Abstract
Cannabis sativa L. is one of the oldest domesticated crops. Hemp-type cultivars, which predominantly produce non-intoxicating cannabidiol (CBD), have been selected for their fast growth, seed, and fibre production, while drug-type chemovars were bred for high accumulation of tetrahydrocannabinol (THC). We investigated how the generation of CBD-dominant chemovars by introgression of hemp- into drug-type Cannabis impacted plant performance. The THC-dominant chemovar showed superior sink strength, higher flower biomass, and demand-driven control of nutrient uptake. By contrast, the CBD-dominant chemovar hyperaccumulated phosphate in sink organs leading to reduced carbon and nitrogen assimilation in leaves, which limited flower biomass and cannabinoid yield. RNA-seq analyses determined organ- and chemovar-specific differences in expression of genes associated with nitrate and phosphate homeostasis as well as growth-regulating transcription factors that were correlated with measured traits. Among these were genes positively selected for during Cannabis domestication encoding an inhibitor of the phosphate starvation response, SPX DOMAIN GENE3, nitrate reductase, and two nitrate transporters. Altered nutrient sensing, acquisition, or distribution are likely a consequence of adaption to growth on marginal, low-nutrient-input lands in hemp. Our data provide evidence that such ancestral traits may become detrimental for female flower development and consequently overall CBD yield in protected cropping environments.
Keywords: Cannabinoids, Cannabis sativa, drug-type, flower development, hemp, hemp-type, medicinal Cannabis, nitrate, nutrient acquisition, nutrient assimilation, phosphate, RNA-seq, source–sink relations, transcriptional regulation
In Cannabis sativa , introgression of hemp- into drug-type chemovars can alter source–sink relations, ion homeostasis, and the transcriptional regulation of flower development and nutrient acquisition.
Introduction
Cannabis sativa L. (hereafter Cannabis) is an ancient crop that has been cultivated in Asia for more than 12 000 years. Cannabis varieties commonly referred to as hemp-type are primarily used for fibre and seed, while drug-type (medicinal) Cannabis varieties are valued for their accumulation of secondary metabolites in female flowers (Clarke and Merlin, 2016). Medicinal use of Cannabis is largely based on the substantial production of secondary metabolites of the cannabinoid family, the most abundant being cannabidiol (CBD), and Δ9-tetrahydrocannabinol (THC), as well as other phenolic meroterpenoids. Research interest, whilst inhibited by the plant’s illegal status in many jurisdictions around the world, has increased with the discovery of the endocannabinoid system in humans about 32 years ago (Lu and Mackie, 2016). The interaction of phytocannabinoids with human cannabinoid receptors has opened new avenues for drug discovery and led to a re-evaluation of Cannabis use in therapeutics (UNODC, 2023).
Hemp-type Cannabis not only features high stem elongation rates and very active cell wall metabolism to produce strong bast fibres, but has also been selected for its seed oil rich in polyunsaturated fatty acids, and for its persistence on marginal lands with low nitrogen input (Struik et al., 2000; Tang et al., 2017). In addition, hemp-type cultivars can contain high levels of CBD, which is recognized for its non-intoxicating pharmacological properties (Russo and Marcu, 2017). The ability of hemp roots to forage for scarce nutrients make it highly nutrient efficient, while its capacity to exclude or redistribute certain soil contaminants, such as heavy metals and sodium, away from growing organs has also fostered its use in phytoremediation (Linger et al., 2002; Husain et al., 2019; Adesina et al., 2020). CBD is often extracted as a by-product from the chaff of hemp fibre or seed crops and hence the quality of CBD-containing products (oil, seeds) varies greatly with soil and seasonal factors as they frequently contain traces of heavy metals (arsenic, cadmium, lead) (Gardener et al., 2022). THC-dominant drug-type Cannabis varieties are in many ways the polar opposite of hemp-types: they were selected for a compact stature, early flowering, and compound racemose female inflorescences with high glandular trichome density and flower biomass production. Their substantial flower value has led to a selection of high-yielding genotypes and beneficial anatomy for protected cropping environments (Barcaccia et al., 2020; Danziger and Bernstein, 2021; Ingvardsen and Brinch-Pedersen, 2023).
Centuries of selection have led to a notable population divergence between the two main genetic groups, and genome-wide analysis of Cannabis germplasm has identified genes associated with traits differentiating hemp- from drug-type chemovars (Ren et al., 2021). Genes encoding strigolactone and brassinosteriod signalling components that suppress branching, as well as cellulose and lignin biosynthetic genes for fibre production have undergone positive selection in hemp-types, while genes associated with flower (JOINTLESS, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE1, EMBRYONIC FLOWER1) and root (SCARECROW-LIKE1, ZINC FINGER PROTEIN1) development, hormone synthesis (AMINOCYCLOPROPANE CARBOXYLASE OXIDASE5, CYTOKININ OXIDASE5, GIBBERELLIN 2-OXIDASE9, JASMONIC ACID CARBOXYL METHYLTRANSFERASE), as well as nitrate (NITRATE TRANSPORTER1.5, NRT1.8) and amino acid transport (AMINO ACID PERMEASE1, AAP3) were prominent selection targets in drug-types (Ren et al., 2021). These findings demonstrate that human selection has resulted in two very distinct genetic groups within Cannabis that differ substantially in their physiology with respect to nutrient acquisition, light response, and the regulation of organ growth and development.
As with other plants, substantial variation in growth conditions used for the growth of hemp- or drug-type Cannabis with respect to light, temperature, day length, or cultivation spaces (field, glasshouse, controlled environments) are expected to determine genotype-specific nutrient requirements, their optimization, and impact on yield (Backer et al., 2019; Jin et al., 2019). For the production of highly regulated phytoceuticals such as cannabinoids, protected cropping systems are ideally suited as they allow for controlled nutrient supply avoiding heavy metals or sodium accumulation, while also allowing integrated pest management and thus avoiding the introduction of unwanted chemical residue or toxins (Stone, 2014). Phosphate (Pi) supply has positive correlations with both dry matter and cannabinoid content (Coffman and Gentner, 1977). However, recent studies also suggest that increased Pi supply negatively affects cannabinoid concentration while improving flower biomass production (Bevan et al., 2021; Shiponi and Bernstein, 2021). Nitrogen (N) has a lesser impact on these agronomic traits in the greenhouse (Coffman and Gentner, 1977), while in more controlled environments increased N input led to higher flower biomass but lower cannabinoid concentration (Bevan et al., 2021; Saloner and Bernstein, 2021). In drug-type Cannabis, low N shifts carbon metabolism towards amino acid and polyamine synthesis (Song et al., 2023) whereas in hemp-type cultivars, it has been suggested that low N supply benefits both fibre quality and the phenylpropanoid pathway (Landi et al., 2019).
With CBD gaining approval from national drug agencies for the treatment of drug-resistant seizures (Farrelly et al., 2021), CBD-dominant drug-types are highly sought after by certified growers as they suit indoor cultivation due to their compact stature and higher flower yield per square metre of growing space. To generate drug-types with the desired cannabinoid profiles, plant breeders have crossed CBD-dominant hemp-type with THC-dominant drug-types, and the resulting high-yielding feminized CBD-dominant progeny have retained about 90% of the genes from drug-type Cannabis (Grassa et al., 2021). However, the gene introgression from hemp-type and the high heterozygosity of Cannabis more generally (Barcaccia et al., 2020) have resulted in mosaic genomes in these chemovars with some gene compositions leading to undesirable traits impeding reproducible product quality and quantity (Ingvardsen and Brinch-Pedersen, 2023). To enable the use of modern breeding techniques such as marker-assisted breeding or genome editing, causal relationships between trait combinations and underlying genes need to be established for effective selection of desired genotypes, and these need to be evaluated in the context of both complex genetic architecture due to the dioecious reproduction of Cannabis and the growth environment (Ingvardsen and Brinch-Pedersen, 2023).
In this study, we compared the performance of female plants of a CBD-dominant with a THC-dominant drug-type Cannabis chemovar in a controlled-environment glasshouse. Quantification of biomass and biochemical composition (nutrients, cannabinoids, anthocyanins, chlorophyll, total protein, and starch) in eight organs (root, basal/youngest internodes, old/mature fan leaves, high/low order reduced leaves, inflorescence) at flower maturity reveal chemovar-specific differences in metabolite distribution and sink–source allocation between the two Cannabis chemovars. These traits were correlated to gene expression patterns determined by RNA-seq analyses using weighted gene co-expression network analysis (WGCNA). The results uncover the potential consequences of hemp trait introgression into drug-type Cannabis and identify gene targets that determine desirable traits with potential for improving the growth performance of Cannabis chemovars.
Materials and methods
Plant material and growth conditions
Two drug-type Cannabis sativa L. chemovars with contrasting cannabinoid profiles were selected for this study: the CBD-dominant chemovar ‘Cannatonic’, which is assumed to be a hybrid derived from a cross between ‘MK Ultra’ and ‘G13 Haze’, and the THC-dominant chemovar ‘Northern Lights’, which is labelled as a hybrid of landraces ‘Afghani’ and ‘Thai’ (Rahn et al., 2016).
Clonal material was derived from nodal cuttings of mother plants. The bases of cuttings were dipped into auxin-enriched gel (Purple Clonex Rooting Hormone Gel, Yates). Cuttings were then transferred into moist propagation cubes (EasyPlug, Goirle, Netherlands) in trays with humidity lids and grown in a controlled environment growth room with 18 h light, a light intensity of 200 µmol m−2 s−1, temperatures of 24 °C (day) and 21 °C (night), and 55% relative humidity. After 2 weeks, rooted cuttings were transferred to 500 ml rectangular pots containing a coconut coir–perlite blend (70:30 (v/v), Nutrifield, Sunshine West, VIC, Australia). Clones were grown in a climate-controlled glasshouse [18 h light, with an average light intensity of 216 µmol m−2 s−1, temperatures of 28 °C (day) and 22 °C (night), and 55% relative humidity]. Pots were watered daily as required and fertilized every third day with vegetative nutrient mix (1.25 ml each of GP3 Grow, GP3 Micro, and 1.0 ml of GP3 Bloom stock solutions per litre of water, EC 1.2, pH 6.0, Green Planet Nutrients/EasyGrow, Epping, VIC, Australia). After 2 weeks, established plants were transferred to 2.5 litre spiral pots (Garden City Plastics, Dandenong South, Australia) with the same substrate. Two weeks later plants were transferred to a flowering compartment [12 h light, with an average light intensity of 216 µmol m−2 s−1, temperatures of 30 °C (day) and 25 °C (night), and 55% relative humidity]. Nutrient mixes were changed to transition bloom (0.75 ml each of GP3 Grow, GP3 Micro and GP3 Bloom stock solutions per litre of water, EC 1.6, pH 6.0, Green Planet Nutrients/EasyGrow) for 1 week, then early bloom for 4 weeks (0.25 ml of GP3 Grow, 1.0 ml of GP3 Micro, and 1.5 ml of GP3 Bloom stock solutions per litre of water, EC 1.8, pH 6.0, Green Planet Nutrients/EasyGrow), and late bloom (1.0 ml of GP3 Micro and 2.0 ml of GP3 Bloom stock solutions per litre of water, EC 1.8, pH 6.0, Green Planet Nutrients/EasyGrow) for another 4 weeks. Twenty-four hours prior to harvest, plants were flushed with water (pH 6.0).
Plant material harvest and processing
At full female flower maturity (brown curly stigmas and swollen carpels; Punja and Holmes, 2020), three representative clones of each chemovar were selected for full destructive harvest. Each plant was separated into female flower (FF), higher order reduced (or so-called ‘sugar’) leaves (RL1), lower (second to fourth) order reduced leaves (RL2), youngest and oldest fully expanded fan leaf pair (YL/OL), youngest and oldest stem internodes (YS/OS), and lateral roots (LR) (Spitzer-Rimon et al., 2019). For root harvest, root balls were carefully removed from pots and loose substrate removed. Roots were then placed on a large plastic mesh and the growth substrate was washed away. Roots were dried between paper towels. Samples of harvested organs were patted dry, wrapped in aluminium foil, snap-frozen in liquid nitrogen, and stored at −80 °C.
Frozen samples were transferred into 15 ml polycarbonate vials with silicone lined caps (OPS Diagnostics, Lebanon, NJ, USA). Prior to transfer, stems were broken down between sheets of aluminium foil under liquid nitrogen using a hammer. One 1 cm diameter metal ball was added to each vial, and frozen samples were ground in a Geno/Grinder automated tissue homogenizer (SPEX SamplePrep, Metuchen, NJ, USA) for 2 × 30 s at 1000 rpm. Ground samples were transferred into 50 ml tubes, snap-frozen in liquid nitrogen, and stored at −80 °C.
Biochemical analyses
One aliquot (50–70 mg) of the ground bulk sample was extracted with 10 volumes of 1% (v/v) acetic acid at 4 °C in the dark. Fresh extracts were used to determine anthocyanin concentration using the pH differential method (Wrolstad et al., 2005). Free phosphate was measured using the ammonium molybdate assay described earlier (Jost et al., 2015). Sulfate ions were quantified from the same extract measuring turbidity after barium chloride precipitation (Coutinho, 1996).
Another tissue aliquot was extracted with 10 volumes at 80 °C in ultra-pure water for 20 min, with supernatant from a 10 min centrifugation at 11 000 g at 4 °C used for colorimetric assays to determine nitrate (Hachiya and Okamoto, 2017) and ammonium (Vega-Mas et al., 2015).
Chlorophyll, protein, and starch were determined successively from a third tissue aliquot. First, chlorophyll was determined in the supernatant after a 30 min extraction in 10 volumes of 80% (v/v) ethanol in 10 mM MES buffer pH 5.9 at 70 °C and a 10 min centrifugation at 11 000 g at 4 °C (Lichtenthaler, 1987). Subsequently, protein was determined in the supernatant at 595 nm using Coomassie Plus Protein Assay Reagent (Thermo Fisher Scientific, Scoresby, VIC, Australia) resulting from 30 min resuspension of the leftover pellet in 0.1 M NaOH at 80 °C and centrifugation as above. The remaining pellet was neutralized in 0.5 M HCl in 0.1 M sodium acetate buffer pH 4.9, and the suspension used for O/N starch hydrolysis at 37 °C. Starch-derived glucose was quantified in a coupled enzymatic assay using the NADH released by ATP-dependent hexokinase as described (Cross et al., 2006).
