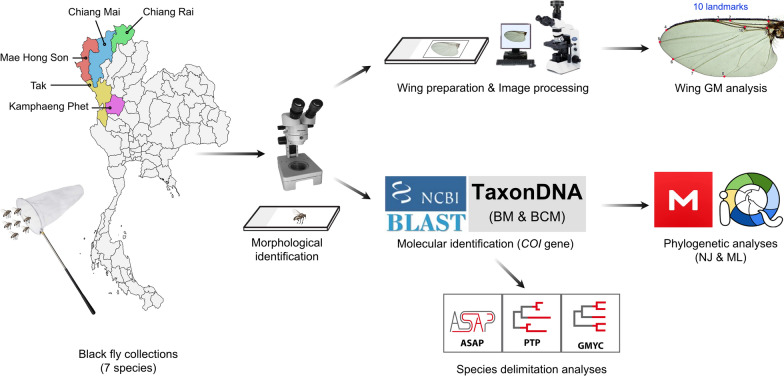
Abstract
Background
Fast and reliable species identification of black flies is essential for research proposes and effective vector control. Besides traditional identification based on morphology, which is usually supplemented with molecular methods, geometric morphometrics (GM) has emerged as a promising tool for identification. Despite its potential, no specific GM techniques have been established for the identification of black fly species.
Methods
Adult female black flies collected using human bait, as well as those reared from pupae, were used in this study. Here, landmark-based GM analysis of wings was assessed for the first time to identify human-biting black fly species in Thailand, comparing this approach with the standard morphological identification method and DNA barcoding based on the mitochondrial cytochrome c oxidase subunit I (COI) gene. To explore genetic relationships between species, maximum likelihood (ML) and neighbor-joining (NJ) phylogenetic trees were built. Additionally, three different methods of species delimitation, i.e., assemble species by automatic partitioning (ASAP), generalized mixed yule coalescent (GMYC), and single Poisson tree processes (PTP), were utilized to identify the morphologically defined species. The effectiveness of a COI barcode in identifying black fly species was further examined through the best match (BM) and best close match (BCM) methods.
Results
Seven black fly species, namely Simulium tenebrosum Takaoka, Srisuka & Saeung, 2018 (complex), S. doipuiense Takaoka & Choochote, 2005 (complex), S. nigrogilvum Summers, 1911, S. nodosum Puri, 1933, S. asakoae Takaoka & Davies, 1995, S. chamlongi Takaoka & Suzuki, 1984, and S. umphangense Takaoka, Srisuka & Saeung, 2017 were morphologically identified. Compared with the standard method, the GM analysis based on wing shape showed high success in separating species, achieving an overall accuracy rate of 88.54%. On the other hand, DNA barcoding surpassed wing GM for species identification with a correct identification rate of 98.57%. Species delimitation analyses confirmed the validity of most nominal species, with an exception for S. tenebrosum complex and S. doipuiense complex, being delimited as a single species. Moreover, the analyses unveiled hidden diversity within S. asakoae, indicating the possible existence of up to four putative species.
Conclusions
This study highlights the potential of wing GM as a promising and reliable complementary tool for species identification of human-biting black flies in Thailand.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-024-06597-8.
Keywords: Morphometric analysis, DNA barcodes, Species delimitation, Hematophagous insect, Medical entomology, Simulium
Background
Black flies are small, dark, humpbacked flies that belong to the family Simuliidae, with more than 2400 species formally recorded worldwide [1]. The female flies usually feed on the blood of birds and mammals, including humans, which can cause irritation, pain, swelling, and redness at the bite site due to an immunoglobulin (Ig)E-mediated reaction to salivary gland proteins [2–9]. During the biting, some species could transmit various pathogens to both animals and humans, especially the filarial worm Onchocerca volvulus, the causative agent of human onchocerciasis or river blindness [9, 10]. This disease is the second leading cause of infectious blindness globally and is one of the 21 neglected tropical diseases (NTDs) that the World Health Organization (WHO) has targeted for elimination by 2030 [11]. In Thailand, a total of 146 species of black flies have been officially documented, with seven species (Simulium asakoae, S. nigrogilvum, S. tenebrosum complex, S. doipuiense complex, S. umphangense, S. chamlongi, and S. nodosum) considered to be human biters [12–15]. Most recently, host blood meal analysis of adult flies based on mitochondrial cytochrome b (cytb) gene revealed that two other species, S. chumpornense Takaoka & Kuvangkadilok, 2000 and S. striatum species-group fed on human blood [16]. However, no human infectious diseases transmitted by these flies have been reported so far in the country [17]. Notably, three human-biting species, namely S. asakoae, S. nigrogilvum and S. nodosum, have been incriminated as natural vectors of various species of filarial or nonfilarial worms, including the genus Onchocerca [17–22]. Furthermore, S. chumpornense, S. khelangense Takaoka, Srisuka & Saeung, 2022, and S. asakoae were recently identified as natural vectors of avian blood protozoa of the genera Leucocytozoon and Trypanosoma [23–26]. Considering the critical role of black flies as vectors for transmitting diverse pathogens, rapid and accurate species identification is crucial for comprehending disease epidemiology and developing effective vector control measures [27].
Traditionally, black fly species identification relies on meticulous examination of morphological characteristics of a large series of larvae, pupae, and adults (males and females), sometimes even requiring dissection [28]. Due to the small size and morphological uniformity of this insect, morphological classification is extremely difficult and often requires trained experts, proving impractical or even impossible for specimens that are damaged or closely related species [28, 29]. To overcome these limitations, other methods (cytogenetics and DNA barcoding) have been developed and used together to assist black fly species identification and confirmation [29–32]. Currently, DNA barcoding based on COI gene sequences plays a significant role in black fly species identification, enabling researchers to distinguish species rapidly and accurately. This method is not only used as an effective identification tool, but can also reveal hidden diversity within nominal species [28, 33–39]. Nevertheless, molecular approaches are relatively expensive, sophisticated, and need to be conducted in well-equipped laboratories [40, 41]. A faster, cost-effective, easy to use, and reliable method for accurately identifying species is necessary.
Over the past decade, geometric morphometrics (GM) analysis has emerged as a potential game changer, being used as an effective complementary tool for the species identification of various insect groups, particularly those with medical and veterinary importance [41–50]. This approach has also proved to be a valuable tool for detecting sexual dimorphism, recognition of cryptic diversity, and studying evolution [48, 51–55]. Presently, the landmark-based GM method of insect wings is the most popular and is a powerful tool used for supplementing and enhancing morphological identification [56, 57]. This method analyzes wing size and shape based on the positions of anatomical landmarks (also known as true landmarks) and the distances between them, which includes both size and shape data [56, 57]. While GM analysis has been employed successfully for distinguishing species of several insect groups, its application for black fly identification remains unexplored. To the best of our knowledge, there is only one study that applied this technique for interpreting evolutionary transitions in the black fly wings [51].
In the present study, landmark-based GM analysis of wings was evaluated, for the first time, for the identification of seven human-biting black fly species of Thailand. For comparison, DNA barcoding based on the COI gene was used to distinguish the species and the effectiveness of this gene for species identification was also tested using the best match (BM) and best close match (BCM) methods. Additionally, three species delimitation methods [assemble species by automatic partitioning (ASAP), generalized mixed yule coalescent (GMYC), and single Poisson tree processes (PTP)] were employed to identify the recognized morphospecies.
Methods
Black fly samples and morphological identification
Most adult female flies were caught with a sweep net while flying around a human bait, while others were reared from pupae collected with fine forceps from available substrates in streams. The wild-caught females were promptly euthanized by submerging them in a 15 ml centrifuge tube containing 80% alcohol. Meanwhile, matured pupae were kept individually in a 15 ml centrifuge tube with minimal water at the bottom until adulthood. The emerged adult females were kept alive in the same tube for at least 24 h to ensure the hardening and coloration of their exoskeleton [15]. All specimens were preserved in 80% ethanol and stored in a freezer (−20 °C) until further analysis. Species identification of the adult flies was based on the examination of their morphological characteristics under a stereomicroscope (Olympus SZ51, Japan) using the standard keys for the black flies of Thailand [58]. In total, 253 adult female black flies (seven species or complex) belonging to the subgenera Gomphostilbia [one species designated here as “S. asakoae” although our specimens probably include not only S. asakoae but also several other species of the S. asakoae species-group, which are morphologically indistinguishable from one another (n = 50)] and Simulium s. str. [six species including S. chamlongi (n = 30), S. doipuiense complex (n = 39), S. nigrogilvum (n = 39), S. nodosum (n = 31), S. tenebrosum complex (n = 31), and S. umphangense (n = 33)] of the genus Simulium Latreille s. l. were utilized in this study as outlined in Table 1.
Table 1.
