Abstract
CRISPR/Cas9-based multiplex genome editing via electroporation is relatively efficient; however, lipofection is versatile because of its ease of use and low cost. Here, we aimed to determine the efficiency of lipofection in CRISPR/Cas9-based multiplex genome editing using growth hormone receptor (GHR) and glycoprotein alpha-galactosyltransferase 1 (GGTA1)-targeting guide RNAs (gRNAs) in pig zygotes. Zona pellucida-free zygotes were collected 10 h after in vitro fertilization and incubated with Cas9, gRNAs, and Lipofectamine 2000 (LP2000) for 5 h. In Experiment 1, we evaluated the mutation efficiency of gRNAs targeting either GHR or GGTA1 in zygotes transfected using LP2000 and cultured in 4-well plates. In Experiment 2, we examined the effects of the culture method on the development, mutation rate, and mutation efficiency of zygotes with simultaneously double-edited GHR and GGTA1, cultured using 4-well (group culture) and 25-well plates (individual culture). In Experiment 3, we assessed the effect of additional GHR-targeted lipofection before and after simultaneous double gRNA-targeted lipofection on the mutation efficiency of edited embryos cultured in 25-well plates. No significant differences in mutation rates were observed between the zygotes edited with either gRNA. Moreover, the formation rate of blastocysts derived from GHR and GGTA1 double-edited zygotes was significantly increased in the 25-well plate culture compared to that in the 4-well plate culture. However, mutations were only observed in GGTA1 when zygotes were transfected with both gRNAs, irrespective of the culture method used. GHR mutations were detected only in blastocysts derived from zygotes subjected to GHR-targeted lipofection before simultaneous double gRNA-targeted lipofection. Overall, our results suggest that additional lipofection before simultaneous double gRNA-targeted lipofection induces additional mutations in the zygotes.
Keywords: CRISPR/Cas9 system, Growth hormone receptor (GHR), Glycoprotein alpha-galactosyltransferase 1 (GGTA1), Lipofection, Porcine zygote
Genetically modified pigs are anatomically and physiologically similar to humans, making them important animal models for biomedical research [1, 2]. Targeted nucleases are powerful tools for high-precision gene modification in pigs [3]. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system, which consists of a guide RNA (gRNA) and a Cas9 nuclease, is widely used for gene editing in various organisms [4]. Genetically modified pigs are generally established via somatic cell nuclear transfer using genetically modified somatic cells or by direct introduction of gene editors into the cytoplasm of zygotes and embryos via microinjection or electroporation [3, 5]. However, specialized equipment is required for these operations. Owing to their flexibility and versatility, various chemical methods such as lipofection are preferred for the delivery of the CRISPR/Cas9 complex. These methods substantially improve the value of pigs as experimental animals, particularly in laboratories lacking specialized equipment.
Liposome-mediated transfection, also known as lipofection, enhances the cellular uptake of polynucleotides using lipophilic reagents, resulting in the delivery of foreign genes into several organisms, including oocytes and embryos [6, 7]. We previously optimized the lipofection system using zona pellucida (ZP)-free embryos to generate mutant embryos with different single-target genes and successfully produced genetically modified piglets with a monoallelic mutation in the myostatin gene [8]. The co-expression of multiple gRNAs in the CRISPR/Cas9 system improves genome editing efficiency by simultaneously disrupting the genomic DNA at multiple sites [9, 10]. In contrast to the establishment of rodent models [11], the establishment of pig models is an expensive and time-consuming process that requires the editing of multiple genes via the breeding of established single-knockout animals and involves a long gestation period and time to reach puberty. Multiplex CRISPR/Cas9-based genome editing and lipofection can greatly increase the efficiency, shorten the duration, and reduce the cost of this process. However, to date, no study has investigated one-step multigene genome editing of mammalian zygotes using lipofection alone. A simple one-step multigene targeting approach can improve the feasibility and efficiency of porcine genome-editing systems.