Elemental composition was quantified using inductively coupled plasma mass spectrometry (ICP-MS) of another aliquot following aqua regia-based acid digest of 10 mg dried sample for 3 h at 70 °C as described (Yi et al., 2021).
Total carbon and nitrogen were determined in 2 mg of dried sample upon 1000 °C combustion followed by chemiluminescence analysis of nitrous oxide in a 2400 Series II CHNS/O Analyser (Perkin Elmer, Glen Waverley, VIC, Australia). For both elemental composition and C/N analyses, NIST1573A tomato leaf (Sigma-Aldrich) was used as reference material.
Cannabinoids were determined from 10 mg of fresh-frozen sample after extraction in 1 ml HPLC-grade methanol spiked with 0.5 ppm triple-deuterated cannabidiol-D3 (Cerilliant) as internal standard. Following incubation in a ThermoMixer (Eppendorf, Macquarie Park, NSW, Australia) at 25 °C and 1800 rpm for 20 min, samples were centrifuged for 15 min at 10 800 g, at room temperature. Supernatants were filtered through a 0.2 µm Titan3 polytetrafluoroethylene filter (Thermo Fisher Scientific) and diluted 1:200 in methanol. Reference standards of cannabidiolic acid (CBDA), CBD, Δ9-tetrahydrocannabinolic acid A (THCA), and THC (Cerilliant, Round Rock, TX, USA) ranged in concentration from 0.001 to 10 ppm. Diluted and undiluted samples and standards were run on a Vanquish Flex UHPLC system (Thermo Fisher Scientific) with a photodiode array detector coupled to an Orbitrap ID-X Tribrid high resolution mass spectrometer (Thermo Fisher Scientific). Chromatographic separation was carried out on a reverse phase C18 column as described (Lu et al., 2023). Positive ion polarity tandem mass spectrometry was carried out using the full scan MS and targeted MS2 acquisition modes with the ID-X Orbitrap mass resolution set at 60 000 and 15 000, respectively. The mass spectrometer made use of a heated electrospray ionization probe with the electrospray voltage set at 3.5 kV, ion transfer tube temperature at 325 °C, vaporize temperature at 350 °C, and S-lens RF level at 35%, as described (Welling et al., 2021). Xcalibur (v4.6) software (Thermo Fisher Scientific) was used for data acquisition. All peak areas were normalized against the internal standard. To avoid peak saturation, the acid form of CBD was quantified using the 1:200 dilutions of all organs of the CBD-dominant chemovar—apart from old stem and fine roots. For both chemovars, 1:200 dilutions of all organs apart from old stem and fine roots were used to quantify acid forms of THC. For neutral cannabinoids, undiluted samples were used to determine peak areas. The slope of the linear range of each standard curve was used to calculate cannabinoid concentration in µmol g−1 fresh weight (FW).
RNA-seq analyses
Total RNA was extracted from three biological replicates using the Spectrum Plant Total RNA kit (Sigma-Aldrich) according to the manufacturer’s protocol. Removal of genomic DNA was performed with an On-Column DNase I (Sigma-Aldrich) digestion kit prior to RNA elution. RNA-seq libraries were generated with a TruSeq Stranded mRNA Library Prep Kit and sequenced on a NextSeq500 instrument (both Illumina) as 84 bp reads with an average quality score (Q30) of above 95% and an average of 25.3 million reads per sample. Quality control was performed using FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Transcript abundances as transcripts per million (TPM) and estimated counts were quantified and summed to the gene level by pseudo-aligning reads against a k-mer index built from the transcript models for the Cannabis sativa CS10 reference genome (Grassa et al., 2021) downloaded from the Ensembl Plants database (https://plants.ensembl.org/Cannabis_sativa_female) using the kallisto program with 100 bootstraps (Bray et al., 2016). Only genes with TPM>1 in at least one of the organs in each chemovar were retained for further analysis.
Bioinformatic analyses
In addition to the CS10 genome annotation, transcripts were also functionally categorized and annotated using the Mercator pipeline (Schwacke et al., 2019). The WGCNA and associated analyses were performed with log transformed TPM values and the corresponding software package for the R programming environment (Langfelder and Horvath, 2008), using a soft thresholding power of 12, a signed-hybrid network type, and Pearson’s correlation together with an automatic module detection and dynamic tree-cutting algorithm. Gene ontology (GO) term enrichment analyses were performed using the GO annotation of the CS10 genome assembly (GCA_900626175.2) downloaded from NCBI with the clusterProfiler tool using the simplify function with the ‘Rel’ option to remove redundancy (Wu et al., 2021). For sequence comparisons, Cannabis protein sequences were obtained for the CS10 genome assembly (https://plants.ensembl.org/Cannabis_sativa_female) and used for BLAST searches against Arabidopsis proteins annotated in TAIR (https://www.Arabidopsis.org/). BLAST hits with a cut-off of E<10−50 were retained for sequence alignments using the Clustal Omega algorithm (Sievers et al., 2011). Subsequent tree reconstruction was performed using the UPGMA method in the Geneious Prime (version 2020.2.5) software (https://www.geneious.com/). Overlaps in gene lists were analysed and visualized by UpSet plots (Conway et al., 2017). Heatmaps were generated with the ComplexHeatmaps package in R (Z. Gu et al., 2016). For comparison of Cannabis transcription factor gene expression with the Arabidopsis homologue(s), a Pearson correlation was calculated and for one-to-many relationships averaged using Fisher’s z-transformation. RNA-seq data for Arabidopsis gene expression in comparable organs (root, stem, leaf, flower) was retrieved from a previous study (Mergner et al., 2020).
Statistical analyses
The statistical analyses were performed using the R software environment (https://www.r-project.org/) by two-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test as indicated.
Results
Differences in cannabinoid concentration, growth habit, and flower yield between a tetrahydrocannabinol- and a cannabidiol-dominant chemovar
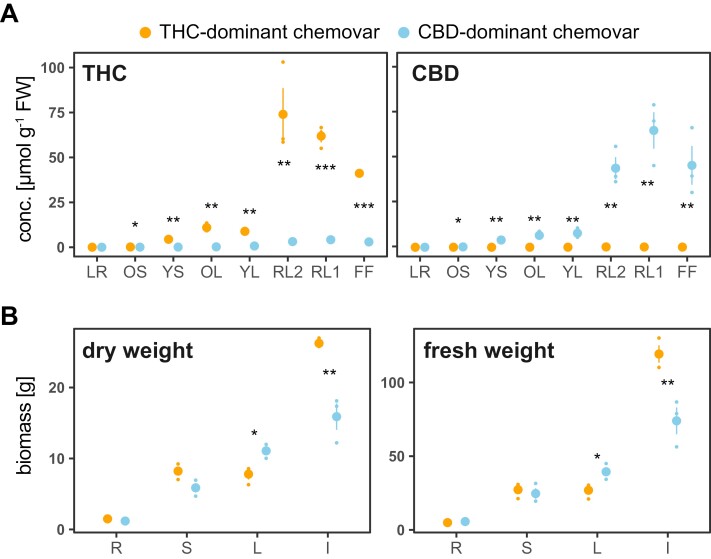
To investigate how the introgression of hemp traits into the genetic background of a high-performing drug-type chemovar to achieve high CBD yield affects growth performance and nutrient use, we analysed two contrasting chemovars with suitable pedigree: one THC-dominant and one CBD-dominant drug-type Cannabis hybrid (from now on referred to as THC- and CBD-dominant chemovars, respectively; see ‘Material and methods’ for details). Analyses were conducted on eight organs of each chemovar at flower maturity: lateral roots (LR), the oldest and youngest stem internodes (OS, YS); the oldest and youngest fully expanded fan leaf pair (OL, YL), lower (second to fourth) order reduced leaves (RL2), higher order reduced (‘sugar’) leaves (RL1), and the female flower (FF). As expected, THC and CBD concentrations determined across organs of the two chemovars differed markedly. Floral organs of the CBD-dominant chemovar contained an average 51 ± 7 µmol total CBD g−1 FW (Fig. 1A). This corresponds to 9 ± 2% total CBD of flower dry weight (DW), with 98.7% present in its acid (CBDA) form. Its flowers also contained some THC (3.4 ± 0.4 µmol total THC g−1 FW). Flowers of the THC-dominant chemovar had an average of 59 ± 10 µmol total THC g−1 FW, which corresponds to 9 ± 1% of flower DW, with 99.9% present in its acid form (THCA). Its flowers contained only trace amounts of total CBD (0.15 ± 0.02 µmol g−1 FW). Across chemovars, reduced leaves had similar (CBD-dominant chemovar) or higher (THC-dominant chemovar) total cannabinoid concentration than the female flowers.
Fig. 1.
Difference in flower biomass and cannabinoid profile between the two chemovars. Shown are total tetrahydrocannabinol (THC) and cannabidiol (CBD) concentrations (A) and fresh and dry weights (B) for the indicated organs of the two chemovars at destructive harvest. Values are means ±SE of three plants per genotype as well as individual data points. Asterisks indicate statistically significant differences between organs across chemovars (two-way ANOVA, Tukey post-hoc test; *P<0.05, **P<0.01, ***P<0.001). Abbreviations in (A): FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves. Abbreviations in (B): I, inflorescence; L, leaves; R, root; S, stem.
At final harvest when flowers had reached maturity, the overall stature of the plants differed significantly, with the THC-dominant chemovar being compact and erect with less than 45° branching angles, while the CBD-dominant chemovar had a weeping habit with greater than 70° branching angles (Supplementary Fig. S1A). Average plant height was 55 ± 2 cm for the THC and 63 ± 1 cm for the CBD-dominant chemovar. The THC-dominant chemovar produced 8.3 ± 0.2 branches, and the CBD-dominant chemovar 11.8 ± 0.2. Racemose inflorescences were densely or loosely stacked in the THC-dominant and CBD-dominant chemovar, respectively (Supplementary Fig. S1B). Mature fan leaves of the former were dark green, while those of the latter turned yellow and purple, indicating early senescence presumably as a consequence of nutrient stress (Supplementary Fig. S1C). The root system featured more fine roots in the THC-dominant chemovar, while the root structure in the CBD-dominant chemovar was poorly developed (Supplementary Fig. S1D). Importantly, biomass data revealed that the CBD-dominant chemovar was sink-limited as it produced significantly higher leaf fresh and dry matter, while the flower yield was 2-fold lower than in the THC-dominant chemovar (Fig. 1B).
Given similar total cannabinoid concentrations in floral organs of the two chemovars (Fig. 1A), differences in the overall yield between them are due to the 2-fold difference in flower biomass (Fig. 1B).
Primary metabolite profiles highlight differences in resource allocation among chemovars
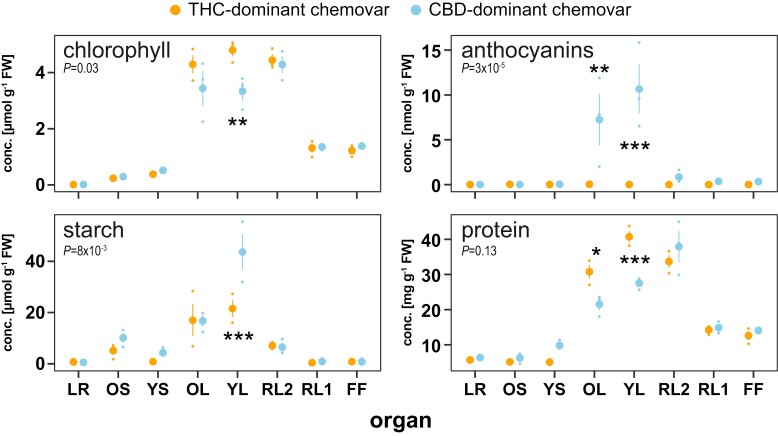
We next analysed biochemical compounds typically associated with contrasting growth characteristics and biomass. Pigment analysis confirmed the visual differences in fan leaf colour (Supplementary Fig. S1C), with an average 34% higher chlorophyll content in the fan leaves of the THC-dominant chemovar (Fig. 2). Young fully expanded and older fan leaves of the CBD-dominant chemovar had high anthocyanin levels, while there was none detected in those of the THC-dominant chemovar (Fig. 2). The anthocyanin accumulation in young leaves was accompanied by 2-fold higher starch levels indicating metabolic restrictions in carbon assimilation due to nutrient stress (Fig. 2) (Jezek et al., 2023). Protein content was on average 46% higher in young and old fan leaves of the THC-dominant chemovar, again supporting its superior performance compared with the sink-limited CBD-dominant chemovar (Fig. 2) (Sulpice et al., 2010).
Fig. 2.
Source leaves differ in key primary metabolite and anthocyanin concentrations. Concentrations of key metabolites chlorophyll, anthocyanins, starch, and protein were determined across organs at final destructive harvest for the two chemovars. Values are means ±SE of three plants. Asterisks indicate statistically significant differences between organs and the indicated P-value between chemovars overall (two-way ANOVA, Tukey post-hoc test; *P<0.05, **P<0.01, ***P<0.001). Abbreviations: CBD, cannabidiol; FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves; THC, tetrahydrocannabinol.
