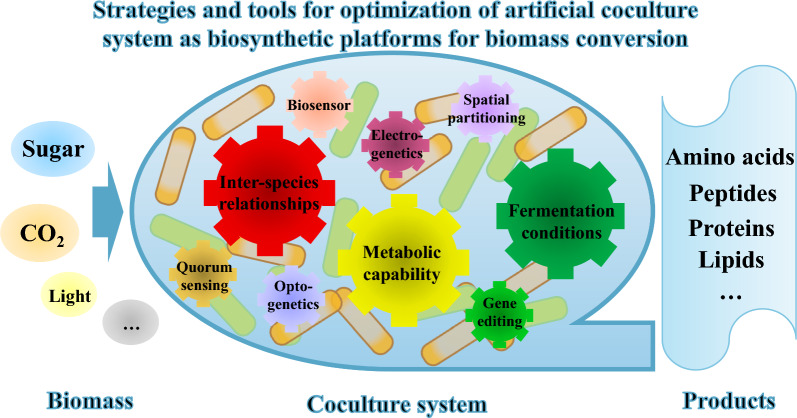
Abstract
Inspired by the natural symbiotic relationships between diverse microbial members, researchers recently focused on modifying microbial chassis to create artificial coculture systems using synthetic biology tools. An increasing number of scientists are now exploring these systems as innovative biosynthetic platforms for biomass conversion. While significant advancements have been achieved, challenges remain in maintaining the stability and productivity of these systems. Sustaining an optimal population ratio over a long time period and balancing anabolism and catabolism during cultivation have proven difficult. Key issues, such as competitive or antagonistic relationships between microbial members, as well as metabolic imbalances and maladaptation, are critical factors affecting the stability and productivity of artificial coculture systems. In this article, we critically review current strategies and methods for improving the stability and productivity of these systems, with a focus on recent progress in biomass conversion. We also provide insights into future research directions, laying the groundwork for further development of artificial coculture biosynthetic platforms.
Graphical Abstract
Keywords: Artificial coculture system, Stability, Productivity, Biomass conversion
Introduction
The production of bio-based chemicals, derived from various biotechnological or chemical processes using organic streams such as non-food lignocellulosic biomass or municipal wastes, as well as CO2, provides environmentally friendly alternatives to fossil fuels and derivatives [1]. Microbial cell factories employ various strategies to convert renewable resources into fermentable sugars. However, converting these substrates for biosynthesis presents challenges for the monoculture, whether natural or engineered. For instance, strains of the Trichoderma genus, one of the predominant genera used in industrial enzyme production, exhibit relatively low β-glucosidase activity, an enzyme crucial for cellulose degradation that works synergistically with cellobiohydrolase and endoglucanase [2]. As a result, the monoculture of Trichoderma is unable to produce the full range of enzymes necessary for the complete breakdown of cell wall components. While CO2 conversion can be performed by cyanobacteria or microalgae, extracting sugars such as sucrose from the culture supernatant is costly, and large-scale sucrose production often leads to contamination issues [3]. In most cases, multiple modifications to cellular metabolic pathways are necessary to enable the synthesis of desired chemicals from CO2 [4].
In nature, most microorganisms interact with microbial communities or complex ecosystems to increase their chances of survival and growth [5]. Natural microbial symbioses have undergone millions of years of evolutionary selection, resulting in intricate yet stable interactions among various strains. Lichens serve as a prime example of such stable, self-sustaining symbiotic organisms, composed of photosynthetic autotrophs and heterotrophic fungi, and are distributed globally [6]. Utilizing naturally occurring microbial communities offers a promising approach for degrading complex substrates, though controlling the behaviour of community members can be challenging [1]. For instance, in cocultures of naturally occurring strains, the excessive accumulation of intermediate products may negatively affect the final yield. Moreover, unlike engineered strains, naturally occurring strains in cocultures lack the ability to precisely fine-tune the metabolic functions of individual bacteria to meet specific needs.
Advances in synthetic biology technologies have enabled researchers to mimic natural symbiotic systems by creating artificial coculture systems with different microbial chassis, serving as next-generation biosynthesis platforms. Artificial microbial coculture involve the collaboration of two or more microbial members to establish a reaction network for chemical production [1]. Using the strategy of coculture, one-pot conversion of renewable resources into fine chemicals can be achieved, offering distinct advantages over cultivating each microorganism independently [7]. By dividing labor, the members of the coculture system reduce metabolic burdens on individual species, especially in lengthy product pathways [8]. Photosynthetic microorganisms, which can harness CO2 and light energy for organic carbon production, can be incorporated into coculture systems to provide carbon sources for heterotrophic species [9, 10]. This approach enables the environmentally friendly synthesis of products from CO2 in systems that combine autotrophic and heterotrophic species. Additionally, many anaerobic bacteria, such as Clostridia, are capable of degrading cellulose through cellulosomes, spurring significant scientific interest in developing coculture systems with cellulose-degrading strains for biofuel production [11, 12].
Considering the advantages of artificial coculture systems, additional efforts have been made to establish various coculture systems for the production of valuable products, such as amino acids, peptides, proteins, lipids, biofuels, polyphenols, alkaloids, terpenoids, and so on. In Tables 1 and 2, we outline the applications of artificial coculture systems comprising two or more species. The stability of an artificial coculture system refers to its ability to sustain consistent microbial interactions and performance over time, while productivity refers to the amount of product generated per unit of time within the coculture. To fully harness artificial microbial coculture systems as innovative platforms for biomass conversion, it is essential to enhance both the stability and productivity of these systems [5, 13].
Table 1.
The applications of artificial two-species coculture systems as biosynthetic platforms
| Products | Coculture systems | Substrate/mid-products | Production before optimization | Optimization method | Coculture production | Refs |
|---|---|---|---|---|---|---|
| Amino acids | ||||||
| L-Lysine | E. coli–C. glutamicum | Starch/ lysine, glucose, amylase | ~ 0.77 g/L with glucose | Establishing cross-feeding interactions between member species | 0.4 g/L without glucose | [128] |
| L-Pipecolic acid | E. coli–C. glutamicum | Starch/ lysine, glucose, amylase | ~ 4 mM* with glucose | Establishing cross-feeding interactions between member species | 3.4 ± 0.1 mM without glucose | [128] |
| γ-Amino butyric acid | C. lacerate–L. plantarum | Glucose, soybean flour, rice bran | — | Adjusting environmental parameters | 15.53 mg/mL | [129] |
| Peptides | ||||||
| Nisin | Y. lipolytica–L. lactis | Sugar beet molasses/ lactic acid | 176 mg/L* | Adjusting environmental parameters | 270 mg/L | [130] |
| Dentigerumycin E | Streptomyces sp. JB5–Bacillus sp. GN1 | Malt extract, glucose, yeast extract | — | Adjusting environmental parameters | 34 mg | [131] |
| Fengycin | B. subtilis–C. glutamicum | Maltodextrin, sucrose, yeast extract / proline | 871.86 mg/L* | Dividing metabolic pathways and enhancing the transport of essential metabolites | 2.31 g/L | [132] |
| Proteins | ||||||
| Alpha amylase | B. cereus–B. thuringiensis | Starch | 14.5 ± 0.1 U/ml/min | Adjusting environmental parameters | 44.0 U/ml/min | [133] |
| Total protein | Microalgae–L. starkeyi | CO2 | 0.3 g/g* | Establishing cross-feeding interactions between member species | 0.15 g/g (lipids and carbohydrates production was higher) | [134] |
| Lipids | ||||||
| Lipid, total fatty acid | C. pyrenoidosa–R. glutinis | CO2, glucose | Lipid: 0.74 ± 0.05 g/L, Total fatty acid: 91.2 ± 2.57 mg/L/day*(C. pyrenoidosa)、70.08 ± 1.97 mg/L/day*(R. glutinis) | Adjusting environmental parameters | Lipid: 2.48 ± 0.09 g/L, total fatty acid: 175.64 ± 2.32 mg/L/day | [135] |
| Lipid | R. glutinis–C. vulgaris | Acetate, CO2 | 1 g/L | Adjusting environmental parameters | 2.6 g/L | [136] |
| Biofuels | ||||||
| H2 | E. aerogenes–C. butyricum | Crude glycerol, apple pomace hydrolysate | 19.46 mmol/L | Adjusting environmental parameters | 26.07 ± 1.57 mmol/L | [137] |
| n-Butanol | E. coli–E. coli | Cellulose hydrolysate of rice straw/ butyrate | 0.093 g/L/h* | Dividing metabolic pathways | 0.163 g/L/h | [138] |
| Ethanol | A. niger–S. cerevisiae | Potato waste/ glucose | 21.58 g/L* | Constructing confined spaces | 37.93 g/L | [71] |
| Methane | C. cellulovorans–methanogens | Sugar beet pulp/ H2, CO2 | — | Adjusting environmental parameters | 34.0 L/kg | [139] |
| Isobutanol | T. reesei–E. coli | Pretreated corn stover | — | Improving the biosynthesis of essential intermediate metabolites | 1.88 g/L | [140] |
| Isopropanol | E. coli–E. coli | Cellobiose/ glucose | — | Utilization of quorum-sensing | 16.0 ± 2.2 mM | [81] |
| Ethanol | A. niger–S. cerevisiae | Tofu waste/ sugar | 7.69 g/L | Adjusting environmental parameters | 11.39 g/L | [141] |
| Fusel alcohol | E. coli–E. coli | Distillers’ grains with solubles hydrolysates | 12 g/L* (from glucose synthetic medium) | Adjusting environmental parameters | 10.3 g/L | [142] |
| Polyphenols | ||||||
| p-Coumaric acid, Caffeic acid | S. cerevisiae–S. cerevisiae | Carboxymethyl-cellulose/ glucose | 0.46 mg/L*, — | Establishing cross-feeding interactions between member species and adjusting environmental parameters | 71.71 mg/L, 8.33 mg/L | [143] |
| Caffeic acid | E. coli–C. glycerinogenes | Sugarcane bagasse hydrolysate/ shikimate | 133.10 mg/L* | Enhancing the transport of essential metabolites | 1943.2 mg/L | [144] |
| Salidroside | E. coli–E. coli | Cellobiose, glucose, xylose/ Glucose, tyrosol | 128.2 mg/L | Utilization of quorum-sensing | 1.18 g/L | [82] |
| Coniferol, chavicol | B. subtilis–E. coli | Corncob slurry/ferulic acid, coumaric acid | — | Application of biosensors | 55 ± 2.5 mg/L, 72 ± 1.3 mg/L | [77] |
| β-carotene | S. elongatus–P. putida | CO2/ sucrose | ~ 0.06 g/L | Hydrogel encapsulation | 1.3 g/L | [145] |
| Others | ||||||
| 3-Hydroxypropionic acid | S. elongatus–E. coli | CO2/ sucrose | ~ 10 mg/L* | Adjusting environmental parameters | 68.29 mg/L | [7] |
| Medium-chain-length polyhydroxyalkanoate | P. putida–E. coli | Lignocellulose/ acetic acid, free fatty acids | — | Reducing competition between species members | 1.64 g/L | [25] |
| Polyhydroxybutyrate | S. elongatus–H. boliviensis | CO2/ sucrose | ~ 0.005 mg/L/day | Hydrogel encapsulation | 28.3 mg/L/day | [9] |
| Polyhydroxyalkanoate | S. elongatus–P. putida | CO2, 2,4-dinitrotoluene/ sucrose | — | Hydrogel encapsulation | 5 mg/L/day | [146] |
| Lactate | T. asperellum–L. paracasei | Cellulose/ glucose | 1.38 g/L | 3D printing | 13.82 g/L | [102] |
—, the production data were not provided in the original reference. *, Data from monoculture production
Table 2.
