Abstract
Background
Enterohemorrhagic Escherichia coli (EHEC), a group of enteric pathogenic bacteria that is a major cause of human diarrheal disease, must interact with the diverse intestinal microbiome during colonization and subsequently overcome the environmental challenges to survive and cause disease. While this relationship, and how the microbiome modulates infection of EHEC, has been studied, it is less understood how the microbiome is impacted during treatment for an EHEC infection. One area that is notably lacking in knowledge is how vaccination can impact the intestinal microbiome composition, and therefore, influence vaccine efficacy. We previously developed vaccine formulations consisting of gold nanoparticles (AuNPs) conjugated to various EHEC antigens and tested them in mice models using both EHEC and its murine counterpart Citrobacter rodentium. The goal of this study was to evaluate the relationship between these EHEC vaccines and their effects on the gut microbiome.
Results
We found that immunization with the vaccines or adjuvant-only control did not lead to major alterations in the composition of the fecal microbiome; however, there were measurable variations in individual mice within the same vaccine group housed in separate cages. Also, immunization with one vaccine (AuNP-EscC) prevented both a decrease in the diversity of the fecal microbiome and an increase in detectable C. rodentium following infection compared to control animals.
Conclusions
Overall, our small study argues in favor of evaluating the intestinal microbiome during vaccine development not just for EHEC, but for other enteric pathogens.
Keywords: EHEC, Citrobacter rodentium, Nanovaccines, Microbiome, Gold nanoparticles
Background
The gut microbiota in mammals profoundly influences metabolic and nutritional processes, as well as the host’s immune responses [1]. There is increasing evidence that links alterations in the microbiota with human diseases, from gastrointestinal to neurological pathologies. Escherichia coli is a commensal, non-pathogenic bacteria in human and other mammals’ intestinal microbiota; however, some pathogroups, such as enterohemorrhagic E. coli (EHEC) can cause intestinal disease [2]. The interplay between EHEC, a human diarrheal illness-causing bacteria, and the intestinal microbiota during infection is an area of active investigation. It has been well reported that to coordinate the expression of its virulence factors, EHEC must interact directly with the intestinal environment and its residents, as well as recognize various microbiota-derived molecules, such as short chain fatty acids (SCFAs) [3, 4]. For example, butyrate, which is a SCFA produced in high concentrations in the large intestine primarily by Firmicutes, can increase expression of genes involved in EHEC adherence to intestinal epithelial cells [4, 5]. This attachment is mediated by enhanced expression of a specialized Type 3 Secretion System (T3SS), a virulence mechanism encoded by a pathogenicity island known as the Locus of Enterocyte Effacement (LEE) [2]. Additionally, certain strains of EHEC are known to produce Shiga toxins (Stx) [2]. Stx acts by inhibiting protein synthesis and inducing apoptosis of endothelial cells [6]. These effects can be local in the intestines or in distant organs such as the kidneys upon bloodstream dissemination. A high-fiber diet approach in mice was demonstrated to increase lethality and disease severity by EHEC following oral infection [7]. This diet not only increased intestinal butyrate and the expression of the Stx cellular binding target Gb3, which increased Stx-coordinated disease outcomes, but it also led to a reduction in commensal E. coli, providing a more favorable niche for EHEC colonization.
While the microbiome is known to promote pathogenesis of EHEC, there is also evidence that it is involved in resistance to infection. These mechanisms can be directly, either through nutrient competition or production of SCFAs or other molecules, or indirectly by modulating the intestinal immune response [4]. Conventional mice are typically resistant to EHEC infection and are only transiently colonized with minimal to no symptoms, despite evidence that the pathogen can adhere to mouse intestinal epithelial cells [8, 9]. This resistance is in part attributed to the composition of the microbiome [9]. Alteration of the mouse gut microbiome, either through antibiotic (streptomycin) treatment or with germ-free mouse strains, increases the susceptibility of mice to EHEC [9]. Additionally, the murine pathogen Citrobacter rodentium, which is an LEE-encoding bacteria that has been used extensively as a surrogate model of infection to study EHEC-related pathogenesis, can disrupt the microbiome and cause dysbiosis to enhance its own colonization by mechanisms such as increasing oxygenation at the intestinal epithelial cell (IEC) interface [10]. These changes result in increased colonization of facultative anaerobes (like itself and other Enterobacteriaceae) and a reduction of commensal obligate anaerobes, further proving the role of commensal organisms in protection against invading pathogens [10].
Considering the ability of the microbiome to both promote and prevent EHEC colonization, which has also been demonstrated with other enteric pathogens [3, 4], prophylactic and post-exposure modulation of the intestinal micro-environment have been explored as potential therapies to limit infections. Examples of these methods include the administration of pre- and pro-biotics and fecal microbiome transplantation [11]. One area that has garnered less attention is the effects that vaccines have on the microbiome, and vice versa. Several studies, both in animals and humans, have indicated that the composition of the microbiome can impact vaccine immunogenicity, mostly due to its role in modulating immune responses [12]. Fewer investigations have examined vaccine-induced changes in the microbiome and if these can influence efficacy. Studies in humans have largely focused on microbiome alterations following vaccinations against HIV-1 and SARS-CoV-2, while those in animals have included Mycobacterium tuberculosis and Lawsonia interacellularis [13]. Only one group has explored this area in the context of E. coli vaccinations, and they found that intranasally immunizing mice with three enterotoxigenic E. coli (ETEC) antigens, which are also conserved in some commensal bacteria, did not significantly alter the fecal microbiome composition [14].