Mutations in the growth hormone receptor (GHR) gene cause Laron syndrome, an autosomal disease involving growth hormone resistance; it is characterized by a slow growth rate and small body size [12]. However, this mutation confers many advantages, such as ease of handling the pigs, the need for a small holding space, and the ability to use low doses of test substances for laboratory tests. Inactivation of the glycoprotein alpha-galactosyltransferase 1 (GGTA1)-encoded enzyme, which mediates xenoantigen generation, is necessary for prolonging organ survival after xenotransplantation [13]. Generation of GHR/GGTA1-knockout pigs is technically challenging because of the need to disrupt these two targets. Our previous study used a group culture system (4-well plate culture) for ZP-free zygotes and embryos after transfection, which may lead to the aggregation of two or more embryos in certain cases, resulting in low blastocyst formation and chimeric blastocysts [14, 15]. Therefore, individual culture systems should be examined for ZP-free embryos after lipofection, for example, using individual microwell cultures, for achieving efficient embryo development and thus addressing the limiting factor of embryo aggregation, which leads to chimerism.
Various CRISPR/Cas9 components, such as expression plasmids, mRNAs, and proteins/nucleases, may affect gene mutation efficiency [16]. The introduction of the CRISPR/Cas9 system closer to the time of pronuclear formation increases mRNA translation, thereby increasing the mutation rate [17, 18]. We hypothesized that an additional lipofection period would increase the mutation rate in pig zygotes, owing to the possibility of re-editing embryos in the mosaic state during different transfection periods.
In this study, we investigated whether lipofection could be used for CRISPR/Cas9-based multiplex genome editing using GHR and GGTA1 in pig zygotes. In the first experiment, we evaluated the mutation efficiencies of gRNAs targeting either GHR or GGTA1 in zygotes transfected using lipofection. In the second experiment, we examined the effects of the culture method on the development and mutation efficiency of GHR and GGTA1 double-edited zygotes in 4- and 25-well plates. In the third experiment, we assessed the effect of an additional 5 h exposure to GHR-targeted lipofection before and after simultaneous double gRNA-targeted lipofection on the mutation efficiency of GHR and GGTA1 double-edited embryos.
Materials and Methods
Ethical approval
All animal experiments were approved by the Institutional Animal Care and Use Committee of Tokushima University (approval number: T2019-11).
Oocyte collection and in vitro maturation (IVM)
Oocyte collection and IVM were performed as previously described [19]. Briefly, the ovaries of prepubertal crossbred gilts (Landrace × Large White × Duroc) were collected from a local slaughterhouse. Cumulus–oocyte complexes (COCs) were collected from follicles with uniformly dark-pigmented ooplasm and intact cumulus cell masses using a surgical blade. Approximately 50 COCs were cultured in 500 µl of maturation medium, consisting of TCM 199 with Earle’s salts (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% (v/v) porcine follicular fluid, 0.6 mM cysteine (Sigma-Aldrich, St. Louis, MO, USA), 50 µg/ml gentamicin (Sigma-Aldrich), 50 µM sodium pyruvate (Sigma-Aldrich), 50 µM β-mercaptoethanol (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan), 2 mg/ml D-sorbitol (FUJIFILM Wako Pure Chemical Corporation), 10 IU/ml equine chorionic gonadotropin (Kyoritsu Seiyaku, Tokyo, Japan), and 10 IU/ml human chorionic gonadotropin (Kyoritsu Seiyaku). Next, COCs were cultured in an IVM medium with hormones for 22 h in 4-well dishes (Nunc A/S, Roskilde, Denmark). Subsequently, they were transferred to an IVM medium without hormones and cultured for an additional 22 h at 39°C in a humidified incubator containing 5% CO2.
In vitro fertilization and culture of embryos
In vitro fertilization (IVF) was performed as previously described [19]. Briefly, frozen-thawed spermatozoa were transferred to 5 ml of porcine fertilization medium (PFM; Research Institute for the Functional Peptides Co., Yamagata, Japan) and washed via centrifugation at 500 × g for 5 min. Approximately 50 matured oocytes were transferred to 500 µl of PFM-containing sperm and co-incubated for 5 h. The final sperm concentration was adjusted to 1 × 106 sperms/ml. After co-incubation, the inseminated zygotes were denuded and cultured for 3 days in porcine zygote medium (PZM-5; Research Institute for the Functional Peptides Co.) overlaid with mineral oil. All cleaved embryos were transferred to a porcine blastocyst medium (Research Institute for the Functional Peptides Co.) and cultured for 4 days to evaluate their ability to develop into blastocysts and to assess the genotype of the resulting blastocysts.
