Abstract
Background
Our previous study demonstrated that temperature-related microwave ablation (MWA) can safely modulate growth plates of piglets’ vertebrae. Therefore, this study is designed to investigate the effects of different temperatures on chondrocyte viability and the underlying molecular mechanisms in vitro.
Methods
Following a 10-minute treatment at different temperatures (37 °C, 40 °C, 42 °C, 44 °C, 46 °C, 48 °C, and 50 °C), CCK-8 assay was used to examine the viability of ATDC5 cells at 12 h. Differentially expressed genes (DEGs) and the hub genes in ATDC5 cells treated at 37 °C, 40 °C and 44 °C were identified using RNA-seq. The expression of hub genes in ATDC5 cells was validated using RT-qPCR.
Results
Compared with 37 °C, exposure to 40 °C significantly increased the viability of ATDC5 cells, while 42 °C had no significant effect. Additionally, exposure to 44 °C, 46 °C, 48 °C, and 50 °C exhibited the opposite pattern, with ATDC5 cells being particularly less than 50% active after treatment at 46 °C, 48 °C, and 50 °C. Differential expression analysis identified 179, 374 and 221 DEGs in the comparisons of 40 °C vs. 37 °C, 44 °C vs. 37 °C, and 44 °C vs. 40 °C, respectively. These DEGs predominantly regulated proliferation, differentiation, necrosis, inflammatory and immune responses, and ECM synthesis/degradation. Furthermore, they were associated with the Ras, PI3K/AKT, mTOR, cAMP, and MAPK pathways. Agt, Hspa1a, Hspb1, and Nlrc4 were identified as hub genes in DEGs, and RT-qPCR confirmed that the mRNA expression patterns of these hub genes in ATDC5 cells were largely consistent with the RNA-seq results.
Conclusion
The regulation of chondrocyte viability by temperature is associated with Ras, PI3K/AKT, mTOR, cAMP, and MAPK pathways. Additionally, Agt, Hspa1a, Hspb1, and Nlrc4 may be the key regulatory genes in this process.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-02070-8.
Keywords: Scoliosis, Growth plate, Chondrocyte, Temperature, Cell viability, RNA sequencing
Introduction
Scoliosis is a prevalent and complex three-dimensional deformity of the spine in children and adolescents, characterized by lateral deviation in the coronal plane, lordosis or kyphosis in the sagittal plane, and vertebral rotation in the transverse plane [1, 2]. Untreated scoliosis can lead to severe deformities of the thoracic cage, impose restrictions on cardiopulmonary growth and development [3, 4]. In a long-term follow-up, untreated children and adolescent patients with spinal scoliosis have a significantly increased risk of death due to respiratory failure by the age of 40–50, with the highest mortality rates observed at 60 years old, when is four times higher than normal [5]. Therefore, early intervention for scoliosis is important to ensure basic cardiorespiratory development and minimize patient mortality rates. Chondrocytes within the vertebral growth plate control the growth of the spine [6, 7]. Thus, an asymmetrical growth pattern of the vertebral growth plate is considered a contributing factor to the development of scoliosis. Chondrocytes within the vertebral growth plate resemble those of long bone epiphyses, and are responsible for the longitudinal growth of the spine through processes such as proliferation, differentiation, and chondral ossification. Therefore, it is crucial to effectively regulate the phenotype of chondrocytes within the growth plate for the prevention of scoliosis-induced cardiopulmonary dysplasia.
Microwave ablation (MWA) is a minimally invasive technique that achieves favorable treatment outcomes with minimal trauma [8–13]. Microwaves possess strong tissue penetration and can cause rapid heating of the tissue within the ablation area, resulting in thermal coagulation and necrosis [8–13]. Continuous application of MWA can generate extremely high temperatures (> 150 °C) in the tissue target area, which enhances the ablation effect through increasing thermal conduction to the surrounding tissue [8–14]. Compared with traditional means such as surgical resection, MWA is minimally invasive, safe, effective, efficient, precise, and widely applicable [8–13]. Currently, MWA is used to treat benign and malignant tumors, malignant central airway obstruction, fractures, varicose veins, and atrial fibrillation [8–13]. Notably, our previous study demonstrated that MWA treatment resulted in dysplasia of the growth plate and the annulus fibrosus on the ablation side, as well as wedge-shaped vertebral bodies, leading to scoliosis with markedly imbalanced spinal growth in piglets [15]. These effects were associated with chondrocyte apoptosis and necrosis in the growth plate. Moreover, no instances of nerve root paralysis, spinal cord injury, or other serious complications were observed in the postoperative period among the piglets. This suggests that MWA has the potential to induce apoptosis and necrosis of chondrocyte within vertebral growth plate, thereby regulating the balance of spinal growth, making it a promising therapeutic approach for scoliosis. Importantly, temperature may be central to MWA-mediated apoptosis and necrosis of chondrocyte within vertebral growth plate.
Notably, 43 °C is the maximum thermal dose for hyperthermia of localized central nervous system in cats, rabbits, minipigs, dogs, and monkeys without causing significant complications [16]. Therefore, 43 °C was established as the safety threshold for MWA at the posterior edge of the vertebral body in our previous study [15]. Additionally, thermal ablation therapies at 50–54 °C for 6 min, including MWA, result in irreversible tissue damage [14]. Chen et al. [17] applied MWA at different temperatures (T1 = 43–45 °C; T2 = 46–48 °C; T3 = 49–51 °C) in the femoral growth plates for animals, and exposed rabbit chondrocytes to temperatures of 37 °C, 44 °C, 47 °C, and 50 °C for 5 min. Therefore, the present study proposes to stimulate chondrocytes at different temperatures (37℃-50℃) in vitro to investigate chondrocyte growth changes, and use RNA sequencing (RNA-seq) to uncover the underlying molecular mechanisms. The aim is to provide a theoretical foundation and molecular insights for the application of temperature-related strategies in the clinical management of scoliosis.
Methods and materials
Cell culture and treatment
Mouse adult chondrocytes (ATDC5) were obtained from Bena Culture Collaboration (Henan, China; Art. No. BNCC350793), inoculated in a medium consisting of 4.5 g/L high glucose DMEM (Gibco, USA; Art. No. 11965118) and 10% FBS (Gibco, USA; Art. No. A5669402), and then incubated in an incubator of 95% air and 5% CO2 at 37 °C.
