Abstract
The episymbiotic Candidatus Saccharibacteria is the most studied lineage of candidate phyla radiation. Living an epiparasitic lifestyle, Saccharibacteria might be associated with human mucosal diseases by modulating the structure of the oral microbiome through interactions with host bacteria. However, the knowledge of Saccharibacterial genomic diversity and the potential underlying their adaptation to a wide range of habitats remains limited. Here, we construct a high-quality genome collection of Saccharibacteria from multiple sources, providing 2041 high-quality genomes and previously unidentified taxa. The comparative genomic analysis shows the widespread metabolic defects of Saccharibacteria. Specific metabolic modules are commonly found in Saccharibacteria of different habitats, suggesting Saccharibacteria might have undergone habitat adaptation during the transition from different environments. We additionally show that Saccharibacteria account for ~1% of the Chinese oral microbiome. A preliminary analysis of rheumatoid arthritis individuals and healthy controls implies that Saccharibacteria might be associated with human systemic disease.
Subject terms: Microbiome, Metagenomics, Metagenomics
Introduction
The Candidatus Saccharibacteria, formerly known as candidate division TM7, cocultured with host bacteria, is the first member of the candidate phyla radiation (CPR)1,2 to be cultured3,4. It took ~20 years from the discovery of Saccharibacteria to uncover their epiparasitic lifestyle4. In the human oral cavity, there are at least G1-G6 six groups of Saccharibacteria3, while all species of Saccharibacteria cultivated from the human oral cavity are from the G1 group5. So far, multiple species of Saccharibacteria have been cultured from human oral cavity4,6–11, wastewater sludge12 and Cicadae periostracum13. Saccharibacteria exhibit diversification and adaptation during the transition from environments to mammals at genome level14.
The successful co-culture of Saccharibacteria with bacterial hosts provide the first insight into the episymbiotic relationship between bacteria4. The host bacteria undergo rapid evolution of decreased susceptibility after continuous co-culture with Saccharibacteria, then the two maintain a stable relationship15. Saccharibacteria may exert negative impacts on host bacteria and lead to their physiological changes such as increased stress responses, reduced cell growth, inhibited cell division, and cell lysis3,6,12,15–17. However, some species of Saccharibacteria may benefit their bacterial hosts by promoting their adaptation ability. For example, the Nanosynbacter lyticus strain TM7x isolated from the human oral cavity is able to promote the biofilm formation of its bacterial host Actinomyces odontolyticus strain XH00118, and facilitate XH001 achieve better survival under acid stress19. Over the past dozen years, the cocultures of host-epibiont pairs demonstrate Saccharibacteria have a limited host range restricted to some species of Actinobacteria7–13, and the type IV pili (T4P) play a part in the adhesion between Saccharibacteria and host bacteria12,13. In the scheme of the epibiotic lifecycle of TM7i, Saccharibacteria adhere to the host bacteria through T4P and proliferate, the progeny cells are dissociative, then adhere to new hosts by motility mechanisms mediated by T4P13.
Saccharibacteria includes members distributed both in mammalian host-associated niches and natural environments14, providing valuable insights into the potential functions of CPR bacteria across various ecological niches. The human oral cavity is one of the main habitats of Saccharibacteria, and the relative abundance of Saccharibacteria was higher in the human oral cavity than in other ecological niches20. It has been reported that an increased abundance of Saccharibacteria is often found in mucosal diseases such as periodontitis and gingivitis, and there are speculations that Saccharibacteria may be periodontal pathogens3,5,21. However, a recent study has reversed the assumption and found that in a ligature-induced periodontitis mouse model, Saccharibacterial species such as HMT346, 356, and 952 from periodontitis patients reduce inflammatory bone loss by modulating the pathogenicity of their host bacteria21. These findings indicate that the associations between Saccharibacteria and human health still need to be further explored22. In activated sludge, the co-culture of Candidatus Mycosynbacter amalyticus strain JR1 and its host bacteria was unexpectedly obtained when isolating bacteriophages from activated sludge. The host bacteria species of JR1, such as Gordonia amarae and Gordonia pseudoamarae, have been reported to be associated with the formation of wastewater foams and hinder the wastewater treatment process, while JR1 can lyse these host bacteria. In the future, JR1 might be used to improve wastewater treatment efficiency.
Considering the mysterious associations between Saccharibacteria with human health5 and the pioneering role of Saccharibacteria in CPR bacteria, it’s important to conduct a comprehensive survey of the whole Saccharibacteria lineage, especially those widespread in different environments that have not been cultured. Here, we construct a genome collection of Saccharibacteria from multiple sources and further investigate their taxonomic diversity, metabolic potential, and survival adaptation. We also investigate the relative abundance of Saccharibacteria in the Chinese oral cavity and preliminarily explore the associations between Saccharibacteria and rheumatoid arthritis (RA). Overall, the genome collection provided in this study will lay the foundation for a more comprehensive understanding of Saccharibacteria and provide novel insights for CPR.
Results
The expanded taxonomic diversity of Saccharibacteria
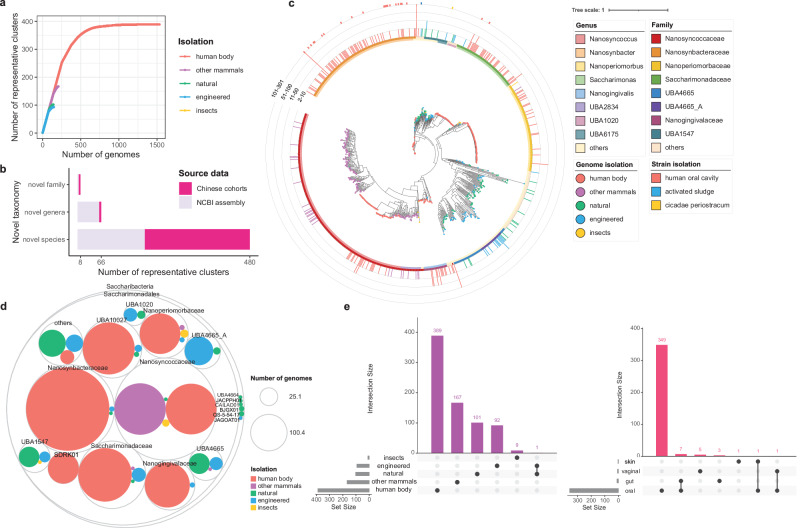
After exhaustive quality control, a Saccharibacteria genome collection comprised of 2041 high-quality genomes was constructed, there were 632 genomes from NCBI assembly23 and 1409 from Chinese cohorts24, all 2041 genomes were used for subsequent analyses (Supplementary Figs. 1–3, and Supplementary Table 1). The genomes were distributed in a variety of niches, including human body (1533; 75.11%), other mammals (208; 10.19%), natural environments (148; 7.25%), engineered environments (142; 6.96%), and insects (10; 0.49%). The majority of the genomes of the human body were from the human oral cavity (1508), and others were from the vagina (12), gut (11), and skin (2). The Saccharibacteria genome collection spanned a wide taxonomic diversity and contained members of Nanosynbacteraceae (538; 26.36%), Nanosyncoccaceae (407; 19.94%), Saccharimonadaceae (238; 11.66%), UBA10027 (214; 10.49%), Nanogingivalaceae (158; 7.74%), Nanoperiomorbaceae (136; 6.66%), SDRK01 (73; 3.58%), and other families (277; 13.57%) (Supplementary Fig. 3b, and Supplementary Table 2), and genomes were taxonomically affiliated to the genera Nanosynbacter (531; 26.02%), Nanosyncoccus (374; 18.32%), Saccharimonas (237; 11.61%), SDRW01 (208; 10.19%), Nanogingivalis (158; 7.74%), Nanoperiomorbus (128; 6.27%), SDRK01 (73; 3.58%) and other genera (332; 16.27%).
