Abstract
To uniformly test and benchmark the secure evaluation of transformer-based models, we designed the iDASH24 homomorphic encryption track dataset. The dataset comprises a protein family classification model with a transformer architecture and an example dataset that is used to build and test the secure evaluation strategies. This dataset was used in the challenge period of iDASH24 Genomic Privacy Competition, where the teams designed secure evaluation of the classification model using a homomorphic encryption scheme. Combined with the benchmarking results and companion methods, iDASH24 dataset is a unique resource that can be used to benchmark secure evaluation of neural network models.
Keywords: Genomic privacy, homomorphic encryption (HE), transformer model
BACKGROUND
Protecting individual privacy has always been a major challenge in health data science, especially when handling sensitive personal information such as genome sequences [1]. The proliferation of large transformer-based architectures has further complicated this issue due to dependency on large individual-level training sets, leading to complex ethical challenges [2], [3]. Due to the large resources needed to evaluate these models, querying is exclusively outsourced to online servers, where the users submit their queries (e.g., ChatGPT questions) via online APIs to servers [4], [5], [6]. When the queries carry sensitive information such as health and medical data, they can cause extensive risk to the users. Furthermore, the queries may be used to tune the models by manual selection and postprocessing, which may further exacerbate the privacy risks to users.
The protection of the queries can be implemented via homomorphic encryption (HE) [7], [8]. In a HE-enabled setup, the queries are first encoded and encrypted via an appropriate HE scheme (e.g., CKKS [9]). Next, the user submits the encrypted query to the server, which securely evaluates the model, obtains encrypted results, and sends the results back to the user (Fig. 1). Results are decrypted to obtain the plaintext results. While HE was deemed impractical after its inception, there is renewed interest in HE-based techniques, thanks to the recent theoretical and practical breakthroughs. These have brought forth orders of magnitude improvement in runtime performance [10], which has led to, for example, generic HE-compilers that can generate HE-enabled code [11], [12]. There is strong industry and academic interest in HE for developing practical applications in machine learning [13], [14], [15]. While there are limitations to the practical usage of HE-enabled systems such as embedded systems, and for processing very large datasets, most HE-schemes are highly parallelizable and can make use of single instruction-multiple data operations optimizations.
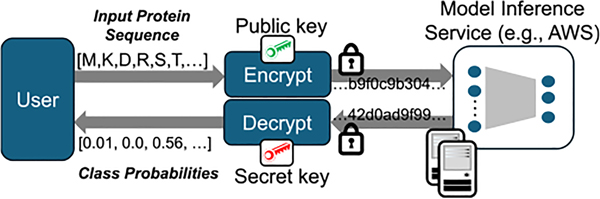
FIG. 1.
Illustration of the secure model inference service. User encrypts and submits the protein sequence. The encrypted data are sent to model inference service, which securely evaluates the classification model and sends the encrypted results back to the user, who decrypts and obtains the results.
There are currently limited benchmarking resources to evaluate secure neural network inference methods effectively. Most benchmark studies focus on comparing the performance of different schemes and libraries [16]. Secure evaluation of the transformer-based models is especially valuable since majority of the outsourced tasks rely on these architectures and they represent the current pinnacle of state-of-the-art in generative tasks [17] with immediate impact on individual privacy.
To address this gap, we present the iDASH24 dataset, to benchmark HE-based evaluation of a transformer model for protein classification for the homomorphic encryption track of iDASH24 genomic privacy challenge [18]. This dataset contains a neural network model for protein sequence classification, and an example dataset that can be used to test and explore the model. The benchmarking metrics and results are included in the dataset, making iDASH24 a unique community contributed resource. In this data descriptor article, we hope to facilitate advancements in privacy-preserving machine learning techniques and promote their adoption in sensitive application domains.
