Abstract
The continuing evolution of SARS-CoV-2 variants challenges the durability of existing spike (S)-based COVID-19 vaccines. We hypothesized that vaccines composed of both S and nucleocapsid (N) antigens would increase the durability of protection by strengthening and broadening cellular immunity compared with S-based vaccines. To test this, we examined the immunogenicity and efficacy of wild-type SARS-CoV-2 S- and N-based DNA vaccines administered individually or together to K18-hACE2 mice. S, N, and S + N vaccines all elicited polyfunctional CD4+ and CD8+ T cell responses and provided short-term cross-protection against Beta and Omicron BA.2 variants, but only co-immunization with S + N vaccines provided long-term protection against Omicron BA.2. Depletion of CD4+ and CD8+ T cells reduced the long-term efficacy, demonstrating a crucial role for T cells in the durability of protection. These findings underscore the potential to enhance long-lived protection against SARS-CoV-2 variants by combining S and N antigens in next-generation COVID-19 vaccines.
Subject terms: Vaccines, Biotechnology
Introduction
In response to the alarming transmissibility of SARS-CoV-2 virus and the resulting COVID-19 pandemic, more than 13 billion doses of COVID-19 vaccines have been administered in the past 3 years1. These vaccines, based on various platforms, have been shown to reduce SARS-CoV-2 disease severity and death; nevertheless, they still face two major challenges that adversely affect their effectiveness: the emergence of mutated variants of concern (VOCs) that evade vaccine-induced immunity2,3, and the rapidity with which vaccine-induced immunity wanes, as supported by studies showing loss of 20%–50% humoral immunity within 4 to 5 months after vaccination4–7. These two factors have driven recommendations for booster doses to maintain vaccine cross-protection against SARS-CoV-2 VOCs and are the main reasons that COVID-19 remains a threat to public health worldwide. Thus, there remains a pressing need to develop vaccines that elicit broader and more durable protective immunity against SARS-CoV-2.
All currently licensed COVID-19 vaccines target the SARS-CoV-2 spike (S) protein with the goal of eliciting neutralizing Abs (nAbs), although they also activate a T cell response8,9. Key VOC mutations are concentrated in the S protein and have been shown to limit the vaccine-induced nAb response10. Several studies suggest that a SARS-CoV-2-specific T cell response could provide broad and durable protection against COVID-1911–16, even in the absence of nAbs17,18, because T cell responses that target multiple viral proteins and conserved epitopes could potentially compensate for the waning nAb response15,19,20. Another study demonstrated that protection elicited by mRNA vaccination against VOCs relied on contributions from both humoral and cellular immunity21. These studies support a vaccine development strategy in which the antigens elicit robust T cell responses in addition to nAbs.
The SARS-CoV-2 nucleocapsid (N) protein, which plays a role in viral RNA genome packaging and assembly into particles, is abundant and much more conserved than S protein across SARS-CoV-2 VOCs22. Antigenically, N protein triggers both Ab and T cell responses23,24 that correlate with control of SARS-CoV-2 infection in humans and K18-hACE2 mouse model17,25,26. Longitudinal studies of SARS-CoV-1–infected patients have shown that N-specific memory T cells are sustained for up to 1127 to 17 years28, suggesting that N-specific cellular immunity against the related SARS-CoV-2 may also be durable. T cell responses to conserved antigens such as the N protein are expected to provide broader cross-protection against SARS-CoV-2 variants compared with responses to S protein alone, and co-immunization with both S and N antigens was thus hypothesized as a means to increase both the durability and breadth of vaccine-induced protection29–37. Accordingly, combined (S + N) mRNA vaccines have been shown to confer improved short-term efficacy against SARS-CoV-2 variants compared with vaccines based on S protein alone38,39. However, these studies have not addressed whether combination vaccines improve the durability of vaccine-induced immunity.
In this study, we compared the efficacy of DNA vaccines encoding wild-type SARS-CoV-2 (D614G) S and N proteins, administered separately or in combination, in protecting against infection with Beta and Omicron BA.2 variants in transgenic K18-hACE2 mice, which express high levels of the human SARS-CoV-2 receptor angiotensin-converting enzyme (ACE2) and are highly susceptible to SARS-CoV-2 infection40. The durability of vaccine-induced cross-protection was evaluated over the short and long term by challenging with virus at ~35 and ~140 days, respectively, post-immunization. The S, N, and S + N DNA vaccination protocols all elicited Ab responses against S and N proteins derived from D614G, Beta, and Omicron BA.2 strains; however, only the S and S + N vaccines induced nAbs against D614G and Beta, and none of the vaccines induced nAbs against Omicron BA.2 or Omicron XBB1.5. Notably, all three vaccines elicited polyfunctional and cytotoxic CD4+ and CD8+ T cell responses against the appropriate immunodominant S and/or N protein epitopes. In the short-term protocol, vaccination with S, N, and S + N protected against infection of the upper and/or lower respiratory tract with Beta or Omicron BA.2 variants; but strikingly, only the S + N vaccination protocol provided complete and sustained long-term (140-day) protection of the lungs against Omicron BA.2, even though cross-reactive anti-Omicron BA.2 nAbs were undetectable well before viral infection. Cell depletion experiments showed that this long-term protection was mediated by T cells, at least in part, suggesting that vaccines targeting both the N and S proteins can elicit T cell immunity that increases the durability of vaccine-induced immunity, as we initially hypothesized. Thus, our data support the development of combined S- and N-based DNA vaccines to elicit more durable protective immunity against SARS-CoV-2 compared with that induced by vaccines targeted to a single protein.
Results
S, N, and S + N DNA vaccines elicit short-term S, N, and RBD protein-binding IgG and nAb responses against homologous and/or heterologous SARS-CoV-2 strains
To evaluate the immunogenicity and protective efficacy of SARS-CoV-2 S and N DNA vaccines, two vaccines were constructed based on optimized sequences encoding full-length S or N proteins from wild-type SARS-CoV-2 (USA/WA-CDC-WA1/2020) cloned into pVAX plasmid vector41. We first assessed the humoral response by priming (day 0) and boosting (day 21) K18-hACE2 transgenic mice with S or N vaccines intramuscularly (ipsilateral quadriceps for prime and boost) or with both S and N vaccines simultaneously (contralateral quadriceps for prime and boost), and collecting blood before (day 20) or after (days 28 and 35) boosting (Fig. 1a). SARS-CoV-2 N-, S- and RBD-binding IgG endpoint titers were then analyzed by ELISA, and production of S-specific nAbs was analyzed with the pVNT assay, which uses recombinant replication-deficient VSV vectors pseudotyped with S proteins from the SARS-CoV-2 variants.
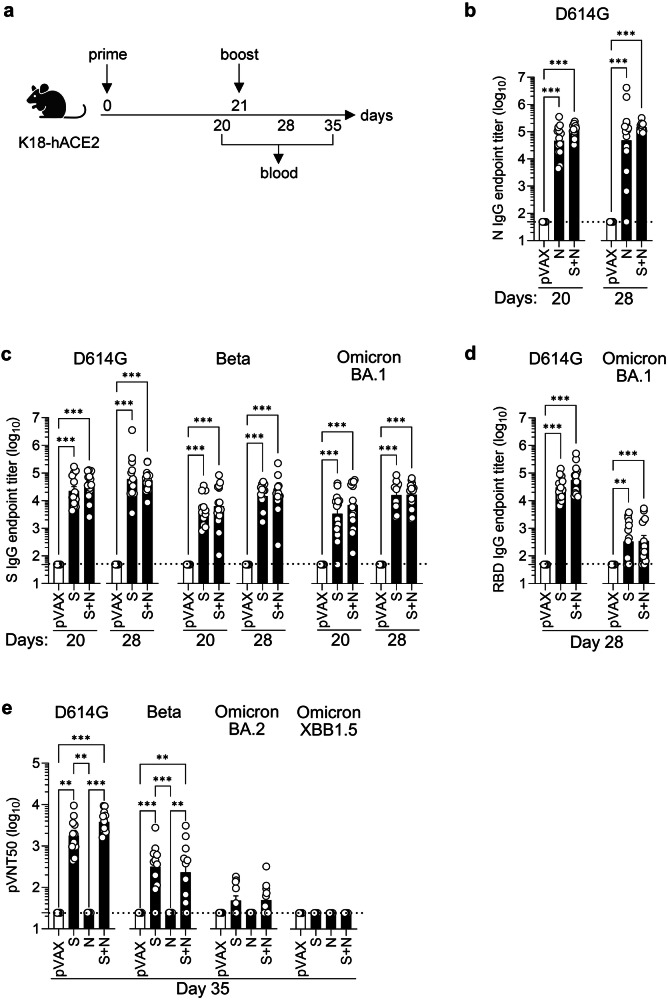
Fig. 1. S and/or N DNA vaccines induce SARS-CoV-2 S- and N-specific IgG and nAb responses in K18-hACE2 mice.
a Experimental protocol: On days 0 and 21, K18-hACE2 mice were inoculated with S and/or N DNA vaccines or pVAX negative control (25 µg, IM), and blood was harvested 1 day pre-boost and 7- and 14-days post-boost. Representation of the experiment timeline was Created with BioRender.com. b–d ELISA assays of serum harvested on days 20 and 28 showing endpoint titers of IgG specific for N, S, or RBD proteins from the indicated SARS-CoV-2 strains. Data are from 3 independent experiments (n = 13–15 mice/group). e Serum harvested on day 35 was subjected to pVNT50 nAb assay using pseudoviruses harboring S proteins from the indicated SARS-CoV-2 strains. Data are from 2 independent experiments (n = 10–11 mice/group). b–e Serum samples were tested in duplicate on the same plates. Group means were compared by the non-parametric Kruskal-Wallis test. *P < 0.05, **P < 0.01, ***P < 0.001. Circles, individual mice; dashed lines, detection limit.
