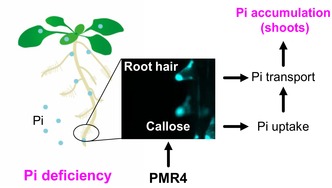
SUMMARY
Plants acquire phosphorus (P) primarily as inorganic phosphate (Pi) from the soil. Under Pi deficiency, plants induce an array of physiological and morphological responses, termed phosphate starvation response (PSR), thereby increasing Pi acquisition and use efficiency. However, the mechanisms by which plants adapt to Pi deficiency remain to be elucidated. Here, we report that deposition of a β‐1,3‐glucan polymer called callose is induced in Arabidopsis thaliana root hairs under Pi deficiency, in a manner independent of PSR‐regulating PHR1/PHL1 transcription factors and LPR1/LPR2 ferroxidases. Genetic studies revealed PMR4 (GSL5) callose synthase being required for the callose deposition in Pi‐depleted root hairs. Loss of PMR4 also reduces Pi acquisition in shoots and plant growth under low Pi conditions. The defects are not recovered by simultaneous disruption of SID2, mediating defense‐associated salicylic acid (SA) biosynthesis, excluding SA defense activation from the cause of the observed pmr4 phenotypes. Grafting experiments and characterization of plants expressing PMR4 specifically in root hair cells suggest that a PMR4 pool in the cell type contributes to shoot growth under Pi deficiency. Our findings thus suggest an important role for PMR4 in plant adaptation to Pi deficiency.
Keywords: root hair, cell wall, Glucan synthase likes (GSLs), phosphate acquisition, phosphate transport, plant nutrition, nutrient deficiency, salicylic acid (SA), pattern‐triggered immunity
Significance Statement
We reveal that PMR4 callose synthase mediates callose deposition in root hairs under phosphate (Pi) deficiency, without requiring Pi starvation response regulators PHR1/PHL1 or LPR1/LPR2. The loss of the callose deposition is accompanied by decreases in Pi acquisition and plant growth in pmr4. Root hair cell‐specific PMR4 expression restores callose deposition in root hairs and shoots growth under Pi deficiency, indicating a critical role for root hair callose in plant adaptation to Pi deficiency.
INTRODUCTION
As an essential macronutrient, plants acquire and distribute P from the rhizosphere to the whole tissues, primarily in the form of inorganic phosphate (Pi). To maintain Pi homeostasis under Pi deficiency, plants induce an array of adaptive responses, for example, increasing Pi uptake, remobilization, and utilization, termed Pi starvation response (PSR), and involve beneficial microbes in mutualistic associations (Chien et al., 2018; Isidra‐Arellano et al., 2021; Péret et al., 2011). PSR is accompanied by extensive transcriptional reprogramming, mainly through the transcription factor PHR1 and related PHL1, which leads to increases in Pi transporter expression, root hair formation and anthocyanin accumulation (Bustos et al., 2010; Rubio et al., 2001). PHR1/PHL1 mediate systemic coordination of these responses according to the internal Pi availability. In addition, Pi depletion leads to remodeling root system architecture, and a decrease in primary root growth while increasing lateral root formation and growth, through the ferroxidases LPR1 and LPR2 (Balzergue et al., 2017; Péret et al., 2014; Svistoonoff et al., 2007). LPR1/LPR2 are required for arresting primary root growth when the external Pi levels are reduced surrounding the root tips (Svistoonoff et al., 2007). Recent studies illuminated an intimate relationship between PSR and biotic interactions, but the molecular basis for their coordination remains poorly understood (Chan et al., 2021).
Callose deposition on the plasma membrane through callose synthases Glucan Synthase Likes (GSLs) reinforces barrier functions of the cell wall and restricts plasmodesmatal permeability in plant development and biotic/abiotic stress responses (Chowdhury et al., 2014; Dong et al., 2005; Ellinger & Voigt, 2014; Enns et al., 2005; Jacobs et al., 2003; Nishikawa et al., 2005; Nishimura et al., 2003; Sager & Lee, 2014; Wu et al., 2018). In Arabidopsis thaliana (hereafter Arabidopsis), callose deposition is induced in the root phloem in response to excess iron, thereby reducing plasmodesmatal permeability (O'Lexy et al., 2018). Of 12 Arabidopsis GSLs, GSL2 (CALS5), and GSL5 (PMR4, CALS12) respectively contribute to basal accumulation and iron‐induced deposition of callose in the root phloem (O'Lexy et al., 2018). Callose also accumulates in the stem cell niche (SCN) surrounding the quiescent center cells, thereby inhibiting cell‐to‐cell movement of the transcription factor SHORT ROOT (SHR) during PSR. This serves to suppress cell division and elongation in the root apical meristem and elongation zone, respectively, through the LPR1/LPR2 pathway (Mora‐Macías et al., 2017; Müller et al., 2015). GSL5 (PMR4) also mediates callose deposition in response to pathogen challenge and microbe/damage‐associated molecular patterns (MAMPs/DAMPs), and promotes fungal penetration resistance (Wang et al., 2021). However, the loss of GSL5 function results in strong resistance to powdery mildew fungi through tightened SA‐based defenses, demonstrated by gsl5 mutants revealed as powdery mildew resistance4 (pmr4) (Nishimura et al., 2003; Vogel & Somerville, 2000). This resistance is thought to represent a backup defense activated when GSL5 (PMR4)‐mediated callose deposition, a key output of MAMP/DAMP‐triggered defense signaling, is disrupted.
Although the significance of PMR4 has been genetically shown, our knowledge is very limited for the mechanisms that involve and regulate PRM4 in callose deposition during defense responses (Wang et al., 2021). PMR4/GSL5 is expressed ubiquitously in unwounded Arabidopsis thaliana plants (Enns et al., 2005). Inoculation with Plectosphaerella cucumerina fungi results in a slight increase in GSL5 expression (García‐Andrade et al., 2011). Downy mildew Hyaloperonospora and the application of SA and flg22 induce GSL5 expression (Dong et al., 2008; Keppler et al., 2018). Whether and if so how GSL5 expression or activity is regulated according to the nutrient status remains poorly understood.
Here, we report that callose deposition is induced in root hairs under Pi deficiency, in a manner independent of PHR1/PHL1 and LPR1/LPR2. Our genetic studies revealed that the Pi depletion‐induced callose deposition is impaired in pmr4 (gsl5) root hairs. The loss of PMR4‐mediated callose deposition is associated with reduced plant growth as well as exaggerated stress symptoms under Pi deficiency, in addition to enhanced powdery mildew resistance. The results indicate PMR4‐mediated callose deposition in root hairs as an important step in plant adaptation to Pi‐limiting conditions.
RESULTS
Callose deposition occurs in root hairs under phosphate deficiency, independently of PHR1/PHL1 and LPR1/LPR2
Under low Pi conditions (Pi at 0 or 50 μM), we noticed callose deposition in the epidermal cells, particularly in root hairs of the differentiation zone (Figure 1A,B; O'Lexy et al., 2018). However, callose deposition was not detected in root hairs under the tested N, Fe, or Fe/Pi deficient conditions (Figure 1A). The results indicate that the callose deposition is specific to Pi deficiency and is dependent on Fe, consistent with the interplay between Fe and Pi (Müller et al., 2015; Abel, 2017; Xue et al., 2022). The absence of callose staining in the epidermis of root hair‐less rhd6 mutants (Figure 1C) validated that callose deposition occurs in root hairs under low Pi. In contrast to previous study (Müller et al., 2015), we did not detect callose deposition in the root SCN and cortex cells of the elongation zone under our experimental conditions (Figure 1A,C).
Figure 1.
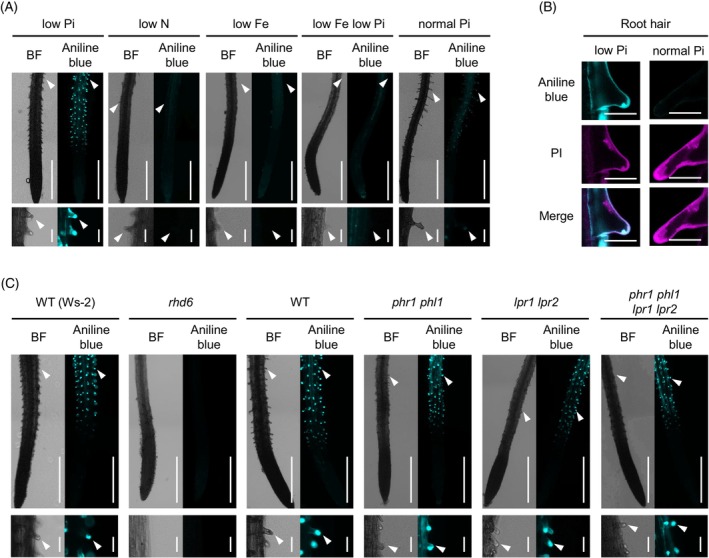
Callose deposition is induced in root hairs under phosphate deficiency, independently of PHR1, PHL1, LPR1, and LPR2. Aniline blue staining of the primary roots 3 days after transfer of 5‐days‐old Arabidopsis thaliana seedlings to low Pi (P 50 μM), low N (N 0 μM), low Fe (Fe 0 μM), low Fe low Pi (Fe 0 μM, P 0 μM), and normal Pi (P 625 μM) liquid media.
(A, C, left). Bright‐field (BF) images. (A, C, right) Aniline blue staining images. White arrowheads indicate root hair tips.
(B) Aniline blue and propidium iodide (PI) staining of root hairs under low Pi and normal Pi conditions.
(C) The wild‐type (WT) used for rhd6 was Ws‐2. Magnified images of the upper panels (A, C, lower panel). Bars, 500 μm (A, C, upper panel), 50 μm (A, C lower panel) and 25 μm (B).
We next assessed whether PHR1/PHL1 and LPR1/LPR2 play a role in the root hair callose deposition. Unlike LPR1/LPR2‐mediated callose deposition in SCN (Müller et al., 2015), the root hair callose deposition was not affected in lpr1 lpr2 (Figure 1C) despite its Fe dependence (Figure 1A). Furthermore, wild‐type (WT)‐like callose deposition was retained in root hairs of phr1 phl1 lpr1 lpr2 under low Pi conditions (Figure 1C). The results indicate that callose deposition is induced in root hairs under Pi deficiency, independently of these PSR regulators, and its separation from callose deposition in the root SCN.