ATDC5 cells were digested with 0.25% trypsin (Gibco, USA; Art. No. 25200-056) for 1 min, and the digestion was terminated using complete medium. After centrifugation at 1000 rpm for 3 min, ATDC5 cells were resuspended as single cell suspensions in complete medium. Subsequently, ATDC5 cells were divided into seven group (N = 6) and exposed for 10 min at different temperatures (37 °C, 40 °C, 42 °C, 44 °C, 46 °C, 48 °C, and 50 °C) in an HWS-12 thermostat water bath (Yiheng, China). ATDC5 cells were collected for subsequent CCK-8, RNA-seq, and RT-qPCR assays.
CCK-8 assay
The viability of ATDC5 cells was assessed using the Cell Counting Kit-8 (CCK-8) (DOJINDO LABORATORIES, Japan; Art. No. CK04). Briefly, ATDC5 cells (2 × 103 cells/well) treated with different temperatures were inoculated in 96-well plates, and incubated in 5% CO2 incubator at 37 °C for 12 h. A blank control was included for each well. The cell supernatant was removed from the 96-well plates and 100 µl of basal medium was added to each well. Subsequently, 100 µl of CCK-8 solution was added to each well and incubated for 2 h. The optical density (OD) of each well were measured using an EL800 fully automated microplate reader (BIO-TEK, USA).
RNA sequencing (RNA-seq) and bioinformatics analysis
RNA-seq was used to identify the expression profiles of ATDC5 cells treated with different temperatures (37 °C, 40 °C, and 44 °C), with four bio-replications in each group. ATDC5 cells (5 × 107) treated with different temperatures were lysed in 1 ml Trizol reagent (Invitrogen, USA; Art. No. 15596018CN) and 0.2 ml chloroform for 5 min and 3 min, respectively. After centrifugation at 12,000 rpm for 15 min, the upper aqueous phase was collected. RNA was added to 0.5 ml of isopropanol and incubated for 10 min at room temperature. Following an additional centrifugation at 12,000 rpm for 10 min, the RNA precipitate was washed with 75% ethanol. After centrifugation at 7500 rpm for 5 min, RNA was added to 30 µl of H2O containing DEPC and dissolved at 65 °C for 15 min. RNA concentration and integrity were characterized using a NanoDrop 2000 spectrophotometer (ThermoScientific, USA) and a 2100 Bioanalyzer (Agilent, USA), respectively.
RNA-seq and analysis were performed by NovoGene (Beijing, China). For RNA-seq, the sequencing platform was Illumina Novaseq™ 6000 with 2 × 150 bp paired-end sequencing (PE150). For bioinformatics analysis, quality control, reference genome comparison, and quantitative analysis of sequencing data were performed using FastQC (V. 0.10.1) [18], hisat2 (V. 2.2.5) [19], and featureCounts (V. 1.5.0-p3) [20]. Principal component analysis (PCA) and expression pattern cluster analysis of expression profiles were performed using R (V. 3.2.0) and Mfuzz package (V. 2.64.0) [21]. Differential expression analysis and enrichment analysis were performed using DESeq2 (V. 1.20.0) [22] and clusterProfiler (V. 3.8.1) [23]. The visualization of the DEG-associated KEGG pathway was performed using the Pathview package (V. 1.44.0) [24]. Differentially expressed genes (DEGs) satisfied P < 0.05 and log2Fold Change (FC) > 1 or log2FC<-1. DEGs with positive log2FC are up-regulated. DEGs with negative log2FC are down-regulated. The STRING database [25] was used to predict the interactions among DEGs. The minimum required interaction score in the STRING database was set to 0.7. The protein-protein interaction (PPI) network was visualized using CytoScape software (V.3.7.2) [26]. Additionally, the MCODE plug-in [27] of CytoScape software (V.3.7.2) was used to perform the clustering analysis of the PPI network.
RNA extraction and RT-qPCR assay
Total RNA from ATDC5 cells treated with different temperatures (37 °C, 40 °C and 44 °C; n = 4) was extracted using the TRIzol™ Plus RNA Purification Kit (Invitrogen, China; Art. No. 12183555), and the concentration was determined using the MEzDrop1000 spectrophotometer (Blue- Ray, China). Reverse transcription and PCR amplification were performed using FastKing RT Kit (TianGen, Germany; Art. No. KR116-03) and Taq Pro Universal SYBR qPCR Master Mix (Vazyme, China; Art. No. Q712-02), to detect mRNA expression of Agt, Bst2, Col3a1, Gbp5, Hspa1a, Hspb1, Irf7, Nlrc4, S100a4, and Tlr7. For thermal parameters, cDNA samples were denatured by heating at 95 °C for 10 min. A total of 40 cycles of annealing and extension were performed, with each cycle exposing the samples to 95 °C and 60 °C for 15 s and 30 s, respectively. Melting curves were analyzed at 95 °C for 15s, 60 °C for 30s and 95 °C for 15s. Primer sequences are listed in Table 1, and the internal reference is β-actin. RT-qPCR assay was conducted using a 7500 Real Time PCR System (ABI, USA). The 2(-Delta Delta C(T)) method [28] was used to calculate the expression of target genes relative to β-actin.
Table 1.
