Abstract
Background and Objectives
Myotonic dystrophy type 2 (DM2) is a multisystemic repeat disorder caused by the expansion of an unstable CCTG tetranucleotide repeat in the noncoding region of the CNBP gene. Standard diagnostic is based on Southern blot analysis or a unidirectional RP-PCR that amplifies the repeat from the downstream end.
Methods
Our study reevaluated 80 patients (cohort 1) with clinical suspicion of DM2 but homozygous negative results using the standard diagnostic repeat-primed PCR (RP-PCR). Reanalysis was performed using a second RP-PCR that amplifies the repeat from the opposite direction. Individual samples were further analyzed by Oxford Nanopore Technology long-read sequencing, Sanger sequencing, and another RP-PCR. In addition, repeat expansions were further characterized in 168 patients with confirmed DM2 (cohort 2).
Results
We identified 5 of the 80 patients (cohort 1) with expanded repeats in CNBP and, as such, reclassified them as positive for DM2. The initial false-negative results were attributed to variants within the primer binding site of the standard RP-PCR in one patient and an additional novel (TCTG)n repeat downstream to the known (CCTG)n repeat in 4 other patients. By analyzing a cohort of 168 patients with confirmed DM2 (cohort 2), we found that the additional (TCTG)n repeat is present in at least 84% of patients.
Discussion
Our study revealed the presence of an additional repeat (TCTG)n in most of the patients living with DM2. Large expansions of this repeat likely hinder sufficient amplification of the disease causing (CCTG)n repeat. Because the (TCTG)n repeat is likely mosaic in length, (CCTG)n repeat expansions are correctly detected in most patients. However, a few patients are at risk of a false-negative result using the standard RP-PCR, which had a false-negative rate of 0.7% (5/674) and a sensitivity of 97.3% in the cohort studied. Based on our findings, we propose (TG)v(TCTG)w(CCTG)n(TCTG’)m as the updated model for the structure of CNBP repeat expansions and recommend adapting the diagnostic guidelines accordingly. The effect of the (TCTG)n repeat on the phenotype remains to be determined but could be key for establishing a phenotype-genotype correlation for DM2 that remained elusive so far.
Introduction
Myotonic dystrophy type 2 (DM2, OMIM 602668) is a slowly progressive multisystemic disorder characterized by predominantly proximal muscle weakness, muscle stiffness, nociceptive pain, and myotonia.1 Common symptoms in patients with DM2 also include cardiac arrhythmias, diabetes, and cataracts.2-4 The clinical description of DM2 originated in 1994 with identification of patients who exhibited symptoms of myotonic dystrophy but lacked a CTG trinucleotide repeat expansion in the DMPK gene, which is known to be causative for DM1.5,6 The CCTG tetranucleotide repeat responsible for DM2 was identified in 2001.7 This repeat is situated within a complex repeat array in intron 1 of the CNBP gene (cellular nucleic acid–binding protein, previously known as zinc finger protein 9, ZNF9) on chromosome 3q21.3. Within the healthy population, the CCTG repeat is typically interrupted by at least 3 tetraplets, contributing to its stability against expansion.7-9
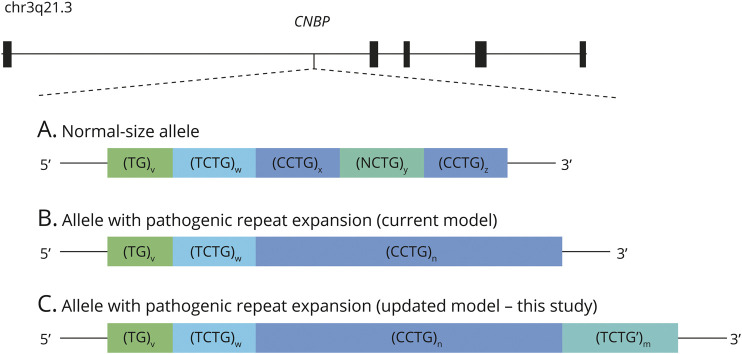
In 2018 normal-size CNBP repeat arrays were characterized in a large cohort and established the current model of their structure as (TG)v(TCTG)w(CCTG)x(NCTG)y(CCTG)z (Figure 1A).10 Pathogenic CCTG repeat expansions, commonly denoted as (CCTG)n, lack interruptions by (NCTG)y repeats and exhibit complete penetrance when exceeding the size of 75 repeat units (Figure 1B).1 Owing to their large size, extreme mosaicism, and high GC content, these pathogenic repeat expansions have a complex structure that is challenging to capture fully. As such, specialized workflows are required for the diagnosis of DM2.
