Abstract
The Hainan muntjac (Muntiacus nigripes) is a wild animal endemic to Hainan, China. Its species distribution and the diversity of muntjac karyotypes have attracted much attention. Although genomic resources have increased in recent years, relevant genome assembly data of Hainan muntjac are still lacking. Meanwhile, molecular evidence for the taxonomic units of this species remains lacking. In this study, we successfully assembled the Hainan muntjac haplotype genome at the chromosome level using Pacbio long read and long sequencing technologies and Hi-C data. The final assembly size was 2.66 Gb, with allele and scaffold N50 values of 29.27 and 700.27 Mb, respectively, and we scaffolded the genome sequence onto three chromosomes. The genome contains a total of 21,451 genes and 10,056 gene families. Phylogenetic analysis using single-copy gene families revealed that Hainan muntjac is most closely related to red muntjac, with a divergence time of 8–11 Ma. This new genomic resource of Hainan muntjac will be crucial for future comparative genomic analyses and genetic evolutionary studies.
Subject terms: Genome evolution, Evolution
Background & Summary
The Hainan muntjac (Muntiacus nigripes), is a small deer species belonging to the genus Muntiacus1,2. This genus is widely distributed across regions such as China, India, Thailand, and Malaysia. The Hainan muntjac is an endemic species of China’s Hainan Province and has garnered attention for its distinctive presence in a geographically unique location3. The Muntiacus species is frequently cited as an excellent model for studying vertebrate evolution due to the unique nature of its chromosome. The range of chromosome numbers varies from 2n = 6 in female Indian muntjac to 2n = 46 in Chinese muntjac(Muntiacus reevesi)4,5.
The chromosome evolution of the Muntiacus genus has been extensively researched. Yang et al.6 and Frönicke et al.7 have provided essential evidence for the homology and occurrence of chromosomal fission-fusion events in Muntiacus species using molecular techniques such as chromosome painting, digital imaging and heterologous fluorescence in situ hybridization (FISH). Mudd et al.8 have substantiated the karyotypic differences and chromosomal evolution in musk deer species through genomic data. In addition, the Red Muntjac wide distribution indicates significant genetic diversity and differentiation among subspecies9,10. At the same time, Hainan muntjac, as an endemic species in Hainan, the time of divergence from its close relatives is still unclear.
High-quality chromosome-level genomes are essential for studying chromosome evolution, species evolution, and population genetic diversity11. A chromosomal-level genome of high quality can improve the detection of chromosomal evolutionary events. It can improve the detection of chromosomal evolutionary events and provide more accurate calculations of population genetic parameters, such as genetic diversity, gene flow, phylogenetic relationships, and genetic load within the genome. Therefore, we have generated the first high-quality chromosome-level haplotype genome of Hainan muntjac by combining Pacbio long-read data, DNBSEQ short-read data, and HiC sequencing data. However, the absence of a Y chromosome in the assembled genome may restrict its utility in investigating male-specific genetics and sex determination. Nevertheless, the female Hainan muntjac genome retains its intrinsic value for examining chromosomal evolution, particularly in light of its distinctive chromosomal characteristics, such as fusions. Overall, the genome enhances the genomic resources of genus Muntiacus, offering crucial support for future research on karyotypic evolution, ecological studies, and species conservation.
Methods
Samples collection and ethics statements
Blood samples were collected from a female Hainan muntjac in Hainan Province, China, for high-molecular-weight DNA extraction. The Hainan muntjac was rescued and anesthetized with an anticoagulant tube before the samples were immediately transferred to liquid nitrogen and stored in a −80 °C refrigerator. The study design, experiments, and sample collection were approved by the Review Board of Hainan University (HNUAUCC-2024-00214).
