Abstract
Glial fibrillary acidic protein (GFAP) is a well‐established biomarker of reactive astrogliosis in the central nervous system because of its elevated levels following brain injury and various neurological disorders. The advent of ultra‐sensitive methods for measuring low‐abundant proteins has significantly enhanced our understanding of GFAP levels in the serum or plasma of patients with diverse neurological diseases. Clinical studies have demonstrated that GFAP holds promise both as a diagnostic and prognostic biomarker, including but not limited to individuals with Alzheimer's disease. GFAP exhibits diverse forms and structures, herein referred to as its proteoform complexity, encompassing conformational dynamics, isoforms and post‐translational modifications (PTMs). In this review, we explore how the proteoform complexity of GFAP influences its detection, which may affect the differential diagnostic performance of GFAP in different biological fluids and can provide valuable insights into underlying biological processes. Additionally, proteoforms are often disease‐specific, and our review provides suggestions and highlights areas to focus on for the development of new assays for measuring GFAP, including isoforms, PTMs, discharge mechanisms, breakdown products, higher‐order species and interacting partners. By addressing the knowledge gaps highlighted in this review, we aim to support the clinical translation and interpretation of GFAP in both CSF and blood and the development of reliable, reproducible and specific prognostic and diagnostic tests. To enhance disease pathology comprehension and optimise GFAP as a biomarker, a thorough understanding of detected proteoforms in biofluids is essential.
Keywords: biology, biomarker, GFAP, immunoassay, proteoform, structure
Glial fibrillary acidic protein (GFAP) is a well‐established biomarker of reactive astrogliosis in the central nervous system. In this review, we explore how the physicochemical characteristics of GFAP may influence its detection, which may underlie the differential diagnostic performance of GFAP in different biological fluids and can provide valuable insights into underlying biological processes. We summarise how knowledge of GFAP biology and proteoforms, including isoforms, post‐translational modifications, discharge mechanisms, breakdown products, higher‐order species and interacting partners, can be utilised to guide future biomarker test development and enhance disease pathology comprehension.
Abbreviations
- 3D
3‐dimensional
- AA
amino acid
- aCSF
artificial cerebrospinal fluid
- AD
Alzheimer's disease
- ALS
amyotrophic lateral sclerosis
- APP
amyloid precursor protein
- AUC‐ROC
area under the curve of the receiver operating characteristic curve
- AxD
alexander disease
- BBB
blood–brain barrier
- BTI
brain trauma indicator
- CNS
central nervous system
- CSF
cerebrospinal fluid
- DLB
dementia with lewy bodies
- EV
extracellular vesicle
- FT
freeze–thaw
- FTD
frontotemporal dementia
- FTLD
frontotemporal lobar degeneration
- GFAP
glial fibrillary acidic protein
- HD
Huntington's disease
- HPA
human protein Atlas
- IF
intermediate filament
- KPBS
potassium phosphate buffer saline
- MS
multiple sclerosis
- MSD
Meso Scale Discovery
- NfL
neurofilament light
- PAD2
peptidylarginine deaminase 2
- PD
Parkinson's disease
- PET
positron emission tomography
- PTM
post‐translational modification
- sHSP
small heat shock protein
- TBI
traumatic brain injury
1. INTRODUCTION
Glial fibrillary acidic protein (GFAP) is an essential component of the cytoplasmic intermediate filament cytoskeleton in astrocytes, facilitating structural integrity, motility, signal transduction and cell homeostasis (Abdelhak et al., 2018; Emirandetti et al., 2006; Kawajiri et al., 2003; Lowery et al., 2015; Messing et al., 1998; Rutka et al., 1994; Yoshida et al., 2007). Following injury, disease or infection of the central nervous system (CNS), levels of GFAP increase (Abdelhak et al., 2022; Heimfarth et al., 2022; Messing & Brenner, 2020; Mondello et al., 2021). In the context of brain injury and various CNS pathologies, astrocytes undergo significant morphological, molecular and functional changes and are termed as ‘reactive astrocytes’ (Escartin et al., 2021). GFAP is, therefore, a widely used biofluid‐ and tissue‐based biomarker of reactive astrogliosis in the CNS since its expression in the brain is astrocyte‐specific and strictly regulated after damage and during disease (Colangelo et al., 2014; Eddleston & Mucke, 1993; Middeldorp & Hol, 2011).
GFAP protein levels can be measured within a detectable range in human biofluids. The measurement of GFAP as a blood‐based biomarker was facilitated by the advent of ultra‐sensitive technologies to detect proteins at biologically relevant concentrations, resulting so far in a large number of studies examining levels of GFAP in clinical samples from patients with different neurological diseases (Abdelhak et al., 2022; Ishiki et al., 2016; Oeckl et al., 2022; Teunissen et al., 2022).
Given that cerebrospinal fluid (CSF) is in direct contact with the brain, it is considered to more accurately and acutely reflect neuropathological changes compared to blood (Aluise et al., 2008). Moreover, peripheral protein sources and transport of brain‐derived proteins across the blood–brain barrier may hamper biomarker detection and result in smaller measurable fold‐changes compared to those measured in CSF (Olsson et al., 2016; Schindler et al., 2019). Thus, brain‐specific proteins, such as GFAP, are generally expected to exhibit better performance as biomarkers in CSF compared to blood (Palmqvist et al., 2020; Simrén et al., 2022). However, for GFAP this is not so clear cut.
The diagnostic value of GFAP varies across biological fluids and neurological diseases. For instance, CSF GFAP shows superior diagnostic performance for Alexander disease (AxD) compared to plasma GFAP (Jany et al., 2015; Kyllerman et al., 2005; Schmidt et al., 2013). A particularly striking finding is that GFAP measured in plasma has a better discriminative performance to distinguish between individuals with and without amyloid pathology across the Alzheimer's disease (AD) clinical continuum compared to GFAP measured in CSF (Baiardi et al., 2022; Benedet et al., 2021; Simrén et al., 2022). Additionally, serum GFAP shows a stronger negative correlation with mini‐mental state examination scores compared to CSF GFAP in a cohort of patients with different types of dementia (Oeckl et al., 2019). These discrepancies between CSF and blood measurements, as well as the secretion mechanism of GFAP from astrocytes to these matrices, are not fully understood.
Despite the increasing scientific interest in GFAP, the implications of its proteoforms are largely unknown. The proteoform properties of GFAP, such as its 3‐dimensional (3D) structure, discharge mechanisms into different body fluids, breakdown products, intermediate filament network and post‐translational modifications (PTMs) may affect its ability to be detected in different matrices. The highly flexible nature of GFAP and the potential impact of its proteoforms on clinical assays emphasise the importance of targeted strategies.
In this article, we aim to provide a comprehensive overview of protein characteristics and highlight knowledge gaps and respective shortcomings of available biomarker tests for GFAP detection and quantification. Based on these, we suggest future directions to improve the understanding of GFAP as a biomarker for various brain disorders, which could improve its clinical utility. To set the stage we first summarise the biology and structural properties of GFAP, and address key findings concerning its diagnostic and prognostic value as a biomarker measured in CSF, serum and plasma. The main section of this review delves into the current understanding and complexity of the protein structure, its physio‐chemical characteristics, and solvent accessibility. We discuss the properties of assays used to detect GFAP, and how antibody attributes translate to measurable levels of GFAP in biofluids. We demonstrate how proteoforms have been successfully utilised for the detection of other neurological biomarkers. Additionally, we provide hypotheses on how these challenges can be overcome and subsequent recommendations for the future development of robust assays targeting specific proteoforms of GFAP.
2. GFAP BIOLOGY
GFAP is a signature intermediate filament (IF) type III protein for astrocytes (Yang & Wang, 2015), but is also expressed in peripheral glia (Kato et al., 1990), enteric glia (Grundmann et al., 2019) and Schwann cells (Hainfellner et al., 2001). The expression of GFAP is higher in white matter compared to grey matter astrocytes, therefore GFAP is highly expressed in regions rich in white matter, such as the medulla oblongata and the hypothalamus (Figure 1a). The amino acid (AA) sequence of GFAP is similar to other IF proteins with a shared central α‐helical (rod) domain flanked by the disordered amino‐ (head) and carboxy‐terminal (tail) domains that largely vary in AA sequence (Chernyatina et al., 2015). GFAP emerged early in the evolution of vertebrates and shows a high degree of conservation across species, with 90% identity between humans and mice and 67% identity between humans and zebrafish (Messing & Brenner, 2020; Nielsen & Jørgensen, 2003). Under physiological conditions, type III IFs assemble into large oligomers that can be visualised by electron microscopy (Parry & Steinert, 1999). A proposed multistep mechanism involves the parallel interactions of monomers through the coiled‐coil region, followed by an antiparallel association of dimers through the core rod domain (composed of four coils; 1A, 1B, 2A, and 2B), leading to the lateral association of tetramers to form octamers. These octamers then aggregate into mature filament structures containing 30–59 monomers in cross‐section (Messing & Brenner, 2020; Parry & Steinert, 1999). A type III IF protein—vimentin, which is a more flexible homologue of GFAP (Kim et al., 2018), serves as a prototypical model for the assembly of other proteins within this family. Similar to GFAP, vimentin expression is up‐regulated in reactive astrocytes (Ridet et al., 1997) and it has similarly been shown to form an antiparallel tetramer structure (Chernyatina et al., 2012). Crystal structures of the GFAP rod 1B domain have revealed a homotetramer architecture, composed of two parallel coiled coils stabilised by salt bridges and hydrophobic interactions (Figure 2a; Kim et al., 2018). However, the rod 1B domain represents only a fraction of the entire GFAP protein, and the native assembly of GFAP remains elusive (Figure 2b). Cryo‐electron tomography experiments have revealed the structure of polymerised vimentin filaments, which are comprised of five protofibrils each having 40 polypeptide chains in cross‐section (Eibauer et al., 2021). Although the 3D structure of GFAP is still not fully understood, a recent study has indicated that GFAP exists in various conformational species and that its dimer structure remains intact under strong denaturing conditions (Gogishvili et al., 2023; Figure 2c). This suggests that GFAP's structural flexibility under different conditions may play a role in its surface accessibility and ultimately function.
FIGURE 1.
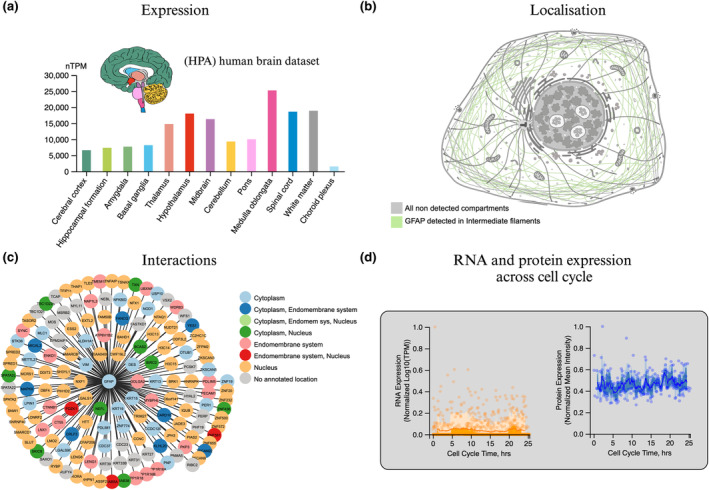
Biology of GFAP curated by Human Protein Atlas (HPA). GFAP is a highly dynamic structural protein involved in a plethora of biological processes, including but not limited to maintaining the integrity of the blood–brain barrier (BBB; Liedtke et al., 1996). (a) Brain‐specific expression of GFAP based on RNA consensus dataset consists of normalised expression levels of 13 brain regions (Uhlén et al., 2015). (b) GFAP localisation characterised by presence in all tested cells (Thul et al., 2017). (c) Interaction summary network of GFAP. The thickness of the edges represents the confidence of the interaction and nodes are coloured according to subcellular location. (d) GFAP RNA and protein expression are highly regulated during the cell cycle as GFAP is essential for the remodelling of glial frameworks in mitosis (Kawajiri et al., 2003; Messing et al., 1998; Rutka et al., 1994; Yoshida et al., 2007). The RNA expression level was determined by single‐cell RNA sequencing of the U‐2 OS FUCCI cell line. This cell line is a variant of the human cervical carcinoma cell line HeLa. Protein expression was determined by indirect immunofluorescence microscopy in the U‐2 OS FUCCI cell line. Normalised RNA and protein expression in individual cells is plotted along a linear representation of cell cycle pseudotime, as determined from the fluorescence intensities of the cell cycle markers (Karlsson et al., 2021).
FIGURE 2.
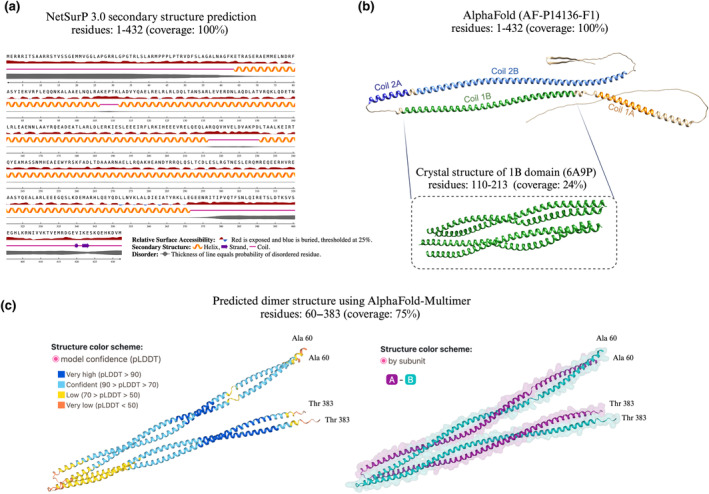
Structural characteristics of mono‐ and dimeric GFAP. Our understanding of the 3‐dimensional (3D) structure of GFAP is limited. (a) The predicted secondary structural components of GFAP using NetSurfP 3.0 covering 100% of the full‐length GFAP AA sequence (Høie et al., 2022; Klausen et al., 2019). (b) The 3D structure of GFAP predicted by AlphaFold covering 100% of the full‐length protein sequence composed of four coils: 1A, 1B, 2A, 2B (Jumper et al., 2021). Below the AlphaFold structure, the X‐ray PDB structure of the 1B domain of GFAP is displayed and determined to form a homotetramer covering 24% of the protein sequence (Kim et al., 2018). (c) The predicted dimer structure of recombinant GFAP using AlphaFold‐Multimer (Evans et al., 2021; Gogishvili et al., 2023) visualised with the PAE viewer tool (Elfmann & Stülke, 2023). Data obtained from hydrogen‐deuterium exchange measurements support the existence of this structure (Gogishvili et al., 2023).
