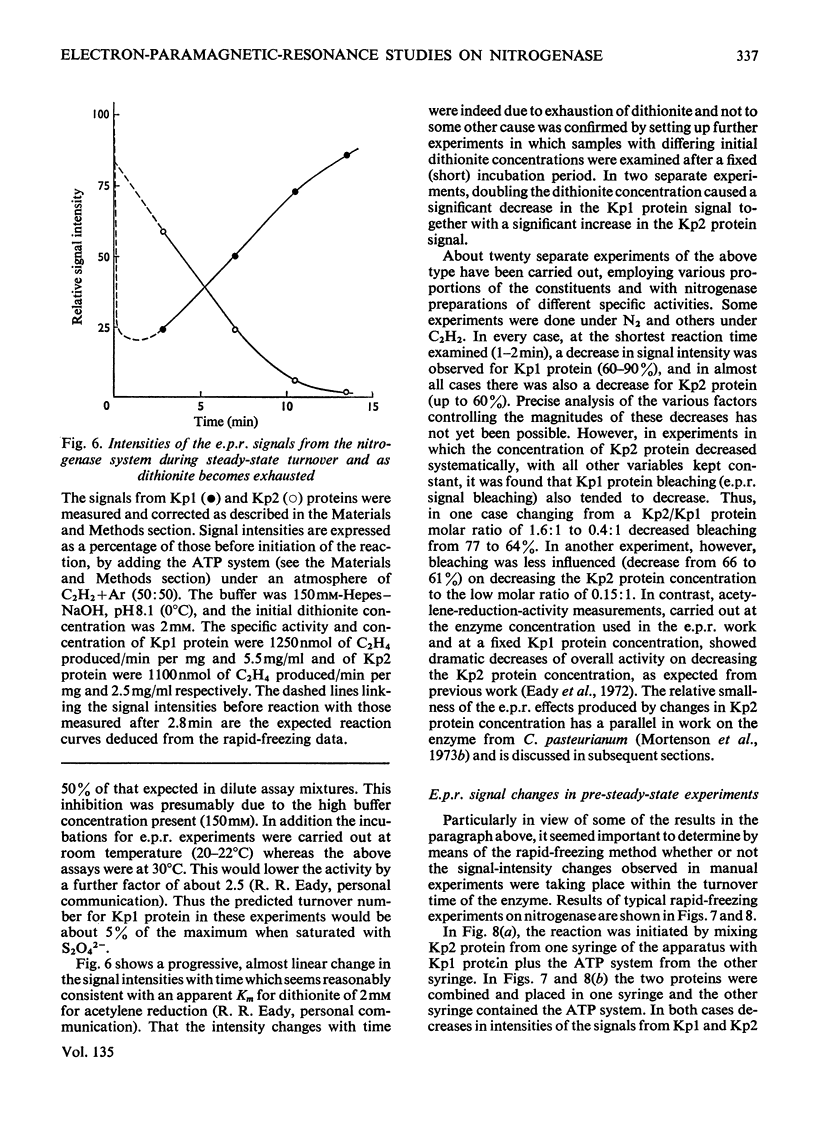
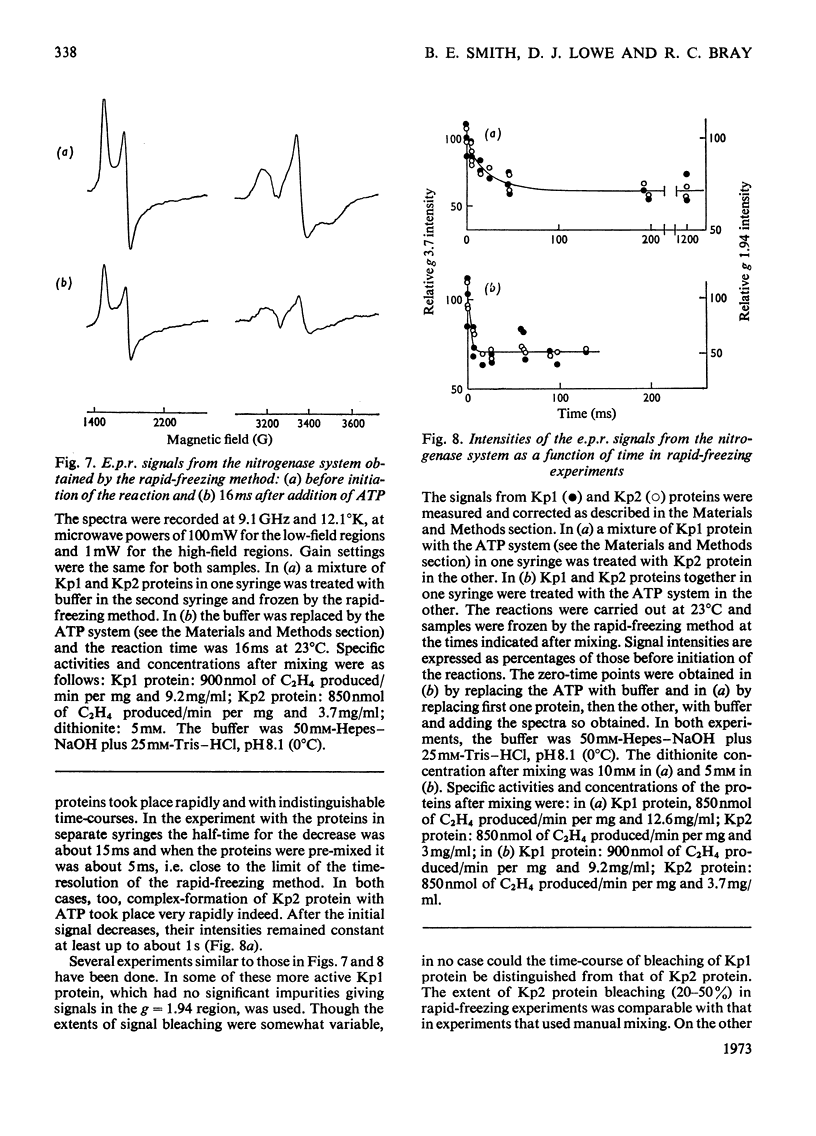
Abstract
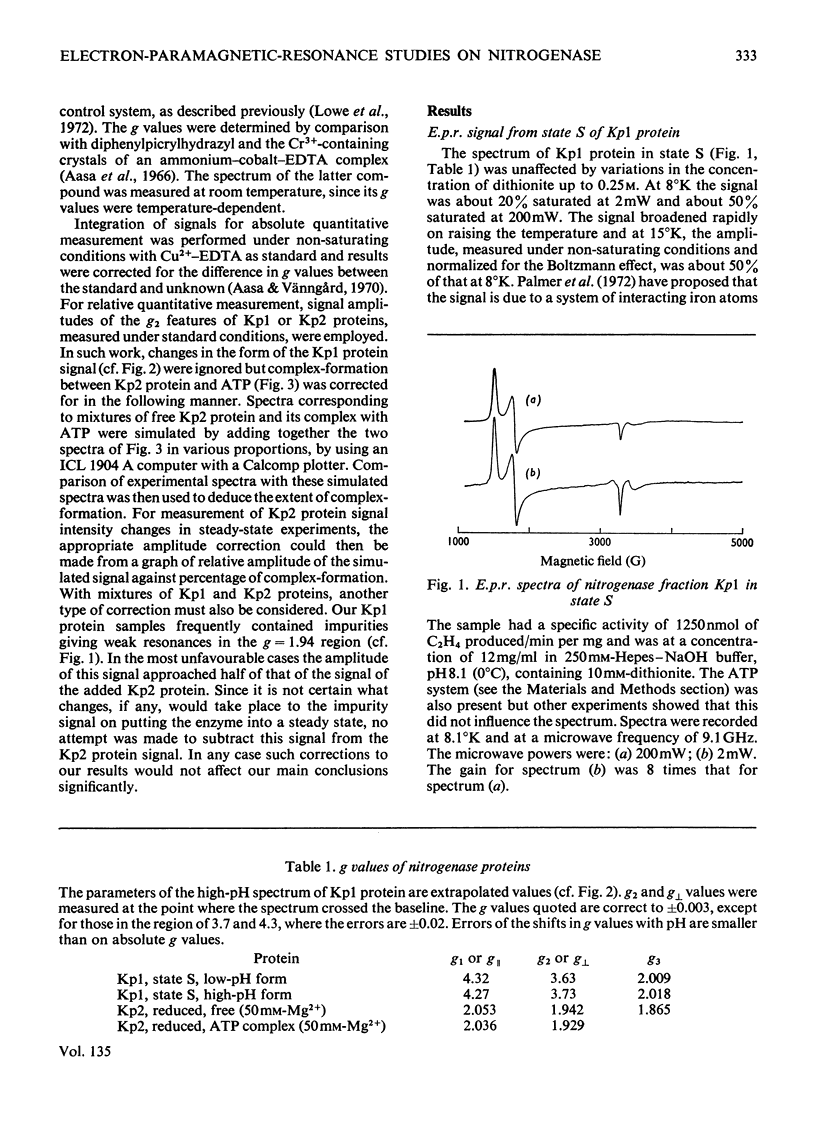
The properties and catalytic reactions of the enzyme nitrogenase purified from Klebsiella pneumoniae were studied by electron-paramagnetic-resonance (e.p.r.) spectroscopy at temperatures down to 8°K. The two protein fractions, Kp1 (the iron–molybdenum protein) and Kp2 (the iron protein), were examined alone and in steady-state mixtures and also in pre-steady-state experiments, by using the rapid-freezing method. Kp1 protein in dithionite solution shows a rhombic type of spectrum with g1 4.32, g2 3.63, g3 2.009 at pH6.8 (0°C). Small changes in the spectrum produced by protons (pK=8.7 at 0°C) or by acetylene indicate binding of these oxidizing substrates to this protein fraction. Kp2 protein shows a rhombic spectrum with g1 2.053, g2 1.942, g3 1.865, which integrates to about 0.45 electron/molecule. Binding of ATP, with a dissociation constant of 4×10−4m, changes the spectrum to an axial form with g∥ 2.036, g⊥ 1.929, thus indicating a conformation change of Kp2 protein. The Kp2 protein spectrum disappears reversibly on cautious oxidation. The signals of both proteins are diminished in their steady-state mixtures, obtained in the presence of ATP and dithionite (with an ATP-generating system and Mg2+ ions) and with protons, N2 or acetylene as oxidizing substrate. At the same time as dithionite is consumed in such reactions, the Kp1 protein signal is gradually restored and the Kp2 protein signal diminishes to zero. In rapid-freezing experiments the signals from the two proteins decreased at indistinguishable rates (t½ about 10ms), then they remained constant. Results are interpreted in terms of a scheme in which reducing equivalents pass from dithionite to Kp2 protein, then, in an ATP-dependent reaction to Kp1 protein, this being finally reoxidized by N2 or another oxidizing substrate. In this scheme Kp1 protein cycles between its signal-giving state and a very highly reduced signal-free state.