Abstract
Female semiochemicals and allyl isothiocyanate (AITC) attract moths, and the moths use odorant-degrading enzymes (ODEs) to break down the excess odor. By identifying antennae-specific ODEs, researchers have established the molecular foundation for odorant degradation and signal inactivation in insects. This enables further exploration of new pest control methods. Currently, the degradation of female semiochemicals and AITC has received limited attention, inspiring this study to identify target ODEs in diamondback moths through transcriptome analysis. Sequencing of antennae from male adults (MA) exposed to female adults (FA) and AITC yielded a substantial 54.18 Gb of clean data, revealing 2276 differentially expressed genes (DEGs) between the MA and MA-FA treatments, and 629 DEGs between MA and MA-AITC treatments. The analysis of MAs exposed to FAs and AITC identified 29 and 17 ODEs, respectively, mainly involving aldehyde dehydrogenases (ALDHs), alcohol dehydrogenases (ADs), cytochrome P450s (CYPs), and UDP-glucuronosyltransferases (UGTs). Pathway analysis revealed primary enrichment in glycolysis/gluconeogenesis and fatty acid degradation in female adult treatments. In contrast, AITC treatments showed major enrichment in pathways related to pentose and glucuronate interconversions, retinol metabolism, and ascorbate and aldarate metabolism. Additionally, qRT-PCR analysis validated the expression patterns of 10 ODE genes in response to these treatments, with varying results observed among the genes. These findings indicate significant changes in ODE expression levels, providing a molecular foundation for identifying potential targets for behavioral inhibitors.
Introduction
The diamondback moth, also known as DMB, or Plutella xylostella L. (Lepidoptera: Plutellidae), is a major agricultural pest. It causes vital damage to brassicaceous vegetables, with annual global economic losses estimated at US$4–5 billion [1,2]. Monitoring, trapping, and mating disruption using semiochemicals are key elements of green pest control and are extensively applied in agricultural practices [3–6]. Semiochemicals are substances or mixtures emitted by an organism that trigger behavioral or physiological responses in individuals of the same or different species, primarily influencing an insect’s behavior with plants or other insects [7]. Host plant volatiles provide critical resources for insects, facilitating feeding, mating, oviposition, and evasion of natural predators. The integration of female semiochemicals and plant volatile constitutes an eco-friendly approach to the monitoring and management of various pest populations [3,7].
The potential applications of pheromones from female semiochemicals are particularly important as they can help reduce the mating rates of female moths by 55% to 75%, hence mitigating crop damage [8]. The main components of DMB’s sex pheromones are (Z)-11-hexadecenal (Z11-16: Ald) and (Z)-11-hexadecenyl acetate (Z11-16: OAc). Using a ratio of 1:10 of sex pheromones and their analogs, specifically (11Z)-hexadec-11-en-1-yl2,2,2-trifluorooate and (11Z)-hexadec-11-en-1-yl 2,2,3,3-pentafluoropropanoate, nearly completely disrupts DMB mating activity in the field [9]. Allyl isothiocyanate (AITC) is a naturally occurring, highly volatile compound found in Brassicaceae family plants. It influences insect behaviors such as feeding and egg-laying and attracts adult moths [10–12]. AITC was shown to effectively control the pest Bradysia odoriphaga in plants both in fields and greenhouses [5]. Furthermore, high concentrations of AITC exhibit anti-insect and anti-bacterial properties, highlighting its potential for agricultural applications [13,14].
Insect antennae are crucial for detecting and distinguishing various semiochemicals [15,16]. In general, odor-binding proteins (OBPs) facilitate the transport of odor molecules to olfactory receptor neurons (ORNs) located in the antennae, where interaction with olfactory receptors (ORs) generates electrical signals, that are subsequently transmitted through axons to the brain [16]. Ultimately, odor-degrading enzymes (ODEs) eliminate excess odors, thereby preparing ORNs for new stimuli [15,17]. Several studies have employed transcriptomic analyses to identify olfactory genes, including research on Dendrolimus houi and Dendrolimus kikuchii [18], Galleria mellonella [19], and Peridroma saucia [20]. However, these investigations have seldom focused on ODEs. ODEs are grouped based on the chain lengths and functional groups present in the target odorant molecules, including key categories like carboxylesterases (CCEs/CXEs), aldehyde oxidases (AOXs) and ALDHs, CYPs, and ADs, etc. [21]. Enzymes such as CCEs /CXEs [22–25] and AOXs [15,26] are associated with the breaking down of sexual signals and plant volatiles. When exposed to strong odors, ODEs act as rapid odor inactivators, suppressing signals and influencing insect behavior by breaking down semiochemicals.
This study identified odor-degrading enzymes in the male adult (MA) antennae of the DMB that are sensitive to female adult (FA) and AITC through transcriptome analysis. Differential gene analysis, along with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, revealed changes in the expression levels of ALDHs, ADs, CYPs, and UGTs. Our findings highlight specific ODEs that provide molecular target for the development of behavioral inhibitors aimed at utilizing female semiochemicals and AITC.
Materials and methods
Insect rearing
Diamondback moths were obtained from a cabbage (Brassica oleracea var. capitata L.) field without using an insecticide disposal in Jiyuan City, He nan province, China (112°60′E, 35°05′N). They were provided with feed and raised in a room at 26 ± 1°C,65 ± 5% humidity, and a 14L: 10 D photoperiod at the Plant Protection Institute of the Fujian Academy of Agricultural Sciences. After about 10 days of growth in the box from the egg stage, the 4th instars were separated by gender [27] and each pupa was placed in a 2 mL glass tube with cotton plugging. Once they emerged, the adults received a 10% (w/v) honey solution.
Insect collection
Male adult antennae were collected from the control group four days post-emergence. For the female adult treatment, male and female adults were housed in a 1:1 ratio in a box with double-layer gauze and male antennae were collected after 24 h. In the allyl isothiocyanate (AITC, Sigma-Aldrich Shanghai Trading Co., Ltd, W203408) treatment group, 0.1 μg/μL of AITC was applied to a 20 μL filter paper strip in the box, and male antennae were collected after 6 h. The dissecting table was disinfected with RnaseZap (Invitrogen, AM9780). Male adults were treated with CO2, immersed in 70% ethanol, and then dissected for antennae collection using forceps under a stereomicroscope. The antennae were immediately frozen in liquid nitrogen at -80°C. A total of 50 pairs of male adult antennae were used in both control and treatment groups, with each treatment consisting of three biological replicates.