Primer sequences applied to RT-qPCR assay
| Target | NCBI accession ID | Forward Sequence | Reverse Sequence |
|---|---|---|---|
| Agt | NM_007428.4/NM_001416312.1 | ACGCTCTCTGGATTTATC | AGTGGCAAGTTCATCTTC |
| S100a4 | NM_011311.3/NM_001410571.1/NM_001410572.1 | CAGAAGGTGATGAGCAACT | AGGACAGGAAGACACAGTA |
| Hspb1 | NM_013560.2 | CAGTGAAGACCAAGGAAG | AGAGATGTAGCCATGTTC |
| Hspa1a | NM_010479.2 | AATCCTATGCCTTCAACAT | CTTCTTCTTGTCAGCCTC |
| Irf7 | XM_006536214.1/NM_001252601.1/NM_016850.3/NM_001252600.1 | GCATAAGGTGTACGAACT | GCTCACTTCTTCCCTATT |
| Nlrc4 | NM_001033367.3 | GTCAAGTGTTATCCAAGTTA | CGCTAATATCATAGTCATCAA |
| Tlr7 | XM_006528713.2/NM_001290758.1/NM_001290755.1/NM_001290757.1/NM_001290756.1/NM_133211.4 | CTTGACCTTGGCACTAAC | ACTTCTCTTGACTCTTCTGA |
| Col3a1 | NM_009930.2 | TGAAGATGTCGTTGATGT | GCAGTGGTATGTAATGTTC |
| Gbp5 | NM_153564.2 | TTCTGCTTGACACTGAAG | CTACTGAGGAGGATTGCTA |
| Bst2 | NM_198095.3 | GCTTGAGAATGAAGTCACGAA | ACTGTGCTAGAAGTCTCCTT |
| β-actin | NM_007393.5 | CTGGAGAAGAGCTATGAG | GATGGAATTGAATGTAGTTTC |
Statistical analysis
All data were expressed as mean ± standard deviation (SD) (N ≥ 4), and statistical analysis and visualization of results were performed using GraphPad Prism software (GraphPad, USA; V.9.0.0). For statistical analysis, CCK-8 and RT-qPCR results were analyzed using one-way ANOVA with Tukey’s multiple comparisons test. P < 0.05 was considered a statistically significant difference.
Results
Effect of different temperatures on the viability of ATDC5 cells and RNA-seq analysis
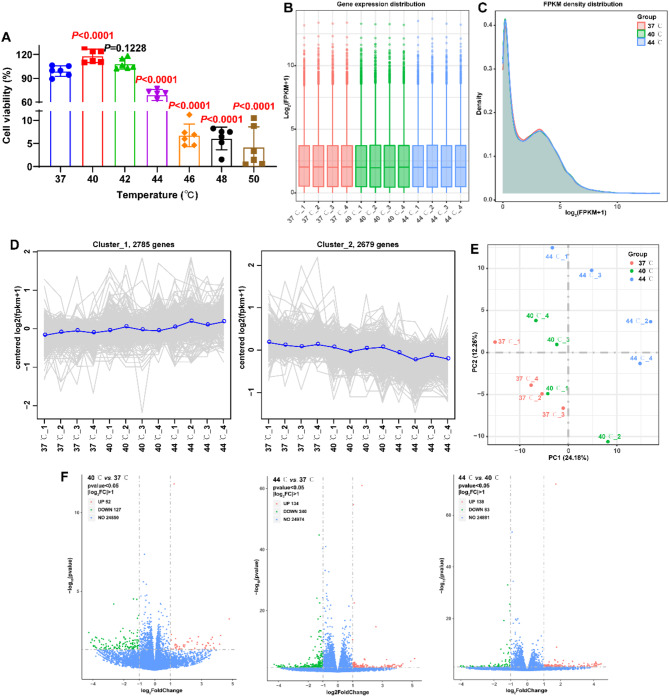
In this study, ATDC5 cells were treated with different temperatures (37 °C, 40 °C, 42 °C, 44 °C, 46 °C, 48 °C, and 50 °C) to investigate the effect of temperature on chondrocyte viability. CCK-8 is a colorimetric assay used to measure cell viability by correlating the production of colored formazan dye to the amount of living cells in culture. In this study, it was used for screening for alternative temperatures for further investigation. CCK-8 assay revealed that stimulation at temperatures of 40 °C, 44 °C, 46 °C, 48 °C and 50 °C significantly modulated the viability of ATDC5 cells compared to 37 °C (Fig. 1A, P < 0.0001).
Fig. 1.
Effects of different temperatures on the viability of ATDC5 cells and differential expression analysis. A: After stimulation for 10 min at different temperatures (37 °C, 40 °C, 42 °C, 44 °C, 46 °C, 48 °C, and 50 °C), CCK-8 assay detected the viability of ATDC5 cells at 12 h (N = 6). Statistical analysis was completed using one-way ANOVA with Tukey’s multiple comparisons test, and P < 0.05 represents a significant difference. B: Box-whisker Plot visualized the distribution of gene with FPKM in the expression profiles of each sample. C: Density Plot characterized the density distribution of the gene with FPKM in the expression profiles of the 37 °C, 40 °C, and 44 °C groups. D: Cluster line plots identified the correlation between the expression patterns of different samples. E: Principal component analysis (PCA) characterized clustering and discretization of expression profiles for different samples. F: Volcano plots visualized the distribution of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C, and 44 °C vs. 40 °C
Temperatures of 40 °C and 44 °C resulted in opposite modulation patterns on the viability of ATDC5 cells, and the viability of ATDC5 cells stimulated by temperatures of 46 °C, 48 °C, and 50 °C was less than 50%. Therefore, the transcriptome expression profiles of ATDC5 cells treated with temperatures of 37 °C, 40 °C, and 44 °C were identified using RNA-seq. The mean values of RNA concentration in the 37 °C, 40 °C and 44 °C groups were 1487.50, 1097.75 and 1947.50 ng/µl, respectively. A total of 569,862,768 raw reads and 85.47 G raw bases were obtained for all the samples, ranging from 44,553,698 to 54,048,196 and 6.68 G to 8.11 G. After quality control, a total of 562,498,458 clean reads and 84.36 G raw bases were obtained for all samples, ranging from 43,912,536 to 53,422,194 and 6.59 G to 8.01 G. The mean values of Q20 and Q30 were 98.10% and 94.68%, respectively, with a range of 97.94–98.25% and 94.30–94.96%. After data normalization, the median and density distributions of FPKM remained consistent across samples, indicating a symmetrical and similar expression profile among the samples (Fig. 1B and C). Further, this study characterized the correlation of expression patterns of different samples using Euclidean Distance. The genes exhibiting similar expression patterns tend to cluster together, resulting in the formation of multiple subclusters based on the gene differences in the expression patterns. The gene expression profiles of the 12 samples were segregated into two subclusters, including 2785 and 2679 genes, respectively. Gene expression in subcluster 1 showed an increasing trend with increasing temperature, and gene expression in subcluster 2 showed the opposite pattern (Fig. 1D). The PCA results revealed that the distances between expression profiles within the 37 °C, 40 °C and 44 °C groups were similar, while significantly different among these groups (Fig. 1E). Differential expression analysis revealed 179 (52 up-regulated and 127 down-regulated DEGs) in the 40 °C group, as well as 374 (134 up-regulated and 240 down-regulated DEGs) DEGs in the 44 °C group, when compared to the 37 °C group (Fig. 1F and Supplementary file 1). Additionally, a total of 221 DEGs were identified in the 44 °C group compared to the 40 °C group, comprising 138 up-regulated and 83 down-regulated DEGs (Fig. 1F and Supplementary file 1). These DEGs may be associated with temperature-mediated chondrocyte viability.
