Abstract
Extrachromosomal circular DNAs (eccDNAs) are a type of circular DNAs originating from but independent of chromosomal DNAs. Nowadays, with the rapid development of sequencing and bioinformatics, the accuracy of eccDNAs detection has significantly improved. This advancement has consequently enhanced the feasibility of exploring the biological characteristics and functions of eccDNAs. This review elucidates the potential mechanisms of eccDNA generation, the existing methods for their detection and analysis, and their basic features. Furthermore, it focuses on the biological functions of eccDNAs in regulating gene expression under both physiological and pathological conditions. Additionally, the review summarizes the clinical implications of eccDNAs in human cancers and health.
Keywords: Extrachromosomal Circular DNA, Circle-Seq, Human Health, Cancer, Generating mechanism
Introduction
Extrachromosomal circular DNAs (eccDNAs) are a type of circular DNAs originating from but independent of chromosomal DNAs, which have been extensively identified in the nuclei and cytoplasms of various eukaryotic organisms, including ciliates, fruit flies, yeast, pigs, and humans [1, 2]. EccDNAs encompass genetic elements from the human genome, ranging from small non-coding regions to entire genes [3, 4]. Unlike the exclusively double-stranded structures observed in mitochondrial DNA, bacterial plasmids, and chloroplast DNA, eccDNAs derived from the nuclei of eukaryotes can exhibit both single-stranded and double-stranded configurations [1].
Despite the initial discovery of eccDNAs in the germ cells of wheat and pigs using electron microscope in 1964 [5, 6], their biological functions still remain largely unclear. This ambiguity is primarily due to their homologous sequences with linear DNA and the limitations of detection methods [7]. Preliminary studies suggest that eccDNAs may regulate gene expression levels by encoding microRNAs or si-like RNAs [8]. Nowadays, with the rapid advancements in sequencing technologies and bioinformatics, the detection accuracy of small DNA fragments, including eccDNAs, has been significantly improved. This progress enhances the feasibility of exploring the biological characteristics and functions of eccDNAs.
This review elucidates the potential mechanisms of eccDNAs generation, along with the existing detection and analysis methods and their biological hallmarks. It also focuses on the molecular functions of eccDNAs in gene expression under both physiological and pathological conditions. Furthermore, the review highlights the promising clinical potential of eccDNAs as innovative biomarkers and therapeutic targets for improving human health and treating diseases.
The categories of eccDNAs
The sizes of eccDNAs vary significantly, ranging from a few hundred base pairs (bp) to several million base pairs (Mb). However, the majority of eccDNAs are less than 1000 bp in length [9] to genomic elements [5]. EccDNAs can be classified into five categories based on their size: microDNAs (200 bp-3 kb) [4], small polydispersed circular DNA (spcDNA; 100 bp-10 kb) [10, 11], telomeric circles (t-circles/c-circles, integral multiples of 738 bp) [12], extrachromosomal ribosomal DNA circle (ERCs, 19.3–40.4 kb) [13] and double minutes/extrachromosomal DNA (ecDNA, 100 kb-10 Mb) [14].
MicroDNA and spcDNA
MicroDNA, derived from unique non-repetitive genomic regions with high density of coding genes and GC contents, are enriched in the 5'-untranslated regions of coding genes, exons, and CpG islands [4]. SpcDNA is significantly more abundant in genetically unstable cells and tissues [15]. Upon its characterization, it displays a heterogeneous size distribution and varied sequence content [11]. The small size of microDNA and spcDNA makes them unable to carry full protein-coding gene sequences or promoter regions. However, microDNA can regulate gene expression through transcription of functional small regulatory RNA, including microRNA (miRNA) and small interfering RNA [8]. In addition, spcDNA can serve as an indicator of genomic instability [16, 17]. For example, spcDNAs carrying Alu elements or LINE-1 sequences occur at a high frequency in the malignant tumors [18].
Telomeric circles
Telomeric circles, consisting only of telomeric repeats, prevent telomere shortening of tumor cells by mediating telomere pruning at the ends of chromosomes [19, 20] and alternative lengthening of telomeres (ALT) [21] to promote the proliferation of tumor cells. Additionally, telomeric circles served as a biomarker for the diagnosis and treatment of telomerase-negative cancers [22]. For example, the telomeric circles showed a significant association with Ki-67 index in gliomas [23]. In high-risk neuroblastoma, a substantial presence of telomere circles is observed, which correlates with the active “telomere trimming” [24, 25]. On the other hand, immature cells lacking telomerase maintain telomere length through the ALT mechanism [26, 27]. In these cells of promyelocytic leukemia, extrachromosomal telomeric DNA circles are present in the nuclei of ALT cells and facilitate telomere elongation via a roll-and-spread mechanism [28].
Extrachromosomal ribosomal circles
Extrachromosomal ribosomal circles (ERCs), generated through homologous recombination (HR), function as transcription templates for ribosomal RNA [13]. They possess self-replicating and amplifying capabilities due to their autonomously replicating sequences [5]. ERCs can also reinsert themselves into the genome in a dosage-dependent manner in response to catastrophic gene loss [29]. Additionally, ERCs tend to accumulate as cells age. During cell division, ERCs can replicate as autonomous replication sequences and are preferentially retained in mother cells, leading to a significant increase in ERCs within senescent mother cells while limiting their number in daughter cells [30]. A typical example is that during the senescence of human lung fibroblasts, it was found that the sizes and copy number of ERCs increased significantly [31], allowing senescent cells to adapt to microenvironmental changes by enriching specific genes through increased copy numbers of ERCs [32].
EcDNA
EcDNA was initially detected in a double-body form, referred to as double minutes [14]. The term ecDNA now refers to those eccDNAs that carry entire or long sequences of coding genes in malignant tumor cells [33], with sizes ranging from 100 kb to 10 Mb [14]. EcDNA lacks centromeres and segregates asymmetrically during mitosis or meiosis. The gene sequences on ecDNA are highly rearranged and amplified, integrating multiple regions that are scattered across different chromosomes in tumors [34].
In this review, we refer to the aforementioned four categories collectively as eccDNAs. EccDNAs are the small-sized circular DNAs carrying small gene segments, including gene sequences, microDNA genes, repetitive or transposable elements, non-coding sequences and so on, which may have diverse regulatory functions [35]. The term ecDNA is specifically used to describe the circular DNA carrying intact genes, especially the oncogenes, ranging from 100 kb to 10 Mb, in malignancies [14, 36].
The biological generating mechanism of eccDNAs
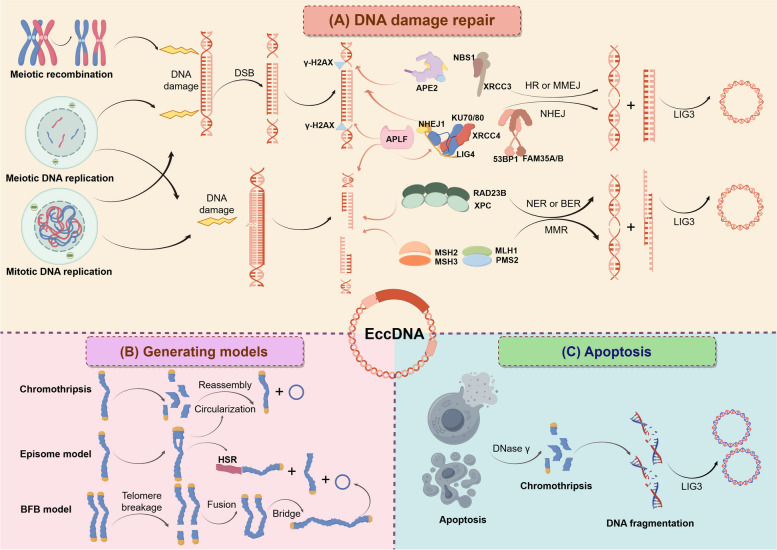
The biological generating mechanisms of eccDNAs vary depending on the type of cell from which they are derived. In human somatic cells, germline cells, and some malignant tumor cells, eccDNAs are primarily generated through processes such as DNA replication, recombination, and damage repair [37]. While in malignant tumor cells, the generation of eccDNAs can also be explained by three models: chromothripsis, the breakage-fusion-bridge (BFB) cycle model and the episome model [1, 2, 9, 38]. Meanwhile, apoptosis and sister chromatid recombination have been demonstrated as the generating mechanisms of eccDNAs (Fig. 1).
Fig. 1.
The biological generating mechanisms of eccDNAs in vivo. A During the process of DNA replication and meiotic recombination, the excised DNA fragments can be circularized into eccDNAs through DNA damage repair mechanisms, including homologous recombination, NHEJ, MMEJ, mismatch repair, nucleotide excision repair and base excision repair. B Chromothripsis, episome model and BFB model were the major generating models of eccDNAs. C Apoptotic DNA fragments can be circularized into eccDNAs under the action of DNA ligase 3 after chromothripsis of apoptotic cells by DNase γ
DNA damage repair mechanisms
The generation of eccDNAs is closely associated with chromosomal DNA damage, functional impairment, and subsequent DNA repair processes [37]. For instance, DNA can undergo excision into smaller fragments through double-strand breaks (DSBs) occurring on the same chromosome. These excised DNA fragments can then circularize and generate eccDNAs [37, 39, 40]. Similarly, damaged DNA fragments may be excised via DSB-activated repair mechanisms such as microhomology-mediated DNA repair, non-homologous end-joining (NHEJ), or mismatch repair processes, subsequently circularizing to generate eccDNAs [37, 39, 40] (Fig. 1A). Thus, the mechanisms underlying eccDNA generation may be determined by the types and locations of chromosomal DNA damage and repair, which can vary across different cell types.
Double-strand breaks
Double-strand breaks (DSBs) commonly occur during mitotic and meiotic DNA replication, as well as in meiotic recombination [41, 42]. High DSB environments, such as those induced by DNA-damaging agents, radiation or stress, are associated with increased eccDNA formation due to heightened repair errors or fragment excision [37]. The circularization of excised or misrepaired DNA fragments often occurs in repetitive sequences [39] (Fig. 1A). As eccDNA usually carry genic fragments, including coding sequences or repetitive sequences, its generation from DSBs introduces genetic variability that may impact gene expression and cellular function [35]. This process has significant implications for both normal cellular activity and diseases like cancer, where eccDNA may carry oncogenes that promote tumor development [37, 39, 43].
Non-homologous end-joining
The DNA sequences flanking the junctional sites of eccDNAs can offer insights into their formation mechanisms. Those eccDNAs originating from the non-homologous regions might be generated via non-homologous end-joining (NHEJ) [44]. This phenomenon has been observed in gliomas [45], cervical cancer cell lines HeLa [46], small cell lung cancer, acute myeloid leukemia [47], and colorectal cancer cell lines [48]. KU70/80 complex can recognize and promote direct ligation of the DSB ends of the excised smaller DNA fragments by recruiting DNA-dependent protein kinase (DNA-PKcs) [49]. And then eccDNAs can be ligated and circularized by interacting with NHEJ regulatory protein such as X-ray repair cross complementing 4 (XRCC4), NHEJ1, LIG4 [50] and APLF [51–53] (Fig. 1A, Table 1). Knockout of the NHEJ regulatory gene KU70/80 heterodimer inhibited eccDNAs generation and subsequently reduced the proliferation of the osteosarcoma cell line SaOS2 [54]. Our research team induced DSBs on the same chromosome to generate endogenous eccDNAs of various sizes in non-homologous regions using spaced clusters of short palindromic repeats (CRISPR) /Cas9 technology, indicating that NHEJ could generate eccDNAs in non-homologous regions [55].
Table 1.
The potential DNA repair genes in generating eccDNAs
| DNA repair | DNA damage types | Genes | Reference |
|---|---|---|---|
| NHEJ | DSB | KU70/80, XRCC4, LIG4, NHEJ4, POLM | [37, 43, 50, 54] |
| APLF | [51–53] | ||
| PRKDC | [37] | ||
| MMEJ | DSB | APE2 | [37, 60] |
| HR | DSB | XRCC3, NBS1 | [63, 64] |
| SHLD2 (FAM35A/B), TP53BP1 | [72] | ||
| POLQ, BRCA1, BRCA2, RAD51 | [37] | ||
| MMR | DNA single-strand break | MSH2, MSH3, MLH1, PMS2 | [37, 40] |
| BLM | [71] | ||
| BER/NER | DNA single-strand break | RAD23B | [73] |
| LIG3 | [43] | ||
| PARP1, PARP2, FEN1, APE1 | [37] |
Microhomology-mediated end-joining
Microhomology-mediated end-joining (MMEJ) is thought to bridge the resulting DNA gap by annealing a 2–25 bp regions of microhomology on each side of the DSB [56, 57]. Previous study has observed that 66.36% of the total eccDNA molecules possessed 4–18 bp direct repeats near eccDNA junction sites [58]. The high frequent occurrence of direct repeats provides paired bases for eccDNA circularization mediated by MMEJ, suggesting MMEJ could be the major generating mechanism of eccDNAs. Meanwhile, the higher GC content of homologous sequences in eccDNAs permitted stronger base-pairing for efficient MMEJ [59]. Additionally, deletion of APE2 genes, which is involved in the excision of damaged DNA fragments and MMEJ, also reduced eccDNAs abundance [37, 60]. Therefore, the sticky ends of DNA fragments with homologous sequences might complement with each other, and then be ligated by DNA ligase 3 to circularize and form eccDNAs [59] (Fig. 1A, Table 1).
Homologous recombination
Regions with high homologous sequences may generate eccDNAs by homologous recombination [5]. Homologous recombination process repairs the missing genetic information in DNA damage using sister chromatids as the homologous templates, a mechanism that can also lead to the generation of eccDNAs. As a result, there are no widespread chromosomal microdeletions found in the hotspots of eccDNA generation [4, 5, 61]. However, due to difficulties in identifying these genomic regions during the bioinformatic analysis of whole genome sequencing data, the role of homologous recombination in generating eccDNAs from repetitive sequence regions is often underestimated. Research indicates that repetitive DNA sequences are disproportionately represented in eccDNAs, accounting for approximately 72% compared with human genomic DNAs [62]. Nonetheless, studies have shown that the abundance of eccDNAs in cells with homologous recombination-related gene defects does not significantly differ from that in wild-type cells [37]. It has been demonstrated that homologous recombination regulatory proteins, such as X-ray repair cross complementing 3 (XRCC3) and nijmegen breakage syndrome 1 (NBS1), can facilitate the generation of eccDNAs, including telomeric circles [63, 64] (Fig. 1A, Table 1).
Mismatch repair
The success rate of DNA repair is influenced by the type and extent of DNA damage [65, 66]. DNA single-strand breaks repair pathways such as mismatch repair have higher success rates compared to pathways for repairing DNA DSBs [67]. Mismatch repair performs DNA repair by identifying and repairing base mismatches and loop structures that occur during annealing of short homologous sequences [68, 69]. It is known that direct repeats on the side of the DNA fragment serve as microhomology for the generation of eccDNAs, indicating mismatch repair may also lead to eccDNA formation based on single-stranded DNA [58, 70]. The potential mechanism is that the formation of eccDNAs occurs when microhomology promotes slippage of the replicative DNA polymerase, leading to looped structures that are excised and ligated into circles through mismatch repair pathways. These single-stranded circles are then converted to double-stranded circles by primed DNA synthesis [39]. In human tumor cells, the deletion of mismatch repair-related genes, including MSH2, MSH3, MLH1 and PMS2, leads to a decrease in the abundance of eccDNAs [37, 39, 40] (Fig. 1A, Table 1). Additionally, BLM proteins, which interact with mismatch repair, also promote the generation of eccDNAs [71].
Breakage-fusion-bridge cycle model
The breakage-fusion-bridge (BFB) cycle model, proposed by McClintock, is a classic model for the generation of eccDNAs [74]. The BFB cycle initiates with the separation of telomeres during the early phase of chromosome DSBs. If these DSBs are not repaired before DNA replication, the chromosome replicates during cell division, resulting in two sister chromatids each lacking a telomere. These chromatids may fuse to form a dicentric chromosome, a chromosome with two centromeres [75]. During anaphase, this dicentric chromosome may break randomly into two chromatids, each still missing a telomere, leading to the formation of a chromatid bridge. The breaking of this bridge results in various chromosomal aberrations, including the generation of eccDNAs [38] (Fig. 1B). The BFB cycle can perpetuate through successive cell cycle, producing reverse repeat sequences in one daughter cell and terminating deletions in another. These unstable reverse repeat sequences are prone to circularization, ultimately generating eccDNAs [76].
Chromothripsis
Chromothripsis is a complex mutational process characterized by a single catastrophic event in which a chromosomal region is shattered into numerous fragments, commonly observed in malignant tumors and congenital diseases [77]. Small-sized eccDNAs are closely associated with late-stage apoptosis during chromosome breakage, with DNA ligase 3 playing a crucial role in the generation of nucleosome-sized circular DNAs [78]. However, the mechanisms underlying the generation of large-sized eccDNAs are intricately linked to chromosome breakage and tumor genome rearrangements [38, 79]. Chromosome breakage resulting from DSBs facilitates the amplification of eccDNAs through the catalytic activity of DNA repair enzymes, including poly ADP-ribose polymerase (PARP) and DNA-dependent protein kinase (DNA-PKc) [38]. Repeated BFB cycles caused by chromosomal fusion is regarded as an initiating factor for a cascade of events such as micronucleation, chromothripsis, and even cell death, thus generating eccDNA [80, 81]. In micronuclei, chromosomes are prone to DNA breakage due to the incomplete nuclear envelope, which can also result in chromothripsis [82]. During mitosis, the reintegration of micronuclei into primary nuclei can facilitate the engagement of DNA fragments in complex chromosomal rearrangements within the genome [38, 77]. Concurrently, some DNA fragments may become circularized, forming eccDNAs through the action of DNA ligase 3 [83] (Fig. 1B). Similar to the generation of eccDNAs, the reintegration of these eccDNAs elements back into the genome may also necessitate processes involving DNA damage and recombination [38, 84, 85].
Episome model
EccDNAs can originate from submicroscopic precursors derived from chromosomal loci and possess the ability to self-replicate. After the small DNA fragments are excised from the chromosome, several non-adjacent chromosomal DNA fragments may fuse and recombine with each other, undergoing cyclical amplification to generate eccDNAs [35, 86, 87]. Alternatively, these small DNA fragments can integrate into chromosomes through the BFB cycle, forming homologous stained regions (HSRs) with gene amplification capabilities [88, 89] (Fig. 1B). For instance, eccDNAs containing the MYC gene have been generated through the excision and amplification of DNA fragments in leukemia cases [90]. Similarly, gradually amplified double minute chromosomes have been detected in CHO cells, accompanied by DNA deletions [91]. Moreover, the CRISPR-CATCH technique has been developed to capture and enrich target ecDNAs from ecDNA containing cells [92].
