Abstract
Background
Postoperative pain is a common complication following surgery, with severity and duration varying between patients. Chronic postoperative pain after inguinal hernia surgery has an incidence rate of approximately 10%. Risk factors for acute and chronic pain following hernia surgery include age, sex, psychosocial factors, and demographic background. Additionally, genetic polymorphisms in enzymes involved in pain mechanisms, as well as the metabolism of analgesics might influence pain perception, pain development, and response to pain medications. Key enzymes include the catechol-o-methyltransferase (COMT), the µ-opioid receptor 1 (OPRM1), and the cytochrome P450 2D6 (CYP2D6).
CYP2D6 plays a crucial role in metabolizing analgesics such as tramadol, codeine, and oxycodone. It is also suspected to be involved in the synthesis of catecholamines and endogenous morphines suggesting a potential role in pathophysiology of pain. We hypothesize that the CYP2D6 activity influences the development of postoperative pain after hernia surgery.
Methods
This study is a prospective, observational, multicenter association study investigating adult patients scheduled for inguinal hernia surgery using a robotic-assisted (rTAPP) approach. Patients are enrolled during the preoperative surgical consultation. A buccal swab is collected for genetic testing at this time. Pain at the site of the hernia is assessed using the validated EuraHSQoL score preoperatively and at 2, 4, and 6 weeks postoperatively. Additionally, information on co-medication and details of the surgery will be collected. The planned number of participants is 350 patients. The primary objective is to analyze the association between different genotype-predicted CYP2D6 phenotypes and patient-reported pain intensity 6 weeks after surgery. Secondary objectives include the association between further genetic variants, such as the COMT rs4680 and OPRM1 rs1799971 genotype, and pain severity. Additionally, the potential of pharmacogenetic panel testing to optimize analgesic therapy in hernia surgery patients will be explored.
Discussion
The findings of this study are expected to provide valuable insights into identifying patients at higher risk for postoperative pain before surgery. This knowledge could pave the way for tailored interventions during and after surgery for these specific patients.
Trial registration
Deutsches Register Klinischer Studien https://www.drks.de/DRKS00034796 Registered on August 07, 2024.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-02064-6.
Keywords: Postoperative pain, Hernia surgery, rTAPP, Pharmacogenetics, CYP2D6, COMT, OPRM1, Analgesics
Background
Postsurgical pain is a common complication following surgery, and its severity and duration can vary from patient to patient. In some instances, the pain may persist and become chronic [1]. Chronic postsurgical pain is characterized by pain that either emerges or intensifies after a surgical procedure and persists for a minimum of three months after the index-procedure [2]. The factors contributing to the onset, progression, and chronicity of postsurgical pain are multifaceted [1]. Research has established that characteristics such as being female, younger age, and the intensity of acute preoperative pain can exert a detrimental influence on the development of postsurgical pain [3]. Furthermore, the type of surgery and the surgical technique employed also hold a pivotal role in pain development, particularly in its chronicity [1]. Notably, chronic postsurgical pain following inguinal hernia repair is not an uncommon occurrence, with an incidence rate of approximately 10% [4]. In a subset of cases, ranging from 0.5% to 6%, the pain may reach a severity level that significantly disrupt daily activities [5]. Systematic reviews indicate that in the context of inguinal hernia repair, endoscopic procedures are linked to a reduced risk of developing chronic inguinal pain in comparison to open procedures [6]. However, it is worth noting that chronic postsurgical pain can still be observed even with endoscopic approaches, including procedures like robotic-assisted transabdominal pre-peritoneal repair (rTAPP) [7].
Prolonged pain persisting after inguinal hernia repair often has a neuropathic nature, originating from nerve damage that can occur during the surgical procedure [8]. However, not all patients with nerve damage develop chronic pain, which indicates the influence of other contributing factors [9, 10]. In addition to age and sex, psychosocial factors and demographic background are assumed to serve as risk factors for experiencing acute and chronic pain following hernia surgery [11]. Moreover, genetic polymorphisms in the enzymes involved in mechanisms underlying pain development or in the metabolism of analgesics may exert an influence on the perception of pain, the development of pain, and the response to analgesics [12, 13].
Numerous genetic variants have been linked to increased postoperative pain sensitivity and the risk of developing chronic pain in genetic association studies. However, the actual significance and relevance of these associations remain uncertain [14]. One of the extensively investigated genes related to pain is the COMT gene, encoding the catechol-O-methyl transferase (COMT) enzyme. The COMT enzyme plays a pivotal role in the degradation of catecholamines like dopamine, epinephrine, and norepinephrine, thereby influencing the physiological pain modulation in the nervous system [15]. The most thoroughly examined single nucleotide polymorphism (SNP) within this context is rs4680 (c.G472A), which is characterized by an amino acid substitution at position 158 (p.Val158Met). The substitution of valine by methionine is assumed to result in the formation of an enzyme with reduced activity [16, 17]. It is hypothesized that the reduced enzyme activity diminishes the degradation of catecholamines, which consequently impacts pain processing by altering dopaminergic, adrenergic, and noradrenergic neurotransmission [15]. Vetterlein et al., described in a meta-analysis, that chronic pain patients who are homozygous for the rs4680 A allele exhibited significantly greater pain sensitivity compared to heterozygotes [15]. However, when it comes to the chronification of postoperative pain, no statistically significant results were obtained from a meta-analysis encompassing studies across various surgical procedures [18]. Notably, to the best of our knowledge, no study has investigated the SNP rs4680 in relation to postoperative pain following inguinal hernia surgery. However, there are data on another polymorphism in the COMT, namely the variant rs4633, which exhibits a high linkage disequilibrium with rs4680. This particular SNP has been shown to be associated with altered “pain-related activity impairment” 6 months after hernia surgery [19].