Sink organs of the CBD-dominant chemovar hyperaccumulate phosphate, which restricts nutrient assimilation
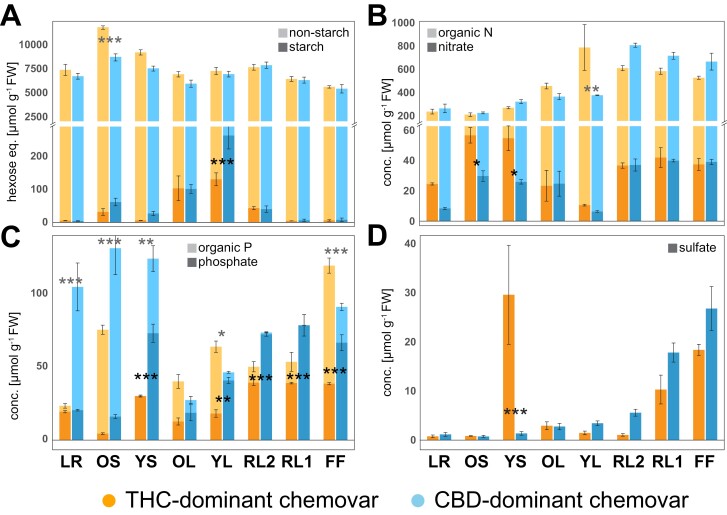
Given the sink limitation of the CBD-dominant chemovar, that is, higher resource investment into leaves and less into flowers when compared with the THC-dominant chemovar, we looked closely at assimilation of the macronutrients nitrogen (N) and phosphorus (P), as well as total carbon (C) content. Apart from significant differences in starch accumulation in young leaves (Figs 2, 3A), overall carbon status was similar between chemovars. The 26% higher total carbon in stems of the THC-dominant chemovar may either indicate higher transient storage in stems to support reproductive structures or investment into root growth (Fig. 3A) (Smith and Stitt, 2007). N assimilation was highly comparable between chemovars, with floral organs containing the highest fraction of organic N with an average 540 ± 24 µmol g−1 FW in the THC- and 696 ± 42 µmol g−1 FW in the CBD-dominant chemovar (Fig. 3B). Free nitrate levels were 7 and 5% of the total N fraction, respectively. The only statistically significant differences between chemovars were higher nitrate levels in stems of the THC-dominant chemovar, and a 52% decrease in the organic N fraction of young leaves of the CBD-dominant chemovar. These hint at reduced xylem loading of nitrate, and reduced N assimilation in source leaves of the CBD-dominant chemovar (Y-Y. Wang et al., 2018). By contrast, phosphorus allocation profiles differed greatly between the two chemovars (Fig. 3C). Apart from roots, all sink organs (i.e. young stems, young fan leaves, and floral organs) of the CBD-dominant chemovar hyperaccumulated phosphate (66 ± 4 µmol g−1 FW) compared with 33 ± 2 µmol g−1 FW of phosphate in sink organs of the THC-dominant chemovar, which is much more in the expected range for growing organs (Cuyas et al., 2023). At the same time, the organic P fraction of young leaves and floral organs was greatly reduced in the CBD- compared with the THC-dominant chemovar. Organic P fractions in lateral roots (21-fold higher) and stems (1.6-fold higher in old stems and more than 50-fold higher in young stems), however, were much larger in the CBD- than the THC-dominant chemovar. This suggests a lack of suppression of phosphate acquisition and translocation in the CBD-dominant chemovar under P-replete conditions, and subsequent localized assimilation of phosphate into the organic P fraction by roots and stem tissues due to exhausted storage capacity for free phosphate in sink organ vacuoles (Shane et al., 2004a; Takagi et al., 2020). Free sulfate levels across organs of both chemovars—apart from flowers—were below a healthy leaf concentration range of 10–30 µmol g−1 FW, which suggest plants were S-limited (Fig. 3D) (Blake-Kalff et al., 2000). Surprisingly, the THC-dominant chemovar accumulated very high sulfate levels in young stems—which could provide local protection against fungal pathogens (Williams et al., 2002).
Fig. 3.
Fractionation of carbon, nitrogen, phosphorus, and concentration of sulfate across organs of the two chemovars. Concentrations of total carbon (A; starch, non-starch), organic and inorganic pools of nitrogen (B; nitrate, organic N) and phosphorus (C; phosphate, organic phosphorus) as well as sulfate (D) across organs at final destructive harvest for the two chemovars. Colours represent the two chemovars (orange, blue) and colour shades (dark, light) the different pools. Given are means ±SE of three plants per genotype. Asterisks indicate statistically significant differences between organs (two-way ANOVA, Tukey post-hoc test; *P<0.05, **P<0.01, ***P<0.001). Abbreviations: CBD, cannabidiol; FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves; THC, tetrahydrocannabinol.
Phosphate hyperaccumulation impairs calcium and potassium homeostasis in source leaves
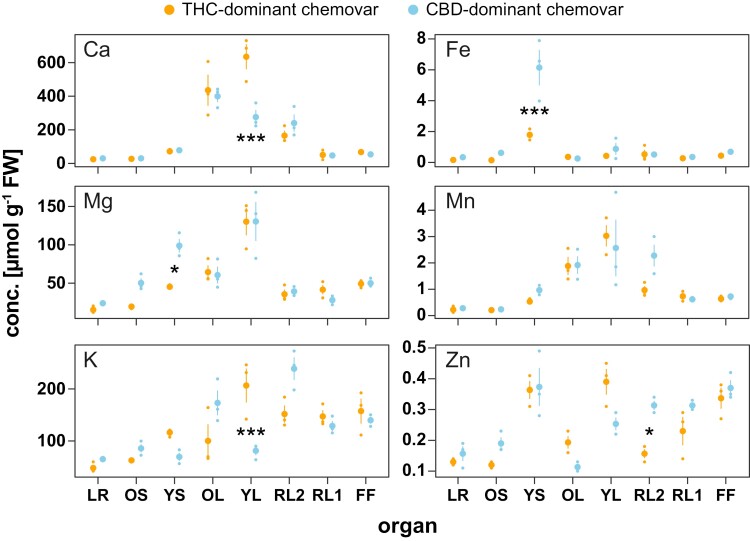
To investigate the effect of unrestricted phosphate acquisition and translocation in the CBD-dominant chemovar further, the elemental composition of all plant organs was determined across the two contrasting genotypes (Fig. 4). While magnesium, iron, and manganese concentrations were similar between leaves of the two chemovars, concentrations of calcium, potassium, and to a lesser degree zinc were reduced by 61%, 57%, and 36%, respectively, in young fully expanded fan leaves of the CBD-dominant chemovar (Fig. 4). These reductions could be a consequence of storage incompatibility between calcium and phosphate with phosphate hyperaccumulation in the CBD-dominant chemovar lowering the calcium concentration in the leaf mesophyll (Hayes et al., 2019). In young stems of the latter, magnesium and iron concentrations were 2- and 3-fold higher, respectively, than those in the THC-dominant chemovar, which could be an early indication of reduced allocation to sink organs because of lower potassium levels and phosphate hyperaccumulation (Sánchez-Rodríguez et al., 2014; Wu et al., 2019).
Fig. 4.
Profiles of macro- and microelements across organs of the chemovars. Organ concentrations of cations calcium (Ca), iron (Fe), magnesium (Mg), manganese (Mn), potassium (K), and zinc (Zn) in the two chemovars were determined at final destructive harvest. Values are means ±SE of three plants per genotype. Asterisks indicate statistically significant differences between organs (two-way ANOVA, Tukey post-hoc test; *P<0.05, **P<0.01, ***P<0.001). Abbreviations: CBD, cannabidiol; FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves; THC, tetrahydrocannabinol.
RNA-seq analyses identifies patterns of gene co-expression in the two chemovars
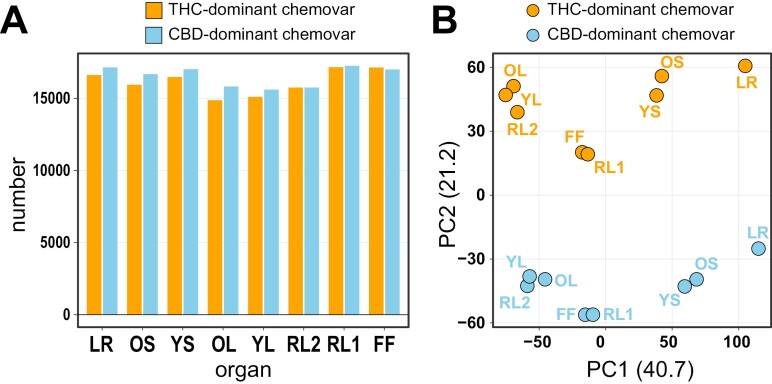
To better understand their physiological and morphological differences, we performed RNA-seq analysis across organs of the two Cannabis chemovars. The number of detectable genes (TPM>1 in at least one organ per chemovar) across organs and chemovars was similar (Fig. 5A), ranging from 14 872 (OL, THC-dominant chemovar) to 17 256 (RL1, CBD-dominant chemovar) and was generally lowest in leaves (OL, YL, RL2), likely because of their primary photosynthetic function. Of 20 625 genes detected in at least one organ of one chemovar across all samples, 12 609 genes were commonly expressed in all organs of both chemovars (Supplementary Table S1). A principal component analysis separated the samples by organ (PC1, 41%) and chemovar (PC2, 21%), with similar organ types (leaf—OL/YL/RL2, flower—RL1/FF, stem—OS/YS) clustering together (Fig. 5B).
Fig. 5.
Gene expression profiles distinguish organs and chemovars. (A) Number of genes detected by RNA-seq in each of the two chemovar organs. A cut-off of TPM>1 in at least one of the organs for each chemovar was used. (B) Principal component analysis of RNA-seq data. Abbreviations: CBD, cannabidiol; FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves; THC, tetrahydrocannabinol; TPM, transcripts per million.
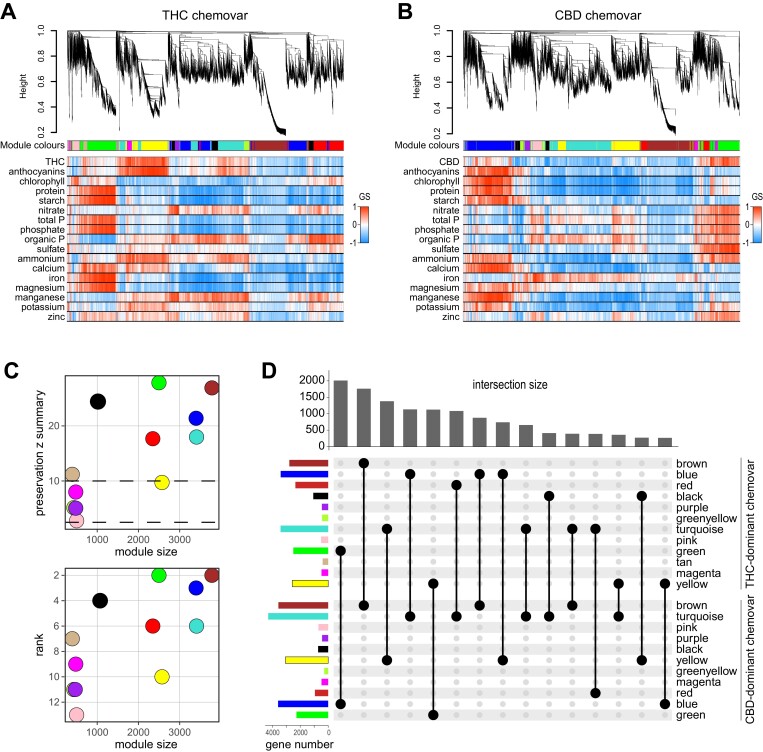
We next performed a WGCNA (Langfelder and Horvath, 2008) on the 20 625 genes detected to identify genes with common expression patterns and to correlate these gene clusters to physiological and biochemical traits shown in Figs 1–4. The WGCNA identified 12 modules of genes with distinct expression patterns for the THC-dominant chemovar and 11 modules for the CBD-dominant chemovar (Fig. 6A, B, upper panels; Supplementary Tables S1–S3). Determination of gene significance (Langfelder and Horvath, 2008), a measure of biological significance of each gene for each quantified trait, indicated several modules with importance for traits such as cannabinoid concentration (yellow and green module) or protein concentration (green and blue modules) in the THC- and CBD-dominant chemovar, respectively (Fig. 6A, B, lower panels). It is important to note that while modules for both chemovars are labelled with colours by the WGCNA algorithm, there is no direct relationship between modules and colour assignment. We therefore determined which modules of the THC-dominant chemovar match those of the CBD-dominant chemovar and the degree of shared genes within each module by analysing module preservation (Langfelder et al., 2011). This showed that the brown, green, blue, black, turquoise, red, and tan modules of the THC-dominant chemovar were strongly preserved (z summary statistics >10) in the CBD-dominant chemovar with median rank statistic confirming this (Fig. 6C). Correspondingly, these modules also shared the highest number of genes, with the most genes common between the green module of the THC-dominant chemovar and the blue module of the CBD-dominant chemovar (Fig. 6D).
Fig. 6.
Weighted gene co-expression network analysis (WGCNA) of RNA-seq data of the two chemovars. RNA-seq data of the two contrasting chemovars were used to determine co-expressed genes across the eight analysed organs using the WGCNA package (Langfelder and Horvath, 2008). (A, B) Gene dendrograms for obtained modules of co-expressed genes and assigned module colours for the THC- (A) and CBD-dominant (B) chemovars are indicated in the top panels. Automatic module detection was based on a Pearson correlation, signed hybrid network, and a manual tree height cut-off of 0.2. The heatmaps in the bottom panels indicate the gene significance (GS; based on the correlation of gene expression profiles with a sample trait; Langfelder and Horvath, 2008) of each gene for the given traits. (C) Preservation of modules for the THC- and CBD-dominant chemovars. The top panel indicates the z summary statistics of preservation for each of the THC-dominant chemovar modules (y-axis) and module sizes (x-axis). The bottom panel shows the median ranking for each THC-dominant chemovar module (y-axis) and module sizes (x-axis). Dotted lines delineate high (upper) and no (lower) statistical significance of module preservation. (D) Overlaps in genes, represented by an UpSet plot, across modules of the two chemovars confirm the module preservation shown in (C). Abbreviations: CBD, cannabidiol; THC, tetrahydrocannabinol.