Enrichment analysis (GO, KEGG, Reactome) for temperature treatment-associated DEGs
Further, this study identified the possible functional and molecular mechanisms underlying temperature treatment-associated DEGs using enrichment analysis (GO, KEGG, Reactome).
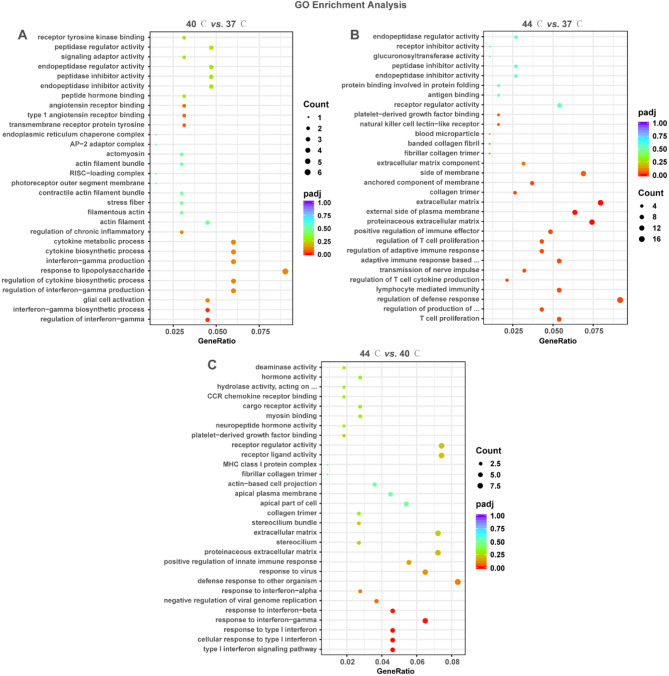
Biological processes, molecular functions, and cellular composition of DEGs were identified using GO enrichment analysis. DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C, and 44 °C vs. 40 °C were enriched to 247, 339, and 98 GO terms, respectively (Supplementary file 2, P < 0.05). As illustrated in Fig. 2A and C, these GO terms mainly included cytokine metabolic process, signaling adaptor activity, glucuronosyltransferase activity, natural killer cell lectin-like receptor, extracellular matrix (ECM) component, T cell proliferation, response to type I interferon. These results suggest that DEGs may regulate glucose metabolism, inflammatory and immune responses, and ECM synthesis/degradation.
Fig. 2.
GO enrichment analysis of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C and 44 °C vs. 40 °C. A-C: Bubble plots displayed the GO enrichment results of DEGs in 40 °C vs. 37 °C (A), 44 °C vs. 37 °C (B), and 44 °C vs. 40 °C (C). The vertical coordinate is the name of the GO term; the horizontal coordinate is the gene ratio. The size and color of the dots represent the count and the adjusted P-value, respectively
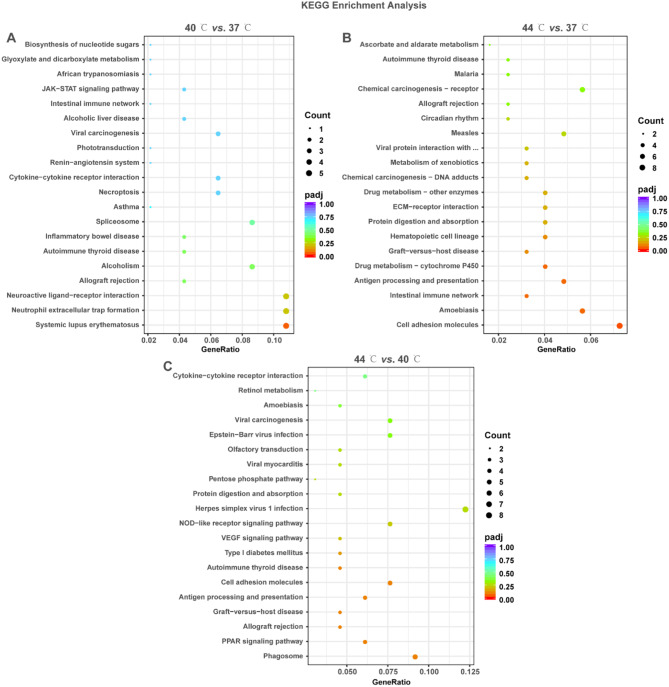
The signaling pathways associated with DEGs were identified using KEGG enrichment analysis. The results demonstrated that DEGs were enriched to 8, 21 and 18 KEGG pathways, respectively (Supplementary file 3, P < 0.05). These KEGG pathways mainly included JAK-STAT signaling pathway, cytokine-cytokine receptor interaction, necroptosis, neutrophil extracellular trap formation, ECM-receptor interaction, biosynthesis of nucleotide sugars, NOD-like receptor signaling pathway, VEGF signaling pathway, PPAR signaling pathway (Fig. 3A and C). These results suggest that DEGs may be involved in the pathway related to nucleotide metabolism, cell proliferation, necroptosis, angiogenesis, lipid metabolism, inflammatory and immune responses, and ECM synthesis/degradation.
Fig. 3.