PMR4 callose synthase contributes to callose deposition under Pi deficiency
To unravel genetic requirements for the callose deposition in root hairs under Pi deficiency, we examined glucan synthase like (gsl) mutants for alternations in callose deposition under phosphate deficiency. Of the tested gsl mutants, gsl5 (hereafter, pmr4) mutants specifically lacked callose deposition in root hairs (Figure S1). We also screened a total of 18 550 ethyl methane sulfonate (EMS)‐mutagenized individuals in Col‐0 and Ler backgrounds. This revealed two mutants in the Col‐0 background, designated callose alteration under Pi starvation 1 and 2 (caps1 and caps2, Figure 2A). caps1 and caps2 mutants were severely defective in callose deposition of root hairs under low Pi conditions (Figure 2A). DNA sequencing on the PMR4 genomic region revealed a DNA substitution within the coding region, resulting in a non‐synonymous amino acid substitution, in both caps1 and caps2 alleles (Figure 2B). The substitution in caps1 converts tryptophan to Stop codon at the 260th amino acid (PMR4W260*), resulting in PMR4 truncation, consistent with the severely impaired callose deposition in caps1 as well as pmr4‐1 (PMR4W687*). In caps2, glutamic acid is substituted with lysine at the 1181st amino acid (PMR4E1181K), located within the cytosolic side of a predicted glucan‐synthase domain (residue 575 to 1558, orange‐marked domain, Figure 2B). The results suggest that dysfunction of PMR4 fails to induce root hair callose deposition under Pi deficiency in these caps alleles. We validated that leaf callose deposition in response to the bacterial MAMP flg22, a PMR4‐dependent output in pattern‐triggered immunity (PTI), is abolished in these caps alleles (Figure S2). Moreover, we showed strong resistance to powdery mildew fungi in these caps plants (Figure 2C), as previously described in pmr4 (Nishimura et al., 2003; Vogel & Somerville, 2000). The results confirm that PMR4 functions are impaired in both caps alleles.
Figure 2.
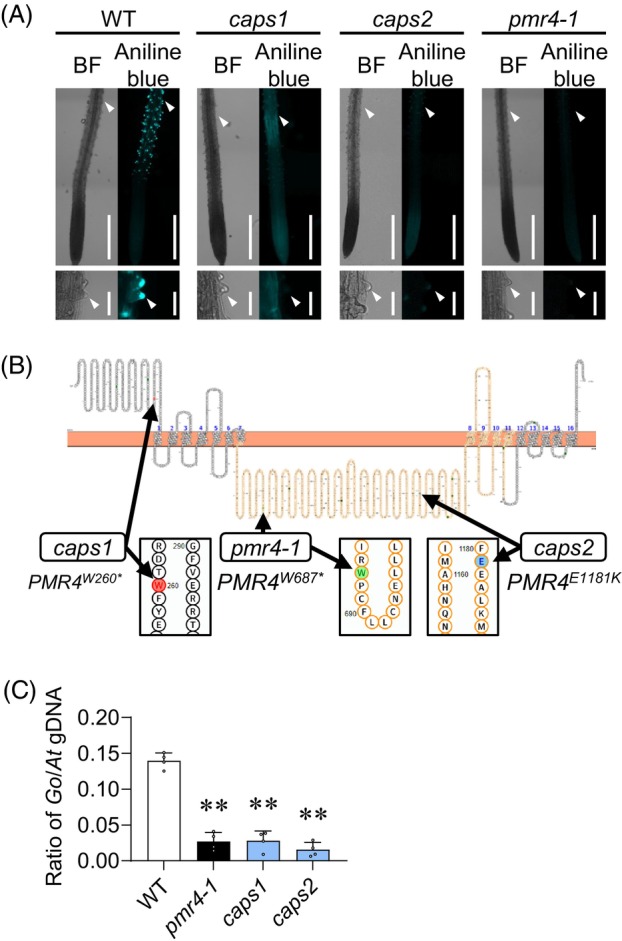
Callose synthase PMR4 is required for callose induction in root hairs under phosphate deficiency.
(A) Aniline blue staining of the primary roots 3 days after transfer of 5‐days‐old Arabidopsis thaliana plants to low Pi (0 μM) liquid media. Left, bright‐field (BF) images. Right, aniline blue staining images. Two caps mutants are defective in inducing root hair callose under phosphate starvation. Lower panels, magnified images of upper panels. White arrowheads indicate root hair tips. Bars, 500 μm (upper panel) and 50 μm (lower panel).
(B) Positions of mutated sites in the PMR4 locus of caps mutants, illustrated with Protter (Omasits et al., 2014). An orange region corresponds to the catalytic domain of PMR4. Asterisks indicate stop codons.
(C) Ratios of G. orontii genomic DNA to Arabidopsis genomic DNA were determined by qPCR with the primers for plasma membrane ATPase 1 (Go_V1_Conting3757) and RNA‐binding family protein (AT3G21215), respectively. Data show means ± SD (n = 3). Asterisks (**P < 0.01) indicate statistically significant differences relative to WT according to Student's t‐test.
PMR4 influences root development traits induced under Pi deficiency
By using pmr4‐1 as a representative mutant allele, we further investigated the biological significance of PMR4 under Pi deficiency. Recent studies noted that some Pi deficiency‐induced root responses are limited to plate‐grown plants whose roots are light irradiated (Gao et al., 2021; Yeh et al., 2020; Zheng et al., 2019). To test this possibility, we examined callose deposition in Pi‐deplete and Pi‐replete plants under darkness. At 3 days after the transfer of seedlings to the different Pi conditions under darkness, we detected callose deposition in non‐irradiated root hairs of WT but not pmr4 plants specifically under low Pi, validating that PMR4‐mediated, root hair callose deposition is not a consequence of light exposure to the root (Figure S3A). Moreover, we also detected callose deposition in the cotyledons of WT but not pmr4 plants under these conditions (Figure S3B). The results strengthen the physiological relevance of PMR4‐mediated callose deposition under low Pi.
The loss of PMR4 leads to an increase in salicylic acid (SA) biosynthesis and defense under biotic stress (Nishimura et al., 2003; Vogel & Somerville, 2000), in a manner requiring SID2 isochorismate synthase that mediates a critical step in SA biosynthesis during defense activation (Wildermuth et al., 2002). Given the negative impact of SA defense activation on plant growth (Mateo et al., 2006; Zhang et al., 2010), we assessed the possible SA dependence of the observed pmr4 phenotypes, by examining genetic interactions between pmr4 and sid2. Callose deposition was induced in sid2 root hairs but not induced in pmr4 sid2 under low Pi conditions (Figure 3A), demonstrating that SID2‐mediated SA biosynthesis is not required for PMR4‐mediated callose deposition in root hairs. The results also exclude over‐activation of SA defense as the cause of the callose defects in pmr4 root hairs.
Figure 3.
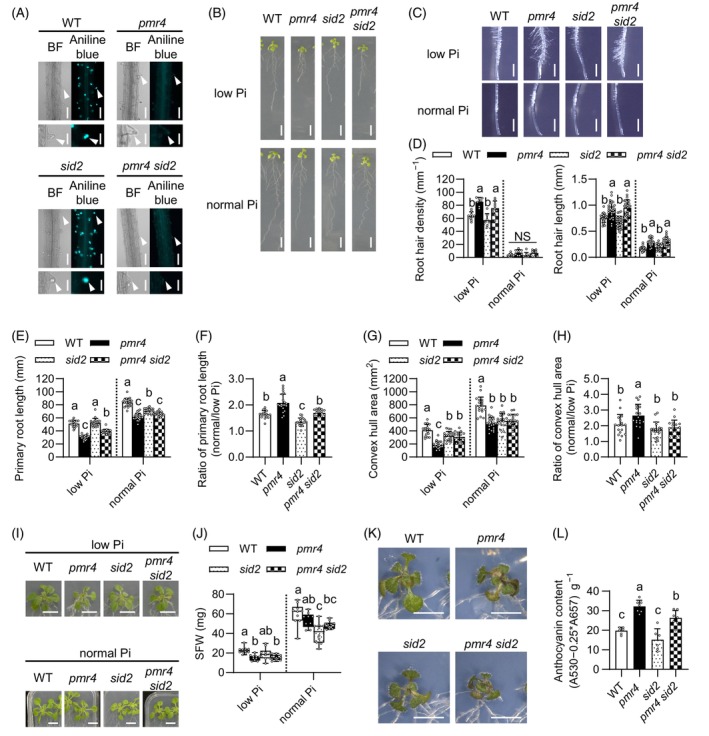
PMR4 modulates root and shoot growth under phosphate deficiency.
(A) Aniline blue staining of the primary roots 3 days after transfer of 5‐days‐old Arabidopsis seedlings to low Pi (50 μM) liquid media. Left, bright‐field (BF) images. Right, aniline blue staining images. Lower panels, magnified images of upper panels. White arrowheads indicate root hair tips. Bars, 100 μm (upper panel) and 50 μm (lower panel).
(B) Six‐days‐old seedlings were exposed to low Pi (50 μM) or normal Pi (625 μM) media for 6 days. Bar, 10 mm.
(C) The primary roots 6 days after transfer of 6‐days‐old seedling to low Pi (50 μM) and normal Pi media. Bar, 1 mm.
(D) Root hair density and root hair length in (C).
(E) Primary root length in (B).
(F) Ratio of the primary root length under normal Pi relative to low Pi of (E).
(G) Convex hull area of the roots in (B).
(H) Ratio of convex hull area under normal Pi relative to low Pi of (G).
(I) Six‐days‐old seedlings were exposed to low Pi (50 μM) or normal Pi for 16 days. Bar, 10 mm.
(J) SFW in (I).
(K) Six‐days‐old seedlings were exposed to low Pi (10 μM) media for 10 days. Bar, 10 mm.
(L) Anthocyanin contents in (K).
(D–H, J, L) Data show means ± SD (n = 7 ~ 30). Statistical significance was assessed by one‐way ANOVA and Tukey honestly significant different (HSD) test for each medium dataset. Different letters indicate significant differences (P < 0.05). NS, not significant.
In Arabidopsis, root hair cells specifically occur at the junction of two underlying cortical cells, whereas non‐hair cells occur in the other epidermal cells (Dolan et al., 1994; Schiefelbein et al., 2009). Root hair density was increased under low Pi conditions compared with normal Pi conditions, and the increase was exaggerated in pmr4 as well as pmr4 sid2, without ectopic root hair formation (Figure 3B–D; Figure S4A). Likewise, root hair length was also increased in pmr4 as well as pmr4 sid2 under both Pi conditions (Figure 3C,D). The results suggest that PMR4 is not required for the differentiation or outgrowth of root hair cells, but that PMR4 contributes to restricting root hair formation and elongation, in the presence or absence of SID2. Conversely, primary root length was reduced in pmr4 compared with WT in both low and normal Pi conditions (Figure 3B,E). We then determined the ratio of the primary root length under normal Pi versus low Pi as an indicator for the primary root growth inhibition under Pi deficiency. The ratio (normal Pi/low Pi) was increased in pmr4 compared with WT, indicating that primary root growth inhibition, a Pi deficiency‐characteristic response, is exaggerated in pmr4 (Figure 3F). We also observed a decrease in the root meristem region in pmr4 compared with WT under Pi deficiency, suggesting that the meristem exhaustion coincides with the inhibition of primary root growth in pmr4 (Figure S4B). The primary root growth inhibition under low Pi was alleviated in sid2, but it was again exaggerated in pmr4 sid2 compared with sid2 (Figure 3E,F), indicating that the loss of PMR4 enhances the Pi deficiency response, with or without SID2.