Genome clustering at 95% average nucleotide identity (ANI) yielded a total of 759 non-redundant representative clusters (Fig. 1a, and Supplementary Table 2). Of the 759 representative clusters, 389 were from humans, and 167, 102, 93, 9 were from other mammals, natural environments, engineered environments, and insects, respectively. There were 389 clusters from body, suggesting that Saccharibacteria have developed a rich diversity during the adaptation to the human body, which further supported the previous conclusion14. Among the 759 non-redundant representative clusters, 480 clusters were unclassified at species level, 66 clusters lacked a genus-level match, and 8 clusters lacked a family-level annotation, underlying that the database revealed an unprecedented diversity of Saccharibacteria (Fig. 1b). Among the unidentified clusters, the Chinese cohorts contributed 293 novel species, 6 novel genera, and 5 novel families. Moreover, it was worth mentioning that compared with cultivation, the culture-independent method revealed a richer diversity of Saccharibacteria (Fig. 1c).
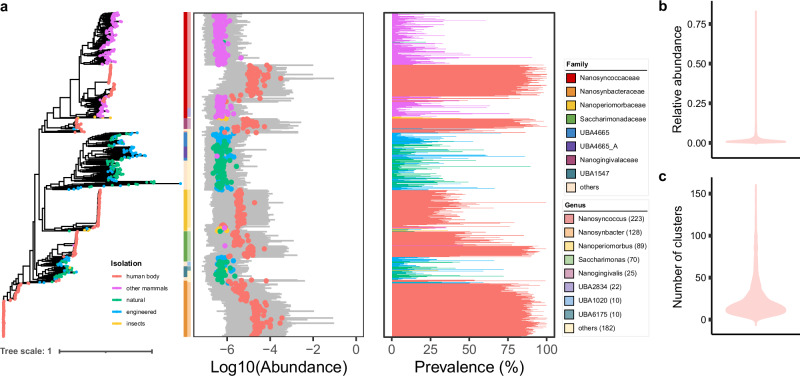
Fig. 1. Taxonomic profile of Saccharibacteria.
a Rarefaction curves of the number of representative clusters obtained as a function of the number of genomes analyzed. Separate colored curves are depicted for clusters from different environments. b Stack plots showing a number of new taxa contributed by Saccharibacteria representative clusters. The numbers of novel taxa at the species, genus, and family levels are shown separately, colored by public data or China cohorts. c Maximum-likelihood phylogenetic tree of the 2041 genomes. The inner dots are colored by the environment for each cluster, with the outer layer depicting the GTDB genus and family annotation. Bar graphs indicate the number of genomes from each cluster. The cluster containing the cultured strains is displayed on the outermost layer. d Circle packing plot, displaying the isolation source of different Saccharibacterial taxa. Number of genomes analyzed per taxon is indicated by the circle size. e Upset plot, displaying the number of clusters found across different types of environments and different niches of the human body, ordered by level of overlap. Vertical bars represent the number of clusters shared between the different environments or human body niches highlighted with dots in the lower panel. Horizontal bars in the lower panel indicate the total number of clusters in each type of environment or human body niche.
Saccharibacteria had a high taxonomic diversity and were widespread in different types of environments (Fig. 1c). We further explore the association between the phylogeny of Saccharibacteria and their isolation sources. The results suggested that each group of Saccharibacteria had a distinctive distribution pattern with respect to others (Fig. 1d, e). At the family level, the members of SDRK01 (73, 100%), Nanosynbacteraceae (535, 99.44%), Nanogingivalaceae (156, 98.73%), Nanoperiomorbaceae (127, 93.38%) and Saccharimonadaceae (221, 92.86%) were mainly distributed in human body, Nanosyncoccaceae were mainly distributed in other mammals (204, 50.12%) and human body (198, 48.65%), UBA1547 (32, 76.19%) and UBA4665 (33, 70.21%) were mainly distributed in natural environments, and UBA4665_A (63, 94.03%) and UBA1020 (14, 73.68%) were mainly distributed in natural and engineered environments (Fig. 1d). In addition, at cluster level, every cluster contained genomes of a specific type of environment, except for one cluster containing 4 genomes from natural environments (groundwater), and 3 genomes from engineered environments (hydrocarbon-contaminated groundwater) (Fig. 1e). Moreover, when we focused on the clusters distributed throughout the human body, we found that several clusters were shared between human oral cavity and other niches of human body, such as human gut, skin and vagina.
A highly reduced genome is a common feature of CPR bacteria1,2, same as other CPR bacteria, Saccharibacteria have reduced genomes3. In our high-quality genome collection, the genome sizes of Saccharibacteria ranged from 4.5 Kbp to 1.5 Mbp, with the smallest genome from the ruminant gastrointestinal tract and the largest from the rhizosphere soil. Notably, Saccharibacteria from natural and engineered environments had larger genome sizes (Supplementary Fig. 4a) and proteome sizes (Supplementary Fig. 4b) than those from mammalian host-associated environments, which could be due to the fact that Saccharibacteria in open environments require a larger stock of functional genes to cope with complex and variable environments, as speculated in a previous study14. Moreover, we also noticed that Saccharibacteria from other mammals had larger genome sizes and proteome sizes than those from the human body.