COLLECTION METHODS AND DESIGN
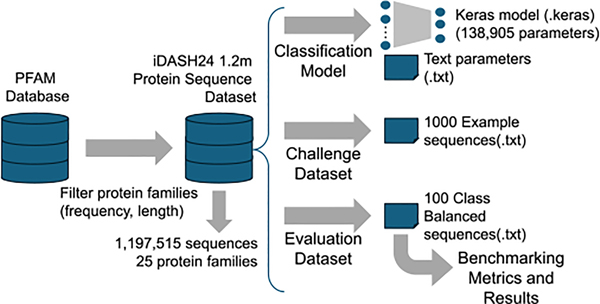
In iDASH24 dataset, we present a 25-class protein sequence dataset that is obtained from PFAM [19] database and a neural network model that is trained using the database (Fig. 2).
FIG. 2.
Processing steps for generating iDASH24 datasets.
iDASH24 1.2 m Protein Sequence Dataset
We downloaded 52786549 FASTA formatted protein sequences from the PFAM database [19], which was accessed on 17 March 2024 (Table I). Each sequence contains an identifier, followed by the aminoacid sequence for the protein. The aminoacids are denoted as single letters out of a 25-letter alphabet. We selected the families that had at least 40000 and at most 60000 examples in the PFAM database. We next sorted the protein classes by decreasing frequency and selected the sequences in the most abundant 25 classes of proteins. For each selected sequence, we extracted a random 50 aminoacid long fragment and saved it with the corresponding family label in [0,24]. Any sequence that is shorter than 50 aminoacids is excluded from the final outputs for the corresponding class. The selected sequences were saved in a text file with 50 aminoacid sequences and the class label separated by a semicolon. Overall, the dataset contains 1197515 sequences for the 25 protein classes. We denote this dataset as the 1.2 m protein sequence dataset.
TABLE I.
Data Source and Filtering Criteria
| Main Data Source | PFAM Database [19] |
|---|---|
| Source Size | 52786549 (17 March 2024) |
| Format | FASTA |
| Selection Criteria | 25 Most Abundant, Frequency between 40000–60000 sequences |
| Exclusion Criteria | Shorter than 50 amino acids |
| Final Dataset | 1197515 sequences with class labels in [0,24] |
iDASH24 Protein Classification Model
The protein classification model uses a transformer architecture comprising a tokenizer, an encoder, a transformer block with a four-head self-attention layer, and a final dense layer that performs classification. The model contains 138905 parameters in total. Model was trained using 1.2 m protein sequence dataset for ten epochs, with categorical cross-entropy loss, and Adam optimizer. Twenty percent of the training dataset (239503 sequences) was used as the validation set to track model fitting. The remaining 958012 sequences were used for training the model. The final model file was saved as a keras file. We also extracted all model parameters as text files that can be loaded and explored in other languages and computing platforms.
iDASH24 Challenge and Evaluation Datasets
One thousand sequences were randomly selected and were distributed to the teams as example sequences. We also extracted 100 class-balanced protein sequences (four sequences per class) that were used for benchmarking the submitted solutions for accuracy and resource requirements.
iDASH24 Benchmarking and Evaluation Results
The participating teams were asked to develop HE-based solutions to evaluate the classification model on encrypted protein sequences. The teams were allowed to tokenize the input sequences and apply one linear scaling to the input data before they were encrypted and used as input to the encoder layer. All layers after encoders were required to process encrypted data. The teams were free to select the encryption library/scheme (e.g., SEAL [20], Lattigo [21], and OpenFHE [22]) with the constraint that the encryption parameters satisfy 128-bit security under HE-standard [23].
Out of 15 registered teams, we received six solutions from five teams. The 100-sequences evaluation dataset was used for benchmarks, which were done on Intel Xeon Platinum 8168 processor. Each submission was run in a Docker container limited to four cores or processor, 2.5 GB of disk storage space, and 128 GB memory (Docker Engine Version 26.1.3). The end-to-end runtime and the microaveraged area under curve (microAUC, using roc_auc_score function in scikit-learn library [24]) was calculated for each solution (Table II). Overall, we observed that the solutions finished in less than 40 min except for one solution, which ran for 12.6 h. One method finished in 7 min and 25 s.