RNA extraction and sequencing
Total RNA was extracted from the male adult antennae using the TRIzol reagent (Invitrogen, 15596026CN) as per the manufacturer’s instructions. RNA concentration and purity were assessed with a Nanodrop2000 (Thermo Scientific), and RNA integrity was evaluated with an Agilent Bioanalyzer 2100 (Agilent Technologies). Sequencing libraries were prepared according to NEB (USA) guidelines, utilizing the NEBNext® Ultra™ RNA Library Prep Kit. Each sample received specific index codes for sequence attribution. The RNA-seq libraries underwent cluster generation with the TruSeq PE Cluster Kit v3-cBot-HS and the cBot Cluster Generation System from Illumina. Following library qualification, sequencing was conducted on the Illumina NovaSeq6000 platform in the PE150 mode, with each sample sequenced to a depth of 6 G. The unigene library for this species was generated through the assembly of clean data.
Quality control
To ensure raw read suitability for analysis, specific quality measures were implemented. Reads containing adapters or showing low quality were removed. Specifically, reads with an N ratio over 10% and those with more than 50% of bases at a quality score of Q ≤ 10 were discarded. An evaluation was made of the Q20, Q30, GC-content, and sequence duplication levels of the clean data. The high-quality clean data were then provided in FASTQ format.
Expression statistics and functional annotation
Reads from sequencing were compared using Bowtie [28], and expression levels were estimated with RSEM [29]. The expression abundance of each unigene was represented by the fragment number per kilobase transcript (FPKM) value [30]. Correlation coefficients between transcript and gene expressions were calculated with DESeq2 software [31]. This analysis identified differentially expressed genes (DEGs) between the comparison groups. DEGs were defined with criteria Fold Change≥2 and a false discovery rate (FDR) of less than 0.01 [32]. Principal Component Analysis (PCA) was utilized to moderate the dimensionality of gene expression datasets, thereby facilitating the examination of sample distribution patterns and the assessment of sample dispersion [33]. Moreover, to assess the reproducibility of biological experimental procedures, the reliability of DEGs, and identify any outlier samples, the relationship between gene expression levels across various samples was evaluated through correlation analyses [34]. Functional enrichment analysis was conducted using topGO [35] for GO analysis and KOBAS [36] for KEGG analysis, offering insights into the DEGs’ linked biological processes and pathways.
Quantitative real-time PCR analysis and data analysis
Dissimilarities in gene expressions were determined by quantitative real-time polymerase chain reaction (qRT-PCR) following the method of Zheng et al. [37]. cDNA was synthesized from 1 μg of RNA using the HiScript® II 1st Strand cDNA Synthesis Kit (Vazyme). qPCR was performed with the Taq Pro Universal SYBR qPCR Master Mix (Vazyme) using Pxylβ-tubulin (GenBank accession number: XM_011550528.1) as an internal control. The primer sequences are provided in S3 Table and Table 3. The reaction comprised 5 μl of the 2×Taq Pro Universal SYBR qPCR Master Mix, 0.3 μl (0.3 μM) of each specific primer, 1 μl of cDNA, and the volume was adjusted to 10 μl with RNase-free water. Thermal cycling conditions included an initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60–64°C for 30 s. Amplicon specificity was confirmed through melting curve analysis. For qRT-PCR, three biological replicates were conducted and gene expression changes were measured using the 2−ΔΔCt method with normalization [38]. Statistical analyses were performed using IBM SPSS Statistics version 21 (SPSS, Chicago, IL, USA), employing Student’s t-tests to compare treatment means.
Table 3. qRT-PCR primers used for odor degradation enzyme in P. xylostella male antenna.
| Gene Name | Gene ID | Sequences (5’-3’) |
|---|---|---|
| Tubulin | XM_011550528.1 |
CAATCAGGCCAATTTACCGC
CTGGGTTTACGCCAGTTACG |
| ALDH | TRINITY_DN29578_c0_g1 |
GCCAAACGGGACCTCTTAT
GGCAACAGCACGGTCAAT |
| ALDH | TRINITY_DN42494_c0_g1 | AGGTTGCCTTCACTGGCTCTA CCACTTGGCGGCTTGTT |
| CYP | TRINITY_DN42642_c0_g1 |
AGAGCGAAGAGGAAGGTGAAA
TGCTTGCGTTATGGATGAATG |
| CYP | TRINITY_DN39031_c0_g1 |
AGATGGTGCTGGGTGGAA
CCTTGTGCTGGCGGATAT |
| AD | TRINITY_DN29028_c0_g1 |
CGGCCATCAATTCGTAAACC
GATTCCGTTGTCAAGACCATTTC |
| ALDH | TRINITY_DN40472_c0_g1 |
GTCTGATTTGGGACCACTCG
CCCTTAGCGAACTTCCCTTT |
| ALDH | TRINITY_DN39018_c0_g1 |
AGAATCCCTTGTCTCCGTGTC
CACCTTCATGGGTGCTCAA |
| CYP | TRINITY_DN7385_c0_g1 |
GACCCTCGATCCACCTTCAC
TGTGCTTAGGCAGGATATAGGC |
| AD | TRINITY_DN74_c0_g1 |
CCAGTTGGGCGAACGTAGTCA
TGGTTTGGGAACTCTTGCTGTG |
| AD | TRINITY_DN74_c0_g2 | GCAGCCTGGACAGCATCA TTGGCCGGTATCAAGTGGT |
Results
Transcriptome assembly and subsequent annotation
Our study analyzed transcriptomes from three groups: female adult treatment, allyl isothiocyanate treatment, and a control group of male adults. The primary aim was to assess the effects of exposure to female semiochemicals and plant-derived volatile compounds on gene expression. Transcriptome sequencing for all nine samples generated 54.18 Gb of clean data. Each sample produced at least 5.70 Gb, with a Q30 base percentage exceeding 91.75%, and an average GC content of 46.4% (Table 1). All clean reads are available in the NCBI database. The distribution of contig lengths, summarized in Table 2, shows 43,405 unigenes with an average length of 983.17 and an N50 of 1875. Notably, 50.73% of the contigs ranged from 200 to 500 bp, while 49.27% exceeded 500 bp, indicating strong assembly accuracy.
Table 1. Quality statistics of reads after raw filtering of P. xylostella transcriptomes from male antennas.
| Sample Names | Read Number | Base Number | GC Content (%) | %≥Q30(%) |
|---|---|---|---|---|
| MA-1 | 20,336,664 | 6,077,635,772 | 46.64 | 94.36 |
| MA-2 | 20,007,849 | 5,969,812,230 | 46.50 | 94.28 |
| MA-3 | 19,807,348 | 5,920,106,086 | 46.34 | 94.26 |
| MA-FA-1 | 20,081,716 | 6,001,452,576 | 46.99 | 94.11 |
| MA-FA-2 | 20,713,808 | 6,184,639,190 | 46.80 | 94.05 |
| MA-FA-3 | 19,099,355 | 5,706,083,310 | 46.92 | 94.40 |
| MA-AITC-1 | 21,360,009 | 6,378,744,478 | 45.94 | 94.20 |
| MA-AITC-2 | 20,796,990 | 6,211,483,148 | 45.52 | 94.21 |
| MA-AITC-3 | 19,179,139 | 5,727,957,678 | 45.93 | 91.75 |
Read Number is the total number of pair-end reads in the clean data. MA, control; MA-FA, female exposed; MA-AITC, AITC exposed.