Another gene that contributes to the pathomechanism of pain and that is linked to the effectiveness of analgesics is the OPMR1 gene, which encodes for the µ-opioid receptor 1 (OPMR1). An extensively studied variant within this gene is rs1799971 (c.A118G), known for an amino acid substitution at position 40 (p.Asn40Asp). The substitution from asparagine to aspartic acid has been linked to diminished responses to both endogenous opiates (e.g. endorphin, etc.) and externally administered synthetic opiates. However, also for this variant, the available data on the association to the chronification of postoperative pain remains inconclusive [18].
Lastly, another enzyme scrutinized for its involvement in both pain development and opiate efficacy is the cytochrome P450 2D6 (CYP2D6) [20]. The role of CYP2D6 in drug metabolism has undergone thorough investigation, being implicated in the metabolism of approximately 25% of all available drugs. Notably, CYP2D6 is highly polymorphic, with various genetic variants affecting the enzymes activity. Based on the enzymes activity four phenotypes are distinguished: ultra-rapid metabolizers (UMs) with increased activity, extensive metabolizers (EMs) with normal activity, intermediate metabolizers (IMs) characterized by decreased CYP2D6 activity, and poor metabolizers (PMs) displaying no CYP2D6 activity [21]. Concerning analgesics, CYP2D6 plays a crucial role in the metabolism of tramadol, codeine, and oxycodone. Notably, there are pharmacogenetic recommendation guidelines for specific phenotypes related to tramadol and codeine. For instance, CYP2D6 PM individuals are anticipated to experience a diminished therapeutic effect of tramadol and codeine due to reduced bioactivation [22, 23]. However, the involvement of CYP2D6 in the development of pain is not as thoroughly researched. It is assumed that CYP2D6 contributes to the production of catecholamines and of endogenous morphine, suggesting its potential role in the pathomechanism of pain [20]. Literature indicates that individuals with CYP2D6 PM status demonstrate enhanced sensitivity to pressure pain [24] and are prone to more severe acute postoperative pain [25, 26]. As far as we know, no study has explored the correlation between CYP2D6 phenotypes and the onset of chronic postoperative pain.
In addition to CYP2D6, several other enzymes are involved in the metabolism of analgesics used to treat postoperative pain. For example, pharmacogenetic variants in CYP2C9 influence the gastrointestinal tolerability of non-steroidal anti-inflammatory drugs (NSAIDs) [27]. Also relevant are genetic variations in CYP2C19, as these affect the efficacy of proton pump inhibitors (PPIs), which are commonly used in the context of NSAID therapy [28].
In conclusion, it can be asserted that several enzymes and receptors play a role in the development of pain, and the interaction of various genetic factors in pain development is intricate and not fully comprehended. Given this gap in knowledge and the significant impact of postoperative pain on patients' quality of life, we aim to enhance the understanding of genetic predisposition in the development of postoperative pain. Specifically, we intend to investigate the association between the genetic makeup of the CYP2D6 enzyme, the COMT enzyme and the OPMR1 receptor with postoperative patient-reported pain intensity after inguinal hernia repair using the rTAPP method. Furthermore, our goal is to assess whether a pharmacogenetic panel test for patients prone to developing chronic pain could be beneficial in optimizing their pain therapy.
Methods
Project objectives and design
Hypothesis and primary objective
We hypothesize that the CYP2D6 phenotype influences the development of postoperative pain after hernia surgery applying the rTAPP methodology. Specifically, we propose that patients with a CYP2D6 PM status will experience more severe pain compared to patients with CYP2D6 EM status 6 weeks postoperatively.
The primary objective of this observational study is to examine the association between different CYP2D6 phenotypes and the patient-reported pain intensity 6 weeks after inguinal hernia surgery with the rTAPP approach. Accordingly, the following null hypothesis results for the primary objective:
H₀: There is no significant difference in pain severity 6 weeks postoperatively after rTAPP inguinal hernia repair between individuals with different CYP2D6 phenotypes.
Patient-reported severity of pain 6 weeks postoperatively will be, determined by the European Hernia Society quality-of-life (EuraHS-QoL) postoperative score [29], considering the domain "Pain" (range 0–30, with an average range of 0–10) (see additional file 1). This specific time point was chosen due to its alignment with clinical practice, where patients routinely undergo postoperative follow-ups with surgeons, and pain is typically assessed during this period. This aims to facilitate the early identification and treatment of patients who may develop potentially chronic pain. Pain management in the initial weeks following surgery appears to be crucial in preventing chronification of postoperative pain [1].
Secondary objectives
Additional objectives will be assessed to further explore the role of the CYP2D6 phenotype, as well as other genetic variants, in the development of postoperative inguinal pain. These objectives encompass:
Association between the genotype predicted CYP2D6 phenotype and patient-reported quality of life 6 weeks postoperatively: Determined by the EuraHS-QoL postoperative score, focusing on the domains of "Pain" and "Restriction of Activities" (range 0–70, with an average range of 0–10).
Association between the genotype predicted CYP2D6 phenotype and patient-reported preoperative pain severity: Determined by the EuraHS-QoL preoperative score, specifically examining the "Pain" domain (range 0–30, with an average range of 0–10).
Association between specific CYP2D6 genotypes and patient-reported postoperative pain severity 6 weeks after inguinal hernia surgery: Determined by the EuraHS-QoL postoperative score, focusing on the "Pain" domain (range 0–30, with an average of 0–10).
Association between the COMT rs4680 genotype and OPMR1 rs1799971 genotype, and patient-reported pain severity before and 6 weeks after inguinal hernia surgery: Determined by the EuraHS-QoL pre- and postoperative scores, considering the "Pain" domain (range 0–30, with an average of 0–10).