Although EHEC poses a significant healthcare burden as a major cause of diarrheal disease in developed countries and South America, as well as being a leading cause of hemolytic-uremic children (HUS) in children due to Stx, there is currently no human vaccine available to protect against EHEC [15]. Previously, our lab developed novel vaccines using gold nanoparticles (AuNPs) conjugated to EHEC antigens and tested them in mice using both EHEC and C. rodentium (including a strain expressing Stx2d toxin) as infection models [16–19]. In these studies, we concluded that some vaccine formulations (AuNP-EscC) could successfully limit colonization of non-Stx2d-producing C. rodentium [19]. Therefore, the purpose of the current study was to determine if these AuNP vaccines induced changes to the murine intestinal microbiome and if AuNP-EscC could affect infection-induced microbiome alterations compared to non-immunized animals.
Methods
Bacterial strains and growth conditions
The non-Stx2d-producing Citrobacter rodentium strain used in this study (ATCC DBS771) was purchased from the American Type Culture Collection (ATCC). It was routinely grown in Luria-Bertani (LB) broth, supplemented with antibiotics for selection (chloramphenicol and kanamycin). For animal infections, a final concentration of 106 CFU per dose was prepared in PBS, as previously described [19].
Vaccine formulation
The cloning and purification of the EHEC antigens EscC and Eae (intimin gamma protein) were performed as described [19, 20]. The synthesis of AuNPs was performed using the Turkevich method [19, 21]. Gold-nanoparticles with the correct size and structure were conjugated with the proteins and finally resuspended in PBS for animal use [19].
Animal studies
Female 5-to-7-week-old C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA), housed in microisolator cages under pathogen-free conditions and maintained on a 12 h light cycle, with food and water available ad libitum. All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of The University of Texas Medical Branch (Protocol #2112077). Mice were immunized subcutaneously (s.c.) three times at 2-week intervals with either AuNP-EscC, AuNP-Eae, or AuNP-EscC + AuNP-Eae (n = 6 per group), along with detoxified cholera toxin B subunit (Sigma, Cream Ridge, NJ, USA) and 2% Alhydrogel® (InvivoGen, San Diego, CA, USA) as adjuvants [19]. Control mice (n = 6) were given unconjugated AuNPs along with the same concentration of both adjuvants. For vaccine efficacy assessment and microbiome evaluation following infection, n = 6 mice from AuNP-EscC or the control group were infected with 106 CFU of C. rodentium DBS771 using a feeding method of infection 3 weeks following the last immunization dose and kept for 21 days, as previously described [19].
Fecal collection and processing
Fecal samples were collected from mice in all groups before vaccine administration (pre) and 2 weeks following the last immunization dose (post). Feces were also collected from AuNP-EscC-immunized and control mice at 3- and 7-days post-infection (dpi). Feces were frozen at -80˚C until processing. To extract microbial DNA, feces were processed using the QIAGEN QIAamp PowerFecal Pro DNA kit according to manufacturer instructions. Purified DNA samples were stored at -20˚C until further use.
Transcriptomic analysis
High-throughput sequencing of the bacterial 16S ribosomal RNA gene was performed using gDNA isolated from each sample. Sequencing libraries for each isolate were generated using universal 16S rRNA V3-V4 region primers [22] by Illumina 16S rRNA metagenomic sequencing library protocols. The samples were barcoded for multiplexing using Nextera XT Index Kit v2. Sequencing was performed on an Illumina MiSeq instrument using a MiSeq Reagent Kit v2 (500-cycles). To identify the presence of known bacteria, sequences were analyzed using the CLC Genomics Workbench 8.0.1 Microbial Genomics Module1. Reads containing nucleotides below the quality threshold of 0.05 (using the modified Richard Mott algorithm) and those with two or more unknown nucleotides or sequencing adapters were trimmed out. All reads were trimmed to 264 bases for subsequent operational taxonomic unit (OTU) classification. Reference-based OTU picking was performed using the SILVA SSU v119 database [23]. Sequences present in more than one copy but not clustered to the database were placed into de novo OTUs (97% similarity) and aligned against the reference database with 80% similarity threshold to assign the “closest” taxonomical name where possible. Chimeras were removed from the dataset if the absolute crossover cost was three using a k-mer size of six. Microbiome Analyst 2.0 (https://www.microbiomeanalyst.ca/) was used for visualization and to perform the statistical analysis of the microbiome data [24, 25]. Alpha diversity was measured using Shannon and Chao1 entropy (OTU level). Beta diversity was calculated using the Bray-Curtis diversity measure (OTU level). The significance was evaluated by PERMANOVA, ANOVA, t-test, or Pearson’s correlation tests, as specified in each case.