Apoptosis and sister chromatid recombination
The sizes of eccDNAs in human cells exhibit a multimodal distribution, with several prominent peak within 600 base pairs [93]. This distribution corresponds to the ladder-like fragmentation pattern of chromosomal linear DNA into nucleosome-sized fragments observed in apoptotic cells. In cells undergoing apoptosis, a significant amount of eccDNAs can be generated by DNase γ and DNA ligase 3, resulting in a multimodal distribution of eccDNA sizes. Consequently, DNA fragments produced during apoptosis can be circularized to form eccDNAs via DNA ligase 3 [78] (Fig. 1C).
Additionally, eccDNAs can be generated through sister chromatid recombination between non-allelic homologous chromosomes [94], replication slippage [39], and tandem duplication of chromosomal recombination [95]. The deletions at recombination hotspots also promote the generation of eccDNAs in germlines [96]. During spermatogenesis, primordial diploid germ cells undergo meiosis to develop into haploid sperm cells, a process that ensures genetic variation through the mechanisms of random segregation and homologous recombination [94]. Within prophase I, recombination between homologous chromosomes is initiated by programmed DSBs that occur during the leptotene stage [97]. DSB sites are located in hotspots on chromatin loops [41], and are subsequently repaired to produce crossovers during the pachytene stage [98]. H3K36me3 and H3K9me3 also play roles in histone replacement during the recombination, and failure in this process may contribute to the generation of eccDNAs [59, 99]. Therefore, the generation of eccDNA occurred in the process of DSB and repair during meiotic recombination.
The detection and analysis of eccDNAs
Imaging
Before the advent of sequencing technology, researchers employed various visual methods such as optical microscope [100], electron microscope [94], fluorescence in situ hybridization [101], neutral two-dimensional gels to detect eccDNAs [7]. Large-sized ecDNAs, known as double minutes, can be observed using optical microscope, but this technique was insufficient for identifying small-sized eccDNAs [100]. Presently, fluorescently labeled eccDNAs can be visualized through advanced techniques such as structured illumination microscope and scanning atomic force microscope [78, 94]. Furthermore, the circular configuration of eccDNAs has been confirmed using ultra-resolution three-dimensional structured illumination microscopy and transmission electron microscope [14]. Currently, the spatio-temporal dynamics of eccDNAs within living cells can be monitored through a DNA labeling system utilizing CRISPR/dCas9 [102]. Additionally, the asymmetric segregation of eccDNAs during mitosis can be tracked using fluorescent RNA probes targeting the junction sites of eccDNAs [102].
The methods of eccDNAs sequencing
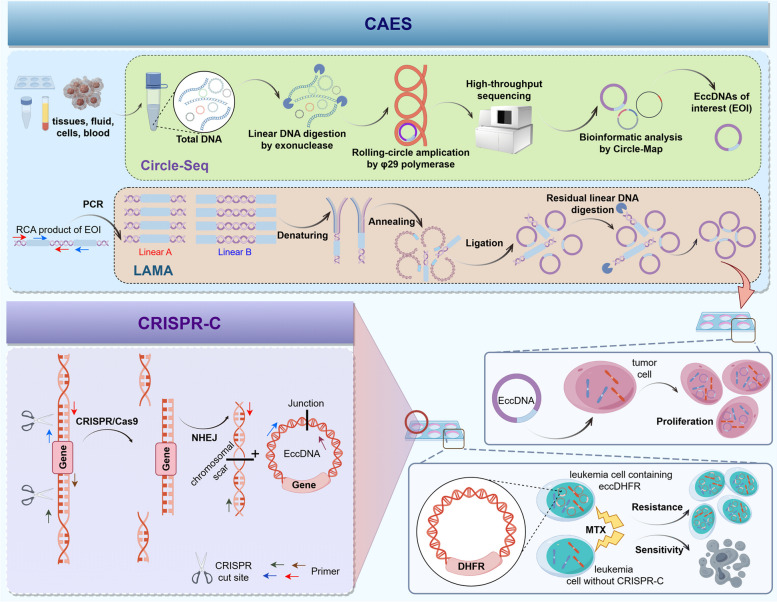
Given the low natural abundance of eccDNAs, their enrichment typically necessitates amplification [6]. Initially, the density gradient centrifugation method involving CsCl-ethidium bromide (CsCl-EtBr) was employed in 1967 to isolate eccDNAs, eventually becoming the standard technique for eccDNA separation [102, 103]. Nonetheless, this method demands a high abundance of eccDNAs for effective detection and results in significant loss of eccDNAs due to their co-migration with linear DNA during CsCl-EtBr density gradient centrifugation [16]. In response to these limitations, Shibata et al. introduced a novel method in 2012 to purify and amplify eccDNAs: nuclei are lysed in an alkaline solution, removing linear DNA through exonuclease digestion. Subsequently, eccDNAs are amplified using rolling-circle amplification (RCA) [4]. Shoura et al. further refined this approach by integrating CsCl-EtBr gradient centrifugation to prevent exonuclease V digestion of linear DNA, utilizing Tn5 transposition fragments for direct labeling of eccDNAs [104]. Additionally, the CRISPR-CATCH technique, in combination with CRISPR-Cas9, facilitates the selective purification of eccDNAs with millions of base pairs (Mb) in sizes from agarose gel electrophoresis in vitro [92].
Circle-Seq
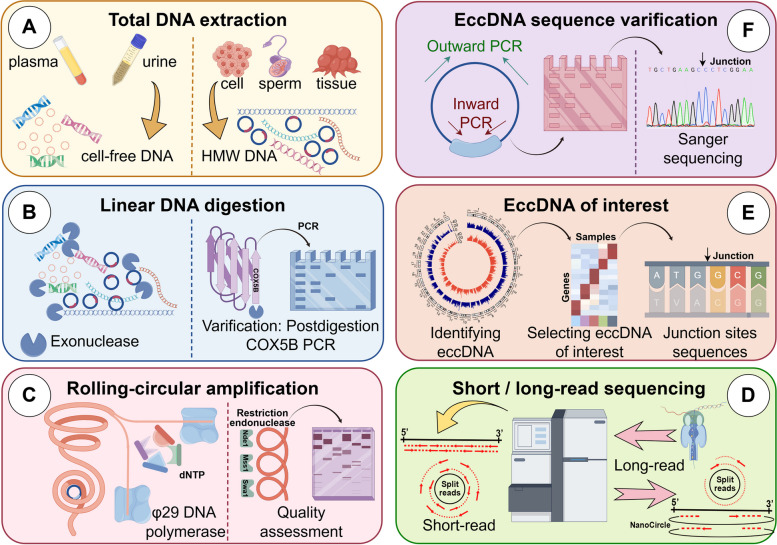
Circle-Seq technology integrates eccDNAs purification with high-throughput sequencing technology to detect novel eccDNAs with low copy number or arbitrary sizes [105]. This technique is characterized by its high sensitivity and wide genomic detection range, leveraging established bacterial plasmid purification and sequencing methods to amplify and identify eccDNAs [40, 106]. The specific process of Circle-Seq involves several steps: (A) The samples are cleaved in alkaline solution and genomic DNAs are collected into elution buffer using ion exchange columns. (B) The linear chromosome DNAs were digested with Plasmid-Safe DNase (PSD). (C) The purified eccDNAs were further amplified by φ29 rolling-circle amplification (RCA). (D) Short-read/long-read sequencing and eccDNA-specific mapping software were employed to identify eccDNAs [7, 94, 106, 107]. (E) Bioinformatic analysis is used to identify the eccDNAs of interest (EOI). (F) The junction site sequences of the EOI were verified through outward and inward PCR, and Sanger sequencing was performed for PCR product (Fig. 2). However, due to RCA's tendency to preferentially amplify smaller eccDNAs, the accuracy of Circle-Seq technique in detecting the copy number of a single eccDNA showed relatively low [7]. During the preparation of NGS libraries, the use of transferase-labeled eccDNAs can enhance the accuracy of eccDNA copy number detection after RCA [78, 108].
Fig. 2.
Flow diagram of the purification and detection of eccDNA by Circle-Seq
Further attention should be paid to the details involved in the purification and amplification of eccDNAs using Circle-Seq. (A) The extraction of total DNA from various samples. As for the traditional high-speed centrifugation can disrupt the double-stranded helix structure of the large-sized eccDNAs, thereby reducing the extraction efficiency [109], magnetic bead method was used for the extraction of total DNA in our researches [107, 110]. Depending on the nature of the body fluid and cell samples, cell-free DNA (cf-DNA) [58, 101, 111, 112] and high-molecular weight DNA (HMW DNA) [7, 59, 94, 107, 110] were identified as the most suitable DNA categories for this step (Fig. 2A). (B) The selection of exonuclease and the verification of digestive efficacy. Exonuclease is a nucleic acid hydrolase that hydrolyzes the phosphodiester bond to form a single nucleotide from the end of a linear DNA strand, which can digest and hydrolyze linear DNA while preserving the closed circular DNA [113]. Plasmid-safe DNA exonuclease has been shown to lack enzymatic activity on double-stranded circular DNA and is therefore a key enzyme for linear DNA digestion before RCA in Circle-Seq technology [107, 110]. Exonucleases V and VIII are not suitable alternatives as they can digest nicked eccDNAs. However, electron microscopy has demonstrated that exonuclease alone cannot completely eliminate linear DNA [4, 39]. Residual linear DNA was verified by COX5B PCR detection to mitigate its impact on Circle-Seq technology (Fig. 2B) [7]. (C) The quality assessment of RCA product using restrictive endonuclease. Restrictive endonuclease is a class of nucleic acid hydrolases that hydrolyze circular DNA by hydrolyzing phosphodiester bonds through specific recognition of DNA palindromic sequences [114]. Specific restrictive endonucleases not only assist exonucleases in removing linear DNA but also target endogenous circular DNA outside the Circle-Seq product [105]. For example, Mss1 recognizes mitochondrial DNA for hydrolysis [7], while Swa1 excises plasmids with a 2 μm diameter [115]. Restrictive endonuclease Nde1, capable of excising both plasmids and circular DNA, is appropriate for evaluating the quality of RCA products (Fig. 2C) [107, 110, 116]. (D) The appropriate sequencing methods for RCA product. Post-enrichment, eccDNA undergoes library construction for sequencing on platforms like short-read high-throughput sequencing or long-read nanopore sequencing techniques. This method can reveal the origin, biological generation and immune stimulation function of eccDNAs [78, 105], in which 104 types of eccDNAs can be detected from 106 myocardial nuclei (Fig. 2D) [105]. (E) The selection of EOI by bioinformatic analysis. The frequency of occurrence [107, 110, 116] and the eccDNA abundance related to genes [111] are the two primary methods for screening EOIs (Fig. 2E). (F) Sanger sequencing of PCR product should be applied to validate the appearance of the target eccDNA [107, 110, 116] (Fig. 2F).
Short-read sequencing
The advancement of high-throughput sequencing technology has significantly facilitated eccDNA research. Researchers can leverage the unique characteristics of eccDNAs mapping reads, such as split reads and discordant paired end reads, to differentiate eccDNAs from linear DNAs using high-throughput sequencing methods like SMOOTH-seq data [117]. In short-read whole genome sequencing (WGS), the copy number of eccDNAs increases with cell proliferation, making short-read WGS more suitable for detecting eccDNAs with higher copy numbers. Since the vast majority of sequencing reads originate from linear DNA, short-read WGS and short-read ATAC-seq are limited in their ability to effectively detect low-abundance eccDNAs [118]. However, defining the nucleotide sequence of large-sized eccDNAs is challenging with short-read sequencing, as it often capture only one breakpoint at a time, limiting comprehensive computational analysis [84]. In addition, paired-end sequencing with short-reads often falls short in accurately reconstructing the full circular structure of eccDNAs.
Long-read sequencing
Currently, long-read sequencing technologies, such as the 5–10 kb sequence reads available from Nanopore, Pacific Biosciences platforms, as well as optical mapping technologies like BioNano [84], enable the capture of multiple breakpoints, providing stronger evidence regarding the frequency and structure of large-sized or chimeric eccDNAs [119, 120]. These methods have been employed to enhance the accuracy of identifying eccDNAs, particularly those with repeat elements [35, 78, 86, 94, 119]. Researchers have demonstrated that long-read Circle-seq by nanopore sequencing showed the highest detection accuracy and low base pair difference for large-sized eccDNAs [118]. Repeat elements are commonly detected in the sequencing data that are used for identifying eccDNAs [121]. Long-read sequencing displayed significantly elevated proportions of reads mapping to LTRs, SINEs, and LINEs compared to short-read counterparts [118]. However, a pattern of redundancy in eccDNAs identification has been observed with long-read sequencing [122]. Upon calculating the duplication rates, nanopore sequencing could identify multiple similar copies from a single eccDNA sequence. These substantial duplication rates present considerable obstacles for the experimental validation of their predictions [118]. Advanced sequencing methods, including CIDER-Seq, ChIP-seq, 4C-seq and PLAC-seq technologies, enable the detection of eccDNAs biological characteristics by integrating their sequence information with insights into chromatin accessibility and epigenetic regulation [14, 78, 105, 123–125].
Single-cell sequencing
Single-cell sequencing can identify copy number variations or single nucleotide variations in eccDNAs, but it struggles to capture the heterogeneity of eccDNAs due to the limited number of recognizable eccDNAs [126]. Single-cell ATAC-seq (scATAC-seq) employs Tn5 transposase to cleave circular DNA, producing chimeric reads in the ATAC-seq library that correspond to circles. Modified ecc_finder, based on bulk sequencing data, is then used to identify circular DNA in scATAC-seq data [127, 128]. Single-cell whole genome sequencing (scWGS-seq) [129] and SMOOTH-seq [117] identified candidate eccDNAs through single-cell DNA sequencing methods.
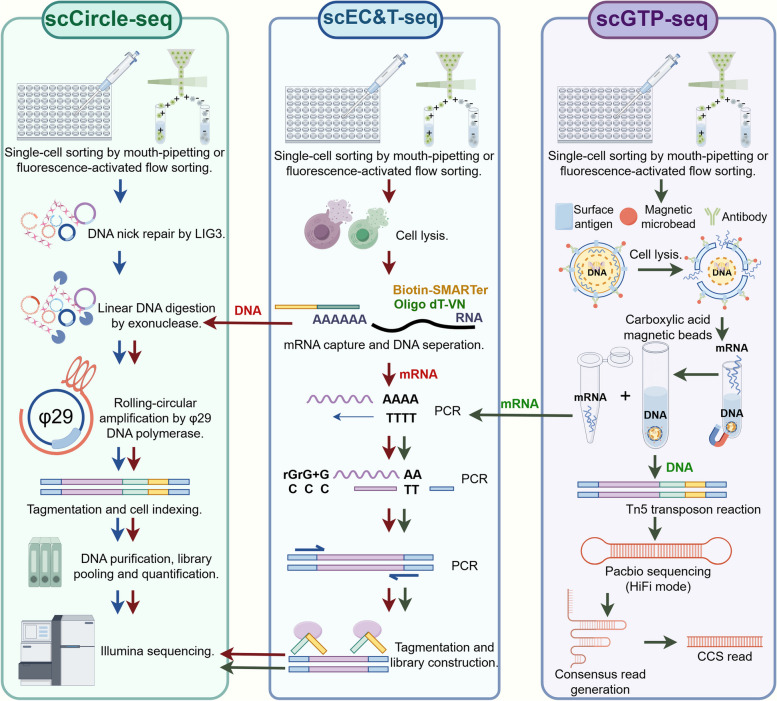
Single-cell Circle-Seq (scCircle-Seq) is a method involving fluorescence-activated flow sorting for single-cell suspension preparation and eccDNA amplification via RCA [130]. Researchers have unveiled cancer-specific eccDNA landscapes and a higher propensity for eccDNAs to form within amplified genomic regions of different cell clusters using scCircle-Seq (Fig. 3) [130]. Single-cell extrachromosomal circular DNA and transcriptome sequencing (scEC&T-seq) was a plate-based method for parallel sequencing of circular DNAs and full-length mRNA from single cells [131, 132]. And scEC&T-seq can be combined with different sequencing technologies, including Strand-seq [133] and single-cell tri-channel processing (Fig. 3) [134]. This method generates a uniform number of reads per cell and produces independent sequencing libraries that can be selected and sequenced, offering a significant advantage when high sequencing coverage is required [132]. Single-cell paralleled genome and transcriptome sequencing(scGTP-seq) on a third-generation platform was a method that physically separates the transcriptome and genome, adapting the SMOOTH-seq procedure [117] by integrating parallel scRNA-seq analysis using Smart-seq2 (Fig. 3) [135]. For instance, researchers have identified hepatic cells, tumor-associated macrophages and tumor-associated endothelial cells in a clinical sample of hepatocellular carcinoma, and obtained eccDNAs from these cells, demonstrating a viable way to monitor the evolution of eccDNAs and structural variations during cancer progression [135].
Fig. 3.
The novel eccDNAs sequencing methods at the single-cell level
However, the relatively low sensitivity, limited throughput, and high cost of long-read sequencing hindered us from conducting long-read deep sequencing for each individual cell [135]. The relatively high sequencing error rate of the Nanopore platform can lead to false positives. This issue becomes particularly problematic with very lengthy sequencing amplicons, as longer reads are more prone to error. For example, U2OS cells sequenced with the Nanopore platform achieved the longest read lengths, but showed a significantly lower read mapping rate [135]. Additionally, mapping the time-resolved eccDNAs copy number distribution by single-cell sequencing of various eccDNAs-carried oncogenes will be essential for future research to better understand more complex fitness models [136].
The bioinformatic analysis of eccDNAs
In recent years, a variety of bioinformatic methods and databases have been established to identify and analyze the biological characteristics of eccDNAs. These methods vary depending on the type of sequencing data and the specific objectives of the analysis. Split and discordant reads within short-read data, and breakpoint reads in long-read data, are primary markers for eccDNA identification [84]. Researchers have developed a Python script to generate simulated eccDNA datasets to evaluate the efficacy of eccDNAs identification [118]. For short-read data analysis, Circle-Map maintained stable base pair differences among different sequencing depth, while the base pair difference of Circle_finder showed significant changes. In the realm of long-read data, eccDNA_RCA_nanopore kept lowest base pair differences across different sequencing depths, but CReSIL showed superior performance at higher depths [118]. The bioinformatic methods and their respective functions for analyzing eccDNAs are detailed in Table 2:
Table 2.