Compared with the pVAX negative control, the N and S + N vaccines elicited high levels of wild-type (D614G) SARS-CoV-2 N protein-binding IgG, with titers that were similar on days 20 (pre-boost) and 28 (post-boost) (Fig. 1b). S and S + N vaccines induced robust production of wild-type SARS-CoV-2 S protein-binding IgGs that were cross-reactive with S protein from Beta and Omicron BA.1 (Fig. 1c), although the anti-D614G S IgG titer was slightly higher compared with the anti-Beta S and anti-Omicron BA.1 S IgG titers. These S-binding IgG titers were only minimally increased post-boost, suggesting that a near-maximal response had been elicited by priming alone. The RBD-binding IgG response was also evaluated because RBD is the dominant target for SARS-CoV-2 nAbs42,43. At 14 days post-boost, S- and S + N-vaccinated mice had similar titers of anti-D614G and anti-Omicron BA.1 RBD IgG titers, but with both vaccination protocols, the response to D614G RBD was much more robust than that to Omicron BA.1 RBD (Fig. 1d).
To evaluate the neutralizing activity of vaccine-elicited anti-SARS-CoV-2 Abs, sera collected at 14 days post-boost were subjected to a pVNT assay. As expected, given that the assay is S protein-specific, neither the pVAX control vaccine nor the N vaccine elicited nAbs (Fig. 1e). In contrast, S and S + N vaccination elicited high nAb titers against homologous SARS-CoV-2 (D614G) and Beta, but only low levels of nAbs against Omicron BA.2 were detectable in sera from 5 of the 11 S-vaccinated mice and 6 of the 11 S + N-vaccinated mice; furthermore, none of the S- or S + N-vaccinated mice exhibited nAbs against Omicron XBB1.5 (Fig. 1e).
These data indicate that the S and S + N vaccination protocols elicited comparable S protein-binding IgG responses against D614G, Beta, and Omicron BA.1 SARS-CoV-2 strains, whereas both vaccination protocols elicited higher titers of RBD-binding IgG and/or nAbs against the homologous D614G strain than against either Beta or Omicron BA.1, BA.2, or XBB1.5. Importantly, all of the Ab responses in the S + N co-vaccinated mice were as robust or better than those observed in the N or S single-vaccinated mice, indicating that co-administration of the N vaccine did not impair the humoral immune response to the S protein, and vice versa.
S, N, and S + N DNA vaccines elicit antigen-specific polyfunctional CD4+ and CD8+ T cell responses
To investigate T cell responses elicited by our S and/or N vaccines, we quantified antigen-specific T cells in spleens and lungs harvested from immunized K18-hACE2 mice at 7 days post-boost (Fig. 2a). Splenocytes and lung leukocytes were stimulated in vitro with SARS-CoV-2 S or N peptides that were previously identified as the immunodominant epitopes for CD8+ and CD4+ T cells in C57BL/6 mice (the genetic background of K18-hACE2 mice) (Table 1)44. Following in vitro stimulation, effector memory CD4+ and CD8+ T cells were phenotyped by intracellular cytokine staining (ICS). Representative flow cytometry plots are shown in Figure S1.
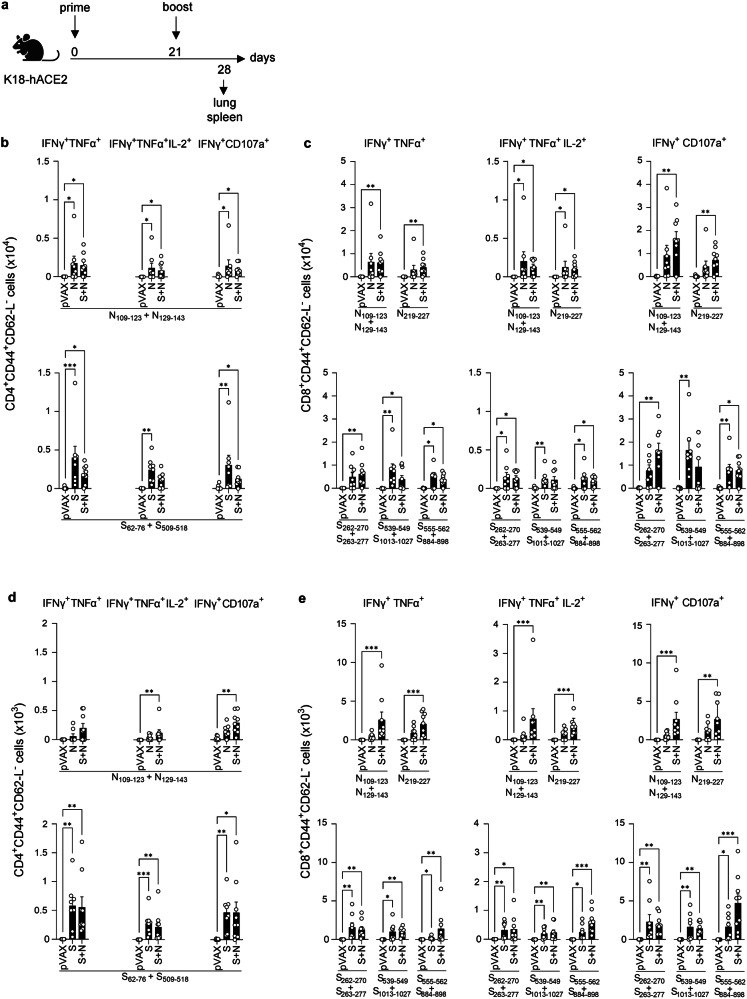
Fig. 2. S and/or N DNA vaccines induce antigen-specific polyfunctional T cell responses in K18-hACE2 mice.
a Experimental protocol: On days 0 and 21, K18-hACE2 mice were inoculated with S and/or N DNA vaccines or pVAX control (25 µg, IM), and tissues were harvested at 7 days post-boost. Representation of the experiment timeline was Created with BioRender.com. b–e Splenocytes (b, c) and lung leukocytes (d, e) were stimulated with the indicated SARS-CoV-2 S or N peptides, and polyfunctional (IFN-γ+TNF-α+ or IFN-γ+TNF-α+IL-2+) or cytotoxic (IFN-γ+CD107a+) CD4+ and CD8+ T cells within the population of live CD3+ cells were quantified by ICS (representative gating strategy is shown in Figure S1). Data are from 2 independent experiments (n = 7–9 mice/group). Group means were compared using the non-parametric Kruskal-Wallis test. *P < 0.05, **P < 0.01, ***P < 0.001. Circles, individual mice.
Table 1.
S and N peptide sequences from SARS-CoV-2 reference strain
| Protein | Peptide | Sequence |
|---|---|---|
| Nucleoprotein (N) | N109-123 | YFYYLGTGPEAGLPY |
| N129-143 | GIIWVATEGALNTPK | |
| N219-227 | LALLLLDRL | |
| Spike (S) | S62-76 | VTWFHAIHVSGTNGT |
| S262-270 | AAAYYVGYL | |
| S263-277 | AAYYVGYLQPRTFLL | |
| S509-518 | RVVVLSFELL | |
| S539-549 | VNFNFNGLTGT | |
| S555-562 | SNKKFLPF | |
| S884-898 | SGWTFGAGAALQIPF | |
| S1013-1027 | IRAAEIRASANLAAT |
As expected, T cells from the pVAX-immunized control mice did not respond to the S or N peptides. In contrast, spleens from N- and S + N-vaccinated mice contained elevated numbers of N-specific CD4+ and CD8+ effector memory T cells with polyfunctional cytokine-secreting (IFN-γ+TNF-α+IL-2+) and cytotoxic (IFN-γ+CD107a+) activities (Fig. 2b, c top). Similarly, spleens from S and S + N-vaccinated mice had higher numbers of S-specific CD4+ and CD8+ effector memory T cells with polyfunctional and cytotoxic activities than did pVAX control mice (Fig. 2b, c bottom). We also evaluated the T cell response in the lungs, the primary target of SARS-CoV-2 infection and pathogenesis. Lungs from S and S + N-vaccinated mice exhibited increased numbers of antigen-specific CD4+ and CD8+ polyfunctional and cytotoxic T cells after stimulation with S or N peptides, respectively, whereas more muted responses were observed in the lungs from N-vaccinated mice (Fig. 2d, e). Thus, the S and S + N vaccines, and to a lesser extent, the N vaccine, elicited SARS-CoV-2 S and N-specific CD4+ and CD8+ T cells with Th1-biased polyfunctionality and cytotoxicity in both spleen and lung, with the S + N vaccine eliciting comparable or slightly superior responses compared with the S or N vaccines.
S, N, and/or S + N DNA vaccines confer short-term cross-protection against SARS-CoV-2 Beta infection and elicit S- and N-specific memory T cell responses
Having shown that the wild-type S, N, and S + N vaccines induce robust IgG, nAb, and/or T cell responses in K18-hACE2 mice (Figs. 1 and 2), we next addressed the main goal of the study, which was to determine whether these next-generation SARS-CoV-2 DNA vaccines can confer protection against infection by heterologous SARS-CoV-2 strains. To this end, vaccinated K18-hACE2 mice were challenged intranasally with SARS-CoV-2 Beta at 14 days post-boost, and tissues were analyzed 3 days later for subgenomic and genomic viral RNA load in nasal turbinate, lung, and brain tissues, as well as CD4+ and CD8+ T cell responses in the spleen (Fig. 3a).
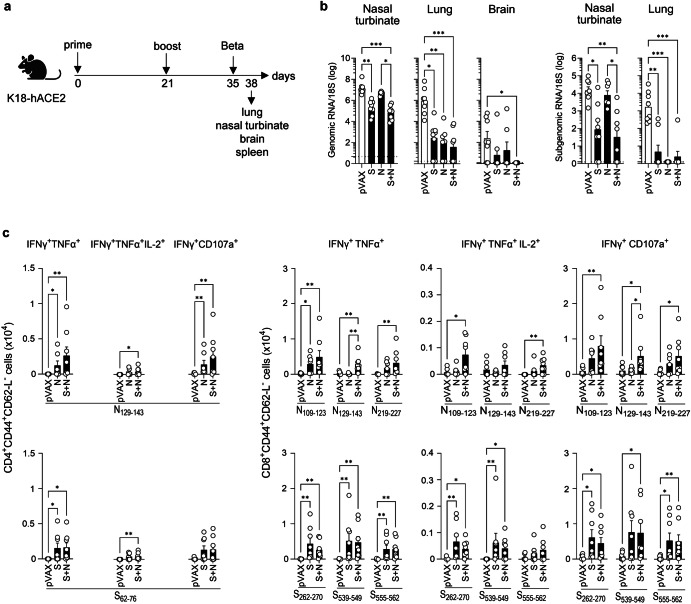
Fig. 3. S and/or N DNA vaccines protect against SARS-CoV-2 Beta infection in K18-hACE2 mice.
a Experimental protocol: On days 0 and 21, K18-hACE2 mice were inoculated with S and/or N DNA vaccines or pVAX control (25 µg, IM), challenged at 2 weeks post-boost with SARS-CoV-2 Beta (B.1.351) variant (103 PFU, IN), and tissues were harvested 3 days later. Representation of the experiment timeline was Created with BioRender.com. b SARS-CoV-2 genomic and subgenomic RNA levels were quantified by RT-qPCR. Dashed lines, detection limit. c Splenocytes were stimulated with the indicated SARS-CoV-2 S or N peptides, and polyfunctional and cytotoxic CD4+ and CD8+ T cells within the population of CD3+ cells were quantified by ICS. b, c Data are from 2 independent experiments (n = 7-8 mice/group). Group means were compared by the non-parametric Kruskal-Wallis test. *P < 0.05, **P < 0.01, ***P < 0.001. Circles, individual mice.
