Abstract
Background
Metagenome-assembled viral genomes have significantly advanced the discovery and characterization of the human gut virome. However, we lack a comparative assessment of assembly tools on the efficacy of viral genome identification, particularly across next-generation sequencing (NGS) and third-generation sequencing (TGS) data.
Results
We evaluated the efficiency of NGS, TGS, and hybrid assemblers for viral genome discovery using 95 viral-like particle (VLP)-enriched fecal samples sequenced on both Illumina and PacBio platforms. MEGAHIT, metaFlye, and hybridSPAdes emerged as the optimal choices for NGS, TGS, and hybrid datasets, respectively. Notably, these assemblers recovered distinct viral genomes, demonstrating a remarkable degree of complementarity. By combining individual assembler results, we expanded the total number of nonredundant high-quality viral genomes by 4.83 ~ 21.7-fold compared to individual assemblers. Among them, viral genomes from NGS and TGS data have the least overlap, indicating the impact of data type on viral genome recovery. We also evaluated four binning methods, finding that CONCOCT incorporated more unrelated contigs into the same bins, while MetaBAT2, AVAMB, and vRhyme balanced inclusiveness and taxonomic consistency within bins.
Conclusions
Our findings highlight the challenges in metagenome-driven viral discovery, underscoring tool limitations. We advocate for combined use of multiple assemblers and sequencing technologies when feasible and highlight the urgent need for specialized tools tailored to gut virome assembly. This study contributes essential insights for advancing viral genome research in the context of gut metagenomics.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01981-z.
Background
The human gut harbors a substantial population of viruses, predominantly featuring double-stranded DNA (dsDNA) phages [1–4]. These phages exert their influence on the ecosystem structure of the intestinal microbiota [5] by modulating bacterial populations within the gut through mechanisms such as predation or lysogeny [6]. Furthermore, phages have shown great promise as precise antibiotic agents, capable of selectively targeting and eliminating their hosts [7]. This holds particular relevance in the context of the alarming surge in antibiotic resistance [8].
In recent years, there has been a notable surge in the detection of viral genomes through metagenomic assemblies, enabling the retrieval of numerous viral genomes from human gut metagenome sequencing data, whether enriched with viral-like particles (VLP) [3, 9, 10] or not [11–13]. Obtaining high-quality assembled genomes is an important prerequisite for downstream analyses such as viral genome detection, host prediction, community composition, or phylogenetic analysis [13].
However, owing to the rapid evolutionary pace of viral genomes and the resulting heightened micro-diversity in their genomic sequences within a sample [14], the development of a dedicated genome assembler for viral metagenomes is an urgent requirement yet one that remains unaddressed. Consequently, the majority of research has resorted to employing assemblers originally designed for assembling single genomes [15, 16] or bulk metagenome sequencing data [13, 17, 18].
In addition to the next-generation sequencing (NGS or short-read) data, third-generation sequencing (TGS or long read) has recently been applied to bulk metagenome [19–21] and gut virome sequencing derived from VLP-enriched samples [10, 22–24]. In response to this growing trend, alternative sequencing and informatics workflows [25, 26] to improve viral metagenomic assemblies designed for second- and third-generation sequencing have been published and widely adopted. Previous results have shown the critical role of assembly software in characterizing the human gut virome using NGS mock viral communities [27] or NGS in silico simulated viral metagenomes [28]. Furthermore, integrating long- and short-read sequencing for the human gut virome (using three samples) [29] and viral mock communities [30] has demonstrated the advantages of long reads in recovering high-quality viral genomes. Despite these findings, a comprehensive evaluation of viral identification methods across both NGS and TGS platforms using a large number of samples has been notably lacking, particularly with paired data—where the same set of samples is sequenced using both NGS and TGS platforms. A particularly critical, yet overlooked, aspect is the overlap and complementarity in gut viral genomes obtained by different methods and sequencing technologies. Additionally, the applicability of binning methods, extensively used in bulk metagenomic analysis, remains untested in the context of VLP metagenome data.
In this study, based on paired long- and short-read sequencing data from 95 VLP-enriched human fecal samples, we assessed the quality and detection efficiency of viral contigs generated by short-read, long-read, and hybrid assemblers. Subsequently, we extensively analyzed the distinctions and complementarity of viral genomes obtained from different assemblers at various taxonomic levels, especially those derived from short- and long-read sequencing data. Finally, we evaluated four binning methods to assess the inclusiveness and taxonomic consistency of binned contigs. Our findings would guide researchers in the selection of the most suitable detection strategy as well as sequencing platforms for their gut virome study and help developers to know the limitations of the current methods and how their performance is affected by the gut virome-specific characteristics.
Methods
Selection of metagenomic assemblers and binners for gut virome analysis
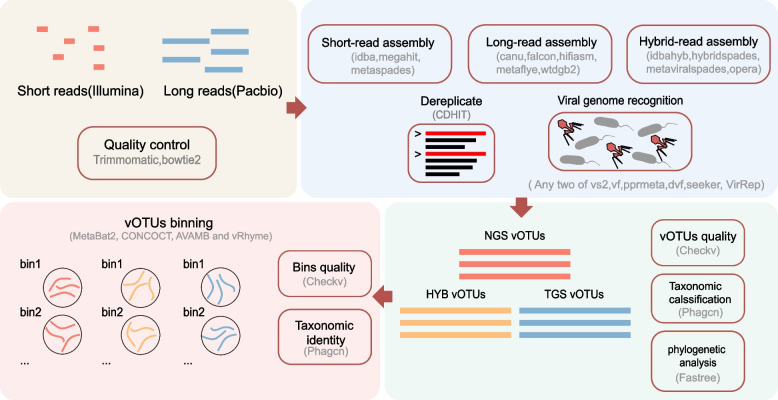
To identify the optimal assemblers and binners within the enterovirus group data, we collected three short-read assemblers, five long-read assemblers, four hybrid assemblers, and four binners into our comprehensive analysis. Tools including its associated information are presented in Table 1 and Fig. 1 for reference.
Table 1.
Metagenomic assemblers and binners used in this study
| Tool | Data type | Version | Algorithms | Last updated | Designed for metagenomics |
|---|---|---|---|---|---|
| IDBA-UD [31] | NGS | v1.1.3 | De Bruijn graph | Dec 31, 2016 | Yes |
| MEGAHIT [18] | NGS | v1.2.9 | De Bruijn graph | Feb 14, 2023 | Yes |
| metaSPAdes [17] | NGS | v3.15.4 | De Bruijn graph | Jul 16, 2022 | Yes |
| Canu [15] | TGS | v2.2 | Overlap-layout consensus | Dec 15, 2023 | No |
| FALCON [32] | TGS | v1.8.1 | Overlap-layout consensus | Sep 11, 2020 | No |
| Hifiasm-meta [33] | TGS | v0.3 | Graph-dependent algorithms | Jun 2, 2023 | Yes |
| metaFlye [34] | TGS | v2.9.1 | Repeat graph | Sep 9, 2023 | Yes |
| wtdbg2 [16] | TGS | v2.5 | Fuzzy Bruijn graph | Dec 11, 2023 | No |
| IDBA-hyb [31] | HYB | v1.1.3 | De Bruijn graph | Dec 31, 2016 | Yes |
| hybridSPAdes [35] | HYB | v3.15.4 | De Bruijn graph | Jul 16, 2022 | Yes |
| metaViralSPAdes [36] | HYB | v3.15.4 | De Bruijn graph | Jul 16, 2022 | Yes |
| OPERA-MS [37] | HYB | v0.83 | De Bruijn graph | Apr 14, 2023 | Yes |
| CONCOCT [38] | - | v1.1.0 | Unsupervised clustering | Nov 11, 2019 | - |
| MetaBAT2 [39] | - | v2.15.2 | Label propagation | Apr 11, 2023 | - |
| AVAMB [40] | - | v4.1.3 | Variational autoencoders | Jun 2, 2023 | - |
| vRhyme [41] | - | v1.1.0 | Supervised machine learning | Jul 13, 2022 | - |
Fig. 1.