KEGG enrichment analysis of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C and 44 °C vs. 40 °C. A-C: Bubble plots displayed the KEGG enrichment results of DEGs in 40 °C vs. 37 °C (A), 44 °C vs. 37 °C (B), and 44 °C vs. 40 °C (C). The vertical coordinate is the name of the KEGG term; the horizontal coordinate is the gene ratio. The size and color of the dots represent the count and the adjusted P-value, respectively
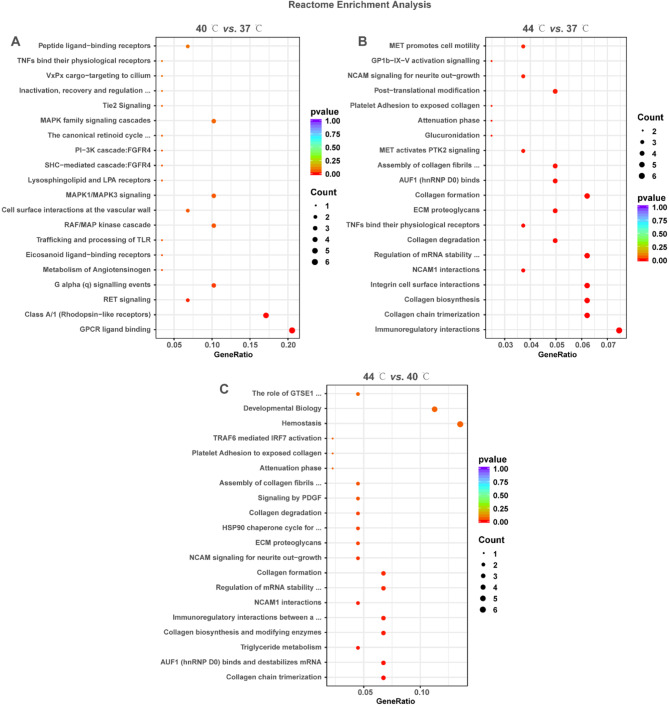
Further, the signaling pathways associated with DEGs were complemented using Reactome enrichment analysis. The results revealed that DEGs in the three comparisons were enriched to 13, 32 and 14 Reactome reactions, respectively (Supplementary file 4, P < 0.05). These Reactome reactions mainly included TNFs bind their physiological receptors, MAPK family signaling cascades, PI3K cascades, RET signaling, trafficking and processing of TLR, immunoregulatory interactions, collagen formation/degradation, ECM proteoglycans, triglyceride metabolism, HSP90 chaperone cycle (Fig. 4A and C). These results suggest that DEGs may be involved in the reaction related to cell proliferation, cell differentiation, lipid metabolism, inflammatory and immune responses, and synthesis/degradation of ECM.
Fig. 4.
Reactome enrichment analysis of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C and 44 °C vs. 40 °C. A-C: Bubble plots displayed the Reactome enrichment results of DEGs in 40 °C vs. 37 °C (A), 44 °C vs. 37 °C (B), and 44 °C vs. 40 °C (C). The vertical coordinate is the name of the KEGG term; the horizontal coordinate is the gene ratio. The size and color of the dots represent the count and the adjusted P-value, respectively
PPI networks for temperature treatment-associated DEGs
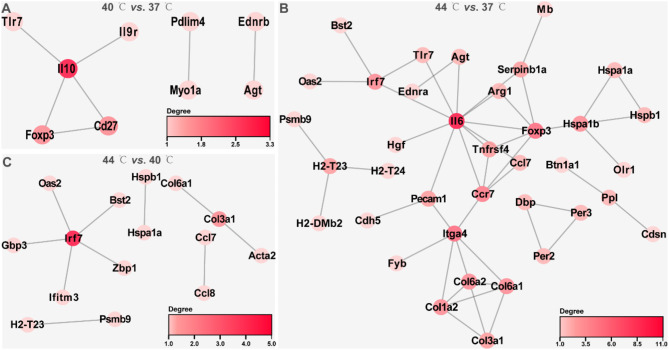
In this study, PPI networks were constructed to identify hub genes of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C and 44 °C vs. 40 °C. In 40 °C vs. 37 °C, the PPI network consisted of 9 nodes and 7 interaction pairs, with Il-10 exhibiting the highest degree centrality (Fig. 5A and Supplementary file 5; minimum required interaction score = 0.7). The PPI network for 44 °C vs. 37 °C comprised of 37 nodes and 48 interaction pairs, with the top 5 genes in terms of degree being Il6, Foxp3, Itga4, Ccr7, and Irf7 (Fig. 5B and Supplementary file 5; minimum required interaction score = 0.7). The PPI network in 44 °C vs. 40 °C contained 15 nodes and 10 interaction pairs, with Irf7 exhibiting the highest degree centrality (Fig. 5C and Supplementary file 5; minimum required interaction score = 0.7).
Fig. 5.
PPI networks of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C and 44 °C vs. 40 °C. A-C: The String database predicted the interactions of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C, and 44 °C vs. 40 °C, and the visualization of PPI networks was performed using Cytoscape software. A, B, and C are the PPI networks of DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C, and 44 °C vs. 40 °C, respectively. The color of a node represents the size of the degree, and the darker the node, the larger the degree
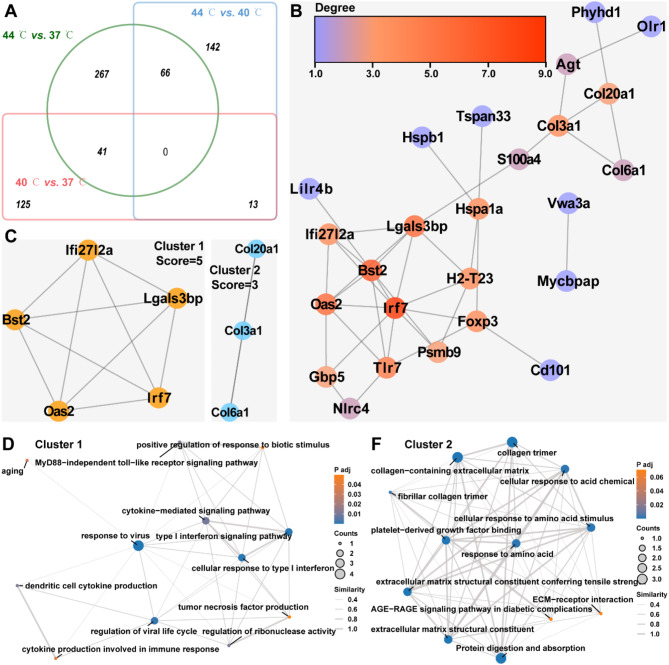
The intersecting DEGs were further utilized to construct PPI networks. As illustrated in Fig. 6A, a total of 280 were found to intersect between at 40 °C vs. 37 °C, 44 °C vs. 37 °C, and 44 °C vs. 40 °C. The PPI network constructed using theseintersecting DEGs comprised of 25 nodes and 39 interaction pairs, with Irf7, Bst2, Lgals3bp, Oas2, and Tlr7 being the top 5 genes based on their degree centrality (Fig. 6B and Supplementary file 6; minimum required interaction score = 0.7). The sub-networks of this PPI network were identified using the MCODE plug-in. The PPI network exhibited two sub-networks with clustering scores of 5 and 3, respectively (Fig. 6C). Importantly, the hub genes in subnetwork 1 mainly regulated the cellular response to type I interferon, aging, positive regulation of response to biotic stimulus, cytokine- mediated signaling pathway and MyD88-independent toll-like receptor signaling pathway, and there was a correlation between them (Fig. 6D). The hub genes in subnetwork 2 mainly regulated collagen-containing extracellular matrix, cellular response to acid chemical, AGE-RAGE signaling pathway in diabetic complications and ECM-receptor interaction (Fig. 6F). Furthermore, a significant correlation was observed among these processes. These results suggest that Bst2, Irf7, Oas2, Tlr7, Lgals3bp, and Foxp3 serve as temperature-mediated hub genes in chondrocytes and regulate chondrocyte proliferation, senescence, inflammatory and immune responses, and ECM synthesis/degradation.