The bioinformatic methods and their functions for analyzing eccDNAs
| Bioinformatic methods | Function | Long-read or short-read |
|---|---|---|
| AmpliconArchitect [36] | Predicting eccDNAs with 83% sensitivity and 85% precision from short-read WGS data | Short |
| AmpliconReconstructor [137] | Improving the resolution of large-sized eccDNAs (> 150 kb) in NGS data | Short |
| Circle-Map [138] | Detecting eccDNAs from Circle-Seq data using split reads and discordant read pairs | Short |
| Circle-Map + + [43] | Detecting eccDNAs from Circle-Seq data using soft-clipped reads and discordant read pairs with higher efficiency and detection accuracy | Short |
| Circle_finder [139] | Identifying eccDNAs in bidirectional sequencing data for short-read ATAC-seq, WGS, and WES | Short |
| ECCsplorer [140] | Detecting candidate eccDNAs from eccDNA-rich data | Short |
| ecc_finder [141] | Identifying eccDNAs from short-read and long-read sequencing profiles without reliance on copy number information | Long and short |
| eccDNA_RCA_nanopore [78] | Detecting eccDNAs from Circle-Seq data using Nanopore sequencing | Long |
| Nanocircle [94] | Detecting eccDNAs from Circle-Seq data using Nanopore sequencing | Long |
| CReSIL [122] | Utilizing coverage depths and breakpoint reads to identify eccDNAs from long-read WGS data | Long |
Meanwhile, there are some eccDNAs databases to investigate the candidate eccDNAs in different tissues and diseases. CircleBase is a human eccDNAs resource and analysis platform featuring a highly interactive eccDNAs visualization function. It integrates sequencing data sets, computational prediction, and manual annotation to identify functional eccDNAs, offering comprehensive eccDNAs annotations [142]. Studies have shown that eccDNAs are primarily enriched in the Ras and PI3K/Akt signaling pathways [142]. eccDNAdb is a database screening potential differential functional eccDNAs carrying genes from a database of eccDNAs profiles in human cancers through WGS [143]. And eccDNA Atlas database contains functional eccDNAs with sequences, diseases, functions, characterization and validation strategy [144]. Specifically, researchers have found that eccDNA carrying FAM48B had the highest abundance in prostate cancer tissues by using the eccDNAdb database. And then the contributions of FAM84BFAM84B carried by eccDNAs was examined in prostate cancer development and related pathways [145].
The basic features of eccDNAs
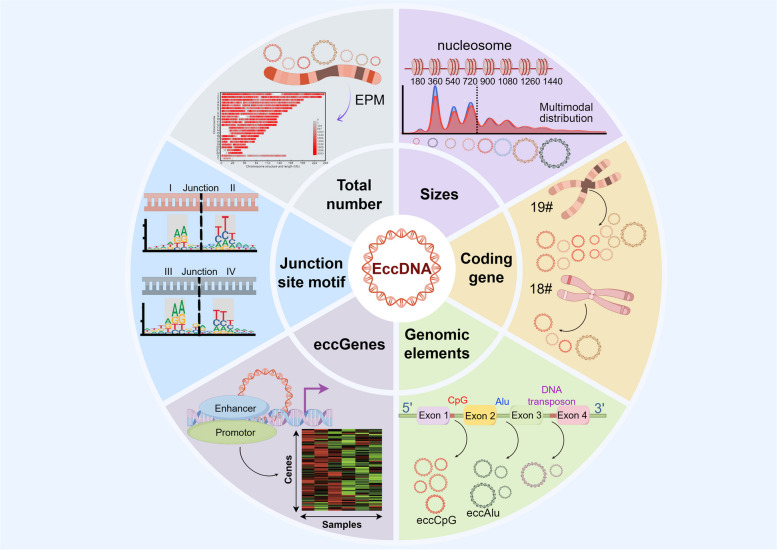
The basic features of eccDNAs were usually located in the total numbers and sizes, the chromosomal origin and their association with coding genes and repetitive sequences, the proportion of eccDNAs from non-repetitive sequences, and the EOI carrying specific genes [7, 59, 94]. These features can provide insights into the mechanisms underlying eccDNA generation. Additionally, the sequence characteristics of genes and the nucleotide motif patterns flanking the junctional sites of eccDNAs can offer insights into the underlying mechanisms driving eccDNAs generation (Fig. 4).
Fig. 4.
The basic features of eccDNAs by Circle-Map
The total numbers and sizes of eccDNAs
EccDNAs per million mappable reads (EPM) is defined as the yield of eccDNA counts in one million sequencing reads. This metric is used in statistical analyses to uncover the biological distribution of eccDNAs while minimizing bias from sequencing depth [7, 94]. Researchers have utilized eccDNA EPM to reflect the dynamic changes in the total number of eccDNAs across various cancers and non-neoplastic diseases [146–148] (Fig. 4).
The size distribution of eccDNAs exhibits a multimodal pattern with a distinct periodicity of approximately 180 bp. Histone synthesis within chromatin is a critical prerequisite for DNA replication [149, 150]. Following DNA replication, DNA double-stranded structures form nucleosomes around histones [151]. Each nucleosome comprises 147 bp of DNA wrapped around a histone core [152, 153], with 20–30 bp linker regions connecting adjacent nucleosomes [154, 155]. Consequently, the 180 bp fragments correspond to the length of a nucleosome [76]. DNA fragment clipping tends to occur at internucleosome junctions, causing eccDNAs to often display sizes that are integer multiples of the molecular weight of nucleosomal DNA [156] (Fig. 4). This size distribution of eccDNAs reflects their nucleosomal origin. Meanwhile, the majority of small-sized eccDNAs, such as microDNAs, are typically between 200 and 3000 bp [5, 70]. Researchers have explored the potential mechanisms behind the formation of both large and small-sized eccDNAs by examining their genomic distribution characteristics and their correlation with normal homologous chromosome meiotic recombination rates, using 3 kb as a cutoff [59]. They found that small-sized eccDNAs are associated with euchromatin, whereas large-sized eccDNAs are preferentially derived from heterochromatin [59].
EccDNAs chromosomal origin and their association with coding genes
The gene sequences found in eccDNAs can serve as predictors for the chromosomal sources from which they originate. In somatic tissues, such as human skeletal muscle and plasma leukocytes, the eccDNA effectively formed in chromosomes 17 and 19, which harbor a higher abundance of coding genes. Additionally, the EPM of eccDNAs shows a significant positive correlation with the presence of coding genes [7] (Fig. 4). This suggests that the generation of eccDNAs in normal tissues might be related to characteristics of gene transcription. Conversely, the EPM of large-sized eccDNAs (≥ 3 kb) derived from spermatozoa is negatively correlated with coding genes [59, 94]. This likely occurs because small-sized eccDNAs (< 3 kb) tend to originate from euchromatin regions, while large-sized eccDNAs are more often derived from heterochromatin regions. Thus, the dense packing of chromatin in heterochromatin regions may impede the formation of small-sized eccDNAs [74].
Association of eccDNAs with repetitive elements
Repetitive elements in the genome can be categorized into tandem repeats and interspersed elements, also known as transposable elements, based on the continuity of their repetitive regions. Tandem repeats are further divided into satellite repeats, microsatellite repeats, and minor satellite repeats. On the other hand, interspersed elements encompass DNA transposons and retrotransposons, which include long interspersed elements (LINEs), Alu elements and other short interspersed elements (SINEs), long terminal repeats (LTRs), and SINE-VNTR-Alus (SVAs) [157] (Fig. 4). The specific mechanisms by which eccDNAs are generated can be analyzed by examining these different source repetitive regions.
Transposons utilize transcribed mRNA as an intermediate to integrate reverse-transcribed complementary DNA into the human genome via a “copy-paste” mechanism [158]. SINEs play a role in recruiting chromatids to gene-rich regions [159], and are linked to early replication regions characterized by high GC content [157], making up approximately 13% of the human genome [160]. Small-sized eccDNAs derived from spermatozoa show a positive correlation with SINEs, suggesting that the generation of eccDNAs during spermatogenesis may originate from gene-rich regions [59].
Alu elements are the most prevalent repetitive elements within SINEs, constituting about 11% of the human genome [161]. Alu elements are associated with early DNA replication and exhibit higher concentration on chromosome 19, which is rich in coding genes. The significant positive correlation between small-sized eccDNAs derived from urine and spermatozoa and Alu elements suggests that Alu elements play a role in eccDNA formation during DNA replication [58, 59, 94]. Additionally, Alu elements are associated with cohesins, which have fundamental roles in meiotic recombination mediating sister chromatid cohesion, chromosome pairing, and synaptonemal complex assembly [162, 163]. Alu elements mediate correct alignment between homologous chromosomes, particularly in gene-rich regions, while suppressing illegitimate intrachromatid recombination that would lead to eccDNA [94]. Previous studies have found that large-sized eccDNAs derived from spermatozoa showed negative association with both the meiotic recombination rate [164–166] and Alu elements [59, 94]. This phenomenon indicates that the abundance of Alu elements [162, 163] has an adaptive significance in maintaining genome integrity by reducing the rate of illegitimate recombination, leading to fewer large-sized eccDNAs from gene-rich regions [59, 94].
Long interspersed elements (LINEs) are associated with late replication regions that have high AT content [157], and recruit chromosomes into gene-poor regions [159], as evidenced by the positive correlation between LINEs and large-sized eccDNAs derived from spermatozoa, and their negative association with coding genes [94]. LTRs are another rich transposition elements, accounting for about 8% of the human genome. LTRs integrated the new copies into the characteristic sequences of the human genome through the "copy-paste" mechanism [167]. The positive correlation between spermatozoa-derived eccDNAs and LTRs further underscores the role of DNA replication in the generation mechanism of eccDNAs [94].
DNA transposons are mobile genetic elements that move within the genome via a "cut-and-paste" mechanism, facilitated by transposon enzymes that integrate their sequences into different genomic locations [168]. Double palindromic repeats may serve as DNA binding sites for these transposons [169], which are then circularized into eccDNAs by transposon enzymes [8, 170]. The spermatozoa-derived eccDNAs EPM shows a positive correlation with DNA transposons [94], and the presence of direct repeats at eccDNAs junction sites further confirms the characteristics of DNA transposons [59, 108]. Microsatellite repeats are related to genomic stability during DNA repair, and microsatellite mutations lead to genomic instability, thus promoting the proliferation and metastasis of malignant tumors [171].
Association of eccDNAs with non-repetitive elements
The ratio of non-repetitive elements in the total number of eccDNAs can indicate the genomic preference generated by eccDNAs [58]. The non-repetitive elements include exon, intron, CpG islands, 5'-untranslated regions (5'UTR), 3'UTR and 2 kb regions upstream/ downstream of genes (Gene2kU/Gene2kD) [7, 58, 111].
CpG islands are abundant with highly condensed cytosine (C)-phosphate (p)-guanine (G) dinucleotides in the promoter and exon regions, playing essential roles in the coding genes in human genome. Methylation of CpG islands in promoters is a key regulatory mechanism of gene expression, typically resulting in gene silencing [172]. It was found that urinary cell-free eccDNAs (ucf-eccDNAs) were enriched in CpG island, suggesting ucf-eccDNAs carrying CpG island may play essential roles in the regulation of coding gene expression [58]. In cases of Type 2 diabetes mellitus (T2DM) following short-term intensive insulin therapy (SIIT), there was a significant decrease in the abundance of eccDNAs in exons, CpG islands, UTRs, and Gene2kU/Gene2kD regions, while enrichment in introns increased. This change suggests the effectiveness of SIIT in managing T2DM [111].
EOI and nucleotide motif patterns flanking eccDNAs junctional sites
The differences of occurrence frequency [65, 68, 78] and the eccDNA abundance for specific genes [70] are the two primary methods used to identify eccDNAs of interest that carry particular genes (Fig. 4). Researchers can utilize synthetic methods to generate eccDNAs containing specific genes in vitro [55, 110, 173], allowing for the exploration of their biological functions in cancer cells and other non-neoplastic diseases. This includes studying their role in cancer proliferation and immunostimulatory activity [78, 107, 116, 136].
The recurrent nucleotide motif patterns flanking the eccDNAs junctional sites shed light on the mechanisms responsible for eccDNAs generation. In order to identify eccDNAs junctional sites precisely, researchers focused on the start site, the 5’-upstream edge in the genome domains, and the end site, the 3’-downstream edge in the genome domains, which were the potential positions where excised DNA segments and gave rise to eccDNAs circularization. Thus, the DNA sequences from 8 bp upstream to 8 bp downstream flanking at eccDNAs start and end sites of each identified eccDNAs for recurrent nucleotide motif signatures were searched to investigate the nucleotide motif patterns [58, 59, 108]. Due to the potential sequence motifs of 3-kb length, the trinucleotide segments were termed as I, II, III, and IV following their genomic orientations. A pair of trinucleotide segments with 4-bp “spacer” was located in between flanking both the start and end sites of eccDNAs [58, 108] (Fig. 4). The double palindromic sequences of eccDNAs junctional sites served as the binding sites for DNA transposon [169]. DNA transposon generated eccDNAs by cutting DNA sequences through the “cut-and-paste” mechanism [168]. The presence of micro-homologous double-stranded direct repeats of eccDNAs junctional sites suggests that the generation of eccDNAs is linked to mechanisms such as microhomologous-mediated HR and MMEJ and other DNA repair pathways [4, 40, 58, 59, 70, 93, 108].
The synthesis methods of eccDNAs in vitro
Genetic engineering is the primary approach for synthesizing eccDNA in vitro. By synthesizing specific sequences of eccDNAs, scientists are able to investigate the roles of endogenous eccDNAs in various biological processes under controlled laboratory conditions. In recent years, our group and other research teams have successfully developed and applied three synthetic methods for in vitro studies: CRISPR-C [55, 174], ligase-assisted mini-circle accumulation (LAMA) [107] and Circle-seq based Artificial EccDNA Synthesis (CAES) [110] (Fig. 5).
Fig. 5.
The synthesis methods of eccDNAs in vitro
CRISPR-C
Our research team first utilized CRISPR-C in 2018 [55] to investigate the biological mechanisms underlying the generation and stability of eccDNAs, as well as their effects on cells in vitro. Chromosomal DSBs can lead to the generation of endogenous eccDNAs [37, 175]. CRISPR/Cas9 genome editing technology simulates this endogenous process by inducing dual DSBs in one chromosome [174]. The core technique of CRISPR-C involves using guide RNA to direct the Cas9 nuclease to specific genetic loci, enabling the simultaneous cleavage of two gene loci on the same chromosome (Fig. 5). This process results in the formation of extrachromosomal DNA fragments that circularize automatically to form eccDNAs, ranging from hundreds to millions of base pairs [55, 136, 174]. Researchers have applied CRISPR-C to generate eccDNAs carrying the dihydrofolate reductase (DHFR) gene and the MYC gene in human chronic myelogenous leukemia cell line HAP1 and human colorectal adenocarcinoma cell lines COLO320-DM, respectively [136]. These eccDNAs, carrying DHFR and MYC genes, can confer resistance to methotrexate and chemotherapy in chronic myelogenous leukemia and colorectal adenocarcinoma [136].
LAMA
Ligase-assisted mini-circle accumulation (LAMA) was first applied in 2007 [173] and enabled the artificial synthesis in vitro by mimicking known microDNAs sequences [78]. LAMA synthesizes small-sized eccDNAs through a process of cyclical annealing, ligation, and denaturation [173] (Fig. 5). Paulsen et al. applied LAMA technology to study in vitro and found that microDNAs not only expressed functional micro-regulatory microRNA (miRNA), but also expressed novel siRNA [8]. Recently, our research team has utilized LAMA technology to synthesize eccDNAs carrying microRNA genes derived from gastric cancer. This research was to explore the role of eccDNAs in the progression and metastasis of gastric cancer in vitro [107]. In addition, LAMA technology was also recently used to synthesize eccDNAs carrying TAOK2 gene fragments in plasma of schizophrenia patients. Their inhibitory effect on mRNA synthesis of the host TAOK2 gene has been verified in glioma and neuroblastoma cell lines U-251MG and SH-SY5Y by our research team [116].
CAES
Circle-seq based Artificial EccDNA Synthesis (CAES) is a rapid and efficient method for synthesizing eccDNAs in vitro, first applied by our research team in 2023. Using this technology, we identified eight types of eccDNAs (eccMIR) containing different microRNA genes in gastric cancer tissues [110]. The CAES process begins with the amplification of eccDNAs using Circle-Seq technology, followed by high-throughput sequencing. Next, Circle-Map is employed to screen for eccDNAs of interest (EOIs). For each eccDNA, two semi-complementary linear DNA fragments are synthesized via PCR, and eccDNAs are subsequently generated through the LAMA reaction. Finally, any residual linear DNA in the LAMA reaction products is removed using restriction exonuclease digestion (Fig. 5). The DNA circularization efficiency of CAES technology ranges from 15.6% to 31.1%, and it is inversely related to the size of the eccDNAs [110].
The mechanisms for biological functions of eccDNAs
Asymmetrical segregation
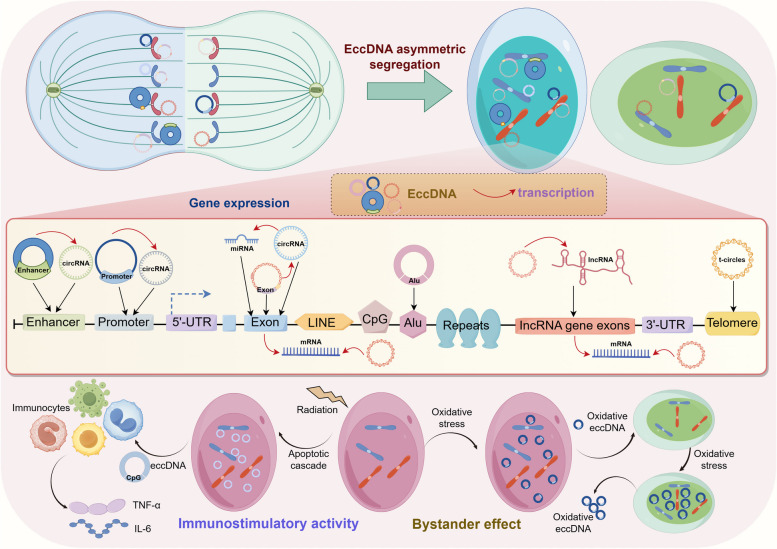
EcDNAs represent the primary types of functional circular DNAs in human neoplastic tissues and cells, characterized by sizes in the millions of base pairs (Mb) [3]. This size is sufficient to carry full-length sequences that promote oncogene amplification. The intact genes carried by ecDNAs exhibit high levels of rearrangement and amplification, integrating sequences from multiple chromosomal regions within tumor cells [137]. The junction sites of ecDNAs are randomly distributed around oncogenes, suggesting that the origin of the ecDNAs genome possesses random characteristics. During tumor evolution, ecDNAs gene fragments undergo fusion, rearrangement, and mutation, increasing gene diversity [36]. However, due to the lack of a recognizable centromere, ecDNAs and eccDNAs are both asymmetrically segregated by attaching to chromosome ends furthest from the mitotic spindle poles, resulting in a binomial random distribution or Gaussian distribution in daughter cells [102, 107]. Consequently, the uneven segregation leads to the accumulation of ecDNAs carrying oncogenes within cells, fostering the formation of heterogeneous tumor cell populations. These cells with an increased number of ecDNAs gain a survival advantage (Fig. 6) [176]. For the asymmetrical segregation of eccDNAs, our research team propose a theoretical hypothesis that eccDNAs cooperate with gastric cancer cells and facilitate gastric cancer progression in the context of genomic instability. The cancer cells produce massive eccDNAs carrying random genomic segments. EccDNAs will be asymmetrical segregated randomly to daughter cells as cell division. The host cells harboring ecDNAs carrying oncogene segments may grow faster or develop tolerance to environmental stress. This cyclic process helps cancer cells to overcome environmental impacts such as target treatment or immune attack, and eventually evolve to malignant tumors [107].