Details of adult female black flies used in this study
| Species | Sampling site | Coordinate (latitude/longitude) | Elevation (m) | Date of collection | n | Total | |
|---|---|---|---|---|---|---|---|
| S. asakoae | Ban Pang Dang, Doi Saket, Chiang Mai | 19°03′35.0″ N/99°21′52.9″ E | 932 | 28 August 2023 | 25 | 50 | |
| Ban Pang Bong, Doi Saket, Chiang Mai | 18°49′04.3″ N/99°20′06.2″ E | 981 | 28 August 2023 | 25 | |||
| S. chamlongi | A-Frame, Doi Pha Hom Pok, Chiang Mai | 20°02′12.9″ N/99°09′55.8″ E | 1529 | 29 November 2012 | 21a | 30 | |
| Mae Kam Pong Waterfall, Mae On, Chiang Mai | 18°51′48.6″ N/99°21′22.9″ E | 1076 | 1 June 2015 | 4a | |||
| Pangkhon, Mueang, Chiang Rai | 19°54′17.6″ N/99°35′54.0″ E | 1386 | 29 October 2016 | 5a | |||
| S. doipuiense complex | Mae Klong Kee, Umphang, Tak | 16°13′33.0″ N/98°58′46.8″ E | 1264 | 24 June 2021 | 2 | 39 | |
| Mae Klong Kee Unit, Umphang, Tak | 16°14′45.8″ N/98°59′52.1″ E | 1188 | 17 August 2021 | 12 | |||
| 16°13′30.9″ N/98°58′47.3″ E | 1237 | 18 August 2021 | 16 | ||||
| Pa Deuk Dum Bun, Umphang, Tak | 16°14′38.0″ N/98°59′55.6″ E | 1263 | 19 August 2021 | 9 | |||
| S. nigrogilvum | Mae Klong Kee, Umphang, Tak | 16°13′29.1″ N/98°58′44.9″ E | 1279 | 24 June 2021 | 12 | 39 | |
| Mae Klong Kee Unit, Umphang, Tak | 16°14′45.8″ N/98°59′52.1″ E | 1188 | 17 August 2021 | 19 | |||
| 16°13′30.9″ N/98°58′47.3″ E | 1237 | 18 August 2021 | 8 | ||||
| S. nodosum | Tham Pla, Mueang, Mae Hong Son | 19°30′09.2″ N/98°00′22.9″ E | 385 | 24 October 2014 | 20 | 31 | |
| Tao Dam Waterfall, Khlonglan, Kamphaeng Phet | 16°18′04.6″ N/99°06′44.3″ E | 539 | 20 December 2016 | 11a | |||
| S. tenebrosum complex | Doi Inthanon, Chom Thong, Chiang Mai | 18°35′12.8″ N/98°29′14.2″ E | 2534 | 19 December 2018 | 31 | 31 | |
| S. umphangense | Ban Lek, Doi Pha Hom Pok, Chiang Mai | 20°04′30.9″ N/99°11′07.4″ E | 1468 | 29 May 2010 | 1 | 33 | |
| Chong Yen, Mae Wong, Kamphaeng Phet | 16°06′02.3″ N/99°06′29.0″ E | 1276 | 23 March 2016 | 3 | |||
| 19 January 2017 | 13 | ||||||
| 4 November 2018 | 3 | ||||||
| Mae Klong Kee Unit, Umphang, Tak | 16°13′34.8″ N/98°58′46.1″ E | 1272 | 1 March 2013 | 6 | |||
| Upstream Mae Klong Yai, Umphang, Tak | 16°18′00.5″ N/99°01′22.6″ E | 1095 | 22 March 2016 | 1 | |||
| Pa Deuk Dum Bun, Umphang, Tak | 16°14′38.0″ N/98°59′55.6″ E | 1263 | 5 March 2020 | 3 | |||
| Mae Klong Kee, Umphang, Tak | 16°13′33.0″ N/98°58′46.8″ E | 1264 | 24 June 2021 | 3 | |||
aAdult female reared from pupa
Wing preparation, image processing, and landmark digitization
After morphological identification of the species, at least 30 specimens of each black fly species with intact wings were selected for geometric morphometric analysis (Table 1). A subset of these fly samples (ten specimens/species) was also randomly chosen for molecular analysis.
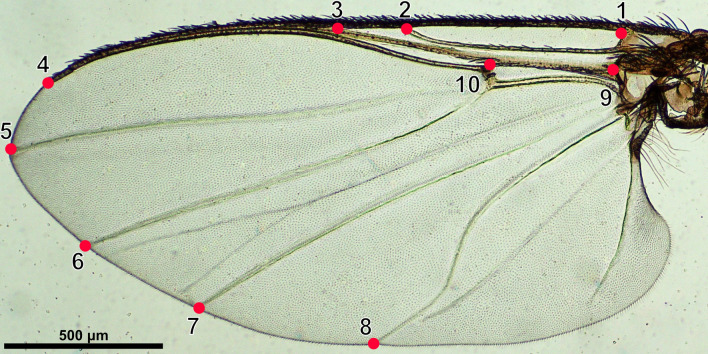
To prepare a semi-permanent slide, the right wing of each individual was first removed from the thorax using an insect needle under a stereomicroscope (Olympus SZ51, Japan). Each wing was then transferred to a new slide, placed on a drop of 80% alcohol, and covered with a coverslip. To prevent leakage and evaporation of the alcohol while observing and photographing the specimens, air-drying nail polish (Revlon, Indonesia) was applied to seal the edges of a coverslip. Digital images of each wing were captured using a DP27 digital camera attached to a 4× magnification light microscope (Olympus CX41, Japan). All images were annotated with a 500 μm reference scale bar. A total of ten landmarks (Fig. 1) slightly modified from [51] were digitized on each wing.
Fig. 1.
A representative image of black fly wing showing ten landmarks used in geometric morphometric analysis
Repeatability
To assess the accuracy of digitizing landmarks, a repeatability test for the shape was performed. Ten wing images of each fly species were randomly selected and digitized twice by the same user for intra-user repeatability. The measurement error, expressed as the repeatability index (R), was calculated by comparing two sets of digitized images using the Procrustes analysis of variance (ANOVA) method [59].
Allometry
As previously reported, wing sizes could influence wing shape variation (allometry) [60, 61]. This correlation was examined before conducting the wing shape analysis. The coefficient of determination (R2), obtained from analyzing the linear regression between the wing centroid size (CS) and the first principal component (PC) of wing shape, was used to assess the extent of the influence.
Wing size and shape analysis
To estimate the global wing size of each species, we employed the centroid size (CS), which was derived from the distances between the centroid point of each configuration and each landmark [62]. The variation of wing CS among Simulium species was illustrated using a violin plot. Statistically significant differences in wing CS between species were assessed using a one-way ANOVA (1000 iterations) with Bonferroni adjustment for significance testing at a p-value < 0.05.
For the wing shape analysis, shape variables were derived through a Procrustes superimposition of landmark configurations using the generalized Procrustes analysis (GPA) [63, 64]. Subsequently, partial wrap scores generated from principal component analysis were used as final shape variables for subsequent analyses. To explore species separation, discriminant analysis (DA) was performed using the final shape variables as an input and was illustrated as a factor map. The Mahalanobis distance, obtained from DA analysis, was calculated to estimate shape divergence between species. Wing shape differences among species based on the Mahalanobis distances were analyzed using a nonparametric permutation test (1000 iterations) with a Bonferroni adjustment for significance testing at a p-value < 0.05. Additionally, a hierarchical clustering tree (UPGMA algorithm) based on Mahalanobis distances was constructed to assess the relationships of wing shape among species [65].
Validated classification
A cross-validated classification (jackknife classification) was performed to test the accuracy of wing size and shape for correct species identification [66]. Each individual was successively excluded from the total sample and then allocated to the most probable group for size and the nearest group for shape using the maximum likelihood method [67] and Mahalanobis distance [68], respectively.
Morphometric software
The geometric morphometrics analysis, including landmark digitization, repeatability test, allometric examination, wing size and shape analyses, and a cross-validated classification test were performed using the online application XY Online Morphometrics (XYOM) version 2 [69]. The software is freely accessible at https://xyom.io/, accessed on 10 March 2024. In addition, the results generated from the XYOM were further beautified in RStudio [70].
Molecular identification based on DNA barcoding
For supplementing the morphological species identification, DNA barcoding using the COI gene was performed on the randomly selected specimens of each black fly species (ten specimens/species). Total DNA was extracted from the thorax of individual flies using the TIANamp Genomic DNA Kit (TIANGEN Biotech, Beijing, China), according to the manufacturer’s protocol. A DNA fragment of the mitochondrial COI gene (658 bp in length) was amplified using the universal primers: LCO1490 (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′) and HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) [71]. Each PCR reaction (20 μl total volume) was composed of 2 μl of DNA template, 1 U of Taq DNA polymerase, 3 mM MgCl2, 0.2 mM dNTPs, and 0.2 μM of each primer. The PCR cycling conditions included: an initial denaturation at 94 °C for 2 min followed by 40 cycles at 94 °C for 30 s, 50 °C for 45 s, and 72 °C for 45 s, with a final extension at 72 °C for 5 min. Subsequently, the PCR products were checked by agarose gel electrophoresis (1.5%), visualized by non-toxic Ultrapower (BioTeke, Beijing, China) dye. All PCR samples with a band of the expected size (658 bp) were sent to First Base Laboratories Sdn Bhd (Malaysia) for purifying and sequencing using the BigDye Terminator v.3.1 cycle sequencing kit on an ABI 3730XL Genetic Analyzer (Applied Biosystems Inc., Foster City, CA, USA). To generate a consensus sequence of each specimen, both forward and reverse sequences were assembled and edited manually in Geneious Prime 2024.0.5 [72]. Calculation of intra- and interspecific genetic distances based on the Kimura two-parameter (K2P) model [73] was conducted in MEGA 11 [74, 75]. The species identities of each black fly specimen were determined by comparing the newly generated sequences with previously published sequences deposited in the GenBank database, using the basic local alignment search tool (BLAST), available at http://blast.ncbi.nlm.nih.gov/Blast.cgi. Success rates of the DNA barcoding in species identification were evaluated based on the BM and BCM methods using the TaxonDNA [76]. All COI sequences obtained in the present study were deposited in the GenBank database under the following accession numbers: PP422429–PP422472.
Multiple sequence alignment and phylogenetic analysis
All sequences obtained in this study and others fetched from GenBank database were aligned with MUSCLE 3.7 [77], performed in MEGA 11 [74, 75]. Subsequently, the COI alignment was used to infer the genetic relationships among seven human-biting black fly species based on the neighbor-joining (NJ) and maximum likelihood (ML) methods. The NJ tree was constructed in MEGA 11 based on the K2P model with 1000 bootstrap iterations [74, 75, 78], while the ML tree was reconstructed using IQ-TREE version 2.3.1 [79] with 10,000 ultrafast bootstrap iterations [80]. The best-fitting substitution model for the ML method, which was K3Pu + F + I + R2, was selected using ModelFinder based on Bayesian Information Criterion (BIC) [81]. The final tree, visualized by FigTree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/), was rooted using the COI sequence of S. khongchiamense Takaoka, Srisuka & Saeung, 2023, a member of the subgenus Asiosimulium.