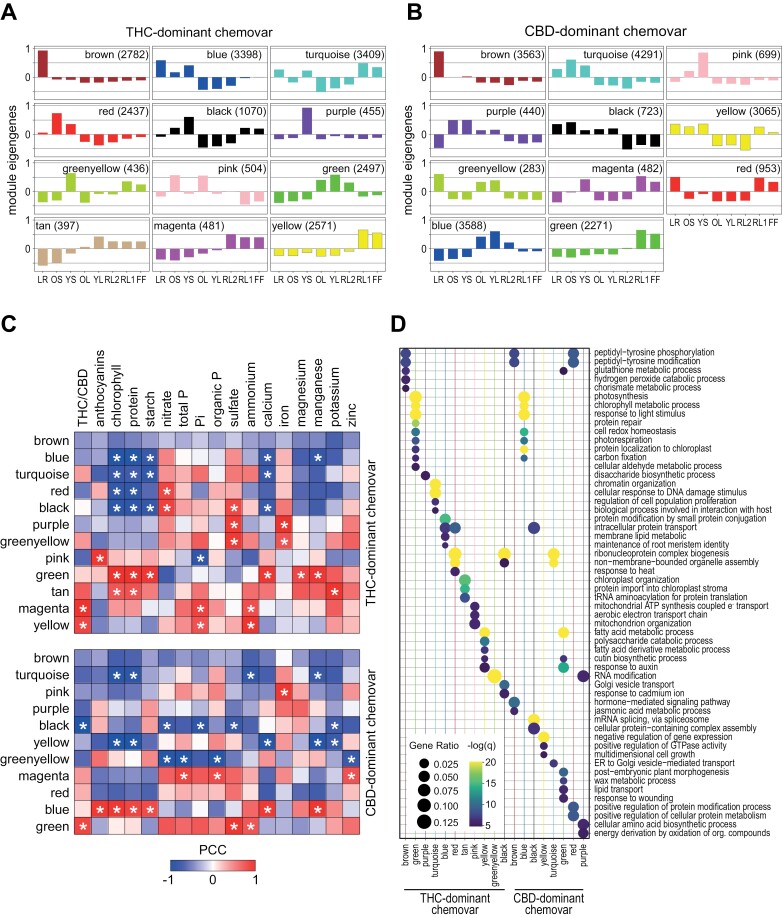
The module eigengene (ME; the first principal component for a given module) summarizes the weighted average expression of genes in each organ across modules (Langfelder and Horvath, 2008). This indicated that the preserved brown modules for the THC- and CBD-dominant chemovar include root-specific genes, while the green and blue modules of the THC- and CBD-dominant chemovar, respectively, are preferentially expressed in photosynthetically active fan leaves (OL, YL, RL2) (Fig. 7A, B). A correlation analysis between module eigengenes and quantified traits showed that cannabinoid concentration had high positive correlation with the inflorescence-specific modules of both chemovars (magenta, yellow—THC-dominant chemovar; green—CBD-dominant chemovar) as expected, while the black module of the CBD-dominant chemovar, containing genes expressed in non-floral organs, was negatively correlated to the CBD concentration. In addition, there was a strong positive correlation between modules with high gene expression in the metabolically active fan leaves (green, tan—THC-dominant chemovar; blue—CBD-dominant chemovar) and key compounds such as chlorophyll, protein, and starch, and also a negative correlation with genes representing modules of organs with low photosynthetic activity such as roots and stems (blue, turquoise, red, black—THC-dominant chemovar; turquoise, yellow—CBD-dominant chemovar).
Fig. 7.
Co-expressed genes are correlated with specific traits and enriched for GO terms. (A, B) Module eigengenes identified across organs of the THC- (A) and CBD-dominant (B) chemovar as proxies for the average expression of genes within each module (Langfelder and Horvath, 2008). Modules across chemovars share organ specific expression of their corresponding genes. (C) Gene expression patterns of each module, represented by their module eigengenes, were correlated with quantified traits for both chemovars. Colour scale indicates the Pearson correlation coefficient and asterisks statistically significant correlations (P<0.05). (D) Comparison of GO term enrichment for genes in the modules of the two chemovars. For the analysis, genes with a module membership value (kME) of above 0.9 were used. Dots sizes indicate the gene ratio (ratio of module genes to all genes associated to GO term) and colour scale the statistical significance (−log10 of the false discovery rate). Abbreviations: CBD, cannabidiol; FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves; THC, tetrahydrocannabinol.
Next, we performed a Gene Ontology (GO) term enrichment analysis for all modules using only genes with a high module membership (kME>0.9, Supplementary Tables S2, S3). This revealed for the leaf-specific green module in the THC-dominant chemovar and the blue module of the CBD-dominant chemovar enrichment of GO terms related to photosynthesis, photorespiration, chlorophyll metabolism, and carbon fixation (Fig. 7D; Supplementary Table S4), with underlying genes having well-documented functions within these processes (Supplementary Table S2). GO terms enriched for the root-specific brown modules were related to peptide phosphorylation largely based on receptor-like kinases. Although many of these are associated with defence responses, receptor kinases also play an important role in the regulation of root development (Ou et al., 2021). One gene in this list encodes a homologue of the Arabidopsis PHYTOSULFOKINE RECEPTOR KINASE1 (PSKR1), which is a receptor for the growth-regulating peptide phytosulfokine (Amano et al., 2007), suggesting a similar role for this peptide in the development of the Cannabis root.
In Cannabis, the cannabinoid-producing floral organs are of particular interest. As would be expected, the modules containing genes with preferential expression in flowers (RL1, FF), that is, the yellow module in the THC-dominant chemovar and the green module of the CBD-dominant chemovar, contained the genes encoding all enzymes of the cannabinoid biosynthetic pathway (Gülck and Møller, 2020), namely ACYL-ACTIVATING ENZYME1, OLIVETOL SYNTHASE, OLIVETOLIC ACID CYCLASE, CANNABIGEROLIC ACID SYNTHASE, CANNABIDIOLIC ACID SYNTHASE, and TETRAHYDROCANNABINOLIC ACID SYNTHASE (Supplementary Tables S1–S3). In addition, enriched GO terms for these modules were associated with fatty acid metabolism and cutin biosynthesis (Fig. 7D; Supplementary Table S4). These are underpinned by expression of genes supporting the precursor supply for cannabinoid synthesis. Many of these fatty-acid-related genes are annotated as lipoxygenases, fatty acid desaturases and CoA reductases, often with predicted chloroplast localization, in agreement with the proposed generation of the cannabinoid precursor hexanoic acid by breakdown of unsaturated fatty acids (Stout et al., 2012). Similarly, cutin biosynthesis-linked genes are expected in these modules to drive cuticle formation in the prolific glandular trichomes (Livingston et al., 2021). Interestingly, for inflorescence-specific modules of the two chemovars ‘response to auxin’ was another enriched GO term (Fig. 7D). Associated genes were mainly from the auxin-induced family of SMALL AUXIN UPREGULATED RNAs (SAUR) and transcriptional repressors of the AUXIN RESPONSE FACTOR/INDOLE-3-ACETIC ACID INDUCIBLE (AUX/IAA) family. Genes of both families are highly up-regulated by auxin, suggesting increased levels of indole-3-acetic acid (IAA) in floral organs when compared with other organs, in line with the critical role of this hormone for floral development (Cucinotta et al., 2021). ‘Polysaccharide catabolism’ was also an enriched GO term for the yellow module of the THC-dominant chemovar, largely made up of genes involved in cell wall and pectin modification (Fig. 7D; Supplementary Table S4).
In summary, gene modules determined for both chemovars were highly correlated with physiological traits, and the enriched GO terms were in agreement with organ-specific morphological and functional differences. The analysis for flower-specific modules of both chemovars identified genes driving the development of abundant floral and trichome structures as well as biosynthesis of cannabinoids and their precursors.
Transcriptional regulators of organ development and plant growth
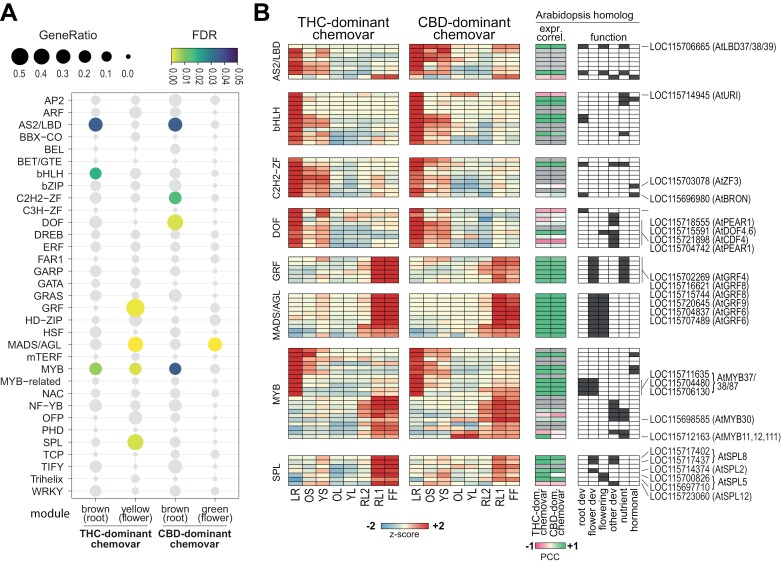
WGCNA enables the identification of ‘hub genes’, which are the most highly connected genes within modules and their expression profile are close proxies for the entire module. To identify possible regulators governing resource allocation and source–sink relations between the two Cannabis chemovars, we focused on transcription factors (TFs) with highest module membership (kME>0.95) in the modules representing fast growing sink organs, i.e., the highly root- and flower-specific modules of both chemovars (brown and yellow for THC-dominant chemovar, brown and green for the CBD-dominant chemovar), respectively (Supplementary Table S5). Investigating the gene list of modules allowed us to identify a number of TF families that were significantly enriched in the corresponding modules (Fisher’s exact test, false discovery rate <0.05; Fig. 8A). For these hub genes, sequence comparisons were performed to identify their closest homologues in Arabidopsis to allow for functional interpretation. For this, Cannabis protein sequences were searched using BLAST (cut-off E<10−50) against sequences of all Arabidopsis TFs downloaded from the Plant Transcription Factor Database (http://planttfdb.gao-lab.org/) (Supplementary Fig. S2).
Fig. 8.
Analysis of genes encoding transcription factors among hub genes identified by weighted gene co-expression network analysis (WGCNA). Transcription factors (TFs) among the hub genes of the flower and root specific modules were defined with a cut-off kME >0.95 for further analysis for TF family enrichment and expression. (A) Bubble plot indicating the enrichment of hub genes in TF families. Only TF families with more than 10 members detected by RNA-seq were included in the analysis. Colour scale specifies statistical significance of enrichment (Fisher’s exact test, false discovery rate (FDR) <0.05) and dot size the gene ratio (number of TFs in hub gene list to total number of family members in Cannabis genome). (B) Heatmaps representing expression of TF hub genes in enriched TF families [left panels; colour scale indicates the z-score of the log2(TPM+1) across organs of each chemovar], the correlation of Cannabis and Arabidopsis homologue expression [middle panel; colour scale indicates the Pearson’s correlation coefficient (PCC) for homologue expression; grey colour: no Arabidopsis data available; Arabidopsis data for root, stem, leaf and flower taken from Mergner et al. (2020)], and function of Arabidopsis homologues [right panel, dark grey boxes indicate genes with a documented function in this category [i.e. root or flower development, flowering time/induction, other development (trichome, cell differentiation)], nutrient metabolism or hormonal signalling; see Supplementary Table S6 for references]. Cannabis genes and their Arabidopsis homologues discussed in the main text are indicated. General abbreviations: CBD, cannabidiol; FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves; THC, tetrahydrocannabinol; TPM, transcripts per million. Abbreviations for transcription factor families: AP2, APETALA 2; ARF, AUXIN RESPONSE FACTOR; AS2/LBD, ASYMMETRIC LEAVES 2/LOB DOMAIN; BBX-CO, B-BOX DOMAIN-CONSTANS; BEL, BELL Homeodomain; BET/GTE, BET1P/SFT1P-LIKE GLOBAL TRANSCRIPTION FACTOR GROUP E; bHLH, basic HELIX-LOOP-HELIX; bZIP, basic leucine-zipper; C2H2-ZF, C2H2-like zinc finger; C3H-ZF, Cys3-His zinc finger domain; DOF, DNA binding with one finger; DREB, dehydration-responsive element-binding protein; ERF ethylene response factor; FAR1, FAR-RED IMPAIRED RESPONSE 1; GARP, GOLDEN2, ARR-B, Psr1-domain; GATA, GATA motif factor; GRAS, GRAS domain; GRF, GROWTH-REGULATING FACTOR; HD-ZIP, homeodomain leucine zipper; HSF, Heat stress factor; MADS/AGL, MADS-box/AGAMOUS-LIKE; mTERF, Mitochondrial Transcription Termination Factor; MYB, MYB domain; MYB-related, MYB domain-related; NAC, NAM/ATAF/CUC transcription factors; NF-YB, Nuclear Factor YB; OFP, ovate family protein; PHD, PHD finger-containing; SPL, SQUAMOSA promoter binding-like; TCP, teosinte branched1/CYCLOIDEA/PROLIFERATING CELL; TIFY, TIFY motif; Trihelix, trihelix structure; WKRY, WRKY domain.
TFs encoding the ASYMMETRIC LEAVES2/LOB DOMAIN-CONTAINING PROTEIN (AS2/LBD), MADS-box AGAMOUS-LIKE (MADS/AGL), and the MYB DOMAIN families were enriched for both chemovars. The HELIX-LOOP-HELIX (bHLH), GROWTH-REGULATING FACTOR (GRF), and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) families were statistically significantly enriched only for the THC-dominant chemovar, while the CYS2–HIS2 ZINC FINGER (C2H2-ZF) and the DNA-BINDING ONE ZINC FINGER (DOF) TFs were enriched only in the CBD-dominant chemovar (Fig. 8A). Organ-specific expression of the corresponding TF-encoding genes agreed with their role as hub genes in the root- and flower-specific modules (Figs 7A, 8B, left panels). These expression patterns were also highly correlated with the identified Arabidopsis homologues (Fig. 8B, middle panel; Supplementary Fig. S2; Supplementary Table S6), further supporting their functional conservation. Generally, the enriched TF families have a role in root (AS2/LBD, bHLH, MYB, and GRF) and flower (MADS/AGL, MYB, and SPL) development, flowering induction/timing (AS2/LBD, DOF, MADS/AGL, and SPL), nutrient or hormonal signalling, and the regulation of interactions between these pathways (Fig. 8B, right panel; for specific references see Supplementary Table S6).