Figure 1. Location and Architecture of the Intronic CNBP Repeat Array Causative for DM2.
(A) Normal-size alleles are usually interrupted by one or more tetranucleotides (NCTG, N = G, T) and comprise less than 26 CCTG repeat units. (B) Pathogenic repeat expansions are typically uninterrupted and carry 75 or more CCTG repeat units. (C) Proposed updated model of the architecture of CNBP repeat expansions.
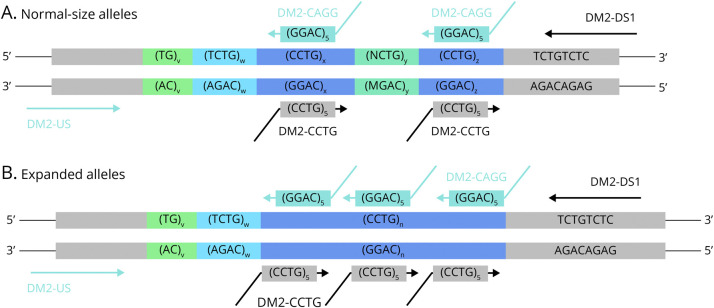
Most laboratories conducting molecular diagnostic analyses for individuals suspected of having DM2 follow the “Best practice guidelines and recommendations on the molecular diagnosis of myotonic dystrophy types 1 and 2.”11 Per these guidelines, the first step involves using a conventional PCR (short-range PCR) combined with fragment-length analysis to determine whether an individual possesses 2 normal-size alleles. If only one allele is detected by short-range PCR, a specific unidirectional repeat-primed PCR (“CCTG-RP-PCR”) based on the protocol of Catalli et al.,12 or Southern blotting, is used to detect possible repeat expansions (Figure 2, eFigure 1). Owing to the laborious and time-consuming nature of Southern blotting, diagnostic laboratories often tend to prefer the alternative RP-PCR method, underscoring the need for further validation of this method. The current unidirectional CCTG-RP-PCR uses a primer combination designed to bind to the (CCTG)n repeat and a region located 4 nucleotides downstream from the repeat array (Figure 2, eTable 1).
Figure 2. Repeat-Primed PCR Methods for the Analysis of the CNBP Repeat Array.
Different repeat-primed PCR methods and their primers used to analyze the CNBP repeat array exemplified for (A) normal-size alleles and (B) repeat expansions. Primers of the standard CCTG-RP-PCR amplifying the repeat from the downstream end are given in black. Primers of the CAGG-RP-PCR amplifying the repeat from the upstream end are given in light blue.
Prompted by a patient who received a false-negative result, we reevaluated 80 patients with homozygous negative results within a cohort of 857 individuals previously tested for DM2 using a bidirectional RP-PCR analysis. Using this approach, we identified 5 patients who initially received false-negative results. Further examination of CNBP repeat expansions in these patients, along with a larger cohort of 168 patients with confirmed DM2, revealed the presence of an additional (TCTG)n repeat in most of the patients with DM2. Based on these results, we propose an updated model for the structure of CNBP repeat expansions and recommend revising the current diagnostic guidelines for DM2.
Methods
Patient DNA Samples and Study Approval
Cohort 1
Within a cohort of 857 patients (eFigure 2) previously diagnosed using the conventional approach, including standard CCTG-RP-PCR, 183 had a positive result, 594 had a negative result showing 2 differently sized normal alleles (heterozygous samples), and 80 had a negative result showing only one repeat size (homozygous samples). Patients were tested for DM2 either because of having symptoms consistent with the disease on clinical or neurologic examination or because a family member had a genetic diagnosis of DM2. False-negative results were assumed to result from the missing amplification of a repeat expansion by the unidirectional CCTG-RP-PCR, leading to an apparent homozygous negative result. As such, these 80 samples were reanalyzed and assembled for cohort 1 (eTable 2).
Cohort 2
Cohort 2 contained 168 patients with a confirmed repeat expansion in CNBP. Of these patients, 53 were diagnosed using the unidirectional RP-PCR and 115 by Southern blot analysis.
Standard Protocol Approvals, Registrations, and Patient Consents
All patients were analyzed as diagnostic samples within our institute. Informed consent was obtained from all patients, and the study was approved by local institutions (Bayerische Landesärztekammer, 2019-210). All genetic analyses and investigations were performed in accordance with the guidelines of the Declaration of Helsinki.
