Abstract
Background
Specific cis-elements and the associated trans-acting factors have been implicated in the post-transcriptional regulation of gene expression. In the era of genome wide analyses identifying novel trans-acting factors and cis-regulatory elements is a step towards understanding coordinated gene expression. UV-crosslink analysis is a standard method used to identify RNA-binding proteins. Uridine is traditionally used to radiolabel substrate RNAs, however, proteins binding to cis-elments particularly uridine poor will be weakly or not detected. We evaluate here the possibility of using UV-crosslinking with RNA substrates radiolabeled with each of the four ribonucleotides as an approach for screening for novel sequence specific RNA-binding proteins.
Results
The radiolabeled RNA substrates were derived from the 3'UTRs of the cloned Eg and c-mos Xenopus laevis maternal mRNAs. Specific, but not identical, uv-crosslinking signals were obtained, some of which corresponded to already identified proteins. A signal for a novel 90 kDa protein was observed with the c-mos 3'UTR radiolabeled with both CTP and GTP but not with UTP. The binding site of the 90 kDa RNA-binding protein was localised to a 59-nucleotide portion of the c-mos 3'UTR.
Conclusion
That the 90 kDa signal was detected with RNAs radiolabeled with CTP or GTP but not UTP illustrates the advantage of radiolabeling all four nucleotides in a UV-crosslink based screen. This method can be used for both long and short RNAs and does not require knowledge of the cis-acting sequence. It should be amenable to high throughput screening for RNA binding proteins.
Background
It is now well documented that gene expression can be regulated post-transcriptionally and a number of RNA sequence elements have been identified that play a role in targeting a mRNA for a particular mode of regulation. For instance, the stability, the translation and the localisation of a mRNA are regulated by cis-elements, many of which are situated in the 3' untranslated region (3'UTR), but such sequence elements are not restricted to the 3'UTR (for review see [1]).
In most cases, regulatory cis-elements are associated with specific RNA-binding proteins (RNA-BP). Several functional pairs of cis-element and trans-acting factor have been identified, for instance the Iron responsive element and the Iron Responsive Protein (reviewed in [2]), the bruno responsive element and bruno [3,4], the Cytoplasmic polyadenylation element (CPE) and the CPE-binding protein [5-7], the Embryo Deadenylation ElemeNt (EDEN) and the EDEN-binding protein [8,9] and lastly, the nanos responsive element and pumilio [10,11]. In at least one instance, the identified cis-element can associate with several factors that confer opposing behaviours: AU-rich elements composed of multiple AUUUA motifs can bind both HuR and AUF1. AUF1 binding is correlated with rapid mRNA degradation [12] whereas the mRNA is more stable when HuR binds [13,14].
Sequence specific RNA-BPs are, in general, part of a functional multimeric complex and their primary role in recognising a specific sequence motif could be to anchor the functional complex onto certain mRNAs; anchoring in this context is probably a dynamic process. Accordingly, we hypothesised that identifying the proteins that exhibit binding specificity for a given mRNA or family of similarly regulated mRNAs would be a useful starting point to characterize both novel regulatory cis-acting sequence elements and the associated multimeric complexes. Within the framework of this hypothesis the experimental approach used should be independent of a prior knowledge of the cis-element and should be amenable to the simultaneous analysis of many mRNAs.
The identification of regulatory cis-elements is commonly addressed by preparing chimeric genes containing different parts of the mRNA under study and analysing the behaviour of the transcribed RNAs either in vivo or in cell extracts. This approach is not readily applicable to the simultaneous analysis of many RNAs. For identification of RNA-BPs, several techniques have been recently developed that allow their direct purification or cloning, including RNA-affinity chromatography [15] and the three-hybrid screen in yeast cells [16]. Again, neither of these methods can be readily used for high-throughput screening. Both are nominally aimed at identifying proteins that bind to a known sequence element; the three-hybrid screen can also be used to identify a sequence element bound by a known protein. In the three-hybrid screen, using long RNAs such as those representing the complete 3'UTRs of certain mRNAs would cause technical problems [16].
With the long-term aim of setting up a screening method to identify novel regulatory cis-acting sequence elements and the associated multimeric complexes, we have evaluated the use of uv-crosslinking. This method can be miniaturised and time saving procedures have been described [17] that should allow its application in a high-throughput protocol.