Overall workflow of this study, including sequencing reads processing, assembly, dereplication, viral genome identification, binning, and quality assessment. Vs2, VirSorter2; vf, VirFinder; dvf, DeepVirFinder
Illumina and PacBio sequencing data of human gut virome samples
Sequencing data in the Chinese Human Gut Virome (CHGV) [10] catalog, containing fecal samples of 95 healthy Chinese residents submitted to both short- and long-read sequencing, were employed for our analysis.
Briefly, human fecal samples (totaling ≈500 g each) were obtained from anonymous healthy volunteers recruited from Wuhan and Shanghai, China. Viral-like particles (VLPs) were obtained by utilizing a virome enrichment protocol adapted from ref. [42–45], as outlined below. A total of 400–500 g of frozen feces from a − 80 °C freezer was added to 5 l of SM buffer (200-mM NaCl, 10-mM MgSO₄, 50-mM Tris–HCl, pH 7.5) and stirred at 120 rpm at room temperature using an automated stirrer (A200plus, OuHor, Shanghai, China) until fully dispersed. The mixture was then filtered through four layers of gauze (21 s × 32 s/28 × 28) and centrifuged at 5000 × g for 45 min at 4 °C. The supernatant was transferred and centrifuged again at 8000 × g for 45 min at 4 °C. The resulting supernatant was concentrated to approximately 300 ml using a 100-kD ultrafiltration membrane (Sartorius, Vivaflow 200). NaCl was added to a final concentration of 0.5 M, and the samples were stored at 4 °C for 1 h. Next, PEG 8000 was added to a final concentration of 10% (w/v), and the samples were incubated overnight at 4 °C. Phage particles were then sedimented the following day by centrifugation at 13,000 × g for 35 min at 4 °C.
Nucleic acid was then extracted using a HiPure HP DNA Maxi Kit (D6322, Magen, Guangzhou, China) according to the manufacturer’s instructions. Double-stranded DNAs extracted were subjected to next-generation sequencing (NGS) using the Illumina HiSeq2000 sequencer (Novogen, Beijing, China) and third-generation sequencing (TGS) using the PacBio RS II sequencer (Pacific Biosciences, Menlo Park, CA, USA).
Preprocessing of VLP sequencing data
For the NGS raw sequencing (short-reads) data, we employed Trimmomatic (v0.39) [46] to perform adaptor removal and eliminate low-quality bases. The parameters used were as follows: LEADING:3, TRAILING:3, SLIDINGWINDOW:15:30, and MINLEN:50. For the correction of third-generation sequencing (TGS; long-reads) data, we utilized the default settings of pbccs (v4.0.0) (https://github.com/nlhepler/pbccs) (Fig. 1).
To identify potential human reads within the trimmed short-reads or CCSed long-reads data, we conducted alignment against the human reference genome hg38 (GCA_000001405.15) employing the Bowtie2 (v2.4.2) [47] (Fig. 1). Subsequently, any human-associated reads were removed from the dataset.
Assembly of VLP sequencing data
The VLP sequencing data were then assembled from the reads of each individual sample using the selected assemblers. Default parameters were used unless otherwise stated. Briefly, for the NGS data, we used IDBA-UD [31], MEGAHIT [18], and metaSPAdes [17]. For the TGS data, we selected Canu [15], FALCON [32], Hifiasm-meta [33], metaFlye [34], and wtdbg2 [16]. For hybrid assembly that combines the NGS and TGS data, we used IDBA-hyb, hybridSPAdes [35], metaViralSPAdes [36], and OPERA-MS [37] (Fig. 1).
Mis-assembly was then identified using metaMIC [48] with default parameters for the contig generated from all assemblers. Mis-assembled contigs were corrected by splitting into fragments at the mis-assembled positions reported by the metaMIC tool; the fragments were considered as contigs and also used for subsequent analysis.
Contig dereplication and viral contig identification
Dereplication was performed on contigs obtained by each tool on each sample or multi-tools on all samples using cd-hit (v4.6.8) [49] with a parameter of -c 0.95 and -aS 0.85 according to a MIUViG [50] (Fig. 1).
Viral contigs were then identified using a similar procedure to human Gut Virome Database (GVD) [9], with modifications (Fig. 1). Briefly, the following virus recognition software were firstly used, including VirSorter2 (v2.2.4) [51], DeepVirFinder (v1.0) [52], VirFinder (v1.1) [53], Seeker [54], PPR-Meta (v1.1) [55], and VirRep [56]. Their parameters were listed as the following.
VirSorter score ≥ 0.7
DeepVirFinder with the default parameter
VirFinder score > 0.6
Seeker with the default parameter
PPR-Meta phage score > 0.7
VirRep with the default parameter
Secondly, a contig was considered as a virus if it passed at least two out of the above six criteria and had sequence length > 1.5 kb.
We referred the viral contigs to viral operational taxonomic units (vOTUs) at strain level, as previously described [10].
Binning of viral contigs
Following the assembly, we performed multi-coverage binning (i.e., when clustering contigs of a sample into bins, the read coverage of these contigs across all samples was also considered) [57] on the identified viral vOTUs from each sample. We used CONCOCT [38], MetaBAT2 [39], AVAMB [40], and vRhyme [41] with default parameters to generate bins (Fig. 1).
Evaluation of the quality of vOTUs and bins
To evaluate the quality of the vOTUs, CheckV (v1.0.1) [4] was used (Fig. 1) with the parameter “end_to_end,” and the vOTUs were assigned into different groups, including “complete,” “high quality,” “medium quality,” “low quality,” and “not determined,” which correspond to completeness scores of 100%, > 90%, 50–90%, 0–50%, and non-determined, respectively. In this study, we referred to the vOTUs with > 90% completeness and no “contig > 1.5 × longer than expected genome length” and “high kmer _ freq may indicate large duplication” warning information as the “the high-quality vOTUs (hq-vOTUs).”
Currently, there is no specific use for evaluating viral bins, and CheckV can only accept contig as input content. Here, we adopted a method from [41], which used 50 consecutive characters Ns (CheckV treats Ns as a gap instead of the shortest length of a sequence) to join all contigs in a bin into a single sequence, and evaluated its quality with CheckV.
Taxonomic annotation and phylogenetic analysis
We employed PhaGCN_newICTV [58] to perform taxonomy annotations at the family-level for all hq-vOTUs (Fig. 1). To ensure the reliability of our annotations, we selected annotations with a PhaGCN_newICTV score equal to 1 (ranging from 0 to 1) as the final results.