Extraction of gDNA
Genomic DNA (gDNA) was obtained from total peripheral EDTA blood samples by extraction of white blood cells with a Biomek FX system (Beckman Coulter) using the NucleoMag Blood 3 mL Kit (Machery-Nagel, #REF 744502.1) or FlexiGene DNA Kit (Qiagen, ID 51206) as per manufacturer's instructions. All DNA samples showed high purity as determined by optical density measurements (A260/A280 > 1.9 and A260/A230 > 2.0).
RP-PCR and Fragment-Length Analysis
The sequence of all primers used in the following is given in eTable 1. Patients were diagnosed as positive for myotonic dystrophy type 2 when a (CCTG)n repeat expansion larger than 75 repeat units is detected.
Short-Range PCR
Short-range PCR was performed according to reference 11.
Standard RP-PCR for Amplifying (CCTG)n Repeat Expansions From the Downstream End (Standard CCTG-RP-PCR)
Following the best practice guidelines, 250 ng of genomic DNA was amplified in a reaction volume of 30 µL, using 0.5x GoTaq Reaction B buffer white (Promega), 0.42 mM MgCl2, 333 µM dNTPs incl. 7-deAZA-GTP (Invitrogen), 0.75 units of GoTaq DNA Polymerase (Promega, Part# 9PIM300), 0.33 µM DM2-DS1 flanking primer, 0.07 µM DM2-CCTG repeat primer, and 0.33 µM anchor primer. The following cycling conditions were applied: initial denaturation at 94°C for 5 minutes followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 2 minutes, with a final extension at 72°C for 10 minutes.
RP-PCR for Amplifying (CCTG)n Repeat Expansions From the Upstream End (CAGG-RP-PCR)
Sixty ng of genomic DNA was amplified in a reaction volume of 10 µL, using 1x HOT FIREPol DNA Buffer B2 (Solis BioDyne), 2 mM MgCl2, 3x GC-rich enhancer solution (Solis BioDyne), 400 µM dNTPs, 1 unit of HOT FIREPol DNA Polymerase (Solis BioDyne), 0.2 µM DM2-US flanking primer, 0.1 µM DM2-CAGG repeat primer, and 0.2 µM anchor primer. The following cycling conditions were applied: initial activation of Hot Fire Polymerase at 95°C for 15 minutes followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 57°C for 1 minute, and extension at 72°C for 5 minutes, with a final extension at 72°C for 15 minutes.
Optimized RP-PCR for Amplifying (CCTG)n Repeat Expansions From the Downstream End (Optimized CCTG-RP-PCR)
Optimized CCTG-RP-PCR was performed as described for the CAGG-RP-PCR by replacing the flanking primer DM2-US by DM2-DS1 and repeat primer DM2-CAGG by DM2-CCTG.
RP-PCR to Detect a (TCTG)n Repeat Expansion Downstream to the (CCTG)n Repeat Expansion (TCTG-RP-PCR)
TCTG-RP-PCR was performed as described for the CAGG-RP-PCR by replacing the repeat flanking primer DM2-US by DM2-DS1 and primer DM2-CAGG by DM2-TCTG; the annealing temperature was adjusted to 50°C.
Fragment Analysis
After dilution with HiDi Formamide (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA) and the addition of GeneScan 500 LIZ dye Size Standard (Applied Biosystems), capillary electrophoresis was performed on ABI Prism Genetic Analyzer 3730 (Applied Biosystems) with an injection time of 5 seconds. The results were assessed using GeneMarker software v2.4.0 (SoftGenetics, State College, PA).
Sanger Sequencing
PCR for Normal-Size Alleles
One hundred ng of genomic DNA was amplified in a reaction volume of 20 µL using 1x HOT FIREPol DNA Buffer B2 (Solis BioDyne), 2 mM MgCl2, 1x GC-rich enhancer solution (Solis BioDyne), 300 µM dNTPs, 1.5 units of HOT FIREPol DNA Polymerase (Solis BioDyne), 0.25 µM DM2-DS2 flanking primer, and 0.25 µM DM2-US flanking primer. The following cycling conditions were applied: initial activation of Hot Fire Polymerase at 95°C for 15 minutes followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 57°C for 30 seconds, and extension at 72°C for 45 seconds, with a final extension at 72°C for 15 minutes.