TABLE II.
Performance Scores and Execution Times for Different Solutions
| Solution | Score (MicroAUC) | End-to-End Time |
|---|---|---|
| Solution-1 | 0.963 | 26m 50.541s |
| Solution-2 | 0.941 | 7m 25.820s |
| Solution-3 | 0.984 | 35m 37.218s |
| Solution-4 | 0.983 | 25m 13.271s |
| Solution-5 | 0.525 | 12.57 h |
VALIDATION AND QUALITY
We chose not to filter sequences based on their amino acid content so as to include rare protein classes with distinct properties, promoting diversity within the dataset. While most classes were well represented, we observed that three classes had fewer sequences due to the sequence length filtering criterion (minimum of 50 amino acids). To address potential class imbalance and maintain inclusivity, we decided to include these smaller classes in the iDASH24 dataset. For model validation, we evaluated the transformer-based classification model on the validation set, achieving an accuracy of 86.95%, which closely matched the training accuracy of 86.84%. This parity suggests that the model generalizes well and does not overfit the training data.
As a separate test, we tested the quality of the dataset and the neural network model using protein classification accuracy from the transformer model. For this, we extracted a separate 1000 random sequences from the 1.2 m protein sequence dataset and evaluated the network model. Overall, we found that the classification accuracy was 89%, which indicates that the model provides high classification accuracy among the 25-protein classes. We further ensured that the released text-based model parameters matched the total number of parameters reported by the model (138905 parameters), confirming the consistency and integrity of the model files. The model was also extensively tested the teams participating in the iDASH24 Challenge. Their independent evaluations help to further validate the model’s reliability and performance.
RECORDS AND STORAGE
We describe the detailed files shared in iDASH24 dataset in Table III.
TABLE III.
Description of Formats and Contents of the iDASH24 Dataset Files
| File Name | Format | Description |
|---|---|---|
| Example Sequences (example_AA_sequences.txt) | Space delimited file. Class | Protein sequences for evaluation and challenge |
| Dashformer.keras | Keras model file | Protein classification model |
| Dashformer_model_parameters | Text-formatted Parameters | Directory contains the parameters of classification model |
| DASHformer_Challenge.py | Python code for the model | The Python code for exploring and evaluating model |
| dashformer_tokenizer.json | Json file | Tokenizer file for processing input sequences |
| PFAM_training_sequences.txt | Space delimited text file | 1.2m protein sequence database |
| DASHformer.requirements | Python requirements list | The list of requirements to run the classification model |
In addition to the classification model and the protein sequence data, iDASH24 contains documentation files including a CHALLENGE.README file and Python model summaries (text and png formatted) that describe the model architecture. We will also release the benchmarking scripts (as Python scripts) and final benchmarking results for all the teams that have completed the challenge. Given the extensive participation from the cryptography research teams in this year’s competition, we foresee that the dataset combined with the benchmarking results will serve as a unique resource for the future development of HE-based neural network evaluation methods.
INSIGHTS AND NOTES
In this work, we have presented the iDASH24 dataset, a comprehensive resource designed to facilitate the benchmarking and development of secure neural network evaluation methods using HE. By providing a large-scale protein sequence dataset and a transformer-based classification model, we aim to bridge the gap between advanced cryptographic techniques and practical machine learning applications. We invite researchers and practitioners to utilize this dataset to advance the field of privacy-preserving machine learning, ultimately contributing to the secure and ethical handling of sensitive biological data.
Previous benchmarking datasets about protein sequence analysis focus onbroad number of tasks for smaller models and may occasionally not satisfy the size requirements for large model training [25], [26], [27]. In comparison, our objective was to train and benchmark a large model on more simple task. Therefore, iDASH24 is specifically selected to be uniformly filtered and processed to include a randomly selected 50-mer fragment from each protein sequence. We also aimed to have a large dataset to ensure the large model can be trained on the sequences.
