Abstract
Background
Loss-of-function mutations of liver kinase B (LKB1, also termed as STK11 (serine/threonine kinase 11)) are frequently detected in patients with non-small cell lung cancer (NSCLC). The LKB1 mutant NSCLC was refractory to almost all the antitumor treatments, including programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) blockade therapy. Unfortunately, mechanisms underlying resistance to immunotherapy are not fully understood. In this study, we deciphered how LKB1 regulated sensitivity to anti-PD-1/PD-L1 immunotherapy.
Methods
We investigated the mutational landscape of LKB1 mutant NSCLC in next generation sequencing (NGS) data sets. Expression of LKB1, PD-L1 and S-phase kinase-associated protein 2 (Skp2) in NSCLC samples were assessed by immunohistochemistry (IHC). The tumor microenvironment (TME) profiling of LKB1 wild type (WT) and mutant NSCLC was performed using fluorescent multiplex IHC. Mass spectrometry and enrichment analysis were used to identify LKB1 interacting proteins. Mechanistic pathways were explored by immunoblotting, ubiquitination assay, cycloheximide chase assay and immunoprecipitation assay.
Results
By using NGS data sets and histological approaches, we demonstrated that LKB1 status was positively associated with PD-L1 protein expression and conferred a T cell-enriched “hot” TME in NSCLC. Patients with good responses to anti-PD-1/PD-L1 immunotherapy possessed a high level of LKB1 and PD-L1. Skp2 emerged as the molecular hub connecting LKB1 and PD-L1, by which Skp2 catalyzed K63-linked polyubiquitination on K136 and K280 residues to stabilize PD-L1 protein. Inhibition of Skp2 expression by short hairpin RNA or its E3 ligase activity by compound #25 abrogated intact expression of PD-L1 in vitro and generated a T cell-excluded “cold” TME in vivo. Thus, the LKB1-Skp2-PD-L1 regulatory loop was crucial for retaining PD-L1 protein expression and manipulation of this pathway would be a feasible approach for TME remodeling.
Conclusion
LKB1 and Skp2 are required for intact PD-L1 protein expression and TME remodeling in NSCLC. Inhibition of Skp2 resulted in a conversion from “hot” TME to “cold” TME and abrogated therapeutic outcomes of immunotherapy. Screening LKB1 and Skp2 status would be helpful to select recipients who may benefit from anti-PD-1/PD-L1 immunotherapy.
Keywords: Immune Checkpoint Inhibitor, Immunotherapy, Lung Cancer, Next generation sequencing - NGS, Tumor infiltrating lymphocyte - TIL
WHAT IS ALREADY KNOWN ON THIS TOPIC
Mutations in liver kinase B (LKB1) indicated poor response to immunotherapy in patients with non-small cell lung cancer (NSCLC) but the underlying mechanism has not been fully understood.
WHAT THIS STUDY ADDS
LKB1 was found to cooperate with S-phase kinase-associated protein 2 (Skp2) to maintain programmed death-ligand 1 expression and created an inflamed “hot” tumor microenvironment.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
LKB1 and Skp2 may be novel biomarkers for the selection of patients with NSCLC before the initiation of immunotherapy.
Introduction
The evasion of immune surveillance primes tumorigenesis and tumor outgrowth. Mechanisms underlying the immune suppressive property of the tumor microenvironment (TME) are quite heterogenous, while one common explanation is that cancer cells could remodel TME through upregulating a panel of negative regulatory immune checkpoints, such as programmed cell death protein 1 (PD-1) and its ligand PD-L1.1 2 Blocking the engagement of PD-1 and PD-L1 removes substantial immune inhibitory potency to enable cytotoxic T lymphocyte infiltration and result in immunologic cancer cell death.3 Higher expression of PD-L1 in tumor cells is associated with a favorable response to PD-1/PD-L1 blockade therapy in patients with advanced cancer, compared with those lacking PD-L1 expression.4,6 Since the expression of PD-L1 protein has been demonstrated to dictate the response to anti-PD-1/PD-L1 immunotherapy, there is increasing interest in exploiting the molecular mechanisms that control the PD-L1 protein level.
Aside from PD-L1 as a dominant factor indicating favorable therapeutic response, there are also biomarkers inversely conferring therapeutic outcomes to PD-1/PD-L1 blockade therapy. In patients with non-small cell lung cancer (NSCLC), genetic alterations in liver kinase B (LKB1, also known as STK11 (serine/threonine kinase 11)) define a distinct subtype of patients, characterized by refractory to almost all the antitumor treatment and poor response to anti-PD-1/PD-L1 immunotherapy.7 For example, in the phase III CheckMate 057 trial evaluating the efficacy of nivolumab (anti-PD-1 immunotherapy) versus docetaxel in previously treated NSCLC, the objective response rate to PD-1 blockade in the LKB1 mutant subgroup was only 7.4%, whereas more than 35% of patients in the LKB1 wild type (WT) subgroup responded to the treatment (p<0.001). The medium overall survival of the LKB1 WT arm reached 16 months, but patient survival declined to 6.4 months if the tumor harbored LKB1 mutations (p=0.0045).8 9 These interesting findings imply the potential involvement of LKB1 in antitumor immunity, although the underlying mechanisms remain to be elucidated. Early studies on LKB1 revealed that it encodes a constitutively activated STK and acts as a tumor suppressor. One of the key components of the LKB1 pathway is activate AMP-activated protein kinase (AMPK), whose function is primarily sensing intracellular energy stress when the ATP level falls down.10 Despite targeting LKB1 mutant tumor through nutrition deprivation, or using AMPK agonists, which have yielded substantial therapeutic efficacy in preclinical models,11 there is still very limited access to an effective treatment for LKB1 mutant NSCLC.
The F-box protein S-phase kinase-associated protein 2 (Skp2) is a prototypical and best-characterized E3 ligase for the proteolytic processing of tumor suppressive proteins, including p27, p21, and FOXO1, through K48-linked polyubiquitination.12 13 Mechanistically, Skp2 integrates with Cullin-1, Skp1 and Rbx1 to resemble the SCF complex, which catalyzes the engagement of the ubiquitin (Ub) chain to substrate proteins for proteasomal degradation. Skp2 also regulates Akt subcellular distribution and augments PI3K/Akt signaling transduction by the non-proteolytic K63-linked ubiquitination.14 Overexpression of Skp2 has been identified in various human malignancies, such as breast cancer, NSCLC, colon cancer, and it has been shown to be associated with poor prognosis as well as increased migrative and metastatic potency.15 16 Thus, Skp2 is prominently recognized as a proto-oncogene and offers a potential druggable target for cancer treatment. The E3 Ub ligase activity of Skp2 is regulated by phosphorylation, by which phosphorylation at Ser72 residue stabilizes Skp2 to increase its oncogenic activity.17 18 Recent studies also showed the Ser256 residue phosphorylated by LKB1/AMPK is highly conserved and contributes to the integrity and E3 ligase activity of Skp2.19 As such, Skp2 may function as a protein adaptor integrating the LKB1-AMPK signaling and downstream oncogenic proteins, as a consequence, facilitating immune surveillance and tumor outgrowth.
In this study, we unravel the mechanism underlying poor response to anti-PD-1/PD-L1 immunotherapy in patients with LKB1 mutant NSCLC. We found that the TME of LKB1 WT NSCLC was inflamed and enriched in cytotoxic T cell infiltration and PD-L1 expression, whereas mutation in LKB1 generated an uninflamed “cold” TME. Using immunoprecipitation in conjunction with mass spectrometry (MS) analysis, we identified Skp2 as a binding partner for LKB1 to maintain PD-L1 protein stability. Skp2 as an E3 ligase triggers K63-linked polyubiquitination of PD-L1 at Lys136 and Lys280 residues to protect it from being degraded by β-TrCP (β-transducin repeats-containing protein). Notably, we found that compound #25, a small molecular inhibitor selectively targets Skp2 E3 ligase activity,20 antagonizes the antitumor efficacy of anti-PD-L1 immunotherapy. Our study revealed novel insights into how the sensitivity to PD-1/PD-L1 blockade therapy is regulated by LKB1, and established the LKB1-Skp2-PD-L1 regulatory axis for interpreting immunotherapy in patients with NSCLC.
Materials and methods
Reagents
MG132 (S2619), compound C (S7840), compound #25 (S9724), and metformin (S1950) were purchased from Selleck Chemicals. Nocodazole (HY-13520) was purchased from MedChemExpress. Cycloheximide (CHX) (2112S) was purchased from Cell Signaling Technology. Puromycin (P8833) was from Sigma. Anti-mouse PD-L1 antibody (BE0101) was purchased from Bio X Cell.
Cell culture
Beas2B, A549, H1299, H292, Lewis lung cancer (LLC) and breast cancer MDA-MB-231 and BT-549 cells have been extensively described and authorized by short tandem repeat analysis. Beas2B, H1299, H292 and BT-549 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. A549, LLC and HEK293 cells were cultured with Dulbecco’s Modified Eagle’s Medium. MDA-MB-231 cells were cultured in Leibovitzs-15 medium. Cells were tested for mycoplasma contamination and used within 3 months after resuscitation.