Lipofection treatment
ZP-free zygotes were prepared before lipofection. First, zygotes were exposed to 0.5% (w/v) actinase-E (Kaken-Seiyaku Corp., Tokyo, Japan) in Dulbecco’s phosphate-buffered saline (Thermo Fisher Scientific) for 20–30 sec and subsequently transferred to PZM-5 without actinase-E. After removing the ZP via gentle pipetting, the zygotes were subjected to lipofection.
We previously successfully performed single gene editing via lipofection in ZP-free porcine zygotes and embryos by Lipofectamine 2000 (LP2000) [7, 20]. To determine whether a lipofection-mediated gene editing system can perform multiple gene edits using the same lipofection system, we used LP2000 (Thermo Fisher Scientific) in this study [20]. The lipofection solution was prepared by adding 2 µl of LP2000 to 8 µl of Nuclease-Free Duplex Buffer (IDT, Integrated DNA Technologies, Coralville, IA, USA) containing Cas9–gRNA ribonucleoprotein complexes (RNPs). Cas9–gRNA RNPs were prepared in Nuclease-Free Duplex Buffer by mixing 300 ng/μl Cas9 protein (Guide-it Recombinant Cas9, Takara Bio, Shiga, Japan) with 100 ng/μl gRNAs (Alt-R CRISPR crRNAs and tracrRNA, chemically modified and length-optimized variants of native gRNAs; Integrated DNA Technologies) targeting GHR (5′-GCTCAAGTGATGCTTTTTCT-3′) or GGTA1 (5′-AGACGCTATAGGCAACGAAA-3′); the final volume was made up to 20 μL. After mixing, the solutions were incubated at 25°C for 15 min to facilitate the formation of CRISPR/Cas RNPs and subsequently added to 180 µl of PZM-5 containing ZP-free zygotes for transfection. Double-editing of GHR and GGTA1 was performed by adding 2 µl of LP2000 to 8 µl of Nuclease-Free Duplex Buffer containing Cas9–gRNA RNP complexes, which were prepared by mixing each gRNA (100 ng/µl) with Cas9 protein (300 ng/µl) to make a final volume of 20 µl.
Mutation analysis
Genomic DNA was isolated from blastocysts by boiling in 50 mM NaOH (FUJIFILM Wako Pure Chemical Corporation). After neutralization, the DNA samples were subjected to polymerase chain reaction (PCR) using the KOD One PCR Master Mix (Toyobo, Osaka, Japan), according to the manufacturer’s instructions, with the following primers: 5′-CCCACCGGAAGTAGCATTTA-3′ (forward) and 5′-ACAACACTCCCGGAAACATC-3′ (reverse) for GHR and 5′-CCTGTCGGGAATGTTCTCAT-3′ (forward) and 5′-AAAAGGGGAGCACTGAACCT-3′ (reverse) for GGTA1. The PCR products were extracted via agarose gel electrophoresis by a Fast Gene Gel/PCR Extraction Kit (Nippon Genetics, Tokyo, Japan). The targeted genomic regions of the PCR products were directly sequenced using Sanger sequencing with the BigDye Terminator Cycle Sequencing Kit version 3.1 (Thermo Fisher Scientific) and an ABI 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The Tracking of Indels by Decomposition (TIDE) bioinformatics package was used for determining the genotype of each blastocyst [21]. The genotypes of the examined blastocysts were categorized as mosaic (carrying more than one type of mutation and the wild-type (WT) sequence) or WT (carrying only the wild-type sequence). Mutation rate was defined as the ratio of the number of mutant blastocysts (mosaics) to the total number of sequenced blastocysts. Mutation efficiency was defined as the proportion of indel mutation events in each mutant blastocyst. The TIDE bioinformatics package was used for estimating the proportion of indel mutation events in each mutant blastocyst, that is, the relative efficiency with which each of the four nucleotides was introduced after the break site, compared with that in the WT sequence [21].