Table 2. Statistics of contigs assembled of P. xylostella transcriptomes from male antennas.
| Length Range(nt) | Transcripts | Unigenes |
|---|---|---|
| 200–300 | 15,438(20.76%) | 13,169(30.34%) |
| 300–500 | 12,837(17.26%) | 8,852(20.39%) |
| 500–1000 | 15,165(20.39%) | 8,536(19.67%) |
| 1000–2000 | 15,674(21.07%) | 6,864(15.81%) |
| 2000+ | 15,260(20.52%) | 5,984(13.79%) |
| Total Number | 74,374 | 43,405 |
| Total Length | 93,290,340 | 42,674,647 |
| N50 Length | 2,215 | 1,875 |
| Mean Length | 1254.34 | 983.17 |
The unigene N50 length denotes the length of the fragment at which the cumulative length reaches 50% of the total length of all unigenes.
Enrichment analysis of DEGs
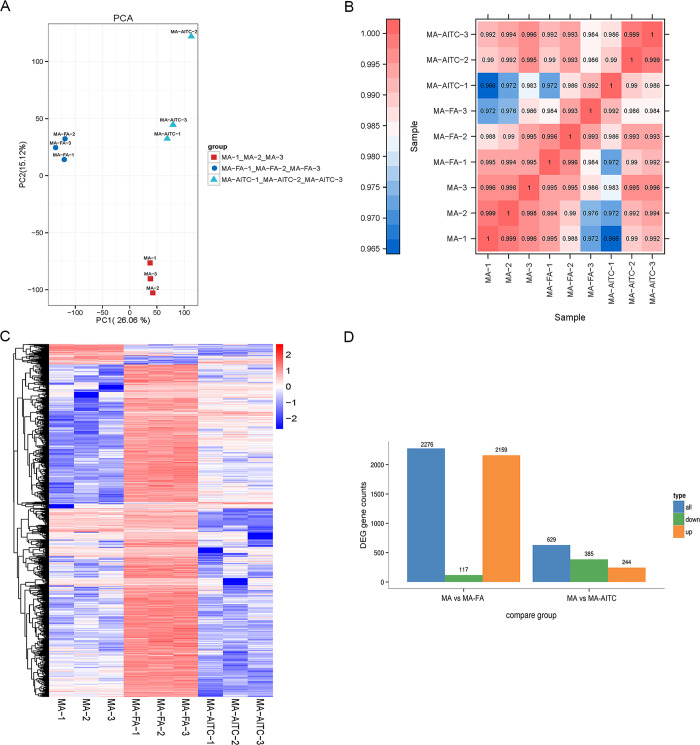
Quality control of RNA-Seq data was assessed using PCA and correlation analyses (Fig 1A and 1B). The MA, MA-FA, and MA-AITC groups were well separated indicating notable variations in their expression levels (Fig 1A). Meanwhile, within the groups, a strong correlation in the levels of gene expression was identified (Fig 1B). Hierarchical clustering detected genes with similar expression patterns across samples, displayed as heatmaps. The comparison revealed a higher percentage of upregulated genes in the MA-FA group and more downregulated genes in the MA-AITC group (Fig 1C). Specifically, 2276 DEGs were identified between the MA and MA-FA treatments, and 629 DEGs between the MA and MA-AITC treatments (Fig 1D). Post-FA treatment, the number of genes upregulated in MA increased to 2159.
Fig 1. Transcriptome quality validation and differential gene analysis of P. xylostella male antennas (MA).
Quality control metrics for transcriptomes are shown as principal components analysis (PCA) (A), correlation analysis (B), and gene cluster heatmap (C) for the MA, MA-FA, and MA-AITC groups. An overview of differentially expressed genes under different treatments (D). MA, control; MA-FA, female exposed; MA-AITC, AITC exposed.
Gene functional annotations of DEGs
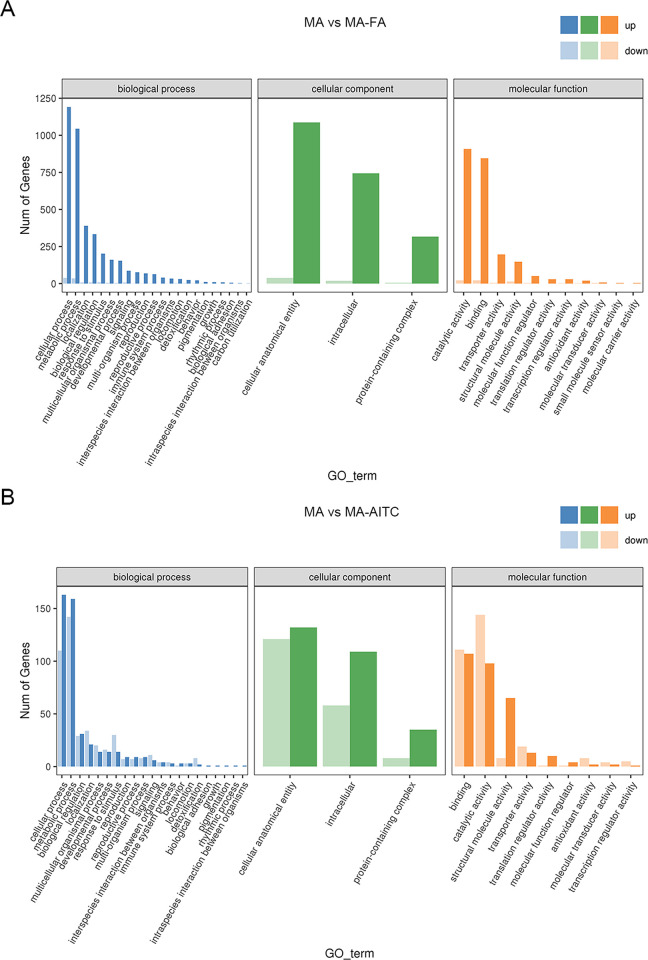
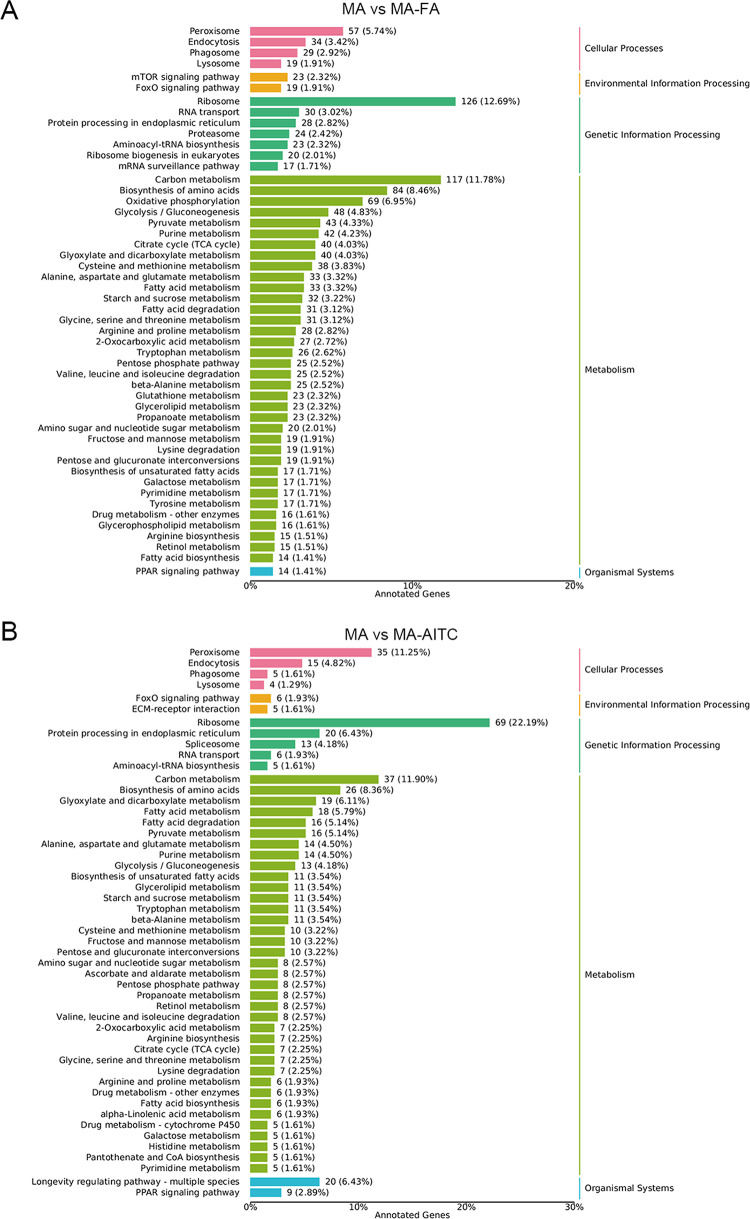