Due to the substantial contrast in resource allocation between chemovars, we focussed on TFs for which expression profiles differed across sink and source organs. The Cannabis LOC115706665 gene encodes a homologue of proteins in the AtLBD37/38/39 clade involved in the repression of key genes in the nitrogen assimilation pathway and also in the N-dependent regulation of anthocyanin accumulation (Rubin et al., 2009). It showed preferential expression in roots, stems, and floral organs of the CBD-dominant chemovar, while it had similar and low expression in all organs of the THC-dominant chemovar (Fig. 8B; Supplementary Tables S1, S6). In the bHLH family, expression of the UPSTREAM REGULATOR OF IRT1 (AtURI) homologue LOC115714945 in young stems and fan leaves of the CBD-dominant chemovar could be linked to its higher iron concentration in stems compared with those of the THC-dominant chemovar (Fig. 4) (Kobayashi and Nishizawa, 2012). For C2H2-ZF family genes with preferential expression in the roots of both chemovars, the expression domain of seven members extends into stems of the THC-dominant chemovar. These genes encode proteins homologous to Arabidopsis TFs involved in hormone-dependent repression of plant growth (LOC115703078, AtZF3; LOC115696980, AtBRON) (Fig. 8B; Supplementary Fig. S2; Supplementary Table S6) (Clark et al., 2021; Zhang et al., 2021). They might therefore control developmental processes contributing to the more compact growth habit of the THC-dominant chemovar. Contrastingly, the expression of DOF TFs encoded by a set of Cannabis genes (LOC115704700, LOC115704742, LOC115718555, LOC115715591, LOC115721898, LOC115718400, LOC115697354) was higher in stems of the CBD- than those of the THC-dominant chemovar (Fig. 8B; Supplementary Fig. S2). Stronger vegetative growth of the CBD-dominant chemovar could be associated with the auxin-modulated function of these genes (Zou and Sun, 2023).
The CBD-dominant chemovar features a less densely stacked inflorescence than the THC-dominant chemovar (Supplementary Fig. S1). Four Cannabis MYB genes (LOC115712024, LOC115711635, LOC115706130, LOC115704480) were more strongly expressed in floral organs of the CBD-dominant chemovar whilst being highly expressed in roots of both chemovars. These genes are homologous to a subgroup of Arabidopsis MYB TFs [AtMYB37/38/87; the former two are also termed REGULATOR OF AXILLARY MERISTEMS1 (RAX1) and RAX2] involved in the regulation of axillary meristem formation (Fig. 8B; Supplementary Fig. S2; Supplementary Table S6). They control the formation of new meristems, which has a major impact on plant architecture (Keller et al., 2006; Müller et al., 2006). LOC115698585 and LOC115712163 had about 10-fold higher expression in leaves of the CBD- than the THC-dominant chemovar (Supplementary Table S1). These genes are homologues of AtMYB30 and ATMYB11/12/111, respectively. The former is a negative regulator of photomorphogenesis by promoting the accumulation of PHYTOCHROME-INTERACTING FACTORS4 (PIF4) and PIF5 (Yan et al., 2020). These two PIFs have also been implicated in light-stimulated activation of phosphate acquisition in some Arabidopsis accessions (Sakuraba et al., 2018). ATMYB11/12/111 control the primary steps of the flavonoid biosynthetic pathway (Stracke et al., 2007), which then branches into anthocyanidin and flavanol synthesis. Therefore, the higher expression of these two Cannabis MYB TFs in leaves of the CBD-dominant chemovar might be related to the accumulation of phosphate and anthocyanins (Fig. 2). The SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family of TFs was enriched among the hub genes of the yellow module for the THC-dominant chemovar only (Fig. 8A). Their 3- to 4-fold higher expression in flowers of the THC-dominant chemovar is likely associated with its complex floral architecture, that is, a highly branched and compact inflorescence, and with higher overall flower biomass (Fig. 8B; Supplementary Tables S1, S6) (Xing et al., 2013; Jung et al., 2016; Chao et al., 2017; Hyun et al., 2017).
In summary, exploration of hub genes defined by our WGCNA identified sets of genes encoding TF families as possible integrators of flowering, developmental transitions, and nutrient homeostasis. Their organ-specific, differential expression might control the expression of downstream pathways leading to the observed physiological and morphological differences in the two analysed chemovars.
Genes involved in nitrogen and phosphorus acquisition, allocation, and homeostasis
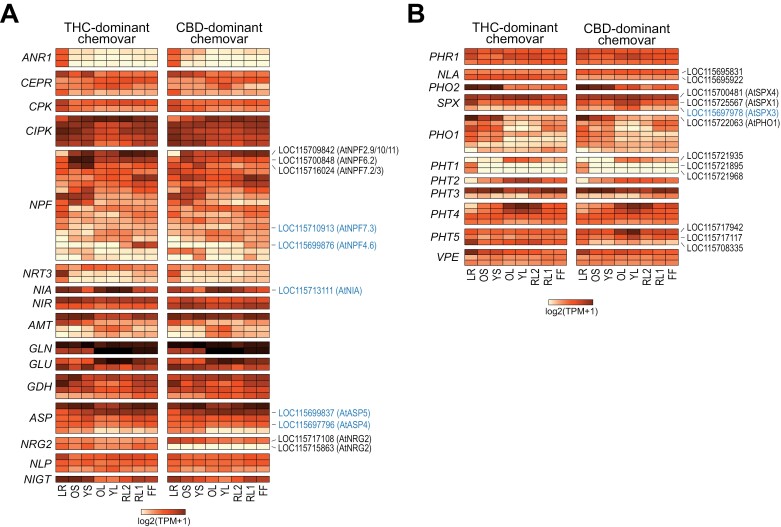
To gain deeper insight into the observed chemovar-specific differences in nutrient status, we analysed the expression of genes involved in the uptake, organ distribution, and signalling of macronutrients nitrogen and phosphorus, which showed the most prominent organ-specific variation in the two chemovars. In Cannabis, both pathways are largely uncharacterized on a molecular or gene level and correspondingly their annotation is limited. Therefore, we first identified all Cannabis homologues of the well-characterized and conserved genes involved in these pathways by sequence homology (BLAST cut-off E<10−50) to Arabidopsis genes and further expert curation (Wang et al., 2021; Paz-Ares et al., 2022). For the expression analysis, only genes with an expression of at least 10 TPM in one organ were included to restrict the analysis to only the major isoforms present (Fig. 9; Supplementary Tables S7, S8).
Fig. 9.
Expression of nitrogen and phosphate homeostasis-related genes across organs of the two chemovars. Shown is the expression of genes involved in the sensing, uptake, distribution, and assimilation of nitrate/ammonium (A) and phosphate (B). Scale of the heatmaps indicates the log2(TPM+1) values determined by RNA-seq (see Supplementary Tables S7, S8 for details). Cannabis genes and their Arabidopsis homologues discussed in the main text are indicated. Genes highlighted in blue have been under selection during Cannabis domestication (Ren et al., 2021). General abbreviations: CBD, cannabidiol; FF, female flower; LR, lateral roots; OL, YL; oldest and youngest fully expanded fan leaf pair; OS, YS; oldest and youngest stem internodes; RL1, higher order reduced (‘sugar’) leaves; RL2, lower (second to fourth) order reduced leaves; THC, tetrahydrocannabinol; TPM, transcripts per million. Gene abbreviations in (A) (in figure order): ANR1, ARABIDOPSIS NITRATE REGULATED 1; CEPR, CEP-peptide receptor; CPK, calcium-dependent protein kinase; CIPK, CBL-interacting serine/threonine-protein kinase; NPF, nitrate and peptide transporter family; NRT3, nitrate transporter 3; NIA, nitrate reductase; NIR, nitrite reductase; AMT, ammonium transporter; GLN, glutamine synthetase; GLU, glutamate synthase; GDH, glutamate dehydrogenase; ASP, aspartate aminotransferase; NRG2, NITRATE REGULATORY GENE2; NLP, NIN-LIKE PROTEIN; NIGT, NITRATE-INDUCIBLE, GARP-TYPE TRANSCRIPTIONAL REPRESSOR. Gene abbreviations in (B) (in figure order): PHR1, PHOSPHATE STARVATION RESPONSE 1; NLA, Nitrogen Limitation Adaptation; PHO2, PHOSPHATE 2; SPX, SPX DOMAIN GENE; PHO1, PHOSPHATE 1; PHT, Phosphate transporter; VPE, vacuolar phosphate efflux.
Nitrogen
Across the two chemovars there were only minor differences in the expression of genes homologous to those involved in the sensing of the nitrogen status in Arabidopsis (Fig. 9A; Supplementary Table S7) (Y-Y. Wang et al., 2018). This matches their overall similar nitrate and organic N profiles shown in Fig. 3B and indicates sufficient N-supply to these Cannabis plants. Indeed, genes of the high-affinity NITRATE TRANSPORTER2 (NRT2) family, which are strongly induced by nitrate starvation, were only very lowly expressed (TPM<2) (Supplementary Table S1). Homologues of the MADS-box transcription factor ANR1 were only expressed in the roots of both chemovars, with one homologue also detected in the stem of the CBD-dominant chemovar (Fig. 9A; Supplementary Table S7; Supplementary Fig. S3). This agrees with its function in Arabidopsis to promote lateral root formation under external nitrate supply and suggests a similar function for ANR1 in the regulation of nitrate-dependent root development in Cannabis.
The main difference between the chemovars was the expression of genes belonging to the NITRATE TRANSPORTER 1 (NRT1)/PEPTIDE TRANSPORTER (PTR) (NPF) family. For our analysis we included only Cannabis homologues with the highest sequence similarity to NPFs with known specificity for nitrate in Arabidopsis (Y-Y. Wang et al., 2018) (Supplementary Fig. S3). These genes showed complex expression patterns across organs of both chemovars, indicating an intricate regulation of nitrate uptake and distribution throughout the plant (Fig. 9A; Supplementary Table S7). Out of 18 genes, three (LOC115716024, LOC115709842, LOC115700848) showed higher expression in the stems of the THC- over the CBD-dominant chemovar suggesting a role in long-distance nitrate transport. Our sequence analysis indicated that LOC115709842 is a homologue of the Arabidopsis NPF2.9/NRT1.9, NPF2.10, and NPF2.11 clade (Supplementary Fig. S3). Its higher expression in the stems of the THC-dominant chemovar might explain higher stem nitrate concentration, which could fuel N assimilation in young fan leaves (Wang and Tsay, 2011; Nour-Eldin et al., 2012) (Figs 3B, 9A). LOC115710913, a homologue of Arabidopsis AtNPF7.3/NRT1.5 involved in root to shoot transport of nitrate (Lin et al., 2008), showed about 7-fold higher expression in roots, but 3-fold lower expression in flowers of the THC- over the CBD-dominant chemovar. Similarly, the LOC11571311 gene encoding nitrate reductase (i.e. the enzyme of the first step in nitrate assimilation) was twice as highly expressed in roots and stems of the THC- than the CBD-dominant chemovar. In both chemovars, LOC115699876, the homologue of the low-affinity Arabidopsis nitrate transporter NFP4.6/NRT1.2 (Huang et al., 1999), was specifically expressed in flowers only, but with about 10-fold higher transcript abundance in the THC-dominant chemovar (Fig. 9A; Supplementary Table S7).
Two Cannabis homologues for a key component of the nitrate regulatory network in Arabidopsis, bZIP TF AtNRG2 (Xu et al., 2016), were detected in the RNA-seq data. The homologue encoded by the LOC115717108 gene was expressed in both chemovars and at similar levels across all organs of the THC-dominant chemovar. For the CBD-dominant chemovar, its expression was higher in the roots (LR) and stem internodes (OS, YS) than in other organs. Interestingly, the second homologue (LOC115715863) was only expressed in the THC-dominant chemovar and at similar levels across all organs, while no expression was detected in the CBD-dominant chemovar.
Phosphorus
Measurements of phosphorus status indicated significant differences in the acquisition, translocation, and assimilation of Pi between the two chemovars (Fig. 3C). Therefore, we investigated the expression of genes involved in Pi homeostasis. Consistent with the constitutive expression of the PHR1-LIKE gene family in Arabidopsis organs (Wang et al., 2022), the three homologues of the major transcriptional regulator of the Pi starvation response MYB-CC transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1) (Rubio et al., 2001) identified by our sequence analyses (Supplementary Fig. S3) showed no difference in expression between the two chemovars, with only minor organ-specific differences in expression between isoforms (Fig. 9B, Supplementary Table S8). Similarly, genes encoding ubiquitin E2 conjugase PHOSPHATE2 (PHO2) and E3 ligase NITROGEN LIMITATION ADAPTATION (NLA), both key post-translational regulators of PHR1 as well as Pi transporting proteins, namely membrane-localized PHOSPHATE TRANSPORTERS (PHTs) and xylem-loading Pi exporter PHOSPHATE1 (PHO1) in Arabidopsis (Aung et al., 2006; Lin et al., 2013; Medici et al., 2019), showed similar expression profiles in the two chemovars (Fig. 9B; Supplementary Fig. S3; Supplementary Table S8). Interestingly, isoform preference switched between chemovars, with NLA-encoding LOC115695922 more highly expressed in organs of the THC-dominant, and LOC115695831 transcripts more abundant in organs of the CBD-dominant chemovar. The PHO2 homologue showed higher expression in roots and stems than other organs. This is consistent with the preferential expression in the vasculature and roots observed for Arabidopsis PHO2 (Aung et al., 2006).
In contrast to the above, Cannabis genes encoding proteins with homology to the central sensors of P status in Arabidopsis and rice, namely the inositol pyrophosphate binding phosphate starvation response co-repressor SPX DOMAIN PROTEIN (SPX), showed more pronounced differences in expression between the chemovars (Lv et al., 2014; Puga et al., 2014; Osorio et al., 2019) (Fig. 9B; Supplementary Table S8). We were able to detect three Cannabis SPX genes and named them here according to their sequence similarity with their Arabidopsis homologues (AtSPX1-4) as CsSPX1 (LOC115725567), CsSPX3 (LOC115697978), and CsSPX4 (LOC115700481) (Supplementary Fig. S3). CsSPX1 and CsSPX3 had varied expression across organs: while both were highly expressed in the root of the THC-dominant chemovar, their transcripts accumulated more strongly (by 2- to 8-fold) in fan leaves of the CBD-dominant chemovar (Fig. 9B; Supplementary Table S3). The differential expression of these two SPX genes might have implications for Pi acquisition and translocation between chemovars. Higher expression in roots than other organs of the THC-dominant chemovar could be a sign of local activation of Pi acquisition and translocation (Hani et al., 2021). Higher expression in leaves of the CBD-dominant chemovar indicates an activation of the phosphate starvation response in shoots to signal higher demand for Pi despite full vacuolar Pi stores (Fig. 3A) (Duan et al., 2008; Wang et al., 2009).