Association between the COMT rs4680 genotype and OPMR1 rs1799971 genotype, and patient-reported quality of life before and 6 weeks after inguinal hernia surgery: Determined by the EuraHS-QoL pre- and postoperative scores, considering the domains of "Pain" and "Restriction of Activities" (range 0–70, with an average of 0–10).
Association between patient-reported pain severity and combinations of the genotype predicted CYP2D6 phenotype, COMT rs4680 genotype, or OPMR1 rs1799971 genotype.
Graphical representation of the development of pain over time for different groups defined by the genotype predicted CYP2D6 phenotypes, COMT rs4680 genotype, and OPMR1 rs1799971 genotype in separate graphs.
Potential of pharmacogenetic panel test in patients undergoing hernia surgery: The potential of the pharmacogenetic panel test will be assessed based on the proportion of patients exhibiting other phenotypes than EM in CYP2D6, CYP2C9, and/or CYP2C19. Those patients might response atypically to pain medications in the future.
Study design
This is a prospective, observational, multicenter association study investigating adult patients scheduled for inguinal hernia surgery employing the rTAPP approach.
Project population and study procedures
Project population, inclusion and exclusion criteria
The project encompasses a total of 350 adult patients. Recruitment is conducted at three Swiss hospitals: Kantonsspital Olten, Olten, Kantonsspital Baselland, Liestal, and Spital Männedorf, Männedorf. Eligibility for inclusion in the trial requires meeting the following criteria.
Inclusion criteria:
Patients with a primary, unilateral, or bilateral inguinal hernia intending to undergo rTAPP inguinal hernia repair.
Age ≥ 18 years.
Exclusion criteria:
History of chronic pain syndrome.
Pain attributable to current malignant disease or chronic infection.
Lumbar Disc Syndrome with current pain.
Disease impairing central or peripheral nerve function.
Previous neurectomy.
Opioid addiction and/or enrollment in an opioid substitution program.
Recurrent inguinal hernia.
Patients with prior operations affecting sensitivity in the inguinal region (e.g., urological surgery).
Pregnancy.
Advanced dementia or other cognitive impairments hindering comprehension of the study protocol.
Study procedures
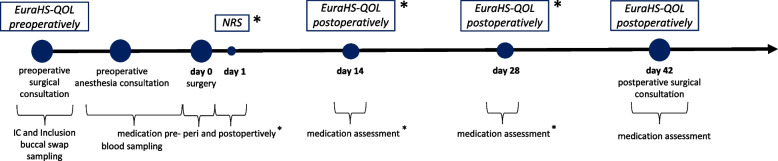
The study procedure is depicted in Fig. 1 and described in the summary table (additional file 2).
Fig. 1.
Study procedures. NRS: Numeric Rating Scale * assessment by email consultation or paper based diary
Preoperative surgical consultation (day > -1)
Patients with an inguinal hernia with a prospect rTAPP inguinal hernia repair typically undergo a standard preoperative consultation with the surgeon, during which they are informed about the operation. At this stage, patients are screened for eligibility and provided information about the study. Furthermore, patients are informed that participation in the study includes a pharmacogenetic analysis conducted by Stratipharm®, humatrix AG. Once the patients have signed the informed consent (IC) and the Stratipharm® laboratory requisition, preoperative hernia pain will be assed using the EuraHS QOL preoperatively score. Additionally, a buccal swab from the patients' oral mucosa will be collected.
The buccal swab is sent to humatrix AG (Pfungstadt, Germany, https://www.stratipharm.de) for Stratipharm® pharmacogenotyping and phenotype prediction. Stratipharm® is an approved in vitro diagnostic (IVD) test according to Article 9 of the European Directive 98/79/EC and consists of a real-time polymerase chain reaction (PCR) primer/probe (assay) using the automated Life Technologies QuantStudio 12k flex system. It is a qualitative genotyping assay for the detection and identification of SNPs in 30 different genes from genomic DNA, including CYP2D6, CYP2C9, CYP2C19, COMT, and OPMR1. TaqMan® GTXpress™ Master Mix is used in combination with the respective primer/probe mix (assay). The IVD product consists of multiple primer/probe combinations (assays) to specifically detect variants of the included genes. For each SNP, an individual PCR reaction is performed with the diluted DNA of the sample, the master mix and the specific assay. Each assay contains two PCR primers and two fluorescence-labelled probes. The probes are short DNA molecules that specifically bind to the sequences of the reference or the variant allele. Performance of this genotyping technique has been validated according to the methods described by Welch et al. [30]. All SNPs included in the Stratipharm® panel are detailed in additional file 3.
Based on the individual polymorphism pattern, a genetic profile is established for each patient highlighting pharmacogenetically relevant variations of each of the analysed genes. Also, Stratipharm® provides a card to the patient containing a personal access code to their genetic data. With this access code, pharmacists or physicians authorized by the patients can excess to the pharmacogenetic data on Stratipharm's web portal. The data are presented in a user-friendly format, enabling physicians and pharmacists to individualize the patient's medication and adjust it to a safer and more effective regimen. Patients are informed that they are allowed to contact the Clinical Pharmacy at Kantonsspital Olten at any time after week 6 (after postoperative surgical consultation) with questions regarding their genetic profile and the selection of future medications.
Preoperative anesthesia consultation (day > -1)
Prior to surgery, patients undergo an informative session with the anesthesiologist. During this consultation, a comprehensive set of patient-specific data, including comorbidities and smoking status, is documented. Additionally, preoperative medications are systematically recorded (generic names, dosage, dosing interval, last intake). Medications that are CYP2D6 or COMT inhibitors or inducers are specifically documented.