Immunization with S, N, or S + N vaccines successfully reduced genomic and subgenomic SARS-CoV-2 Beta RNA levels in all three tissues examined, albeit with differing effectiveness, compared with the pVAX control vaccine (Fig. 3b). Notably, the S and/or S + N vaccines, but not the N vaccine, significantly reduced levels of genomic and subgenomic RNA in the nasal turbinates (S and S + N) and genomic RNA in the brain (S + N), whereas all three vaccines virtually eliminated genomic and subgenomic RNA from the lungs (Fig. 3b). Thus, all three SARS-CoV-2 D614G DNA-based vaccines cross-protected against Beta infection of the lungs; both S and S + N vaccines were cross-protective in the upper respiratory tract; and only the S + N vaccine provided significant protection of the brain.
We next assessed CD4+ and CD8+ T cell responses from the immunized and challenged mice by stimulating splenocytes in vitro with N or S peptides, on day 3 post-infection, as indicated above (Table 1 and Fig. 3c). Splenocytes from all three vaccinated/infected mouse groups contained increased numbers of Th1-biased polyfunctional (IFN-γ+TNF-α+ and IFN-γ+TNF-α+IL-2+) and cytotoxic (IFN-γ+CD107a+) T cells specific for multiple S and N epitopes compared with the cells from pVAX-immunized and infected control mice (Fig. 3c). Note that the low response in the pVAX group was expected because a primary T cell response is typically not detected by day 3 post-infection. Interestingly, the S + N vaccine was consistently more effective than the N vaccine in increasing the mean number of N-specific CD4+ and CD8+ T cells, whereas the S and S + N vaccines evoked comparable numbers of S-specific CD4+ and CD8+ T cell responses (Fig. 3c). Taken together, these data indicate that D614G-based S, N, and S + N vaccines elicit S- and N-specific polyfunctional T cell responses in Beta-challenged mice, with S + N-vaccinated mice exhibiting slightly superior responses. Moreover, the unique efficacy of the S + N vaccine in preventing Beta infection of the brain may suggest a potential role for S + N vaccine-elicited T cell immunity in conferring brain protection.
S, N, and/or S + N DNA vaccines confer short-term cross-protection against SARS-CoV-2 Omicron BA.2 infection, reduce lung damage, and elicit S- and N-specific memory T cell responses
We next performed similar experiments to evaluate the short-term cross-protective capacity of the D614G-based vaccines against SARS-CoV-2 Omicron BA.2, which harbors more mutations, particularly in S protein, than Beta. K18-hACE2 mice were vaccinated, challenged with Omicron BA.2 at day 14 post-boost, and tissues were collected 3 days later for analysis (Fig. 4a). Vaccination with S or S + N significantly reduced genomic and subgenomic Omicron BA.2 RNA in the nasal turbinates and lungs of virtually all mice, when compared with the pVAX-vaccinated mice (Fig. 4b). However, N-vaccinated mice had little to no effect on genomic and subgenomic RNA levels in the lung and nasal turbinates of Omicron BA.2-infected mice, which contrasts with the N vaccine effect on Beta protection (compare Figs. 4b and 3b). To determine whether S, N, or S + N vaccination could reduce Omicron BA.2-induced lung disease, we performed histopathological analysis and quantified the scores for several key features of SARS-CoV-2-induced lung damage45. Compared with the pVAX control, S + N vaccination significantly reduced 4 of the 5 parameters evaluated: necrosis of bronchiolar epithelial cells (BEC), cellular debris in bronchioles, perivascular lymphocytic cuffing and bronchointerstitial pneumonia; whereas N vaccination significantly reduced perivascular lymphocytic cuffing; and S vaccination did not significantly reduce any parameter analyzed compared with the pVAX control (Fig. 4c). Representative images of these findings are illustrated in Fig. 4d. Overall, mice immunized with the S + N vaccine exhibited healthier lungs compared with mice immunized with S alone, with significantly lower scores for necrosis of BEC and inducible bronchus-associated lymphoid tissue (iBALT) hyperplasia. Moreover, despite the relatively poor efficacy of N vaccination in reducing Omicron BA.2 RNA levels in the respiratory tract, there was a significant beneficial effect of vaccination in the histopathology lung scores in these mice.
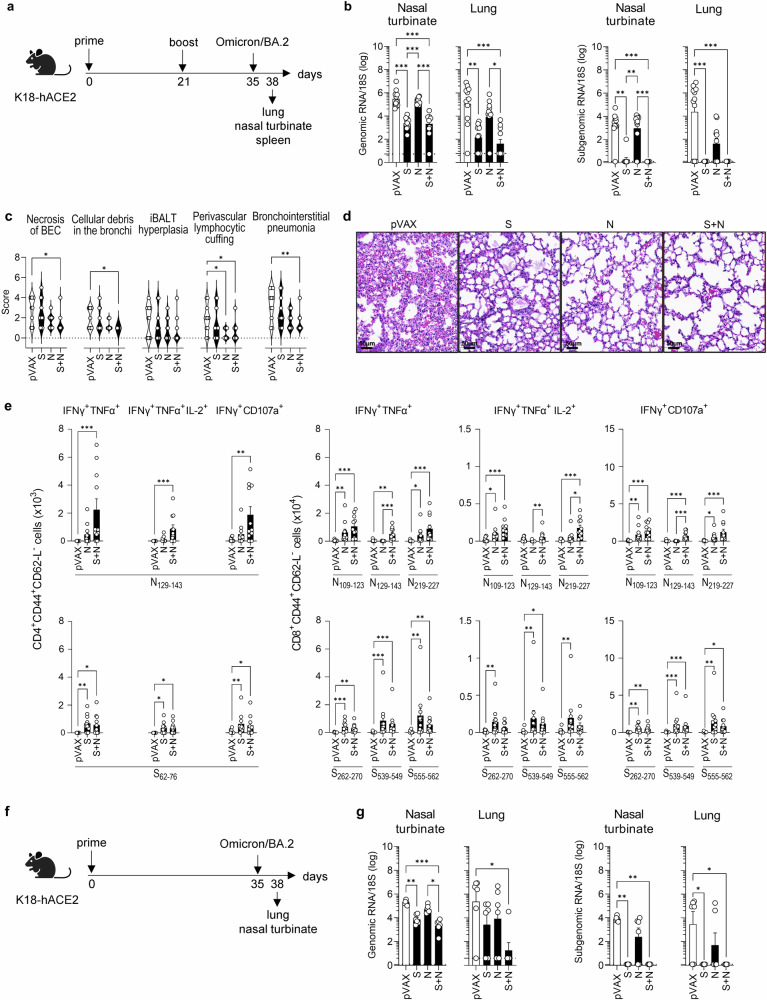
Fig. 4. S and S + N DNA vaccines protect against SARS-CoV-2 Omicron BA.2 infection in K18-hACE2 mice.
a Experimental protocol: On days 0 and 21, K18-hACE2 mice were inoculated with S and/or N DNA vaccines or pVAX control (25 µg, IM), challenged at 2 weeks post-boost with SARS-CoV-2 Omicron BA.2 variant (104 PFU, IN), and tissues were harvested 3 days later. Representation of the experiment timeline was Created with BioRender.com. b SARS-CoV-2 genomic and subgenomic RNA were quantified by RT-qPCR. Dashed lines, detection limit. c Histopathology of SARS-CoV-2-induced lung disease. c Five histopathological parameters (necrosis of bronchiolar epithelial cells [BEC], cellular debris in bronchioles, inducible bronchus-associated lymphoid tissue [iBALT] hyperplasia, perivascular lymphocytic cuffing, and bronchointerstitial pneumonia) were scored from 0 (least severe) to 5 (most severe). d Representative bronchointerstitial pneumonia images, H&E-stain. e Splenocytes were stimulated with the indicated SARS-CoV-2 S or N peptides, and polyfunctional (IFN-γ+TNF-α+ or IFN-γ+TNF-α+IL-2+) and cytotoxic (IFN-γ+CD107a+) CD4+ and CD8+ T cells within the population of CD3+ cells were quantified by ICS. f Experimental protocol for one-dose vaccination regimen: On day 0, K18-hACE2 mice were inoculated with S and/or N DNA vaccines or pVAX control (25 µg, IM), challenged on day 35 with SARS-CoV-2 Omicron BA.2 (104 PFU, IN), and tissues were harvested 3 days later. Representation of the experiment timeline was Created with Biorender.com. g SARS-CoV-2 genomic and subgenomic RNA quantified by RT-qPCR; dashed lines, detection limit. b–d Data are from 2 independent experiments (n = 10–11 mice/group). f, g Data is from 1 experiment (n = 6 mice/group). Group means were compared by the non-parametric Kruskal-Wallis test. *P < 0.05, **P < 0.01, ***P < 0.001. Circles, individual mice.
We next assessed CD4+ and CD8+ T cell responses to S and N peptides in splenocytes from vaccinated Omicron BA.2-challenged mice (Fig. 4e) using the same protocol as described above (Fig. 3c). Splenocytes from mice immunized with S + N vaccine contained higher numbers of N-specific polyfunctional/Th1-biased CD4+ and CD8+ T cells than splenocytes from N-vaccinated mice, whereas similar numbers of S-specific CD4+ and CD8+ T cells were observed among splenocytes from S-vaccinated and S + N-vaccinated mice (Fig. 4e), as also observed in Beta-challenged mice.