Experimental design
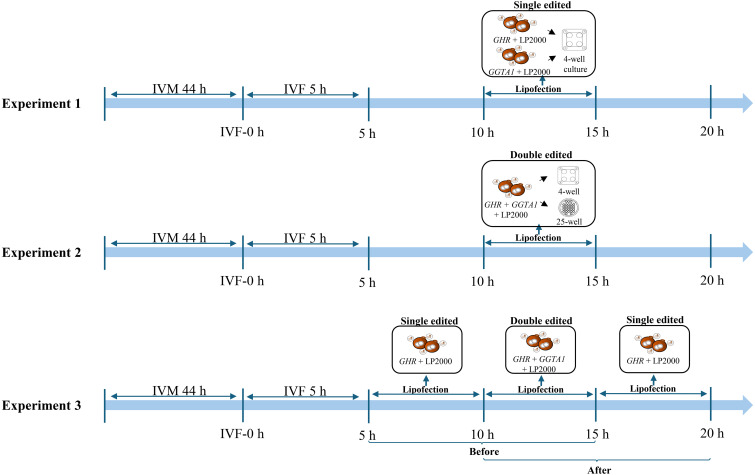
The experimental design is shown schematically in Fig. 1.
Fig. 1.
A schematic of the experimental design. Zona pellucida-free zygotes collected after in vitro fertilization (IVF) were co-incubated with Lipofectamine 2000 (LP2000) and guide RNAs targeting growth hormone receptor (GHR) and glycoprotein alpha-galactosyltransferase 1 (GGTA1) for 5 h. Subsequently, they were cultured in 4-well plates (group culture) or 25-well plates (individual culture). IVM, in vitro maturation.
Experiment 1: To assess the development and mutation of embryos edited with gRNAs targeting GHR and GGTA1, ZP-free zygotes collected 10 h after the start of IVF were co-incubated with RNPs containing gRNAs targeting either GHR or GGTA1 and LP2000 for 5 h and subsequently cultured in 4-well plates (50 embryos per 500 µl of culture medium) for 7 days.
Experiment 2: To examine the effects of the culture method on the development and mutation of zygotes double-edited with GHR and GGTA1, ZP-free zygotes collected 10 h after the start of IVF were co-incubated with RNPs containing two gRNAs targeting GHR and GGTA1 and LP2000 for 5 h. After lipofection, the zygotes were cultured in 4-well plates (50 embryos per 500 µl of culture medium) and 25-well plates (one embryo per 15 µl of culture medium; ART Culture Dish, Nipro, Osaka, Japan) for 7 days.
Experiment 3: Because Experiment 2 revealed that GHR was not mutated in the edited zygotes, we evaluated the effects of additional GHR-targeted lipofection (5 h) on the development and mutation of GHR and GGTA1 double-edited embryos. Before and after simultaneous double-targeted lipofection of GHR and GGTA1, ZP-free zygotes were co-incubated for 5 h with gRNA targeting GHR and LP2000. In this experiment, 25-well plates were used for culturing the edited zygotes because, in Experiment 2, the 25-well plate culture showed improved blastocyst formation and mutation rate in the edited zygotes compared with the 4-well plate culture. As a control, the zygotes were simultaneously edited with gRNAs targeting GHR and GGTA1 10 h after the start of IVF.
Statistical analyses
The percentage data for embryo development and mutation efficiency in Experiments 1 and 2 were evaluated using an independent Student’s t-test with STATVIEW (Abacus Concepts, Inc., Berkeley, CA, USA). The percentage data in Experiment 3 were evaluated using analysis of variance followed by Fisher’s least significant difference test. The data were subjected to arcsine transformation before statistical analysis. The percentage of mutant blastocysts was evaluated using the chi-square test with Yates correction. Statistical significance was set at P < 0.05.
Results
Experiment 1
No significant differences were observed in the blastocyst formation rates, mutation rates, or mutation efficiencies between the GHR- and GGTA1-gRNA-targeting groups (12.5 vs. 15.8%, 11.8 vs. 25.0%, and 9.1 vs. 16.4%, respectively; Table 1).