Like other type III IF proteins, GFAP is a highly dynamic structural protein involved in the formation of the cytoskeleton (Figure 1b). Having a large interactome (Figure 1c), GFAP is involved in various cellular processes, including (i) cell motility and migration (Yoshida et al., 2007), (ii) remodelling glial frameworks in mitosis, essential for cell proliferation, during which GFAP RNA and protein expression are highly regulated (Figure 1d; Kawajiri et al., 2003; Messing et al., 1998; Rutka et al., 1994; Yoshida et al., 2007), (iii) exocytosis and vesicle mobility (Potokar et al., 2007), (iv) synapse formation (Emirandetti et al., 2006), neuronal plasticity (Emirandetti et al., 2006), neurite outgrowth (Rozovsky et al., 2002) and neuronal sprouting (Finch, 2003), (v) the maintenance of CNS myelination (Giménez y Ribotta et al., 2000; Liedtke et al., 1996), and (vi) maintaining the integrity of the blood–brain barrier (BBB; Liedtke et al., 1996). Multiple studies have demonstrated that GFAP and vimentin knockout mice are more susceptible to severe long‐term consequences following brain injury, such as ischemic brain damage (Nawashiro et al., 1998, 2000), demonstrating the protective astrocytic function related to GFAP.
3. GFAP AS A BIOMARKER IN BRAIN DISORDERS
The activation of common inflammatory pathways is linked to early stages of neuropathological processes (Colangelo et al., 2014). Astrogliosis—glial activation, proliferation (present in acute damage), and increased GFAP expression were shown to be important to recover from initial damage during CNS injury. Nevertheless, such processes can become harmful in severe stress conditions (Kumar et al., 2021). Following acute damage such as spinal cord and traumatic brain injury (TBI), GFAP is up‐regulated immediately, and has been shown to reach a peak in the blood at 20 h following TBI (Papa et al., 2016, 2023), and a peak in both CSF and blood during the first 24–36 h after spinal cord injury (Kwon et al., 2010; Leister et al., 2023), subsequent to which levels decrease. In the context of chronic CNS injury, such as dementia, elevation of plasma GFAP levels begins 10–20 years prior to the onset of symptoms and neurodegeneration, and this rise in GFAP concentration continues throughout the dementia continuum (Chatterjee et al., 2023; Guo et al., 2024; Montoliu‐Gaya et al., 2023). Because of this early and sustained increase in GFAP levels, plasma GFAP has excellent prognostic value for conversion to dementia (Verberk et al., 2020). This difference in the time course of GFAP in acute compared to chronic events may be a reflection of the molecular and functional astrocytic changes in response to acute neuronal injury compared to chronic neurodegenerative disease pathology.
Different aspects of reactive astrogliosis and distinct subtypes of astrocytes may also underlie the difference in measurement of astrocytes in the living brain by positron emission tomography (PET) compared to using GFAP as a fluid biomarker. 11C‐DED is the gold‐standard PET radiotracer for imaging reactive astrogliosis and is a selective inhibitor of monoamine oxidase type B, expression of which increases in reactive astrocytes (Ekblom et al., 1993). A negative association was observed between plasma GFAP and 11C‐DED binding in autosomal dominant and sporadic AD brains (Chiotis et al., 2023). As such, it is proposed that 11C‐DED binding may reflect a ‘first‐wave’ of reactive astrogliosis, potentially in response to pre‐plaque soluble amyloid, whereas GFAP measured in biofluids may reflect more advanced amyloid pathology in AD progression, and thus a later reactive astrogliosis process (Fontana et al., 2023). Measurement of plasma GFAP and 11C‐DED binding in the context of other neurodegenerative diseases can help elucidate the relationship between these measures, functional astrocytic changes and other types of neuropathology.
AxD is a rare disorder specific to astrocytes neuropathologically defined by Rosenthal fibres, which are aggregates of GFAP. This disease is caused by de novo mutations in the gene encoding GFAP, the majority of which are coding for regions located in the central rod domain (69–377 AA) of the protein (Messing, 2018; Figure 3b). CSF GFAP levels are increased in AxD patients compared to controls (Jany et al., 2015; Kyllerman et al., 2005; Schmidt et al., 2013). The same effect has not been demonstrated for blood‐based GFAP, which showed no significant difference between controls, infantile‐, juvenile‐ and adult‐onset AxD patients, although this has only been investigated in one study to date (Jany et al., 2015). Reactive astrogliosis has been linked to many other CNS diseases, such as AD, Parkinson's disease (PD), frontotemporal dementia (FTD), amyotrophic lateral sclerosis (ALS), dementia with Lewy bodies (DLB), multiple sclerosis (MS), Huntington's disease (HD), and glioma (Glass et al., 2010; Jiwaji & Hardingham, 2022; van Asperen, Fedorushkova, et al., 2022). Elevated GFAP levels have, therefore, been found in the CSF of patients with various neurodegenerative diseases compared to controls (Axelsson et al., 2011; Oeckl et al., 2019). Blood‐based GFAP generally displays a similar pattern, with increases shown in AD, PDD, DLB and FTD cases compared to controls (Tang et al., 2023; Thijssen et al., 2022), and serum GFAP has been shown to distinguish between MS phenotypes (Ayrignac et al., 2020; Högel et al., 2020). In addition to being a promising diagnostic biomarker for various neurodegenerative diseases, GFAP can also be utilised for prognostic applications: rate of cognitive decline and higher risk of conversion to dementia (Benedet et al., 2021; Cicognola et al., 2021; Cullen et al., 2021; Verberk et al., 2020).
FIGURE 3.
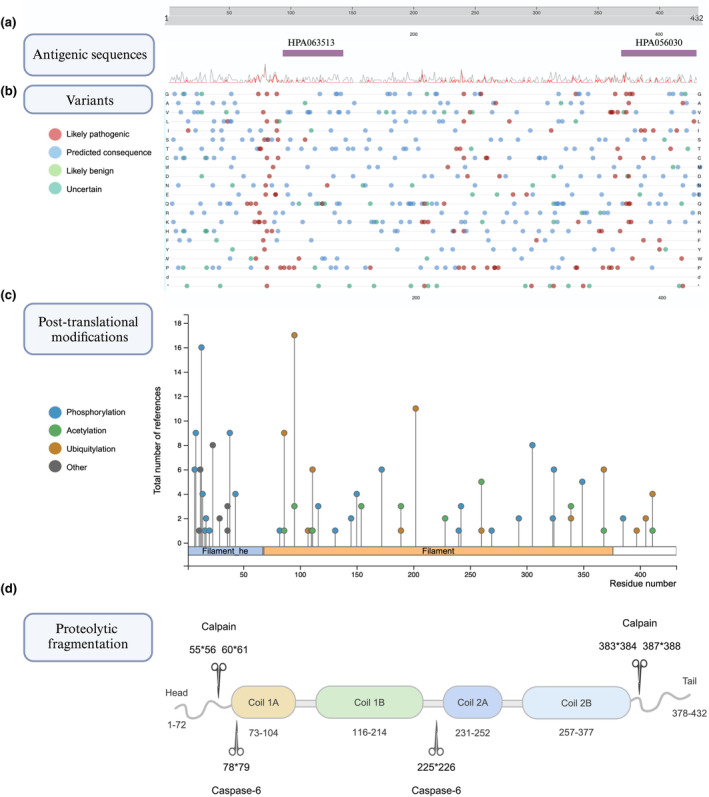
GFAP feature overview and breakdown products. GFAP and its modified or cleaved products play a key role in various cellular processes. GFAP epitopes which are targeted by commercially available immunoassays are mostly poorly characterised. (a) Demonstrates known antigenic sequences along the full‐length GFAP (antibodies targeting GFAP: HPA063513; HPA056030) and (b) mutations curated by UniProt (Consortium, T. U, 2023). Mutations and types of modifications are colour‐coded. (c) Protein post‐translational modifications (PTMs) functionally regulate the localisation, activity and assembly of GFAP. The visualisation of PTMs is based on PhosphoSitePlus (Hornbeck et al., 2014). (d) Full‐length GFAP is susceptible to proteolysis by calpain and caspase enzymes. A schematic representation of the proteolytic fragmentation of GFAP is shown. GFAP is shown as a linear model and major calpain and caspase 6 cleavage sites are indicated with scissors, whereby asterisks show between which amino acids the cleavage sites are. Adapted from Yang et al. (2022).
As evidenced, the measurements of GFAP in different biological fluids are not always equivalent across different CNS diseases. For instance, CSF GFAP is a better diagnostic biomarker in AxD whereas plasma GFAP has been demonstrated to outperform CSF GFAP for differentiation of amyloid‐positive and amyloid‐negative individuals in the context of AD. These distinctions emphasise the importance of context‐specific evaluation of GFAP levels, highlighting the need for tailored diagnostic strategies.
4. COMMERCIALLY AVAILABLE GFAP IMMUNOASSAYS
Before delving into GFAP's proteoform complexity, it should be disclosed that the epitopes which are targeted by commercially available immunoassays are mostly unknown or poorly characterised (Figure 3a; Waury et al., 2022). The assay with the most evidence concerning the antibodies used is the Quanterix Simoa singleplex or multiplex GFAP assay. This assay is widely used in clinical research and utilises antibodies from Banyan Biomarkers. The capture antibody is a mouse monoclonal IgG antibody (clone 2H12) and the detector antibody is a rabbit polyclonal antibody raised against the midsection of full‐length GFAP (Papa et al., 2012). Two epitopes for the capture antibody within human GFAP are reported, neither of which are entirely conserved between rat and human GFAP, or mouse and human GFAP (Zoltewicz et al., 2012). Both antibodies have been shown to recognise full‐length GFAP and a range of GFAP breakdown products varying in size from 48 to 38 kDa (Zoltewicz et al., 2012). Since both full‐length GFAP and various breakdown products are recognised with this assay (Zoltewicz et al., 2012), the antibodies likely bind to epitopes within the central rod domain (69–377 AA).
The Banyan Biomarkers' Brain Trauma Indicator (BTI) is an in‐vitro diagnostic test for the measurement of GFAP in the serum of suspected mild patients with traumatic brain injury. The BTI received a breakthrough device marketing authorisation from the FDA in 2018 (US Food and Drug Administration, 2018). In the decision memorandum from the FDA, it is demonstrated that the assay shows cross‐reactivity to NfL, but to no other proteins with similar homology to GFAP (US Food and Drug Administration, 2018). As such, a limitation of the procedure listed on the Banyan BTI Package Insert describes that because of the cross‐reactivity of neurofilament light (NfL) with the antibodies in the Banyan GFAP Kit, patients with neurodegenerative diseases such as Guillain‐Barré syndrome, ALS, PD, AD, or Creutzfeldt‐Jakob disease may have erroneously high Banyan GFAP, hence a false‐positive result.
Another GFAP biomarker test includes the NeuroToolKit from Roche, in which several biomarkers including GFAP can be measured in CSF or blood using a panel of automated exploratory prototype sandwich immunoassays (Johnson et al., 2023). Roche has developed a research‐use‐only GFAP electrochemiluminescence immunoassay to be used on the cobas e 801 and cobas e 402 immunoassay analysers. This assay uses monoclonal recombinant capture and detector antibodies; however, it is unknown which GFAP epitopes these antibodies target (Mayer et al., 2013). Other commercial GFAP immunoassays are the R‐Plex and S‐plex assays from Meso Scale Discovery (MSD; Kivisäkk et al., 2023; Spanos et al., 2022). These MSD assays utilise mouse monoclonal antibodies as both capture and detector antibodies, which were raised against the full‐length GFAP protein and show cross‐reactivity to mouse and rat GFAP protein (Kivisäkk et al., 2023; Spanos et al., 2022). No other information concerning these assays is publicly available.
To summarise, relatively little is known about which GFAP proteoforms are being targeted in commercially available immunoassays. The lack of detailed information about antibodies makes it challenging to compare studies and interpret discrepancies. This leaves plenty of room for improving our strategies to accurately measure GFAP to first unravel its function in brain pathologies and take advantage of this knowledge for developing specific biomarker tests.
5. PROTEOFORM COMPLEXITY
Given the major potential of GFAP as a biomarker of reactive astrogliosis for neurodegenerative and neurological diseases, as well as glioma, and its implementation in clinical settings (Abdelhak et al., 2022; Glass et al., 2010; Heimfarth et al., 2022; Jiwaji & Hardingham, 2022; Messing & Brenner, 2020; Mondello et al., 2021; van Asperen, Fedorushkova, et al., 2022), it is paramount to understand how the physio‐chemical characteristics of GFAP may influence its detection which may underlie the differential diagnostic performance of GFAP in different matrices. Although GFAP can be quantified well with commercially available immunoassays, it is unclear which GFAP proteoforms are being targeted with the available antibodies. Proteoforms of GFAP are altered in disease (Battaglia et al., 2019; Herskowitz et al., 2010; Ishigami et al., 2015; Kamphuis et al., 2014; Korolainen et al., 2005; Lin et al., 2021; Nicholas et al., 2004; Porchet et al., 2003) and could underly discrepancies between CSF and blood GFAP performance as a biomarker across various CNS diseases. Furthermore, targeting specific GFAP proteoforms could therefore result in disease‐specific biomarker tests. In the following section, we delve into the proteoform complexity of GFAP, which covers GFAP isoforms, PTMs, half‐life and breakdown products, surface accessibility, including structural flexibility and aggregation patterns, and protein–protein interactions.