Plasmids, mutagenesis and lentivirus production
His-Ub, His-Ub K48-only, His-Ub K63-only, Flag-PD-L1, Myc-PD-L1, Flag-PD-1, Flag-Skp2 and HA-β-TrCP overexpression plasmids were preserved in our in-house plasmid bank as previously described. Human HA-LKB1 in pCMV3 vector was purchased from Sino Biological (China). Mouse LKB1 (mLKB1) plasmid was from HanBio (China). The HA-LKB1 K78I (kinase dead), Myc-PD-L1 K136R, Myc-PD-L1 K280R, Myc-PD-L1 K136/K280R, Flag-Skp2 ΔLRR constructs were generated by mutagenesis following manufacturer’s instruction. All the plasmids used in this study have been verified by sequencing and were available on reasonable request.
The TRCN pLKO.1 short hairpin RNA (shRNA) plasmids targeting the coding sequence (CDS) of LKB1 and Skp2 were purchased from Sigma. To generate shRNAs lentivirus, pLKO.1 shRNA was transfected into HEK293 cells, together with psPAX2 and pMD2.G packing plasmids using Lipofectamine 3000 (Invitrogen). The cell culture supernatants containing indicated shRNA lentivirus were collected 48 hours after transfection and stored at −80°C. Cells were infected with shRNAs lentivirus and treated with 8 µg/mL polybrene. After infection, the stable cells were selected with 2∼5 µg/mL puromycin.
Real-time PCR assay
The RNA was extracted using TRIzol reagents (Takara) following standard protocol. The complementary DNA (cDNA) was synthesized using the reverse transcription kit (Takara). Quantitative real-time PCR was performed with transcribed cDNA as the template. Relative expression of indicated genes was normalized to β-actin or GAPDH (Bio-Rad).
Nocodazole synchronization
Cells were seeded at 2×106 cells/10 cm dish and grew overnight to allow cell attachment. The cell culture medium was replaced with a fresh medium containing nocodazole (200 ng/mL). After treatment with nocodazole for 16 hours, cells were then cultured in drug-free RPMI-1640 media to release cell cycle transition and harvested at selected time points.
Western blot and immunoprecipitation
After the indicated treatment, cells were collected and centrifugated. Cell pellets were lyzed on ice in RIPA lysis buffer (Beyotime) containing phosphatase/proteasome inhibitors cocktail (Roche). The protein concentration was measured using a BCA reagent kit (Thermo). An equal amount of lysate (20∼35 µg) was separated on SDS-PAGE gel and transferred to a nitrocellulose membrane (Millipore). The membrane was blocked in 5% skim milk and incubated with primary antibody overnight at 4°C. On the next day, the membrane was washed with TBST buffer and incubated with horseradish peroxidase (HRP)-conjugated anti-Rabbit or anti-mouse immunoglobulin G (IgG) at room temperature for an additional 2 hours. Protein bands were visualized by the chemiluminescence analysis system. Antibody source was available in online supplemental table S1).
For the immunoprecipitation experiment, a total amount of 500 µg cell lysate was mixed with 1 µg primary antibody, or isotype IgG as a negative control, with gentle rotation overnight at 4°C. The immunocomplex was captured and precipitated by Protein A/G agarose beads (Santa Cruz). After extensive washing with phosphate-buffered saline, the deposition was suspended in 2×loading buffer and then boiled for Western blot analysis.
MS analysis
Whole-cell lysates of H292 and H1299 cells were prepared as described. To pull down proteins that interacted with LKB1, 1 µg antibody against LKB1 was added into cellular extracts followed by gentle rotation overnight at 4°C. An equal amount of isotype IgG was added as a negative control. The LKB1 and its interaction proteins were precipitated with Protein A/G agarose beads and subjected to electrophoresis, protein bands were visualized using Coomassie blue staining. The bands of interest were carefully excised, extensively destained and in-gel digested with sequencing-grade trypsin. The peptides were then extracted from the gel matrix and prepared for MS analysis.
Animal experiments
The immunocompetent, specific pathogen-free male C57BL/6J mice aged 6–8 weeks (n=32) were purchased from GemPharmatech. LLC cells stably expressed mLKB1 or negative control (NC) were resuspended in 80 µL of matrigel and injected into the right upper limbs of C57BL/6J mice. Four days later, the tumor volume was measured and recorded using the following formula: Tumor volume=L (length)×W (width)2/2. When the tumor volume reached 50 mm3, treatment was initiated. Mice were treated with anti-PD-L1 antibody or compound #25 separately, or in their combination. Specifically, anti-mouse PD-L1 antibody was given intraperitoneally at a dose of 200 µg per mouse once weekly. The compound #25 was intraperitoneally injected at a dose of 40 mg/kg per mouse once daily. An equal amount of vehicle was given in the same manner as treatment controls. At the end of the experiment, the mice were humanistically sacrificed to isolated subcutaneous tumors and proceeded for pathological evaluation. All the animal experiments were conducted in compliance with institutional guidelines and approved by the Ethical Review Committee of Jinling Hospital.
Immunohistochemistry
The paraffin-embedded sections were subjected to antigen retrieval and incubation with indicated primary antibody at 4°C overnight, followed by incubation with HRP-conjugated secondary antibody at room temperature for 1 hour. The expression of the indicated protein was visualized by DAB (Dako) and hematoxylin counterstaining. The immunohistochemistry (IHC) density was scored as negative (score 0), weakly positive (score 1), moderately positive (score 2) and strongly positive (score 3). The percentage of positive cells was also scored (<5%, score 0; 6–25%, score 1; 26–50%, score 2; 51–75%, score 3; and>75%, score 4). The intensity score was multiplied by the proportion score to yield the IHC H-score as we have described previously.
Characterization of TME by fluorescent multiplex IHC
The PerkinElmer/Akoya Biosciences Phenoptics system was applied to characterize TME as described. Briefly, the FFPE slides were subjected to antigen retrieval, antibody incubation, tyramine signal amplification, antibody stripping, and a new round of IHC up to five times. The primary antibodies targeting CD8, Foxp3, CD68 or F4/80, granzyme B (GZMB) and PD-L1, and Opal fluorescent IHC kit (PerkinElmer) were used. We used Opal 480 channel for CD8 (cyan), Opal 520 channel for Foxp3 (green), Opal 690 channel for CD68 or F4/80 (red), Opal 780 channel for GZMB (white), and Opal 570 channel for PD-L1 (yellow), respectively. DAPI was used to stain the nucleus. The slides were then scanned by the Olympus FV1000 confocal system and images were reviewed using Phenochart software (PerkinElmer). A selection of at least three representative regions of interest was used to analyze TME.
Patient characteristics and clinical information
Paired paraffin-embedded LKB1 WT and LKB1 mutant NSCLC samples were collected from the Affiliated Hospital of Nantong University and Jinling Hospital Affiliated with Nanjing University. A total number of 40 patients with LKB1 WT NSCLC who received anti-PD-1/PD-L1 immunotherapy in our institution from 2018 to 2022 were evaluated for PD-L1 expression and TME. The patients’ clinical information, including age, gender, smoking state, tumor location, pathological type, differentiation degree, and tumor, node, metastases stage were collected and analyzed (table 1). The experiment using human specimens was approved by the Institutional Ethics Committee of Jinling Hospital (DZGZRDW2400194). Written informed consent was obtained from each participant.
Table 1. Baseline characteristics of patients with NSCLC receiving anti-PD-1/PD-L1 immunotherapy.
| NDB | DCB | Total | ||
| Age | ≤60 | 6 | 11 | 17 |
| >60 | 11 | 12 | 23 | |
| Gender | Male | 11 | 19 | 30 |
| Female | 6 | 4 | 10 | |
| Smoking status | Smoker | 11 | 17 | 28 |
| Former/never smoker | 6 | 6 | 12 | |
| Histology | Adenocarcinoma | 7 | 9 | 16 |
| squamous carcinoma | 10 | 14 | 24 | |
| Differentiation grade | High and moderate | 6 | 6 | 12 |
| Low | 7 | 12 | 19 | |
| NA | 4 | 5 | 9 | |
| TNM stage | III stage | 7 | 14 | 20 |
| IV stage | 10 | 9 | 20 | |
| Total | 17 | 23 | 40 |
DCBdurable clinical benefitNDBno durable benefitNSCLCnon-small cell lung cancerPD-1programmed cell death protein-1PD-L1programmed death-ligand 1TNMtumor, node, metastases
Statistical analysis
Statistical analysis was performed using SPSS V.25.0 software, with Student’s t-test, χ² test or Mann-Whitney U test. The data were calculated as the mean±SEM. *p<0.05 was considered statistically significant.