The GO annotation analysis showed that the MA-FA treatment affected 1785 genes, with 1726 upregulated and 59 downregulated genes. In contrast, the MA-AITC treatment influenced 455 genes, with 210 upregulated and 245 downregulated genes. In biological processes, the cellular process pathway saw the most upregulated genes, with 1191 from the MA-FA treatment and 163 from the MA-AITC treatment. The metabolic process pathway also had significant upregulation, with 1044 and 159 genes from the MA-FA and MA-AITC treatments, respectively. Additionally, the cellular components analysis indicated 1087 upregulated genes in the cellular anatomical entity category from the MA-FA treatment and 132 from the MA-AITC treatment. In the molecular function category, the catalytic activity pathway had the highest upregulation, with 908 and 98 genes from the MA-FA and MA-AITC treatments, respectively. Following this, the binding pathway had 846 upregulated genes from the MA-FA treatment and 107 from the MA-AITC treatment (Fig 2A and 2B). The KEGG pathway analysis showed both treatments had enriched pathways like the ribosome, carbon metabolism, and amino acid biosynthesis (Fig 3A and 3B). The key difference was that MA-FA treatment enriched pathways were oxidative phosphorylation, mTOR signaling, and Proteasome (Fig 3A), while the MA-AITC treatment pathways were specific to longevity regulation and spliceosome (Fig 3B).
Fig 2.
Gene Ontology (GO) enrichment of differentially expressed genes in MA vs MA-FA (A) and MA vs MA-AITC (B). The X-axis represents the GO classification, while the Y-axis represents the number of genes; colors indicate different primary classifications.
Fig 3.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed genes in MA vs MA-FA (A) and MA vs MA-AITC (B). The left axis shows the annotated KEGG metabolic pathway names and the right axis shows the corresponding primary classification names. The X-axis represents the number of genes from the annotated pathway and their proportion to the total genes annotated.