Of note, we also examined the efficacy of a single dose of the vaccines in Omicron BA.2-challenged mice in preventing infection of the upper and lower respiratory tract (Fig. 4f) and found similar results to the two-dose vaccination protocol (Fig. 4g). Thus, our results indicate that the S + N vaccine provides robust cross-protection against Omicron BA.2 in the upper and lower respiratory tracts; reduces Omicron BA.2-induced lung damage; and elicits similar or superior antigen-specific CD4+ and CD8+ T cell responses compared with the single-protein vaccines following viral challenge.
S, N, and S + N DNA vaccines elicit long-term S, N, and RBD protein-binding IgG and nAb responses against homologous and/or heterologous SARS-CoV-2 strains
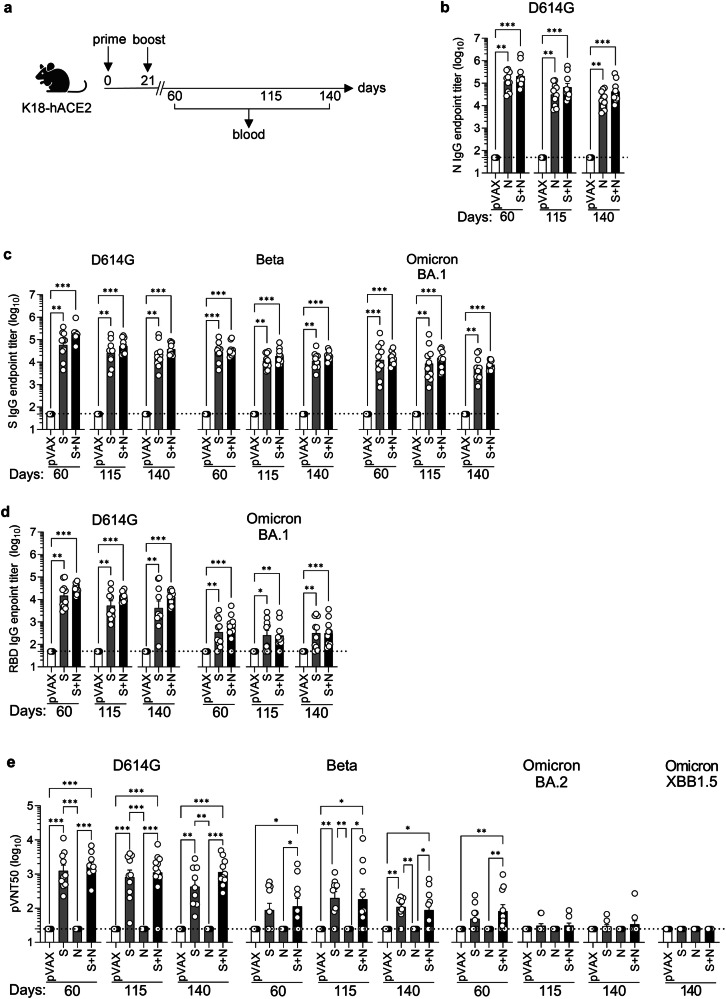
In addition to providing broad protection against different variants, next-generation SARS-CoV-2 vaccines need to confer long-lived protective immunity compared with current vaccines. To evaluate the durability of the Ab responses to our vaccines, K18-hACE2 mice were primed and boosted on days 0 and 21, respectively, with S, N, and S + N vaccines, and blood samples were collected at day 60, 115, and 140 ( ~ 5, 13, and 17 weeks) post-boost (Fig. 5a). Endpoint titers of N-, S- and RBD-specific IgG were determined by ELISA and neutralizing activities against D614G, Beta, Omicron BA.2, and Omicron XBB1.5 S proteins were evaluated using pVNT assays.
Fig. 5. S and N DNA vaccines elicit long-term IgG and nAb responses in a SARS-CoV-2 variant-specific manner in K18-hACE2 mice.
a Experimental protocol: On days 0 and 21, K18-hACE2 mice were inoculated with S and/or N DNA vaccines or pVAX control (25 µg, IM) and blood was harvested on days 60, 115, and 140. Representation of the experiment timeline was Created with BioRender.com. b–d ELISA assays showing endpoint titers of IgG against N, S, or RBD proteins from the indicated SARS-CoV-2 strains. Data are from 2 independent experiments (n = 9–10 mice/group). e Neutralization assay (pVNT50) using pseudoviruses harboring S proteins from the indicated SARS-CoV-2 strains. Data are from 2 independent experiments (n = 9-11 mice/group). b-e Serum samples were tested in duplicate on the same plates. Group means were compared by the non-parametric Kruskal-Wallis test. *P < 0.05, **P < 0.01, ***P < 0.001. Circles, individual mice; dashed lines, detection limit.
Sera from mice immunized with N, S, or S + N vaccines contained comparably high levels of N- and/or S-specific IgG at all three time points evaluated (Fig. 5b, c), whereas the RBD-specific IgG response against Omicron BA.1 protein was much weaker than that against the homologous protein at all three time points (Fig. 5d). Notably, all of the IgG responses evaluated were sustained for at least 17 weeks, demonstrating highly durable Ab responses to all vaccine protocols. Examination of the neutralizing capacity of the S and S + N-elicited Ab responses revealed strong responses against the homologous (D614G) S protein that remained at relatively constant levels for 17 weeks post-boost, and a weaker but cross-neutralizing response against Beta that was also durable (Fig. 5e). However, only a weak and transient (day 60) cross-neutralizing response against Omicron BA.2 was detected and no cross-neutralizing activity against either Omicron BA.2 or Omicron XBB1.5 was observed at day 140 (Fig. 5e). As expected, the N vaccine did not elicit an S protein-specific nAb response.
These data indicate that SARS-CoV-2 D614G S and S + N vaccines elicited IgG responses that cross-reacted with heterologous N, S, and/or RBD proteins and were sustained for at least 140 days post-vaccination. However, only the robust homologous anti-D614G nAb response and weaker anti-Beta nAb response were sustained long-term, whereas the anti-Omicron BA.2 nAb response was weak and transient, becoming undetectable by day 140.
Only the S + N DNA vaccine confers long-term protection against SARS-CoV-2 Omicron BA.2 infection in the lungs and does so in a CD4+ and CD8+ T cell-dependent manner
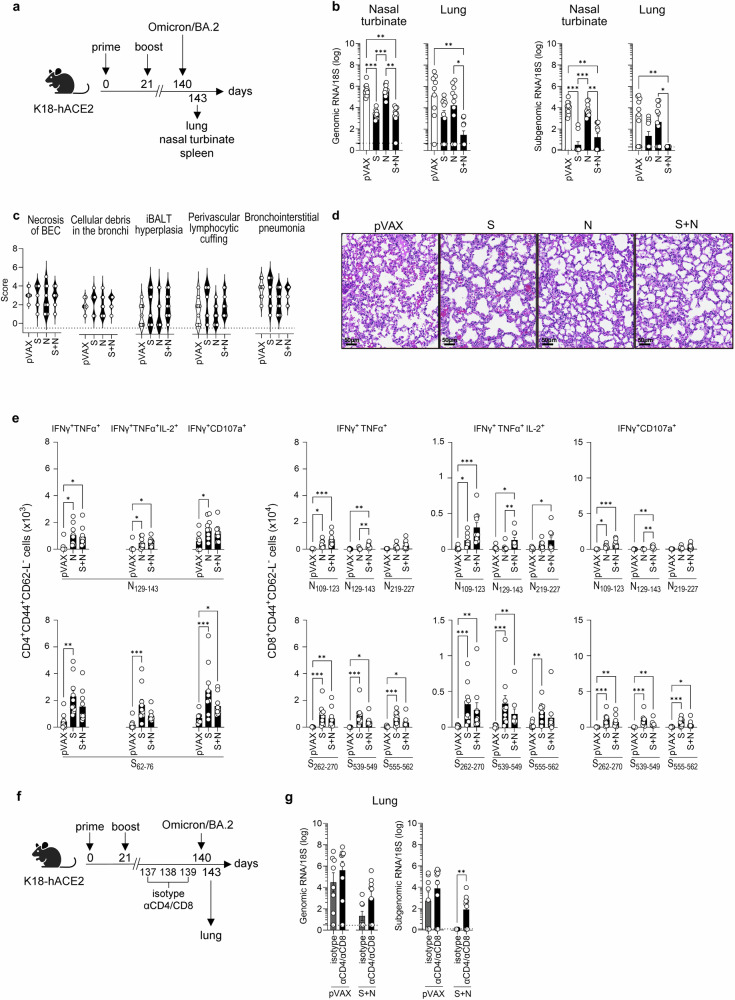
Having shown that S and S + N vaccines elicit durable cross-reactive IgG, but not nAb, responses against Omicron BA.2 (Fig. 5), we next determined whether the S, N, and S + N vaccines are able to induce cross-reactive T cell responses or confer long-term protection against Omicron BA.2 infection. Accordingly, mice were primed and boosted at days 0 and 21, respectively; challenged with Omicron BA.2 at ~17 weeks post-boost, and tissues were collected 3 days later for analysis of viral RNA load, lung pathology, and T cell responses (Fig. 6a).