Results
Mutational landscape of LKB1 mutant NSCLC
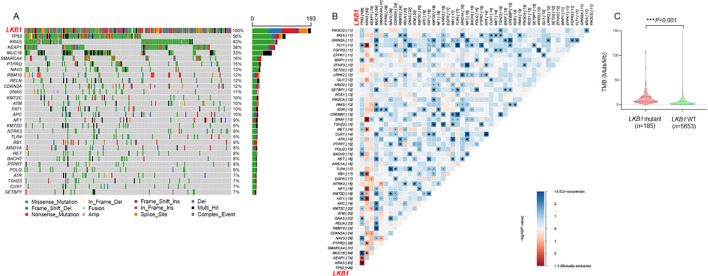
In order to understand the mechanism accounting for poor response to anti-PD-1/PD-L1 immunotherapy in patients with LKB1 mutant NSCLC, we first analyzed LKB1-associated co-mutations based on the next generation sequencing (NGS) database. According to a patient cohort consisting of 193 cases of NSCLC harboring LKB1 mutation, we found various co-mutations that were implicated in apoptosis, DNA damage and cell cycle transition (figure 1A). The most prominent mutations included TP53 (56%), KRAS (43%) and KEAP1 (38%), which was consistent with previous studies showing a prevalence of these genetic mutations in patients with LKB1 mutant NSCLC.21 Moreover, an unbiased mutation enrichment analysis was performed to unveil the co-mutation gene patterns of the LKB1 mutant cohort. We selected the top 50 LKB1 co-mutant genes and analyzed their probability of co-mutation or mutual exclusivity by using the R package maftools (online supplemental table S2). As shown in figure 1B, the gene pairs in blue color indicated the probability of co-mutation, whereas the red color indicated the probability of mutually exclusive. Strikingly, we found that 35 cases of LKB1 mutant NSCLC harbored concomitant TP53 and KRAS mutation, whereas they were mutually exclusive from each other in 122 cases. It was concluded that TP53/KRAS co-mutation was not a common event in patients with LKB1 mutant NSCLC. There were also gene pairs simultaneously mutated in our study. For example, MUC16 and RBM10, TLR4 and FLT1, were two pairs of highly probable co-mutant genes. These genes were found to either enhance or suppress tumorigenesis and therapeutic outcomes of NSCLC. Thus, the genetic landscape of LKB1 mutant NSCLC is quite complicated, and NGS testing may not be a reliable approach to direct anti-PD-1/PD-L1 immunotherapy for patients with LKB1 mutant NSCLC.
Figure 1. The mutational landscape of LKB1-mutant NSCLC. (A) A patient cohort consisting of 193 cases of NSCLC harboring LKB1 mutations was analyzed. Tumor samples were arranged from left to right. Alterations of LKB1 co-occurring genes were annotated for each sample according to the color panel below the image. The somatic mutation frequencies for each candidate gene were plotted on the right panel. (B) Co-mutation gene pattern analysis of the LKB1 mutant cohort. The gene pairs in blue color indicated the probability of co-mutation, and the red color indicated the probability of mutually exclusive. (C) The distribution of TMB in LKB1 mutant and LKB1 WT cohorts. LKB1, liver kinase B; NSCLC, non-small cell lung cancer; TMB, tumor mutation burden; WT, wild type.
Mutations in LKB1 were also accompanied by alterations in gene copy number variations. As shown in online supplemental table S3, the frequency of MYC and MCL-1 (myeloid cell leukemia-1) proto-oncogene amplification was around 5% in the LKB1 WT cohort, however, there was a 3-fold increase in the incidence of MYC and MCL-1 copy number gain. Therefore, it was reasonable to see an increased tumor mutation burden (TMB) when LKB1 was mutated. In our LKB1 mutant patient cohort, TMB data was available in 185 patients and we found that 45.95% (85/185) of patients elicited a high TMB state (≥10 Muts/Mb). In contrast, only 16.24% (918/5,653) of LKB1 WT patients possessed a TMB value over 10 Muts/Mb (figure 1C). When setting 20 Mut/Mb as the threshold to definite high TMB tumor, we found that only 4.14% of LKB1 WT NSCLC were defined as TMBhigh tumor, whereas this proportion increased to 14.59% in the LKB1 mutant cohort (online supplemental table S4). The TMBhigh state is generally believed to confer more favorable responses to PD-1/PD-L1 inhibitors, however, mutations in LKB1 may disrupt antitumor immunological response and result in impaired efficacy of anti-PD-1/PD-L1 immunotherapy. We thus explored the mechanism underlying how LKB1 manipulated antitumor immunity in patients with NSCLC.
LKB1 altered TME and sensitivity to immunotherapy
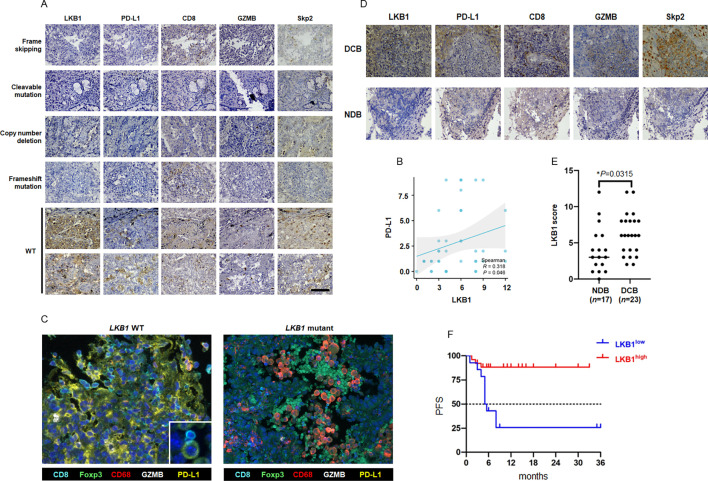
To study the effect of LKB1 on TME, we first evaluated the expression of PD-L1. Representative IHC images of LKB1 and PD-L1 staining in tumor specimens are shown in figure 2A. In NSCLC without LKB1 mutation, the tumor cells were found to express abundant LKB1 protein, along with positive staining of cell membrane-bounded PD-L1. The LKB1 WT NSCLC manifested as an inflamed tumor because a considerable amount of CD8+T cells that were also active for GZMB were found to infiltrate TME. PD-L1 expression was not detected in the LKB1 mutant tumor, regardless of mutational patterns, suggesting a positive correlation between LKB1 and PD-L1 (figure 2B). Notably, the LKB1 negative tumors lacked T cell infiltration and GZMB production (uninflamed TME). These findings partially reflected a “hot”/“cold” status of TME and highlighted LKB1 as a potential biomarker to mirror TME. To gain a comprehensive understanding of the spatial context of TME in the presence or absence of LKB1, we performed fluorescent multiplex IHC that enabled the visualization of multiple TME components at a much better resolution on FFPE samples. The TME panel consisted of markers of tumor-infiltrating immune cells, including CD8, Foxp3, CD68 or F4/80, and GZMB for the identification of active cytotoxic T cells, regulatory T cells (Tregs) and macrophages. In surgical resected LKB1 WT NSCLC, abundant CD8+T cells invade TME and these Opal 480-labeled T cells were found to abut Opal 570-labeled PD-L1+tumor cells (figure 2C). An influx of inhibitory Foxp3+Tregs and CD56+macrophages was noticed in the LKB1 mutant tumor, along with loss of PD-L1 expression on tumor cells. These results indicated a “hot” (PD-L1+/CD8+T cell-enriched) to “cold” (PD-L1-enriched/inhibitory Tregs and macrophages-enriched) TME switch on LKB1 mutation, suggesting that LKB1 is a determinant factor for PD-L1 expression and TME remodeling. We therefore speculated that LKB1 may act as a reliable biomarker to mirror TME and dictate sensitivity to immunotherapy for patients with NSCLC.
Figure 2. Effect of LKB1 on TME and sensitivity to anti-PD-1/PD-L1 immunotherapy in patients with NSCLC. (A) A total number of 40 cases of surgical resected NSCLC were analyzed for LKB1, PD-L1, CD8, GZMB and Skp2 expression by IHC staining. Representative IHC images of LKB1 mutant tumor (the top four panels) and LKB1 WT (the bottom two panels) tumor were shown. The distinct alterations in LKB1 were listed in the left side of images. (B) The linear regression analysis of IHC H-score to determine the correlation between LKB1 and PD-L1. (C) Analysis of TME components in LKB1-WT and LKB1-mutant NSCLC by fluorescent mIHC. Each fluorescent channel indicated a specific biomarker for immune cells or tumor cells. Cell nucleus was visualized by DAPI staining. (D) Representative IHC images of LKB1, PD-L1, CD8, GZMB and Skp2 protein expression in patients with NSCLC receiving anti-PD-1/PD-L1 immunotherapy with different therapeutic outcomes. (E) The distribution of LKB1 IHC-H score in patients with NSCLC with favorable response (DCB) and unfavorable response (NDB) to anti-PD-1/PD-L1 immunotherapy. ((F) Progression-free survival analysis of patients with NSCLC receiving anti-PD-1/PD-L1 immunotherapy with different LKB1 status. DCB, durable clinical benefit; GZMB, granzyme B; IHC, immunohistochemistry; LKB1, liver kinase B; mIHC, multiplex immunohistochemistry; NDB, no durable benefit; NSCLC, non-small cell lung cancer; PD-1, programmed cell death protein-1; PD-L1, programmed death-ligand 1; PFS, progression-free survival; Skp2, S-phase kinase-associated protein 2; TME, tumor microenvironment; WT, wild type.
To collaborate with this notion, we evaluated LKB1 and PD-L1 expression in a separate NSCLC cohort consisting of 40 LKB1 WT patients who received anti-PD-1/PD-L1 immunotherapy and more than 3 years follow-up data were available (table 1, (online supplemental table S5), figure 2D). Patients with complete response and partial response to treatment were classified as responders (durable clinical benefit, DCB), whereas patients with progressive disease or stable disease were classified as non-responders (no durable benefit, NDB). LKB1 could be detected in both groups, in which 18 out of 23 patients in the DCB group showed a high expression of LKB1, whereas only 8 patients with a modest level of LKB1 expression were detected in the NDB group (table 2, figure 2E). In the 26 patients with an LKB1 high expression state, 18 cases (69%) revealed a positive staining of PD-L1, while the percentage of PD-L1 positive cases was 35% in the LKB1 low expression group (table 3). Moreover, the infiltration of active CD8+T cells tended to be more evident in biopsy samples of the DCB group, but not in the NDB group. Log-rank survival analysis also showed the LKB1high cohort benefited more from anti-PD-1/PD-L1 immunotherapy than the LKB1low cohort (modified progression-free survival (mPFS): not reached vs 5.25 months, χ2=12.52, p=0.0004, figure 2F). Taken together, these clinical evidence strongly suggested that intact LKB1 was required for PD-L1 expression and dictated a “hot” TME. The expression of LKB1 therefore conferred a favorable therapeutic response to anti-PD-1/PD-L1 immunotherapy in patients with NSCLC.