The applications of artificial more species coculture systems as biosynthetic platforms
| Products | Coculture systems | Carbon source, precursor/ mid-products | Production before optimization | Optimization method | Coculture production | Refs |
|---|---|---|---|---|---|---|
| Caffeic acid | S. cerevisiae–S. cerevisiae–S. cerevisiae | Carboxymethyl-cellulose/ glucose, p-Coumaric acid | 8.33 mg/L | Adjusting environmental parameters | 16.91 mg/L | [143] |
| Lipopeptide | C. glutamicum–B. amyloliquefaciens–P. pastoris | Kitchen waste/ L-proline, amylase | ~ 29.77 mg/L* | Dividing metabolic pathways | 74.13 mg/L | [147] |
| H2 | Enterococcus–Enterococcus–Enterococcus | Wheat-straw xylan | — | Dividing metabolic pathways | 79.54 mL/g wheat-straw xylan | [148] |
| Lipid | R. opacus–R. jostii–R. jostii | Lignin, glucose/ acetyl-CoA, β-ketoadipyl-CoA | — | Reducing competition between species members | 0.08 g/g cell dry weight | [149] |
| Lipopeptides | B. amyloliquefaciens–C. glutamicum–C. glutamicum–P. pastoris | Kitchen waste, glucose/ proline, serline, amylase | ~ 193.47 mg/L* | Dividing metabolic pathways and enhancing the transport of essential metabolites | 269.17 mg/L | [150] |
| Butyl Butyrate | C. acetobutylicum–C. tyrobutyricum-E. coli–T. asperellum | Microcrystalline cellulose/ lipase, acetate, butyrate, butanol | 13.52 g/L with glucose | Dividing metabolic pathways | 2.94 g/L without glucose | [151] |
| Butyric acid | T. reesei–L. pentosus–C. tyrobutyricum–L. brevis | Cellulose, xylose/ acetate, lactate, butyrate, O2 | 9.5 g/L | Membrane separation | 10.2 g/L | [103] |
| Electricity | S. elongatus-E. coli–S. oneidensis–G. sulfurreducens | CO2/ sucrose, lactate, acetate | ~ 0.6 W·m−2 | Dividing metabolic pathways and adjusting environmental parameters | 1.7 W·m−2 | [152] |
| Iturin A | B. amyloliquefaciens–B. subtilis–B. subtilis | Food waste/ amylase, lipase, glucose, fatty acid chain | 7.66 mg/L | Dividing metabolic pathways | 8.12 mg/L | [153] |
—, the production data were not provided in the original reference. *, data from monoculture production
In this article, we review recent advances in utilizing coculture systems as biosynthetic platforms for biomass conversion. We also discussed the factors that influence stability and productivity in coculture systems, along with strategies and tools to address these challenges. Additionally, we proposed further perspectives on optimizing coculture systems through the development of novel tools, analysis of interspecies relationships, and the regulation of metabolic balance. These insights provide a foundation for future efforts aimed at production of bio-based chemicals from renewable resources using coculture systems.
Enhancing stability through the regulation of interspecies relationships
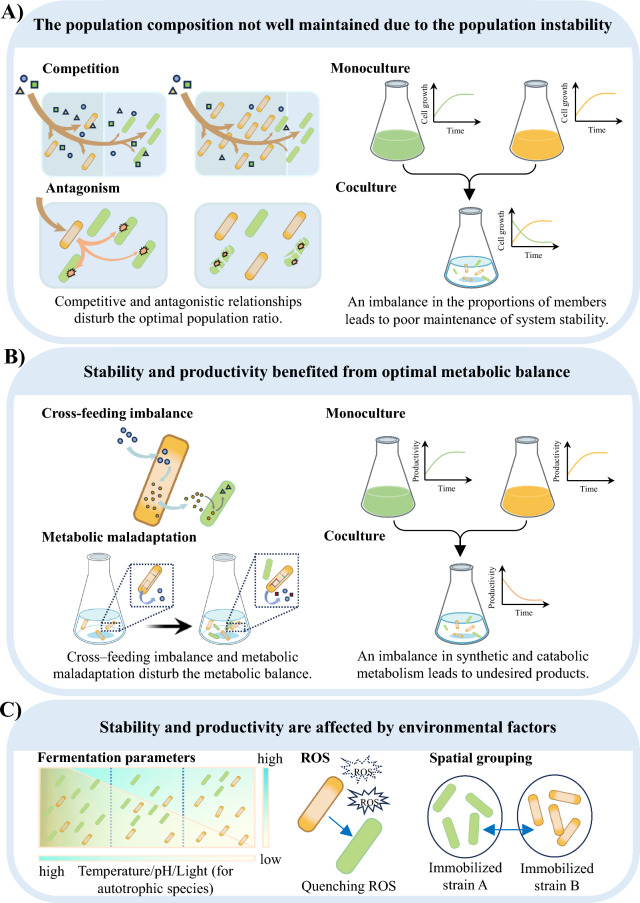
The population composition not well maintained due to the population instability
In coculture systems, the development of competitive and antagonistic relationships among microbial members can disrupt the intended optimal population ratio, leading to a shortened period of system stability (Fig. 1A). In competitive relationships, both microbes vie for limited resources, which suppresses their growth, survival, or reproduction [14]. For example, in an artificial autotrophic–heterotrophic coculture system comprising Synechococcus elongatus cscB+ and engineered E. coli, the growth of E. coli significantly reduced compared to its growth in LB medium [7, 15]. Subsequently, Ma et al., from the same research group identified the competitive consumption of phosphate and nitrogen between microbial members in this system, as revealed through multi-omics analyses [16]. Similarly, the construction of a tri-member coculture system consisting of Azotobacter vinelandii, Bacillus licheniformis and Paenibacillus curdlanolyticus often results in system instability due to the “winner-takes-all” dynamic, driven by competition for nitrogen and substrates [17]. Antagonism is another biological interaction where one organism inhibits the growth of another by releasing toxins, inhibitory compounds, or other means [18]. For example, plant microbiomes containing Bacillus cereus and Pseudomonas fluorescens can protect plants from fungal and bacterial pathogens, either through direct antagonism or by activating plant defense mechanisms [19]. However, under iron-deficient conditions, P. fluorescens inhibited the vegetative growth of B. cereus by producing iron-chelating molecules [20]. Antagonistic interactions have also been observed among nitrogen-fixing bacteria that promote plant growth. Notably, strains of Gluconacetobacter diazotrophicus exhibit antagonistic activity against other strains of the same species, as well as closely related species like Gluconacetobacter johannae, Gluconacetobacter azotocaptans and Gluconacetobacter liquefaciens [21].
Fig. 1.
Challenges in enhancing the stability and productivity of coculture systems. A The population composition not well maintained due to the population instability. Left, competitive and antagonistic relationships disturb the optimal population ratio. Moreover, an imbalance in the proportions of members leads to poor maintenance of system stability. B Stability and productivity benefited from optimal metabolic balance. Left, cross-feeding imbalance and metabolic maladaptation disturb the metabolic balance. Right, an imbalance in synthetic and catabolic metabolism leads to undesired products. C Stability and productivity are affected by environmental factors. The environmental factors included fermentation parameters (such as temperature/pH/light (for autotrophic species)), ROS and spatial grouping between the member species
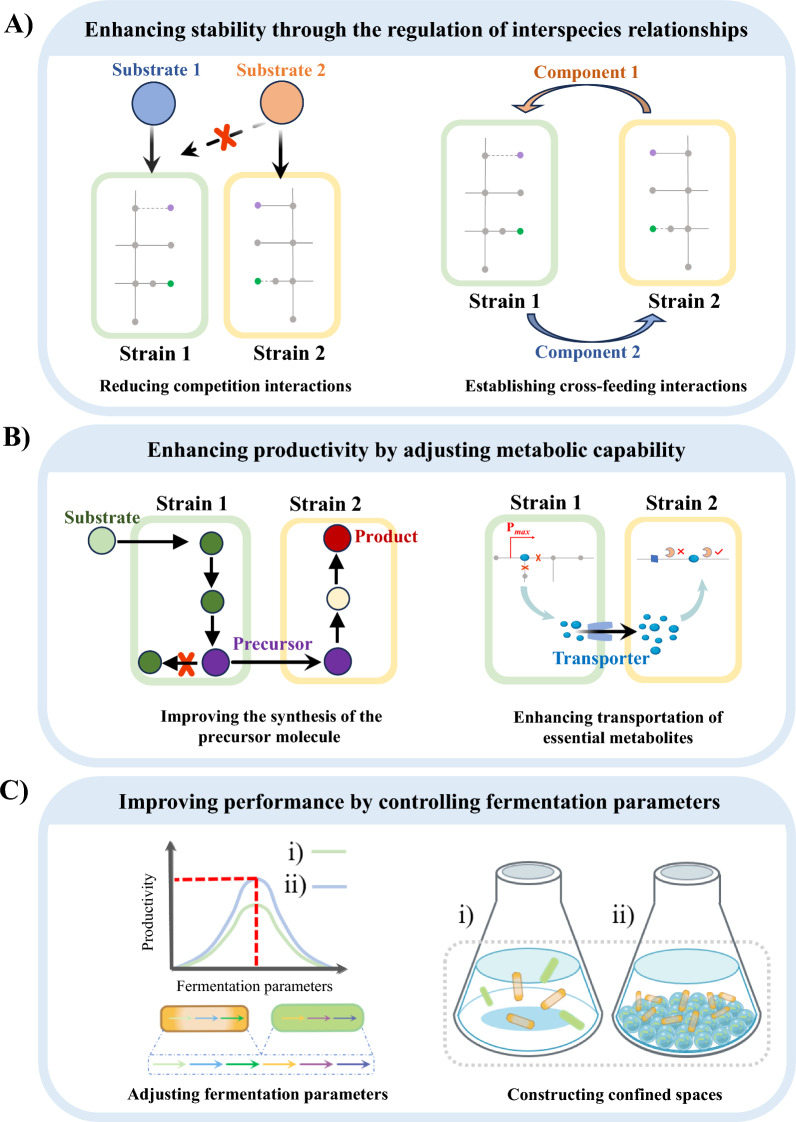
Optimization of population relationships via reducing competition and establishing cross-feeding interactions between species members
Since interactions among microbial members in artificially designed coculture systems may not occur naturally, unexpected behaviours can arise during cocultivation. The nutritional patterns of microbial members, designed according to specific requirements, can potentially alter original interactions [22, 23]. Thus, regulating these interactive relationships, such as minimizing competition and establishing cross-feeding between species, is anticipated to become an effective optimization strategy (Fig. 2A).
Fig. 2.