For phylogenetic analysis of selected vOTUs, we first annotated their protein coding genes using Prokka (v1.14.6) [59], from which we selected gene and protein sequences belonging to the large terminases. Subsequently, we conducted multiple sequence comparisons on the protein sequences using MUSCLE (v3.8.1551) [60]. The resulting multiple-sequence alignments were analyzed by FastTree (v2.1.11) [61] to construct phylogenetic trees using the maximum-likelihood algorithm (Fig. 1). Finally, we employed iTOL (v6.8) [62] and Evolview v3 [63] for visualization and annotation of the phylogenetic trees.
Results
Identifying the optimal assemblers for vOTU detection using short-, long-, and hybrid-sequencing data
To comprehensively evaluate the effect of different assembly and binning tools on viral genome discovery, we used 95 viral-like particle (VLP)-enriched human fecal samples sequenced on both Illumina (next-generation sequencing, NGS, or short reads) and PacBio (third-generation sequencing, TGS, or long-reads) platforms from our previous study [10] (“ Methods”).
Our evaluation is shown in Fig. 1. First is genome assembly. We selected a total of 12 state-of-the-art assemblers for (meta)-genome analysis, including 3 NGS, 5 TGS, and 4 hybrid assemblers (“ Methods” and Table 1). Secondly, we performed an in-sample dereplication of the contigs assembled from all samples for each tool. Thirdly, we identified viral contigs using a customized bioinformatics pipeline and clustered them into the nonredundant species-level viral contigs referred to as vOTUs (“ Methods”). Fourthly, for the viral contigs generated by each assembler, we used CONCOCT [38], MetaBAT2 [39], AVAMB [40], and vRhyme [41] for multi-coverage binning [57] (“ Methods”). Then, we conducted a systematic evaluation of the tools at the assembly level and the binning level. The quality metrics of viral contigs and bins, taxonomy classification analysis, and phylogenetic status were included in the process (Fig. 1).
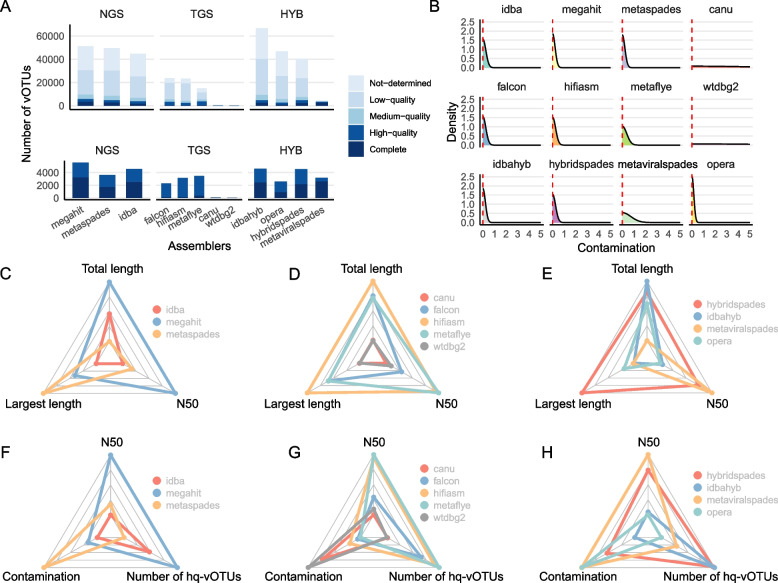
After assembly and viral genome identification, we first compared the numbers of vOTUs obtained from the assemblers. We found that MEGAHIT, FALCON, and IDBA-hyb generated the highest number of vOTUs among the NGS, TGS, and hybrid assembler groups, respectively. However, when considering only the high-quality vOTUs (hq-vOTUs) with > 90% genome completeness and no “contig > 1.5 × longer than expected genome length” and “high kmer _ freq may indicate large duplication” warning information according to CheckV [4] (“ Methods”), MEGAHIT, metaFlye, and hybridSPAdes performed the best within their respective assembler categories (Fig. 2A). Notably, assemblers that were not optimized for metagenomic data, such as canu and wtdgb2, generated significantly less vOTUs (Fig. 2A).
Fig. 2.
Evaluation of the identification and quality of vOTU across assemblers. A Stacked barplots showing numbers of vOTUs derived from the assembly tools, color-coded according to the CheckV quality scores. The tools are stratified by the sequencing data types they could handle such as NGS (for short reads), TGS (for long reads), and hybrid (for both reads types; HYB). Upper panel, all vOTUs; lower panel, high-quality vOTUs (hq-vOTUs) with > 90% completeness and no “contig > 1.5 × longer than expected genome length” and “high kmer _ freq may indicate large duplication” warning information. B Density plots showing the distribution of vOTU contaminations according to CheckV, with the vertical dashed lines indicating the median contamination of each tool. C, D, E Radar plots showing the strength and weakness of the assemblers in length metrics including total contig length, N50 length, and maximum vOTU length of the vOTUs derived from the NGS (C), TGS (D), and HYB assemblers (E). F, J, H Similar to C, D, and E but with different evaluation metrics such as vOTU N50 length, the proportion of vOTUs with 0 contamination, and the number of hq-vOTUs
We observed that > 97% of the vOTUs by all assemblers contained < 5% contaminations (Figs. 2B, S1). This is because CheckV only counted bacterial genes at the end of the assembled contigs as contaminations [4]. We thus did not consider the contamination levels as a key measurement of the vOTUs.
Finally, we compared the assembly length metrics of the vOTUs, including the lengths of the longest contig, total contigs, and N50. For the NGS assemblers, MEGAHIT generated contigs with the longest total length and the highest N50, while metaSPAdes achieved the longest contigs (Fig. 2C). Among the TGS assemblers, Hifiasm-meta had the largest total length and the largest contig length. However, it is noteworthy that metaFlye, despite having the highest N50, did not significantly lag behind Hifiasm-meta in terms of total length and the largest contig length (Fig. 2D). Among the hybrid assemblers, hybridSPAdes achieved the largest contig length and was comparable to IDBA-hyb and metaViralSPAdes in terms of total lengths and N50 values, with only marginal differences in these metrics (Fig. 2E).
Overall, our results suggest that MEGAHIT, metaFlye, and hybridSPAdes stand out as the best tools in the NGS, TGS, and hybrid assembler categories, respectively, featuring the identification of more and longer vOTUs with higher quality (Fig. 2F, G, H).