Table 2. IHC evaluation of LKB1 and Skp2 expression and their association with therapeutic response to anti-PD-1/PD-L1 immunotherapy.
| NDB | DCB | Total | P value | ||
| LKB1 | Low (IHC score<4) | 9 | 5 | 14 | 0.044 |
| High (IHC score≥4) | 8 | 18 | 26 | ||
| Skp2 | Low (IHC score<4) | 14 | 10 | 24 | 0.014 |
| High (IHC score≥4) | 3 | 13 | 16 | ||
| Total | 17 | 23 | 40 |
DCBdurable clinical benefitIHCimmunohistochemistryLKB1liver kinase BNDBno durable benefitPD-1programmed cell death protein-1PD-L1programmed death-ligand 1Skp2S-phase kinase-associated protein 2
Table 3. IHC evaluation of LKB1 and Skp2 and the association with PD-L1 expression.
| PD-L1 (−) | PD-L1 (+) | Total | ||
| LKB1 | Low (IHC score<4) | 5 | 9 | 14 |
| High (IHC score≥4) | 8 | 18 | 26 | |
| Skp2 | Low (IHC score<4) | 10 | 14 | 24 |
| High (IHC score≥4) | 3 | 13 | 16 | |
| Total | 13 | 27 | 40 |
IHCimmunohistochemistryLKB1liver kinase BPD-L1programmed death-ligand 1Skp2S-phase kinase-associated protein 2
LKB1 selectively manipulates PD-L1 expression at protein level
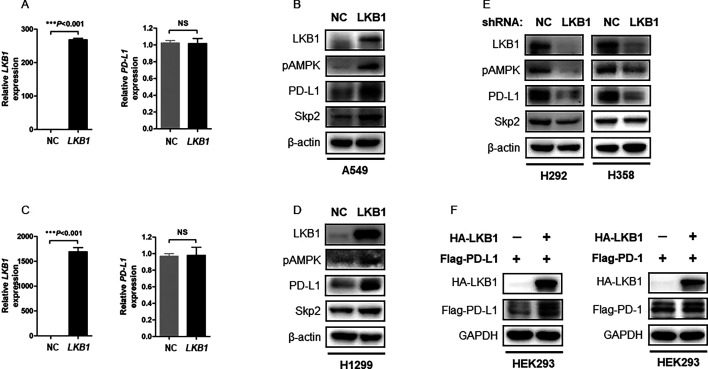
We next explored how LKB1 preserved PD-L1 expression in NSCLC. To do this, we overexpressed LKB1 in a panel of NSCLC cell lines and measured PD-L1 expression at messenger RNA (mRNA) and protein levels, respectively. We found that induction of LKB1 in the LKB1-null A549 cells had minimal effect on PD-L1 transcripts (figure 3A), while immunoblotting assay showed LKB1 readily increased PD-L1 protein abundance (figure 3B). Similarly, overexpression of LKB1 did not alter PD-L1 mRNA level (figure 3C), but increased PD-L1 protein level in H1299 cells (figure 3D). Knockdown of endogenous LKB1 by shRNA selectively reduced PD-L1 protein abundance in H292 and H358 cells (figure 3E). As such, we concluded that LKB1 predominantly regulated PD-L1 at the protein level. The interaction between LKB1 and PD-L1 was highly selective and specific, since we failed to detect an impact of LKB1 on PD-1 expression in HEK293 cells transfected with plasmids encoding HA-LKB1 and Flag-PD-1 (figure 3F).
Figure 3. LKB1 selectively regulates PD-L1 protein expression. (A) qPCR assay of the LKB1-null A549 cells transfected with NC or LKB1 plasmids. Expression of LKB1 and PD-L1 mRNA was determined by qPCR analysis (NS: not significant). (B) Immunoblotting of A549 cell lysates transfected with NC or LKB1 plasmids. β-actin was used as equal loading control. (C) Measurement of PD-L1 mRNA expression in H1299 cells transfected with NC or LKB1 plasmids (NS: not significant). (D) Immunoblotting of H1299 cell lysates transfected with NC or LKB1 plasmids. (E) Immunoblotting of cellular extracts from H292 and H358 stable cells infected with NC shRNA or LKB1 shRNA lentivirus. (F) HEK293 cells were transfected with HA-LKB1, together with Flag-PD-L1 or Flag-PD-1 plasmids. 48 hours after transfection, cells were lyzed for immunoblotting analysis of Flag-tagged PD-L1 or PD-1 protein expression. GAPDH was used as equal loading control. LKB1, liver kinase B; mRNA, messenger RNA; NC, negative control; PD-1, programmed cell death protein-1; PD-L1, programmed death-ligand 1; qPCR, quantitative real-time PCR; shRNA, short hairpin RNA; Skp2, S-phase kinase-associated protein 2.
LKB1-AMPK signaling governs PD-L1 protein abundance
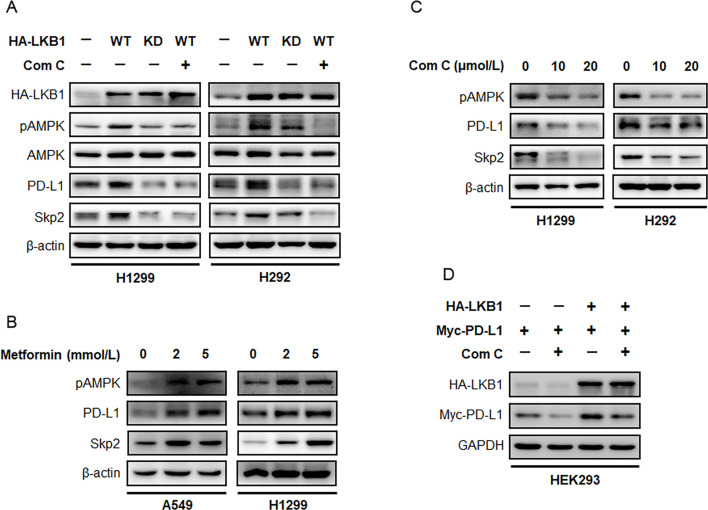
Given that LKB1 is a STK, we investigated whether its kinase activity is required for maintaining a high expression state of PD-L1. We generated a HA-tagged full-length LKB1 (WT) construct and its kinase dead (K78I KD) mutant and expressed these plasmids in H292 and H1299 NSCLC cells. We found overexpression of LKB1 WT markedly increased PD-L1 protein abundance in both cell lines, however, this effect was attenuated when cells were engineered to express the LKB1 mutant defective in kinase activity (figure 4A). We also noticed an augmentation in the phosphorylation level of AMPK in cells ectopically expressing HA-LKB1 WT, but not HA-LKB1 KD plasmid. Because AMPK is a major signaling output of the LKB1 pathway, we thought to exploit the effect of manipulating AMPK on PD-L1 expression. A549 and H1299 cells were treated with increasing concentrations of metformin, an AMPK agonist, and it was found that metformin treatment readily exacerbated the phosphorylation level of AMPK. This effect was accompanied by a dose-dependent increase in the expression of PD-L1 protein (figure 4B, online supplemental figure S1A). In contrast, pharmacological blockade of AMPK activity by compound C suppressed PD-L1 protein level. These findings partially indicated that AMPK is involved in LKB1-regulated PD-L1 protein expression in NSCLC. To confirm whether AMPK is indeed required for LKB1 to maintain PD-L1 protein level, we transfected H292 and H1299 cells with the HA-LKB1 construct and treated the resultant cells with or without compound C. It was noted that ectopic expression of HA-LKB1 augmented AMPK phosphorylation and increased PD-L1 protein level, whereas concurrently blocking AMPK activity by compound C efficiently repressed this augmentation (figure 4C).
Figure 4. LKB1/AMPK pathway determines PD-L1 protein expression. (A) H1299 and H292 cells were transfected with HA-LKB1 WT or dominant negative HA-LKB1 KD mutant, and treated with 20 µmol/L compound C for 24 hours. Immunoblotting assay was performed to detect the expression indicated proteins after treatment. (B) A549 and H1299 cells were treated with metformin, an AMPK agonist, for 48 hours. Expression of pAMPK, PD-L1 and Skp2 was determined by immunoblotting. (C) H1299 and H292 cells were treated with increasing concentrations of compound C and evaluated for pAMPK, PD-L1 and Skp2 expression by Western blot. (D) HEK293 cells were transfected with HA-LKB1 and Myc-PD-L1, treated with or without compound C to block LKB1/AMPK pathway. Protein level of ectopically expressed Myc-PD-L1 was assessed by immunoblotting. AMPK, AMP-activated protein kinase; KD, kinase dead; LKB1, liver kinase B; PD-L1, programmed death-ligand 1; Skp2, S-phase kinase-associated protein 2; WT, wild type.
In HEK293 cells transfected with Myc-PD-L1, we noticed compound C repressed Myc-tagged PD-L1 protein level despite the absence of ectopic HA-LKB1, probably due to the inhibition of endogenous phosphorylated AMPK. When HA-LKB1 was simultaneously introduced, we found a dramatic increase in Myc-PD-L1 protein level, which could be effectively abrogated by compound C treatment (figure 4D). These results thus demonstrated a causal relation between LKB1/AMPK in governing PD-L1 protein abundance, and an active state of AMPK is required for this regulation.