In uv-crosslinking a covalent bond is formed by UV irradiation between the RNA and closely interacting proteins. However, this reaction is relatively inefficient and the formation of a photo-induced product is strongly dependent on structural parameters such as the proximity of reactive amino acids and base. Not all amino acids and bases are equivalent with respect to this photo-induced reaction [18] and references therein). After UV irradiation, the RNA-protein complex is digested by an RNase. This produces a protein with an associated short oligo ribonucleotide that corresponds to the protected RNA fragment. RNAs containing radiolabeled nucleotides are often used in uv-crosslinking experiments. In this case the resulting protein-oligonucleotide product will be radiolabeled only if the oligonucleotide contains a radiolabeled nucleotide. Accordingly the probability of detecting an RNA-BP that binds to a sequence motif rich in a particular base will be increased if this ribonucleotide is radiolabeled. It is important to note that the radiolabeled nucleotide is not necessarily in contact with or covalently linked to the protein.
The most commonly used radiolabeled ribonucleotide is uridine. Other ribonucleotides have been used when the sequence element studied was composed exclusively or predominantly of another base. [19,20]). In order to improve the ability of uv-crosslinking to reveal RNA-BPs we have used all four ribonucleotides to radiolabel RNA substrates; we have called this method "comprehensive uv-crosslinking". We describe here experiments that validate this approach as a method to screen for novel RNA-BPs even with no prior knowledge of the regulatory element.
Results and Discussion
Comparison of the UV-crosslink patterns obtained with different radiolabeled nucleotides
To evaluate comprehensive uv-crosslinking as a screening method for novel RNA-BPs 32P-labeled UTP, GTP, CTP and ATP were used to radiolabel the RNA substrates corresponding to the 3'UTRs of Eg1, Eg2, Eg5 and c-mos. These RNAs were chosen for an evaluation of the screening method as the pattern of UV-crosslinked proteins obtained with 32P UTP has been described [21]. Also, these RNAs undergo the same post-transcriptional modifications during oocyte maturation and after fertilisation. One would expect therefore that some of the UV-crosslinked proteins would be common to these RNAs. As controls of specificity, similarly labeled RNAs corresponding to the 3'UTRs of the Xenopus laevis mRNAs encoding ornithine decarboxylase (ODC), phosphatase 2A catalytic subunit (PP2Ac), elongation factor 1β (EF1β) and elongation factor 1γ (EF1γ) were also synthesised. The 32P-labeled transcripts were incubated in cytoplasmic extracts made from 4h embryos and processed for UV-crosslink analysis as described in the Materials and Methods. The UV-crosslink patterns obtained with the four radiolabeled nucleotides are shown in Figure 1. For simplicity the radiolabeled RNA-protein complexes produced by UV-irradiation will be called radiolabeled proteins.
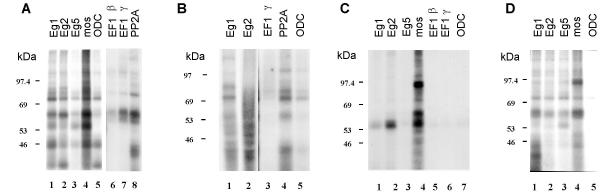
Figure 1.
Comparative analysis of proteins that UV crosslink to Eg1, Eg2, Eg5 and c-mos 3'UTRs.32P-labeled transcripts corresponding to the 3' untranslated regions of the RNAs indicated above the lanes were incubated in extracts made from 4 hour (pre-transcription) Xenopus embryos, irradiated with UV light and digested with RNase A. The proteins rendered radioactive by crosslinking to the labeled transcripts were analysed by electrophoresis on 10% polyacrylamide-SDS gels followed by autoradiography. A32P-UTP labeled RNAs; B32P-ATP labeled RNAs; C32P-GTP labeled RNAs; D32P-CTP labeled RNAs. In each panel the sizes of the molecular weight markers are indicated on the left.
When using 32P-labeled UTP the Eg and c-mos derived RNAs produced a number of radiolabeled proteins (Figure 1A lanes 1–4). We have already shown that these RNAs specifically bind a 53/55 kDa protein (EDEN-BP) [9]. With the exception of the signal produced by this protein, most of the bands were also present in the patterns produced with at least one of the control RNAs (compare lanes 1–4 with 5–8).