Complementarity of different assemblers in recovering high-quality viral genomes
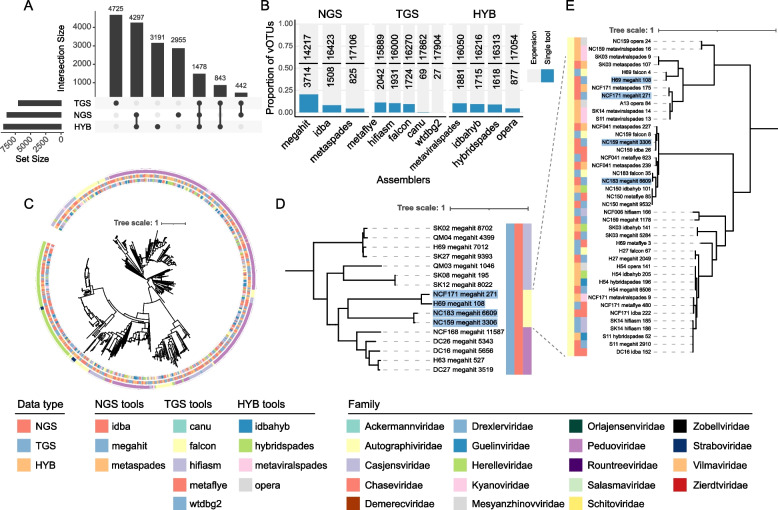
We next examined the overlaps and differences in the detected vOTUs across assemblers. We focused on the hq-vOTUs with CheckV completeness > 90% and no “contig > 1.5 × longer than expected genome length” and “high kmer _ freq may indicate large duplication” warning information to avoid misevaluation due to genome incompleteness. Combining all such vOTUs from all assemblers and dereplicated at a 95% threshold using cd-hit (“ Methods”), we obtained a combined set of 17,931 nonredundant hq-vOTUs (Table S1). Surprisingly, we found that more than half (54.5%, 9771) of them were assembler specific (Fig. S2). We also examined the overlaps among the three assembler groups (NGS, TGS, HYB) and found that few hq-vOTUs were recovered by all three groups (n = 1478, 8.24% out of 17,931) or by two groups (i.e., n = 442 between TGS and NGS, n = 843 between TGS and HYB). We did find a significant overlap between the NGS and HYB groups (4297), likely because the pre-assembly step of these hybrid assemblers is using NGS reads during the assembly [35, 37]. Additionally, the TGS group derived the highest number of unique hq-vOTUs (n = 4725, 26.4%), followed by the hybrid (n = 3191, 17.8%) and NGS (n = 2955, 16.5%) (Fig. 3A). These results suggest that in addition to the choice of tools, the type of sequencing data, i.e., short vs long reads, also significantly influences the vOTU identification results, highlighting the necessity of using both the long and short reads for a complete gut virome characterization.
Fig. 3.
Assembler-specific vOTUs accounted most of the total vOTUs and were of high quality. A Upset plot showing the number of hq-vOTUs derived from the three assembler groups. B Stacked barplots showing the number of high-quality vOTUs (hq-vOTUs) assembled by individual assemblers (dark blue and lower number) compared to those expanded by other assemblers (light gray and upper number). C Phylogenetic tree of the 1026 hq-vOTUs encoding the large terminase gene. The annotation ring next to the leaf labels consists of three circles, with the inner, mid, and outer circles color-coded according to the assembly tools, the data types, and family-level taxonomical annotations, respectively. D A phylogenetic tree constructed from selected hq-vOTUs derived from the megahit tool on NGS data, with leaves forming a sub-branch belonging to the Autographiviridae family highlighted by blue. E A phylogenetic tree constructed from all hq-vOTUs belonging to the Autographiviridae in the nonredundant combined set. Branch leaves highlighted with blue background are the megahit-derived hq-vOTUs. The three columns of the heatmap on the left indicate phylum annotation according to the PhagCN_newICTV tool, data type (i.e., NGS, TGS, and hybrid), and assembly tool (from left to right)
When compared with individual assemblers, we found that the combined set significantly expanded the numbers of the hq-vOTUs compared with individual assemblers, from 4.83-fold increase for MEGAHIT to 21.7-fold increase for metaSPAdes (Fig. 3B). These results indicate significant complementarity of different assemblers in recovering high-quality viral genomes.
Assembler-specific metagenome-assembled genomes can be error-prone, and we thus adopted a phylogenetic approach to further validate the quality of these hq-vOTUs from different assemblers. We annotated the large terminase genes in hq-vOTUs and used the protein sequences for phylogenetic analysis. The dsDNA virus terminal enzyme gene, often employed as a marker gene for phylogenetic analysis, encodes a crucial enzyme involved in DNA replication and repair processes [64]. About 16% of the hq-vOTUs encoded the large terminase (Fig. S3). We built multiple sequence alignments using the large terminase proteins and constructed a maximum-likelihood tree (“ Methods”). As shown in Fig. 3C, we observed a significant concordance between the tree clades and the phage families annotated by PhaGCN_newICTV ( [58]; see also the “ Methods”). Specifically, genomes belonging to different phage families formed discrete clades on the phylogenetic tree, each with well-defined boundaries (Fig. 3C; outer ring). Notably, within each clade (family), we often found nonredundant vOTUs derived from multiple assemblers (Fig. 3C). For example, the Autographiviridae family contained 4 hq-vOTUs from the NGS assembler MEGAHIT (Fig. 3D), while other assemblers contributed 37 more hq-vOTUs to this family (Fig. 3E). More importantly, the terminase proteins from these genomes showed significant sequence divergence (Fig. S4), which was also evident from the long branch lengths on the phylogenetic tree (Fig. 3E). These results together indicate that our multi-assembler approach could indeed expand the gut virome identification by contributing assembler-specific and high-quality viral genomes.
Biases of different assemblers in recovering vOTUs at higher taxonomic levels
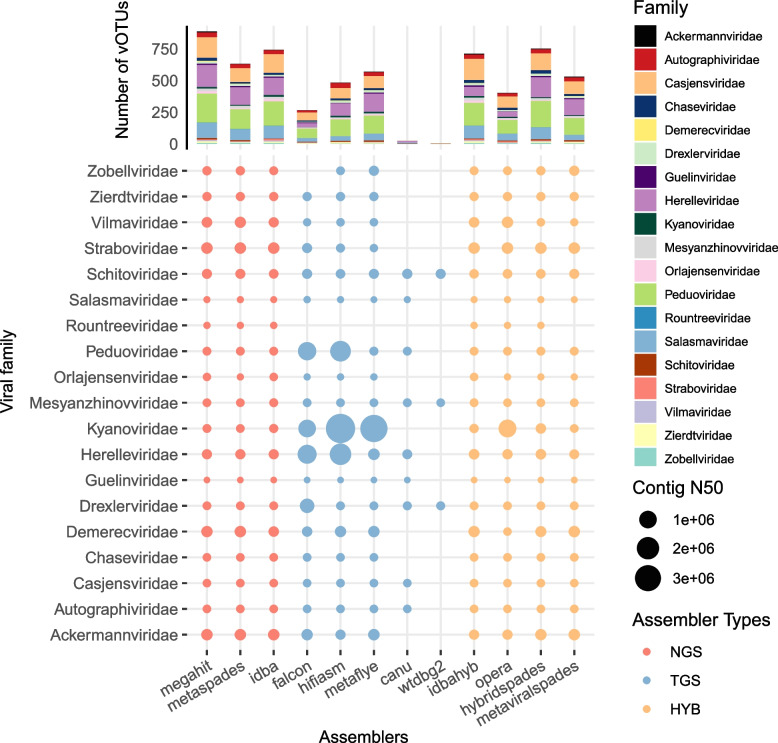
Next, we examined the overlaps in the identified viral contigs at higher taxonomic levels among all the assemblers. We annotated the hq-vOTUs into known viral families using PhaGCN_newICTV [58], resulting in 8 ~ 43% of annotation rates across the assemblers, with an average of ~ 16% (Fig. S5). A total of 19 viral families were annotated. All NGS assemblers were able to detect members of all families and so were all the hybrid assemblers except the metaViralSPAdes, which did not detect any members of the Rountreeviridae family (Fig. 4). Conversely, we observed significant performance variations among the TGS assemblers. Specifically, metaFlye and Hifiasm-meta could recover all families except the Rountreeviridae, while falcon additionally did not recover the Zobellviridae. Furthermore, wtdbg2 and canu missed majority of the families and recovered fewer family members when they did. Interestingly, all the TGS assemblers did not recover any members of the Rountreeviridae family; further study should be implemented to determine whether it is because of the fewer members presented in the human gut or its unique sequence and/or abundance characteristics.