DNA/RNA extraction, libraries preparation and sequencing
The high molecular weight genomic DNA for PacBio HiFi sequencing was isolated using 3.5 ml blood sample and the cetyltrimethylammonium bromide (CATB) method, and purified with the Blood & Cell Culture DNA Midi Kit (QIAGEN). DNA quality was assessed using a 1% agarose gel and a NanoDrop One UV-Vis spectrophotometer (Thermo Fisher Scientific, USA), while DNA quantity was measured using a Qubit 4.0 (Invitrogen, USA). The PacBio HiFi library was constructed at the Genome Center of NextOmics Bioscience Co. (Wuhan, China). The genome center of Ltd. in Wuhan, China followed the standard protocol of PacBio (Pacific Biosciences, USA) using SMRTbell 15 kb preparation solution to prepare the libraries. The libraries were then sequenced on a PacBio Sequel II sequencer using Sequencing Primer V2 and Sequel II Binding Kit 2.0. Total RNA was extracted from a 2 ml blood sample using the TRNzol Universal Kit (TIANGEN). RNA concentration and purity were assessed using NanoDrop One (Thermo Scientific, USA) and Qubit 4 Fluorometer-1 (Thermo Scientific, USA), while RNA integrity was evaluated with the Agilent 2100 Bioanalyzer. For short-threaded RNA sequencing, libraries were prepared using the TruSeq mRNA Library Kit (Illumina, USA) and sequenced on an Illumina HiSeq X Ten Sequencer (Illumina, USA) or the MGIEasy RNA Library Preparation Kit (MGI, China). Hi-C libraries were prepared for Hainan muntjac using the dnpII restriction endonuclease. The libraries were constructed on the DNBSEQ platform following the manufacturer’s instructions (MGIEasy Universal DNA Library Preparation Kit, BGI) and sequenced on a DNBSEQ-T1 sequencer (MGI, China).
Genome assembly and assessment
The study sequenced and assembled the genome of Hainan muntjac using a combination of Pacbio (HIFI) technology, HiC chromosome conformation sequencing, and short read length sequencing. The heterozygous genome was assembled using hifiasm software12 with the raw HIFI data (Table S1), and then combined with the HIC data to decompose the hybrid genome into chromosome-level haplotypes. The genome was improved using GapCloser13 (v. 1.12) to intercombine short read data and gap-fill the genomes, resulting in improved assembly.
The completeness of the final genome assembly was assessed using BUSCO v514 and compared to the mammalian database (mammal_odb10). The Burrows-Wheeler aligner was used to map reads to the genome assemblies, and coverage, depth, and alignment were calculated to assess the completeness and uniformity of the assembly.
A chromosome-level and haplotype-resolved genome for the Hainan muntjac was generated by combining ~23x PacBio HiFi long-read data, DNBSEQ short-read data, and Hi-C data. The Hainan muntjac genome, referred to as Haimjac, was assembled with a size of 2.66 Gb and a scaffold N50 of 700.27 Mb (Fig. 1, Table 1). Approximately 99.34% of the sequence bridges, equivalent to over 2.40 Gb, were mapped to three chromosome-scale pseudomolecule (Figure S1, Table S2). Our assembly results and the specific karyotype composition of this species are very similar to that of the closely related Muntiacus muntjak species8.
Fig. 1.
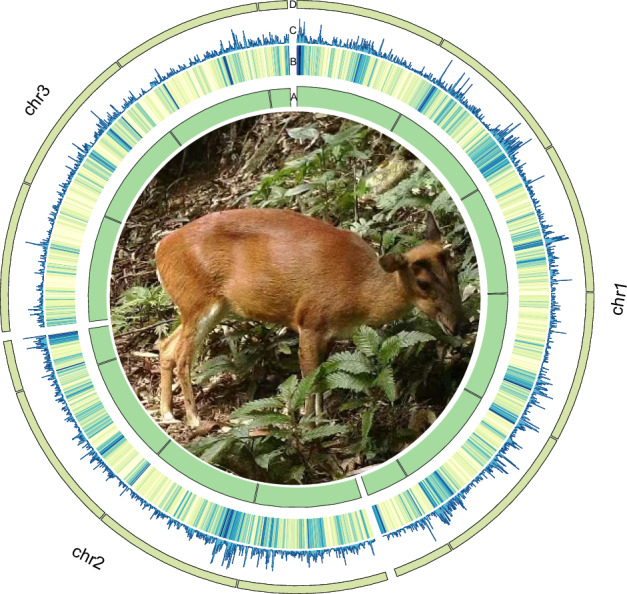
Genomic landscapes of the Hainan muntjac. A. sequencing depth; B. GC contents; C. gene density; D. Schematic diagram of chromosomes; The photo of a female Hainan muntjac taken with a normal color camera that was infrared controlled in the field.
Table 1.
