Abstract
Cannabis sativa L. is an ancient crop, but its agricultural adoption has been interrupted to prevent the use of marijuana as a psychoactive drug. Nevertheless, hemp—the C. sativa type with low concentrations of intoxicating Δ9-tetrahydrocannabinoid—is experiencing a resurgence in interest due to loosened cultivation restrictions and its potential as a multipurpose bio-based crop. Hemp has valuable applications, including production of medicines from its non-intoxicating cannabinoids, food, medical, and industrial uses of its seed oil rich in polyunsaturated fatty acids, and production of fibers for textiles and industry from its stems. Recently, several hemp genomic and genetic resources have been developed, allowing significant expansion of our knowledge of major hemp traits, such as synthesis of cannabinoids, oil, and fibers, and regulation of flowering and sex determination. Still, hemp is an underimproved crop, and its development will depend on the ability to expand and collectively use the novel resources arising from fast advancements in bioinformatics and plant phenotyping. This review discusses current genetic and genomic knowledge of the most important hemp traits, and provides a perspective on how to further expand such knowledge and tackle hemp improvement with the most up-to-date tools for plant and hemp research.
Keywords: Breeding tools, cannabinoids, fiber genetics, flowering genetics, hemp breeding, hemp genetics, hemp genomics, terpenes
Advances in hemp genetics allow its potential to be unlocked through integrative bioinformatic, molecular, and breeding approaches, but questions on the genetic control of crucial hemp traits are also still open.
Introduction
Cannabis sativa L. is the only plant species within the Cannabis genus from the Cannabaceae family (Ren et al., 2021). This species is an annual, herbaceous, diploid plant, with a genome of ~830 Mb made up of 10 chromosome pairs (2n=20) (Van Bakel et al., 2011; Divashuk et al., 2014). Of these, nine pairs are homomorphic autosomes, while the 10th pair groups semi-heteromorphic XX/XY sex chromosomes (Moliterni et al., 2004; Petit et al., 2020a; Prentout et al., 2020). Cannabis sativa is naturally dioecious, with male (XY) and female (XX) plants producing male and female flowers, respectively, which are fertilized through wind pollination (Salentijn et al., 2019; Smart et al., 2022). However, monoecious plants also occur, which are generally genetic females (XX) producing a variable number of both male and female flowers (Faux et al., 2014; Salentijn et al., 2019). Cannabis sativa has its center of origin in central Asia (Devkota, 2022), where it has been domesticated by humans since at least 6000 years ago (Hillig, 2005; Ren et al., 2021). The initial use of this plant was for medicinal purposes (Hillig, 2005; Liu et al., 2017), but since ~3000 years ago it has also been used for fiber production (Liu et al., 2017; Ren et al., 2021). Nowadays, C. sativa is abundant worldwide, mostly in temperate regions, and is a promising but underimproved multipurpose crop that can be used to produce diverse products such as fibers, oil, cannabinoids, and terpenes (Andre et al., 2016; Kovalchuk et al., 2020; Devkota, 2022). Specifically, multipurpose hemp is the C. sativa type that can be cultivated and used for the aforementioned applications, due to the low amount of the intoxicating Δ9-tetrahydrocannabinoid (THC; <0.3% of inflorescence dry weight) (Small, 2015; Petit, 2020). Still, hemp plants can synthesize >100 types of cannabinoids, among which cannabidiolic acid (CBDA) is prevalent (Johnson, 2019; Lange and Zager, 2022). Hemp cannabinoids—especially CBD—have valuable medical applications in formulations to treat epilepsy, chronic pain, disorders from drugs abuse, anxiety, inflammation, and cancer (Blessing et al., 2015; Überall, 2020; Morel et al., 2021; O’Brien, 2022). Thus, these molecules constitute the basis of the modern use of hemp as a medicinal plant. Apart from cannabinoids, hemp seeds contain ~28–35% oil with high amounts of α-linolenic acid, γ-linolenic acid, tocopherols, and antioxidant polyphenols, which are all compounds with high value for use in food and cosmetic products (Cerino et al., 2021; Devkota, 2022). Moreover, hemp oil can also be used to produce biofuels (Cerino et al., 2021; Devkota, 2022). Finally, hemp biomass is also an excellent source of natural fibers, which mostly consist of bast fibers from hemp stems, and can be used to replace glass fibers, producing textiles, and synthesizing bioplastics and building materials, such as ‘hempcrete’ (Ranalli and Venturi, 2004; Andre et al., 2016; Novakova and Sal, 2019).
Despite the potential of hemp as a multipurpose crop to produce several bio-based products, this species is currently underimproved and far from expressing its genetic potential for diverse applications (Salentijn et al., 2015; Schluttenhofer and Yuan, 2017). This is mainly due to the restrictions to C. sativa cultivation to prevent illegal production of marijuana (i.e. C. sativa types with >0.3% THC) that for more than 80 years have also extensively blocked hemp research and breeding (Smart et al., 2022). As such, the major available industrial hemp cultivars have been bred through conventional basic breeding methods, such as mass selection, without the use of any marker or genomic technology (Salentijn et al., 2015; Schluttenhofer and Yuan, 2017). Still, large genetic variation for the content and quality of the major products extractable from hemp—fibers, cannabinoids, terpenes, and oil—has been recorded in multiple hemp populations (Kriese et al., 2004; Galasso et al., 2016; Petit et al., 2020c; Johnson and Wallace, 2021; Stack et al., 2021). Moreover, the resurgence of hemp research that has taken place over the last decades has produced a wealth of genomic and genetic resources—including multiple hemp genome assemblies (Van Bakel et al., 2011; Braich et al., 2020; Grassa et al., 2021; Wei et al., 2024), transcriptomic resources (Guerriero et al., 2017; Braich et al., 2019; Adal et al., 2021; Tang et al., 2023), and molecular markers (Petit et al., 2020a, b; Borin et al., 2021)—that are extremely valuable to perform technology-driven breeding research on this crop. Finally, the increasing availability of genetic, genomic, and functional data on specific traits across all plant species, as well as of bioinformatic tools for large-scale comparative analyses, opens up novel scenarios for crop improvement through the translation of genetic information between species.
In the context just discussed, this paper reviews the advancements achieved in understanding the genetic basis of major target traits for the utilization of hemp in a bio-based economy. Moreover, a perspective is offered on how the further development of multipurpose hemp varieties can flourish by integrating those research advancements with new genomic and bioinformatic technologies. Major target hemp traits for a multipurpose use of hemp in a bio-based economy include the content and quality of hemp cannabinoids, terpenes, oil, and fibers. Moreover, regulation of sex determination and flowering behavior are also important traits, as they directly influence the expression of major industrial hemp characters, while in parallel allowing for specific breeding schemes (i.e. hybrid breeding) (Salentijn et al., 2019). In the future, hemp improvement will integrate conventional breeding methods with novel genomic technologies, bioinformatic tools, and biotechnological approaches based on the genetics of target traits from both hemp and other crops.
The genetics of relevant hemp traits from the perspective of crop improvement
Cannabinoids
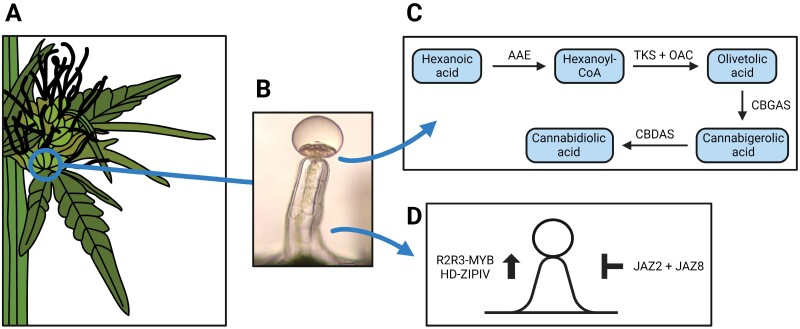
Maximizing the yield of non-intoxicating cannabinoids is a important objective in hemp research (Smart et al., 2022), as these secondary metabolites—particularly CBDA—have several pharmaceutical applications with high economic value. Cannabis sativa synthesizes cannabinoids as phytoprotectans (Stack et al., 2023), and the biosynthetic pathways of both THCA and CBDA largely overlap (Fig. 1) (Tahir et al., 2021). Specifically, the common part of the THCA and CBDA biosynthetic pathway starts with the conversion of hexanoic acid into hexanoyl-CoA by an acyl-activating enzyme (AAE) (Laverty et al., 2019; Kovalchuk et al., 2020). Hexanoyl-CoA is then converted into cannabigerolic acid (CBGA) through a series of condensations, cyclations, and aromatizations catalyzed by the enzymes tetraketide synthase [TKS; also known as olivetol synthase (OLS)], olivetolic acid cyclase (OAC), and cannabigerolic acid synthase (CBGAS, an aromatic prenyltransferase) [reviewed by Tahir et al., 2021 and Innes and Vergara, 2023)]. CBGA is the precursor of both CBDA and THCA (Innes and Vergara, 2023), whose final synthesis is carried out by the oxidocylase enzymes CBDA and THCA synthases (CBDAS and THCAS), respectively (Sirikantaramas et al., 2004; Taura et al., 2007).
Fig. 1.
Genes taking part in biosynthesis of cannabidiolic acid and establishment of glandular trichomes in the context of hemp female flowers and glandular trichome development. (A) Drawing of a typical hemp female flower. (B) Microscopic structure of a typical hemp glandular trichome [adapted from Andre et al. (2016)]. (C) The biosynthetic pathway of cannabidiolic acid, with the main molecules (blue boxes) and genes/enzymes (arrows) involved, which takes place in the disk cells of glandular trichomes. AAE, acyl-activating enzyme; TKS, tetraketide synthase; OAC, olivetolic acid cyclase; CBGAS, cannabigerolic acid synthase; CBDAS, cannabidiolic acid synthase. (D) The major genes involved in the promotion (left) and repression (right) of glandular trichome development based on research on Arabidopsis and tomato, as described in the text.
Over the years, several genes encoding the enzymes of the cannabinoid pathway have been identified in hemp [see the works of Van Bakel et al. (2011), Laverty et al. (2019), McGarvey et al. (2020), Grassa et al. (2021), van Velzen and Schranz (2021), Fulvio et al. (2021), Innes and Vergara (2023), and Fulvio et al. (2023) for an overview of the results in this area over the years]. A major achievement was the identification of the genomic location of the oxidocyclase genes synthesizing either CBDA or THCA within a long terminal repeat (LTR)-rich region on C. sativa chromosome 7 (26–31 Mb) (Grassa et al., 2021). At this locus, the presence of either CBDAS or THCAS functional genes underlies the production of either CBDA or THCA (Laverty et al., 2019; McKernan et al., 2020, Preprint; Grassa et al., 2021). Specifically, CBDAS functionality appears pivotal for regulating CBDA production (McKernan et al., 2020, Preprint) and, even though recent studies showed that both CBDAS and THCAS loci display little sequence variation in a large panel of C. sativa accessions (Lynch et al., 2024, Preprint), the relationship between CBDAS genetic diversity and CBDA levels deserves further attention, as induced CBDAS mutations can significantly affect CBDAS catalytic activity (Dai et al., 2024). In addition to CBDAS, relevant genetic patterns were recently uncovered for some of the genes acting upstream of cannabinoid oxidocyclases within the cannabinoid pathway, including OLS and OAC (Innes and Vergara, 2023). Specifically, some OLS and OAC tandem paralogs display copy number variation across different hemp accessions, and future research could investigate functional redundancy of variable gene copies, as well as their possible gene dosage effect on the cannabinoid pathway (Innes and Vergara, 2023).