Results
AuNP-protein immunization and adjuvant-only treatment led to some differences in the composition of the fecal microbiome of mice, with only minimal changes in the overall diversity
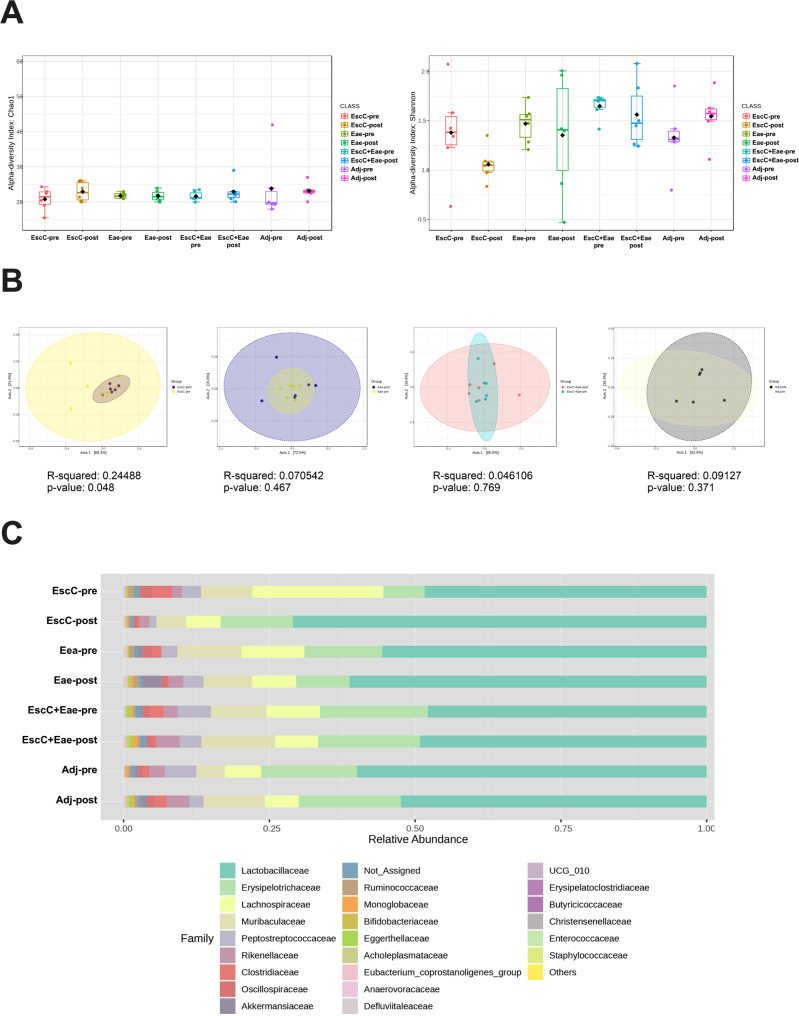
Although EHEC is pathogenic to humans, there are commensal strains of E. coli that reside as normal members of the microbiota. To guarantee that vaccines formulated against EHEC do not affect these commensals, antigens that are not shared between pathogenic and commensal strains are preferable. Both proteins used for this vaccine study – EscC and Eae – are encoded by the LEE [26], which is only known to be present in pathogenic strains. However, we confirmed that the AuNP-proteins and the adjuvants did not alter the mouse microbiome composition. For this study, C57BL/6 mice were subcutaneously immunized with AuNPs conjugated to either EscC, Eae, or a combination of the antigens, along with the adjuvants. Mice immunized with unconjugated AuNPs mixed with adjuvants were used as controls. The full regimen used, and the antigen selection, were described in our previous publication [19]. Feces were collected before immunization and 2 weeks following the last dose. Fecal DNA was used for 16S rRNA gene sequencing and analysis. Chao1 and Shannon indices revealed there were no significant changes in alpha diversity within any of the vaccine groups before (pre) and after vaccination (post). However, there was a downward trend in the Shannon diversity index in AuNP-EscC immunized mice (Fig. 1A). The beta diversity within each group pre- and post-vaccination did not vary significantly, except in the AuNP-EscC vaccinated animals (p = 0.048) (Fig. 1B).
Fig. 1.
Alterations in mice fecal microbiome before and after immunization with either AuNP-proteins or AuNP-unconjugated. (A) Alpha-diversity of bacterial transcripts at the family level -pre and after -post immunization with AuNP-proteins or AuNP + adjuvant-only with Chao1 and Shannon indices. No significant differences were found by Weltch T-test/ANOVA analysis. (B) Beta-diversity of bacterial transcripts using the Bray-Curtis dissimilarity index and PERMANOVA statistical method. Each graph is a comparison of the -pre and -post diversity at the family level within each treatment group. From left to right: EscC, Eae, EscC + Eae, and adjuvant-only. (C) Relative abundances at the family level from -pre and -post treatment groups. -pre, baseline samples before immunization; -post, post-immunization sample
Taxonomic analysis of each vaccine group showed some differences in the relative abundances of various bacterial families, including in the adjuvant-only group (Fig. 1C). A list of the families that were significantly differentially abundant pre- and post-immunization are listed in Table 1, along with the associated p-value and if the family was increased (I) or decreased (D) following immunization. Lactobacillaceae was the most abundant family in all animal groups, with a noticeable, but not statistically significant, increase seen after AuNP-EscC immunization (pre: 53.4%; post: 71.5%) (Fig. 1C). This change was associated with a significant decrease in Lachnospiraceae species (Fig. 1C; Table 1). Furthermore, the relative abundance of Clostridiaceae was significantly reduced following immunization with each antigen alone [EscC (pre: 3.4%; post: 0.3%); Eae (pre: 1.7%; post: 0.1%)] and EscC + Eae together (pre: 2.4%; post: 0.5%), with this being the only family differentially abundant in the combo group (Fig. 1C; Table 1). Animals receiving the adjuvant-only mostly had an increase in the abundance of certain families, except for Peptostreptococaceae, which decreased (pre: 5.49%; post: 2.7%) (Fig. 1C; Table 1).
Table 1.