Fig. 6.
PPI Networks for Temperature treatment-associated DEGs. A: Venn diagrams showed intersecting and concatenated DEGs in 40 °C vs. 37 °C, 44 °C vs. 37 °C, and 44 °C vs. 40 °C. B: String database predicted interactions of intersecting DEGs and the visualization of PPI networks was conducted by Cytoscape software. The color of the node represents the size of the degree, and the redder the node, the larger the degree. C: Cluster analysis of PPI networks associated with intersecting DEGs was performed using the MCODE plug-in of Cytoscape software to identify sub-networks. D-F: EMAP plots exhibited the GO term and KEGG pathways associated with DEGs in cluster 1 (D) and cluster 2 (E), and the correlation between them
Validation of hub genes in temperature treatment-associated DEGs
This study confirmed differential expression of 10 hub genes (Agt, Bst2, Col3a1, Gbp5, Hspa1a, Hspb1, Irf7, Nlrc4, S100a4, and Tlr7) in ATDC5 cells. The mean values of RNA concentration in the 37 °C, 40 °C and 44 °C groups were 792.90, 762.74 and 795.90 ng/µl, respectively. Treatment at 40 °C resulted in a significant upregulation in Agt and Hspa1a mRNA, while there was a significant downregulation in Hspb1, Nlrc4, S100a4, and Tlr7 (Fig. 7, P < 0.05). Treatment at 44 °C resulted in the upregulation of Agt, Hspa1a, Hspb1, and Nlrc4 mRNA levels, while downregulating the levels of Col3a1, Gbp5, Irf7, S100a4, and Tlr7 mRNAs (Fig. 7A, P < 0.05). The expression pattern of mRNAs was consistent with the RNA-seq results, except for Bst2 and Hspb1 (Supplementary file 1). Additionally, significant positive correlations were observed between the levels of Agt, Bst2, Col3a1, Gbp5, Hspa1a, Hspb1, Irf7, Nlrc4, S100a4, and Tlr7 in both RNA-seq and RT-qPCR results (Fig. 7B, P < 0.05).
Fig. 7.
Validation of Hub Genes in Temperature treatment-associated DEGs. A: RT-qPCR was used to detect the level differences of AGT, BST2, COL3A1, GBP5, HSPA1A, HSPB1, IRF7, NLRC4, S100A4, and TLR7 mRNA in the 37 °C, 40 °C, and 44 °C groups (N = 4). Statistical analysis was completed using one-way ANOVA with Tukey’s multiple comparisons test, and P < 0.05 represents a significant difference. B: Correlation of RNA-seq and RT-qPCR results for hub genes (AGT, BST2, COL3A1, GBP5, HSPA1A, HSPB1, IRF7, NLRC4, S100A4, and TLR7) (N = 4). Statistical analysis was completed using Pearson correlation analysis, and P < 0.05 represents a significant difference
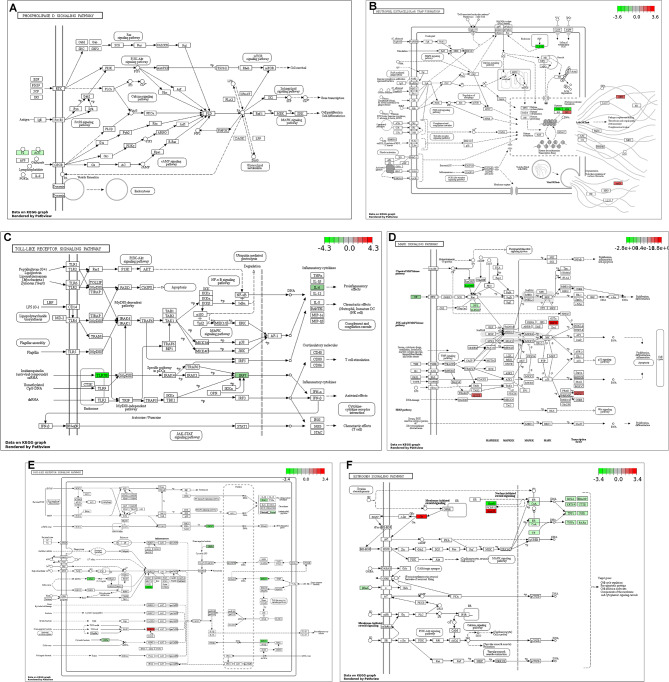
Further, this study visualized the possible molecular mechanisms underlying the regulation of these hub genes using Pathview. As indicated in Fig. 8A, AGT may promote gene transcription and cell viability, survival as well as differentiation of ATDC5 cells through Ras, PI3K/AKT, mTOR, cAMP and MAPK pathways (Fig. 8A). TLR7 and IRF7 may regulate neutrophil extracellular trap formation by activating the Toll-like receptor pathway (Fig. 8B and C). In addition, GBP5, HSPA1A, and HSPB1 were associated with MAPK, NOD-like receptor and ESTROGEN pathway (Fig. 8D and F).
Fig. 8.