Species delimitation analysis
Three different methods of DNA sequence-based species delimitation, including ASAP, GMYC, and single PTP were performed to estimate the number of molecular operational taxonomic units (MOTUs). ASAP, a distance-based method, was conducted online using the ASAP webserver, available at https://bioinfo.mnhn.fr/abi/ public/asap/ , with default parameters [82]. The PTP analysis was run through mPTP webserver (https://mptp.h-its.org/#/tree) using the maximum likelihood implementation with a single Poisson distribution [83]. The ML tree generated from IQ-TREE was used as the input tree in the PTP analysis with a default p-value of 0.001. For the GMYC analysis, an ultrametric tree was generated with BEAUti2 software under the Yule process speciation model and the relaxed clock log-normal model [84, 85]. The best fitting substitution model (TrN + I + G) based on the BIC was determined using jModelTest 2.1.7 [86]. The MCMC chain was run in BEAST v2.6.7 for 20 million iterations with sampling frequency of 1000 iterations each. As previously recommended [87], the output file was checked using Tracer v1.7 software to ensure that all the effective sample size (ESS) values were greater than 200. To infer a maximum clade credibility tree from the set of posterior trees, the output tree was subjected to TreeAnnotator v2.6.7 with a burn-in of 20% [85]. The resulting tree was then analyzed under the single-threshold operation using the R package “splits” on the R platform [70, 88].
Results
Repeatability
The repeatability test for the wing shape revealed a very low measurement error score (1%) and a high repeatability score (99%), suggesting a high level of accuracy in landmark placement on the tested wing image set.
Allometric effect
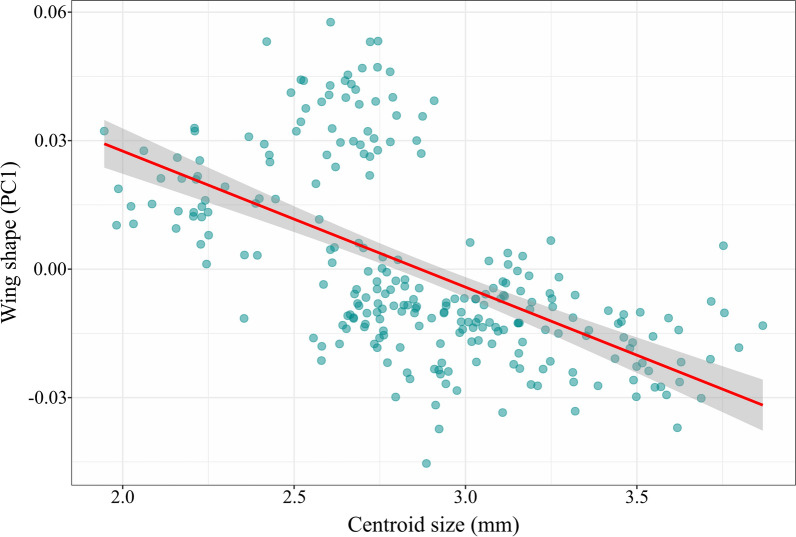
Assessment of the allometric effect revealed an apparent negative relationship between wing size and wing shape (R2 = 33.7%) with statistical significance (p < 0.05) (Fig. 2). This indicated that wing size variation influences wing shape divergence between species.
Fig. 2.
Scatter plot showing the allometric relationship between the wing shape (the first principal component, PC1) and wing size (centroid size) of seven black fly species. The red line indicates the linear regression prediction with 95% confidence intervals (shaded areas), while the sapphire dots represent individual samples
Wing size variation
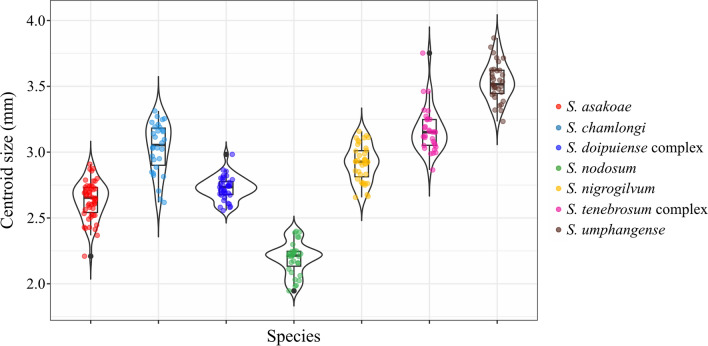
The variation in wing size (CS) among seven human-biting black fly species is depicted in Fig. 3. Simulium umphangense displayed the largest wing size of 3.53 ± 0.15 mm (mean ± S.D.), while S. nodosum exhibited the smallest wing size at 2.19 ± 0.12 mm (Table 2). The comparisons of wing size based on a nonparametric permutation test (1000 iterations) with Bonferroni adjustment revealed significant differences (p < 0.05) between almost all species pairs, with the exception of S. asakoae versus S. doipuiense complex and S. chamlongi versus S. nigrogilvum and S. tenobrosum complex.
Fig. 3.
Violin plot overlaid with a box plot showing the distribution of wing centroid sizes (CS) in millimeters (mm) of seven black fly species
Table 2.
Average wing centroid sizes (CS) in millimeters and statistical differences in wing size among seven human-biting black fly species
| Species | n | Wing size (mm) | ||
|---|---|---|---|---|
| Mean ± SD | Minimum | Maximum | ||
| S. asakoae | 50 | 2.64 ± 0.14a | 2.21 | 2.91 |
| S. chamlongi | 30 | 3.03 ± 0.19b | 2.62 | 3.31 |
| S. doipuiense complex | 39 | 2.72 ± 0.09a | 2.56 | 2.98 |
| S. nodosum | 31 | 2.19 ± 0.12c | 1.95 | 2.40 |
| S. nigrogilvum | 39 | 2.92 ± 0.14b,d | 2.66 | 3.16 |
| S. tenebrosum complex | 31 | 3.17 ± 0.17b,e | 2.86 | 3.75 |
| S. umphangense | 33 | 3.53 ± 0.15f | 3.23 | 3.87 |
Different superscript letters denote statistically significant differences at p < 0.05
Wing shape variation
The visual comparisons of superimposition of the mean landmark configurations among seven black fly species showed the most noticeable landmark displacement in the lower part of the wing, particularly in landmark positions 3 and 4 (Fig. 4). The analysis also indicated that S. asakoae exhibited the most different wing shape, as its landmark position 4 clearly separated it from the other six species (Fig. 4).
Fig. 4.
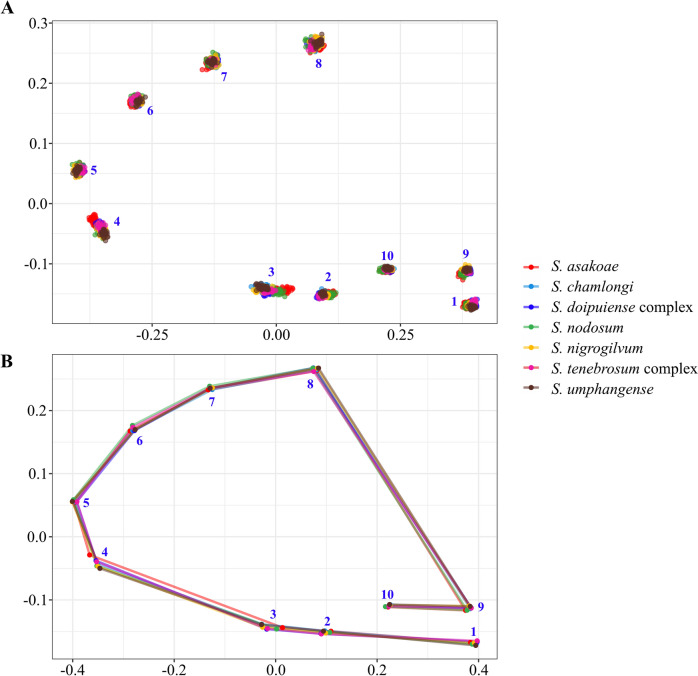
Shape differences in wing venation based on anatomical landmarks of seven black fly species. (A) Scatter plot showing residual coordinates of ten landmarks aligned by Procrustes analysis and (B) a wireframe graph showing the superposition of the overall mean shape
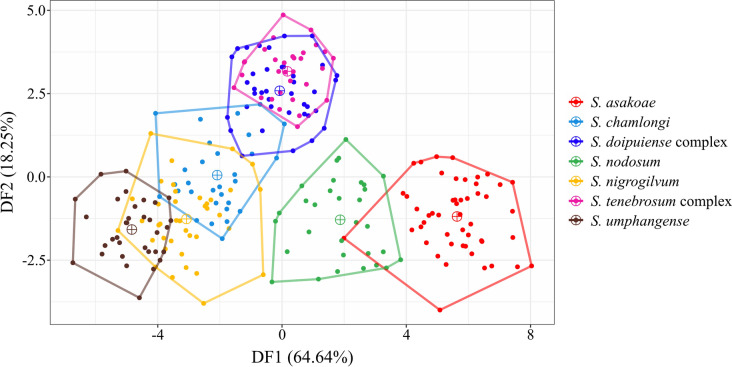
Discriminant analysis of the wing shape variables among the seven black fly species showed clear separation, with a small overlap for two species, namely S. asakoae and S. nodosum. Conversely, the five other species revealed large overlapping among species, especially the two closely related species complexes, S. tenebrosum complex and S. doipuiense complex (Fig. 5). Despite the large overlap among species, significant differences (p < 0.05) in wing shape were observed among all black fly species based on the pairwise Mahalanobis distances (Table 3).