PCR for Expanded Alleles
One hundred and twenty ng of genomic DNA was amplified in a reaction volume of 20 µL, using 1x HOT FIREPol DNA Buffer B2 (Solis BioDyne), 2.5 mM MgCl2, 3x GC-rich enhancer solution (Solis BioDyne), 400 µM dNTPs, 1.5 units of HOT FIREPol DNA Polymerase (Solis BioDyne), 0.25 µM DM2-DS2 flanking primer, and 0.3 µM DM2-CCTGa repeat primer or DM2-TCTGa repeat primer, respectively. The following cycling conditions were applied: initial activation of Hot Fire Polymerase at 95°C for 15 minutes followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 57°C for 1 minute for DM2-CCTGa or 50°C for 1 minute for DM2-TCTGa repeat primer, respectively, and extension at 72°C for 5 minutes, with a final extension at 72°C for 15 minutes.
Sanger Sequencing Step
Each cleaned-up PCR product (by Exo-SAP-IT, Applied Biosystems) was used for Sanger sequencing in a 10-µL mix of BigDye Terminator Sequencing Kit (Applied Biosystems). For normal-size alleles, the following reagents were used: 1x Q-Solution (Qiagen), 1x sequencing buffer, 0.3 µM DM2-DS3 sequencing primer, and 200 µM BigDye v1.1. For expanded alleles, the following reagents were used: 1x Q-Solution, 0.5 µM DM2-DS3, and 400 µM BigDye v3.1.
The following conditions were used for sequencing: pre-denaturation for 2 minutes at 96°C followed by 26 cycles of denaturation at 96°C for 30 seconds, 4 minutes of extension at 60°C for normal alleles or at 68°C for expanded alleles, and final extension steps for 10 minutes.
Products were cleaned up with Sephadex G-50 Superfine (Sigma Aldrich, Merck, St. Louis, MO), diluted with 30 µL of nuclease-free water (Qiagen), and sequenced on ABI Prism Genetic Analyzer 3730. Data were assessed with Mutation Surveyor software v3.10 (SoftGenetics).
Oxford Nanopore Technology Long-Read Sequencing
One patient (patient 33, cohort 1) was analyzed by Oxford Nanopore Technology (ONT) long-read sequencing to identify possible alterations within the CNBP repeat and adjunct region explaining the false-negative result. Library preparation and flow cell loading were performed according to the ONT Cas9-targeted sequencing protocol using ONT's SQK-CS9109 kit and 7 µg of input gDNA. CRISPR RNAs (crRNAs) to enrich the CNBP repeat array were designed using CHOPCHOP 8 (eTable 1). Sequencing was performed with ONT FLO-MIN106D R9 flow cells on the GridION X5 sequencer for 48 hours. Base calling from electrical data was performed using Guppy (v5.0.16).13 The generated FASTQ files were aligned to the human reference genome (GRCh38/hg38) using Minimap2 (v2.17).14 For quality control of the aligned reads, we used NanoPlot (v1.29.1).15 The CNBP locus was analyzed manually by visual inspection of all reads mapped to that region in the Integrative Genome Viewer (IGV).
Data Availability
Anonymized data not published within this article will be made available by request from any qualified investigator.
Results
Reevaluation of Patients With Homozygous Negative Results for DM2
The reevaluation of patients who tested negative for DM2 based on standard unidirectional CCTG-RP-PCR was prompted by a patient with an apparent DM2 phenotype and positive family history (patient 1) but missed by this analysis. The short-range PCR showed only one normal-size allele in this patient. Despite testing in 3 diagnostic laboratories, the unidirectional standard CCTG-RP-PCR did not detect a repeat expansion, resulting in an initial negative DM2 diagnosis. However, owing to strong clinical suspicion, Southern blotting was further performed, which detected a CNBP repeat expansion, thus confirming DM2 in this patient. This led us to suspect a failure of the standard CCTG-RP-PCR as the cause of the false-negative result. Reanalysis of this patient using a second RP-PCR (CAGG-RP-PCR, Figure 2, eFigure 3), which amplifies the (CCTG)n repeat from the opposite direction using a reverse CAGG repeat primer, detected a large (CCTG)n repeat expansion (>75 repeat units), in line with the Southern blotting results.
