Abstract
Background
The fatal diffuse midline gliomas (DMG) are characterized by an undruggable H3K27M mutation in H3.1 or H3.3. K27M impairs normal development by stalling differentiation. The identification of targetable pathways remains very poorly explored. Toward this goal, we undertake a multi-omics approach to evaluate replication timing profiles, transcriptomics, and cell cycle features in DMG cells from both H3.1K27M and H3.3K27M subgroups and perform a comparative, integrative data analysis with healthy brain tissue.
Results
DMG cells present differential replication timing in each subgroup, which, in turn, correlates with significant differential gene expression. Differentially expressed genes in S phase are involved in various pathways related to DNA replication. We detect increased expression of DNA replication genes earlier in the cell cycle in DMG cell lines compared to normal brain cells. Furthermore, the distance between origins of replication in DMG cells is smaller than in normal brain cells and their fork speed is slower, a read-out of replication stress. Consistent with these findings, DMG tumors present high replication stress signatures in comparison to normal brain cells. Finally, DMG cells are specifically sensitive to replication stress therapy.
Conclusions
This whole genome multi-omics approach provides insights into the cell cycle regulation of DMG via the H3K27M mutations and establishes a pharmacologic vulnerability in DNA replication, which resolves a potentially novel therapeutic strategy for this non-curable disease.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-024-03460-y.
Keywords: Diffuse midline gliomas H3 K27-altered, H3K27M, Cell cycle, Replication timing, Replication stress
Background
The temporal control of DNA replication, i.e., replication timing, is highly regulated. This is necessary to ensure that the genome is entirely and correctly duplicated during S phase. Replication timing is associated with the epigenetic landscape [1], gene expression [2], and cell fate [3]. Replication timing is highly dysregulated in diseases, including cancers [4, 5]. Alteration of the progression of the replication fork leads to replication stress, DNA damage, and associates with genomic instability [6, 7]. Replication stress creates vulnerabilities that can be therapeutically targeted in cancers [8]. Despite these observations, the relationship between specific cancer alterations, replication timing, and associated cellular processes remains largely unexplored.
Replication timing exhibits distinct features: initiation zones (origin of replication from which the replication forks diverge in opposite directions), timing transition regions (replication forks moving away from initiation sites), constant timing regions (early and late plateaus of replication), and termination sites (replication forks converging and terminating replication) [9]. Replication timing and its different features associate with distinct epigenetic patterns [1]. One such example is H3K27me3, which is enriched in regions replicating during mid to late S phase and associates with timing transition regions and termination sites [1]. The H3K27me3 landscape is reshaped in many cancers due to alterations in the chromatin modifiers writing or erasing this mark or in the histone H3 itself [10].
Diffuse midline gliomas (DMG), H3 K27-altered, are non-curable brain cancers primarily diagnosed in children. More than 80% of DMG feature a K27M mutation in either HIST1H3B/C, coding for the replicative histone H3.1, or H3F3A, coding for the variant H3.3 [11, 12]. These mutations define two DMG subgroups presenting differences in phenotypes, response to therapy, and prognosis [13]. Despite these differences, commonalities and dissimilarities between these subgroups remain largely unexplored outside of gene regulation.
While the H3.1 replicative histone is inserted in the entire genome as the replication fork progresses, the histone variant H3.3 is systematically deposited where it was before DNA replication, displacing nascent H3.1 and creating boundaries between H3.3 and H3.1 [14]. These H3.1/H3.3 boundaries demarcate the initiation zones of the early origins of replication [14]. In Drosophila, the genomic deposition of H3.1K27M and H3.3K27M is consistent with the DNA replication-dependent deposition of H3.1 and the predominantly replication-independent deposition of H3.3 [15]. In C. elegans, incorporation of the H3.3K27M mutation in one of the five H3.3 genes leads to germline defects: ectopic activation of DNA replication, accumulation of DNA damage, and aberrant progression of the cell cycle [16]. Corroborating the importance of cell cycle progression, oncohistones need to be incorporated in the chromatin via cell cycling for the K27M mutations to propagate a pathogenic effect [15]. While the relationship between oncohistones, cell cycle, and DNA replication was demonstrated in model organisms, replication timing within the DMG subgroups, its association with gene expression, their impact on measurable features of replication stress, and potential associated therapeutic vulnerabilities have yet to be described.