Fig. 4.
Evaluation of taxonomic annotation of vOTUs assembled by different assemblers. The performance of each assembler in assembling nonredundant contigs of each virus family, the size of the dots represents the N50 of nonredundant contigs of that family of viruses assembled by that type of tool, the color of the dots represents the classification of the assembler, and a bar in the above representation represents the number of the contigs of each virus family assembled by that tool
Within each assembler category, we observed little difference in the performance of the three NGS assemblers in recovering viral families (Fig. 4). Hifiasm-meta and metaFlye, as TGS assemblers, assembled a broader range of viral families and increase the N50 values of several families. HybridSPAdes enabled the assembly of all families as well as being the most numerous in terms of contigs within the hybrid assemblers.
Together, our results indicate biases of different assemblers in recovering viral contigs at higher taxonomic levels, especially those of the TGS assemblers.
Different binners exhibit markedly distinct behaviors in the binning of vOTUs
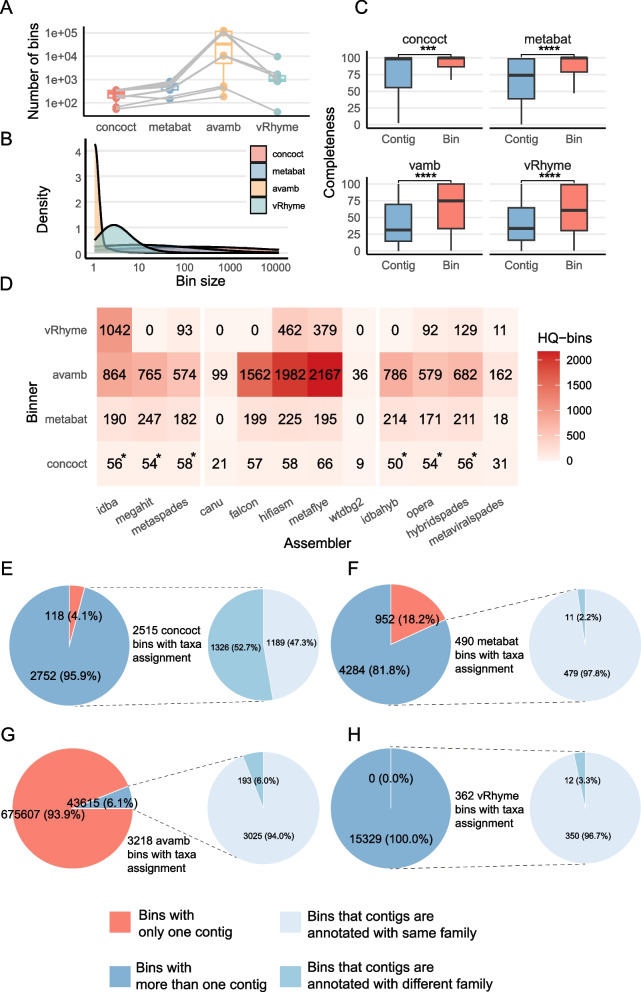
We also evaluated the performance of four binning tools on vOTUs, namely CONCOCT [38], MetaBAT2 [39], AVAMB [40], and vRhyme [41]. AVAMB consistently produced a greatest number of bins on all assemblers (Fig. 5A). Consequently, we found that bins created by CONCOCT contained a significantly high number of contigs (median 154) than those by other binners (MetaBAT2: median 8, AVAMB: median 1, vRhyme: median 2; p < 0.0001, Wilcoxon test; Fig. 5B and Fig. S6).
Fig. 5.
Evaluation of binning results generated by binners selected. A Boxplots showing the number of bins generated by four binning approaches using vOTUs recovered from the assemblers. Each dot represents an assembler. B Density plot showing the distribution of bin size (i.e., number of vOTUs in each bin) by four binning approaches. C Boxplot showing completeness improvement by the four binners. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon rank-sum test; ***p < 0.001, ****p < 0.0001. D Heatmaps showing the numbers of high-quality bins (i.e., CheckV completeness > 90% and no “contig > 1.5 × longer than expected genome length” and “high kmer _ freq may indicate large duplication” warning information) obtained from the assembler groups by the two binners, * indicates the number of bins after removal of oversized bins (i.e., concatenated length > 30 million base pairs) that cannot be analyzed by CheckV. E, F, G, H Pie charts showing the proportion of multi-vOTU bins obtained by four binners in which all member vOTUs are annotated to the same virus family
Subsequently, we applied the CheckV tool to assess the completeness and quality of the bins derived from the 12 assemblers and the 4 binners (“ Methods”). Of note, CONCOCT produced 95 oversized bins comprising thousands of contigs, which exceeded the capacity of CheckV for completeness evaluation. We thus excluded these oversized bins from further analysis.
Overall, we observed that all binning methods significantly improved the completeness of viral genomes when we compared the completeness of the bins to the member contigs with the highest completeness values (Fig. 5C; p < 0.01 in CONCOCT and < 0.0001 in others, Wilcoxon rank-sum test). AVAMB achieved the greatest improvement in completeness among all the binning tools (the average increased completeness per bin for AVAMB, CONCOCT, MetaBAT2, and vRhyme was 17.6, 3.04, 10.7, and 17.3 respectively, Table S2). We observed the same trends across almost all assemblers (Figs. S7, S8, S9, S10). Additionally, AVAMB consistently generated a greater number of HQ bins (i.e., those having > 90% completeness and no “contig > 1.5 × longer than expected genome length” and “high kmer _ freq may indicate large duplication” warning information) compared to other binners (Fig. 5D).
We proceeded to compare the consistency of taxonomy annotation results for contigs within bins. Strikingly, among the 2515 multi-contig bins generated by CONCOCT and were taxonomically annotated, more than half (52.7%, 1326) contained vOTUs that were annotated to different viral families (Fig. 5E). In contrast, 97.8% of the multi-vOTU bins by MetaBAT2 showed consistent annotations results within the same family (Fig. 5F). Notably, only 6.1% of AVAMB-generated bins contained more than one vOTUs. However, within these multi-bins, a high level of taxonomic annotation consistency was observed, with 94.0% (3025 multi-bins) displaying consistent taxonomic classifications (Fig. 5G). Conversely, all bins generated by vRhyme contained multiple vOTUs and exhibited high consistency (350 bins, 96.7%) in taxonomy annotations (Fig. 5H). These findings indicate that MetaBAT2, AVAMB, and vRhyme exhibits superior taxonomy annotations consistency than CONCOCT, while the latter tended to be more inclusive and cluster vOTUs from varying taxonomic levels.