We next tested whether and if so how PMR4 affects the expression of Pi deficiency‐inducible genes. Quantitative RT‐PCR analyses on WT and pmr4 roots revealed that the expression of Pi starvation‐induced genes, AT4 and IPS1 long non‐coding RNAs (Franco‐Zorrilla et al., 2007), PHT1;5 Pi transporter (Nagarajan et al., 2011), and SPX1 Pi‐binding suppressor of PHR1 (Puga et al., 2014), was not affected in pmr4 under the normal (625 μM) and low Pi (50 μM and 10 μM) conditions (Figure S5). The results are consistent with WT‐like callose deposition in the root hairs of phr1 phl1 and phr1 phl1 lpr1 lpr2 (Figure 1C) and indicate that PMR4 is not required for the expression of these PSR‐related genes.
The primary root growth inhibition under Pi deficiency relies on LPR1 and LPR2 (Müller et al., 2015; Svistoonoff et al., 2007). We examined the possible relationship between LPR1/LPR2 and PMR4 in the regulation of primary root growth. The primary root length was reduced in pmr4 lpr1 lpr2 compared with lpr1 lpr2 under both Pi conditions, whereas it was increased in pmr4 lpr1 lpr2 compared with pmr4 specifically under low Pi (Figure S6A,B). The results indicate that PMR4 serves to sustain the primary root growth irrespective of the Pi availability or LPR1/LPR2 activity, whereas the LPR1/LPR2 inhibition is specific under low Pi with or without PMR4. The ratio of the primary root length under normal Pi versus low Pi was lowered to a similar extent in lpr1 lpr2 and pmr4 lpr1 lpr2 compared with WT (Figure S6C), despite the decrease in the primary root length of pmr4 (Figure S6B), indicating that lpr1 lpr2 is epistatic to pmr4. Consistently, under Pi deficiency, PMR4 expression was reduced in lpr1 lpr2, but not in phr1 phl1, compared with WT roots (Figure S7). Together with the WT‐like callose deposition in lpr1 lpr2 root hairs (Figure 1C), the data suggest that PMR4 expression is dependent on LPR1/LPR2 but that the residual PMR4 expression is sufficient for the callose deposition in lpr1 lpr2 root hairs.
We next determined the convex hull root area as a proxy of overall root system growth, which was correlated with Pi availability (Figure 3G). It was reduced in pmr4 and sid2 compared with WT under both normal and low Pi conditions, albeit to a lesser extent in sid2 than pmr4 under low Pi (Figure 3G), suggesting that PMR4 and SID2 both contribute to expanding the root area. The ratio of the root area under normal Pi versus low Pi conditions was increased in pmr4 relative to WT in a manner dependent on SID2 (Figure 3H), suggesting that SID2 contributes to low Pi‐induced root growth inhibition in pmr4. Consistently, the root area was greater in pmr4 sid2 compared with pmr4 under low Pi (Figure 3G). The results suggest that SID2 negatively impacts root expansion specifically in the absence of PMR4 under Pi deficiency. The results also imply that SID2‐mediated SA biosynthesis promotes root growth expansion in Pi‐depleted plants when PMR4 is present. The latter seems to be consistent with a previously described uncoupling of SA induction from defense activation under Pi deficiency (Gulabani et al., 2021).
PMR4 serves to sustain shoot growth and restrict anthocyanin accumulation under Pi deficiency
We next examined whether PMR4 influences shoot growth by determining shoot fresh weight (FW) under Pi deficiency and sufficiency. Shoot FW was lowered in pmr4 compared with WT, and it was not even partially restored by the simultaneous loss of SID2 in pmr4 sid2 under low‐Pi media or soil (Figure 3I,J; Figure S8). Together with the root phenotypes described above, the results exclude SID2 as the major cause of the growth inhibition in pmr4. Although high SA accumulation was previously described to reduce plant growth (Miura et al., 2010), we did not detect the negative impacts of SID2 on shoot growth under low Pi conditions (Figure 3J). SID2 even positively influenced shoot growth under the normal Pi conditions, indicated by the reduced shoot biomass in sid2 compared with WT (Figure 3J).
Anthocyanin content was increased in pmr4 compared with WT, in the presence or absence of SID2 (Figure 3K,L). Although SA was described to enhance anthocyanin accumulation under Pi deficiency (Morcillo et al., 2020), sid2 was largely indistinguishable from WT in the anthocyanin contents under our low Pi conditions (Figure 3K,L). The results imply that PMR4‐mediated Pi acquisition and allocation serves to alleviate Pi starvation causing anthocyanin accumulation under Pi deficiency.
PMR4 promotes the acquisition and root‐to‐shoot translocation of Pi
The PMR4 effects on shoot and root growth prompted us to examine its possible role in Pi acquisition from the rhizosphere and Pi mobilization within the plant. We found that shoot Pi concentrations were greatly reduced in pmr4 compared with WT, without alterations in root Pi concentrations, under Pi deficiency (Figure 4A,B). This indicates that PMR4 contributes to shoot Pi accumulation in Pi‐depleted plants. It was described that PHR1/PHL1 promotes shoot Pi accumulation (Wang et al., 2018) and that PHO1 Pi transporter mediates Pi mobilization from roots to shoots through the root xylem vessels (Stefanovic et al., 2007). We validated that Pi contents were lowered in the shoots but increased in the roots, in phr1 phl1 and pho1 compared with WT under low Pi (Figure 5A). A decrease in shoot Pi contents was also observed in phr1 phl1 and pho1 under normal Pi (Figure 5B). Since root hairs contribute to Pi accumulation under Pi deficiency (Holz et al., 2018; Robertson‐Albertyn et al., 2017; Tanaka et al., 2014), we next determined Pi contents in a root hair‐less mutant, rhd6. As seen in pmr4, phr1 phl1 and pho1, Pi contents were lowered in the shoots but not in the roots of rhd6 compared with WT, specifically under Pi deficiency (Figure 6A,B). As illustrated in Figures 5C and 6C, all these mutants displayed a decrease in shoot Pi contents without reducing root Pi contents under Pi deficiency. We inferred from these results that PMR4‐mediated callose deposition in root hairs contributes to shoot Pi accumulation, possibly through increasing the uptake or root‐to‐shoot translocation of Pi. To test the hypothesis, we determined the rates of Pi uptake and Pi translocation by applying and tracing radiolabeled P. The rate of 32P uptake per time and the ratio of 32P in the shoot to whole seedling, standing for Pi translocation rate, were both lowered in pmr4 and pmr4 sid2 compared with WT and sid2, respectively, under low Pi (Figures 5D, 6D). In the presence or absence of SID2, PMR4 contribution to the two proxies was significant only under low Pi (Figures 5D and 6D). These results suggest that PMR4 promotes Pi uptake and root‐to‐shoot Pi translocation under Pi deficiency. Moreover, the 32P uptake rate was reduced in sid2 compared with WT, whereas pmr4 sid2 was not distinguishable from pmr4 under low Pi (Figure 6D), suggesting that SID2 positively influences Pi uptake under Pi deficiency in the presence of PMR4.
Figure 4.
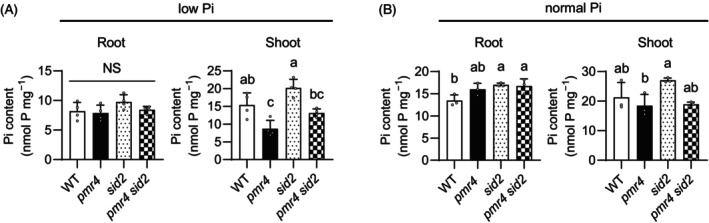
PMR4 contributes to shoot Pi accumulation under phosphate deficiency.
(A, B) Pi contents per root or shoot fresh weight (FW) of 6‐days‐old Arabidopsis seedlings exposed to low Pi (150 μM) or normal Pi (625 μM) for 9 days. Data show means ± SD (n = 3 ~ 4) in (A, B). Statistical significance was assessed by one‐way ANOVA and Tukey HSD test for each medium dataset (A, B). Different letters indicate significant differences (P < 0.05). NS, not significant.
Figure 5.
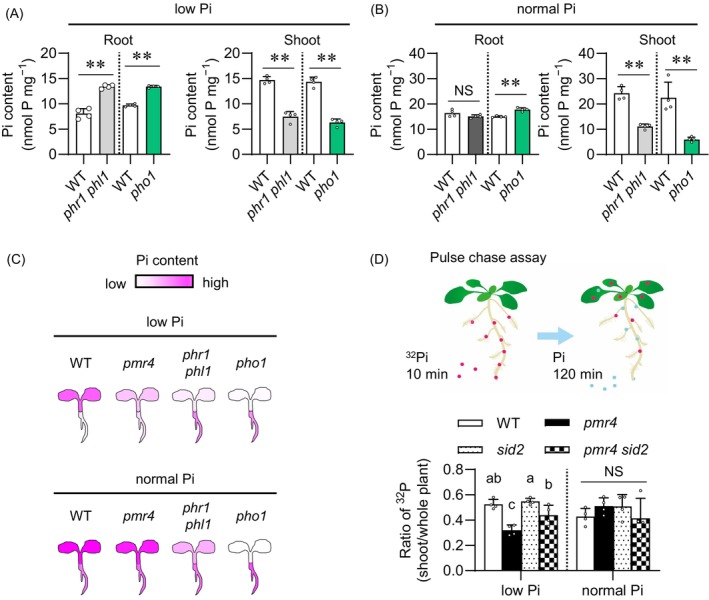
PMR4 contributes to Pi translocation under phosphate deficiency.
(A, B) Pi contents per root or shoot fresh weight (FW) of 6‐days‐old Arabidopsis seedlings exposed to low Pi (150 μM) or normal Pi (625 μM) for 9 days.
(C) Illustration of Pi accumulation in the indicated genotypes, referred to (Figures 4A, 5A,B).
(D) Ratio of 32P in the shoots relative to the whole plants after pulse‐chase labeling. Seedlings grown on low and normal Pi media (50 and 625 μM, respectively) were exposed to 32P‐labeled Pi for 10 min, and then transferred to non‐labeled Pi media for 120 min (pulse chase). The 32P ratio of the shoots to the whole seedlings was calculated for the translocation rate of Pi. Data show means ± SD (n = 3 ~ 4) in (A, B, D). Asterisks (**P < 0.01) indicate statistically significant differences relative to WT according to Student's t‐test (A, B). Statistical significance was assessed by one‐way ANOVA and Tukey HSD test for each medium dataset (D). Different letters indicate significant differences (P < 0.05). NS, not significant.
Figure 6.