Fig. 5.
Factor map based on discriminant analysis (DA) showing the shape divergence of seven black fly species. Each polygon represents a different species, with dots indicating individual specimens and a sun cross marking the mean values for each species
Table 3.
Pairwise Mahalanobis distances and significant differences in wing shape of seven black fly species
| Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| (1) S. asakoae | – | ||||||
| (2) S. chamlongi | 8.13* | – | |||||
| (3) S. doipuiense complex | 7.03* | 4.36* | – | ||||
| (4) S. nodosum | 5.79* | 6.13* | 5.69* | – | |||
| (5) S. nigrogilvum | 8.86* | 3.41* | 5.13* | 6.25* | – | ||
| (6) S. tenebrosum complex | 7.29* | 4.77* | 2.69* | 5.65* | 5.87* | – | |
| (7) S. umphangense | 10.56* | 4.24* | 6.56* | 7.68* | 2.84* | 7.11* | – |
The superscript asterisks (*) after each Mahalanobis distance values denote statistically significant differences between black fly species at p < 0.05
Phenetic relationships of wing shape among black fly species
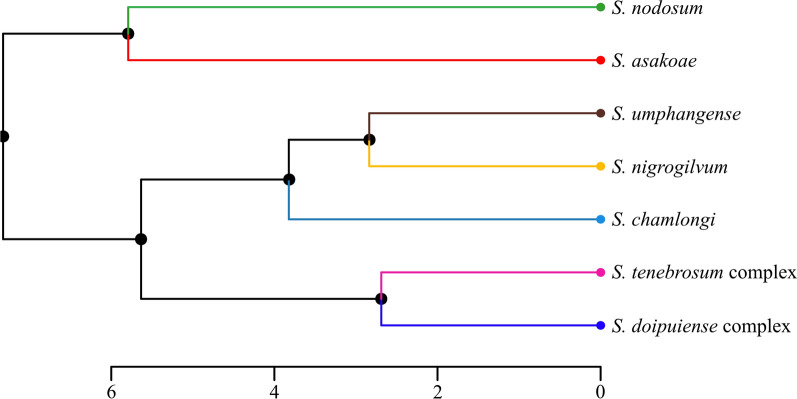
The UPGMA dendrogram based on Mahalanobis distances demonstrated that the seven black fly species were separated into two distinct groups (Fig. 6). Simulium nodosum and S. asakoae formed one group, while the other five species formed another group, which was further divided into two subgroups: (1) S. chamlongi + (S. umphangense + S. nigrogilvum) and (2) S. tenebrosum complex + S. doipuiense complex.
Fig. 6.
UPGMA dendrogram based on the Mahalanobis distances between average group shapes showing the phenetic relationship of wing shape among seven black fly species. The scale bar represents the Mahalanobis distance
Validated classification
The success rate of landmark-based GM analysis of the wings for identifying seven black fly species is summarized in Tables 4 and 5. The size-based cross-validated classification revealed a low level of correct identification, with an overall accuracy score of 47.43% (120/253), while the shape-based cross-validated classification yielded a higher level of successful identification, with a total classification score of 88.54% (224/253). Furthermore, the shape-based classification showed a perfect result (100% accuracy score) for species identification of S. asakoae.
Table 4.
Percentage of correct identification based on cross-validated classification of the wing size (centroid size) and wing shape (Mahalanobis distance values) of seven black fly species
| Species | Percentage of correct classification (assigned/observed) | |
|---|---|---|
| Based on wing size | Based on wing shape | |
| S. asakoae | 56.00% (28/50) | 100.00% (50/50) |
| S. chamlongi | 23.33% (7/30) | 80.00% (24/30) |
| S. doipuiense complex | 46.15% (18/39) | 89.74% (35/39) |
| S. nodosum | 29.03% (9/31) | 96.77% (30/31) |
| S. nigrogilvum | 41.03% (16/39) | 76.92% (30/39) |
| S. tenebrosum complex | 45.16% (14/31) | 87.10% (27/31) |
| S. umphangense | 84.85% (28/33) | 84.85% (28/33) |
| Total performance | 47.43% (120/253) | 88.54% (224/253) |
Table 5.
Percentage of correctly assigned individuals based on the cross-validated classification of the wing shape of seven black fly species
| Species | Classified as | Total | Correct identification | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| (1) S. asakoae | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 100.00% |
| (2) S. chamlongi | 0 | 24 | 1 | 0 | 4 | 1 | 0 | 30 | 80.00% |
| (3) S. doipuiense complex | 0 | 1 | 35 | 0 | 1 | 2 | 0 | 39 | 89.74% |
| (4) S. nodosum | 0 | 1 | 0 | 30 | 0 | 0 | 0 | 31 | 96.77% |
| (5) S. nigrogilvum | 0 | 4 | 1 | 0 | 30 | 0 | 4 | 39 | 76.92% |
| (6) S. tenebrosum complex | 0 | 0 | 4 | 0 | 0 | 27 | 0 | 31 | 87.10% |
| (7) S. umphangense | 0 | 0 | 0 | 0 | 5 | 0 | 28 | 33 | 84.85% |
| Correctly assigned/total individual | 224/253 | 88.54% | |||||||
The rows represent given species, while columns represent predicted species
Sequence variation and genetic distance
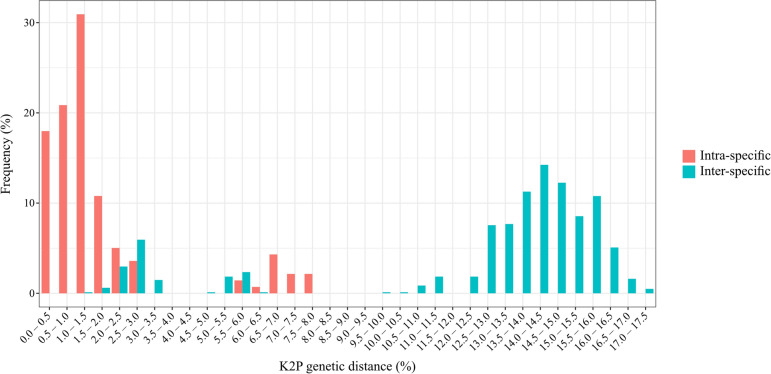
In total, 70 COI sequences (658 bp long) of seven human-biting black fly species were obtained in this study, with 44 sequences identified as unique haplotypes. Sequence analysis revealed a high AT content, with an average base composition of A = 0.275, C = 0.178, G = 0.168, and T = 0.379. An overlap between the maximum intraspecific and minimum interspecific divergences was also noted, suggesting the absence of a barcoding gap (Fig. 7).
Fig. 7.
Frequency distribution of intraspecific and interspecific K2P genetic distances based on the COI gene of seven human-biting black fly species
The maximum intraspecific genetic distance based on the K2P model ranged from 0.15% (S. chamlongi) to 7.92% (S. asakoae). Most species showed low intraspecific divergences (maximum value < 3%). The exception was S. asakoae, which displayed the highest intraspecific divergence (maximum value > 7%) (Table 6).
Table 6.
Mean intraspecific (in bold) and interspecific genetic distances (%) with maximum values indicated in parentheses among seven human-biting black fly species
| Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| (1) S. asakoae | 5.49 (7.92) | ||||||
| (2) S. chamlongi | 15.69 (17.13) | 0.15 (0.15) | |||||
| (3) S. doipuiense complex | 14.97 (16.09) | 13.01 (13.53) | 1.11 (1.86) | ||||
| (4) S. nodosum | 16.08 (17.00) | 14.46 (14.79) | 15.46 (16.14) | 0.23 (0.31) | |||
| (5) S. nigrogilvum | 15.9 (17.07) | 11.07 (11.38) | 12.83 (13.75) | 14.12 (14.42) | 0.73 (1.23) | ||
| (6) S. tenebrosum complex | 14.91 (15.71) | 13.22 (13.71) | 2.64 (3.30) | 14.61 (15.93) | 13.21 (13.93) | 1.48 (2.81) | |
| (7) S. umphangense | 15.72 (16.48) | 10.82 (11.24) | 14.18 (14.89) | 14.29 (14.67) | 5.512 (6.06) | 13.81 (14.11) | 0.58 (1.07) |
The mean interspecific genetic divergences ranged from 2.64% (S. tenebrosum complex versus S. doipuiense complex) to 16.08% (S. asakoae versus S. nodosum), with the highest value of 17.13% (S. asakoae versus S. chamlongi). Low interspecific genetic divergence values were observed in two species pairs: S. tenebrosum complex and S. doipuiense complex, and S. nigrogilvum and S. umphangense, with maximum values of 3.30% and 6.06%, respectively. Meanwhile the remaining species pairs showed very high levels of interspecific divergence, with maximum values exceeding 11% (Table 6).
Species determination through a BLAST search
Using a BLAST search, all haplotypes could be identified to the species level (Table S1). Four out of seven species, including S. nodosum, S. tenebrosum complex, S. nigrogilvum, and S. umphangense were correctly classified to their respective species or complex with high sequence similarity (98–100%). On the other hand, seven haplotypes of S. asakoae displayed the highest similarity (> 99%) with several different species of the S. asakoae species-group as follows: S. nanthaburiense (H1), S. chaowaense (H4), S. pitasawatae (H5), S. tamdaoense (H6), and S. asakoae (H3, H7), whereby the H2 haplotype was identical to three different species, namely S. asakoae, S. vinhphucense, and S. hongthaii. S. chamlongi and S. doipuiense complex were also ambiguously assigned to the species level. One haplotype (H9) of S. chamlongi matched perfectly to S. phuluense, while the other (H8) was identical to both S. phuluense and S. chamlongi. Additionally, the majority of haplotypes (9/10) of the S. doipuiense complex showed the highest similarity (> 98%) to the S. tenebrosum complex, with only one (H14) being accurately identified as the S. doipuiense complex with a sequence similarity of 98.63%.