5.1. Isoforms
Twelve different human and seven murine GFAP isoforms have been described to this day (de Reus et al., 2024; van Asperen, Robe, & Hol, 2022): α, β, γ, δ /ε, κ, ζ, λ, μ, ∆135, ∆164, ∆exon6, ∆exon7 (de Reus et al., 2024; Kamphuis et al., 2012; van Asperen, Robe, & Hol, 2022; Yang & Wang, 2015; Figure 4). Some of the isoforms have an alternate head (β γ), tail (δ /ε, κ, μ), rod (ζ, λ), or shortened (GFAP+1) rod domains affecting filament assembly. Notably, for many of the isoforms, full‐length RNAs have not been described and it is not always known where the transcripts start and end (indicated with question marks in Figure 4). The most predominant in the brain and spinal cord and most often studied isoform is 432 AA long GFAPα synthesised from 9 exons of the GFAP gene (Middeldorp & Hol, 2011). GFAPβ (>432 AA) is expressed in Schwann cells and includes a sequence before exon 1 originating in the 5′ untranslated region. The levels of GFAPβ were shown to be associated with neuronal injury (Condorelli et al., 1999). GFAPγ (<432 AA) similarly includes a sequence before exon 1 and has an intron instead of exon 1 (Zelenika et al., 1995). GFAPδ /ε is the second most common 431 AA long isoform that includes an extra exon 7a and lacks exons 8 and 9 (de Reus et al., 2024).
FIGURE 4.
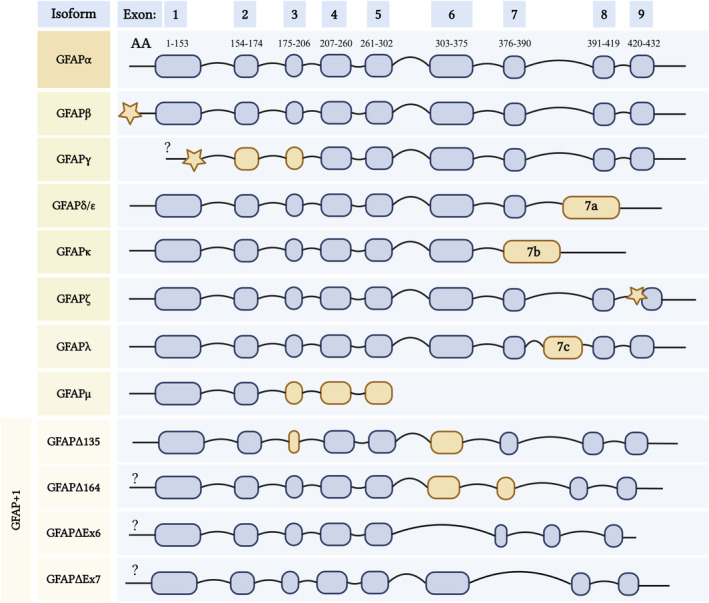
Twelve known isoforms of human GFAP. GFAPα (canonical isoform) comprises 9 exons represented as rounded rectangles. Respective exons and splice junctions were mapped to the amino acid sequence of GFAPα using the CCDS (NCBI) database (Pujar et al., 2018). GFAPβ and γ isoforms originate from alternative transcription start sites and have a varied N‐terminal (head). The rest of the isoforms result from alternative splicing: δ /ε, κ, and μ have shortened C‐terminal (tail), λ and ζ have alternate rod domains and less common GFAP +1 isoforms have shortened rod domains. Stars represent introns and yellow rounded rectangles indicate alternate regions. Longer linkers in the last two cases of GFAP+1 isoforms indicate that GFAP∆Ex6 lacks exon 6 and GFAP∆Ex7 lacks exon 7. Question marks indicate that the exact start sites of these isoforms are not yet known. Adapted from Middeldorp & Hol (2011), de Reus et al. (2024), and Yang & Wang (2015).
AA long isoform that includes an extra exon 7a and lacks exons 8 and 9 (de Reus et al., 2024). Increased GFAPδ /ε expression was detected in human astrocytic tumours reporting a direct correlation between the tumour malignancy and the isoform levels (Choi et al., 2009). GFAPκ is the third most commonly investigated isoform, which is 328 AA long and is enriched in the Rosenthal fibres of post‐mortem brains of AxD patients (Lin et al., 2021). GFAPκ lacks exons 8 and 9 but contains exon 7b, which consists of exon 7 and intron 7a (de Reus et al., 2024; Yang & Wang, 2015). GFAPζ (>438 AA) includes an intron between exon 8–9 (Kamphuis et al., 2012). AxD mutations result in over‐expression of GFAPλ (472 AA), which contains an altered exon 7c (de Reus et al., 2024; Helman et al., 2020). GFAPμ (179 AA) has the shortest coding sequence among the known isoforms and is expressed in healthy brain tissue, glioma cell lines, and primary glioma cells (van Bodegraven et al., 2021). Skipping exon 2 results in an out‐of‐frame transcript with a premature termination codon in exon 3 (van Bodegraven et al., 2021). Four less common GFAP isoforms—GFAP+1 collectively refer to variants caused by a single frame‐shift: ∆135 with shortened exon 6 lacking Coil 2B (374 AA), ∆164 with shortened exon 6, 7 and Coil 2B (366 AA), ∆exon6 lacking exon 6 and Coil 2B (347 AA), and ∆exon7 lacking exon 7 (418 AA; Yang & Wang, 2015). The role and abundance of different isoforms in neurodegenerative diseases have not yet been investigated, other than GFAPα and GFAPδ being elevated in AD brains (Kamphuis et al., 2014). A focused proteomic analysis of GFAP isoforms in neurodegenerative diseases could aid in understanding the role of these different isoforms per disease and potentially pinpoint discovering disease‐specific GFAP isoforms to develop into novel biomarkers.
5.2. Post‐translational modifications
PTMs functionally regulate intermediate filament formation (Snider & Omary, 2014) and GFAP is no exception. As shown in Figure 3c, GFAP is heavily modified throughout its sequence, and these PTMs are key in determining the localisation, activity and assembly of the protein. PTMs that lie in epitope regions can potentially hinder the binding of antibodies or the pairs thereof. To this end, the impact of PTMs on the analytical performance and clinical use of different GFAP immunoassays has been poorly characterised (Abdelhak et al., 2022).
Phosphorylation is one of the major PTM types involved in the (dis)assembly of GFAP polymers. Phosphorylation of GFAP in the head domain (Thr‐7, Ser‐8, Ser‐13, Ser‐17 and Ser‐34) regulates the filament disassembly during mitosis (Battaglia et al., 2019). Phosphorylation of Ser‐8 is thought to affect binding to 13‐3‐3γ (Li et al., 2006). Increased Ser‐13 phosphorylation is implicated in several pathologies, including disease severity in AxD (Battaglia et al., 2019), disease progression in frontotemporal lobar degeneration (FTLD; Herskowitz et al., 2010), and associated with hypoxic–ischemic brain injury (Sullivan et al., 2012). Furthermore, proteoforms of GFAP which are phosphorylated and N‐glycosylated, are increased in the frontal cortices of AD patients compared to age‐matched controls, whereas isoforms which are O‐glycosylated, showed no such difference (Korolainen et al., 2005). Moreover, GFAP in Rosenthal fibres of AxD patients and rodent models was shown to be ubiquitylated suggesting its critical role in GFAP aggregation (Lin et al., 2024). Another interesting PTM is citrullination, representing an enzymatic deimination forming citrulline from arginine. GFAP is believed to be one of the major deiminated proteins in both health and disease (Brenner & Nicholas, 2017). Citrullination was proposed to influence GFAP filament formation (Inagaki et al., 1989). Peptidylargenine deaminase 2 (PAD2) is an enzyme responsible for the citrullination of GFAP and the amount of PAD2 and citrullinated GFAP is increased in the hippocampi of AD patients compared to non‐AD controls (Ishigami et al., 2015). Additionally, GFAP citrullination was suggested to be a result of an immune response to inflammation in MS (Faigle et al., 2019). As such, citrullinated GFAP is increased in the brains of secondary progressive multiple sclerosis patients compared to controls (Nicholas et al., 2004). Lastly, lipoxidation is another PTM that can potentially affect GFAP polymerisation. The only cysteine at position 294 is susceptible to lipoxidation and is believed to be involved in filament formation (Viedma‐Poyatos et al., 2018). Its mutation to serine has been shown to affect the formation of the cytoskeletal network, suggesting that lipoxidation of this cysteine residue might lead to a similar outcome (Messing & Brenner, 2020; Viedma‐Poyatos et al., 2018). Furthermore, in vitro and cell‐based studies demonstrate that cystine‐generating mutations promote GFAP crosslinking by cysteine‐dependent oxidation, resulting in defective GFAP assembly and decreased filament solubility (Lin et al., 2024). Cys‐291 (mouse GFAP) is palmitoylated in vitro and in vivo and hyper‐palmitoylation was shown to accelerate astrogliosis and neurodegenerative pathology in PPT1‐deficient mice (Yuan et al., 2021). To enhance the reliability of GFAP detection strategies, it is key to consider PTMs during antibody selection. Using a combination of antibodies that target different sites, including those less likely to be affected by known modifications, may improve assay sensitivity and specificity.
5.3. Discharge mechanisms
The discharge of GFAP from the brain to the CSF and blood could occur via multiple pathways, which may underlie the established difference in diagnostic performance between plasma and CSF GFAP (Benedet et al., 2021). One study has demonstrated that GFAP efflux into the blood occurs via the glymphatic system in murine models (Plog et al., 2015). Within the glymphatic system, there is first an influx of CSF through AQP4 channels on astrocytes to the interstitial space. This influx of CSF and interstitial fluid in the brain parenchyma then drives a fluid efflux to the perivascular space and venous system (Jessen et al., 2015).
Other hypothesised discharge mechanisms of GFAP, which have yet to be proven, include direct secretion of GFAP from reactive astrocytes to the bloodstream since astrocytic end‐feet surrounds blood capillaries in the brain (Giannoni et al., 2018). Another proposed mechanism is that GFAP may diffuse from the cytosol of injured astrocytes across the blood–brain barrier which is altered and can become ‘leaky’ or damaged in the context of many types of dementia (Hussain et al., 2021), traumatic brain injury (Plog et al., 2015) and stroke (Dvorak et al., 2009). This hypothesis is supported by evidence comparing serum GFAP levels of intracerebral haemorrhage patients, who experience rapid blood–brain barrier disruption, to ischaemic stroke patients, where the opening of the blood–brain barrier occurs more gradually (Dvorak et al., 2009). From 2 to 6 h following stroke onset, serum GFAP was significantly increased in intracerebral haemorrhage patients compared to ischaemic stroke patients. Elevation of GFAP occurred at a much later time‐point of 48 h in ischaemic stroke patients (Dvorak et al., 2009). Another potential mechanism of GFAP release could occur via extracellular vesicles (EVs) since GFAP has been previously detected and quantified in EVs (Flynn et al., 2021). Moreover, GFAP has a high probability of being EV‐associated based on various physio‐chemical properties and PTMs according to a recently developed machine learning model (Waury et al., 2024), suggesting that GFAP is likely to be actively transported through vesicles.
5.4. Breakdown products
The full‐length intact 50 kDa GFAP is highly susceptible to proteolysis by calpain and caspase enzymes, by which it is mainly processed to 42 and 38 kDa breakdown products (Escartin et al., 2021; Figure 3d). These generated fragments have different stabilities, ranging from seconds to 20 h, depending on the amino acids that are exposed during cleavage (Phillips et al., 2023). Following calpain enzyme proteolysis, predicted GFAP cleavage sites expose residues, such as serine and alanine, which stabilise the product and cause it to be long‐lived; this suggests these breakdown products may have an important functional role (Phillips et al., 2023).
In a clinical context, two independent studies demonstrated the presence of 36–44 kDa GFAP breakdown products in AD brains (Korolainen et al., 2005; Porchet et al., 2003). GFAP fragments have also been detected in human biofluids, specifically a 38 kDa breakdown product was detected in the CSF (Yang et al., 2022) and plasma (Okonkwo et al., 2013) of patients with TBI within the first 24 h post‐incident. Measurement of total GFAP and its breakdown products in the serum of TBI patients aided in the diagnosis of intracranial injury compared to clinical screening alone (McMahon et al., 2015). A comparison of the 38 kDa GFAP proteolytic fragment versus intact GFAP measured in CSF to distinguish between TBI patients versus controls showed better discriminative performance for the 38 kDa fragment compared to full‐length GFAP (AUC‐ROC of 0.944 versus 0.909; Yang et al., 2022). Interestingly, phosphorylation of GFAP in AxD leads to proteolytic cleavage by caspase 6, resulting in fragments of varying molecular weight compared to the 38 kDa product, which is only produced following acute injury (Battaglia et al., 2019). Different GFAP breakdown products may therefore define acute astrocyte injury, in the context of TBI, versus chronic injury in the context of neurodegenerative conditions.