The limited metabolic potential of yet-uncultured Saccharibacteria
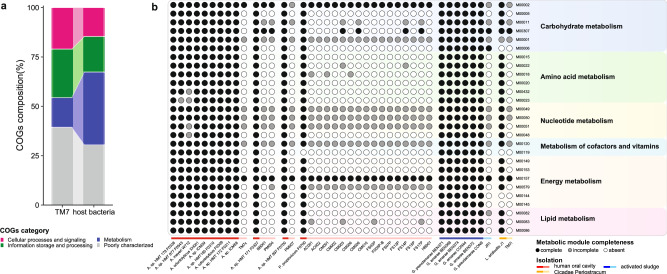
Although it has been reported that Saccharibacteria are metabolically restrained, and depend on host bacteria for survival4,7–13, the conclusion is based on cultivation studies and a limited number of genomes. With the acquisition of more Saccharibacterial genomes, whether the ubiquitous yet-uncultured Saccharibacteria possesses the potential for an independent living warrants further investigation. Here, to further explore this question, we compared the functional gene composition of the host-epibiont pairs4,7–13, and subsequently evaluated the metabolic potential of uncultivated Saccharibacteria in our genome collection. In genomes of Saccharibacteria, ~15% of the genes are related to metabolism, while in genomes of the bacterial hosts, metabolism-related genes account for ~37% (Fig. 2a). Using a Python script to calculate the completeness of metabolic modules, we found similar metabolic dependency patterns between different host-epibiont pairs (Fig. 2b, and Supplementary Tables 3a, b). Species of Saccharibacteria lacked the potential to synthesize biomacromolecules such as amino acids (proline, threonine, serine, leucine, tryptophan, etc.), nucleotides (adenine ribonucleotide and guanine ribonucleotide biosynthesis, etc.), vitamins (coenzyme A, pantothenate, etc.), and fatty acids, while host strains had corresponding complete metabolic modules, which is consistent with previous research findings (Fig. 2b).
Fig. 2. The metabolic dependency between Saccharibacteria and their bacterial hosts.
a COG function composition of genomes of cultured Saccharibacteria and their host bacteria, colored by COGs category. b Heatmap showing metabolic module completeness of genomes of Saccharibacteria and their bacterial hosts. Rows indicate metabolic modules. Columns indicate genomes of Saccharibacteria and their host bacteria. The color of the circles represents the completeness of metabolic modules in each genome, black circles represent complete metabolic modules, gray circles represent incomplete metabolic modules, white circles represent the metabolic module is absent. Specific parasitic pairs are represented by horizontal lines, colored by the isolation source of organisms, darker colors represent host bacteria, lighter colors represent Saccharibacteria.
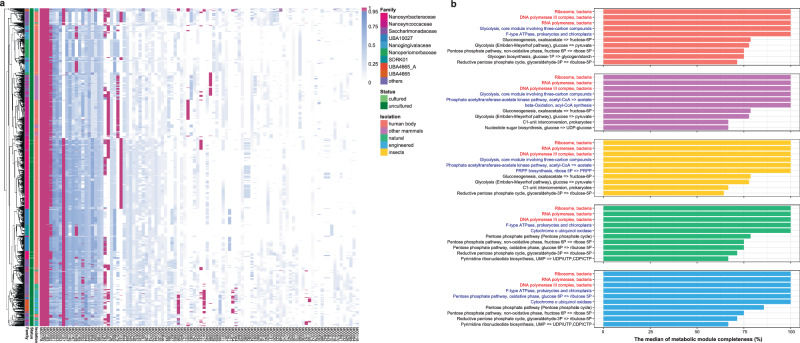
Further investigation of the metabolic potential of the whole Saccharibacterial genome collection revealed a common feature of metabolic deficiency, namely, the inability to synthesize biological macromolecules such as amino acids, nucleotides, and vitamins (Fig. 3a, b). For Saccharibacteria from different habitats, there were few metabolic modules with a median of completeness of 100%, and only essential modules for information storage and processing, such as DNA polymerase III complex, RNA polymerase, and ribosome were found complete in all genomes (Fig. 3b). Moreover, it was worth mentioning that the metabolic deficiencies of Saccharibacteria appeared to be independent of genome size. Although several genomes in the genome collection were larger, such as >1.25 Mbp, these genomes also exhibited metabolic deficiencies (Supplementary Fig. 5). As the genome size increased, metabolic modules for the biosynthesis of amino acids, nucleotides, cofactors, and vitamins were still incomplete, suggesting Saccharibacteria with a larger genome size may not be able to live independently. Thus, our results suggested that the ubiquitous yet-uncultured Saccharibacteria rely on other bacteria for survival.
Fig. 3. The metabolic potential of Saccharibacteria is limited.
a Heatmap of the metabolic module completeness of the 2041 genomes of Saccharibacteria. Rows indicate genomes of Saccharibacteria. Columns indicate metabolic modules, only the 100 modules with the highest sum of module completeness in 2041 genomes are displayed. The color of the cell represents the completeness of the metabolic module. Color of the cell indicates the completeness of the metabolic module, purple and blue of the cell represents metabolic modules above and below 95%, respectively. The family annotation, isolation source and culture status are indicated by column annotation. b Bar plot showing the median of metabolic module completeness of genomes of Saccharibacteria from different habitats. Only the top ten modules with the highest median of each habitat were shown. The red color of the module text indicates that the module is complete in all genomes of Saccharibacteria, and the blue color indicates that the median of the completeness of the metabolic module in the genomes from a certain habitat is 100%. Annotation of genome isolation source as in (a).
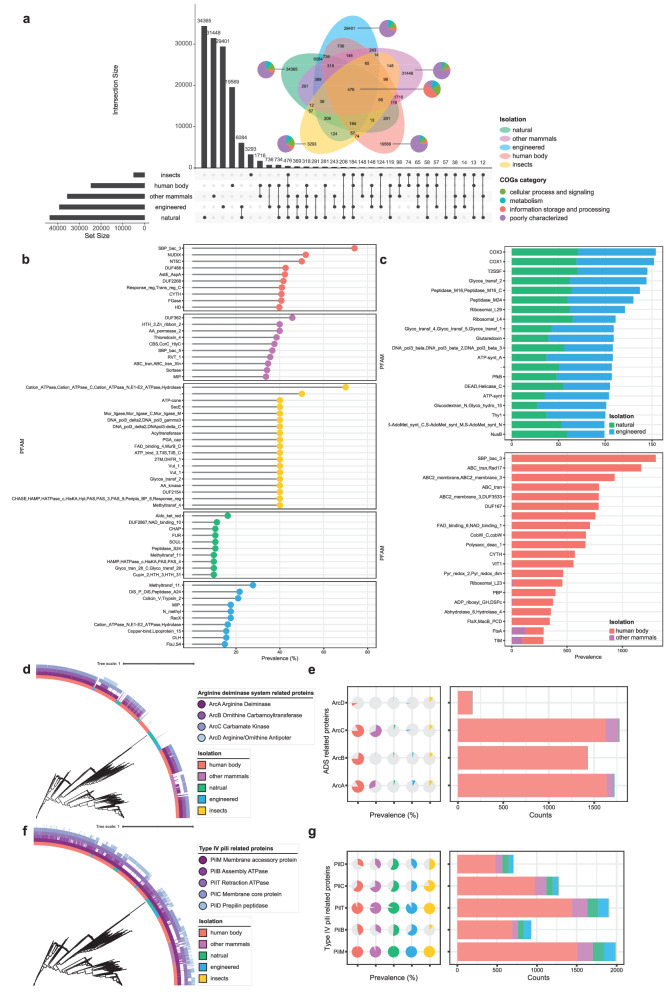
Fig. 4. Comparative genomic analysis of Saccharibacteria.