The greatest proportion of hemp cannabinoids is synthesized by stalked glandular trichomes located on hemp female flowers, which are the hemp plant structures richest in CBDA (Fig. 1) (Mahlberg and Kim, 2004; Livingston et al., 2020). These trichomes are formed by multicellular, large stalks supporting 12–16 disk cells and a secretory cavity, and their occurrence is highest on calyces and bracts of hemp female flowers (Livingston et al., 2020; Tanney et al., 2021). The disk cells synthesize CBDA by forming polarized ‘supercells’ that contain non-photosynthetic metabolic plastids (Livingston et al., 2022). CBDA is then accumulated in the secretory cavities of the trichomes (Livingston et al., 2022; Xie et al., 2023). Because of the importance of glandular trichomes for CBDA production, elucidation of the factors influencing their number, size, and metabolic activity is a relevant research area (Tanney et al., 2021), particularly because these properties are genetically controlled in hemp (Punja et al., 2023). Bassolino et al. (2020) recently identified three C. sativa MYB transcription factor genes likely to be involved in trichome development regulation based on homology with Arabidopsis thaliana. This study highlights the relevance of knowledge about glandular trichomes obtained from other dicot species, given the overall scarce information on the genetics of hemp glandular trichome development (Tanney et al., 2021). In this context, both tomato (Solanum lycopersicum) and sweet wormwood (Artemisia annua) are models to study the genetics underlying trichome development, and two transcription factor gene families—R2R3-MYB and HD-ZIP IV—regulate the initiation of glandular trichome formation in these species (Chalvin et al., 2020). Specifically, the genes AaMYB1, AaMIXTA1, and SlSIMX1 from the R2R3-MYB family, as well as the genes AaHD1, AaHD8, SlCD2, and SlWO from the HD-ZIP IV family all positively influence glandular trichome development in A. annua (Matías‐Hernández et al., 2017; Yan et al., 2017; Shi et al., 2018) and tomato (Nadakuduti et al., 2012; Yang et al., 2015; Ewas et al., 2016, 2017). Moreover, their overexpression leads to increased yield of glandular trichome metabolites in A. annua (Yan et al., 2017; Shi et al., 2018). Conversely, jasmonate signaling genes such as AaJAZ8 or SlJAZ2 are known to repress glandular trichome formation by down-regulating R2R3-MYB and HD-ZIP IV transcription factor genes (Yan et al., 2017; Yu et al., 2018). Finally, quantitative trait loci (QTLs) associated with variability in shape and density of glandular trichomes in tomato have also been mapped (Momotaz et al., 2010; Bennewitz et al., 2018). Overall, these genetic resources can serve as a starting point for translational genomics studies aimed at studying the regulation of glandular trichome development in hemp, by identifying hemp gene orthologs or genomic regions that share conserved blocks of genes in the same relative positions as compared with known QTLs (syntenic QTL regions). Such gene orthologs and syntenic QTL regions can in turn be targeted for functional studies or screenings of favorable allelic diversity for both trichome- and metabolic-related traits. In this regard, the recent works of Kundan et al. (2022) and Haiden et al. (2022) appear relevant, as they respectively identified 99 R2R3-MYB genes in the hemp genome, and showed the functionality of a C. sativa MIXTA gene homolog to A. annua to increase trichome density upon overexpression in tobacco leaves.
Terpenes
After cannabinoids, terpenes are another important class of hemp secondary metabolites. Terpenes determine the typical C. sativa aroma and, like cannabinoids, are synthesized and accumulated in stalked glandular trichomes (Sommano et al., 2020). Terpenes can be used for multiple applications, including as aroma regulators and bioactive compounds in cosmetics, flavor additives in food, or bioactive molecules in pharmaceuticals thanks their antimicrobial, antioxidant, anti-inflammatory, and antidiabetic properties (Nuutinen, 2018; Chen and Pan, 2021). Furthermore, terpenes can be blended with CBD to enhance the medicinal properties of CBD itself—a phenomenon known as the ‘entourage effect’ (Ferber et al., 2020; Anand et al., 2021). At a chemical level, terpenes are a diverse group of hydrocarbon molecules made up of isoprene chains (Hanuš and Hod, 2020; Sommano et al., 2020). Hemp trichomes can synthesize monoterpenes (two isoprene subunits), sesquiterpenes (three isoprene subunits), diterpenes (four isoprene subunits), and triterpenes (six isoprene subunits), with the vast majority of C. sativa terpenes being mono- and sesquiterpenes (Booth et al., 2020; Hanuš and Hod, 2020; Sommano et al., 2020).
Terpene synthases (TPSs) are responsible for terpene synthesis in all land plants (Chen et al., 2011). In hemp, TPSs cyclize the last intermediates of the mevalonate (MEV) and non-mevalonate (MEP) pathways—geranyl diphosphate and farnesyl diphosphate, respectively—into the variety of mono- and sesquiterpenes found in hemp (Booth et al., 2020). The TPS gene family is large and diversified across all land plants, accounting for eight known TPS subfamilies—TPS-a to TPS-h (Chen et al., 2011; Jiang et al., 2019). Among these, the TPS-a and TPS-b genes are of particular relevance for hemp, as they synthesize mono- and sesquiterpenes, respectively (Booth et al., 2020). Research showed that TPS genes form a large gene family in hemp, with a variable total number of genes across different accessions. Specifically, Booth et al. (2017, 2020) identified 13 and 19 TPS genes in the transcriptome of the hemp cultivar ‘Finola’ and in the draft genome of the cultivar ‘Purple Kush’, respectively (Van Bakel et al., 2011). More recently, Allen et al. (2019) and Xu et al. (2024) mapped 55 and 41 distinct TPS genes in the complete C. sativa genomes from cultivars CBDRx (Grassa et al., 2021) and ‘Jamaica Lion’ (McKernan et al., 2020, Preprint), respectively. Finally, McKernan et al. (2020, Preprint) found extensive copy number variation for TPS genes potentially associated with variability in terpene profiles across 40 re-sequenced C. sativa accessions. As TPS genes are organized in genomic clusters, (proximal), gene duplications have driven the expansion and diversification of these genes in hemp (Booth et al., 2020; Xu et al., 2024). Most of the hemp TPS genes belong to the TPS-a and TPS-b subfamilies, in line with the prevalence of mono- and sesquiterpenes among hemp terpenes (Booth et al., 2017, 2020; Xu et al., 2024). Nevertheless, the genomic differentiation of TPS genes, reflected in their expression variability across different hemp accessions, seems key to determine the specific blends of hemp terpenes—including α-pinene, limonene, myrcene, and β-caryophyllene (Zager et al., 2019; Booth et al., 2020). As such, a further sampling of the genomic and transcriptomic diversity of hemp TPS genes, coupled with functional analyses on individual TPS copies, seems a promising strategy to acquire the genetic knowledge needed to modify hemp terpenes toward specific blends.
In addition to TPS genes, the other genes within the MEV and MEP pathways are also important to understand the genetics of hemp terpene biosynthesis, as well as to modify the synthesis of these molecules. These genes encode multiple enzymes that globally catalyze the conversion of pyruvate into geranyl diphosphate (MEP pathway) and of acetoacetyl-CoA into farnesyl diphosphate (MEV pathway) [see Booth et al. (2020) for an overview]. Notably, most of the MEP and MEV pathway genes are present in multiple copies within the hemp genome, often arranged in tandem clusters (Booth et al., 2020). Furthermore, MEP and MEV pathway genes display co-expression with specific TPS genes during terpene synthesis in hemp (Zager et al., 2019). As such, it would be relevant to study the variability in copy number and in the coordinated regulation of the MEP and MEV pathway genes in panels of multiple hemp accessions, and to correlate the results with overall terpene production and profiles. This could identify targets to increase terpene production through biotechnology.
Oil
Seed oil is another important product of industrial hemp, with several high-value applications in nutraceutical, cosmetic, and pharmaceutical industries (Devkota, 2022; Smart et al., 2022). This is due to the hemp seed oil composition, which includes 50–70% linoleic acid (18:2n-6), 15–25% α-linolenic acid (18:3n-3), and ~9% γ-linolenic (18:3n-6) and stearidonic acids (18:4n-3) (Matthäus and Brühl, 2008; Rezvankhah et al., 2019). These are all polyunsaturated fatty acids (PUFAs), which deliver health benefits upon consumption or when used in medicines against diseases such as glaucoma, cancer, inflammation, arthritis, and allergies (Rezvankhah et al., 2019). Moreover, tocopherols (i.e. vitamin E) and phytosterols—which are antioxidants and can reduce low-density lipoprotein (LDL)-cholesterol—are also found in high amounts in hemp seed oil (up to 800 mg kg–1 tocopherols and ~5 g kg–1 phytosterols) (Oomah et al., 2002; Matthäus and Brühl, 2008). Industrial hemp varieties have typically not been bred for oil content and composition, which is the reason why hemp oil yield is often unstable, while oil quality is heterogeneous, even within cultivars (Matthäus and Brühl, 2008). As such, pursuing an increased and stable oil yield, as well as an optimized and stable oil composition, is a major target for research on industrial hemp.
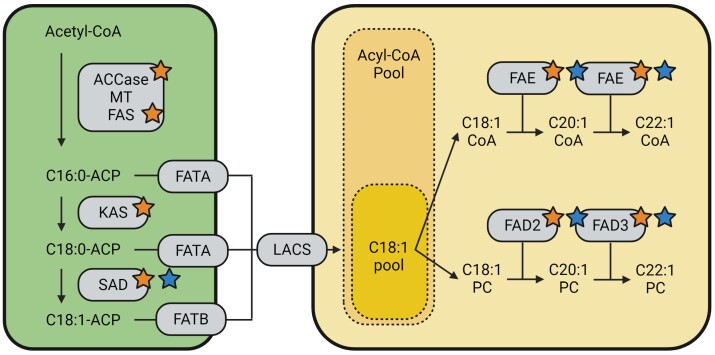
So far, improvement of hemp oil-related traits has followed conventional methods of selective breeding, as in the case of the development of the ‘Finola’ variety (Callaway and Laakkonen, 1997). As plant oil yield and quality are typically highly heritable traits (Schierholt and Becker, 2001; Bachlava et al., 2008; Khan et al., 2010), selective breeding can lead to significant improvements. Still, research in different oilseed crops showed that genetic manipulation of the oil pathway is also very effective both to increase seed oil content and yield, and to tailor oil composition to specific uses (Savadi et al., 2017; Porokhovinova et al., 2022). In this context, the work of Bielecka et al. (2014) pioneered this research area in hemp, by showing that the silencing of two FATTY ACID DESATURASE (FAD) genes identified through homology with other oilseed dicots greatly changed the oil fatty acid profile of hemp (Fig. 2). More recently, Wei et al. (2024) released the reference genome of a major Chinese seed hemp cultivar, ‘Yushe’, which was used to identify 36 genes underlying hemp oil synthesis and covering the conventional plant oil biosynthetic pathway (Fig. 2). Moreover, it was shown that seed hemp experienced an expansion of the STEAROYL-ACP DESATURASE (SAD) and FAD gene families relative to other species, which might underpin its high seed oil content and high amounts of oil PUFAs (Wei et al., 2024). Further research could explore variability in SAD and FAD copy number across diverse hemp accessions and verify correlations with variability in seed oil content and composition. Moreover, reverse genetics studies such as that of Bielecka et al. (2014) could be replicated for other hemp oil genes. In this regard, FATTY ACID ELONGASE (FAE) represents an interesting target, as six FAE copies have been found in seed hemp (Wei et al., 2024) and its down-regulation increased the levels of oleic acid in Brassicaceae (Tian et al., 2011; Shi et al., 2015). This, coupled with the overexpression of endogenous FAD genes, could lead to increased PUFAs levels (Liu et al., 2024), which can be a valuable target for seed hemp improvement.