The molecules containing 32P-ATP yielded patterns with a large number of radiolabeled proteins (Figure 1B). A protein migrating just above the 69 kDa marker was present for all the transcripts tested except EF1γ. With the exception of EF1γ, all the transcripts terminated by a 65-nt poly(A) tail. Hence this protein may correspond to the 70 kDa embryonic Poly(A) binding protein [20]. Several proteins migrating between the 46 and 53 kDa markers are labeled in the presence of the Eg1 and Eg2 3'UTR RNAs but not with the PP2Ac or ODC derived RNAs (Figure 2B compare lanes 1 and 2 with 4 and 5).
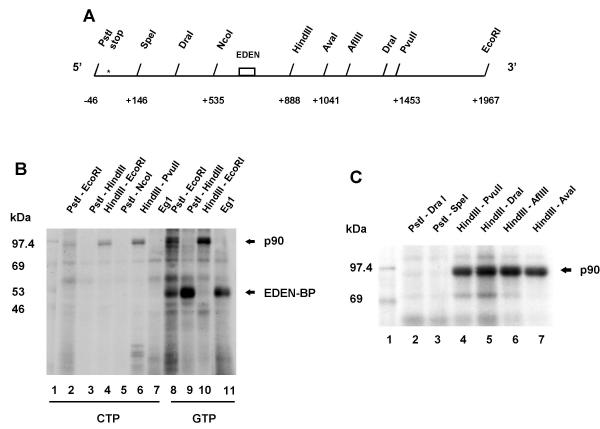
Figure 2.
p90 is a sequence specific RNA-binding protein.A Restriction map of c-mos 3'UTR. Numbering is from the stop codon which is indicated by a star (*), EDEN represents the EDEN sequence present in the 3'UTR of c-mos. B and C Mapping of the binding sites of p90 in the c-mos 3'UTR. 32P-CTP (panel B: lanes 2–7) and 32P-GTP (panel B: lanes 8–11 and panel C: lanes 2–7) labeled transcripts, that correspond to different regions of the c-mos 3'UTR, were synthesised and used in UV crosslinking analyses as described in the legend to Figure 1. The portions of the c-mos 3'UTR cDNA used as template to synthesise the different RNAs is indicated above the lanes. The positions of p90 and EDEN-BP are indicated on the right of the panels and the sizes of the molecular weight markers (lane 1) are indicated on the left.
The 32P-GTP labeled Eg and c-mos RNAs produced much simpler crosslink patterns (Figure 1C). The Eg1 and Eg2 derived RNAs labeled with 32P-GTP produced one predominant band migrating at the position of EDEN-BP (Figure 1C lanes 1 and 2). Three predominant signals (90, 60 and 53 kDa) were observed with the c-mos RNA (lane 4), one of which (53 kDa) also migrated at the position of EDEN-BP. Only faint, non-sequence specific, UV-crosslink patterns were obtained with all the other RNAs (lanes 3 and 5–7). Interestingly, the Eg5 RNA labeled with 32P-GTP did not produce a signal at the position of EDEN-BP.
When labeled with 32P-CTP, the Eg and c-mos derived RNAs produced two strong signals at 60 and 72 kDa (Figure 1D, lanes 1–4). The 72 kDa protein was also faintly seen with the ODC derived RNA (lane 5). Similar to the GTP-labeled RNA, a 90 kDa protein was also present when the c-mos RNA was labeled with 32P-CTP.
Several interesting features are highlighted by a comparison between the patterns obtained with the different 32P-labeled ribonucleotides; assuming that radiolabeled proteins with the same molecular weight are identical. Firstly, a protein with the binding specificity of EDEN-BP [21] is revealed when UTP and GTP but not by CTP are used to synthesis the RNA. This is coherent with the sequences of the characterised EDEN elements [9,22] that are rich in U/purine dinucleotides and contain few or no cytidines. A 60 kDa protein is observed with all the RNAs labeled with 32P-UTP or CTP except that derived from ODC mRNA. When using the 32P-GTP labeled transcripts, this protein only becomes radiolabeled in the presence of c-mos RNA. A 72 kDa protein appears to be bound to all of the RNAs. It is detected with the 32P-UTP and CTP labeled molecules but not with the 32P-GTP label. Lastly, binding of a 90 kDa protein detected with 32P-GTP and CTP but not with the 32P UTP, was only observed with the c-mos RNA.