Changes in the expression of ODEs
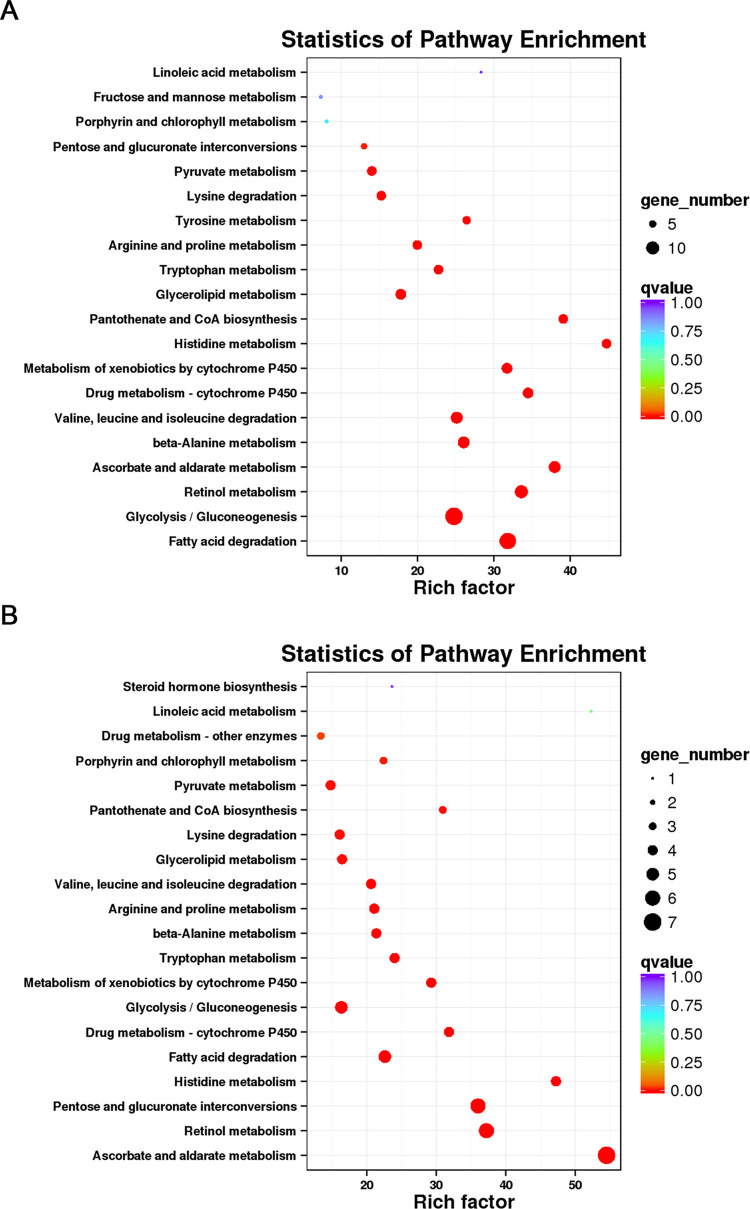
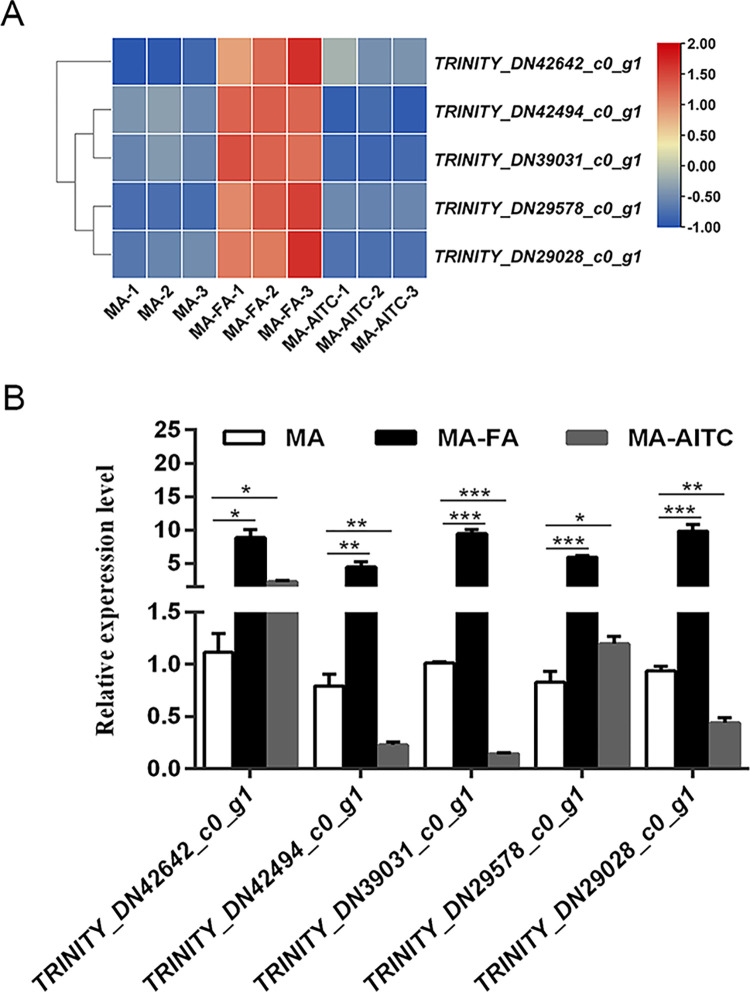
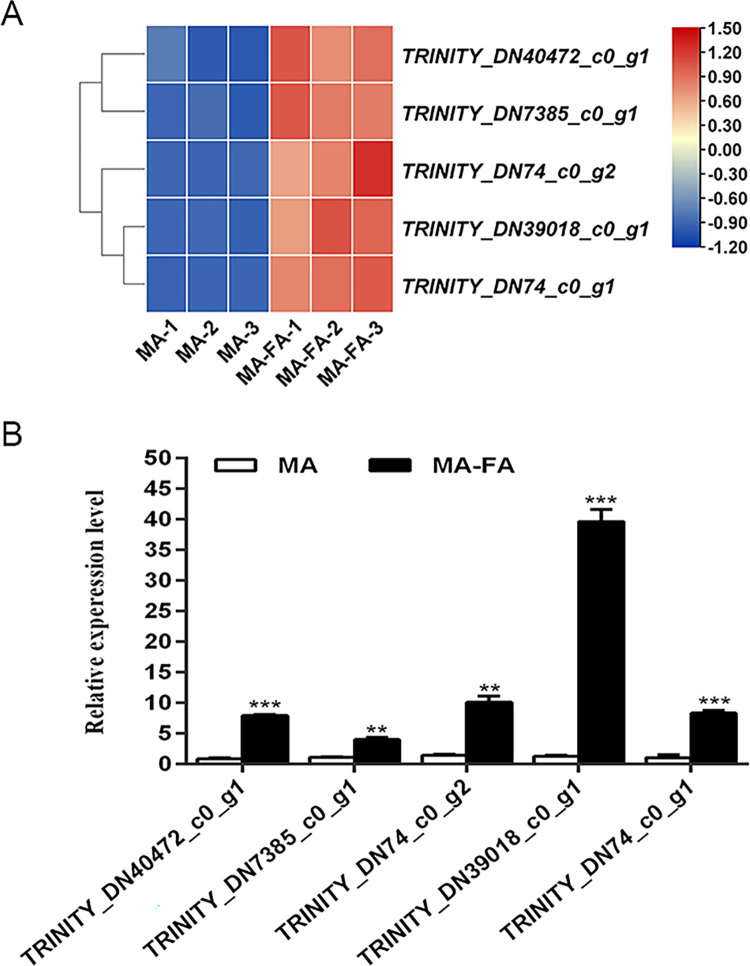
This study found differential expression of 29 ODE genes in the MA vs MA-FA comparison (S1 Table) and 17 in the MA vs MA-AITC comparison (S2 Table). The MA vs MA-FA group showed primary pathway enrichment in glycolysis/gluconeogenesis and fatty acid degradation (Fig 4A), while the MA vs MA-AITC group was major enriched in pentose and glucuronate interconversions, retinol metabolism, and ascorbate and aldarate metabolism pathways (Fig 4B). Comparison among MA, MA-FA, and MA-AITC groups identified five genes with higher expression for further investigation. TRINITY_DN42642_c0_g1 and TRINITY_DN29578_c0_g1 were upregulated in both MA-FA and MA-AITC groups compared to MA. The remaining three genes were downregulated in the MA-AITC group but significantly upregulated in the MA-FA group (Table 3 and Fig 5). Five genes were notably upregulated in the MA-FA group, with TRINITY_DN39018_c0_g1 showing a significant 30.0-fold upregulation in the MA-FA group compared to MA (Table 3 and Fig 6). Additionally, a decrease in the expression of five heat shock protein (Hsp) genes was noted in the MA-AITC group compared to MA (S3 Table and S1 Fig). The qRT-PCR analysis validated the reliability of the DEG data by confirming the RNA-seq data.