Discussion
The human gut virome is an essential component of the human microbiome due to its significant impact on modulation of gut microbial structure and function [5, 65]. Metagenomic approaches are crucial for comprehensively studying the diverse and complex human gut virome, enabling the identification of novel viruses and understanding their functional roles [3, 9–11, 13]. Studies using both short-read and long-read assemblies of viral genomes have found that Illumina is preferable when using a single data type to recovering complete genomes [30]. However, the addition of long reads can improve the assembly of higher-quality genomes [29]. There are similar benchmarks for approaches to recovering the human gut viral genomes, but they used either only short-reads assemblers [27, 28] or only mock [27, 30] or in silico simulated [28] communities. Additionally, the number of real samples used in the benchmarking has been very small (e.g., n = 3 in ref [29]). Therefore, we lack a comparative evaluation of assembly tools on the efficacy of viral genome identification, especially for both next-generation sequencing (NGS) and third-generation sequencing (TGS) data from large number of samples. Here, we systematically evaluated the performance of 12 assemblers and 2 binners on a paired long- and short-read sequencing dataset consisting of 95 human fecal viral-like particle-enriched samples.
We first evaluated the number of contigs, completeness, contamination, and long-read metrics at the assembly level. We determined the MEGAHIT, metaFlye, and hybridSPAdes as the best metagenomic assemblers for short-read, long-read, and hybrid assemblies. We also found that third-generation sequencing (TGS) assemblers could enhance the N50 of Straboviridae, Peduoviridae, Kyanoviridae, and Herelleviridae viral family genomes, but they were not able to recover the genomes of some viral families, in particular canu and wtdbg2, which may due to the fact that they are not specifically designed to be applied to metagenomic data. In addition, the number of virus families depends on the type of virus family itself rather than the choice of tool.
We then found that contigs assembled using short-read and long-read data have little overlap, while the assembly results for short-read and hybrid data have considerable overlap, suggesting that the assembly of viral genomes is heavily influenced by the type of sequencing approaches. It is worth noting that the results from different tools are highly complementary to each other. Regardless of the categories of tools (i.e., NGS, TGS, or hybrid assemblers), the viral genomes identified by multiple assemblers significantly expand those of the individual tools. And we confirm that it is not mis-assembly that causes the difference between nonredundant contigs. Therefore, we suggest that when assembling metagenomic data from human gut virome, it is best to use multiple tools and merge the nonredundant results after making mis-assembly corrections. We also advocate the development of new tools and software suitable for the assembly of viral metagenomic data.
Of the four binners, we found that AVAMB outperformed others in terms of the number of high-quality bins and MetaBAT2 demonstrated the highest taxonomic consistency within bins. However, vRhyme exhibited well-balanced performance across all evaluated metrics. In conclusion, our findings suggest that future researchers can select different binning tools based on their specific requirements.
Despite our efforts, some genome fragment reassembly (assembly improving) tools (Phables [66], COBRA [67]) were ultimately not included in our study, because they only improve the length of 1 ~ 2% of the contigs assembled from randomly selected samples through reassembly (Table S3). The potential reasons for this outcome are the low quality of viral assemblies (with only about 10% of contigs being high quality) (Table S1) and the low viral abundance. Recently, studies [29, 66, 67] have shown that Phables and COBRA outperform binning tools in terms of genome completeness, contamination, and contiguity. This suggests that these tools may be more suitable for reassembling viral vOTUs in low-quality viral bins. PHAMB was not included in our evaluation as it is designed for selecting viral bins from metagenomic bins, which is not applicable to our VLP data. Moreover, our workflow already incorporates viral sequence identification. Additionally, the impact of trying different parameters during assembly was not tested due to the widespread use of default parameters in existing studies [9, 12] and vast time consumption. However, it is essential to adjust assembler parameters to accommodate specific data or situations. In future research, such attempts may help identify the most suitable parameters for optimal performance of different assemblers on various datasets.
In summary, our analysis pipeline, including both the dataset and performance evaluation matrices, could be easily adapted to test any new tools.
Conclusions
Based on a dataset comprising 95 paired long-read and short-read sequenced human fecal enriched virus-like particles, we conducted a comprehensive array of analyses encompassing raw data quality control, assembly, binning, viral sequence identification, and taxonomic annotation. In our examination of 12 assemblers and 4 binners, we observed that MEGAHIT, metaFlye, and hybridSPAdes exhibited superior performance within their respective categories grouped by data type. The various binners exhibited substantial differences in performance across multiple aspects. Furthermore, our findings indicate that vOTUs (viral operational taxonomic units) generated from diverse assemblers and data types demonstrated high complementarity and differentiation. This underscores the imperative of employing a multi-tool approach and encompassing multiple data types for the proficient recovery of viral genomes from virome data.
Supplementary Information
Supplementary Material 1: Figure S1. Pie chart showing the proportion of contamination greater than or equal to 5 and less than 5 for all vOTUs generated from the 12 assemblers. Figure S2. Upset plot shows assembler-specific hq-vOTUs of all assemblers. The connecting lines indicate the overlaps (hq-vOTUs present in a cluster when using a parameter of 95% sequence similarity for de-redundancy in our study) situation between the connected assemblers. Figure S3. Barplot of the number and proportion of hq-vOTUs generated from each assembler that carry Large terminase gene or not. Figure S4. Heatmap showing the results of a sequence-by-sequence comparison of large terminase proteins from hq-vOTUs annotated as the Autographivirid family, with color shades indicating sequence similarity. The identity between 0 and 95 is represented in blue, while that above 95 is shown in red. Figure S5. Barplot of the number and proportion of hq-vOTUs generated from each assembler that can be annotated by PhagCN to family level or not. Figure S6. Boxplots showing the size of bins (number of vOTUs contained in the bin) generated by CONCOCT and MetaBAT2 using vOTUs recovered from the assemblers. Wilcoxon test; ****: p < 0.0001. Figure S7. Boxplot showing completeness improvement of each assembler by CONCOCT. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, **: p < 0.01, ****: p < 0.0001. Figure S8. Boxplot showing completeness improvement of each assembler by MetaBAT2. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, **: p < 0.01, ****: p < 0.0001. Figure S9. Boxplot showing completeness improvement of each assembler by AVAMB. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, ****: p < 0.0001. Figure S10. Boxplot showing completeness improvement of each assembler by vRhyme. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, ****: p < 0.0001. Table S1. A list of basic information about all vOTUs, hq-vOTUs, non-redundant hq-vOTUs and hq-vOTUs with phagCN taxonomic annotations. Table S2. A list of completeness of all bins formed by binner selected and the vOTUs with the highest completeness in each bin. Table S3. A list of the original number of contigs input for the two assembly improvement tools and the number of contigs that showed improved assembly results.
Acknowledgements
We thank all members of the Chen, Zhao labs for their help related to this work.
Abbreviations
- NGS
Next-generation sequencing
- TGS
Third-generation sequencing
- VLP
Viral-like particle
- dsDNA
Double-stranded DNA
- CHGV
Chinese Human Gut Virome
- GVD
Gut Virome Database
- vOTUs
Viral operational taxonomic units
- hq-vOTUs
High-quality vOTUs
Authors’ contributions
WHC and XMZ designed the study. JC managed the sampling and did some of the experiments. CS and YL carried out quality control, preprocessing of the raw data and some data analysis. HW analyzed the data and wrote the draft manuscript. WHC and HW revised the manuscript through multiple rounds of discussions. All authors read and commented on the manuscript.
Funding
This research is supported by NNSF-VR Sino-Swedish Joint Research Programme (82161138017 to W. H. C.), National Natural Science Foundation of China (32070660 to W. H. C.; T2225015 and 61932008 to X. M. Z.), and National Key Research and Development Program of China (2020YFA0712403 to X. M. Z.; 2019YFA0905600 to W. H. C.).