Fig. 6.
The mechanisms of action for eccDNAs in the invasion of neoplasms
Regulatory role of gene expression
EcDNAs increase the transcription level and promote overexpression of oncogenes by amplifying the copy number of oncogenes and carrying oncogenes with more accessible chromatin [136]. Meanwhile, ecDNAs carrying oncogenes have the ability to acting as trans-acting factors such as mobile super-enhancer [87, 177, 178]. In turn, this contributes to poor prognosis of patients with malignant tumors [178–180]. The dynamic changes of ecDNAs copy number will affect the expression level of the specific oncogenes, allowing adaptation to changing external environments [175]. EcDNAs promote tumor heterogeneity and accelerate genomic evolution by carrying and regulating drug-resistance-related genes, leading to drug resistance in tumor cells [181].
For eccDNAs carrying specific gene segments, they also regulate gene expression by transcribed into microRNAs and si-like RNAs [8]. Furthermore, the large amounts of eccDNAs generated during genome rearrangement can serve as templates for the transcription of long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs), thus regulating oncogene expression (Fig. 6) [182]. For example, ecDNAs can significantly increase the copy number of EGFR, MDM2, CDK4 and BRAF in tumor cells, participating in the regulation of PI3K/AKT pathway, p53 pathway and MAPK/ERK pathway [183, 184].
However, some eccDNAs carrying specific gene segments may also reduce the expression level of target genes [8]. For instance, eccDNAs enhance the expression of proto-oncogenes such as TERT by integrating with these genes, but the expression of tumor suppressor DCLK1 is inhibited by eccDNA integration [35]. This phenomenon indicated that eccDNA-derived rearrangements lead to the opposite effect of specific genes through oncogenic genome remodeling [35]. Therefore, although the precise mechanisms by which eccDNAs carrying specific gene segments enhancing or suppressing the expression levels of target genes are not yet fully understood, this issue warrants further attention.
Bystander effect
EccDNAs can also influence neoplasms development through bystander effect [185] and tumor progression signaling pathway. The bystander effect of eccDNAs is intricately linked to oxidative stress and the apoptotic cascade [185]: When patients with malignant tumors receive ionizing radiation, the radiated tumor cells undergo apoptosis and release oxidizing eccDNAs, which in turn activate the oxidative signaling pathways. These eccDNAs interact with adjacent cells, inducing secondary oxidative stress and a bystander effect. This interaction initiates an apoptotic cascade, resulting in the release of additional oxidizing eccDNAs (Fig. 6) [185].
Immunostimulatory activity
EccDNAs can regulate the secretion of proinflammatory cytokines through immunostimulatory activity [78]. EccDNAs that are rich in CpG islands can function as ligands for TLR9, thereby promoting the secretion of IL-6 and TNF-α by activating TLR9/MyD88/NF-κB signal pathway [78, 186]. In contrast, eccDNAs lacking CpG islands do not stimulate the secretion of pro-inflammatory cytokines [78, 186]. It's suggested that eccDNAs may act as a type of immunostimulatory DNA, triggering innate immunity via the activation of the cyclic GMP-AMP synthase-cytoplasmic DNA sensor STING pathway [187, 188]. Moreover, eccDNAs can induce higher levels of cytokine secretion compared to linear genomic DNA fragments of the same size. When eccDNAs are converted into linear genomic DNA fragments, they lose their ability to stimulate an immune response, indicating that their potent immunostimulatory capability is dependent on their circular structure (Fig. 6) [78].
The clinical implications of eccDNAs in human cancers and health
EccDNAs are widely identified in various human tissues and body fluids, including tumor tissues [107, 189], skeletal muscle, leukocytes [7], plasma [108, 111], urine [58], spermatozoa [59, 94] and other normal tissues. They play important roles in aging [190], genomic diversity [94], malignant tumorigenesis and drug resistance [189]. Previous studies have mainly focused on the roles of eccDNAs in malignant tumors. This review not only highlights the latest research progress of eccDNAs in neoplastic diseases, but also focuses on their clinical relevance in human health and non-neoplastic diseases. The aim is to inspire innovative approaches for the clinical application of eccDNAs in the prevention, diagnosis, and therapy of human health and diseases.
The biological functions of eccDNAs in human cancers
As is mentioned above, ecDNAs carrying intact oncogenes regulate gene expression by amplifying the copy number of oncogenes [136]. While eccDNAs carrying enhancer elements or genic segments regulate the expression of oncogenes by acting as a mobile super-enhancer [178–180] or transcribing into microRNAs, si-like RNAs [8], circRNAs and lncRNAs [182]. The biological functions of eccDNAs and ecDNAs lie in mediating neoplastic progression, serving as biomarkers and therapeutic targets, and promoting drug-resistance (Table 3).
Table 3.
The eccDNA/ecDNA-related genes and their functions in different cancers
| Neoplasms | EccDNA/ecDNA-related genes | Function |
|---|---|---|
| Bladder cancer | APOBEC3 [43] | Progression |
| Breast cancer | HER2 [222, 223]; ERBB2 [87]; MIR6748 [224] | Progression & Therapeutic target |
| FAT2, CTNNB1, CACNA2D2, CACNA1D [225] | Prognostic biomarker | |
| Cervical cancer | RAD54 [221] | Drug resistance |
| Colorectal cancer | DHFR [48, 218, 226]; HER2, EGFR [227, 228]; c-myb [229]; DNA-PKcs [217, 218] | Drug resistance |
| Gastric cancer | MIR433, MIR548w, MIR1206 [107] | Progression |
| Glioblastoma | EGFRvIII [87, 176, 230] | Progression & drug resistance |
| Hepatocellular carcinoma | SLC16A3, BAIAP2L2 [231]; MIR1792 [232] | Prognostic biomarker |
| Leukaemia | MYC, MYCN [195–197]; ATRX, DAXX [233]; APB [28] | Progression |
| Melanoma | BRAF [183]; LIG4 [17] | Drug resistance |
| Neuroblastoma | MYCN [177] | Progression |
| Non-small cell lung cancer | PLCG2 [203]; PDZRN3, LGR6 [204, 205] | Prognostic biomarker |
| Oral squamous cell carcinoma | RAB3B [189] | Drug resistance |
| Ovarian cancer | PIK3CA, MCL1, MYCN, DHFR, eIF-5A2 [10, 213, 234–236]; MAR [237] | Progression |
| Perihilar cholangiocarcinoma (pCCA) | ALDH3B2, RACGAP1, PDE4D, SCAPER, STX18 [147] | Prognostic biomarker |
| Prostate cancer | FAM84B [145] | Progression |
| Thyroid cancer | MIR1203 [238] | Non-invasive diagnostic Biomarker |
EccDNAs in neoplastic progression
EccDNAs and ecDNAs promote oncogene expansion and neoplastic progression by mediating oncogene transcription, altering the structure of regulatory elements, enhancing chromatin accessibility and inducing genome remodeling [35, 85]. Specifically, the MYC family of transcription factors, comprised of MYC and MYCN, are together the most commonly altered oncogenes [191] and play critical roles in tumorigenesis and progression in cancer [192]. EcDNAs actively regulate the expression level of MYC gene and the balance between oncogenes and tumor suppressor genes through genomic rearrangement and remodeling [193]. After the elimination of ecDNAs carrying segments of MYCN, the proliferation of tumor cells is significantly inhibited [194]. For instance, ecDNAs drives the amplification of MYCN copy number in neuroblastoma tissues and cell line IMR-32 [177]. While in acute myeloid leukemia, a large number of MYCN genes are amplified in the form of ecDNAs and circular chromosomes [195–197]. EcDNAs containing MYC family genes have also been detected in glioblastoma, colon cancer and ovarian cancer [198]. And they can promote MYC overexpression through the interaction of enhancer and gene through binding with exodomain protein BRD4 in colon cancer [198]. In addition, eccDNAs carrying segments of MYCN also drive the amplification of MYCN by regulating genome remodeling [35]. EccDNAs carrying promoter of MYCN can be integrated into MYCN gene sequences, which mediate the amplification of MYCN gene by genomic rearrangements in neuroblastoma [35] (Table 3).
EccDNAs as biomarkers for monitoring neoplasms
The level of extracellular nucleic acids in body fluids reflects the pathological processes of benign and malignant tumors and can be used as biomarkers for non-invasive diagnosis of malignant tumors, thus replacing classical invasive biopsy of tumor tissues in the future [199]. Due to the stable circular structure and the ability of resistance to digestion by exonuclease, eccDNAs have the potential to be used as biomarkers for monitoring malignant tumors [93]. For instance, there are significant differences in eccDNAs types between primary and metastatic high-grade serous ovarian cancer. The reduced copy number of eccDNA DNMT1Circle10302690−10302961 is significantly positively correlated with poor prognosis of patients with high-grade ovarian cancer, and has the potential to be used as a biomarker for monitoring this neoplasm [200]. Meanwhile, eccDNAs in preoperative plasma exhibit larger sizes compared with those in the plasma of patients who have undergone surgery for ovarian cancer [200]. Therefore, plasma eccDNAs can be used as a biomarker for non-invasive diagnosis and prognosis prediction of ovarian cancer [201]. However, due to the low abundance and the high heterogeneity of eccDNAs in plasma, the clinical application of plasma eccDNAs as biomarkers for gynaecological tumors is still challenging [200].
The abundance of eccDNAs have also potential as biomarkers of neoplastic prognosis and survival. Studies have found that the survival rate of patients with a high number of eccDNAs is significantly lower than that of patients without eccDNAs, and the total number of eccDNAs in malignant tumors is also significantly higher than that in benign tumors or non-neoplastic tissues [202]. Our research team confirmed that the total number of eccDNAs in gastric cancer tissues was significantly higher than that in adjacent normal tissues, and eccDNAs had functions of promoting tumor proliferation and invasion through transcription into functional miRNA to monitor the prognosis of patients with gastric cancer [107]. The eccDNAs carrying gene segments of PLCG2 [203], PDZRN3 and LGR6 [204, 205] in plasma and eccDNAs abundance in peripheral blood lymphocytes [206] has been proven to be an independent prognostic factor of lung cancer.
EccDNAs as therapeutic targets for malignant tumors
Some drug treatments can reduce the total number of eccDNAs or ecDNAs originating from malignant tumors. The development of eccDNAs-targeted drugs for reducing the expansion and expression of eccDNA/ecDNA-carrying oncogenes can provide a new strategy for the targeted treatment of malignant tumors, making eccDNAs or ecDNAs potential therapeutic targets for malignant tumors. Researchers have discovered that non-cytotoxic doses of hydroxyurea can reduce the total number of ecDNAs carrying oncogenes in some advanced ovarian cancer patients, thereby enhancing the efficacy of conventional chemotherapy [207]. In vitro therapeutic strategies that inhibit DNA replication, such as Hydroxyurea (HU), gecitabine and radiotherapy, can induce the generation of micronucleus and excrete amplified genes such as MYC and MYCN, demonstrating anti-cancer effectiveness in various malignancies, including leukemia [208, 209]. In neuroblastoma cells, exposure to low doses of HU results in morphological changes such as increased size, flattening, and granulation of cell lines containing eccDNAs, along with elevated expression of senescence-associated β-galactosidase, leading to reduced cell activity [210]. In addition, HU can eliminate the ecDNAs carrying oncogenes in the S phase of the mitotic interphase in colon cancer, but cannot eliminate the copy number of amplified genes in HSR [211]. Thus, the efficacy of HU is limited by the dynamic changes of ecDNAs and HSR.
Gemcitabine can facilitate the transport of the circular structure of ecDNAs into the micronucleus, where they are subsequently degraded in the ovarian cancer cell line UACC-1598. This process leads to the down-regulation of ecDNAs carrying oncogenes such as EIF5A2, MYCN, and MCL1, thereby reducing the proliferation and invasion capabilities of the UACC-1598 cell line [212]. In addition, ERK1/2 inhibitors significantly reduce the abundance of ecDNAs and the expression of oncogenes by inhibiting ERK1/2 activation in ovarian cancer cells [213] (Table 3). However, since the pharmacodynamic characteristics and anti-tumor efficacy of ecDNAs-targeted therapy are still not well understood, new strategies are needed to further reduce the occurrence of eccDNAs in malignant tumors during treatment.
EccDNAs in promoting drug-resistance of malignant tumors
Drug resistance poses a major challenge in the treatment of malignant tumors, often leading to the recurrence and metastasis [214]. EccDNAs and ecDNAs promote tumor resistance mechanisms by regulating drug resistance-related genes, asymmetric segregation, or metastasis. Schimke et al. first identified the association of ecDNAs with unstable amplification of the DHFR gene in methotrexate (MTX)-resistant cells [215, 216]. EcDNAs were generated by tumor cells exposed to MTX, while MTX deficiency reduced the copy number of DHFR gene in ecDNAs of drug-resistant cell lines, suggesting that the dynamic changes in ecDNAs influence chemotherapy response [48]. The dynamic conversion between ecDNAs carrying the oncogenic mutation BRAFV600E and HSR in melanoma cells also reflects the dynamic change of drug concentration, with ecDNAs carrying the BRAF gene increasing resistance to BRAF and MEK inhibitors in melanoma cells [183]. In addition, inhibition of NHEJ key protein DNA-PKcs can inhibit the generation of ecDNAs carrying DHFR, thereby increasing the sensitivity of colon cancer cells to MTX [217, 218]. HU can promote the degradation of ecDNAs carrying resistance genes in oral squamous cell carcinoma cells resistant to vincristine and MTX [219]. Therefore, eliminating ecDNAs carrying drug resistance genes is a crucial strategy for overcoming colon cancer resistance.
The total number of ecDNAs has a significant positive correlation with drug resistance of human cervical cancer cell line, and ecDNAs in peripheral blood can also predict the sensitivity of ovarian cancer to chemotherapy [220]. EcDNA promotes malignant tumor progression and drug resistance gene expression by increasing the intratumor heterogeneity of ovarian cancer and cervical cancer cells [220]. Excessive chromatin disruption can lead to continuous circular structural changes in ecDNAs, which increases the drug resistance of tumors [136]. The absence of HR key protein RAD54 leads to a significant increase in eccDNA content in MTX-resistant cervical cancer cells, and eccDNA-induced DHFR amplification may contribute to the generation of MTX drug resistance [221]. In pharyngeal squamous cell cancer cells, eccDNA carrying RAB3B gene induces autophagy through RAB3B amplification to promote the resistance of pharyngeal squamous cell carcinoma to cisplatin [189] (Table 3). Collectively, these studies suggest that eccDNAs and ecDNAs play a crucial role in tumor progression and drug resistance.
The clinical relevance of eccDNAs in different systems of human body
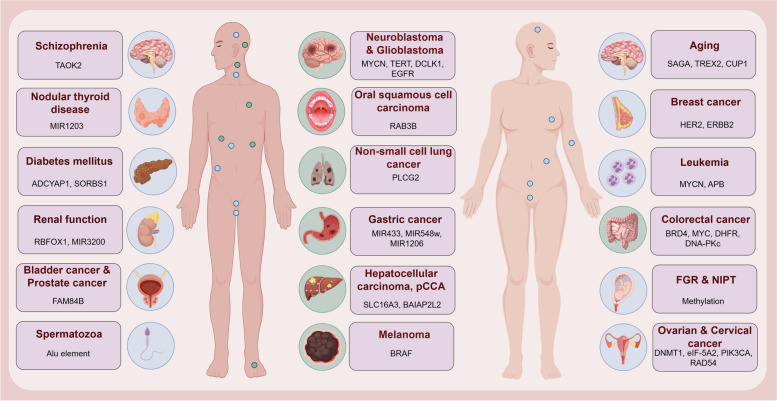
Recently, eccDNAs and ecDNAs have been shown to play essential roles in 16 types of tumor tissues and cells, such as gynecological tumors [234, 237], digestive system neoplasms [107], nervous system neoplasms [176]. Additionally, eccDNAs derived from spermatozoa and urine play essential roles in male reproductive system [59, 94] and human renal function health [58], respectively. Meanwhile, serum or plasma-derived eccDNAs not only have the potential as a biomarker of non-invasive prenatal examination, but also play essential roles in type 2 diabetes [111], gout [112] and bone immune inflammatory diseases [239]. Considering that human body as a complex system consisting of various subsystems, this review focused on the advanced research progress of eccDNAs’ roles in human cancers and health, categorized according to classifications of human systems (Fig. 7).
Fig. 7.
Schematic diagram of the biological functions and differentially expressed genes of eccDNAs or ecDNAs in different cancers and non-neoplastic diseases
Gynecologic cancers and reproductive health
Ovarian cancer ranks as the eighth most lethal gynecologic malignancy worldwide [240]. Unfortunately, approximately 70–80% of ovarian cancer patients are diagnosed with metastatic disease at the outset [241], commonly referred to as high-grade serous ovarian cancer [242]. Cervical cancer is among the most common cancers and is a leading cause of cancer-related deaths in women [243]. Notably, 87% of cervical cancer cases occur in developing countries, where it ranks as the third leading cause of cancer death among women [244]. In primary ovarian cancer tissue, ovarian cancer cell line UACC-1598 and cervical cancer cell line HeLa, eccDNAs carrying gene segments, such as PIK3CA, MCL1, MYCN, DHFR and eIF-5A2, can lead to the amplification of targeted oncogenes, resulting in the tumorigenesis and progression of gynecologic cancers [10, 213, 234–236]. Meanwhile, eukaryotic initiation factor eIF-5A2 is amplified by eccDNAs carrying enhancer in the ovarian cancer cell line UACC-1598, thereby promoting tumor proliferation and metastasis [234]. In addition, research has identified five matrix attachment regions (MARs) within a 682 kb eccDNA. These MARs enhance oncogene expression in ovarian cancer by binding to the nuclear matrix in vivo, as demonstrated by luciferase assays. Therefore, the overexpression of oncogenes is not solely achieved through oncogene amplification but also requires the co-amplification of MAR elements in the non-coding regions of eccDNAs [237].
Breast cancer is currently the most common cancer among women, and its prognosis depends not only on the stage at which the disease is detected but also on the specific type of breast cancer [245, 246]. In human epidermal growth factor receptor 2 (HER2) positive breast adenocarcinoma tissues, the amplification ratio of HER2 in eccDNAs and HSR were 30% and 60%, respectively [222, 223]. This suggests that eccDNAs promote breast cancer progression by amplifying HER2 copy numbers, thereby activating the MAPK and PI3K oncogene pathways [247–249]. EcDNAs carrying ERBB2 and enhancer also enhance the adaptability of breast cancer cells to the tumor microenvironment and promote the progression of breast cancer by amplifying this oncogene and its related enhancers [87]. Meanwhile, researchers have showed the potential of eccDNAs carrying gene segments of FAT2, CTNNB1, CACNA2D2 and CACNA1D as biomarkers of prognosis and overall survival in breast invasive carcinoma tissues, which might assist in clinical medical treatment [225]. EccDNA carrying MIR6748 gene could upregulate miR-6748 to promote the invasion and progression of breast cancer, which suppress the expression of tumor-suppressor gene TUSC5 [224]. EccDNAs containing functional genomic segments play a role in the initiation and progression of breast cancer [224]. These eccDNAs or ecDNAs offer a dynamic source of genomic plasticity and hold potential as biomarkers and therapeutic targets.