In this study, we leverage whole genome multi-omics approaches to evaluate DMG cells (H3.1K27M and H3.3K27M subgroups), normal astrocytes, tumors, and normal brain cells for novel cellular control that allows therapeutic vulnerabilities to be established for targetable pathways in DMG. Our studies resolved a critical role for H3K27M in replication timing genome-wide, while establishing an opportunity to therapeutically intervene with replication stress therapies.
Results
H3.1K27M DMG, H3.3K27M DMG, and normal astrocytes harbor differential DNA replication timing
To evaluate DMG H3 K27-altered in the context of the cell cycle, we explored replication timing throughout the S phase and gene expression profiles over the entire cell cycle in DMG and non-cancer brain cells. We leveraged a Fluorescence Activated Cell Sorting (FACS)-based method for these studies to avoid chemical-induced alterations and remove such confounders in the assessment [1]. With the goal of identifying new therapeutic opportunities that would not affect non-cancer cells in the same environment, we used three patient-derived pediatric DMG cell lines harboring the H3.1K27M mutation (DIPG4, DIPG21, DIPG33), three DMG cell lines harboring the H3.3K27M mutation (DIPG19, DIPG24, DIPG29), and two normal astrocyte cell lines (NHA, HA-bs). Oligodendrocyte precursor cells, the likely cell of origin of DMG [17], have a very low doubling capacity (2 weeks), which precludes their use for cell cycle-centric studies. However, astrocytes are abundant and have a comparable proliferative capacity in the normal brain, making them ideal to compare to the DMG cells. Each of these primary cell lines was sorted based on DNA content in four equal phases across S phase (S1, S2, S3, S4) for replication timing sequencing [1, 18], and in four phases across the entire cell cycle (G1, early S (ES), late S (LS), G2/M) for RNA sequencing [1] (Fig. 1A).
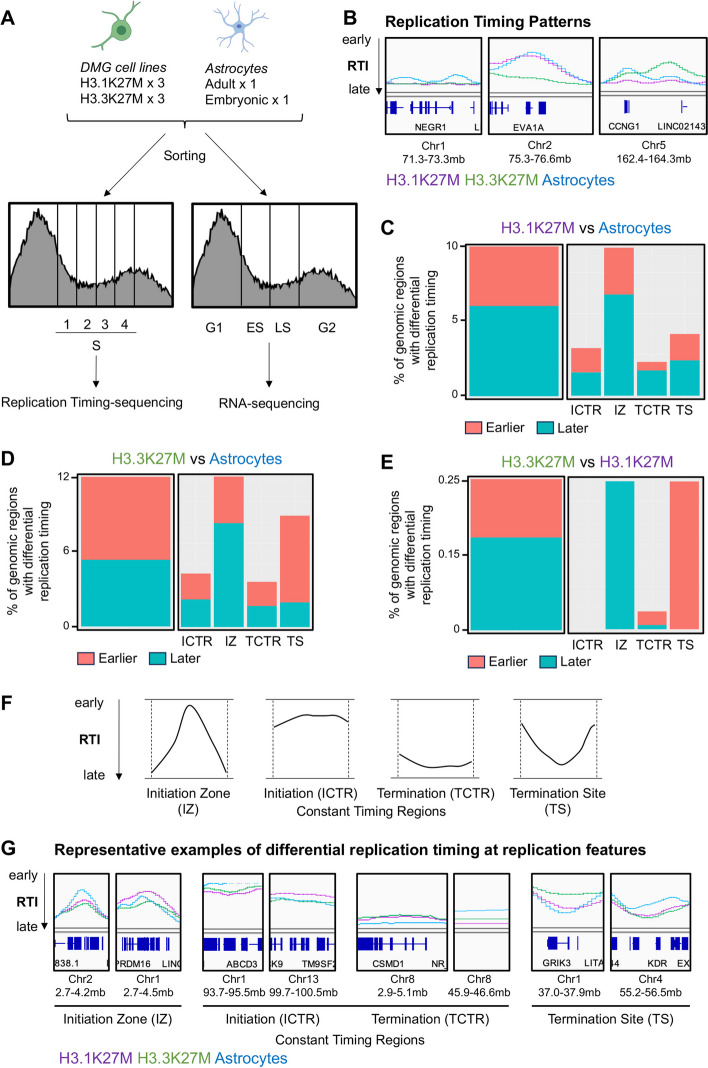
Fig. 1.