Pathways involved in Temperature treatment-associated DEGs. A-F: The pathview package of R software was used to visualize temperature treatment-associated DEGs (AGT, TLR7, IRF7, GBP5, HSPA1A, and HSPB1) in the phospholipase D signaling pathway (A), neutrophil extracellular trap formation (B), Toll-like receptor pathway (C), MAPK signaling pathway (D), NOD-like receptor signaling pathway (E), and ESTROGEN signaling pathway (F)
Discussion
In the present study, we explored the effects of different temperature stimuli (37 °C, 40 °C, 42 °C, 44 °C, 46 °C, 48 °C, and 50 °C) on chondrocyte viability. The present study demonstrated that compared to the physiological temperature of 37 °C, stimulation at 40 °C significantly enhanced chondrocyte viability. Conversely, temperatures of 44 °C and above exhibited a contrasting effect on cell survival. Notably, Voss et al. [29] have indicated that thermal stimulation at 45 °C, 50 °C, 55 °C, 60 °C, and 65 °C resulted in a significant decrease in live cells and a significant increase in dead cells in chondrocytes as compared to 37 °C. This is consistent with the findings of this study, which supports the conclusions. However, the evaluation of chondrocyte viability under 40 °C conditions was not conducted by Voss et al. We believe that stimulation at 40 °C contributes to chondrocyte viability. Interestingly, Serrat et al. [30] have found that stimulation at 39 °C resulted in enhanced chondrocyte proliferation and extracellular matrix synthesis in metatarsal growth plates compared to temperatures of 32 °C and 37 °C in vitro, indicating that a temperature of 39 °C was more conducive to limb growth compared to temperatures of 32 °C and 37 °C. This finding is in general agreement with the results of the present study. It is suggested that temperature of 39 °C/40°C may serve as a critical point for regulating the growth of vertebral growth plates in scoliosis. However, this remains to be verified at both the animal and clinical levels.
Further, this study identified potential molecular mechanisms underlying temperature-mediated chondrocyte viability using RNA-seq. In this study, a total of 654 DEGs were identified, primarily regulated proliferation, differentiation, necrosis, inflammatory and immune responses and ECM synthesis/degradation. Additionally, these DEGs were associated with Ras, PI3K/AKT, mTOR, cAMP and MAPK pathways. These phenotypes and pathways have been demonstrated to be associated with chondrocyte proliferation and growth plate development [31–33]. Notably, Agt, Bst2, Col3a1, Gbp5, Hspa1a, Hspb1, Irf7, Nlrc4, S100a4, and Tlr7 were identified as hub genes among the DEGs. A bioinformatics analysis revealed differential expression of AGT in seven chondrocyte clusters, implicating its involvement in cartilage metabolism and development [34]. Moreover, Wang et al. [35] demonstrated that AGT exerts regulatory effects on the inflammatory response of chondrocytes in osteoarthritis by modulation of the JAK2/STAT3 pathway. The present study revealed that stimulation at 40 °C resulted in an upregulation of Agt mRNA level in chondrocytes, potentially activating the cAMP and MAPK pathways. The cAMP and MAPK pathways are essential for chondrocyte proliferation and growth plate development [36, 37]. Therefore, we hypothesize that stimulation at 40 °C may enhance chondrocyte viability by the upregulation of AGT, thereby activating the cAMP and MAPK pathways. NLRC4, as an inflammasome proteins, facilitates chondrocyte pyroptosis and inflammation in osteoarthritis [38, 39]. The present study found that stimulation at 40 °C resulted in downregulation of Nlrc4 mRNA level in chondrocytes and may be associated with the NOD-like receptor pathway. Activation of the NOD-like receptor pathway contributes to dysplasia of cartilage and growth plates [40], and is facilitated by NLRC4 [41]. Therefore, we hypothesize that stimulation at 40 °C may facilitate chondrocyte viability by downregulation of Nlrc4 mRNA level, leading to a reduction in the activity of NOD-like receptor pathway. However, these temperature-related potential molecular mechanisms remain to be validated by cellular and animal experiments.
In conclusion, the present study found that chondrocyte viability is enhanced by stimulation at 40 °C, while temperatures of 44 °C and above exhibit the opposite pattern in vitro. Additionally, Agt, Bst2, Col3a1, Gbp5, Hspa1a, Hspb1, Irf7, Nlrc4, S100a4 and Tlr7 are likely to be the key regulatory genes for this process. The findings will provide a molecularly relevant theoretical foundation for temperature-related therapies, such as MWA, and will facilitate the advancement of temperature-related therapeutic applications for cartilage-related disorders, including scoliosis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- CCK-8
Cell Counting Kit-8
- DEGs
Differentially Expressed Genes
- ECM
Extracellular Matrix
- FC
Fold Change
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MWA
Microwave Ablation
Author contributions
W.Z., Y.W., J.X., J.Z., Z.Z., T.L., Z.S., and J.X. conceived and designed the experiments; W.Z., Y.W., J.X., J.Z., Z.Z., T.L., Z.S., and J.X. performed the experiments; W.Z. and Y.W. analyzed the data; J.Z. and Z.Z. contribute to resources and data curation; W.Z. contribute to writing-Original Draft; W.Z., J.X., J.Z., Z.Z., T.L., Z.S., and J.X. contribute to writing-Review & Editing. All authors have read and agreed to the published version of the manuscript.
Funding
National Natural Science Foundation of China (Grant No.82260447).