At this time point, at Kantonsspital Olten, blood samples (4 ml EDTA whole blood and 6 ml serum) are collected for subsequent analysis, which may arise from further research questions that may emerge after the completion of this study. These analyses may include, the identification of other variants in pharmacogenes or measurement of substance concentrations in whole blood or serum. Standardized and established methods, such as TaqMan® genotyping assays, direct DNA Sanger sequencing, or LC–MS/MS, are employed for these analyses. Blood samples are only taken if the patient consents to the further use of the biological samples and genetic data. The biological material is stored in the Clinical Sample Archive of the Biopharmacy at the University of Basel.
Surgery (day 0)
On Day 0, patients undergo surgery. During the operation, standard information about the surgery and anesthesia is routinely recorded. This includes the duration of the surgery and anesthesia, details of the used anesthetics and analgesics, as well as potential perioperative complications. Information about the hernia, like side of hernia or classification of hernia, are routinely recorded in the electronic patient record.
Postoperative assessment (day 1, day 14, and day 28, ± 4 days)
Following surgery, patients undergo hospital discharge based on the surgeon's risk assessment: Immediate release or lasting hospitalization. For inpatients, postoperative pain medication and acute pain levels (Numeric Rating Scale (NRS) on Day 1) are systematically documented daily in the electronic patient record, as part of routine hospital practice.
Upon discharge patients are provided, according to their preference, with either a paper-based pain diary for manual completion or the option to choose email notifications for online surveys on days 1, 14, and 28. In the pain diary or email survey, patients record the following assessments: NRS for acute postoperative pain on day 1, and the EuraHS QOL postoperatively score for assessing postoperative pain in the inguinal region at two weeks (day 14) and four weeks (day 28) postoperatively. Furthermore, patients document their actual pain medication on days 1, 14, and 28. Patients who have been provided with a paper-based diary are requested to hand it in at the postoperative surgical consultation at day 42.
Postoperative surgical consultation (day 42, ± 7 days)
Approximately 6 weeks postoperatively (day 42, ± 7 days), patients routinely have an appointment with the surgeon for a surgical follow-up. At this time, the postoperative pain in the inguinal region will be assessed using the EuraHS QOL postoperatively score, along with the current medication. Additionally, postoperative complications such as neuropathic pain, seroma, hematoma, wound healing disorder, thrombosis, and recurrence of hernia are recorded. If the collection of buccal swabs has not yet been done, it will be carried out at that time.
Statistics and methodology
Statistical analysis plan
The sample size was calculated for a two-sample t-test comparing CYP2D6 PMs with CYP2D6 EMs, as the strongest effect is expected in this comparison. In the calculation, we considered that the groups would be of different sizes. We assumed that CYP2D6 PMs would constitute 6.5% of the patient population, while CYP2D6 EMs would account for 48%. The frequency was chosen based on the CYP2D6 phenotype frequency for Caucasians according to the Pharmacogenomics Knowledgebase (PharmGKB) [31]. Additionally, we took into account the following criteria: power = 80%, α = 5%, mean difference = 1, and standard deviation of 1.6. The mean difference was chosen based on the consideration that a clinically relevant difference in pain score rating from 0 to 10 must be at least 1 [32]. The standard deviation was calculated based on the EuraHS QOL score of approximately 70 patients previously operated on at Kantonsspital Olten. Taking all these factors into account, we calculated a sample size of 340 patients. It is important to note that not all CYP2D6 genotypes can be clearly assigned to a specific phenotype by Stratipharm®. In rare cases (2.55%), we won't be able to clearly assign CYP2D6 phenotype (see section on Handling of Missing Data). We have accounted for these potential missing data in the sample size calculation, resulting in a total of 350 patients.
To compare the various CYP2D6 phenotypes regarding postoperative pain, we will employ univariate ANCOVA for statistical analysis. As a covariate, we will consider pain medication at week 6, expressed as an analgesic score (rating of analgesic strength from 0–5 according to Widder et al.) [7]. Post hoc tests (Tukey–Kramer Test) will be conducted to identify group differences, such as comparing CYP2D6 PMs with CYP2D6 EMs.
For secondary objectives, we will also use univariate ANCOVA and the Post hoc tests, noting that the sample size calculation is targeted at the pairwise comparison of CYP2D6 PMs to CYP2D6 EMs at the 6-week postoperative time point, making the secondary endpoints purely exploratory in nature.
To assess the Hardy–Weinberg Equilibrium, we will use Pearson’s X2 test. For CYP2D6, rs3892097 will be selected for calculations as it represents the most frequent SNP resulting in a CYP2D6 enzyme without function. Additionally, the Hardy–Weinberg Equilibrium for COMT rs4680 and OPMR1 rs1799971 will be calculated.
The possible potential of the pharmacogenetic panel test in patients undergoing hernia surgery will be evaluated based on the proportion of patients exhibiting other phenotypes than EM in CYP2D6, CYP2C9, and/or CYP2C19. As those patients might response atypically to pain medications in the future.
Statistical analyses will be performed using the software “IBM SPSS Statistics” and “GraphPad Software”. An interim analysis will be conducted once 170 patients have been enrolled in the study. This milestone is predetermined based on a sample size calculation for a two-sample t-test comparing patients with COMT A/A genotype to those with G/G genotype in rs4680. Any deviation from the initial statistical plan will be detailed and justified in the conclusive trial report.