For genes encoding proteins directly involved in the uptake and distribution of Pi, (i.e. PHT and PHO1 transporter families; Supplementary Fig. S2), expression patterns were complex across chemovars and organs owing to their diverse tissue-specific and subcellular functions (Wang et al., 2021). Overall, expression of the CsPHO1 genes was lowest in the leafy organs (OL, YL, RL1), suggesting that Cannabis PHO1 proteins might have similar functions as in Arabidopsis such as loading the xylem with Pi for its translocation to aerial parts (AtPHO1) or the integration with developmental processes such as photoperiod-sensitive regulation of flowering (AtPHO1;H4/SHB1) (Kang and Ni, 2006; Zhou and Ni, 2009; Arpat et al., 2012; Liu et al., 2016). The expression of genes encoding PHT1 to PHT4 transporters, which localize to various membranes within the cell (plasma membrane, PHT1; plastid envelope, PHT2/4; outer mitochondrial membrane, PHT3; M. Gu et al., 2016), and those encoding the vacuolar Pi efflux transporters (tonoplast, PHT5/SPX-MFS and VPTs; Liu et al., 2016; Xu et al., 2019) was largely very similar in the two chemovars with some organ-specific expression (Fig. 9B; Supplementary Fig. S3; Supplementary Table S8). For example, two CsPHT1 genes (LOC115721895, LOC115721968) were root-specific, while the other CsPHT1 gene (LOC115721935) was only expressed in the leaf and floral organs, indicating specific functions of PHT1 family members in Cannabis mediating uptake of Pi by different organs (Mudge et al., 2002; Lapis-Gaza et al., 2014). The Cannabis PHT2 isoform was more strongly expressed in leaves of the THC-dominant chemovar, which coincides with higher chlorophyll and lower starch levels (Fig. 2). Members of the PHT5 family also had chemovar-specific expression patterns: LOC115717117 and LOC115717942 were more highly expressed in fan leaves (YL, OL) of the CBD- than those of the THC-dominant chemovar. One Cannabis PHT5 gene (LOC115708335) showed higher expression in the root than in other organs, with strongest expression in roots of the THC-dominant chemovar (Fig. 9B; Supplementary Table S8). Higher expression of PHT5 family genes in organs of the CBD-dominant chemovar might be a consequence of higher Pi influx requiring increased vacuolar sequestration to maintain cytosolic Pi homeostasis (Liu et al., 2016; Luan et al., 2022).
In conclusion, strong expression of genes encoding NPF nitrate transporter family members in stems of the THC-dominant chemovar could explain higher xylem loading and transient accumulation of nitrate in this organ (Fig. 3B). Organ- and chemovar-specific differences in the expression of SPX and PHT5 genes are in agreement with the observed lack of suppression of Pi acquisition in the CBD-dominant chemovar. However, which mechanisms lead to the loss of this important regulatory function and subsequent Pi hyperaccumulation in sink organs requires further investigation.
Candidate genes with drug- and hemp-type selection history
In the list of genes connecting physiological traits with organ development and differential expression in the two chemovars described above (Figs 8, 9), four genes overlapped with genes associated with the domestication of hemp-type, drug-type, and/or ancestral Cannabis (Ren et al., 2021). The two nitrate transporter genes, LOC115710913 and LOC115699876, were putatively selected in drug-type versus hemp-type Cannabis, while the nitrate reductase-encoding gene LOC115713111 had a positive selection signature in drug-type versus basal Cannabis accessions (Fig. 9, highlighted in blue). LOC115697978 encoding SPX3, a key regulator of phosphate homeostasis (Fig. 9, highlighted in blue), has been selected for in both drug- and hemp-types during their domestication (Ren et al., 2021). Our analyses further support a role of these genes as drivers of drug- and hemp-type divergence or early Cannabis domestication. These may have utility as selective expression markers for genotype improvement.
Discussion
Development of high-yielding plant varieties often leads to genetic erosion due to human selection and mitigation of unfavourable environmental conditions, such as irrigation or fertilizer application, leading to increasing genetic homogeneity and potential loss of important resilience traits (Khoury et al., 2022). Plants from P-impoverished landscapes carefully balance availability of N and P with those of other resources such as water, light, and other macro- and micronutrients (Prodhan et al., 2019). These plants have acquired P efficiency traits to enhance Pi acquisition by roots, remobilize nutrients from older leaves, and remodel membrane lipids to reduce the ratio of phospho- over sulfolipids. In addition, they display greater allocation of P to the leaf mesophyll of mature leaves facilitated by delayed greening of young leaves and tight control of nitrate and sulfate uptake, leaf protein and other N/S pools. However, some plant species have lost tight control over Pi uptake due to its scarcity in the natural environment, to the point where relatively low Pi supply results in hyperaccumulation in leaf mesophyll cells and toxicity symptoms such as necrotic leaf margins and ultimately plant death (Shane et al., 2004a, b; Takagi et al., 2020). By contrast, plants from N-impoverished landscapes take up nitrate and ammonium as they become available and only activate Pi uptake when N pools are in surplus (Gojon et al., 2009; DeLoose et al., 2024). In agricultural systems, plants often have ample nutrient supply and suppress Pi uptake to match N and C assimilation rates as well as water availability. This is especially the case in protected cropping systems—such as hydro-, aero- or aquaponics, where nutrient supply is never limiting and rather nutrient overaccumulation may impact product quality (Colla et al., 2018; Bian et al., 2020).
Cannabis represents an interesting case study, given its two-pronged domestication history: hemp-type Cannabis has been bred predominantly for vegetative, stem biomass and/or seed production, while drug-type Cannabis was selected for maximum female flower biomass and secondary metabolite (cannabinoid) content (Petit et al., 2020; Ren et al., 2021; Schilling et al., 2021). Hemp-type Cannabis is nutrient-efficient as it is often grown on marginal land, whilst drug-type Cannabis is often grown in well-fertilized soil or hydroponically (Small, 2017; McPartland et al., 2019; Zheng et al., 2021). Due to recent interest by the pharmaceutical industry, high CBD content in female drug-type flowers was selected for after introgressive hybridization of hemp- into drug-type Cannabis (Grassa et al., 2021).
In our study we investigated a THC- and a CBD-dominant drug-type chemovar with contrasting trait expressions. The THC dominant cultivar has large compact inflorescences and high cannabinoid yield (Fig. 1). Genes encoding select GRF, MYB, and SPL TFs are strongly and specifically expressed in floral organs, which correlates with a larger and more compact inflorescence (Fig. 8) (Noda et al., 1994; Li et al., 2009; Oshima et al., 2013; Xing et al., 2013; Jung et al., 2016; Wang et al., 2016; Chao et al., 2017; Li et al., 2018). The THC-dominant chemovar also adjusts nutrient uptake to the demand of growing organs, in particular flowers; its leaves are photosynthetically active with high chlorophyll and protein concentration and very low starch levels (Fig. 2). Pools of C, N, P, and other macro- and microelements are well balanced, with higher levels of free nitrate and phosphate in roots and stems indicating sink-driven translocation to leaves and flowers (Figs 3, 4). Expression of genes encoding members of the AS2/LBD, bHLH, DOF, and MYB TF families was mainly restricted to roots where they regulate root architecture in response to nutrient supply (Fig. 8) (Keller et al., 2006; Müller et al., 2006; Kobayashi and Nishizawa, 2012; Zhang et al., 2021; Schulten et al., 2022). Higher expression of, for example, CsNPF2.9 in stems of the THC-dominant chemovar alongside higher overall expression of a second CsNRG2 isoform across organs is consistent with higher nitrate concentrations in translocating tissues (Figs 3, 9A) (Xu et al., 2016). High expression of CsPHO2 in roots and stems suggests effective negative regulation of Pi uptake and translocation through ubiquitin-mediated turnover of PHT1 and PHO1 proteins (Liu et al., 2012; Huang et al., 2013; Xu et al., 2016). Transient storage of Pi in root cell vacuoles is reflected in the high expression of PHT5 and VPE isoforms in roots of the THC-dominant chemovar (Fig. 9B) (Liu et al., 2016; Luan et al., 2022). Strong expression of CsPHT2 in leaves reflects Pi demand of plastids for carbon shuttling due to high photosynthetic activity (Figs 2, 9B) (Carstensen et al., 2018).
By contrast, the CBD-dominant chemovar has limited ability to regulate high nutrient availability, indicating retention of hemp-type features associated with efficient nutrient uptake, which appears to coincide with hyperaccumulation of phosphate observed in other species (Shane et al., 2004a; Tang et al., 2017; Takagi et al., 2020; Anderson et al., 2021). High nutrient supply triggers poor transitioning to flowering—and hence a ‘leafy’ growth habit (Fujita et al., 2014; Lin and Tsay, 2017; Olas et al., 2019). Low expression of genes encoding flower-specific GRF and SPL TFs also indicates weaker commitment to flowering in the CBD-dominant chemovar. Hyperaccumulation of phosphate in sink organs may contribute to reduced photosynthetic activity highlighted by reduced chlorophyll and protein concentration as well as starch accumulation in leaves (Figs 2, 3) (Shane et al., 2004a; Chiou et al., 2005; Zhou et al., 2008; Schlüter et al., 2012; Takagi et al., 2020). High transcript abundance of a MYB11/12/111 homologue and resulting anthocyanin accumulation in leaves of the CBD-dominant chemovar is also indicative of metabolic stress (Figs 2, 8B) (Jezek et al., 2023). Lack of expression of a second CsNRG2 isoform across CBD-dominant chemovar organs suggests reduced nitrate acquisition and translocation, which further exacerbate leaf P toxicity (Figs 3B, 9A) (Xu et al., 2016; Medici et al., 2019), while offloading Pi into mesophyll cell vacuoles is supported by increased expression of two PHT5 isogenes in leaves (Liu et al., 2016; Xu et al., 2019). In the CBD-dominant chemovar, there is a lack of significant down-regulation of PHT1 and PHO1 genes in response to excess Pi in leaves, in conjunction with the high transcript abundance of P status sensors and transcriptional co-repressors CsSPX1 and CsSPX3. In Arabidopsis and rice, these two genes are induced by Pi starvation (Duan et al., 2008; Wang et al., 2009). Their misregulation in leaves of the CBD-dominant chemovar therefore hints at a reduced ability to sense cellular P status, reminiscent of the Arabidopsis pho2-1 mutant that also accumulates toxic levels of Pi in leaves and shows high SPX1 expression under combined low Pi and nitrate supply (Aung et al., 2006; Medici et al., 2019). Misregulation may either be a cause or a consequence of perturbed regulatory networks and further investigations may help to answer the question as to why the CBD-dominant chemovar continues to accumulate Pi to toxic levels in leaves. In this respect, it is of note that expression of one AtLBD37/38/39 homologue (LOC115706665) is very low across organs of the THC-dominant chemovar, while its expression is strong in roots, stems, and floral organs of the CBD-dominant chemovar (Fig. 8B). The three class II LBD isoforms in Arabidopsis are strongly induced by nitrate and inhibit nitrate uptake and assimilation as well as anthocyanin biosynthesis as transcriptional repressors of PAP1/MYB75 and PAP2/MYB90 (Rubin et al., 2009). Similar to the CBD-dominant chemovar, LBD overexpression in Arabidopsis results in less nitrate and protein, as well as higher starch levels compared with wild-type plants. They also show increased basal rosette branching under low nitrogen supply, similar to the shorter internodes and leafier growth habit of the CBD-dominant chemovar (Supplementary Fig. S1) (Rubin et al., 2009). In summary, our results suggest that hemp traits for optimized vegetative growth were retained in the CBD-dominant chemovar and limit flower production and thus cannabinoid yield.
Recent analysis of genomic data from 110 Cannabis sativa accessions determined genomic regions associated with positive selection in either hemp- or drug-type chemovars during domestication (Ren et al., 2021). Remarkably, and similarly to our study, this identified genes linking the regulation of nutrient signalling, and in particular phosphate homeostasis (e.g. SPX proteins, purple acid phosphatases) and nitrogen assimilation (e.g. NRG2, NRT1, NITRATE REDUCTASE, NITRITE REDUCTASE), with flower development (e.g. AGLs, SPLs, LBDs, FT) as key contributors to superior performance in drug-type Cannabis (Ren et al., 2021). In Arabidopsis and rice, knockout of nitrate sensor/transporter NRT1.1 orthologs impairs N utilization and transition to reproductive growth, and the nrt1.1 mutant phenotype in Arabidopsis is rescued by FLC knockdown (Guo et al., 2001; W. Wang et al., 2018; Teng et al., 2019). Nitrate responsive TFs AtNLP6 and AtNLP7 have been implicated in promoting flowering through binding to nitrate responsive elements in the promoters of SPL3 and SPL5, which then modify SOC1 expression (Olas et al., 2019). Interestingly, loss of PHO2 function promotes flowering in warmer conditions through increase expression of TWIN SISTER OF FT (TSF)—consistent with Pi deficiency delaying floral meristem transition and low temperature inhibiting Pi translocation to shoots (Hurry et al., 2000; Kim et al., 2011). In rice, OsPHO2 interacts with OsGIGANTEA (OsGI), and Osgi mutants hyperaccumulated Pi in leaves featuring leaf tip necrosis similar to Ospho2 mutants, with flowering delayed in both rice mutants (Li et al., 2017).
Given these links between nutrient signalling and plant development, it will be paramount to understand how their genetic determinants can be either manipulated or selected for in breeding programmes to suit protected indoor cropping. In Cannabis this is especially important given that the supply of N and P is a driver of growth and reproductive investment and there is an economic trade-off between flower biomass production and fertiliser cost (Elser et al., 2000; Fujita et al., 2014). A key to cultivation of drug-type Cannabis will be to consider the origin of parental lines as this impacts sensitivity to environmental factors such as nutrient supply, day length, or light. While investigating a broader set of genotypes will be necessary to generalise, the phosphate sensitivity of the CBD-dominant chemovar observed in this study is a good example of evolutionary constraints having an impact on performance and yield. From recent research, it is emerging that excessive and environmentally damaging phosphate supply does not improve cannabinoid yield or product quality (Westmoreland and Bugbee, 2022). For future cultivar selection, it will be essential to understand nutritional requirements and to identify underlying genetic markers as this will help reduce fertilizer inputs and create a more sustainable, cost-effective cultivation strategy without affecting yield.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Anatomy of the two analysed chemovars.