Significant differentially abundant families pre- and post-immunization
| Group | Family | p-value | I or D From Pre-Vaccination |
|---|---|---|---|
| EscC | Bifidobacteriaceae | 0.015152 | D |
| Clostridiaceae | 0.004329 | D | |
| Eubacterium_coprostanoligenes_group | 0.025974 | I | |
| Lachnospiraceae | 0.025974 | D | |
| Oscillospiraceae | 0.008658 | D | |
| Eae | Butyricicoccaceae | 0.015152 | D |
| Clostridiaceae | 0.002165 | D | |
| Eggerthellaceae | 0.041126 | I | |
| Erysipelatoclostridiaceae | 0.002165 | I | |
| Monoglobaceae | 0.004329 | I | |
| Rikenellaceae | 0.002165 | I | |
| EscC + Eae | Clostridiaceae | 0.008658 | D |
| Adj | Acholeplasmataceae | 0.025974 | I |
| Anaerovoracaceae | 0.015152 | I | |
| Bifidobacteriaceae | 0.015152 | I | |
| Defluviitaleaceae | 0.062771 | I | |
| Eggerthellaceae | 0.008658 | I | |
| Muribaculaceae | 0.041126 | I | |
| Peptostreptococcaceae | 0.041126 | D |
I, increased in post-immunization compared with pre-vaccination; D, decreased in post-immunization compared with pre-vaccination. Significance was evaluated with t-test
AuNP-EscC immunized mice had no significant cage-to-cage differences in microbiota composition
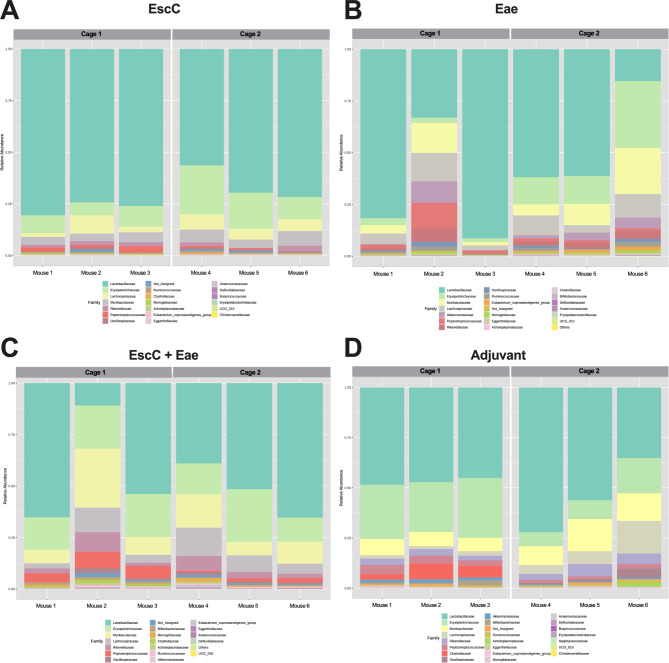
During our vaccine study, AuNP-protein and adjuvant-only treated animals were subsequently infected with 106 CFU of C. rodentium The infection resulted in cage-to-cage variations in pathogen shedding within the same group, i.e., all mice in one cage were highly colonized while mice in the other cage shed much lower concentrations, as we have previously shown [19]. This outcome was not completely surprising, considering it is well-known that C57BL/6 mice display inconsistent colonization by C. rodentium [27]. However, due to this observed phenomenon, we sought to examine the potential differences in the microbiome before infection in immunized animals within the same group but housed in different cages. It is important to note that AuNP-EscC immunized mice consistently shed C. rodentium at lower concentrations than the adjuvant-only controls and had significantly less organ burden at the end of the study [19]. Both AuNP-Eae immunized mice cages shed similar levels while mice in Cage 1 in both the AuNP-EscC + Eae immunized and adjuvant-only treated groups consistently shed much lower concentrations of C. rodentium than Cage 2 [19]. Interestingly, following vaccination with AuNP-EscC, only the relative abundance of Defluviitaleaceae was significantly different between Cage 1 and Cage 2 (Fig. 2A), whereas there were multiple differences between the two cages in the other groups, such as Clostridiaceae or Peptostreptococcaceae (Fig. 2B-D). The average relative abundances for the significantly differentially abundant families for each group and the corresponding p-values are given in Table 2. Overall, this data indicates that mice within the same treatment group can have differences in their microbiota composition, which appeared to be, in our conditions, cage dependent. We then evaluated changes in the fecal microbiome following infection.
Fig. 2.
Fecal microbiome compositions of individual mice treated with AuNP vaccine formulations. Relative abundances at the family level in (A) AuNP-EscC, (B) AuNP-Eae, (C) EscC + Eae, and (D) adjuvant-only vaccinated animals. Each chart displays a comparison between Cage 1 and Cage 2 (each housing 3 mice) within each treatment group
Table 2.
Significant differentially abundant families post-immunization
| Average Relative Abundance (%) | ||||
|---|---|---|---|---|
| Group | Family | Cage 1 | Cage 2 | p-value |
| EscC | Defluviitaleaceae | 0.01 | 0.03 | 0.03351 |
| Eae | Clostridiaceae | 0.02 | 0.16 | 0.007693 |
| Eubacterium_coprostanoligenes_group | 0.15 | 1.02 | 0.027702 | |
| EscC + Eae | Bifidobacteriaceae | 1.03 | 0.30 | 0.010424 |
| Clostridiaceae | 1.00 | 0.04 | 0.018588 | |
| Peptostreptococcaceae | 6.18 | 1.50 | 0.013972 | |
| Adj | Akkermansiaceae | 1.43 | 0.69 | 0.012242 |