5.5. Aggregation, dynamics and sample stability
Several GFAP‐isoforms have a high propensity to form Rosenthal fibre‐like aggregates and a high percentage of such isoforms can lead to an IF‐network collapse (de Reus et al., 2024; Lin et al., 2021). In AxD, a specific mutation in GFAP alters splicing, leading to an increase in aggregation‐prone isoforms: GFAP‐δ, −κ and ‐λ (Lin et al., 2021). These isoforms are less soluble compared to non‐pathological GFAP, making them prone to form rod‐shaped proteinaceous aggregates inside astrocytes called Rosenthal fibres. The fibres interfere with cell mitosis, alter the morphology of aggregate‐bearing astrocytes and are the pathological hallmark of AxD (Lin et al., 2021). They are also sometimes present in MS (Wippold et al., 2006) and glioma (Gullotta et al., 1985). Rosenthal fibre‐like aggregates can be extraordinarily stable and this may limit the detection of GFAP in blood in AxD (Abdelhak et al., 2022). Aggregation or de‐aggregation of GFAP may occur in different matrices and/or in response to temperature changes; GFAP levels in blood were shown to increase with storage or freeze–thaw (FT) cycles at −20°C (Gouda et. al, in prep) or at −80°C (Verberk et al., 2022). However, there is no direct evidence that this increase in the level of GFAP monomers occurs because of protein de‐aggregation. Conversely, CSF GFAP levels have been shown to decrease with FT cycles, and CSF GFAP was shown to be more susceptible to FT cycles compared to blood GFAP measured in the same individual (Simrén et al., 2022). However, even in fresh samples, GFAP measured in blood was superior to CSF measurements in discriminating between amyloid‐positive and amyloid‐negative individuals (thereby reflecting AD pathology), suggesting that the matrix discrepancy is likely not solely because of sample stability and alterations in GFAP aggregation. A recent study has highlighted structural heterogeneity and the existence of multiple conformational forms of GFAP (Gogishvili et al., 2023). The study explored the structural dynamics of recombinant GFAP under three conditions, namely potassium phosphate buffer saline (KPBS) and two setups in artificial CSF (aCSF) using hydrogen‐deuterium exchange mass spectrometry. Under aCSF conditions, recombinant GFAP showed an overall increase in solvent accessibility and simultaneously displayed hotspots of aggregation, suggesting the existence of multiple conformations of GFAP (Gogishvili et al., 2023).
5.6. Interaction partners
Protein‐antibody accessibility can also be hampered because of interacting partners. Intermediate filament‐associated proteins play an important role in filament stability and facilitate links to other structures within the cell (Middeldorp & Hol, 2011). Plectin is a widely expressed IF‐binding protein, which is thought to provide mechanical strength to cells by cross‐linking to microtubules and the actin cytoskeleton, and was shown to bind to GFAP in the rod domain (Tian et al., 2006). Additionally, decreased plectin levels lead to the formation of a disorganised aggregate of GFAP (severe type of AxD mutation, R239C (RC); Tian et al., 2006). GFAP was shown to interact with the family of regulatory proteins 14‐3‐3 and that the interaction is influenced by the phosphorylation of GFAP in a cell‐cycle dependent manner (Li et al., 2006). Small Heat shock proteins (sHSPs) are a group of ATP‐independent chaperones expressed ubiquitously in all kingdoms of life (Haslbeck et al., 2019) and play a key role in preventing protein misfolding and aggregation (Haslbeck & Vierling, 2015). Upon stress, HSP27 has a phosphorylation‐activated role in actin filament regulation and prevents filament disruption and degeneration (Graceffa, 2011; Guay et al., 1997). Moreover, sHSPs are involved in cytoskeletal rearrangement. HSP27 along with αB‐crystallin was shown to interact with GFAP regulating filament assembly (Perng et al., 1999). Increasing GFAPδ levels by transient transfection in astrocyte‐derived cell lines were shown to have deleterious effects, causing the increased association of αB‐crystallin and the disruption of IF network (Perng et al., 2008). Moreover, HSP27 was found in Rosenthal fibres (GFAP inclusions) both in the brains of patients suffering from AxD (Tomokane et al., 1991), as well as in mice overexpressing GFAP (Eng et al., 1998).
6. DISCUSSION
The comprehensive evaluation of GFAP has the potential to enable longitudinal evaluation of the astrocyte response in brain and spinal cord disorders. A better understanding of GFAP proteoforms can ultimately assist with the development of accurate, early and discriminative diagnosis. There is much to discover about the implications of GFAP proteoforms and accessibility in the context of neurodegeneration. To better understand disease pathology, improve the utilisation of GFAP as a biomarker and unlock various biological insights, we need to have a comprehensive understanding of what we are detecting and quantifying in biomarker tests.
GFAP (post‐)transcriptional regulation has a key role in glial cell physiology and pathology, as GFAP isoforms vary in cellular localisation and determine mechanical properties of the IF‐network (de Reus et al., 2024). There are numerous open questions regarding the implications of distinct GFAP isoforms, concerning the function of GFAP isoforms in ageing, brain injury and disease. In more detail, using isoform‐specific antibodies may hold promise for staging AD in terms of inflammation; the shortened rod isoforms (GFAP+1) are associated with disease progression as GFAP+1 positive astrocytes have been shown to increase in number over the course of AD (Kamphuis et al., 2014). Another interesting avenue to investigate is the GFAP isoform ratio. Amyloid precursor protein (APP)‐derived peptides exemplify the case where the ratio of Aβ 42 to Aβ 40 (Aβ 42/40 ratio) is superior to the concentration of Aβ 42 alone in discriminating patients with AD from controls (Shoji et al., 1998). A lower ratio is indicative of disrupted amyloid metabolism and is used as a diagnostic tool for AD (Dumurgier et al., 2015; Perez‐Grijalba et al., 2019). For GFAP isoform ratios, a change in the GFAPα/GFAPδ ratio has been shown to alter cell‐environment interactions and cell migration in the context of glioma cell invasion (van Asperen, Robe, & Hol, 2022). Importantly, glioma does not directly translate to dementia and there are major differences in pathological processes. Yet, it may be valuable to investigate the role of the GFAPα/GFAPδ ratio to reflect different states of astrocyte activation in the context of dementias. The interplay between these isoforms and their differential effects on protein aggregation and neurotoxicity highlights the importance of understanding the GFAP proteoform landscape for unravelling the complexity of neurodegenerative disorders and advancing biomarker research.
PTMs play a pivotal role in regulating the functional properties of GFAP (Snider & Omary, 2014). Among others, phosphorylation regulates the assembly and disassembly of GFAP polymers, binding to other proteins and disease severity (Battaglia et al., 2019; Herskowitz et al., 2010; Li et al., 2006; Sullivan et al., 2012). PTMs affecting filament formation can impact the accessibility of GFAP to antibodies used in assays, through obscuring or exposing epitopes leading to variability in assay results. Understanding the complex cross‐talk and regulatory mechanisms of these PTMs is crucial for unravelling their functional significance and developing PTM‐specific biomarker tests for differential diagnosis and disease staging (Battaglia et al., 2019; Herskowitz et al., 2010). The importance of detecting specific PTMs for differential dementia diagnosis can be highlighted with the example of phosphorylated tau. Phosphorylated tau has long been established to reflect abnormal tau metabolism in the brain. The identification and quantification of specific phospho‐tau epitopes have proven instrumental in elucidating disease diagnosis, progression and severity. For instance, CSF p‐tau181 is one of the core biomarkers for AD diagnosis (Olsson et al., 2016). Recently, CSF p‐tau217 has been shown to perform better for diagnostic workup in AD (Janelidze et al., 2020). Both plasma p‐tau231 and p‐tau217 were shown to associate with the earliest cerebral Aβ pathologies (Milà‐Alomà et al., 2022), implicating their role in early diagnosis. The potential of phospho‐GFAP or other PTMs as biomarkers for differential dementia diagnosis has not yet been explored, but based on research summarised above, phospho‐GFAP Serine 13 could be a potential FTLD‐specific biomarker test, and citrullinated GFAP may hold promise for AD and/or MS.
GFAP clearance mechanisms and transport from the brain to the CSF and blood could underly differences in diagnostic performance between plasma and CSF GFAP. The contribution of each of the hypothesised mechanisms and their disruption could impact the performance of GFAP assays in different biological matrices. For example, traumatic brain injury reduces clearance via the glymphatic system which has been shown to suppress TBI‐induced increases of GFAP in the blood, which negatively impacts its clinical utility (Plog et al., 2015). Contrarily, a disrupted blood–brain barrier in AD could enhance the discharge of GFAP to the blood, underlying its superior performance compared to CSF. Further work studying GFAP dynamics in both CSF and blood matrices, in combination with MRI scans of blood–brain barrier quality in the context of various diseases with different dynamics of astrocyte injury can help elucidate the contribution of different proposed pathways.
Given the susceptibility of full‐length GFAP to proteolysis, it is crucial to understand its cleavage products. It is yet to be determined exactly how GFAP fragments are released from astrocytes for detection in CSF and blood, whether they are related to the pathogenesis and progression of the disease, and whether they outperform full‐length GFAP as stand‐alone biomarkers. Breakdown products have different stabilities compared to the native full‐length protein depending on the residues exposed on the breakdown products (Phillips et al., 2023), and potentially the matrix they are in. Investigating the differences in the abundance of specific GFAP breakdown products among biological matrices in various disease contexts could elucidate this. Additionally, breakdown products may have distinct functions compared to native proteins and could act to serve as stand‐alone biomarkers, as is the case for TBI (Yang et al., 2022), and they may reflect different cellular processes and disease pathologies. The development of novel breakdown product‐specific immunoassays would help answer these questions. In the case of NfL, which is another intermediate filament protein that is widely used as a biomarker for axonal damage, characterisation of the Uman antibodies used in commercially available immunoassays revealed their neurodegeneration‐specific staining properties (Shaw et al., 2023). Surprisingly, neither antibody stains neurofilaments in healthy cells but rather recognises degenerated neuronal NfL. This study highlights the importance of targeting protein products which are specifically produced in the context of neurodegeneration rather than constitutively produced.
GFAP is a highly dynamic and flexible protein co‐existing in multiple conformational species. Focusing on specific oligomeric or aggregated GFAP species can provide valuable insights into the severity of the disease. Under specific conditions, GFAP may become more disordered and floppy (as shown in the case of artificial CSF; Gogishvili et al., 2023), which increases solvent accessibility, especially in the interface regions, possibly leading to amorphous aggregation. The structural dynamics of GFAP can affect the performance of immunoassays potentially in the context of both different matrices or buffers used in the assay protocol.
In future studies, cross‐linking mass spectrometry and hydrogen‐deuterium exchange mass spectrometry techniques can be explored for mapping the binding interface of GFAP and to better understand GFAP solvent accessibility. This can help elucidate optimal epitope sites for antibody binding when generating novel immunoassays. Furthermore, understanding which proteins GFAP binds to in the context of different diseases can help us gain insight as to the exact biological process the biomarker is reflecting.
7. CONCLUSION
GFAP has been proven to be a highly valuable addition to the biomarker toolbox for early and discriminative diagnosis of brain and spinal cord disorders. By taking an example of other successful biomarkers developed for various neurological diseases, we can harness the GFAP proteoform diversity to develop more accurate and disease‐specific biomarkers (see Concluding remarks). Proteoform‐specific targeting (including isoforms, PTMs, discharge mechanisms, breakdown products, and higher‐order species) could reveal novel biological insights, which might lead to more reliable and reproducible tests and improved diagnostic performance.
7.1. Concluding remarks
Potential avenues to explore for advancing GFAP as a biomarker:
Isoforms can provide valuable information about the underlying CNS pathology. For instance, GFAP+1 isoforms may hold promise for staging AD in terms of inflammation, while GFAPα/GFAPδ ratio may reflect different states of astrocyte activation.
Post‐translational modifications (PTMs) of GFAP can be exploited by developing PTM‐specific biomarker tests that could aid in differential diagnosis and/or disease staging. For example, phosphorylated GFAP may be useful for identifying AxD, AD, or FTLD, while citrullinated GFAP could help diagnose AD and MS.
Discharge mechanisms of GFAP from the brain to the CSF and blood could underlie differences in diagnostic performance between plasma and CSF GFAP. Additionally, clearance mechanisms have clinical relevance for utilising GFAP as a biomarker of acute astrocytic injury, such as for TBI.
Breakdown products of GFAP may exhibit distinct functions compared to full‐length GFAP. Furthermore, different cleavage products could serve as markers for acute versus chronic astroglial injury.
Aggregates or higher‐order species of GFAP are associated with disease progression and may serve as useful biomarkers for AxD and potentially MS patients. Moreover, detecting specific oligomeric or aggregated GFAP species could provide valuable insights into the severity of the disease.
Interaction partners of GFAP can influence protein‐antibody accessibility, potentially affecting biomarker test performance in specific matrices. Understanding the GFAP interactome in different pathological conditions can provide crucial biological insights into the mechanisms underlying disease progression.
AUTHOR CONTRIBUTIONS
Dea Gogishvili: Conceptualization; investigation; visualization; writing – original draft; writing – review and editing. Madison I. J. Honey: Conceptualization; investigation; writing – original draft; writing – review and editing. Inge M. W. Verberk: Conceptualization; writing – original draft; writing – review and editing. Lisa Vermunt: Conceptualization; writing – original draft; writing – review and editing. Elly M. Hol: Writing – original draft; writing – review and editing. Charlotte E. Teunissen: Conceptualization; funding acquisition; writing – original draft; writing – review and editing. Sanne Abeln: Conceptualization; funding acquisition; writing – original draft; writing – review and editing.
CONFLICT OF INTEREST STATEMENT
CT is the recipient of ABOARD, which is a public‐private partnership receiving funding from ZonMW (#73305095007) and HealthHolland, Topsector Life Sciences & Health (PPP‐allowance; #LSHM20106). More than 30 partners participate in ABOARD. ABOARD also receives funding from Edwin Bouw Fonds and Gieskes‐Strijbisfonds. CT has a collaboration contract with ADx Neurosciences, Quanterix and Eli Lilly, performed contract research or received grants from AC‐Immune, Axon Neurosciences, Biogen, Brainstorm Therapeutics, Celgene, EIP Pharma, Eisai, Olink, PeopleBio, Roche, Toyama, Vivoryon. CT serves on editorial boards of Medidact Neurologie/Springer, Alzheimer Research and Therapy, Neurology: Neuroimmunology & Neuroinflammation, and is editor of a Neuromethods book Springer. LV is supported by research grants from Amsterdam UMC, ZonMw, Stichting Diorapthe, and Olink and consultancy/speaking fees from Roche and Eli Lilly, all paid to her institution. Outside the submitted work: SA reports a patent pending; SA is in a consortium agreement with Cergentis BV as part of the TargetSV project; SA is in a consortium agreement with Olink and Quanterix as part of the NORMAL project. The rest of the authors do not have any competing interests to declare.
PEER REVIEW
The peer review history for this article is available at https://www.webofscience.com/api/gateway/wos/peer‐review/10.1111/jnc.16226.