Statistics of the sequencing data, assembly, and annotation results of the Hainan muntjac genome.
| Category | Haimuj | |
|---|---|---|
| Sequencing data | PacBio (Gb) | 63.24 |
| WGS (Gb) | 195.26 | |
| Hi-C (Gb) | 224.39 | |
| RNA-seq (Gb) | 22.25 | |
| Assembly | Assembled genome size (Gb) | 2.66 |
| Contig N50 (Mb) | 29.27 | |
| Scaffold N50 (Mb) | 700.27 | |
| Longest scaffold (Mb) | 1,132 | |
| GC content (%) | 42.19 | |
| Annotation | Repeat sequences (%) | 36.20 |
| Number of protein-coding genes | 21,451 | |
| Number of functionally annotated genes | 21,139 |
Through the utilization of (Benchmarking Universal Single-Copy Orthologs) to evaluate genome quality, we achieved a score of 95.9%, demonstrating a high level of genomic integrity and quality (Table S3).
Genome annotation
Firstly, we annotated repetitive elements using a combination of de novo and homologous methods. To identify novel repetitive sequences, we used RepeatModeler215 (v2.0.1) and LTR finder16 (v1.0.6), and merged them with known elements in the RepBase database. We then performed a conserved BLASTN search on the RepBase library using RepeatMasker17 (v4.0.5) to classify transposable elements. The RepeatProteinMask program from RepeatMasker17 (v4.0.5) was utilized to identify homologous repeat proteins. Furthermore, the Finder software18 (v4.07) was employed to annotate tandem repeats. Additionally, blastall19 (v2.2.26), tRNAscan-SE20 (v2.0.9), and INFERNAL v1.1.121 were used to characterize ribosomal RNA (rRNA), transfer RNA (tRNA), and microRNA (miRNA), respectively, with the Rfam database.
The results based on the above methodology show that genome contains 963.24 Mb (36.7%) of repetitive elements, with LINE being the most prevalent at 35.24% (Table S4, S5). The LTR is the next largest, occupying 11.03% of the genome. In the Hainan muntjac genome, we predicted 250 rRNA, 386 miRNA, 339 tRNA, and 352 snRNA (Table S6).
For protein-coding gene annotation, we masked all repetitive elements in the genome and combined transcripts, homology evidence, and de novo evidence to construct the final gene set. To annotate genes based on homology, we used blastall19 (v2.2). To conduct comparisons, set the E-value truncation to 1e-5 and include the following species: Cervus canadensis, Bos taurus, Ovis aries, Homo sapiens, Cervus elaphus. For de novo gene annotation, we used Augustus22 (v3.0.3), GlimmerHMM23 (v3.0.1), and SNAP24 (v11/29/2013) for prediction. Long-read and short-read RNA sequencing data were employed for transcript-based gene prediction. Transcripts were identified using the IsoSeq (v3) pipeline. Short-read mapping was performed using HISAT225 (v2.1.0). The final gene set was generated by the MAKER pipeline26 (v3.01.03) through the combination of genes predicted through RNA-seq, homology, and novel approaches.
Using de novo prediction, homology-based protein alignment, and RNA-seq mapping methods, we identified 21,451 gene models in the genome (Figure S2). These gene regions span over 732.68 Mb, accounting for 27.53% of the Haimjac genome (Table S7). The final integrated gene set was subjected to BUSCO analysis, and the gene set integrity of the Hainan muntjac genome was high with a score of 91.5% (Table S8). Notably, 98.55% of the genes in the Hainan muntjac genome received functional annotations (Table S9 and Figure S3).
The genome was functionally annotated by conducting BLAST searches against the SwissProt, TrEMBL, and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, using E-value cutoffs of 1e-5. To predict motifs, domains, and gene ontology (GO) terms, InterProScan27 (v5.52–86.0) was utilized.
