Abstract
Hematological malignancies are typically treated with chemotherapy and radiotherapy as the first-line conventional therapies. However, non-coding RNAs (ncRNAs) are a rapidly expanding field of study in cancer biology that influences the growth, differentiation, and proliferation of tumors by targeting immunological checkpoints. This study reviews the results of studies (from 2012 to 2024) that consider the immune checkpoints and ncRNAs in relation to hematological malignancies receiving immunotherapy. This article provides a summary of the latest advancements in immunotherapy for treating hematological malignancies, focusing on the role of immune checkpoints and ncRNAs in the immune response and their capacity for innovative strategies. The paper also discusses the function of immune checkpoints in maintaining immune homeostasis and how their dysregulation can contribute to developing leukemia and lymphoma. Finally, this research concludes with a discussion on the obstacles and future directions in this rapidly evolving field, emphasizing the need for continued research to fully harness the capacity of immune checkpoints and ncRNAs in immunotherapy for hematological malignancies.
Keywords: miRNA, lncRNA, Leukemia, Immune checkpoints, Immunotherapy
Introduction
Anti-programmed death-1 (PD-1) and anti-cytotoxic T-lymphocyte antigen-4 (CTLA-4) immune checkpoint inhibition have drastically changed the solid tumor cancer therapy landscape [1, 2]. However, their efficacy in hematological malignancies has been less pronounced. It is recognized that cancerous cells can circumvent immune surveillance by activating immune checkpoints, which serve as inhibitors of the immune system. Immune checkpoint inhibitors (ICIs) have revolutionized solid tumor treatment by reactivating the anti-tumor effect mediated by T-lymphocytes [3–5]. A number of T cell immune checkpoint molecules, such as CTLA-4 and PD-1, have been shown to be viable targets for therapy because of their non-redundant control over T cell responses in malignancies that are hematological in nature [6]. Hematological malignancies have responded better to immunotherapeutic approaches such as stem cell transplantation and anti-cancer monoclonal antibodies; nevertheless, the clinical advantages of immune checkpoint blockade (ICB) are restricted to tumor forms with significant levels of immune cell infiltration [7, 8]. Long-term disease management is a common challenge for ICB therapy, underscoring the necessity for a thorough understanding of immunological microenvironments unique to each illness in order to improve effectiveness [9–11].
Non-coding RNAs (ncRNAs) play a pivotal role in leukemia therapy, especially in relation to immune checkpoints. ncRNAs are implicated in the primary mechanisms underlying the development of drug resistance [12, 13]. They regulate physiological and pathological processes. In cases of chemo-resistant leukemia, ncRNAs are known to modulate the onset and progression of drug resistance [12, 14].
According to recent studies, leukemia and other malignancies are closely associated with the dysregulated expression of non-coding RNA genes and their mutational range [15, 16]. Thus, ncRNAs could serve as therapeutic targets and be used as biomarkers for diagnosis or prognosis [17]. ncRNAs may change how immune cells and cancer stem cells interact with the tumor microenvironment, which might lead to immunotherapy resistance [18, 19]. Immunologically associated ncRNAs have the ability to directly or indirectly affect immune responses by their effects on nearby protein-coding genes or their ability to sponge microRNAs (miRNAs) through a variety of methods [20–22]. However, more investigation is necessary to fully understand their potential in leukemia therapy explicitly. This paper primarily discusses (from 2012 to 2024) the function of ncRNAs in immunological checkpoints and their unique processes in the immunotherapy of hematological malignancies.
Characteristics and functions of non-coding RNAs in hematological malignancies
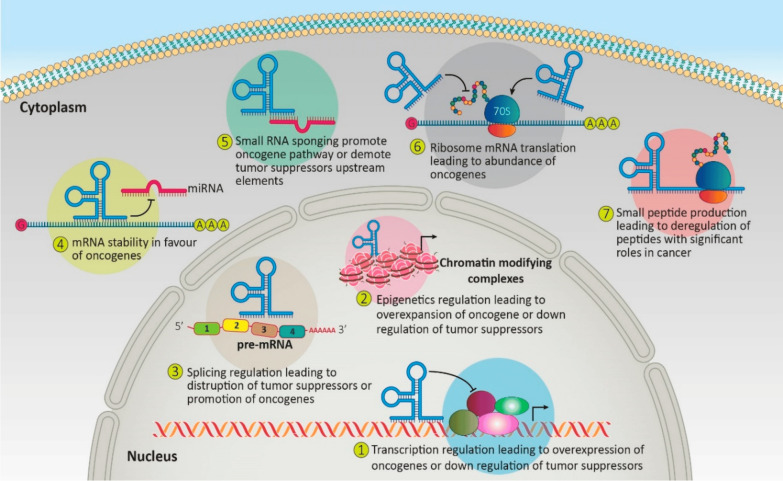
The eukaryotic genome contains around 98% ncRNAs in its transcription, with around 2% translated into proteins [23, 24]. NcRNAs are functional RNA molecules that control biological functions by focusing on certain key biochemical pathways. MiRNAs, which bind to messenger RNAs (mRNAs) of target genes, can lead to translational repression or mRNA degradation. They can act as oncogenes or tumor suppressors depending on their targets. Long non-coding RNAs (LncRNAs) can influence gene expression through interactions with chromatin-modifying complexes, acting as scaffolds for transcription factors or interfering with their binding. They can also influence mRNA stability, splicing, or translation. Some RNAs, like lncRNAs and circular RNAs, can act as miRNA sponges, sequestering miRNAs and increasing the expression of oncogenes or tumor suppressors (Fig. 1) [17, 25, 26]. They are pivotal in gene expression, RNA maturation, and protein synthesis [27]. Recent evidence indicates that non-coding RNAs, including protein-coding mutations and variations within the non-coding genome, contribute to various cancer etiologies [28, 29]. Compelling evidence shows that transcriptional, post-transcriptional, and translational controls, mediated by multiple non-coding RNAs, exert pleiotropic effects on several aspects of leukemia biology [12]. This has led to identifying and characterizing ncRNAs as biomarkers in hematological malignancies, sparking numerous studies in this field over the past decade [13, 30, 31].
Fig. 1.
The process by which non-coding RNAs influence the regulation of oncogenes and the network of tumor suppressors [17]
MicroRNAs
Non-coding RNAs with a distinct hairpin secondary structure, measuring 19–20 nucleotides in length, are known as miRNAs (Fig. 2) [12, 32]. Since its discovery in Caenorhabditis elegans, the species has been detected in the majority of eukaryotic creatures, including humans [33]. At least 30% of the genes that code for proteins are regulated by miRNAs, which comprise 1–5% of the human genome [34]. There are 940 unique miRNA molecules discovered. Encoded at distinct chromosomal loci, miRNAs are translated by RNA polymerase II into primary miRNA transcripts (pri-miRNAs) and then transformed by DROSHA and DiGeorge Critical Region 8 (DGCR8) into pre-miRNA transcripts [32, 35, 36]. Pre-miRNAs produced in the nucleus are exported into the cytoplasm, and DICER and TRBP (transactivation response element RNA-binding protein) may handle them. MiRNAs have the ability to upregulate target translation and suppress gene expression by attaching to untranslated regions of target mRNAs [37, 38]. Blood, plasma, and urine all contain miRNAs, which function as signaling molecules that move cargo between different cells or organs [39]. MiRNAs are crucial in gene expression regulation, acting as fine-tuners rather than switches [40]. They can target hundreds of different mRNAs and multiple mRNAs, creating complex regulatory networks. This allows miRNAs to modulate entire biological pathways and cellular processes [41]. They can also be involved in regulatory circuits like negative feedback loops, feed-forward loops, and double-negative feedback loops, where two miRNAs reciprocally repress each other's expression [42]. MiRNAs are essential for various cellular processes such as development, differentiation, proliferation, apoptosis, and stress responses [43]. They can be regulated through their own turnover, such as enzymatic degradation, target-directed miRNA degradation, or modifications like uridylation or adenylation [44, 45]. They can also be packaged into extracellular vesicles or bound to proteins, influencing gene expression in distant tissues [46, 47]. Their dysregulation has been linked to diseases like cancer, cardiovascular disorders, and neurodegenerative conditions, making them crucial targets for therapeutic interventions and biomarker development [32].
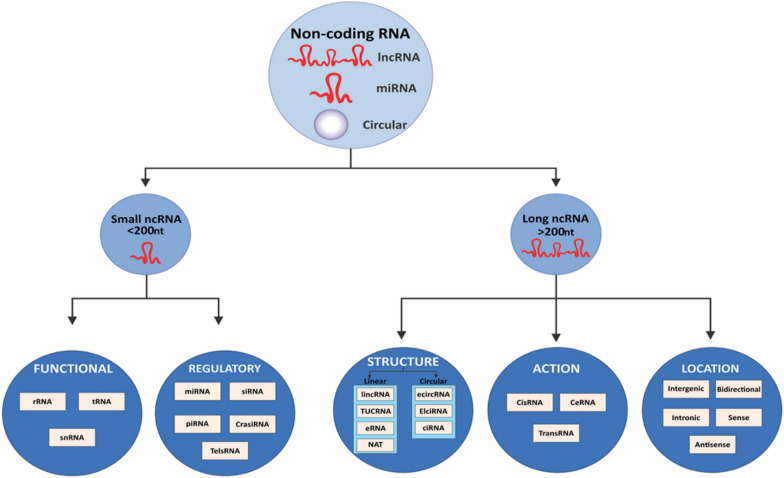
Fig. 2.
Categorization of noncoding RNAs (ncRNAs) [12]
MicroRNAs can act as oncogenes or tumor suppressors in leukemia [48]. They can be dysregulated, affecting gene expression profiles and cellular processes [12]. Oncogenic miRNAs promote cell proliferation and survival by targeting tumor suppressor genes, while tumor-suppressive miRNAs downregulate, increasing oncogene expression and anti-apoptotic proteins [49, 50]. MiRNAs also influence leukemia stem cell behavior, modulating signaling pathways and impacting the bone marrow microenvironment [51, 52]. Their ability to regulate multiple target genes simultaneously allows them to have significant effects on leukemia cell biology and disease progression [53]. However, there isn’t much research on miRNA molecules' involvement in leukemia, and our knowledge of their precise targets and biological activities is still incomplete [53–55].
Chronic myeloid leukemia (CML)
Resistance to Imatinib has been identified as a significant challenge in treating CML [56]. MiRNAs play a major role in the development of drug resistance and tumors, including CML [57]. According to recent research, the miR-221-STAT5 pathway is critical in regulating how sensitive CML cells are to the medication imatinib [58, 59]. MiR-214 alters ABCB1 expression, which has been connected to imatinib resistance in CML patients [60]. MiR-30e inhibits the translation of ABL protein by targeting ABL mRNA. In K562 cells, elevated expression causes apoptosis, suppresses growth, and heightens susceptibility to imatinib therapy. Although miR-203 expression is downregulated in bone marrow, it increases the susceptibility of CML patients to imatinib [61, 62].
Chronic lymphocytic leukemia (CLL)
Venetoclax is a prime example of ncRNAs' crucial role in creating new anti-cancer drugs [63]. This medication makes up for the lack of miR-15a/miR-16-1 targeting B-cell lymphoma-2 (BCL-2) in CLL. It is strong, selective, and well-tolerated [64]. In order to address the absence of miR-15a/miR-16-1 targeting of receptor tyrosine kinase-like orphan receptor 1 (ROR1) in CLL, cercozumab was created [65]. As seen by the synergistic impact on patient-treated CLL cells, the medicines can be used to target distinct pathways in CLL dysregulated by the same driving event, miR-15a/miR-16-1 loss [53]. According to the study, leukemic cells characterized by overexpression of BCL-2 and ROR1 genes, coupled with reduced expression of microRNAs miR-15a and miR-16-1, demonstrate enhanced therapeutic efficacy when subjected to combination treatment utilizing venetoclax, a BCL-2 inhibitor, and cirtuzumab, an anti-ROR1 monoclonal antibody [66]. Drug development relies heavily on microRNAs, as seen by the discovery of the monoclonal antibody ianalumab, which inhibits the production of miR-155 to prevent cancer [67]. In CLL, miR-155 overexpression is carcinogenic, and signals from the microenvironment, such as B-cell activating factor (BAFF) binding to its receptor on the cell surface of the disease, can cause dysregulation of miR-155 [68, 69]. The miR-17/92 polycistronic miRNA cluster, which includes miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a, is overexpressed in lymphoid malignancies such as CLL [70, 71]. A study using transgenic mice overexpressed miR-17/92 in B-cells showed it can function as an oncogene in leukemia development. 80% of these mice developed a B-cell malignancy with increased CD19+ B cells [72]. The mechanism triggering miR-17/92 expression is not fully understood. Still, Studies show that MYC induction occurs prior to miRNA up-regulation in unmutated immunoglobulin heavy-chain variable region gene (IGHV) CLL, suggesting a relationship between MYC, breakpoint cluster region protein (BCR) activation, and miR-transcription in CLL [31].
Acute myeloid leukemia (AML)
MicroRNAs have the ability to function as tumor suppressors or oncogenes. BCL-2-associated transcription factor 1 is the target of miR-194-5p, and imbalances between the two are commonly seen in AML patients [73]. Relapsed instances of AML have been reported to have overexpressed MiR-10a-5p [74, 75]. MiR-96 expression is associated with the leukemic load and declines in newly diagnosed AML patients [76, 77]. Tumor-suppressive miRNAs are downregulated in the majority of cancers, whereas oncogenic miRNAs are increased [78]. Hematopoietic stem cells from AML patients with FLT3 internal tandem duplication (FLT3-ITD) and nucleophosmin (NPM1) gene mutations had elevated levels of MiR-155 [79, 80]. It has been proposed that aberrant expression of miR-155 targets two important factors in granulopoiesis, SHIP1 and CEBPB, in individuals with AML [81, 82]. MiR-133 objectives leukemic cells with ecotropic viral integration site 1 exhibit increased drug sensitivity, indicating possible therapeutic targets for leukemias overexpressing ecotropic viral integration site-1 (EVI1) [83, 84]. According to the study, elevated miR-223 expression in AML cell lines both promotes and inhibits cell death and inhibits cell motility and proliferation [85]. In pediatric AML patients, there has been a decrease in MiR-370 expression, which may act as a non-invasive diagnostic and prognostic indicator for the course of the illness [86]. According to the study, by inhibiting ATM, miR-181a upregulation can improve cell cycling and proliferation in the leukemic cell lines HL60 and NB4 [87]. In HL60 cell lines, transfection of MiR-128 has been shown to increase drug sensitivity and apoptosis [88]. While MiR-128 is elevated and overexpressed in a number of cancers, AML cells with NPM1 mutations exhibit a reduction in its expression [12].
Acute lymphoblastic leukemia (ALL)
In patients with high-risk early T-cell precursor ALL (ETP-ALL), miR-221 and miR-222 are increased, which may block ETS1 expression and contribute to the myeloid nature of ETP-ALL [89]. In a mouse model of neurogenic locus notch homolog protein 1 (Notch1)-induced T-cell ALL (T-ALL), miR-19 is the most highly expressed component, and it enhances lymphocyte survival and starts leukemogenesis [90]. In ALL, miR-223, which targets forkhead box protein O1 (FOXO1), increases cell apoptosis while reducing cell invasion, migration, and proliferation [91]. Significant upregulation of the E3 ligase FBXW7 (F-Box And WD Repeat Domain Containing 7) has been seen in T-ALL, which may have a role in the development of Notch1-driven leukemia [92]. MYC (mostly referred to as c-Myc) transcriptionally suppresses the expression of MiR-30a, a microRNA linked to T-ALL, which prevents NOTCH1 and NOTCH2. T-cells are also impacted by the tumor suppressor miR-146b-5p, which is suppressed by TAL1 [93]. The microRNA miR-142-3p, which plays a crucial role in hematopoietic stem cell function, exhibits significantly elevated expression levels in ALL samples. This overexpression is particularly pronounced in pediatric T-cell ALL cases, where patients typically face poorer prognoses compared to healthy donor T-cells [94, 95]. By encouraging leukemic cell proliferation, miR-142-3p causes glucocorticoid treatment resistance in T-ALL. This miRNA is essential for both proliferation and chemo-resistant characteristics [96]. When overexpressed in mouse bone marrow cells relative to B-ALL patient samples, miR-196b, a miRNA overexpressed in T-ALL patient samples, co-expresses with HOXA cluster genes and improves proliferative potential and survival [97, 98]. A tumor suppressor called miR-26b targets PIK3CD (Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Delta), which in turn suppresses the PI3K/AKT pathway [99]. Phosphatase and Tensin Homolog deleted on Chromosome 10 (PTEN) plays a role in controlling Ikaros isoforms, which in turn promotes miR-26b expression [100]. MiR-181a increases pre-T cell receptor (pre-TCR) signals and decreases Notch's negative feedback to augment Notch’s oncogenic signals in T-ALL [101]. Comparing patients with multiple medication resistance to those without resistance, the expression levels of miR-326 are considerably lower in the former group [102]. According to recent studies, leukemia patients with the ETV6-RUNX1+ fusion gene had higher cell survival and lower apoptosis when the miR-125b-2 cluster was overexpressed. This finding raises the possibility of treating pediatric ALL [103]. By improving our understanding of the processes behind leukemias, the work helps physicians understand the function of miRNA and inform treatment plans.