ACKNOWLEDGEMENTS
This study is supported by the European Commission (Marie Curie International Training Network, grant agreement No 860197 (MIRIADE). MH and CT are recipients of ABOARD, which is a public‐private partnership receiving funding from ZonMW (#73305095007) and Health Holland, Topsector Life Sciences & Health (PPP‐allowance; #LSHM20106). IV is supported by research grants from Amsterdam UMC and HealthHolland (a public‐private partnership grant executed in collaboration with Olink and Quanterix). EH is a recipient of the following grants related to GFAP and astrocytes: Alzheimer NL (#WE.03‐ 2022‐04), ZonMW MODEM dementia project (#10510032120006), EJP RD JTC 2019 (#463002004),’la Caixa’ Foundation (#LCF/PR/HR21/52410002). The research of SA is supported by Health‐Holland).
Gogishvili, D. , Honey, M. I. J. , Verberk, I. M. W. , Vermunt, L. , Hol, E. M. , Teunissen, C. E. , & Abeln, S. (2025). The GFAP proteoform puzzle: How to advance GFAP as a fluid biomarker in neurological diseases. Journal of Neurochemistry, 169, e16226. 10.1111/jnc.16226
Dea Gogishvili and Madison I. J. Honey contributed equally to this work.
Contributor Information
Dea Gogishvili, Email: d.gogishvili@vu.nl.
Madison I. J. Honey, Email: m.i.j.honey@amsterdamumc.nl.
DATA AVAILABILITY STATEMENT
The data that support this study are openly available. Several tools were employed to analyse the structure of GFAP, namely, NetSurfP 3.0 (Høie et al., 2022; Klausen et al., 2019), AlphaFold‐Multimer (Evans et al., 2021; Gogishvili et al., 2023), and the PAE viewer tool (Elfmann & Stülke, 2023). The Human Protein Atlas (HPA) was utilised for the analysis of GFAP expression, interaction network, and cell localisation (Karlsson et al., 2021; Kawajiri et al., 2003; Thul et al., 2017; Uhlén et al., 2015; Yoshida et al., 2007). The feature overview of GFAP was obtained from curated UniProt annotations (Consortium, T. U, 2023). For mapping exons and splice junctions to the amino acid sequence of GFAPβ, the NCBI‐CCDS database was used (Pujar et al., 2018). Figures 3d and 4 were created with Biorender.com.
REFERENCES
- Abdelhak, A. , Foschi, M. , Abu‐Rumeileh, S. , Yue, J. K. , D'Anna, L. , Huss, A. , Oeckl, P. , Ludolph, A. C. , Kuhle, J. , Petzold, A. , Manley, G. T. , Green, A. J. , Otto, M. , & Tumani, H. (2022). Blood gfap as an emerging biomarker in brain and spinal cord disorders. Nature Reviews Neurology, 18, 158–172. [DOI] [PubMed] [Google Scholar]
- Abdelhak, A. , Huss, A. , Kassubek, J. , Tumani, H. , & Otto, M. (2018). Serum gfap as a biomarker for disease severity in multiple sclerosis. Scientific Reports, 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluise, C. D. , Sowell, R. A. , & Butterfield, D. A. (2008). Peptides and proteins in plasma and cerebrospinal fluid as biomarkers for the prediction, diagnosis, and monitoring of therapeutic efficacy of alzheimer's disease. Biochimica et Biophysica Acta (BBA) ‐ Molecular Basis of Disease, 1782, 549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson, M. , Malmeström, C. , Nilsson, S. , Haghighi, S. , Rosengren, L. , & Lycke, J. (2011). Glial fibrillary acidic protein: A potential biomarker for progression in multiple sclerosis. Journal of Neurology, 258, 882–888. [DOI] [PubMed] [Google Scholar]
- Ayrignac, X. , Le Bars, E. , Duflos, C. , Hirtz, C. , Maleska Maceski, A. , Carra‐Dallière, C. , Charif, M. , Pinna, F. , Prin, P. , Menjot de Champfleur, N. , Deverdun, J. , Kober, T. , Marechal, B. , Fartaria, M. J. , Corredor Jerez, R. , Labauge, P. , & Lehmann, S. (2020). Serum GFAP in multiple sclerosis: Correlation with disease type and MRI markers of disease severity. Scientific Reports, 10(1), 10923. 10.1038/s41598-020-67934-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiardi, S. , Quadalti, C. , Mammana, A. , Dellavalle, S. , Zenesini, C. , Sambati, L. , Pantieri, R. , Polischi, B. , Romano, L. , Suffritti, M. , Bentivenga, G. M. , Randi, V. , Stanzani‐Maserati, M. , Capellari, S. , & Parchi, P. (2022). Diagnostic value of plasma p‐tau181, nfl, and gfap in a clinical setting cohort of prevalent neurodegener‐ ative dementias. Alzheimer's Research & Therapy, 14, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia, R. A. , Beltran, A. S. , Delic, S. , Dumitru, R. , Robinson, J. A. , Kabiraj, P. , Herring, L. E. , Madden, V. J. , Ravinder, N. , Willems, E. , Newman, R. A. , Quinlan, R. A. , Goldman, J. E. , Perng, M. D. , Inagaki, M. , & Snider, N. T. (2019). Site‐specific phosphorylation and caspase cleavage of gfap are new markers of alexander disease severity. eLife, 8, e47789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedet, A. L. , Milà‐Alomà, M. , Vrillon, A. , Ashton, N. J. , Pascoal, T. A. , Lussier, F. , Karikari, T. K. , Hourregue, C. , Cognat, E. , Dumurgier, J. , Stevenson, J. , Rahmouni, N. , Pallen, V. , Poltronetti, N. M. , Salvadó, G. , Shekari, M. , Operto, G. , Gispert, J. D. , Minguillon, C. , … Stevensson, A. (2021). Differences between plasma and cerebrospinal fluid glial fibrillary acidic protein levels across the alzheimer disease continuum. JAMA Neurology, 78, 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, M. , & Nicholas, A. P. (2017). The significance of deiminated gfap in neurodegenerative diseases with special emphasis on alexander disease. In Protein deimination in human health and disease (pp. 391–412). Springer. [Google Scholar]
- Chatterjee, P. , Vermunt, L. , Gordon, B. A. , Pedrini, S. , Boonkamp, L. , Armstrong, N. J. , Xiong, C. , Singh, A. K. , Li, Y. , Sohrabi, H. R. , Taddei, K. , Molloy, M. , Benzinger, T. L. S. , Morris, J. C. , Karch, C. , Berman, S. , Chhatwal, J. , Cruchaga, C. , Graff‐Radford, N. R. , … and the Dominantly Inherited Alzheimer Network . (2023). Plasma glial fibrillary acidic protein in autosomal dominant alzheimer's disease: Associations with aβ ‐pet, neurodegeneration, and cognition. Alzheimer's & Dementia, 19, 2790–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyatina, A. A. , Guzenko, D. , & Strelkov, S. V. (2015). Intermediate filament structure: The bottom‐up approach. Current Opinion in Cell Biology, 32, 65–72. [DOI] [PubMed] [Google Scholar]
- Chernyatina, A. A. , Nicolet, S. , Aebi, U. , Herrmann, H. , & Strelkov, S. V. (2012). Atomic structure of the vimentin central α‐helical domain and its implications for intermediate filament assembly. Proceedings of the National Academy of Sciences, 109, 13620–13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiotis, K. , Johansson, C. , Rodriguez‐Vieitez, E. , Ashton, N. J. , Blennow, K. , Zetterberg, H. , Graff, C. , & Nordberg, A. (2023). Tracking reactive astrogliosis in autosomal dominant and sporadic alzheimer's disease with multi‐modal pet and plasma gfap. Molecular Neurodegeneration, 18, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.‐C. , Kwak, S.‐E. , Kim, J.‐E. , Sheen, S. H. , & Kang, T.‐C. (2009). Enhanced glial fibrillary acidic protein‐δ expression in human astrocytic tumor. Neuroscience Letters, 463, 182–187. [DOI] [PubMed] [Google Scholar]
- Cicognola, C. , Janelidze, S. , Hertze, J. , Zetterberg, H. , Blennow, K. , Mattsson‐Carlgren, N. , & Hansson, O. (2021). Plasma glial fibrillary acidic protein detects alzheimer pathology and predicts future conversion to alzheimer dementia in patients with mild cognitive impairment. Alzheimer's Research & Therapy, 13, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo, A. M. , Alberghina, L. , & Papa, M. (2014). Astrogliosis as a therapeutic target for neurodegenerative diseases. Neuroscience Letters, 565, 59–64. [DOI] [PubMed] [Google Scholar]
- Condorelli, D. F. , Nicoletti, V. G. , Dell'Albani, P. , Barresi, V. , Caruso, A. , Conticello, S. G. , Belluardo, N. , & Giuffrida Stella, A. M. (1999). GFAPbeta mRNA expression in the normal rat brain and after neuronal injury. Neurochemical Research, 24(5), 709–714. 10.1023/a:1021016828704 [DOI] [PubMed] [Google Scholar]
- Consortium, T. U . (2023). Uniprot: The universal protein knowledgebase in 2023. Nucleic Acids Research, 51, D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, N. C. , Leuzy, A. , Palmqvist, S. , Janelidze, S. , Stomrud, E. , Pesini, P. , Sarasa, L. , Allué, J. A. , Proctor, N. K. , Zetterberg, H. , Dage, J. L. , Blennow, K. , Mattsson‐Carlgren, N. , & Hansson, O. (2021). Individualized prognosis of cognitive decline and dementia in mild cognitive impairment based on plasma biomarker combinations. Nature Aging, 1, 114–123. [DOI] [PubMed] [Google Scholar]
- de Reus, A. J. E. M. , Basak, O. , Dykstra, W. , van Asperen, J. V. , van Bodegraven, E. J. , & Hol, E. M. (2024). GFAP‐isoforms in the nervous system: Understanding the need for diversity. Current Opinion in Cell Biology, 87, 102340. 10.1016/j.ceb.2024.102340 [DOI] [PubMed] [Google Scholar]
- Dumurgier, J. , Schraen, S. , Gabelle, A. , Vercruysse, O. , Bombois, S. , Laplanche, J. L. , Peoc'h, K. , Sablonnière, B. , Kastanenka, K. V. , Delaby, C. , Pasquier, F. , Touchon, J. , Hugon, J. , Paquet, C. , & Lehmann, S. (2015). Cerebrospinal fluid amyloid‐β 42/40 ratio in clinical setting of memory centers: A multicentric study. Alzheimer's Research & Therapy, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, F. , Haberer, I. , Sitzer, M. , & Foerch, C. (2009). Characterisation of the diagnostic window of serum glial fibrillary acidic protein for the differentiation of intracerebral haemorrhage and ischaemic stroke. Cerebrovascular Diseases, 27, 37–41. [DOI] [PubMed] [Google Scholar]
- Eddleston, M. , & Mucke, L. (1993). Molecular profile of reactive astrocytes—Implications for their role in neurologic disease. Neuroscience, 54, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibauer, M. , Weber, M. S. , Turgay, Y. , Sivagurunathan, S. , Goldman, R. D. , & Medalia, O. (2021). The molecular architecture of vimentin filaments. bioRxiv, 2021‐07. 10.1101/2021.07.15.452584 [DOI] [Google Scholar]
- Ekblom, J. , Jossan, S. S. , Bergstrüm, M. , Oreland, L. , Walum, E. , & Aquilonius, S. M. (1993). Monoamine oxidase‐b in astrocytes. Glia, 8, 122–132. [DOI] [PubMed] [Google Scholar]
- Elfmann, C. , & Stülke, J. (2023). PAE viewer: A webserver for the interactive visualization of the predicted aligned error for multimer structure predictions and crosslinks. Nucleic Acids Research, 51, W404–W410. 10.1093/nar/gkad350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emirandetti, A. , Zanon, R. G. , Sabha, M., Jr. , & de Oliveira, A. L. R. (2006). Astrocyte reactivity influences the number of presynaptic terminals apposed to spinal motoneurons after axotomy. Brain Research, 1095, 35–42. [DOI] [PubMed] [Google Scholar]
- Eng, L. F. , Lee, Y. L. , Kwan, H. , Brenner, M. , & Messing, A. (1998). Astrocytes cultured from transgenic mice carrying the added human glial fibrillary acidic protein gene contain rosenthal fibers. Journal of Neuroscience Research, 53, 353–360. [DOI] [PubMed] [Google Scholar]
- Escartin, C. , Galea, E. , Lakatos, A. , O'Callaghan, J. P. , Petzold, G. C. , Serrano‐Pozo, A. , Steinhäuser, C. , Volterra, A. , Carmignoto, G. , Agarwal, A. , Allen, N. J. , Araque, A. , Barbeito, L. , Barzilai, A. , Bergles, D. E. , Bonvento, G. , Butt, A. M. , Chen, W. T. , Cohen‐Salmon, M. , … Verkhratsky, A. (2021). Reactive astrocyte nomenclature, definitions, and future directions. Nature Neuroscience, 24, 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, R. , O'Neill, M. , Pritzel, A. , Antropova, N. , Senior, A. , Green, T. , Žídek, A. , Bates, R. , Blackwell, S. , Yim, J. , Ronneberger, O. , Bodenstein, S. , Zielinski, M. , Bridgland, A. , Potapenko, A. , Cowie, A. , Tunyasuvunakool, K. , Jain, R. , Clancy, E. , … Hassabis, D. (2021). Protein complex prediction with AlphaFold‐Multimer. bioRxiv, 2021‐10. 10.1101/2021.10.04.463034 [DOI] [Google Scholar]
- Faigle, W. , Cruciani, C. , Wolski, W. , Roschitzki, B. , Puthenparampil, M. , Tomas‐Ojer, P. , Sellés‐Moreno, C. , Zeis, T. , Jelcic, I. , Schaeren‐Wiemers, N. , Sospedra, M. , & Martin, R. (2019). Brain citrullination patterns and T cell reactivity of cerebrospinal fluid‐derived cd4+ T cells in multiple sclerosis. Frontiers in Immunology, 10, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch, C. E. (2003). Neurons, glia, and plasticity in normal brain aging. Neurobiology of Aging, 24, S123–S127. [DOI] [PubMed] [Google Scholar]