Non-invasive prenatal testing (NIPT) has been widely utilized to detect fetal chromatin and genetic abnormalities during pregnancy [250]. As the diversity and number of fetal DNA detection methods expand, the diagnostic applications of NIPT are progressively increasing. Researchers from Dennis Lo's group, known for pioneering NIPT, were the first to identify maternal and fetal eccDNAs in maternal plasma. They found that fetal eccDNAs are smaller in size compared to maternal eccDNAs, and the closed circular structure of eccDNAs provides greater resistance to exonuclease digestion [108]. Additionally, fetal eccDNAs exhibit significantly lower methylation levels than maternal eccDNAs, and this methylation level positively correlates with eccDNA size. Notably, fetal eccDNAs are rapidly cleared from the maternal body after delivery. Therefore, eccDNAs hold promise as diagnostic biomarkers for pregnancy-related conditions such as preeclampsia via NIPT [251].
Newborns with fetal growth restriction (FGR) have higher mortality. Early diagnosis of FGR during pregnancy is of great significance to the life and health of fetuses and newborns. Yang et al. have found that the total number of eccDNAs in the placenta of patients with FGR was significantly higher than that of normal individuals. These eccDNAs may contribute to FGR through interactions between immune signaling pathways and non-coding RNAs [252]. Therefore, further research is needed to explore the relationship between eccDNAs and prenatal-related diseases, as well as their application in NIPT.
Male urogenital cancers and health
Urologic cancers include cancers of the kidney, bladder, prostate and testes, with prostate cancer remaining a leading cause of male cancer death worldwide [253]. Researchers have found that eccDNAs carrying gene segments of FAM84B could significantly upregulated the expression of FAM84B in prostate cancer cells, thereby developing the growth and metastasis of prostate cancer [145]. FAM84B promoted by eccDNA have an ability to accelerate prostate cancer progression through mediating degradation of CDKN1B via MYC/WWP1 [145].
Urothelial carcinomas of the bladder are among the ten most prevalent malignancies worldwide, accounting for over 90% of all bladder cancer cases [254]. The invasive cystoscope and following biopsy pathology are the major methods for accurate diagnosis of bladder cancer [255]. The high detection rate (74%) of identifying eccDNAs in urine samples underscores the potential for non-invasive cancer-specific eccDNAs monitoring [43]. APOBEC3A and APOBEC3B are regarded as the two predominant APOBEC enzymes related to chromosomal mutagenesis. The increase of these 2 genes expression showed their roles in the clustered mutagenesis of eccDNAs in bladder cancer tissues [43]. Additionally, the expression of DNA repair genes, LIG3, POLQ and BRCA1/2, was significantly correlated with the eccDNAs levels, suggesting DNA repair mechanisms play an essential role in the generation of eccDNAs in bladder cancer [43].
Urinary cell-free eccDNAs (ucf-eccDNAs) serve as essential biomarker of renal function. The total number of ucf-eccDNAs is significantly elevated in advanced (grade 3–5) chronic kidney diseases (CKD). These ucf-eccDNAs carrying genic segments from gene-rich and LINE regions, typically less than 1000 base pairs in size, may significantly impact renal function through interactions with miRNA [58]. In addition, the total number of eccDNAs in peripheral blood of patients with end-stage renal disease (stage 5 CKD) was also markedly increased [256]. Therefore, eccDNAs derived from peripheral blood and urine have an important effect on renal function health.
Genetic diversity is mainly related to the mutation and recombination of germline cells, whereas eccDNAs contribute to genetic variation and genomic instability by amplifying or inserting into the genome [257]. Studies have identified a large number of eccDNAs in human spermatozoa, significantly impacting male reproductive health and genetic diversity [59, 94]. EccDNAs in germline cells mainly arise from gene fragment replication, leading to copy number variation and subsequent human genetic variation [258]. The generation of eccDNAs in spermatozoa is also linked to MMEJ [59]. Additionally, the production of large-sized eccDNAs in spermatozoa is inversely correlated with the rate of meiotic recombination [59]. These eccDNAs may integrate into the genome of gamete or zygote and become stably inherited by the offspring, thereby enhancing genomic diversity and potentially contributing to the rapid adaptive evolution of mammals [94]. However, for lack of transcription, translation, and DNA repair activity in spermatozoa [259], eccDNAs carrying gene segments or transposons may transiently transcribe and function in the zygote, potentially influencing early embryonic development [260]. Furthermore, the total number of spermatozoa-derived eccDNAs is significantly positively correlated with sperm motility [94], highlighting the need for further research into the role of eccDNAs in male infertility, such as asthenozoospermia.
Endocrine cancers and health
Nodular thyroid diseases are divided into benign nodular thyroid goitre and malignant papillary thyroid carcinoma. Preoperative ultrasonography was the major non-invasive diagnostic methods for the differentiation of malignant from benign goitrous nodules, but the accuracy should be improved by novel methods [261]. The total number of eccDNAs was elevated in papillary thyroid carcinoma than nodular thyroid goitre, and MIR1203 was the differential genes in malignant thyroid nodules [238]. The plasma eccDNAs might be the potential non-invasive biomarker for distinguishing benign or malignant thyroid tumor.
Type 2 diabetes mellitus (T2DM) is the most common endocrine disease in human beings, with the subcutaneous or intravenous insulin being the major therapeutic strategy [262]. It was found that the total number of eccDNAs in serum of patients with T2DM increased significantly after SIIT. This increase significantly affects the gene distribution of plasma eccDNAs, potentially influencing the therapeutic efficacy of SIIT through metabolic genes such as ADCYAP1 [111]. Therefore, eccDNAs carrying gene segments of ADCYAP1 in plasma have the potential as a biomarker to monitor the T2DM progression and therapeutic efficacy of SIIT in patients with T2DM [111]. In addition, eccDNA SORBS1circle97206791−97208025, derived from the insulin resistance gene SORBS1, shows a notable increase in copy number in the plasma of T2DM patients with insulin resistance and in HepG2 hepatocytes induced by high glucose and palmitate (HG/PA). These studies proved that eccDNAs carrying specific genes played crucial roles in disease diagnosis and surveillance [101].
Digestive cancers and health
Gastric cancer is the third most common cause of cancer-related death worldwide [263], accounting for almost 10% of all cancer deaths [264, 265]. Our research team have found that eccDNAs carrying MIR433, MIR548w and MIR1206 were the eccDNAs of interest in gastric cancer tissues [107]. Interestingly, these eccDNAs may contribute to the down-regulation of the Differentially Expressed Genes in gastric cancer. However, eccDNAs carrying functional genomic segments have been associated with the carcinogenesis of gastric cancer and show the potential to facilitate cancer progression. This implies that cancerous eccDNAs could function as a flexible reservoir for genomic plasticity and the swift adaptive evolution of cancer. Consequently, inhibiting the pathways involved in eccDNA generation might offer a new therapeutic approach for treating gastric cancer [107].
Hepatocellular carcinoma is the second leading cause of cancer-related death globally, accounts for approximately 90% of primary liver cancer cases [232]. EccDNA carrying MIR1792 suppress the expression of E-cadherin, PTEN, GSK3β, P21 and BIM through regulating MIR-17–92 in hepatocellular carcinoma. This type of eccDNA can significantly accelerate the proliferation and migration of hepatocellular carcinoma cell lines [232]. The copy number of eccDNAs carrying MIR1792 is an independent risk factor for overall survival and poor survival of patients with hepatocellular carcinoma [232]. Additionally, SLC16A3 and BAIAP2L2, which exhibit higher transcription levels in tumor tissues, were associated with eccDNAs and negatively correlated with survival rates in hepatocellular carcinoma [231]. EccDNA could be also detected in bile and plasma of perihilar cholangiocarcinoma (pCCA) patients, with a higher concentration. A prognostic model based on eccDNA-related genes, ALDH3B2, RACGAP1, PDE4D, SCAPER, STX18, showed the potential to predict the survival and immune microenvironment of patients with cholangiocarcinoma [147].
Acute liver failure is a rapidly progressive, life-threatening deterioration of liver function that results in altered neurological state and coagulopathy [266]. Our research team demonstrated that eccDNAs showed higher abundance and shorter in liver failure than healthy individuals. Additionally, eccDNA carrying ZMYM1 primarily regulated the PI3K-Akt and HIF-1 signaling pathways, thus mediating acute liver failure [267]. These findings suggest that liver failure-specific circulating extracellular vesicle-associated eccDNAs may exert diverse molecular effects on cells, indicating that some may accelerate the progression of liver failure by exerting systemic impacts on various organs.
Nervous system cancers and mental health
Glioblastoma is the most aggressive and deadliest form of glioma with a very limited prognosis, accounting for over 50% of all glioma cases [268]. EcDNAs carrying gene segments of specific oncogenes and their enhancers can encode and synergistically amplify epidermal growth factor receptor (EGFR) and other top 1% oncogenic genes, such as MYC, CDK4 and MDM2 [45], to promote the proliferation of glioblastoma [87], proving that ecDNAs can amplify oncogenes with higher copy number [14]. Although alterations in the EGFR are common in glioblastoma, numerous agents targeting EGFR or its signaling pathways have failed in glioblastoma clinical trials [269, 270]. EcDNAs carrying EGFR can also promote drug-resistance for targeted inhibitor and radiation therapy in glioblastoma. For instance, EGFRvIII mutated tumor cells are sensitive to EGFR tyrosine kinase inhibitors (TKIs). EccDNA-mediated expansion of oncogene EGFRvIII can eliminate mutated EGFR in exposure to TKIs in glioblastoma, thereby adapting to environmental changes and generating resistance to TKIs targeted therapy [176]. Researchers have found that the resistance to TKIs in glioblastoma patients was associated with a reduction in the proportion of EGFRvIIIHigh/ EGFRvIIILow tumor cells. The rapid increasing number of ecDNAs carrying EGFRvIII in the intervals of treatment provided a theoretical basis for high-dose EGFR TKI pulse intermittent therapy in glioblastoma patients [176]. Additionally, neoplastic cells harboring ecDNAs with the EGFR gene exhibit significantly increased radiation tolerance. Upon exposure to radiation, these neoplastic cells can shift their mode of respiration to oxidative phosphorylation and enhance neovascularization within the tumor microenvironment, thereby increasing their aggressiveness. Consequently, glioblastoma cells with ecDNAs demonstrate higher levels of aggressiveness and resistance to radiotherapy, potentially leading to tumor recurrence [230].
Plasma eccDNAs hold potential as a non-invasive biomarker for the diagnosis and monitoring of schizophrenia. Our research team discovered that the total number of eccDNAs in schizophrenia is significantly lower than in healthy individuals. What is more interesting is that eccDNAs carrying TAOK2 intronic fragments appear to inhibit the production of TAOK2 mRNA, thereby exerting a negative effect on schizophrenia [116]. However, whether eccDNAs carrying gene segments promote or suppress gene amplification are not fully understood, further studies will shed light on this issue in the future.
The deficiency and future prospect of eccDNAs
The clinical application of eccDNAs in human body fluids is under investigation to elucidate their biological characteristics in malignant tumors and overall human health. However, several challenges remain. Firstly, there are significant differences in the abundance of eccDNAs across different tissues. In certain diseases, eccDNA abundance is too low to be detected, and the difficulty in collecting clinical tissue samples limits their clinical application [2]. Secondly, eccDNAs data analysis methods require further optimization. Although Circle-Seq, based on RCA with random primers, is the most commonly used method for eccDNA detection at present, there is still a lack of reliable analytical techniques to accurately quantify eccDNA abundance [105]. The application of long-read sequencing presents considerable obstacles as a result of identifying multiple similar copies from a single eccDNA sequence [118]. Additionally, while HU therapy can eliminate ecDNAs and their associated oncogenes, it does not reduce oncogene amplification in HSR. This underscores the potential of ecDNA elimination as a therapeutic strategy for drug-resistant malignancies, yet effective methods to specifically target and eliminate ecDNAs are still lacking [181].
Besides, several important issues remain to be addressed: Firstly, transposons are ancestral traces of transposable elements past invasions, occupying nearly half of the human genome [271]. It has been proved that retrotransposons exploit the alternative end-joining DNA repair process for circularization and second-strand synthesis, essential for their replication and new insertions, with 90% resulting in the generation of eccDNAs [121]. Similarly, the biological roles of the tremendous amount of eccDNAs carrying transposable elements, such as Alu element, SINE, LINE, LTR and DNA transposons, are needed to be urgently addressed [87, 177]. Secondly, despite the biological functions of ecDNAs in malignant tumors have been extensively studied, the molecular function of small-sized eccDNAs carry gene segments still remain elusive. EccDNAs carrying segments of MYCN also drive the amplification of MYCN in neuroblastoma by regulating genome remodeling [35]. However, eccDNAs carrying TAOK2 intronic fragments suppress the expression of TAOK2 [116]. Therefore, further studies are needed to confirm the precise roles of eccDNAs carrying specific gene segments in either promoting or suppressing target gene expression. Moreover, the role of other types of eccDNAs, such as eccDNAs carrying the enhancer or repetitive elements, in human health and non-neoplastic diseases requires further exploration [272]. Additionally, the interactions between eccDNAs and other biological processes, such as epigenetic changes, DNA modifications, three-dimensional genome remodeling and histone modifications, need to be understood in greater detail [252]. Apart from these areas, it is also noteworthy to consider the potential interaction between piRNAs and eccDNAs containing transposable elements in germline cells. PiRNAs are a very large class of small RNAs present in germ line cells, particularly spermatozoa, and are believed to partial function to silence retrotransposons [273]. Recently, in has been proved that retrotransposon achieve its propagation by hijacking the alt-EJ DNA repair process of the host cells to form eccDNA [121]. Hence, the roles of eccDNAs carrying transposable elements and their methylation in interaction with piRNA is worthy of particular attention in the future. In summary, future research could benefit from a comprehensive mapping of the genomic characteristics of eccDNAs, which could advance our understanding of their roles in human health and disease.
Acknowledgements
The authors thank the OE Biotech CO. Ltd. (Shanghai, China) for supporting. The figures were created using Figdraw 2.0 on Home for Researchers. We thank the investigators whose studies have not yet been cited.
List of abbreviations
- eccDNA
Extrachromosomal circular DNAs
- bp
Base pairs
- Mb
Million base pairs
- spcDNA
Small polydispersed circular DNA
- ERC
Extrachromosomal ribosomal DNA circle
- ecDNA
Extrachromosomal DNA
- miRNA
MicroRNA
- ALT
Alternative lengthening of telomeres
- APB
ALT-associated promyelocytic leukemia body
- HR
Homologous recombination
- DSB
Double-strand breaks
- MMEJ
Microhomology-mediated end-joining
- NHEJ
Non-homologous end-joining
- MMR
Mismatch repair
- BER
Base excision repair
- NER
Nucleotide excision repair
- HU
Hydroxyurea
- CRISPR
Spaced clusters of short palindromic repeats
- XRCC
X-ray repair cross complementing
- DNA-PKc
DNA-dependent protein kinase
- BFB
Breakage-fusion-bridge
- PARP
Poly ADP-ribose polymerase
- HSR
Homologous stained regions
- LIG3
DNA ligase 3
- CsCl-EtBr
CsCl-ethidium bromide
- PSD
Plasmid-Safe DNase
- RCA
Rolling-circle amplification
- EOI
EccDNAs of interest
- cfDNA
Cell-free DNA
- HMW DNA
High-molecular weight DNA
- WGS
Whole genome sequencing
- NGS
Next-generation sequencing
- scEC&T-seq
Single-cell extrachromosomal circular DNA and transcriptome sequencing
- scGTP-seq
Single-cell paralleled genome and transcriptome sequencing
- EPM
Per million mappable reads
- LINE
Long interspersed elements
- SINE
Short interspersed elements
- LTR
Long terminal repeats
- SVA
SINE-VNTR-Alus
- UTR
Untranslated regions
- ucf-eccDNA
Urinary cell-free eccDNA
- T2DM
Type 2 diabetes mellitus
- SIIT
Short-term intensive insulin therapy
- LAMA
Ligase-assisted mini-circle accumulation
- CAES
Circle-seq based Artificial EccDNA Synthesis
- DHFR
Dihydrofolate reductase
- MTX
Methotrexate
- MAR
Matrix attachment regions
- HER2
Human epidermal growth factor receptor 2
- NIPT
Non-invasive prenatal testing
- FGR
Fetal growth restriction
- EGFR
Epidermal growth factor receptor
- TKI
Tyrosine kinase inhibitor
- pCCA
Perihilar cholangiocarcinoma
Authors’ contributions
Chunxiao He, Xi Xiang and Changze Song designed and conceptualized the study and wrote the article. Zilong Wang, Jiaying Yu and Wenli Zhu collected the documents and reviewed the manuscript. Zilong Wang, Xiaoning Hong, Zhen Xu, Lei Huang and Peng Han illustrated the images. Xi Xiang and Changze Song designed and conceptualized the study and wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the grants from Clinical Research Fund of the Seventh Affiliated Hospital, Sun Yat-sen University (ZSQYLCKYJJ202317), Shenzhen Medical Research Special Fund Project of Shenzhen Medical Academy of Research and Translation (A2301001), Shenzhen Science and Technology Innovation Commission (JCYJ20220530145014033 and JCYJ20230807110401003), the Research Start-up Fund of the Seventh Affiliated Hospital, Sun Yat-sen University (592026), Guangdong Provincial Key Laboratory of Digestive Cancer Research (2021B1212040006), the Open Fund of Guangdong Provincial Key Laboratory of Digestive Cancer Research (GPKLDCR202206M), Fundamental Research Funds for the Central Universities, Sun Yat-sen University (2023KYPT02) and the Lateral Research Project of Sun Yat-sen University (ZSQY-XQ-2309–0884).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zilong Wang, Jiaying Yu and Wenli Zhu contributed equally to this work and share first authorship.
Contributor Information
Chunxiao He, Email: hechx26@mail.sysu.edu.cn.
Changze Song, Email: songchz@mail.sysu.edu.cn.
Xi Xiang, Email: xiangx25@mail.sysu.edu.cn.