This comparison shows that when performing an initial screen for proteins bound to a particular RNA or a family of RNA molecules, using several radiolabeled nucleotides should be particularly informative. In contrast to an analysis using only one radionucleotide, it allows the identification of the full complement of proteins that are susceptible to uv-crosslinking. The 90 kDa protein which shows binding specificity to the c-mos 3'UTR is an example of a protein that is not detected with the widely used 32P-labeled UTP.
Localisation of the p90 binding site in c-mos 3'UTR
The 90 kDa protein detected in the UV-crosslink experiments described above appears to exhibit sequence specific binding as this signal was only observed with the c-mos derived RNA. To confirm specificity we mapped the p90 binding site, again using UV crosslinking. The 1.8 kb cDNA corresponding to the c-mos 3'UTR is partitioned in approximately two equal parts by a HindIII restriction site (see Figure 2A). This site was used to subclone the 5' and 3' portions of this cDNA. Templates for in vitro transcription were made by linearizing the resulting plasmids with various restriction enzymes (see Figure 2A). Both 32P-GTP and CTP were used to make in vitro transcripts for UV-crosslink analysis. The Eg1 3'UTR labeled with 32P-CTP was included as a negative control as it does not bind a 90 kDa protein.
The data shown in Figure 2B demonstrate that p90 binding, detected with both radiolabels (lanes 2 and 8), is not observed with the 5' half of the c-mos 3'UTR (lanes 3, 5, and 9). Binding is observed with the 3' half of the c-mos 3'UTR (lanes 4 and 10) and it is conserved with the 565-nt portion of the 3' half situated between the central HindIII site and the PvuII site in the cDNA (Figure 2B lane 6). The p90 signal detected with both the 32P-GTP and CTP labeled RNAs appears as a doublet (Figure 2B lanes 2, 4, 6 and 8). Due to this similarity in the form of the signal and the sequence requirements shown by localisation, we assume that the same protein is being detected with the 32P-GTP and CTP labeled RNAs. Hence, for finer mapping of the p90 binding site only 32P-GTP was used. Further truncation of the cDNA containing the sequence of the 3' half of c-mos 3'UTR was done using the endogenous restriction sites (Figure 2A). The UV crosslink patterns obtained with the corresponding 32P-GTP labeled RNAs (Figure 2C) localised the p90 binding site to within 153 nt transcribed from the HindIII-AvaI portion of the cDNA (Figure 2C lanes 4–7). RNAs transcribed from the 5' half of the c-mos 3'UTR were included as negative controls (lanes 2 and 3).
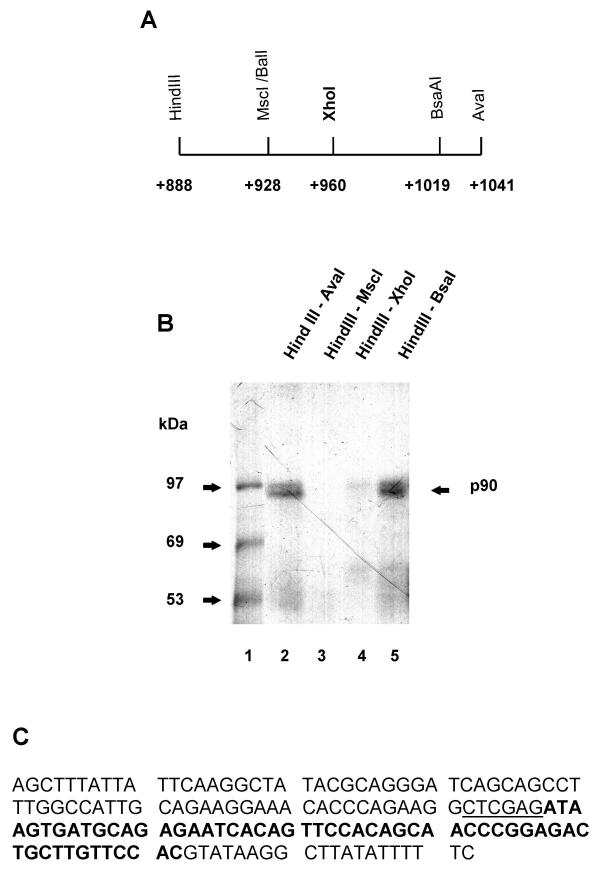
Finally, a XhoI restriction site was introduced, by site directed mutagenesis, midway between the HindIII and AvaI restriction sites in the c-mos cDNA (Figure 3A). The resulting plasmid was cut by different restriction enzymes to make in vitro transcription templates. The modified RNA corresponding to the HindIII-AvaI portion of the cDNA to c-mos 3'UTR still binds p90 with a distribution of signal between the two bands of the doublet that is identical to that of the wild-type RNA (compare Figure 3B lane 2 with Figure 2B lane 8). Hence, we did not disrupt sequence information needed for p90 binding by inserting the XhoI site into the cDNA. The 131 nt RNA transcribed from templates cut with BsaAI also retained p90 binding (Figure 3B lane 4) but p90 binding was not observed when the RNA only contained sequence upstream of the introduced XhoI site (lanes 3 and 4). Therefore, sequence information (Figure 3C, bold type) between nucleotides 960 (XhoI) and 1019 (BsaAI) of c-mos 3'UTR are required for p90 binding. At present we cannot formally exclude that sequence information upstream of the introduced XhoI site also affects p90 binding.