- Flynn, S. , Leete, J. , Shahim, P. , Pattinson, C. , Guedes, V. A. , Lai, C. , Devoto, C. , Qu, B. X. , Greer, K. , Moore, B. , van der Merwe, A. , Ekanayake, V. , Gill, J. , & Chan, L. (2021). Extracellular vesicle concentrations of glial fibrillary acidic protein and neurofilament light measured 1 year after traumatic brain injury. Scientific Reports, 11, 3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana, I. C. , Scarpa, M. , Malarte, M. L. , Rocha, F. M. , Ausellé‐Bosch, S. , Bluma, M. , Bucci, M. , Chiotis, K. , Kumar, A. , & Nordberg, A. (2023). Astrocyte signature in alzheimer's disease continuum through a multi‐pet tracer imaging perspective. Cells, 12, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoni, P. , Badaut, J. , Dargazanli, C. , de Maudave, A. F.’. H. , Klement, W. , Costalat, V. , & Marchi, N. (2018). The pericyte–glia interface at the blood–brain barrier. Clinical Science, 132, 361–374. [DOI] [PubMed] [Google Scholar]
- Giménez y Ribotta, M. , Langa, F. , Menet, V. , & Privat, A. (2000). Comparative anatomy of the cerebellar cortex in mice lacking vimentin, gfap, and both vimentin and gfap. Glia, 31, 69–83. [DOI] [PubMed] [Google Scholar]
- Glass, C. K. , Saijo, K. , Winner, B. , Marchetto, M. C. , & Gage, F. H. (2010). Mechanisms underlying inflammation in neurodegen‐ eration. Cell, 140, 918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogishvili, D. , Illes‐Toth, E. , Harris, M. J. , Hopley, C. , Teunissen, C. E. , & Abeln, S. (2023). Structural flexibility and heterogeneity of recombinant human glial fibrillary acidic protein (gfap). Proteins: Structure, Function, and Bioinformatics, 92(5), 649–664. [DOI] [PubMed] [Google Scholar]
- Graceffa, P. (2011). Hsp27‐actin interaction. Biochemistry Research International, 2011, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann, D. , Loris, E. , Maas‐Omlor, S. , Huang, W. , Scheller, A. , Kirchhoff, F. , & Schäfer, K. H. (2019). Enteric glia: S100, gfap, and beyond. The Anatomical Record, 302, 1333–1344. [DOI] [PubMed] [Google Scholar]
- Guay, J. , Lambert, H. , Gingras‐Breton, G. , Lavoie, J. N. , Huot, J. , & Landry, J. (1997). Regulation of actin filament dynamics by p38 map kinase‐mediated phosphorylation of heat shock protein 27. Journal of Cell Science, 110, 357–368. [DOI] [PubMed] [Google Scholar]
- Gullotta, F. , Schindler, F. , Schmutzler, R. , & Weeks‐Seifert, A. (1985). Gfap in brain tumor diagnosis: Possibilities and limitations. Pathology Practice, 180, 54–60. [DOI] [PubMed] [Google Scholar]
- Guo, Y. , You, J. , Zhang, Y. , Liu, W. S. , Huang, Y. Y. , Zhang, Y. R. , Zhang, W. , Dong, Q. , Feng, J. F. , Cheng, W. , & Yu, J. T. (2024). Plasma proteomic profiles predict future dementia in healthy adults. Nature Aging, 4, 247–260. [DOI] [PubMed] [Google Scholar]
- Hainfellner, J. A. , Voigtländer, T. , Ströbel, T. , Mazal, P. R. , Maddalena, A. S. , Aguzzi, A. , & Budka, H. (2001). Fibroblasts can express glial fibrillary acidic protein (gfap) in vivo. Journal of Neuropathology and Experimental Neurology, 60, 449–461. [DOI] [PubMed] [Google Scholar]
- Haslbeck, M. , & Vierling, E. (2015). A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. Journal of Molecular Biology, 427, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck, M. , Weinkauf, S. , & Buchner, J. (2019). Small heat shock proteins: Simplicity meets complexity. The Journal of Biological Chemistry, 294, 2121–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimfarth, L. , Passos, F. R. S. , Monteiro, B. S. , Araújo, A. A. S. , Quintans Júnior, L. J. , & Quintans, J. S. S. (2022). Serum glial fibrillary acidic protein is a body fluid biomarker: A valuable prognostic for neurological disease–a systematic review. International Immunopharmacology, 107, 108624. [DOI] [PubMed] [Google Scholar]
- Helman, G. , Takanohashi, A. , Hagemann, T. L. , Perng, M. D. , Walkiewicz, M. , Woidill, S. , Sase, S. , Cross, Z. , du, Y. , Zhao, L. , Waldman, A. , Haake, B. C. , Fatemi, A. , Brenner, M. , Sherbini, O. , Messing, A. , Vanderver, A. , & Simons, C. (2020). Type ii alexander disease caused by splicing errors and aberrant overexpression of an uncharacterized gfap isoform. Human Mutation, 41, 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, J. H. , Seyfried, N. T. , Duong, D. M. , Xia, Q. , Rees, H. D. , Gearing, M. , Peng, J. , Lah, J. J. , & Levey, A. I. (2010). Phosphoproteomic analysis reveals site‐specific changes in gfap and ndrg2 phosphorylation in frontotemporal lobar degeneration. Journal of Proteome Research, 9, 6368–6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högel, H. , Rissanen, E. , Barro, C. , Matilainen, M. , Nylund, M. , Kuhle, J. , & Airas, L. (2020). Serum glial fibrillary acidic protein correlates with multiple sclerosis disease severity. Multiple Sclerosis Journal, 26, 210–219. [DOI] [PubMed] [Google Scholar]
- Høie, M. H. , Kiehl, E. N. , Petersen, B. , Nielsen, M. , Winther, O. , Nielsen, H. , Hallgren, J. , & Marcatili, P. (2022). Netsurfp‐3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Research, 50, W510–W515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck, P. V. , Zhang, B. , Murray, B. , Kornhauser, J. M. , Latham, V. , & Skrzypek, E. (2014). PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Research, 43, D512–D520. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, B. , Fang, C. , & Chang, J. (2021). Blood–brain barrier breakdown: An emerging biomarker of cognitive impairment in normal aging and dementia. Frontiers in Neuroscience, 15, 688090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki, M. , Takahara, H. , Nishi, Y. , Sugawara, K. , & Sato, C. (1989). Ca2+−dependent deimination‐induced disassembly of intermediate filaments involves specific modification of the amino‐terminal head domain. The Journal of Biological Chemistry, 264, 18119–18127. [PubMed] [Google Scholar]
- Ishigami, A. , Masutomi, H. , Handa, S. , Nakamura, M. , Nakaya, S. , Uchida, Y. , Saito, Y. , Murayama, S. , Jang, B. , Jeon, Y. C. , Choi, E. K. , Kim, Y. S. , Kasahara, Y. , Maruyama, N. , & Toda, T. (2015). Mass spectrometric identification of citrullination sites and immunohistochemical detection of citrullinated glial fibrillary acidic protein in alzheimer's disease brains. Journal of Neuroscience Research, 93, 1664–1674. [DOI] [PubMed] [Google Scholar]
- Ishiki, A. , Kamada, M. , Kawamura, Y. , Terao, C. , Shimoda, F. , Tomita, N. , Arai, H. , & Furukawa, K. (2016). Glial fibrillar acidic protein in the cerebrospinal fluid of Alzheimer's disease, dementia with lewy bodies, and frontotemporal lobar degeneration. Journal of Neurochemistry, 136, 258–261. [DOI] [PubMed] [Google Scholar]
- Janelidze, S. , Stomrud, E. , Smith, R. , Palmqvist, S. , Mattsson, N. , Airey, D. C. , Proctor, N. K. , Chai, X. , Shcherbinin, S. , Sims, J. R. , Triana‐Baltzer, G. , Theunis, C. , Slemmon, R. , Mercken, M. , Kolb, H. , Dage, J. L. , & Hansson, O. (2020). Cerebrospinal fluid p‐tau217 performs better than p‐tau181 as a biomarker of alzheimer's disease. Nature Communications, 11, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jany, P. L. , Agosta, G. E. , Benko, W. S. , Eickhoff, J. C. , Keller, S. R. , Köehler, W. , Koeller, D. , Mar, S. , Naidu, S. , Marie Ness, J. , Pareyson, D. , Renaud, D. L. , Salsano, E. , Schiffmann, R. , Simon, J. , Vanderver, A. , Eichler, F. , van der Knaap, M. S. , & Messing, A. (2015). Csf and blood levels of gfap in alexander disease. Eneuro, 2, ENEURO.0080–ENEU15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen, N. A. , Munk, A. S. F. , Lundgaard, I. , & Nedergaard, M. (2015). The glymphatic system: A beginner's guide. Neurochemical Research, 40, 2583–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiwaji, Z. , & Hardingham, G. E. (2022). Good, bad, and neglectful: Astrocyte changes in neurodegenerative disease. Free Radical Biology & Medicine, 182, 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S. C. , Suárez‐Calvet, M. , Suridjan, I. , Minguillón, C. , Gispert, J. D. , Jonaitis, E. , Michna, A. , Carboni, M. , Bittner, T. , Rabe, C. , Kollmorgen, G. , Zetterberg, H. , & Blennow, K. (2023). Identifying clinically useful biomarkers in neurodegenerative disease through a collaborative approach: The neurotoolkit. Alzheimer's Research & Therapy, 15, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper, J. , Evans, R. , Pritzel, A. , Green, T. , Figurnov, M. , Ronneberger, O. , Tunyasuvunakool, K. , Bates, R. , Žídek, A. , Potapenko, A. , Bridgland, A. , Meyer, C. , Kohl, S. A. A. , Ballard, A. J. , Cowie, A. , Romera‐Paredes, B. , Nikolov, S. , Jain, R. , Adler, J. , … Hassabis, D. (2021). Highly accurate protein structure prediction with alphafold. Nature, 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis, W. , Mamber, C. , Moeton, M. , Kooijman, L. , Sluijs, J. A. , Jansen, A. H. P. , Verveer, M. , de Groot, L. R. , Smith, V. D. , Rangarajan, S. , Rodríguez, J. J. , Orre, M. , & Hol, E. M. (2012). Gfap isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of alzheimer disease. PLoS One, 7, e42823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis, W. , Middeldorp, J. , Kooijman, L. , Sluijs, J. A. , Kooi, E. J. , Moeton, M. , Freriks, M. , Mizee, M. R. , & Hol, E. M. (2014). Glial fibrillary acidic protein isoform expression in plaque related astrogliosis in alzheimer's disease. Neurobiology of Aging, 35, 492–510. [DOI] [PubMed] [Google Scholar]
- Karlsson, M. , Zhang, C. , Méar, L. , Zhong, W. , Digre, A. , Katona, B. , Sjöstedt, E. , Butler, L. , Odeberg, J. , Dusart, P. , Edfors, F. , Oksvold, P. , von Feilitzen, K. , Zwahlen, M. , Arif, M. , Altay, O. , Li, X. , Ozcan, M. , Mardinoglu, A. , … Lindskog, C. (2021). A single–cell type transcriptomics map of human tissues. Science Advances, 7, eabh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, H. , Yamamoto, T. , Yamamoto, H. , Ohi, R. , So, N. , & Iwasaki, Y. (1990). Immunocytochemical characterization of supporting cells in the enteric nervous system in hirschsprung's disease. Journal of Pediatric Surgery, 25, 514–519. [DOI] [PubMed] [Google Scholar]
- Kawajiri, A. , Yasui, Y. , Goto, H. , Tatsuka, M. , Takahashi, M. , Nagata, K. I. , & Inagaki, M. (2003). Functional significance of the specific sites phosphorylated in desmin at cleavage furrow: Aurora‐b may phosphorylate and regulate type iii intermediate filaments during cytokinesis coordinatedly with rho‐kinase. Molecular Biology of the Cell, 14, 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B. , Kim, S. , & Jin, M. S. (2018). Crystal structure of the human glial fibrillary acidic protein 1b domain. Biochemical and Biophysical Research Communications, 503, 2899–2905. [DOI] [PubMed] [Google Scholar]
- Kivisäkk, P. , Carlyle, B. C. , Sweeney, T. , Trombetta, B. A. , LaCasse, K. , el‐Mufti, L. , Tuncali, I. , Chibnik, L. B. , das, S. , Scherzer, C. R. , Johnson, K. A. , Dickerson, B. C. , Gomez‐Isla, T. , Blacker, D. , Oakley, D. H. , Frosch, M. P. , Hyman, B. T. , Aghvanyan, A. , Bathala, P. , … Arnold, S. E. (2023). Plasma biomarkers for diagnosis of alzheimer's disease and prediction of cognitive decline in individuals with mild cognitive impairment. Frontiers in Neurology, 14, 1069411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen, M. S. , Jespersen, M. C. , Nielsen, H. , Jensen, K. K. , Jurtz, V. I. , Sønderby, C. K. , Sommer, M. O. A. , Winther, O. , Nielsen, M. , Petersen, B. , & Marcatili, P. (2019). Netsurfp‐2.0: Improved prediction of protein structural features by integrated deep learning. Proteins: Structure, Function, and Bioinformatics, 87, 520–527. [DOI] [PubMed] [Google Scholar]
- Korolainen, M. A. , Auriola, S. , Nyman, T. A. , Alafuzoff, I. , & Pirttilä, T. (2005). Proteomic analysis of glial fibrillary acidic protein in alzheimer's disease and aging brain. Neurobiology of Disease, 20, 858–870. [DOI] [PubMed] [Google Scholar]
- Kumar, A. , Fontana, I. C. , & Nordberg, A. (2021). Reactive astrogliosis: A friend or foe in the pathogenesis of alzheimer's disease. Journal of Neurochemistry, 164, 309–324. [DOI] [PubMed] [Google Scholar]