Table 1. Development and mutation of zygotes edited with guide RNAs (gRNAs) targeting the growth hormone receptor (GHR) and glycoprotein alpha-galactosyltransferase 1 (GGTA1).
| Target | No. of zygotes | No. (%) of zygotes developed to blastocysts | No. of blastocysts examined | No. (%) of gene-edited blastocysts * |
||
|---|---|---|---|---|---|---|
| WT | Mosaic | Average efficiency | ||||
| GHR | 200 | 25 (12.5 ± 2.5) | 17 | 15 (88.2) | 2 (11.8) | 9.1 |
| GGTA1 | 196 | 31 (15.8 ± 3.3) | 16 | 12 (75.0) | 4 (25.0) | 16.4 |
Zona pellucida-free zygotes collected 10 h after in vitro fertilization were co-incubated with guide RNAs (gRNAs) and Lipofectamine 2000 (LP2000) for 5 h and cultured in 4-well culture plates. Four replicates were performed. * Percentages were calculated by dividing the number of mutant blastocysts by the number of blastocysts examined. Average efficiency indicates the proportion of indel mutation events in mutant blastocysts, as determined using the Tracking of Indels by Decomposition (TIDE) analysis.
Experiment 2
When the effects of the culture method on the development and mutation of GHR and GGTA1 double-edited zygotes were examined, the 25-well plate showed a significantly increased blastocyst formation and mutation of zygotes compared to those in the 4-well plate (32.6 vs. 16.5% and 45.0 vs. 11.1%, respectively; P < 0.05). Neither the 4-well nor 25-well plates showed GHR-targeted mutant blastocysts derived from zygotes edited with the two gRNAs. Moreover, no differences were observed in the mutation efficiency of the edited embryos between the two culture plates (Table 2).
Table 2. Development and mutation of zygotes cultured in different culture plates after simultaneous growth hormone receptor (GHR) and glycoprotein alpha-galactosyltransferase 1 (GGTA1) double-targeted lipofection.
| Culture plate | No. of zygotes | No. (%) of zygotes developed to blastocysts | No. of blastocysts examined | No. (%) of GHR-edited blastocysts * |
No. (%) of GGTA1-edited blastocysts * |
||||
|---|---|---|---|---|---|---|---|---|---|
| WT | Mosaic | Average efficiency | WT | Mosaic | Average efficiency | ||||
| 4-well | 200 | 33 (16.5 ± 4.6) a | 18 | 18 (100) | 0 (0.0) | – | 16 (88.9) a | 2 (11.1) a | 16.3 |
| 25-well | 197 | 64 (32.6 ± 2.3) b | 20 | 20 (100) | 0 (0.0) | – | 11 (55.0) b | 9 (45.0) b | 9.8 |
Zona pellucida-free zygotes collected 10 h after in vitro fertilization were co-incubated with two guide RNAs (gRNAs) targeting GHR, GGTA1, and Lipofectamine 2000 (LP2000) for 5 h and cultured in 4-well or 25-well plates. Four replicates were performed. * Percentages were calculated by dividing the number of mutant blastocysts by the number of blastocysts examined. Average efficiency indicates the proportion of indel mutation events in mutant blastocysts, as determined using the Tracking of Indels by Decomposition (TIDE) analysis. a,b Values with different superscript letters are significantly different (P <0.05).
Experiment 3
Evaluation of the additional lipofection treatment with GHR before and after simultaneous double gRNA-targeted lipofection revealed no significant differences in the development and mutation rates of the edited zygotes, regardless of pre- or post-GHR-targeted lipofection treatments. However, GHR-targeted mutations were detected only in blastocysts derived from zygotes obtained via GHR-targeted lipofection before simultaneous double gRNA-targeted lipofection (Table 3).
Table 3. Effects of additional lipofection of the growth hormone receptor (GHR) before and after simultaneous GHR and glycoprotein alpha-galactosyltransferase 1 (GGTA1) double-targeted lipofection on the development and mutation of double-edited zygotes.
| Additional lipofection treatment | No. of zygotes | No. (%) of zygotes developed to blastocysts | No. of blastocysts examined | No. (%) of GHR-edited blastocysts * |
No. (%) of GGTA1-edited blastocysts * |
||||
|---|---|---|---|---|---|---|---|---|---|
| WT | Mosaic | Average efficiency | WT | Mosaic | Average efficiency | ||||
| Control | 250 | 67 (26.8 ± 3.6) | 20 | 20 (100.0) | 0 (0.0) | – | 8 (40.0) | 12 (60.0) | 16.6 |
| Before | 250 | 56 (22.4 ± 3.9) | 19 | 17 (89.5) | 2 (10.5) | 6.2 | 8 (42.1) | 11 (57.9) | 17.9 |
| After | 250 | 63 (25.2 ± 3.6) | 20 | 20 (100.0) | 0 (0.0) | – | 11 (55.0) | 9 (45.0) | 14.5 |
Before and after simultaneous GHR and GGTA1 double-targeted lipofection, zona pellucida-free zygotes were co-incubated for 5 h with Lipofectamine 2000 (LP2000) and guide RNAs (gRNAs) targeting GHR. As a control, zygotes were simultaneously edited with gRNAs targeting GHR and GGTA1 10 h after the start of in vitro fertilization. Five replicates were performed. * Percentages were calculated by dividing the number of mutant blastocysts by the number of blastocysts examined. Average efficiency indicates the proportion of indel mutation events in mutant blastocysts, as determined using the Tracking of Indels by Decomposition (TIDE) analysis.