Handling of missing data
When determining the CYP2D6 phenotype using Stratipharm®, there will be certain genotypes for which the phenotype cannot clearly be defined. This primarily impacts patients with copy number variations in CYP2D6, as the multiplied gene cannot be accurately stratified, leaving uncertainty regarding whether the polymorphism is present within the multiplied gene. According to retrospective data from Stratipharm®, this is expected to occur in 2.55% of the patient cases, which was taken into account during the sample size calculation. Additionally, these missing data will be excluded for the determination of the primary endpoint. To assess the impact of the missing data, we will perform sensitivity analyses. For this purpose, all unknown CYP2D6 phenotypes will be assigned to either the PM, IM, EM, or UM CYP2D6 status, based on this the primary endpoint (pain 6 weeks postoperatively) will be recalculated, comparing the scenarios.
If patients omit individual values while completing the EuraHS QOL score, these "missing" values will be handled as described elsewhere [33]. In short, if 1 value is missing in the domain “Pain”, it will be substituted with the mean of the values for the other two Pain questions; if 2 or 3 values are missing, the domain value will be rated as missing. If 1 or 2 values are missing in the domain "Restriction of Activities”, the value(s) will be substituted with the mean of the values for the 2 or 3 other questions in the domain; if 3 or 4 values are missing, the domain value will be rated as missing. If a value is missing in the domain “Cosmetic”, it will be substituted with the value for the other question in this domain; if 2 values are missing, the domain value will be rated as missing.
Any other missing data will be retrospectively retrieved from medical records if possible. Study dropouts will be replaced to achieve the final calculated study size of 350 patients.
Quality control and data protection
Data recording and source data
Study data is recorded in an electronic Case Report Form using the REDCap® software (2024 Vanderbilt University, version 14.0.42). Some data are routinely collected during internal procedures in the hospital (preoperative consultation with the surgeon or anesthetist, surgery, postoperative follow-up with the surgeon) as part of daily practice and documented in the hospital's electronic patient dossier. These data are transferred from the electronic patient dossier to REDCap® and include the following information:
Age, sex
Body height, body weight, BMI
Comorbidities: ASA Score, Diabetes Mellitus, Chronic Obstructive Pulmonary Disease, Chronic Kidney Disease, Psychiatric Disease
Nicotine abuse
Dates of preoperative surgical consultation, clinic entry, operation, postoperative surgical consultation
Duration of operation (start time and stop time of operation), duration of anesthesia (start time and stop time of anesthesia)
Perioperative complications (urinary retention, postoperative bleeding, perioperative nausea and vomiting, other)
Inpatient or outpatient status, duration of hospital stay for inpatient patients (day of entry and day of discharge)
Medication preoperatively (generic names, ATC Code, daily dose, unit, route of administration) and perioperatively (generic names, ATC Code, cumulative dose during operation, unit, route of administration)
Side of hernia, EHS groin hernia classification [34]
Data not routinely collected are obtained through paper CRFs and later transferred to REDCap®.
self-reported origin
EuraHS-QOL preoperatively and postoperatively at day 42
qualitative description of postoperative pain (nociceptive, neuropathic) at day 42
analgesic medication at day 42
Pain diary (NRS at day 1, EuraHS-QOL postoperatively at day 14 and 28, analgesic medication at day 1, 14, and 28)
Postoperative complications (seroma, hematoma, wound healing disorder, thrombosis, primary recurrence of hernia)
For patients who prefer electronic pain diary management, they can provide their email address, which will be used in REDCap® for sending REDCap surveys. Pain assessment (NRS at day 1, EuraHS-QOL postoperatively at day 14 and 28) and pain medication recording (at day 1, 14, and 28) will be directly captured by patients via REDCap surveys. For subsequent analyses, patient data will be used in coded form. The email address is marked as an identifier in REDCap® and will not be extracted from REDCap® and cannot be later associated with patient data.
The genetic data for CYP2D6, CYP2C9, CYP2C19, COMT, and OPRM1 provided by Stratipharm®, as well as the predicted genotype and phenotype are also captured in REDCap®.
Confidentiality and coding
Project data will be handled with uttermost discretion and is only accessible to authorized personnel who require the data to fulfil their duties within the scope of the research project. On the CRFs and further project specific documents, participants are only identified by a unique participant number.1 The corresponding patient identification list is securely kept at the study site, accessible only to the study personnel authorized by the principal investigator.
Buccal swab and genetic data
The DNA extracted from the buccal swab sent to humatrix AG in Germany (Stratipharm®) will be destroyed three weeks after genotyping, leaving only the generated genetic data. These data are protected with a sample-specific password and assigned to a database (server without internet access), where they are stored until revoked. Only Stratipharm® humatrix AG employees have access to this database, and these employees must adhere to strict data protection regulations. Humatrix AG adheres to standards recognized and equivalent in Switzerland. Once the genetic analysis is available, the project leader is informed by email notification. From that point on, the results will be visible on Stratipharm's web portal. The web portal is password-protected, accessible only to pharmacists or physicians authorized by the patient. As part of the study, the patient will provide written consent for the research team to use the password and thus access the genetic data. Genetic data required for the analyses outlined in the study protocol will be extracted from Stratipharm® and stored in REDCap®. For the final analysis, genetic data will be extracted from REDCap® in a coded format and cannot be subsequently linked to patient data.
Blood samples
The blood samples collected in this study are labeled with the study-specific patient ID. The participant's name will not be visible on any of the samples. Blood samples will be collected at Kantonsspital Olten and temporarily stored in Kantonsspital Olten's laboratory at -80 °C. The access is regulated and badge-secured and the freezer temperature is monitored. For storage and further analyses, the biological material is transferred to the Clinical Sample Archive in the Laboratory for Biopharmacy at the University of Basel. Transportation is carried out, if necessary, with dry ice. In the Laboratory for Biopharmacy at the University of Basel, the blood samples, isolated DNA, and processed blood serum are appropriately stored at -20 °C or -80 °C and are accessible only to authorized laboratory personnel. Laboratory personnel handling the biological material do not have access to the patient identification list or any clinical data. Analyses are only conducted upon instruction by the Project Leader. Details regarding the handling, storage and transfer of biological samples between the study center and the Clinical Sample Archive are detailed in an MTA project agreement.