Data availability
The raw sequencing data used in this study are available in the CNCB GSA database under accession code PRJCA008836 (accessible via either the GSA link https://ngdc.cncb.ac.cn/gsa/browse/CRA006494 or the BioProject page https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA008836).
The fasta files containing viral contigs generated by each assembler have been deposited to https://figshare.com/articles/dataset/Viral_contigs_of_Virome_Benchmark/25060193.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Tongji Medical College of Huazhong University of Science and Technology (No. S1241) and the Human Ethics Committee of the School of Life Sciences of Fudan University (No. BE1940).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xing-Ming Zhao, Email: xmzhao@fudan.edu.cn.
Wei-Hua Chen, Email: weihuachen@hust.edu.cn.
References
- 1.Shkoporov AN, Hill C. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe. 2019;25(2):195–209. [DOI] [PubMed] [Google Scholar]
- 2.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21(10):1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185(20):6220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol. 2021;39(5):578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen J, Zhang J, Mo L, Li Y, Li Y, Li C, Kuang X, Tao Z, Qu Z, Wu L, et al. Large-scale phage cultivation for commensal human gut bacteria. Cell Host Microbe. 2023;31(4):665-677 e667. [DOI] [PubMed] [Google Scholar]
- 6.Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4(1):4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin M, Chen J, Zhao X, Hu G, Wang H, Liu Z, Chen WH. An Engineered lambda Phage Enables Enhanced and Strain-Specific Killing of Enterohemorrhagic Escherichia coli. Microbiol Spectr. 2022;10(4):e0127122. [DOI] [PMC free article] [PubMed]
- 8.Ferri M, Ranucci E, Romagnoli P, Giaccone V. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57(13):2857–76. [DOI] [PubMed] [Google Scholar]
- 9.Gregory AC, Zablocki O, Zayed AA, Howell A, Bolduc B, Sullivan MB. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe. 2020;28(5):724–40 e728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Sun C, Dong Y, Jin M, Lai S, Jia L, Zhao X, Wang H, Gao NL, Bork P, et al. Efficient Recovery of Complete Gut Viral Genomes by Combined Short- and Long-Read Sequencing. Adv Sci (Weinh). 2024;11(13):e2305818. [DOI] [PMC free article] [PubMed]
- 11.Nishijima S, Nagata N, Kiguchi Y, Kojima Y, Miyoshi-Akiyama T, Kimura M, Ohsugi M, Ueki K, Oka S, Mizokami M, et al. Extensive gut virome variation and its associations with host and environmental factors in a population-level cohort. Nat Commun. 2022;13(1):5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camarillo-Guerrero LF, Almeida A, Rangel-Pineros G, Finn RD, Lawley TD. Massive expansion of human gut bacteriophage diversity. Cell. 2021;184(4):1098-1109 e1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayfach S, Paez-Espino D, Call L, Low SJ, Sberro H, Ivanova NN, Proal AD, Fischbach MA, Bhatt AS, Hugenholtz P, et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat Microbiol. 2021;6(7):960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung P, Eltahla AA, Lloyd AR, Bull RA, Luciani F. Understanding the complex evolution of rapidly mutating viruses with deep sequencing: beyond the analysis of viral diversity. Virus Res. 2017;239:43–54. [DOI] [PubMed] [Google Scholar]
- 15.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruan J, Li H. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 2020;17(2):155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–6. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Zhao N, Cao J, Liu X, Xu J, Ma Y, Yu Y, Zhang X, Zhang W, Guan X, et al. Short- and long-read metagenomics expand individualized structural variations in gut microbiomes. Nat Commun. 2022;13(1):3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin H, Quan K, He Q, Kwok L-Y, Ma T, Li Y, Zhao F, You L, Zhang H, Sun Z. A high-quality genome compendium of the human gut microbiome of Inner Mongolians. Nat Microbiol. 2023;8(1):150–61. [DOI] [PubMed] [Google Scholar]
- 21.Warwick-Dugdale J, Tian F, Michelsen ML, Cronin DR, Moore K, Farbos A, Chittick L, Bell A, Zayed AA, Buchholz HH, et al. Long-read powered viral metagenomics in the oligotrophic Sargasso Sea. Nat Commun. 2024;15(1):4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao L, Shi Y, Lau HC, Liu W, Luo G, Wang G, Liu C, Pan Y, Zhou Q, Ding Y, et al. Uncovering 1058 Novel Human Enteric DNA Viruses Through Deep Long-Read Third-Generation Sequencing and Their Clinical Impact. Gastroenterol. 2022;163(3):699-711. [DOI] [PubMed]
- 23.Cook R, Hooton S, Trivedi U, King L, Dodd CER, Hobman JL, Stekel DJ, Jones MA, Millard AD. Hybrid assembly of an agricultural slurry virome reveals a diverse and stable community with the potential to alter the metabolism and virulence of veterinary pathogens. Microbiome. 2021;9(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaulaurier J, Luo E, Eppley JM, Uyl PD, Dai X, Burger A, Turner DJ, Pendelton M, Juul S, Harrington E, et al. Assembly-free single-molecule sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 2020;30(3):437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warwick-Dugdale J, Solonenko N, Moore K, Chittick L, Gregory AC, Allen MJ, Sullivan MB, Temperton B. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ. 2019;7: e6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zablocki O, Michelsen M, Burris M, Solonenko N, Warwick-Dugdale J, Ghosh R, Pett-Ridge J, Sullivan MB, Temperton B. VirION2: a short- and long-read sequencing and informatics workflow to study the genomic diversity of viruses in nature. PeerJ. 2021;9:e11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutton TDS, Clooney AG, Ryan FJ, Ross RP, Hill C. Choice of assembly software has a critical impact on virome characterisation. Microbiome. 2019;7(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux S, Emerson JB, Eloe-Fadrosh EA, Sullivan MB. Benchmarking viromics: an in silico evaluation of metagenome-enabled estimates of viral community composition and diversity. PeerJ. 2017;5: e3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook R, Telatin A, Hsieh SY, Newberry F, Tariq MA, Baker DJ, Carding SR, Adriaenssens EM. Nanopore and Illumina sequencing reveal different viral populations from human gut samples. Microb Genom 2024;10(4):001236. [DOI] [PMC free article] [PubMed]
- 30.Cook R, Brown N, Rihtman B, Michniewski S, Redgwell T, Clokie M, Stekel DJ, Chen Y, Scanlan DJ, Hobman JL, et al. The long and short of it: benchmarking viromics using illumina, nanopore and PacBio sequencing technologies. Microb Genom. 2024;10(2):001198. [DOI] [PMC free article] [PubMed]
- 31.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–8. [DOI] [PubMed] [Google Scholar]
- 32.Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13(12):1050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng X, Cheng H, Portik D, Li H. Metagenome assembly of high-fidelity long reads with hifiasm-meta. Nat Methods. 2022;19(6):671–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolmogorov M, Bickhart DM, Behsaz B, Gurevich A, Rayko M, Shin SB, Kuhn K, Yuan J, Polevikov E, Smith TPL, et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods. 2020;17(11):1103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antipov D, Korobeynikov A, McLean JS, Pevzner PA. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 2016;32(7):1009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antipov D, Raiko M, Lapidus A, Pevzner PA. Metaviral SPAdes: assembly of viruses from metagenomic data. Bioinformatics. 2020;36(14):4126–9. [DOI] [PubMed] [Google Scholar]
- 37.Bertrand D, Shaw J, Kalathiyappan M, Ng AHQ, Kumar MS, Li C, Dvornicic M, Soldo JP, Koh JY, Tong C, et al. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat Biotechnol. 2019;37(8):937–44. [DOI] [PubMed] [Google Scholar]