Fig. S2. Sequence comparisons for Cannabis and Arabidopsis transcription factors.
Fig. S3. Sequence comparisons for Cannabis and Arabidopsis genes involved in nitrate and phosphate assimilation.
Table S1. TPM values for genes detected by RNA-seq across organs in the two chemovars. Modules assignment by WGCNA is also indicated.
Table S2. Module assignment and module membership (kME) for all genes of the THC-dominant chemovar after WGCNA.
Table S3. Module assignment and module membership (kME) for all genes of the CBD-dominant chemovar after WGCNA.
Table S4. Results of GO term enrichment analyses.
Table S5. Hub genes (kME>0.95) in the flower- and root-specific modules of both chemovars.
Table S6. z-Scored expression of transcription factors in the flower- and root-specific enriched TF families (Fig. 8B).
Table S7. TPM values for genes involved in the nitrate/ammonium sensing, uptake, distribution, or assimilation (Fig. 9).
Table S8. TPM values for genes involved in the phosphate sensing, uptake, distribution, or assimilation.
Acknowledgements
We would like to thank Dr Thy Truong (La Trobe University Proteomics and Metabolomics Platform) for method development in cannabinoid profiling as well as Asha Haslem and the La Trobe University Genomics Platform for next-generation transcriptome sequencing. We also thank Dr Damien L. Callahan (Deakin University) for ICP-MS analyses. The authors acknowledge the generosity of the Ian Potter Foundation (Grant #31110299), the ARC LIEF scheme grant (LE200100117), and La Trobe University infrastructure funds for contributing to the purchase of the mass spectrometers in the La Trobe University Proteomics and Metabolomics Research Platform. We wish to acknowledge support from La Trobe University and industry partners, particularly Cann Group for access to their cultivation facilities, that have made this work possible.
Contributor Information
Ricarda Jost, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Oliver Berkowitz, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Amelia Pegg, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Bhavna Hurgobin, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Muluneh Tamiru-Oli, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Matthew T Welling, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Myrna A Deseo, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Hannah Noorda, Cann Group Limited, Port Melbourne, VIC 3207, Australia.
Filippa Brugliera, Cann Group Limited, Port Melbourne, VIC 3207, Australia.
Mathew G Lewsey, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia; Australian Research Council Centre of Excellence in Plants for Space, La Trobe University, Bundoora, VIC, Australia.
Monika S Doblin, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
Antony Bacic, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, La Trobe University, Bundoora, VIC 3086, Australia.
James Whelan, Australian Research Council Research Hub for Medicinal Agriculture, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia.
Rainer Melzer, University College Dublin, Ireland.
Author contributions
RJ, OB, JW, MGL, MSD, and AB conceptualized the project. AP, RJ, OB, MO, MW, and MAD developed methodologies and carried out experimental work, with support and resources from HN and FB. OB, RJ, AP, and BH performed the data curation, formal analyses, and validation; RJ and OB wrote the original draft with revision by all authors.
Conflict of interest
The authors declare no conflict of interest. HN was an employee and FB is an employee of Cann Group Limited, which provided Cannabis chemovars, expertise, and growth facilities.
Funding
This work was funded by the Australian Research Council Industrial Transformation Research Hub for Medicinal Agriculture (grant ID IH180100006) of which Cann Group Limited is key industry partner.
Data availability
RNA-seq read data of the two chemovars were deposited at the NCBI SRA database under project ID PRJNA884161 and PRJNA884162.
References
- Adesina I, Bhowmik A, Sharma H, Shahbazi A.. 2020. A review on the current state of knowledge of growing conditions, agronomic soil health practices and utilities of hemp in the United States. Agriculture 10, 129. [Google Scholar]
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y.. 2007. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proceedings of the National Academy of Sciences, USA 104, 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL 2nd, Pearson B, Kjelgren R, Brym Z.. 2021. Response of essential oil hemp (Cannabis sativa L.) growth, biomass, and cannabinoid profiles to varying fertigation rates. PLoS One 16, e0252985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpat AB, Magliano P, Wege S, Rouached H, Stefanovic A, Poirier Y.. 2012. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. The Plant Journal 71, 479–491. [DOI] [PubMed] [Google Scholar]
- Aung K, Lin S-I, Wu C-C, Huang Y-T, C-l S, Chiou T-J.. 2006. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology 141, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer R, Schwinghamer T, Rosenbaum P, et al. 2019. Closing the yield gap for cannabis: a meta-analysis of factors determining cannabis yield. Frontiers in Plant Science 10, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaccia G, Palumbo F, Scariolo F, Vannozzi A, Borin M, Bona S.. 2020. Potentials and challenges of genomics for breeding cannabis cultivars. Frontiers in Plant Science 11, 573299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan L, Jones M, Zheng Y.. 2021. Optimisation of nitrogen, phosphorus, and potassium for soilless production of Cannabis sativa in the flowering stage using response surface analysis. Frontiers in Plant Science 12, 764103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Wang Y, Zhang X, Li T, Grundy S, Yang Q, Cheng R.. 2020. A review of environment effects on nitrate accumulation in leafy vegetables grown in controlled environments. Foods 9, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake-Kalff MMA, Hawkesford MJ, Zhao FJ, McGrath SP.. 2000. Diagnosing sulfur deficiency in field-grown oilseed rape (Brassica napus L.) and wheat (Triticum aestivum L.). Plant and Soil 225, 95–107. [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Carstensen A, Herdean A, Schmidt SB, Sharma A, Spetea C, Pribil M, Husted S.. 2018. The impacts of phosphorus deficiency on the photosynthetic electron transport chain. Plant Physiology 177, 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L-M, Liu Y-Q, Chen D-Y, Xue X-Y, Mao Y-B, Chen X-Y.. 2017. Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Molecular Plant 10, 735–748. [DOI] [PubMed] [Google Scholar]
- Chiou T-J, Aung K, Lin S-I, Wu C-C, Chiang S-F, C-l S.. 2005. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. The Plant Cell 18, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Nolan TM, Wang P, Song G, Montes C, Valentine CT, Guo H, Sozzani R, Yin Y, Walley JW.. 2021. Integrated omics networks reveal the temporal signaling events of brassinosteroid response in Arabidopsis. Nature Communications 12, 5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Merlin M.. 2016. Cannabis: evolution and ethnobotany. Berkeley, Los Angeles: University of California Press. [Google Scholar]
- Coffman CB, Gentner WA.. 1977. Responses of greenhouse-grown Cannabis sativa L. to nitrogen, phosphorus, and potassium1. Agronomy Journal 69, 832–836. [Google Scholar]
- Colla G, Kim H-J, Kyriacou MC, Rouphael Y.. 2018. Nitrate in fruits and vegetables. Scientia Horticulturae 237, 221–238. [Google Scholar]
- Conway JR, Lex A, Gehlenborg N.. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho J. 1996. Automated method for sulphate determination in soil‐plant extracts and waters. Communications in Soil Science and Plant Analysis 27, 727–740. [Google Scholar]
- Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, Palacios N, Stitt M.. 2006. Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiology 142, 1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta M, Cavalleri A, Chandler JW, Colombo L.. 2021. Auxin and flower development: a blossoming field. Cold Spring Harbor Perspectives in Biology 13, a039974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuyas L, David P, de Craieye D, et al. 2023. Identification and interest of molecular markers to monitor plant Pi status. BMC Plant Biology 23, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danziger N, Bernstein N.. 2021. Plant architecture manipulation increases cannabinoid standardization in ‘drug-type’ medical cannabis. Industrial Crops and Products 167, 113528. [Google Scholar]
- DeLoose M, Clúa J, Cho H, Zheng L, Masmoudi K, Desnos T, Krouk G, Nussaume L, Poirier Y, Rouached H.. 2024. Recent advances in unraveling the mystery of combined nutrient stress in plants. The Plant Journal 117, 1764–1780. [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P.. 2008. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. The Plant Journal 54, 965–975. [DOI] [PubMed] [Google Scholar]
- Elser JJ, Sterner RW, Gorokhova E, Fagan WF, Markow TA, Cotner JB, Harrison JF, Hobbie SE, Odell GM, Weider LW.. 2000. Biological stoichiometry from genes to ecosystems. Ecology Letters 3, 540–550. [Google Scholar]
- Farrelly AM, Vlachou S, Grintzalis K.. 2021. Efficacy of phytocannabinoids in epilepsy treatment: novel approaches and recent advances. International Journal of Environmental Research and Public Health 18, 3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Venterink HO, van Bodegom PM, et al. 2014. Low investment in sexual reproduction threatens plants adapted to phosphorus limitation. Nature 505, 82–86. [DOI] [PubMed] [Google Scholar]
- Gardener H, Wallin C, Bowen J.. 2022. Heavy metal and phthalate contamination and labeling integrity in a large sample of US commercially available cannabidiol (CBD) products. The Science of the Total Environment 851, 158110. [DOI] [PubMed] [Google Scholar]
- Gojon A, Nacry P, Davidian J-C.. 2009. Root uptake regulation: a central process for NPS homeostasis in plants. Current Opinion in Plant Biology 12, 328–338. [DOI] [PubMed] [Google Scholar]
- Grassa CJ, Weiblen GD, Wenger JP, Dabney C, Poplawski SG, Timothy Motley S, Michael TP, Schwartz CJ.. 2021. A new Cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytologist 230, 1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Chen A, Sun S, Xu G.. 2016. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Molecular Plant 9, 396–416. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Gülck T, Møller BL.. 2020. Phytocannabinoids: Origins and biosynthesis. Trends in Plant Science 25, 985–1004. [DOI] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM.. 2001. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. The Plant Cell 13, 1761–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Okamoto Y.. 2017. Simple spectroscopic determination of nitrate, nitrite, and ammonium in Arabidopsis thaliana. Bio-Protocol 7, e2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hani S, Cuyas L, David P, et al. 2021. Live single-cell transcriptional dynamics via RNA labelling during the phosphate response in plants. Nature Plants 7, 1050–1064. [DOI] [PubMed] [Google Scholar]
- Hayes PE, Clode PL, Guilherme Pereira C, Lambers H.. 2019. Calcium modulates leaf cell-specific phosphorus allocation in Proteaceae from south-western Australia. Journal of Experimental Botany 70, 3995–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF.. 1999. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. The Plant Cell 11, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI, et al. 2013. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. The Plant Cell 25, 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurry V, Strand A, Furbank R, Stitt M.. 2000. The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of Arabidopsis thaliana. The Plant Journal 24, 383–396. [DOI] [PubMed] [Google Scholar]
- Husain R, Weeden H, Bogush D, Deguchi M, Soliman M, Potlakayala S, Katam R, Goldman S, Rudrabhatla S.. 2019. Enhanced tolerance of industrial hemp (Cannabis sativa L.) plants on abandoned mine land soil leads to overexpression of cannabinoids. PLoS One 14, e0221570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Coupland G.. 2017. Competence to flower: age-controlled sensitivity to environmental cues. Plant Physiology 173, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvardsen CR, Brinch-Pedersen H.. 2023. Challenges and potentials of new breeding techniques in Cannabis sativa. Frontiers in Plant Science 14, 1154332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezek M, Allan AC, Jones JJ, Geilfus C-M.. 2023. Why do plants blush when they are hungry? New Phytologist 239, 494–505. [DOI] [PubMed] [Google Scholar]
- Jin D, Jin S, Chen J.. 2019. Cannabis indoor growing conditions, management practices, and post-harvest treatment: a review. American Journal of Plant Sciences 10, 925–946. [Google Scholar]
- Jost R, Pharmawati M, Lapis-Gaza HR, Rossig C, Berkowitz O, Lambers H, Finnegan PM.. 2015. Differentiating phosphate-dependent and phosphate-independent systemic phosphate-starvation response networks in Arabidopsis thaliana through the application of phosphite. Journal of Experimental Botany 66, 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-H, Lee H-J, Ryu Jae Y, Park C-M.. 2016. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Molecular Plant 9, 1647–1659. [DOI] [PubMed] [Google Scholar]
- Kang X, Ni M.. 2006. Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. The Plant Cell 18, 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P.. 2006. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. The Plant Cell 18, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury CK, Brush S, Costich DE, et al. 2022. Crop genetic erosion: understanding and responding to loss of crop diversity. New Phytologist 233, 84–118. [DOI] [PubMed] [Google Scholar]
- Kim W, Ahn HJ, Chiou T-J, Ahn JH.. 2011. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Molecules and Cells 32, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK.. 2012. Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology 63, 131–152. [DOI] [PubMed] [Google Scholar]
- Landi S, Berni R, Capasso G, Hausman J-F, Guerriero G, Esposito S.. 2019. Impact of nitrogen nutrition on Cannabis sativa: an update on the current knowledge and future prospects. International Journal of Molecular Sciences 20, 5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S.. 2011. Is my network module preserved and reproducible? PLoS Computational Biology 7, e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapis-Gaza HR, Jost R, Finnegan PM.. 2014. Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biology 14, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, et al. 2018. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ying Y, Secco D, Wang C, Narsai R, Whelan J, Shou H.. 2017. Molecular interaction between PHO2 and GIGANTEA reveals a new crosstalk between flowering time and phosphate homeostasis in Oryza sativa. Plant, Cell & Environment 40, 1487–1499. [DOI] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW.. 2009. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. The Plant Cell 21, 72–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. [34] Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382. [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, et al. 2008. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. The Plant Cell 20, 2514–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-Y, Huang T-K, Chiou T-J.. 2013. NITROGEN LIMITATION ADAPTATION, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. The Plant Cell 25, 4061–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Tsay YF.. 2017. Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. Journal of Experimental Botany 68, 2603–2609. [DOI] [PubMed] [Google Scholar]
