Abstract
Background
Osteosarcoma (OS) is the most common primary malignant bone neoplasm. Growing researches have highlighted the tumor promoting role of miR-615-3p in various cancers. Notwithstanding, the biological function and underlying mechanisms of miR-615-3p in OS development still unclear.
Methods
Quantitative Real-Time PCR analysis (qRT-PCR) and RNA fluorescence in situ hybridization (FISH) staining were performed to measure miR-615-3p expression in OS. CCK-8 assay, colony formation assay and EdU assay were applied to analyze the OS cell proliferation activity. Cell metastasis abilities were evaluated using Transwell assays. Analysis of apoptosis was performed based on flow cytometric detection. The potential mechanisms of miR-615-3p in OS progression were investigated through RNA immunoprecipitation (RIP) assays, dual-luciferase reporter assays, qRT-PCR and western blotting. In vivo experiments, mouse xenograft model was carried out to assess the tumorigenicity of miR-615-3p.
Results
This study demonstrated a significant upregulation of miR-615-3p in OS. In addition, miR-615-3p knockdown suppressed OS proliferation, invasion, metastasis and EMT. Mechanistically, miR-615-3p regulated sestrin 2 (SESN2) expression negatively by targeting its 3’UTR. Moreover, silencing SESN2 facilitated OS progression and activated mTOR pathway. Noteworthy, the anticancer functions of miR-615-3p knockdown were partially recovered by SESN2 silencing. Taken together, the miR-615-3p/SESN2/mTOR pathway is critical for regulating OS progression.
Conclusion
Our results revealed that miR-615-3p modulated mTOR signaling, thus influencing the progression of OS. For OS treatment, molecular strategies that target the miR-615-3p/SESN2/mTOR pathway is promising.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03604-x.
Keywords: miR-615-3p, SESN2, mTOR, EMT, OS
Introduction
Osteosarcoma (OS) is the most prevalent and aggressive bone tumor that usually arises in children and adolescents [1]. Originating from interstitial tissue, OS is characterized by heterogeneity, rapid progression and early metastasis, primarily to the lungs [2]. Due to the development of various therapeutic strategies, including surgery, neoadjuvant radiotherapy and chemotherapy, the 5-year survival rate of patients without metastasis has reached 60%[3, 4]. However, the molecular mechanisms driving OS pathogenesis are not yet fully understood. Hence, there is an urgent need to explore the underlying mechanism and develop new effective targets for OS treatment.
MicroRNAs (miRNAs), evolutionarily conserved non-coding RNAs, exert their biological functions via accelerating mRNA degradation or translational inhibition posttranscriptionally [5]. Substantial evidence indicates that miRNAs and their biogenesis machinery play a crucial role in cancer development [6–8]. Accumulating studies has showed that several miRNAs may be used as diagnostic and prognostic biomarkers in the future [9–12]. miR-615-3p situates within the intron of the Hoxc5 gene at human chromosome 12q13.13[13]. As reported, miR-615-3p affects the progression of numerous tumors as an oncogene. miR-615-3p is highly expressed and leads to an inferior prognosis in prostate cancer [14]. Another study demonstrated that aberrant highly expressed miR-615-3p facilitated proliferation and metastasis of gastric cancer via targeting CELF2[15]. Nevertheless, whether miR-615-3p is involved in osteosarcoma procession and the underlying regulatory mechanisms are not well known.
The activation of mTOR often occurs in various diseases, including cancers [16, 17]. mTOR influences protein synthesis, cell growth and metabolism by phosphorylating 70S6K and 4E-BP1[10–20]. Notably, in addition to enhancing cell proliferation, Numerous reports indicate that the mTOR pathway can promotes metastasis of cancers through activating and maintaining the epithelial-mesenchymal transition (EMT) progression [21–23]. EMT, which is a crucial mechanism resulting in tumor metastasis, is a phenomenon in which epithelial cells transform into mesenchymal cells through a complex process. During EMT, a lower expression of E-cadherin, which is an epithelial marker, was measured, while the expression of mesenchymal markers (N-cadherin and vimentin) increase [24]. It is reported that EMT attenuated the interaction of tumor cells, thus contributing to distant metastasis [25, 26].
In this study, our data elucidated that miR-615-3p was upmodulated in OS and inhibition of miR-615-3p hindered OS development via suppressing mTOR pathway. Mechanistically, we found that SESN2, involved in the mTOR pathway as a key regulator, was suppressed by miR-615-3p, thereby leading to the activation of mTOR. Collectively, our study indicated that molecular strategy targeting the miR-615-3p/SESN2/mTOR pathway may be hopeful for OS treatment.
Materials and methods
Cell culture
The human OS cell lines U-2OS, 143B, MG63, and MNNG-HOS, as well as the human osteoblast cell line hFOB1.19, were obtained from the Cell Bank of the Chinese Academy of Sciences. Incubating OS cells at 37 °C in humidified air with 5% CO2, while maintaining hFOB1.19 cells at 34.5 °C with 5% CO2 humidified air. All cells were grown in a medium consisting of 10% fetal bovine serum (FBS) along with 1% penicillin-streptomycin.
Cell transfection
Control inhibitor and miR-615-3p inhibitor, obtained from Gene-Pharma (Shanghai, China), were introduced into the OS cells using Lipofectamine 3000 (Invitrogen). Transfection was performed meticulously following the detailed instructions provided by the supplier to ensure optimal delivery and functioning of the inhibitors within the cellular environment. Lentiviral vector to knockdown SESN2 was purchased from Sigma (catalog numbers: TRCN0000087791). Each lentiviral vector and packaging plasmids were introduced into HEK293T cells with Lipofectamine 3000. Cell supernatants gathered at 24, 48, and 72 h following transfection, filtered (0.45 μm) and then introduced to the target cells along with 4 µg/µl of polybrene. Plasmids, including SESN2 and negative controls, were provided by OBiO Technology (Shanghai, China). SESN2-overexpressing cell lines were constructed via the plasmids constructed above. Puromycin was used to select infected cells after 72 h. qRT-PCR were used for verification of transfection efficiency.
Quantitative real-time PCR analysis (qRT-PCR)
Total RNA was meticulously extracted from all cell lines. This process was followed by the reverse transcription of the isolated RNA into cDNA. All operations were performed following the instructions provided by manufacturer (Vazyme, Nanjing, China). qRT‒PCR analysis was performed as outlined in earlier studies. Normalization was achieved using β-Actin and U6 expression.
Cell proliferation assays and cell apoptosis assay
Cell counting kit-8 (CCK-8) assays, colony formation assays and cell apoptosis assays were executed following the established protocols [27].
Western blotting
We used RIPA buffer (Sigma, USA) to extract protein, which contained protease inhibitors (Roche, Mannheim, Germany). Electrophoresis was carried out with 12% SDS–PAGE gels to separate proteins. Following transfer, primary antibodies were incubated overnight at 4 °C following a blocking step with 5% skim milk for 1 h. TBST was used to wash the membranes before secondary antibodies were added. The membranes were then washed once more in TBST, and luminescence was measured with an ECL detection kit (Share-bio, Shanghai, China).
EdU assay
Assays of cell proliferation were performed using a EdU kit (Beyotime, Shanghai, China). Briefly, target cells were placed in 12-well chambers and treated with 200 µl EdU each well for 2 h. Next, the cells were fixed using 4% paraformaldehyde and treated with 0.3% Triton X-100 to allow for permeabilization. Following washing, each well received 5 µg/ml of Hoechst 33,342, which was then incubated for 30 min. Images were acquired under the inverted microscope (Olympus).