Fig. 2.
Representation of the plant oil biosynthesis pathway and the genes involved. Orange stars next to genes indicate that hemp orthologs have been identified in the seed hemp genome by Wei et al. (2024). Blue stars indicate genes that are interesting hemp targets for screenings of allelic diversity and copy number variation, or genetic engineering. ACCase, acetyl-CoA carboxylase; MT, malonyl-CoA-ACP transferase; FAS, fatty acid synthase; KAS, ketoacyl-ACP synthase; SAD, stearoyl-ACP desaturase; FATA, acyl-ACP thioesterase A; FATB, acyl-ACP thioesterase B; LACS, long-chain acyl-CoA synthetase; FAE, fatty acid elongase; FAD2, fatty acid desaturase 2; FAD3, fatty acid desaturase 3.
Despite being valuable for food and industrial applications, PUFA-rich plant oils are prone to lipid peroxidation by reactive oxygen species (Ali et al., 2022), making the oil unstable. This issue can be controlled by decreasing the PUFA content and raising the content of the oxidatively stable oleic acid (C18:1) in the oil, for example by down-regulating FAD genes (Kinney, 1998; Bielecka et al., 2014). However, this approach also decreases the yield of valuable PUFAs that are naturally prevalent in hemp oil. An alternative approach can aim at increasing tocopherols in hemp oil, which act as antioxidants against PUFA peroxidation (Mène-Saffrané and DellaPenna, 2010; Boonnoy et al., 2017; Ali et al., 2022), have several food and industrial applications, and are naturally present in relatively high amounts in hemp oil. Several key genes underlying tocopherol biosynthesis have been found in different plants, including A. thaliana, carrot, maize, and tobacco [see Fritsche et al. (2017) for a detailed review]. Specifically, the genes p-HYDROXYPHENYLPYRUVATE DIOXYGENASE 1 (PSD1) and several VITAMIN E (VTE) loci deeply affect the synthesis and content of tocopherols in multiple plant organs, including seeds, and are targets for engineering these plant traits (Fritsche et al., 2017). Wei et al. (2024) identified 16 genes involved in tocopherol biosynthesis in the seed hemp genome, including six homologs of the A. thaliana VTE loci. This finding is a valuable starting point for reverse genetics research aimed at understanding the synthesis of hemp tocopherols, and modulating their content in seed oil.
Fibers
The utilization of hemp for the production of fibers is one of the most ancient uses of this crop (Ren et al., 2021; Smart et al., 2022). Hemp fibers are essentially made up of cell walls and are located in the stems of the plants (Petit, 2020; Smart et al., 2022). They comprise bast fibers—originating from the cambium, rich in crystalline cellulose and low in lignin—and hurd fibers (or shives)—forming the woody core of the hemp stems, with a high content of lignin and xylan-rich hemicellulosic polysaccharides (Van der Werf et al., 1994; Salentijn et al., 2015). Bast fibers are the most valuable hemp fibers, and the maximization of their yield and quality are pre-eminent targets of industrial hemp breeding (Salentijn et al., 2015). The quality of bast fibers involves the composition of the cell walls constituting the fibers themselves, and is maximized when fibers are rich in crystalline cellulose with a low angle of cellulose microfiber deposition, and display a reduced content of lignin and xylan hemicelluloses (Salentijn et al., 2015; Petit, 2020).
The yield and quality of hemp bast fibers have a strong genetic basis, as shown by the high heritability of these traits (Hennink, 1994; Petit et al., 2020c). Nevertheless, the study of their genetic architecture is difficult, as both fiber yield and fiber quality are complex traits, controlled by several genomic loci and genes with intricated pleiotropic effects (Petit et al., 2020b). The study of the genetics underlying complex traits typically involves genetic mapping to identify regions and genes associated with patterns of trait variability in the progenies of crossings of phenotypically contrasting lines, or in phenotypically diverse panels of accessions. In this context, the work of Petit et al. (2020b) pioneered genetic mapping in hemp aimed at uncovering the genetic architecture of fiber yield and quality. These analyses uncovered 16 QTLs associated with hemp fiber yield and quality variability across diverse environments, as well as multiple candidate genes underlying the QTLs (Petit et al., 2020b). Notably, a large proportion of these candidate genes is involved in the synthesis of lignin and pectins (Petit et al., 2020b). As such, selection to reduce the content of these cell wall constituents may represent a promising strategy to improve the quality of bast fibers. Interestingly, protocols to screen these phenotypes in hemp at the scale of breeding programs have been developed (Petit et al., 2019), and can thus be used in selective breeding schemes.
In the absence of further studies on the genetic basis of fiber yield and quality in hemp, information on this trait from other plant species can be highly valuable. As hemp bast fibers are essentially constituted of cell walls, research on the genetics underlying variability in cell wall composition conducted in grasses, forage crops, and model species such as A. thaliana is particularly relevant. Notably, these studies have largely elucidated the biosynthetic pathway of lignin (Bonawitz and Chapple, 2010; Yoon et al., 2015) and have shown its amenability to genetic engineering (Ralph et al., 2019; Carpita and McCann, 2020). Specifically, knockouts of key lignin genes such as CINNAMATE 4-HYDROXYLASE (C4H), 4-COUMARATE:CoA LIGASE (4CL), CINNAMATE 3-HYDROXYLASE (C3H), CINNAMOYL REDUCTASE (CCR), and CAFFEOYL O-METHYLTRANSFERASE (COMT) all determine considerable reductions of lignin content in the cell walls and tissues of diverse plant species (Leple et al., 2007; Sattler et al., 2009, 2012; Bjurhager et al., 2010; Ralph et al., 2012; Saballos et al., 2012; Xiang et al., 2017). Therefore, the mining of these genes in the hemp genome and their knockout could represent valuable strategies to reduce lignin content in hemp bast fibers. However, tissue-specific patterns of gene expression should probably be studied, as lignin genes such as those just mentioned typically form multigene families whose members take part in lignin deposition in specific tissues or cells (Chantreau et al., 2014; Le Roy et al., 2017; MacMillan et al., 2017). Accordingly, in hemp it would be important to ensure that reduction of lignin content is confined to bast fibers, while extensive crop growth sustained by the woody core of hemp stems can still take place.
Regarding the possibility of increasing the crystalline cellulose content in hemp bast fibers, research in other crops showed that the genetics underlying this trait is more complex and less understood than those of lignin biosynthesis. Given the current status of plant cell wall research, the most likely relevant target genes to modify these traits are the CELLULOSE SYNTHASE (CESA) genes, whose down-regulation and overexpression affect both the total cellulose content and cellulose crystallinity in different plant species (Harris et al., 2009; Joshi et al., 2011; Jayawardhane et al., 2020). Moreover, multiple CESA genes are specifically responsible for the deposition of plant fibers, for example in cotton (Li et al., 2013, 2016), making these genes relevant in the context of fiber improvement. Research demonstrated that cotton-specific patterns in the genomic diversification and physical organization of multiple CESA gene copies probably underly the massive deposition of fibers observed in cotton (Pancaldi et al., 2022a). In this respect, mining the hemp homologs of functionally studied CESA genes could identify gene targets for screenings of allelic diversity and copy number variation, or for genetic engineering approaches to modify the cellulose properties of fibers. To conclude, it is noteworthy that the genomic organization of cell wall genes has co-evolved with their differential functionalization across both different members of multigene families and different plant species (Pancaldi et al., 2022a, 2023a). Therefore, it is important to consider the ‘genomic contexts’ (Dewey, 2011) of specific gene copies when mining homologs of target genes between diverse plant clades.
Flowering behavior and sex determination
The regulation of flowering time and the mechanisms underlying sex determination are crucial hemp traits affecting plant growth and the yield and quality of the major harvestable hemp products (Faux et al., 2013; Salentijn et al., 2019). Regarding flowering, hemp is a short-day crop with a quantitative flowering phenology resulting from the interaction of genetic and metabolic factors with environmental variables such as specific photoperiod length and temperature (Salentijn et al., 2019; Petit et al., 2020a). The onset of flowering marks the end of vegetative growth, the start of bast fiber formation, and an intensified lignin deposition in stems (Keller et al., 2001; Faux et al., 2013; Liu et al., 2015). In parallel, the time between flowering onset and hemp harvest is crucial to optimize the content and composition of seed oil and proteins (Amaducci et al., 2015). Hemp has an XY sex chromosomes system, which underlies the prevalence of dioecious female (XX) and male (XY) plants (Salentijn et al., 2019; Smart et al., 2022). However, monoecious accessions also exist and are generally genetic females (XX) with quantitative variation in the proportion of female and male flowers, indicating that sex regulation is not entirely controlled by sex chromosomes (Faux et al., 2016; Petit et al., 2020a). Notably, monoecious cultivars display more uniform plant height and total stem and seed production (which are all traits suitable for dual harvest of stems and seeds), while the total production and quality of bast fibers are generally higher in dioecious accessions (Salentijn et al., 2019). Controlling sex is therefore crucial to obtain hemp cultivars fitted to specific applications (Moliterni et al., 2004; Salentijn et al., 2019).