a Lollipop chart showing the PFAMs corresponding to the top ten unique protein clusters of each habitat. b Cleveland dot plots showing the PFAMs composition corresponding to top 20 shared protein clusters of Saccharibacteria from different environments. c Upset plot and Venn diagram displaying the number of protein clusters found across different types of environments. Vertical bars of the upset plot represent the number of protein clusters shared between the different environments. Horizontal bars of the upset plot in the lower panel indicate the total number of protein clusters in each type of environment. A Venn diagram was added with the COG composition of protein clusters. d Phylogenetic tree of 2041 genomes combined with the heatmap on the outermost layer indicating the isolation source of genomes and the presence or absence of ADS-related proteins. e Distribution patterns of ADS-related proteins in the genome collection, colored by the same annotation of isolation source of genomes as in (d). f Phylogenetic tree of 2041 genomes combined with the heatmap on the outermost layer indicating the isolation source of genomes and the presence or absence of T4P-related proteins. g Distribution patterns of T4P-related proteins in the genome collection, colored by the same annotation of isolation source of genomes as in (f).
Insights into Saccharibacteria habitat adaptation
Even though metabolically restrained, the ultra-small Saccharibacteria are able to live in a variety of environments14, impelling us to investigate their functional potential, which might help them adapt to different niches. The comparative genomic analysis of Saccharibacteria was conducted on the basis of eggNOG annotations. Most of the unique protein clusters of each habitat were poorly characterized, others were involved in biological processes such as metabolism, cellular process and signaling, and information storage and processing (Fig. 4a). When we focused on unique protein clusters that might be associated with the habitat adaptation of Saccharibacteria, a protein cluster with PFAM SBP_bac_3 was found, the protein cluster had a high prevalence (74%) in the human body-derived Saccharibacteria (Fig. 4b, and Supplementary Table 4a). SBP_bac_3 belongs to the bacterial solute-binding proteins (SBP) family, and usually binds to specific substrates, and plays an important role in the nutrient uptake and metabolism of bacteria25. Thus, we speculated that the habitat adaptation of Saccharibacteria to the human body may be associated with the unique nutrient absorption mechanism mediated by SBP. We also found a protein cluster with PFAMs Cation_ATPase, Cation_ATPase_C, Cation_ATPase_N, E1-E2_ATPase, Hydrolase had a high prevalence (70%) in the insect-derived Saccharibacteria (Fig. 4b). The protein was corresponding to the P-type Mg2+ transporter that mediates magnesium influx into the cytosol. Moreover, we noticed that Saccharibacteria of natural and engineered environments had a large number of protein clusters in common (Supplementary Table 4c). Of note, the top two protein clusters unique to Saccharibacteria of natural and engineered environments were annotated as COX3 and COX1, corresponding to subunit 3 and subunit 1 of the cytochrome o ubiquinol oxidase, which might support the aerobic respiration of Saccharibacteria (Fig. 4c). Saccharibacteria from human body and other mammals also shared many protein clusters, although these protein clusters had different prevalence (Fig. 4c, Supplementary Table 4b). Actually, for the key functions of habitat adaptation of mammal-associated Saccharibacteria, the arginine deiminase system (ADS) is often found complete in mammal-associated Saccharibacteria but missing in environmental counterparts, allows metabolically restrained Saccharibacteria to maintain higher viability and infectivity when separated from the host bacteria, and protects Saccharibacteria and its host bacterium from acid stress5,19. Here, we found that the arginine deiminase, ornithine carbamoyl transferase, and carbamate kinase had high prevalence (83%, 73%, 82%) in Saccharibacteria from the human body (Fig. 4d, e).
We also examined the metabolic modules of Saccharibacteria across different habitats. (Fig. 3b, and Supplementary Tables 3c, d). For example, the cytochrome o ubiquinol oxidase complex (M00417) was found in more than 50% of Saccharibacteria from both natural and engineered environments. This complex, which is composed of cytochrome o ubiquinol oxidase subunits I, II, and III, and the cytochrome o ubiquinol oxidase operon protein cyoD, functions as the terminal oxidase in the electron transport chain. It catalyzes the oxidation of ubiquinol to ubiquinone and the reduction of O2 to H2O, creating a transmembrane proton gradient that provides the energy for ATP synthesis. This suggested that some Saccharibacteria in natural and engineered environments might respire aerobically, which has been reported in previous studies26–28. The acyl-CoA synthesis of the beta-Oxidation (M00086), which plays a crucial role in fatty-acid metabolism, was commonly identified in Saccharibacteria across other mammals. It requires a long-chain fatty-acid-CoA ligase which activates long-chain fatty acids by converting them into long-chain acyl-CoA. This process is essential for both the degradation and synthesis of fatty acids. Through this activation, long-chain fatty acids can be efficiently utilized for energy production and other metabolic processes. This implied that some gastrointestinal-derived Saccharibacteria in mammals might be able to use the host-derived fatty acids. On the other hand, several specific metabolic modules were consistently identified in Saccharibacteria from various environments, which might also be related to the adaptation of Saccharibacteria. For example, the F-type ATPase (M00157) was commonly identified in Saccharibacteria from the human body, natural and engineered environments. It uses a proton gradient to drive ATP synthesis. Protons passively flow across the membrane down their electrochemical gradient, and the energy released from this transport reaction is then used to release newly formed ATP from the active site of the F-ATPase. Moreover, the core module involving three-carbon compounds of glycolysis (M00002) was commonly found in Saccharibacteria from the human body, other mammals, and insects. The acetyl-CoA => acetate of phosphate acetyltransferase-acetate kinase module (M00579) was commonly found in Saccharibacteria of other mammals and insects.
The type IV pili (T4P) are surface-associated filamentous appendages of bacteria, participating in twitching motility, cellular adherence, and DNA uptake29. Previous studies have identified five protein-coding genes related to T4P in genomes of Saccharibacteria from different environments, including assembly ATPase, retraction ATPase, membrane core protein, membrane accessory protein, and prepilin peptidase13. Here, we investigated the distribution of related protein-coding genes of T4P in our genome collection and confirmed that two primary genes of T4P had high prevalence in Saccharibacteria genomes of all habitat types (Fig. 4f, g). The pilM gene, which encodes the membrane accessory protein, was annotated in 97.84% of the genomes, and the pilT gene, encoding the retraction ATPase, was annotated in 91.47% of the genomes (Fig. 4g). Although the other three T4P-related genes were not annotated in the majority of genomes in our collection, we believe the consistent detection of the two key genes of T4P, pilM and pilT could support the routinely detected T4P and facilitate Saccharibacteria the maintenance of an epiparasitic lifestyle13.