Strategies for enhancing the stability and productivity of coculture systems. A Enhancing stability through the regulation of interspecies relationships; B enhancing productivity by adjusting metabolic capability; and C improving performance by controlling fermentation parameters
Reducing competition between species members. Similar to the principle of survival of the fittest, microbial members with less capacity to utilize available resources may be outcompeted. In artificial coculture systems, the poor growth of one member can lead to the collapse of the entire system [22]. One approach to mitigate resources competition in coculture systems is to allocate carbon sources to different species [24]. For instance, to minimize glucose competition between E. coli and Pseudomonas putida, the ptsG and manZ genes were knocked out in E. coli to enable preferentially xylose utilization. This create a coculture of xylose-utilizing E. coli and glucose-utilizing P. putida, resulting in an efficient production of medium-chain-length polyhydroxyalkanoate, with a yield of 1.64 g/L [25]. Similarly, Li et al. improved the stability of a three-strain coculture system by using different carbon substrates for three metabolically engineered E. coli strains, achieving 172 mg/L rosmarinic acid—38-fold more than the parent strain in monoculture [26].
Establishing cross-feeding interactions between member species. More artificial coculture systems still rely on the transfer of a single metabolite as a linker between upstream and downstream strains [27, 28]. In such setups, the growth of upstream strains is minimally affected by downstream strains, while downstream strains are heavily dependent on upstream strains, placing them in a disadvantaged ecological niche [29]. Establishing metabolite cross-feeding interactions within coculture systems shifts the relationship toward mutualistic symbiosis, where the growth and production of strains are influenced by environmental conditions and the regulatory behavior of partner strains. This mutualism enhances system stability [1]. Key metabolites like amino acids, vitamins and ATP are often used to establish cross-feeding relationships, a strategy widely adopted to improve performance in an artificial coculture systems [30]. For example, Konstantinidis et al. used WT lactic acid bacteria capable of secreting B-group vitamins, such as riboflavin and folate, to cocultivate with vitamin-deficient S. cerevisiae. Through adaptive laboratory evolution, they increased riboflavin production 14.4-fold (up to 144 ± 55.1 ng/mL) and enhanced folate secretion from 48.3 ± 10.5 ng/mL to 190 ± 35.6 ng/mL in the evolved strain [31]. Nutritional co-dependence, or syntrophy, also shows potential for enhancing biotechnological processes by leveraging cooperation between cell types. For example, Losoi et al. developed a carbon cross-feeding system using E. coli and Acinetobacter baylyi ADP1 strains, both engineered to be incapable of growing on glucose independently. When cultured together with glucose as the sole carbon source, their growth was supported by the exchange of gluconate and acetate, enabling intrinsic control over carbon availability and population balance [32]. Similarly, Peng et al. engineered a cross-feeding relationship using nutrient-deficient and metabolite-overexpressing yeast strains to efficiently synthesize resveratrol [33].
Enhancing biosynthetic performance by adjusting metabolic capability
Stability and productivity benefited from optimal metabolic balance
Disruptions in metabolic balance or maladaptation during the transition from monoculture to coculture can negatively impact the productivity of coculture systems (Fig. 1B). In monoculture, the metabolic pool is unified within a single strain, but in coculture, it is divided between multiple strains, introducing complexity and uncertainty into the fermentation process [34]. Specially, intermediate metabolites are excreted into the extracellular environment and transferred from donor to recipient cells through synthetic metabolic pathways during cocultivation. Since the extracellular volume typically exceeds the intracellular volume, metabolites become diluted, reducing the efficiency of synthetic metabolism in recipient strains, ultimately affecting the productivity of coculture systems [34]. For example, Kawai et al. divided the glucose-to-isoprenol synthesis pathway into two modules, assigning them to two E. coli strains [34]. The upstream strain converted glucose to mevalonate, while the downstream strain converted mevalonate to isoprenol. However, the conversion efficiency of mevalonate to isoprenol in the downstream strain was initially low. Through adaptive laboratory evolution and rational metabolic pathway design of the metabolic pathway in the downstream chassis, the conversion efficiency in the downstream strain, conversion efficiency improved, ultimately enhancing overall productivity. The shift from monoculture to coculture introduced complex ecological and metabolic dynamics. In microbial cell factories, key metabolite pathways are optimized by enhancing key enzymes and silencing competitive pathways to increase product yield [35, 36]. However, changes in cultivation patterns can alter metabolic behavior. For instance, Seo et al. developed a coculture system with engineered E. coli strains for the simultaneous consumption of mixed sugars, including glucose and xylose [37]. They isolated a mutant strain (HSEC0415xyl) with mutations in the xylR gene (R121C and P363S) to overcome glucose-driven carbon catabolite repression [37]. Despite these efforts, 0.6 g/L xylose remained in the culture when glucose consumption began, indicating that the strain still preferentially consumed glucose [37]. This preferential glucose consumption, also known as carbon catabolite repression, has been observed in other studies. Take an example, Shin et al. designed a coculture system with strains specialized in either glucose or xylose utilization for ethanol production. However, the fermentation rate of xylose specialist in mixed sugar is significantly lower than that in xylose alone [38].
Adjusting metabolic capability by improving the biosynthesis or transport of essential intermediate metabolites
Microbial interactions can reduce the metabolic burden on individual strains, benefiting overall metabolic productivity [39]. In coculture systems, cooperative relationships are typically established by designing specific metabolites to synthesize target products. Optimizing both anabolic and catabolic processes—by enhancing the biosynthesis of crucial metabolites and improving their transport—can significantly increase system productivity (Fig. 2B).
Improving the biosynthesis of essential intermediate metabolites. An unequal distribution of metabolite “production” and “consumption” is a key factor limiting efficient production in coculture systems [40]. For instance, Wang et al. developed an E. coli–E. coli coculture system for sakuranetin biosynthesis, but low concentration of the malonyl-CoA, a crucial precursor, limited production [41]. By adding malonate (the precursor of malonyl-CoA) to the medium and overexpressing of two heterologous enzymes involved in malonyl-CoA synthesis, they increased sakuranetin yield from lower than 10 mg/L in monoculture to 29.7 mg/L in coculture [41]. Similarly, Thuan et al. used an E. coli–E. coli coculture system to produce apigetrin [42]. They knocked out the gene such as ushA (encoding UDP-glucose hydrolase), zwf (glucose-6-phosphate 1-dehydrogenase), and pgi (glucose-6-phosphate isomerase) to redirect carbon flux, enhancing the synthesis of UDP-glucose, a key precursor. This optimization resulted in an apigenin yield of 16.6 mg/L [42]. Additionally, metabolic pathway can be optimized by selecting enzymes from different sources for cocultured strains. For instance, Liu et al. employed a three-strain E. coli coculture system for the biosynthesis of genistein, a natural plant product with various plant-derived biological activities [29]. In the study, six different sources of chalcone synthase (CHS) and four sources of chalcone isomerase-like proteins (CHILs) were tested to optimize genistein yield. The combination of EbCHS from Erigeron breviscapus and PhCHIL from a Petunia hybrid result in the highest production of genistein and the lowest byproducts formation [29]. Additionally, for target products of eukaryotic origin, coculture systems incorporating prokaryotic–eukaryotic or eukaryotic–eukaryotic interactions may represent a more promising strategy for optimizing key enzymes functionality [43].
Enhancing the transport of essential metabolites. Metabolites are typically transferred between intracellular and extracellular environments to maintain cellular homeostasis [44]. Engineering transport proteins can improve the use of low-cost alternative substrates, reduce losses of pathway intermediates, and enhance the efficacy and yield of target products [45]. Designing methods for transporting small molecules is an effective strategy for improving the performance of coculture systems. For example, Gargatte et al. combined the amino acid exporter protein PhpCAT with biosensor-assisted cell selection to increase the biosynthesis of tyrosine, a key pathway intermediate [46]. This strategy led to a 96% increase in the production of 4-hydroxystyrene in an E. coli–E. coli coculture system compared to a control coculture without the exporter tyrosine [46]. In another study involving cis, cis-muconic acid (MA) production, Zhang et al. observed that the most of the precursor of 3-dehydroshikimic acid (DHS) accumulated outside the cell, suggesting that the intracellular availability of DHS for conversion to MA was a limiting step [47]. To improve MA production, they engineered a membrane-bound transporter, ShiA, to transport DHS into the cell, significantly increasing the yield of MA [47].
Enhancing performance by modulating the internal and external environment
Stability and productivity are affected by environmental factors
The parameters of a coculture systems, such as inoculation timing, carbon sources, temperature, pH, light intensity (only for autotrophy), and other inducers, can significantly impact the production of individual metabolites (Fig. 1C) [48]. Even small changes in these factors can dramatically influence system performance. Typically, to accommodate the growth conditions of all members in a coculture system, individual strains often must compromise on their optimal requirements. For example, E. coli grows best at 37 °C, but in a coculture with phototrophic cyanobacteria—producers of carbon sources for E. coli—the temperature is adjusted to 30 °C to support cyanobacterial growth. As a result, E. coli’s productivity is expected to decrease [7, 49]. Similarly, oxygen levels in the medium affect the growth and production of strains, especially in systems involving microalgae and bacteria for H2 production [50].
In autotrophic–heterotrophic coculture systems, reactive oxygen species (ROS) also play a critical role in affecting cell growth and productivity. While phototrophs provide organic carbon to heterotrophs, the heterotrophs help phototrophs from oxidative stress, contributing to their survival and robust growth. This mutualistic interaction is essential for maintaining system stability. Similar findings have been observed in other autotrophic–heterotrophic cocultures [7, 51, 52].
Spatially structuring within coculture systems can further stabilize mutualistic cross-feeding [53]. Spatial organization strengthens local interactions, prevents exogenous microbes from outcompeting functional strains, and improves system resilience to environmental stresses [54, 55]. Kim et al. used mathematical models to highlight the significance of spatial structure in bacterial communities, demonstrating its influence on both symbiotic and competitive interactions [17]. For example, in tri-member coculture of A. vinelandii, B. licheniformis and P. curdlanolyticus, separating into different compartments transformed their relationship from competitive to mutualistic [17].
Improving performance by optimizing environmental parameters and space organization
To achieve controlled fermentation in coculture systems, as in monoculture, it is essential to establish optimal environmental parameters (e.g., pH, temperature, oxygen demand) and define acceptable ranges of substrate and product concentrations [56, 57]. Moreover, the complexity of microbial—whether positive or negative—can be influenced by the spatial distribution of bacteria [58]. Therefore, adjusting environmental parameters and constructing confined spaces to optimize system performance are key strategies for improving the stability and productivity of coculture systems (Fig. 2C).
Adjusting environmental parameters. Early studies have shown that optimizing fermentation parameters such as temperature, pH, inoculum ratio, medium components, inoculation time, and anaerobic conditions plays a crucial role in enhancing the stability and productivity of coculture systems [59, 60]. pH, in particular, is a critical factor affecting cell growth and inducing metabolic shifts [61]. Strategies for controlling pH—such as adjusting the phosphate buffer concentrations, maintaining a fixed pH, or adding exogenous regulators like eggshell biowaste, have also been shown to improve the stability and productivity of many other coculture systems [62–64]. Temperature is another important factor, especially when coculture members have different optimal temperature ranges. For example, high temperatures favour E. coli, while low temperatures favor P. putida. To address this, enable coexistence and program the community composition of such systems, Krieger et al. developed a cycling regime that alternates between temperature conditions, allowing the two strains to coexist and enabling tunable control over community composition [65].
Redox levels in the culture medium also impact coculture stability and productivity. Microbial interactions can alter the redox conditions, which in turn affects growth and metabolite production. For instance, E. coli was found to reduce ROS levels under cocultivation conditions, enhancing the growth of autotrophs in a coculture system with S. elongatus 2973 [7]. Similarly, Li et al. observed a significant reduction in ROS levels in a coculture system involving S. elongatus 7942 and S. cerevisiae [66]. These studies revealed that well-designed coculture systems, particularly those involving photoautotrophs and heterotrophs, can establish interactions that naturally reduce ROS, contributing to the system stability and productivity.