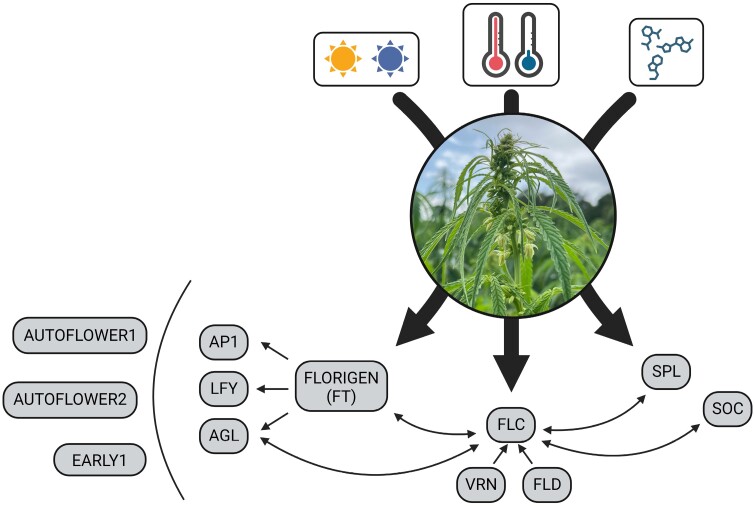
Current understanding of the genetics underlying hemp flowering behavior is not complete. Nevertheless, it is clear that this trait has a strong genetic basis and is highly heritable (Petit et al., 2020c). Moreover, studies on separate species identified several genes involved in flowering time regulation, highlighting the complex nature of this trait in plants [see Freytes et al. (2021) for an updated review]. Flowering-related genes belong to different, interacting, signaling pathways, including the photoperiod-dependent flowering pathway, the temperature-dependent pathway, and the endogenous pathway that incorporates stimuli from internal hormones, metabolites, and aging factors (Zhang et al., 2014; Salentijn et al., 2019) (Fig. 3). Some of the genes in these pathways are well studied and conserved across plant species, such as FLOWERING LOCUS T (FT or FLORIGEN; a major flowering promoter gene responsive to photoperiod and highly conserved across plants) (Wickland and Hanzawa, 2015), FLOWERING LOCUS C (FLC; a central flowering repressor responsive to temperature and regulated by multiple transcription factors) (Ruelens et al., 2013; Cheng et al., 2017), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC; a conserved gene responsive to multiple flowering pathways and promoting floral meristem) (Immink et al., 2012), and SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE genes (SPL genes; involved in the endogenous flowering pathway and conserved across angiosperms) (Chen et al., 2010; Preston and Hileman, 2013). Moreover, these primary flowering genes interact with several other genes that can be extensively conserved across plants as well, including APETALA1, LEAFY, AGAMOUS-LIKE PROTEIN, VERNALIZATION (VRN), and FLOWERING LOCUS-D (Thomson and Wellmer, 2019; Sharma et al., 2020) (Fig. 3). The increased availability of hemp genomic resources allowed investigations of the genetics of flowering regulation also in hemp. Petit et al. (2020a) performed the first genome-wide association study (GWAS) on this trait, anchored to the draft hemp genome of Van Bakel et al. (2011). More recently, other genetic mapping studies were performed by Woods et al. (2021), Toth et al. (2022), and Dowling et al. (2024). Overall, these studies found several hemp QTLs associated with variability in flowering time, with candidate genes often corresponding to known critical flowering regulators. For example, the flowering-related QTLs of Petit et al. (2020a) contain homologs of FT, FLC, FLD, SOC1, SPL, and VRN genes, highlighting the likely importance of these flowering genes also in hemp (Fig. 3). More recently, Toth et al. (2022) successfully mapped on hemp chromosome 1 two major loci—AUTOFLOWER1 and EARLY1—controlling, respectively, photoperiod insensitivity and flowering time across multiple hemp populations. Importantly, molecular markers were also developed for both loci (Toth et al., 2022), allowing high-throughput screening of hemp material for key flowering alleles to tailor novel hemp varieties to specific latitudes and agricultural rotations. Another locus—AUTOFLOWER2—was identified by Dowling et al. (2024) on hemp chromosome 8, most probably caused by a hemp FT gene. Finally, Steel et al. (2023) performed comparative mapping of flowering-related loci from several hemp mapping studies, finding ample co-localizations for some QTLs and genes identified across them. Similarly, co-localization between QTLs for flowering, agronomic, and morphological hemp traits was also found in a separate QTL mapping study (Woods et al., 2021). Overall, these observations suggest that translating the ample knowledge on flowering time regulation available in model species to hemp can represent a promising strategy to unveil the complete genetic architecture of hemp flowering time. Moreover, the reported co-localizations between QTLs for flowering time and other traits indicate opportunities for the parallel improvement of multiple traits in hemp, by careful selection of the diversity in initial material.
Fig. 3.
Representation of the major hemp flowering pathways (photoperiod-dependent, temperature-dependent, and endogenous) and the main genes involved in those pathways, based on genetic knowledge on flowering regulation for both hemp and other plant species. These genes represent interesting targets for hemp improvement. Arrows between genes indicate known genetic interactions among them. AP1, Apetala1, LFY, Leafy, AGL, Agamous-like protein; FT, Flowering locus T (or Florigen); FLC, Flowering Locus C; VRN, Vernalization; FLD, Flowering Locus D; SPL, Squamosa Protein-like; SOC, Suppressor of Overexpression of Constans 1.
Similarly to flowering behavior, the genetic factors underlying hemp sex determination are also not fully understood. Still, this trait is also highly heritable, meaning that it has a strong genetic basis, whose core lies in the XY hemp sex chromosomes (Faux et al., 2014; Petit et al., 2020c). The hemp system of XY chromosomes is one of the oldest among plants, firstly evolved in the hemp–hop ancestor 20–25 million years ago (Prentout et al., 2020, 2021). The X and Y chromosomes of hemp differ in size, with the Y chromosome being ~40 Mbp larger than the X chromosome due to a loading of long interspersed nuclear element (LINE)-like retrotransposons (Sakamoto et al., 1998; Van Bakel et al., 2011; Divashuk et al., 2014). The accumulation of LINE-like retrotransposons in the hemp Y chromosome underlies the suppression of recombination over a portion of the X–Y chromosomes, as well as their still-evolving structural heteromorphism (Moliterni et al., 2004; Vergara et al., 2016; Kovalchuk et al., 2020), despite the occurrence of a relatively large pseudo-autosomal recombining region (Peil et al., 2003; Prentout et al., 2021). Nevertheless, hemp sex determination seems to rely on the X-to-autosomes balance rather than on a purely Y-active mechanism (Faux et al., 2014). Moreover, the occurrence of variable proportions of male and female flowers in female (XX) monoecious hemp cultivars suggests interaction between sex chromosomes and other regions of the hemp genome to determine the phenotypic expression of hemp sex (Faux et al., 2016; Salentijn et al., 2019; Petit et al., 2020a). These observation explain why the mapping of stable male-linked sex markers is a notoriously difficult task in hemp (Kovalchuk et al., 2020), even if high-throughput genotyping methods have recently allowed the development of effective assays (Toth et al., 2020; Torres et al., 2022). Moreover, studying the genes located on the hemp sex chromosomes both on their own and in relation to other targets on the autosomal chromosomes could be promising to depict the mechanisms underlying hemp sex determination. In this direction, Prentout and co-workers applied comparative RNA-seq analysis to identify a common set of 112 sex-linked genes across hemp and hop (Prentout et al., 2020, 2021), which appear particularly relevant to identify targets underlying the X and Y chromosome-dependent components of sex determination in hemp. An RNA-seq approach was also used by Adal et al. (2021), who identified ~200 genes displaying up-regulation upon chemical induction of male flowers in genetically female C. sativa plants, including genes involved in anther and pollen development, as well as hormone signaling. Genes related to hormone signaling, including auxin- and gibberellin-responsive transcription factors and bZIP genes regulating auxin and gibberellin balances, were also found to be related to hemp sex determination by Petit et al. (2020a), who performed a GWAS on this trait in a hemp panel including monoecious cultivars. Overall, these studies show that integrating genomic, transcriptomic, and genetic mapping approaches at the whole-genome level retains the potential to further dissect the genetic architecture of sex determination in hemp. As such, the combined profiling of the expression of hemp genes from sex and autosomal chromosomes, along with their comparative functional analysis in monoecious male and female accessions, could be performed to reveal interactive networks underlying hemp sex determination.
The future of hemp improvement guided by novel tools
Following the resurgence in interest toward hemp as a multipurpose crop and the loosened restrictions to hemp cultivation in some countries (Devkota, 2022; Rathi et al., 2022), the research conducted over the last decades have been of paramount importance in supporting hemp improvement. However, hemp currently remains underimproved, far from reaching its potential as a multipurpose crop (Salentijn et al., 2015; Smart et al., 2022). Specifically, great efforts in the development of genetic and genomic hemp resources have been made, but the genetic knowledge on major traits is still incomplete. Moreover, the translation of fundamental hemp research into superior marketed varieties has been limited so far (Salentijn et al., 2015; Smart et al., 2022).
Further study and improvement of different hemp traits will require different strategies, as the genetic factors underlying different characters differ significantly in complexity, genomic structure, and level of understanding. For example, the application of genomic selection coupled with high-throughput CBDA phenotyping could be explored to attain increased CBDA yields, by selecting favorable alleles at genomic loci involved in cannabinoid biosynthesis (McKernan et al., 2020, Preprint; Innes and Vergara, 2023; Lynch et al., 2024, Preprint). So far, genomic selection has not been performed in hemp, but the availability of multiple high-quality genome assemblies (Grassa et al., 2021; Wei et al., 2024) and of ever cheaper genotyping technologies allows for pursuing this goal. Alternatively, methods for targeted sequencing [see, for example, Scaglione et al. (2019)] could also be applied by restricting genotyping to CBDA-associated loci while increasing the accuracy of detecting and characterizing diverse alleles with a positive effect on CBDA synthesis. Regarding phenotyping, near infrared spectroscopy is emerging as a valuable high-throughput methodology to analyze the content of different cannabinoids—including CBDA and THCA—in diverse types of hemp material (Callado et al., 2018; Birenboim et al., 2022; Tran et al., 2023), allowing the screening of large populations for the content of these compounds. Finally, hemp polyploidization through in vitro approaches or crossing of genotypes with different ploidy levels can also be investigated further as a strategy to increase hemp CBDA yield. In principle, polyploid hemp accessions are expected to display increased CBDA concentrations and/or CBDA yield (Crawford et al., 2021; Suchoff et al., 2024). However, the studies performed are not conclusive in this respect, deserving further research (Mansouri and Bagheri, 2017; Crawford et al., 2021). Nevertheless, the creation of sterile triploid hemp genotypes appears to be a good strategy to prevent reduction of CBDA yield due to uncontrolled cross-pollination (Kurtz et al., 2020).
In addition to improving CBDA yield, the modulation of the content and fatty acid composition of hemp seed oil is also an essential trait, particularly to fit hemp oil to specific uses, such as food and cosmetic applications (for which high amounts of PUFAs, particularly linoleic and α-linolenic fatty acids, is desirable) or biofuel production (for which a high amount of oxidative stable fatty acids, such as oleic acid, represents an important target) (Callaway, 2004; Vogl et al., 2004; Carlsson et al., 2011). In this regard, similarly to what was discussed for cannabinoids, the screening of sequence and structural genetic variation through genotyping-based or whole-genome resequencing methods can also be applied to the loci underlying hemp seed oil synthesis (Wei et al., 2024). Specifically, favorable alleles, copy number variation, or presence/absence variation could be searched for genes such as SAD, FAD, and FAE, which are key to regulate the oil content of oleic acid, PUFAs, and elongated fatty acids, respectively (Bates et al., 2013; Bielecka et al., 2014; Wei et al., 2024). For example, copy number expansion or selection for more active alleles at the SAD loci should favor the synthesis of oleic acid for biofuel applications (Wang et al., 2024). The same would be expected by selecting for reduced copy number and non-functional alleles at the hemp FAD and FAE loci (Carlsson et al., 2011; Bielecka et al., 2014). Conversely, boosting the activity of FAD genes can promote the accumulation of PUFAs, which would be beneficial for food or cosmetic applications (Wang et al., 2021). The achievement of these goals will be facilitated by using high-throughput methods for phenotyping seed oil content and composition in large plant populations. For this purpose, both NMR and infrared spectroscopy are well-established methodologies across different plant species (Jasinski et al., 2016; Melchinger et al., 2018; Anderson et al., 2019), and a protocol for their use in hemp has recently been developed (Siudem et al., 2019). Finally, the generally high heritability of seed oil content and composition (Schierholt and Becker, 2001; Bachlava et al., 2008; Khan et al., 2010) allows for crossing superior accessions identified through genetic screening and directly operates genomically informed selection of superior material from progenies. To this aim, it would also be worth developing seed hemp mapping populations both to evaluate genes in the oil pathway that are of particular relevance to determine desired patterns of oil content and quality variation, as well as to mine molecular markers that can be used in selective breeding to deal with linkage drag. These types of studies are currently missing for hemp seed oil traits.