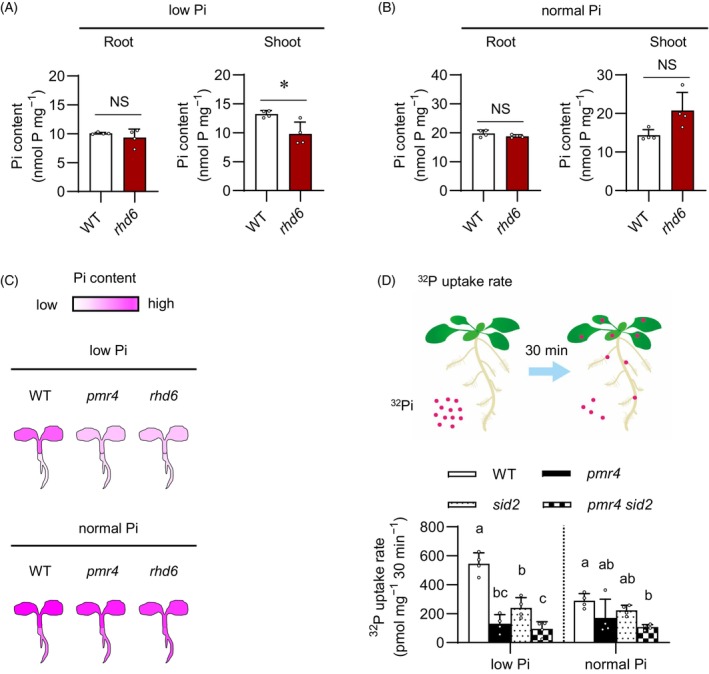
PMR4 contributes to Pi uptake under phosphate deficiency.
(A, B) Pi contents per root or shoot fresh weight (FW) of 6‐days‐old Arabidopsis seedlings exposed to low Pi (150 μM) or normal Pi (625 μM) for 9 days. The WT used for rhd6 was Ws‐2 (C) Illustration of Pi accumulation in the indicated genotypes, referred to (Figures 4A, 6A,B).
(D) 32P uptake rate determined in the whole seedlings exposed to the indicated media (low Pi and normal Pi, 50 and 625 μM Pi, respectively) supplemented with 32P‐labeled Pi for 30 min. Data show means ± SD (n = 3 ~ 4) in (A, B, D). Asterisks (*P < 0.05) indicate statistically significant differences relative to WT according to Student's t‐test (A, B). Statistical significance was assessed by one‐way ANOVA and Tukey HSD test for each medium dataset (D). Different letters indicate significant differences (P < 0.05). NS, not significant.
We further assessed the possible PMR4 contributions to the acquisition and tissue mobilization of Pi and other elements, by inductively coupled plasma mass spectrometry (ICP‐MS) analyses for P, K, Mg, Ca, Mn, Fe, Zn, Mo, and B contents in Pi‐depleted and Pi‐replete plants. Under Pi sufficiency, although the contents of these elements were not distinguishable between WT and pmr4 roots, the contents of K and the rest were increased and lowered, respectively, in pmr4 compared with WT shoots (Figure S9). Under low Pi, Mn, Zn, and Mo contents were greater in pmr4 compared with WT roots, whereas the contents of all the tested elements except K were lowered in pmr4 compared with WT shoots (Figure 7). The results were consistent with PMR4‐mediated shoot Pi accumulation without affecting root Pi contents under low Pi conditions (Figure 4A). Additionally, the results suggest that PMR4 also positively influences shoot accumulation of Mg, Ca, Mn, Fe, Zn, Mo, and B (Figure 7; Figure S9). WT‐like and increased K accumulation in pmr4 shoots compared with WT under Pi deficiency and sufficiency, respectively, show that the root‐to‐shoot transport is not entirely reduced for all elements in pmr4, pointing to a certain degree of element specificity in PMR4 dependence (Figure 7; Figure S9).
Figure 7.
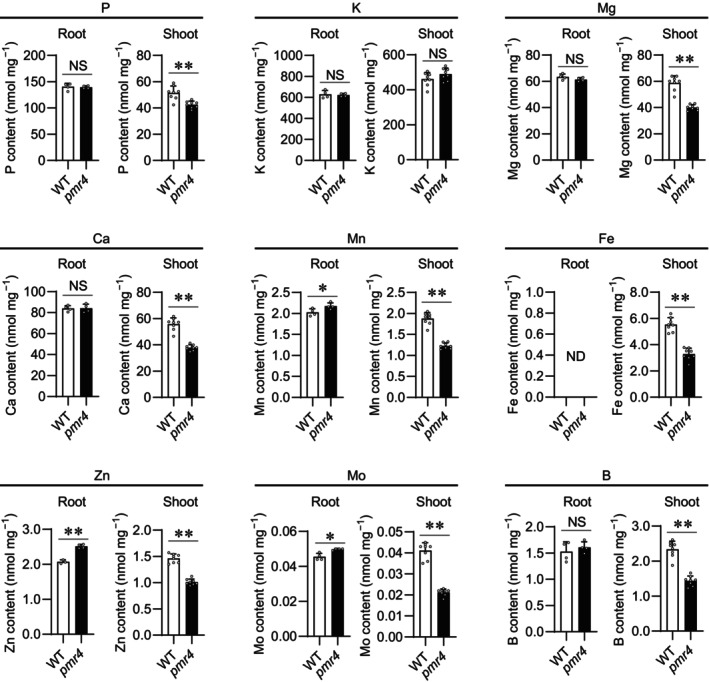
Plant element contents in shoot and root under low phosphate conditions.
Element contents (P, K, Mg, Ca, Mn, Fe, Zn, Mo, and B) per root or shoot dry weight (DW) of 5‐days‐old Arabidopsis thaliana seedlings exposed to low Pi (10 μM) for 4 days. Data show means ± SD (n = 4–8). Asterisks (**P < 0.01, *P < 0.05) indicate statistically significant differences relative to WT according to Student's t‐test. ND, no data; NS, not significant.
The role of PMR4 in the shoot accumulation of different elements prompted us to examine whether PMR4 influences plant responses to different nutrient deficiencies. We examined pmr4 phenotypes under B scarcity, as B scarcity induces inhibition of the primary root and transcriptional reprogramming related to ROS and defense in roots (Das & Purkait, 2020; Miwa et al., 2013). Although the primary root length was reduced in pmr4 compared with WT over the tested range of B conditions, its ratio under B sufficiency (100 μM) versus the examined low B conditions was not affected in pmr4 (Figure S10). The results point to the specific contribution of PMR4 to plant growth under Pi deficiency.
Root PMR4 contributes to shoot growth under Pi deficiency
We next examined the possible significance of the root PMR4 pool in promoting shoot growth, through micrografting between WT and pmr4 plants. We validated that the micrografted plants of WT/WT (shoot/root genotypes) and pmr4/pmr4 showed the corresponding WT and pmr4 phenotypes in root growth and Shoot fresh weight (SFW) (Figure 8A–C). The root hair callose was detected only when PMR4 was present in the roots, although the callose signals were not as strong as in intact plants, possibly reflecting the stresses imposed by the grafting procedure (Figure 8D). Remarkably, SFW was greatly reduced in WT/pmr4 whereas it was less affected in pmr4/WT (Figure 8B), pointing to a critical role for a root PMR4 pool in shoot growth under Pi deficiency. However, the root growth was partially restored in WT/pmr4 (Figure 8C), pointing to a role for a shoot PMR4 pool in sustaining the primary root growth in Pi‐depleted plants.
Figure 8.
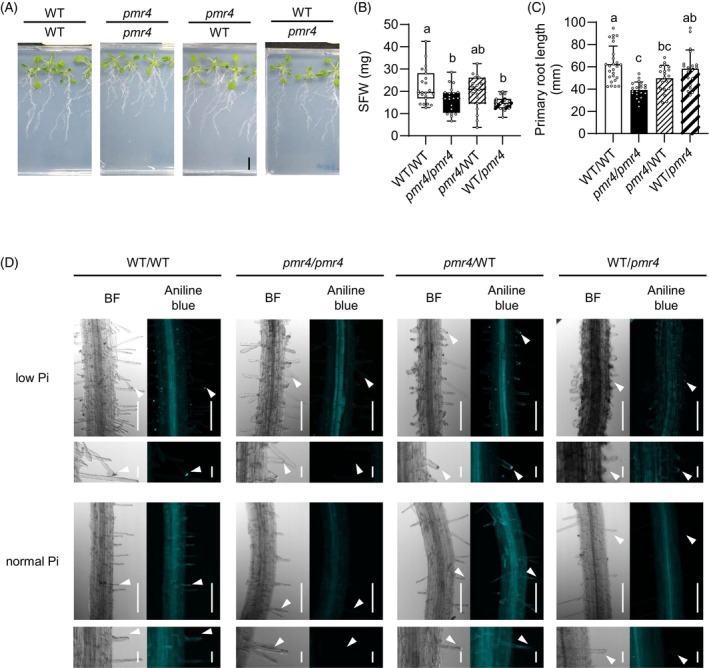
Root PMR4 contributes to shoot growth under phosphate deficiency.
(A) Reciprocal micrografting of Arabidopsis WT and pmr4 mutants. Images were obtained at 14 days after transfer to low Pi (150 μM) media. Bar, 10 mm.
(B) Shoot fresh weight (SFW) of (A).
(C) The primary root length of (A).
(D) Aniline blue staining of grafted‐seedlings at 9 days after transfer to low Pi (50 μM). (D, left) Bright filed (BF) images. (D, right) Aniline blue staining images. (D, lower panel) Magnified images of (D, upper panel). White arrows indicate root hair tips. Bar, 100 μm (C, upper panel) or 20 μm (C, lower panel).
(B, C) Data show means ± SD (n = 19–26). Statistical significance was assessed by one‐way ANOVA and Tukey HSD test for each medium dataset. Different letters indicate significant differences (P < 0.05).
To further examine the role of a PMR4 pool in root hairs, we generated transgenic plants expressing PMR4 under the control of a root hair‐specific LRX1 promoter (pLRX1, Baumberger et al., 2001) in the pmr4‐1 background (Figure 9A,B). pLRX1::PMR4 complemented pmr4 in terms of the callose deposition in root hairs and shoot growth under low Pi conditions (Figure 9A–C). By contrast, the primary root growth remained suppressed in the pLRX1::PMR4 transgenic lines as well as in pmr4 (Figure 9B,D). The results indicate that PMR4 in root hairs is sufficient to restore shoot growth, but not the primary root growth, under Pi deficiency.
Figure 9.
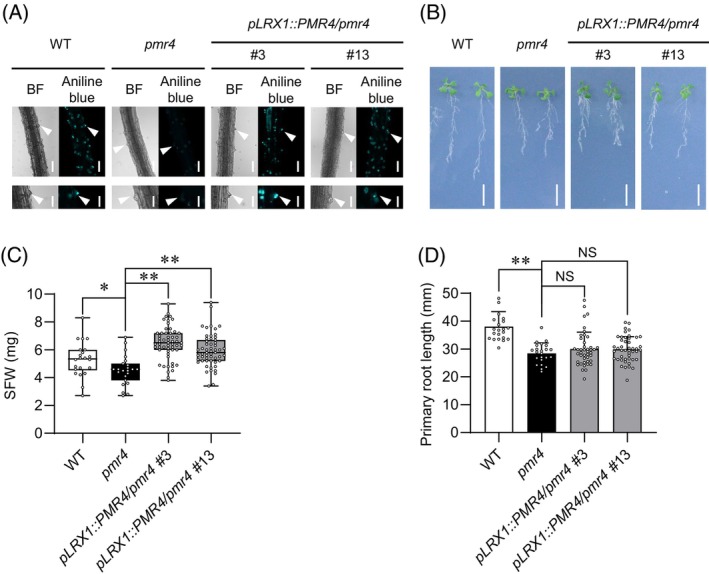
PMR4 in root hairs contributes to shoot growth under phosphate deficiency.