References
- 1.Wu N, Wei L, Zhu Z, Liu Q, Li K, Mao F, Qiao J, Zhao X. Innovative insights into extrachromosomal circular DNAs in gynecologic tumors and reproduction. Protein Cell. 2023;15(1):6-20. [DOI] [PMC free article] [PubMed]
- 2.Yang L, Jia R, Ge T, Ge S, Zhuang A, Chai P, Fan X. Extrachromosomal circular DNA: biogenesis, structure, functions and diseases. Signal Transduct Target Ther. 2022;7:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noer JB, Horsdal OK, Xiang X, Luo Y, Regenberg B. Extrachromosomal circular DNA in cancer: history, current knowledge, and methods. Trends Genet. 2022;38:766–81. [DOI] [PubMed] [Google Scholar]
- 6.Hotta Y. Bassel a: molecular size and circularity of dna in cells of mammals and higher plants. Proc Natl Acad Sci U S A. 1965;53:356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Halling JF, Plomgaard P, Maretty L, Hansen AJ, Snyder MP, Pilegaard H, et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. 2018;9:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulsen T, Shibata Y, Kumar P, Dillon L, Dutta A. Small extrachromosomal circular DNAs, microDNA, produce short regulatory RNAs that suppress gene expression independent of canonical promoters. Nucleic Acids Res. 2019;47:4586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling X, Han Y, Meng J, Zhong B, Chen J, Zhang H, Qin J, Pang J, Liu L. Small extrachromosomal circular DNA (eccDNA): major functions in evolution and cancer. Mol Cancer. 2021;20:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CA, Vinograd J. Small polydisperse circular DNA of HeLa cells. J Mol Biol. 1972;69:163–78. [DOI] [PubMed] [Google Scholar]
- 11.Gaubatz JW. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat Res. 1990;237:271–92. [DOI] [PubMed] [Google Scholar]
- 12.Basenko EY, Cesare AJ, Iyer S, Griffith JD, McEachern MJ. Telomeric circles are abundant in the stn1-M1 mutant that maintains its telomeres through recombination. Nucleic Acids Res. 2010;38:182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinclair DA, Guarente L. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell. 1997;91:1033–42. [DOI] [PubMed] [Google Scholar]
- 14.Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, Luebeck J, Rajkumar U, Diao Y, Li B, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575:699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen S, Regev A, Lavi S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene. 1997;14:977–85. [DOI] [PubMed] [Google Scholar]
- 16.Cohen S, Lavi S. Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol Cell Biol. 1996;16:2002–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen Z, Bacharach E, Lavi S. Mouse major satellite DNA is prone to eccDNA formation via DNA Ligase IV-dependent pathway. Oncogene. 2006;25:4515–24. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt H, Taubert H, Lange H, Kriese K, Schmitt WD, Hoffmann S, Bartel F, Hauptmann S. Small polydispersed circular DNA contains strains of mobile genetic elements and occurs more frequently in permanent cell lines of malignant tumors than in normal lymphocytes. Oncol Rep. 2009;22:393–400. [PubMed] [Google Scholar]
- 19.Xu J, McEachern MJ. Maintenance of very long telomeres by recombination in the Kluyveromyces lactis stn1-M1 mutant involves extreme telomeric turnover, telomeric circles, and concerted telomeric amplification. Mol Cell Biol. 2012;32:2992–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. 2009;16:1010–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 2003;194:155–62. [DOI] [PubMed] [Google Scholar]
- 22.Yu EY, Perez-Martin J, Holloman WK, Lue NF. Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient Ustilago maydis. Plos Genet. 2015;11:e1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fogli A, Demattei MV, Corset L, Vaurs-Barriere C, Chautard E, Biau J, Kemeny JL, Godfraind C, Pereira B, Khalil T, et al. Detection of the alternative lengthening of telomeres pathway in malignant gliomas for improved molecular diagnosis. J Neurooncol. 2017;135:381–90. [DOI] [PubMed] [Google Scholar]
- 24.Yu EY, Cheung IY, Feng Y, Rabie MO, Roboz GJ, Guzman ML, Cheung NV, Lue NF. Telomere trimming and DNA damage as signatures of high risk neuroblastoma. Neoplasia. 2019;21:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dagg RA, Pickett HA, Neumann AA, Napier CE, Henson JD, Teber ET, Arthur JW, Reynolds CP, Murray J, Haber M, et al. Extensive proliferation of human cancer cells with ever-shorter telomeres. Cell Rep. 2017;19:2544–56. [DOI] [PubMed] [Google Scholar]
- 26.Plantinga MJ, Pascarelli KM, Merkel AS, Lazar AJ, von Mehren M, Lev D, Broccoli D. Telomerase suppresses formation of ALT-associated single-stranded telomeric C-circles. Mol Cancer Res. 2013;11:557–67. [DOI] [PubMed] [Google Scholar]
- 27.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–9. [PubMed] [Google Scholar]
- 28.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansisidor A, Molinar TJ, Srivastava P, Dartis DD, Pino DA, Blitzblau HG, Klein H, Hochwagen A. Genomic copy-number loss is rescued by self-limiting production of DNA circles. Mol Cell. 2018;72:583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denoth-Lippuner A, Krzyzanowski MK, Stober C, Barral Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. Elife. 2014;3:e03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunisada T, Yamagishi H, Ogita Z, Kirakawa T, Mitsui Y. Appearance of extrachromosomal circular DNAs during in vivo and in vitro ageing of mammalian cells. Mech Ageing Dev. 1985;29:89–99. [DOI] [PubMed] [Google Scholar]
- 32.Hull RM, Houseley J. The adaptive potential of circular DNA accumulation in ageing cells. Curr Genet. 2020;66:889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karami FM, Akbari OM, Ramezani A, Barjoie MF, Khalesi B, Delazar S, Anjomrooz M, Taghizadeh A, Taghizadeh S, Payandeh Z, Pourzardosht N. Extra chromosomal DNA in different cancers: individual genome with important biological functions. Crit Rev Oncol Hematol. 2021;166:103477. [DOI] [PubMed] [Google Scholar]
- 34.Sanborn JZ, Salama SR, Grifford M, Brennan CW, Mikkelsen T, Jhanwar S, Katzman S, Chin L, Haussler D. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013;73:6036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koche RP, Rodriguez-Fos E, Helmsauer K, Burkert M, MacArthur IC, Maag J, Chamorro R, Munoz-Perez N, Puiggros M, Dorado GH, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat Genet. 2020;52:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, Nguyen NP, Turner K, Wu S, Gujar AD, Luebeck J, Liu J, Deshpande V, Rajkumar U, Namburi S, et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet. 2020;52:891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsen T, Malapati P, Shibata Y, Wilson B, Eki R, Benamar M, Abbas T, Dutta A. MicroDNA levels are dependent on MMEJ, repressed by c-NHEJ pathway, and stimulated by DNA damage. Nucleic Acids Res. 2021;49:11787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoshani O, Brunner SF, Yaeger R, Ly P, Nechemia-Arbely Y, Kim DH, Fang R, Castillon GA, Yu M, Li J, et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature. 2021;591:137–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dillon LW, Kumar P, Shibata Y, Wang YH, Willcox S, Griffith JD, Pommier Y, Takeda S, Dutta A. Production of extrachromosomal microDNAs is linked to mismatch repair pathways and transcriptional activity. Cell Rep. 2015;11:1749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moller HD, Parsons L, Jorgensen TS, Botstein D, Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci U S A. 2015;112:E3114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie C, Wang W, Tu C, Meng L, Lu G, Lin G, Lu LY, Tan YQ. Meiotic recombination: insights into its mechanisms and its role in human reproduction with a special focus on non-obstructive azoospermia. Hum Reprod Update. 2022;28:763–97. [DOI] [PubMed] [Google Scholar]
- 42.Olivieri M, Cho T, Alvarez-Quilon A, Li K, Schellenberg MJ, Zimmermann M, Hustedt N, Rossi SE, Adam S, Melo H, et al. A genetic map of the response to DNA damage in human cells. Cell. 2020;182:481–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lv W. Extrachromosomal circular DNA orchestrates genome heterogeneity in urothelial bladder carcinoma. Theranostics. 2024;14:5102–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–24. [DOI] [PubMed] [Google Scholar]
- 45.Vogt N, Lefevre SH, Apiou F, Dutrillaux AM, Cor A, Leuraud P, Poupon MF, Dutrillaux B, Debatisse M, Malfoy B. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci U S A. 2004;101:11368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Loon N, Miller D, Murnane JP. Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res. 1994;22:2447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.L’Abbate A, Macchia G, D’Addabbo P, Lonoce A, Tolomeo D, Trombetta D, Kok K, Bartenhagen C, Whelan CW, Palumbo O, et al. Genomic organization and evolution of double minutes/homogeneously staining regions with MYC amplification in human cancer. Nucleic Acids Res. 2014;42:9131–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng X, Qi X, Guo H, Cai M, Li C, Zhu J, Chen F, Guo H, Li J, Zhao Y, et al. Novel role for non-homologous end joining in the formation of double minutes in methotrexate-resistant colon cancer cells. J Med Genet. 2015;52:135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manjon E, Edreira T, Munoz S, Sanchez Y. Rgf1p (Rho1p GEF) is required for double-strand break repair in fission yeast. Nucleic Acids Res. 2017;45:5269–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao K, Ma Y, Li Y, Shen X, Zhao J, Tian S, Zhang C, Liang C, Zhao Z, Yang Y, et al. A di-acetyl-decorated chromatin signature couples liquid condensation to suppress DNA end synapsis. Mol Cell. 2024;84:1206–23. [DOI] [PubMed] [Google Scholar]
- 51.Corbeski I, Guo X, Eckhardt BV, Fasci D, Wiegant W, Graewert MA, Vreeken K, Wienk H, Svergun DI, Heck A, et al. Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF. Sci Adv. 2022;8:o517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corbeski I, Dolinar K, Wienk H, Boelens R, van Ingen H. DNA repair factor APLF acts as a H2A–H2B histone chaperone through binding its DNA interaction surface. Nucleic Acids Res. 2018;46:7138–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nemoz C, Ropars V, Frit P, Gontier A, Drevet P, Yu J, Guerois R, Pitois A, Comte A, Delteil C, et al. XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat Struct Mol Biol. 2018;25:971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B, Reddy S, Comai L. Depletion of Ku70/80 reduces the levels of extrachromosomal telomeric circles and inhibits proliferation of ALT cells. Aging (Albany NY). 2011;3:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moller HD, Lin L, Xiang X, Petersen TS, Huang J, Yang L, Kjeldsen E, Jensen UB, Zhang X, Liu X, et al. CRISPR-C: circularization of genes and chromosome by CRISPR in human cells. Nucleic Acids Res. 2018;46:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iyer S, Suresh S, Guo D, Daman K, Chen J, Liu P, Zieger M, Luk K, Roscoe BP, Mueller C, et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature. 2019;568:561–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez-Galvez G, Joshi P, Friedberg I, Manduca A, Ekker SC. Deploying MMEJ using MENdel in precision gene editing applications for gene therapy and functional genomics. Nucleic Acids Res. 2021;49:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lv W, Pan X, Han P, Wang Z, Feng W, Xing X, Wang Q, Qu K, Zeng Y, Zhang C, et al. Circle-Seq reveals genomic and disease-specific hallmarks in urinary cell-free extrachromosomal circular DNAs. Clin Transl Med. 2022;12:e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu J, Zhang Z, Xiao S, Cao Y, Chen Y, Weng J, Jiang H, Li W, Chen JY, Liu C. Microhomology-mediated circular DNA formation from oligonucleosomal fragments during spermatogenesis. Elife. 2023;12:RP87115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fleury H, MacEachern MK, Stiefel CM, Anand R, Sempeck C, Nebenfuehr B, Maurer-Alcala K, Ball K, Proctor BR, Belan O, et al. The APE2 nuclease is essential for DNA double-strand break repair by microhomology-mediated end joining. Mol Cell. 2023;83:1429–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hull RM, King M, Pizza G, Krueger F, Vergara X, Houseley J. Transcription-induced formation of extrachromosomal DNA during yeast ageing. Plos Biol. 2019;17:e3000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moller HD, Ramos-Madrigal J, Prada-Luengo I, Gilbert M, Regenberg B. Near-random distribution of chromosome-derived circular DNA in the condensed genome of pigeons and the larger, more repeat-rich human genome. Genome Biol Evol. 2020;12:3762–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rivera T, Haggblom C, Cosconati S, Karlseder J. A balance between elongation and trimming regulates telomere stability in stem cells. Nat Struct Mol Biol. 2017;24:30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Compton SA, Choi JH, Cesare AJ, Ozgur S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–9. [DOI] [PubMed] [Google Scholar]
- 65.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakkas D, Alvarez JG. Sperm DNA fragmentation: mechanisms of origin, impact on reproductive outcome, and analysis. Fertil Steril. 2010;93:1027–36. [DOI] [PubMed] [Google Scholar]
- 67.Coban O, Serdarogullari M, Yarkiner Z, Serakinci N. Investigating the level of DNA double-strand break in human spermatozoa and its relation to semen characteristics and IVF outcome using phospho-histone H2AX antibody as a biomarker. Andrology-Us. 2020;8:421–6. [DOI] [PubMed] [Google Scholar]
- 68.Fujimori H, Hyodo M, Matsuno Y, Shimizu A, Minakawa Y, Atsumi Y, Nakatsu Y, Tsuzuki T, Murakami Y, Yoshioka KI. Mismatch repair dependence of replication stress-associated DSB recognition and repair. Heliyon. 2019;5:e3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen W, Jinks-Robertson S. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol Cell Biol. 1998;18:6525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paulsen T, Kumar P, Koseoglu MM, Dutta A. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 2018;34:270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Min J, Wright WE, Shay JW. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019;33:814–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noordermeer SM, Adam S, Setiaputra D, Barazas M, Pettitt SJ, Ling AK, Olivieri M, Alvarez-Quilon A, Moatti N, Zimmermann M, et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao X, Wang X, Hu X, Liu Z, Liu J, Zhou H, Shen X, Wei Y, Huang Z, Ying W, et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017;27:801–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murnane JP. Telomere dysfunction and chromosome instability. Mutat Res. 2012;730:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murnane JP, Sabatier L. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. BioEssays. 2004;26:1164–74. [DOI] [PubMed] [Google Scholar]
- 77.Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Wang M, Djekidel MN, Chen H, Liu D, Alt FW, Zhang Y. eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature. 2021;599:308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cortes-Ciriano I, Lee JJ, Xi R, Jain D, Jung YL, Yang L, Gordenin D, Klimczak LJ, Zhang CZ, Pellman DS, Park PJ. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 2020;52:331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Umbreit NT, Zhang CZ, Lynch LD, Blaine LJ, Cheng AM, Tourdot R, Sun L, Almubarak HF, Judge K, Mitchell TJ, et al. Mechanisms generating cancer genome complexity from a single cell division error. Science. 2020;368:eaba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ly P, Cleveland DW. Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol. 2017;27:917–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo T, Chen GQ, Li XF, Wang M, Liu KM, Yang XY, Liu SC, Feng YL, Liu PY, Lin H, Xie AY. Small extrachromosomal circular DNA harboring targeted tumor suppressor gene mutations supports intratumor heterogeneity in mouse liver cancer induced by multiplexed CRISPR/Cas9. Genome Med. 2023;15:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verhaak R, Bafna V, Mischel PS. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer. 2019;19:283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yi E, Chamorro GR, Henssen AG, Verhaak R. Extrachromosomal DNA amplifications in cancer. Nat Rev Genet. 2022;23:760–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Helmsauer K, Valieva ME, Ali S, Chamorro GR, Schopflin R, Roefzaad C, Bei Y, Dorado GH, Rodriguez-Fos E, Puiggros M, et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat Commun. 2020;11:5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, Allan KC, Mack SC, Wang X, Gimple RC, et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell. 2019;179:1330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, Vermeesch JR, Stul M, Dutta B, Boeckx N, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–9. [DOI] [PubMed] [Google Scholar]
- 89.Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol Cell. 1998;2:259–65. [DOI] [PubMed] [Google Scholar]
- 90.Storlazzi CT, Lonoce A, Guastadisegni MC, Trombetta D, D’Addabbo P, Daniele G, L’Abbate A, Macchia G, Surace C, Kok K, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20:1198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carroll SM, DeRose ML, Gaudray P, Moore CM, Needham-Vandevanter DR, Von Hoff DD, Wahl GM. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol Cell Biol. 1988;8:1525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hung KL, Luebeck J, Dehkordi SR, Colon CI, Li R, Wong IT, Coruh C, Dharanipragada P, Lomeli SH, Weiser NE, et al. Targeted profiling of human extrachromosomal DNA by CRISPR-CATCH. Nat Genet. 2022;54:1746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar P, Dillon LW, Shibata Y, Jazaeri AA, Jones DR, Dutta A. Normal and cancerous tissues release extrachromosomal circular DNA (eccDNA) into the circulation. Mol Cancer Res. 2017;15:1197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Henriksen RA, Jenjaroenpun P, Sjostrom IB, Jensen KR, Prada-Luengo I, Wongsurawat T, Nookaew I, Regenberg B. Circular DNA in the human germline and its association with recombination. Mol Cell. 2022;82:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cohen S, Segal D. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res. 2009;124:327–38. [DOI] [PubMed] [Google Scholar]
- 96.Lukaszewicz A, Lange J, Keeney S, Jasin M. De novo deletions and duplications at recombination hotspots in mouse germlines. Cell. 2021;184:5970–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev. 2013;23:147–55. [DOI] [PubMed] [Google Scholar]
- 98.Henderson IR, Bomblies K. Evolution and plasticity of genome-wide meiotic recombination rates. Annu Rev Genet. 2021;55:23–43. [DOI] [PubMed] [Google Scholar]
- 99.Udugama M, Vinod B, Chan FL, Hii L, Garvie A, Collas P, Kalitsis P, Steer D, Das PP, Tripathi P, et al. Histone H3.3 phosphorylation promotes heterochromatin formation by inhibiting H3K9/K36 histone demethylase. Nucleic Acids Res. 2022;50:4500–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cox D, Yuncken C. Spriggs AI: minute chromatin bodies in malignant tumours of childhood. Lancet. 1965;1:55–8. [DOI] [PubMed] [Google Scholar]