- Linger P, Müssig J, Fischer H, Kobert J.. 2002. Industrial hemp (Cannabis sativa L.) growing on heavy metal contaminated soil: fibre quality and phytoremediation potential. Industrial Crops and Products 16, 33–42. [Google Scholar]
- Liu T-Y, Huang T-K, Tseng C-Y, Lai Y-S, Lin S-I, Lin W-Y, Chen J-W, Chiou T-J.. 2012. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. The Plant Cell 24, 2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Huang T-K, Yang S-Y, Hong Y-T, Huang S-M, Wang F-N, Chiang S-F, Tsai S-Y, Lu W-C, Chiou T-J.. 2016. Identification of plant vacuolar transporters mediating phosphate storage. Nature Communications 7, 11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston SJ, Bae EJ, Unda F, Hahn MG, Mansfield SD, Page JE, Samuels AL.. 2021. Cannabis glandular trichome cell walls undergo remodeling to store specialized metabolites. Plant and Cell Physiology 62, 1944–1962. [DOI] [PubMed] [Google Scholar]
- Lu HC, Mackie K.. 2016. An introduction to the endogenous cannabinoid system. Biological Psychiatry 79, 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HT, Li W, Deseo MA, Stevens GW, Bacic A, Doblin MS, Mumford KA.. 2023. Green method for recovery of cannabinoids from Cannabis sativa flowers: pH-controlled aqueous leaching. Separation and Purification Technology 326, 124754. [Google Scholar]
- Luan M, Zhao F, Sun G, Xu M, Fu A, Lan W, Luan S.. 2022. A SPX domain vacuolar transporter links phosphate sensing to homeostasis in Arabidopsis. Molecular Plant 15, 1590–1601. [DOI] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, et al. 2014. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. The Plant Cell 26, 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPartland JM, Hegman W, Long T.. 2019. Cannabis in Asia: its center of origin and early cultivation, based on a synthesis of subfossil pollen and archaeobotanical studies. Vegetation History and Archaeobotany 28, 691–702. [Google Scholar]
- Medici A, Szponarski W, Dangeville P, et al. 2019. Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. The Plant Cell 31, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergner J, Frejno M, Messerer M, Lang D, Samaras P, Wilhelm M, Mayer KFX, Schwechheimer C, Kuster B.. 2020. Proteomic and transcriptomic profiling of aerial organ development in Arabidopsis. Scientific Data 7, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW.. 2002. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. The Plant Journal 31, 341–353. [DOI] [PubMed] [Google Scholar]
- Müller D, Schmitz G, Theres K.. 2006. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. The Plant Cell 18, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K-i, Glover BJ, Linstead P, Martin C.. 1994. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369, 661–664. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jørgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA.. 2012. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534. [DOI] [PubMed] [Google Scholar]
- Olas JJ, Van Dingenen J, Abel C, Działo MA, Feil R, Krapp A, Schlereth A, Wahl V.. 2019. Nitrate acts at the Arabidopsis thaliana shoot apical meristem to regulate flowering time. New Phytologist 223, 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M.. 2013. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. The Plant Cell 25, 1609–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio MB, Ng S, Berkowitz O, De Clercq I, Mao C, Shou H, Whelan J, Jost R.. 2019. SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiology 181, 332–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Kui H, Li J.. 2021. Receptor-like kinases in root development: current progress and future directions. Molecular Plant 14, 166–185. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Puga MI, Rojas-Triana M, Martinez-Hevia I, Diaz S, Poza-Carrión C, Miñambres M, Leyva A.. 2022. Plant adaptation to low phosphorus availability: Core signaling, crosstalks, and applied implications. Molecular Plant 15, 104–124. [DOI] [PubMed] [Google Scholar]
- Petit J, Salentijn EMJ, Paulo M-J, Denneboom C, van Loo EN, Trindade LM.. 2020. Elucidating the genetic architecture of fiber quality in hemp (Cannabis sativa L.) using a genome-wide association study. Frontiers in Genetics 11, 566314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodhan MA, Finnegan PM, Lambers H.. 2019. How does evolution in phosphorus-impoverished landscapes impact plant nitrogen and sulfur assimilation? Trends in Plant Science 24, 69–82. [DOI] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, et al. 2014. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punja ZK, Holmes JE.. 2020. Hermaphroditism in marijuana (Cannabis sativa L.) inflorescences – impact on floral morphology, seed formation, progeny sex ratios, and genetic variation. Frontiers in Plant Science 11, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn B, Pearson BJ, Trigiano RN, Gray DJ.. 2016. The derivation of modern cannabis varieties. Critical Reviews in Plant Sciences 35, 328–348. [Google Scholar]
- Ren G, Zhang X, Li Y, et al. 2021. Large-scale whole-genome resequencing unravels the domestication history of Cannabis sativa. Science Advances 7, eabg2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible W-R.. 2009. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. The Plant Cell 21, 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J.. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo EB, Marcu J.. 2017. Cannabis pharmacology: the usual suspects and a few promising leads. In: Kendall D, Alexander SPH, eds. Advances in pharmacology. Vol. 80. Academic Press, 67–134. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Kanno S, Mabuchi A, Monda K, Iba K, Yanagisawa S.. 2018. A phytochrome-B-mediated regulatory mechanism of phosphorus acquisition. Nature Plants 4, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Saloner A, Bernstein N.. 2021. Nitrogen supply affects cannabinoid and terpenoid profile in medical cannabis (Cannabis sativa L.). Industrial Crops and Products 167, 113516. [Google Scholar]
- Sánchez-Rodríguez AR, del Campillo MC, Torrent J.. 2014. The severity of iron chlorosis in sensitive plants is related to soil phosphorus levels. Journal of the Science of Food and Agriculture 94, 2766–2773. [DOI] [PubMed] [Google Scholar]
- Schilling S, Dowling CA, Shi J, Ryan L, Hunt DJL, O’Reilly E, Perry AS, Kinnane O, McCabe PF, Melzer R.. 2021. The cream of the crop: biology, breeding, and applications of Cannabis sativa. In: Roberts JA, ed. Annual plant reviews online. Vol. 4. John Wiley & Sons, 471–528. [Google Scholar]
- Schlüter U, Mascher M, Colmsee C, Scholz U, Bräutigam A, Fahnenstich H, Sonnewald U.. 2012. Maize source leaf adaptation to nitrogen deficiency affects not only nitrogen and carbon metabolism but also control of phosphate homeostasis. Plant Physiology 160, 1384–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten A, Pietzenuk B, Quintana J, et al. 2022. Energy status-promoted growth and development of Arabidopsis require copper deficiency response transcriptional regulator SPL7. The Plant Cell 34, 3873–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Ponce-Soto GY, Krause K, et al. 2019. MapMan4: A refined protein classification and annotation framework applicable to multi-omics data analysis. Molecular Plant 12, 879–892. [DOI] [PubMed] [Google Scholar]
- Shane MW, McCully ME, Lambers H.. 2004a. Tissue and cellular phosphorus storage during development of phosphorus toxicity in Hakea prostrata (Proteaceae). Journal of Experimental Botany 55, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Shane MW, Szota C, Lambers H.. 2004b. A root trait accounting for the extreme phosphorus sensitivity of Hakea prostrata (Proteaceae). Plant, Cell & Environment 27, 991–1004. [Google Scholar]
- Shiponi S, Bernstein N.. 2021. Response of medical cannabis (Cannabis sativa L.) genotypes to P supply under long photoperiod: functional phenotyping and the ionome. Industrial Crops and Products 161, 113154. [Google Scholar]
- Sievers F, Wilm A, Dineen D, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E. 2017. Cannabis: a complete guide. Boca Raton, London, New York: CRC Press. [Google Scholar]
- Smith AM, Stitt M.. 2007. Coordination of carbon supply and plant growth. Plant, Cell & Environment 30, 1126–1149. [DOI] [PubMed] [Google Scholar]
- Song C, Saloner A, Fait A, Bernstein N.. 2023. Nitrogen deficiency stimulates cannabinoid biosynthesis in medical cannabis plants by inducing a metabolic shift towards production of low-N metabolites. Industrial Crops and Products 202, 116969. [Google Scholar]
- Spitzer-Rimon B, Duchin S, Bernstein N, Kamenetsky R.. 2019. Architecture and florogenesis in female Cannabis sativa plants. Frontiers in Plant Science 10, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D. 2014. Cannabis, pesticides and conflicting laws: the dilemma for legalized States and implications for public health. Regulatory Toxicology and Pharmacology 69, 284–288. [DOI] [PubMed] [Google Scholar]
- Stout JM, Boubakir Z, Ambrose SJ, Purves RW, Page JE.. 2012. The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. The Plant Journal 71, 353–365. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B.. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal 50, 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struik PC, Amaducci S, Bullard MJ, Stutterheim NC, Venturi G, Cromack HTH.. 2000. Agronomy of fibre hemp (Cannabis sativa L.) in Europe. Industrial Crops and Products 11, 107–118. [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, et al. 2010. Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. The Plant Cell 22, 2872–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi D, Miyagi A, Tazoe Y, Suganami M, Kawai-Yamada M, Ueda A, Suzuki Y, Noguchi K, Hirotsu N, Makino A.. 2020. Phosphorus toxicity disrupts Rubisco activation and reactive oxygen species defence systems by phytic acid accumulation in leaves. Plant, Cell & Environment 43, 2033–2053. [DOI] [PubMed] [Google Scholar]
- Tang K, Struik PC, Amaducci S, Stomph T-J, Yin X.. 2017. Hemp (Cannabis sativa L.) leaf photosynthesis in relation to nitrogen content and temperature: implications for hemp as a bio-economically sustainable crop. GCB Bioenergy 9, 1573–1587. [Google Scholar]
- Teng Y, Liang Y, Wang M, Mai H, Ke L.. 2019. Nitrate Transporter 1.1 is involved in regulating flowering time via transcriptional regulation of FLOWERING LOCUS C in Arabidopsis thaliana. Plant Science 284, 30–36. [DOI] [PubMed] [Google Scholar]
- UNDOC. 2023. UN World Drug Report 2023. Vienna: United Nations Office on Drugs and Crime (UNODC). [Google Scholar]
- Vega-Mas I, Marino D, Sánchez-Zabala J, González-Murua C, Estavillo JM, González-Moro MB.. 2015. CO2 enrichment modulates ammonium nutrition in tomato adjusting carbon and nitrogen metabolism to stomatal conductance. Plant Science 241, 32–44. [DOI] [PubMed] [Google Scholar]
- Wang W, Hu B, Yuan D, et al. 2018. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. The Plant Cell 30, 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang F, Lu H, Liu Y, Mao C.. 2021. Phosphate uptake and transport in plants: an elaborate regulatory system. Plant and Cell Physiology 62, 564–572. [DOI] [PubMed] [Google Scholar]
- Wang Y-Y, Cheng Y-H, Chen K-E, Tsay Y-F.. 2018. Nitrate transport, signaling, and use efficiency. Annual Review of Plant Biology 69, 85–122. [DOI] [PubMed] [Google Scholar]
- Wang Y-Y, Tsay Y-F.. 2011. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. The Plant Cell 23, 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P.. 2009. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. Journal of Integrative Plant Biology 51, 663–674. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang Y, Kohalmi SE, Amyot L, Hannoufa A.. 2016. SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2 controls floral organ development and plant fertility by activating ASYMMETRIC LEAVES 2 in Arabidopsis thaliana. Plant Molecular Biology 92, 661–674. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zheng Z, Zhu Y, Kong S, Liu D.. 2022. PHOSPHATE RESPONSE 1 family members act distinctly to regulate transcriptional responses to phosphate starvation. Plant Physiology 191, 1324–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welling MT, Deseo MA, Bacic A, Doblin MS.. 2021. Untargeted metabolomic analyses reveal chemical complexity of dioecious Cannabis flowers. Australian Journal of Chemistry 74, 463–479. [Google Scholar]
- Westmoreland FM, Bugbee B.. 2022. Sustainable Cannabis nutrition: elevated root-zone phosphorus significantly increases leachate P and does not improve yield or quality. Frontiers in Plant Science 13, 1015652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hall SA, Hawkesford MJ, Beale MH, Cooper RM.. 2002. Elemental sulfur and thiol accumulation in tomato and defense against a fungal vascular pathogen. Plant Physiology 128, 150–159. [PMC free article] [PubMed] [Google Scholar]
- Wrolstad RE, Durst RW, Lee J.. 2005. Tracking color and pigment changes in anthocyanin products. Trends in Food Science & Technology 16, 423–428. [Google Scholar]
- Wu L-B, Holtkamp F, Wairich A, Frei M.. 2019. Potassium ion channel gene OsAKT1 affects iron translocation in rice plants exposed to iron toxicity. Frontiers in Plant Science 10, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, et al. 2021. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Salinas M, Garcia-Molina A, Höhmann S, Berndtgen R, Huijser P.. 2013. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. The Plant Journal 75, 566–577. [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao H, Wan R, et al. 2019. Identification of vacuolar phosphate efflux transporters in land plants. Nature Plants 5, 84–94. [DOI] [PubMed] [Google Scholar]
- Xu N, Wang R, Zhao L, et al. 2016. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. The Plant Cell 28, 485–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Li C, Dong X, et al. 2020. MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. The Plant Cell 32, 2196–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Wang X, Chen Q, Callahan DL, Fournier-Level A, Whelan J, Jost R.. 2021. Diverse phosphate and auxin transport loci distinguish phosphate tolerant from sensitive Arabidopsis accessions. Plant Physiology 187, 2656–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mitsuda N, Yoshizumi T, Horii Y, Oshima Y, Ohme-Takagi M, Matsui M, Kakimoto T.. 2021. Two types of bHLH transcription factor determine the competence of the pericycle for lateral root initiation. Nature Plants 7, 633–643. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Fiddes K, Yang L.. 2021. A narrative review on environmental impacts of cannabis cultivation. Journal of Cannabis Research 3, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P.. 2008. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146, 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ni M.. 2009. SHB1 plays dual roles in photoperiodic and autonomous flowering. Developmental Biology 331, 50–57. [DOI] [PubMed] [Google Scholar]
- Zou X, Sun H.. 2023. DOF transcription factors: specific regulators of plant biological processes. Frontiers in Plant Science 14, 1044918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq read data of the two chemovars were deposited at the NCBI SRA database under project ID PRJNA884161 and PRJNA884162.