Fig. 6. Co-immunization with S and N DNA vaccines provides long-term protection against SARS-CoV-2 Omicron BA.2 infection in K18hACE2 mice.
a Experimental protocol: On days 0 and 21, K18-hACE2 mice were inoculated with S and/or N DNA vaccines or pVAX control (25 µg, IM), challenged 17 weeks later with SARS-CoV-2 Omicron BA.2 (104 PFU, IN), and tissues were harvested at 3 days post-challenge. Representation of the experiment timeline was Created with BioRender.com. b SARS-CoV-2 genomic and subgenomic RNA were quantified by RT-qPCR. Data are from 2 independent experiments (n = 9–10 mice/group). c Histopathology of SARS-CoV-2-induced lung disease. Five histopathological parameters (necrosis of bronchiolar epithelial cells [BEC], cellular debris in bronchioles, inducible bronchus-associated lymphoid tissue [iBALT] hyperplasia, perivascular lymphocytic cuffing, and bronchointerstitial pneumonia) were scored from 0 (least severe) to 5 (most severe). Data are from 2 independent experiments (n = 9–10 mice/group). d Representative bronchointerstitial pneumonia images, H&E-stain. e Splenocytes were stimulated with the indicated SARS-CoV-2 S or N peptides and polyfunctional (IFN-γ+TNF-α+ or IFN-γ+TNF-αIL-2+) and cytotoxic (IFN-γ+CD107a+) CD4+ and CD8+ T cells within the population of CD3+ cells were quantified by ICS. Data are from 2 independent experiments (n = 8–10 mice/group). f In vivo T cell depletion protocol (Created with BioRender.com): On days 0 and 21, K18-hACE2 mice were inoculated with S + N DNA vaccines or pVAX control (25 µg, IM). On days 137, 138, and 139, mice were injected with anti-CD4 and anti-CD8 or isotype control monoclonal Abs (250 µg each, IP), and then challenged on day 140 with SARS-CoV-2 Omicron BA.2 (104 PFU, IN). Tissues were harvested 3 days later. Representation of the experiment timeline was Created with Biorender.com. g SARS-CoV-2 genomic and subgenomic RNA were quantified by RT-qPCR. Data are from 3 independent experiments (n = 8–9 mice/group). b, e, g Group means were compared by the non-parametric Kruskal-Wallis test (b, e) or non-parametric Mann–Whitney test (g). *P < 0.05, **P < 0.01. ***P < 0.001. Circles, individual mice; dashed lines (b, e), detection limit.
As noted in the short-term experiments, neither genomic nor subgenomic Omicron BA.2 RNA levels were reduced in the nasal turbinates or lungs of mice immunized with the N vaccine compared with pVAX control vaccine (Fig. 6b). In contrast, genomic Omicron BA.2 RNA was significantly reduced in the nasal turbinates and lungs of both S- and S + N-immunized mice, whereas subgenomic RNA was only significantly decreased in the lungs of S + N-immunized mice; indeed, subgenomic RNA was undetectable in all animals in this mouse group (Fig. 6b). Thus, co-vaccination with S and N antigens elicited long-lived cross-protective immunity against Omicron BA.2 infection of the upper and lower respiratory tract (Fig. 6b) despite the absence of Omicron BA.2 nAbs (Fig. 5e), suggesting that T cells may play a dominant role in the durability of cross-protection.
Histopathological analysis of lung tissue from mice challenged with Omicron BA.2 on day 140 did not reveal a clear protective effect of any of the DNA vaccines in reducing lung damage (Fig. 6c, d), which contrasts with our observations using the short-term challenge protocol (Fig. 4c, d). Examination of splenic CD4+ and CD8+ T cell responses to S and N peptides at day 143 (3 days post-challenge) revealed a generally similar pattern of response by splenocytes to that observed in splenocytes from mice following challenge on day 35 (compare Fig. 6eand Fig. 4e), irrespective of the vaccination protocol. Thus, the S, N, and S + N vaccines elicited S- and N-specific polyfunctional T cell responses that persist for at least 17 weeks (Fig. 6e).
To directly test the contribution of T cells to the long-term protection against Omicron BA.2 elicited by S + N vaccination, we repeated the experiment with mice that had been treated with depleting anti-CD4 and anti-CD8 mAbs for 3 days prior to the Omicron BA.2 challenge on day 140 (Fig. 6f), and we verified the efficacy of T cell depletion by flow cytometry (Figure S2). As expected, Omicron BA.2 genomic and subgenomic RNA were reduced or undetectable, respectively, in the lungs of S + N-immunized mice pretreated with isotype control mAb (Fig. 6g). However, genomic and subgenomic RNA were readily detectable in the lungs of S + N-immunized mice pretreated with T cell-depleting mAbs (Fig. 6g), demonstrating a crucial role for long-lasting S + N-elicited T cell immunity in controlling infection of the respiratory tract with the heterologous Omicron BA.2 strain.
Collectively, these data indicate that although both S and S + N vaccinations elicited long-term protection against Omicron BA.2 in the upper and lower respiratory tracts, only the S + N vaccine completely inhibited viral replication in the lungs and did so in a manner at least partly dependent on long-lived vaccine-elicited T cell responses.
Discussion
While licensed COVID-19 vaccines have been successful in reducing the incidence of serious illness and death following SARS-CoV-2 infection, they have two major short-comings. First, the durability of the responses is poor, even with booster doses, resulting from a sharp drop in Ab-induced immunity at about 4 to 6 months after vaccination6,7,46. Second, the current vaccines exhibit poor cross-reactivity to variants, allowing escape from existing immunity. In this study, we explored the potential benefit of combining SARS-CoV-2 S and N antigens in DNA vaccines as a strategy to achieve long-lasting broadly cross-reactive immunity against SARS-CoV-2 variants. Our rationale for using DNA vaccines was that they can be manufactured at low cost, are highly stable, have plasmid sequences that can be easily modified, and they induce robust T cell responses. The main finding of our study is that vaccination with wild-type SARS-CoV-2 (D614G) DNA vaccines can confer short-term protection against infection of the respiratory tract by Beta (S, N, and S + N vaccines) and Omicron BA.2 (S and S + N vaccines), but only S + N co-vaccination induces robust long-term ( ≥ 4 months) cross-protection against Omicron BA.2 infection of the lungs. In addition, we show that this durable cross-protection is T cell dependent. To our knowledge, this is the first study to evaluate long-term protection against Omicron infection using a vaccine strategy that combined SARS-CoV-2 N and S antigens.
Other groups have investigated the combined immunogenicity of S and N protein vaccines against SARS-CoV-2. In one study, a self-amplifying RNA vaccine encoding both S-RBD and N antigens also showed improved control of Beta and Omicron BA.1 compared with single-protein vaccines39. A second study showed that combined S + N mRNA vaccines provided better control of Delta and Omicron variants in lungs and enhanced protection in the upper respiratory tract of hamsters compared with S mRNA vaccine alone38. A third study showed that mice immunized with an RBD + N protein subunit recombinant vaccine were highly protected against Delta and Omicron in survival analysis36. Although other studies have also shown protection with vaccines composed of both S and N antigens, they lacked comparison with single S-based vaccines, making it impossible to predict the potential advantages of adding N antigen to current vaccines32,34,35. Most importantly, none of the cited studies conducted long-term efficacy experiments, and thus did not examine the possible benefit of S + N vaccines in extending the durability of protection. Our data demonstrate that co-vaccination with both S and N antigens is superior to single-protein vaccination in promoting durable and cross-protective immunity to Omicron BA.2.
SARS-CoV-2 S and N proteins are important targets of the humoral immune response47,48. Neutralizing Abs against S protein, especially its RBD domain, are the primary mechanism of vaccine-induced defense against SARS-CoV-249,50. The RBD encompasses a large number of epitope targets for SARS-CoV-2 neutralization51,52. Our S and S + N vaccines induced anti-S-binding IgG capable of recognizing the S proteins of SARS-CoV-2 Beta and Omicron, but less able to recognize Omicron RBD, compared with the D614G proteins, which might explain why we observed robust neutralization of D614G and Beta, but little or no neutralization of Omicron BA.2 and Omicron XBB1.5. Future experiments should assess neutralizing activities against contemporaneous SARS-CoV-2 variants such as XEC, KP.3.1.1, and MC.1.
Despite Omicron BA.2 nAbs being virtually undetectable at day 35 or 140 post-immunization, all S + N-vaccinated mice and all except one S-vaccinated mouse were fully protected against Omicron BA.2 infection in the short-term experiments, with no detectable levels of viral replication in the nasal turbinates or lungs. Our data are consistent with human cohort studies showing that VOCs, including Beta, Delta and, in particular, Omicron, are resistant to anti-S nAbs2,10,53, and some convalescent individuals with low or no nAbs nevertheless resolve SARS-CoV-2 infection54,55. Thus, our data and these human studies suggest a key role for T cell immunity and/or effector functions of Abs other than neutralization in controlling SARS-CoV-2 variant infection.
Several published studies point to the possible effector functions of non-neutralizing Abs. In humans, COVID-19 vaccination induces Abs that bind S protein on the surface of infected cells, leading to Ab-dependent cellular cytotoxicity56,57. Indeed, in BNT162b2-vaccinated individuals, despite the loss of Omicron anti-RBD Abs (with probable neutralizing potential), anti-S Abs maintain their avidity in Fc-mediated effector functions58. Further, one study in K18-hACE2 mice showed that an anti-N monoclonal Ab protected against SARS-CoV-2 infection by Ab-dependent cellular cytotoxicity, and passive immunization from mice immunized with an N adenovirus vaccine reduced SARS-CoV-2 infection in lungs26.
In addition to Abs, COVID-19 vaccines elicit T cell responses that appear to control viral spread at later stages of infection and generate long-lasting, cross-reactive immune memory8,59. In fact, recent data has shown that SARS-CoV-2 T cell immunity elicited after infection or vaccination can compensate for the absence of Abs in B cell-deficient patients, reducing the risk of severe COVID-19 outcomes60. Most virus-specific T cells from wild-type SARS-CoV-2-infected and/or -vaccinated individuals cross-react with variants due to their ability to recognize conserved epitopes8,61,62. Although both S and N proteins are major targets of the T cell response to SARS-CoV-224,63, N-specific are more likely than S-specific T cell responses to provide broader protection against variants, since the N protein is the more conserved of the two. Indeed, several human studies point to the potential for N-specific T cell immunity to improve vaccine durability and breadth. For example, SARS-CoV-2 N-specific CD8+ T cells suppressed viral replication of the original SARS-CoV-2 strain and multiple variants in vitro for at least 6 months after infection64; N-specific CD4+ and CD8+ T cells were shown to be associated with control of SARS-CoV-2 replication in the upper airways before seroconversion of patients with mild COVID-19 disease17; and, in SARS-CoV-1–infected patients, N-specific memory T cells are preserved for up to 1127 or 17 years28. In line with these human studies, we showed that addition of the N antigen improved the durability of the wild-type SARS-CoV-2 S DNA vaccine-induced cross-protection against Omicron in mice. The S, N, and S + N DNA vaccines induced epitope-specific CD4+ and CD8+ T cells with polyfunctional, cytotoxic, and Th1-biased cytokine production phenotypes in the spleens and lungs of K18-hACE2 mice. These phenotypes are relevant in the context of antiviral T cell responses in COVID-19 vaccinated individuals65,66. Our in vivo T cell depletion experiment suggests that vaccine-induced antigen-specific CD4+ and CD8+ T cells play a major role in conferring long-term protection. Although our experiment did not differentiate between contributions by CD4+ and CD8+ subpopulations, two other studies examining short-term protection against SARS-CoV-2 showed that depletion of either CD4+ or CD8+ T cells could reduce protection provided by an RBD + N protein vaccine36, and depletion of CD8+ T cells reduced the ability of an N + S mRNA vaccine to control replication of variants in hamsters38. Moreover, there is evidence to support an important role for coronavirus-cross-reactive T cell-mediated immunity in mitigating COVID-19 disease and improving vaccination response in humans67–69. T cell epitopes have been identified that are highly conserved between SARS-CoV-2 and other human endemic coronaviruses (HCoV-OC43, HCoV-229E, HCoV-HKU1, and HCoV-NL63), including S and N epitopes28,70–72. Thus, the broad and durable T cell response specific for highly conserved epitopes induced by our S + N vaccine suggests that a similar strategy might be effective for inducing pan-CoV protective immunity in the future.