Genomic engineering aimed at boosting or constraining specific branches of the hemp oil pathway is also a valid strategy to improve seed oil content or composition. This approach would be particularly suited to optimize hemp seed oil properties in plant material already improved for other traits such as CBDA profile, which could be difficult to keep genetically stable while breeding for other traits (Ingvardsen and Brinch-Pedersen, 2023). It is noteworthy that different tools to enable genomic engineering in hemp have recently been developed. These include the achievement of successful transformation and regeneration of hemp plants from hypocotyls (Galán-Ávila et al., 2021), despite hemp’s notorious recalcitrance to these techniques (Ingvardsen and Brinch-Pedersen, 2023). Moreover, Zhang et al. (2021) recently reported the first successful example of CRISPR/Cas9-mediated genome editing in hemp, consisting of the knockout of the PHYTOENE DESATURASE gene. Altogether, these methods could be applied in cis-genesis or gene editing experiments to overexpress one or more genes participating in oleic acid synthesis in plastids, from ACETYL-Coa CARBOXYLASE to SAD genes, as their overexpression typically translates to higher seed oil content. In parallel, FAD genes could be either down-regulated or overexpressed, to aim at higher oleic acid or PUFAs in hemp oil. This type of research could also make the oil pathway a model for the development of genome editing techniques in hemp.
Compared with cannabinoids and oil, knowledge of the genetics underlying fiber yield and quality, flowering behavior, and sex determination in hemp is more fragmented. In this sense, further genetic mapping studies should be performed, as they have proven to be valuable tools to identify genes and markers for these traits in hemp (Faux et al., 2016; Petit et al., 2020a, b; Toth et al., 2022). Moreover, in parallel to ‘conventional’ QTL mapping and genome-wide association approaches, bulk segregant analysis (BSA) should also be considered as an approach for variable hemp traits that probably rely on a limited number of loci, such as sex determination in monoecious hemp accessions (Salentijn et al., 2019). For this trait, BSA between pools of plants displaying extremely high proportions of male or female flowers, respectively, could reveal relevant underlying loci. In turn, comparative genomics could be used to further characterize those loci based on functional information from other species. More broadly, the translation of characterized genomic loci from (model) plants to a crop such as hemp is also a promising approach to uncover the genetics of complex traits in C. sativa. In this respect, pipelines have recently been developed for projecting multiple genomic loci underlying quantitative traits in diverse model species to target crops, as well as for analyzing allelic variation of the projected loci in the target crops (Pancaldi et al., 2022b, 2023b). These strategies exploit large-scale and high-throughput analysis of genome synteny (Zhao and Schranz, 2017) to translate entire syntenically conserved QTLs across species (Pancaldi et al., 2023b) and identify underlying functionally conserved genes based on conserved ‘genomic contexts’ (Dewey, 2011; Zhao and Schranz, 2017; Pancaldi et al., 2023b). Collectively, these methods can quickly identify conserved loci and genes underlying traits of interest in understudied crops, and characterize (favorable) variation at such loci by using, for example, whole-genome re-sequencing data. In turn, accessions carrying superior alleles can be selected for by targeted genotyping or sequencing of diverse populations and included in breeding programs, accelerating crop improvement. Breeding gain can be maximized if recurrent breeding schemes are applied, with the crossing of novel superior material with well-performing lines (Smart et al., 2022). In this respect, speed breeding can also be considered to maximize the number of crosses and generations per year, and a method to apply speed breeding in hemp has recently been developed (Schilling et al., 2023).
Conclusion
Hemp is a multipurpose crop with high potential to fuel diverse valuable industrial applications in a developing bio-based economy. From this perspective, the recent loosening of the restrictions on hemp cultivation in different countries has acted as a substantial boost to research and commercial interest in this crop. Accordingly, the last decades have already seen unprecedented development of genetic, genomic, and breeding resources for hemp. These developments have provided many new insights into important traits for successfully adopting hemp in agriculture and its products in industries. This review has highlighted future research areas, and proposed approaches in fundamental and applied genetics to further advance hemp. Unlocking the potential of this crop will require the integration of resources and tools in creative research efforts to deliver new insights in an effective, rapid, and cost-effective manner. For this purpose, all the diverse resources and tools available on key traits for hemp and other crops need to be collectively analyzed with innovative bioinformatic approaches, and the insights obtained should be translated into applied achievements by using the most cutting-edge breeding strategies and tools—from precise genomic engineering to speed breeding.
Contributor Information
Francesco Pancaldi, Plant Breeding, Wageningen University & Research, Droevendaalsesteeg 1, 6708PB, Wageningen, The Netherlands.
Elma M J Salentijn, Plant Breeding, Wageningen University & Research, Droevendaalsesteeg 1, 6708PB, Wageningen, The Netherlands.
Luisa M Trindade, Plant Breeding, Wageningen University & Research, Droevendaalsesteeg 1, 6708PB, Wageningen, The Netherlands.
Ricarda Jost, La Trobe University, Australia.
Author contributions
FP: writing, with input from EMJS and LMT. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflict of interest.
Funding
This research was part of a project that received funding from the European Union’s Horizon 2020 Research and Innovation Program MIDAS (Marginal lands, industrial crops and innovative bio-based value chains) project, under grant agreement no. 101082070.
References
- Adal AM, Doshi K, Holbrook L, Mahmoud SS.. 2021. Comparative RNA-Seq analysis reveals genes associated with masculinization in female Cannabis sativa. Planta 253, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali E, Hussain S, Hussain N, Kakar KU, Shah JM, Zaidi SHR, Jan M, Zhang K, Khan MA, Imtiaz M.. 2022. Tocopherol as plant protector: an overview of tocopherol biosynthesis enzymes and their role as antioxidant and signaling molecules. Acta Physiologiae Plantarum 44, 20. [Google Scholar]
- Allen KD, McKernan K, Pauli C, Roe J, Torres A, Gaudino R.. 2019. Genomic characterization of the complete terpene synthase gene family from Cannabis sativa. PLoS One 14, e0222363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaducci S, Scordia D, Liu F, Zhang Q, Guo H, Testa G, Cosentino S.. 2015. Key cultivation techniques for hemp in Europe and China. Industrial Crops and Products 68, 2–16. [Google Scholar]
- Anand U, Pacchetti B, Anand P, Sodergren MH.. 2021. Cannabis-based medicines and pain: a review of potential synergistic and entourage effects. Pain Management 11, 395–403. [DOI] [PubMed] [Google Scholar]
- Anderson JV, Wittenberg A, Li H, Berti MT.. 2019. High throughput phenotyping of Camelina sativa seeds for crude protein, total oil, and fatty acids profile by near infrared spectroscopy. Industrial Crops and Products 137, 501–507. [Google Scholar]
- Andre CM, Hausman J-F, Guerriero G.. 2016. Cannabis sativa: the plant of the thousand and one molecules. Frontiers in Plant Science 7, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachlava E, Burton JW, Brownie C, Wang S, Auclair J, Cardinal AJ.. 2008. Heritability of oleic acid content in soybean seed oil and its genetic correlation with fatty acid and agronomic traits. Crop Science 48, 1764–1772. [Google Scholar]
- Bassolino L, Buti M, Fulvio F, Pennesi A, Mandolino G, Milc J, Francia E, Paris R.. 2020. In silico identification of MYB and bHLH families reveals candidate transcription factors for secondary metabolic pathways in Cannabis sativa L. Plants 9, 1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Stymne S, Ohlrogge J.. 2013. Biochemical pathways in seed oil synthesis. Current Opinion in Plant Biology 16, 358–364. [DOI] [PubMed] [Google Scholar]
- Bennewitz S, Bergau N, Tissier A.. 2018. QTL mapping of the shape of type VI glandular trichomes in tomato. Frontiers in Plant Science 9, 1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecka M, Kaminski F, Adams I, Poulson H, Sloan R, Li Y, Larson TR, Winzer T, Graham IA.. 2014. Targeted mutation of Δ12 and Δ15 desaturase genes in hemp produce major alterations in seed fatty acid composition including a high oleic hemp oil. Plant Biotechnology Journal 12, 613–623. [DOI] [PubMed] [Google Scholar]
- Birenboim M, Kengisbuch D, Chalupowicz D, Maurer D, Barel S, Chen Y, Fallik E, Paz-Kagan T, Shimshoni JA.. 2022. Use of near-infrared spectroscopy for the classification of medicinal cannabis cultivars and the prediction of their cannabinoid and terpene contents. Phytochemistry 204, 113445. [DOI] [PubMed] [Google Scholar]
- Bjurhager I, Olsson A-M, Zhang B, Gerber L, Kumar M, Berglund LA, Burgert I, Sundberg B, Salmén L.. 2010. Ultrastructure and mechanical properties of Populus wood with reduced lignin content caused by transgenic down-regulation of cinnamate 4-hydroxylase. Biomacromolecules 11, 2359–2365. [DOI] [PubMed] [Google Scholar]
- Blessing EM, Steenkamp MM, Manzanares J, Marmar CR.. 2015. Cannabidiol as a potential treatment for anxiety disorders. Neurotherapeutics 12, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C.. 2010. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annual Review of Genetics 44, 337–363. [DOI] [PubMed] [Google Scholar]
- Boonnoy P, Karttunen M, Wong-Ekkabut J.. 2017. Alpha-tocopherol inhibits pore formation in oxidized bilayers. Physical Chemistry Chemical Physics 19, 5699–5704. [DOI] [PubMed] [Google Scholar]
- Booth JK, Page JE, Bohlmann J.. 2017. Terpene synthases from Cannabis sativa. PLoS One 12, e0173911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth JK, Yuen MM, Jancsik S, Madilao LL, Page JE, Bohlmann J.. 2020. Terpene synthases and terpene variation in Cannabis sativa. Plant Physiology 184, 130–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borin M, Palumbo F, Vannozzi A, Scariolo F, Sacilotto GB, Gazzola M, Barcaccia G.. 2021. Developing and testing molecular markers in Cannabis sativa (hemp) for their use in variety and dioecy assessments. Plants 10, 2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braich S, Baillie RC, Jewell LS, Spangenberg GC, Cogan NO.. 2019. Generation of a comprehensive transcriptome atlas and transcriptome dynamics in medicinal cannabis. Scientific Reports 9, 16583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braich S, Baillie RC, Spangenberg GC, Cogan NO.. 2020. A new and improved genome sequence of Cannabis sativa. Gigabyte 2020, gigabyte10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callado CS-C, Núñez-Sánchez N, Casano S, Ferreiro-Vera C.. 2018. The potential of near infrared spectroscopy to estimate the content of cannabinoids in Cannabis sativa L.: a comparative study. Talanta 190, 147–157. [DOI] [PubMed] [Google Scholar]