Phylogeny construction and gene family expansion
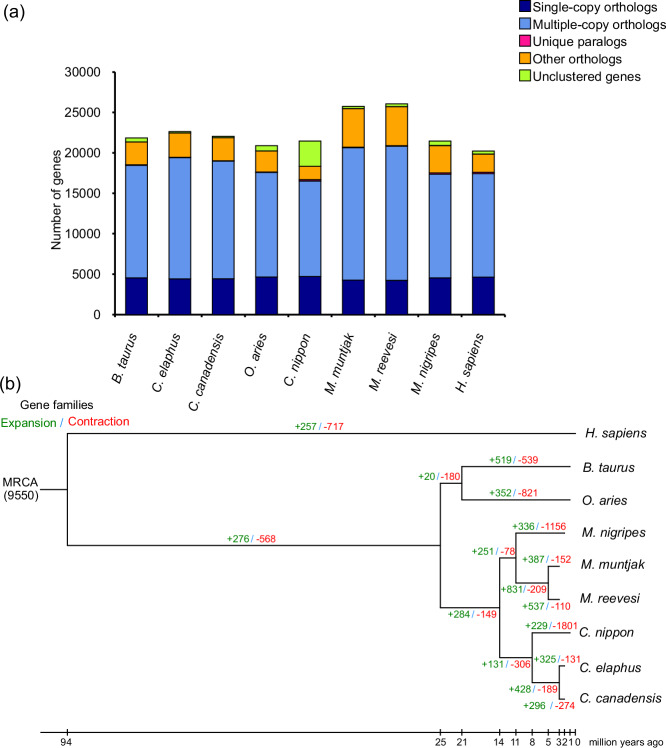
The protein sequences of single-copy gene families were compared using the MUSCLE algorithm28 to perform sequence alignment. RAxML29 was used to construct phylogenetic trees, and the corresponding dendrogram files were generated. To enhance clarity, we visualized the dendrograms using MEGA software. To estimate the divergence time of species, we utilized the software packages PAML 430 and MCMCTREE31. This step aimed to provide a more precise estimate of the evolutionary timeline of the species. To cluster gene families, we utilized the BLASTP tool to compare protein sequences among all species, estimating similarities and differences between genes. Setting the E-value threshold at 1e-7 ensured the fidelity of our outcomes. Subsequently, gene clustering was executed via H-cluster software, enhancing our comprehension of intra-family correlations. Employing the TreeFam methodology, we elucidated homologous and paralogous associations across species, thereby unveiling the evolutionary trajectory of single-copy gene families within the phylogenetic framework. The analysis of gene family clustering in Hainan muntjac identified 10,056 gene families, which included 4,538 single-copy orthologs, 12822 multiple-copy orthologs 173 unique paralogs and 3364 other orthologs (Fig. 2a, Table S10). Additionally, the analysis of gene family expansion and contraction (CAFE) revealed 336 expanded and 1156 contracted gene families (P < 0.05; Fig. 2b).
Fig. 2.
(a) Comparison of the number of homologous genes. (b) The phylogenetic relationship of 9 species and the estimated divergence time. Numbers on the branch of the phylogenetic tree represent the number of significantly expanded (green) and contracted (red) gene families.
Data Records
The Hainan muntjac genome assembly has been deposited in the NCBI BioProject database under accession number PRJNA1090610. The genomic RNA sequencing data are available in the NCBI Sequence Read Archive (SRA) under accession number SRR2881045932. The Hi-C sequencing data are deposited under SRA accession numbers SRR2881046133 and SRR2881046234, while the genomic Pacbio sequencing data are available under accession number SRR2881046335. The assembled genome has been deposited in GenBank under accession number GCA_039877825.136. The annotation of the genome, including information on repetitive sequences, gene structure and functional predictions, is available in the Figshare database37.
Technical Validation
Genomic integrity, fragmentation, and potential loss rates were measured using BUSCO V5. Among 9226 prospective conserved core genes in the mammalian database, 95.9% and 1% were identified as complete BUSCOs and fragment BUSCOs, respectively, indicating that the assembled genome had high integrity and validity and could be used for further analysis.
Supplementary information
Acknowledgements
This study was supported by 2022 Central Finance Forestry and Grassland Ecological Protection and Restoration Fund (National Park Subsidy)-Project of Impact of human interference on wildlife and their habitats.
Author contributions
Y.C., Y.L. and H.L. wrote the manuscript. Y.L., J.L. and M.C. collected the samples. Y.C., M.C., X.L. and Z.X. performed the RNA/DNA isolation. Y.C., Y.L., Z.X. performed the data analysis. X.L. and H.L. reviewed the manuscript. H.L. provided the supervision of this project All authors read and approved the final manuscript.
Code availability
No specific scripts were used in this work. All codes and pipelines for data processing were executed following the manuals and protocols of the respective bioinformatics software. The specific software versions are detailed in the Methods section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yilin Cui, Yakui Lv.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04167-2.