- Kwon, B. K. , Stammers, A. M. T. , Belanger, L. M. , Bernardo, A. , Chan, D. , Bishop, C. M. , Slobogean, G. P. , Zhang, H. , Umedaly, H. , Giffin, M. , Street, J. , Boyd, M. C. , Paquette, S. J. , Fisher, C. G. , & Dvorak, M. F. (2010). Cerebrospinal fluid inflammatory cytokines and biomarkers of injury severity in acute human spinal cord injury. Journal of Neurotrauma, 27, 669–682. [DOI] [PubMed] [Google Scholar]
- Kyllerman, M. , Rosengren, L. , Wiklund, L.‐M. , & Holmberg, E. (2005). Increased levels of gfap in the cerebrospinal fluid in three subtypes of genetically confirmed alexander disease. Neuropediatrics, 36, 319–323. [DOI] [PubMed] [Google Scholar]
- Leister, I. , Altendorfer, B. , Maier, D. , Mach, O. , Wutte, C. , Grillhösl, A. , Arevalo‐Martin, A. , Garcia‐Ovejero, D. , Aigner, L. , & Grassner, L. (2023). Trajectory of serum levels of glial fibrillary acidic protein within four weeks post‐injury is related to neurological recovery during the transition from acute to chronic spinal cord injury. Journal of Neurotrauma, 40, 999–1006. [DOI] [PubMed] [Google Scholar]
- Li, H. , Guo, Y. , Teng, J. , Ding, M. , Yu, A. C. H. , & Chen, J. (2006). 14‐3‐3γ affects dynamics and integrity of glial filaments by binding to phosphorylated gfap. Journal of Cell Science, 119, 4452–4461. [DOI] [PubMed] [Google Scholar]
- Liedtke, W. , Edelmann, W. , Bieri, P. L. , Chiu, F. C. , Cowan, N. J. , Kucherlapati, R. , & Raine, C. S. (1996). Gfap is necessary for the integrity of cns white matter architecture and long‐term maintenance of myelination. Neuron, 17, 607–615. [DOI] [PubMed] [Google Scholar]
- Lin, N. H. , Jian, W. S. , Snider, N. , & Perng, M. D. (2024). Glial fibrillary acidic protein is pathologically modified in Alexander disease. The Journal of Biological Chemistry, 107402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, N.‐H. , Yang, A.‐W. , Chang, C.‐H. , & Perng, M.‐D. (2021). Elevated gfap isoform expression promotes protein aggregation and compromises astrocyte function. The FASEB Journal, 35, e21614. [DOI] [PubMed] [Google Scholar]
- Lowery, J. , Kuczmarski, E. R. , Herrmann, H. , & Goldman, R. D. (2015). Intermediate filaments play a pivotal role in regulating cell architecture and function. The Journal of Biological Chemistry, 290, 17145–17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, C. A. , Brunkhorst, R. , Niessner, M. , Pfeilschifter, W. , Steinmetz, H. , & Foerch, C. (2013). Blood levels of glial fibrillary acidic protein (gfap) in patients with neurological diseases. PLoS One, 8, e62101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, P. J. , Panczykowski, D. M. , Yue, J. K. , Puccio, A. M. , Inoue, T. , Sorani, M. D. , Lingsma, H. F. , Maas, A. I. R. , Valadka, A. B. , Yuh, E. L. , Mukherjee, P. , Manley, G. T. , Okonkwo, D. O. , TRACK‐TBI investigators including , Casey, S. S. , Cheong, M. , Cooper, S. R. , Dams‐O'Connor, K. , Gordon, W. A. , … Vassar, M. J. (2015). Measurement of the glial fibrillary acidic protein and its breakdown products gfap‐bdp biomarker for the detection of traumatic brain injury compared to computed tomography and magnetic resonance imaging. Journal of Neurotrauma, 32, 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing, A. (2018). Alexander disease. Handbook of Clinical Neurology, 148, 693–700. [DOI] [PubMed] [Google Scholar]
- Messing, A. , & Brenner, M. (2020). Gfap at 50. ASN Neuro, 12, 1759091420949680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing, A. , Head, M. W. , Galles, K. , Galbreath, E. J. , Goldman, J. E. , & Brenner, M. (1998). Fatal encephalopathy with astrocyte inclusions in gfap transgenic mice. The American Journal of Pathology, 152, 391–398. [PMC free article] [PubMed] [Google Scholar]
- Middeldorp, J. , & Hol, E. (2011). Gfap in health and disease. Progress in Neurobiology, 93, 421–443. [DOI] [PubMed] [Google Scholar]
- Milà‐Alomà, M. , Ashton, N. J. , Shekari, M. , Salvadó, G. , Ortiz‐Romero, P. , Montoliu‐Gaya, L. , Benedet, A. L. , Karikari, T. K. , Lantero‐Rodriguez, J. , Vanmechelen, E. , Day, T. A. , González‐Escalante, A. , Sánchez‐Benavides, G. , Minguillon, C. , Fauria, K. , Molinuevo, J. L. , Dage, J. L. , Zetterberg, H. , Gispert, J. D. , … Blennow, K. (2022). Plasma p‐tau231 and p‐tau217 as state markers of amyloid‐β pathology in preclinical Alzheimer's disease. Nature Medicine, 28(9), 1797–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello, S. , Sorinola, A. , Czeiter, E. , Vámos, Z. , Amrein, K. , Synnot, A. , Donoghue, E. , Sándor, J. , Wang, K. K. W. , Diaz‐Arrastia, R. , Steyerberg, E. W. , Menon, D. K. , Maas, A. I. R. , & Buki, A. (2021). Blood‐based protein biomarkers for the management of traumatic brain injuries in adults presenting to emergency departments with mild brain injury: A living systematic review and meta‐analysis. Journal of Neurotrauma, 38, 1086–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoliu‐Gaya, L. , Alcolea, D. , Ashton, N. J. , Pegueroles, J. , Levin, J. , Bosch, B. , Lantero‐Rodriguez, J. , Carmona‐Iragui, M. , Wagemann, O. , Balasa, M. , Kac, P. R. , Barroeta, I. , Lladó, A. , Brum, W. S. , Videla, L. , Gonzalez‐Ortiz, F. , Benejam, B. , Arranz Martínez, J. J. , Karikari, T. K. , … Fortea, J. (2023). Plasma and cerebrospinal fluid glial fibrillary acidic protein levels in adults with down syndrome: A longitudinal cohort study. eBioMedicine, 90, 104547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawashiro, H. , Brenner, M. , Fukui, S. , Shima, K. , & Hallenbeck, J. M. (2000). High susceptibility to cerebral ischemia in gfap‐null mice. Journal of Cerebral Blood Flow and Metabolism, 20, 1040–1044. [DOI] [PubMed] [Google Scholar]
- Nawashiro, H. , Messing, A. , Azzam, N. , & Brenner, M. (1998). Mice lacking gfap are hypersensitive to traumatic cerebrospinal injury. Neuroreport, 9, 1691–1696. [DOI] [PubMed] [Google Scholar]
- Nicholas, A. P. , Sambandam, T. , Echols, J. D. , & Tourtellotte, W. W. (2004). Increased citrullinated glial fibrillary acidic protein in secondary progressive multiple sclerosis. The Journal of Comparative Neurology, 473, 128–136. [DOI] [PubMed] [Google Scholar]
- Nielsen, A. L. , & Jørgensen, A. L. (2003). Structural and functional characterization of the zebrafish gene for glial fibrillary acidic protein, gfap. Gene, 310, 123–132. [DOI] [PubMed] [Google Scholar]
- Oeckl, P. , Anderl‐Straub, S. , von Arnim, C. A. F. , Baldeiras, I. , Diehl‐Schmid, J. , Grimmer, T. , Halbgebauer, S. , Kort, A. M. , Lima, M. , Marques, T. M. , Ortner, M. , Santana, I. , Steinacker, P. , Verbeek, M. M. , Volk, A. E. , Ludolph, A. C. , & Otto, M. (2022). Serum gfap differentiates alzheimer's disease from frontotemporal dementia and predicts mci‐to‐dementia conversion. Journal of Neurology, Neurosurgery, and Psychiatry, 93, 659–667. [DOI] [PubMed] [Google Scholar]
- Oeckl, P. , Halbgebauer, S. , Anderl‐Straub, S. , Steinacker, P. , Huss, A. M. , Neugebauer, H. , von Arnim, C. A. , Diehl‐Schmid, J. , Grimmer, T. , Kornhuber, J. , Lewczuk, P. , Danek, A. , Consortium for Frontotemporal Lobar Degeneration German , Ludolph, A. C. , & Otto, M. (2019). Glial fibrillary acidic protein in serum is increased in Alzheimer's disease and correlates with cognitive impairment. Journal of Alzheimer's Disease, 67(2), 481–488. [DOI] [PubMed] [Google Scholar]
- Okonkwo, D. O. , Yue, J. K. , Puccio, A. M. , Panczykowski, D. M. , Inoue, T. , McMahon, P. J. , Sorani, M. D. , Yuh, E. L. , Lingsma, H. F. , Maas, A. I. R. , Valadka, A. B. , Manley and Transforming Research an, G. T. , Casey, S. S. , Cheong, M. , Cooper, S. R. , Dams‐O'Connor, K. , Gordon, W. A. , Hricik, A. J. , Hochberger, K. , … Vassar, M. J. (2013). Gfap‐bdp as an acute diagnostic marker in traumatic brain injury: Results from the prospective transforming research and clinical knowledge in traumatic brain injury study. Journal of Neurotrauma, 30, 1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, B. , Lautner, R. , Andreasson, U. , Öhrfelt, A. , Portelius, E. , Bjerke, M. , Hölttä, M. , Rosén, C. , Olsson, C. , Strobel, G. , Wu, E. , Dakin, K. , Petzold, M. , Blennow, K. , & Zetterberg, H. (2016). Csf and blood biomarkers for the diagnosis of alzheimer's disease: A systematic review and meta‐analysis. The Lancet Neurology, 15, 673–684. [DOI] [PubMed] [Google Scholar]
- Palmqvist, S. , Janelidze, S. , Quiroz, Y. T. , Zetterberg, H. , Lopera, F. , Stomrud, E. , Su, Y. , Chen, Y. , Serrano, G. E. , Leuzy, A. , Mattsson‐Carlgren, N. , Strandberg, O. , Smith, R. , Villegas, A. , Sepulveda‐Falla, D. , Chai, X. , Proctor, N. K. , Beach, T. G. , Blennow, K. , … Hansson, O. (2020). Discriminative accuracy of plasma phospho‐tau217 for alzheimer disease vs other neurodegenerative disorders. JAMA, 324, 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, L. , Brophy, G. M. , Alvarez, W. , Hirschl, R. , Cress, M. , Weber, K. , & Giordano, P. (2023). Sex differences in time course and diagnostic accuracy of gfap and uch‐l1 in trauma patients with mild traumatic brain injury. Scientific Reports, 13, 11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, L. , Brophy, G. M. , Welch, R. D. , Lewis, L. M. , Braga, C. F. , Tan, C. N. , Ameli, N. J. , Lopez, M. A. , Haeussler, C. A. , Mendez Giordano, D. I. , Silvestri, S. , Giordano, P. , Weber, K. D. , Hill‐Pryor, C. , & Hack, D. C. (2016). Time course and diagnostic accuracy of glial and neuronal blood biomarkers gfap and uch‐l1 in a large cohort of trauma patients with and without mild traumatic brain injury. JAMA Neurology, 73, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, L. , Lewis, L. M. , Falk, J. L. , Zhang, Z. , Silvestri, S. , Giordano, P. , Brophy, G. M. , Demery, J. A. , Dixit, N. K. , Ferguson, I. , Liu, M. C. , Mo, J. , Akinyi, L. , Schmid, K. , Mondello, S. , Robertson, C. S. , Tortella, F. C. , Hayes, R. L. , & Wang, K. K. W. (2012). Elevated levels of serum glial fibrillary acidic protein breakdown products in mild and moderate traumatic brain injury are associated with intracranial lesions and neurosurgical intervention. Annals of Emergency Medicine, 59, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, D. A. , & Steinert, P. M. (1999). Intermediate filaments: Molecular architecture, assembly, dynamics and polymorphism. Quarterly Reviews of Biophysics, 32, 99–187. [DOI] [PubMed] [Google Scholar]
- Pérez‐Grijalba, V. , Romero, J. , Pesini, P. , Sarasa, L. , Monleón, I. , San‐José, I. , Arbizu, J. , Martínez‐Lage, P. , Munuera, J. , Ruiz, A. , Tárraga, L. , Boada, M. , Sarasa, M. , & AB255 Study Group . (2019). Plasma Aβ42/40 ratio detects early stages of Alzheimer's disease and correlates with CSF and neuroimaging biomarkers in the AB255 study. The Journal of Prevention of Alzheimer's Disease, 6, 34–41. [DOI] [PubMed] [Google Scholar]
- Perng, M. D. , Cairns, L. , van den Ijssel, P. , Prescott, A. , Hutcheson, A. M. , & Quinlan, R. A. (1999). Intermediate filament interactions can be altered by hsp27 and alphab‐crystallin. Journal of Cell Science, 112, 2099–2112. [DOI] [PubMed] [Google Scholar]
- Perng, M.‐D. , Wen, S. F. , Gibbon, T. , Middeldorp, J. , Sluijs, J. , Hol, E. M. , & Quinlan, R. A. (2008). Glial fibrillary acidic protein filaments can tolerate the incorporation of assembly‐compromised gfap‐δ, but with consequences for filament organization and αb‐crystallin association. Molecular Biology of the Cell, 19, 4521–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C. L. , Faridounnia, M. , Armao, D. , & Snider, N. T. (2023). Stability dynamics of neurofilament and gfap networks and protein fragments. Current Opinion in Cell Biology, 85, 102266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plog, B. A. , Dashnaw, M. L. , Hitomi, E. , Peng, W. , Liao, Y. , Lou, N. , Deane, R. , & Nedergaard, M. (2015). Biomarkers of traumatic injury are transported from brain to blood via the glymphatic system. The Journal of Neuroscience, 35, 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchet, R. , Probst, A. , Bouras, C. , Dráberová, E. , Dráber, P. , & Riederer, B. M. (2003). Analysis of gial acidic fibrillary protein in the human entorhinal cortex during aging and in Alzheimer's disease. PROTEOMICS: International Edition, 3(8), 1476–1485. [DOI] [PubMed] [Google Scholar]