Figure 3.
Fine mapping of the p90 binding site.A Restriction map of the HindIII – AvaI region of c-mos 3'UTR cDNA. The position of the XhoI site introduced by mutagenesis is also indicated. Numbering is from the translation stop codon in the complete mRNA. B UV crosslink analysis of 32P-GTP-labeled transcripts that contain sequence information from different portions of the HindIII-AvaI regions of the c-mos 3'UTR cDNA, as indicated above the lanes. The position of the p90 protein is indicated on the right and the sizes of the molecular weight markers (lane 1) are indicated on the left. C The sequence of c-mos 3'UTR contained between the HindIII – AvaI restriction sites in the cDNA. The introduced XhoI site is underlined. The portion required for p90 binding is indicated in bold type.
The potential role of a cis-acting element can sometimes be inferred from knowing in which mRNAs it occurs. Therefore a blast search of the non-redundant 3'UTR database http://bighost.area.ba.cnr.it was made using the 59nt region of c-mos 3'UTR containing the p90 binding site. This search did not identify any regions of extended homology (above 25nt) in known 3'UTR sequences. It did identify 7 mRNAs, including c-mos that contained a common eight nucleotide sequence (ATGCAGAG). The Zuker m-fold program [23]http://bioinfo.math.rpi.edu/~mfold calculated that in the c-mos 3'UTR the last 6-nt of this sequence are presented as a loop on a 5 base pair stem. However this structural presentation is not conserved amongst the other six 3'UTRs. Therefore, the pertinence of this sequence with respect to p90 binding is not known.
Conclusions
Our data show that when performing UV-crosslink analysis to screen for RNA binding proteins, using several radiolabeled nucleotides allows a larger population of proteins to be identified. It should be emphasised that this method, like all others, cannot identify all the RNA-BPs associated with an RNA. Only those proteins that are structurally able to form a UV-induced product with the RNA molecule can be detected. In this study, a 90 kDa protein that shows binding specificity to a 59 nt region of the c-mos 3'UTR was detected with 32P-labeled CTP and GTP, but not with the widely used 32P-labeled UTP. The delimited 59 nt region is not particularly GC rich (Figure 3C) which emphasises the utility of screening with substrate RNAs radiolabeled with several nucleotides as opposed to only one.
Comprehensively screening for proteins that produce UV-induced RNA-protein adducts should result in the identification of novel and specific RNA binding proteins. Simultaneously screening several or many RNAs will give an immediate indication of specificity as observed in the present study. We consider that such screens will constitute a beneficial starting point for the localization of regulatory elements and the characterization of associated multimeric complexes involved in post-transcriptional regulation. At least some of the modifications in global gene expression observed when the physiological states of cells or organelles change are controlled at a post-transcriptional level. It would be interesting therefore to couple screens such as that proposed here to microarray based analyses of the genes expressed. In order for this to be informative a high throughput of samples should be achieved. Technical modifications that reduce the time for uv-crosslinking analyses have been described [17] and the major part of a uv-crosslinking analysis could probably be performed by the automated systems required for a high throughput of samples. Furthermore in vitro transcription using PCR produced templates could be used to considerably reduce the workload required to sufficiently delimit the binding site within the RNA. Once this has been achieved cloning of the identified factor could be addressed either by RNA affinity chromatography or by the three-hybrid approach.