Efficiency of the COI gene for species identification
Species identification based on the COI sequences demonstrated high success rate, with 98.57% (69/70) correct identification for both the BM and BCM methods. The only exception was the misidentification of one specimen (TN5) of S. tenebrosum complex as S. doipuiense complex.
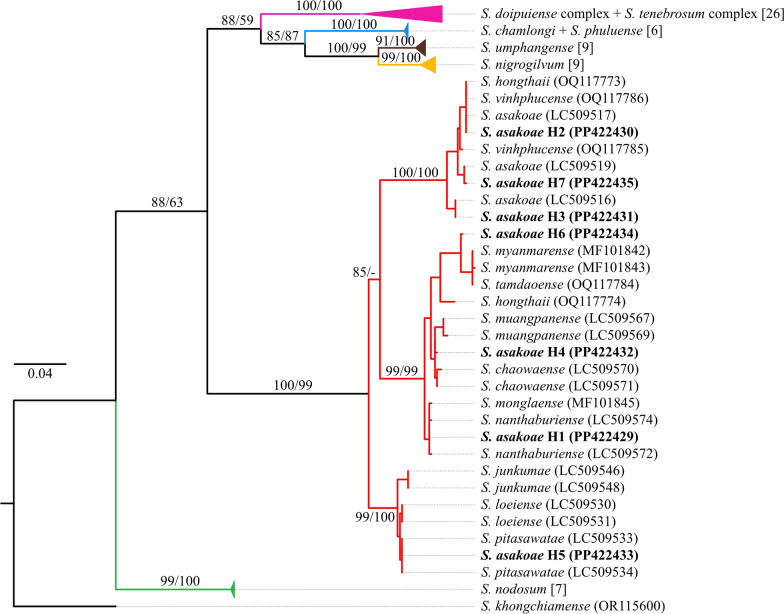
Phylogenetic analysis
Both NJ and ML phylogenetic trees based on the COI sequences yielded similar tree topologies. Thus, only the ML tree is demonstrated (Fig. 8). Three species, namely S. nigrogilvum, S. nodosum, and S. umphangense were identified as distinct species, forming their own monophyletic groups with strong bootstrap support. In contrast, the remaining four species—S. asakoae, S. tenebrosum complex, S. doipuiense complex, and S. chamlongi—were not monophyletic. Sequences of S. chamlongi were clustered with S. phuluense from Vietnam, while the sequences of S. tenebrosum complex were intermixed with S. doipuiense complex. Notably, seven haplotypes of S. asakoae were divided into three subclades and placed into different species members of the S. asakoae species-group as follows: (1) S. asakoae/S. vinhphucense/S. hongthaii for H2, H3, and H7; (2) S. pitasawatae for H5; (3) S. tamdaoense/S. myanmarense for H6; (4) S. nanthaburiense/S. monglaense for H1; (5) S. chaowaense for H4.
Fig. 8.
Maximum likelihood phylogenetic tree based on 585 bp COI gene of seven human-biting black fly species and their related species. Bootstrap support values (ML/NJ) greater than 50% are indicated near the branches. Some distinct clades were collapsed for clearer presentation, and the number of sequences falling within those clades is indicated in square brackets. Sequences obtained in this study are highlighted in bold type. All sequences used for constructing the tree are detailed in Table S2
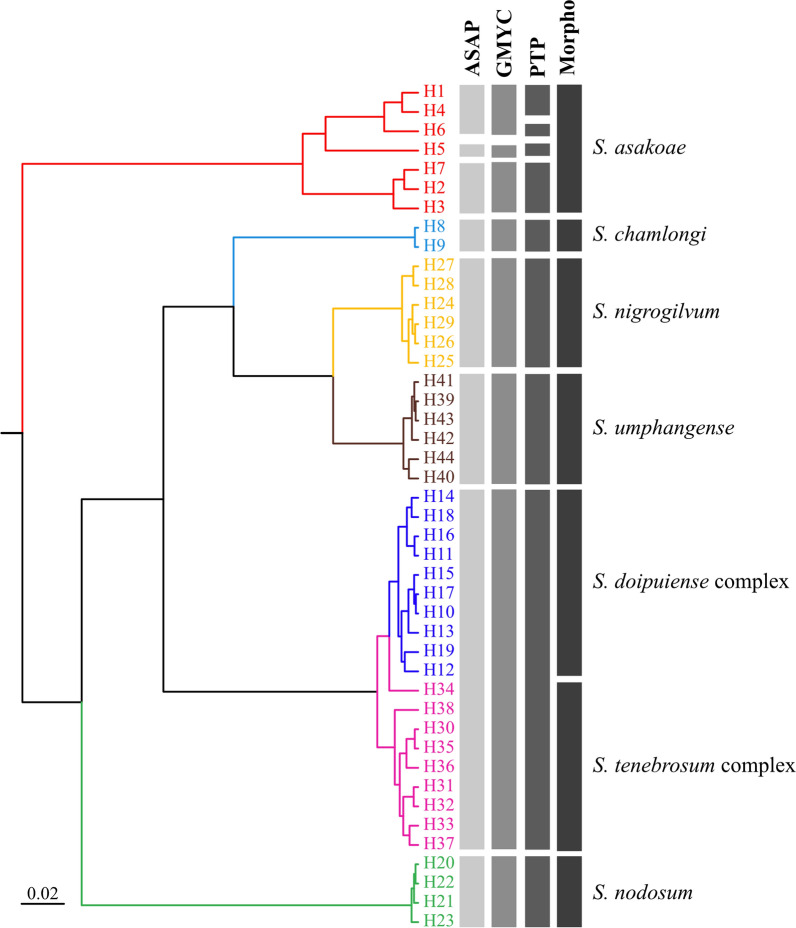
Species delimitation analysis
Three species delimitation methods (ASAP, PTP, and GMYC) mostly supported seven morphospecies, as summarized in Fig. 9. Both ASAP and PTP methods identified eight putative species, while GMYC method recovered more putative species (nine species). All species delimitation methods merged S. tenebrosum complex and S. doipuiense complex into a single species, disagreeing with their morphologically defined species. Regarding S. asakoae, seven haplotypes were divided into three (ASAP and PTP) up to four (GMYC) putative species, suggesting cryptic diversity in this species.
Fig. 9.
Summary of the three species delimitation analyses (ASAP, GMYC, and PTP) based on the COI haplotypes from seven different nominal species of black flies. The maximum clade credibility tree built from BEAST is colored according to morphospecies. The vertical bars at the tips of the tree correspond to the result of each species delimitation method and morphological identification, respectively. Detailed information of each haplotype is provided in Table S2
Discussion
Here, we demonstrate for the first time that landmark-based GM of wing shape is a promising tool for the identification of the adult human-biting black flies in Thailand on a species level, although not with perfection. As expected, wing shape analysis is superior to wing size analysis in distinguishing black fly species, consistent with findings in numerous previous reports across various insect groups [46–48, 89, 90]. Unlike wing size, which is susceptible to environmental influences and is not a suitable parameter for distinguishing species [57, 91], wing shape appears to be a fixed trait that preserves genetic information, making it a desirable species-specific indicator [92, 93]. Our allometric analysis also indicated that one variable (wing size) tends to increase, while the other (wing shape) tends to decrease (shows a negative correlation), as has been observed in several medically and forensically important insects, e.g., Anopheles barbirostris mosquitoes [46], Armigeres mosquitoes [50], Culex vishnui subgroup [94], and Stomoxys flies [90]. Therefore, this study excluded wing size data before wing shape analysis. Noticeably, our GM analysis based on wing shape was often unable to correctly assign the closely related species to their respective species. This result aligns with expectations, given the high morphological similarities between S. umphangense and S. nigrogilvum, where species separation relies mainly on subtle difference in the leg color of the fore tibia [14]. Similarly, the distinction of the two closely related species complexes, S. tenebrosum complex and S. doipuiense complex, is feasible based on certain features of the legs, specifically the coloration of the hind tibia and basitarsus [95].
The phenotypic relationships among seven black fly species mostly align with the phylogenetic tree based on the COI gene, suggesting that landmark-based GM analysis of wing shape could be used as an alternative tool for evolutionary studies, taxonomy, and systematics [51, 61, 94]. However, both phenotypic and phylogenetic analyses erroneously placed S. nodosum in the wrong group, contradicting its morphological classification and previous molecular studies [58, 96]. Instead of assigning all seven species to their respective subgenus, S. nodosum of the subgenus Simulium s.l. was placed in a distinct group or clustered as a sister species with S. asakoae of the subgenus Gomphostilbia. The incorrect assignment of S. nodosum seems to be due to the lack of intermediate taxa in the analyses. Therefore, incorporating additional black fly specimens of different species and subgenera into the analyses is recommended to improve both phenotypic and phylogenetic accuracy [97, 98].