We strongly recommend any users of iDASH24 data refer to other papers published with this dataset. The users can refer to these publications and announcements to perform future benchmarks. Additionally, the users of iDASH24 dataset can make use of the 1.2 m dataset to generate new benchmarking datasets with similar characteristics and do more extensive tests. This is possible because the classification model was trained on a large portion of this dataset and should classify this dataset well.
ACKNOWLEDGMENT
A.H., M.K., and X.J. conceived the conceptual development of iDASH24 homomorphic encryption challenge and details. A.H. performed data processing and model building, training. A.H., M.K., and X.J. worked on documentation updates and communication with the teams over the challenge period. A.H., L.C., and X.J. performed evaluation and benchmarking of solutions. A.H., L.C., M.K., and X.J. wrote the manuscript. All authors read and approved the final manuscript.
The authors acknowledge the contributions of the iDASH organizing committee.
This work was supported by the NIH under Grant R13HG012902. The work of Arif Harmanci was supported by the NIH under Grant R01HG012604.
Footnotes
SOURCE CODE AND SCRIPTS
The authors released the scripts that were used to download and extract the PFAM dataset, the iDASH24 protein classification model source code, and the model file, with the documentation on IEEE DataPort (doi: 10.21227/9fdg-pz55) and on Zenodo (doi: 10.5281/zenodo.13922565).
REFERENCES
- [1].Wan Z, Hazel JW, Clayton EW, Vorobeychik Y, Kantarcioglu M, and Malin BA, “Sociotechnical safeguards for genomic data privacy,” Nature Re. Genetics, vol. 23, no. 7, pp. 429–445, Jul. 2022. [Google Scholar]
- [2].Li H et al. , “Privacy in large language models: Attacks, defenses and future directions,” Oct. 2023, arXiv:2310.08888. [Google Scholar]
- [3].Feretzakis G and Verykios VS, “Trustworthy AI: Securing sensitive data in large language models,” Sep. 2024, arXiv:2409.12345. [Google Scholar]
- [4].Gm H, Gourisaria MK, Pandey M, and Rautaray SS, “A comprehensive survey and analysis of generative models in machine learning,” Comput. Sci. Rev, vol. 38, Nov. 2020, Art. no. 100285. [Google Scholar]
- [5].Samsi S et al. , “From words to watts: Benchmarking the energy costs of large language model inference,” Oct. 2023, arXiv:2310.04123. [Google Scholar]
- [6].Zhao WX et al. , “A survey of large language models,” Mar. 2023, arXiv:2303.18223. [Google Scholar]
- [7].Gentry C, “Computing arbitrary functions of encrypted data,” Commun. ACM, vol. 53, no. 3, pp. 97–105, Mar. 2010. [Google Scholar]
- [8].Albrecht M et al. , “Homomorphic encryption standard,” 2018. Accessed: Apr. 18, 2022. [Online]. Available: http://homomorphicencryption.org/wp-content/uploads/2018/11/HomomorphicEncryptionStandardv1.1.pdf [Google Scholar]
- [9].Cheon JH, Kim A, Kim M, and Song Y, “Homomorphic encryption for arithmetic of approximate numbers,” in Proc. Adv. Crypt, ASIACRYPT, Takagi T and Peyrin T, Eds., vol. 10624. Cham, Switzerland: Springer, 2017, pp. 409–437. [Google Scholar]
- [10].Jiang L and Ju L, “FHEBench: Benchmarking fully homomorphic encryption schemes,” Mar. 2022, arXiv:2203.00345. [Google Scholar]
- [11].Boemer F, Lao Y, Cammarota R, and Wierzynski C, “nGraph-HE: A graph compiler for deep learning on homomorphically encrypted data,” 2019. [Online]. Available: https://arxiv.org/abs/1810.10121 [Google Scholar]