Materials and Methods
Plasmids and binding site mapping
Eg5, ODC and PP2Ac
The plasmids containing the cDNAs corresponding to the complete 3'UTRs of Eg5, ODC and PP2Ac have been described previously [21].
Eg1
The 900 bp BglII – EcoRI cDNA fragment containing the complete Eg1 3'UTR [24] was cloned into pBluscript previously restricted by BamHI and EcoRI enzymes.
EF1γ
The 180 bp EcoRI cDNA fragment containing sequence of the complete 3'UTR of EF1γ mRNA [25] was isolated, blunt ended and cloned into pBluscript restricted by SmaI and EcoRV. Plasmids in which the 5' extremity of the insert was oriented towards the T7 promoter were selected by restriction analysis.
EF1β and c-mos
The PstI – Eco RI cDNA fragments that correspond to the compete 3'UTRs of EF1β (200 bp) [26] and c-mos (2000 bp) [27] mRNAs were isolated and cloned into pBluscript vectors restricted by these same enzymes.
To map protein binding sites in the c-mos 3'UTR, the cloned cDNA corresponding to the full length 3'UTR was first divided into two almost equal parts by digestion with HindIII (Figure 2A). Digestion with this enzyme excised the 3' portion of the c-mos 3'UTR cDNA. The HindIII digested plasmid, that retained the 5' portion of the c-mos 3'UTR, was closed by ligation. The excised 3' half of the 3'UTR was isolated and cloned into pT7/T3α19 (Gibco-BRL) previously restricted by HindIII. The plasmids in which the 5' end of this insert was oriented towards the T7 promoter were selected by restriction analysis. Finally, a XhoI site was introduced by PCR at position 960 of the c-mos 3'UTR, in the context of the HindIII – EcoRI terminal portion. The 5' part of this region was amplified with the primers: FHindIIImos 5'GCAAAGCTTTATTATTCAAGGCTATACGC and RXhoImos 5' CATCACTTATCTCGAGCCTTCTGGG. The 3' part was amplified with the primers: FXhoImos 5' GAAGGCTCGAGATAAGTGATGCAGA and REcoRImos 5' CGACGGCCAGTGAATTAACCCTC. The introduced XhoI site is underlined. The two PCR products were digested with XhoI and ligated together. After further restriction by HindIII and EcoRI the ligated product was cloned into pT7/T3α19 previously digested by the same enzymes. All constructions were verified by sequencing.
In vitro transcription and UV crosslinking
For in vitro transcription, the plasmids containing the cDNAs corresponding to the complete 3'UTRs of Eg1, Eg2, c-mos and EF1β were linearized by EcoRV. The Eg5 and PP2Ac plasmids were linearized by BamHI and the EF1γ plasmid by HindIII. The plasmid containing the modified c-mos cDNA was linearized by the enzyme indicated as the 3' terminus. 32P-labeled uncapped transcripts were made using the Promega Riboprobe transcription kit as previously described [21]. For the Eg5 and PP2Ac plasmids, transcription was from the T3 promoter. For the other plasmids transcription was from the T7 promoter.
Crosslinking of proteins to the variously labeled RNAs was performed as previously described [21]. Briefly, 50–100 femtomoles of 32P-labeled transcript were incubated in 10 μl of cytoplasmic extract (prepared from 4 hour Xenopus embryos) for 30 min, exposed to UV light, and digested with RNase A for 45 min at 37°C. Reactions were stopped with 2X SDS loading buffer and the entire reaction resolved by SDS-PAGE on a 10% Laemmli gel. Gels were fixed, dried and visualised by autoradiography.
Authors' contributions
Author 1 (RH) performed the initial UV crosslinking experiments. Author 2 (VM) did the fine mapping of p90 binding site. Author 3 (HBO) conceived of the study and participated in its design and co-ordination.
Acknowledgments
Acknowledgments
Support was obtained from the Centre National de la Recherche Scientifique, the European Union Biotechnology Program (BIO4-CT95-0045) and the Association pour la Recherche sur le Cancer (ARC n°9529). RH. was supported by a fellowship from the Human Frontier Science Program (LT0095/97-M).
Contributor Information
Rebecca Hartley, Email: rebecca-hartley@uiowa.edu.
Valerie Le Meuth-Metzinger, Email: lemeuth@roazhon.inra.fr.