In this study, we also assessed the performance of wing GM analysis in comparison to DNA barcoding based on the COI gene for distinguishing morphologically defined species. DNA barcoding clearly outperformed wing GM analysis, demonstrating an almost perfect efficiency (> 98%) in species identification using both BM and BCM methods. The high accuracy (> 90%) of DNA barcoding in distinguishing Thai black fly species was also observed in previous studies, even when analyzing a larger number of species (41–89 nominal species) [35, 96]. Further, our phylogenetic analysis of the COI gene coupled with species delimitation analysis revealed hidden diversity within S. asakoae, suggesting the presence of up to four putative species. A previous molecular study using the COI gene also indicated that S. asakoae in Thailand consisted of at least seven groups [34]. Subsequent investigations, which extensively analyzed a large number of morphologically defined S. asakoae specimens further confirmed this observation, with a total of 23 new species described in recent years [99–101]. The high morphological variability and the high similarity of the COI sequences of several members of the S. asakoae species-group hampers the correct assignment of our S. asakoae specimens to the defined morphospecies [99, 102]. Initially, DNA barcoding was considered highly effective in distinguishing the true S. asakoae from other members in the S. asakoae species-group [102]. However, a recent study in Vietnam [39], as well as this study, demonstrated that the COI gene is no longer a suitable barcoding marker. In these studies, two related species from Vietnam, namely S. hongthaii and S. vinhphucense, were placed in the same clade as the true S. asakoae, leading to their delimitation as a single species. Some previous reports also suggested that the COI gene, or even the rapidly evolving nuclear BZF gene provide insufficient signal to distinguish most members of the S. asakoae species-group [35, 39, 99, 103]. To overcome this problem, further studies using more variable gene markers, such as the elongation complex protein 1 (ECP1), the 5-intron gene (5intG) [104], or molybdenum cofactor sulfurase (MCS) [105], are required to enhance species differentiation within the S. asakoae species-group. Additionally, we found that DNA barcoding was ineffective in identifying S. tenebrosum complex and S. doipuiense complex, and three species delimitation methods further supported this result by merging them as a single species [35, 39]. A more recent study examining the efficiency of two rapidly evolving nuclear genes suggested that the ECP1 gene is a promising barcoding marker for the successful identification of the two species complexes [106]. The molecular analysis in this study also revealed a similar result that S. chamlongi is not monophyletic as its clade including S. phuluense from Vietnam [39]. It may be presumed that S. chamlongi and S. phuluense are morphologically distinct but molecularly homosequential, since they are readily distinguished by the color of female legs, the number of male upper-eye (large) facets and the presence or absence of tubercles on the pupal head [107, 108. This suggests the need for additional genetic markers beyond the COI gene to resolve their phylogenetic relationships.
Although the accuracy of landmark-based GM of wing shape for black fly species identification in this study does not reach that of DNA barcoding, the results suggest that it is a promising complementary method to traditional and molecular methods. In addition, wing GM analysis is simple, reliable, cost-effective, rapid, and only needs nondamaged wings, in contrast to molecular identification [47, 61, 109]. Since this is the first study that applies wing GM analysis for black fly species identification, we focus on the most important species (anthropophilic species) and use the most popular method, landmark-based GM [56, 57]. Further studies that include more species of different subgenera and employ both landmark- and outline-based GM approaches are needed to determine the extent to which wing GM analysis can serve as a tool for identifying adults of black fly species.
Conclusions
In this research, we demonstrate that landmark-based GM analysis of wing shape, which achieves greater accuracy than wing size analysis in species identification, is a promising, reliable tool for supplementing and enhancing morphological identification of black fly adults. Our molecular analysis suggests that there may be up to four putative species within the morphologically defined S. asakoae.
Supplementary Information
Additional file 1: Table S1. Results from the NCBI BLAST search of black fly samples based on COI gene
Additional file 2: Table S2. Details of COI sequences generated from the present study and retrieved from NCBI GenBank databased used for molecular analysis
Acknowledgements
We would like to thank Wanida Kanta for her assistance with part of the GM analysis.
Abbreviations
- ASAP
Assemble species by automatic partitioning
- BCM
Best close match
- BM
Best match
- COI
Cytochrome c oxidase subunit I
- GM
Geometric morphometrics
- GMYC
Generalized mixed yule coalescent
- PCR
Polymerase chain reaction
- PTP
Poisson tree processes
Author contributions
KA conceptualized the idea of the study, designed the experiments, performed laboratory investigations, analyzed the data and wrote the paper. KA and WS collected specimens. WS, KL, SS, HT, and AS gave advice and revised the paper. KA, KL, and AS interpreted the results. AS managed the project and provided resources.
Funding
This work was supported by Faculty of Medicine, Chiang Mai University, grant no. 106–2565 to Kittipat Aupalee.
Availability of data and materials
The authors confirm that all data supporting the findings of this study are available within the article. All sequences generated from the study have been deposited in the GenBank database with assigned accession numbers PP422429–PP422472.
Declarations
Ethics approval and consent to participate
The procedures and research methodology used in this study were approved by the Research Ethics Committee (Institutional Animal Care and Use Committee) (protocol number: 28/2564) of the Faculty of Medicine, Chiang Mai University, Chiang Mai Province, Thailand.
Consent for publication
All authors have read and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adler PH. World blackflies (Diptera: Simuliidae): a comprehensive revision of the taxonomic and geographical inventory [2024]. 2024. https://biomia.sites.clemson.edu/pdfs/blackflyinventory.pdf.
- 2.Orange JS, Song LA, Twarog FJ, Schneider LC. A patient with severe black fly (Simuliidae) hypersensitivity referred for evaluation of suspected immunodeficiency. Ann Allergy Asthma Immunol. 2004;92:276–80. [DOI] [PubMed] [Google Scholar]
- 3.Hellberg W, Mellor PS, Torsteinsdottir S, Marti E. Insect bite hypersensitivity in the horse: comparison of IgE-binding proteins in salivary gland extracts from Simuliumvittatum and Culicoidesnubeculosus. Vet Immunol Immunopathol. 2009;132:62–7. [DOI] [PubMed] [Google Scholar]
- 4.Schaffartzik A, Weichel M, Crameri R, Bjornsdottir TS, Prisi C, Rhyner C, et al. Cloning of IgE-binding proteins from Simuliumvittatum and their potential significance as allergens for equine insect bite hypersensitivity. Vet Immunol Immunopathol. 2009;132:68–77. [DOI] [PubMed] [Google Scholar]
- 5.Schaffartzik A, Marti E, Crameri R, Rhyner C. Cloning, production and characterization of antigen 5 like proteins from Simuliumvittatum and Culicoidesnubeculosus, the first cross-reactive allergen associated with equine insect bite hypersensitivity. Vet Immunol Immunopathol. 2010;137:76–83. [DOI] [PubMed] [Google Scholar]
- 6.Schaffartzik A, Hamza E, Janda J, Crameri R, Marti E, Rhyner C. Equine insect bite hypersensitivity: what do we know? Vet Immunol Immunopathol. 2012;147:113–26. [DOI] [PubMed] [Google Scholar]
- 7.Chattopadhyay P, Goyary D, Dhiman S, Rabha B, Hazarika S, Veer V. Immunomodulating effects and hypersensitivity reactions caused by Northeast Indian black fly salivary gland extract. J Immunotoxicol. 2014;11:126–32. [DOI] [PubMed] [Google Scholar]
- 8.Chiriac A, Brzezinski P, Miron L, Moldovan C, Podoleanu C, Stolnicu S. Simuliosis–a dermatosis caused by black flies. Allergol Int. 2016;65:217–8. [DOI] [PubMed] [Google Scholar]
- 9.Adler PH, McCreadie JW. Black flies (Simuliidae). In: Mullen GR, Durden LA, editors. Medical and veterinary entomology. 3rd ed. London: Academic Press; 2019. p. 237–59. [Google Scholar]
- 10.Basanez MG, Boussinesq M. Population biology of human onchocerciasis. Philos Trans R Soc Lond B Biol Sci. 1999;354:809–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization, Department of Neglected Tropical Disease. Ending the neglect to attain the sustainable development goals: a road map for neglected tropical diseases 2021–2030. 2020. https://iris.who.int/bitstream/handle/10665/338565/9789240010352-eng.pdf?sequence=1
- 12.Choochote W, Takaoka H, Fukuda M, Otsuka Y, Aoki C, Eshima N. Seasonal abundance and daily flying activity of black flies (Diptera: Simuliidae) attracted to human baits in Doi Inthanon National Park, northern Thailand. Med Entomol Zool. 2005;56:335–48. [Google Scholar]
- 13.Pramual P, Thaijarern J, Wongpakam K. DNA barcoding of human-biting black flies (Diptera: Simuliidae) in Thailand. Acta Trop. 2016;164:33–40. [DOI] [PubMed] [Google Scholar]
- 14.Takaoka H, Srisuka W, Saeung A. A new human-biting black fly species of Simulium (Simulium) (Diptera: Simuliidae) from Thailand. J Med Entomol. 2017;54:945–8. [DOI] [PubMed] [Google Scholar]
- 15.Srisuka W, Takaoka H, Taai K, Maleewong W, Aupalee K, Saeung A. Morphological description and genetic analysis of a new black fly species (Diptera: Simuliidae) in the subgenus Asiosimulium from central Thailand. Parasit Vectors. 2024;17:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomontean B, Jumpato W, Wongpakam K, Tangkawanit U, Wannasingha W, Thanee I, et al. Diversity, distribution and host blood meal analysis of adult black flies (Diptera: Simuliidae) from Thailand. Insects. 2024;15:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aupalee K, Saeung A, Srisuka W, Fukuda M, Streit A, Takaoka H. Seasonal filarial infections and their black fly vectors in Chiang Mai province, northern Thailand. Pathogens. 2020;9:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda M, Choochote WET, Bain O, Aoki C, Takaoka H. Natural infections with filarial larvae in two species of black flies (Diptera: Simuliidae) in northern Thailand. Jpn J Trop Med Hyg. 2003;31:99–102. [Google Scholar]