Discussion
As a method for introducing the CRISPR/Cas9 system into porcine embryos without the use of specialized equipment, lipofection shows great potential for RNP delivery and can substantially improve the value of pig resources as experimental animals, particularly in ill-equipped laboratories. In this study, we investigated whether lipofection could be used for CRISPR/Cas9-based multiplex genome editing targeting GHR and GGTA1 in pig zygotes. When ZP-free zygotes were co-incubated with individual gRNAs targeting GHR or GGTA1, no differences were observed in embryo development rates, mutation rates, or mutation efficiencies between the two gRNA groups. These findings are consistent with those of our previous study, which showed that lipofection-mediated gene editing can be performed at various target sites during embryogenesis [8]. However, when zygotes were simultaneously transfected with the two gRNAs, the resulting blastocysts showed no GHR-targeted mutations and only GGTA1-targeted mutations, regardless of the culture method used. In this study, the concentrations of gRNA (10 ng/µl at final concentration) and Cas9 protein (30 ng/µl at final concentration) used for the double gRNA-targeted lipofection were the same as those used for the single-gene-targeting experiment. This indicates crosstalk among multiple gRNAs, which affects the editing efficiency. Therefore, potential crosstalk interference should be avoided when CRISPR systems are used to regulate multiple genes [22]. In contrast to our findings, the introduction of the CRISPR/Cas9 system via microinjection exhibited high efficiency in the one-step disruption of multiple target genes in the embryos of several mammalian species, including mice [23], monkeys [24], and rabbits [25]. One possible explanation for this discrepancy is that the limiting factors in the endocytosis delivery method (lipofection) are the sizes of the gRNAs and Cas9 protein; the simultaneous transfection of multiple small-sized gRNAs may reduce the ability to mutate.
A previous study demonstrated that differences in editing efficiency during multiple edits depend on differences in the target genes in the embryo [26]. Moreover, the co-delivery of multiple smaller-sized gRNAs has been shown to reduce the efficiency of lipofection [27]. In the current study, we observed that when zygotes were simultaneously transfected with two gRNAs, no GHR-targeted mutations were detected in the resulting blastocysts. Therefore, to induce GHR-targeted mutations by lipofection in double-edited embryos, we performed additional lipofection of GHR before and after simultaneous lipofection with double gRNA treatment. GHR-targeted mutations were detected only in blastocysts derived from zygotes subjected to GHR-targeted lipofection before double gRNA-targeted lipofection. Introduction of the CRISPR/Cas9 system close to the time of pronuclear formation increases mRNA translation, thereby increasing the mutation rate [17, 18]. In pigs, male and female pronucleus formation in IVF zygotes begins 6–10 h and 8 h after fertilization, respectively [28]. In this study, the mutation rate increased when the zygotes were transfected 5–15 h after IVF. This finding suggests that additional lipofection treatment at the beginning of pronuclear formation facilitates the re-editing of non-mutated or inefficiently mutated embryos. Moreover, examination of the development and mutation of the desired GHR and GGTA1 double-edited zygotes revealed increased blastocyst formation and mutations in the 25-well plates compared with that in the 4-well plates. Since the volume of culture medium per embryo was similar in the 4-well plates (10 μl per embryo) and 25-well plates (15 μl per embryo), the higher blastocyst formation rate in the 25-well plates may be attributed to the single-embryo culture conditions. Although single-embryo culture in 4-well plates via the well-of-the-well (WOW) system avoids embryo aggregation, the WOW system results in significantly lower rates of blastocyst formation in single-embryo cultures than that in group-embryo cultures [29]. Therefore, 25-well plates may be more effective for achieving single-embryo culture conditions than 4-well plates. The aggregation of ZP-free embryos in 4-well plates may lead to genetic mosaicism [8]. However, in this study, the resulting blastocysts showed no GHR-targeted mutations. Therefore, GHR-mutant blastocysts may be derived from single-gene editing rather than double-gene editing with GHR and GGTA1, indicating that the introduction of multiple genes results in a larger RNP insertion size, which affects gene editing efficiency. Moreover, the variable editing efficiencies between GHR and GGTA1 during double editing might be associated with the difference in locations of the targeted site in genes because editing efficiencies during multiple editing depend on the target genes in the embryo [26].