Long non-coding RNAs
Transcripts longer than 200 nucleotides that are present in differentiated tissues or particular cancer types are known as lncRNAs (Fig. 2) [12, 29]. RNA polymerase II or RNAP III and single-polypeptide nuclear RNA polymerase IV are the transcription factors for lncRNAs, which control gene expression at several cellular levels. They can be bidirectional, sense- or antisense-oriented, intronic, and intergenic [104]. They conduct RNA editing processes, show transcriptional activation patterns, and have traits like those of mRNA transcription [105]. With some functioning as oncogenes and others as tumor suppressors, lncRNAs are essential for RNA processing, nuclear organization, and the control of gene expression. They also regulate gene expression at different levels and are involved in cellular processes such as cell cycle, tumor invasion, and metastasis [105, 106]. By promoting DNA methylation and histone modification, as well as the recruitment of chromatin remodeling complexes to certain genomic regions, they play a critical role in epigenetic modifications [107, 108]. Numerous human illnesses, including several cancers, have been connected to the dysregulation of lncRNAs [109–111]. Even though the activities of miRNAs have been well studied, research on lncRNAs is still developing, and their involvement in hematological malignancies remains mysterious [112, 113]. Leukemia has been shown to exhibit distinct lncRNA expression patterns, suggesting that lncRNAs may be useful as new biomarkers and therapeutic targets [114–116].
CML
Nuclear-enriched autosomal transcript1 (NEAT1) expression is reduced in CML, and its suppression increases imatinib-induced apoptosis [117]. LncRNA-IUR1, a key counteractive regulator of Bcr-Abl-induced carcinogenesis, was barely produced in Bcr-Abl-positive CML cells [118]. The expression of HOTAIR has been associated with the clinical-pathological prognostic classification in AML, indicating that it may be a useful marker and prospective target for future treatment of AML and CML [119]. AML protein HOTAIR reduces the number of colony-forming cells, triggers apoptosis, and stops cell division, all of which lead to CML's imatinib resistance, particularly in those with high MRP1 expression and K562-imatinib-resistant cells. Reducing HOTAIR increases the body's susceptibility to imatinib treatment [120]. LncRNA CCAT2 has been found to be a putative marker for CML diagnosis that may also be used to predict how well a patient would respond to imatinib [121]. Compared to normal cells and CML patients, K562-imatinib-resistant cells had higher levels of the LncRNA SNHG5. In K562-imatinib-resistant cells, overexpression promotes resistance, whereas knockdown decreases resistance [122]. It was discovered that K562 cells had an increased level of metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) and that suppressing it inhibited cell division and the cell cycle by focusing on miR-328 [123]. According to recent studies, the lncRNA MALAT1/miR-328 pathway may enhance CML cell growth and resistance to imatinib, suggesting new options for CML treatment [124]. According to the study, chromatin interaction allows lncRNA human meiotic recombination hot spot locus (Hmrhl) to control the expression of genes associated with malignancy in chronic myelogenous leukemia [125].
AML
ZNF571-AS1, a lncRNA, has been identified as a significant player in AML, potentially influencing Janus kinase (JAK), Signal transducer and activator of transcription 5A (STAT 5A), and KIT according to correlation studies [126]. Another lncRNA, LNC00899, has shown substantial promise as a predictive and diagnostic marker for AML [127]. The tumor suppressor MEG3 (Maternally Expressed 3) is associated with a notably poor survival rate in AML and is linked to human cancers [128, 129]. Three lncRNAs have been found as predictive variables for AML risk: lncRNAs AC008753.6, CES1P1, and RP11-342 M1.7. This research used complete lncRNA expression profiling [130]. AML progression is mostly controlled by the LncRNA LINP1 via the HNF4alpha/AMPK/WNT5A signaling pathway [131]. It has been discovered that the lncRNA CRNDE promotes cell cycle progression, proliferation, and apoptosis suppression in the U937 cell line, especially in AML patients [132]. LncRNA PVT1 has been connected to tumor stage, poor prognosis, and carcinogenesis in a number of malignancies [133] and was found to be upregulated in the bone marrow and peripheral blood mononuclear cells of ALL and AML patients compared to healthy individuals [134, 135]. Elevated apoptosis, a G0/G1 arrest in the cell cycle, reduced proliferation, and lower c-Myc protein stability were the outcomes of PVT1 (Plasmacytoma Variant Translocation 1) suppression in Jurkat cells [136]. It has been discovered that lncRNA TUG1, a gene that is overexpressed in AML cells, modulates cell proliferation by decreasing apoptosis and promoting cell proliferation in vitro [137]. As an endogenous RNA that competes with miR155, CCAT1 promotes cell division and suppresses myeloid cell differentiation [138]. LncRNA NR-104098 suppressed EZH2 (Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit) transcription by interacting with E2F1, substantially reducing AML proliferation and promoting differentiation [139]. LncRNA CCD26 was elevated in ALL and AML patients [140]. Higher expression of LINC00265 in the blood and bone marrow of AML patients suggests that this gene may serve as an AML prognostic biomarker. Suppression increased apoptosis and decreased cell invasion, migration, and proliferation [141]. The lncRNA NEAT1, which plays a crucial role in the formation of paraspeckles within the nucleus, was observed to be significantly downregulated in de novo acute promyelocytic leukemia (APL) samples when compared to samples from healthy donors. Further investigation provided evidence that this repression of NEAT1 expression was mediated by the PML-RARα fusion protein. Moreover, during the process of all-trans retinoic acid (ATRA)-induced differentiation of NB4 cells, a substantial upregulation of NEAT1 was noted. Importantly, when NEAT1 expression was reduced through the application of small interfering RNA (siRNA), the ATRA-induced differentiation process was inhibited. These findings collectively suggest a potential regulatory role for NEAT1 in APL pathogenesis and treatment response [142].
ALL
In pre-B ALL patients, overexpression of the genes BALR-1, BRL-6, and LINC0098 is associated with cytogenetic abnormalities and survival rates in B-ALL [143]. In comparison to pre-B-cells (CD10+CD19+) derived from human cord blood, it was observed that BALR-1 and LINC00958 exhibited heightened expression in ETV6-RUNX1 subtypes. Notably, an increase in BALR-2 expression was detected in patients with MLL rearrangements and those carrying either the t(4;11) or t(9;11) translocations. To explore the functional implications of newly discovered B-ALL lncRNAs, Ouimet and colleagues employed siRNA to reduce the levels of 5 candidate lncRNAs identified in their study [144, 145]. The study discovered that in human NALM-6 leukemia cells, lowering lncRNA RP11-137H2.4 reduced cell migration and proliferation and restored glucocorticoid sensitivity in previously resistant cells. Using the non-leukemic cell line GM12878 as a comparison, Gioia and colleagues examined three downregulated lncRNAs (RP-11-624C23.1, RP11-203E8, and RP11-446E9) in B-ALL further [146]. The lncRNA RP11-137H2.4 has a major effect on cell migration, proliferation, and apoptosis. By inhibiting it, resistant B-ALL cells can undergo apoptosis by regaining an NR3C1-independent glucocorticoid response [145]. Both T-ALL and B-ALL patients showed a decrease in linc-PINT expression [147]. Differential expression of wild-type and mutant NOTCH1 was seen in T-ALL cells for LUNAR1 and lnc-FAM120AOS-1 [148]. Through the HOXA3/EGFR/Ras/Raf/MEK/ERK pathway, the LncRNA HOXA cluster antisense RNA2 (HOXA-AS2) has been shown to reduce glucocorticoid sensitivity in acute lymphoblastic leukemia [149, 150]. In children with ALL, dysregulation of miR-335-3p, which is controlled by the lncRNAs NEAT1 and MALAT1, is associated with a poor prognosis [151].
Circular RNAs
Circular RNAs (circRNAs) constitute a significant category of regulatory transcripts, predominantly originating from protein-coding exons [152]. CircRNAs are produced in a tissue- or developmental stage-specific way, form closed-loop structures, and show relative stability in the cytoplasm [153]. CircRNAs are RNA viruses that are usually created at low levels by co-transcription from mRNA. They have several exons in their structure and are controlled by cis and trans-acting elements. Through alternative splicing, a single gene can produce several circRNA isoforms by including or deleting internal introns [154]. CircRNAs play critical physiological and functional roles in regulating gene expression, as evidenced by a recent study. By serving as spies for microRNA binding, decreasing their cellular availability, and upregulating target mRNAs, circRNAs contribute to post-transcriptional control [155, 156]. It has been discovered that circular RNAs are changed in a number of clinical situations, suggesting a possible function for them in human illnesses, including cancer [157]. A miRNA intermediate is frequently involved in the connection between circRNAs and a number of human disorders, including cancer, according to recent studies [158–160]. CircRNAs, which are more prevalent in blood than linear mRNAs, have been shown in research to have potential use as new biomarkers in routine clinical blood samples [12, 161].
CML
Researchers Pan and colleagues discovered a link between resistance to Imatinib, a second-generation tyrosine kinase inhibitor (TKI), and f-circRNA circBA9.3, which is derived from BCR-ABL1 mRNA. In TKI-resistant patients, they discovered higher circBA9.3 levels; in BCR-ABL-negative cell lines, they discovered enhanced cell proliferation and cancer development. Additionally, they discovered a positive association between BCR-ABL1 expression and circBA9.3 levels, with high circBA9.3 expression correlated with lower apoptosis [162]. Hsa_circ_100053 levels were discovered to be higher in CML patient cells and serum by Ping et al.'s investigation, indicating that it may be a possible biomarker for CML. Elevated expression was linked to BCR/ABL1 mutant status, advanced clinical stage, and imatinib resistance. Furthermore, it was suggested that elevated hsa_circ_100053 levels were a poor predictor of overall survival in CML patients [163]. In a separate study, Liu and colleagues [164] identified hsa_circ_0080145 as up-regulated in CML patients cells and in K562 and KU812 cell lines through an RNA-sequencing screen. The research discovered that by acting as a sponge for miR-29b, hsa_circ_0080145 silencing inhibited leukemic cell growth. These genes were implicated in the biosynthesis of heterocycles, the cAMP signaling pathway, and the systemic lupus erythematosus pathway. This is consistent with a prior work that discovered that overexpressing miR-29b in K562 cells and downregulating it in CML prevented leukemic cell proliferation and encouraged apoptosis [165]. Recent investigations have revealed circRNAs as putative drug resistance indicators, prospective therapeutic targets, and potential biomarkers for CML.
CLL
It has been discovered that the circRNA circulating factor beta (circCBFB) is up-regulated in untreated CLL cells, indicating that it may serve as a prognostic and diagnostic marker for CLL patients. The Wnt/ß-catenin pathway is activated by circulating factor beta, and this can result in a proliferative and anti-apoptotic phenotype that aids in the development of CLL. Low overall survival and a shorter survival duration are linked to high levels of circCBFB and circRPL15, respectively [166]. Because circ_0132266 may bind to miR-337-3p in CLL cell lines, Wu et al. suggested it as a possible tumor-suppressor in CLL. MiR-337-3p's primary target, PML, controls both gene expression and cell survival. MiR-337-3p levels may rise in response to circ_0132266 reductions, perhaps displaying tumor-suppressive characteristics in CLL [167]. Tests have been conducted on the circular RNA circRPL15 as a possible diagnostic biomarker in CLL patients, particularly those without an IGHV mutation. Increased RAF1 protein levels brought on by circRPL15 upregulation trigger mitogen‑activated protein kinase (MAPK) signaling and encourage cell proliferation. This theory was supported by the reduction in mitogenic factor phosphorylation seen in human cell lines following circRPL15 knockdown [168].
AML
Over the past few years, a growing number of dysregulated circRNAs have been identified in AML. For instance, Chen et al. [169] discovered that via sponging microRNAs in the miR-181 family, which control hematopoietic differentiation, circANAPC7, an up-regulated circRNA, may be linked to AML. Moreover, they discovered that circ_0009910 is a circRNA that increases the development of cancer by sponging up the tumor-suppressor miR-20a-5p [170]. Fan et al. [171] found that circ_100290 increased AML cell proliferation and inhibited apoptosis by sponging miR-293, thereby increasing Rab10 expression, a member of the oncogenic RAS family. Circ_0004277 levels were discovered to vary across AML patients by Li et al. [172]; low levels were observed in initially diagnosed patients, but they were restored during induction treatment. Nevertheless, during relapse, levels dropped, indicating promise as a predictive and diagnostic biomarker. Circ 0004277 may be a component of a complex network comprising mRNAs and miRNAs, according to a bioinformatic study. In cytogenetically normal AML (CN-AML) cell lines, Hirsch and colleagues found numerous circRNAs originating from NPM1, with circRNA hsa_circ_0075001 being more abundant in AML cells [173]. Forty-six undifferentiated blast patients had larger quantities of circular RNA, which had an inverse relationship with the expression of genes influencing the differentiation of hematopoietic cells [174–176]. Patients with high hsa_circ_0075001 levels showed a drop in the abundance of the miR-181 target gene, which may be related to circRNA sequestration by NPM1, which has miR-181 binding sites [173]. It's significant to remember that miR-181 is essential for both cellular differentiation and the emergence of hematological cancers [177]. In pediatric Acute Myeloid Leukemia patients, Yuan and colleagues detected a rise in circ_0004136, indicating that it may promote cell proliferation by binding to and suppressing miR-142, a microRNA associated with pediatric AML [178]. In another investigation, Yi et al. found that circVIM, which is produced from the VIM (vimentin) gene, was present in 113 AML patients [179]. Elevated circVIM levels in AML are associated with reduced leukemia-free survival and overall survival, indicating that it may serve as a prognostic indicator. Circulated circRNA, circDLEU2, inhibits the action of miR-496 and encourages the growth of tumors in mice. Elevated circDLEU2 levels have been shown to upregulate PRKACB expression, which may alter cell apoptosis and proliferation [180]. Elevated levels of the circular RNA circKLHL8 have been associated with better overall outcomes, longer survival times between events, and a decreased proportion of malignant blasts in bone marrow and blood [181]. It was proposed that two more circRNAs, circFOXO3 and circFBXW7, function as tumor suppressors in AML [181, 182]. APL-derived NB4 cells treated with ATRA showed differential expression of a number of circRNAs, including circHIPK2, circHIPK3, circPVT1, circRELL1, and circSMARCA5. Li and colleagues discovered these circRNAs. CircHIPK2 rose with complete remission and fell in those who had just received a diagnosis. Its association with cell maturation was important because it upregulated the transcription factor CEBPA, which is involved in hematopoiesis, and sponged miR-124-3p [183]. Up-regulated circRNAs in extramedullary infiltration (EMI) bone marrow samples may affect signal transduction, migration, and cell adhesion, according to research by Lv et al. [184]. According to the study, hsa_circ_0004520 could control VEGFA expression, which might encourage angiogenesis in AML-EMI. The design of AML treatment is complicated by the intricacy of cytogenetic and molecular abnormalities. Resistance may eventually arise even with cytarabine and anthracycline induction treatment [185]. Sun et al. discovered that circMYBL2 knockdown reduced cell proliferation in animals in culture while increasing the susceptibility of human FLT3-ITD+ cells to TKI quizartinib. CircMYBL2 improved FLT3 translation by enabling PTBP1-mediated mRNA binding [186]. Shang et al. discovered that circPAN3 plays a significant role in AML cell lines' doxorubicin resistance. By binding miR-153-3p and miR-183-5p, CircPAN3 modifies the expression levels of XIAP. Drug sensitivity was restored upon downregulation of circPAN3, indicating circPAN3's involvement in AML resistance to traditional chemotherapies [187].