Fig 4.
KEGG enrichment analysis of odorant-degrading enzymes in MA vs MA-FA (A) and MA vs MA-AITC (B). The enriched pathways were visualized with q-values (marked in represented colors); the enrichment factor is on the X-axis, and the number of involved differentially expressed genes is indicated by the corresponding circle size.
Fig 5.
Transcriptomic (A) and qRT-PCR (B) expression levels of five odorant-degrading enzyme genes were compared between control (MA) and female exposed (MA-FA), as well as between control (MA) and AITC exposed (MA-AITC). Values are the mean ± SE (n = 3). Statistical significance was determined by Student’s t-test: * P < 0.05, ** P < 0.01, or *** P < 0.001.
Fig 6.
Transcriptomic (A) and qRT-PCR (B) expression levels of five odorant-degrading enzyme genes were compared between control (MA) and female exposed (MA-FA). Values are the mean ± SE (n = 3). Statistical significance was determined by Student’s t-test: ** P < 0.01, or *** P < 0.001.
Discussion
Past research has mostly concentrated on the screening and identification of olfactory proteins, such as OBPs, ORs, and chemosensory proteins (CSPs), while neglecting ODEs. In this study, we explored the response of male DBMs to the female semiochemicals and plant volatile compound AITC. We mainly aimed to screen for ODEs capable of degrading the female semiochemicals and AITC. By identifying ODEs that respond to specific chemical signals, this research offers insights into potential novel strategies for pest control.
Previous research has indicated that exposure to various odors alters the expression of ODEs in insect antennae [21,22,39]. In a transcriptome analysis of Grapholita molesta, 23 candidate CXEs were identified. Notably, GmolCXE1 and GmolCXE5 showed significant upregulation in the antennae of male moths in response to female adults, while GmolCXE14 and GmolCXE 21 displayed approximately two-fold upregulation in larval heads in response to fresh mature fruit odors. These genes may play a role in degrading sex pheromones or fruit odors, potentially affecting mating and foraging behaviors [22]. Additionally, research on DBM showed that two homologous carboxylesterases, CCE16a and CCE16c, can degrade sex pheromones and plant volatiles. CCE16c exhibited greater hydrolysis activity than CCE16a, indicating its potential role in odorant degradation [21]. Using exposure to female semiochemicals and AITC, we also screened several ODEs. In the MA vs MA-FA comparison, 29 ODEs were upregulated in male antennae (S1 Table) that are primarily enriched in glycolysis/glycogenesis and fatty acid degradation pathways (Fig 4A). This suggests that male insects may upregulate these genes to degrade excess female semiochemicals (S1 Table and Figs 5 and 6). After exposure to AITC, 17 ODEs were identified. These are majorly enriched in pentose and glucuronate interconversions, retinol metabolism, and ascorbate and aldarate metabolism pathways. The insect’s response to AITC is concentration-dependent, where moderate AITC levels are promoting [40] while high levels are toxic [41,42]. During a 6 h AITC exposure, 14 ODE genes were downregulated and 3 were upregulated (S2 Table and Fig 5), indicating a subtle response based on treatment duration and concentration. Notably, in our preliminary experiments, known ODE expressions exhibited an increase following a 6 h treatment with AITC. This initial finding guided our decision to focus on the 6 h time point for subsequent transcriptomic analysis. Nevertheless, the findings from the detailed study indicate that 6 h treatment with AITC may not be the optimal time for identifying the upregulated ODE expressions effectively.
Furthermore, we identified four classes of ODEs (ALDHs, ADs, CYPs, and UGTs), particularly ALDHs and CYPs showing the most significant expression changes. Previous studies demonstrated that ALDHs metabolize volatile compounds such as acetaldehyde [43], ethanol [44], farnesal [45], and verbenone [46]. Insect cytochrome P450 monooxygenases are involved in metabolizing and detoxifying external substances, including plant chemicals, insecticides, and environmental pollutants [47–49]. Knockout experiments targeting the CYP6AE subfamily have shown increased sensitivity to insecticides and plant chemicals, highlighting the importance of CYPs in insect physiology [50,51]. This suggests that these target genes hold potential for further research.
In this study, we identified ODEs associated with the degradation of female semiochemicals and AITC through transcriptome screening. These can be categorized into four groups: ALDHs, ADs, CYPs, and UGTs. Future studies should prioritize genes with increased ALDHs and CYPs expression levels. To facilitate this, we should conduct bioinformatics assessments, cloning, prokaryotic expression, and protein assays to evaluate the degradation of female semiochemicals and AITC in DBM. Given the fact that even minimal quantities of volatiles can be lethal to insects that are incapable of decomposing scents, the application of dsODEs in the use of semiochemicals can markedly lower the cost of pest control.
Supporting information
Transcriptomic (A) and qRT-PCR (B) expression levels of five heat shock protein genes were compared between control (MA) and AITC exposed (MA-AITC). Values are the mean ± SE (n = 3). Statistical significance was determined by Student’s t-test: ** P < 0.01, or *** P < 0.001.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
The article contains data along with Supplementary Materials. The RNA-seq raw data has been uploaded to the NCBI Short Read Archive (SRA) and is accessible under the accession number PRJNA1106633.