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRAY R. C., PETTERSSON R. Electron-spin-resonance measurements. Biochem J. 1961 Oct;81:194–195. doi: 10.1042/bj0810194. [DOI] [PubMed] [Google Scholar]
- BRAY R. C. Sudden freezing as a technique for the study of rapid reactions. Biochem J. 1961 Oct;81:189–193. doi: 10.1042/bj0810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray G. A., Gallagher T. F., Jr Suppression of appetite by bile acids. Lancet. 1968 May 18;1(7551):1066–1067. doi: 10.1016/s0140-6736(68)91415-3. [DOI] [PubMed] [Google Scholar]
- Davis L. C., Shah V. K., Brill W. J., Orme-Johnson W. H. Nitrogenase. II. Changes in the EPR signal of component I (iron-molybdenum protein) of Azotobacter vinelandii nitrogenase during repression and derepression. Biochim Biophys Acta. 1972 Feb 28;256(2):512–523. doi: 10.1016/0005-2728(72)90079-5. [DOI] [PubMed] [Google Scholar]
- Eady R. R., Smith B. E., Cook K. A., Postgate J. R. Nitrogenase of Klebsiella pneumoniae. Purification and properties of the component proteins. Biochem J. 1972 Jul;128(3):655–675. doi: 10.1042/bj1280655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good N. E., Winget G. D., Winter W., Connolly T. N., Izawa S., Singh R. M. Hydrogen ion buffers for biological research. Biochemistry. 1966 Feb;5(2):467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- Lowe D. J., Lynden-Bell R. M., Bray R. C. Spin-spin interaction between molybdenum and one of the iron-sulphur systems of xanthine oxidase and its relevance to the enzymic mechanism. Biochem J. 1972 Nov;130(1):239–249. doi: 10.1042/bj1300239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortenson L. E., Zumpft W. G., Palmer G. Electron paramagnetic resonance studies on nitrogenase. 3. Function of magnesium adenosine 5'-triphosphate and adenosine 5'-diphosphate in catalysis by nitrogenase. Biochim Biophys Acta. 1973 Feb 22;292(2):422–435. doi: 10.1016/0005-2728(73)90048-0. [DOI] [PubMed] [Google Scholar]