ALL
Studies on circRNAs revealed that circRNAs accounted for more than 10% of transcripts in hematopoietic stem cells and naïve B cells, suggesting that circRNAs express differently in acute lymphoblastic leukemia [188]. Various circRNAs produced from the partner fusion gene AF4 of Mixed Lineage Leukemia (MLL) were found in a 2019 research by Huang et al. [189]. Elevated circAF4 levels were discovered in leukemia cell lines and in patients less than eight years old. The degree of the illness was shown to be correlated with Circaf4 levels, and in cells that had the MLL-AF4 translocation, silencing Circaf4 enhanced apoptosis. In mice, circAF4 knockdown decreased spleen infiltration and increased survival. Circaf4 may bind to miR-128-3p and sequester microRNA from the fusion MLL-AF4 mRNA, allowing MLL-AF4 to be expressed. The overexpression of miR-128-3p in vivo and the silencing of circAF4 supported this regulatory axis. According to a 2019 study by Dal Molin et al., [190] some rearrangements between MLL and other genes may also lead to the synthesis of disease-associated aberrant circRNAs in addition to producing alternate isoforms of circRNAs in various leukemia subtypes. According to Hu et al.'s 2018 study, [191] dysregulation of circPVT1 in ALL cell lines increased the expression of MYC and BCL2, sponging miR-125 and let-7 and controlling the let-7 family members' ability to operate as tumor suppressors. These effects were seen to enhance cell proliferation and prevent apoptosis [192]. Using bioinformatics, Gaffo et al. detected and measured circRNAs in T cells, B cells, and monocytes. Depending on the cell type and differentiation stage, the signatures changed. Examination of circRNAs that were differently expressed in pediatric ALL patients' B-cell progenitors revealed that the transcription factors BSAP are encoded by circPVT1, circHIPK3, and circPAX5. These circRNAs are up-regulated. CircPAX5 and CircHIPK3 binding to miR-124-5p may work in concert to obstruct B cell development and advance the course of the illness [193].
Immune checkpoints and hematological malignancies
Programmed death-1 (PD-1)
The transmembrane protein PD-1, also known as CD279, was initially identified by Ishida et al. in 1992 during a search for apoptosis-inducing genes [194]. The 288 amino acid protein, which is produced by the PDCD1 gene, has an intracellular domain with an ITSM and ITIM immunoreceptor tyrosine-based motif, a transmembrane domain, and a single V-like domain [195, 196]. Not found in resting T cells, PD-1 is expressed in a variety of immune cells such as effector T cells, regulatory T cells, naïve and activated B cells, natural killer cells, myeloid dendritic cells, and monocytes [197]. These immunological checkpoints are frequently used to cover a variety of tumors, including hematologic cancers [198, 199]. Tumor-infiltrating lymphocytes (TILs) can provide extrinsic or intrinsic signals that activate PD-L1 or PD-L2 in tumor cells, which in turn induce immunological escape signals [200, 201]. Large B cell lymphomas that are rich in T cells and histiocytes (TCHRBCLs) are distinguished by a dense population of CD8+ T cells and histiocytes, together with a small number of malignant B cells [202]. The expression of PD-L1 in TCHRBCL is diverse at the interface between malignant B cells and the inflammatory background. It is particularly robust in the histiocytes that are in close proximity to lymphoma cells, indicating that immune escape signals are influenced by both tumor and background inflammatory cells [203]. Four mechanisms have been reported for the expression of PD-L1 in lymphoid neoplasms: copy number alterations, translocations involving 9p24.1/PD-L1/PD-L2, and overexpression in tumor cells of classical Hodgkin lymphoma, primary mediastinal large B cell lymphoma, Epstein-Barr virus-negative PCNSL, primary testicular lymphoma, and a subset of diffuse large B cell lymphoma. These mechanisms account for the majority of PD-L1 expression in lymphoid neoplasms [204–208]. According to a study, all cases of EBV-positive diffuse large B-cell lymphoma (DLBCL) and DLBCL linked to EBV immunodeficiency express PD-L1 [203]. Other EBV-related lymphoproliferative illnesses, such as plasmablastic lymphoma, primary effusion lymphoma, extranodal NK/T cell lymphoma, and EBV+ post-transplant lymphoproliferative disorder, all express PD-L1 [203, 209, 210]. Preclinical research indicates that the PD-1 pathway is dysregulated in AML. The murine leukemic cell C1498 exhibits increased PD-L1 expression in vivo, indicating that the leukemic cells' milieu supports PD-L1 expression [211]. After being inoculated with C1498, regulatory T cells (Tregs) and CD8+ T cells expressing PD-1 increased in the liver, where C1498 leukemic cells spread [212]. This finding is also observed in the bone marrow of AML patients [213]. Tregs limit IFN-γ release and CD8+ T cell proliferation; nevertheless, in animals lacking PD-1 or in mice given anti-PD-L1 antibody injections, their suppressive function is compromised [212]. When compared to wild-type mice, PD-1 KO animals that were implanted with C1498 leukemia cells had a greater anti-tumor response and a longer life time [211, 212]. The in vivo administration of anti-PD-L1 antibody to C1498-challenged wild-type mice demonstrated similar anti-tumor activity [211, 212]. Clinical evidence shows that AML patients have a dysregulated PD-1 pathway, with much greater T cell PD-1 expression than in healthy persons [214].
Cytotoxic T-lymphocyte antigen-4 (CTLA-4)
CTLA-4, also known as CD152, is a protein that was initially discovered by Brunet et al. [215]. This protein is encoded by the CTLA-4 gene, which is located on chromosome 2q33.2 and consists of four exons. The protein has a single V-like domain with ligand binding sites and is a member of the immunoglobulin superfamily [216, 217]. The CTLA-4 protein is composed of 223 amino acids and has a calculated molecular weight of 24.6 kDa. After activation, one to two days later, naïve resting T cells will exhibit surface expression of CTLA-4, which is mostly located in the cytoplasm [218, 219]. Conversely, memory T cells exhibit a rapid induction of CTLA-4 expression upon activation, and this expression persists longer than in naïve resting T cells [220]. Notably, CTLA-4 is constitutively expressed in regulatory T cells [221]. In patients with peripheral T-cell lymphoma, mycosis fungoides, and Sézary syndrome, there is an observed upregulation of CTLA-4 expression. However, this is not the case in B-cell lymphoma [222–224]. A rearrangement between CTLA4 and CD28 is evident in a subset of patients with various conditions, including angioimmunoblastic T-cell lymphoma, extranodal NK/T-cell lymphoma, peripheral T-cell lymphoma (not otherwise specified), Sézary syndrome, and adult T-cell leukemia/lymphoma [225–228]. A fusion protein produced by the rearrangement of CTLA4 and CD28 enhances T-cell signaling through the MAPK and AKT pathways [226]. Research has indicated that CTLA-4 contributes to the immune evasion of AML and that CTLA-4 inhibition improves the ability of cytotoxic T-lymphocytes to destroy any remaining leukemic cells [229]. The CTLA-4 polymorphism CT60 AA genotype, found in the 3′-UTR of the CTLA-4 gene, has been linked to relapse in AML patients [230].
Lymphocyte activation gene-3 (LAG-3)
In the wake of the clinical success achieved by targeting co-inhibitory molecules CTLA-4 and PD-1, other molecules of the same class, namely LAG-3, have garnered increased attention. The lymphocyte activation gene-3 (LAG-3, also known as CD223) was first identified by Triebel and colleagues in 1990 [231]. This gene, located at 12p13.31, is composed of eight exons and encodes a protein of 498 amino acids. Structurally, LAG-3 bears a resemblance to CD4, comprising one immunoglobulin-like V-type domain and three immunoglobulin-like C2-type domains. The intracellular domain of LAG-3 contains a unique KIEELE motif, which plays a crucial role in modulating T-cell activity [232]. LAG-3 expression is observed in activated T cells, NK cells, activated B cells, and plasmacytoid dendritic cells [231, 233, 234]. LAG-3 acts as a negative regulator of CD4 and CD8 T cell expansion both in vitro and in vivo [235]. The exact mechanisms behind this regulation remain unclear, but the co-expression of LAG-3 and PD-1 in TILs in mouse models and human tissue suggests a similar role for LAG-3 [236–238]. Research has indicated that when both PD-1 and LAG-3 are simultaneously inhibited, CD8+ T cells' anti-tumor activity is increased as opposed to when only one molecule is targeted [236, 237]. In the context of Hodgkin lymphoma (HL), it has been observed that an elevated presence of LAG-3 on TILs and peripheral blood lymphocytes is linked to the suppression of Epstein-Barr Virus (EBV)-specific T-cell-mediated immunity. Notably, compared to HL patients in remission, the number of CD4+ LAG-3 circulating T-cells was considerably greater in patients with active illness [239]. A particular study highlighted that in the Tumor Microenvironment (TME) of classical HL, LAG-3 is almost invariably co-expressed [240]. LAG-3, which is mostly expressed on PD-1+ T-cells, is overexpressed in follicular lymphoma (FL) and is associated with worse outcomes. IL-12 causes this upregulation, which eventually wears down T cells. The enhancement of CD8+ T-cell activity via blocking both PD-1 and LAG-3 leads to a rise in IL-2 and IFN-γ production [241].
T cell immunoglobulin and mucin domain-containing protein-3 (TIM-3)
Hepatitis A virus cellular receptor 2 (HAVCR2), also known as TIM-3, was initially discovered by Monney et al. in 2002 [242]. The HAVCR2 gene, located at 5q33.3, encodes TIM-3 and comprises seven exons. The transmembrane protein TIM-3 has a cytoplasmic tail, immunoglobulin-like V-type domain, mucin domain, and signal peptide sequence [242]. It is expressed in various immune cells, including cytotoxic T cells, T helper 1 cells, regulatory T cells, NK cells, monocytes, and dendritic cells. TIM-3 has multiple ligands, such as galectin-9, high mobility group protein B1 (HMGB1), and phosphatidyl serine [243, 244]. It is still unclear how exactly the tyrosine residues in TIM-3’s cytoplasmic domain interact with downstream signaling pathways. Studies have shown that targeting TIM-3 can significantly enhance anti-tumor activity in tumor mouse models [245]. Moreover, it has been demonstrated that concurrent suppression of TIM-3 and PD-1 increases the anti-tumor efficacy of CD8+ TILs [246].
B7-H3/CD276
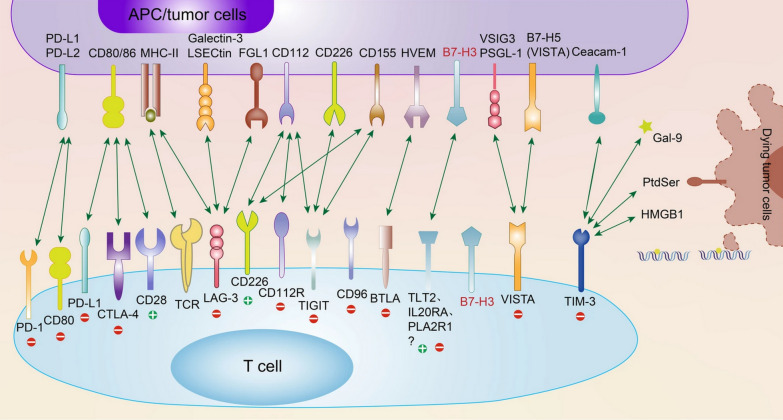
The B7 family, recognized as a collection of immune regulatory ligands, plays a crucial role in the modulation of T lymphocyte activation and differentiation. They are widely distributed in both innate and adaptive immune cells, as well as different cancer tissues. They are closely related to the CD28 superfamily (Fig. 3) [247]. This enhances cancer's capacity to elude the immune system [248]. The B7 family is composed of ten members, including the well-studied B7-H1 (PD-L1), B7-1 (CD80), B7-2 (CD86), B7-DC (PD-L2), B7-H2 (CD275), B7-H3 (CD276), B7-H4, B7-H5, B7-H6, and B7-H7 (HHLA2) [249]. B7-H3, a member of the B7 family, has been the focus of considerable attention since its discovery in 2001 [250]. Through a variety of pathways, B7-H3 has a paradoxical function in T cell activation, contributing to carcinogenesis, metastasis, and malignant behaviors. A worse prognosis is associated with its expression. A thorough analysis of B7-H3 is required to further our knowledge of cancer immunotherapy and spur clinical development. The function, underlying mechanisms, expression, involvement in various cancer types, and advancements in B7-H3 immunotherapy in clinical trials are all outlined in this review of current research [247]. The type 1 transmembrane glycoprotein human B7-H3 gene, located at 15q24.1, encodes 316 amino acids through 12 exons and has two isoforms: 2IgB7-H3 (B7-H3 VC) and 4IgB7-H3 [250, 251]. In humans, 4IgB7-H3 is the primary isoform expressed on immunocytes and malignant cells [252]. The 93% amino acid similarity between the structure of the human 2IgB7-H3 and the mouse B7-H3 gene, which is found on chromosome 9, is noteworthy [253]. The FG loop of the IgV domain is essential to the inhibitory activity of the murine B7-H3 protein, according to its crystal structure [254]. In addition to its transmembrane form, soluble B7-H3 (sB7-H3) has been detected in normal human blood [255]. sB7-H3 is produced by alternative splicing from the fourth intron of B7-H3 [256] or matrix metallopeptidase (MMP) [255], and serum levels of sB7-H3 have been linked to prognosis in a number of cancers [257, 258]. Primary AML blasts and monocytic AML cell lines both exhibit significant levels of B7-H3 expression. Its CAR-T has shown strong anti-AML properties both in vitro and in a xenograft form [259].
Fig. 3.
Presently recognized immune checkpoint receptors along with their corresponding ligands [247]
Immune checkpoints and ncRNAs
NcRNAs are essential for controlling immunological checkpoints and other cellular processes. Trans-membrane CTLA-4, which is expressed on CD4+ and CD8+ T cells, binds to CD80 and CD86 to block T-cell signaling. It has been determined that there are two miRNAs that target CTLA-4, with MiR-138 acting as a tumor suppressor in cancer. Through its binding to PD-1 and CTLA-4, MiR-138 inhibits the development of glioma cells in vivo [260]. Conversely, by preventing CTLA-4 expression on T cells, MiR-155 stimulates anti-cancer immune responses; its overexpression may improve immunotherapy [261].
miR-155
MicroRNA-155 (miR-155) is linked to B-cell lymphoma progression by enhancing the interaction between B-lymphoma cells and CD8+ T cells in the tumor microenvironment, potentially inhibiting the PD-1/PD-L1 pathway [262]. MiR-155 transgenic mice displayed pre-B-cell proliferation in the spleen and bone marrow and exhibited malignant B-cell transformation [263]. MiR-155 enhances PD-L1 expression in B-lymphoma cells by binding to the 3′-UTR, promoting CD8+ T-cell apoptosis and maintaining tumor immunity in a PD-1/PD-L1-dependent manner [262]. The AKT and ERK pathways are crucial regulators of PD-1-mediated CD8+ T-cell function [264]. Overexpression of miR-155 in DB cells increased their sensitivity to anti-PD-L1 and anti-PD-1 antibodies [262]. In nude mice, downregulation of miR-155 induced apoptosis in B-lymphoma cells and delayed the formation of xenograft tumors [265]. Two inhibitory receptors, miR-155 and BTLA (B- and T-lymphocyte attenuator), may be suitable targets for anti-PD-L1 protein therapy in malignant B-cell neoplasms. When BTLA is coupled to SHP-1 and SHP-2 phosphatase, it becomes phosphorylated and decreases the production of IL-2. Activated Th1 cells produce BTLA [266]. According to Liu J. et al., the elevated protein miR-155 selectively targets the 3′UTR of BTLA, resulting in a 60% reduction in the protein's surface expression in T-activated cells [267].