- 19.Takaoka H, Choochote W, Aoki C, Fukuda M, Bain O. Black flies (Diptera: Simuliidae) attracted to humans and water buffalos and natural infections with filarial larvae, probably Onchocerca sp., in northern Thailand. Parasite. 2003;10:3–8. [DOI] [PubMed] [Google Scholar]
- 20.Ishii Y, Choochote W, Bain O, Fukuda M, Otsuka Y, Takaoka H. Seasonal and diurnal biting activities and zoonotic filarial infections of two Simulium species (Diptera: Simuliidae) in northern Thailand. Parasite. 2008;15:121–9. [DOI] [PubMed] [Google Scholar]
- 21.Saeung A, Srisuka W, Aupalee K, Fukuda M, Otsuka Y, Taai K, et al. Natural infections with larvae of Onchocerca species type I in the human-biting black fly, Simuliumnigrogilvum (Diptera: Simuliidae), in western Thailand. Acta Trop. 2020;204:105344. [DOI] [PubMed] [Google Scholar]
- 22.Huang F, Srisuka W, Aupalee K, Streit A, Fukuda M, Pitasawat B, et al. Diversity of nematodes infecting the human-biting black fly species, Simuliumnigrogilvum (Diptera: Simuliidae) in central Thailand. Acta Trop. 2021;224:106140. [DOI] [PubMed] [Google Scholar]
- 23.Jumpato W, Tangkawanit U, Wongpakam K, Pramual P. Molecular detection of Leucocytozoon (Apicomplexa: Haemosporida) in black flies (Diptera: Simuliidae) from Thailand. Acta Trop. 2019;190:228–34. [DOI] [PubMed] [Google Scholar]
- 24.Thaijarern J, Tangkawanit U, Wongpakam K, Pramual P. Molecular detection of Trypanosoma (Kinetoplastida: Trypanosomatidae) in black flies (Diptera: Simuliidae) from Thailand. Acta Trop. 2019;200:105196. [DOI] [PubMed] [Google Scholar]
- 25.Pramual P, Thaijarern J, Tangkawanit U, Wongpakam K. Molecular identification of blood meal sources in black flies (Diptera: Simuliidae) suspected as Leucocytozoon vectors. Acta Trop. 2020;205:105383. [DOI] [PubMed] [Google Scholar]
- 26.Stangarm J, Mintara R, Jumpato W, Gomontean B, Thanee I, Wongpakam K, et al. Molecular detection of blood protozoa and identification of black flies of the Simuliumvaricorne species group (Diptera: Simuliidae) in Thailand. Acta Trop. 2024;254:107207. [DOI] [PubMed] [Google Scholar]
- 27.Yssouf A, Almeras L, Raoult D, Parola P. Emerging tools for identification of arthropod vectors. Future Microbiol. 2016;11:549–66. [DOI] [PubMed] [Google Scholar]
- 28.Rivera J, Currie DC. Identification of Nearctic black flies using DNA barcodes (Diptera: Simuliidae). Mol Ecol Resour. 2009;9:224–36. [DOI] [PubMed] [Google Scholar]
- 29.Adler PH, Huang S. Chromosomes as Barcodes: Discovery of a new species of black fly (Diptera: Simuliidae) from California, USA. Insects. 2022;13:903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thapa S, Adler PH, Chhetri S, Varma R, Henry W. Chromosomal evidence for a new cryptic species of black fly in the Simulium praelargum complex (Diptera: Simuliidae) from West Bengal, India. Zootaxa. 2017;4244:137–44. [DOI] [PubMed] [Google Scholar]
- 31.Nascimento J, Hamada N, Andrade-Souza V, Adler PH. A new anthropophilic species of Simulium (Trichodagmia) (Diptera: Simuliidae) from Amazonia: Morphology, chromosomes, and DNA sequences. J Med Entomol. 2018;55:90–111. [DOI] [PubMed] [Google Scholar]
- 32.Adler PH, Fukuda M, Takaoka H, Reeves WK, Kim SK, Otsuka Y. Revision of Simuliumrufibasis (Diptera: Simuliidae) in Japan and Korea: Chromosomes, DNA, and Morphology. J Med Entomol. 2020;57:388–403. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez-Triana LM, Montes De Oca F, Prosser SW, Hebert PD, Gregory TR, McMurtrie S. DNA barcoding as an aid for species identification in austral black flies (Insecta: Diptera: Simuliidae). Genome. 2017;60:348–57. [DOI] [PubMed] [Google Scholar]
- 34.Jomkumsing P, Tangkawanit U, Wongpakam K, Pramual P. Who is biting you? DNA barcodes reveal cryptic diversity in human-biting black flies (Diptera: Simuliidae). Acta Trop. 2019;196:22–9. [DOI] [PubMed] [Google Scholar]
- 35.Pramual P, Jomkumsing P, Wongpakam K, Wongwian P. DNA barcoding of tropical black flies (Diptera: Simuliidae) in Thailand: one decade of progress. Acta Trop. 2021;224:106116. [DOI] [PubMed] [Google Scholar]
- 36.Hew YX, Ya’cob Z, Adler PH, Chen CD, Lau KW, Sofian-Azirun M, et al. DNA barcoding of black flies (Diptera: Simuliidae) in Indonesia. Parasit Vectors. 2023;16:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izwan-Anas N, Low VL, Ya’cob Z, Lourdes EY, Halim MRA, Sofian-Azirun M, et al. DNA barcodes and species boundaries of black flies (Diptera: Simuliidae) in Malaysia. Arthropod Syst Phylogeny. 2023;81:931–43. [Google Scholar]
- 38.Kúdelová T, Krčmárik S, Lužáková I, Bujačková B, Matická K, Kúdela M. DNA barcoding of black flies (Diptera: Simuliidae) in Slovakia and its utility for species identification. Diversity. 2023;15:661. [Google Scholar]
- 39.Putt QY, Ya’cob Z, Adler PH, Chen CD, Hew YX, Izwan-Anas N, et al. From bites to barcodes: uncovering the hidden diversity of black flies (Diptera: Simuliidae) in Vietnam. Parasit Vectors. 2023;16:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garros C, Dujardin J-P. Genetic and phenetic approaches to Anopheles systematics. London: InTech; 2013. [Google Scholar]
- 41.Sumruayphol S, Apiwathnasorn C, Ruangsittichai J, Sriwichai P, Attrapadung S, Samung Y, et al. DNA barcoding and wing morphometrics to distinguish three Aedes vectors in Thailand. Acta Trop. 2016;159:1–10. [DOI] [PubMed] [Google Scholar]
- 42.Jaramillo ON, Dujardin JP, Calle-Londono D, Fonseca-Gonzalez I. Geometric morphometrics for the taxonomy of 11 species of Anopheles (Nyssorhynchus) mosquitoes. Med Vet Entomol. 2015;29:26–36. [DOI] [PubMed] [Google Scholar]
- 43.Sontigun N, Samerjai C, Sukontason K, Wannasan A, Amendt J, Tomberlin JK, et al. Wing morphometric analysis of forensically important flesh flies (Diptera: Sarcophagidae) in Thailand. Acta Trop. 2019;190:312–9. [DOI] [PubMed] [Google Scholar]
- 44.Champakaew D, Junkum A, Sontigun N, Sanit S, Limsopatham K, Saeung A, et al. Geometric morphometric wing analysis as a tool to discriminate female mosquitoes from different suburban areas of Chiang Mai province. Thailand PloS one. 2021;16:e0260333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Limsopatham K, Klong-Klaew T, Fufuang N, Sanit S, Sukontason KL, Sukontason K, et al. Wing morphometrics of medically and forensically important muscid flies (Diptera: Muscidae). Acta Trop. 2021;222:106062. [DOI] [PubMed] [Google Scholar]
- 46.Chaiphongpachara T, Changbunjong T, Laojun S, Sumruayphol S, Suwandittakul N, Kuntawong K, et al. Geometric morphometric and molecular techniques for discriminating among three cryptic species of the Anophelesbarbirostris complex (diptera: culicidae) in Thailand. Heliyon. 2022;8:e11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hadj-Henni L, Millot C, Lehrter V, Augot D. Wing morphometrics of biting midges (Diptera: Culicoides) of veterinary importance in Madagascar. Infect Genet Evol. 2023;114:105494. [DOI] [PubMed] [Google Scholar]
- 48.Ardkhongharn N, Ravichotikul R, Aksornchai P, Weluwanarak T, Chaiphongpachara T, Changbunjong T. Wing geometric morphometrics to distinguish and identify Haematobosca flies (Diptera: Muscidae) from Thailand. Int J Parasitol Parasites Wildl. 2023;21:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laojun S, Changbunjong T, Sumruayphol S, Chaiphongpachara T. Outline-based geometric morphometrics: wing cell differences for mosquito vector classification in the Tanaosri mountain range, Thailand. Acta Trop. 2024;250:107093. [DOI] [PubMed] [Google Scholar]
- 50.Laojun S, Changbunjong T, Sumruayphol S, Pimsuka S, Chaiphongpachara T. Wing geometric morphometrics and DNA barcoding to distinguish three closely related species of Armigeres mosquitoes (Diptera: Culicidae) in Thailand. Vet Parasitol. 2024;325:110092. [DOI] [PubMed] [Google Scholar]
- 51.Pepinelli M, Spironello M, Currie DC. Geometric morphometrics as a tool for interpreting evolutionary transitions in the black fly wing (Diptera: Simuliidae). Zool J Linn Soc. 2013;169:377–88. [Google Scholar]
- 52.Gérard M, Martinet B, Dehon M, Rasmont P, Williams PH, Michez D. The utility of wing morphometrics for assigning type specimens to cryptic bumblebee species. Syst Entomol. 2020;45:849–56. [Google Scholar]
- 53.Boumaza M, Merabti B, Adjami Y, Ouakid ML, Carvajal TM. Geometric morphometric wing analysis of avian malaria vector, Culisetalongiareolata, from two locations in Algeria. Insects. 2022;13:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaiphongpachara T, Duvallet G, Changbunjong T. Wing phenotypic variation among Stomoxyscalcitrans (Diptera: Muscidae) populations in Thailand. Insects. 2022;13:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santoni MM, Magri LA, de Oliveira SV, da Silva BH, Del Lama MA. Sex ratio, sexual eimorphism, and wing geometric morphometrics in species of Trypoxylon Latreille, 1796 (Hymenoptera: Crabronidae). Neotrop Entomol. 2023;52:1005–17. [DOI] [PubMed] [Google Scholar]
- 56.Dujardin J-P. Morphometrics applied to medical entomology. Infect Genet Evol. 2008;8:875–90. [DOI] [PubMed] [Google Scholar]
- 57.Lorenz C, Almeida F, Almeida-Lopes F, Louise C, Pereira SN, Petersen V, et al. Geometric morphometrics in mosquitoes: what has been measured? Infect Genet Evol. 2017;54:205–15. [DOI] [PubMed] [Google Scholar]
- 58.Takaoka H, Srisuka W, Saeung A. Checklist and keys for the black flies (Diptera: Simuliidae) of Thailand. Med Entomol Zool. 2019;70:53–77. [Google Scholar]
- 59.Arnqvist G, Martensson T. Measurement error in geometric morphometrics: empirical strategies to assess and reduce its impact on measures of shape. Acta Zool Acad Sci Hung. 1998;44:73–96. [Google Scholar]
- 60.Gidaszewski NA, Baylac M, Klingenberg CP. Evolution of sexual dimorphism of wing shape in the Drosophilamelanogaster subgroup. BMC Evol Biol. 2009;9:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sontigun N, Sukontason KL, Zajac BK, Zehner R, Sukontason K, Wannasan A, et al. Wing morphometrics as a tool in species identification of forensically important blow flies of Thailand. Parasit Vectors. 2017;10:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bookstein FL. Morphometric tools for landmark data: geometry and biology. New York: Cambridge University Press; 1991. [Google Scholar]
- 63.Gower JC. Generalized procrustes analysis. Psychometrika. 1975;40:33–51. [Google Scholar]
- 64.Rohlf FJ. Rotational fit (Procrustes) methods. In: Rohlf F, Bookstein F, (eds). Proceedings of the Michigan Morphometrics Workshop Special Pu Ann Arbor. The University of Michigan Museum of Zoology. 1990. p. 227–36.