Constructing confined spaces. Compartmentalization is a key feature of natural systems and a promising tool for engineering biochemical pathways in both cells and cell-free environments [67]. By creating confined spaces through immobilization, the natural microenvironment of enzymes can be simulated, reducing mass transfer limitations and enhancing system performance [68]. For instance, immobilization can boost hydrogen production, acclimatize bacteria more effectively, and reduce the lag phase in bacterial cultivation [69]. In a study by Kao et al., biohydrogen production increased in an immobilized coculture system consisting of Clostridium butyricum and Rhodopseudomonas palustris compared to free coculture. Immobilized cell beads can achieved high cumulative hydrogen production [70]. Similarly, Izmirlioglu et al. used biofilm reactors to coculture Aspergillus niger and Saccharomyces cerevisiae for ethanol production from potato waste through simultaneous saccharification and fermentation [71]. A plastic composite scaffold to supported biofilm formation, while the hyphae of A. niger provided additional attachment sites for S. cerevisiae [71]. Under optimal conditions, ethanol production reached 37.93 g/L [71].
Tools for enhancing the stability and productivity of coculture systems
Researchers have developed numerous to enhance the stability and productivity of coculture systems by focusing on intercellular communication and the environmental conditions. These strategies include the use of genetic circuits, spatial partitioning, and advanced biotechnological tools to optimize coculture system.
Engineering coculture systems via novel genetic approaches
Strategies to reduce competition between species, establish cross-feeding interactions, and enhance the synthesis of key metabolites were focused on individual chassis and typically lacked considerations of the interactions between microorganisms, which could result in less than optimal performance in coculture systems. To address this, genetic circuits that enable microbial partners to collaborate and respond dynamically to environmental changes have been developed to optimize coculture systems [72].
Application of biosensors
Biosensors detect and convert biological signals into measurable outputs, serving as regulatory tools that respond to metabolite concentrations and guide coculture system performance (Fig. 3A) [73]. In a coculture system, biosensors can serve as regulatory components that respond to the concentration of target metabolites and, depending on the specific function of the sensor, provide different guidelines to regulate the performance of the coculture system [74, 75]. For instance, Kang et al. constructed a coculture system using Vibrio sp. dhg and E. coli to produce 3-hydroxypropionic acid (3-HP) from alginate [76]. A biosensor sensitive to 3-HP was introduced into E. coli, enabling it to degrade ampicillin in response to 3-HP production. This modification led to a 20-fold increase in 3-HP production compared to an unmodified control system [76]. Similarly, Chacόn et al. developed a hydroxy cinnamic acid-inducible biosensor that responds to ferulic acid and coumaric acid in a coculture system of Bacillus subtilis and E. coli for the synthesis coniferol and chavicol from corncob lignocellulose [77]. The biosensor induced enzyme production in response to these acids, resulting in 173 ± 9.3 mg/L of coniferol and chavicol without external inducers [77].
Fig. 3.
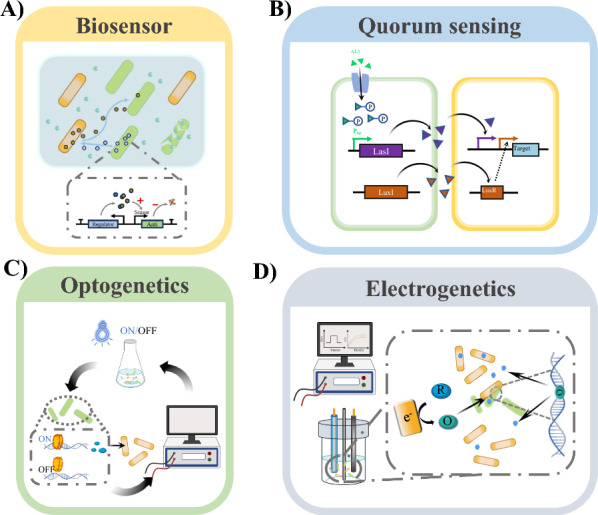
Genetic tools for enhancing the stability and productivity of coculture systems. A The population proportions were balanced using biosensor tools; B the expression level of the target gene was adjusted by the QS signal to the cell density; C the balance between growth and production was adjusted using computer monitoring and photosensitivity; D cell growth and production were regulated using electrical signals
Utilization of quorum-sensing
Quorum sensing (QS) is a common phenomenon in the cell–cell communication process that involves the detection of and responses to changes in cell population density caused by specific signalling molecules, such as autoinducer 1 (AI-1), autoinducer 2 (AI-2), and acylated homoserine lactone (AHL), which regulate gene expression (Fig. 3B) [78–80]. Several components of the QS system were employed to enhance coculture system performance by autonomously regulating population density. Honjo et al. designed a synthetic QS system to cascade task execution through cell–cell communication within the coculture system [81]. When the cell density of the β-glucosidase-secreting strain reached a threshold, cells lysed and released β-glucosidase. Cellobiose was then broken down into glucose in the medium, and the production strain initiated the isopropanol production pathway by recognizing AHL signalling. The isopropanol yield of the coculture system was nearly three times more than that of the single-strain system [81]. Wu et al. employed a series of combinations of four AHL-based QS systems (lux, rpa, tra, las) to determine the feasibility and optimality of these systems, which simultaneously involved cell growth competition and cooperative isopropanol production; subsequently, they constructed a coculture system for the production of salidroside from glucose, xylose, and cellobiose using the selected lux and rpa systems, with a yield nine times higher than that of the control group [82].
Application of optogenetics technology
Optogenetics is a novel technique in which light is utilized to control specific genes and proteins by employing natural and engineered photoreceptors [83]. Since light can be easily adjusted in both time and space, it can dynamically regulate microbial metabolic processes (Fig. 3C) [84–86]. In coculture systems, regardless of the population ratio and the composition of the culture medium, engineered strains carrying optogenetic components can be dynamically controlled based on the flexibility of light usage [87]. This characteristic also facilitates easy integration of the coculture system with computers, enabling visual monitoring and the automatic adjustment of cultivation processes [88]. Lalwani et al. employed an optogenetic circuit combined with the MazEF toxin–antitoxin (TA) system in a coculture system consisting of E. coli and S. cerevisiae, in which E. coli growth could be tuned using only blue light to control the population composition of E. coli and S. cerevisiae [87]. E. coli growth was suppressed in the dark through PFixK2 expression of mazF leading to mRNA degradation. E. coli growth was enabled in blue light through PR expression of mazE, which ceases expression of mazF and inhibits MazF. As a result, this coculture system exhibited an 83% greater isobutyl acetate yield than the control [87].
Application of electrogenetics tools
In electrogenetics, selective chemicals are utilized to convert electrical signals into biological signals, initiating oxidative stress mechanisms within cells to regulate the population density of a single strain in a coculture system to control the strain-to-strain ratio or the biosynthetic ability of the strains (Fig. 3D) [89]. By leveraging electrogenetic principles, genetic tools can be developed that allow for the modulation of interactions and behaviours among cocultured organisms through electrical signalling or the manipulation of cellular activities. Like in optogenetics, the controllability and monitorability of electrical signals can also be integrated with computers or various sensors to achieve autonomous and automated processes [90]. For instance, VanArsdale et al. combined electrogenetics with QS to modulate the production of tryptophan in a coculture system [91]. The researchers used electrogenetics to transform redox signals into the quorum-sensing autoinducer AI-1, which in turn, induced a tyrosine biosynthesis in a second population [91]. An electrogenetic method was used to stimulate AI-1 to actuate the expression of ptsH, increasing the growth rate of tyrosine-producing cells, and finally achieving approximately 0.15 g/L of tyrosine [91]. Similarly, VanArsdale et al. employed a combination of electrogenetics and QS to convert electrical signals into AI-1 and control cell lysis through the oxidative stress transcription factor OxyR to regulate population density in a coculture system [92]. Tschirhart et al. presented a simple electrogenetic device that employs the native transcriptional regulator SoxR and the promoter PsoxS to control cell responses quickly and reversibly and is dependent on the amplitude and frequency of the imposed electronic signals [93].
Optimizing coculture systems by spatial partitioning
Beyond genetic modifications, physical methods such as adjusting inoculation ratios, medium composition, and environmental conditions (e.g., temperature, pH) are often applied to optimize coculture systems [94–96]. However, manual adjustments alone are insufficient for maintaining optimal conditions throughout the process. Spatial partitioning, which divides microbial populations into distinct compartments, enhances resource utilization, alleviates feedbacks inhibition, and reduces competition, thereby promoting system synergy and performance [1, 97].
Hydrogel encapsulation
Hydrogels, composed of polymers that allow the diffusion of proteins, nutrients, and metabolites while confining microbes, have been used to construct spatially segregated microbial consortia (Fig. 4A) [98]. Wang et al. developed a microcapsule-based method to arrange 34 strains in a stable and controllable coculture system [99]. Weiss et al. encapsulated S. elongatus 7942 in alginate to increase sucrose export rates and enhance the stability of its coculture with Halomonas boliviensis, achieving stable coculture performance for over 5 months [9].
Fig. 4.
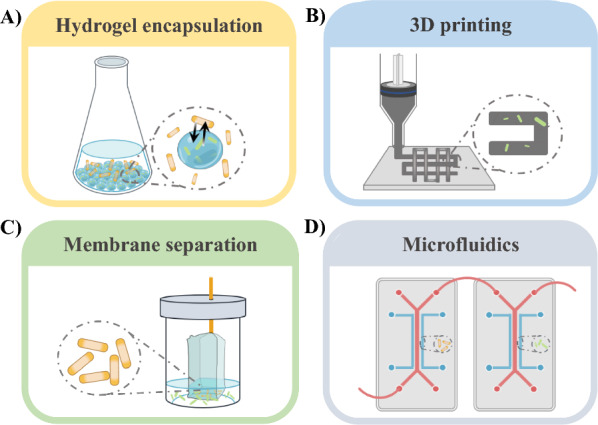
Optimizing coculture systems by spatial partitioning. A Microcapsule-based spatial arrangement using hydrogels; B 3D printing technology to form a spatial isolation structure; C separating different strains using a semipermeable membrane; D using microfluidics technology to control the contact frequency of strains
3D printing
More recently, combined 3D printing technology and material encapsulation for spatial isolation has been considered a feasible approach for optimizing coculture systems. 3D printing enables the precise customization of spatial requirements for any coculture system, which provides researchers with the flexibility to effectively adjust microbial population density (Fig. 4B) [100]. Sun et al. used 3D printing to encapsulate cocultured algae, achieving heterotrophic growth and flexible spatial distribution [101]. Gao et al. constructed a 3D-printed bioreactor for lactate production, achieving 13.82 g/L lactate from 40 g/L microcrystalline cellulose in a coculture system [102].
Membrane separation
Membrane separation is typically achieved by using physical membranes, such as dialysis bags or other materials, to directly isolate cells, permitting the passage of metabolites while constraining the movement of cells. Another approach involves utilizing biofilms to establish spatial ecological niche separation under specific conditions (Fig. 4C). Xie et al. employed membrane separation to coculture Cellvibrio pealriver with microalgae, increasing lipid production by 1.7 to 1.9 times [97]. Similarly, Shahab et al. used a membrane-aerated biofilm reactor to create an oxygen-replete niche for cross-kingdom microbial coculture system, demonstrating the advantages of spatial segregation in lignocellulose processing, such as regulating oxygen, temperature, light, or pH [103].