| Anaerovoracaceae | 0.05 | 0.12 | 0.018279 | |
| Butyricicoccaceae | 0.00 | 0.02 | 0.02549 | |
| Clostridiaceae | 5.33 | 0.03 | 0.023607 | |
| Erysipelotrichaceae | 27.23 | 11.31 | 0.010853 | |
| Muribaculaceae | 7.22 | 13.04 | 0.042468 | |
| Peptostreptococcaceae | 3.79 | 1.65 | 0.023874 | |
Comparison of the relative abundances of families between both cages in which animals in the same group were housed. Significance was evaluated with t-test
AuNP-EscC immunization protected mice from a reduction in diversity and colonization by C. rodentium
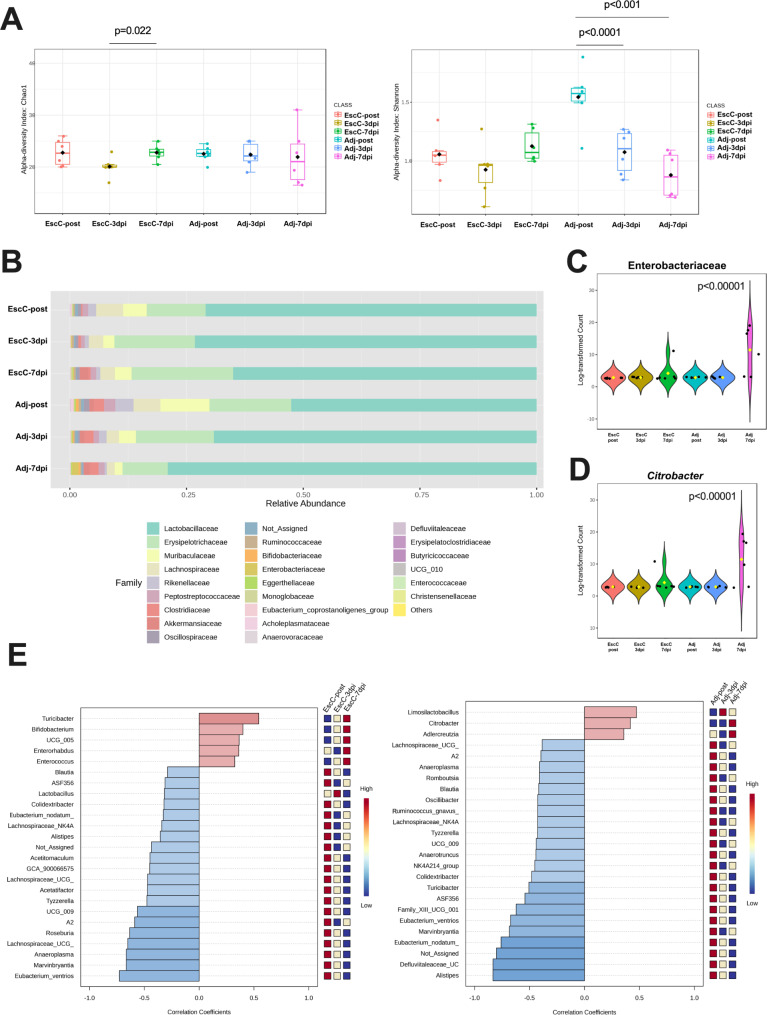
As described above, mice immunized with AuNP-EscC were less colonized by C. rodentium and had significantly less organ burden at 14 days post-infection (dpi) compared to the adjuvant-only treated animals. Because C. rodentium causes intestinal dysbiosis to aid in its own colonization [10], we wanted to determine if the vaccine could prevent intestinal microbiota imbalance. The fecal microbiome was analyzed in AuNP-EscC immunized and adjuvant-only treated animals before infection (post-treatment) and at 3dpi and 7dpi with 106 CFU C. rodentium. Although both Chao1 and Shannon indices showed changes in the alpha diversity in AuNP-EscC immunized animals following infection, this was only significant between 3dpi and 7dpi (Chao 1 [p = 0.022] and Shannon index [p = 0.09]) (Fig. 3A). These changes were not significant when comparing EscC-post to either dpi. There was a significant reduction in alpha diversity throughout the infection in adjuvant-only treated animals according to the Shannon index (Adj-post versus Adj-3dpi p < 0.0001; Adj-post versus Adj-7dpi p < 0.001) (Fig. 3A). Furthermore, both “-post” groups and “-7dpi” differed significantly in the Shannon index as well (EscC-post versus Adj-post p = 0.0036; EscC-7dpi versus Adj-7dpi p = 0.0317). This was also reflected in the taxonomic analyses. The AuNP-EscC vaccinated animals had few changes in the relative abundances of identified families, with only a noticeable increase in Akkermansiaceae after the infection (from 0.008% in EscC-post, to 0.078% in EscC-3dpi, and up to 1.9% in EscC-7dpi), in Bifidobacteriaceae (from 0.007% in EscC-post, to 0.11% in EscC-3dpi, and 0.15% in EscC-7dpi), and in Erysipelotrichaceae (from 12.62% in EscC-post, to 17.25% in EscC-3 dpi, and to 21.76% in EscC-7dpi). Adjuvant-only treated animals had a bloom of Lactobacillaceae (52.58% in Adj-post, 69.15% in Adj-3dpi, and 79% in Adj-7dpi) and concurrent reductions of other families such as Erysipelotrichaceae (17.53% in Adj-post, 16.68% in Adj-3dpi, and 9.69% in Adj-7dpi), Muribaculaceae (10.5% in Adj-post, 3.64% in Adj-3dpi, and 1.65% in Adj-7dpi), and Lachnospiraceae (5.75% in Adj-post, 2.65% in Adj-3dpi, and 1.70% in Adj-7dpi) throughout the infectious process (Fig. 3B). Lachnospiraceae was also reduced in the vaccinated group after the infection (5.8% in EscC-post, 3.09% in EscC-3dpi, and 3.14% in Adj-7dpi). There was an increase in Enterobacteriaceae in adjuvant-only treated animals (Fig. 3C), as well as a measured increase in C. rodentium (Fig. 3D), with statistical significance between Adj-pre and Adj-7dpi in both cases. We further explored these differences using the pattern search analysis in the correlation network of the top 25 genera associated with each group before and after infection. Following infection, both AuNP-EscC immunized and adjuvant-only treated animals were associated with increases and decreases of certain genera (Fig. 3E). For example, Turicibacter increased with time in the vaccinated group, while it decreased over the infection course in the control animals. Others like Alistipes, Blautia, and members of the Lachnospiraceae family exhibited a reduction over time in both treatment groups (Fig. 3E). Most importantly, however, an increase in Citrobacter was only associated with adjuvant-only treated animals, confirming the reduction in colonization by the pathogen in the immunized mice (Fig. 3D-E).
Fig. 3.