Based on this case, we reanalyzed 79 additional patients (in total, 80 patients (cohort 1), results are provided in eTable 2) with a negative result and who appeared homozygous for one normal-size allele. In 4 patients (4, 24, 33, 56), the CNBP repeat expansion was detected using the alternative RP-PCR, reclassifying these patients as DM2 positive. All 5 patients with initial false-negative result showed symptoms compatible with DM2 and could clinically not be differentiated from the other symptomatic patients with homozygous negative result.
Among the 674 patients diagnosed as negative for DM2 in this cohort and 188 true-positive patients with DM2, the standard method recommended in the current guidelines shows a false-negative rate of 0.7% (5/674) and a sensitivity of 97.3% (183/188). Extensive optimization of all parameters of the standard CCTG-RP-PCR (optimized CCTG-RP-PCR), as performed for establishing the CAGG-RP-PCR protocol, resulted in insufficient amplification of the (CCTG)n repeat in all 5 initially false-negative patients. Although a resulting stuttering pattern with very low intensity was observed, the electropherogram interpretation remained susceptible to false-negative results.
Characterization of the Repeat Expansions in Patients With False-Negative Results in the Unidirectional RP-PCR
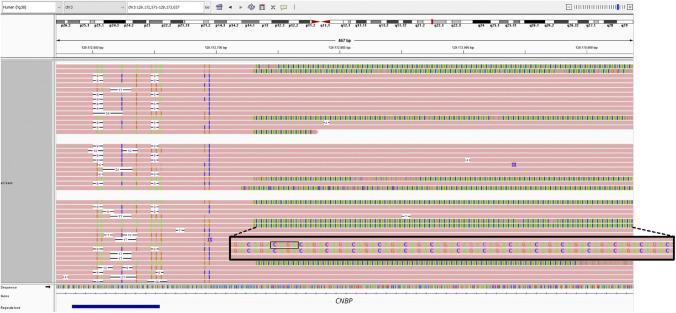
We hypothesized that the allelic dropout in these 5 patients might be due to variants within the primer binding site or the CNBP repeat array itself. To investigate this further, we performed Oxford Nanopore long-read sequencing on one sample (patient 33) with sufficient DNA quality. This analysis revealed an additional (TCTG)n repeat downstream of the known (CCTG)n repeat (Figure 3). Although the exact length of both repeats could not be determined because of the borderline DNA fragment lengths of the input DNA, we confirmed the presence of this additional (TCTG)n repeat in 4 of the 5 false-negative samples (patients 1, 24, 33, 56) using Sanger sequencing of the downstream end of the repeat array (eFigure 4). The fifth patient (patient 4) showed a complex structure of an additional downstream repeat corresponding to the sequence (CCTG-TCTG)n. Sanger sequencing further revealed a deletion-insertion of ‘GAGA’ to ‘CAGG’ in the binding site of the primer (eFigure 4).
Figure 3. Long-Read Sequencing of the Downstream End of the CNBP Repeat Array.
Integrative Genome Viewer (IGV) screenshot of reads mapped to the downstream end of the expanded and nonexpanded CNBP repeat array of patient 33 after ONT long-read sequencing presented as a reverse complement with a subset of the reads carrying the additional downstream (TCTG)n repeat motif.
Because Sanger sequencing is only capable of capturing a limited number of repeat units, we implemented an additional RP-PCR (TCTG-RP-PCR, eFigure 5) to estimate the size of the additional repeat element (results are provided in eTable 2). This analysis showed an expansion (>75 repeat units) of the additional (TCTG)n repeat in all 4 patients carrying this additional repeat element. The fifth sample showed only a weak stutter pattern, likely due to the complex (CCTG-TCTG)n repeat pattern of the additional downstream repeat and the variations within the primer binding site. Based on these findings, we propose that the false-negative result in patients 1, 24, 33, and 56 primarily results from a large expansion of the additional (TCTG)n repeat between the CCTG repeat primer and the downstream flanking primer DM2-DS1, which hinders sufficient amplification. In patient 4, variations within the primer binding site likely cause the false-negative result.