In conclusion, our study provides proof-of-concept for a vaccination strategy that could increase both the durability and breadth of coverage of the current SARS-CoV-2 vaccines by concomitantly immunizing with both N and S antigens. Abs and T cells specific for S and N antigens offer a “one-two punch” strategy against SARS-CoV-2 that should be taken into account in the design and development of next-generation vaccines.
Methods
Study design
The goal of this study was to investigate the immunogenicity and efficacy of S and N DNA vaccines (alone or combined) against SARS-CoV-2 VOCs. In vitro S and N antigen expression driven by S and N DNA vaccines was confirmed in transfected cells. Immunogenicity and protection were assessed in single- or co-vaccinated K18-hACE2 mice later infected with Beta or Omicron BA.2 VOCs. Short- and long-term protocols were conducted to assess durability. Similar numbers of male and female mice were assigned to experimental groups (N = 3 to 6). Sample sizes for each mouse group were estimated based on previous efficacy and viral challenge experiments in our laboratory and in the literature, aiming to balance the numbers required for statistical rigor while minimizing animal use. Each animal experiment was conducted at least twice. Studies were not randomized or blinded. The animal experiments were performed in strict accordance with recommendations outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at the La Jolla Institute for Immunology ABSL2 and ABSL3 facilities (protocol number AP00001242). Tissue samples were collected from immunized and/or infected mice to evaluate Ab and T cell immunity in addition to SARS-CoV-2 RNA levels. All in vitro assays (ELISA, pVNT50, qRT-PCR) included experimental samples and controls in duplicate.
Mice
K18-hACE2 transgenic mice were obtained from The Jackson Laboratory or bred at the La Jolla Institute for Immunology and maintained under pathogen-free conditions. K18-hACE2 transgenic mice express human ACE2 (primary cell receptor for SARS-CoV-2) under the control of the human keratin-18 promoter. In K18-hACE2 mice, human ACE2 is expressed in epithelial cells, including the mucosal epithelium lining the airways; as a result, these mice are highly susceptible to SARS-CoV-2 infection40.
S and N DNA vaccines
S and N DNA vaccines41 were designed to express full-length wild-type SARS-CoV-2 S and N proteins with sequences from SARS-CoV-2/human/USA/WA-CDC-WA1/2020 (GenBank accession: MN985325.1). Coding sequences were codon-optimized for human expression by GenScript and cloned into pVAX1 plasmid vector (Thermo Fisher Scientific) under the control of human cytomegalovirus immediate-early promoter. To enhance translational initiation and performance, optimized DNA sequences were preceded by Kozak and IgE leader sequences. In vitro antigen expression driven by S and N DNA vaccines was confirmed as previously described41.
Immunization and tissue collection for immunogenicity experiments
K18-hACE2 mice were immunized via the right or left quadriceps (see below) with 25 µg of S and/or N DNA vaccines diluted in 50 µL of Tris-EDTA pH 8.0 (Invitrogen, 15568025), followed by minimally invasive electroporation73. The intramuscular (IM) injections used 30-gauge ultra-fine insulin syringes (BD Bioscience, BD-25150) and electroporation used the BTX AgilePulse IM System (47-0500 N) with a 4 x 4 x 5 mm needle array (47-0045). At 21 days post-priming, mice were boosted in the same manner (boost omitted for the one-dose vaccination protocol). For single immunization with either S or N vaccines, mice were primed in one muscle and boosted ipsilaterally. For co-immunization with both vaccines, mice were primed with S vaccine in the right hind limb and with N vaccine in the left hind limb, and then boosted in the contralateral limb. In all experiments, control mice received commercial vector pVAX1.
For tissue collection, mice were either anesthetized with isoflurane or euthanized with CO2. In short-term experiments, blood was collected on days 20 (1 day before boost) and 28 (7 days post-boost) into serum-gel collection tubes (Sarstedt, 41.1378.005) from the facial vein or via cardiac puncture, and lungs and spleens were harvested on day 28 (7 days after the boost) in sterile RPMI 1640 media (Gibco, 1187519) supplemented with 10% fetal bovine serum (FBS, Gibco, 1187519), 1% penicillin–streptomycin (Gibco, 15140-122), 1% HEPES (Gibco, 15630130). In long-term experiments, blood samples were collected on days 60, 115, and 140.
SARS-CoV-2 S and N IgG ELISA
SARS-CoV-2-binding Abs were detected by endpoint ELISA assays using recombinant wild-type S, RBD, and N proteins, and commercial RBD from the Omicron BA.1 (B.1.1.529.1) variant (Sino Biological, 40592-V08H121) as coating antigens. Recombinant wild-type SARS-CoV-2 S, N, and RBD proteins were generated from synthetic codon-optimized DNA of Wuhan-Hu-1 isolate (GenBank accession MN908947.3) by sub-cloning into the pHCMV3 expression vector. Site-directed mutagenesis was performed to generate sequences of the Beta (B.1.351) and Omicron BA.1 (B.1.1.529.1) variants, and positive clones were fully sequenced to ensure that no additional mutations were introduced. Protein purification was previously described74.
High-binding flat-bottom plates (Corning, 9018) were coated overnight (4 °C) with 0.1 µg/mL of S, RBD, or N proteins in phosphate buffered saline (PBS, Corning, 21040). The following steps were carried out at room temperature. Plates were washed 3 times with 1% BSA (Sigma-Aldrich, A3059) in PBS (PBS/BSA), and blocked with 5% Blotting Grade Blocker (Bio-Rad, 1706404) in PBS for 2 h. Serial dilutions of mouse sera were prepared in PBS/BSA (5-fold dilutions; 1:50 to 1:156.250) and added to the plates for 1.5 h. After 3 washes, plates were incubated with HRP-labeled anti-mouse IgG (Jackson ImmunoResearch, 115-035-008) in PBS/BSA for 1.5 h, washed, developed in the dark for 10 min with 1 Step TMB (Thermo Fisher Scientific, 34028), and the reaction was then stopped by addition of an equal volume of 2 N sulfuric acid (Fisher Chemical, A468-2). Control wells were treated identically except without the serum incubation. Optical density was immediately measured at 450 nm using the SpectraMax M2 microplate reader (Molecular Devices). The universal cut-off value for all ELISA assays was 0.2 (3 standard deviations above the mean of the control wells). Endpoint titers were calculated based on the interpolation from the cut-off value in a 4-parameter logistic curve fit of each test sample.
Pseudovirus neutralization (pVNT) assay
SARS-CoV-2-specific nAbs in mouse sera were measured using a pVNT assay based on recombinant replication-deficient vesicular stomatitis virus (VSV) vectors encoding GFP pseudotyped with SARS-CoV-2 S protein derived from the Wuhan-Hu-1 isolate reference strain (GenBank: MN908947.3), or the Beta (B.1.351), Omicron BA.2 (B.1.1.529.2), or Omicron XBB1.5 variants generated by site-directed mutagenesis, as previously described in ref. 74. Briefly, Vero cells (ATCC CCL81) were seeded (2.5 × 104 cells/well) in flat-bottom 96-well black plates (Corning, CLS3603) to achieve 80% to 90% confluence at the time of infection. Mouse sera were heat-inactivated (30 min, 56 °C), serially diluted in PBS (3-fold dilutions; 1:25 to 1:54,675), incubated with pre-titrated amounts of rVSV-SARS-CoV-2 (1.5 h, 5% CO2), and then added to confluent Vero monolayers for 16 to 18 h (37 °C, 5% CO2). After infection, cells were washed with PBS, fixed with 4% paraformaldehyde (Electron Microscopy Science, 15700), stained in the dark with 1 µg/mL of Hoechst (Thermo Fisher Scientific, 62249) in PBS for (30 min, room temperature), and washed twice with PBS. Pseudovirus titers were quantified as the number of focus forming units (FFU/mL) using a Cell Insight CX5 imager (Thermo Fisher Scientific). Neutralizing Ab titers were computed using a 4-parameter logistic curve fitting regression.