- Callaway J. 2004. Hempseed as a nutritional resource: an overview. Euphytica 140, 65–72. [Google Scholar]
- Callaway J, Laakkonen T.. 1997. Cultivation of Cannabis oil seed varieties in Finland. Journal of the International Hemp Association 3, 32–34. [Google Scholar]
- Carlsson AS, Yilmaz JL, Green AG, Stymne S, Hofvander P.. 2011. Replacing fossil oil with fresh oil—with what and for what? European Journal of Lipid Science and Technology 113, 812–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, McCann MC.. 2020. Redesigning plant cell walls for the biomass-based bioeconomy. Journal of Biological Chemistry 295, 15144–15157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerino P, Buonerba C, Cannazza G, D’Auria J, Ottoni E, Fulgione A, Di Stasio A, Pierri B, Gallo A.. 2021. A review of hemp as food and nutritional supplement. Cannabis and Cannabinoid Research 6, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalvin C, Drevensek S, Dron M, Bendahmane A, Boualem A.. 2020. Genetic control of glandular trichome development. Trends in Plant Science 25, 477–487. [DOI] [PubMed] [Google Scholar]
- Chantreau M, Portelette A, Dauwe R, et al. 2014. Ectopic lignification in the flax lignified bast fiber1 mutant stem is associated with tissue-specific modifications in gene expression and cell wall composition. The Plant Cell 26, 4462–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Pan Z.. 2021. Cannabidiol and terpenes from hemp—ingredients for future foods and processing technologies. Journal of Future Foods 1, 113–127. [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E.. 2011. The family of terpene synthases in plants: a mid‐size family of genes for specialized metabolism that is highly diversified throughout the kingdom. The Plant Journal 66, 212–229. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang Z, Liu D, Zhang K, Li A, Mao L.. 2010. SQUAMOSA promoter‐binding protein‐like transcription factors: star players for plant growth and development. Journal of Integrative Plant Biology 52, 946–951. [DOI] [PubMed] [Google Scholar]
- Cheng J-Z, Zhou Y-P, Lv T-X, Xie C-P, Tian C-E.. 2017. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiology and Molecular Biology of Plants 23, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S, Rojas BM, Crawford E, Otten M, Schoenenberger TA, Garfinkel AR, Chen H.. 2021. Characteristics of the diploid, triploid, and tetraploid versions of a cannabigerol-dominant F1 hybrid industrial hemp cultivar, Cannabis sativa ‘Stem Cell CBG’. Genes 12, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Niu T, Luo R, et al. 2024. Improvement of cannabidiolic acid synthetase activity through molecular docking and site-directed mutagenesis. Industrial Crops and Products 208, 117860. [Google Scholar]
- Devkota HP. 2022. Hemp (Cannabis sativa L.)—taxonomy, distribution and uses. In: Belwal T, Belwal NC, eds. Revolutionizing the potential of hemp and its products in changing the global economy. Cham: Springer, 1–10. [Google Scholar]
- Dewey CN. 2011. Positional orthology: putting genomic evolutionary relationships into context. Briefings in Bioinformatics 12, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divashuk MG, Alexandrov OS, Razumova OV, Kirov IV, Karlov GI.. 2014. Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS One 9, e85118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling CA, Shi J, Toth JA, Quade MA, Smart LB, McCabe PF, Melzer R, Schilling S.. 2024. A FLOWERING LOCUS T ortholog is associated with photoperiod-insensitive flowering in hemp (Cannabis sativa L.). The Plant Journal 119, 383–403. [DOI] [PubMed] [Google Scholar]
- Ewas M, Gao Y, Ali F, Nishawy EM, Shahzad R, Subthain H, Amar M, Martin C, Luo J.. 2017. RNA-seq reveals mechanisms of SlMX1 for enhanced carotenoids and terpenoids accumulation along with stress resistance in tomato. Science Bulletin 62, 476–485. [DOI] [PubMed] [Google Scholar]
- Ewas M, Gao Y, Wang S, et al. 2016. Manipulation of SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Science Bulletin 61, 1413–1418. [DOI] [PubMed] [Google Scholar]
- Faux A-M, Berhin A, Dauguet N, Bertin P.. 2014. Sex chromosomes and quantitative sex expression in monoecious hemp (Cannabis sativa L.). Euphytica 196, 183–197. [Google Scholar]
- Faux A-M, Draye X, Flamand M-C, Occre A, Bertin P.. 2016. Identification of QTLs for sex expression in dioecious and monoecious hemp (Cannabis sativa L.). Euphytica 209, 357–376. [Google Scholar]
- Faux A-M, Draye X, Lambert R, D’Andrimont R, Raulier P, Bertin P.. 2013. The relationship of stem and seed yields to flowering phenology and sex expression in monoecious hemp (Cannabis sativa L.). European Journal of Agronomy 47, 11–22. [Google Scholar]
- Ferber SG, Namdar D, Hen-Shoval D, Eger G, Koltai H, Shoval G, Shbiro L, Weller A.. 2020. The ‘entourage effect’: terpenes coupled with cannabinoids for the treatment of mood disorders and anxiety disorders. Current Neuropharmacology 18, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytes SN, Canelo M, Cerdán PD.. 2021. Regulation of flowering time: when and where? Current Opinion in Plant Biology 63, 102049. [DOI] [PubMed] [Google Scholar]
- Fritsche S, Wang X, Jung C.. 2017. Recent advances in our understanding of tocopherol biosynthesis in plants: an overview of key genes, functions, and breeding of vitamin E improved crops. Antioxidants 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulvio F, Paris R, Montanari M, et al. 2021. Analysis of sequence variability and transcriptional profile of cannabinoid synthase genes in Cannabis sativa L. chemotypes with a focus on cannabichromenic acid synthase. Plants 10, 1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulvio F, Righetti L, Minervini M, Moschella A, Paris R.. 2023. The B1080/B1192 molecular marker identifies hemp plants with functional THCA synthase and total THC content above legal limit. Gene 858, 147198. [DOI] [PubMed] [Google Scholar]
- Galán-Ávila A, Gramazio P, Ron M, Prohens J, Herraiz FJ.. 2021. A novel and rapid method for Agrobacterium-mediated production of stably transformed Cannabis sativa L. plants. Industrial Crops and Products 170, 113691. [Google Scholar]
- Galasso I, Russo R, Mapelli S, Ponzoni E, Brambilla IM, Battelli G, Reggiani R.. 2016. Variability in seed traits in a collection of Cannabis sativa L. genotypes. Frontiers in Plant Science 7, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassa CJ, Weiblen GD, Wenger JP, Dabney C, Poplawski SG, Motley TS, Michael TP, Schwartz CJ.. 2021. A new cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytologist 230, 1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero G, Behr M, Legay S, Mangeot-Peter L, Zorzan S, Ghoniem M, Hausman J-F.. 2017. Transcriptomic profiling of hemp bast fibres at different developmental stages. Scientific Reports 7, 4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiden SR, Apicella PV, Ma Y, Berkowitz GA.. 2022. Overexpression of CsMIXTA, a transcription factor from Cannabis sativa, increases glandular trichome density in tobacco leaves. Plants 11, 1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanuš LO, Hod Y.. 2020. Terpenes/terpenoids in cannabis: are they important? Medical Cannabis and Cannabinoids 3, 25–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D, Stork J, Debolt S.. 2009. Genetic modification in cellulose‐synthase reduces crystallinity and improves biochemical conversion to fermentable sugar. GCB Bioenergy 1, 51–61. [Google Scholar]
- Hennink S. 1994. Optimisation of breeding for agronomic traits in fibre hemp (Cannabis sativa L.) by study of parent–offspring relationships. Euphytica 78, 69–76. [Google Scholar]
- Hillig KW. 2005. Genetic evidence for speciation in Cannabis (Cannabaceae). Genetic Resources and Crop Evolution 52, 161–180. [Google Scholar]
- Immink RG, Posé D, Ferrario S, et al. 2012. Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiology 160, 433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvardsen CR, Brinch-Pedersen H.. 2023. Challenges and potentials of new breeding techniques in Cannabis sativa. Frontiers in Plant Science 14, 1154332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes PA, Vergara D.. 2023. Genomic description of critical cannabinoid biosynthesis genes. Botany 101, 270–283. [Google Scholar]
- Jasinski S, Lécureuil A, Durandet M, Bernard-Moulin P, Guerche P.. 2016. Arabidopsis seed content QTL mapping using high-throughput phenotyping: the assets of near infrared spectroscopy. Frontiers in Plant Science 7, 1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardhane KN, Singer SD, Ozga JA, Rizvi SM, Weselake RJ, Chen G.. 2020. Seed-specific down-regulation of Arabidopsis CELLULOSE SYNTHASE 1 or 9 reduces seed cellulose content and differentially affects carbon partitioning. Plant Cell Reports 39, 953–969. [DOI] [PubMed] [Google Scholar]
- Jiang S-Y, Jin J, Sarojam R, Ramachandran S.. 2019. A comprehensive survey on the terpene synthase gene family provides new insight into its evolutionary patterns. Genome Biology and Evolution 11, 2078–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MS, Wallace JG.. 2021. Genomic and chemical diversity of commercially available high-CBD industrial hemp accessions. Frontiers in Genetics 12, 682475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. 2019. Defining hemp: a fact sheet. Washington, DC: Congressional Research Service. [Google Scholar]
- Joshi CP, Thammannagowda S, Fujino T, et al. 2011. Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic aspen. Molecular Plant 4, 331–345. [DOI] [PubMed] [Google Scholar]
- Keller A, Leupin M, Mediavilla V, Wintermantel E.. 2001. Influence of the growth stage of industrial hemp on chemical and physical properties of the fibres. Industrial Crops and Products 13, 35–48. [Google Scholar]
- Khan NU, Marwat KB, Hassan G, Farhatullah SB, Makhdoom K, Ahmad W, Khan HU.. 2010. Genetic variation and heritability for cotton seed, fiber and oil traits in Gossypium hirsutum L. Pakistan Journal of Botany 42, 615–625. [Google Scholar]
- Kinney A. 1998. Production of specialised oils for industry. In: Harwood JL, ed. Plant lipid biosynthesis: fundamentals and agricultural applications. Cambridge: Cambridge University Press, 273–285. [Google Scholar]
- Kovalchuk I, Pellino M, Rigault P, Van Velzen R, Ebersbach J, Ashnest J, Mau M, Schranz M, Alcorn J, Laprairie R.. 2020. The genomics of cannabis and its close relatives. Annual Review of Plant Biology 71, 713–739. [DOI] [PubMed] [Google Scholar]
- Kriese U, Schumann E, Weber W, Beyer M, Brühl L, Matthäus B.. 2004. Oil content, tocopherol composition and fatty acid patterns of the seeds of 51 Cannabis sativa L. genotypes. Euphytica 137, 339–351. [Google Scholar]
- Kundan M, Gani U, Fayaz M, Angmo T, Kesari R, Rahul VP, Gairola S, Misra P.. 2022. Two R2R3-MYB transcription factors, CsMYB33 and CsMYB78 are involved in the regulation of anthocyanin biosynthesis in Cannabis sativa L. Industrial Crops and Products 188, 115546. [Google Scholar]
- Kurtz LE, Brand MH, Lubell-Brand JD.. 2020. Production of tetraploid and triploid hemp. HortScience 55, 1703–1707. [Google Scholar]
- Lange BM, Zager JJ.. 2022. Comprehensive inventory of cannabinoids in Cannabis sativa L.: can we connect genotype and chemotype? Phytochemistry Reviews 21, 1273–1313. [Google Scholar]