Data availability
Data available on request from the authors. RNA sequencing data has been stored in GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE269585).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang YB, Zhang JG. Treatment of early-onset scoliosis: techniques, indications, and complications. Chin Med J. 2020;133(3):351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marya S, Tambe AD, Millner PA, Tsirikos AI. Adolescent idiopathic scoliosis: a review of aetiological theories of a multifactorial disease. Bone Joint J 2022;104–B(8):915–21. [DOI] [PubMed]
- 3.Koumbourlis AC. Chest wall abnormalities and their clinical significance in childhood. Paediatr Respir Rev. 2014;15(3):246–54; quiz 254–5. [DOI] [PubMed] [Google Scholar]
- 4.Redding GJ, Hurn H, White KK, Bompadre V, Emerson J, Garza RZ, Anigian K, Waldhausen J, Krengel W, Joshi A. Persistence and Progression of Airway Obstruction in Children with Early Onset Scoliosis. J Pediatr Orthop. 2020;40(4):190–5. [DOI] [PubMed] [Google Scholar]
- 5.Pehrsson K, Larsson S, Oden A, Nachemson A. Long-term follow-up of patients with untreated scoliosis. A study of mortality, causes of death, and symptoms. Spine. 1992;17(9):1091–6. [DOI] [PubMed] [Google Scholar]
- 6.Bylski-Austrow DI, Glos DL, Wall EJ, Crawford AH. Scoliosis vertebral growth plate histomorphometry: comparisons to controls, growth rates, and compressive stresses. J Orthop Res. 2018;36(9):2450–9. [DOI] [PubMed] [Google Scholar]
- 7.Villemure I, Stokes IA. Growth plate mechanics and mechanobiology. A survey of present understanding. J Biomech. 2009;42(12):1793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gartshore A, Kidd M, Joshi LT. Applications of Microwave Energy in Medicine. Biosensors. 2021;11(4):96. [DOI] [PMC free article] [PubMed]
- 9.Senitko M, Oberg CL, Abraham GE, Hillegass WB, Akhtar I, Folch E. Microwave ablation for malignant central Airway obstruction: a pilot study. Respir Int Rev Thorac Dis. 2022;101(7):666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin L, Li XY, Zhu LL, Chen GL, Xiang Z, Wang QQ, Bi JW, Wang Q. Clinical application status and prospect of the combined anti-tumor strategy of ablation and immunotherapy. Front Immunol. 2022;13:965120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pusceddu C, Dessì G, Melis L, Fancellu A, Ruggiu G, Sailis P, Congia S, Derudas D, Cau R, Senis I et al. Combined microwave ablation and osteosynthesis for long bone metastases. Medicina (Kaunas). 2021;57(8):825. [DOI] [PMC free article] [PubMed]
- 12.Zhao N, Guo H, Zhang Y, Hu X, He JN, Wang D, Huang W, Gan H, Pang PF. Comparison of endovenous microwave ablation versus radiofrequency ablation for lower limb varicose veins. J Vasc Surg Venous Lymphat Disord. 2024;12(1):101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaynor SL, Byrd GD, Diodato MD, Ishii Y, Lee AM, Prasad SM, Gopal J, Schuessler RB, Damiano RJ Jr. Microwave ablation for atrial fibrillation: dose-response curves in the cardioplegia-arrested and beating heart. Ann Thorac Surg. 2006;81(1):72–6. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed M, Brace CL, Lee FT Jr., Goldberg SN. Principles of and advances in percutaneous ablation. Radiology. 2011;258(2):351–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du ZS, Wang YS, Xie JM, Li T, Shi ZY, Lu QA, Zhang Y, Zhao Z, Bi N, Song ZB, et al. Feasibility of microwave ablation of the vertebral growth plate for spine growth regulation: a preliminary study. Int J Hyperthermia. 2021;38(1):1233–41. [DOI] [PubMed] [Google Scholar]
- 16.Haveman J, Sminia P, Wondergem J, van der Zee J, Hulshof MC. Effects of hyperthermia on the central nervous system: what was learnt from animal studies? Int J Hyperthermia. 2005;21(5):473–87. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, Yan F, Zhong A, Cai L. Effect of thermal ablation on growth plates: a study to explore the thermal threshold of rabbit growth plates during microwave ablation. Cardiovasc Interv Radiol. 2023;46(1):112–9. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 21.Kumar L. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Cambridge (Mass). 2021;2(3):100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinf (Oxford England). 2013;29(14):1830–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego Calif). 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 29.Voss JR, Lu Y, Edwards RB, Bogdanske JJ, Markel MD. Effects of thermal energy on chondrocyte viability. Am J Vet Res. 2006;67(10):1708–12. [DOI] [PubMed] [Google Scholar]
- 30.Serrat MA, King D, Lovejoy CO. Temperature regulates limb length in homeotherms by directly modulating cartilage growth. Proc Natl Acad Sci USA. 2008;105(49):19348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun K, Luo J, Guo J, Yao X, Jing X, Guo F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthr Cartil. 2020;28(4):400–9. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Dai A, Yang M, Chen S, Deng Z, Li L. p38MAPK Signaling Pathway in Osteoarthritis: pathological and therapeutic aspects. J Inflamm Res. 2022;15:723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Y, Peng T, Yao X, Sun C, Wang X. KLF2 reduces dexamethasone-induced injury to growth plate chondrocytes by inhibiting the Runx2-mediated PI3K/AKT and ERK signalling pathways. Autoimmunity. 2023;56(1):1–7. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Zhang Y, Ge Y, Ren H. Integrated single-cell and bulk RNA sequencing analysis identified pyroptosis-related signature for diagnosis and prognosis in osteoarthritis. Sci Rep. 2023;13(1):17757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Han X, Zhao T, Zhang X, Qu P, Zhao H. AGT, targeted by miR-149-5p, promotes IL-6-induced inflammatory responses of chondrocytes in osteoarthritis via activating JAK2/STAT3 pathway. Clin Exp Rheumatol. 2020;38(6):1088–95. [PubMed] [Google Scholar]
- 36.Sakamoto A, Chen M, Kobayashi T, Kronenberg HM, Weinstein LS. Chondrocyte-specific knockout of the G protein G(s)alpha leads to epiphyseal and growth plate abnormalities and ectopic chondrocyte formation. J bone Mineral Research: Official J Am Soc Bone Mineral Res. 2005;20(4):663–71. [DOI] [PubMed] [Google Scholar]
- 37.Thouverey C, Caverzasio J. Focus on the p38 MAPK signaling pathway in bone development and maintenance. BoneKEy Rep. 2015;4:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, He Z, Guo J, Li F, Huang Z, Zheng W, Xing W, Li M, Zhu Y, Yang X. Sesamin-mediated high expression of BECN2 ameliorates cartilage endplate degeneration by reducing autophagy and inflammation. Aging. 2024;16(2):1145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundaram B, Kanneganti TD. Advances in understanding activation and function of the NLRC4 inflammasome. Int J Mol Sci 2021, 22(3):1048. [DOI] [PMC free article] [PubMed]
- 40.Wang C, Xu CX, Alippe Y, Qu C, Xiao J, Schipani E, Civitelli R, Abu-Amer Y, Mbalaviele G. Chronic inflammation triggered by the NLRP3 inflammasome in myeloid cells promotes growth plate dysplasia by mesenchymal cells. Sci Rep. 2017;7(1):4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnett KC, Li S, Liang K, Ting JP. A 360° view of the inflammasome: mechanisms of activation, cell death, and diseases. Cell. 2023;186(11):2288–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request from the authors. RNA sequencing data has been stored in GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE269585).