Fecal microbiome composition in immunized or control mice before and following challenge with C. rodentium. (A) Alpha-diversity of bacterial transcripts at the family level from -post, -3dpi, and -7dpi of AuNP-EscC and adjuvant-only vaccinated groups with Chao1 and Shannon index. (B) Relative abundances of bacterial families from -post, -3dpi, and -7dpi of AuNP-EscC and adjuvant-only groups. (C) The relative abundance of the Enterobacteriaceae family in each treatment group before and after infection. Significance of p < 0.00001 evaluated between Adj-7dpi and all other groups through Pearson’s correlation. (D) The relative abundance of the Citrobacter genus in each treatment group before and after infection. Significance of p < 0.00001 evaluated between Adj-7dpi and all other groups through Pearson’s correlation. (E) The top 25 bacterial genera exhibiting the most significant differences throughout the infection timeline, in AuNP-EscC vaccinated (left) and adjuvant-only treated animals (right). -post, post-immunization fecal sample; -3dpi, fecal samples from 3 days after infection; -7dpi, fecal samples from 7 days after infection
Discussion
Pathogenic E. coli remains an important cause of human food-borne diarrheal disease, with some strains able to cause more severe sequalae like HUS or even death in susceptible populations (reviewed in [28]). In cases in which antimicrobial treatment is available because of a sensitivity profile of the bacterium, it is contraindicated when EHEC are the source of the infection since antibiotics are known to increase the production of the Stx [29–31]. Development of a prophylactic vaccine for human use to protect against EHEC is associated with a myriad of challenges. These include the lack of reliable animal models for testing [32] and the antigen similarity between pathogenic and commensal strains, which limits antigenic options. There is also a gap in knowledge regarding how these vaccines impact the gut microbiota, despite its known role in modulating EHEC pathogenesis. A further limitation is age-related fluctuations in the microbiome, which has been proven to affect susceptibility of the host to enteric infections, and how this could affect long-term vaccine studies [33].
Previously, our lab designed a vaccination protocol to test the efficacy of a AuNPs coupled with different EHEC antigenic proteins that are not present in commensal E. coli. Although we demonstrated varying levels of protection with these formulations after animal immunization and challenges [16–19], all combinations and routes tested elicited a strong humoral response. Therefore, we questioned if the individual and collective microbiomes of the different groups in our most recent vaccine trial had a significant impact on infection clearance.
The alpha diversity of the animal groups both before and after vaccination showed no significant differences, indicating that the richness (Chao1) and the evenness (Shannon) of the groups are comparable (Fig. 1A) This is similar to what has been demonstrated by other researchers [14]. However, the beta diversity, used as a measurement of the similarity between groups, showed significant differences only in the AuNP-EscC vaccinated group (Fig. 1B). Based on this data, we can conclude that there are no differences attributed to the injection route or adjuvants used but only to the antigen, making the chosen vaccination regimen appropriate in those terms. Additionally, all vaccinated animals exhibited a significant reduction in the abundance of Clostridiaceae after the vaccination, which was not observed in the control (adjuvant-only) group (Table 1). A higher presence of Clostridium spp. is commonly associated with adverse effects on normal gastrointestinal (GI) activity [34] and linked with proinflammatory cytokines [35]. However, it has been shown that its prevalence differs from tissue and feces [35].
During our previous study, we tested our EHEC vaccines against both high (109) and low (106) infectious doses of C. rodentium [19]. Following the low dose, we observed cage-to-cage variation in the shedding and colonization of C. rodentium, even in mice within the same treatment group. However, animals in the same group infected with the high dose shed comparable concentrations of bacteria, regardless of cage. As a result, we hypothesize that the number of bacteria that colonized the GI tract was too elevated to be outcompeted by the commensal microorganisms. As described earlier, natural C. rodentium infection leads to inconsistent disease kinetics and outcomes in C57BL/6 mice, which could explain the pronounced colonization differences seen in the low dose infected animals. Additionally, it has been reported that the intestinal microbiome contributes to the susceptibility of mice to this pathogen infection [36]. Therefore, we compared the individual mouse’s fecal microbiome within each group prior to infection with the low dose to help determine if differences in the commensal composition could explain the variability in C. rodentium colonization, considering all vaccine formulations elicited similar antibody responses in all immunized animals. The animals vaccinated with EscC and Eae antigens alone exhibited similar shedding and organ burden among animals in the same group [19] and were also the groups that exhibited fewer microbiome differences between both cages (Fig. 2A-B; Table 2). In the control groups and AuNP-EscC+Eae vaccinated animals, cages 1 exhibited lower bacterial burden than cages 2, and in both cases, they shared a higher abundance of families Clostridiaceae and Peptostreptococcaceae (Fig. 2C-D; Table 2). Some species of Clostridium have been reported as potential probiotics that can control the gut inflammatory response, so they might be able to have preventive effects on EHEC infections [28]. Also, Peptostreptococcaceae is usually more abundant in healthy individuals’ gut and helps in maintaining homeostasis [37]. Overall, our data indicates that variations in the gut microbiota from one mouse to the next could have implications in vaccine effectiveness and should be explored further when testing vaccines for enteric pathogens. Limitations to this approach, however, include the coprophagy behavior of mice, which has been demonstrated to horizontally transmit the intestinal microbiota from one mouse to another during cohousing [38]. It is unclear if this contributed to the cage-to-cage variations in both C. rodentium colonization and microbiome composition seen in our studies, but it could be explored in future vaccine experiments. Interestingly, the intestinal microbiome has been extensively proven to be variable among individuals, and factors such as age, diet, and other environmental factors, including geographical location, are known to effect the composition [39]. It remains to be seen how these variations could affect future vaccine trials in humans.