- Potokar, M. , Kreft, M. , Li, L. , Daniel Andersson, J. , Pangršič, T. , Chowdhury, H. H. , Pekny, M. , & Zorec, R. (2007). Cytoskeleton and vesicle mobility in astrocytes. Traffic, 8, 12–20. [DOI] [PubMed] [Google Scholar]
- Pujar, S. , O'Leary, N. A. , Farrell, C. M. , Loveland, J. E. , Mudge, J. M. , Wallin, C. , Girón, C. G. , Diekhans, M. , Barnes, I. , Bennett, R. , Berry, A. E. , Cox, E. , Davidson, C. , Goldfarb, T. , Gonzalez, J. M. , Hunt, T. , Jackson, J. , Joardar, V. , Kay, M. P. , … Pruitt, K. D. (2018). Consensus coding sequence (ccds) database: A standardized set of human and mouse protein‐coding regions supported by expert curation. Nucleic Acids Research, 46, D221–D228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridet, J. , Privat, A. , Malhotra, S. , & Gage, F. (1997). Reactive astrocytes: Cellular and molecular cues to biological function. Trends in Neurosciences, 20, 570–577. [DOI] [PubMed] [Google Scholar]
- Rozovsky, I. , Wei, M. , Stone, D. J. , Zanjani, H. , Anderson, C. P. , Morgan, T. E. , & Finch, C. E. (2002). Estradiol (e2) enhances neurite outgrowth by repressing glial fibrillary acidic protein expression and reorganizing laminin. Endocrinology, 143, 636–646. [DOI] [PubMed] [Google Scholar]
- Rutka, J. T. , Hubbard, S. L. , Fukuyama, K. , Matsuzawa, K. , Dirks, P. B. , & Becker, L. E. (1994). Effects of antisense glial fibrillary acidic protein complementary dna on the growth, invasion, and adhesion of human astrocytoma cells. Cancer Research, 54, 3267–3272. [PubMed] [Google Scholar]
- Schindler, S. E. , Bollinger, J. G. , Ovod, V. , Mawuenyega, K. G. , Li, Y. , Gordon, B. A. , Holtzman, D. M. , Morris, J. C. , Benzinger, T. L. S. , Xiong, C. , Fagan, A. M. , & Bateman, R. J. (2019). High‐precision plasma β ‐amyloid 42/40 predicts current and future brain amyloidosis. Neurology, 93, e1647–e1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H. , Kretzschmar, B. , Lingor, P. , Pauli, S. , Schramm, P. , Otto, M. , Ohlenbusch, A. , & Brockmann, K. (2013). Acute onset of adult alexander disease. Journal of the Neurological Sciences, 331, 152–154. [DOI] [PubMed] [Google Scholar]
- Shaw, G. , Madorsky, I. , Li, Y. , Wang, Y. S. , Jorgensen, M. , Rana, S. , & Fuller, D. D. (2023). Uman‐type neurofilament light antibodies are effective reagents for the imaging of neurodegeneration. Brain Communications, 5, fcad067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, M. , Matsubara, E. , Kanai, M. , Watanabe, M. , Nakamura, T. , Tomidokoro, Y. , Shizuka, M. , Wakabayashi, K. , Igeta, Y. , Ikeda, Y. , Mizushima, K. , Amari, M. , Ishiguro, K. , Kawarabayashi, T. , Harigaya, Y. , Okamoto, K. , & Hirai, S. (1998). Combination assay of csf tau, aβ 1‐40 and aβ 1‐42 (43) as a biochemical marker of alzheimer's disease. J. Neurological Sciences, 158, 134–140. [DOI] [PubMed] [Google Scholar]
- Simrén, J. , Weninger, H. , Brum, W. S. , Khalil, S. , Benedet, A. L. , Blennow, K. , Zetterberg, H. , & Ashton, N. J. (2022). Differences between blood and cerebrospinal fluid glial fibrillary acidic protein levels: The effect of sample stability. Alzheimer's & Dementia, 18, 1988–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider, N. T. , & Omary, M. B. (2014). Post‐translational modifications of intermediate filament proteins: Mechanisms and functions. Nature Reviews. Molecular Cell Biology, 15, 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanos, M. , Shachar, S. , Sweeney, T. , Lehmann, H. I. , Gokulnath, P. , Li, G. , Sigal, G. B. , Nagaraj, R. , Bathala, P. , Rana, F. , Shah, R. V. , Routenberg, D. A. , & das, S. (2022). Elevation of neural injury markers in patients with neurologic sequelae after hospitalization for sars‐cov‐2 infection. Iscience, 25, 104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, S. M. , Sullivan, R. K. P. , Miller, S. M. , Ireland, Z. , Björkman, S. T. , Pow, D. V. , & Colditz, P. B. (2012). Phosphorylation of gfap is associated with injury in the neonatal pig hypoxic‐ischemic brain. Neurochemical Research, 37, 2364–2378. [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Han, L. , Li, S. , Hu, T. , Xu, Z. , Fan, Y. , Liang, X. , Yu, H. , Wu, J. , & Wang, J. (2023). Plasma GFAP in Parkinson's disease with cognitive impairment and its potential to predict conversion to dementia. npj . Parkinson's Disease, 9(1), 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen, C. E. , Verberk, I. M. W. , Thijssen, E. H. , Vermunt, L. , Hansson, O. , Zetterberg, H. , van der Flier, W. M. , Mielke, M. M. , & del Campo, M. (2022). Blood‐based biomarkers for alzheimer's disease: Towards clinical implementation. The Lancet Neurology, 21, 66–77. [DOI] [PubMed] [Google Scholar]
- Thijssen, E. H. , Verberk, I. M. W. , Kindermans, J. , Abramian, A. , Vanbrabant, J. , Ball, A. J. , Pijnenburg, Y. , Lemstra, A. W. , van der Flier, W. M. , Stoops, E. , Hirtz, C. , & Teunissen, C. E. (2022). Differential diagnostic performance of a panel of plasma biomarkers for different types of dementia. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring, 14(1), e12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul, P. J. , Åkesson, L. , Wiking, M. , Mahdessian, D. , Geladaki, A. , Ait Blal, H. , Alm, T. , Asplund, A. , Björk, L. , Breckels, L. M. , Bäckström, A. , Danielsson, F. , Fagerberg, L. , Fall, J. , Gatto, L. , Gnann, C. , Hober, S. , Hjelmare, M. , Johansson, F. , … Lundberg, E. (2017). A subcellular map of the human proteome. Science, 356, eaal3321. [DOI] [PubMed] [Google Scholar]
- Tian, R. , Gregor, M. , Wiche, G. , & Goldman, J. E. (2006). Plectin regulates the organization of glial fibrillary acidic protein in alexander disease. The American Journal of Pathology, 168, 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomokane, N. , Iwaki, T. , Tateishi, J. , Iwaki, A. , & Goldman, J. (1991). Rosenthal fibers share epitopes with alpha b‐crystallin, glial fibrillary acidic protein, and ubiquitin, but not with vimentin. Immunoelectron microscopy with colloidal gold. The American Journal of Pathology, 138, 875. [PMC free article] [PubMed] [Google Scholar]
- Uhlén, M. , Fagerberg, L. , Hallström, B. M. , Lindskog, C. , Oksvold, P. , Mardinoglu, A. , Sivertsson, Å. , Kampf, C. , Sjöstedt, E. , Asplund, A. , Olsson, I. M. , Edlund, K. , Lundberg, E. , Navani, S. , Szigyarto, C. A. K. , Odeberg, J. , Djureinovic, D. , Takanen, J. O. , Hober, S. , … Pontén, F. (2015). Tissue‐based map of the human proteome. Science, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- US Food and Drug Administration . (2018). Evaluation of automatic Class III designation for Banyan Brain Trauma Indicator: Decision memorandum (pp. 1–32). US Food and Drug Administration. [Google Scholar]
- van Asperen, J. V. , Fedorushkova, D. M. , Robe, P. A. , & Hol, E. M. (2022). Investigation of glial fibrillary acidic protein (gfap) in body fluids as a potential biomarker for glioma: A systematic review and meta‐analysis. Biomarkers, 27, 1–12. [DOI] [PubMed] [Google Scholar]
- van Asperen, J. V. , Robe, P. A. , & Hol, E. M. (2022). Gfap alternative splicing and the relevance for disease–a focus on diffuse gliomas. ASN Neuro, 14, 17590914221102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bodegraven, E. J. , Sluijs, J. A. , Tan, A. K. , Robe, P. A. , & Hol, E. M. (2021). New gfap splice isoform (gfapμ) differentially expressed in glioma translates into 21 kda n‐terminal gfap protein. The FASEB Journal, 35, e21389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberk, I. M. W. , Thijssen, E. , Koelewijn, J. , Mauroo, K. , Vanbrabant, J. , de Wilde, A. , Zwan, M. D. , Verfaillie, S. C. J. , Ossenkoppele, R. , Barkhof, F. , van Berckel, B. N. M. , Scheltens, P. , van der Flier, W. M. , Stoops, E. , Vanderstichele, H. M. , & Teunissen, C. E. (2020). Combination of plasma amyloid beta (1‐42/1‐40) and glial fibrillary acidic protein strongly associates with cerebral amyloid pathology. Alzheimer's Research & Therapy, 12, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberk, I. M. W. , Misdorp, E. O. , Koelewijn, J. , Ball, A. J. , Blennow, K. , Dage, J. L. , Fandos, N. , Hansson, O. , Hirtz, C. , Janelidze, S. , Kang, S. , Kirmess, K. , Kindermans, J. , Lee, R. , Meyer, M. R. , Shan, D. , Shaw, L. M. , Waligorska, T. , West, T. , … Teunissen, C. E. (2022). Characterization of pre‐analytical sample handling effects on a panel of Alzheimer's disease–related blood‐based biomarkers: Results from the Standardization of Alzheimer's Blood Biomarkers (SABB) working group. Alzheimer's & Dementia, 18(8), 1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viedma‐Poyatos, Á. , de Pablo, Y. , Pekny, M. , & Pérez‐Sala, D. (2018). The cysteine residue of glial fibrillary acidic protein is a critical target for lipoxidation and required for efficient network organization. Free Radical Biology & Medicine, 120, 380–394. [DOI] [PubMed] [Google Scholar]
- Waury, K. , Gogishvili, D. , Nieuwland, R. , Chatterjee, M. , Teunissen, C. E. , & Abeln, S. (2024). Proteome encoded determinants of protein sorting into extracellular vesicles. Journal of Extracellular Biology, 3, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waury, K. , Willemse, E. A. J. , Vanmechelen, E. , Zetterberg, H. , Teunissen, C. E. , & Abeln, S. (2022). Bioinformatics tools and data resources for assay development of fluid protein biomarkers. Biomarker Research, 10, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippold, F. , Perry, A. , & Lennerz, J. (2006). Neuropathology for the neuroradiologist: Rosenthal fibers. Am. Journal Neuroradiol‐ Ogy, 27, 958–961. [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Arja, R. D. , Zhu, T. , Sarkis, G. A. , Patterson, R. L. , Romo, P. , Rathore, D. S. , Moghieb, A. , Abbatiello, S. , Robertson, C. S. , Haskins, W. E. , Kobeissy, F. , & Wang, K. K. W. (2022). Characterization of calpain and caspase‐6‐generated glial fibrillary acidic protein breakdown products following traumatic brain injury and astroglial cell injury. International Journal of Molecular Sciences, 23, 8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , & Wang, K. K. (2015). Glial fibrillary acidic protein: From intermediate filament assembly and gliosis to neurobiomarker. Trends in Neurosciences, 38, 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T. , Tomozawa, Y. , Arisato, T. , Okamoto, Y. , Hirano, H. , & Nakagawa, M. (2007). The functional alteration of mutant gfap depends on the location of the domain: Morphological and functional studies using astrocytoma‐derived cells. Journal of Human Genetics, 52, 362–369. [DOI] [PubMed] [Google Scholar]
- Yuan, W. , Lu, L. , Rao, M. , Huang, Y. , Liu, C. E. , Liu, S. , Zhao, Y. , Liu, H. , Zhu, J. , Chao, T. , Wu, C. , Ren, J. , Lv, L. , Li, W. , Qi, S. , Liang, Y. , Yue, S. , Gao, J. , Zhang, Z. , & Kong, E. (2021). Gfap hyperpalmitoylation exacerbates astrogliosis and neurodegenerative pathology in ppt1‐deficient mice. Proceedings of the National Academy of Sciences, 118, e2022261118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenika, D. , Grima, B. , Brenner, M. , & Pessac, B. (1995). A novel glial fibrillary acidic protein mrna lacking exon 1. Molecular Brain Research, 30, 251–258. [DOI] [PubMed] [Google Scholar]
- Zoltewicz, J. S. , Scharf, D. , Yang, B. , Chawla, A. , Newsom, K. J. , & Fang, L. (2012). Characterization of antibodies that detect human gfap after traumatic brain injury. Biomarker Insights, 7, BMI–S9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support this study are openly available. Several tools were employed to analyse the structure of GFAP, namely, NetSurfP 3.0 (Høie et al., 2022; Klausen et al., 2019), AlphaFold‐Multimer (Evans et al., 2021; Gogishvili et al., 2023), and the PAE viewer tool (Elfmann & Stülke, 2023). The Human Protein Atlas (HPA) was utilised for the analysis of GFAP expression, interaction network, and cell localisation (Karlsson et al., 2021; Kawajiri et al., 2003; Thul et al., 2017; Uhlén et al., 2015; Yoshida et al., 2007). The feature overview of GFAP was obtained from curated UniProt annotations (Consortium, T. U, 2023). For mapping exons and splice junctions to the amino acid sequence of GFAPβ, the NCBI‐CCDS database was used (Pujar et al., 2018). Figures 3d and 4 were created with Biorender.com.