- 65.Murtagh F, Contreras P. Algorithms for hierarchical clustering: an overview, II. WIREs Data Min Knowl Discovery. 2017;7:e1219. [Google Scholar]
- 66.Manly BFJ. Multivariate statistical methods: a primer. 3rd ed. USA: Chapman and Hall/CRC; 2004. [Google Scholar]
- 67.Dujardin J-P, Dujardin S, Kaba D, Santill An-Guayasamín S, Villacís AG, Piyaselakul S, et al. The maximum likelihood identification method applied to insect morphometric data. Zool Syst. 2017;42:46–58. [Google Scholar]
- 68.Mahalanobis PC. On the generalized distance in statistics. Proc Natl Inst Sci (Calcutta). 1936;2:49–55. [Google Scholar]
- 69.Dujardin S, Dujardin J-P. Geometric morphometrics in the cloud. Infect Genet Evol. 2019;70:189–96. [DOI] [PubMed] [Google Scholar]
- 70.Team R: RStudio: integrated development for R. Boston, MA; 2020.
- 71.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9. [PubMed] [Google Scholar]
- 72.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. [DOI] [PubMed] [Google Scholar]
- 74.Stecher G, Tamura K, Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol. 2020;37:1237–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meier R, Shiyang K, Vaidya G, Ng PK. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol. 2006;55:715–28. [DOI] [PubMed] [Google Scholar]
- 77.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Puillandre N, Brouillet S, Achaz G. ASAP: assemble species by automatic partitioning. Mol Ecol Resour. 2021;21:609–20. [DOI] [PubMed] [Google Scholar]
- 83.Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, et al. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol. 2006;55:595–609. [DOI] [PubMed] [Google Scholar]
- 85.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ezard T, Fujisawa T, Barraclough T. splits: SPecies’ LImits by Threshold Statistics. R package version 1.0–14/r31. 2009.
- 89.Changbunjong T, Prakaikowit N, Maneephan P, Kaewwiset T, Weluwanarak T, Chaiphongpachara T, et al. Landmark data to distinguish and identify morphologically close Tabanus spp. (Diptera: Tabanidae). Insects. 2021;12:974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Changbunjong T, Chaiphongpachara T, Weluwanarak T. Species discrimination of Stomoxys flies S. bengalensis, S. calcitrans, and S. sitiens (Diptera: Muscidae) using wing geometric morphometrics. Animals. 2023;13:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gomez GF, Marquez EJ, Gutierrez LA, Conn JE, Correa MM. Geometric morphometric analysis of Colombian Anophelesalbimanus (Diptera: Culicidae) reveals significant effect of environmental factors on wing traits and presence of a metapopulation. Acta Trop. 2014;135:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lorenz C, Suesdek L. The use of wing shape for characterising macroevolution in mosquitoes (Diptera: Culicidae). Infect Genet Evol. 2020;77:104052. [DOI] [PubMed] [Google Scholar]
- 93.Chonephetsarath S, Raksakoon C, Sumruayphol S, Dujardin JP, Potiwat R. The unequal taxonomic signal of mosquito wing cells. Insects. 2021;12:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saiwichai T, Laojun S, Chaiphongpachara T, Sumruayphol S. Species identification of the major Japanese Encephalitis vectors within the Culexvishnui Subgroup (Diptera: Culicidae) in Thailand using geometric morphometrics and DNA barcoding. Insects. 2023;14:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takaoka H, Srisuka W, Low VL, Saeung A. Morphological and molecular analyses of Simuliumrufibasis (Diptera: Simuliidae) in Thailand. J Med Entomol. 2019;56:408–15. [DOI] [PubMed] [Google Scholar]
- 96.Pramual P, Adler PH. DNA barcoding of tropical black flies (Diptera: Simuliidae) of Thailand. Mol Ecol Resour. 2014;14:262–71. [DOI] [PubMed] [Google Scholar]
- 97.Pollock DD, Zwickl DJ, McGuire JA, Hillis DM. Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol. 2002;51:664–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong Z, Qu S, Landrein S, Yu WB, Xin J, Zhao W, et al. Increasing taxa sampling provides new insights on the phylogenetic relationship between Eriobotrya and Rhaphiolepis. Front Genet. 2022;13:831206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takaoka H, Srisuka W, Fukuda M, Saeung A. Twenty-one new species of the Simulium (Gomphostilbia) asakoae species group (Diptera, Simuliidae) in Thailand, with their genetic relationships. Zookeys. 2020;950:51–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Srisuka W, Aupalee K, Low VL, Yacob Z, Fukuda M, Saeung A, et al. A new species of Simulium (Gomphostilbia) (Diptera: Simuliidae) from Northern Thailand, with its genetic relationship in the S.asakoae species-group. Acta Trop. 2021;218:105889. [DOI] [PubMed] [Google Scholar]
- 101.Srisuka W, Aupalee K, Fukuda M, Saeung A, Takaoka H. Morphological description and phylogenetic position of a new cryptic species in the Simuliumasakoae species-group, Simulium (Gomphostilbia) mokroense, from Western Thailand. Acta Trop. 2022;228:106313. [DOI] [PubMed] [Google Scholar]
- 102.Low VL, Srisuka W, Saeung A, Tan TK, Ya’cob Z, Yeong YS, et al. DNA barcoding of Simuliumasakoae (Diptera: Simuliidae) from northern Thailand. J Med Entomol. 2020;57:1675–8. [DOI] [PubMed] [Google Scholar]
- 103.Low VL, Takaoka H, Adler PH, Ya’cob Z, Norma-Rashid Y, Chen CD, et al. A multi-locus approach resolves the phylogenetic relationships of the Simuliumasakoae and Simuliumceylonicum species groups in Malaysia: evidence for distinct evolutionary lineages. Med Vet Entomol. 2015;29:330–7. [DOI] [PubMed] [Google Scholar]
- 104.Senatore GL, Alexander EA, Adler PH, Moulton JK. Molecular systematics of the Simuliumjenningsi species group (Diptera: Simuliidae), with three new fast-evolving nuclear genes for phylogenetic inference. Mol Phylogenet Evol. 2014;75:138–48. [DOI] [PubMed] [Google Scholar]
- 105.Winkler IS, Blaschke JD, Davis DJ, Stireman JO 3rd, O’Hara JE, Cerretti P, et al. Explosive radiation or uninformative genes? origin and early diversification of tachinid flies (Diptera: Tachinidae). Mol Phylogenet Evol. 2015;88:38–54. [DOI] [PubMed] [Google Scholar]
- 106.Pramual P, Jumpato W, Adler PH. Fast-evolving nuclear genes as barcoding markers for black flies (Diptera: Simuliidae) in Thailand. Acta Trop. 2023;246:106988. [DOI] [PubMed] [Google Scholar]
- 107.Takaoka H, Sofian-Azirun M, Ya’cob Z, Chen CD, Lau KW, Low VL, et al. The black flies (Diptera: Simuliidae) of Vietnam. Zootaxa. 2017;4261:1–165. [DOI] [PubMed]
- 108.Takaoka H, Suzuki H. The blackflies (Diptera: Simuliidae) from Thailand. Med Entomol Zool. 1984;35:7–45.
- 109.Sauer FG, Pfitzner WP, Jost H, Rauhoft L, Kliemke K, Lange U, et al. Using geometric wing morphometrics to distinguish Aedes japonicusjaponicus and Aedes koreicus. Parasit Vectors. 2023;16:418. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Results from the NCBI BLAST search of black fly samples based on COI gene
Additional file 2: Table S2. Details of COI sequences generated from the present study and retrieved from NCBI GenBank databased used for molecular analysis
Data Availability Statement
The authors confirm that all data supporting the findings of this study are available within the article. All sequences generated from the study have been deposited in the GenBank database with assigned accession numbers PP422429–PP422472.