SNHG14/ZEB1
Zinc finger E-box-binding homeobox gene 1 (ZEB1), a transcription factor, is associated with gene regulation in various cancer cells, influencing invasion, migration, epithelial-mesenchymal transition (EMT), and proliferation [268–270]. The SNHG14 (Small Nucleolar RNA Host Gene 14) regulates invasion, migration, and proliferation, which gives many illnesses chemo-resistance. This function is critical in the development of cancer [271]. Prior research has demonstrated that ZEB1 can improve immune evasion by upregulating PD-L1 expression in cancer cells [272, 273]. When SNHG14 was silenced, ZEB1's RNA expression decreased, which in turn affected ZEB1 and PD-L1's protein expressions in DLBCL cells [271]. SNHG14/ZEB1 promoted DLBCL cells' interaction with CD8+ T lymphocytes and triggered death via the PD-1/PD-L1 pathway [271]. This shows that PD-1/PD-L1 and SNHG14/ZEB1 may combine to provide a viable target for DLBCL therapy.
miR-340-5p
KMT5A, a miR-340-5p target, regulates CD8+ T cell activity and enhances DLBCL cells' immune system inhibition, thereby influencing tumor development control [274]. Tumor cells expressing CD73 negatively regulate the anti-tumor T-cell response and can also increase T-cell apoptosis [275]. KMT5A silencing boosted CD73's ubiquitination in LY-1 cells, which was subsequently decreased by COP1 knockdown, resulting in CD73's downregulation [276]. In DLBCL cells, the miR-340-5p/KMT5A axis functions as an antitumor mediator independently of immunological modulation [276]. This presents a fresh viewpoint on the management of DLBCL.
miR-21
MicroRNA-21 (miR-21) is a key regulator in the disease progression of B-cell lymphoma [277, 278]. An experiment revealed a pre-B malignant lymphoid-like phenotype caused by miR-21 overexpression [279]. Through the regulation of p-STAT3, MiR-21 upregulates ICOS on Tregs, boosting their interaction with epithelial cells and encouraging tumor formation and chemo-resistance in B-cell lymphoma [280]. These results could offer new approaches to treating B-cell lymphoma.
miR-28
According to recent studies, miR-28 modulates T-cell fatigue by upregulating the expression of checkpoint inhibitor receptors such as PD-1, TIM-3, and LAG3, as well as lowering T-cell secretions of TNF-a and IL-2 [281]. In addition, miR-28 serves as a protective agent against Burkitt lymphoma (BL), and its suppression by MYC plays a role in the development of B-cell lymphoma [12]. By focusing on immunological checkpoints, RNA therapies are thought to be a viable approach to cancer immunotherapy. Nevertheless, more research into the molecular processes and advances in medication development is required to completely comprehend the connection between ncRNA and immunological checkpoints.
Role of checkpoint inhibition in hematological malignancies
ICIs, a type of monoclonal antibody, target immune checkpoints that are frequently overexpressed on cancer cells and cells in the surrounding immune and stromal microenvironment [282]. ICIs have been licensed for use in conjunction with chemotherapy, targeted cancer treatment, and other immunotherapeutic drugs; they have dramatically improved patient prognosis [283–285]. Some cancer types have shown long-term remission with ICI treatment; however, not all patients benefit from this because of tumor immunogenicity and immune microenvironment phenotypic reductions [286–288]. Because of ncRNA's diverse roles and ability to simultaneously block several checkpoint receptors, RNA-based therapies hold great promise for advancements in cancer immunotherapy [289]. Developing treatment approaches, identifying predictive biomarkers of ICI response, and comprehending basic and acquired resistance mechanisms are important obstacles in the field of cancer immunotherapy. Numerous ncRNAs linked to the ICI response have been discovered recently [290, 291]. Classical Hodgkin Lymphoma (CHL) is the most extensively researched lymphoid neoplasm in the context of PD-1 blockade. Nivolumab, a fully humanized IgG4 anti-PD-1 monoclonal antibody, has been shown to have a satisfactory safety profile and significant clinical activity in patients with relapsed or refractory CHL, as evidenced by a phase 1b study (NCT01592370) [292]. Pembrolizumab, a different, completely humanized IgG4 anti-PD-1 monoclonal antibody, has equivalent clinical efficacy to nivolumab and a tolerable safety profile in patients with relapsed or refractory CHL (NCT01953692, KEYNOTE-013) [293, 294]. The clinical activity of pembrolizumab was further substantiated with a multicohort phase 2 study (KEYNOTE-087, NCT02453594), which observed an overall response rate (ORR) of 65–72% with a CR rate of 22% across all cohorts [295, 296]. Primary mediastinal large B cell lymphoma (PMBL), primary central nervous system lymphoma (PCNSL), and primary testicular lymphoma (PTL) are thought to be strong candidates for PD-1 blocking based on the underlying genetic abnormalities. Nineteen patients with relapsed or refractory PMBL were enrolled in an independent cohort in phase 1b research using pembrolizumab (NCT01953692, KEYNOTE-013). The ORR was 41% with a median follow-up of 11.3 months, and 2 and 5 patients, respectively, achieved CR and PR. A global multi-center phase 2 experiment (KEYNOTE-170, NCT02576990) is presently being conducted in response to these findings [297]. Patients with varying ORR (30–40%), such as those with mycosis fungoides/Sézary syndrome (MF/SS), follicular lymphoma, T cell lymphoma, or DLBCL have also tried PD-1 blocking (NCT01592370 and NCT02243579) [298, 299]. Numerous anti-PD-1 antibodies, including as nivolumab, pembrolizumab, AMP-224, BGB-A317, MEDI0680, PDR001, PF-06801591, and REGN2810, are being studied in the context of immunotherapy. The US Food and Drug Administration (FDA) has approved atezolizumab (Tecentriq®, Genentech), a completely humanized IgG1 anti-PD-L1 monoclonal antibody, to treat metastatic non-small cell lung cancer. According to preliminary findings, individuals with relapsed/refractory DLBCL or FL who receive atezolizumab in addition to obinutumab (an anti-CD20 antibody) show good tolerance and clinical effectiveness (NCT02220842) [300]. Two completely human monoclonal anti-CTLA-4 antibodies are available: tremelimumab (Pfizer) and imipomumab (Yervoy®, Bristol-Myers Squibb). In patients with relapsed or resistant B cell lymphoma, an early pilot study using ipilimumab monotherapy showed a poor overall response rate (ORR) of 11% [301]. The three forms of LAG-3 therapeutics include LAG-3 fusion proteins, LAG-3-targeting monoclonal antibodies, and bispecific LAG-3 antibodies. The majority of LAG-3 therapies are completely humanized IgG4 monoclonal antibodies [302]. It has been shown that LAG-3-targeting monoclonal antibodies decrease the synthesis of both IL-12 and IFN-γ. Additionally, these antibodies block the positive signal that MHC-II sends to monocytes and the T-cell response to IL-12 [303]. Anti-LAG-3 monotherapy might not be the optimal course of action; combination therapies, especially those including PD-1 inhibitors, are being researched. The quantity and functionality of antigen-specific CD8+ T cells are increased by blocking PD-1 and LAG-3 [304]. Other monoclonal antibodies targeting LAG-3, such as HLX26 (NCT05078593 and NCT05400265), IBI110 in diffuse large B cell lymphoma (NCT05039658), INCAGN02385 (NCT03538028, NCT04370704, NCT05287113, NCT04586244), Sym022 (NCT03489369, NCT03311412, NCT04641871), and TSR-033 (NCT03250832, NCT02817633), are also under clinical investigation [305]. Ipilimumab, an immunotherapeutic agent, has demonstrated efficacy in patients with AML who have relapsed following allogeneic stem cell transplantation. A phase I/Ib study (NCT01822509) administered ipilimumab at a dosage of 10 mg/kg to patients with relapsed hematologic malignancies post-allogeneic stem cell transplant. This cohort comprised 16 patients with AML, 2 with Myelodysplastic Syndromes (MDS), and 1 with myeloproliferative neoplasm. Out of the 22 patients who received the 10 mg/kg dosage, 5 patients (23%) exhibited a complete response. This included 3 patients with leukemia cutis, 1 with myeloid sarcoma, and 1 with AML. Furthermore, 4 additional AML patients, although not achieving an objective response, displayed a reduction in tumor burden [306]. In a separate phase I study, ipilimumab monotherapy was administered to high-risk MDS patients (n = 11) who had not responded to hypomethylating agents. While no objective responses were reported, disease stabilization was observed in five patients (45%) [307]. Numerous clinical trials are currently investigating the use of anti-CTLA-4 antibodies, either as monotherapy or in combination with other treatments, in patients with MDS or AML (NCT01757639, NCT02117219, NCT02846376, and NCT02890329). Preliminary results from a phase 2 study (NCT02530463) investigating various combinations of nivolumab, ipilimumab, and azacitidine in MDS patients have also been reported [308].
Non-coding RNA-based immunotherapy
The potential of ncRNAs as therapeutic targets for cancer treatment is being explored due to their link with resistance to immunotherapy. A range of immunoregulatory miRNA mimics/antagonists, including miR-26, miR-33a, miR-34, miR-101, miR-125, miR-21, miR-31, miR-32, miR-100, miR-192, and miR-211, are currently undergoing pre-clinical and clinical trials for cancer treatment. However, none have achieved a significant therapeutic breakthrough [309–312]. The application of miRNA mimics/antagonists in cancer treatment is hindered by their physiochemical properties, such as their vulnerability to nuclease-mediated degradation, potential for off-target side effects, and low cellular uptake [313]. Various strategies are being investigated to overcome these challenges. For instance, chemical modifications like locked nucleic acid (LNA), phosphorothioate-containing oligonucleotides, and peptide nucleic acids (PNAs) have been shown to enhance the stability, cellular targeting, uptake, and delivery efficacy of miRNA-based drugs [314]. Furthermore, developing targeted delivery formulations, such as liposomal and polymeric-based delivery platforms, offers a significant opportunity to apply ncRNA-based therapeutics [315].
Drug resistance has been a significant hurdle in the effectiveness of treatment, leading to disease relapse/progression and impacting prognosis. Clinical studies have shown that approximately 30–50% of cancers develop primary or secondary resistance after an initial response to T-cell-based immunotherapy [316–318]. This resistance is potentially due to immune evasion from immune-surveillance, facilitated by alterations in tumor cells and the TME at various levels [319]. Immunomodulatory treatments have shown efficacy in promoting a balanced anti-tumor immune response, activating cytotoxic T lymphocytes (CTLs), and inhibiting tumor growth [320]. However, these approaches are not without limitations, including adverse events and resistance observed in certain types of cancer [321, 322]. Recent focus has shifted towards the role of immunoregulatory ncRNAs in eliciting and monitoring specific immune responses in the context of cancer immunotherapy [323, 324]. Previous studies have highlighted the crucial role of miR-491 in regulating the proliferation and apoptosis of CD8+ and CD4+ T-cells. This regulation is achieved by reducing the expression of IFN-γ by targeting cyclin-dependent kinase 4, the transcription factor T-cell factor 1, and the anti-apoptotic protein B-cell lymphoma 2-like 1[325]. Circ_0009910 has the potential to inhibit proliferation, sphere formation, and autophagy while promoting apoptosis in AML cells. This is achieved by regulating B4GALT5 expression and activating the PI3K/AKT signaling pathway through the absorption of miR-491-5p. These findings suggest that circ_0009910 could serve as a potential biomarker for AML treatment [326]. Consequently, miR-491 could serve as a potential immunomodulatory biomarker in cancer immunotherapy. Furthermore, miR-381-3p has been shown to induce T-cell differentiation by targeting FOXO1, activating the transcription factors T-bet and RORγt [327]. Also, the reestablishment of ROCK1 (Rho Associated Coiled-Coil Containing Protein Kinase 1) expression negates the suppressive effect of miR-381-3p on cell proliferation, invasion, and migration. This suggests that miR-381-3p functions as a tumor suppressor in pediatric AML by targeting ROCK1 [328]. MiR-381-3p, a key immunomodulatory ncRNA, has the potential to be a therapeutic target for pediatric AML treatment, potentially serving as a predictive biomarker for patient response to such treatments.
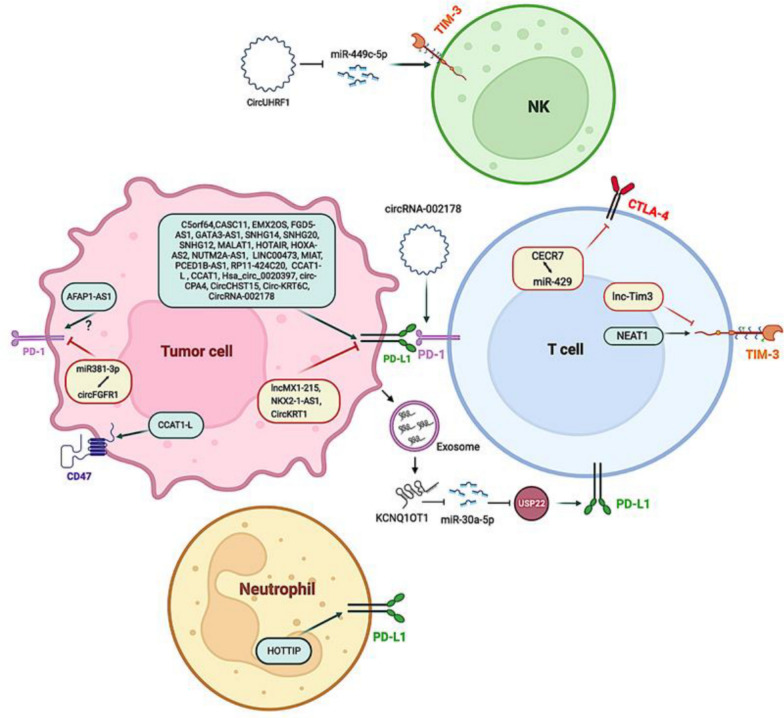
Certain lncRNAs, referred to as immune-related lncRNAs, play a crucial role in modulating the T-cell-mediated immune response and the release of inflammatory cytokines. This modulation results in an immunosuppressive TME, which leverages immune checkpoint pathways (Fig. 4) [329]. Studies reveal that different lncRNAs attract and activate cells that suppress the immune system, such as Tregs and myeloid-derived suppressor cells (MDSCs), and these cells are associated with a worse prognosis and resistance to immunological treatment in the TME [330]. In the context of tumor biology, the lncRNA known as lnc-chop plays a crucial role in the regulation of myeloid-derived suppressor cells (MDSCs). Specifically, the suppression of lnc-chop within MDSCs enhances the release of Interferon-gamma (IFN-γ) by both CD4+ and CD8+ T cells, thereby fostering an immunosuppressive environment. This process is further modulated by the activation of the transcription factor CCAAT-enhancer-binding protein β (C/EBPβ) and the subsequent upregulation of Arginase-1 (Arg-1), Nitric Oxide Synthase 2 (NOS2), NADPH Oxidase 2 (NOX2), and Cyclooxygenase-2 (COX2). This upregulation occurs via the binding of both the C/EBPβ homologous protein (CHOP) and the liver-enriched inhibitory protein (LIP), which are integral to this process. Moreover, lnc-chop augments the production of Nitric Oxide (NO), Hydrogen Peroxide (H2O2), and Reactive Oxygen Species (ROS), as well as the expression of Arg-1. This is achieved by promoting the enrichment of histone H3 lysine 4 trimethylation (H3K4me3) in the promoter regions of Arg-1, NOS2, NOX2, and COX2 [331].
Fig. 4.
The control of immune checkpoint molecules in cancer by long non-coding RNAs [373]
According to recent research, modifying lncRNAs may function as an immune sensitizer to counteract the resistance of immunotherapy. For example, lncRNAs that suppress NEAT1 have been shown to decrease CD8+ T-cell apoptosis and boost cytolytic activity through the miR-155/Tim-3 pathway [332]. Among the lncRNAs that affect antigen presentation, long intergenic non-coding RNA for kinase activation (LINK-A) is found to be highly expressed in a subset (25%) of triple-negative breast cancer patients. LINK-A appears to negatively regulate the recruitment of Antigen-Presenting Cells (APCs) and CD8+ T cells, resulting in decreased infiltration of APCs and activated CD8+ T cells, as well as reduced β-2 M and MHC-I expression [333, 334]. LINK-A’s prognostic role may be attributed to its impact on the degradation of TPSN, TAP1, TAP2, and CALR proteins of the peptide-loading complex (PLC), affecting the loading and editing of MHC-I. Consequently, LINK-A inhibitors could enhance the effect of ICIs by increasing the infiltration of hyperactivated CD8+ T cells at the tumor level [333]. In DLBCL, MALAT1 upregulates the expression of PD-L1 through miR-195, promoting migration and immune escape mediated by CD8+ T cells. Inhibiting MALAT1 could reverse this effect [335]. However, additional clinical validation is required to establish immunoregulatory ncRNAs as viable therapeutic targets for improved leukemia management.