Migration and invasion assays
Transwell assays were applied for cell metastasis ability assessment. In the migration assay, 3 × 104 treated OS cells were placed in the upper chamber filled with serum-free medium, while the lower chamber was supplied with medium containing 10% FBS to serve as a chemo-attractant. For invasion assay, diluted Matrigel (BD Biosciences, USA) was used to coat transwell inserts (8-µm pore size, 24-well inserts; Corning, NY, USA). 4% formaldehyde was used to fix the cells after 48 h of incubation. After staining with crystal violet, inverted microscope (Olympus) was used to count and photograph the cells that passed through the membrane.
Mouse xenograft model
Approval for all animal experiments was granted by the Animal Protection and Use Committee of the Affiliated Hospital of Nanjing Medical University. 5-week-old female BALB/C nude mice received subcutaneous injections of 1.5 × 106 transfected cells (n = 5 per group). We assessed tumor volume every five days and calculated them as follows: Width2 × Length × 0.5. All mice were euthanized after 20 days, and then tumors were isolated and weighed.
Immunohistochemistry (IHC) staining
Cell proliferation was tested by the anti-Ki67 antibody (GB13030; Servicebio, Wuhan, China). Tunel kit (Roche, Basel, Switzerland) was employed for cell apoptosis of xenograft tumor tissues assessment. Fluorescence images were captured using a microscope.
Fluorescence in situ hybridization (FISH)
Alena Biotechnology Co., Ltd. (Xi’an, China) provide the OS tissue microarray (n = 40). The miR-615-3p and SESN2 probes were used to hybridize OS tissue sections (Servicebio, Wuhan, China). FISH assay was executed as mentioned previously [28].
Luciferase activity assays
The dual-luciferase reporter assays were executed to verify the association between miR-615-3p and SESN2 as previously described [29].
Database analysis
Potential target genes of miR-615-3p were examined through the online tools StarBase (http://starbase.sysu.edu.cn) and TargetScan (http://www.targetscan.org/).
Bioinformatic Analysis
Data from the GSE28423 microarray dataset, which includes 18 disease samples and 3 healthy controls, was sourced from Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). The normalizeBetweenArrays function in ‘limma’ package of R was used to normalize the gene expression. The analysis of differential expression was conducted in R employing both the limma as well as DESeq2 packages.
Survival analysis
Metastasisfree survival analysis of 88 OS patients was performed via Kaplan-Meier method by the online database (http://hgserver1.amc.nl).
Statistical analyses
All statistical results are presented as means ± SD. Two-tailed Student’s t-test was performed to compare differences between groups (GraphPad Prism, version 9.3.1). A p value of < 0.05 was regarded as indicative of a statistically significant difference.
Results
Mir-615-3p is remarkably upregulated in OS
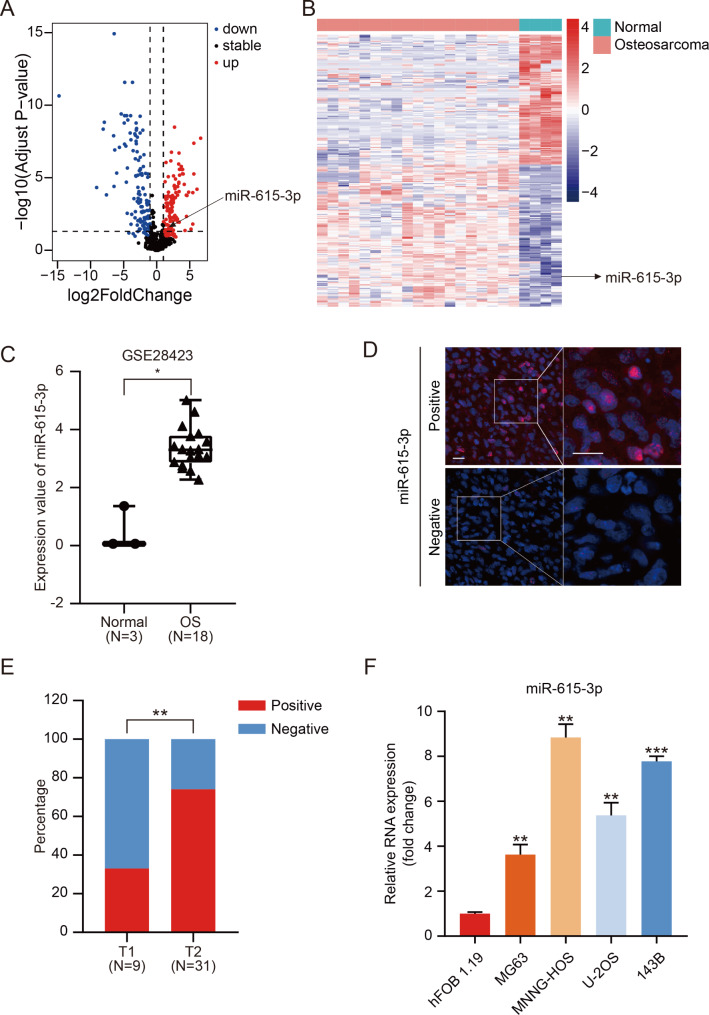
To identify the miRNAs that are differentially expressed in OS, we applied the R package to dissect the miRNA expression profiles of GSE28423 from GEO database. As depicted in volcano plot and heatmap (Fig. 1A, B), differential expression miRNAs between normal tissues and OS were generated using R package limma (logFC > 1 and adj.P.Value < 0.05). Among these differentially expressed miRNAs, we noted that miR-615-3p was obviously upregulated in OS (Fig. 1C). To further verify these results, we assessed miR-615-3p expression by FISH assay using a specific miR-615-3p-probe on OS tissue microarray (n = 40). A positive correlation between miR-615-3p expression and OS pathological staging was found (Fig. 1D, E). Moreover, miR-615-3p expression in hFOB1.19 cells and in OS cell lines (MG63, U-2OS, MNNG-HOS and 143B) was detected through qRT-PCR. As presented in Fig. 1F, miR-615-3p was obviously upregulated in OS, especially in 143B and MNNG-HOS cells. In conclusion, our data elucidated that miR-615-3p was high-expression in OS and may be associated with osteosarcoma progression.
Fig. 1.