H3K27-altered DMG feature differential replication timing with H3.1K27M/H3.3K27M-subgroup specificities. A Schematic of the experimental setup. Three DMG cell lines of each subgroup (H3.1K27M and H3.3K27M) and two normal astrocyte cell lines were used. For replication timing sequencing S phase was divided into four equal cell populations. For cell-cycle RNA sequencing cells were divided into four groups consisting of G1, early S (ES), late S (LS), and G2/M. B Representative genomic tracks at the regions with differential replication timing between cell types (H3.1K27M in purple, H3.3K27M in green, astrocytes in blue), RTI = replication timing index (from top to bottom, replication earlier to later during S phase). Each line represents an average of biological replicates. C–E Differential replication timing of H3.1K27M (C) and H3.3K27M (D) compared to astrocytes and of H3.3K27M compared to H3.1K27M (E) (earlier and later replication represented in coral and teal, respectively). Left panels: percentage of genomic regions presenting differential replication timing between respective cell types. Right panels: differential replication timing at specific replication features. H3.1K27M/Astrocytes: 2224 regions replicating earlier and 3379 regions replicating later; H3.3K27M/Astrocytes: 3780 regions replicating earlier and 3038 regions replicating later; H3.3K27M/H3.1K27M: 60 regions replicating earlier and 96 regions replicating later. F Schematic representation of replication features: IZ = initiation zones, TCTR = termination constant timing regions, ICTR = initiation constant timing regions, TS = termination sites. G Representative examples of genomic regions with differential replication timing at replication features
Replication timing sequencing characterizes the time during S phase at which specific genomic regions duplicate. Cells are pulse-labeled with BrdU and FACS sorted in four phases across S phase based on their DNA content. The BrdU-labeled DNA is then immunoprecipitated and prepared for sequencing [1, 18, 19]. This method maps nascent DNA across S phase, therefore determining which regions of the genome replicate “early” or “late” during the DNA replication process. This information is translated into a replication timing index (RTI): quantification of replication timing at a given genomic region as one continuous value between 0 and 1, the strongest read density in early (S1) and late (S4) phase resulting in RTI = 0.10 or 0.91, respectively (Fig. 1B) [1, 20].
Using this approach, we observed that H3.1K27M, H3.3K27M, and astrocytes presented differential replication timing patterns at various genomic loci (Fig. 1B and Additional file 1: Fig. S1). Some regions presented differential replication timing for astrocytes (Fig. 1B, left panel), while others presented differential replication timing for only one subgroup of DMG (Fig. 1B, middle panel), or both DMG subgroups and astrocytes presented unique patterns (Fig. 1B, right panel). Overall, H3.1K27M and H3.3K27M presented differential replication timing of 10% and 12% of the genome when compared to astrocytes, respectively (Fig. 1C–D). Over half of the regions presenting differential timing replicated later in H3.1K27M DMG than in astrocytes (2224 regions replicating earlier and 3379 regions replicating later) (Fig. 1C, left panel), whereas over half of the regions presenting differential timing replicated earlier in H3.3K27M DMG than in astrocytes (3780 regions replicating earlier and 3038 regions replicating later) (Fig. 1D, left panel). While comparing directly H3.3K27M to H3.1K27M presented a much higher variability, which is anticipated from cell lines established post-mortem from patients that underwent different treatment regimens, these genotypes presented differential replication timing of 0.25% of the genome (60 regions replicating earlier and 96 regions replicating later) (Fig. 1E).