(A) Aniline blue staining of the primary roots 3 days after transfer of 5‐days‐old Arabidopsis seedlings to low Pi (10 μM). White arrowheads indicate root hair chips. Bars, 100 μm (upper panel) and 50 μm (lower panel).
(B) Five‐days‐old seedlings were exposed to low Pi (50 μM) for 10 days. Bar, 10 mm.
(C) SFW of (B).
(D) Primary root length of (B).
(C, D) Data show means ± SD (n = 22–48). Asterisks (**P < 0.01, *P < 0.05) indicate statistically significant differences relative to pmr4 according to Student's t‐test. NS, not significant.
DISCUSSION
Our studies on callose deposition in root hairs induced under Pi deficiency revealed PMR4 as a positive regulator of plant adaptation to Pi deficiency (Figure 10). PMR4 mediates defense‐induced callose deposition in PTI and pre‐invasion fungal resistance (Chowdhury et al., 2014; Jacobs et al., 2003), and its loss is linked to enhanced SA‐based pathogen resistance (Nishimura et al., 2003). Therefore, PMR4 seems to play a critical role in an array of biotic and abiotic stress responses, possibly through callose deposition at different sites.
Figure 10.
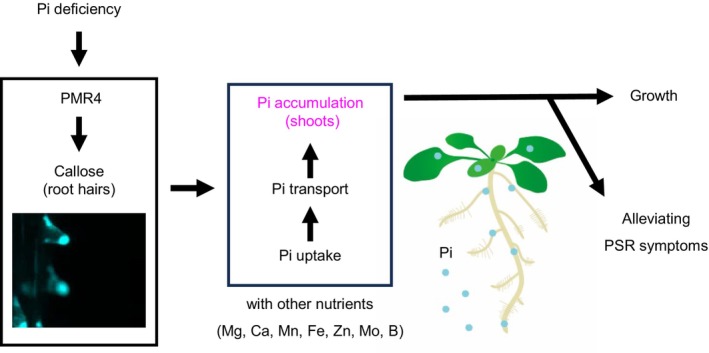
Model for PMR4‐mediated root hair callose deposition in plant adaptation to Pi deficiency.
PMR4 induces callose deposition in root hairs under phosphate deficiency, and thus contributes to shoot Pi accumulation and growth through enhancing Pi uptake and root‐to‐shoot transfer. This contribution alleviates phosphate starvation symptoms, such as root system area reduction and anthocyanin accumulation.
The present study suggests that the root hair callose contributes to root Pi uptake and shoot Pi accumulation under Pi deficiency. Importantly, the loss of PMR4 affects both PHR1‐dependent and LPR1‐dependent processes (Figures 3 and 4) (Bates & Lynch, 1996; Thibaud et al., 2010), which have been genetically uncoupled in PSR (Balzergue et al., 2017; Wang et al., 2010). Pi accumulation and anthocyanin accumulation are suppressed in phr1 phl1 (Bustos et al., 2010; Rubio et al., 2001; Wang et al., 2018). Shoot Pi contents and plant biomass are also reduced in pmr4, implying that PMR4 positively influences PHR1/PHL1‐mediated Pi acquisition and plant growth under internal Pi deficiency. However, over‐accumulation of anthocyanin in pmr4 (Figure 3K,L) in contrast to phr1 phl1 (Bustos et al., 2010; Rubio et al., 2001; Wang et al., 2018), suggests that PMR4 contribution is specific to the former two outputs of PHR1/PHL1 and that its loss results in exaggeration of Pi starvation symptom. The loss of PMR4 also strengthens inhibition of the primary root growth under Pi deficiency, which is pronounced in the presence of LPR1/LPR2 (Figure S6), consistent with LPR1/LPR2‐dependent PMR4 expression (Figure S7). The LPR1 pathway mediates callose deposition at the root SCN to inhibit the cell‐to‐cell movement of the transcription factor SHR, thereby suppressing the cell division and subsequent root elongation (Mora‐Macías et al., 2017; Müller et al., 2015). Our results suggest a role for PMR4 in antagonizing LPR1/LPR2‐dependent root growth inhibition.
The failure of a root hair PMR4 pool to restore primary root growth (Figure 9B,D) implies the involvement of PMR4 from another cell type(s) than root hairs in this regulation. Consistently, shoot Pi accumulation is partially and essentially retained in rhd6 under low and normal Pi conditions, respectively, despite the lack of root hairs (Figure 6A–C). It is conceivable that PMR4‐mediated callose deposition in different cell types serves to sustain the primary root growth against LPR1/LPR2‐mediated attenuation under Pi deficiency. In line with this, we observed low Pi‐induced, PMR4‐mediated callose deposition in cotyledons as well under darkness (Figure S3), strengthening its critical role in plant adaptation to Pi deficiency. Previous studies suggest that the inhibition of primary root growth in Pi‐depleted plants is particularly pronounced when roots are exposed to light (Gao et al., 2021; Yeh et al., 2020; Zheng et al., 2019). The mechanisms underlying antagonistic interactions between PMR4 and LPR1/LPR2 require further studies including the full identification of critical callose deposition sites.
Shoot Pi accumulation depends on root Pi uptake and subsequent Pi translocation. PHT1;1 and PHT1;4 provide two major Pi transporters in roots, contributing greatly to Pi acquisition under Pi deficiency (Shin et al., 2004). PHO1 mediates Pi translocation from roots to shoots through Pi loading into the root xylem (Hamburger et al., 2002; Liu et al., 2012; Poirier et al., 1991). Notably, the loss of PMR4 results in a decrease in the shoot Pi contents without affecting the root Pi contents (Figures 4A,B and 7). Likewise, the loss of PHR1/PHL1 and PHO1 also lowers shoot Pi contents without reducing root Pi contents (Figure 5A,B; Liu et al., 2012; Wang et al., 2018). In pht1;1 pht1;4, a decrease of Pi uptake in roots is associated with that of Pi translocation to the shoots and shoot Pi accumulation (Shin et al., 2004). An intimate linkage between root uptake and root‐to‐shoot translocation is also seen for K+ (Nieves‐Cordones et al., 2019). It is thus conceivable that the lowered Pi accumulation in pmr4 shoots is caused by a decrease in the rates of the root Pi uptake and the root‐to‐shoot Pi translocation, possibly through the compromised root hair function in the absence of PMR4. Indeed, 32P tracer analyses indicate that PMR4 promotes both root Pi uptake and root‐to‐shoot Pi translocation, without affecting the root Pi contents (Figures 4, 5D, and 6D). This is consistent with the view that the root‐to‐shoot Pi translocation is driven by the Pi uptake in the roots (Shin et al., 2004). Leaf transpiration affects root‐to‐shoot solute translocation (Nieves‐Cordones et al., 2019), but it might be ignorable under constitutive high humidity in the closed plate assays used in the present study. Notably, PMR4 also contributes to the accumulation of different elements except K in shoots under Pi deficiency (Figure 7; Figure S9). This might largely be attributed to secondary effects of PMR4‐dependent root growth and expansion (Figure 3E–H), but the separate regulation of K (Figure 7; Figure S9) cannot be explained by this model. Notably, the disruption of multiple PHT1 members (PHT1;1/1;2/1;3 silencing in the phf1 pht1;4 mutant background, designated mut5) results in a decrease in Na as well as P contents compared with WT (Ayadi et al., 2015). PHR1 mediates cross‐regulation among Fe, Zn, and Pi homeostasis (Briat et al., 2015). Further studies will be required to unravel the mechanisms underlying the complex coordination of the acquisition and tissue distribution among different elements and the observed role of PMR4 in the regulation (Figure 7; Figure S9).
Root hairs increase the root surface area to promote root anchorage, nutrient acquisition, and metabolite exudation, thereby facilitating the interactions with the soil environments (Datta et al., 2011; Han et al., 2023; Poole et al., 2018; Robe et al., 2021; Vissenberg et al., 2020). The elongation and increased densities of root hairs in pmr4 (Figure 3C,D) can be inferred that the root hairs formed in the absence of callose deposition are less functional in Pi acquisition or Pi allocation to the other tissues, thereby leading to further formation of root hairs.
How does PMR4‐mediated callose deposition contribute to the root hair functions under low Pi? Our findings are reminiscent of the nutrient transport through the phloem that requires callose deposition in sieve plates (Barratt et al., 2010, Xie & Hong, 2011). The loss of callose from sieve plate pores reduces the phloem conductivity and thus the phloem transport (Barratt et al., 2010). Therefore, it is tempting to speculate that the root hair callose plays a role in cell wall remodeling to reinforce the root hairs for effective transport of water and nutrients and for providing a driving force for their root‐to‐shoot transport. The precise mechanisms by which callose deposited in the root hairs promotes Pi acquisition require further investigation.
Root hairs also provide a key interface to soil‐borne microbes and are intimately associated with root defense regulation. Root hair formation and elongation are facilitated by jasmonate (JA) (Zhu et al., 2006), which mediates the expression of key transcription factors for root hair differentiation and the de‐repression of RHD6 from the interference by JAZ proteins (Han et al., 2020). In beneficial interactions with the epiphytic bacterium Pseudomonas simiae WCS417, activation of defense‐related genes is increased in hair cell lineages compared with non‐hair cell lineages (Verbon et al., 2023). The endogenous elicitor‐active Pep peptides induce root hair formation (Jing et al., 2024; Okada et al., 2021), and Pep receptor signaling from the root hairs can activate defense responses without root growth inhibition (Okada et al., 2021). These findings suggest a specific capacity of root hair cells in root immunity. How these properties of root hairs are influenced by PMR4‐mediated callose deposition under Pi deficiency also represents an interesting topic for future studies.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
The Arabidopsis thaliana accessions used were Col‐0 and Ws‐2, including the mutants examined (Table S1) and the wild‐type used was Col‐0 unless otherwise stated. We generated ethyl methanesulfonate (EMS)‐mutagenesis (M2) seeds for screening in both Col‐0 and Ler backgrounds. All seeds were sterilized using 6% sodium hypochlorite and 0.2% Trion X‐100 for 15 min, and then kept at 4°C for 2 days before sowing a half‐strength Murashige and Skoog (MS) medium with 25 mM sucrose and 0.8% Agar (WAKO, Japan). If not stated otherwise, the normal Pi media contained 625 μM KH2PO4, whereas the low Pi media contained 0 μM, 10 μM, 50 μM or 150 μM KH2PO4 as indicated. The low N and low Fe media used contained 0 μM N and 0 μM Fe, respectively. Plants were grown in a growth chamber under 12 h: 12 h, white light: dark cycles at 22°C unless otherwise stated. The white light intensity was 80 μmol m−2 s−1. Six‐day (d)‐old seedlings were transferred to the indicated Pi liquid media or agar media with 1% Difco Agar, Bacteriological (BD, Franklin Lakes, NJ, USA).