Characterization of the Repeat Expansions in a Larger Cohort of Patients With DM2
To investigate whether the additional (TCTG)n repeat is a rare variation of a few repeat expansions that are predisposed to have a false-negative result in the standard CCTG-RP-PCR or whether it is a more general component of CNBP repeat expansions, we analyzed an additional cohort of 168 patients with confirmed DM2 (cohort 2) using the TCTG-RP-PCR method. We identified clear signals for downstream (TCTG)n repeat expansions (>75 units) in 83% of the samples (139/168). Two samples (1%, 2/168) showed short (TCTG)n expansions, quantified at 19 and 23 repeat units, respectively. In 10% of samples (16/168), a low-intensity repeat stutter pattern indicated the potential presence of an additional (TCTG)n repeat, although a more complex repeat pattern cannot be excluded. Alternatively, a large expansion of the pure (TCTG)n repeat or variants in the primer binding site of primer DM2-DS1 could account for the reduced signal. In 7% of samples (11/168), no (TCTG)n repeat was detected. Clinically, no obvious difference between the groups of patients with different results in the TCTG-RP-PCR could be identified.
Discussion
Molecular testing for DM2 typically involves a two-step workflow: initial short-range PCR and, if only one allele is detected, additional RP-PCR based on the protocol of Catalli et al., followed by fragment-length analyses.11,12 While this approach resolves most cases, a small proportion of DM2-positive samples are missed because of low-to-barely visible signals in the standard unidirectional RP-PCR–based fragment analyses, resulting from an insufficient amplification of the expanded (CCTG)n repeat.
We introduced a second RP-PCR analysis (CAGG-RP-PCR) that amplifies the repeat from the downstream end to identify potential false-negative cases within our previously analyzed cohort. This approach identified 5 false-negative patients with DM2, corresponding to a sensitivity of 97.3% for the recommended standard CCTG-RP-PCR in our cohort. This is considerably lower than the 100% reported by Catalli et al. and carries a risk of missing patients, especially when PCR methods are not carefully optimized.11,12 Several RP-PCR methods for analyzing the CNBP repeat from the upstream end, as in our CAGG-RP-PCR, have been reported.2,9,16,17 Amplification of the repeat in this direction includes the polymorphic (TG)n(TCTG)n part of the repeat array from the normal-size and expanded alleles. Owing to the variable sizes of the (TG)v(TCTG)w regions, the resulting amplicons start at different positions in the electropherogram, leading to a complex repeat pattern that complicates interpretation.16 Consequently, this method has not been widely adopted.
Based on the results of our study and known discrepancies between Southern blotting and standard CCTG-RP-PCR, we advocate for performing the molecular diagnosis of DM2 exclusively using a bidirectional RP-PCR, combining both the CCTG-RP-PCR and CAGG-RP-PCR with optimized PCR conditions as reported in this study. Laboratories using the standard unidirectional CCTG-RP-PCR can easily adopt their procedures because no additional equipment is required and both RP-PCRs can be performed in parallel for time efficiency.
In 4 of the 5 initial false-negative samples, we identified an additional (TCTG)n repeat expansion downstream of the known (CCTG)n repeat. We hypothesized its large expansion to be the cause of the false-negative result. In the fifth sample, a variant within the primer binding site and a complex additional downstream (CCTG-TCTG)n repeat could be identified as likely causative for the false-negative result. The analysis of a larger cohort of 168 patients with confirmed DM2 (cohort 2) showed that the additional downstream (TCTG)n repeat is a general feature of CNBP repeat expansions, present in at least 84% of the patients. Based on these findings, we propose an updated model of the complex DM2 repeat locus, including the (TCTG)n downstream repeat: (TG)v(TCTG)w(CCTG)n(TCTG’)m (Figure 1C). Because RP-PCR is limited to detecting the presence of an expanded allele that carries a specific motive, the size and variability of (TCTG)n repeat expansions remain to be determined. An evident absence of the additional (TCTG)n repeat was observed in only 7% of patients while 10% had a TCTG-RP-PCR result that was difficult to interpret because of the low intensity of repeat stuttering. This low signal intensity could result from variations in the (TCTG)n downstream repeat motif, as in patient 4, or its presence in only a small proportion of the cells.
Our finding of an additional downstream (TCTG)n repeat aligns with recent independent findings of Alfano et al.,18 who analyzed 9 patients with confirmed DM2 using CRISPR/Cas9-targeted ONT long-read sequencing and detected the downstream (TCTG)n repeat in 7 of 9 patients. While their study was limited to only a few patients, primarily from one family, we could confirm the presence of this additional repeat element in a large cohort of unrelated patients with DM2. In contrast to the (CCTG)n repeat, Alfano et al. did not observe the (TCTG)n repeat in all reads, with proportions varying widely (11–86%). Whether this variation is an artifact resulting from CRISPR/Cas9 enrichment, a sequencing bias, or whether it reflects the naturally occurring heterogeneity of CNBP repeat expansions remains to be determined. Furthermore, it is not yet possible to distinguish whether the (TCTG)n repeat is present in the germline or merely arises somatically during life, as would be conceivable given the pronounced instability of the CNBP repeat array.