As previously mentioned, the AuNP-EscC immunized mice showed the lowest burden following infection [19] and was also the only group that exhibited greater differences in the microbiome after vaccination (Fig. 1B). These differences, however, were more homogeneous among the animals (Fig. 2A). Because of the significant reduction in colonization in AuNP-EscC immunized mice compared to the control group that was not seen in the other vaccine groups, we only compared the microbiome between those groups before (-post immunization) and after (-3dpi, -7dpi) infection. There was a clear and significant difference in bacterial richness in the fecal microbiota after infection in both groups, but this was more noteworthy in the control rather than the vaccinated group (Fig. 3A). There was also a difference in the evenness, probably due to the high concentrations of Citrobacter (Fig. 3D). Specifically, differences in the abundance of families were seen between the AuNP-EscC vaccinated and the control animals (Fig. 3B). AuNP-EscC immunized animals exhibited an increase of Akkermansiaceae and Bifidobacteriaceae relative abundances after the infection. However, the adjuvant group showed only an increase in Lactobacillaceae with reductions in Maribaculaceae and Lachnospiraceae. Some in vivo studies have shown a protective effect of probiotics in EHEC-challenged animals, which they linked with higher levels of Akkermansiaceae and Lachnospiraceae in the treated animals [40]. In our study, when we analyzed the trends at the genus level, there was a clear increase of Bifidobacterium in AuNP-EscC vaccinated animals after the infection, which has been reported to inhibit the growth of EHEC and reduce the production of Stx in animal models [28]. Nevertheless, the microbiome is a complex community, and our evidence suggests that it should be considered during vaccination due to its proven roles in protection against enteric infection.
Given the observed heterogeneity in microbiome composition and its potential influence on vaccine efficacy, subsequent research should focus on further elucidating the interaction between vaccination, microbiome diversity, and immunological responses across varying host demographics and environmental conditions. Such efforts could ultimately support the development of more universally effective vaccines and enhance our understanding of microbiome-mediated immunological protection.
Conclusion
With this work, we have evaluated the gut microbiota modifications in animals that were vaccinated with AuNPs conjugated to EHEC antigens and then subsequently challenged with the mouse pathogen C. rodentium. Although there are limitations to this work, in terms of the number of animals used and that only the fecal microbiota was analyzed, it can pave the way for future studies and does highlight the importance of commensal microbiota in resisting colonization of bacterial pathogens. Our study reinforces the complex role that the gut microbiota plays in modulating the immune response to pathogens and suggests its critical impact on the effectiveness of vaccines targeting enteric infections such as those caused by EHEC. However, as our lab and others have already demonstrated, robust antibody production is not always a direct indication of EHEC protection. While our AuNP-EscC-based vaccine demonstrated promising protective effects, particularly in reducing bacterial colonization and influencing microbiota composition, variability among individuals points to the microbiome as a significant factor in vaccine response. This finding underlines the necessity of incorporating microbiome analysis into vaccine trials, especially since the gut microbiota has shown potential for either promoting or inhibiting colonization resistance and influencing immune responses. Future work is focused on optimizing the vaccines to maximize their efficacy against enteric pathogens and understanding how microbiome maintenance and alterations can apply to humans in protection against EHEC.
Acknowledgements
The authors would like to thank the UTMB Microbiome and Bioinformatics Analysis core for their contribution in 16S rRNA sequencing of extracted DNA samples.
Author contributions
Conceptualization, S.B., I.C.-G. and A.G.T.; methodology, S.B., I.C.-G.; validation, S.B. and I.C.-G.; formal analysis, S.B. and I.C.-G.; investigation, S.B. and I.C.-G.; data curation, S.B. and I.C.-G.; writing—original draft preparation, S.B. and I.C.-G.; writing—review and editing, S.B., I.C.-G. and A.G.T.; supervision, A.G.T.; project administration, A.G.T.; funding acquisition, A.G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by NIH NIAID grant AI155844 and UTMB SIVS funds awarded to A.G.T. S.B. was further supported by the UTMB Sealy Institute for Vaccine Sciences Pre-Doctoral Fellowship. The opinions and assertions contained herein are those of the authors and do not reflect the official policy or position of the National Institutes of Health.
Data availability
All data generated or analyzed during this study is available upon request from the authors.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sarah Bowser and Itziar Chapartegui-González equal contribution.