- Orme-Johnson W. H., Hamilton W. D., Jones T. L., Tso M. Y., Burris R. H., Shah V. K., Brill W. J. Electron paramagnetic resonance of nitrogenase and nitrogenase components from Clostridium pasteurianum W5 and Azotobacter vinelandii OP. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3142–3145. doi: 10.1073/pnas.69.11.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer G., Multani J. S., Cretney W. C., Zumft W. G., Mortenson L. E. Electron paramagnetic resonance studies on nitrogenase. I. The properties of molybdoferredoxin and azoferredoxin. Arch Biochem Biophys. 1972 Nov;153(1):325–332. doi: 10.1016/0003-9861(72)90452-3. [DOI] [PubMed] [Google Scholar]
- Smith B. E., Lowe D. J., Bray R. C. Nitrogenase of Klebsiella pneumoniae: electron-paramagnetic-resonance studies on the catalytic mechanism. Biochem J. 1972 Nov;130(2):641–643. doi: 10.1042/bj1300641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneley R. N., Eady R. R. Nitrogenase of Klebsiella pneumoniae: evidence for an adenosine triphosphate-induced association of the iron-sulphur protein. Biochem J. 1973 Jun;133(2):405–408. doi: 10.1042/bj1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G., Cretney W. C., Huang T. C., Mortenson L. E., Palmer G. On the structure and function of nitrogenase from Clostridium pasteurianum W5. Biochem Biophys Res Commun. 1972 Sep 26;48(6):1525–1532. doi: 10.1016/0006-291x(72)90887-x. [DOI] [PubMed] [Google Scholar]
- Zumft W. G., Palmer G., Mortenson L. E. Electron paramagnetic resonance studies on nitrogenase. II. Interaction of adenosine 5'-triphosphate with azoferredoxin. Biochim Biophys Acta. 1973 Feb 22;292(2):413–421. doi: 10.1016/0005-2728(73)90047-9. [DOI] [PubMed] [Google Scholar]