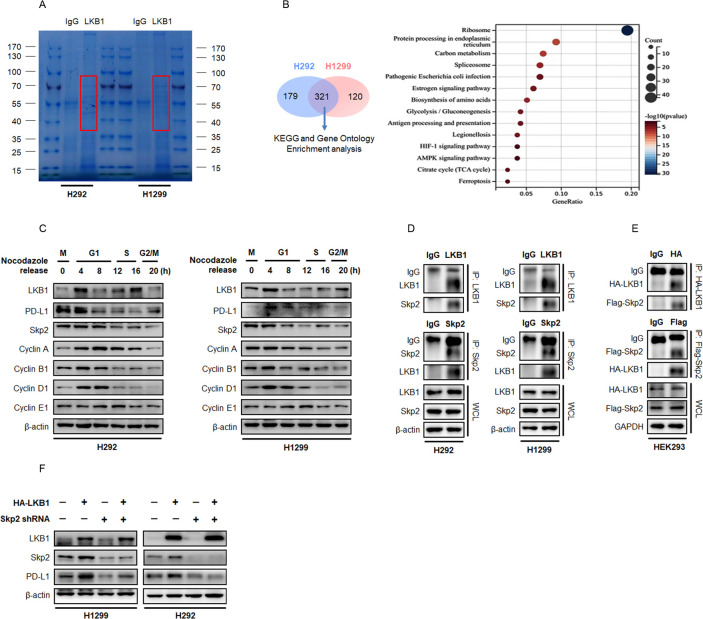
Identification of Skp2 as the LKB1/PD-L1 interacting protein
In order to identify the regulatory factor controlling PD-L1 expression in the presence of LKB1, we first analyzed the LKB1 interacting proteins using MS and ingenuity pathway analysis. We pulled down endogenous LKB1 and its interacting proteins from H292 and H1299 cells with an antibody targeting LKB1. The LKB1 immunocomplex was captured by Protein A/G agarose beads and separated on an SDS-PAGE gel. As shown in figure 4A, protein bands that were captured by LKB1 (∼52 kDa) migrated around the position of the IgG heavy chain were subjected to MS analysis (figure 5A). Using this approach, we identified a total number of 441 peptides from H1299 cell extract and 500 peptides from the H292 cell extract, with 321 peptides overlapping the two tested cell lines (figure 5B). The KEGG and Gene Ontology gene set analysis suggested that the overlapping peptides captured by LKB1 were enriched in the protein processing pathway and Skp2 emerged as an interacting protein of interest. Skp2 has been well documented to play a crucial role in driving cell cycle G1/S phases progression.22 Notably, PD-L1 protein abundance fluctuated during the cell cycle transition in NSCLC cells synchronized by nocodazole, featuring the highest abundance in M and early G1 phases, followed by a sharp reduction in late G1/S phases (figure 5C). The fluctuation of LKB1 and Skp2 proteins also behaved in a similar manner when cells were arrested in the M phase by nocodazole block, indicating potential functional interactions of these proteins.
Figure 5. LKB1 regulates PD-L1 expression through Skp2 in NSCLC. (A) Representative image of Coomassie blue stained SDS-PAGE gel. Proteins that interacted with LKB1 were pulled down by anti-LKB1 antibody, separated by electrophoresis and visualized by Coomassie blue staining. Proteins potentially interacted with LKB1 were highlighted in red box and the gels were excised and proceed for MS analysis. (B) Venn diagram showing the overlap of LKB1 interacting proteins between H292 (n=500) and H1299 (n=441) cells. Enrichment analysis on overlap proteins using clusterProfiler in GO terms. Ranking of the interacting proteins among these enriched signatures according to their frequency. (C) Immunoblotting of whole cell lysates (WCL) derived from H292 and H1299 cells synchronized in M phase by nocodazole followed by releasing back into cell cycle. (D) Reciprocal immunoprecipitation of endogenous LKB1 and Skp2 in H292 and H1299 cells. β-actin was used as equal loading control. (E) Reciprocal immunoprecipitation of ectopically expressed HA-tagged LKB1 and Flag-tagged Skp2 in HEK293 cells. GAPDH was used as equal loading control. (F) H1299 and H292 cells were stably expressed HA-LKB1 with or without shRNA targeting Skp2. Expression of LKB1, Skp2 and PD-L1 was determined by immunoblotting. GO, Gene Ontology; IgG, immunoglobulin G; LKB1, liver kinase B; MS, mass spectrometry; NSCLC, non-small cell lung cancer; PD-L1, programmed death-ligand 1; shRNA, short hairpin RNA; Skp2, S-phase kinase-associated protein 2.
To verify the specific association between LKB1 and Skp2, we first precipitated endogenous LKB1 in NSCLC cell lines and immunoblotting analysis showed Skp2 was indeed captured. When the reciprocal experiment was performed with an antibody targeting Skp2, LKB1 was also found to bind Skp2 (figure 5D). To collaborate with these findings, we used an anti-HA antibody to pull down HA-tagged LKB1 protein ectopically expressed in HEK293 cells and probed the precipitates with an anti-Flag tag antibody. The result of the immunoblotting assay also yielded protein interaction between HA-LKB1 and Flag-Skp2, and vice versa (figure 5E). Thus, the LKB1-Skp2 complex was universally present in living cells, raising the possibility that Skp2 executed the PD-L1 regulatory efficacy of LKB1. In support of this notion, transfection of Flag-Skp2 plasmid into the LKB1-null H1299 cells or H292 cells could promote PD-L1 expression, whereas LKB1-induced PD-L1 expression was antagonized by shRNAs-mediated knockdown of Skp2 (figure 5F). In surgical resected NSCLC, Skp2 tended to be detected in tumors without LKB1 mutation (figure 2A), and there was a tendency for increased survival benefits to anti-PD-1/PD-L1 immunotherapy in patients with high levels of Skp2 (mPFS: not reached vs 5.5 months, χ2=5.879, p=0.0153, (online supplemental figure S2A–B), table 2). Spearman logistic regressive analysis also yielded a positive correlation between Skp2 and PD-L1 expression (R=0.314, p=0.049, (online supplemental figure S2C), table 3). Importantly, these Skp2-positive NSCLCs were susceptible to be positive for LKB1 expression and displayed features of inflamed “hot” tumors (figure 2A). We therefore speculated that Skp2 acts as a molecular hub integrating LKB1 and PD-L1 to shape TME and dictates therapeutic outcomes of immunotherapy in patients with NSCLC.
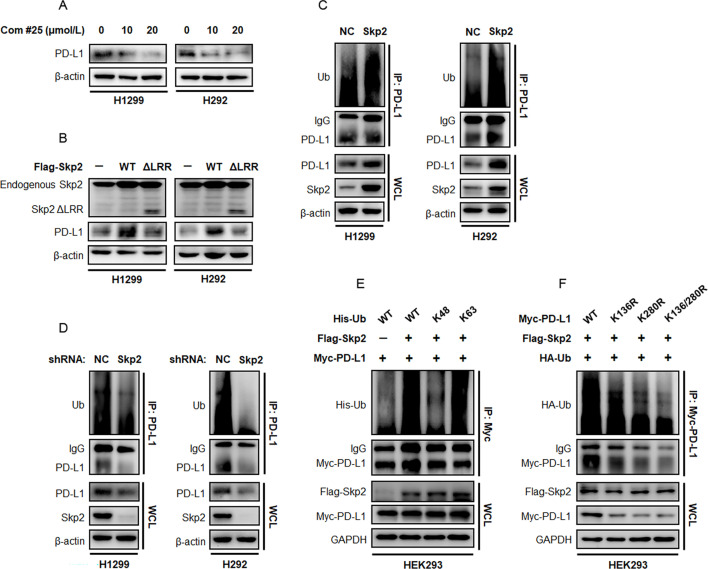
Skp2 stabilizes PD-L1 protein by promoting K63-linked polyubiquitination
Skp2 has been described as an E3 ligase to promote substrate protein ubiquitination. To determine if ubiquitination processing is involved in PD-L1 expression, we used compound #25 that specifically inhibited Skp2 E3 ligase activity.20 Immunoblotting experiment showed a brief treatment with compound #25 suppressed both baseline and LKB1-induced PD-L1 protein expression in NSCLC and HEK293 cells (figure 6A, (online supplemental figure S3A–C). Moreover, ectopic expression of a Skp2 mutant defective in the LRR domain (Skp2 ΔLRR) failed to increase PD-L1 protein expression in comparison to Skp2 WT (figure 6B, (online supplemental figure S3B). The K48-linked polyubiquitin chain impended targeted protein for proteasomal degradation, whereas non-K48 polyubiquitin linkages, particularly K63-linked Ub chains, regulate protein distribution, activity and interaction with other proteins. To determine whether and how Skp2 augmented PD-L1 protein ubiquitination, we transfected plasmids encoding Skp2 with an N-terminal Flag tag in H292 and H1299 cells and precipitated endogenous PD-L1 with an antibody targeting PD-L1. The ubiquitination state of PD-L1 protein was probed with anti-Ub antibody and the result showed ectopic expression of Flag-Skp2 markedly promoted Ub chain engaged to PD-L1 protein (figure 6C). In contrast, inhibition of Skp2 expression by shRNAs or its E3 ligase activity by compound #25 efficiently removed the higher molecular weight smearing pattern of PD-L1 on the SDS-PAGE gel (figure 6D, (online supplemental figure S3D). These findings impended the idea that PD-L1 was regulated by Skp2 E3 ligase, by which PD-L1 protein is modified by Ub adducts in response to active Skp2.