Owing to the high complexity of the CNBP repeat expansions, extensive variability in size, and somatic instability, no significant phenotype-genotype correlation has been established.2,7,19,20 Such analysis is further complicated by technical limitations in determining the repeat motif and size of repeat expansions. Despite these challenges, long-read sequencing such as ONT or Pacific Biosciences offers novel opportunities to overcome these limitations and has been successfully implemented for other repeat disorders.21,22 In addition to deciphering the detailed architecture of CNBP repeat expansions, the degree of somatic mosaicism and the instability dynamics can be accessed using long-read sequencing. Understanding these parameters may lead to a more precise molecular basis of DM2. Based on a detailed phenotyping of patients with DM2, these studies should also elucidate the biological effect of (TCTG)n repeat expansions on the clinical phenotype because their previously unknown presence might contribute to the lack of a phenotype-genotype correlation.
Our study revealed a significant gap for DM2 under current diagnostic recommendations. This limitation can be overcome by implementing a bidirectional RP-PCR together with optimizing PCR conditions as demonstrated in our study. In addition, we propose an updated CNBP repeat expansion architecture model and reveal that most patients with DM2 carry an additional downstream (TCTG)n repeat. A pronounced (TCTG)n expansion may cause false-negative results in the standard unidirectional CCTG-RP-PCR. To fully understand the full complexity of this repeat locus, further characterization using long-read sequencing methods is necessary. Comprehensive genotyping could establish a crucial phenotype-genotype relationship needed for genetic counseling, drug development, and clinical trials in patients living with DM2.
Glossary
- DM1
myotonic dystrophy type 1
- DM2
myotonic dystrophy type 2
- gDNA
genomic DNA
- IGV
Integrative Genome Viewer
- ONT
Oxford Nanopore Technology
- RP-PCR
repeat-primed PCR
Author Contributions
M. Wendlandt: drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; study concept or design; analysis or interpretation of data. H. Erdmann: drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data. S. Rost: drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data. M.C. Lucas: drafting/revision of the manuscript for content, including medical writing for content. K. Becker: drafting/revision of the manuscript for content, including medical writing for content. S. Kleinle: drafting/revision of the manuscript for content, including medical writing for content. M. Timmer: drafting/revision of the manuscript for content, including medical writing for content. A. Bier: drafting/revision of the manuscript for content, including medical writing for content. G. Wunderlich: drafting/revision of the manuscript for content, including medical writing for content. S. Wenninger: drafting/revision of the manuscript for content, including medical writing for content. M.C. Walter: drafting/revision of the manuscript for content, including medical writing for content. T. Neuhann: drafting/revision of the manuscript for content, including medical writing for content. B. Schoser: drafting/revision of the manuscript for content, including medical writing for content; analysis or interpretation of data. E. Holinski-Feder: drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data. A. Abicht: drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data.
Study Funding
Financial support for this study was given to H. Erdmann by the Deutsche Gesellschaft für Muskelkranke e.V.
Disclosure
The authors report no relevant disclosures. Go to Neurology.org/NG for full disclosures.