miR-615-3p was upregulated in OS cells and tissue. A. Volcano plot representing the differentially expressed genes in GEO datasets (GSE28423). B. Heatmap showed the differentially expressed genes in GSE28423 downloaded from the GEO database. C. The expression of miR-615-3p in normal and osteosarcoma samples of GSE28423 dataset. D. Representative FISH photographs of the expression patterns of miR-615-3p in human OS tissues. Scale bars = 20 μm. E. Statistical analysis of FISH results based on the expression level of miR-615-3p in T1 stage (n = 9) and T2 stage (n = 31) OS tissues. F. Relative miR-615-3p mRNA levels of MG63, MNNG-HOS, U-2OS and 143B relative to hFOB1.19 cells were determined using qRT-PCR. Results are displayed as mean ± SD, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
Mir-615-3p promotes proliferation and suppresses apoptosis in OS cell
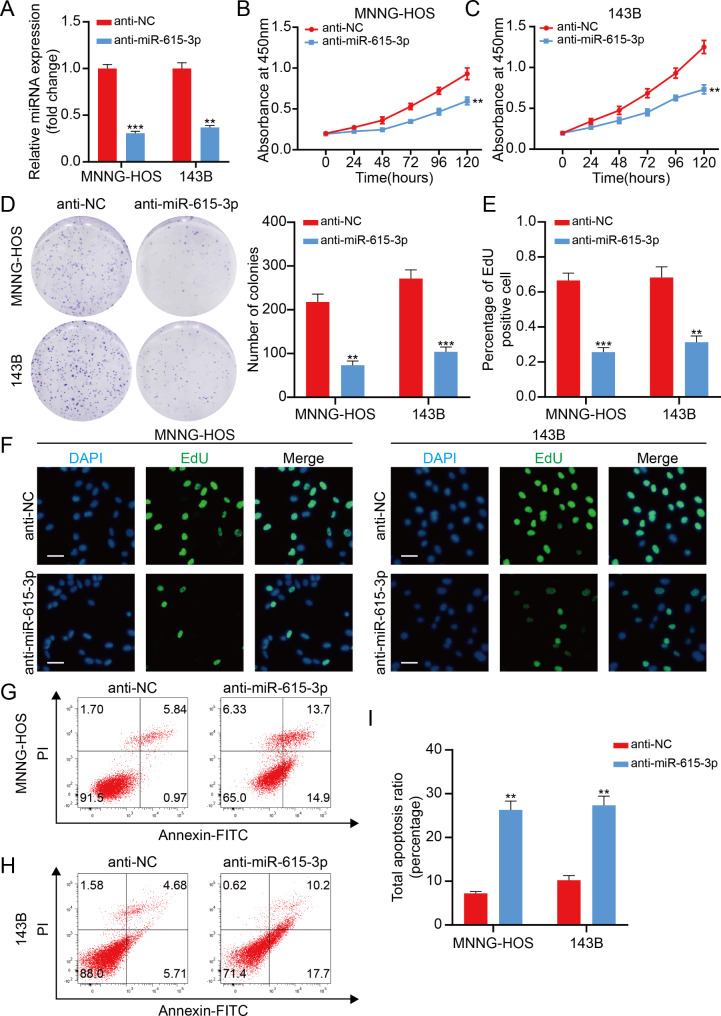
To investigate the function of miR-615-3p in OS, we silenced miR-615-3p in MNNG-HOS and 143B cells. Silencing efficiency of miR-615-3p was measured using qRT-PCR (Fig. 2A). As detected by CCK-8 assay, knockdown of miR-615-3p remarkably attenuated OS cell proliferative capacity (Fig. 2B, C). Meanwhile, the number of clones significantly reduced in miR-615-3p knockdown group (Fig. 2D). Subsequently, we performed the EdU assay and got the same result (Fig. 2E, F). Furthermore, we found that miR-615-3p silencing greatly stimulated OS cell apoptosis (Fig. 2G-I). In general, these data clearly clarified that miR-615-3p stimulated OS cell proliferation and weakened cell apoptosis.
Fig. 2.
miR-615-3p promotes proliferation and inhibited apoptosis in OS cell. A. Interference efficacy of sh-RNA targeting of miR-615-3p in 143B and MNNG-HOS cells was determined by qRT-PCR. B and C. Knockdown of miR-615-3p suppressed proliferation capability of 143B and MNNG-HOS cells using the CCK-8 assay. D. Knockdown of miR-615-3p suppressed the proliferation of OS cells (143B and MNNG-HOS) using the colony formation assay. E. miR-615-3p inhibitor decreased the percentage of EdU-positive OS cells. F. Representative photographs of the EdU incorporation assay. Scale bars = 20 μm. G-I. The knockdown of miR-615-3p significantly induces apoptosis of 143B and MNNG-HOS cells. Results are displayed as mean ± SD, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
MiR-615-3p facilitates OS cell metastasis through stimulating EMT in vitro
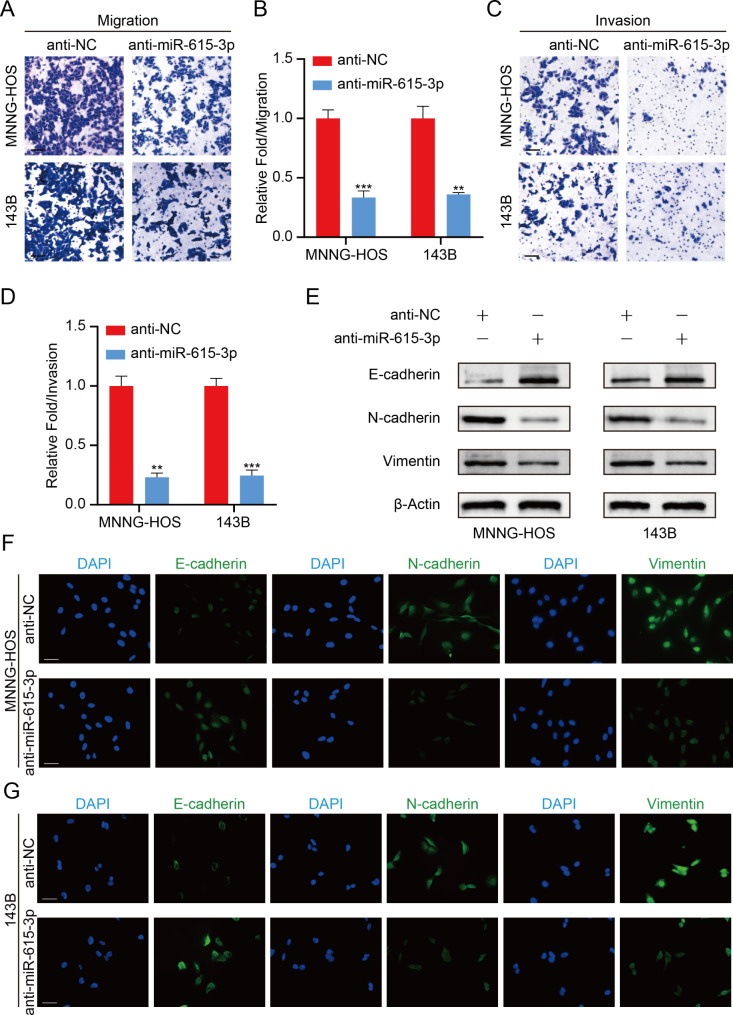
Next, we examined the effect of miR-615-3p on OS cell metastasis by transwell assays. As shown in Fig. 3A-D, the migration and invasion abilities of OS cell were substantially suppressed due to the inhibition of miR-615-3p.
Fig. 3.
miR-615-3p promotes OS cell metastasis via stimulating EMT in vitro. A. Cell migration ability was detected in OS cells with miR-615-3p knockdown or not. Scale bars = 50 μm. B. Quantification of the cell migration ability from the Fig. 3A. C. Cell invasion ability was detected in OS cells with miR-615-3p knockdown or not. Scale bars = 50 μm. D. Quantification of the cell invasion ability from the Fig. 3C. E. Protein levels of EMT markers was determined in different groups (anti-NC and anti-miR-615-3p). F and G. Immunofluorescence staining showed the changes in the expression of EMT markers (green) in OS cells. Nuclei were counterstained with DAPI (blue). Scale bars = 20 μm. Results are displayed as mean ± SD, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
Recently, growing researches demonstrated that EMT exerts a crucial function in tumor metastasis. Therefore, we carried out western blotting to measure EMT markers expression. The results demonstrated that miR-615-3p knockdown led to an increase in E-cadherin expression, which was a marker of epithelial phenotype, while the expression of mesenchymal markers was significantly inhibited after miR-615-3p silencing (Fig. 3E). To further validate the above results, we carried out Immunofluorescence assay. We found that inhibition of miR-615-3p induced a sharp decline in the fluorescence intensity of E-cadherin, while a significant increase in the fluorescence intensity of N-cadherin and Vimentin was observed in miR-615-3p knockdown group (Fig. 3F, G). Together, our research revealed that miR-615-3p accelerated metastasis of OS via stimulating EMT.
Mir-615-3p directly targets SESN2 in OS cells
Growing studies reported that the mTOR pathway exerted a crucial impact on proliferation, EMT process and distant metastasis [30, 31]. Therefore, we sought to explore whether miR-615-3p exerts its biological function in OS as an oncogene by stimulating the mTOR pathway.