Construction and plant transformation
The genomic DNA sequences of the POWDERY MILDEW RESISTANCE 4 (PMR4, AT4G03550) loci, together with a regulatory DNA sequence for root hair‐specific expression (below), were cloned into the Gateway binary vector pGWB504 (Nakagawa et al., 2007; primer information Table S2) to express a C‐terminally GFP‐fused PMR4 under its native regulatory DNA sequence. The root hair‐specific promoter sequence used was derived from LRR‐EXTENSIN 1 (LRX1, AT1G12040) (Baumberger et al., 2003). Transgenic lines were generated by Agrobacterium‐mediated transformation of pmr4‐1 with the DNA construct and subsequently selected on hygromycin resistance and DNA genotyping.
Aniline blue and PI staining
Callose staining with aniline blue was conducted as described in Schenk and Schikora (2015). Plants were examined at 3 days after the transfer of 5‐days‐old seedlings to the indicated Pi, N, or Fe liquid media (Figure 1A,C). To test callose deposition under darkness, plants were grown in a growth chamber on a 12‐h light and 12‐h dark cycle at 22°C for 5 days. After transferring the seedlings to low phosphate (Pi) conditions (10 μM) or normal phosphate (Pi) conditions (625 μM), the plates were covered with aluminum foil for an additional 3 days (Figure S3). Plant tissues were fixed in a solution containing acetic acid and ethanol (1:3 v/v) overnight. The fixation solution was exchanged to a 150 mM K2HPO4 solution before callose staining with 0.1% aniline blue in the K2HPO4 solution for 2 h. The data presented are representative of three independent experiments (Figures 1A,C, 2A, and 3A), two independent experiments (Figures 1B and 6D; Figures S1, S2) and one experiment (Figure 7A; Figure S3). To localize the root meristematic regions, the primary roots were stained with 20 μg/mL propidium iodide (PI). Callose or PI signals were examined under microscopy using a fluorescence microscope DMI 6000B (Leica Microsystems, Wetzlar, Germany) and a confocal microscope SP8 (Leica Microsystems, Wetzlar, Germany). A 405 nm excitation laser and collection emission 430–515 nm spectrum were used for detecting Aniline blue fluorescence. A 561 nm excitation laser and collection emission spectrum 600–640 nm were used for detecting PI fluorescence.
Quantitative real‐time PCR analysis
Three‐day‐old seedlings were exposed to low Pi or normal Pi media for 4 days. Total RNA was then extracted from the root tissues by NucleoSpin® RNA Plant kit (Machete‐Nagel, Dueren, Germany). Total RNA was reverse‐transcribed using the PrimeScript RT reagent kit including a genomic DNA eraser (Perfect Real Time, Takara Bio, Ohtsu, Japan). Quantitative PCR was performed with SYBR Green (Life Technologies, Carlsbad, CA, USA) and the Thermal Cycler Dice® Real‐Time System (TakaraBio). Expression levels of the examined genes (Table S2) were normalized relative to a reference gene, ACTIN 2 (ACT2). PSR‐inducible markers examined were AT4 (At5g03545), IPS1 (AT3g09922), PHT1;5 (AT2g32830), and SPX1 (AT5g20150).
Data are representative of two independent experiments.
Identification of mutants altered in the root hair callose deposition
We examined callose deposition in gsl mutants (Table S1) after aniline blue staining of the roots transferred to the low Pi liquid media (50 μM) for 3 days. In the EMS mutagenic screening, we subjected a region ~5 cm from the root tip to aniline blue staining, after cutting out of the M2 individuals transferred to the Pi ‐depleted liquid media (0 μM) for 3 days. This enabled us to keep the plant alive for subsequent characterization and seed propagation. The PMR4 genomic region was sequenced in the selected mutant candidates.
Inoculation analyses with powdery mildew fungi
Inoculation of Golovinomyces orontii and determination of fungal versus plant DNA ratio was conducted essentially by following Weßling and Panstruga (2012). Eighteen‐days‐old plants grown in 10 h: 14 h, light: dark cycles at 22°C were inoculated with G. orontii spores at ~ 750 spores cm−2. The inoculated leaves were harvested and frozen in liquid nitrogen before determining fungal biomass at 10 days after inoculation. Genomic DNA was extracted essentially by following Brouwer et al. (2003). Quantitative real‐time PCR was performed with primers (Table S2) and Brilliant SYBR Green QPCR Reagent Kit (Stratagene, Waldbronn, Germany). The ratio of G. orontii to Arabidopsis genomic DNA was calculated using the ΔΔCt method (Pfaffl, 2001). Data are representative of two independent experiments (Figure 2C).
Determination of root hair density, primary root length and root area
Six‐day‐old seedlings were transferred and further grown in the low (50 μM) or normal Pi media (625 μM) for 6 days, and then root hairs, primary root, and root system area were examined under stereomicroscope M80 (Leica Microsystems, Wetzlar, Germany). The number of root hairs was determined in a region 2.0–3.0 mm from the root tip using Image J software (National Institutes of Health, Bethesda, MD, USA). Data are representative of similar results from three independent experiments (Figure 3C,D) and two independent experiments (Figure S4). The primary root length and convex hull area were determined using ImageJ software and Root nav software (Pound et al., 2013). Data are representative of three independent experiments (Figure 3E–H). For growth under different Boron (B) conditions, plants were grown in MGRL solid media containing 1% sucrose and 1% gellan gum (WAKO, Japan). B concentrations were adjusted with boric acid. Plants were grown under 16 h: 8 h, light: dark cycles at 22°C. The data presented are from one independent experiment (Figure S10).
Determination of shoot fresh weight, anthocyanin and pi contents
In plant growth assays on media, 6‐day‐old seedlings were transferred to the indicated media (10 μM, 50 μM, or 625 μM Pi) and then further grown for the indicated days. SFW was determined 16 days after transfer to the indicated media (50 μM or 625 μM Pi; Figure 3I,J). In plant growth assays with pLRX1::PMR4 lines, SFW was determined 10 days after the transfer to the low Pi media (10 μM:Figure 9A,C). In plant growth assay on soil, organic black soil (Maruishi Engei, Osaka, Japan) and Ezo sand (Kobayashi Sangyo, Kita‐Nagoya, Japan) mixed at 1:1 (v/v) were used as low Pi soil, and Sakata‐Super‐Mix‐A (Sakata Seed, Kyoto, Japan) and Vermiculite G20 (Maruishi Engei, Osaka, Japan) mixed at 1:1 (v/v) were used as normal Pi soil (Figure S8). SFW of seedlings grown on soil was determined 14 days after the transfer of 10‐days‐old seedlings to the low and normal Pi soils. Data are representative of two independent experiments (Figure 3I,J; Figure S5) and one independent experiment (Figure 9B,C).
In the determination of anthocyanin content, 6‐day‐old seedlings were transferred to and further grown on low Pi media (10 μM) for 10 days (Figure 3K,L). The shoots were subject to the procedure described in Mita et al. (1997). In brief, a HCl: methanol mixture at 1:99 (v/v) was added to the ground tissues with 1 mL per sample, and then the resultant homogenates were incubated at 4°C overnight. A 530 and A 657 of the supernatants were determined with a spectrometer UV‐1800 (Shimadzu, Kyoto, Japan). Anthocyanin content was calculated following the formula: (A530−0.25 × A657) × fresh weight (FW)−1. Data are representative of two independent experiments with the same conclusions (Figure 3K,L).
In the determination of Pi content, 6‐days‐old seedlings were transferred to and further grown on the low Pi (150 μM) or normal Pi (625 μM) media for 9 days (Figures 4, 5A,B, and 6A,B). Pi content was determined by following Ames (1966). In brief, the extraction buffer (10 mM Tris, 1 mM EDTA, 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride pH 8.0) was added with 10 μL per sample mg FW. The homogenate 100 μL was mixed with 1% glacial acetic acid 900 μL, and then incubated at 42°C for 30 min. The supernatant was recovered to be incubated in a solution (0.35% NH4MoO4, 0.86 N H2SO4, and 1.4% ascorbic acid) at 42°C for 30 min. A 820 of the aliquot was measured under Eppendorf BioSpectrometer® basic (Eppendorf, Hamburg, Germany). Then, Pi content was calculated by a P calibration curve prepared with a P standard solution. Data are representative of three independent experiments (Figure 4A,B) and two independent experiments (Figures 5D and 6D) with the same conclusions.
Scanning electron microscope
The maturation zone of primary roots 10 days after transfer of 5‐days‐old seedlings to low Pi (50 μM) liquid media was fixed in half‐Karnovsky's fixative overnight at 4°C, and then postfixed in 1% osmium tetroxide for 1 h at room temperatures (Karnovsky, 1964). The specimens were dehydrated in an ethanol series (30%, 50%, 70%, 80%, and 90%, and 100% at three times) for 10 min each at RT, and then the specimens were dipped in t‐butyl alcohol and dried with a vacuum evaporator. They were coated with vapor of 1% osmium tetroxide and then observed with a scanning electron microscope (JIB‐4601F, JEOL, Tokyo, Japan).
Inductively coupled plasma mass spectrometer (ICP‐MS) analyses
Five‐day‐old seedlings were transferred to and further grown on the growth media (10 μM or 625 μM KH2PO4) for 4 days (Figure 7; Figure S9). Shoots and roots were harvested and dried at 60°C. After determining the dry weight, the dried tissues were digested with 2 mL nitric acid and 0.5 mL hydrogen peroxide (Wako Pure Chemicals) at 110°C. Digested samples were then dissociated with 5 mL of 2% (w/v) nitric acid and subjected to inductively coupled plasma mass spectrometry (NexION 2000, Perkin‐Elmer). The contents of P, K, Mg, Ca, Mn, Fe, Zn, Mo, and B were determined. The data were obtained from one experiment (Figure 7; Figure S9).
Determination of 32P uptake rate and translocation ratio
Seven‐day‐old seedlings were transferred to and further grown on the growth media (50 or 625 μM KH2PO4) for 10 days. The seedlings were exposed to the liquid media containing 32P‐labeled Pi (32PO4 3− 20 kBq/mL) for 30 min. The plant tissues were fixed to an Imaging Plate (GE Healthcare co., Chicago, USA) for 3 h and then scanned by Typhoon FLA 9000 (GE Healthcare Co., Chicago, USA). 32P uptake rate was calculated based on a calibration curve for 32P. For determining the Pi translocation rate into shoots, the seedlings were exposed to the Pi‐containing liquid media containing 32P‐labeled Pi for 10 min, and then transferred to non‐labeled agar media for 120 min. The translocation rate of Pi was determined by calculating 32P ratio of the shoot to the whole seedlings. The data presented are representative of two independent experiments with the same conclusions (Figures 5D and 6D).