Funding Statement
This project was supported by the National Natural Science Foundation under grant number 32072425, and the Basic Research Projects of Fujian Academy of Agricultural Sciences under grant numbers GJYS202205 and XTCXGC2021017. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Furlong MJ, Wright DJ, Dosdall LM. Diamondback moth ecology and management: problems, progress, and prospects. Annu Rev Entomol. 2013; 58(1):517–541. doi: 10.1146/annurev-ento-120811-153605 [DOI] [PubMed] [Google Scholar]
- 2.You MS, Ke FS, You SJ, Wu ZY, Liu QF, He WY, et al. Variation among 532 genomes unveils the origin and evolutionary history of a global insect herbivore. Nat Commun. 2020; 11(1):2321. doi: 10.1038/s41467-020-16178-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witzgall P, Kirsch P, Cork A. Sex pheromones and their impact on pest management. J Chem Ecol. 2010; 36(1):80–100. doi: 10.1007/s10886-009-9737-y [DOI] [PubMed] [Google Scholar]
- 4.Schroeder PC, Shelton AM, Ferguson CS, Hoffmann MP, Petzoldt CH. Application of synthetic sex pheromone for management of diamondback moth, Plutella xylostella, in cabbage. Entomol Exp Appl. 2010; 94(3): 243–248. 10.1046/j.1570-7458.2000.00626.x [DOI] [Google Scholar]
- 5.Shi CH, Hu JR, Xie W, Yang YT, Wang SL, Zhang YJ. Control of Bradysia odoriphaga (Diptera: Sciaridae) with allyl isothiocyanate under field and greenhouse conditions. J Econ Entomol. 2017; 110(3):1127–1132. doi: 10.1093/jee/tow303 [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Zheng XB, Yuan JJ, Wang SY, Xu BY, Wang SL, et al. Insecticide resistance monitoring of the diamondback moth (Lepidoptera: Plutellidae) populations in China. J Econ Entomol. 2021; 114(3): 1282–1290. doi: 10.1093/jee/toab027 [DOI] [PubMed] [Google Scholar]
- 7.El-Ghany NMA. Semiochemicals for controlling insect pests. J Plant Protect Res. 2019; 59(1):1–11. 10.24425/jppr.2019.126036 [DOI] [Google Scholar]
- 8.Cui GZ, Zhu JJ. Pheromone-based pest management in China: past, present, and future prospects. J Chem Ecol. 2016; 42(7):557–570. doi: 10.1007/s10886-016-0731-x [DOI] [PubMed] [Google Scholar]
- 9.Wang AJ, Zhang KX, Gao YL, Weng AZ, Wang LY, Zhang YH, et al. Synthesis and bioactivity studies of sex pheromone analogs of the diamond back moth, Plutella xylostella. Pest Manag Sci. 2019; 75(4):1045–1055. doi: 10.1002/ps.5214 [DOI] [PubMed] [Google Scholar]
- 10.Pivnick KA, Lamb RJ, Reed D. Response of flea beetles, Phyllotreta spp., to mustard oils and nitriles in field trapping experiments. J Chem Ecol. 1992; 18(6):863–873. doi: 10.1007/BF00988327 [DOI] [PubMed] [Google Scholar]
- 11.Tian HJ, Lin S, Chen Y, Chen YX, Zhao JW, Gu XJ, et al. Electroantennogram responses to plant volatiles associated with fenvalerate resistance in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J Econ Entomol. 2018; 111(3):1354–1360. doi: 10.1093/jee/toy022 [DOI] [PubMed] [Google Scholar]
- 12.Xiao Y, Sun L, Wu YH, Wang Q, Zhang YJ, Jing XF, et al. The larvae of Phyllotreta striolata share the same olfactory cues for locating Brassicaceae plant with conspecific adults. J Pest Sci. 2024; 97(2):979–992. 10.1007/s10340-023-01690-w [DOI] [Google Scholar]
- 13.Lin CM, Preston JF 3rd, Wei CI. Antibacterial mechanism of allyl isothiocyanate. J Food Prot. 2000; 63(6):727–734. doi: 10.4315/0362-028x-63.6.727 [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Zhang GA, Zeng SY, Liu KC. Extraction of allyl isothiocyanate from horseradish (Armoracia rusticana) and its fumigant insecticidal activity on four stored-product pests of paddy. Pest Manag Sci. 2009; 65(9):1003–1008. doi: 10.1002/ps.1786 [DOI] [PubMed] [Google Scholar]
- 15.Wang MM, He M, Wang H, Ma YF, Dewer Y, Zhang F, et al. A candidate aldehyde oxidase in the antennae of the diamondback moth, Plutella xylostella (L.), is potentially involved in the degradation of pheromones, plant-derived volatiles and the detoxification of xenobiotics. Pestic Biochem Physiol. 2021; 171:104726. doi: 10.1016/j.pestbp.2020.104726 [DOI] [PubMed] [Google Scholar]
- 16.Gadenne C, Barrozo RB, Anton S. Plasticity in insect olfaction: to smell or not to smell? Annu Rev Entomol. 2016; 61(1):317–333. doi: 10.1146/annurev-ento-010715-023523 [DOI] [PubMed] [Google Scholar]
- 17.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013; 58(1):373–391. doi: 10.1146/annurev-ento-120811-153635 [DOI] [PubMed] [Google Scholar]
- 18.Zhang SF, Zhang Z, Wang HB, Kong XB. Antennal transcriptome analysis and comparison of olfactory genes in two sympatric defoliators, Dendrolimus houi and Dendrolimus kikuchii (Lepidoptera: Lasiocampidae). Insect Biochem Mol Biol. 2014; 52: 69–81. doi: 10.1016/j.ibmb.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 19.Jiang XC, Liu S, Jiang XY, Wang ZW, Xiao JJ, Gao Q, et al. Identification of olfactory genes from the greater wax moth by antennal transcriptome analysis. Front Physiol. 2021; 12:663040. doi: 10.3389/fphys.2021.663040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong JF, Wang K, Sun YL, Tian CH, Wang SL. Antennal transcriptome analysis of odorant-binding proteins and characterization of GOBP2 in the variegated cutworm Peridroma saucia. Front Physiol. 2023; 14:1241324. doi: 10.3389/fphys.2023.1241324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang MM, Long GJ, Guo H, Liu XZ, Wang H, Dewer Y, et al. Two carboxylesterase genes in Plutella xylostella associated with sex pheromones and plant volatiles degradation. Pest Manag Sci. 2021; 77(6):2737–2746. doi: 10.1002/ps.6302 [DOI] [PubMed] [Google Scholar]
- 22.Wei HS, Tan SQ, Li Z, Li JC, Moural TW, Zhu F, et al. Odorant degrading carboxylesterases modulate foraging and mating behaviors of Grapholita molesta. Chemosphere. 2021; 270:128647. doi: 10.1016/j.chemosphere.2020.128647 [DOI] [PubMed] [Google Scholar]
- 23.He P, Zhang YN, Li ZQ, Yang K, Zhu JY, Liu SJ, et al. An antennae-enriched carboxylesterase from Spodoptera exigua displays degradation activity in both plant volatiles and female sex pheromones. Insect Mol Biol. 2014; 23(4):475–486. doi: 10.1111/imb.12095 [DOI] [PubMed] [Google Scholar]