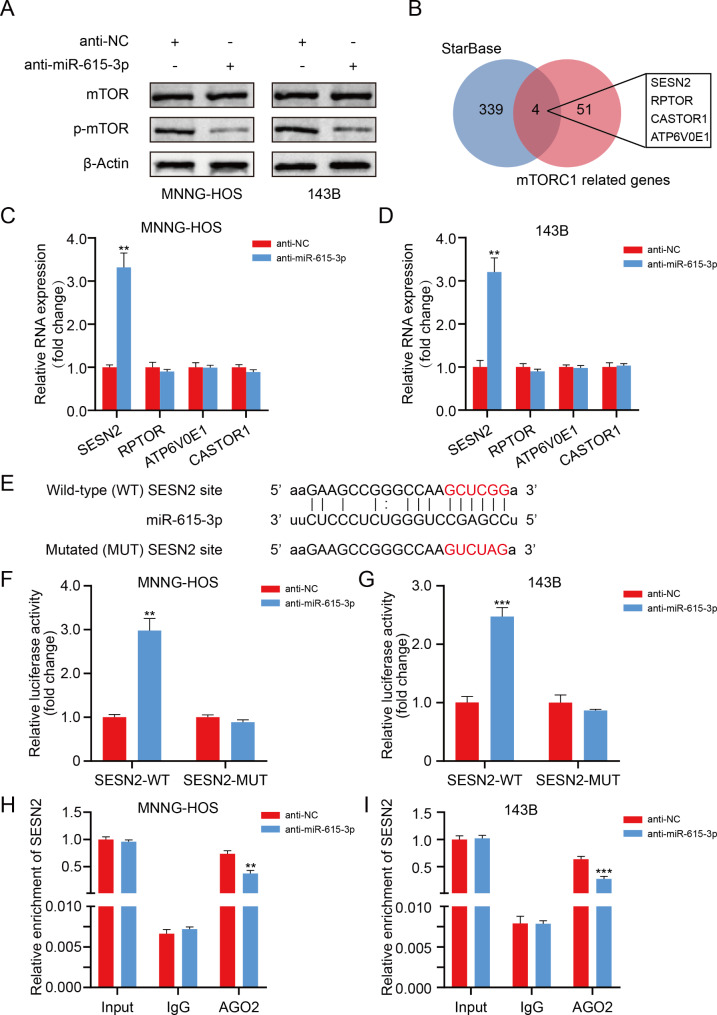
Afterwards, western blotting was performed to clarify the connection between miR-615-3p and the mTOR pathway in OS cells. We found that the phosphorylation degree of mTOR markedly decreased after interfering with miR-615-3p (Fig. 4A).
Fig. 4.
miR-615-3p directly targets SESN2 in OS cells. A. Protein levels of total and phosphorylated mTOR in different groups (anti-NC and anti-miR-615-3p). B. Venn diagram showing the predicted mTORC1-related target genes of miR-615-3p obtained from StarBase databases. C and D. The mRNA expression patterns of the predicted target genes in different groups (anti-NC and anti-miR-615-3p). E. The wild-type and the mutated sequences of the SESN2 mRNA 3’-UTR (mutation site: red). F and G. The luciferase activity of the OS cells (143B and MNNG-HOS) in luciferase reporter plasmid containing wild-type SESN2 3’-UTR (SESN2-WT) and mutant SESN2 3’-UTR (SESN2-MUT) co-transfected with miR-615b-3p inhibitors or negative control was assessed. H and I. RIP assays using antibodies against AGO2 or IgG were performed in cellular lysates from 143B and MNNG-HOS cells. qRT-PCR showed the relative enrichment of SESN2 in different groups (anti-NC and anti-miR-615-3p). Results are displayed as mean ± SD, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
To elucidate the underlying mechanisms of miR-615-3p in regulating mTOR pathway in OS, potential miR-615-3p target mRNA candidates were identified through two target gene prediction algorithms (StarBase and TargetScan). We found that only SESN2, RPTOR, CASTOR1 and ATP6V0E1 were related to both miR-615-3p and mTOR pathway (Fig. 4B). Subsequently, we discovered that the miR-615-3p inhibition stimulated SESN2 expression remarkably in OS cells, while no noticeable changes in the expression of RPTOR, CASTOR1 and ATP6V0E1 were observed (Fig. 4C, D). Moreover, an assessment of the connection between SESN2 expression and the prognosis in OS was carried out via R2 database. As shown in Fig. S1A, low SESN2 expression predicts a poor prognosis for OS patients. (p = 0.020). The potential binding site of miR-615-3p on SESN2 was displayed in Fig. 4E. To verify in depth the interaction between miR-615-3p and SESN2, luciferase reporter assays were carried out. As expected, the miR-615-3p knockdown induced a visible enhancement in the luciferase activity of SESN2-3’-UTR-wildtype reporter, while the luciferase intensity of SESN2-3’-UTR-mutation reporter did not change obviously (Fig. 4F, G). In addition, the RNA immunoprecipitation (RIP) assay illustrated that inhibition of miR-615-3p led to a lower enrichment level of SESN2 in the anti-Argonaute2 (Ago2) group (Fig. 4H, I). Collectively, our data demonstrated that miR-615-3p interacts directly with SESN2, thus regulating SESN2 expression in OS.
SESN2 attenuates OS cell progression through activating the mTOR pathway
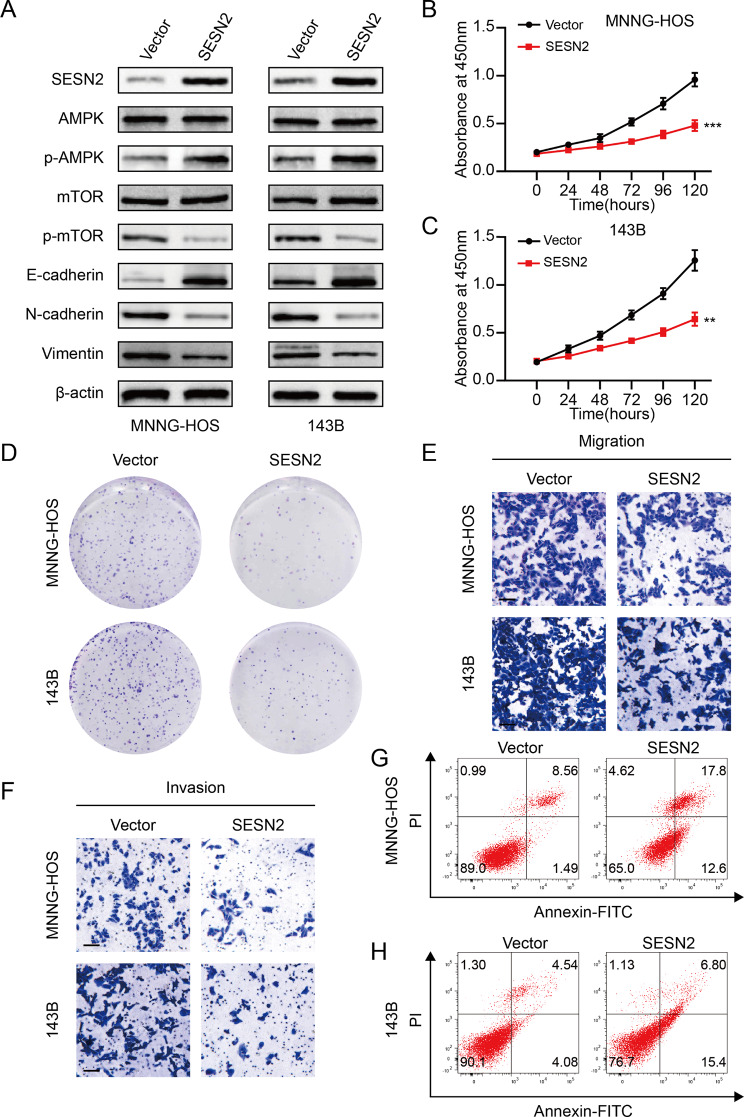
SESN2, a key regulator of mTORC1, exerts its biological function by mediating AMPK activation [32, 33]. It has been reported that SESN2 was involved in processes of numerous cancers [34, 35]. Nevertheless, the function of SESN2 in OS progression requires further exploration. We firstly overexpressed SESN2 in OS cells and overexpression efficiency was detected through western blotting (Fig. 5A). Additionally, western blotting analysis indicated that SESN2 substantially suppressed the activation of mTOR pathway as well as the expression of mesenchymal markers (Fig. 5A). As a result, CCK-8 assays revealed that the proliferative activity of OS cells was remarkably suppressed in the SESN2 overexpression group (Fig. 5B, C). The same results were observed in colony formation assays (Fig. 5D and Fig. S2A). Meanwhile, the transwell assays confirmed that SESN2 notably reduced the metastasis capacity of OS cells (Fig. 5E, F and Fig. S2B, C). Additionally, a higher apoptosis rate of OS cells was induced by the overexpression of SESN2 (Fig. 5G, H and Fig. S2D). These observations suggested that SESN2, at least to some extent, inhibited OS progression via regulating mTOR pathway.