Microfluidics
Microfluidics, a high-throughput platform capable of precise manipulation of microbial communities, has been employed to optimize coculture interactions (Fig. 4D) [104, 105]. Wei et al. constructed a coculture system consisting of Synechocystis sp. PCC 6803 and Shewanella oneidensis MR-1 in a microfluidic chip, in which Synechocystis 6803 utilizing solar energy to produce O2 and organic substrates in the upper layer, while the S. oneidensis MR-1 utilize O2 and organic substrates to discharge electricity in the lower layer [106]. Without additional organic substrates, the coculture system generated self-sustaining current at a density of 6 μA/cm2, approximately 400 times higher than that achieved using only photosynthetic autotrophs [106]. Liu et al. utilized microfluidic droplet generators to encapsulate Chlorella vulgaris and Bacillus licheniformis, resulting in a 62.91% increase in lipid content in C. vulgaris compared to monoculture [107].
Perspectives and future directions
Developing novel tools for coculture system studies
Due to the presence of two or more different engineered strains in coculture systems, sometimes involving multiple species, favourable interactions should be considered, and beneficial metabolic sharing among various member strains should be established in the optimization of coculture systems. The development of novel tools provides necessary technical support for the construction of stable and efficient coculture systems and is expected to offer promising solutions for addressing the challenges associated with the industrial application of these systems.
Multiomics technologies. Multiomics analysis could provide a “global view” of various microbial family members in microbial coculture systems, increasing the understanding of the interactions between microbial members [108–110]. The development of multi-omics technologies contributes to a greater understanding of the diversity, functions, and interactions within microbial communities in coculture systems and is crucial for designing more complex and efficient coculture systems.
High-throughput screening. The “switch”-controlled fermentation processes used in manufacturing and in laboratory-scale models can significantly impact the metabolic state of strains, potentially through interactions with designed or random genetic modifications [111]. High-throughput screening techniques provide opportunities for establishing ideal screening models, which can contribute to the identification of engineered strains with robust metabolic capabilities [111, 112]. The development of high-throughput screening methods that allow for coculture systems to rapidly assess the synergistic effects and product synthesis capabilities of different microbial combinations could expedite the design process of coculture systems.
Genetic tools. Using synthetic biology and gene editing tools and methods, fine-tuning the expression levels of genes, introducing new metabolic pathways, or optimizing existing pathways can achieve more precise metabolic engineering of host strains [113, 114]. To further optimize coculture systems, it is necessary to continuously improve gene editing and regulation tools to precisely design and adjust the gene expression of microbial communities, achieving specific cooperative relationships and product syntheses.
Analysing interspecies relationships between microbial members
Inspection of failed mechanisms can also help inform the development of future strategies. However, researchers have not systematically examined why some coculture systems did not function in the past. Liu et al. established a coculture system consisting of S. elongatus–E. coli for isoprene production, achieving an eightfold increase in isoprene production compared to that of an axenic culture [115]. The authors also demonstrated that oxidative stress mitigation pathways might contribute to a long-lasting fermentation process, which lasts longer than 400 h, as revealed through analysis of differential omics profiles [115]. Similarly, Ma et al. employed an integrated proteomics, transcriptomics and metabolomics approach to analyse the metabolic responses of cyanobacteria to a heterotrophic partner in an artificial coculture system consisting of sucrose-secreting cyanobacteria and sucrose-utilizing E. coli and demonstrated that the improved cell growth of cultured cyanobacteria may be due to synergistic effects, including oxidative stress alleviation, enhanced CO2 availability and increased sucrose sink capacity [16]. According to omics analyses, the supply of phosphate and nitrogen in the coculture system was insufficient, which informed the optimization of the performance of the coculture system [16]. The mechanisms operating within coculture systems have been analysed using multi-omics techniques, alongside strategies like genetic circuits construction and spatial segregation. Inspired by these studies, this combined approach may offer a novel and precise method for optimizing coculture systems.
Moreover, despite the application of numerous genetic circuit tools in coculture systems, an understanding of the potential benefits of dynamically adjusting these components over time is lacking, which could limit the development of reliable gene circuits, especially in large-scale cultivation scenarios. The integration of AI, high-throughput screening and synthetic biology technologies holds great promise for advancing the ability of researchers to engineer biological systems with precision and efficiency [116, 117]. Researchers have initiated preliminary attempts to analyse the effect of gene(s) burden on coculture systems by constructing a mechanistic mathematical model describing the three modules and their impact on the growth of two bacterial species sharing a single growth compartment [118]. The use of computer simulation and artificial intelligence (AI) technology allows for real-time modelling and prediction of microbial behaviour in coculture systems. Researchers could construct a metabolic network model of a coculture system to guide the reconstruction of coculture systems by integrating and analysing information from metabolic pathways, metabolic reactions, and cross-feeding relationships of multiple microbial consortia.
Improving the metabolic balance of microbial members
The microbial population of a coculture system with different physiological attributes can rarely be controlled simply by using initial conditions because the growth of each cell is significantly affected by the medium composition, which changes dynamically throughout the cultivation period. The general optimization process depends on predetermined empirical parameters, which must be determined through labour-intensive experiments. However, this approach poses numerous challenges in industrial applications, especially for coculture systems. Adaptive laboratory evolution is a crucial approach for enhancing microbial growth, stress resistance, and production efficiency [119]. Synthetic acclimation is proposed to serve as an efficient strategy for improving the production of diverse value-added biochemicals via artificial coculture systems. A representative example is the coculture system consisting of alginate-utilizing Vibrio sp. dhg and an engineered E. coli strain for the direct production of 3-HP constructed by Kang et al. [76]. Via systematic design, 3-HP production increased up to 4.3-fold with minimal accumulation of acetate, which was attributed to changes in the population composition of cocultures based on the concentrations of growth-inhibiting compounds [76]. All these results provide us with the insight that constructing gene circuits, combined with domestication methods, is an effective means of improving the performance of coculture systems.
Designing specific bioreactors for coculture cultivation
Microbial growth necessitates suitable conditions in terms of factors, such as temperature, pH, and gas supply. For instance, ensuring that a reactor maintains stable temperature and pH is crucial for optimizing microbial growth and metabolism. An adequate oxygen supply must be ensured to meet microbial respiratory needs. Photoautotrophic microorganisms typically require light and CO2 for photosynthesis, and factors such as the design of appropriate light sources, light intensity, light cycling, and ventilation are especially critical for photoautotrophic–heterotrophic coculture systems. This complexity in designing bioreactors is heightened in coculture systems, as different member microorganisms have different requirements. Bioreactors offer precise control over production conditions such as temperature, pH, and gas composition, enabling better regulation of microbial growth processes and consequently improving the quality and stability of the products [120]. Currently, there is limited research on the design of bioreactors specifically for coculture systems. To meet the industrial application requirements of coculture systems, it is essential to design bioreactors that are specifically tailored to the characteristics of coculture systems to ensure the stable and efficient operation of these systems.
Immobilized fermentation is a process in which selected microorganisms are cultivated on a moist, solid, nonsoluble organic material that serves as both a support and nutrient source, allowing growth in the absence or near-absence of free-flowing water [121]. Immobilized fermentation allows a system to maintain aseptic conditions and meet oxygen requirements for aerobic fermentation and allows for the monitoring of heat transfer and mass transfer effects and regulation of environmental conditions to increase growth and yield [56]. Moreover, immobilized fermentation has become a crucial strategy for enhancing the performance of coculture systems [122]. This includes optimizing the spatial distribution of different microbial populations to reduce competition and antagonistic relationships and promoting collaborative cooperation, thereby improving the productivity of coculture systems [48]. Additionally, immobilized fermentation contributes to simplifying the separation process, mitigating mutual interference from substances in the mixture [48]. Wang et al. developed a microcapsule-based spatial arrangement method using polymeric microcapsules to construct coculture systems [99]. In this study, the immobilization of member species within microcapsules was employed to create coculture systems, which included a single-species E. coli system, a two-species consortium containing E. coli and S. cerevisiae or E. coli and P. pastoris; a multispecies consortium containing E. coli, S. cerevisiae and C. glutamicum; and a phototrophic–heterotrophic consortium containing S. elongatus and E. coli [99]. This study inspires us to believe that designing biological materials for immobilized bioreactors may become a strategy to expand the industrial application of coculture systems.
Although some researchers have employed omics approaches to explore interactions among member microorganisms, there have been limited or no efforts to comprehend the dynamic interactions between member microbial species. A significant constraint in such studies is the lack of real-time monitoring systems for coculture systems. Kim et al. developed a membrane-separated bioreactor and associated protocols and control strategies to facilitate the quantitative study of coculture systems [123]. Through the in-house developed cell retention modules and gas mixing apparatuses, as well as dual-mode continuous/pseudocontinuous operation, Kim et al. were able to not only independently monitor and control the biomass development of each strain but also independently control the different oxygen utilization rate conditions for each strain [123]. Establishing a real-time monitoring system in artificial coculture systems to track the growth of member microorganisms, the accumulation of metabolic products, and other key parameters will facilitate timely adjustments to operational conditions, enhancing the stability and yield of systems.
Conclusions
Artificial microbial coculture systems have gained increasing attention due to their unique characteristics and functionalities, often surpassing those of individual microbial populations. In coculture systems, members can engage in one-way, two-way, or even multi-way communication through the exchange of signalling molecules, detection, and mutual responses [124, 125]. This mutual coordination ensures the stability of the community’s structure and function [126, 127]. By specializing in distinct tasks, each member enables the system to perform complex functions that are beyond the capabilities of a single strain. However, constructing stable and productive coculture systems is far more challenging than cultivating individual strains, primarily due to the difficulty in maintaining optimal population ratios and striking a balancing anabolism and catabolism over long-term cultivation. In this review, we summarized the issues challenging the stability and productivity of coculture systems. We also introduce strategies and tools for optimizing these systems for biomass conversion. Finally, we provided perspectives and future research directions for coculture systems as novel biosynthetic platforms, laying a foundation for the development of more sophisticated coculture systems in the future.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2019YFA0904600) and the National Natural Science Foundation of China (No. 32270091).