References
- 1.Schoser B. Myotonic Dystrophy Type 2. GeneReviews®; 2006. Accessed January 29, 2024. ncbi.nlm.nih.gov/books/NBK1466/ [Google Scholar]
- 2.Day JW, Ricker K, Jacobsen JF, et al. Myotonic dystrophy type 2: molecular, diagnostic and clinical spectrum. Neurology. 2003;60(4):657-664. doi: 10.1212/01.wnl.0000054481.84978.f9 [DOI] [PubMed] [Google Scholar]
- 3.Ricker K, Koch MC, Lehmann-Horn F, et al. Proximal myotonic myopathy. Clinical features of a multisystem disorder similar to myotonic dystrophy. Arch Neurol. 1995;52(1):25-31. doi: 10.1001/archneur.1995.00540250029009 [DOI] [PubMed] [Google Scholar]
- 4.Ricker K. Myotonic dystrophy and proximal myotonic myophathy. J Neurol. 1999;246(5):334-338. doi: 10.1007/s004150050359 [DOI] [PubMed] [Google Scholar]
- 5.Ricker K, Koch MC, Lehmann-Horn F, et al. Proximal myotonic myopathy: a new dominant disorder with myotonia, muscle weakness, and cataracts. Neurology. 1994;44(8):1448-1452. doi: 10.1212/wnl.44.8.1448 [DOI] [PubMed] [Google Scholar]
- 6.Thornton CA, Griggs RC, Moxley RT. Myotonic dystrophy with no trinucleotide repeat expansion. Ann Neurol. 1994;35(3):269-272. doi: 10.1002/ana.410350305 [DOI] [PubMed] [Google Scholar]
- 7.Liquori CL, Ricker K, Moseley ML, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293(5531):864-867. doi: 10.1126/science.1062125 [DOI] [PubMed] [Google Scholar]
- 8.Radvanszky J, Surovy M, Polak E, Kadasi L. Uninterrupted CCTG tracts in the myotonic dystrophy type 2 associated locus. Neuromuscul Disord. 2013;23(7):591-598. doi: 10.1016/j.nmd.2013.02.013 [DOI] [PubMed] [Google Scholar]
- 9.Liquori CL, Ikeda Y, Weatherspoon M, et al. Myotonic dystrophy type 2: human founder haplotype and evolutionary conservation of the repeat tract. Am J Hum Genet. 2003;73(4):849-862. doi: 10.1086/378720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahyera AS, Schneider T, Halliger-Keller B, et al. Distribution and structure of DM2 repeat tract alleles in the German population. Front Neurol. 2018;9:463. doi: 10.3389/fneur.2018.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamsteeg EJ, Kress W, Catalli C, et al. Best practice guidelines and recommendations on the molecular diagnosis of myotonic dystrophy types 1 and 2. Eur J Hum Genet. 2012;20(12):1203-1208. doi: 10.1038/ejhg.2012.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catalli C, Morgante A, Iraci R, Rinaldi F, Botta A, Novelli G. Validation of sensitivity and specificity of tetraplet-primed PCR (TP-PCR) in the molecular diagnosis of myotonic dystrophy type 2 (DM2). J Mol Diagn. 2010;12(5):601-606. doi: 10.2353/jmoldx.2010.090239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oxford Nanopore Technologies. Guppy Protocol: Modified Base Calling. Accessed February 2, 2022. community.nanoporetech.com/protocols/Guppy-protocol/v/gpb_2003_v1_revz_14dec2018/modified-base-calling [Google Scholar]
- 14.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094-3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34(15):2666-2669. doi: 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radvansky J, Ficek A, Kadasi L. Repeat-primed polymerase chain reaction in myotonic dystrophy type 2 testing. Genet Test Mol Biomarkers. 2011;15(3):133-136. doi: 10.1089/gtmb.2010.0127 [DOI] [PubMed] [Google Scholar]
- 17.Bachinski LL, Udd B, Meola G, et al. Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different European origins: a single shared haplotype indicates an ancestral founder effect. Am J Hum Genet. 2003;73(4):835-848. doi: 10.1086/378566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alfano M, De Antoni L, Centofanti F, et al. Characterization of full-length CNBP expanded alleles in myotonic dystrophy type 2 patients by Cas9-mediated enrichment and nanopore sequencing. eLife. 2022;11:e80229. doi: 10.7554/eLife.80229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoser BGH, Kress W, Walter MC, Halliger-Keller B, Lochmüller H, Ricker K. Homozygosity for CCTG mutation in myotonic dystrophy type 2. Brain. 2004;127(Pt 8):1868-1877. doi: 10.1093/brain/awh210 [DOI] [PubMed] [Google Scholar]
- 20.Udd B, Meola G, Krahe R, et al. Report of the 115th ENMC workshop: DM2/PROMM and other myotonic dystrophies. 3rd Workshop, 14-16 February 2003, Naarden, The Netherlands. Neuromuscul Disord. 2003;13(7-8):589-596. doi: 10.1016/s0960-8966(03)00092-0 [DOI] [PubMed] [Google Scholar]
- 21.Erdmann H, Schöberl F, Giurgiu M, et al. Parallel in-depth analysis of repeat expansions in ataxia patients by long-read sequencing. Brain. 2023;146(5):1831-1843. doi: 10.1093/brain/awac377 [DOI] [PubMed] [Google Scholar]
- 22.Stevanovski I, Chintalaphani SR, Gamaarachchi H, et al. Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Sci Adv. 2022;8(9):eabm5386. doi: 10.1126/sciadv.abm5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data not published within this article will be made available by request from any qualified investigator.