References
- 1.Afzaal M, Saeed F, Shah YA, Hussain M, Rabail R, Socol CT, et al. Human gut microbiota in health and disease: unveiling the relationship. Front Microbiol. 2022;13:999001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. [DOI] [PubMed] [Google Scholar]
- 3.Iacob S, Iacob DG, Luminos LM. Intestinal microbiota as a host defense mechanism to infectious threats. Front Microbiol. 2019;10:426119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan I, Bai Y, Zha L, Ullah N, Ullah H, Shah SRH, et al. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen infection. Front Cell Infect Microbiol. 2021;11:716299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takao M, Yen H, Tobe T. LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2014;93:1302–13. [DOI] [PubMed] [Google Scholar]
- 6.Melton-Celsa AR. Shiga Toxin (Stx) classification, structure, and function. Microbiol Spectr. 2014;2:EHEC–0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DS, O’Brien AD. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc Natl Acad Sci U S A. 2013;110:E2126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roxas JL, Koutsouris A, Bellmeyer A, Tesfay S, Royan S, Falzari K, et al. Enterohemorrhagic E. Coli alters murine intestinal epithelial tight junction protein expression and barrier function in a Shiga toxin independent manner. Lab Invest. 2010;90:1152–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien AD, Mohawk KL. Mouse models of Escherichia coli O157:H7 Infection and Shiga Toxin Injection. J Biomed Biotechnol. 2011; 2011:258185. [DOI] [PMC free article] [PubMed]
- 10.Hopkins EGD, Roumeliotis TI, Mullineaux-Sanders C, Choudhary JS, Frankel G. Intestinal epithelial cells and the microbiome undergo swift reprogramming at the inception of colonic Citrobacter rodentium infection. mBio. 2019;10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yadav M, Chauhan NS. Microbiome therapeutics: exploring the present scenario and challenges. Gastroenterol Rep (Oxf). 2022;10:goab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynn DJ, Benson SC, Lynn MA, Pulendran B. Modulation of immune responses to vaccination by the microbiota: implications and potential mechanisms. Nat Rev Immunol. 2021;22:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann P. The immunological interplay between vaccination and the intestinal microbiota. NPJ Vaccines. 2023;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hays MP, Ericsson AC, Yang Y, Hardwidge PR. Vaccinating with conserved Escherichia coli antigens does not alter the mouse intestinal microbiome. BMC Res Notes. 2016;9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojas-Lopez M, Monterio R, Pizza M, Desvaux M, Rosini R. Intestinal pathogenic Escherichia coli: insights for vaccine development. Front Microbiol. 2018;9:333650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Villamil JI, Tapia D, Torres AG. Optimization of multivalent gold nanoparticle vaccines eliciting Humoral and Cellular Immunity in an in vivo model of Enterohemorrhagic Escherichia coli O157:H7 colonization. mSphere. 2022;7:e0093421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez-Villamil JI, Tapia D, Torres AG. Development of a gold nanoparticle vaccine against Enterohemorrhagic Escherichia coli O157:H7. mBio. 2019;10:e01869–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowser S, Melton-Celsa A, Chapartegui-González I, Torres AG. Efficacy of EHEC gold nanoparticle vaccines evaluated with the Shiga toxin-producing Citrobacter rodentium mouse model. Microbiol Spectr. 2023;12:e02261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowser S, Melton-Celsa A, Chapartegui-González I, Torres AG. Further evaluation of Enterohemorrhagic Escherichia coli Gold Nanoparticle vaccines utilizing Citrobacter rodentium as the Model Organism. Vaccines (Basel). 2024;12:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gansheroff LJ, Wachtel MR, O’Brien AD. Decreased adherence of enterohemorrhagic Escherichia coli to HEp-2 cells in the presence of antibodies that recognize the C-terminal region of intimin. Infect Immun. 1999;67:6409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John Turkevich B, Cooper Stevenson P, Hillier J. A study of the nucleation and growth processes in the synthesis of Colloidal Gold. Discuss Faraday Soc. 1951;11:55–75. [Google Scholar]
- 22.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15:799–821. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Zhou G, Ewald J, Pang Z, Shiri T, Xia J. MicrobiomeAnalyst 2.0: comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 2023;51:W310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elliott SJ, Sperandio V, Giron JA, Shin S, Mellies JL, Wainwright L, et al. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in Enteropathogenic and Enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mundy R, Macdonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–706. [DOI] [PubMed] [Google Scholar]
- 28.Lee KS, Jeong YJ, Lee MS. Escherichia coli Shiga Toxins and Gut Microinteractionsctions. Toxins (Basel). 2021;13:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freedman SB, Xie J, Neufeld MS, Hamilton WL, Hartling L, Tarr PI, et al. Shiga Toxin–Producing Escherichia coli infection, antibiotics, and risk of developing hemolytic uremic syndrome: a Meta-analysis. Clin Infect Dis. 2016;62:1251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong CS, Jelacic S, Habeen RL, Watkins SL, Tarr PI. The risk of hemolytic-uremic syndrome after Antibiotic Treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;342:1930–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kakoullis L, Papachristodoulou E, Chra P, Panos G. Shiga toxin-induced haemolytic uraemic syndrome and the role of antibiotics: a global overview. J Infect. 2019;79:75–94. [DOI] [PubMed] [Google Scholar]
- 32.Ritchie JM. Animal models of Enterohemorrhagic Escherichia coli infection. Microbiol Spectr. 2014;2:EHEC–0022. [DOI] [PubMed] [Google Scholar]
- 33.Walrath T, Dyamenahalli KU, Hulsebus HJ, McCullough RL, Idrovo JP, Boe DM, et al. Age-related changes in intestinal immunity and the microbiome. J Leukoc Biol. 2021;109:1045–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh R, Zogg H, Wei L, Bartlett A, Ghoshal UC, Rajender S, et al. Gut Microbial Dysbiosis in the Pathogenesis of Gastrointestinal Dysmotility and Metabolic disorders. J Neurogastroenterol Motil. 2021;27:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borgognone A, Elizalde-Torrent A, Casadellà M, Romero L, Escribà T, Parera M, et al. Vaccination with an HIV T-cell immunogen induces alterations in the mouse gut microbiota. NPJ Biofilms Microbiomes. 2022;8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osbelt L, Thiemann S, Smit N, Lesker TR, Schröter M, Gálvez EJC, et al. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog. 2020;16:e1008448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho S, Kumar SS, Ramirez S, Valientes R, Kim IH. Dietary eubiotics of microbial muramidase and glycan improve intestinal villi, ileum microbiota composition and production trait of broiler. J Anim Sci Biotechnol. 2024;15:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caruso R, Ono M, Bunker ME, Núñez G, Inohara N. Dynamic and asymmetric changes of the Microbial communities after Cohousing in Laboratory mice. Cell Rep. 2019;27:3401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 2017;18:690–9. [DOI] [PubMed] [Google Scholar]
- 40.Bao CL, Liu SZ, Shang ZD, Liu YJ, Wang J, Zhang WX, et al. Bacillus amyloliquefaciens TL106 protects mice against enterohaemorrhagic Escherichia coli O157:H7-induced intestinal disease through improving immune response, intestinal barrier function and gut microbiota. J Appl Microbiol. 2021;131:470–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study is available upon request from the authors.