- 24.He P, Mang DZ, Wang H, Wang MM, Ma YF, Wang J, et al. Molecular characterization and functional analysis of a novel candidate of cuticle carboxylesterase in Spodoptera exigua degradating sex pheromones and plant volatile esters. Pestic Biochem Physiol. 2020; 163:227–234. doi: 10.1016/j.pestbp.2019.11.022 [DOI] [PubMed] [Google Scholar]
- 25.Ma YF, Gong LL, Zhang MQ, Liu XZ, Gao H, Hull JJ, et al. Two antenna-enriched carboxylesterases mediate olfactory responses and degradation of ester volatiles in the German cockroach Blattella germanica. J Agric Food Chem. 2023; 71(12):4789–4801. doi: 10.1021/acs.jafc.2c08488 [DOI] [PubMed] [Google Scholar]
- 26.Pelletier J, Bozzolan F, Solvar M, François MC, Jacquin-Joly E, Maïbèche-Coisne M. Identification of candidate aldehyde oxidases from the silkworm Bombyx mori potentially involved in antennal pheromone degradation. Gene. 2007; 404(1–2):31–40. doi: 10.1016/j.gene.2007.08.022 [DOI] [PubMed] [Google Scholar]
- 27.Liu YB, Tabashnik BE. Visual determination of sex of diamondback moth larvae. Can Entomol. 1997; 129(3):585–586. 10.4039/Ent129585-3 [DOI] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10(3):R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Colin ND. RSEM: accurate transcript quantification from RNA Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts andisoform switching during cell differentiation. Nat Biotechnol. 2010; 28(5):511–515. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995; 57(1):289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 33.Greenacre M, Groenen PJF, Hastie T, d’Enza AI, Markos A, Tuzhilina E. Principal component analysis. Nat Rev Method Prime. 2022; 2(1): 100. 10.1038/s43586-022-00184-w [DOI] [Google Scholar]
- 34.Liu MY, Ma ZT, Zheng TR, Sun WJ, Zhang YJ, Jin WQ, et al. Insights into the correlation between Physiological changes in and seed development of tartary buckwheat (Fagopyrum tataricum Gaertn.). BMC Genomics. 2018; 19(1):648. doi: 10.1186/s12864-018-5036-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. Bioconductor; 2010. 10.18129/B9.bioc.topGO [DOI] [Google Scholar]
- 36.Mao XZ, Cai T, Olyarchuk JG, Wei LP. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005; 21(19):3787–3793. doi: 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 37.Zheng YQ, Zhang XY, Liu X, Qin NN, Xu KF, Zeng RS, et al. Nitrogen supply alters rice defenses against the striped stem borer Chilo suppressalis. Front Plant Sci. 2021; 12:691292. doi: 10.3389/fpls.2021.691292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001; 25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 39.Gao SS, Zhang KP, Wei LT, Wei GY, Xiong WF, Lu YY, et al. Insecticidal activity of Artemisia vulgaris essential oil and transcriptome analysis of Tribolium castaneum in response to oil exposure. Front Genet. 2020; 11:589. doi: 10.3389/fgene.2020.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raina AK, Kingan TG, Mattoo AK. Chemical signals from host plant and sexual behavior in a moth. Science. 1992; 255(5044):592–594. doi: 10.1126/science.255.5044.592 [DOI] [PubMed] [Google Scholar]
- 41.de Souza LP, Faroni LRD, Lopes LM, de Sousa AH, Prates LHF. Toxicity and sublethal effects of allyl isothiocyanate to Sitophilus zeamais on population development and walking behavior. J Pest Sci. 2018; 91(2):761–770. 10.1007/s10340-017-0950-0 [DOI] [Google Scholar]
- 42.Sun Y, Jiang YJ, Wu H, Xu N, Ma ZQ, Zhang C. Function of four mitochondrial genes in fumigation lethal mechanisms of allyl isothiocyanate against Sitophilus zeamais adults. Pestic Biochem Physiol. 2021; 179:104947. doi: 10.1016/j.pestbp.2021.104947 [DOI] [PubMed] [Google Scholar]
- 43.Leal JFM, Barbancho M. Acetaldehyde detoxification mechanisms in Drosophila melanogaster adults involving aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH) enzymes. Insect Biochem Mol Biol. 1992; 22(8):885–892. 10.1016/0965-1748(92)90115-U [DOI] [Google Scholar]
- 44.Fry JD, Saweikis M. Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet Res. 2006; 87(2):87–92. doi: 10.1017/S0016672306008032 [DOI] [PubMed] [Google Scholar]
- 45.Rivera-Perez C, Nouzova M, Clifton ME, Garcia EM, LeBlanc E, Noriega FG. Aldehyde dehydrogenase 3 converts farnesal into farnesoic acid in the corpora allata of mosquitoes. Insect Biochem Mol Biol. 2013; 43(8):675–682. doi: 10.1016/j.ibmb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao QJ, Koski TM, Li HP, Zhang C, Sun JH. The effect of inactivation of aldehyde dehydrogenase on pheromone production by a gut bacterium of an invasive bark beetle, Dendroctonus valens. Insect Sci. 2023; 30(2):459–472. doi: 10.1111/1744-7917.13101 [DOI] [PubMed] [Google Scholar]
- 47.Snyder MJ, Glendinning JI. Causal connection between detoxification enzyme activity and consumption of a toxic plant compound. J Comp Physiol A. 1996; 179(2):255–261. doi: 10.1007/BF00222792 [DOI] [PubMed] [Google Scholar]
- 48.Brattsten LB, Wilkinson CF, Eisner T. Herbivore-plant interactions: mixed-function oxidases and secondary plant substances. Science. 1977; 196(4296):1349–1352. doi: 10.1126/science.196.4296.1349 [DOI] [PubMed] [Google Scholar]
- 49.Lu K, Song YY, Zeng RS. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr Opin Insect Sci. 2021; 43:103–107. doi: 10.1016/j.cois.2020.11.004 [DOI] [PubMed] [Google Scholar]
- 50.Wang HD, Shi Y, Wang L, Liu S, Wu SW, Yang YH, et al. CYP6AE gene cluster knockout in Helicoverpa armigera reveals role in detoxification of phytochemicals and insecticides. Nat Commun. 2018; 9(1):4820. doi: 10.1038/s41467-018-07226-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nauen R, Bass C, Feyereisen R, Vontas J. The role of cytochrome P450s in insect toxicology and resistance. Annu Rev Entomol. 2022; 67(1):105–124. doi: 10.1146/annurev-ento-070621-061328 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptomic (A) and qRT-PCR (B) expression levels of five heat shock protein genes were compared between control (MA) and AITC exposed (MA-AITC). Values are the mean ± SE (n = 3). Statistical significance was determined by Student’s t-test: ** P < 0.01, or *** P < 0.001.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The article contains data along with Supplementary Materials. The RNA-seq raw data has been uploaded to the NCBI Short Read Archive (SRA) and is accessible under the accession number PRJNA1106633.