Retention and destruction of project data and biological material
The archiving of study data is intended for 10 years after the completion or premature termination of the study. This includes baseline data and all study materials, which will be stored in the study clinic's archive. After 10 years they will be irreversibly anonymized by destruction of the key file linking the unique patient identifier with personal contact details. Buccal swabs and the DNA extracted from them will be destroyed by humatrix AG three weeks after the laboratory analysis. Individual patient and genetic data will be retained by humatrix AG until revoked by the patient. If participants consent to the further use of the analyzed and coded genetic data and biological material, the collected blood samples and the processed samples (DNA and blood serum) will be stored for 10 years in the Clinical Sample Archive of the Biopharmacy at the University of Basel. After 10 years they will be destroyed. Additional data obtained through the analysis of blood and serum samples will be irreversibly anonymized and retained indefinitely.
Ethical considerations
Patients treatment will not be affected whether the patient participates in the study or not. Participation in the study is optional, and participants have the right to withdraw their consent at any time without the need for explanation, and it will not result in any negative impact on their ongoing medical care.
The expected risk from study-related procedures is not greater than that of routine clinical practices. Blood sampling by vein puncture may lead to inflamed blood vessels, bruising, or nerve injuries.
The study falls under the category of "Research involving human subjects with the exception of clinical trials," according to the local research legislation, and is classified as Risk Category A (low risk). It has been approved by the local ethics committees (Ethikkommission Nordwest- und Zentralschweiz, Basel, Switzerland, and Kantonale Ethikkommission Zürich, Zürich, Switzerland; reference number 2024–01080).
Discussion
Given the high incidence of chronic postsurgical pain following inguinal hernia repair and the significant impact of postoperative pain on patients' quality of life [5], it is crucial to identify patients at risk for postoperative pain. In the clinical practice of the study hospitals, a postoperative surgical consultation is conducted 6 weeks after surgery to identify patients who are still experiencing pain and who are at risk of developing chronic pain.
Multiple factors influence chronification of postoperative inguinal pain, including age, sex, surgical techniques, nerve damage, psychosocial influences, and demographic background. Acute postoperative pain and delays in pain treatment are recognized as additional risk factors [11]. Moreover, genetic factors are assumed to affect both the pathophysiology of pain and the efficacy and safety of drugs used in pain management, whereby influencing postoperative pain [12, 13].
One aim of this project is to investigate the association between the genetic makeup of genes assumed to be involved in pain development and in the metabolism of analgesics with post-operative pain. Specifically, the genes CYP2D6, COMT, and OPRM1 have been selected for analysis. This study will examine how specific genetic variants within these genes are linked to the patient-reported intensity of postoperative pain 6 weeks after inguinal hernia repair using the rTAPP method.
Considering all possible risk factors, the insights from genetic variants could be of help in identifying patients at higher risk for postoperative pain, even before they undergo surgery. This knowledge could pave the way for tailored interventions during and after surgery for these patients. Such interventions might include structured follow-ups and closer monitoring of postoperative pain, enabling early therapeutic intervention to prevent chronic pain and restore quality of life.
Additionally, we aim to identify which medications are actually prescribed to patients in our study for pain management and to determine the proportion of patients who exhibit altered enzyme activity, leading to atypically metabolism of analgesics in the future. Currently, official pharmacogenetic dosing guidelines exist for tramadol and codeine [22], NSAIDs [27], and PPIs [28] when used concomitantly with NSAIDs. For tricyclic antidepressants (TCAs), there are pharmacogenetic guidelines for their use as antidepressants [35].
For the pharmacological treatment of chronic postoperative inguinal pain guidelines recommend primarily, gabapentin, duloxetine, and TZAs due to the frequent neuropathic pain component. However, NSAIDs or cyclooxygenase-II (COX-II) inhibitors combined with paracetamol should not be excluded, given the often present inflammation. Capsaicin patches can also be used. As a second-line treatment, tramadol is recommended due to its serotonin-norepinephrine reuptake inhibitor effect, which could be advantageous for neuropathic pain [36].
In summary, for some of the analgesics recommended after hernia surgery, there is evidence that pharmacogenetic testing could be beneficial. Clinical studies have shown that using pharmacogenetic panel tests and pharmacogenetically guided selection of postoperative analgesia after various surgeries can lead to lower pain scores [37], reduced postoperative opioid consumption [38], or both [39, 40]. For our specific study population of patients undergoing hernia surgery, these endpoints would need to be further investigated in additional clinical trials. Pharmacogenetic panel tests could potentially be beneficial for patients undergoing rTAPP hernia surgery.
In conclusion, our study could contribute to identifying patients at an increased risk of postoperative pain and potentially offer a solution through pharmacogenetic testing, which would enable the provision of optimized analgesic treatment for these patients. Personalized treatments are likely to represent the future of medicine, with particular emphasis on individualization in the management of pain conditions. This emphasis is underscored by the well-documented substantial inter-individual variability in pain sensitivity and pain experience.
Supplementary Information
Additional file 2: Schedule of assessments.
Additional file 3: Stratipharm® SNPs and annotation.
Acknowledgements
We used Deepl Translator (DeepL SE, Köln, Germany, version 24.6.1) for translation of specific text passages from German to English and ChatGPT (OpenAI, San Fracisco, CA, USA, version 3.5) for language editing to improve clarity and readability.