Intracellular cytokine staining (ICS) assay
Single-cell suspensions of leukocytes and splenocytes were prepared from the lungs and spleens, respectively, and then seeded into 96-well round-bottom plates (Corning, 38018) in complete RPMI media (2 × 106 cells/well). Cells were stimulated with individual or pooled SARS-CoV-2 S- or N-derived peptides (2 µg of each peptide/well, 5 h, 37 °C, 5% CO2; Table 1). Prediction of peptide-MHC class I or II binding affinity was performed using tools from the Immune Epitope Data Base website (www.iedb.org) and selection of the “IEDB-recommended” method, as described previously44. The selected peptides were identified as immunodominant in previous INF-γ -ELISPOT assays (data not shown) for CD8+ and CD4+ T cells in C57BL/6 mice (the genetic background of K18-hACE2 mice). After 1 h incubation, brefeldin A (BioLegend, 420601) and anti-CD107a PE (clone eBio1D4B, eBioscience, 12-1071-83) were added and the incubation was continued for 4 h. Positive and negative control cells were incubated with a commercial stimulation cocktail containing PMA and ionomycin (eBioscience, 00-4970-93) or RPMI 1640 medium alone, respectively. Peptide-stimulated cells were then labeled with viability dye efluor 455 UV, (eBioscience, 65-0868-18), blocked with Fc Block (CD16/CD32 mAb 2.4G2; BD Biosciences, 553142) and stained with fluorophore-conjugated Abs: anti-CD3e PE-Cy7 (clone 145-2C11, eBioscience, 25-0031-82), anti-CD4 BUV395 (clone GK1.5, BD Bioscience, 565974), anti-CD8a BV510 (clone 53-6.7, BioLegend, 100751), anti-CD44 BV785 (clone IM7, BioLegend, 103041) and anti-CD62-L APC efluor 780 (clone MEL-14, eBioscience, 47-0621-82). Cells were then fixed, permeabilized with Cytofix/Cytoperm commercial kit (BD Bioscience, 554722), and stained with anti-IFN-γ FITC (clone XMG1.2, Tonbo Bioscience, 35-7311-U100), anti-TNF-α APC (clone MP6-XT22, eBioscience, 17-7321-82), and anti-IL-2 BV711 (clone JES6-5H4, BioLegend, 503837). Data were acquired on an LSRFortessa flow cytometer (BD Bioscience) and analyzed using FlowJo software v10.8.1 (Tree Star).
Viruses and in vivo challenge
Vaccine efficacy was assessed against SARS-CoV-2 Beta (hCoV-19/South Africa/KRISP-K005325/2020, BEI NR-54009) and Omicron BA.2 (CoV-19/Japan/UT-NCD1288-2N/2022, NCD1288), generously provided by Ralph Baric (University of North Carolina) and Yoshihiro Kawaoka (University of Wisconsin; via Michael Diamond at Washington University in St. Louis), respectively. The SARS-CoV-2 Beta strain was propagated in Vero cells (ATCC, CCL81), and the Omicron BA.2 strain in Vero/TMPRSS2 cells (kindly donated by Michael Diamond, Washington University) for 3 days in DMEM (Corning, 10-013-CV) supplemented with 10% FBS, 1% penicillin–streptomycin, 1% HEPES, and 1% non-essential amino acids (Gibco, 11140050), and the supernatants were then harvested and frozen. The genetic homogeneity of both virus stocks was confirmed by deep-sequencing analysis (La Jolla Institute for Immunology Sequencing Core). The viral stocks were titrated by plaque assay. Briefly, viral supernatants were serially diluted 10-fold and added to confluent Vero E6 cells (ATCC, CRL-1587) in 24-well plates for 1 h (8 × 104 cells/well); the medium was then switched (DMEM, 1% carboxymethylcellulose, 2% FBS) and the cells incubated for 3 days (all incubations were at 37 °C, 5% CO2). Cells were then fixed with 10% formaldehyde (1 h, room temperature), washed, and stained with 0.1% crystal violet in methanol (20 min, room temperature), and plaque-forming units (PFU) per mL quantified. All SARS-CoV-2 propagation and titration were performed in a BSL-3 facility.
For the viral challenge protocol, immunized K18-hACE2 mice were inoculated intranasally (IN, 15 µL per nostril, 30 µL total) with Beta (103 PFU) or Omicron BA.2 (104 PFU) variants on days 35 or 140 (14- or 119-days post-boost, respectively). At 3 days post-challenge, mice were euthanized by CO2 inhalation. Lungs and nasal turbinates were harvested in 1 mL RNA/DNA shield (ZYMO Research, R1100-250) and spleens were harvested in complete RPMI medium for isolation of splenocytes used in ICS assays. All SARS-CoV-2 infections and tissue harvesting were performed in an ABSL-3 facility.
In vivo T cell depletion and viral challenge
For in vivo T cell depletion, pVAX and S + N-immunized K18-hACE2 mice were inoculated intraperitoneally (IP, 200 µL total) with 250 µg anti-CD4 (clone GK1.5, Bio X Cell, BE0003-1) and anti-CD8 (clone 2.43, Brand, BE0061) monoclonal Abs or isotype control Abs on days 137, 138, and 139. On day 140, mice were inoculated with Omicron BA.2 variant as described above, and at 3 days post-challenge, lungs and nasal turbinates were harvested.
To assess T cell depletion efficiency, mice were anesthetized with isoflurane inhalation and blood collected via the facial vein just before viral challenge. Blood cells were labeled with viability dye efluor 455 UV (eBioscience, 65-0868-18), blocked with Fc Block (CD16/CD32 mAb 2.4G2; BD Biosciences, 553142), and stained with the following 3 fluorophore-conjugated Abs: anti-CD3e PE-Cy7 (clone 145-2C11, eBioscience, 25-0031-82), anti-CD4 BUV395 (clone GK1.5, BD Bioscience, 565974), anti-CD8a BV510 (clone 53-6.7, BioLegend, 100751). Data were acquired on an LSRFortessa flow cytometer (BD Bioscience) and analyzed using FlowJo software v10.8.1 (Tree Star).
Lung histopathology
Histopathological analysis was conducted according to previous methods41. Briefly, lungs from immunized/challenged mice were harvested and fixed in zinc formalin for 24–48 h at room temperature with gentle agitation. After fixation, samples were transferred to 70% alcohol. Lung tissues were then embedded in paraffin using standard procedures, sectioned into 4-μm slices, stained with H&E using a Leica ST5020 autostainer, and imaged with a Zeiss AxioScan Z1 (40 × 0.95 NA objective). Histopathological evaluation was performed by a certified veterinary pathologist who was blinded to group identities. Sections were scored on a scale of 0–5 based on 10 criteria for SARS-CoV-2-induced lung pathology, as observed in hamsters, monkeys, and COVID-19 patients45. Scores for 5 parameters are shown: necrosis of bronchiolar epithelial cells (BEC), cellular debris in bronchioles, inducible bronchus-associated lymphoid tissue (iBALT) hyperplasia, perivascular lymphocytic cuffing, and bronchointerstitial pneumonia.
SARS-CoV-2 quantification in tissues
Lungs and nasal turbinates were harvested from immunized/challenged mice into 1 mL RNA/DNA shield (Zymo Research, R1100-250), and SARS-CoV-2 RNA was extracted using RNeasy Mini Kits (Qiagen, 52904) and then stored at −80 °C. Total SARS-CoV-2 genomic and subgenomic RNA copies were quantified using the qScript One-Step qRT-PCR Kit (Quanta BioSciences, 76047-080). Genomic RNA was quantified using the envelope gene as a target and the following primer sets75: Fwd, 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′; Rev, 5′-ATATTGCAGCAGTACGCACACA-3′; and Probe, FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ. Subgenomic RNA copies were quantified using the Orf7a gene as a target and the following primer sets76: Fwd, 5′-TCCCAGGTAACAAACCAACCAACT-3′; Rev, 5′-AAATGGTGAATTGCCCTCGT-3′; and Probe, FAM-CAGTACTTTTAAAAGAACCTTGCTCTTCTGGAAC-Tamra-Q. Amplification for genomic and subgenomic RNA was performed using the CFX Real-Time PCR System and the following program: 48 °C for 30 min, 95 °C for 10 min, and then 40 cycles of 95 °C for 15 s and 55 °C for 1 min. Viral RNA concentration was calculated using a standard curve composed of four 100-fold serial dilutions of in vitro-transcribed RNA from SARS-CoV-2 (RNA/human/USA/WA-CDC-WA1/2020, ATCC NR-52347).
Statistical analysis
Statistical analyses were performed using GraphPad Prism v10.0.2 software. Outliers were identified using GraphPad Prism outlier calculator. All data are presented as the mean ± SEM. Differences between group means were analyzed by the non-parametric Kruskal-Wallis test for more than 2 groups, or the non-parametric Mann–Whitney test for 2 groups; P < 0.05 was considered significant.
Supplementary information
Acknowledgements
We would like to thank the Department of Laboratory Animal Care (DLAC; Morag Mackay, Pascual Barajas, and Joseph Garza), Department of Environmental Health and Safety (Dr. Laurence Cagnon and David Hall), and Flow Cytometry Core (Cheryl Kim) at the La Jolla Institute for Immunology for their assistance with husbandry and experiments. This study was supported by the Coordination of Improvement of Higher Education Personnel (CAPES) from the Oswaldo Cruz Institute (IOC-Fiocruz), grant 88887.472772/2019-00 and 88887.583319/2020-00 (P.B.A.P.) and NIH U19 AI142790-02S1, the GHR Foundation, and the Overton family (S.S. and E.O.S.), the Arvin Gottleib Foundation and the Prebys Foundation (to S.S.). The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author contributions
Revised: Conceptualization, P.B.A.P., A.E.N., and S.S.; Methodology, P.B.A.P., A.E.N., J.T., K.C., Q.H.L., H.H.L., E.M., M.N.N., R.P.S.A., S.K.V., F.A., K.V., S.L.B., and K.H.; Investigation, P.B.A.P., A.E.N., J.T., K.C., Q.H.L., H.H.L., M.N.N., R.P.S.A., K.V., S.L.B., K.K., and K.H.; Writing – Original Draft, P.B.A.P., A.E.N., and S.S.; Writing – Review & Editing, A.E.N., K.K., and S.S.; Visualization, P.B.A.P., and A.E.N.; Supervision, S.S., and A.A.; Project administration, A.E.N., and S.S.; Funding Acquisition, S.S., and E.O.S.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Annie Elong Ngono, Email: aelong@lji.org.
Sujan Shresta, Email: sujan@lji.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-01043-3.
References
- 1.Organization, W. H. COVID-19 vaccine tracker and landscape. (2023).