Fig. 5.
SESN2 attenuates OS cell progression through activating the mTOR pathway. A. Protein levels of SESN2, total and phosphorylated AMPK, total and phosphorylated mTOR and EMT markers in 143B and MNNG-HOS cells transfected with SESN2 plasmid or negative control. B and C. SESN2 overexpression suppressed the cell viability of 143B and MNNG-HOS cells using the CCK- 8 assay. D. SESN2 overexpression suppressed the colony formation capability of OS cells (143B and MNNG-HOS). E and F. SESN2 overexpression suppressed the cell migration and invasion ability of 143B and MNNG-HOS cells. G and I. SESN2 overexpression induced the apoptosis of OS cells (143B and MNNG-HOS). Results are displayed as mean ± SD, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
Mir-615-3p promotes OS development via SESN2-mediated mTOR pathway
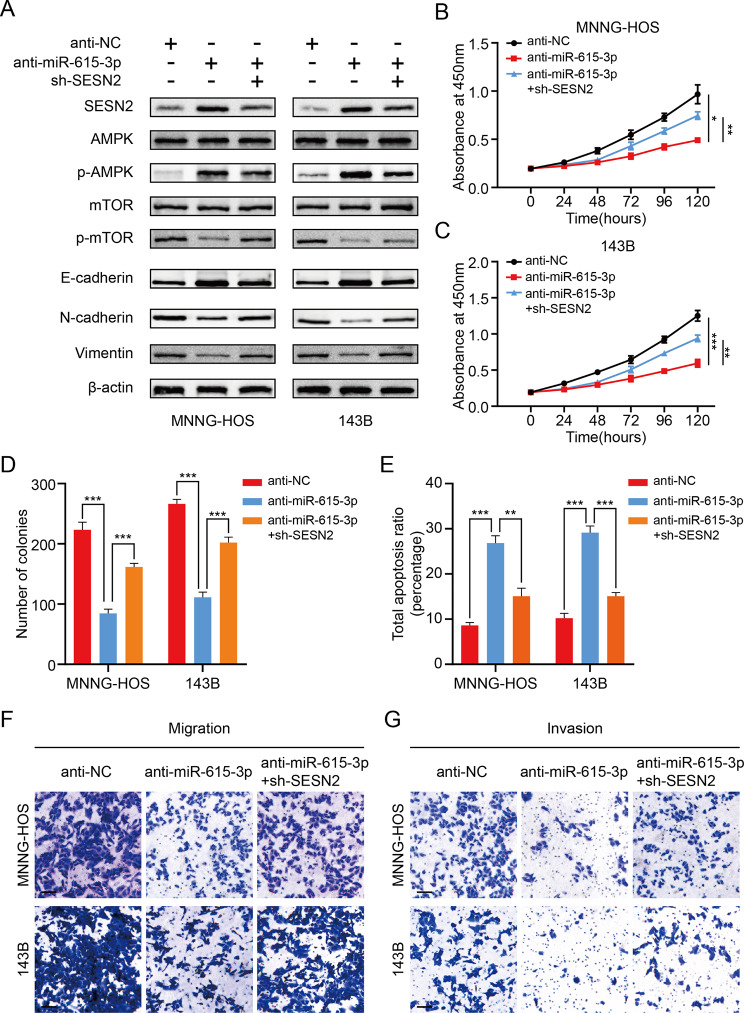
To assess whether miR-615-3p is involved in OS development via SESN2-mediated mTOR pathway in depth, a set of rescue experiments were conducted. We treated OS cells using miR-615-3p inhibitors plus sh-SESN2. As shown in Fig. 6A, the influence of silencing miR-615-3p on SESN2, p-AMPK, p-mTOR and EMT biomarkers was partly reversed by SESN2 knockdown. Next, CCK-8 assays demonstrated that miR-615-3p knockdown markedly suppressed OS cell proliferation, which was partially restored through downregulating SESN2 (Fig. 6B, C). The same results were confirmed by colony formation assays (Fig. 6D and Fig. S3A). Meanwhile, the results of apoptosis revealed that silencing SESN2 dramatically weaken the positive impact of miR-615-3p knockdown on apoptosis (Fig. 6E and Fig. S3B, C). Additionally, we found that the inhibition of migration and invasion abilities was partly rescued in the miR-615-3p knockdown group after SESN2 silencing (Fig. 6F, G and Fig.S3D, E). These data demonstrated that miR-615-3p exerted its tumor-promoting function by regulating the activity of SESN2-mediated mTOR pathway.
Fig. 6.