Saccharibacteria in Chinese oral cavity
The human oral cavity is one of the main habitats of Saccharibacteria3,20. Although accumulating evidence has shown that Saccharibacteria are part of the core microbiome of the human oral cavity14,30, the distribution patterns of Saccharibacteria in the Chinese oral cavity have been poorly characterized. Here, we investigated the relative abundance of Saccharibacteria in the Chinese oral cavity (Fig. 5a) utilizing the 759 species-level representative genomes of Saccharibacteria and 445 presentative genomes of cultivated oral bacteria (Supplementary Table 5). Overall, the oral microbiome of Chinese harbored Saccharibacteria at a low relative abundance (Fig. 5b), the median of Saccharibacterial relative abundance across all 4362 samples was 1.2% and the mean was 1.89%, which was slightly higher than the previously reported relative abundance of 1% of Saccharibacteria in non-disease states3. The oral-derived Saccharibacteria had higher relative abundances and occurrence in the Chinese oral cavity than Saccharibacteria from other habitats (Fig. 5a). We further focused on representative clusters with higher relative abundance, with a relative abundance of 0.01% in the metagenome selected as a threshold, we found the median and mean of Saccharibacteria clusters in each sample were 16 and 23, respectively (Fig. 5c). Of note, there were three clusters (RSZYD18187466_A_saliva.metaspades.bin.39, RSZYD18078343_A_saliva.metaspades.bi-n.34, RSZYD18078188_A_saliva.metaspades.bin.16) present in over 99% of the samples, and the median of relative abundance was 0.035%, 0.031, 0.029%, respectively, indicating these three clusters represented the most prevalent and abundant taxa of Saccharibacteria in Chinese oral cavity.
Fig. 5. The distribution of Saccharibacteria in Chinese oral cavity.
a Abundance and prevalence of Saccharibacteria in Chinese oral cavity. Gray box, Log10 (relative abundance); Dot, median of log10 (relative abundance); Bar, prevalence; color, isolation of clusters. Columns, genus, and family of clusters. b Violin plot displaying a relative abundance of Saccharibacteria in each sample. c Violin plot displaying the number of clusters of Saccharibacteria in each sample. Only the number of clusters with relative abundances above 0.01% in each sample is shown.
Associations between Saccharibacteria and RA
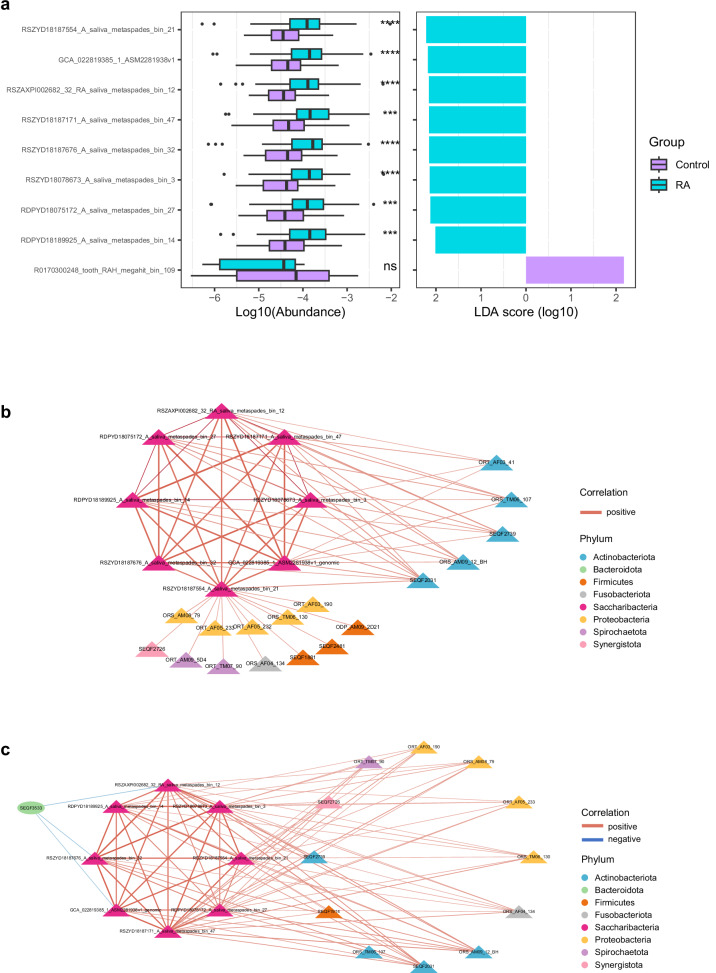
The connections of the human oral cavity to the rest of the body have been increasingly explored and understood, and dysbiosis in the oral microbiome is often implicated in the pathogenesis of both oral and systemic diseases5. As a part of human commensal bacteria, Saccharibacteria has been reported to be associated with human oral mucosal diseases such as periodontitis and gingivitis3,30, yet the associations between Saccharibacteria and human systemic diseases remain understudied. RA is a long-term autoimmune disorder that primarily affects joints. Previous studies have reported different compositions of the oral microbiome between healthy individuals and RA individuals31,32. Here, we downloaded 47 metagenomes of healthy controls (HC) and 50 metagenomes of RA patients (RA) from a public database31, and initially investigated the potential associations between Saccharibacteria and RA. Utilizing a total of 1204 species-level bacterial clusters as the database, the abundance profiles were conducted. The linear discriminant analysis (LDA) effect size (LEfSe)33 was then performed to identify clusters with significantly different abundance between the HC and RA groups. There were 104 clusters significantly enriched in HC, and 53 clusters significantly enriched in RA (Supplementary Fig. 7, Supplementary Table 6a). Among the clusters, eight clusters of Saccharibacteria were found to be significantly enriched in RA, and one cluster of Saccharibacteria was significantly enriched in HCs (Fig. 6a and Supplementary Fig. 7). Subsequently, the correlation network of Saccharibacterial clusters and related bacterial clusters differing in abundance in HC (Fig. 6b, Supplementary Table 6b) and RA (Fig. 6c, Supplementary Table 6c) was conducted, respectively. Result indicated that, in HC, apart from one Saccharibacterial cluster was significantly positively correlated with bacterial clusters from multiple phyla, most Saccharibacterial clusters were significantly positively correlated with bacterial clusters within Actinobacteriota. Conversely, in RA patients, the network was more complex. Several clusters of Saccharibacteria showed a significant positive correlation with bacterial clusters from multiple phyla in RA patients, such as clusters within Actinobacteria, Proteobacteria, Fusobacteriota, etc. The above preliminary results implied that species of Saccharibacteria might play an important role in the changes of the microbiome during disease conditions. Given the currently limited sample size, the associations between Saccharibacteria and RA disease require validation with a larger cohort and further in-depth research.