In conclusion, our findings suggest that additional lipofection before simultaneous double gRNA-targeted lipofection induces additional mutations in the zygotes. However, the resultant mutant blastocysts may be derived from single-gene editing rather than double-gene editing, even if the culture conditions are changed. Therefore, extensive technical improvements are necessary for maximizing the efficiency of multiplex gene editing using lipofection.
Conflict of interests
The authors declare no conflicts of interest.
Acknowledgments
This study was partially supported by KAKENHI (grant numbers JP22H02499 and JP22K19896) from the Japan Society for the Promotion of Science (JSPS) and the Uzushio Program of Tokushima University. We would like to thank Nippon Food Packer and K. K. Shikoku (Tokushima, Japan) for supplying pig ovaries for this study.
Availability of data and materials
The authors declare that the data supporting the findings of this study are available in the manuscript and its supplementary information files.
References
- 1.Lossi L, D’Angelo L, De Girolamo P, Merighi A. Anatomical features for an adequate choice of experimental animal model in biomedicine: II. Small laboratory rodents, rabbit, and pig. Ann Anat 2016; 204: 11–28. [DOI] [PubMed] [Google Scholar]
- 2.Hryhorowicz M, Lipiński D, Hryhorowicz S, Nowak-Terpiłowska A, Ryczek N, Zeyland J. Application of genetically engineered pigs in biomedical research. Genes (Basel) 2020; 11: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan W, Proudfoot C, Lillico SG, Whitelaw CBA. Gene targeting, genome editing: from Dolly to editors. Transgenic Res 2016; 25: 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanihara F, Takemoto T, Kitagawa E, Rao S, Do LTK, Onishi A, Yamashita Y, Kosugi C, Suzuki H, Sembon S, Suzuki S, Nakai M, Hashimoto M, Yasue A, Matsuhisa M, Noji S, Fujimura T, Fuchimoto D, Otoi T. Somatic cell reprogramming-free generation of genetically modified pigs. Sci Adv 2016; 2: e1600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe A, Miyanohara A, Friedmann T. Polybrene increases the efficiency of gene transfer by lipofection. Gene Ther 1998; 5: 708–711. [DOI] [PubMed] [Google Scholar]
- 7.Hirata M, Wittayarat M, Namula Z, Anh Le Q, Lin Q, Takebayashi K, Thongkittidilok C, Tanihara F, Otoi T. Lipofection-Mediated Introduction of CRISPR/Cas9 System into Porcine Oocytes and Embryos. Animals (Basel) 2021; 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirata M, Wittayarat M, Namula Z, Le QA, Lin Q, Takebayashi K, Thongkittidilok C, Mito T, Tomonari S, Tanihara F, Otoi T. Generation of mutant pigs by lipofection-mediated genome editing in embryos. Sci Rep 2021; 11: 23806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J, Wu L, Zhang S-M, Lu M, Cheung WKC, Cai W, Gale M, Xu Q, Yan Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res 2016; 44: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minkenberg B, Wheatley M, Yang Y. CRISPR/Cas9-Enabled Multiplex Genome Editing and Its Application. Prog Mol Biol Transl Sci 2017; 149: 111–132. [DOI] [PubMed] [Google Scholar]
- 11.Colucci F, Soudais C, Rosmaraki E, Vanes L, Tybulewicz VLJ, Di Santo JP. Dissecting NK cell development using a novel alymphoid mouse model: investigating the role of the c-abl proto-oncogene in murine NK cell differentiation. J Immunol 1999; 162: 2761–2765. [PubMed] [Google Scholar]