Figure 6. Skp2 promotes ubiquitination of PD-L1 protein. (A) H1299 and H292 cells were treated with increasing concentrations of compound #25 for 48 hours and evaluated for endogenous PD-L1 protein expression. (B) H1299 and H292 cells were transfected with Flag-tagged Skp2 WT or its E3 ligase deficient ΔLRR mutant. The effect of Skp2 on endogenous PD-L1 protein expression was assessed by immunoblotting. (C) Endogenous PD-L1 protein in H1299 and H292 cells was pulled down by an anti-PD-L1 antibody and PD-L1 ubiquitination status was determined with an anti-Ub antibody. (D) Effect of shRNA-mediated Skp2 inhibition on PD-L1 protein ubiquitination in H1299 and H292 cells. (E) HEK293 cells were transfected with Myc-PD-L1, Flag-Skp2, together with His-Ub WT, His-Ub K48-only or His-Ub K63-only mutants. PD-L1 protein was precipitated by an anti-Myc tag antibody and its ubiquitination status was probed with an antibody targeting His-tag protein. (F) HEK293 cells were transfected with HA-Ub, Flag-Skp2, together with Myc-PD-L1 WT, Myc-PD-L1 K136R mutant, Myc-PD-L1 K280R mutant, or K136/280R double mutant. Recombinant Myc-PD-L1 protein was precipitated by an anti-Myc tag antibody and its ubiquitination state was probed with an antibody targeting HA tag protein. IgG, immunoglobulin G; NC, negative control; PD-L1, programmed death-ligand 1; shRNA, short hairpin RNA; Skp2, S-phase kinase-associated protein 2; Ub, ubiquitin; WCL, whole cell lysates; WT, wild type.
To further decipher the Skp2-mediated Ub processing of PD-L1, we expressed Flag-tagged Skp2, together with Myc-tagged PD-L1 and His-tagged Ub, in HEK293 cells. Using an antibody targeting Myc tag, the ectopically expressed PD-L1 protein was pulled down and probed with His antibody to determine Ub-linkage specific PD-L1 protein. We noticed profound polyubiquitination of Myc-PD-L1 in HEK293 cells transfected with Flag-Skp2 and His-Ub WT plasmids. Skp2 prominently led to K63-linked polyubiquitination, since Skp2 failed to yield PD-L1 protein ubiquitination when His-tagged Ub WT or K63-only constructs were replaced by K48-only mutant (figure 6E), indicating Skp2-mediated ubiquitination on PD-L1 as a non-proteolytic modification.
In order to study the role of PD-L1 ubiquitylation, we next identified the Ub acceptor lysine residues of PD-L1. We analyzed lysine evolutionary profiles and functional domain on PD-L1 protein by computational prediction and found the K136 and K280 residues composed of putative ubiquitination sites (online supplemental figure S3E). To this end, we mutagenized each lysine to arginine and transfected the KR mutant into HEK293 cells along with Flag-tagged Skp2 and HA-tagged Ub plasmids. We found that mutation of the two evolutionarily conserved lysine residues of PD-L1, K136 and K280, consistently led to a substantial decrease in Skp2-mediated ubiquitination, with a mutation in K280 residue rendered more potent suppression in ubiquitination. Notably, mutation of both lysine residues reduced ubiquitination status more drastically, suggesting that K136 and K280 were major ubiquitination sites for Skp2 on PD-L1 (figure 6F). Aside from the ubiquitination state, the total amount of PD-L1 protein also declined to different degrees when these K63 ubiquitination sites were replaced by arginine. The K163/280R double mutant yielded the strongest inhibitory potency on the PD-L1 protein level, raising a possibility that ubiquitination at K136 and K280 might stabilize PD-L1 protein.
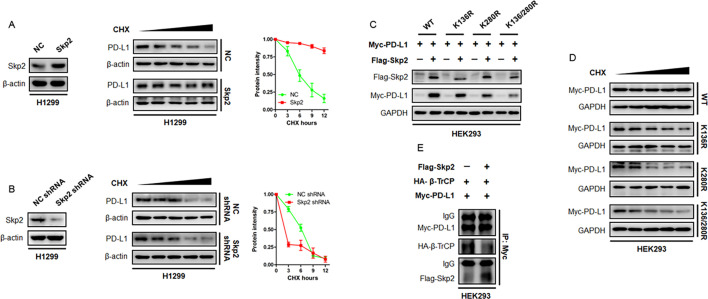
Skp2 is required for maintaining PD-L1 expression
To examine the importance of PD-L1 ubiquitination, we explored PD-L1 protein expression and stability. Skp2 was overexpressed in H1299 and H292 cells and endogenous PD-L1 protein synthesis was stalled by CHX treatment. We found that PD-L1 protein intensity declined to half of its baseline level in approximately 6 hours, while Skp2 was noticed to suppress PD-L1 destruction and stabilize PD-L1 protein (figure 7A). Consistently, the knockdown of endogenous Skp2 by shRNA accelerated PD-L1 protein breakdown and reduced its half-life (figure 7B). Thus, Skp2 was essential for PD-L1 expression and stability.
Figure 7. Effect of Skp2 on PD-L1 protein stability. (A) Protein synthesis was stalled by CHX treatment. The half-life of PD-L1 protein in H1299 cells with overexpression of Skp2 or NC was measured by CHX chase assay. Representative PD-L1 protein intensity curve following CHX treatment was shown. (B) The endogenous Skp2 in H1299 cells were knockdown by shRNA. The effect of Skp2 inhibition on PD-L1 protein stability was evaluated by CHX chase assay. Representative PD-L1 protein intensity curve following CHX treatment was shown. (C) Myc-PD-L1 WT or its KR mutants, together with or without Flag-Skp2, were expressed in HEK293 cells. Expression of Myc-PD-L1 WT or recombinant Myc-PD-L1 KR protein was evaluated by immunoblotting. (D) Myc-PD-L1 WT or its KR mutants was expressed in HEK293 cells. PD-L1 protein stability was determined by CHX chase assay. (E) Myc-PD-L1, HA-β-TrCP plasmids, with or without Flag-Skp2, were expressed into HEK293 cells. The PD-L1 binding to HA-β-TrCP or Flag-Skp2 was determined by immunoprecipitation assay. β-TrCP, β-transducin repeats-containing protein; CHX, cycloheximide; IgG, immunoglobulin G; NC, negative control; PD-L1, programmed death-ligand 1; shRNA, short hairpin RNA; Skp2, S-phase kinase-associated protein 2; WT, wild type.
To exploit the significance of Skp2-mediated ubiquitination sites on PD-L1 dynamics, HEK293 cells were transfected with WT PD-L1, K136R, K280R, or K136/280R double mutant, together with or without Skp2. Accordingly, co-expression of Skp2 readily increased PD-L1 protein abundance, however, this augmentation was compromised when the indicated lysine residues were substituted by arginine. It was noted that the abundance of protein product encoded by the K136/280R double mutant was the lowest compared with HEK293 cells transfected with WT PD-L1 or KR mutant on a single lysine residue (figure 7C). Aside from affecting PD-L1 protein level, these ubiquitination sites were found to contribute to PD-L1 protein stability. As shown in figure 7D, Myc-tagged PD-L1 protein was quite stable, but the recombinant PD-L1 protein products defective in K63-Ub engagement became unstable and their half-life times dropped to different degrees with the K136/280R double mutant being the most prominent.
Having demonstrated the K63-linked non-proteolytic ubiquitination by Skp2 augmented PD-L1 protein stability, we proposed an E3 ligase competition model in which Skp2 antagonized PD-L1 binding to other pro-proteolytic E3 ligases. To validate this model, we assessed PD-L1 affinity to β-TrCP, a potent E3 ligase that promoted PD-L1 protein undergoing K48-linked polyubiquitination and degradation,23 in the absence or presence of Skp2. The Myc-PD-L1, HA-β-TrCP and Flag-Skp2 plasmids were expressed in HEK293 cells and the anti-Myc-PD-L1 precipitates were probed with antibodies against HA tag and Flag tag. Strikingly, the binding partner of PD-L1 switched from β-TrCP E3 ligase to Skp2 E3 ligase when Skp2 was overexpressed (figure 7E). Skp2 thus competes with β-TrCP to protect PD-L1 protein from undergoing proteolytic K48-linked poly-ubiquitination and subsequent degradation by β-TrCP.