RNA interference approaches
RNA interference (RNAi), a cellular regulatory mechanism found in most eukaryotes, has been harnessed as a method of drug action in the creation of RNAi-based therapies [336]. This natural process, known to researchers for over two decades, involves short RNA strands, specifically siRNAs, leading to targeted gene silencing [337]. Short, double-stranded RNAs known as siRNAs attach to certain messenger RNA sequences to start a chain reaction that cleaves and degrades the target mRNA, thus blocking the production and function of the target gene [338]. siRNAs offer significant therapeutic potential, providing a means to selectively target and silence the mRNA products of genes, which were previously deemed “undruggable” targets [339]. The majority of protein-coding genes can now be decoded and annotated thanks to the human genome, which facilitates the production of complementary siRNA molecules for subsequent protein silencing. However, because they function at the protein level, conventional small-molecule medications demand more structural accuracy and a more involved development procedure [340, 341]. The journey from theoretical to practical drug-related knowledge in the development of siRNA-based drugs has spanned nearly two decades. In August 2018, two decades after the discovery of RNAi, the FDA approved the first siRNA drug, patisiran [342, 343]. This was followed by the approval of givosiran in November 2019 [344] and lumasiran in November 2020 [345]. At present, seven siRNA drugs, including vutrisiran, nedosiran, inclisiran, fitusiran, teprasiran, cosdosiran, and tivanisiran, are in the late stages of Phase 3 clinical trials, with some nearing FDA approval [346, 347]. The difficulty of site-specific delivery, in which large anionic siRNA molecules must cross physiological barriers to enter the cytoplasm of target cells, has slowed the development of siRNA therapeutics. To overcome the delay in realizing the therapeutic potential of RNA interference, stability and specificity have been improved through the use of chemical modifications and delivery methods [347].
Antibodies that bind to either CTLA-4 or PD-1 thereby relieving immune inhibition, have shown promising clinical results in patients [348]. However, only a small percentage of patients had a lasting response to the therapy. Moreover, a high percentage of patients experienced severe side effects, especially those treated with anti-CTLA-4 therapy compared to anti-PD1 therapy [349]. These effects are likely due to the systemic administration of the antibodies causing polyclonal activation of autoreactive T cells. Therefore, additional targeted strategies to inhibit the expression of these co-inhibitory molecules are needed. In this regard, Hobo et al. utilized RNAi to reduce the expression of PD-1 ligands in Dendritic cells (DCs) [350]. The expansion of MiHA-specific CD8+ T cells in mice was also boosted by siRNA silencing of PD-1 ligands on DC vaccines [351]. Recently, a new cationic lipid formulation, referred to as SAINT-18, which is compatible with GMP manufacturing, has been used to deliver PD-L1 and PD-L2 siRNAs to DCs [352, 353]. DCs silenced for PD-L and loaded with mRNA encoding for MiHA demonstrated a greater potential for activating MiHA-specific T-cells than control DCs. The data suggests that the immunogenic function of DCs can be enhanced by silencing PD-1 ligands, leading to stronger antigen-specific CTL responses in vitro models and anti-cancer immunity in various mouse cancer models [354]. SiRNAs elicit potent CTL responses by inhibiting certain inhibitory molecules. Chemically produced siRNAs or shRNAs may be added to DCs at no further expense through electroporation. Chemical alterations and lipid-based siRNA delivery techniques provide an alternative to electroporation. The successful targeting of siRNAs to T cells in the bloodstream implies that they may be developed for use as cancer immunotherapies in clinical settings [348]. Malignancies and autoimmune diseases can result from B-cell malfunction. Inadequate siRNA delivery techniques have impeded the development of RNAi-based therapies. Utilizing αCD38 antibody-LNPs encapsulating CycD1 siRNA, an effective and non-immunogenic method has been devised to suppress CycD1 expression in a mouse model of human Mantle Cell Lymphoma [355]. The study showed that encapsulating siRNA into layer-by-layer nanoparticles coated with a targeting antibody allows for the targeted delivery of siRNA to B cells. Unlike the failed use of siRNA therapies in hematologic malignancies because of cell resistance, this approach provides a viable treatment plan for B-cell malignancies [356]. The LbL-np is a siRNA delivery device that encloses the target gene in polyelectrolyte layers to shield it from bloodstream nucleases. Its outer layer, which is dual-targeting (CD20/CD44), guarantees accurate binding to blood cancer cells. The pro-survival protein BCL-2 was silenced both in vivo and in vitro using this method. The dual-targeted nanoparticle's systemic injection caused apoptosis and reduced blood cancer cells' ability to proliferate, indicating that LbL nano assemblies are a potentially effective way to deliver therapeutic siRNA [357].
Conclusions and future perspectives
NcRNAs play a pivotal role in gene regulatory networks, and advancements in genomics and biotechnology have positioned them as promising therapeutic targets in hematological malignancies [15, 30]. Dysregulation of small non-coding RNAs is often linked to altered gene expression in cancer cells [358, 359]. This seminal discovery has paved the way for significant strides in the development of innovative and more potent cancer drugs [360]. lncRNAs operate through different mechanisms compared to miRNAs in cancer, suggesting that their targeting could unveil key tumorigenesis mechanisms [361]. Artificial modulation of ncRNA expression can also restore sensitivity to conventional chemotherapy [362]. Because miRNAs regulate post-transcriptional processes and have dual roles as oncogenes and tumor suppressors in a variety of malignancies, including leukemia and lymphoma, they are the subject of much research in the field of leukemia [313, 363].
In recent times, ncRNAs have been identified as predictive biomarkers for cancer immunotherapy [364]. These biomarkers could offer early evaluation of immunotherapeutic responses, patient prognoses, and cancer recurrence. Various ncRNAs regulate distinct pathways within the cancer microenvironment. However, molecular biomarker indicators are still in their infancy, and no studies have elucidated technical constraints associated with using ncRNA biomarkers for monitoring immunotherapy response [365]. Further research is needed to ensure the reliability of clinical applications. The combination of ncRNA biomarker analysis could reveal novel immunomodulatory agents and therapeutic targets, bolstering the clinical application of cancer immunotherapy [366, 367]. The concurrent use of ncRNAs to modulate specific tumor cell signaling pathways and immunotherapeutic interventions could enhance treatment responses and improve patient prognoses. The high concentration of miRNA in liquid biopsies and tumor tissue may be the reason for the restricted screening of short and lncRNAs as biomarkers in the circulatory systems of cancer patients [113].
The precision of ncRNA-based treatments could be enhanced by directing efforts toward the precursors of ncRNAs. This would involve the use of nucleic acid oligonucleotides or peptides to inhibit their synthesis and maturation [33]. Despite promising results from pre-clinical studies and initial clinical trials, the effective clinical application of ncRNA-based immunotherapy remains unrealized [368]. However, as our understanding of the role of ncRNA in tumor immunity and immunotherapy expands, there is optimism that ncRNA-based treatments could emerge as innovative cancer therapies [20]. Advanced genomic techniques are employed to identify functionally relevant miRNA-mRNA target pairs that regulate the growth of leukemia, which will likely be advantageous for pre-clinical models [369, 370]. The need for better miRNA delivery vehicles for successful therapy is highlighted by the ability of miRNA sponges and anti-miRNA oligonucleotides to silence aberrant miRNAs and the ability of nanoparticle vectors to target oncogenic lncRNAs through the effective delivery of small interfering RNAs [371]. It is possible to modify high-affinity antisense oligonucleotides to decrease carcinogenic lncRNAs via degradation, altering RNA and protein interactions, or alternative splicing. By enabling steady transfection of RNA products into tissues, lentiviral vectors efficiently introduce the siRNA sequence into the intended cell type [372].
The interaction between ncRNAs and immune checkpoints is intricate. The interplay between lncRNAs and immune checkpoints contributes to cancer progression. The ncRNA types play specific roles in advancing the research of tumor resistance and the creation of new drug targets or immunotherapy alternatives. However, the precise roles of lncRNAs on immune checkpoints are not fully understood and necessitate further investigation. The targeted influence of ncRNAs on immune checkpoints suggests potential advancements in leukemia immunotherapy. Notwithstanding their wide range of uses in cancer treatment, ncRNAs in leukemia pose some obstacles to their therapeutic application, such as the intricate cellular milieu and the requirement for a safe and effective delivery mechanism. Furthermore, innovative strategies must be developed to mitigate RNA degradation and thereby increase their bioavailability. While the field of ncRNAs is extensively researched, the exploration of their role as biomarkers and therapeutic targets in leukemia is still in progress.
Acknowledgements
The authors would like to express their gratitude for the support provided by the Blood Transfusion Research Center, High Institute for Research and Education in Transfusion Medicine, Tehran, Iran. It must be noted that we utilized advanced AI tools to enhance the fluidity and grammar of our manuscript. After using these services, we accepted full responsibility for the publication's content and undertook necessary reviews and edits.
Author contributions
Samira Anvari: Methodology, Investigation, Visualization, Writing-Original draft preparation, Data Curation, Software, Visualization, Resources, Formal analysis, Writing- Reviewing and Editing; Mohsen Nikbakht: Software, Writing- Reviewing and Editing; Mohammad Vaezi: Software, Writing- Reviewing and Editing; Sedigheh Amini Kafiabad: Funding acquisition, Conceptualization, Project, Supervision; Mohammad Ahmadvand: Funding acquisition, Conceptualization, Project, Supervision.
Availability of data and materials
By ordering from the corresponding author.
Declarations
Ethical approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sedigheh Amini-Kafiabad, Email: dr.amini@gmail.com.
Mohammad Ahmadvand, Email: Ahmadvand.mohamad64@yahoo.com.
References
- 1.Ganesan S, Mehnert J. Biomarkers for response to immune checkpoint blockade. Ann Rev Cancer Biol. 2020;4:331–51. [Google Scholar]
- 2.Lee J, Kim EH. Mechanisms underlying response and resistance to immune checkpoint blockade in cancer immunotherapy. Front Oncol. 2023;13:1233376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salik B, Smyth MJ, Nakamura K. Targeting immune checkpoints in hematological malignancies. J Hematol Oncol. 2020;13(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aru B, et al. Comparison of laboratory methods for the clinical follow up of checkpoint blockade therapies in leukemia: current status and challenges ahead. Front Oncol. 2022;12: 789728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hradska K, Hajek R, Jelinek T. Toxicity of immune-checkpoint inhibitors in hematological malignancies. Front Pharmacol. 2021;12: 733890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bewersdorf JP, Shallis RM, Zeidan AM. Immune checkpoint inhibition in myeloid malignancies: moving beyond the PD-1/PD-L1 and CTLA-4 pathways. Blood Rev. 2021;45: 100709. [DOI] [PubMed] [Google Scholar]
- 7.Abaza Y, Zeidan AM. Immune checkpoint inhibition in acute myeloid leukemia and myelodysplastic syndromes. Cells. 2022;11(14):2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aru B, et al. A potential area of use for immune checkpoint inhibitors: targeting bone marrow microenvironment in acute myeloid leukemia. Front Immunol. 2023;14:1108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroll MH, Rojas-Hernandez C, Yee C. Hematologic complications of immune checkpoint inhibitors. Blood. 2022;139(25):3594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marin-Acevedo JA, Kimbrough EO, Lou Y. Next generation of immune checkpoint inhibitors and beyond. J Hematol Oncol. 2021;14(1):1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köhler N, et al. The role of immune checkpoint molecules for relapse after allogeneic hematopoietic cell transplantation. Front Immunol. 2021;12: 634435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat AA, et al. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol Cancer. 2020;19:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahmati A, et al. Non-coding RNAs in leukemia drug resistance: new perspectives on molecular mechanisms and signaling pathways. Ann Hematolx. 2023;103:1–28. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharya M, Gutti RK. Non-coding RNAs: are they the protagonist or antagonist in the regulation of leukemia? Am J Transl Res. 2022;14(3):1406. [PMC free article] [PubMed] [Google Scholar]
- 15.Ghafouri-Fard S, Esmaeili M, Taheri M. Expression of non-coding RNAs in hematological malignancies. Eur J Pharmacol. 2020;875: 172976. [DOI] [PubMed] [Google Scholar]
- 16.Balatti V, Croce CM. Small non-coding RNAs in leukemia. Cancers. 2022;14(3):509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghazimoradi MH, Karimpour-Fard N, Babashah S. The promising role of non-coding RNAs as biomarkers and therapeutic targets for leukemia. Genes. 2023;14(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen C, et al. Long non-coding RNAs: emerging regulators for chemo/immunotherapy resistance in cancer stem cells. Cancer Lett. 2021;500:244–52. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, et al. Noncoding RNAs: the shot callers in tumor immune escape. Signal Transduct Target Ther. 2020;5(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vishnubalaji R, et al. Noncoding RNAs as potential mediators of resistance to cancer immunotherapy. Semin Cancer Biol. 2020;65:65–79. 10.1016/j.semcancer.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Yu F, Li P. Noncoding RNAs as an emerging resistance mechanism to immunotherapies in cancer: basic evidence and therapeutic implications. Front Immunol. 2023;14:1268745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taeb S, et al. Role of Tumor microenvironment in cancer stem cells resistance to radiotherapy. Curr Cancer Drug Targets. 2022;22(1):18–30. [DOI] [PubMed] [Google Scholar]
- 23.Nemeth K, et al. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genetics. 2023;25:1–22. [DOI] [PubMed] [Google Scholar]
- 24.Loganathan T, Doss T. Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets. Funct Integr Genom. 2023;23(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, et al. Non-coding RNAs/DNMT3B axis in human cancers: from pathogenesis to clinical significance. J Transl Med. 2023;21(1):621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashrafizadeh M, et al. Noncoding RNAs as regulators of STAT3 pathway in gastrointestinal cancers: roles in cancer progression and therapeutic response. Med Res Rev. 2023. 10.1002/med.21950. [DOI] [PubMed] [Google Scholar]
- 27.Ala U. Competing endogenous RNAs, non-coding RNAs and diseases: an intertwined story. Cells. 2020;9(7):1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkle M, et al. Noncoding RNA therapeutics—challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu SJ, et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dieter C, Lourenco ED, Lemos NE. Association of long non-coding RNA and leukemia: a systematic review. Gene. 2020;735: 144405. [DOI] [PubMed] [Google Scholar]
- 31.Fabris L, Juracek J, Calin G. Non-coding RNAs as cancer hallmarks in chronic lymphocytic leukemia. Int J Mol Sci. 2020;21(18):6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. 2021;14(4):dmm047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratti M, et al. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target Oncol. 2020;15(3):261–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dexheimer PJ, Cochella L. MicroRNAs: from mechanism to organism. Front Cell Dev Biol. 2020;8:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smolarz B, et al. miRNAs in cancer (review of literature). Int J Mol Sci. 2022;23(5):2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussen BM, et al. MicroRNA: A signature for cancer progression. Biomed Pharmacother. 2021;138: 111528. [DOI] [PubMed] [Google Scholar]
- 37.Shademan B, et al. MicroRNAs as targets for cancer diagnosis: interests and limitations. Adv Pharm Bull. 2023;13(3):435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Brien J, et al. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salido-Guadarrama I, et al. MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther. 2014;7:1327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rani V, Sengar RS. Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol Bioeng. 2022;119(3):685–92. [DOI] [PubMed] [Google Scholar]
- 41.Ali Syeda Z, et al. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci. 2020;21(5):1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferro E, et al. From endogenous to synthetic microRNA-mediated regulatory circuits: an overview. Cells. 2019;8(12):1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivey KN, Srivastava D. microRNAs as developmental regulators. Cold Spring Harb Perspect Biol. 2015;7(7): a008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiers NM, et al. Target-directed microRNA degradation: mechanisms, significance, and functional implications. Wiley Interdiscip Rev RNA. 2024;15(2): e1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Rooij LA, et al. The microRNA lifecycle in health and cancer. Cancers (Basel). 2022;14(23):5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groot M, Lee H. Sorting mechanisms for MicroRNAs into extracellular vesicles and their associated diseases. Cells. 2020;9(4):1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashoub MH, et al. Extracellular microvesicles: biologic properties, biogenesis, and applications in leukemia. Mol Cell Biochem. 2024;479(2):419–30. [DOI] [PubMed] [Google Scholar]