H Beverley Osborne, Email: beverley.osborne@univ-rennes1.fr.
References
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Henderson BR, Kühn LC. Interactions between iron-regulatory porteins and their RNA target sequences, iron-responsive elements. Prog Mol Subcel Biol. 1997;18:117–140. doi: 10.1007/978-3-642-60471-3_6. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. Translational repressor bruno plays mutilple roles in development and is widely conserved. Genes Dev. 1997;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CA, Sheets MD, Wickens M. Poly(A) addition during maturation of frog oocytes: Distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Omilli F, Arlot-Bonnemains Y, Legagneux V, Roghi C, Bassez T, Osborne HB. The deadenylation conferred by the 3' untranslated region of a developmentally controlled mRNA in Xenopus embryos is switched to polyadenylation by deletion of a short sequence element. Mol Cell Biol. 1994;14:1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediates sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. RNA Regulatory Elements Mediate Control of Drosophila Body Pattern by the Posterior Morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol. 1999;62:257–91. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. Embo J. 1998;17:3461–70. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. Embo J. 2000;19:2340–50. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski D, Ostareck N, Standart N, Jackson R. Affinity methods for isolating RNA binding proteins. In: Smith C, editor. RNA:Protein Interactions: A pratical Approach. New York: Oxford University Press; 1998. pp. 137–158. [Google Scholar]
- SenGupta D, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci U S A. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A, Rogers J, Walker C, Staton J, Leedman P. Optimizer RNA Gel-shift and UV Cross-linking Assays for Characterization of Cytopmasmic RNA-Protein Interactions. BioTechniques. 1999;27:1032–1040. doi: 10.2144/99275rr03. [DOI] [PubMed] [Google Scholar]
- Hockensmith JW, Kubasek WL, Vorachek WR, Von Hippel PH. Laser cross-linking of proteins to nucleic acids. I. Examining physical parameters of protein-nucleic acid complexs. J Biol Chem. 1993;268:15712–15720. [PubMed] [Google Scholar]
- Murray KE, Roberts AW, Barton DJ. Poly(rC) binding proteins mediate poliovirus mRNA stability. Rna. 2001;7:1126–41. doi: 10.1017/S1355838201010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G, Ongkasuwan J, Standart N, Steitz J. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–88. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legagneux V, Bouvet P, Omilli F, Chevalier S, Osborne HB. Identification of RNA-binding proteins specific to Xenopus Eg maternal mRNAs: association with the portion of Eg2 mRNA that promotes deadenylation in embryos. Development. 1992;116:1193–1202. doi: 10.1242/dev.116.4.1193. [DOI] [PubMed] [Google Scholar]
- Audic Y, Omilli F, Osborne HB. Embryo deadenylation element-dependent deadenylation is enhanced by a cis element containing AUU repeats. Mol Cell Biol. 1998;18:6879–84. doi: 10.1128/mcb.18.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH. In: Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide In RNA Biochemistry and Biotechnology. J Barciszewski & BFC Clark, editor. Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
- Paris J, LeGuellec R, Couturier A, LeGuellec K, Omilli F, Camonis J, MacNeil S, Philippe M. Cloning by differential screening of a Xenopus cDNA coding for a protein highly homologous to cdc2. Proc Natl Acad Sci U S A. 1991;88:1039–1043. doi: 10.1073/pnas.88.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier P, Osborne HB, Morales J, Bassez T, Poulhe R, Mazabraud A, Mulner-Lorillon O, Bell R. Molecular cloning of Xenopus elongation factor 1 ç, major M-phase promoting factor substrate. Nucleic Acids Res. 1991;19:6644. doi: 10.1093/nar/19.23.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier P, Osborne HB, Morales J, Bassez T, Minella O, Pouhle R, Bellé R, Mulner-Lorillon O. Elongation factor 1 contains two homologous guanine-nucleotide exchange proteins as shown from the molecular cloning of beta and delta subunits. Nucleic Acids Res. 1993;21:743. doi: 10.1093/nar/21.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N, Oskarsson M, Copeland T, Brumbaugh J, Vande Woude GF. Function of c-mos proto-oncogene product in meiotic maturation in Xenopus oocytes. Nature (Lond) 1988;335:519–525. doi: 10.1038/335519a0. [DOI] [PubMed] [Google Scholar]