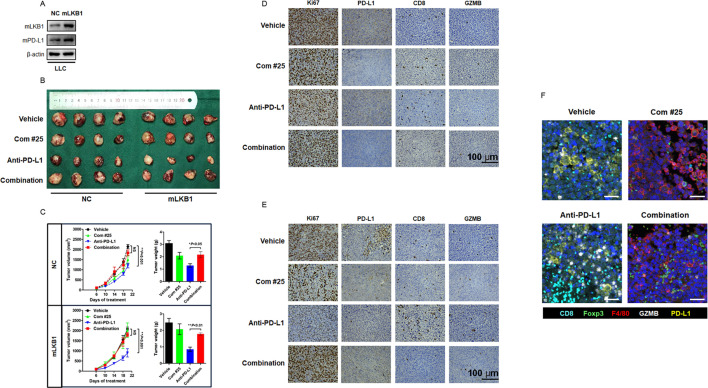
Inhibiting Skp2 generated a “cold” TME and impaired therapeutic efficacy of anti-PD-L1 immunotherapy
To extend our findings for in vivo settings, we stably expressed mLKB1 in LLC cells and transplanted the resultant cells into the right flank of immune-competent C57/BL6 mice (figure 8A). The mice were treated with anti-mouse PD-L1 antibody, compound #25, or their combination, and the syngeneic subcutaneous tumors were monitored every 3 days. We noticed treatment with an anti-mouse PD-L1 antibody yielded profound antitumor activity in both NC and mLKB1 syngeneic tumors, while overexpression of ectopic mLKB1 resulted in a more remarkable response to immunotherapy, as the reduction in size and weight of mLKB1 tumors were more significant than NC tumors. Treatment with compound #25 to inhibit Skp2 activity seemed to suppress tumor outgrowth to a modest degree, however, anti-PD-L1 antibody-induced tumor regression was retarded with the addition of compound #25 (figure 8B,C). As expected from our earlier observations, LLC-mLKB1 cells yielded a high level of PD-L1 expression. Treatment with anti-PD-L1 antibody provoked active CD8+T cell infiltration and created an inflamed “hot” TME. Once Skp2 was inhibited by compound #25, cancer cells lost sufficient expression of PD-L1 and failed to recruit CD8+T cells (figure 8D,E). Importantly, compound #25 was found to turn the inflamed “hot” tumor into an uninflamed “cold” tumor, and the dominant cells invading TME were F4/80+macrophages. The anti-PD-L1 antibody and compound #25 combination also generated an immune suppressive TME, characterized by infiltration of Foxp3+Tregs and F4/80+macrophages. The changes in the TME component therefore underlie differential responses to immunotherapy in the LLC orthotopic tumor model (figure 8F). Restoration of PD-L1 expression by LKB1/AMPK agonists, but not Skp2 inhibitors, is expected to enhance the clinical efficacy of immunotherapy in patients with NSCLC.
Figure 8. Inhibition of Skp2 antagonized PD-L1 blockade therapy and generated a “cold” TME. (A) Mouse LKB1 was stably expressed in LLC cells and its expression was confirmed by Western blot analysis. (B) Representative image of the gross observation of isolated subcutaneous tumor in mice treated with Vehicle, compound #25 (40 mg/kg daily), anti-mouse PD-L1 antibody (200 µg per week), or their combination. (C) The syngeneic tumor volumes were calculated and recorded at indicated time points. NS, not significant (combination vs vehicle). **p<0.01 (anti-PD-L1 vs vehicle). At the end of experiment, tumor nodules were carefully isolated and weighted. *p<0.05 and **p<0.01 (anti-PD-L1 vs combination). (D) Representative IHC staining of Ki67, PD-L1, CD8 and GZMB for NC syngeneic tumor after indicated treatment. (E) Representative IHC staining of Ki67, PD-L1, CD8 and GZMB for mLKB1 syngeneic tumor after indicated treatment. (F) mIHC staining of TME component after indicated treatment in vivo. Each fluorescent channel indicated a specific biomarker for immune cells or tumor cells. Cell nucleus was visualized by DAPI staining. GZMB, granzyme B; IHC, immunohistochemistry; LKB1, liver kinase B; LLC, Lewis lung cancer; mIHC, multiplex immunohistochemistry; NC, negative control; PD-L1, programmed death-ligand 1; Skp2, S-phase kinase-associated protein 2; TME, tumor microenvironment.
Discussion
Despite poor response to anti-PD-1/PD-L1 immunotherapy in LKB1 mutant NSCLC has been extensively recognized in the clinic, surprisingly, little is known about the underlying machinery. In our study, we provided preliminary evidence showing how PD-L1 protein abundance and TME are regulated by LKB1. We demonstrated a direct correlation between LKB1 mutation and loss of PD-L1 expression, which shapes a CD8+T cell-excluded and uninflamed “cold” TME. Accordingly, induction of functional LKB1 or activation of AMPK, downstream of LKB1, thus serves as alternative signals for retaining PD-L1 expression and TME remodeling. We also demonstrated that PD-L1 is ubiquitinated by Skp2, and ubiquitination at K136 and K280 residues is crucial for preserving PD-L1 protein expression. Mechanistically, Skp2 acts as a protein adaptor integrating functional LKB1 and PD-L1, Skp2-mediated K63-linked polyubiquitination on PD-L1 augments protein stability and prompts a T cell-enriched and inflamed “hot” TME. Therefore, LKB1-Skp2 permits persistent expression of PD-L1 and recruitment of cytotoxic T cells, which enables durable response to immunotherapy in patients with LKB1 WT NSCLC.
Converting a “cold” TME into a “hot” TME to enhance the therapeutic efficacy of immune checkpoint blockade therapy is an emerging field of great interest. Hot TME is characterized by elevated T cell infiltration, heightened GZMB production, expression of PD-L1, and suppression of inhibitory immune cells. There are considerable amounts of factors affecting TME, such as PD-L1 expression level, transcriptional factors, protein kinases, cytokines and metabolites.24 In our study, we reported a direct association between the status of LKB1 and the landscape of TME. Under such circumstances, introducing functional LKB1 or reactivation of LKB1/AMPK signaling is expected to boost antitumor immunity. Metformin has been showed to activate AMPK activity and sensitize immune checkpoint inhibitors in preclinical models.25 26 In breast cancer, metformin treatment was found to reduce PD-L1 protein levels, which has been further confirmed in the present study.27 While metformin promoted PD-L1 protein expression in NSCLC, presumably through Skp2-mediated K63 Ub chain engagement. These conflicting PD-L1 responses to metformin treatment might be attributed to the discrepancy in LKB1 expression because the incidence of LKB1 deficiency is more frequent in breast cancer (https://www.proteinatlas.org/ENSG00000118046-STK11/pathology). Thus, cancer cell lines derived from breast cancer are less likely to depend on the LKB1/AMPK pathway to manipulate PD-L1 expression. Another explanation is distinct Skp2 expression patterns in NSCLC. Treatment with metformin resulted in Skp2 overexpression in all tested NSCLC cell lines (figure 4B, (online supplemental figure S1A), whereas it suppressed Skp2 expression in breast cancer BT-549 and MDA-MB-231 cells (online supplemental figure S1B). Hence, Skp2 cooperates with LKB1 to permit high expression of PD-L1 protein and creates a “hot” TME in NSCLC, but the LKB1-Skp2-PD-L1 axis is not working in breast cancer. These results may account for the limited survival benefits of anti-PD-1/PD-L1 immunotherapy over chemotherapy in breast cancer (Keynote 355, ΔOS=6.9 months) when compared with that in NSCLC (Keynote 189, ΔOS=11.4 months).28 29 Future studies aiming to decipher the regulatory machinery of PD-L1 in breast cancer hold the promise of improving therapeutic outcomes in these patients.
The most interesting finding in our study is the discovery of Skp2 as a core component of the LKB1-AMPK-PD-L1 regulatory loop in NSCLC. In fact, Skp2 has been well established as an E3 ligase to promote S phase entry through targeted degradation of cyclin-dependent kinase (CDK) inhibitors p21 and p27.30 Meanwhile, Skp2 regulates PI3K/Akt signaling transduction via K63 Ub chain engagement.14 31 It is anticipated that Skp2 also enhances PD-L1 expression through its E3 Ub ligase activity. Although a panel of E3 ligases for PD-L1, including β-TrCP, SPOP, FBXO22 and ITCH, have already been reported,2332,34 these ubiquitination-promoting enzymes prominently add K48 Ub chains and catalyze PD-L1 protein for destruction. In our study, we found Skp2 adds K63-linked, but not K48-linked, Ub chains to PD-L1 protein in NSCLC. PD-L1 ubiquitination by Skp2 is not associated with protein destruction, instead, ubiquitination on K136 and K280 residues stabilizes PD-L1 protein. This model is consistent with the observation that inhibition of Skp2 by compound #25 or shRNAs accelerated PD-L1 protein turnover and abolished its expression. Therefore, Skp2 together with other E3 ligases regulates PD-L1 at multiple magnitudes of ubiquitination, and the balance between K48-Ub promoting enzymes (destruction) and K63-Ub promoting enzymes (stabilization) dictates the final ubiquitination status of PD-L1 protein, TME profiling and sensitivity to anti-PD-1/PD-L1 immunotherapy in NSCLC. It is possible that Skp2 serves as a higher affinity binding protein for PD-L1 over other pro-proteolytic ubiquitinating enzymes to protect PD-L1 protein from being degraded, but this notion needs to be validated by further studies.