- 48.Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1(1):15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcucci G, et al. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117(4):1121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doghish AS, et al. miRNAs as cornerstones in chronic lymphocytic leukemia pathogenesis and therapeutic resistance—an emphasis on the interaction of signaling pathways. Pathol Res Pract. 2023;243: 154363. [DOI] [PubMed] [Google Scholar]
- 51.Gębarowska K, et al. MicroRNA as a prognostic and diagnostic marker in T-cell acute lymphoblastic leukemia. Int J Mol Sci. 2021;22(10):5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cariello M, et al. Drug resistance: the role of exosomal miRNA in the microenvironment of hematopoietic tumors. Molecules. 2022;28(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anelli L, et al. Dysregulation of miRNA in leukemia: exploiting miRNA expression profiles as biomarkers. Int J Mol Sci. 2021;22(13):7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Cui X, Guan H. MicroRNAs: pivotal regulators in acute myeloid leukemia. Ann Hematol. 2020;99:399–412. [DOI] [PubMed] [Google Scholar]
- 55.Katsaraki K, et al. MicroRNAs: tiny regulators of gene expression with pivotal roles in normal B-cell development and B-cell chronic lymphocytic leukemia. Cancers. 2021;13(4):593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alves R, et al. Resistance to tyrosine kinase inhibitors in chronic myeloid leukemia—from molecular mechanisms to clinical relevance. Cancers. 2021;13(19):4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Navabi A, et al. The role of microRNAs in the development, progression and drug resistance of chronic myeloid leukemia and their potential clinical significance. Life Sci. 2022;296: 120437. [DOI] [PubMed] [Google Scholar]
- 58.Jiang X, et al. MicroRNA-221 sensitizes chronic myeloid leukemia cells to imatinib by targeting STAT5. Leuk Lymphoma. 2018. 10.1080/10428194.2018.1543875. [DOI] [PubMed] [Google Scholar]
- 59.Xu X Sr, Jiang X, Xia L. MiR-221 sensitizes chronic myeloid leukemia cells to imatinib by targeting STAT5. Washington, DC: American Society of Hematology; 2018. [Google Scholar]
- 60.Jin J, et al. Decreased expression of microRNA-214 contributes to imatinib mesylate resistance of chronic myeloid leukemia patients by upregulating ABCB1 gene expression. Exp Ther Med. 2018;16(3):1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hershkovitz-Rokah O, et al. MiR-30e induces apoptosis and sensitizes K562 cells to imatinib treatment via regulation of the BCR–ABL protein. Cancer Lett. 2015;356(2):597–605. [DOI] [PubMed] [Google Scholar]
- 62.Ge Y, et al. miR-30e-5p regulates leukemia stem cell self-renewal through the Cyb561/ROS signaling pathway. Haematologica. 2024;109(2):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lasica M, Anderson MA. Review of venetoclax in CLL, AML and multiple myeloma. J Pers Med. 2021;11(6):463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ. 2015;22(1):6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi MY, et al. Pre-clinical specificity and safety of UC-961, a first-in-class monoclonal antibody targeting ROR1. Clin Lymphoma Myeloma Leuk. 2015;15:S167–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pepe F, Balatti V. Role of non-coding RNAs in the development of targeted therapy and immunotherapy approaches for chronic lymphocytic leukemia. J Clin Med. 2020;9(2):593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rogers KA, et al. Investigating the addition of ianalumab (VAY736) to ibrutinib in patients with chronic lymphocytic leukemia (CLL) on ibrutinib therapy: results from a phase Ib study. Blood. 2021;138:2631. [Google Scholar]
- 68.Cui B, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124(4):546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raeisi F, et al. Differential expression profile of miR-27b, miR-29a, and miR-155 in chronic lymphocytic leukemia and breast cancer patients. Mol Ther Oncolytics. 2020;16:230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khalifa M, et al. Prognostic significance of microRNA 17–92 cluster expression in Egyptian chronic lymphocytic leukemia patients. J Egypt Natl Canc Inst. 2021;33:1–14. [DOI] [PubMed] [Google Scholar]
- 71.Zhao W, et al. The miR-17–92 cluster: Yin and Yang in human cancers. Cancer Treat Res Commun. 2022;33:100647. [DOI] [PubMed] [Google Scholar]
- 72.Sandhu SK, et al. B-cell malignancies in microRNA Eμ-miR-17∼ 92 transgenic mice. Proc Natl Acad Sci USA. 2013;110(45):18208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dell’Aversana C, et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia. 2017;31(11):2315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhi Y, et al. Serum level of miR-10-5p as a prognostic biomarker for acute myeloid leukemia. Int J Hematol. 2015;102:296–303. [DOI] [PubMed] [Google Scholar]
- 75.Zhang T-J, et al. Bone marrow miR-10a overexpression is associated with genetic events but not affects clinical outcome in acute myeloid leukemia. Pathol Res Pract. 2018;214(1):169–73. [DOI] [PubMed] [Google Scholar]
- 76.Zhao J, et al. Prognostic value of miR-96 in patients with acute myeloid leukemia. Diagn Pathol. 2014;9:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X-L, Wang W-F, Hao J-M. Effect of MiR-96 on cell invasion and apoptosis in pediatric acute myeloid leukemia via regulating MYB. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2021;29(2):439–44. [DOI] [PubMed] [Google Scholar]
- 78.Barbato S, Solaini G, Fabbri M. MicroRNAs in oncogenesis and tumor suppression. Int Rev Cell Mol Biol. 2017;333:229–68. [DOI] [PubMed] [Google Scholar]
- 79.Faraoni I, et al. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012;5:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palma CA, et al. MicroRNA-155 as an inducer of apoptosis and cell differentiation in acute myeloid leukaemia. Mol Cancer. 2014;13(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khalife J. Targeting the oncomicroRNA miR-155 in acute myeloid leukemia with the NEDD8-activating enzyme inhibitor MLN4924. Columbus: The Ohio State University; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neaga A, et al. MicroRNAs associated with a good prognosis of acute myeloid leukemia and their effect on macrophage polarization. Front Immunol. 2021;11: 582915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamamoto H, et al. miR-133 regulates Evi1 expression in AML cells as a potential therapeutic target. Sci Rep. 2016;6(1):19204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng Z-Z, et al. Serum miR-133 as a novel biomarker for predicting treatment response and survival in acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2020;24(2):777. [DOI] [PubMed] [Google Scholar]
- 85.Xiao Y, Su C, Deng T. miR-223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7. Oncol Lett. 2016;12(5):3531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu G, et al. Low serum miR-223 expression predicts poor outcome in patients with acute myeloid leukemia. J Clin Lab Anal. 2020;34(3): e23096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu X, et al. miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J Cancer Res Clin Oncol. 2016;142:77–87. [DOI] [PubMed] [Google Scholar]
- 88.Seca H, et al. Effect of miR-128 in DNA damage of HL-60 acute myeloid leukemia cells. Curr Pharm Biotechnol. 2014;15(5):492–502. [DOI] [PubMed] [Google Scholar]
- 89.Coskun E, et al. MicroRNA profiling reveals aberrant microRNA expression in adult ETP-ALL and functional studies implicate a role for miR-222 in acute leukemia. Leuk Res. 2013;37(6):647–56. [DOI] [PubMed] [Google Scholar]
- 90.Mavrakis KJ, et al. Genome-wide RNAi screen identifies miR-19 targets in notch-induced acute T-cell leukaemia (T-ALL). Nat Cell Biol. 2010;12(4):372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li C, et al. MicroRNA-223 decreases cell proliferation, migration, invasion, and enhances cell apoptosis in childhood acute lymphoblastic leukemia via targeting Forkhead box O 1. Biosci Rep. 2020;40(10):BSR20200485. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Liu Z, et al. MicroRNA-223-induced inhibition of the FBXW7 gene affects the proliferation and apoptosis of colorectal cancer cells via the Notch and Akt/mTOR pathways. Mol Med Rep. 2021;23(2):1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Correia NC, et al. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Sci Rep. 2016;6(1):31894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu X, et al. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 2013;23(12):1356–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lv M, et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-α and cAMP/PKA pathways. Leukemia. 2012;26(4):769–77. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Y, Liu Y, Xu X. Upregulation of miR-142-3p improves drug sensitivity of acute myelogenous leukemia through reducing P-glycoprotein and repressing autophagy by targeting HMGB1. Transl Oncol. 2017;10(3):410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Correia NC, Barata JT. MicroRNAs and their involvement in T-ALL: a brief overview. Adv Biol Regul. 2019;74: 100650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Z, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3(1):688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yuan T, et al. Regulation of PI3K signaling in T-cell acute lymphoblastic leukemia: a novel PTEN/Ikaros/miR-26b mechanism reveals a critical targetable role for PIK3CD. Leukemia. 2017;31(11):2355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kogut S, et al. Ikaros regulates microRNA networks in acute lymphoblastic leukemia. Epigenomes. 2022;6(4):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim C, et al. miR-181a-regulated pathways in T-cell differentiation and aging. Immun Ageing. 2021;18(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ghodousi ES, Aberuyi N, Rahgozar S. Simultaneous changes in expression levels of BAALC and miR-326: a novel prognostic biomarker for childhood ALL. Jpn J Clin Oncol. 2020;50(6):671–8. [DOI] [PubMed] [Google Scholar]
- 103.Gefen N, et al. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia. 2010;24(1):89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mattick JS, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24(6):430–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Statello L, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rinn JL, Chang HY. Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem. 2020;89:283–308. [DOI] [PubMed] [Google Scholar]
- 107.Kazimierczyk M, Wrzesinski J. Long non-coding RNA epigenetics. Int J Mol Sci. 2021;22(11):6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Razavi ZS, et al. Gynecologic cancers and non-coding RNAs: epigenetic regulators with emerging roles. Crit Rev Oncol Hematol. 2021;157: 103192. [DOI] [PubMed] [Google Scholar]
- 109.Ruffo P, et al. Deregulation of ncRNA in neurodegenerative disease: focus on circRNA, lncRNA and miRNA in amyotrophic lateral sclerosis. Front Genet. 2021;12: 784996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ahmadpour ST, et al. Breast cancer chemoresistance: insights into the regulatory role of lncRNA. Int J Mol Sci. 2023;24(21):15897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fernandes M, et al. Competitive endogenous RNA network involving miRNA and lncRNA in non-hodgkin lymphoma: current advances and clinical perspectives. Biomedicines. 2021;9(12):1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Illarregi U, et al. lncRNA deregulation in childhood acute lymphoblastic leukemia: a systematic review. Int J Oncol. 2022;60(5):1–9. [DOI] [PubMed] [Google Scholar]
- 113.Di Martino MT, et al. miRNAs and lncRNAs as novel therapeutic targets to improve cancer immunotherapy. Cancers. 2021;13(7):1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen S, et al. Long non-coding RNAs: the novel diagnostic biomarkers for leukemia. Environ Toxicol Pharmacol. 2017;55:81–6. [DOI] [PubMed] [Google Scholar]
- 115.Morlando M, Ballarino M, Fatica A. Long non-coding RNAs: new players in hematopoiesis and leukemia. Front Med. 2015;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kirtonia A, et al. Long noncoding RNAs: a novel insight in the leukemogenesis and drug resistance in acute myeloid leukemia. J Cell Physiol. 2022;237(1):450–65. [DOI] [PubMed] [Google Scholar]
- 117.Zeng C, et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol Cancer. 2018;17(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo G, et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34(14):1768–79. [DOI] [PubMed] [Google Scholar]
- 119.Eldin RAA, et al. HOTAIR expression and prognostic impact in acute myeloid leukemia patients. Egypt J Med Hum Genetics. 2021;22(1):1–11.38624675 [Google Scholar]
- 120.Wang H, et al. The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology. 2017;22(4):208–16. [DOI] [PubMed] [Google Scholar]
- 121.Shehata AMF, et al. LncRNA CCAT2 expression at diagnosis predicts imatinib response in chronic phase chronic myeloid leukemia patients. Leuk Res. 2022;116: 106838. [DOI] [PubMed] [Google Scholar]
- 122.He B, et al. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res. 2017;7(8):1704. [PMC free article] [PubMed] [Google Scholar]
- 123.Ahmadi A, et al. Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood research. 2018;53(4):320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wen F, et al. LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem Biophys Res Commun. 2018;507(1–4):1–8. [DOI] [PubMed] [Google Scholar]
- 125.Choudhury SR, et al. LncRNA Hmrhl regulates expression of cancer related genes in chronic myelogenous leukemia through chromatin association. NAR Cancer. 2021;3(4):zcab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pan J-Q, et al. lncRNA co-expression network model for the prognostic analysis of acute myeloid leukemia. Int J Mol Med. 2017;39(3):663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Y, et al. Long non-coding RNA LINC00899 as a novel serum biomarker for diagnosis and prognosis prediction of acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2018;22(21):7364–70. [DOI] [PubMed] [Google Scholar]
- 128.He C, et al. Long Noncoding RNA maternally expressed gene 3 is downregulated, and its insufficiency correlates with poor-risk stratification, worse treatment response, as well as unfavorable survival data in patients with acute myeloid leukemia. Technol Cancer Res Treat. 2020;19:1533033820945815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu Y, et al. LncRNA MEG3 contributes to drug resistance in acute myeloid leukemia by positively regulating ALG9 through sponging miR-155. Int J Lab Hematol. 2020;42(4):464–72. [DOI] [PubMed] [Google Scholar]
- 130.Wang Y. Comprehensive long non-coding RNA expression profiling by RNA sequencing reveals potential biomarkers for acute myeloid leukemia risk. Cancer Biomark. 2019;26(1):93–108. [DOI] [PubMed] [Google Scholar]
- 131.Shi J, et al. LncRNA LINP1 regulates acute myeloid leukemia progression via HNF4α/AMPK/WNT5A signaling pathway. Hematol Oncol. 2019;37(4):474–82. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y, Zhou Q, Ma J-J. High expression of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and promotes the malignant progression in acute myeloid leukemia cell line U937. Eur Rev Med Pharmacol Sci. 2018;22(3):763. [DOI] [PubMed] [Google Scholar]
- 133.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ju J-K, Han W-N, Shi C-L. Long non-coding RNA (lncRNA) plasmacytoma variant translocation 1 gene (PVT1) modulates the proliferation and apoptosis of acute lymphoblastic leukemia cells by sponging miR-486-5p. Bioengineered. 2022;13(2):4587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cheng J, et al. LncRNA PVT1 promotes the malignant progression of acute myeloid leukaemia via sponging miR-29 family to increase WAVE1 expression. Pathology. 2021;53(5):613–22. [DOI] [PubMed] [Google Scholar]
- 136.Yazdi N, et al. Long noncoding RNA PVT1: potential oncogene in the development of acutelymphoblastic leukemia. Turk J Biol. 2018;42(5):405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li G, et al. Long non-coding RNA TUG1 modulates proliferation, migration, and invasion of acute myeloid leukemia cells via regulating miR-370-3p/MAPK1/ERK. Onco Targets Ther. 2019;12:10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen L, et al. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39(4):330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Feng Y, et al. LncRNA NR-104098 inhibits AML proliferation and induces differentiation through repressing EZH2 transcription by interacting with E2F1. Front Cell Dev Biol. 2020;8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hirano T, et al. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol Cancer. 2015;14:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ma L, et al. Long noncoding RNA LINC00265 predicts the prognosis of acute myeloid leukemia patients and functions as a promoter by activating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22(22):7867. [DOI] [PubMed] [Google Scholar]
- 142.Zeng C, et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cruz-Miranda GM, et al. Long non-coding RNA and acute leukemia. Int J Mol Sci. 2019;20(3):735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lajoie M, et al. Specific expression of novel long non-coding RNAs in high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS ONE. 2017;12(3): e0174124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ouimet M, et al. A childhood acute lymphoblastic leukemia-specific lncRNA implicated in prednisolone resistance, cell proliferation, and migration. Oncotarget. 2017;8(5):7477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gioia R, et al. LncRNAs downregulated in childhood acute lymphoblastic leukemia modulate apoptosis, cell migration, and DNA damage response. Oncotarget. 2017;8(46):80645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Garitano-Trojaola A, et al. Deregulation of linc-PINT in acute lymphoblastic leukemia is implicated in abnormal proliferation of leukemic cells. Oncotarget. 2018;9(16):12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Durinck K, et al. The Notch driven long non-coding RNA repertoire in T-cell acute lymphoblastic leukemia. Haematologica. 2014;99(12):1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao Q, et al. TCF7L2 activated HOXA-AS2 decreased the glucocorticoid sensitivity in acute lymphoblastic leukemia through regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway. Biomed Pharmacother. 2019;109:1640–9. [DOI] [PubMed] [Google Scholar]