- 12.Yu H, Long W, Zhang X, Xu K, Guo J, Zhao H, Li H, Qing Y, Pan W, Jia B, Zhao H-Y, Huang X, Wei H-J. Generation of GHR-modified pigs as Laron syndrome models via a dual-sgRNAs/Cas9 system and somatic cell nuclear transfer. J Transl Med 2018; 16: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurh S, Kang B, Choi I, Cho B, Lee EM, Kim H, Kim YJ, Chung YS, Jeong JC, Hwang J-I, Kim JY, Lee BC, Surh CD, Yang J, Ahn C. Human antibody reactivity against xenogeneic N-glycolylneuraminic acid and galactose-α-1,3-galactose antigen. Xenotransplantation 2016; 23: 279–292. [DOI] [PubMed] [Google Scholar]
- 14.Lin Q, Takebayashi K, Torigoe N, Liu B, Namula Z, Hirata M, Tanihara F, Nagahara M, Otoi T. Comparison of chemically mediated CRISPR/Cas9 gene editing systems using different nonviral vectors in porcine embryos. Anim Sci J 2023; 94: e13878. [DOI] [PubMed] [Google Scholar]
- 15.Saadeldin IM, Kim SJ, Lee BC. Blastomeres aggregation as an efficient alternative for trophoblast culture from porcine parthenogenetic embryos. Dev Growth Differ 2015; 57: 362–368. [DOI] [PubMed] [Google Scholar]
- 16.Tanihara F, Hirata M, Otoi T. Current status of the application of gene editing in pigs. J Reprod Dev 2021; 67: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao L, Yang M, Wang X, Zhang Z, Wu Z, Tian J, An L, Wang S. Efficient biallelic mutation in porcine parthenotes using a CRISPR-Cas9 system. Biochem Biophys Res Commun 2016; 476: 225–229. [DOI] [PubMed] [Google Scholar]
- 18.Tanihara F, Hirata M, Nguyen NT, Le QA, Hirano T, Otoi T. Effects of concentration of CRISPR/Cas9 components on genetic mosaicism in cytoplasmic microinjected porcine embryos. J Reprod Dev 2019; 65: 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Q, Aihara M, Shirai A, Tanaka A, Takebayashi K, Yoshimura N, Torigoe N, Nagahara M, Minamikawa T, Otoi T. Porcine embryo development and inactivation of microorganisms after ultraviolet-C irradiation at 228 nm. Theriogenology 2023; 197: 252–258. [DOI] [PubMed] [Google Scholar]
- 20.Lin Q, Le QA, Takebayashi K, Thongkittidilok C, Wittayarat M, Hirata M, Tanihara F, Otoi T. Timing and duration of lipofection-mediated CRISPR/Cas9 delivery into porcine zygotes affect gene-editing events. BMC Res Notes 2021; 14: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 2014; 42: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, Zhang N, Zhu K, Xu J, Hu B, Leng Q, Huang X. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 2014; 46: 49–55. [DOI] [PubMed] [Google Scholar]
- 24.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014; 156: 836–843. [DOI] [PubMed] [Google Scholar]
- 25.Song J, Yang D, Ruan J, Zhang J, Chen YE, Xu J. Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Sci Rep 2017; 7: 12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song R, Wang Y, Zheng Q, Yao J, Cao C, Wang Y, Zhao J. One-step multiple site-specific base editing by direct embryo injection for precision and pyramid pig breeding. bioRxiv 2020: 2020.08.26.267948. [Google Scholar]
- 27.Khan FJ, Yuen G, Luo J. Multiplexed CRISPR/Cas9 gene knockout with simple crRNA:tracrRNA co-transfection. Cell Biosci 2019; 9: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurincik J, Rath D, Niemann H. Differences in pronucleus formation and first cleavage following in vitro fertilization between pig oocytes matured in vivo and in vitro. J Reprod Fertil 1994; 102: 277–284. [DOI] [PubMed] [Google Scholar]
- 29.Dai SJ, Xu CL, Wang J, Sun YP, Chian RC. Effect of culture medium volume and embryo density on early mouse embryonic development: tracking the development of the individual embryo. J Assist Reprod Genet 2012; 29: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available in the manuscript and its supplementary information files.