Abbreviations
- COMT
Catechol-O-methyl transferase
- COX-II
Cyclooxygenase-II
- CYP
Cytochrome P450
- DNA
Deoxyribonucleic acid
- EMs
Extensive metabolizers
- EuraHS-QoL
European Hernia Society quality-of-life
- IM
Intermediate metabolizer
- IVD
In vitro diagnostic
- NRS
Numeric Rating Scale
- NSAID
Non-Steroidal Anti-Inflammatory Drug
- OPMR1
µ-Opioid receptor 1
- PCR
Polymerase chain reaction
- PharmGKB
Pharmacogenomics Knowledgebase
- PM
Poor metabolizer
- PPI
Proton Pump Inhibitors
- rTAPP
Robotic-assisted transabdominal pre-peritoneal repair
- SNP
Single Nucleotide Polymorphism
- TZA
Tricyclic antidepressants
- UM
Ultra-rapid metabolizer
Authors’ contributions
FW, ML, and UD contributed equally in the writing of this manuscript. SA is the sponsor and led the protocol development together with ML as the project leader. SA, HMzS, AT, SL, and RR were involved in the study design, contributed to the additional content, and were responsible for the critical revision of the manuscript. AB was responsible for the critical revision of the manuscript. The authors read and approved the final manuscript.
Funding
The study is fully funded by the study centers and the University of Basel. No external funding was received for this research.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The local ethics committees (Ethikkommission Nordwest- und Zentralschweiz, Basel, Switzerland, and Kantonale Ethikkommission Zürich, Zürich, Switzerland) approved the described study protocol version 2, dated on July 24, 2024, as of October 01, 2024 (reference number 2024–01080). We will obtain consent from all trial participants.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Mandated by the Swiss Association of Research Ethics Committees—Project plan template for human research other than clinical trials, Version 2.5, 31.08.2022.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rosenberger DC, Pogatzki-Zahn EM. Chronic post-surgical pain – update on incidence, risk factors and preventive treatment options. BJA Educ. 2022;22(5):190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schug SA, Lavand’homme P, Barke A, Korwisi B, Rief W, Treede RD, et al. The IASP classification of chronic pain for ICD-11: chronic postsurgical or posttraumatic pain. Pain. 2019;160(1):45. [DOI] [PubMed] [Google Scholar]
- 3.Gerbershagen HJ, Pogatzki-Zahn E, Aduckathil S, Peelen LM, Kappen TH, van Wijck AJM, et al. Procedure-specific risk factor analysis for the development of severe postoperative pain. Anesthesiology. 2014;120(5):1237–45. [DOI] [PubMed] [Google Scholar]
- 4.Aasvang EK, Gmaehle E, Hansen JB, Gmaehle B, Forman JL, Schwarz J, et al. Predictive risk factors for persistent postherniotomy pain. Anesthesiology. 2010Apr;112(4):957–69. [DOI] [PubMed] [Google Scholar]
- 5.van Veenendaal N, Simons MP, Bonjer HJ. Summary for patients: International guidelines for groin hernia management. Hernia. 2018;22(1):167–8. [DOI] [PubMed] [Google Scholar]
- 6.Haladu N, Alabi A, Brazzelli M, Imamura M, Ahmed I, Ramsay G, et al. Open versus laparoscopic repair of inguinal hernia: an overview of systematic reviews of randomised controlled trials. Surg Endosc. 2022;36(7):4685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Widder A, Reese L, Lock JF, Wiegering A, Germer CT, Kindl GK, et al. Postoperative Analgesics Score as a Predictor of Chronic Postoperative Inguinal Pain After Inguinal Hernia Repair: Lessons Learned From a Retrospective Analysis. World J Surg. 2023;47(10):2436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haroutiunian S, Nikolajsen L, Finnerup NB, Jensen TS. The neuropathic component in persistent postsurgical pain: a systematic literature review. Pain. 2013;154(1):95–102. [DOI] [PubMed] [Google Scholar]
- 9.Kehlet H, Jensen TS, Woolf CJ. Persistent postsurgical pain: risk factors and prevention. Lancet Lond Engl. 2006;367(9522):1618–25. [DOI] [PubMed] [Google Scholar]
- 10.Costigan M, Scholz J, Woolf CJ. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci. 2009;32:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen MB, Andersen KG, Crawford ME. Pain following the repair of an abdominal hernia. Surg Today. 2010;40(1):8–21. [DOI] [PubMed] [Google Scholar]
- 12.Lötsch J, Geisslinger G, Tegeder I. Genetic modulation of the pharmacological treatment of pain. Pharmacol Ther. 2009Nov 1;124(2):168–84. [DOI] [PubMed] [Google Scholar]
- 13.Palada V, Kaunisto MA, Kalso E. Genetics and genomics in postoperative pain and analgesia. Curr Opin Anaesthesiol. 2018;31(5):569–74. [DOI] [PubMed] [Google Scholar]
- 14.Young EE, Lariviere WR, Belfer I. Genetic Basis of Pain Variability: Recent Advances. J Med Genet. 2012 Jan;49(1):10.1136/jmedgenet-2011-100386. [DOI] [PMC free article] [PubMed]
- 15.Vetterlein A, Monzel M, Reuter M. Are catechol-O-methyltransferase gene polymorphisms genetic markers for pain sensitivity after all? – A review and meta-analysis. Neurosci Biobehav Rev. 2023;1(148):105112. [DOI] [PubMed] [Google Scholar]
- 16.Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melén K, Julkunen I, et al. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995;34(13):4202–10. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75(5):807–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chidambaran V, Gang Y, Pilipenko V, Ashton M, Ding L. Systematic Review and Meta-Analysis of Genetic Risk of Developing Chronic Postsurgical Pain. J Pain. 2020;21(1–2):2–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belfer I, Dai F, Kehlet H, Finelli P, Qin L, Bittner R, et al. Association of functional variations in COMT and GCH1 genes with postherniotomy pain and related impairment. Pain. 2015;156(2):273–9. [DOI] [PubMed] [Google Scholar]
- 20.Zahari Z, Ismail R. Influence of Cytochrome P450, Family 2, Subfamily D, Polypeptide 6 (CYP2D6) Polymorphisms on Pain Sensitivity and Clinical Response to Weak Opioid Analgesics. Drug Metab Pharmacokinet. 2014;29(1):29–43. [DOI] [PubMed] [Google Scholar]
- 21.Schwabedissen MZ, HE. The role of pharmacogenomics in individualized medicine. In: Fischer T, Langanke M, Marschall P, Michl S, editors. Individualized medicine–ethical, economical and historical perspectives. Springer: Cham; 2014. p. 93–112.