- Laverty KU, Stout JM, Sullivan MJ, et al. 2019. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Research 29, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leple J-C, Dauwe R, Morreel K, et al. 2007. Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. The Plant Cell 19, 3669–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy J, Blervacq A-S, Créach A, Huss B, Hawkins S, Neutelings G.. 2017. Spatial regulation of monolignol biosynthesis and laccase genes control developmental and stress-related lignin in flax. BMC Plant Biology 17, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Wang R, Li X, et al. 2016. Proteomic profiling of cellulase-aid-extracted membrane proteins for functional identification of cellulose synthase complexes and their potential associated-components in cotton fibers. Scientific Reports 6, 26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Xia T, Xu W, et al. 2013. An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense. Planta 237, 1585–1597. [DOI] [PubMed] [Google Scholar]
- Liu F-H, Hu H-R, Du G-H, Deng G, Yang Y.. 2017. Ethnobotanical research on origin, cultivation, distribution and utilization of hemp (Cannabis sativa L.) in China. Indian Journal of Traditional Knowledge 16, 235–242. [Google Scholar]
- Liu M, Fernando D, Daniel G, Madsen B, Meyer AS, Ale MT, Thygesen A.. 2015. Effect of harvest time and field retting duration on the chemical composition, morphology and mechanical properties of hemp fibers. Industrial Crops and Products 69, 29–39. [Google Scholar]
- Liu Y, Du Z, Li Y, Lu S, Tang S, Guo L.. 2024. Improving linolenic acid content in rapeseed oil by overexpression of CsFAD2 and CsFAD3 genes. Molecular Breeding 44, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston SJ, Quilichini TD, Booth JK, Wong DC, Rensing KH, Laflamme‐Yonkman J, Castellarin SD, Bohlmann J, Page JE, Samuels AL.. 2020. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. The Plant Journal 101, 37–56. [DOI] [PubMed] [Google Scholar]
- Livingston SJ, Rensing KH, Page JE, Samuels AL.. 2022. A polarized supercell produces specialized metabolites in cannabis trichomes. Current Biology 32, 4040–4047.e4. [DOI] [PubMed] [Google Scholar]
- Lynch RC, Padgitt-Cobb LK, Garfinkel AR, Knaus BJ, Hartwick NT, Allsing N, Aylward A, Mamerto A, Kitony JK, Colt K.. 2024. Domesticated cannabinoid synthases amid a wild mosaic cannabis pangenome. bioRxiv 2024.05. 21.595196. [Preprint]. [Google Scholar]
- MacMillan CP, Birke H, Chuah A, Brill E, Tsuji Y, Ralph J, Dennis ES, Llewellyn D, Pettolino FA.. 2017. Tissue and cell-specific transcriptomes in cotton reveal the subtleties of gene regulation underlying the diversity of plant secondary cell walls. BMC Genomics 18, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlberg PG, Kim ES.. 2004. Accumulation of cannabinoids in glandular trichomes of Cannabis (Cannabaceae). Journal of Industrial Hemp 9, 15–36. [Google Scholar]
- Mansouri H, Bagheri M.. 2017. Induction of polyploidy and its effect on Cannabis sativa L.In: Chandra S, Lata H, ElSohly M, eds. Cannabis sativa L.—Botany and biotechnology. Cham: Springer, 365–383. [Google Scholar]
- Matías‐Hernández L, Jiang W, Yang K, Tang K, Brodelius PE, Pelaz S.. 2017. AaMYB 1 and its orthologue AtMYB 61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. The Plant Journal 90, 520–534. [DOI] [PubMed] [Google Scholar]
- Matthäus B, Brühl L.. 2008. Virgin hemp seed oil: an interesting niche product. European Journal of Lipid Science and Technology 110, 655–661. [Google Scholar]
- McGarvey P, Huang J, McCoy M, et al. 2020. De novo assembly and annotation of transcriptomes from two cultivars of Cannabis sativa with different cannabinoid profiles. Gene 762, 145026. [DOI] [PubMed] [Google Scholar]
- McKernan KJ, Helbert Y, Kane LT, Ebling H, Zhang L, Liu B, Eaton Z, McLaughlin S, Kingan S, Baybayan P.. 2020. Sequence and annotation of 42 cannabis genomes reveals extensive copy number variation in cannabinoid synthesis and pathogen resistance genes. bioRxiv 2020.01.03.894428. [Preprint]. [Google Scholar]
- Melchinger A, Böhm J, Utz H, Müller J, Munder S, Mauch F.. 2018. High‐throughput precision phenotyping of the oil content of single seeds of various oilseed crops. Crop Science 58, 670–678. [Google Scholar]
- Mène-Saffrané L, Dellapenna D.. 2010. Biosynthesis, regulation and functions of tocochromanols in plants. Plant Physiology and Biochemistry 48, 301–309. [DOI] [PubMed] [Google Scholar]
- Moliterni VC, Cattivelli L, Ranalli P, Mandolino G.. 2004. The sexual differentiation of Cannabis sativa L.: a morphological and molecular study. Euphytica 140, 95–106. [Google Scholar]
- Momotaz A, Scott JW, Schuster DJ.. 2010. Identification of quantitative trait loci conferring resistance to Bemisia tabaci in an F2 population of Solanum lycopersicum× Solanum habrochaites accession LA1777. Journal of the American Society for Horticultural Science 135, 134–142. [Google Scholar]
- Morel A, Lebard P, Dereux A, Azuar J, Questel F, Bellivier F, Marie-Claire C, Fatséas M, Vorspan F, Bloch V.. 2021. Clinical trials of cannabidiol for substance use disorders: outcome measures, surrogate endpoints, and biomarkers. Frontiers in Psychiatry 12, 565617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti SS, Pollard M, Kosma DK, Allen C Jr, Ohlrogge JB, Barry CS.. 2012. Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiology 159, 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova P, Sal J.. 2019. Use of technical hemp for concrete—hempcrete. IOP Conference Series: Materials Science and Engineering 603, 052095. [Google Scholar]
- Nuutinen T. 2018. Medicinal properties of terpenes found in Cannabis sativa and Humulus lupulus. European Journal of Medicinal Chemistry 157, 198–228. [DOI] [PubMed] [Google Scholar]
- O’Brien K. 2022. Cannabidiol (CBD) in cancer management. Cancers 14, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomah BD, Busson M, Godfrey DV, Drover JC.. 2002. Characteristics of hemp (Cannabis sativa L.) seed oil. Food Chemistry 76, 33–43. [Google Scholar]
- Pancaldi F, Schranz ME, Van Loo EN, Trindade LM.. 2023a. Highly differentiated genomic properties underpin the different cell walls of Poaceae and eudicots. Plant Physiology 194, 274–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancaldi F, Van Loo EN, Schranz ME, Trindade LM.. 2022a. Genomic architecture and evolution of the cellulose synthase gene superfamily as revealed by phylogenomic analysis. Frontiers in Plant Science 13, 870818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancaldi F, Van Loo EN, Senio S, Al Hassan M, Van Der Cruijsen K, Paulo M-J, Dolstra O, Schranz ME, Trindade LM.. 2023b. Syntenic cell wall QTLs as versatile breeding tools: intraspecific allelic variability and predictability of biomass quality loci in target plant species. Plants 12, 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancaldi F, Vlegels D, Rijken H, Van Loo EN, Trindade LM.. 2022b. Detection and analysis of syntenic quantitative trait loci controlling cell wall quality in angiosperms. Frontiers in Plant Science 13, 855093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil A, Flachowsky H, Schumann E, Weber W.. 2003. Sex-linked AFLP markers indicate a pseudoautosomal region in hemp (Cannabis sativa L.). Theoretical and Applied Genetics 107, 102–109. [DOI] [PubMed] [Google Scholar]
- Petit J. 2020. Novel molecular tools to uncover the genetic architecture of hemp fibre quality. PhD thesis. Wageningen University.
- Petit J, Gulisano A, Dechesne A, Trindade LM.. 2019. Phenotypic variation of cell wall composition and stem morphology in hemp (Cannabis sativa L.): optimization of methods. Frontiers in Plant Science 10, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J, Salentijn EM, Paulo M-J, Denneboom C, Trindade LM.. 2020a. Genetic architecture of flowering time and sex determination in hemp (Cannabis sativa L.): a genome-wide association study. Frontiers in Plant Science 11, 569958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J, Salentijn EM, Paulo M-J, Denneboom C, Van Loo EN, Trindade LM.. 2020b. Elucidating the genetic architecture of fiber quality in hemp (Cannabis sativa L.) using a genome-wide association study. Frontiers in Genetics 11, 566314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J, Salentijn EM, Paulo M-J, Thouminot C, Van Dinter BJ, Magagnini G, Gusovius H-J, Tang K, Amaducci S, Wang S.. 2020c. Genetic variability of morphological, flowering, and biomass quality traits in hemp (Cannabis sativa L.). Frontiers in Plant Science 11, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porokhovinova E, Matveeva T, Khafizova G, Bemova V, Doubovskaya A, Kishlyan N, Podolnaya L, Gavrilova V.. 2022. Fatty acid composition of oil crops: genetics and genetic engineering. Genetic Resources and Crop Evolution 69, 2029–2045. [Google Scholar]
- Prentout D, Razumova O, Rhoné B, Badouin H, Henri H, Feng C, Käfer J, Karlov G, Marais GA.. 2020. An efficient RNA-seq-based segregation analysis identifies the sex chromosomes of Cannabis sativa. Genome Research 30, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentout D, Stajner N, Cerenak A, Tricou T, Brochier‐Armanet C, Jakse J, Käfer J, Marais GA.. 2021. Plant genera Cannabis and Humulus share the same pair of well‐differentiated sex chromosomes. New Phytologist 231, 1599–1611. [DOI] [PubMed] [Google Scholar]
- Preston JC, Hileman LC.. 2013. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Frontiers in Plant Science 4, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punja ZK, Sutton DB, Kim T.. 2023. Glandular trichome development, morphology, and maturation are influenced by plant age and genotype in high THC-containing cannabis (Cannabis sativa L.) inflorescences. Journal of Cannabis Research 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Akiyama T, Coleman HD, Mansfield SD.. 2012. Effects on lignin structure of coumarate 3-hydroxylase downregulation in poplar. Bioenergy Research 5, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Boerjan W.. 2019. Lignin structure and its engineering. Current Opinion in Biotechnology 56, 240–249. [DOI] [PubMed] [Google Scholar]
- Ranalli P, Venturi G.. 2004. Hemp as a raw material for industrial applications. Euphytica 140, 1–6. [Google Scholar]
- Rathi V, Singh G, Kumar P, Chaudhary M, Singh P, Mishra M.. 2022. Legality of worldwide cannabis use and associated economic benefits. In: Belwal T, Belwal NC, eds. Revolutionizing the potential of hemp and its products in changing the global economy. Cham: Springer, 27–68. [Google Scholar]
- Ren G, Zhang X, Li Y, et al. 2021. Large-scale whole-genome resequencing unravels the domestication history of Cannabis sativa. Science Advances 7, eabg2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvankhah A, Emam-Djomeh Z, Safari M, Askari G, Salami M.. 2019. Microwave-assisted extraction of hempseed oil: studying and comparing of fatty acid composition, antioxidant activity, physiochemical and thermal properties with Soxhlet extraction. Journal of Food Science and Technology 56, 4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelens P, De Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K.. 2013. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nature Communications 4, 2280. [DOI] [PubMed] [Google Scholar]
- Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C, Pedersen JF, Vermerris W.. 2012. Brown midrib2 (Bmr2) encodes the major 4‐coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor L. Moench). The Plant Journal 70, 818–830. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Akiyama Y, Fukui K, Kamada H, Satoh S.. 1998. Characterization; genome sizes and morphology of sex chromosomes in hemp (Cannabis sativa L.). Cytologia 63, 459–464. [Google Scholar]