miR-615-3p promotes OS development via SESN2-mediated mTOR pathway. A. Protein levels of SESN2, total and phosphorylated AMPK, total and phosphorylated mTOR and EMT markers in 143B and MNNG-HOS cells transfected with miR-615b-3p inhibitor, sh-SESN2, or negative control. B and C. SESN2-knockdown partly reversed the suppressed effects of miR-615-3p-knockdown on the cell viability of 143B and MNNG-HOS cells using the CCK- 8 assay. D. SESN2-knockdown partly reversed the suppressed effects of miR-615-3p-knockdown on the colony formation capability of 143B and MNNG-HOS cells. E. SESN2-knockdown partly reversed the induced effect of miR-615-3p-knockdown on the apoptosis of OS cells (143B and MNNG-HOS). F and G. SESN2-knockdown partly reversed the suppressed effects of miR-615-3p-knockdown on the cell migration and invasion ability of OS cells (143B or MNNG-HOS) in different groups (anti-NC, anti-miR-615-3p and anti-miR-615-3p + sh-SESN2). Results are displayed as mean ± SD, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
MiR-615-3p promotes OS development by modulating SESN2 in vivo
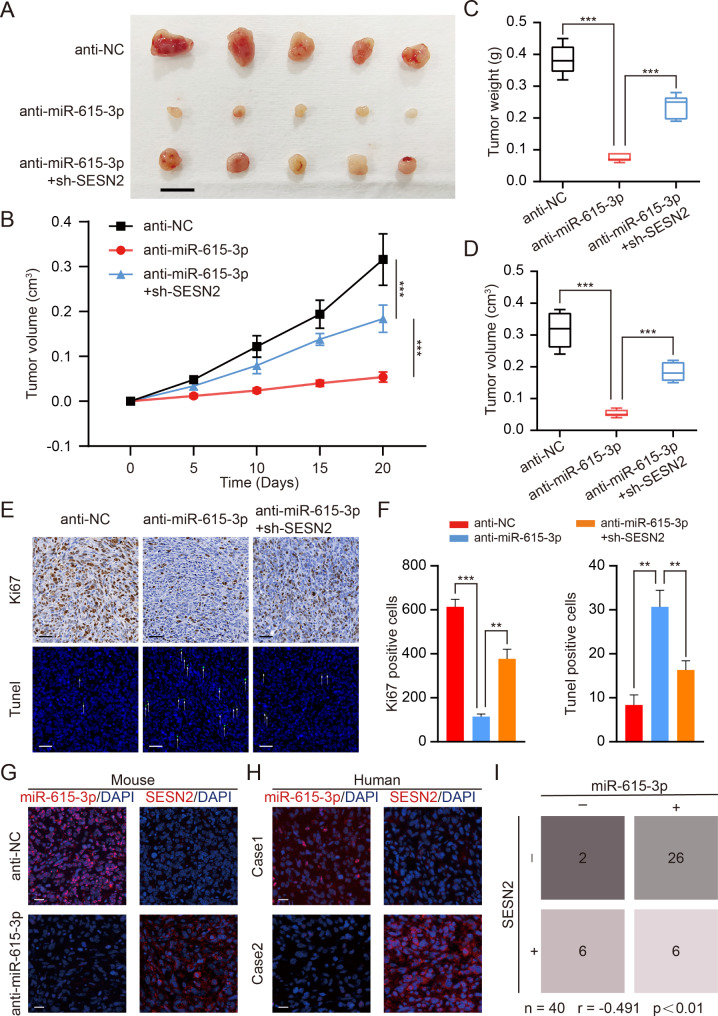
Next, to investigate whether miR-615-3p promotes the viability of OS cells in vivo, we constructed xenograft models by performing subcutaneous injections with treated cells in nude mice. As displayed in Fig. 7A-D, compared to the control conditions, miR-615-3p knockdown greatly diminished the tumor load and suppressed the development of xenograft tumors. However, the partial restoration of the tumor suppressor effect from miR-615-3p silencing was achieved by inhibition of SESN2. Afterwards, we used Ki67 staining and TUNEL assay to assess proliferation and apoptosis of xenograft tumor, respectively. As a result of miR-615-3p knockdown, cell viability decreased and apoptosis was activated in vivo (Fig. 7E, F). This effect can be reversed, however, by SESN2 silencing. FISH and IF staining were used to establish the expression patterns of miR-615-3p as well as SESN2 in xenograft tumors (Fig. 7G). Our data were further confirmed in OS samples (Fig. 7H). The negative relation between miR-615-3p and SESN2 was apparent (Fig. 7I). In summary, our data clarified that miR-615-3p stimulated OS development through targeting SESN2 in vivo.
Fig. 7.
miR-615-3p promotes OS growth by targeting SESN2 in vivo. A. Morphologic characteristics of xenograft tumors from different groups (anti-NC, anti-miR-615-3p, anti-miR-615-3p + sh-SESN2) (n = 5). Scale bars = 1 cm. B. The tumor volumes were measured with calipers every 5 days. C. Tumor weights at 20 days were measured in each group (anti-NC, anti-miR-615-3p, anti-miR-615-3p + sh-SESN2). The median, upper, and lower quartiles were plotted, and the whiskers that extend from each box indicate the range of values that were outside of the intra-quartile range (n = 5). D. Tumor volumes at 20 days were measured in each group (anti-NC, anti-miR-615-3p, anti-miR-615-3p + sh-SESN2). The median, upper, and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range (n = 5). E. Representative images of Ki67 and TUNEL staining in the xenograft tumors from each experimental group of nude mice. A TUNEL positive cell is indicated with an arrow. Scale bars = 50 μm. F. Quantification of positive cells from the Fig. 7E. G. Representative photographs of the expression patterns of miR-615-3p and SESN2 in tumor tissues from subcutaneous xenograft mouse model by IF and FISH. Scale bars = 20 μm. H. Representative photographs of the expression patterns of miR-615-3p and SESN2 in huamn OS tissues by IF and FISH. Scale bars = 20 μm. I. A negative correlation between the expression pattern of miR-615-3p and SESN2 (n = 40, r = -0.491, p < 0.01). Results are displayed as mean ± SD, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
Discussion
So far, the treatment of OS remains a huge clinical challenge. Emerging researches have indicated that many microRNAs are related to the development of multitudinous tumors including OS. miRNAs exert a vital role in a series of biological processes through regulating downstream genes expression [36, 37]. Liang et al. demonstrated that miR-744-5p could regulate MAPK signaling by targeting TGF-β1, thus inhibiting the growth and metastasis of OS [38]. Liu et al. revealed that miR-140 negatively modulated OS proliferation and metastasis via USP22/LSD1/p21 axis [39]. Liang et al. clarified that miR-23b-3p, which was upmodulated in OS, activated the PI3K/AKT pathway by interacting with the 3’-UTR of VEPH1, thus promoting OS progression [40]. Through our analysis of data from GEO database, we discovered an increase in miR-615-3p expression was observed in OS compared to control group. Previous study revealed that miR-615-3p exerted tumor-promoting effect in various cancers. Lei et al. found that miR-615-3p facilitated the metastasis via PICK1/TGFBRI axis mediated EMT in breast cancer [41]. Another report clarified that miR-615-3p, which is upmodulated in prostate cancer, may serve as a prognostic biomarker [42]. In this study, our results revealed that miR-615-3p, which was upregulated in OS, enhanced OS proliferative activity and migration ability and suppressed apoptosis of OS.
SESN2, which belongs to the sestrin family, is induced by various stress signals and is highly conserved in most eukaryotes [43]. SESN2 executes its biological function as a key inhibitor of mTORC1. The effect of SESN2 on mTORC1 activity can be mediated by phosphorylation of AMPK [44]. Growing evidences reported that SESN2/AMPK/mTOR pathway was associated with the development of different diseases. According to previous report, SESN2 played a crucial role in protecting hair cells from gentamicin [45]. Han et al. indicated that hepatic SESN2, which was upregulated during cholestatic liver injury, ameliorated bile acid-induced ER stress via AMPK/mTOR pathway, thus preventing liver damage [46]. In addition, SESN2 mediated anti-tumor effect has been reported. Liang et al. revealed that SESN2 inhibited the growth of bladder cancer by activating autophagy [35]. Shin et al. found that low-expressed SESN2 could stimulate endometrial cancer progression via activating mTOR pathway [47]. It was reported that Tanshinone IIA (TIIA) exerted its anti-cancer effect by SESN2-induced autophagy in OS [48]. In our study, we elucidated that SESN2 could enhance apoptosis and attenuate viability and metastasis of OS. Furthermore, miR-615-3p repressed SESN2 expression through interacting with the 3’-UTR of SESN2, therefore promoting OS progression.
EMT is a process which plays a vital part in progression of various tumors [49]. Interestingly, growing reports illuminated that mTOR pathway was involved in the activation of the EMT. Luo et al. found that YTHDF1 promoted metastasis of hepatocellular carcinoma via PI3K/AKT/mTOR-mediated EMT [50]. Wang et al. demonstrated that APOC2-induced mTOR activation promoted cell metastasis by enhancing the EMT process in gastric cancer [30]. Our research elucidated that miR-615-3p facilitated EMT through SESN2/AMPK/mTOR pathway, thus promoting metastasis of OS.
Conclusion
Generally, these findings demonstrated that miR-615-3p is highly expressed in OS. Silencing miR-615-3p inhibits OS development in vitro and in vivo. Subsequent researches indicated that miR-615-3p functioned as an oncogene by SESN2/AMPK/mTOR axis. These findings underscore the prospect of focusing on miR-615-3p and SESN2 as targets for OS treatment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Yifei Shen and Dr. Yunkun Zhang (Department of Orthopedics, The Affiliated Changzhou Second People’s Hospital of Nanjing Medical University, Changzhou Medical Center, Nanjing Medical University, Changzhou) for technical assistance.