Author contributions
XYS and YJ wrote the draft of the manuscript; LC and WWZ drafted and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2019YFA0904600) and the National Natural Science Foundation of China (No. 32270091).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
We have no conflicts of interest to disclose and all the authors gave their consent for publications.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Diender M, Parera Olm I, Sousa DZ. Synthetic co-cultures: novel avenues for bio-based processes. Curr Opin Biotechnol. 2021;67:72–9. [DOI] [PubMed] [Google Scholar]
- 2.Okeke BC. Cellulolytic and xylanolytic potential of high β-glucosidase-producing Trichoderma from decaying biomass. Appl Biochem Biotechnol. 2014;174:1581–98. [DOI] [PubMed] [Google Scholar]
- 3.Chisti Y. Constraints to commercialization of algal fuels. J Biotechnol. 2013;167(3):201–14. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z, Wang K, Chen Y, Tan T, Nielsen J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat Catal. 2020;3(3):274–88. [Google Scholar]
- 5.Jiang Y, Wu R, Zhang W, Xin F, Jiang M. Construction of stable microbial consortia for effective biochemical synthesis. Trends Biotechnol. 2023;41(11):1430–41. [DOI] [PubMed] [Google Scholar]
- 6.Li T, Jiang L, Hu Y, Paul JT, Zuniga C, Zengler K, et al. Creating a synthetic lichen: mutualistic co-culture of fungi and extracellular polysaccharide-secreting cyanobacterium Nostoc PCC 7413. Algal Res. 2020;45: 101755. [Google Scholar]
- 7.Zhang L, Chen L, Diao J, Song X, Shi M, Zhang W. Construction and analysis of an artificial consortium based on the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 to produce the platform chemical 3-hydroxypropionic acid from CO2. Biotechnol Biofuels. 2020;13(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Xue B, Liu H, Wang S, Su H. Rational construction of synthetic consortia: Key considerations and model-based methods for guiding the development of a novel biosynthesis platform. Biotechnol Adv. 2024;24:108348. [DOI] [PubMed] [Google Scholar]
- 9.Weiss TL, Young EJ, Ducat DC. A synthetic, light-driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production. Metab Eng. 2017;44:236–45. [DOI] [PubMed] [Google Scholar]
- 10.Gao X, Sun T, Pei G, Chen L, Zhang W. Cyanobacterial chassis engineering for enhancing production of biofuels and chemicals. Appl Microbiol Biotechnol. 2016;100(8):3401–13. [DOI] [PubMed] [Google Scholar]
- 11.An X, Chen X, Wang Y, Zhao X, Xiao X, Long H, et al. Cellulolytic bacterium characterization and genome functional analysis: an attempt to lay the foundation for waste management. Bioresour Technol. 2021;321: 124462. [DOI] [PubMed] [Google Scholar]
- 12.Tang H, Ou J, Zhu M. Development of a quantitative real-time PCR assay for direct detection of growth of cellulose-degrading bacterium Clostridium thermocellum in lignocellulosic degradation. J Appl Microbiol. 2015;118(6):1333–44. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Yu H. Insights into constructing a stable and efficient microbial consortium. Chin J Chem Eng. 2021;30:112–20. [Google Scholar]
- 14.Pekkonen M, Ketola T, Laakso JT. Resource availability and competition shape the evolution of survival and growth ability in a bacterial community. PLoS ONE. 2013;8(9): e76471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Pugh S, Nielsen DR, Zhang W, Meldrum DR. Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab Eng. 2013;16:68–77. [DOI] [PubMed] [Google Scholar]
- 16.Ma J, Guo T, Ren M, Chen L, Song X, Zhang W. Cross-feeding between cyanobacterium Synechococcus and Escherichia coli in an artificial autotrophic-heterotrophic coculture system revealed by integrated omics analysis. Biotechnol Biofuels Bioprod. 2022;15(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A. 2008;105(47):18188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feichtmayer J, Deng L, Griebler C. Antagonistic microbial interactions: contributions and potential applications for controlling pathogens in the aquatic systems. Front Microbiol. 2017;8:2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Z, Gu Y, Friman VP, Kowalchuk GA, Xu Y, Shen Q, et al. Initial soil microbiome composition and functioning predetermine future plant health. Sci Adv. 2019;5(9):eaaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simões M, Simoes LC, Pereira MO, Vieira MJ. Antagonism between Bacillus cereus and Pseudomonas fluorescens in planktonic systems and in biofilms. Biofouling. 2008;24(5):339–49. [DOI] [PubMed] [Google Scholar]
- 21.Munoz-Rojas J, Fuentes-Ramirez LE, Caballero-Mellado J. Antagonism among Gluconacetobacter diazotrophicus strains in culture media and in endophytic association. FEMS Microbiol Ecol. 2005;54(1):57–66. [DOI] [PubMed] [Google Scholar]
- 22.Khan N, Maezato Y, McClure RS, Brislawn CJ, Mobberley JM, Isern N, et al. Phenotypic responses to interspecies competition and commensalism in a naturally-derived microbial co-culture. Sci Rep. 2018;8(1):297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein HC, McClure RS, Thiel V, Sadler NC, Kim YM, Chrisler WB, et al. Indirect interspecies regulation: transcriptional and physiological responses of a cyanobacterium to heterotrophic partnership. Msystems. 2017. 10.1128/msystems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim M, Raajaraam L, Raman K. Modelling microbial communities: Harnessing consortia for biotechnological applications. Comp Struct Biotechnol J. 2021;19:3892–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin R, Zhu Y, Ai M, Jia X. Reconstruction and optimization of a Pseudomonas putida-Escherichia coli microbial consortium for mcl-PHA production from lignocellulosic biomass. Front Bioeng Biotechnol. 2022;10:1023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Wang X, Zhang H. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab Eng. 2019;54:1–11. [DOI] [PubMed] [Google Scholar]
- 27.Qiu Z, Liu X, Li J, Qiao B, Zhao G. Metabolic division in an Escherichia coli coculture system for efficient production of kaempferide. ACS Synth Biol. 2022;11(3):1213–27. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Song D, Hu H, Yang R, Lyu X. De novo production of hydroxytyrosol by Saccharomyces cerevisiae-Escherichia coli coculture engineering. ACS Synth Biol. 2022;11(9):3067–77. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Li L, Zhao G. Systems metabolic engineering of Escherichia coli coculture for de novo production of genistein. ACS Synth Biol. 2022;11(5):1746–57. [DOI] [PubMed] [Google Scholar]
- 30.Fritts RK, McCully AL, McKinlay JB. Extracellular metabolism sets the table for microbial cross-feeding. Microbiol Mol Biol Rev. 2021;85(1):e00135-e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinidis D, Pereira F, Geissen EM, Grkovska K, Kafkia E, Jouhten P, et al. Adaptive laboratory evolution of microbial co-cultures for improved metabolite secretion. Mol Syst Biol. 2021;17(8): e10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Losoi PS, Santala VP, Santala SM. Enhanced population control in a synthetic bacterial consortium by interconnected carbon cross-feeding. ACS Synth Biol. 2019;8(12):2642–50. [DOI] [PubMed] [Google Scholar]
- 33.Peng H, Darlington AP, South EJ, Chen HH, Jiang W, Ledesma-Amaro R. A molecular toolkit of cross-feeding strains for engineering synthetic yeast communities. Nat Microbiol. 2024. 10.1038/s41564-023-01596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai R, Toya Y, Miyoshi K, Murakami M, Niide T, Horinouchi T, et al. Acceleration of target production in co-culture by enhancing intermediate consumption through adaptive laboratory evolution. Biotechnol Bioeng. 2022;119(3):936–45. [DOI] [PubMed] [Google Scholar]
- 35.Zhou K, Qiao K, Edgar S, Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33(4):377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R, Zhao S, Wang Z, Koffas MA. Recent advances in modular co-culture engineering for synthesis of natural products. Curr Opin Biotechnol. 2020;62:65–71. [DOI] [PubMed] [Google Scholar]
- 37.Seo H, Castro G, Trinh CT. Engineering a synthetic Escherichia coli coculture for compartmentalized de novo biosynthesis of isobutyl butyrate from mixed sugars. ACS Synth Biol. 2023;13:259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin J, Liao S, Kuanyshev N, Xin Y, Kim C, Lu T, et al. Compositional and temporal division of labor modulates mixed sugar fermentation by an engineered yeast consortium. Nat Commun. 2024;15(1):781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulmer A, Veit S, Erdemann F, Freund A, Loesch M, Teleki A, et al. A two-compartment fermentation system to quantify strain-specific interactions in microbial co-cultures. Bioengineering. 2023;10(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang EX, Ding MZ, Ma Q, Dong XT, Yuan YJ. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation. Microb Cell Fact. 2016;15(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Li Z, Policarpio L, Koffas MAG, Zhang H. De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering. Appl Microbiol Biotechnol. 2020;104(11):4849–61. [DOI] [PubMed] [Google Scholar]
- 42.Thuan NH, Chaudhary AK, Van Cuong D, Cuong NX. Engineering co-culture system for production of apigetrin in Escherichia coli. J Ind Microbiol Biotechnol. 2018;45(3):175–85. [DOI] [PubMed] [Google Scholar]
- 43.Thuan NH, Tatipamula VB, Canh NX, Van Giang N. Recent advances in microbial co-culture for production of value-added compounds. 3 Biotech. 2022;12(5):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katsube S, Ando T, Yoneyama H. L-alanine exporter, AlaE, of Escherichia coli functions as a safety valve to enhance survival under feast conditions. Int J Mol Sci. 2019;20(19):4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Hoek SA, Borodina I. Transporter engineering in microbial cell factories: the ins, the outs, and the in-betweens. Curr Opin Biotechnol. 2020;66:186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gargatte S, Li Z, Zhou Y, Wang X, Zhuang L, Zhang H. Utilizing a tyrosine exporter to facilitate 4-hydroxystyrene biosynthesis in an E coli-E coli co-culture. Biochem Eng J. 2021. 10.1016/j.bej.2021.108178. [Google Scholar]
- 47.Zhang H, Pereira B, Li Z, Stephanopoulos G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci U S A. 2015;112(27):8266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao W, Nokes SE. The use of co-culturing in solid substrate cultivation and possible solutions to scientific challenges. Biofuels Bioprod Biorefining. 2013;7(4):361–72. [Google Scholar]
- 49.Hays SG, Yan LLW, Silver PA, Ducat DC. Synthetic photosynthetic consortia define interactions leading to robustness and photoproduction. J Biol Eng. 2017;11(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Javed MA, Zafar AM, Hassan AA. Regulate oxygen concentration using a co-culture of activated sludge bacteria and Chlorella vulgaris to maximize biophotolytic hydrogen production. Algal Res. 2022;63: 102649. [Google Scholar]
- 51.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol. 2008;74(14):4530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS ONE. 2011;6(2): e16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pande S, Kaftan F, Lang S, Svatos A, Germerodt S, Kost C. Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 2016;10(6):1413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacLean RC, Gudelj I. Resource competition and social conflict in experimental populations of yeast. Nature. 2006;441(7092):498–501. [DOI] [PubMed] [Google Scholar]