Grafting experiments
Grafting of WT and pmr4 was performed with a supportive micro‐device as described in Tsutsui et al. (2020). Four‐day‐old seedlings grown on a 1/2 MS medium containing 0.5% sucrose and 1% agar under constant light at 22°C were subject to grafting. Both hypocotyls of stock and scion were cut horizontally with a knife and assembled on the device using forceps. The grafted plants were grown in a 1/2 MS medium containing 0.5% sucrose and 2% agar at 27°C for 6 days. In callose staining, the grafted plants transferred and grown on low Pi media (50 μM) under constant light at 22°C for 9 days were examined. In plant growth assays, the grafted plants transferred and grown on low Pi media (150 μM) under constant light at 22°C for 14 days were examined. The data presented are representative of two independent experiments with the same conclusions (Figure 8).
Author Contributions
YS conceived the study. KO and YS designed the experiments. KO, KY, TANN, SK, SY, HT, CT, THL, UN, KI, NT, TI, KM, TM and MN developed the methods, performed the experiments, and/or analyzed the data. SM, MN and KH advised on the experiments. KO and YS wrote the manuscript with contributions from the other authors.
Supporting information
Figure S1. Callose deposition in gsl mutants under phosphate deficiency.
Figure S2. flg22‐induced callose deposition is impaired in the caps and pmr4 alleles.
Figure S3. Pi deficiency‐induced callose deposition under dark conditions.
Figure S4. Root hair and meristem morphology in pmr4 under Pi deficiency.
Figure S5. PSR marker gene expression in pmr4 roots.
Figure S6. PMR4 restricts LPR1/LPR2‐mediated inhibition of the primary root growth under low Pi.
Figure S7. PMR4 expression in phr1 phl1 and lpr1 lpr2 roots.
Figure S8. Plant growth is lowered in pmr4 on soil.
Figure S9. Plant element contents in shoot and root under phosphate sufficiency.
Figure S10. Primary root growth inhibition under B deficiency.
Table S1. Arabidopsis thaliana mutants used in this study.
Table S2. DNA Primers used in DNA vector construction and qPCR analyses.
Acknowledgements
We thank the Arabidopsis Biological Research Center for published plant materials, Yaichi Kawakatsu for advice on micrografting, and Michiko Tsukamoto and Mie Matsubara for technical assistance. This work was supported in part by the grants from Mitsubishi Foundation (YS), the Japan Science and Technology Agency (JST, JPMJPR13B6 and JPMJSC1702 (YS) and JPMJPR16Q7 (KH)), and MEXT of Japan 18H02467 and 21H02507 (YS).
References
- Abel, S. (2017) Phosphate scouting by root tips. Current Opinion in Plant Biology, 39, 168–177. [DOI] [PubMed] [Google Scholar]
- Ames, B.N. (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods in Enzymology, 8, 115–118. [Google Scholar]
- Ayadi, A. , David, P. , Arrighi, J.F. , Chiarenza, S. , Thibaud, M.C. , Nussaume, L. et al. (2015) Reducing the genetic redundancy of Arabidopsis phosphate transporter1 transporters to study phosphate uptake and signaling. Plant Physiology, 167, 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzergue, C. , Dartevelle, T. , Godon, C. , Laugier, E. , Meisrimler, C. , Teulon, J.M. et al. (2017) Low phosphate activates STOP1‐ALMT1 to rapidly inhibit root cell elongation. Nature Communications, 8, 15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt, D.H.P. , Kölling, K. , Graf, A. , Pike, M. , Calder, G. , Findlay, K. et al. (2010) Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol., 155(1), 328–341. Available from: 10.1104/pp.110.166330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, T.R. & Lynch, J.P. (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant, Cell & Environment, 19, 529–538. [Google Scholar]
- Baumberger, N. , Ringli, C. & Keller, B. (2001) The chimeric leucine‐rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana . Genes & Development, 15, 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat, J.F. , Rouached, H. , Tissot, N. , Gaymard, F. & Dubos, C. (2015) Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Frontiers in Plant Science, 6, 290. Available from: 10.3389/fpls.2015.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, M. , Lievens, B. , Van Hemelrijck, W. , Van Den Ackerveken, G. , Cammue, B.P.A. & Thomma, B.P.H.J. (2003) Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real‐time fluorescence PCR. FEMS Microbiology Letters, 228, 241–248. [DOI] [PubMed] [Google Scholar]
- Bustos, R. , Castrillo, G. , Linhares, F. , Puga, M.I. , Rubio, V. , Pérez‐Pérez, J. et al. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genetics, 6, e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C. , Liao, Y.Y. & Chiou, T.J. (2021) The impact of phosphorus on plant immunity. Plant & Cell Physiology, 62, 582–589. [DOI] [PubMed] [Google Scholar]
- Chien, P.S. , Chiang, C.P. , Leong, S.J. & Chiou, T.J. (2018) Sensing and signaling of phosphate starvation: from local to long distance. Plant & Cell Physiology, 59, 1714–1722. [DOI] [PubMed] [Google Scholar]
- Chowdhury, J. , Henderson, M. , Schweizer, P. , Burton, R.A. , Fincher, G.B. & Little, A. (2014) Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei . New Phytologist, 204, 650–660. [DOI] [PubMed] [Google Scholar]
- Das, A.K. & Purkait, A. (2020) Boron dynamics in soil: classification, sources, factors, fractions, and kinetics. Communications in Soil Science and Plant Analysis, 51, 2778–2790. [Google Scholar]
- Datta, S. , Kim, C.M. , Pernas, M. , Pires, N.D. , Proust, H. , Tam, T. et al. (2011) Root hairs: development, growth and evolution at the plant‐soil interface. Plant and Soil, 346, 1–14. [Google Scholar]
- Dolan, L. , Duckett, C.M. , Grierson, C. , Linstead, P. , Schneider, K. , Lawson, E. et al. (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis . Development, 120, 2465–2474. [Google Scholar]
- Dong, X. , Hong, Z. , Chatterjee, J. , Kim, S. & Verma, D.P.S. (2008) Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta, 229, 87–98. Available from: 10.1007/s00425-008-0812-3 [DOI] [PubMed] [Google Scholar]
- Dong, X. , Hong, Z. , Sivaramakrishnan, M. , Mahfouz, M. & Verma, D.P.S. (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant Journal, 42, 315–328. [DOI] [PubMed] [Google Scholar]
- Ellinger, D. & Voigt, C.A. (2014) Callose biosynthesis in arabidopsis with a focus on pathogen response: what we have learned within the last decade. Annals of Botany, 114, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns, L.C. , Kanaoka, M.M. , Torii, K.U. , Comai, L. , Okada, K. & Cleland, R.E. (2005) Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Molecular Biology, 58, 333–349. [DOI] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J.M. , Valli, A. , Todesco, M. , Mateos, I. , Puga, M.I. , Rubio‐Somoza, I. et al. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics, 39, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gao, Y.Q. , Bu, L.H. , Han, M.L. , Wang, Y.L. , Li, Z.Y. , Liu, H.T. et al. (2021) Long‐distance blue light signalling regulates phosphate deficiency‐induced primary root growth inhibition. Molecular Plant, 14, 1539–1553. [DOI] [PubMed] [Google Scholar]
- García‐Andrade, J. , Ramírez, V. , Flors, V. & Vera, P. (2011) Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen‐induced callose deposition. Plant Journal, 67, 783–794. [DOI] [PubMed] [Google Scholar]
- Gulabani, H. , Goswami, K. , Walia, Y. , Roy, A. , Noor, J.J. , Ingole, K.D. et al. (2021) Arabidopsis inositol polyphosphate kinases IPK1 and ITPK1 modulate crosstalk between SA‐dependent immunity and phosphate‐starvation responses. Plant Cell Reports, 41, 347–363. [DOI] [PubMed] [Google Scholar]
- Hamburger, D. , Rezzonico, E. , Petétot, J.M.D.C. , Somerville, C. & Poirier, Y. (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell, 14, 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, G. , Wei, X. , Dong, X. , Wang, C. , Sui, N. , Guo, J. et al. (2020) Arabidopsis ZINC FINGER PROTEIN1 acts downstream of GL2 to repress root hair initiation and elongation by directly suppressing bHLH genes. Plant Cell, 32, 206–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X. , Kui, M. , He, K. , Yang, M. , Du, J. , Jiang, Y. et al. (2023) Jasmonate‐regulated root growth inhibition and root hair elongation. Journal of Experimental Botany, 74, 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz, M. , Zarebanadkouki, M. , Kuzyakov, Y. , Pausch, J. & Carminati, A. (2018) Root hairs increase rhizosphere extension and carbon input to soil. Annals of Botany, 121, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidra‐Arellano, M.C. , Delaux, P.M. & Vald°s‐L°pez, O. (2021) The phosphate starvation response system: its role in the regulation of plant–microbe interactions. Plant & Cell Physiology, 62, 392–400. [DOI] [PubMed] [Google Scholar]
- Jacobs, A.K. , Lipka, V. , Burton, R.A. , Panstruga, R. , Strizhov, N. , Schulze‐Lefert, P. et al. (2003) An Arabidopsis Callose synthase, GSL5, is required for wound and papillary Callose formation. Plant Cell, 15, 2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, Y. , Zhao, F. , Lai, K. , Sun, F. , Sun, C. , Zou, X. et al. (2024) Plant elicitor peptides regulate root hair development in Arabidopsis . Frontiers in Plant Science, 15, 1336129. Available from: 10.3389/fpls.2024.1336129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky, M.J. (1964) A formaldehyde‐glutaraldehyde fixative of high osmolality for use in electron microscopy. The Journal of Cell Biology, 27, 1A–149A. Available from: https://www.researchgate.net/publication/244955881 [Google Scholar]
- Keppler, B.D. , Song, J. , Nyman, J. , Voigt, C.A. & Bent, A.F. (2018) 3‐aminobenzamide blocks MAMP‐induced callose deposition independently of its poly(ADPribosyl)ation inhibiting activity. Frontiers in Plant Science, 871, 1907. Available from: 10.3389/fpls.2018.01907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T.Y. , Huang, T.K. , Tseng, C.Y. , Lai, Y.S. , Lin, S.I. , Lin, W.Y. et al. (2012) PHO2‐dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis . Plant Cell, 24, 2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo, A. , Funck, D. , Mühlenbock, P. , Kular, B. , Mullineaux, P.M. & Karpinski, S. (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. Journal of Experimental Botany, 57, 1795–1807. [DOI] [PubMed] [Google Scholar]
- Mita, S. , Murano, N. , Akaike, M. & Nakamura, K. (1997) Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for β‐amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant Journal, 11, 841–851. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Lee, J. , Miura, T. & Hasegawa, P.M. (2010) SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant & Cell Physiology, 51, 103–113. [DOI] [PubMed] [Google Scholar]