Author contributions
D.Z., Y.Q.J. and X.C.Y. were responsible for the concept and experimental design. X.C.Y., X.W., F.X. and X.Y.Z. performed the experiments, data analysis and statistical analysis. M.Y.W., R.K.Z., Z.Y.S., X.H.P., L.F., W.C.Z. and Y.S. technical and material support. X.C.Y., X.W., F.X. and X.Y.Z. were involved in drafting and revision of the manuscript. W.T.Z. performed the animal experiments. Y.Q.J. and D.Z. supervised this study.All authors discussed the results and commented on the manuscript.
Funding
This study was supported from the National Science Foundation Committee (NSFC) of China (Grant number: No. 82160555), Natural Science Foundation of Xinjiang Uygur Autonomous Region (2022D01A317), Zhenjiang Social Development project (FZ2021059).
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Declarations
Ethics Statement
This study was approved by the research medical ethics committee of the Affiliated Hospital of the Nanjing Medical University Animal Protection and Use Committee. Informed Consent: N/A. Registry and the Registration No. of the study/trial: N/A. Animal Studies: N/A.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenting Zhang, Email: wenting_zhang@ntu.edu.cn.
Dong Zhou, Email: zhoudong1012@163.com.
Yuqing Jiang, Email: jyqbaba@126.com.
References
- 1.Park JA, Cheung NV. GD2 or HER2 targeting T cell engaging bispecific antibodies to treat osteosarcoma. J Hematol Oncol. 2020;13(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z, Li Y, Kuang M, Wang X, Jia Q, Cao J, Hu J, Wu S, Wang Z, Xiao J. The CD24(+) cell subset promotes invasion and metastasis in human osteosarcoma. EBioMedicine. 2020;51:102598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu XK, Rao SS, Tan YJ, Yin H, Luo MJ, Wang ZX, Zhou JH, Hong CG, Luo ZW, Du W, Wu B, Yan ZQ, He ZH, Liu ZZ, Cao J, Wang Y, Situ WY, Liu HM, Huang J, Wang YY, Xia K, Qian YX, Zhang Y, Yue T, Liu YW, Zhang HQ, Tang SY, Chen CY, Xie H. Fructose-coated Angstrom silver inhibits osteosarcoma growth and metastasis via promoting ROS-dependent apoptosis through the alteration of glucose metabolism by inhibiting PDK. Theranostics. 2020;10(17):7710–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang YH, Wu SH, Kuo YC, Chen HF, Chiou A, Lee OK. Raman spectroscopy for grading of live osteosarcoma cells. Stem Cell Res Ther. 2015;6(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xing S, Tian Z, Zheng W, Yang W, Du N, Gu Y, Yin J, Liu H, Jia X, Huang D, Liu W, Deng M. Hypoxia downregulated miR-4521 suppresses gastric carcinoma progression through regulation of IGF2 and FOXM1. Mol Cancer. 2021;20(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM, Zhang JF. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the wnt pathway. Genome Biol. 2019;20(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy JW, Zhang Y, Sheehan C, D’Souza-Schorey C. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat Cell Biol. 2019;21(7):856–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Y, Murray-Stewart T, Wang Y, Yu F, Li J, Marton LJ, Casero RA Jr., Oupický D. Self-immolative nanoparticles for simultaneous delivery of microRNA and targeting of polyamine metabolism in combination cancer therapy. J Control Release. 2017;246:110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu Y, Liu S, Zhang X, Chen G, Liang H, Yu M, Liao Z, Zhou Y, Zhang CY, Wang T, Wang C, Zhang J, Chen X. Oncogenic miR-19a and miR-19b co-regulate tumor suppressor MTUS1 to promote cell proliferation and migration in lung cancer. Protein Cell. 2017;8(6):455–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao CC, Ho MY, Liang SM, Liang CM. Autophagic degradation of SQSTM1 inhibits ovarian cancer motility by decreasing DICER1 and AGO2 to induce MIRLET7A-3P. Autophagy. 2018;14(12):2065–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Li R, Zhang J, Meng C, Zhang J, Song X, Lv C. MicroRNA-708-3p as a potential therapeutic target via the ADAM17-GATA/STAT3 axis in idiopathic pulmonary fibrosis. Exp Mol Med 2018, 50 (3), e465. [DOI] [PMC free article] [PubMed]
- 12.Seo Y, Kim SS, Kim N, Cho S, Park JB, Kim JH. Development of a miRNA-controlled dual-sensing system and its application for targeting miR-21 signaling in tumorigenesis. Exp Mol Med. 2020;52(12):1989–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan T, Ooi WF, Qamra A, Cheung A, Ma D, Sundaram GM, Xu C, Xing M, Poon L, Wang J, Loh YP, Ho JHJ, Ng JJQ, Ramlee MK, Aswad L, Rozen SG, Ghosh S, Bard FA, Sampath P, Tergaonkar V, Davies JOJ, Hughes JR, Goh E, Bi X, Fullwood MJ, Tan P, Li S. HoxC5 and mir-615-3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis. Nat Commun. 2018;9(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laursen EB, Fredsøe J, Schmidt L, Strand SH, Kristensen H, Rasmussen AKI, Daugaard TF, Mouritzen P, Høyer S, Kristensen G, Stroomberg HV, Brasso K, Røder MA, Borre M, Sørensen KD. Elevated mir-615-3p expression predicts adverse clinical outcome and promotes Proliferation and Migration of prostate Cancer cells. Am J Pathol. 2019;189(12):2377–88. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Liu L, Sun Y, Xue Y, Qu J, Pan S, Li H, Qu H, Wang J, Zhang J. Mir-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed Pharmacother. 2018;101:406–13. [DOI] [PubMed] [Google Scholar]
- 16.Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer. 2018;18(12):744–57. [DOI] [PubMed] [Google Scholar]
- 17.Marques-Ramos A, Cervantes R. Expression of mTOR in normal and pathological conditions. Mol Cancer. 2023;22(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beauchamp EM, Abedin SM, Radecki SG, Fischietti M, Arslan AD, Blyth GT, Yang A, Lantz C, Nelson A, Goo YA, Akpan I, Eklund EA, Frankfurt O, Fish EN, Thomas PM, Altman JK, Platanias LC. Identification and targeting of novel CDK9 complexes in acute myeloid leukemia. Blood. 2019;133(11):1171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arenas DJ, Floess K, Kobrin D, Pai RL, Srkalovic MB, Tamakloe MA, Rasheed R, Ziglar J, Khor J, Parente SAT, Pierson SK, Martinez D, Wertheim GB, Kambayashi T, Baur J, Teachey DT, Fajgenbaum DC. Increased mTOR activation in idiopathic multicentric Castleman disease. Blood. 2020;135(19):1673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue F, Cheng J, Liu Y, Cheng C, Zhang M, Sui W, Chen W, Hao P, Zhang Y, Zhang C. Cardiomyocyte-specific knockout of ADAM17 ameliorates left ventricular remodeling and function in diabetic cardiomyopathy of mice. Signal Transduct Target Ther. 2022;7(1):259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan S, Huang W, Liu X, Liu X, Chen N, Xu Q, Hu Y, Song W, Zhou J. IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. J Exp Clin Cancer Res. 2018;37(1):304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res. 2019;38(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Meng L, Li X, Li D, Liu Q, Chen Y, Li X, Bu W, Sun H. Regulation of FN1 degradation by the p62/SQSTM1-dependent autophagy-lysosome pathway in HNSCC. Int J Oral Sci. 2020;12(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang W, Cai F, Xu H, Lu Y, Chen J, Liu J, Cao N, Zhang X, Chen X, Huang Q, Zhuang H, Hua ZC. Extracellular signal regulated kinase 5 promotes cell migration, invasion and lung metastasis in a FAK-dependent manner. Protein Cell. 2020;11(11):825–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du X, Zhang Z, Zheng X, Zhang H, Dong D, Zhang Z, Liu M, Zhou J. An electrochemical biosensor for the detection of epithelial-mesenchymal transition. Nat Commun. 