- 38.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11(11):1144–6. [DOI] [PubMed] [Google Scholar]
- 39.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7: e7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindez PP, Johansen J, Kutuzova S, Sigurdsson AI, Nissen JN, Rasmussen S. Adversarial and variational autoencoders improve metagenomic binning. Commun Biol. 2023;6(1):1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kieft K, Adams A, Salamzade R, Kalan L, Anantharaman K. vRhyme enables binning of viral genomes from metagenomes. Nucleic Acids Res. 2022;50(14): e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangalea MR, Paez-Espino D, Kieft K, Chatterjee A, Chriswell ME, Seifert JA, Feser ML, Demoruelle MK, Sakatos A, Anantharaman K, et al. Individuals at risk for rheumatoid arthritis harbor differential intestinal bacteriophage communities with distinct metabolic potential. Cell Host Microbe. 2021;29(5):726-739 e725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shkoporov AN, Ryan FJ, Draper LA, Forde A, Stockdale SR, Daly KM, McDonnell SA, Nolan JA, Sutton TDS, Dalmasso M, et al. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome. 2018;6(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleiner M, Hooper LV, Duerkop BA. Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genomics. 2015;16(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d’Humieres C, Touchon M, Dion S, Cury J, Ghozlane A, Garcia-Garcera M, Bouchier C, Ma L, Denamur E. E PCR: a simple, reproducible and cost-effective procedure to analyse gut phageome: from phage isolation to bioinformatic approach. Sci Rep. 2019;9(1):11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai S, Pan S, Sun C, Coelho LP, Chen WH, Zhao XM. metaMIC: reference-free misassembly identification and correction of de novo metagenomic assemblies. Genome Biol. 2022;23(1):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–9. [DOI] [PubMed] [Google Scholar]
- 50.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, Kuhn JH, Lavigne R, Brister JR, Varsani A, et al. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat Biotechnol. 2019;37(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, Pratama AA, Gazitua MC, Vik D, Sullivan MB, et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren J, Song K, Deng C, Ahlgren NA, Fuhrman JA, Li Y, Xie X, Poplin R, Sun F. Identifying viruses from metagenomic data using deep learning. Quant Biol. 2020;8(1):64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren J, Ahlgren NA, Lu YY, Fuhrman JA, Sun F. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome. 2017;5(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Auslander N, Gussow AB, Benler S, Wolf YI, Koonin EV. Seeker: alignment-free identification of bacteriophage genomes by deep learning. Nucleic Acids Res. 2020;48(21): e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang Z, Tan J, Wu S, Li M, Xu C, Xie Z, Zhu H. PPR-Meta: a tool for identifying phages and plasmids from metagenomic fragments using deep learning. GigaScience. 2019;8(6):giz066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong Y, Chen WH, Zhao XM. VirRep: a hybrid language representation learning framework for identifying viruses from human gut metagenomes. Genome Biol. 2024;25(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mattock J, Watson M. A comparison of single-coverage and multi-coverage metagenomic binning reveals extensive hidden contamination. Nat Methods. 2023;20(8):1170–3. [DOI] [PubMed] [Google Scholar]
- 58.Shang J, Jiang J, Sun Y. Bacteriophage classification for assembled contigs using graph convolutional network. Bioinformatics. 2021;37(Suppl_1):i25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. [DOI] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian B, Gao S, Lercher MJ, Hu S, Chen WH. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47(W1):W270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hilbert BJ, Hayes JA, Stone NP, Xu RG, Kelch BA. The large terminase DNA packaging motor grips DNA with its ATPase domain for cleavage by the flexible nuclease domain. Nucleic Acids Res. 2017;45(6):3591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pargin E, Roach MJ, Skye A, Papudeshi B, Inglis LK, Mallawaarachchi V, Grigson SR, Harker C, Edwards RA, Giles SK. The human gut virome: composition, colonization, interactions, and impacts on human health. Front Microbiol. 2023;14: 963173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mallawaarachchi V, Roach MJ, Decewicz P, Papudeshi B, Giles SK, Grigson SR, Bouras G, Hesse RD, Inglis LK, Hutton ALK, et al. Phables: from fragmented assemblies to high-quality bacteriophage genomes. Bioinformatics. 2023;39(10):btad586. [DOI] [PMC free article] [PubMed]
- 67.Chen L, Banfield JF. COBRA improves the completeness and contiguity of viral genomes assembled from metagenomes. Nat Microbiol. 2024;9(3):737–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Figure S1. Pie chart showing the proportion of contamination greater than or equal to 5 and less than 5 for all vOTUs generated from the 12 assemblers. Figure S2. Upset plot shows assembler-specific hq-vOTUs of all assemblers. The connecting lines indicate the overlaps (hq-vOTUs present in a cluster when using a parameter of 95% sequence similarity for de-redundancy in our study) situation between the connected assemblers. Figure S3. Barplot of the number and proportion of hq-vOTUs generated from each assembler that carry Large terminase gene or not. Figure S4. Heatmap showing the results of a sequence-by-sequence comparison of large terminase proteins from hq-vOTUs annotated as the Autographivirid family, with color shades indicating sequence similarity. The identity between 0 and 95 is represented in blue, while that above 95 is shown in red. Figure S5. Barplot of the number and proportion of hq-vOTUs generated from each assembler that can be annotated by PhagCN to family level or not. Figure S6. Boxplots showing the size of bins (number of vOTUs contained in the bin) generated by CONCOCT and MetaBAT2 using vOTUs recovered from the assemblers. Wilcoxon test; ****: p < 0.0001. Figure S7. Boxplot showing completeness improvement of each assembler by CONCOCT. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, **: p < 0.01, ****: p < 0.0001. Figure S8. Boxplot showing completeness improvement of each assembler by MetaBAT2. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, **: p < 0.01, ****: p < 0.0001. Figure S9. Boxplot showing completeness improvement of each assembler by AVAMB. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, ****: p < 0.0001. Figure S10. Boxplot showing completeness improvement of each assembler by vRhyme. (The vOTU with the highest completeness in each bin is compared to the completeness of the entire bin). Pairwise Wilcoxon Rank Sum test; *: p < 0.05, ****: p < 0.0001. Table S1. A list of basic information about all vOTUs, hq-vOTUs, non-redundant hq-vOTUs and hq-vOTUs with phagCN taxonomic annotations. Table S2. A list of completeness of all bins formed by binner selected and the vOTUs with the highest completeness in each bin. Table S3. A list of the original number of contigs input for the two assembly improvement tools and the number of contigs that showed improved assembly results.
Data Availability Statement
The raw sequencing data used in this study are available in the CNCB GSA database under accession code PRJCA008836 (accessible via either the GSA link https://ngdc.cncb.ac.cn/gsa/browse/CRA006494 or the BioProject page https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA008836).
The fasta files containing viral contigs generated by each assembler have been deposited to https://figshare.com/articles/dataset/Viral_contigs_of_Virome_Benchmark/25060193.