Elevated Skp2 expression has been shown to correlate with an increased risk of lymph node metastasis and poor prognosis in various malignancies. Inhibiting oncogenic Skp2 is thought to be a feasible approach for the treatment of advanced cancer. Consistently, inhibition of Skp2 restores p27 and sensitizes tumor cells to cisplatin, transuzumab and other cytotoxic agents in nasopharyngeal carcinoma and HER2-positive breast cancer.14 35 However, we reported a challenging finding in our study that the Skp2 inhibitor compound #25 failed to sensitize PD-L1 blockade therapy in the LLC syngeneic mouse model. Instead, the addition of compound #25 was found to antagonize antitumor efficacy of the anti-PD-L1 inhibitor. A rational explanation for this controversial observation might be Skp2 inhibitor destabilized the PD-L1 protein. On Skp2 inactivation, the protein protective K63-Ub signals were compromised and consequently led to PD-L1 downregulation and generated an uninflamed “cold” TME. It is proposed that manipulating upstream regulators of PD-L1, including LKB1-Skp2, CDK4-SPOP and CMTM4-CMTM6, to retain PD-L1 expression holds the promise to synergize with immunotherapy.32 36 In line with this model, there are successful clinical attempts of combining metformin (LKB1/AMPK agonist) or palbociclib (CDK4/6 inhibitor) with anti-PD-1 antibodies to enhance therapeutic efficacy and overcome resistance to immunotherapy.37,39 It could be speculated that other strategies aiming to preserve PD-L1 expression may also be beneficial to boost antitumor immunity. In summary, we revealed a novel LKB1-Skp2-PD-L1 regulatory loop that underlies poor response to anti-PD-1/PD-L1 immunotherapy in LKB1 mutant NSCLC. By interacting with Skp2 E3 ligase, LKB1 increases PD-L1 protein expression and activates antitumor immunity. The LKB1 mutant NSCLC manifests as uninflamed “cold” tumors and targeting the LKB1-Skp2 pathway would be a promising strategy to convert “cold” TME into “hot” TME, and therefore enhancing the efficacy of anti-PD-1/PD-L1 immunotherapy in patients with NSCLC.
Conclusion
In this study, we elucidated how LKB1 status dictated therapeutic outcomes of anti-PD-1/PD-L1 immunotherapy in patients with NSCLC, which largely attributed to inducible PD-L1 protein expression through Skp2-dependent K63-linked polyubiquitination machinery. Activation of the LKB1-Skp2 pathway generated a “hot” TME and enhanced the efficacy of immunotherapy, whereas loss-of-function in LKB1 abrogated PD-L1 expression and remodeled a “cold” TME that impaired therapeutic response to anti-PD-1/PD-L1 immunotherapy.
supplementary material
Acknowledgements
We would like to thank Wenyi Wei (Beth Israel Deaconess Medical Center), Hui-Kuan Lin (Duke University) and Yang Gao (Xi’an Jiao Tong University) for their helpful suggestions and technical advice. We also express gratitude to all patients and their families who participated in our study.
Footnotes
Funding: This study was sponsored in part by grants National Natural Science Foundation of China (#82002456 to DW, #82172728 and #82370096 to TL) and Natural Science Foundation of Shandong Province (#ZR2022MH196 to QM).
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Consent obtained directly from patient(s).
Ethics approval: Not applicable.
Contributor Information
Liting Lv, Email: 645829671@qq.com.
Qing Miao, Email: miaoqing971@163.com.
Sutong Zhan, Email: zst_njumed@foxmail.com.
Peilin Chen, Email: 18796598687@163.com.
Wei Liu, Email: liuwei@biokanghui.com.
Jiawen Lv, Email: jiawenlv@126.com.
Wenjie Yan, Email: yanwenjie200531@qq.com.
Dong Wang, Email: doyle_wd@126.com.
Hongbing Liu, Email: netlhb@126.com.
Jie Yin, Email: yinjie506@163.com.
Jian Feng, Email: Jfeng68@126.com.
Yong Song, Email: yong.song@nju.edu.cn.
Mingxiang Ye, Email: mingxiangye88@163.com.
Tangfeng Lv, Email: bairoushui@163.com.
Data availability statement
Data are available upon reasonable request.
References
- 1.Iwai Y, Ishida M, Tanaka Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A. 2002;99:12293–7. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Errico A. Immunotherapy: PD-1-PD-L1 axis: efficient checkpoint blockade against cancer. Nat Rev Clin Oncol. 2015;12:63. doi: 10.1038/nrclinonc.2014.221. [DOI] [PubMed] [Google Scholar]
- 3.Clift R, Souratha J, Garrovillo SA, et al. Remodeling the Tumor Microenvironment Sensitizes Breast Tumors to Anti-Programmed Death-Ligand 1 Immunotherapy. Cancer Res. 2019;79:4149–59. doi: 10.1158/0008-5472.CAN-18-3060. [DOI] [PubMed] [Google Scholar]
- 4.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14:847–56. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 5.Lin H, Wei S, Hurt EM, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest. 2018;128:805–15.:96113. doi: 10.1172/JCI96113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–74. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Federico A, De Giglio A, Parisi C, et al. STK11/LKB1 and KEAP1 mutations in non-small cell lung cancer: Prognostic rather than predictive? Eur J Cancer. 2021;157:108–13. doi: 10.1016/j.ejca.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8:822–35. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borghaei H, Gettinger S, Vokes EE, et al. Five-Year Outcomes From the Randomized, Phase III Trials CheckMate 017 and 057: Nivolumab Versus Docetaxel in Previously Treated Non-Small-Cell Lung Cancer. J Clin Oncol. 2021;39:723–33. doi: 10.1200/JCO.20.01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shackelford DB, Abt E, Gerken L, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–58. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji P, Jiang H, Rekhtman K, et al. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol Cell. 2004;16:47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Regan KM, Wang F, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–54. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan C-H, Li C-F, Yang W-L, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu CQ, Blackhall FH, Pintilie M, et al. Skp2 gene copy number aberrations are common in non-small cell lung carcinoma, and its overexpression in tumors with ras mutation is a poor prognostic marker. Clin Cancer Res. 2004;10:1984–91. doi: 10.1158/1078-0432.ccr-03-0470. [DOI] [PubMed] [Google Scholar]
- 16.Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–24. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- 17.Lin H-K, Wang G, Chen Z, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–32. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D, Inuzuka H, Tseng A, et al. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009;11:397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han F, Li C-F, Cai Z, et al. The critical role of AMPK in driving Akt activation under stress, tumorigenesis and drug resistance. Nat Commun. 2018;9:4728. doi: 10.1038/s41467-018-07188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan C-H, Morrow JK, Li C-F, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–68. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–77. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kossatz U, Dietrich N, Zender L, et al. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 2004;18:2602–7. doi: 10.1101/gad.321004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C-W, Lim S-O, Xia W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Jiang CC, Jin L, et al. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27:409–16. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 25.Cha J-H, Yang W-H, Xia W, et al. Metformin Promotes Antitumor Immunity via Endoplasmic-Reticulum-Associated Degradation of PD-L1. Mol Cell. 2018;71:606–20. doi: 10.1016/j.molcel.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Lu C, Zhang K, et al. Metformin Combining PD-1 Inhibitor Enhanced Anti-Tumor Efficacy in STK11 Mutant Lung Cancer Through AXIN-1-Dependent Inhibition of STING Ubiquitination. Front Mol Biosci. 2022;9:780200. doi: 10.3389/fmolb.2022.780200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai X, Bu X, Gao Y, et al. Energy status dictates PD-L1 protein abundance and anti-tumor immunity to enable checkpoint blockade. Mol Cell. 2021;81:2317–31. doi: 10.1016/j.molcel.2021.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortes J, Rugo HS, Cescon DW, et al. Pembrolizumab plus Chemotherapy in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2022;387:217–26. doi: 10.1056/NEJMoa2202809. [DOI] [PubMed] [Google Scholar]
- 29.Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378:2078–92. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 30.Carrano AC, Eytan E, Hershko A, et al. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–9. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 31.Yao F, Zhou Z, Kim J, et al. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat Commun. 2018;9:2269. doi: 10.1038/s41467-018-04620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Bu X, Wang H, et al. Cyclin D–CDK4 kinase destabilizes PD-L1 via cullin 3–SPOP to control cancer immune surveillance. Nature New Biol. 2018;553:91–5. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De S, Holvey-Bates EG, Mahen K, et al. The ubiquitin E3 ligase FBXO22 degrades PD-L1 and sensitizes cancer cells to DNA damage. Proc Natl Acad Sci U S A. 2021;118:e2112674118. doi: 10.1073/pnas.2112674118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Wang Y, Liu S, et al. Enhancing PD-L1 Degradation by ITCH during MAPK Inhibitor Therapy Suppresses Acquired Resistance. Cancer Discov. 2022;12:1942–59. doi: 10.1158/2159-8290.CD-21-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Wang R, Zhang Y, et al. Skp2-mediated ubiquitination and mitochondrial localization of Akt drive tumor growth and chemoresistance to cisplatin. Oncogene. 2019;38:7457–72. doi: 10.1038/s41388-019-0955-7. [DOI] [PubMed] [Google Scholar]
- 36.Mezzadra R, Sun C, Jae LT, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature New Biol. 2017;549:106–10. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Lin Y, Xiong X, et al. Low-Dose Metformin Reprograms the Tumor Immune Microenvironment in Human Esophageal Cancer: Results of a Phase II Clinical Trial. Clin Cancer Res. 2020;26:4921–32. doi: 10.1158/1078-0432.CCR-20-0113. [DOI] [PubMed] [Google Scholar]
- 38.Yuan Y, Lee JS, Yost SE, et al. Phase I/II trial of palbociclib, pembrolizumab and letrozole in patients with hormone receptor-positive metastatic breast cancer. Eur J Cancer. 2021;154:11–20. doi: 10.1016/j.ejca.2021.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang H-J, Truong CY, Lo EM, et al. Inhibition of Cyclin Dependent Kinase 4/6 Overcomes Primary Resistance to Programmed Cell Death 1 Blockade in Malignant Mesothelioma. Ann Thorac Surg. 2022;114:1842–52. doi: 10.1016/j.athoracsur.2021.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request.