- 22.Crews KR, Monte AA, Huddart R, Caudle KE, Kharasch ED, Gaedigk A, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin Pharmacol Ther. 2021;110(4):888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matic M, Nijenhuis M, Soree B, de Boer-Veger NJ, Buunk AM, Houwink EJF, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2D6 and opioids (codeine, tramadol and oxycodone). Eur J Hum Genet. 2021;30(10):1105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sindrup SH, Poulsen L, Brøsen K, Arendt-Nielsen L, Gram LF. Are poor metabolisers of sparteine/debrisoquine less pain tolerant than extensive metabolisers? Pain. 1993;53(3):335. [DOI] [PubMed] [Google Scholar]
- 25.Candiotti KA, Yang Z, Rodriguez Y, Crescimone A, Sanchez GC, Takacs P, et al. The Impact of CYP2D6 Genetic Polymorphisms on Postoperative Morphine Consumption. Pain Med. 2009;10(5):799–805. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Yang Z, Arheart KL, Morris R, Zhang Y, Rodriguez Y, et al. CYP2D6 Poor Metabolizer Genotype and Smoking Predict Severe Postoperative Pain in Female Patients on Arrival to the Recovery Room. Pain Med. 2012;13(4):604–9. [DOI] [PubMed] [Google Scholar]
- 27.Theken KN, Lee CR, Gong L, Caudle KE, Formea CM, Gaedigk A, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin Pharmacol Ther. 2020;108(2):191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lima JJ, Thomas CD, Barbarino J, Desta Z, Van Driest SL, El Rouby N, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clin Pharmacol Ther. 2021;109(6):1417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muysoms F, Campanelli G, Champault GG, DeBeaux AC, Dietz UA, Jeekel J, et al. EuraHS: the development of an international online platform for registration and outcome measurement of ventral abdominal wall hernia repair. Hernia J Hernias Abdom Wall Surg. 2012;16(3):239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch RA, Lazaruk K, Haque KA, Hyland F, Xiao N, Wronka L, et al. Validation of the performance of a comprehensive genotyping assay panel of single nucleotide polymorphisms in drug metabolism enzyme genes. Hum Mutat. 2008;29(5):750–6. [DOI] [PubMed] [Google Scholar]
- 31.PharmGKB. Gene-specific Information Tables for CYP2D6. https://www.pharmgkb.org/page/cyp2d6RefMaterials Accessed 20 Aug 2024.
- 32.Hilfiker R. Schmerzintensität messen. Physiopraxis. 2008;6(11):46–7. [Google Scholar]
- 33.Muysoms FE, Vanlander A, Ceulemans R, Kyle-Leinhase I, Michiels M, Jacobs I, et al. A prospective, multicenter, observational study on quality of life after laparoscopic inguinal hernia repair with ProGrip laparoscopic, self-fixating mesh according to the European Registry for Abdominal Wall Hernias Quality of Life Instrument. Surgery. 2016;160(5):1344–57. [DOI] [PubMed] [Google Scholar]
- 34.Miserez M, Alexandre JH, Campanelli G, Corcione F, Cuccurullo D, Pascual MH, et al. The European hernia society groin hernia classication: simple and easy to remember. Hernia. 2007;11(2):113–6. [DOI] [PubMed] [Google Scholar]
- 35.Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Müller DJ, Shimoda K, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Veenendaal N, Foss NB, Miserez M, Pawlak M, Zwaans WAR, Aasvang EKA, narrative review on the non-surgical treatment of chronic postoperative inguinal pain: a challenge for both surgeon and anaesthesiologist. Hernia. cited 2022 Nov 11. Available from: 2022Oct 31. 10.1007/s10029-022-02693-9. [DOI] [PMC free article] [PubMed]
- 37.Hamilton WG, Gargiulo JM, Parks NL. Using pharmacogenetics to structure individual pain management protocols in total knee arthroplasty. Bone Joint J. 2020;102-b(6_Supple_A):73–8. [DOI] [PubMed] [Google Scholar]
- 38.Thomas CD, Parvataneni HK, Gray CF, Deen JT, Prieto HA, Pulido LF, et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genet Med. 2021;23(4):621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamilton WG, Gargiulo JM, Reynolds TR, Parks NL. Prospective Randomized Study Using Pharmacogenetics to Customize Postoperative Pain Medication Following Hip and Knee Arthroplasty. J Arthroplasty. 2022;37(6, Supplement):S76–81. [DOI] [PubMed] [Google Scholar]
- 40.Senagore AJ, Champagne BJ, Dosokey E, Brady J, Steele SR, Reynolds HL, et al. Pharmacogenetics-guided analgesics in major abdominal surgery: Further benefits within an enhanced recovery protocol. Am J Surg. 2017;213(3):467–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2: Schedule of assessments.
Additional file 3: Stratipharm® SNPs and annotation.
Data Availability Statement
No datasets were generated or analysed during the current study.