- 55.Lee KW, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8(4):894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prabhu G, Bhat D, Bhat RM, Selvaraj S. A critical look at bioproducts co-cultured under solid state fermentation and their challenges and industrial applications. Waste Biomass Valorization. 2022;13(7):3095–111. [Google Scholar]
- 57.Diao J, Song X, Cui J, Liu L, Shi M, Wang F, et al. Rewiring metabolic network by chemical modulator based laboratory evolution doubles lipid production in Crypthecodinium cohnii. Metab Eng. 2019;51:88–98. [DOI] [PubMed] [Google Scholar]
- 58.Liu W, Røder HL, Madsen JS, Bjarnsholt T, Sørensen SJ, Burmølle M. Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front Microbiol. 2016;7:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pei G, Li X, Liu L, Liu J, Wang F, Chen L, et al. De novo transcriptomic and metabolomic analysis of docosahexaenoic acid (DHA)-producing Crypthecodinium cohnii during fed-batch fermentation. Algal Res. 2017;26:380–91. [Google Scholar]
- 60.Liu L, Diao J, Bi Y, Zeng L, Wang F, Chen L, et al. Rewiring the metabolic network to increase docosahexaenoic acid productivity in Crypthecodinium cohnii by fermentation supernatant-based adaptive laboratory evolution. Front Microbiol. 2022;13: 824189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabemanolontsoa H, Kuninori Y, Saka S. High conversion efficiency of Japanese cedar hydrolyzates into acetic acid by co-culture of Clostridium thermoaceticum and Clostridium thermocellum. J Chem Technol Biotechnol. 2016;91(4):1040–7. [Google Scholar]
- 62.Laurinavichene T, Tsygankov A. Different types of H2 photoproduction by starch-utilizing co-cultures of Clostridium butyricum and Rhodobacter sphaeroides. Int J Hydrog Energy. 2016;41(31):13419–25. [Google Scholar]
- 63.Zagrodnik R, Laniecki M. The role of pH control on biohydrogen production by single stage hybrid dark- and photo-fermentation. Bioresour Technol. 2015;194:187–95. [DOI] [PubMed] [Google Scholar]
- 64.Pachapur VL, Das RK, Brar SK, Le Bihan Y, Buelna G. Valorization of crude glycerol and eggshell biowaste as media components for hydrogen production: A scale-up study using co-culture system. Bioresour Technol. 2017;225:386–94. [DOI] [PubMed] [Google Scholar]
- 65.Krieger AG, Zhang J, Lin XN. Temperature regulation as a tool to program synthetic microbial community composition. Biotechnol Bioeng. 2021;118(3):1381–92. [DOI] [PubMed] [Google Scholar]
- 66.Li T, Li CT, Butler K, Hays SG, Guarnieri MT, Oyler GA, et al. Mimicking lichens: incorporation of yeast strains together with sucrose-secreting cyanobacteria improves survival, growth, ROS removal, and lipid production in a stable mutualistic co-culture production platform. Biotechnol Biofuels. 2017;10(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menon G, Okeke C, Krishnan J. Modelling compartmentalization towards elucidation and engineering of spatial organization in biochemical pathways. Sci Rep. 2017;7(1):12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hwang ET, Lee S. Multienzymatic cascade reactions via enzyme complex by immobilization. ACS Catal. 2019;9(5):4402–25. [Google Scholar]
- 69.Seol E, Manimaran A, Jang Y, Kim S, Oh YK, Park S. Sustained hydrogen production from formate using immobilized recombinant Escherichia coli SH5. Int J Hydrog Energy. 2011;36(14):8681–6. [Google Scholar]
- 70.Kao PM, Hsu BM, Huang KH, Tao CW, Chang CM, Ji W-T. Biohydrogen production by immobilized co-culture of Clostridium Butyricum and Rhodopseudomonas Palustris. Energy Procedia. 2014;61:834–7. [Google Scholar]
- 71.Izmirlioglu G, Demirci A. Simultaneous saccharification and fermentation of ethanol from potato waste by co-cultures of Aspergillus niger and Saccharomyces cerevisiae in biofilm reactors. Fuel. 2017;202:260–70. [Google Scholar]
- 72.McCarty NS, Ledesma-Amaro R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 2019;37(2):181–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yuan J, Zhao K, Tan X, Xue R, Zeng Y, Ratti C, et al. Perspective on the development of synthetic microbial community (SynCom) biosensors. Trends Biotechnol. 2023;41(10):1227–36. [DOI] [PubMed] [Google Scholar]
- 74.Sun T, Li Z, Li S, Chen L, Zhang W. Exploring and validating key factors limiting cyanobacteria-based CO2 bioconversion: case study to maximize myo-inositol biosynthesis. Chem Eng J. 2023;452: 139158. [Google Scholar]
- 75.Sun X, Li S, Zhang F, Sun T, Chen L, Zhang W. Development of a N-acetylneuraminic acid-based sensing and responding switch for orthogonal gene regulation in cyanobacterial Synechococcus strains. ACS Synth Biol. 2021;10(8):1920–30. [DOI] [PubMed] [Google Scholar]
- 76.Kang CW, Lim HG, Won J, Cha S, Shin G, Yang J, et al. Circuit-guided population acclimation of a synthetic microbial consortium for improved biochemical production. Nat Commun. 2022;13(1):6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chacόn M, Percival E, Bugg TDH, Dixon N. Engineered co-culture for consolidated production of phenylpropanoids directly from aromatic-rich biomass. Bioresour Technol. 2024;391: 129935. [DOI] [PubMed] [Google Scholar]
- 78.Scott SR, Hasty J. Quorum sensing communication modules for microbial consortia. ACS Synth Biol. 2016;5(9):969–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shong J, Jimenez Diaz MR, Collins CH. Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol. 2012;23(5):798–802. [DOI] [PubMed] [Google Scholar]
- 80.Wu S, Liu J, Liu C, Yang A, Qiao J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell Mol Life Sci. 2019;77(7):1319–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Honjo H, Iwasaki K, Soma Y, Tsuruno K, Hamada H, Hanai T. Synthetic microbial consortium with specific roles designated by genetic circuits for cooperative chemical production. Metab Eng. 2019;55:268–75. [DOI] [PubMed] [Google Scholar]
- 82.Wu S, Xue Y, Yang S, Xu C, Liu C, Liu X, et al. Combinational quorum sensing devices for dynamic control in cross-feeding cocultivation. Metab Eng. 2021;67:186–97. [DOI] [PubMed] [Google Scholar]
- 83.Wegner SA, Barocio-Galindo RM, Avalos JL. The bright frontiers of microbial metabolic optogenetics. Curr Opin Chem Biol. 2022;71: 102207. [DOI] [PubMed] [Google Scholar]
- 84.Morales NM, Patel Michael T, Stewart Cameron J, Sweeney K, McClean MN. Optogenetic tools for control of public goods in Saccharomyces cerevisiae. mSphere. 2021;6(4):e00581-e621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zou X, Pan T, Chen L, Tian Y, Zhang W. Luminescence materials for pH and oxygen sensing in microbial cells-structures, optical properties, and biological applications. Crit Rev Biotechnol. 2017;37(6):723–38. [DOI] [PubMed] [Google Scholar]
- 86.Wang X, Chen L, Liu J, Sun T, Zhang W. Light-driven biosynthesis of myo-inositol directly from CO2 in Synechocystis sp PCC 6803. Front Microbiol. 2020;11:566117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lalwani MA, Kawabe H, Mays RL, Hoffman SM, Avalos JL. Optogenetic control of microbial consortia populations for chemical production. ACS Synth Biol. 2021;10(8):2015–29. [DOI] [PubMed] [Google Scholar]
- 88.Gutiérrez Mena J, Kumar S, Khammash M. Dynamic cybergenetic control of bacterial co-culture composition via optogenetic feedback. Nat Commun. 2022;13(1):4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Y, Li J, Tschirhart T, Terrell JL, Kim E, Tsao CY, et al. Connecting biology to electronics: Molecular communication via redox modality. Adv Healthc Mater. 2017;6(24):1700789. [DOI] [PubMed] [Google Scholar]
- 90.Terrell JL, Tschirhart T, Jahnke JP, Stephens K, Liu Y, Dong H, et al. Bioelectronic control of a microbial community using surface-assembled electrogenetic cells to route signals. Nat Nanotechnol. 2021;16(6):688–97. [DOI] [PubMed] [Google Scholar]
- 91.VanArsdale E, Pitzer J, Wang S, Stephens K, Chen C, Payne GF, et al. Electrogenetic signal transmission and propagation in coculture to guide production of a small molecule, tyrosine. ACS Synth Biol. 2022;11(2):877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.VanArsdale E, Navid A, Chu MJ, Halvorsen TM, Payne GF, Jiao Y, et al. Electrogenetic signaling and information propagation for controlling microbial consortia via programmed lysis. Biotechnol Bioeng. 2023;120(5):1366–81. [DOI] [PubMed] [Google Scholar]
- 93.Tschirhart T, Kim E, McKay R, Ueda H, Wu HC, Pottash AE, et al. Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nat Commun. 2017;8(1):14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Metreveli E, Kachlishvili E, Singer SW, Elisashvili V. Alteration of white-rot basidiomycetes cellulase and xylanase activities in the submerged co-cultivation and optimization of enzyme production by Irpex lacteus and Schizophyllum commune. Bioresour Technol. 2017;241:652–60. [DOI] [PubMed] [Google Scholar]
- 95.Tizazu S, Tesfaye G, Andualem B, Wang A, Guadie A. Evaluating the potential of thermo-alkaliphilic microbial consortia for azo dye biodegradation under anaerobic-aerobic conditions: Optimization and microbial diversity analysis. J Environ Manage. 2022;323: 116235. [DOI] [PubMed] [Google Scholar]
- 96.Ergal İ, Gräf O, Hasibar B, Steiner M, Vukotić S, Bochmann G, et al. Biohydrogen production beyond the Thauer limit by precision design of artificial microbial consortia. Commun Biol. 2020;3(1):443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie Z, Lin W, Luo J. Co-cultivation of microalga and xylanolytic bacterium by a continuous two-step strategy to enhance algal lipid production. Bioresour Technol. 2021;330: 124953. [DOI] [PubMed] [Google Scholar]
- 98.Balasubramanian S, Yu K, Meyer AS, Karana E, Aubin-Tam ME. Bioprinting of regenerative photosynthetic living materials. Adv Funct Mater. 2021;31(31):2011162. [Google Scholar]
- 99.Wang L, Zhang X, Tang C, Li P, Zhu R, Sun J, et al. Engineering consortia by polymeric microbial swarmbots. Nat Commun. 2022;13(1):3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao H, Zhang W, Yu Z, Xin F, Jiang M. Emerging applications of 3D printing in biomanufacturing. Trends Biotechnol. 2021;39(11):1114–6. [DOI] [PubMed] [Google Scholar]
- 101.Sun Z, Wen H, Di Z, Zhang Y, Zhang S, Zhang Z, et al. Photosynthetic living fiber fabrication from algal–bacterial consortia with controlled spatial distribution. ACS Biomater Sci Eng. 2023;9(11):6481–9. [DOI] [PubMed] [Google Scholar]
- 102.Gao H, Li M, Yang L, Jiang Y, Jiang W, Yu Z, et al. Spatial niche construction of a consortium-based consolidated bioprocessing system. Green Chem. 2022;24(20):7941–50. [Google Scholar]
- 103.Shahab RL, Brethauer S, Davey MP, Smith AG, Vignolini S, Luterbacher JS, et al. A heterogeneous microbial consortium producing short-chain fatty acids from lignocellulose. Science. 2020;369:6507. [DOI] [PubMed] [Google Scholar]
- 104.Burmeister A, Grünberger A. Microfluidic cultivation and analysis tools for interaction studies of microbial co-cultures. Curr Opin Biotechnol. 2020;62:106–15. [DOI] [PubMed] [Google Scholar]
- 105.Wondraczek L, Pohnert G, Schacher FH, Köhler A, Gottschaldt M, Schubert US, et al. Artificial microbial arenas: materials for observing and manipulating microbial consortia. Adv Mater. 2019;31(24):1900284. [DOI] [PubMed] [Google Scholar]
- 106.Wei X, Yang W, Choi S. A high power-density, self-sustained hybrid bio-solar cell with co-culture of heterotrophic and photosynthetic bacteria. Solid-State Sensors, Actuators and Microsystems Workshop; June; South Carolina; 2016. 110–4.