- 150.Baghdadi H, et al. Long non-coding RNA signatures in lymphopoiesis and lymphoid malignancies. Non-coding RNA. 2023;9(4):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen L, et al. LncRNA CDKN2B-AS1 contributes to tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia through miR-335–3p/TRAF5 axis. Anti Cancer Drugs. 2020. 10.1097/CAD.0000000000001001. [DOI] [PubMed] [Google Scholar]
- 152.Liu C-X, Chen L-L. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185:2016. [DOI] [PubMed] [Google Scholar]
- 153.Chen L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–90. [DOI] [PubMed] [Google Scholar]
- 154.Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29(3):481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Prats A-C, et al. Circular RNA, the key for translation. Int J Mol Sci. 2020;21(22):8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.He Z, Zhu Q. A Circular RNAs: Emerging roles and new insights in human cancers. Biomed Pharmacother. 2023;165: 115217. [DOI] [PubMed] [Google Scholar]
- 157.Tang X, et al. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li J, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends in cancer. 2020;6(4):319–36. [DOI] [PubMed] [Google Scholar]
- 159.He AT, et al. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Memczak S, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE. 2015;10(10): e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pan Y, et al. CircBA9.3 supports the survival of leukaemic cells by up-regulating c-ABL1 or BCR-ABL1 protein levels. Blood Cells Mol Dis. 2018;73:38–44. [DOI] [PubMed] [Google Scholar]
- 163.Ping L, et al. High circ_100053 predicts a poor outcome for chronic myeloid leukemia and is involved in imatinib resistance. Oncol Res. 2019. 10.3727/096504018X15412701483326. [DOI] [PubMed] [Google Scholar]
- 164.Liu J, et al. Global identification of circular RNAs in chronic myeloid leukemia reveals hsa_circ_0080145 regulates cell proliferation by sponging miR-29b. Biochem Biophys Res Commun. 2018;504(4):660–5. [DOI] [PubMed] [Google Scholar]
- 165.Li Y, et al. miR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp Cell Res. 2013;319(8):1094–101. [DOI] [PubMed] [Google Scholar]
- 166.Xia L, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem Biophys Res Commun. 2018;503(1):385–90. [DOI] [PubMed] [Google Scholar]
- 167.Wu W, et al. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging (Albany NY). 2019;11(11):3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu Z, et al. Circ-RPL15: a plasma circular RNA as novel oncogenic driver to promote progression of chronic lymphocytic leukemia. Leukemia. 2020;34(3):919–23. [DOI] [PubMed] [Google Scholar]
- 169.Chen H, et al. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family. Cell Physiol Biochem. 2018;47(5):1998–2007. [DOI] [PubMed] [Google Scholar]
- 170.Ping L, et al. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis. 2019;75:41–7. [DOI] [PubMed] [Google Scholar]
- 171.Fan H, et al. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem Biophys Res Commun. 2018;507(1–4):178–84. [DOI] [PubMed] [Google Scholar]
- 172.Li W, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18(3):597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hirsch S, et al. Circular RNAs of the nucleophosmin (NPM1) gene in acute myeloid leukemia. Haematologica. 2017;102(12):2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nagai Y, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24(6):801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Okamoto M, et al. Toll-like receptors (TLRs) are expressed by myeloid leukaemia cell lines, but fail to trigger differentiation in response to the respective TLR ligands. Br J Haematol. 2009;147(4):585–7. [DOI] [PubMed] [Google Scholar]
- 176.Eriksson M, et al. Agonistic targeting of TLR1/TLR2 induces p38 MAPK-dependent apoptosis and NFκB-dependent differentiation of AML cells. Blood Adv. 2017;1(23):2046–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Su R, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 2015;34(25):3226–39. [DOI] [PubMed] [Google Scholar]
- 178.Yuan D-M, Ma J, Fang WB. Identification of non-coding RNA regulatory networks in pediatric acute myeloid leukemia reveals circ-0004136 could promote cell proliferation by sponging miR-142. Eur Rev Med Pharmacol Sci. 2019;23(21):9251. [DOI] [PubMed] [Google Scholar]
- 179.Yi YY, et al. Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J Cell Physiol. 2019;234(4):3711–9. [DOI] [PubMed] [Google Scholar]
- 180.Wu D-M, et al. Role of circular RNA DLEU2 in human acute myeloid leukemia. Mol Cell Biol. 2018. 10.1128/mcb.00259-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Papaioannou D, et al. Clinical and functional significance of circular RNAs in cytogenetically normal AML. Blood Adv. 2020;4(2):239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhou J, et al. Circ-Foxo3 is positively associated with the Foxo3 gene and leads to better prognosis of acute myeloid leukemia patients. BMC Cancer. 2019;19(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li S, et al. Profiling and functional analysis of circular RNAs in acute promyelocytic leukemia and their dynamic regulation during all-trans retinoic acid treatment. Cell Death Dis. 2018;9(6):651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lv C, et al. Circular RNA regulatory network reveals cell–cell crosstalk in acute myeloid leukemia extramedullary infiltration. J Transl Med. 2018;16(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127(1):53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sun Y-M, et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134(18):1533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shang J, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. 2019;85: 106198. [DOI] [PubMed] [Google Scholar]
- 188.Salzman J, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7(2): e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huang W, et al. circRNA circAF4 functions as an oncogene to regulate MLL-AF4 fusion protein expression and inhibit MLL leukemia progression. J Hematol Oncol. 2019;12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dal Molin A, et al. CircRNAs are here to stay: a perspective on the MLL recombinome. Front Genet. 2019;10:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hu J, et al. Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics. 2018;10(6):723–32. [DOI] [PubMed] [Google Scholar]
- 192.Panda AC, et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017;45(7):4021–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gaffo E, et al. Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci Rep. 2019;9(1):14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ishida Y, et al. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11(11):3887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Daëron M, et al. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224(1):11–43. [DOI] [PubMed] [Google Scholar]
- 196.El Firar A, et al. Discovery of a functional immunoreceptor tyrosine-based switch motif in a 7-transmembrane-spanning receptor: role in the orexin receptor OX1R-driven apoptosis. FASEB J. 2009;23(12):4069–80. [DOI] [PubMed] [Google Scholar]
- 197.Keir ME, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Köhnke T, et al. Increase of PD-L1 expressing B-precursor ALL cells in a patient resistant to the CD19/CD3-bispecific T cell engager antibody blinatumomab. J Hematol Oncol. 2015;8:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schnorfeil FM, et al. T cells are functionally not impaired in AML: increased PD-1 expression is only seen at time of relapse and correlates with a shift towards the memory T cell compartment. J Hematol Oncol. 2015;8(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Spranger S, et al. Up-regulation of PD-L1, IDO, and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci Transl Med. 2013;5(200):200ra116-200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Garon EB, et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28. [DOI] [PubMed] [Google Scholar]
- 202.Blank C, et al. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Can Res. 2004;64(3):1140–5. [DOI] [PubMed] [Google Scholar]
- 203.Chen BJ, et al. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res. 2013;19(13):3462–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Green MR, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116(17):3268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chapuy B, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127(7):869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Twa DD, et al. Genomic rearrangements involving programmed death ligands are recurrent in primary mediastinal large B-cell lymphoma. Blood. 2014;123(13):2062–5. [DOI] [PubMed] [Google Scholar]
- 207.Georgiou K, et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 2016;127(24):3026–34. [DOI] [PubMed] [Google Scholar]
- 208.Kiyasu J, et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood. 2015;126(19):2193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Green MR, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18(6):1611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bi X-W, et al. PD-L1 is upregulated by EBV-driven LMP1 through NF-κB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J Hematol Oncol. 2016;9(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhang L, Gajewski TF, Kline J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood. 2009;114(8):1545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhou Q, et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood. 2010;116(14):2484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Goswami M, et al. Increased frequencies of PD-1+ CD8+ marrow-infiltrating lymphocytes associated with highly clonal t-lymphocyte expansions in relapsed and refractory aml patients but not healthy adults. Blood. 2016;128(22):1644. [Google Scholar]
- 214.Daver N, et al. Defining the immune checkpoint landscape in patients (pts) with acute myeloid leukemia (AML). Blood. 2016;128(22):2900. [Google Scholar]
- 215.Brunet J-F, et al. A new member of the immunoglobulin superfamily—CTLA-4. Nature. 1987;328(6127):267–70. [DOI] [PubMed] [Google Scholar]
- 216.Van Coillie S, Wiernicki B, Xu J. Molecular and cellular functions of CTLA-4. Regul Cancer Immune Checkp Mol Cell Mech Ther. 2020. 10.1007/978-981-15-3266-5_2. [DOI] [PubMed] [Google Scholar]
- 217.Hosseini A, et al. CTLA-4: From mechanism to autoimmune therapy. Int Immunopharmacol. 2020;80: 106221. [DOI] [PubMed] [Google Scholar]
- 218.Perkins D, et al. Regulation of CTLA-4 expression during T cell activation. J Immunol (Baltimore, Md: 1950). 1996;156(11):4154–9. [PubMed] [Google Scholar]
- 219.Sobhani N, et al. CTLA-4 in regulatory T cells for cancer immunotherapy. Cancers. 2021;13(6):1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jago C, et al. Differential expression of CTLA-4 among T cell subsets. Clin Exp Immunol. 2004;136(3):463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Takahashi T, et al. Immunologic self-tolerance maintained by CD25+ CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte–associated antigen 4. J Exp Med. 2000;192(2):303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wong HK, et al. Increased expression of CTLA-4 in malignant T cells from patients with mycosis fungoides–cutaneous T-cell lymphoma. J Investig Dermatol. 2006;126(1):212–9. [DOI] [PubMed] [Google Scholar]
- 223.Gibson HM, et al. Impaired proteasome function activates GATA3 in T cells and upregulates CTLA-4: relevance for Sezary syndrome. J Investig Dermatol. 2013;133(1):249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xerri L, et al. In vivo expression of the CTLA4 inhibitory receptor in malignant and reactive cells from human lymphomas. J Pathol. 1997;183(2):182–7. [DOI] [PubMed] [Google Scholar]
- 225.Sekulic A, et al. Personalized treatment of Sézary syndrome by targeting a novel CTLA 4: CD 28 fusion. Mol Genet Genomic Med. 2015;3(2):130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yoo HY, et al. Frequent CTLA4-CD28 gene fusion in diverse types of T-cell lymphoma. Haematologica. 2016;101(6):757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kataoka K, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–15. [DOI] [PubMed] [Google Scholar]
- 228.Ungewickell A, et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47(9):1056–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Saudemont A, Quesnel B. In a model of tumor dormancy, long-term persistent leukemic cells have increased B7–H1 and B7.1 expression and resist CTL-mediated lysis. Blood. 2004;104(7):2124–33. [DOI] [PubMed] [Google Scholar]
- 230.Perez-Garcia A, et al. CTLA-4 genotype and relapse incidence in patients with acute myeloid leukemia in first complete remission after induction chemotherapy. Leukemia. 2009;23(3):486–91. [DOI] [PubMed] [Google Scholar]
- 231.Triebel F, et al. LAG-3, a novel lymphocyte activation gene closely related to CD4. J Exp Med. 1990;171(5):1393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Workman CJ, Vignali DA. The CD4-related molecule, LAG-3 (CD223), regulates the expansion of activated T cells. Eur J Immunol. 2003;33(4):970–9. [DOI] [PubMed] [Google Scholar]
- 233.Kisielow M, et al. Expression of lymphocyte activation gene 3 (LAG-3) on B cells is induced by T cells. Eur J Immunol. 2005;35(7):2081–8. [DOI] [PubMed] [Google Scholar]
- 234.Workman CJ, et al. LAG-3 regulates plasmacytoid dendritic cell homeostasis. J Immunol. 2009;182(4):1885–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Workman CJ, Dugger KJ, Vignali DA. Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J Immunol. 2002;169(10):5392–5. [DOI] [PubMed] [Google Scholar]
- 236.Matsuzaki J, et al. Tumor-infiltrating NY-ESO-1–specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci. 2010;107(17):7875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Woo S-R, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Can Res. 2012;72(4):917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Grosso JF, et al. Functionally distinct LAG-3 and PD-1 subsets on activated and chronically stimulated CD8 T cells. J Immunol. 2009;182(11):6659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gandhi MK, et al. Expression of LAG-3 by tumor-infiltrating lymphocytes is coincident with the suppression of latent membrane antigen–specific CD8+ T-cell function in Hodgkin lymphoma patients. Blood. 2006;108(7):2280–9. [DOI] [PubMed] [Google Scholar]
- 240.El Halabi L, et al. Expression of the immune checkpoint regulators LAG-3 and TIM-3 in classical Hodgkin lymphoma. Clin Lymphoma Myeloma Leuk. 2021;21(4):257-266.e3. [DOI] [PubMed] [Google Scholar]
- 241.Yang Z-Z, et al. Expression of LAG-3 defines exhaustion of intratumoral PD-1+ T cells and correlates with poor outcome in follicular lymphoma. Oncotarget. 2017;8(37):61425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Monney L, et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415(6871):536–41. [DOI] [PubMed] [Google Scholar]
- 243.Zhu C, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6(12):1245–52. [DOI] [PubMed] [Google Scholar]
- 244.Chiba S, et al. Tumor-infiltrating DCs suppress nucleic acid–mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat Immunol. 2012;13(9):832–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ngiow SF, et al. Anti-TIM3 antibody promotes T cell IFN-γ–mediated antitumor immunity and suppresses established tumors. Can Res. 2011;71(10):3540–51. [DOI] [PubMed] [Google Scholar]
- 246.Sakuishi K, et al. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207(10):2187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhao B, et al. Immune checkpoint of B7–H3 in cancer: from immunology to clinical immunotherapy. J Hematol Oncol. 2022;15(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Feng R, et al. The role of B7–H3 in tumors and its potential in clinical application. Int Immunopharmacol. 2021;101: 108153. [DOI] [PubMed] [Google Scholar]
- 249.Liu S, et al. The role of CD276 in cancers. Front Oncol. 2021;11: 654684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chapoval AI, et al. B7–H3: a costimulatory molecule for T cell activation and IFN-γ production. Nat Immunol. 2001;2(3):269–74. [DOI] [PubMed] [Google Scholar]
- 251.Steinberger P, et al. Molecular characterization of human 4Ig-B7-H3, a member of the B7 family with four Ig-like domains. J Immunol. 2004;172(4):2352–9. [DOI] [PubMed] [Google Scholar]
- 252.Zhou YH, et al. 4IgB7-H3 is the major isoform expressed on immunocytes as well as malignant cells. Tissue Antigens. 2007;70(2):96–104. [DOI] [PubMed] [Google Scholar]
- 253.Sun M, et al. Characterization of mouse and human B7–H3 genes. J Immunol. 2002;168(12):6294–7. [DOI] [PubMed] [Google Scholar]
- 254.Vigdorovich V, et al. Structure and T cell inhibition properties of B7 family member, B7–H3. Structure. 2013;21(5):707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhang G, et al. Soluble CD276 (B7–H3) is released from monocytes, dendritic cells and activated T cells and is detectable in normal human serum. Immunology. 2008;123(4):538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Chen W, et al. Characterization of a soluble B7–H3 (sB7-H3) spliced from the intron and analysis of sB7-H3 in the sera of patients with hepatocellular carcinoma. PLoS ONE. 2013;8(10): e76965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Huang L, et al. Evaluation of the role of soluble B7–H3 in association with membrane B7–H3 expression in gastric adenocarcinoma. Cancer Biomark. 2022;33(1):123–9. [DOI] [PubMed] [Google Scholar]
- 258.Kovaleva O, et al. Soluble B7–H3 in ovarian cancer and its predictive value. Bull Exp Biol Med. 2021;171:472–4. [DOI] [PubMed] [Google Scholar]