- 101.Kong X, Wan SJ, Chen TB, Jiang L, Xing YJ, Bai YP, Hua Q, Yao XM, Zhao YL, Zhang HM, et al. Increased serum extrachromosomal circular DNA SORBS1(circle) level is associated with insulin resistance in patients with newly diagnosed type 2 diabetes mellitus. Cell Mol Biol Lett. 2024;29:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yi E, Gujar AD, Guthrie M, Kim H, Zhao D, Johnson KC, Amin SB, Costa ML, Yu Q, Das S, et al. Live-cell imaging shows uneven segregation of extrachromosomal DNA elements and transcriptionally active extrachromosomal DNA hubs in cancer. Cancer Discov. 2022;12:468–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Radloff R, Bauer W, Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967;57:1514–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shoura MJ, Gabdank I, Hansen L, Merker J, Gotlib J, Levene SD, Fire AZ. Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 (Bethesda). 2017;7:3295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moller HD. Circle-Seq: Isolation and sequencing of chromosome-derived circular DNA elements in cells. Methods Mol Biol. 2020;2119:165–81. [DOI] [PubMed] [Google Scholar]
- 106.Wang Y, Wang M, Zhang Y. Purification, full-length sequencing and genomic origin mapping of eccDNA. Nat Protoc. 2023;18:683–99. [DOI] [PubMed] [Google Scholar]
- 107.Jiang X, Pan X, Li W, Han P, Yu J, Li J, Zhang H, Lv W, Zhang Y, He Y, Xiang X. Genome-wide characterization of extrachromosomal circular DNA in gastric cancer and its potential role in carcinogenesis and cancer progression. Cell Mol Life Sci. 2023;80:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sin S, Jiang P, Deng J, Ji L, Cheng SH, Dutta A, Leung TY, Chan K, Chiu R, Lo Y. Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proc Natl Acad Sci U S A. 2020;117:1658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moller HD, Bojsen RK, Tachibana C, Parsons L, Botstein D, Regenberg B. Genome-wide purification of extrachromosomal circular DNA from Eukaryotic Cells. J Vis Exp. 2016;110:e54239. [DOI] [PMC free article] [PubMed]
- 110.Yu J, Zhang H, Han P, Jiang X, Li J, Li B, Yang S, He C, Mao S, Dang Y, Xiang X. Circle-seq based method for eccDNA synthesis and its application as a canonical promoter independent vector for robust microRNA overexpression. Comput Struct Biotechnol J. 2024;23:358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu Z, He J, Han P, Dai P, Lv W, Liu N, Liu L, Liu L, Pan X, Xiang X, et al. Plasma extrachromosomal circular DNA is a pathophysiological hallmark of short-term intensive insulin therapy for type 2 diabetes. Clin Transl Med. 2023;13:e1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pang J, Pan X, Lin L, Li L, Yuan S, Han P, Ji X, Li H, Wang C, Chu Z, et al. Characterization of plasma extrachromosomal circular DNA in gouty arthritis. Front Genet. 2022;13:859513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Matelska D, Steczkiewicz K, Ginalski K. Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res. 2017;45:6995–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miyazono KI, Wang D, Ito T, Tanokura M. Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI. Nucleic Acids Res. 2020;48:5106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moller HD, Larsen CE, Parsons L, Hansen AJ, Regenberg B, Mourier T. Formation of extrachromosomal circular DNA from long terminal repeats of retrotransposons in saccharomyces cerevisiae. Ge (Bethesda). 2015;6:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xiang X, Pan X, Lv W, Chen S, Li J, Zhang H, Liao Y, Yu J, Li J, Dang Y, et al. Identification and functional analysis of circulating extrachromosomal circular DNA in schizophrenia implicate its negative effect on the disorder. Clin Transl Med. 2023;13:e1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan X, Yang C, Li W, Bai X, Zhou X, Xie H, Wen L, Tang F. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021;22:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gao X, Liu K, Luo S, Tang M, Liu N, Jiang C, Fang J, Li S, Hou Y, Guo C, Qu K. Comparative analysis of methodologies for detecting extrachromosomal circular DNA. Nat Commun. 2024;15:9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.DeCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, Koeman J, Seth S, Protopopov A, Felicella M, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet. 2018;50:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu K, Ding L, Chang TC, Shao Y, Chiang J, Mulder H, Wang S, Shaw TI, Wen J, Hover L, et al. Structure and evolution of double minutes in diagnosis and relapse brain tumors. Acta Neuropathol. 2019;137:123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang F, Su W, Chung OW, Tracy L, Wang L, Ramsden DA, Zhang Z. Retrotransposons hijack alt-EJ for DNA replication and eccDNA biogenesis. Nature. 2023;620:218–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wanchai V, Jenjaroenpun P, Leangapichart T, Arrey G, Burnham CM, Tummler MC, Delgado-Calle J, Regenberg B, Nookaew I. CReSIL: accurate identification of extrachromosomal circular DNA from long-read sequences. Brief Bioinform. 2022;23:bbac422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kumar P, Kiran S, Saha S, Su Z, Paulsen T, Chatrath A, Shibata Y, Shibata E, Dutta A. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv. 2020;6:a2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mehta D, Cornet L, Hirsch-Hoffmann M, Zaidi SS, Vanderschuren H. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq. Nat Protoc. 2020;15:1673–89. [DOI] [PubMed] [Google Scholar]
- 125.Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, Satpathy AT, Carter AC, Ghosh RP, East-Seletsky A, et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods. 2016;13:1013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lahnemann D, Koster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, Vallejos CA, Campbell KR, Beerenwinkel N, Mahfouz A, et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kang J, Dai Y, Li J, Fan H, Zhao Z. Investigating cellular heterogeneity at the single-cell level by the flexible and mobile extrachromosomal circular DNA. Comput Struct Biotechnol J. 2023;21:1115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang R, Yang M, Zhang S, Huang M. Advances in sequencing-based studies of microDNA and ecDNA: Databases, identification methods, and integration with single-cell analysis. Comput Struct Biotechnol J. 2023;21:3073–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Spain L, Coulton A, Lobon I, Rowan A, Schnidrig D, Shepherd S, Shum B, Byrne F, Goicoechea M, Piperni E, et al. Late-stage metastatic melanoma emerges through a diversity of evolutionary pathways. Cancer Discov. 2023;13:1364–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen JP, Diekmann C, Wu H, Chen C, Della CG, Berrino E, Georgiadis KL, Bouwman B, Virdi M, Harbers L, et al. scCircle-seq unveils the diversity and complexity of extrachromosomal circular DNAs in single cells. Nat Commun. 2024;15:1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Disentangling extrachromosomal circular DNA heterogeneity in single cells with scEC&T-seq. Nat Genet. 2023;55:740–1. 10.1038/s41588-023-01385-z [DOI] [PubMed]
- 132.Chamorro GR, Conrad T, Stober MC, Xu R, Giurgiu M, Rodriguez-Fos E, Kasack K, Bruckner L, van Leen E, Helmsauer K, et al. Parallel sequencing of extrachromosomal circular DNAs and transcriptomes in single cancer cells. Nat Genet. 2023;55:880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sanders AD, Falconer E, Hills M, Spierings D, Lansdorp PM. Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat Protoc. 2017;12:1151–76. [DOI] [PubMed] [Google Scholar]
- 134.Sanders AD, Meiers S, Ghareghani M, Porubsky D, Jeong H, van Vliet M, Rausch T, Richter-Pechanska P, Kunz JB, Jenni S, et al. Single-cell analysis of structural variations and complex rearrangements with tri-channel processing. Nat Biotechnol. 2020;38:343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chang L, Deng E, Wang J, Zhou W, Ao J, Liu R, Su D, Fan X. Single-cell third-generation sequencing-based multi-omics uncovers gene expression changes governed by ecDNA and structural variants in cancer cells. Clin Transl Med. 2023;13: e1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lange JT, Rose JC, Chen CY, Pichugin Y, Xie L, Tang J, Hung KL, Yost KE, Shi Q, Erb ML, et al. The evolutionary dynamics of extrachromosomal DNA in human cancers. Nat Genet. 2022;54:1527–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Deshpande V, Luebeck J, Nguyen ND, Bakhtiari M, Turner KM, Schwab R, Carter H, Mischel PS, Bafna V. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun. 2019;10:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Prada-Luengo I, Krogh A, Maretty L, Regenberg B. Sensitive detection of circular DNAs at single-nucleotide resolution using guided realignment of partially aligned reads. BMC Bioinformatics. 2019;20:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Su Z, Saha S, Paulsen T, Kumar P, Dutta A. ATAC-Seq-based identification of extrachromosomal circular DNA in Mammalian cells and its validation using inverse PCR and FISH. Bio Protoc. 2021;11:e4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mann L, Seibt KM, Weber B, Heitkam T. ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data. BMC Bioinformatics. 2022;23:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang P, Peng H, Llauro C, Bucher E, Mirouze M. ecc_finder: a robust and accurate tool for detecting extrachromosomal circular DNA from sequencing data. Front Plant Sci. 2021;12:743742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhao X, Shi L, Ruan S, Bi W, Chen Y, Chen L, Liu Y, Li M, Qiao J, Mao F. CircleBase: an integrated resource and analysis platform for human eccDNAs. Nucleic Acids Res. 2022;50:D72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peng L, Zhou N, Zhang CY, Li GC, Yuan XQ. eccDNAdb: a database of extrachromosomal circular DNA profiles in human cancers. Oncogene. 2022;41:2696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhong T, Wang W, Liu H, Zeng M, Zhao X, Guo Z. eccDNA Atlas: a comprehensive resource of eccDNA catalog. Brief Bioinform. 2023;24:bbad037. [DOI] [PubMed] [Google Scholar]
- 145.Jin W, Xu Z, Song Y, Chen F. Extrachromosomal circular DNA promotes prostate cancer progression through the FAM84B/CDKN1B/MYC/WWP1 axis. Cell Mol Biol Lett. 2024;29:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bao Y, Sui X, Wang X, Qu N, Xie Y, Cong Y, Cao X. Extrachromosomal circular DNA landscape of breast cancer with lymph node metastasis. Int J Cancer. 2024;155:756–65. [DOI] [PubMed] [Google Scholar]
- 147.Fu S, Dai Y, Zhang P, Zheng K, Cao G, Xu L, Zhong Y, Niu C, Wang X. Extrachromosomal circular DNA (eccDNA) characteristics in the bile and plasma of advanced perihilar cholangiocarcinoma patients and the construction of an eccDNA-related gene prognosis model. Front Cell Dev Biol. 2024;12:1379435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Luo X, Zhang L, Cui J, An Q, Li H, Zhang Z, Sun G, Huang W, Li Y, Li C, et al. Small extrachromosomal circular DNAs as biomarkers for multi-cancer diagnosis and monitoring. Clin Transl Med. 2023;13:e1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Escobar TM, Oksuz O, Saldana-Meyer R, Descostes N, Bonasio R, Reinberg D. Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA replication. Cell. 2019;179:953–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zasadzinska E, Huang J, Bailey AO, Guo LY, Lee NS, Srivastava S, Wong KA, French BT, Black BE, Foltz DR. Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP. Dev Cell. 2018;47:348–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tekel SJ, Haynes KA. Molecular structures guide the engineering of chromatin. Nucleic Acids Res. 2017;45:7555–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.De Braganca S, Aicart-Ramos C, Arribas-Bosacoma R, Rivera-Calzada A, Unfried JP, Prats-Mari L, Marin-Baquero M, Fortes P, Llorca O, Moreno-Herrero F. APLF and long non-coding RNA NIHCOLE promote stable DNA synapsis in non-homologous end joining. Cell Rep. 2023;42:111917. [DOI] [PubMed] [Google Scholar]
- 153.Talbert PB, Henikoff S. Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Biol. 2017;18:115–26. [DOI] [PubMed] [Google Scholar]
- 154.Hao F, Murphy KJ, Kujirai T, Kamo N, Kato J, Koyama M, Okamato A, Hayashi G, Kurumizaka H, Hayes JJ. Acetylation-modulated communication between the H3 N-terminal tail domain and the intrinsically disordered H1 C-terminal domain. Nucleic Acids Res. 2020;48:11510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Adhireksan Z, Sharma D, Lee PL, Davey CA. Near-atomic resolution structures of interdigitated nucleosome fibres. Nat Commun. 2020;11:4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Navratilova A, Koblizkova A, Macas J. Survey of extrachromosomal circular DNA derived from plant satellite repeats. Bmc Plant Biol. 2008;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yehuda Y, Blumenfeld B, Mayorek N, Makedonski K, Vardi O, Cohen-Daniel L, Mansour Y, Baror-Sebban S, Masika H, Farago M, et al. Germline DNA replication timing shapes mammalian genome composition. Nucleic Acids Res. 2018;46:8299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tristan-Ramos P, Rubio-Roldan A, Peris G, Sanchez L, Amador-Cubero S, Viollet S, Cristofari G, Heras SR. The tumor suppressor microRNA let-7 inhibits human LINE-1 retrotransposition. Nat Commun. 2020;11:5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lu JY, Chang L, Li T, Wang T, Yin Y, Zhan G, Han X, Zhang K, Tao Y, Percharde M, et al. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. 2021;31:613–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weltner J, Balboa D, Katayama S, Bespalov M, Krjutskov K, Jouhilahti EM, Trokovic R, Kere J, Otonkoski T. Human pluripotent reprogramming with CRISPR activators. Nat Commun. 2018;9:2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee CH, Shih YP, Ho MR, Wang AH. The C-terminal D/E-rich domain of MBD3 is a putative Z-DNA mimic that competes for Zalpha DNA-binding activity. Nucleic Acids Res. 2018;46:11806–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brar GA, Hochwagen A, Ee LS, Amon A. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol Biol Cell. 2009;20:1030–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–8. [DOI] [PubMed] [Google Scholar]
- 164.Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen CF, Thomas MA, Haussler D, Jacob HJ. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–7. [DOI] [PubMed] [Google Scholar]
- 166.Morton NE. Parameters of the human genome. Proc Natl Acad Sci U S A. 1991;88:7474–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kesselring L, Miskey C, Zuliani C, Querques I, Kapitonov V, Lauko A, Feher A, Palazzo A, Diem T, Lustig J, et al. A single amino acid switch converts the Sleeping Beauty transposase into an efficient unidirectional excisionase with utility in stem cell reprogramming. Nucleic Acids Res. 2020;48:316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pace JN, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Blundell-Hunter G, Tellier M, Chalmers R. Transposase subunit architecture and its relationship to genome size and the rate of transposition in prokaryotes and eukaryotes. Nucleic Acids Res. 2018;46:9637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cortes-Ciriano I, Lee S, Park WY, Kim TM, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017;8:15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Du Q, Kotlyar A, Vologodskii A. Kinking the double helix by bending deformation. Nucleic Acids Res. 2008;36:1120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yu J, Xiang X, Huang J, Liang X, Pan X, Dong Z, Petersen TS, Qu K, Yang L, Zhao X, et al. Haplotyping by CRISPR-mediated DNA circularization (CRISPR-hapC) broadens allele-specific gene editing. Nucleic Acids Res. 2020;48:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A. 2010;107:18551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Karami FM, Karimfar N, Fazlollahpour NA, Shafa S, Ghasemi SM, Ataei M, Dehghanzadeh H, Nabi AM, Ghadiri T, Payandeh Z, Tarhriz V. Revisiting characteristics of oncogenic extrachromosomal DNA as mobile enhancers on neuroblastoma and glioma cancers. Cancer Cell Int. 2022;22:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhu Y, Gujar AD, Wong CH, Tjong H, Ngan CY, Gong L, Chen YA, Kim H, Liu J, Li M, et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell. 2021;39:694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhu Y, Gong L, Wei CL. Guilt by association: EcDNA as a mobile transactivator in cancer. Trends Cancer. 2022;8:747–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shimizu N. Gene amplification and the extrachromosomal circular DNA. Genes (Basel). 2021;12:1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gu X, Yu J, Chai P, Ge S, Fan X. Novel insights into extrachromosomal DNA: redefining the onco-drivers of tumor progression. J Exp Clin Cancer Res. 2020;39:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yerlici VT, Lu MW, Hoge CR, Miller RV, Neme R, Khurana JS, Bracht JR, Landweber LF. Programmed genome rearrangements in Oxytricha produce transcriptionally active extrachromosomal circular DNA. Nucleic Acids Res. 2019;47:9741–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Song K, Minami JK, Huang A, Dehkordi SR, Lomeli SH, Luebeck J, Goodman MH, Moriceau G, Krijgsman O, Dharanipragada P, et al. Plasticity of extrachromosomal and intrachromosomal BRAF amplifications in overcoming targeted therapy dosage challenges. Cancer Discov. 2022;12:1046–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xia W, Jie W. ZEB1-AS1/miR-133a-3p/LPAR3/EGFR axis promotes the progression of thyroid cancer by regulating PI3K/AKT/mTOR pathway. Cancer Cell Int. 2020;20:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ermakov AV, Konkova MS, Kostyuk SV, Egolina NA, Efremova LV, Veiko NN. Oxidative stress as a significant factor for development of an adaptive response in irradiated and nonirradiated human lymphocytes after inducing the bystander effect by low-dose X-radiation. Mutat Res. 2009;669:155–61. [DOI] [PubMed] [Google Scholar]
- 186.Kostjuk S, Loseva P, Chvartatskaya O, Ershova E, Smirnova T, Malinovskaya E, Roginko O, Kuzmin V, Izhevskaia V, Baranova A, et al. Extracellular GC-rich DNA activates TLR9- and NF-kB-dependent signaling pathways in human adipose-derived mesenchymal stem cells (haMSCs). Expert Opin Biol Ther. 2012;12(Suppl 1):S99–111. [DOI] [PubMed] [Google Scholar]
- 187.Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.de Oliveira MC, Kranzusch PJ. cGAS Conducts Micronuclei DNA Surveillance. Trends Cell Biol. 2017;27:697–8. [DOI] [PubMed] [Google Scholar]
- 189.Lin C, Chen Y, Zhang F, Liu B, Xie C, Song Y. Encoding gene RAB3B exists in linear chromosomal and circular extrachromosomal DNA and contributes to cisplatin resistance of hypopharyngeal squamous cell carcinoma via inducing autophagy. Cell Death Dis. 2022;13:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yuksel A, Altungoz O. Gene amplifications and extrachromosomal circular DNAs: function and biogenesis. Mol Biol Rep. 2023;50:7693–703. [DOI] [PubMed] [Google Scholar]