References
- 1.Groves, C. & Grubb, P. Ungulate taxonomy. (2011).
- 2.Groves, C. Systematics of the Artiodactyla of China in the 21st century. (2016). [DOI] [PMC free article] [PubMed]
- 3.Ohtaishi, N. & Gao, Y. A review of the distribution of all species of deer (Tragulidae, Moschidae and Cervidae) in China. Mammal Review20, 125–144, 10.1111/j.1365-2907.1990.tb00108.x (1990). [Google Scholar]
- 4.Wurster, D. H. & Benirschke, K. Indian Muntjac, Muntiacus muntjak: A Deer with a Low Diploid Chromosome Number. Science168, 1364–1366, 10.1126/science.168.3937.1364 (1970). [DOI] [PubMed] [Google Scholar]
- 5.Wurster, D. H. & Benirschke, K. Chromosome Studies in Some Deer, the Springbok, and the Pronghorn, with Notes on Placentation in Deer. CYTOLOGIA32, 273–285, 10.1508/cytologia.32.273 (1967). [DOI] [PubMed] [Google Scholar]
- 6.Yang, F., Carter, N. P., Shi, L. & Ferguson-Smith, M. A. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma103, 642–652, 10.1007/BF00357691 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Frönicke, L., Chowdhary, B. P. & Scherthan, H. Segmental homology among cattle (Bos taurus), Indian muntjac (Muntiacus muntjak vaginalis), and Chinese muntjac (M. reevesi) karyotypes. Cytogenetics and Cell Genetics77, 223–227, 10.1159/000134581 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Mudd, A. B., Bredeson, J. V., Baum, R., Hockemeyer, D. & Rokhsar, D. S. Analysis of muntjac deer genome and chromatin architecture reveals rapid karyotype evolution. Communications biology3, 480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martins, R. F. et al. Phylogeography of red muntjacs reveals three distinct mitochondrial lineages. BMC Evolutionary Biology17, 34, 10.1186/s12862-017-0888-0 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh, V. K. et al. Genetic diversity and population structure of the northern red muntjac (Muntiacus vaginalis) in Indian Himalayan region. Mammalian Biology102, 537–544, 10.1007/s42991-022-00254-2 (2022). [Google Scholar]
- 11.Fan, H. et al. Chromosome-level genome assembly for giant panda provides novel insights into Carnivora chromosome evolution. Genome Biology20, 267, 10.1186/s13059-019-1889-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature methods18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, M. et al. TGS-GapCloser: a fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience9, giaa094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manni, M., Berkeley, M. R., Seppey, M. & Zdobnov, E. M. BUSCO: assessing genomic data quality and beyond. Current Protocols1, e323 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proceedings of the National Academy of Sciences117, 9451–9457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic acids research35, W265–W268 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, N. Using Repeat Masker to identify repetitive elements in genomic sequences. Current protocols in bioinformatics5, 4–10 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic acids research27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mount, D. W. Using the basic local alignment search tool (BLAST). Cold Spring Harbor Protocols2007, pdb–top17 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic acids research34, W435–W439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics20, 2878–2879 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Korf, I. Gene finding in novel genomes. BMC bioinformatics5, 1–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nature methods12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell, M. S. et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant physiology164, 513–524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics5, 1–19 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics17, 754–755 (2001). [DOI] [PubMed] [Google Scholar]
- 32.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810459 (2024).
- 33.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810461 (2024).
- 34.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810462 (2024).
- 35.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810463 (2024).
- 36.Cui, Y. L. H. GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_039877825.1 (2024).
- 37.Cui, Y. A haplotype-resolved and chromosome-scale genome assembly of Hainan muntjac (Muntiacus nigripes). figshare10.6084/m9.figshare.26993131 (2024). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810459 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810461 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810462 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR28810463 (2024).
- Cui, Y. L. H. GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_039877825.1 (2024).
- Cui, Y. A haplotype-resolved and chromosome-scale genome assembly of Hainan muntjac (Muntiacus nigripes). figshare10.6084/m9.figshare.26993131 (2024). [DOI] [PubMed]
Supplementary Materials
Data Availability Statement
No specific scripts were used in this work. All codes and pipelines for data processing were executed following the manuals and protocols of the respective bioinformatics software. The specific software versions are detailed in the Methods section.