- [12].Viand A, Jattke P, and Hithnawi A, “SoK: Fully homomorphic encryption compilers,” in Proc. IEEE Symp. Secur. Privacy (SP), 2021, pp. 1092–1108. [Google Scholar]
- [13].Song C and Huang R, “Secure convolution neural network inference based on homomorphic encryption,” Appl. Sci, vol. 13, no. 10, 2023, Art. no. 6117. [Online]. Available: https://www.mdpi.com/2076-3417/13/10/6117 [Google Scholar]
- [14].Maloney V, Obrecht RF, Saraph V, Rama P, and Tallaksen K, “High-resolution convolutional neural networks on homomorphically encrypted data via sharding Ciphertexts,” 2024. [Online]. Available: https://arxiv.org/abs/2306.09189 [Google Scholar]
- [15].van Elsloo T, Patrini G, and Ivey-Law H, “SEALion: A framework for neural network inference on encrypted data,” 2019. [Online]. Available: https://arxiv.org/abs/1904.12840 [Google Scholar]
- [16].Gouert C, Mouris D, and Tsoutsos NG, “SoK: New insights into fully homomorphic encryption libraries via standardized benchmarks,” Cryptol. ePrint Arch, Paper 2022/425, 2022. Accessed: Sep. 10, 2024. [Online]. Available: https://eprint.iacr.org/2022/425 [Google Scholar]
- [17].Chillotti I, Joye M, and Paillier P, “New challenges for fully homomorphic encryption,” 2020. Accessed: Sep. 10, 2024. [Online]. Available: https://ppml-workshop.github.io/ppml20/pdfs/Chillotti_et_al.pdf [Google Scholar]
- [18].Kuo T-T et al. , “The evolving privacy and security concerns for genomic data analysis and sharing as observed from the idash competition,” J. Amer. Med. Inform. Assoc, vol. 29, no. 12, pp. 2182–2190, Nov. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mistry J et al. , “Pfam: The protein families database in 2021,” Nucleic Acids Res, vol. 49, no. D1, Jan. 2021, pp. D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Benaissa A, Retiat B, Cebere B, and Belfedhal AE, “TenSEAL: A library for encrypted tensor operations using homomorphic encryption,” 2021. [Online]. Available: https://arxiv.org/abs/2104.03152 [Google Scholar]
- [21].Mouchet C, Troncoso-Pastoriza J, Bossuat J-P, and Hubaux J-P, “Multiparty homomorphic encryption from ring-learning-with-errors,” Cryptol. ePrint Arch, Paper 2020/304, 2020. Accessed: Sep. 10, 2024. [Online]. Available: https://eprint.iacr.org/2020/304 [Google Scholar]
- [22].Badawi AA et al. , “OpenFHE: Open-source fully homomorphic encryption library,” Cryptol. ePrint Arch, Paper 2022/915, 2022. Accessed: Sep. 10, 2024. [Online]. Available: https://eprint.iacr.org/2022/915 [Google Scholar]
- [23].Albrecht M et al. , “Homomorphic encryption security standard,” HomomorphicEncryption.org, Toronto, ON, Canada, Tech. Rep., Nov. 2018. Available: https://eprint.iacr.org/2019/939 [Google Scholar]
- [24].Pedregosa F et al. , “Scikit-learn: Machine learning in Python,” J. Mach. Learn. Res, vol. 12, pp. 2825–2830, 2011. [Google Scholar]
- [25].Sonego P et al. , “A protein classification benchmark collection for machine learning,” Nucleic Acids Res, vol. 35, no. Database issue, pp. D232–D236, Jan. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kertész-Farkas A et al. , “Benchmarking protein classification algorithms via supervised cross-validation,” J. Biochem. Biophys. Methods, vol. 70, pp. 1215–1223, 2008. [DOI] [PubMed] [Google Scholar]
- [27].Yang J et al. , “CARE: A benchmark suite for the classification and retrieval of enzymes,” 2024. [Online]. Available: http://arxiv.org/abs/2406.15669 [Google Scholar]