- Miwa, K. , Wakuta, S. , Takada, S. , Ide, K. , Takano, J. , Naito, S. et al. (2013) Roles of BOR2, a boron exporter, in cross linking of rhamnogalacturonan II and root elongation under boron limitation in Arabidopsis. Plant Physiology, 163, 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora‐Macías, J. , Ojeda‐Rivera, J.O. , Gutiérrez‐Alanís, D. , Yong‐Villalobos, L. , Oropeza‐Aburto, A. , Raya‐González, J. et al. (2017) Malate‐dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proceedings of the National Academy of Sciences of the United States of America, 114, E3563–E3572. Available from: 10.1073/pnas.1701952114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo, R.J. , Singh, S.K. , He, D. , An, G. , Vílchez, J.I. , Tang, K. et al. (2020) Rhizobacterium‐derived diacetyl modulates plant immunity in a phosphate‐dependent manner. The EMBO Journal, 39, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J. , Toev, T. , Heisters, M. , Teller, J. , Moore, K.L. , Hause, G. et al. (2015) Iron‐dependent Callose deposition adjusts root meristem maintenance to phosphate availability. Developmental Cell, 33, 216–230. [DOI] [PubMed] [Google Scholar]
- Nagarajan, V.K. , Jain, A. , Poling, M.D. , Lewis, A.J. , Raghothama, K.G. & Smith, A.P. (2011) Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiology, 156, 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. et al. (2007) Improved gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, Biotechnology, and Biochemistry, 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nieves‐Cordones, M. , Lara, A. , Ródenas, R. , Amo, J. , Rivero, R.M. , Martínez, V. et al. (2019) Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants. Plant, Cell & Environment, 42, 2357–2371. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S.I. , Zinkl, G.M. , Swanson, R.J. , Maruyama, D. & Preuss, D. (2005) Callose (β‐1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biology, 5, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T. , Stein, M. , Hou, B.H. , Vogel, J.P. , Edwards, H. & Somerville, S.C. (2003) Loss of a callose synthase results in salicylic acid‐dependent disease resistance. Science, 301, 969–972. [DOI] [PubMed] [Google Scholar]
- Okada, K. , Kubota, Y. , Hirase, T. , Otani, K. , Goh, T. , Hiruma, K. et al. (2021) Uncoupling root hair formation and defence activation from growth inhibition in response to damage‐associated pep peptides in Arabidopsis thaliana . New Phytologist, 229, 2844–2858. [DOI] [PubMed] [Google Scholar]
- O'Lexy, R. , Kasai, K. , Clark, N. , Fujiwara, T. , Sozzani, R. & Gallagher, K.L. (2018) Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. Journal of Experimental Botany, 69, 3715–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits, U. , Ahrens, C.H. , Müller, S. & Wollscheid, B. (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics, 30, 884–886. [DOI] [PubMed] [Google Scholar]
- Péret, B. , Clément, M. , Nussaume, L. & Desnos, T. (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science, 16, 442–450. [DOI] [PubMed] [Google Scholar]
- Péret, B. , Desnos, T. , Jost, R. , Kanno, S. , Berkowitz, O. & Nussaume, L. (2014) Root architecture responses: in search of phosphate. Plant Physiology, 166, 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research, 29, 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier, Y. , Thoma, S. , Somerville, C. & Schiefelbein, J. (1991) A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiology, 97, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, J. , Day, C.J. , Von Itzstein, M. , Paton, J.C. & Jennings, M.P. (2018) Glycointeractions in bacterial pathogenesis. Nature Reviews. Microbiology, 16, 440–452. Available from: 10.1038/s41579-018-0007-2 [DOI] [PubMed] [Google Scholar]
- Pound, M.P. , French, A.P. , Atkinson, J.A. , Wells, D.M. , Bennett, M.J. & Pridmore, T. (2013) RootNav: navigating images of complex root architectures. Plant Physiology, 162, 1802–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga, M.I. , Mateos, I. , Charukesi, R. , Wang, Z. , Franco‐Zorrilla, J.M. , De Lorenzo, L. et al. (2014) SPX1 is a phosphate‐dependent inhibitor of phosphate starvation response 1 in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 111, 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robe, K. , Conejero, G. , Gao, F. , Lefebvre‐Legendre, L. , Sylvestre‐Gonon, E. , Rofidal, V. et al. (2021) Coumarin accumulation and trafficking in Arabidopsis thaliana: a complex and dynamic process. New Phytologist, 229, 2062–2079. [DOI] [PubMed] [Google Scholar]
- Robertson‐Albertyn, S. , Terrazas, R.A. , Balbirnie, K. et al. (2017) Root hair mutations displace the barley rhizosphere microbiota. Frontiers in Plant Science, 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, V. , Linhares, F. , Solano, R. , Martín, A.C. , Iglesias, J. , Leyva, A. et al. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development, 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager, R. & Lee, J.Y. (2014) Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. Journal of Experimental Botany, 65, 6337–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, S. & Schikora, A. (2015) Staining of callose depositions in root and leaf tissues. Bio‐Protocol, 5(6), e1429. Available from: 10.21769/bioprotoc.1429 [DOI] [Google Scholar]
- Schiefelbein, J. , Kwak, S.H. , Wieckowski, Y. , Barron, C. & Bruex, A. (2009) The gene regulatory network for root epidermal cell‐type pattern formation in Arabidopsis . Journal of Experimental Botany, 60, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H. , Shin, H.S. , Dewbre, G.R. & Harrison, M.J. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low‐ and high‐phosphate environments. Plant Journal, 39, 629–642. [DOI] [PubMed] [Google Scholar]
- Stefanovic, A. , Ribot, C. , Rouached, H. , Wang, Y. , Chong, J. , Belbahri, L. et al. (2007) Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant Journal, 50, 982–994. [DOI] [PubMed] [Google Scholar]
- Svistoonoff, S. , Creff, A. , Reymond, M. , Sigoillot‐Claude, C. , Ricaud, L. , Blanchet, A. et al. (2007) Root tip contact with low‐phosphate media reprograms plant root architecture. Nature Genetics, 39, 792–796. [DOI] [PubMed] [Google Scholar]
- Tanaka, N. , Kato, M. , Tomioka, R. , Kurata, R. , Fukao, Y. , Aoyama, T. et al. (2014) Characteristics of a root hair‐less line of Arabidopsis thaliana under physiological stresses. Journal of Experimental Botany, 65, 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud, M.C. , Arrighi, J.F. , Bayle, V. , Chiarenza, S. , Creff, A. , Bustos, R. et al. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant Journal, 64, 775–789. [DOI] [PubMed] [Google Scholar]
- Tsutsui, H. , Yanagisawa, N. , Kawakatsu, Y. , Ikematsu, S. , Sawai, Y. , Tabata, R. et al. (2020) Micrografting device for testing systemic signaling in Arabidopsis. Plant J., 103, 918–929. Available from: 10.1111/tpj.14768 [DOI] [PubMed] [Google Scholar]
- Verbon, E.H. , Liberman, L.M. , Zhou, J. , Yin, J. , Pieterse, C.M.J. , Benfey, N. et al. (2023) Cell‐type‐specific transcriptomics reveals that root hairs and endodermal barriers play important roles in beneficial plant‐rhizobacterium interactions. Molecular Plant, 16, 1160–1177. Available from: 10.1016/j.molp.2023.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg, K. , Claeijs, N. , Balcerowicz, D. & Schoenaers, S. (2020) Hormonal regulation of root hair growth and responses to the environment in Arabidopsis. Journal of Experimental Botany, 71, 2412–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J. & Somerville, S. (2000) Isolation and characterization of powdery mildew‐resistant Arabidopsis mutants. Proceedings of the National Academy of Sciences of the United States of America, 97, 1897–1902. Available from: 10.1073/pnas.030531997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Du, G. , Wang, X. , Meng, Y. , Li, Y. , Wu, P. et al. (2010) The function of LPR1 is controlled by an element in the promoter and is independent of SUMO E3 ligase SIZ1 in response to low pi stress in Arabidopsis thaliana . Plant & Cell Physiology, 51, 380–394. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Li, X. , Fan, B. , Zhu, C. & Chen, Z. (2021) Regulation and function of defense‐related callose deposition in plants. International Journal of Molecular Sciences, 22, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Zheng, Z. , Song, L. & Liu, D. (2018) Functional characterization of Arabidopsis PHL4 in plant response to phosphate starvation. Frontiers in Plant Science, 9, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling, R. & Panstruga, R. (2012) Rapid quantification of plant‐powdery mildew interactions by qPCR and conidiospore counts. Plant Methods, 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. & Ausubel, F.M. (2002) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 417, 571. [DOI] [PubMed] [Google Scholar]
- Wu, S.W. , Kumar, R. , Iswanto, A.B.B. & Kim, J.Y. (2018) Callose balancing at plasmodesmata. Journal of Experimental Botany, 69, 5325–5339. [DOI] [PubMed] [Google Scholar]
- Xie, B. & Hong, Z. (2011) Unplugging the callose plug from sieve pores. Plant Signaling & Behavior, 6, 491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, C. , Li, W. , Shen, R. & Lan, P. (2022) Impacts of iron on phosphate starvation‐induced root hair growth in Arabidopsis. Plant, Cell & Environ., 46(1), 215–238. Available from: 10.1111/pce.14451 [DOI] [PubMed] [Google Scholar]
- Yeh, C.M. , Kobayashi, K. , Fujii, S. , Fukaki, H. , Mitsuda, N. & Ohme‐Takagi, M. (2020) Blue light regulates phosphate deficiency‐dependent primary root growth inhibition in Arabidopsis. Frontiers in Plant Science, 10, 1803. Available from: 10.3389/fpls.2019.01803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Chen, S. & Mou, Z. (2010) Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis . Journal of Plant Physiology, 167, 144–148. Available from: 10.1016/j.jplph.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Zheng, Z. , Wang, Z. , Wang, X. & Liu, D. (2019) Blue light‐triggered chemical reactions underlie phosphate deficiency‐induced inhibition of root elongation of Arabidopsis seedlings grown in petri dishes. Molecular Plant, 12, 1515–1523. Available from: 10.1016/j.molp.2019.08.001 [DOI] [PubMed] [Google Scholar]
- Zhu, C. , Gan, L. , Shen, Z. & Xia, K. (2006) Interactions between jasmonates and ethylene in the regulation of root hair development in Arabidopsis . Journal of Experimental Botany, 57, 1299–1308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Callose deposition in gsl mutants under phosphate deficiency.
Figure S2. flg22‐induced callose deposition is impaired in the caps and pmr4 alleles.
Figure S3. Pi deficiency‐induced callose deposition under dark conditions.
Figure S4. Root hair and meristem morphology in pmr4 under Pi deficiency.
Figure S5. PSR marker gene expression in pmr4 roots.
Figure S6. PMR4 restricts LPR1/LPR2‐mediated inhibition of the primary root growth under low Pi.
Figure S7. PMR4 expression in phr1 phl1 and lpr1 lpr2 roots.
Figure S8. Plant growth is lowered in pmr4 on soil.
Figure S9. Plant element contents in shoot and root under phosphate sufficiency.
Figure S10. Primary root growth inhibition under B deficiency.
Table S1. Arabidopsis thaliana mutants used in this study.
Table S2. DNA Primers used in DNA vector construction and qPCR analyses.