Fig. 6. Differential patterns of Saccharibacterial clusters in 47 healthy controls (HC) and 50 patients with rheumatoid arthritis (RA).
a The logarithm of abundance (base 10) in each group of differentially abundant Saccharibacterial clusters and the LDA scores of differentially abundant Saccharibacterial clusters analyzed by Linear discriminant analysis Effect Size (LEfSe) analysis (P < 0.05, LDA > 2), colored according to enrichment group. P values are from Wilcoxon rank-sum test (two-sided), and adjusted using the Benjamini–Hochberg procedure, ****P < 0.0001, ** P < 0.01, ns, not significant. b Correlation network of Saccharibacterial clusters and related bacterial clusters differing in abundance in each group in HC. The nodes are colored by phylum. Triangle nodes reflect clusters enriched in RA. c Correlation network of Saccharibacterial clusters and related bacterial clusters differing in abundance in each group in RA. The nodes are colored by phylum. Triangle nodes reflect clusters enriched in RA, and oval nodes reflect clusters enriched in healthy controls. b, c Positive correlations are indicated by red edges and negative correlations by blue edges. Spearman’s correlation is used to calculate correlation, adjusted P values (P < 0.05) are determined using the Benjamini–Hochberg procedure.
Discussion
As the first and most extensively cultured group of CPR, Saccharibacteria has provided profound insights into the morphology characteristics, physiological properties, and ecological functions of CPR bacteria3–13,15–19,21. However, these findings were primarily based on cultured Saccharibacteria, especially from the human oral cavity, and thus may represent only a minority of Saccharibacteria taxa. Alternatively, although previous studies have conducted comparative genomic analyses of existing genomes of Saccharibacteria, the amount of data is very small, and the preliminary analysis results cannot represent the entire Saccharibacterial lineage14,34. With an increasing number of Saccharibacterial genomes obtained through culture-independent methods, several questions remain understudied. For instance, what is the taxonomic diversity of the whole Saccharibacteria lineage, and what are the potential differences in lifestyle and metabolic potential between yet-uncultured Saccharibacteria and their cultured counterparts14. In order to further explore these questions, we collected Saccharibacterial genomes from multiple source data and conducted a comprehensive genomic analysis.
The 2041 Saccharibacterial genomes, which passed rigorous quality control, spanned various habitats and encompassed diverse taxa. Covering a large range of genome size, most of the genomes (1935; 94.80%) ranged from 0.45 Mbp to 1 Mbp, and a small portion of the genomes (106; 5.20%) ranged from 1 Mbp to 1.5 Mbp. Generally, genomes of Saccharibacteria from engineered and natural environments were larger than those from the human body and other mammals, implying that there were potential biological differences between Saccharibacteria derived from different habitats. On the other hand, Saccharibacterial genomes were reduced compared to bacteria outside the CPR lineage, yet the genomes exhibited high diversity in both phylogeny and gene content (Fig. 4a, b, Supplementary Tables 4a–d), indicating the evolution of Saccharibacteria might be very complex. Currently, the understanding of the evolutionary driving force of CPR bacteria is essential in the field of CPR. Gene loss and lateral gene transfer might play a role in shaping the genomic evolution of CPR bacteria1,2,35,36. We envisage our genome collection will be used in future studies on the evolution of CPR bacteria.
Through comparative genomic analysis, genomic traits related to the habitat adaptation of Saccharibacteria were identified. For example, the SBP proteins commonly identified in Saccharibacteria derived from the human body might facilitate nutrient absorption within the human host, and the cytochrome o ubiquinol oxidase complex in Saccharibacteria in natural and engineered environments suggested the potential for aerobic respiration. However, due to the relatively low abundance of Saccharibacteria in various habitats and the challenge in recover genomes in metagenomes, the currently genomes may not represent the full diversity of Saccharibacteria in a given habitat, thus the results need further validation using more datasets. Alternatively, genomes in our collection contained a large proportion of genes that lack annotation, which might also increase the limitations of current results. Thus, understanding the function of the large proportion of unannotated genes is warranted for a deeper understanding of CPR bacteria1,2. Techniques for the genetic manipulation of Saccharibacteria37 are likely to enhance our knowledge of uncharacterized genes of Saccharibacteria and other CPR bacteria in the future.
The host specificity is another key remaining question in the field of CPR bacteria. Even though our analysis implied the ubiquitous uncultured Saccharibacteria might rely on other bacteria to survive, the question of the host specificity of Saccharibacteria still remained. Although several studies have speculated on the potential hosts of Saccharibacteria based on horizontal gene transfer34 and abundance correlations20, these speculations have not been experimentally validated. Given the intricate interactions between Saccharibacteria and their host bacteria, their potential roles in the human body and other niches such as groundwater, rhizosphere soil polluted environments, etc., remain for further analysis. With more cocultures of CPR-host bacteria obtained, a broader host range for CPR might be uncovered, consequently, more dynamic and complex microbial interactions might be unveiled. By then, the functions of CPR bacteria in broader ecosystems will be better elucidated.
To summarize, our study compiles a comprehensive genome collection of Saccharibacteria from diverse sources. We provide new insights into the phylogeny, metabolic potential, and habitat adaptation of Saccharibacteria across various habitats. Additionally, our analysis initially investigates the associations between Saccharibacteria and RA. We envisage our work will foster a more comprehensive understanding of Saccharibacteria and support further research on other CPR bacteria.
Methods
Genome assembly and quality assessment
To better explore the biodiversity of Saccharibacteria, we collected assemblies of Saccharibacteria from NCBI Assembly23 (01-2023) and assembled assemblies of Saccharibacteria from 5427 oral metagenomic samples of multiple Chinese cohorts24. The Chinese cohorts were derived from both our sequencing data and public datasets, including the following: (1) 3953 samples from the Chinese Shenzhen cohort, including 2678 samples from the SZ-4D (Disease, Drug, Diet, Daily life) cohort and 1275 new sequencing samples24; (2) 671 samples from Chinese Yunnan cohort24; (3) 294 RA samples from Chinese Beijing cohort31. The method of assembling from metagenomic shotgun reads was described by Zhu et al.24. In brief, in each sample, contigs were assembled using metaSPAdes38, contigs longer than 1500 bp were binned by MetaBAT239. The CheckM (v.1.2.1)40 custom workflow with a set of 43 marker genes for CPR bacteria20,41 was used to evaluate Saccharibacterial genome completeness and contamination. GUNC (v1.0.5)42 was used to detect chimerism. 4019 Saccharibacterial genomes with 50–90% completeness and 5–10% contamination were considered as medium-quality genomes. Quality filtration at >90% completeness and <5% contamination, plus subsequent removal of chimeric genomes, yielded a non-redundant set of 2041 high-quality genomes for downstream analysis.