2020;11(1):192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao SJ, Shen YF, Li Q, He YJ, Zhang YK, Hu LP, Jiang YQ, Xu NW, Wang YJ, Li J, Wang YH, Liu F, Zhang R, Yin GY, Tang JH, Zhou D, Zhang ZG. SLIT2/ROBO1 axis contributes to the Warburg effect in osteosarcoma through activation of SRC/ERK/c-MYC/PFKFB2 pathway. Cell Death Dis. 2018;9(3):390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Y, Xu J, Pan X, Zhang Y, Weng Y, Zhou D, He S. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020;11(4):278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X, Tan J, Tao T, Zhang X, Weng Y, Weng X, Xu J, Li H, Jiang Y, Zhou D, Shen Y. LINC01123 enhances osteosarcoma cell growth by activating the hedgehog pathway via the miR-516b-5p/Gli1 axis. Cancer Sci. 2021;112(6):2260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Yang Z, Xu E, Shen X, Wang X, Li Z, Yu H, Chen K, Hu Q, Xia X, Liu S, Guan W. Apolipoprotein C-II induces EMT to promote gastric cancer peritoneal metastasis via PI3K/AKT/mTOR pathway. Clin Transl Med 2021, 11 (8), e522. [DOI] [PMC free article] [PubMed]
- 31.Yang C, Dou R, Wei C, Liu K, Shi D, Zhang C, Liu Q, Wang S, Xiong B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther. 2021;29(6):2088–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang AJ, Tang Y, Zhang J, Wang BJ, Xiao M, Lu G, Li J, Liu Q, Guo Y, Gu J. Cardiac SIRT1 ameliorates doxorubicin-induced cardiotoxicity by targeting sestrin 2. Redox Biol. 2022;52:102310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin HR, Du CH, Wang CZ, Yuan CS, Du W. Ginseng metabolite Protopanaxadiol induces Sestrin2 expression and AMPK activation through GCN2 and PERK. Cell Death Dis. 2019;10(4):311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang D, Xu C, Yang W, Chen J, Ou Y, Guan Y, Guan J, Liu Y. E3 ligase RNF167 and deubiquitinase STAMBPL1 modulate mTOR and cancer progression. Mol Cell. 2022;82(4):770–e7849. [DOI] [PubMed] [Google Scholar]
- 35.Liang Y, Zhu J, Huang H, Xiang D, Li Y, Zhang D, Li J, Wang Y, Jin H, Jiang G, Liu Z, Huang C. SESN2/sestrin 2 induction-mediated autophagy and inhibitory effect of isorhapontigenin (ISO) on human bladder cancers. Autophagy. 2016;12(8):1229–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cesarini V, Silvestris DA, Tassinari V, Tomaselli S, Alon S, Eisenberg E, Locatelli F, Gallo A. ADAR2/miR-589-3p axis controls Glioblastoma cell migration/invasion. Nucleic Acids Res. 2018;46(4):2045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clausse V, Zheng H, Amarasekara H, Kruhlak M, Appella DH. Thyclotides, tetrahydrofuran-modified peptide nucleic acids that efficiently penetrate cells and inhibit microRNA-21. Nucleic Acids Res. 2022;50(19):10839–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang H, Li L, Zhu S, Tan J, Yang B, Wang X, Wu G, Xie C, Li L, Liu Z, Li Y, Song H, Chen G, Lin L. MicroRNA-744-5p suppresses tumorigenesis and metastasis of osteosarcoma through the p38 mitogen-activated protein kinases pathway by targeting transforming growth factor-beta 1. Bioengineered. 2022;13(5):12309–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Wang D, Liu L, Wang L, Yan M. miR-140 inhibits osteosarcoma progression by impairing USP22-mediated LSD1 stabilization and promoting p21 expression. Mol Ther Nucleic Acids. 2021;24:436–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan L, Cao X, Lei Y. MicroRNA miR-23b-3p promotes osteosarcoma by targeting ventricular zone expressed PH domain-containing 1 (VEPH1)/phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) pathway. Bioengineered. 2021;12(2):12568–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei B, Wang D, Zhang M, Deng Y, Jiang H, Li Y. Mir-615-3p promotes the epithelial-mesenchymal transition and metastasis of breast cancer by targeting PICK1/TGFBRI axis. J Exp Clin Cancer Res. 2020;39(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pudova EA, Kobelyatskaya AA, Katunina IV, Snezhkina AV, Fedorova MS, Pavlov VS, Bakhtogarimov IR, Lantsova MS, Kokin SP, Nyushko KM, Alekseev BY, Kalinin DV, Melnikova NV, Dmitriev AA, Krasnov GS, Kudryavtseva AV. Lymphatic dissemination in prostate Cancer: features of the Transcriptomic Profile and Prognostic models. Int J Mol Sci 2023, 24 (3). [DOI] [PMC free article] [PubMed]
- 43.Song S, Shi C, Bian Y, Yang Z, Mu L, Wu H, Duan H, Shi Y. Sestrin2 remedies podocyte injury via orchestrating TSP-1/TGF-β1/Smad3 axis in diabetic kidney disease. Cell Death Dis. 2022;13(7):663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng W, Cha J, Yuan J, Haraguchi H, Bartos A, Leishman E, Viollet B, Bradshaw HB, Hirota Y, Dey SK. p53 coordinates decidual sestrin 2/AMPK/mTORC1 signaling to govern parturition timing. J Clin Invest. 2016;126(8):2941–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebnoether E, Ramseier A, Cortada M, Bodmer D, Levano-Huaman S. Sesn2 gene ablation enhances susceptibility to gentamicin-induced hair cell death via modulation of AMPK/mTOR signaling. Cell Death Discov. 2017;3:17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han D, Kim H, Kim S, Le QA, Han SY, Bae J, Shin HW, Kang HG, Han KH, Shin J, Park HW. Sestrin2 protects against cholestatic liver injury by inhibiting endoplasmic reticulum stress and NLRP3 inflammasome-mediated pyroptosis. Exp Mol Med. 2022;54(3):239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin J, Bae J, Park S, Kang HG, Shin SM, Won G, Kim JS, Cho SG, Choi Y, Oh SM, Shin J, Kim JS, Park HW. mTOR-Dependent Role of Sestrin2 in Regulating Tumor Progression of Human Endometrial Cancer. Cancers (Basel) 2020, 12 (9). [DOI] [PMC free article] [PubMed]
- 48.Yen JH, Huang ST, Huang HS, Fong YC, Wu YY, Chiang JH, Su YC. HGK-sestrin 2 signaling-mediated autophagy contributes to antitumor efficacy of Tanshinone IIA in human osteosarcoma cells. Cell Death Dis. 2018;9(10):1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taparra K, Wang H, Malek R, Lafargue A, Barbhuiya MA, Wang X, Simons BW, Ballew M, Nugent K, Groves J, Williams RD, Shiraishi T, Verdone J, Yildirir G, Henry R, Zhang B, Wong J, Wang KK, Nelkin BD, Pienta KJ, Felsher D, Zachara NE, Tran PT. O-GlcNAcylation is required for mutant KRAS-induced lung tumorigenesis. J Clin Invest. 2018;128(11):4924–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo X, Cao M, Gao F, He X. YTHDF1 promotes hepatocellular carcinoma progression via activating PI3K/AKT/mTOR signaling pathway and inducing epithelial-mesenchymal transition. Exp Hematol Oncol. 2021;10(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.