- 259.Lichtman EI, et al. Preclinical evaluation of B7–H3–specific chimeric antigen receptor T cells for the treatment of acute myeloid leukemia. Clin Cancer Res. 2021;27(11):3141–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wei J, et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. 2016;18(5):639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Huffaker TB, et al. Antitumor immunity is defective in T cell–specific microRNA-155–deficient mice and is rescued by immune checkpoint blockade. J Biol Chem. 2017;292(45):18530–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zheng Z, et al. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+ T cells. Mol Cancer. 2019;18(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103(18):7024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. 2016;375(18):1767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Zhu F-Q, et al. MicroRNA-155 downregulation promotes cell cycle arrest and apoptosis in diffuse large B-cell lymphoma. Oncol Res Featur Preclin Clin Cancer Therapeutics. 2016;24(6):415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Watanabe N, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol. 2003;4(7):670–9. [DOI] [PubMed] [Google Scholar]
- 267.Liu Y, et al. B and T lymphocyte attenuator is a target of miR-155 during naive CD4+ T cell activation. Iran J Immunol. 2016;13(2):89–99. [PubMed] [Google Scholar]
- 268.Su W, et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Craene BD, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. [DOI] [PubMed] [Google Scholar]
- 270.Katsura A, et al. ZEB 1-regulated inflammatory phenotype in breast cancer cells. Mol Oncol. 2017;11(9):1241–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zhao L, et al. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019;10(10):731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Chen L, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5(1):5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Noman MZ, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology. 2017;6(1): e1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bobisse S, et al. Sensitive and frequent identification of high avidity neo-epitope specific CD8+ T cells in immunotherapy-naive ovarian cancer. Nat Commun. 2018;9(1):1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chen S, et al. CD73: an emerging checkpoint for cancer immunotherapy. Immunotherapy. 2019;11(11):983–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Xu Y, et al. MiRNA-340-5p mediates the functional and infiltrative promotion of tumor-infiltrating CD8+ T lymphocytes in human diffuse large B cell lymphoma. J Exp Clin Cancer Res. 2020;39:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lawrie CH. Micro RNA s and lymphomagenesis: a functional review. Br J Haematol. 2013;160(5):571–81. [DOI] [PubMed] [Google Scholar]
- 278.Baraniskin A, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117(11):3140–6. [DOI] [PubMed] [Google Scholar]
- 279.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. [DOI] [PubMed] [Google Scholar]
- 280.Zheng Z, et al. MiR21 sensitized B-lymphoma cells to ABT-199 via ICOS/ICOSL-mediated interaction of Treg cells with endothelial cells. J Exp Clin Cancer Res. 2017;36(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Li Q, et al. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget. 2016;7(33):53735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Alturki NA. Review of the immune checkpoint inhibitors in the context of cancer treatment. J Clin Med. 2023;12(13):4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Naimi A, et al. Tumor immunotherapies by immune checkpoint inhibitors (ICIs); the pros and cons. Cell Communication and Signaling. 2022;20(1):1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Vafaei S, et al. Combination therapy with immune checkpoint inhibitors (ICIs); a new frontier. Cancer Cell Int. 2022;22:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–49. [DOI] [PubMed] [Google Scholar]
- 286.Shiravand Y, et al. Immune checkpoint inhibitors in cancer therapy. Curr Oncol. 2022;29(5):3044–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Singh S, et al. Immune checkpoint inhibitors: a promising anticancer therapy. Drug Discovery Today. 2020;25(1):223–9. [DOI] [PubMed] [Google Scholar]
- 288.Johnson DB, et al. Immune-checkpoint inhibitors: long-term implications of toxicity. Nat Rev Clin Oncol. 2022;19(4):254–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Xu S, et al. miR-424 (322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7(1):11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Shek D, et al. Non-coding RNA and immune-checkpoint inhibitors: friends or foes? Immunotherapy. 2020;12(7):513–29. [DOI] [PubMed] [Google Scholar]
- 291.García-Giménez JL, et al. miRNAs related to immune checkpoint inhibitor response: a systematic review. Int J Mol Sci. 2024;25(3):1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ansell SM, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Armand P, et al. Programmed death-1 blockade with pembrolizumab in patients with classical Hodgkin lymphoma after brentuximab vedotin failure. J Clin Oncol. 2016;34(31):3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Armand P, et al. Pembrolizumab in patients with classical Hodgkin lymphoma after brentuximab vedotin failure: long-term efficacy from the phase 1b keynote-013 study. Blood. 2016;128(22):1108. [Google Scholar]
- 295.Chen RW, et al. Pembrolizumab for relapsed/refractory classical Hodgkin lymphoma (R/R cHL): phase 2 KEYNOTE-087 study. Am Soc Clin Oncol. 2016. 10.1200/JCO.2016.34.15_suppl.7555. [Google Scholar]
- 296.Moskowitz CH, et al. Pembrolizumab in relapsed/refractory classical Hodgkin lymphoma: primary end point analysis of the phase 2 Keynote-087 study. Blood. 2016;128(22):1107. [Google Scholar]
- 297.Zinzani PL, et al. Phase 1b study of pembrolizumab in patients with relapsed/refractory primary mediastinal large B-cell lymphoma: results from the ongoing keynote-013 trial. Blood. 2016;128(22):619. [Google Scholar]
- 298.Khodadoust M, et al. Pembrolizumab for treatment of relapsed/refractory mycosis fungoides and Sezary syndrome: clinical efficacy in a CITN multicenter phase 2 study. Blood. 2016;128(22):181. [Google Scholar]
- 299.Lesokhin AM, et al. Nivolumab in patients with relapsed or refractory hematologic malignancy: preliminary results of a phase Ib study. J Clin Oncol. 2016;34(23):2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Till BG, et al. Safety and clinical activity of atezolizumab (anti-PDL1) in combination with obinutuzumab in patients with relapsed or refractory non-Hodgkin lymphoma. Washington DC: American Society of Hematology; 2015. [Google Scholar]
- 301.Ansell SM, et al. Phase I study of ipilimumab, an anti–CTLA-4 monoclonal antibody, in patients with relapsed and refractory B-cell non–Hodgkin lymphoma. Clin Cancer Res. 2009;15(20):6446–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sauer N, et al. LAG-3 as a potent target for novel anticancer therapies of a wide range of tumors. Int J Mol Sci. 2022;23(17):9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Avice M-N, et al. Lymphocyte activation gene-3, a MHC class II ligand expressed on activated T cells, stimulates TNF-α and IL-12 production by monocytes and dendritic cells. J Immunol. 1999;162(5):2748–53. [PubMed] [Google Scholar]
- 304.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ibrahim R, et al. LAG-3 inhibitors: novel immune checkpoint inhibitors changing the landscape of immunotherapy. Biomedicines. 2023;11(7):1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Davids MS, et al. Ipilimumab for patients with relapse after allogeneic transplantation. N Engl J Med. 2016;375(2):143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Zeidan AM, et al. Stabilization of myelodysplastic syndromes (MDS) following hypomethylating agent (HMAs) failure using the immune checkpoint inhibitor ipilimumab: a phase I trial. Blood. 2015;126(23):1666. [Google Scholar]
- 308.Garcia-Manero G, et al. A phase II study evaluating the combination of nivolumab (Nivo) or ipilimumab (Ipi) with azacitidine in Pts with previously treated or untreated myelodysplastic syndromes (MDS). Washington DC: American Society of Hematology; 2016. [Google Scholar]
- 309.Bonneau E, et al. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Ejifcc. 2019;30(2):114. [PMC free article] [PubMed] [Google Scholar]
- 310.Ishida M, Selaru FM. miRNA-based therapeutic strategies. Curr Pathobiol Rep. 2013;1:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Mondal P, et al. Progress and promises of epigenetic drugs and epigenetic diets in cancer prevention and therapy: a clinical update. Semin Cancer Biol. 2022. 10.1016/j.semcancer.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 312.Abd-Aziz N, Kamaruzman NI, Poh CL. Development of microRNAs as potential therapeutics against cancer. J Oncol. 2020;2020:8029721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Segal M, Slack FJ. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin Drug Discov. 2020;15(9):987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Baumann V, Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Fut Med Chem. 2014;6(17):1967–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Mondal P, et al. The emerging role of miRNA in the perturbation of tumor immune microenvironment in chemoresistance: therapeutic implications. Semin Cell Dev Biol. 2022. 10.1016/j.semcdb.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 316.Shin DS, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7(2):188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Sade-Feldman M, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8(1):1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Salavati MS, et al. CAR-NK cells as promising immune therapeutics: platforms and current progress. Int J Cancer Manag. 2024;17(1): e145431. [Google Scholar]
- 319.Kim J, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol. 2016;27(8):1492–504. [DOI] [PubMed] [Google Scholar]
- 320.Matsushita M, Kawaguchi M. Immunomodulatory effects of drugs for effective cancer immunotherapy. J Oncol. 2018;2018:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Taefehshokr S, et al. Cancer immunotherapy: challenges and limitations. Pathol Res Pract. 2022;229: 153723. [DOI] [PubMed] [Google Scholar]
- 322.Abakushina EV, et al. The advantages and challenges of anticancer dendritic cell vaccines and NK cells in adoptive cell immunotherapy. Vaccines. 2021;9(11):1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Wang W-T, et al. Noncoding RNAs in cancer therapy resistance and targeted drug development. J Hematol Oncol. 2019;12(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Papaioannou E, et al. Regulation of adaptive tumor immunity by non-coding RNAs. Cancers. 2021;13(22):5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yu T, et al. MicroRNA-491 regulates the proliferation and apoptosis of CD8+ T cells. Sci Rep. 2016;6(1):30923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Wu Y, et al. Circ_0009910 sponges miR-491-5p to promote acute myeloid leukemia progression through modulating B4GALT5 expression and PI3K/AKT signaling pathway. Int J Lab Hematol. 2022;44(2):320–32. [DOI] [PubMed] [Google Scholar]
- 327.Jiang M, et al. Small extracellular vesicles containing miR-381-3p from keratinocytes promote T helper type 1 and T helper type 17 polarization in psoriasis. J Investig Dermatol. 2021;141(3):563–74. [DOI] [PubMed] [Google Scholar]
- 328.Ye Q, et al. Tumor-suppressing effects of miR-381-3p in pediatric acute myeloid leukemia via ROCK1 downregulation. Funct Integr Genom. 2023;23(1):43. [DOI] [PubMed] [Google Scholar]
- 329.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Gao Q, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25(18):2586–93. [DOI] [PubMed] [Google Scholar]
- 331.Gao Y, et al. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. J Immunol. 2018;200(8):2603–14. [DOI] [PubMed] [Google Scholar]
- 332.Yan K, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8+ T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol. 2019;110:1–8. [DOI] [PubMed] [Google Scholar]
- 333.Hu Q, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. 2019;20(7):835–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Blees A, et al. Structure of the human MHC-I peptide-loading complex. Nature. 2017;551(7681):525–8. [DOI] [PubMed] [Google Scholar]
- 335.Wang R, et al. Downregulation of miRNA-214 in cancer-associated fibroblasts contributes to migration and invasion of gastric cancer cells through targeting FGF9 and inducing EMT. J Exp Clin Cancer Res. 2019;38(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Bobbin ML, Rossi JJ. RNA interference (RNAi)-based therapeutics: delivering on the promise? Annu Rev Pharmacol Toxicol. 2016;56:103–22. [DOI] [PubMed] [Google Scholar]
- 337.Ahn I, Kang CS, Han J. Where should siRNAs go: applicable organs for siRNA drugs. Exp Mol Med. 2023;55(7):1283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Hu B, et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Paul A, et al. siRNA therapeutics and its challenges: recent advances in effective delivery for cancer therapy. OpenNano. 2022;7: 100063. [Google Scholar]
- 340.Alshaer W, et al. siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur J Pharmacol. 2021;905: 174178. [DOI] [PubMed] [Google Scholar]
- 341.Sajid MI, et al. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharmaceuticals. 2020;13(10):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Hoy SM. Patisiran: first global approval. Drugs. 2018;78:1625–31. [DOI] [PubMed] [Google Scholar]
- 343.Wood H. FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat Rev Neurol. 2018;14(10):570–570. [DOI] [PubMed] [Google Scholar]
- 344.Scott LJ. Givosiran: first approval. Drugs. 2020;80(3):335–9. [DOI] [PubMed] [Google Scholar]
- 345.Scott LJ, Keam SJ. Lumasiran: first approval. Drugs. 2021;81:277–82. [DOI] [PubMed] [Google Scholar]
- 346.Yamada Y. Nucleic acid drugs—current status, issues, and expectations for exosomes. Cancers. 2021;13(19):5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Zhang MM, et al. The growth of siRNA-based therapeutics: updated clinical studies. Biochem Pharmacol. 2021;189: 114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Sioud M. Releasing the immune system brakes using siRNAs enhances cancer immunotherapy. Cancers. 2019;11(2):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Boussiotis VA. Somatic mutations and immunotherapy outcome with CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2230–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Hobo W, et al. Improving dendritic cell vaccine immunogenicity by silencing PD-1 ligands using siRNA-lipid nanoparticles combined with antigen mRNA electroporation. Cancer Immunol Immunother. 2013;62(2):285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Hobo W, et al. siRNA silencing of PD-L1 and PD-L2 on dendritic cells augments expansion and function of minor histocompatibility antigen–specific CD8+ T cells. Blood. 2010;116(22):4501–11. [DOI] [PubMed] [Google Scholar]
- 352.Roeven MW, et al. Efficient nontoxic delivery of PD-L1 and PD-L2 siRNA into dendritic cell vaccines using the cationic lipid SAINT-18. J Immunother. 2015;38(4):145–54. [DOI] [PubMed] [Google Scholar]
- 353.Van den Bergh JM, et al. Monocyte-derived dendritic cells with silenced PD-1 ligands and transpresenting interleukin-15 stimulate strong tumor-reactive T-cell expansion. Cancer Immunol Res. 2017;5(8):710–5. [DOI] [PubMed] [Google Scholar]
- 354.Versteven M, et al. Dendritic cells and programmed death-1 blockade: a joint venture to combat cancer. Front Immunol. 2018;9: 351494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Choi KY, et al. Binary targeting of siRNA to hematologic cancer cells in vivo using layer-by-layer nanoparticles. Adv Func Mater. 2019;29(20):1900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Subhan MA, Attia SA, Torchilin VP. Advances in siRNA delivery strategies for the treatment of MDR cancer. Life Sci. 2021;274: 119337. [DOI] [PubMed] [Google Scholar]
- 358.Kyriazi AA, et al. Dual effects of non-coding RNAs (ncRNAs) in cancer stem cell biology. Int J Mol Sci. 2020;21(18):6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65(4):625–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta BBA Rev Cancer. 2021;1: 188491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Zhang X-Z, Liu H, Chen S-R. Mechanisms of long non-coding RNAs in cancers and their dynamic regulations. Cancers. 2020;12(5):1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Zhang X, et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol Cancer. 2020;19:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Uzuner E, et al. The role of MiRNA in cancer: pathogenesis, diagnosis, and treatment. In: Allmer J, Yousef M, editors., et al., miRNomics: MicroRNA biology and computational analysis. New York: Springer; 2022. p. 375–422. [DOI] [PubMed] [Google Scholar]
- 364.Alahdal M, Elkord E. Non-coding RNAs in cancer immunotherapy: Predictive biomarkers and targets. Clin Transl Med. 2023;13(9): e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–67. [DOI] [PubMed] [Google Scholar]
- 366.Martinez-Castillo M, et al. An overview of the immune modulatory properties of long non-coding RNAs and their potential use as therapeutic targets in cancer. Non-Coding RNA. 2023;9(6):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Shakhpazyan NK, et al. Long non-coding RNAs in colorectal cancer: navigating the intersections of immunity, intercellular communication, and therapeutic potential. Biomedicines. 2023;11(9):2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Kaur M, et al. Chapter four-noncoding RNAs as novel immunotherapeutic tools against cancer. Adv Protein Chem Struct Biol. 2022;129:135–61. [DOI] [PubMed] [Google Scholar]
- 369.Wallace JA, O’Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130(11):1290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Salavatipour MS, et al. CRISPR-Cas9 in basic and translational aspects of cancer therapy. BioImpacts. 2024. 10.34172/bi.2024.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Jin SJ, et al. Long non-coding RNA DANCR as an emerging therapeutic target in human cancers. Front Oncol. 2019;9:1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Chen Y, et al. Long non-coding RNAs: From disease code to drug role. Acta Pharm Sin B. 2021;11(2):340–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Saadi W, et al. Long non-coding RNAs as epigenetic regulators of immune checkpoints in cancer immunity. Cancers. 2023;15(1):184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
By ordering from the corresponding author.