Genome clustering and taxonomic analyses
The full set of 2041 genomes was clustered on the basis of whole-genome ANI using dRep (v.3.4.0)43 with a 95% ANI threshold (-sa 0.95 -nc 0.3), yielding a set of 759 representative clusters. All genomes were taxonomically annotated using GTDB-Tk (v.2.1.0)44 ‘classify_wf’ function with default parameters against the GTDB r207 release, defining Saccharibacteria as the class Saccharimonadia of Patescibacteria phylum. The genome maximum-likelihood phylogenetic tree was constructed using GTDB-Tk based on 120 bacterial marker genes and visualized by iTOL45. Information on genome isolation sources was gathered through NCBI. The “human body” includes the human oral cavity, gut, vagina, and skin. The “other mammals” included the ruminant gastrointestinal tract and intestines of mice, baboons, pigs, etc. The “natural” included natural environments such as groundwater, lake, sediment, soil, rhizosphere, marine water, etc. The “insects” included termite gut and cicada slough. The “engineered” isolation category was defined to include human-made or industrial systems as summarized by Jaffe et al.20. A Circle packing plot was generated with RAWGraphs (https://app.rawgraphs.io). An upset plot was generated using Intervene (https://asntech.shinyapps.io/intervene)46.
Genome annotation, protein catalog, and comparative genomic analysis
Genes were predicted using Prokka v.1.14.647 with default parameters and further characterized using eggNOG-mapper v.2.048 and the eggNOG v.5.0 database49. The non-redundant gene catalog was generated by CD-HIT v4.8.150 (parameters: -d 0 -c 0.95 -aS 0.9 -G 0 -M 0). Genes were clustered at different percentage identities, and the number of unique genes resulting per clustering for each isolation source was visualized (Supplementary Fig. 6a). The protein catalog was generated by combining all predicted CDSs derived from the 2041 genomes (Supplementary Fig. 6b). Protein clustering of the concatenated proteins dataset was performed with the ‘linclust’ function of MMseqs2 (Version 14.7e284)51 with options ‘--cov-mode 1 -c 0.8’ and ‘--kmer-per-seq 80’. Comparative genomic analysis was carried out based on a protein catalog with 50% amino-acid sequences similarity. Venn diagram was performed with EVenn52. The metabolic module completeness was calculated with a python script, briefly, the script calculates the completeness of the metabolic modules in each Saccharibacterial genome by counting the annotation of KOs in the eggNOG output and comparing it to the KOs required for the metabolic module in the KEGG database. The T4P-related genes and ADS-related genes were identified by prokka as well as eggNOG, and manually curated.
Metagenomic analysis
For investigating the distribution of Saccharibacteria in a larger Chinese population, human oral metagenome sequencing data of Chinese cohorts were downloaded from the CNGB Sequence Archive (CNSA)53 (https://db.cngb.org/cnsa/) of China National GeneBank DataBase (CNGBdb)54. 3691 metagenomic samples were obtained from the 4D-SZ cohort under the accession code CNP0000687, and 671 metagenomic samples were obtained from the Yunnan cohort under the accession code CNP0001221. For investigating the associations between Saccharibacteria and RA, 47 metagenomic samples from healthy individuals and 50 metagenomics samples from RA individuals31 were downloaded from https://www.ebi.ac.uk/ena/browser/view/PRJEB6997. Reads were quality-filtered and trimmed using fastp v0.19.455, and human contamination was removed using Bowtie2 v2.3.556 and SeqKit v1.357 as described by Zhu et al.24. In order to explore the relative abundance of bacteria in metagenomes, clean reads were mapped to 1204 reference genomes, which included 445 reference genomes of oral bacteria and 759 representative genomes of Saccharibacteria. The 445 reference genomes of oral bacteria were selected from 1019 genomes of the expanded Human Oral Microbiome Database (eHOMD)58 and 1089 genomes of a Cultivated Oral Bacteria Genome Reference (COGR)32, using dRep (v3.4.0)43 with the criterion of 95% ANI as the threshold for distinction at the species level. The relative abundance of representative clusters was estimated using Kraken v2.1.259 and calibrated using Bracken v2.560. Samples or genomes without any reads mapped were filtered out. In order to calculate the coverage and depth of reference genomes, clean reads of each sample were mapped against reference genomes using bowtie2 v2.5.456, the mapped reads were sorted using samtools v1.2061. Subsequently, for contigs of each representative cluster, the coverage of contigs was calculated using bedtools v2.31.162, and the depth of contigs was calculated using pandepth v2.2563. Finally, the average of the coverage and depth within the same representative cluster was utilized as the mean coverage and depth of each representative genome, respectively.
Statistical analysis
Statistical tests were performed using R v4.2.2. P values of differences in genome size and proteome size of Saccharibacteria from different habitats were calculated using the Wilcoxon rank-sum test (two-sided). LDA Effect Size (LEfSe) analyses33 were performed to estimate the clusters that differed significantly among the HCs and RA individuals. LEfSe was calculated using P value < 0.05 for the Wilcoxon test, a LDA score >2 was used as the threshold cutoff. Clusters with differential abundance in HC and RA groups were used for the network analysis in HC and RA groups, respectively. The “rcorr” function of the R package Hmisc was used to compute the Spearman correlations between clusters based on the relative abundances in HC and RA groups, respectively. The strong (r < −0.5 or >0.5) and statistically significant (P value < 0.05 adjusted using the Benjamini–Hochberg method) correlations were retained for the visualization in Cytoscape (v3.9.1)64.
Supplementary information
Supplementary Figures and Supplementary tables.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 32100009), the Natural Science Foundation of Guangdong Province, China (No. 2019B020230001), and Shenzhen Municipal Government of China (No. XMHT20220104017). We thank the colleagues at BGI Research for discussions, and China National GeneBank (CNGB) Shenzhen for DNA extraction, library construction, and sequencing.
Author contributions
Conceived the study: Y.Z., L.X., and W.H. Collected the genomes: H.L. and W.H. Analyzed the data: W.H. and W.L. Contributed reagents/materials/analysis tools: H.L., T.H., X.L., Z.W., J.S., X.L., M.W., X.H, Z.J., and X.T. Wrote the paper: W.H. Revised the paper: X.G., Y.Z., L.X., and X.J. All authors commented on the manuscript.
Data availability
All genomes in the 2041 high-quality genome collection of Saccharibacteria have been deposited into CNSA53 of CNGBdb54 under accession number CNP0004645.
Code availability
Scripts of this work can be downloaded form https://github.com/WenxinHe616/Supplementary-Data-for-reviewers/tree/main.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liang Xiao, Email: xiaoliang@genomics.cn.
Yuanqiang Zou, Email: zouyuanqiang@genomics.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00617-2.