- 107.Liu Y, Chen L, Yu L, Zhu J, Wang F, Jiang F, et al. Benefited wastewater utilization via configurable, spatialized, and microorganisms-integrated biophotonic systems. Chem Eng J. 2023;466: 143250. [Google Scholar]
- 108.Nie L, Wu G, Culley DE, Scholten JC, Zhang W. Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit Rev Biotechnol. 2007;27(2):63–75. [DOI] [PubMed] [Google Scholar]
- 109.Zhang W, Li F, Nie L. Integrating multiple “omics” analysis for microbial biology: application and methodologies. Microbiology. 2010;156(2):287–301. [DOI] [PubMed] [Google Scholar]
- 110.Tian X, Chen L, Wang J, Qiao J, Zhang W. Quantitative proteomics reveals dynamic responses of Synechocystis sp PCC 6803 to next-generation biofuel butanol. J Proteomics. 2013;78:326–45. [DOI] [PubMed] [Google Scholar]
- 111.Rienzo M, Jackson SJ, Chao LK, Leaf T, Schmidt TJ, Navidi AH, et al. High-throughput screening for high-efficiency small-molecule biosynthesis. Metab Eng. 2021;63:102–25. [DOI] [PubMed] [Google Scholar]
- 112.Lv M, Wang F, Zeng L, Bi Y, Cui J, Liu L, et al. Identification and metabolomic analysis of a starch-deficient Crypthecodinium cohnii mutant reveals multiple mechanisms relevant to enhanced growth and lipid accumulation. Algal Res. 2020;50: 102001. [Google Scholar]
- 113.Sun T, Li S, Song X, Diao J, Chen L, Zhang W. Toolboxes for cyanobacteria: Recent advances and future direction. Biotechnol Adv. 2018;36(4):1293–307. [DOI] [PubMed] [Google Scholar]
- 114.Li S, Sun T, Xu C, Chen L, Zhang W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab Eng. 2018;48:163–74. [DOI] [PubMed] [Google Scholar]
- 115.Liu H, Cao Y, Guo J, Xu X, Long Q, Song L, et al. Study on the isoprene-producing co-culture system of Synechococcus elongates-Escherichia coli through omics analysis. Microb Cell Fact. 2021;20(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Radivojevic T, Costello Z, Workman K, Garcia MH. A machine learning automated recommendation tool for synthetic biology. Nat Commun. 2020;11(1):4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Long B, Fischer B, Zeng Y, Amerigian Z, Li Q, Bryant H, et al. Machine learning-informed and synthetic biology-enabled semi-continuous algal cultivation to unleash renewable fuel productivity. Nat Commun. 2022;13(1):541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boo AL, Mehta H, Ledesma-Amaro R, Stan GBV. Host-aware RNA-based control of synthetic microbial consortia. Biorxiv. 2023;15:5. [Google Scholar]
- 119.Dragosits M, Mattanovich D. Adaptive laboratory evolution-principles and applications for biotechnology. Microb Cell Fact. 2013;12(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spier MR, Vandenberghe L, Medeiros ABP, Soccol CR. Application of different types of bioreactors in bioprocesses. In: Antolli PG, Liu Z, editors. Bioreactors: Design, Properties and Applications. New York: Nova Science Publishers; 2011. p. 53–87. [Google Scholar]
- 121.Manan MA, Webb C. Modern microbial solid state fermentation technology for future biorefineries for the production of added-value products. Biofuel Res J. 2017;4(4):730–40. [Google Scholar]
- 122.Lu J, Peng W, Lv Y, Jiang Y, Xu B, Zhang W, et al. Application of cell immobilization technology in microbial cocultivation systems for biochemicals production. Ind Eng Chem Res. 2020;59(39):17026–34. [Google Scholar]
- 123.Kim MH, Liang M, He QP, Wang J. A novel bioreactor to study the dynamics of co-culture systems. Biochem Eng J. 2016;107:52–60. [Google Scholar]
- 124.Du P, Zhao H, Zhang H, Wang R, Huang J, Tian Y, et al. De novo design of an intercellular signaling toolbox for multi-channel cell-cell communication and biological computation. Nat Commun. 2020;11(1):4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dinh CV, Chen X, Prather KLJ. Development of a quorum-sensing based circuit for control of coculture population composition in a naringenin production system. ACS Synth Biol. 2020;9(3):590–7. [DOI] [PubMed] [Google Scholar]
- 126.Lindemann SR, Bernstein HC, Song HS, Fredrickson JK, Fields MW, Shou W, et al. Engineering microbial consortia for controllable outputs. ISME J. 2016;10(9):2077–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jia X, He Y, Jiang D, Liu C, Lu W. Construction and analysis of an engineered Escherichia coli-Pseudomonas aeruginosa co-culture consortium for phenanthrene bioremoval. Biochem Eng J. 2019;148:214–23. [Google Scholar]
- 128.Sgobba E, Stumpf AK, Vortmann M, Jagmann N, Krehenbrink M, Dirks-Hofmeister ME, et al. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Bioresour Technol. 2018;260:302–10. [DOI] [PubMed] [Google Scholar]
- 129.Lee EJ, Lee SP. Novel bioconversion of sodium glutamate to γ-amino butyric acid by co-culture of Lactobacillus plantarum K154 in Ceriporia lacerata culture broth. Food Sci Biotechnol. 2014;23(6):1997–2005. [Google Scholar]
- 130.Ariana M, Hamedi J. Enhanced production of nisin by co-culture of Lactococcus lactis sub sp lactis and Yarrowia lipolytica in molasses based medium. J Biotechnol. 2017;256:21–6. [DOI] [PubMed] [Google Scholar]
- 131.Shin D, Byun WS, Moon K, Kwon Y, Bae M, Um S, et al. Coculture of marine Streptomyces sp with Bacillus sp produces a new piperazic acid-bearing cyclic peptide. Front Chem. 2018;6:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao GR, Wei SY, Ding MZ, Hou ZJ, Wang DJ, Xu QM, et al. Enhancing fengycin production in the co-culture of Bacillus subtilis and Corynebacterium glutamicum by engineering proline transporter. Bioresour Technol. 2023;383: 129229. [DOI] [PubMed] [Google Scholar]
- 133.Abdullah R, Naeem N, Aftab M, Kaleem A, Iqtedar M, Iftikhar T, et al. Enhanced production of alpha amylase by exploiting novel bacterial co-culture technique employing solid state fermentation. Iran J Sci Technol Trans A-Sci. 2016;42(2):305–12. [Google Scholar]
- 134.Winasti NMS, Yulyanita DA, Naser AS, Suyono EA. Enhacment of biomass, carbohydrates, lipids, and proteins content using co-culture of Glagah consortium and Lipomyces starkeyi. J Appl Nat Sci. 2023;15(1):15–22. [Google Scholar]
- 135.Liu L, Chen J, Lim PE, Wei D. Dual-species cultivation of microalgae and yeast for enhanced biomass and microbial lipid production. J Appl Phycol. 2018;30(6):2997–3007. [Google Scholar]
- 136.Gong G, Wu B, Liu L, Li J, He M, Hu G. Enhanced biomass and lipid production by light exposure with mixed culture of Rhodotorula glutinis and Chlorella vulgaris using acetate as sole carbon source. Bioresour Technol. 2022;364: 128139. [DOI] [PubMed] [Google Scholar]
- 137.Pachapur VL, Sarma SJ, Brar SK, Le Bihan Y, Buelna G, Verma M. Biohydrogen production by co-fermentation of crude glycerol and apple pomace hydrolysate using co-culture of Enterobacter aerogenes and Clostridium butyricum. Bioresour Technol. 2015;193:297–306. [DOI] [PubMed] [Google Scholar]
- 138.Saini M, Chiang CJ, Li SY, Chao YP. Production of biobutanol from cellulose hydrolysate by the Escherichia coli co-culture system. FEMS Microbiol Lett. 2016. 10.1093/femsle/fnw008. [DOI] [PubMed] [Google Scholar]
- 139.Tomita H, Okazaki F, Tamaru Y. Biomethane production from sugar beet pulp under cocultivation with Clostridium cellulovorans and methanogens. AMB Express. 2019;9(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Su Y, Zhang W, Zhang A, Shao W. Biorefinery: the production of isobutanol from biomass feedstocks. Appl Sci. 2020;10(22):8222. [Google Scholar]
- 141.Rahayuningsih M, Febrianti F, Syamsu K. Enhancement of bioethanol production from tofu waste by engineered simultaneous saccharification and fermentation (SSF) using co-culture of mold and yeast. IOP Conference Series: Earth and Environmental Science; July; Indonesia: IOP Publishing; 2022. 012004.
- 142.Liu F, Wu W, Tran-Gyamfi MB, Jaryenneh JD, Zhuang X, Davis RW. Bioconversion of distillers’ grains hydrolysates to advanced biofuels by an Escherichia coli co-culture. Microb Cell Fact. 2017;16(1):192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cai M, Liu J, Song X, Qi H, Li Y, Wu Z, et al. De novo biosynthesis of p-coumaric acid and caffeic acid from carboxymethyl-cellulose by microbial co-culture strategy. Microb Cell Fact. 2022;21(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang X, Zhao C, Lu X, Zong H, Zhuge B. Development of a co-culture system for green production of caffeic acid from sugarcane bagasse hydrolysate. Front Microbiol. 2024;15:1379688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao R, Sengupta A, Tan AX, Whelan R, Pinkerton T, Menasalvas J, et al. Photobiological production of high-value pigments via compartmentalized co-cultures using Ca-alginate hydrogels. Sci Rep. 2022;12(1):22163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fedeson DT, Saake P, Calero P, Nikel PI, Ducat DC. Biotransformation of 2, 4-dinitrotoluene in a phototrophic co-culture of engineered Synechococcus elongatus and Pseudomonas putida. Microb Biotechnol. 2020;13(4):997–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen XY, Sun HZ, Qiao B, Miao CH, Hou ZJ, Xu SJ, et al. Improved the lipopeptide production of Bacillus amyloliquefaciens HM618 under co-culture with the recombinant Corynebacterium glutamicum producing high-level proline. Bioresour Technol. 2022;349: 126863. [DOI] [PubMed] [Google Scholar]
- 148.Valdez-Vazquez I, Pérez-Rangel M, Tapia A, Buitrón G, Molina C, Hernández G, et al. Hydrogen and butanol production from native wheat straw by synthetic microbial consortia integrated by species of Enterococcus and Clostridium. Fuel. 2015;159:214–22. [Google Scholar]
- 149.Li X, He Y, Zhang L, Xu Z, Ben H, Gaffrey MJ, et al. Discovery of potential pathways for biological conversion of poplar wood into lipids by co-fermentation of Rhodococci strains. Biotechnol Biofuels. 2019;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun HZ, Chen XY, Zhang YM, Qiao B, Xu QM, Cheng JS, et al. Construction of multi-strain microbial consortia producing amylase, serine and proline for enhanced bioconversion of food waste into lipopeptides. Biochem Eng J. 2022;188: 108682. [Google Scholar]
- 151.Lu J, Jiang W, Dong W, Zhou J, Zhang W, Jiang Y, et al. Construction of a microbial consortium for the de novo synthesis of butyl butyrate from renewable resources. J Agric Food Chem. 2023;71(7):3350–61. [DOI] [PubMed] [Google Scholar]
- 152.Zhu H, Xu L, Luan G, Zhan T, Kang Z, Li C, et al. A miniaturized bionic ocean-battery mimicking the structure of marine microbial ecosystems. Nat Commun. 2022;13(1):5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miao C, Wang X, Qiao B, Xu Q, Cao C, Cheng J. Artificial consortia of Bacillus amyloliquefaciens HM618 and Bacillus subtilis for utilizing food waste to synthetize iturin A. Environ Sci Pollut Res. 2022;29(48):72628–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.