- 2.Andrews, N. et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med.386, 1532–1546 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Risk, M. et al. Comparative Effectiveness of Coronavirus Disease 2019 (COVID-19) Vaccines Against the Delta Variant. Clin. Infect. Dis.75, e623–e629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tartof, S. Y. et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet398, 1407–1416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemaitelly, H. et al. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 Infection in Qatar. N. Engl. J. Med.385, e83 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suah, J. L. et al. Waning COVID-19 Vaccine Effectiveness for BNT162b2 and CoronaVac in Malaysia: An Observational Study. Int J. Infect. Dis.119, 69–76 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews, N. et al. Duration of Protection against Mild and Severe Disease by Covid-19 Vaccines. N. Engl. J. Med.386, 340–350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarke, A. et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell185, 847–859.e811 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GeurtsvanKessel, C. H. et al. Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci. Immunol.7, eabo2202 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, C. et al. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat. Commun.12, 1715 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rydyznski Moderbacher, C. et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell183, 996–1012.e1019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng, Y. et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol.21, 1336–1345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan, A. T. et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep.34, 108728 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, J. et al. CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Sci. Immunol.7, eabq7647 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarke, A. et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med.2, 100204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rümke, L. W. et al. Impaired SARS-CoV-2 specific T-cell response in patients with severe COVID-19. Front Immunol.14, 1046639 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eser, T. M. et al. Nucleocapsid-specific T cell responses associate with control of SARS-CoV-2 in the upper airways before seroconversion. Nat. Commun.14, 2952 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connor, M. A. et al. A replicon RNA vaccine can induce durable protective immunity from SARS-CoV-2 in nonhuman primates after neutralizing antibodies have waned. PLoS Pathog.19, e1011298 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi, S. J. et al. T cell epitopes in SARS-CoV-2 proteins are substantially conserved in the Omicron variant. Cell Mol. Immunol.19, 447–448 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrotri, M. et al. Spike-antibody waning after second dose of BNT162b2 or ChAdOx1. Lancet398, 385–387 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brasu, N. et al. Memory CD8+ T cell diversity and B cell responses correlate with protection against SARS-CoV-2 following mRNA vaccination. Nat. Immunol.23, 1445–1456 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Wu, W., Cheng, Y., Zhou, H., Sun, C. & Zhang, S. The SARS-CoV-2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol. J.20, 6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Algaissi, A. A., Alharbi, N. K., Hassanain, M. & Hashem, A. M. Preparedness and response to COVID-19 in Saudi Arabia: Building on MERS experience. J. Infect. Public Health13, 834–838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grifoni, A. et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell181, 1489–1501.e1415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manfredi, F. et al. Antiviral effect of SARS-CoV-2 N-specific CD8(+) T cells induced in lungs by engineered extracellular vesicles. NPJ Vaccines8, 83 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dangi, T. et al. Improved control of SARS-CoV-2 by treatment with a nucleocapsid-specific monoclonal antibody. J. Clin. Invest.132, 10.1172/jci162282 (2022). [DOI] [PMC free article] [PubMed]
- 27.Ng, O. W. et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine34, 2008–2014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bert, N. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature584, 457–462 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Chiuppesi, F. et al. Vaccine-induced spike- and nucleocapsid-specific cellular responses maintain potent cross-reactivity to SARS-CoV-2 Delta and Omicron variants. iScience25, 104745 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dangi, T., Class, J., Palacio, N., Richner, J. M. & Penaloza MacMaster, P. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep.36, 109664 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn, J. Y. et al. Safety and immunogenicity of two recombinant DNA COVID-19 vaccines containing the coding regions of the spike or spike and nucleocapsid proteins: an interim analysis of two open-label, non-randomised, phase 1 trials in healthy adults. Lancet Microbe3, e173–e183 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, A. et al. Intranasal plus subcutaneous prime vaccination with a dual antigen COVID-19 vaccine elicits T-cell and antibody responses in mice. Sci. Rep.11, 14917 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afkhami, S. et al. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell185, 896–915.e819 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Routhu, N. K. et al. A modified vaccinia Ankara vaccine expressing spike and nucleocapsid protects rhesus macaques against SARS-CoV-2 Delta infection. Sci. Immunol.7, eabo0226 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alcolea, P. J. et al. Non-replicative antibiotic resistance-free DNA vaccine encoding S and N proteins induces full protection in mice against SARS-CoV-2. Front Immunol.13, 1023255 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castro, J. T. et al. Promotion of neutralizing antibody-independent immunity to wild-type and SARS-CoV-2 variants of concern using an RBD-Nucleocapsid fusion protein. Nat. Commun.13, 4831 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Moor, W. R. J. et al. LSDV-Vectored SARS-CoV-2 S and N Vaccine Protects against Severe Clinical Disease in Hamsters. Viruses15, 10.3390/v15071409 (2023). [DOI] [PMC free article] [PubMed]
- 38.Hajnik, R. L. et al. Dual spike and nucleocapsid mRNA vaccination confer protection against SARS-CoV-2 Omicron and Delta variants in preclinical models. Sci. Transl. Med14, eabq1945 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCafferty, S. et al. A dual-antigen self-amplifying RNA SARS-CoV-2 vaccine induces potent humoral and cellular immune responses and protects against SARS-CoV-2 variants through T cell-mediated immunity. Mol. Ther.30, 2968–2983 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarrés-Freixas, F. et al. Heterogeneous Infectivity and Pathogenesis of SARS-CoV-2 Variants Beta, Delta and Omicron in Transgenic K18-hACE2 and Wildtype Mice. Front Microbiol13, 840757 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dos Santos Alves, R. P. et al. Human coronavirus OC43-elicited CD4(+) T cells protect against SARS-CoV-2 in HLA transgenic mice. Nat. Commun.15, 787 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zost, S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature584, 443–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Premkumar, L. et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol.5, 10.1126/sciimmunol.abc8413 (2020). [DOI] [PMC free article] [PubMed]
- 44.Shailendra K. V, et al. Influence of Th1 versus Th2 immune bias on viral, pathological, and immunological dynamics in SARS-CoV-2 variant-infected human ACE2 knock-in mice. eBioMed.In revision (2024). [DOI] [PMC free article] [PubMed]
- 45.Gruber, A. D. et al. Standardization of Reporting Criteria for Lung Pathology in SARS-CoV-2-infected Hamsters: What Matters? Am. J. Respir. Cell Mol. Biol.63, 856–859 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheutlin, A. et al. Durability of Protection Post–Primary COVID-19 Vaccination in the United States. Vaccines10, 1458 (2022). [DOI] [PMC free article] [PubMed]
- 47.Suthar, M. S. et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med.1, 100040 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koerber, N. et al. Dynamics of spike-and nucleocapsid specific immunity during long-term follow-up and vaccination of SARS-CoV-2 convalescents. Nat. Commun.13, 153 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long, Q. X. et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med26, 845–848 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Ma, M. L. et al. Systematic profiling of SARS-CoV-2-specific IgG responses elicited by an inactivated virus vaccine identifies peptides and proteins for predicting vaccination efficacy. Cell Discov.7, 67 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piccoli, L. et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell183, 1024–1042.e1021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Min, L. & Sun, Q. Antibodies and Vaccines Target RBD of SARS-CoV-2. Front Mol. Biosci.8, 671633 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tauzin, A. et al. Humoral Responses against BQ.1.1 Elicited after Breakthrough Infection and SARS-CoV-2 mRNA Vaccination. Vaccines (Basel)11, 10.3390/vaccines11020242 (2023). [DOI] [PMC free article] [PubMed]
- 54.Wu, F. et al. Evaluating the Association of Clinical Characteristics With Neutralizing Antibody Levels in Patients Who Have Recovered From Mild COVID-19 in Shanghai, China. JAMA Intern Med180, 1356–1362, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muecksch, F. et al. Longitudinal Serological Analysis and Neutralizing Antibody Levels in Coronavirus Disease 2019 Convalescent Patients. J. Infect. Dis.223, 389–398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding, S. et al. SARS-CoV-2 Spike Expression at the Surface of Infected Primary Human Airway Epithelial Cells. Viruses14, 10.3390/v14010005 (2021). [DOI] [PMC free article] [PubMed]
- 57.Beaudoin-Bussières, G. et al. A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep.38, 110368 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartsch, Y. C. et al. Omicron variant Spike-specific antibody binding and Fc activity are preserved in recipients of mRNA or inactivated COVID-19 vaccines. Sci. Transl. Med14, eabn9243 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu, J. et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature603, 493–496 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zonozi, R. et al. T cell responses to SARS-CoV-2 infection and vaccination are elevated in B cell deficiency and reduce risk of severe COVID-19. Sci. Transl. Med15, eadh4529 (2023). [DOI] [PubMed] [Google Scholar]
- 61.Naranbhai, V. et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell185, 1041–1051.e1046 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazzoni, A. et al. SARS-CoV-2 Spike-Specific CD4+ T Cell Response Is Conserved Against Variants of Concern, Including Omicron. Front Immunol.13, 801431 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohen, K. W. et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep. Med.2, 100354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng, Y. et al. An immunodominant NP(105-113)-B*07:02 cytotoxic T cell response controls viral replication and is associated with less severe COVID-19 disease. Nat. Immunol.23, 50–61 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sahin, U. et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature595, 572–577 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Phan, J. M. et al. Cytotoxic T Cells Targeting Spike Glycoprotein Are Associated with Hybrid Immunity to SARS-CoV-2. J. Immunol.210, 1236–1246 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kundu, R. et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun.13, 80 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sagar, M. et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J Clin Invest131, 10.1172/jci143380 (2021). [DOI] [PMC free article] [PubMed]
- 69.Loyal, L. et al. Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Sci. (N. Y., N. Y.)374, eabh1823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nelde, A. et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol.22, 74–85 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Mateus, J. et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Sci. (N. Y., N. Y.)370, 89–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prakash, S. et al. Genome-Wide B Cell, CD4(+), and CD8(+) T Cell Epitopes That Are Highly Conserved between Human and Animal Coronaviruses, Identified from SARS-CoV-2 as Targets for Preemptive Pan-Coronavirus Vaccines. J. Immunol.206, 2566–2582 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi, H. et al. Protective immunity by an engineered DNA vaccine for Mayaro virus. PLoS Negl. Trop. Dis.13, e0007042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hastie, K. M. et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Sci. (N. Y., N. Y.)374, 472–478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill25, 10.2807/1560-7917.Es.2020.25.3.2000045 (2020). [DOI] [PMC free article] [PubMed]
- 76.Alexandersen, S., Chamings, A. & Bhatta, T. R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun.11, 6059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.