References
- 1.Castelle, C. J. & Banfield, J. F. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell172, 1181–1197 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Castelle, C. J. et al. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat. Rev. Microbiol.16, 629–645 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Bor, B., Bedree, J. K., Shi, W., McLean, J. S. & He, X. Saccharibacteria (TM7) in the human oral microbiome. J. Dent. Res.98, 500–509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He, X. et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. USA112, 244–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, J. L., Mark Welch, J. L., Kauffman, K. M., McLean, J. S. & He, X. The oral microbiome: diversity, biogeography and human health. Nat. Rev. Microbiol.22, 89–104 (2023). [DOI] [PMC free article] [PubMed]
- 6.Utter, D. R., He, X., Cavanaugh, C. M., McLean, J. S. & Bor, B. The saccharibacterium TM7x elicits differential responses across its host range. ISME J.14, 3054–3067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross, K. L. et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol.37, 1314–1321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bor, B. et al. Insights obtained by culturing saccharibacteria with their bacterial hosts. J. Dent. Res.99, 685–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murugkar, P. P., Collins, A. J., Chen, T. & Dewhirst, F. E. Isolation and cultivation of candidate phyla radiation Saccharibacteria (TM7) bacteria in coculture with bacterial hosts. J. Oral. Microbiol.12, 1814666 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim, A. et al. Adapted protocol for saccharibacteria cocultivation: two new members join the club of candidate phyla radiation. Microbiol. Spectr.9, e0106921 (2021). [DOI] [PMC free article] [PubMed]
- 11.Nie, J. et al. Strain-level variation and diverse host bacterial responses in episymbiotic saccharibacteria. mSystems7, e0148821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batinovic, S., Rose, J. J. A., Ratcliffe, J., Seviour, R. J. & Petrovski, S. Cocultivation of an ultrasmall environmental parasitic bacterium with lytic ability against bacteria associated with wastewater foams. Nat. Microbiol.6, 703–711 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Xie, B. et al. Type IV pili trigger episymbiotic association of Saccharibacteria with its bacterial host. Proc. Natl. Acad. Sci. USA119, e2215990119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLean, J. S. et al. Acquisition and adaptation of ultra-small parasitic reduced genome bacteria to mammalian hosts. Cell Rep.32, 107939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bor, B. et al. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc. Natl. Acad. Sci. USA115, 12277–12282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen, P. H. & Singleton, C. M. Parasitic bacteria control foam formation. Nat. Microbiol.6, 701–702 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Bor, B. et al. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont actinomyces. Micro. Ecol.71, 243–255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedree, J. K. et al. Quorum sensing modulates the epibiotic-parasitic relationship between actinomyces odontolyticus and its saccharibacteria epibiont, a nanosynbacter lyticus strain, TM7x. Front. Microbiol.9, 2049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian, J. et al. Acquisition of the arginine deiminase system benefits epiparasitic Saccharibacteria and their host bacteria in a mammalian niche environment. Proc. Natl. Acad. Sci. USA119, e2114909119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffe, A. L. et al. Patterns of gene content and co-occurrence constrain the evolutionary path toward animal association in candidate phyla radiation bacteria. mBio.12, e0052121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chipashvili, O. et al. Episymbiotic Saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe29, 1649–1662.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker, J. L., Bor, B., Agnello, M., Shi, W. & He, X. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol.25, 362–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitts, P. A. et al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res.44, D73–D80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu, J. et al. Over 50,000 metagenomically assembled draft genomes for the human oral microbiome reveal new taxa. Genomics Proteomics Bioinformatics20, 246–259 (2021). [DOI] [PMC free article] [PubMed]
- 25.Tam, R. & Saier, M. H. Jr. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev.57, 320–346 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantor, R. S. et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio4, e00708–e00713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr, E. P. et al. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome6, 122 (2018). [DOI] [PMC free article] [PubMed]
- 28.Nicolas, A. M. et al. Soil candidate phyla radiation bacteria encode components of aerobic metabolism and co-occur with nanoarchaea in the rare biosphere of rhizosphere grassland communities. Msystems6, e0120520 (2021). [DOI] [PMC free article] [PubMed]
- 29.Craig, L., Forest, K. T. & Maier, B. Type IV pili: dynamics, biophysics and functional consequences. Nat. Rev. Microbiol.17, 429–440 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Naud, S. et al. Candidate phyla radiation, an underappreciated division of the human microbiome, and its impact on health and disease. Clin. Microbiol. Rev.35, e0014021 (2022). [DOI] [PMC free article] [PubMed]
- 31.Zhang, X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med.21, 895–905 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Li, W. et al. A catalog of bacterial reference genomes from cultivated human oral bacteria. npj Biofilms Microbiomes9, 45 (2023). [DOI] [PMC free article] [PubMed]
- 33.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol.12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker, J. L. Complete genomes of clade G6 saccharibacteria suggest a divergent ecological niche and lifestyle. mSphere6, e0053021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffe, A. L. & Banfield, J. F. Candidate phyla radiation bacteria. Curr. Biol.34, R80–R81 (2024). [DOI] [PubMed] [Google Scholar]
- 36.Meheust, R., Burstein, D., Castelle, C. J. & Banfield, J. F. The distinction of CPR bacteria from other bacteria based on protein family content. Nat. Commun.10, 4173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y. et al. Genetic manipulation of Patescibacteria provides mechanistic insights into microbial dark matter and the epibiotic lifestyle. Cell186, 4803–4817.e13 (2023). [DOI] [PMC free article] [PubMed]
- 38.Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res.27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ.7, e7359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown, C. T. et al. Unusual biology across a group comprising more than 15% of domain bacteria. Nature523, 208–211 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Orakov, A. et al. GUNC: detection of chimerism and contamination in prokaryotic genomes. Genome Biol.22, 178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J.11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics36, 1925–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan, A. & Mathelier, A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform.18, 287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Cantalapiedra, C. P., Hernandez-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol.38, 5825–5829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huerta-Cepas, J. et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res.47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinegger, M. & Soding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol.35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Chen, T., Zhang, H., Liu, Y., Liu, Y. X. & Huang, L. EVenn: easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genomics48, 863–866 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Guo, X. et al. CNSA: a data repository for archiving omics data. Database (Oxford)2020, baaa055 (2020). [DOI] [PMC free article] [PubMed]
- 54.Chen, F. Z. et al. CNGBdb: China National GeneBank DataBase. Hereditas42, 799–809 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One11, e0163962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Escapa, I. F. et al. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a Resource for the microbiome of the human aerodigestive tract. mSystems3, e00187-18 (2018). [DOI] [PMC free article] [PubMed]
- 59.Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol.20, 257 (2019). [DOI] [PMC free article] [PubMed]
- 60.Lu, J., Breitwieser, F. P., Thielen, P. & Salzberg, S. L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci.3, e104 (2017).
- 61.Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu, H., Shi, C., He, W., Li, F. & Ouyang, B. PanDepth, an ultrafast and efficient genomic tool for coverage calculation. Brief. Bioinform.25, bbae197 (2024). [DOI] [PMC free article] [PubMed]
- 64.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Supplementary tables.
Data Availability Statement
All genomes in the 2041 high-quality genome collection of Saccharibacteria have been deposited into CNSA53 of CNGBdb54 under accession number CNP0004645.
Scripts of this work can be downloaded form https://github.com/WenxinHe616/Supplementary-Data-for-reviewers/tree/main.