- 191.Zeid R, Lawlor MA, Poon E, Reyes JM, Fulciniti M, Lopez MA, Scott TG, Nabet B, Erb MA, Winter GE, et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat Genet. 2018;50:515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, Sagar V, Luan Y, Chalmers ZR, Unno K, et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell. 2019;36:483–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Von Hoff DD, Forseth B, Clare CN, Hansen KL, VanDevanter D. Double minutes arise from circular extrachromosomal DNA intermediates which integrate into chromosomal sites in human HL-60 leukemia cells. J Clin Invest. 1990;85:1887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Von Hoff DD, McGill JR, Forseth BJ, Davidson KK, Bradley TP, Van Devanter DR, Wahl GM. Elimination of extrachromosomally amplified MYC genes from human tumor cells reduces their tumorigenicity. Proc Natl Acad Sci U S A. 1992;89:8165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Koduru P, Chen W, Haley B, Ho K, Oliver D, Wilson K. Cytogenomic characterization of double minute heterogeneity in therapy related acute myeloid leukemia. Cancer Genet. 2019;238:69–75. [DOI] [PubMed] [Google Scholar]
- 196.Abbate LA, Tolomeo D, Cifola I, Severgnini M, Turchiano A, Augello B, Squeo G, Addabbo PD, Traversa D, Daniele G, et al. MYC-containing amplicons in acute myeloid leukemia: genomic structures, evolution, and transcriptional consequences. Leukemia. 2018;32:2152–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Huh YO, Tang G, Talwalkar SS, Khoury JD, Ohanian M, Bueso-Ramos CE, Abruzzo LV. Double minute chromosomes in acute myeloid leukemia, myelodysplastic syndromes, and chronic myelomonocytic leukemia are associated with micronuclei, MYC or MLL amplification, and complex karyotype. Cancer Genet. 2016;209:313–20. [DOI] [PubMed] [Google Scholar]
- 198.Hung KL, Yost KE, Xie L, Shi Q, Helmsauer K, Luebeck J, Schopflin R, Lange JT, Chamorro GR, Weiser NE, et al. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature. 2021;600:731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. [DOI] [PubMed] [Google Scholar]
- 200.Cen Y, Fang Y, Ren Y, Hong S, Lu W, Xu J. Global characterization of extrachromosomal circular DNAs in advanced high grade serous ovarian cancer. Cell Death Dis. 2022;13:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang Y, Dong K, Jia X, Du S, Wang D, Wang L, Qu H, Zhu S, Wang Y, Wang Z, et al. A novel extrachromosomal circular DNA related genes signature for overall survival prediction in patients with ovarian cancer. Bmc Med Genomics. 2023;16:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kanada M, Bachmann MH, Contag CH. Signaling by extracellular vesicles advances cancer hallmarks. Trends Cancer. 2016;2:84–94. [DOI] [PubMed] [Google Scholar]
- 203.Yang Y, Yang Y, Huang H, Song T, Mao S, Liu D, Zhang L, Li W. PLCG2 can exist in eccDNA and contribute to the metastasis of non-small cell lung cancer by regulating mitochondrial respiration. Cell Death Dis. 2023;14:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xu G, Shi W, Ling L, Li C, Shao F, Chen J, Wang Y. Differential expression and analysis of extrachromosomal circular DNAs as serum biomarkers in lung adenocarcinoma. J Clin Lab Anal. 2022;36:e24425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yang Y, Song T, Liu S, Liu Z, Wang X, Li Y, Liu D. Circle-map profiling of extrachromosomal circular DNA as diagnostic biomarkers for lung cancer. Precis Clin Med. 2024;7:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Minina VI, Sinitsky MY, Druzhinin VG, Fucic A, Bakanova ML, Ryzhkova AV, Savchenko YA, Timofeeva AA, Titov RA, Voronina EN, et al. Chromosome aberrations in peripheral blood lymphocytes of lung cancer patients exposed to radon and air pollution. Eur J Cancer Prev. 2018;27:6–12. [DOI] [PubMed] [Google Scholar]
- 207.Raymond E, Faivre S, Weiss G, McGill J, Davidson K, Izbicka E, Kuhn JG, Allred C, Clark GM, Von Hoff DD. Effects of hydroxyurea on extrachromosomal DNA in patients with advanced ovarian carcinomas. Clin Cancer Res. 2001;7:1171–80. [PubMed] [Google Scholar]
- 208.Ambros IM, Rumpler S, Luegmayr A, Hattinger CM, Strehl S, Kovar H, Gadner H, Ambros PF. Neuroblastoma cells can actively eliminate supernumerary MYCN gene copies by micronucleus formation–sign of tumour cell revertance? Eur J Cancer. 1997;33:2043–9. [DOI] [PubMed] [Google Scholar]
- 209.Eckhardt SG, Dai A, Davidson KK, Forseth BJ, Wahl GM, Von Hoff DD. Induction of differentiation in HL60 cells by the reduction of extrachromosomally amplified c-myc. Proc Natl Acad Sci U S A. 1994;91:6674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Narath R, Ambros IM, Kowalska A, Bozsaky E, Boukamp P, Ambros PF. Induction of senescence in MYCN amplified neuroblastoma cell lines by hydroxyurea. Genes Chromosomes Cancer. 2007;46:130–42. [DOI] [PubMed] [Google Scholar]
- 211.Shimizu N, Itoh N, Utiyama H, Wahl GM. Selective entrapment of extrachromosomally amplified DNA by nuclear budding and micronucleation during S phase. J Cell Biol. 1998;140:1307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yu L, Zhao Y, Quan C, Ji W, Zhu J, Huang Y, Guan R, Sun D, Jin Y, Meng X, et al. Gemcitabine eliminates double minute chromosomes from human ovarian cancer cells. PLoS ONE. 2013;8:e71988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sun W, Quan C, Huang Y, Ji W, Yu L, Li X, Zhang Y, Zheng Z, Zou H, Li Q, et al. Constitutive ERK1/2 activation contributes to production of double minute chromosomes in tumour cells. J Pathol. 2015;235:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7:339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kaufman RJ, Brown PC, Schimke RT. Amplified dihydrofolate reductase genes in unstably methotrexate-resistant cells are associated with double minute chromosomes. Proc Natl Acad Sci U S A. 1979;76:5669–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Alt FW, Kellems RE, Bertino JR, Schimke RT. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978;253:1357–70. [PubMed] [Google Scholar]
- 217.Cai M, Zhang H, Hou L, Gao W, Song Y, Cui X, Li C, Guan R, Ma J, Wang X, et al. Inhibiting homologous recombination decreases extrachromosomal amplification but has no effect on intrachromosomal amplification in methotrexate-resistant colon cancer cells. Int J Cancer. 2019;144:1037–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Morales C, Garcia MJ, Ribas M, Miro R, Munoz M, Caldas C, Peinado MA. Dihydrofolate reductase amplification and sensitization to methotrexate of methotrexate-resistant colon cancer cells. Mol Cancer Ther. 2009;8:424–32. [DOI] [PubMed] [Google Scholar]
- 219.Von Hoff DD, Waddelow T, Forseth B, Davidson K, Scott J, Wahl G. Hydroxyurea accelerates loss of extrachromosomally amplified genes from tumor cells. Cancer Res. 1991;51:6273–9. [PubMed] [Google Scholar]
- 220.Kalavska K, Minarik T, Vlkova B, Manasova D, Kubickova M, Jurik A, Mardiak J, Sufliarsky J, Celec P, Mego M. Prognostic value of various subtypes of extracellular DNA in ovarian cancer patients. J Ovarian Res. 2018;11:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ruiz-Herrera A, Smirnova A, Khoriauli L, Nergadze SG, Mondello C, Giulotto E. Gene amplification in human cells knocked down for RAD54. Genome Integr. 2011;2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Baig M, Lai WF, Mikrani R, Jabeen M, Naveed M, Abbas M, Farooq MA, Ahsan A, Kassim SA, Khan GJ, Ansari MT. Synthetic NRG-1 functionalized DNA nanospindels towards HER2/neu targets for in vitro anti-cancer activity assessment against breast cancer MCF-7 cells. J Pharm Biomed Anal. 2020;182:113133. [DOI] [PubMed] [Google Scholar]
- 223.Shen C, Liu S, Li X, Zhao D, Yang M. Immunoelectrochemical detection of the human epidermal growth factor receptor 2 (HER2) via gold nanoparticle-based rolling circle amplification. Mikrochim Acta. 2018;185:547. [DOI] [PubMed] [Google Scholar]
- 224.Sheng Z, Wang X, Zheng Y, Duan W, Cui J, Gu L, Gao X, Ma J, Cui M, Luo H, et al. Genome-wide characterization of extrachromosomal circular DNA in breast cancer and its potential role in carcinogenesis and cancer progression. Cell Rep. 2024;43:114845. [DOI] [PubMed] [Google Scholar]
- 225.Ouyang Y, Lu W, Wang Y, Wang B, Li F, Li X, Bai Y, Wang Y. Integrated analysis of mRNA and extrachromosomal circular DNA profiles to identify the potential mRNA biomarkers in breast cancer. Gene. 2023;857:147174. [DOI] [PubMed] [Google Scholar]
- 226.Morales C, Ribas M, Aiza G, Peinado MA. Genetic determinants of methotrexate responsiveness and resistance in colon cancer cells. Oncogene. 2005;24:6842–7. [DOI] [PubMed] [Google Scholar]
- 227.Ooi A, Takehana T, Li X, Suzuki S, Kunitomo K, Iino H, Fujii H, Takeda Y, Dobashi Y. Protein overexpression and gene amplification of HER-2 and EGFR in colorectal cancers: an immunohistochemical and fluorescent in situ hybridization study. Mod Pathol. 2004;17:895–904. [DOI] [PubMed] [Google Scholar]
- 228.Chen Y, Qiu Q, She J, Yu J. Extrachromosomal circular DNA in colorectal cancer: biogenesis, function and potential as therapeutic target. Oncogene. 2023;42:941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Alitalo K, Winqvist R, Lin CC, de la Chapelle A, Schwab M, Bishop JM. Aberrant expression of an amplified c-myb oncogene in two cell lines from a colon carcinoma. Proc Natl Acad Sci U S A. 1984;81:4534–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zeng X, Wan M, Wu J. ecDNA within tumors: a new mechanism that drives tumor heterogeneity and drug resistance. Signal Transduct Target Ther. 2020;5:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ye J, Huang P, Ma K, Zhao Z, Hua T, Zai W, Chen J, Fu X. Genome-wide extrachromosomal circular DNA profiling of paired hepatocellular carcinoma and adjacent liver tissues. Cancers (Basel). 2023;15:5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.McCulloch K, Romero N, MacLachlan J, Allard N, Cowie B. Modeling progress toward elimination of hepatitis B in Australia. Hepatology. 2020;71:1170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hu Y, Shi G, Zhang L, Li F, Jiang Y, Jiang S, Ma W, Zhao Y, Songyang Z, Huang J. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci Rep. 2016;6:32280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 2001;61:3806–9. [PubMed] [Google Scholar]
- 235.Kunisada T, Yamagishi H. Sequence organization of repetitive sequences enriched in small polydisperse circular DNAs from HeLa cells. J Mol Biol. 1987;198:557–65. [DOI] [PubMed] [Google Scholar]
- 236.Kunisada T, Yamagishi H. Sequence repetition and genomic distribution of small polydisperse circular DNA purified from HeLa cells. Gene. 1984;31:213–23. [DOI] [PubMed] [Google Scholar]
- 237.Jin Y, Liu Z, Cao W, Ma X, Fan Y, Yu Y, Bai J, Chen F, Rosales J, Lee KY, Fu S. Novel functional MAR elements of double minute chromosomes in human ovarian cells capable of enhancing gene expression. PLoS ONE. 2012;7:e30419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhou M, Lv W, Han P, Sun K, Hao Z, Gao L, Xu Y, Xu Z, Shao S, Ma S, et al. Plasma extrachromosomal circular DNA as a potential diagnostic biomarker for nodular thyroid disease. Clin Transl Med. 2024;14:e1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhou T, Ma S, Zhao Y, Guo D, Wang H, Kuang M, Li X. Identification and characterization of extrachromosomal circular DNA in alcohol induced osteonecrosis of femoral head. Front Genet. 2022;13:918379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Coleman RL, Oza AM, Lorusso D, Aghajanian C, Oaknin A, Dean A, Colombo N, Weberpals JI, Clamp A, Scambia G, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wang S, Blois A, El RT, Liu JF, Hirsch MS, Gravdal K, Palakurthi S, Bielenberg DR, Akslen LA, Drapkin R, et al. Development of a prosaposin-derived therapeutic cyclic peptide that targets ovarian cancer via the tumor microenvironment. Sci Transl Med. 2016;8:329r–34r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yang S, Lheureux S, Karakasis K, Burnier JV, Bruce JP, Clouthier DL, Danesh A, Quevedo R, Dowar M, Hanna Y, et al. Landscape of genomic alterations in high-grade serous ovarian cancer from exceptional long- and short-term survivors. Genome Med. 2018;10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Shrivastava S, Mahantshetty U, Engineer R, Chopra S, Hawaldar R, Hande V, Kerkar RA, Maheshwari A, Shylasree TS, Ghosh J, et al. Cisplatin chemoradiotherapy vs radiotherapy in FIGO stage IIIB squamous cell carcinoma of the uterine cervix: a randomized clinical trial. Jama Oncol. 2018;4:506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zeng X, Zhang X, Li C, Wang X, Jerwick J, Xu T, Ning Y, Wang Y, Zhang L, Zhang Z, et al. Ultrahigh-resolution optical coherence microscopy accurately classifies precancerous and cancerous human cervix free of labeling. Theranostics. 2018;8:3099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Allaoui R, Bergenfelz C, Mohlin S, Hagerling C, Salari K, Werb Z, Anderson RL, Ethier SP, Jirstrom K, Pahlman S, et al. Cancer-associated fibroblast-secreted CXCL16 attracts monocytes to promote stroma activation in triple-negative breast cancers. Nat Commun. 2016;7:13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liao C, Zhang Y, Fan C, Herring LE, Liu J, Locasale JW, Takada M, Zhou J, Zurlo G, Hu L, et al. Identification of BBOX1 as a therapeutic target in triple-negative breast cancer. Cancer Discov. 2020;10:1706–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ruiz-Saenz A, Dreyer C, Campbell MR, Steri V, Gulizia N, Moasser MM. HER2 Amplification in tumors activates PI3K/Akt signaling independent of HER3. Cancer Res. 2018;78:3645–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bardia A, Hurvitz S. Targeted therapy for premenopausal women with HR(+), HER2(-) advanced breast cancer: focus on special considerations and latest advances. Clin Cancer Res. 2018;24:5206–18. [DOI] [PubMed] [Google Scholar]
- 249.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Cernat A, De Freitas C, Majid U, Trivedi F, Higgins C, Vanstone M. Facilitating informed choice about non-invasive prenatal testing (NIPT): a systematic review and qualitative meta-synthesis of women’s experiences. BMC Pregnancy Childbirth. 2019;19:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sin S, Ji L, Deng J, Jiang P, Cheng SH, Heung M, Lau C, Leung TY, Chan K, Chiu R, Lo Y. Characteristics of fetal extrachromosomal circular DNA in maternal plasma: methylation status and clearance. Clin Chem. 2021;67:788–96. [DOI] [PubMed] [Google Scholar]
- 252.Yang H, He J, Huang S, Yang H, Yi Q, Tao Y, Chen M, Zhang X, Qi H. Identification and characterization of extrachromosomal circular DNA in human placentas with fetal growth restriction. Front Immunol. 2021;12:780779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Schaeffer EM, Srinivas S, Adra N, An Y, Barocas D, Bitting R, Bryce A, Chapin B, Cheng HH, D’Amico AV, et al. Prostate cancer, version 4.2023, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2023;21:1067–96. [DOI] [PubMed] [Google Scholar]
- 254.Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49. [DOI] [PubMed] [Google Scholar]
- 255.Cathomas R, Lorch A, Bruins HM, Comperat EM, Cowan NC, Efstathiou JA, Fietkau R, Gakis G, Hernandez V, Espinos EL, et al. The 2021 updated european association of urology guidelines on metastatic urothelial carcinoma. Eur Urol. 2022;81:95–103. [DOI] [PubMed] [Google Scholar]
- 256.Peng Y, Li Y, Zhang W, ShangGuan Y, Xie T, Wang K, Qiu J, Pu W, Hu B, Zhang X, et al. The characteristics of extrachromosomal circular DNA in patients with end-stage renal disease. Eur J Med Res. 2023;28:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wang T, Zhang H, Zhou Y, Shi J. Extrachromosomal circular DNA: a new potential role in cancer progression. J Transl Med. 2021;19:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mouakkad-Montoya L, Murata MM, Sulovari A, Suzuki R, Osia B, Malkova A, Katsumata M, Giuliano AE, Eichler EE, Tanaka H. Quantitative assessment reveals the dominance of duplicated sequences in germline-derived extrachromosomal circular DNA. Proc Natl Acad Sci U S A. 2021;118:e2102842118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Middelkamp S, van Tol H, Spierings D, Boymans S, Guryev V, Roelen B, Lansdorp PM, Cuppen E, Kuijk EW. Sperm DNA damage causes genomic instability in early embryonic development. Sci Adv. 2020;6:z7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wang L, Tracy L, Su W, Yang F, Feng Y, Silverman N, Zhang Z. Retrotransposon activation during Drosophila metamorphosis conditions adult antiviral responses. Nat Genet. 2022;54:1933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lowenstein LM, Basourakos SP, Williams MD, Troncoso P, Gregg JR, Thompson TC, Kim J. Active surveillance for prostate and thyroid cancers: evolution in clinical paradigms and lessons learned. Nat Rev Clin Oncol. 2019;16:168–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, Stein C, Basit A, Chan J, Mbanya JC, et al. IDF diabetes atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract. 2022;183:109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Catenacci D, Tebbutt NC, Davidenko I, Murad AM, Al-Batran SE, Ilson DH, Tjulandin S, Gotovkin E, Karaszewska B, Bondarenko I, et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1467–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Huang YK, Wang M, Sun Y, Di Costanzo N, Mitchell C, Achuthan A, Hamilton JA, Busuttil RA, Boussioutas A. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat Commun. 2019;10:3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Suzuki A, Katoh H, Komura D, Kakiuchi M, Tagashira A, Yamamoto S, Tatsuno K, Ueda H, Nagae G, Fukuda S, et al. Defined lifestyle and germline factors predispose Asian populations to gastric cancer. Sci Adv. 2020;6:v9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ferriero R, Nusco E, De Cegli R, Carissimo A, Manco G, Brunetti-Pierri N. Pyruvate dehydrogenase complex and lactate dehydrogenase are targets for therapy of acute liver failure. J Hepatol. 2018;69:325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Qian Y, Hong X, Yu Y, Du C, Li J, Yu J, Xiao W, Chen C, Huang D, Zhong T, et al. Characterization and functional analysis of extrachromosomal circular DNA discovered from circulating extracellular vesicles in liver failure. Clin Transl Med. 2024;14: e70059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14:92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Van Den Bent M, Eoli M, Sepulveda JM, Smits M, Walenkamp A, Frenel JS, Franceschi E, Clement PM, Chinot O, De Vos F, et al. INTELLANCE 2/EORTC 1410 randomized phase II study of Depatux-M alone and with temozolomide vs temozolomide or lomustine in recurrent EGFR amplified glioblastoma. Neuro Oncol. 2020;22:684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Gan HK, Reardon DA, Lassman AB, Merrell R, van den Bent M, Butowski N, Lwin Z, Wheeler H, Fichtel L, Scott AM, et al. Safety, pharmacokinetics, and antitumor response of depatuxizumab mafodotin as monotherapy or in combination with temozolomide in patients with glioblastoma. Neuro Oncol. 2018;20:838–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Chenais B. Transposable elements and human diseases: mechanisms and implication in the response to environmental pollutants. Int J Mol Sci. 2022;23:2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Chiu R, Dutta A, Hensson AG, Lo Y, Mischel P, Regenberg B. What is extrachromosomal circular DNA and what does it do? Clin Chem. 2020;66:754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Zhang L, Yang M, Marks P, White LM, Hurtig M, Mi QS, Divine G, Gibson G. Serum non-coding RNAs as biomarkers for osteoarthritis progression after ACL injury. Osteoarthritis Cartilage. 2012;20:1631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.