- Salentijn EM, Petit J, Trindade LM.. 2019. The complex interactions between flowering behavior and fiber quality in hemp. Frontiers in Plant Science 10, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentijn EM, Zhang Q, Amaducci S, Yang M, Trindade LM.. 2015. New developments in fiber hemp (Cannabis sativa L.) breeding. Industrial Crops and Products 68, 32–41. [Google Scholar]
- Sattler SE, Palmer NA, Saballos A, Greene AM, Xin Z, Sarath G, Vermerris W, Pedersen JF.. 2012. Identification and characterization of four missense mutations in brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenergy Research 5, 855–865. [Google Scholar]
- Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF.. 2009. A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiology 150, 584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savadi S, Lambani N, Kashyap PL, Bisht DS.. 2017. Genetic engineering approaches to enhance oil content in oilseed crops. Plant Growth Regulation 83, 207–222. [Google Scholar]
- Scaglione D, Pinosio S, Marroni F, Di Centa E, Fornasiero A, Magris G, Scalabrin S, Cattonaro F, Taylor G, Morgante M.. 2019. Single primer enrichment technology as a tool for massive genotyping: a benchmark on black poplar and maize. Annals of Botany 124, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierholt A, Becker H.. 2001. Environmental variability and heritability of high oleic acid content in winter oilseed rape. Plant Breeding 120, 63–66. [Google Scholar]
- Schilling S, Melzer R, Dowling CA, Shi J, Muldoon S, McCabe PF.. 2023. A protocol for rapid generation cycling (speed breeding) of hemp (Cannabis sativa) for research and agriculture. The Plant Journal 113, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluttenhofer C, Yuan L.. 2017. Challenges towards revitalizing hemp: a multifaceted crop. Trends in Plant Science 22, 917–929. [DOI] [PubMed] [Google Scholar]
- Sharma N, Geuten K, Giri BS, Varma A.. 2020. The molecular mechanism of vernalization in Arabidopsis and cereals: role of Flowering Locus C and its homologs. Physiologia Plantarum 170, 373–383. [DOI] [PubMed] [Google Scholar]
- Shi J, Lang C, Wu X, Liu R, Zheng T, Zhang D, Chen J, Wu G.. 2015. RNAi knockdown of fatty acid elongase1 alters fatty acid composition in Brassica napus. Biochemical and Biophysical Research Communications 466, 518–522. [DOI] [PubMed] [Google Scholar]
- Shi P, Fu X, Shen Q, Liu M, Pan Q, Tang Y, Jiang W, Lv Z, Yan T, Ma Y.. 2018. The roles of AaMIXTA 1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytologist 217, 261–276. [DOI] [PubMed] [Google Scholar]
- Sirikantaramas S, Morimoto S, Shoyama Y, Ishikawa Y, Wada Y, Shoyama Y, Taura F.. 2004. The gene controlling marijuana psychoactivity: molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. Journal of Biological Chemistry 279, 39767–39774. [DOI] [PubMed] [Google Scholar]
- Siudem P, Wawer I, Paradowska K.. 2019. Rapid evaluation of edible hemp oil quality using NMR and FT-IR spectroscopy. Journal of Molecular Structure 1177, 204–208. [Google Scholar]
- Small E. 2015. Evolution and classification of Cannabis sativa (marijuana, hemp) in relation to human utilization. The Botanical Review 81, 189–294. [Google Scholar]
- Smart LB, Toth JA, Stack GM, Monserrate LA, Smart CD.. 2022. Breeding of hemp (Cannabis sativa). Plant Breeding Reviews 46, 239–288. [Google Scholar]
- Sommano SR, Chittasupho C, Ruksiriwanich W, Jantrawut P.. 2020. The cannabis terpenes. Molecules 25, 5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack GM, Snyder SI, Toth JA, et al. 2023. Cannabinoids function in defense against chewing herbivores in Cannabis sativa L. Horticulture Research 10, uhad207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack GM, Toth JA, Carlson CH, Cala AR, Marrero‐González MI, Wilk RL, Gentner DR, Crawford JL, Philippe G, Rose JK.. 2021. Season‐long characterization of high‐cannabinoid hemp (Cannabis sativa L.) reveals variation in cannabinoid accumulation, flowering time, and disease resistance. GCB Bioenergy 13, 546–561. [Google Scholar]
- Steel L, Welling M, Ristevski N, Johnson K, Gendall A.. 2023. Comparative genomics of flowering behavior in Cannabis sativa. Frontiers in Plant Science 14, 1227898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchoff DH, Inoa SH, Stack GM, Wares AJ, Snyder SI, Murdock MJ, Rose JK, Smart LB, Caton TA, Pearce RC.. 2024. Characterization of agronomic performance and sterility in triploid and diploid cannabinoid hemp. Agronomy Journal 116, 2470–2482. [Google Scholar]
- Tahir MN, Shahbazi F, Rondeau-Gagné S, Trant JF.. 2021. The biosynthesis of the cannabinoids. Journal of Cannabis Research 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Xu Y, Gao F, Cheng C, Deng C, Chen J, Yuan X, Zhang X, Su J.. 2023. Transcriptomic and metabolomic analyses reveal the differential accumulation of phenylpropanoids and terpenoids in hemp autotetraploid and its diploid progenitor. BMC Plant Biology 23, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney CA, Backer R, Geitmann A, Smith DL.. 2021. Cannabis glandular trichomes: a cellular metabolite factory. Frontiers in Plant Science 12, 721986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura F, Sirikantaramas S, Shoyama Y, Yoshikai K, Shoyama Y, Morimoto S.. 2007. Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Letters 581, 2929–2934. [DOI] [PubMed] [Google Scholar]
- Thomson B, Wellmer F.. 2019. Molecular regulation of flower development. Current Topics in Developmental Biology 131, 185–210. [DOI] [PubMed] [Google Scholar]
- Tian B, Wei F, Shu H, Zhang Q, Zang X, Lian Y.. 2011. Decreasing erucic acid level by RNAi-mediated silencing of fatty acid elongase 1 (BnFAE1.1) in rapeseeds (Brassica napus L.). African Journal of Biotechnology 10, 13194–13201. [Google Scholar]
- Torres A, Pauli C, Givens R, Argyris J, Allen K, Monfort A, Gaudino RJ.. 2022. High-throughput methods to identify male Cannabis sativa using various genotyping methods. Journal of Cannabis Research 4, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth JA, Stack GM, Cala AR, Carlson CH, Wilk RL, Crawford JL, Viands DR, Philippe G, Smart CD, Rose JK.. 2020. Development and validation of genetic markers for sex and cannabinoid chemotype in Cannabis sativa L. GCB Bioenergy 12, 213–222. [Google Scholar]
- Toth JA, Stack GM, Carlson CH, Smart LB.. 2022. Identification and mapping of major-effect flowering time loci Autoflower1 and Early1 in Cannabis sativa L. Frontiers in Plant Science 13, 991680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran J, Vassiliadis S, Elkins AC, Cogan NO, Rochfort SJ.. 2023. Developing prediction models using near-infrared spectroscopy to quantify cannabinoid content in Cannabis sativa. Sensors 23, 2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Überall MA. 2020. A review of scientific evidence for THC: CBD oromucosal spray (nabiximols) in the management of chronic pain. Journal of Pain Research 13, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE.. 2011. The draft genome and transcriptome of Cannabis sativa. Genome Biology 12, R102–R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Werf HM, Van Der Veen JH, Bouma A, Ten Cate M.. 1994. Quality of hemp (Cannabis sativa L.) stems as a raw material for paper. Industrial Crops and Products 2, 219–227. [Google Scholar]
- van Velzen R, Schranz ME.. 2021. Origin and evolution of the cannabinoid oxidocyclase gene family. Genome Biology and Evolution 13, evab130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara D, Baker H, Clancy K, Keepers KG, Mendieta JP, Pauli CS, Tittes SB, White KH, Kane NC.. 2016. Genetic and genomic tools for Cannabis sativa. Critical Reviews in Plant Sciences 35, 364–377. [Google Scholar]
- Vogl CR, Mölleken H, Lissek-Wolf G, Surböck A, Kobert J.. 2004. Hemp (Cannabis sativa L.) as a resource for green cosmetics: yield of seed and fatty acid compositions of 20 varieties under the growing conditions of organic farming in Austria. Journal of Industrial Hemp 9, 51–68. [Google Scholar]
- Wang J, Liu Z, Liu H, Peng D, Zhang J, Chen M.. 2021. Linum usitatissimum FAD2A and FAD3A enhance seed polyunsaturated fatty acid accumulation and seedling cold tolerance in Arabidopsis thaliana. Plant Science 311, 111014. [DOI] [PubMed] [Google Scholar]
- Wang J-J, Shao Y-N, Xin Y, Zhang C, Yuan G, Liu Z-J, Chen M-X.. 2024. Heterogeneous expression of stearoyl-acyl carrier protein desaturase genes SAD1 and SAD2 from Linum usitatissimum enhances seed oleic acid accumulation and seedling cold and drought tolerance capability in Brassica napus. Journal of Integrative Agriculture 23, 1864–1878. [Google Scholar]
- Wei H, Yang Z, Niyitanga S, et al. 2024. The reference genome of seed hemp (Cannabis sativa) provides new insights into fatty acid and vitamin E synthesis. Plant Communications 5, 100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickland DP, Hanzawa Y.. 2015. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Molecular Plant 8, 983–997. [DOI] [PubMed] [Google Scholar]
- Woods P, Campbell BJ, Nicodemus TJ, Cahoon EB, Mullen JL, McKay JK.. 2021. Quantitative trait loci controlling agronomic and biochemical traits in Cannabis sativa. Genetics 219, iyab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Sen SK, Min D, Savithri D, Lu F, Jameel H, Chiang V, Chang H-M.. 2017. Field-grown transgenic hybrid poplar with modified lignin biosynthesis to improve enzymatic saccharification efficiency. ACS Sustainable Chemistry & Engineering 5, 2407–2414. [Google Scholar]
- Xie Z, Mi Y, Kong L, Gao M, Chen S, Chen W, Meng X, Sun W, Chen S, Xu Z.. 2023. Cannabis sativa: origin and history, glandular trichome development, and cannabinoid biosynthesis. Horticulture Research 10, uhad150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Kong L, Ren W, Wang Z, Tang L, Wu W, Liu X, Ma W, Zhang S.. 2024. Identification and expression analysis of TPS family gene in Cannabis sativa L. Heliyon 10, e27817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Chen M, Shen Q, et al. 2017. HOMEODOMAIN PROTEIN 1 is required for jasmonate‐mediated glandular trichome initiation in Artemisia annua. New Phytologist 213, 1145–1155. [DOI] [PubMed] [Google Scholar]
- Yang C, Gao Y, Gao S, Yu G, Xiong C, Chang J, Li H, Ye Z.. 2015. Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genomics 6, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Choi H, An G.. 2015. Roles of lignin biosynthesis and regulatory genes in plant development. Journal of Integrative Plant Biology 57, 902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen G, Tang B, Zhang J, Zhou S, Hu Z.. 2018. The jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Science 267, 65–73. [DOI] [PubMed] [Google Scholar]
- Zager JJ, Lange I, Srividya N, Smith A, Lange BM.. 2019. Gene networks underlying cannabinoid and terpenoid accumulation in cannabis. Plant Physiology 180, 1877–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu G, Cheng C, et al. 2021. Establishment of an Agrobacterium‐mediated genetic transformation and CRISPR/Cas9‐mediated targeted mutagenesis in hemp (Cannabis sativa L.). Plant Biotechnology Journal 19, 1979–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhou Y, Chen Q, Huang X, Tian CE.. 2014. Molecular basis of flowering time regulation in Arabidopsis. Chinese Bulletin of Botany 49, 469. [Google Scholar]
- Zhao T, Schranz ME.. 2017. Network approaches for plant phylogenomic synteny analysis. Current Opinion in Plant Biology 36, 129–134. [DOI] [PubMed] [Google Scholar]