DNA replication exhibits local replication features (Fig. 1F–G). These defined features include initiation zones (IZ, clusters of origin of replication where the replication forks diverge in opposite directions from the same point, Fig. 1F left panel), constant timing regions (ICTR and TCTR, early and late plateaus of replication, Fig. 1F middle panels), and termination sites (TS, replication forks converging and terminating replication, Fig. 1F right panel) [9]. In comparison to normal astrocytes, H3.1K27M cell lines presented genomic regions exhibiting local DNA replication features replicating earlier or later, with a higher proportion of differential replication at initiation zones (Fig. 1C, right panel). H3.3K27M cell lines also presented genomic regions exhibiting local DNA replication features replicating earlier or later, but with a higher proportion of differential replication at initiation zones and termination sites (Fig. 1D, right panel). H3.1K27M and H3.3K27M differed mainly at initiation zones and termination sites, with H3.3K27M cell lines presenting later replication at initiation zones and earlier replication at termination sites (Fig. 1E). These results indicate that H3.1K27M DMG cell lines, H3.3K27M DMG cell lines, and astrocyte cell lines present distinct replication timing profiles.
Differential replication timing correlates with differential gene expression
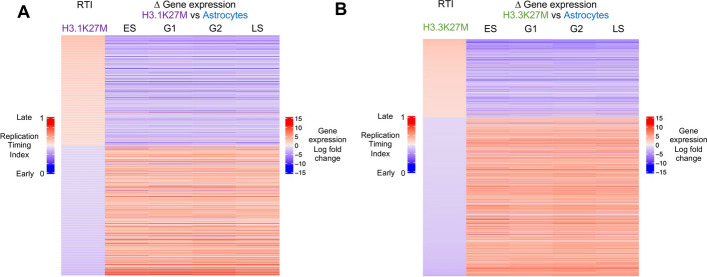
We then assessed whether altered replication timing was connected to differential gene expression in DMG. Regions presenting differential replication timing between DMG cell lines and astrocytes were compared to the differential gene expression within these regions in any given cell cycle phase (Fig. 2A–B, Additional file 1: Fig. S2A, Additional file 2: Tables S1–S3). Regions replicating later in DMG cell lines (RTI > 0.5, red) were associated with downregulated gene expression (log fold change < 0, blue) and vice versa (Fig. 2A–B). This correlation was also observed between DMG subgroups (Additional file 1: Fig. S2A). These regions were enriched in initiation zones when comparing H3.1K27M to astrocytes (Additional file 1: Fig. S2B), in initiation zones and termination sites when comparing H3.3K27M to astrocytes (Additional file 1: Fig. S2C), and in termination sites when comparing H3.3K27M to H3.1K27M (Additional file 1: Fig. S2D). These data are consistent with studies linking earlier replication timing to gene expression and later replication timing with gene suppression [2].
Fig. 2.
Differential replication timing correlates with differential gene expression within the same genomic regions. A–B Heatmap correlating the replication timing index (RTI, the value between 0 for early and 1 for late replication) with gene expression changes in the same regions (downregulated and upregulated in blue and red, respectively) for the genomic regions presenting differential replication timing between H3.1K27M DMG cell lines and astrocytes (872 regions, A) and between H3.3K27M and astrocytes (1369 regions, B)
H3.1K27M and H3.3K27M DMG exhibit changes in gene expression in all phases of the cell cycle when compared to normal astrocytes
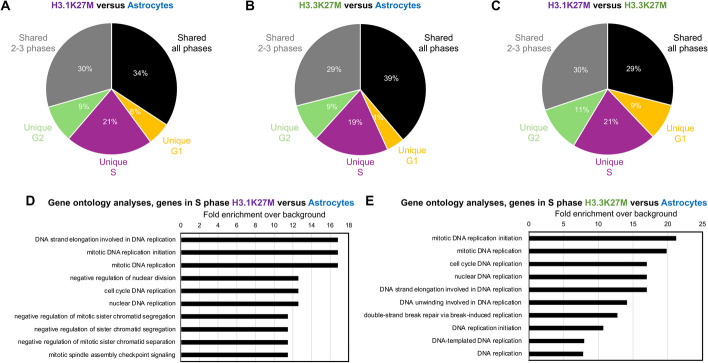
Given the patterns of differential gene expression and replication timing, we further investigated gene expression in different cell cycle phases (G1, ES, LS, G2/M) in H3.1K27M DMG cell lines, H3.3K27M DMG cell lines, and astrocytes (Fig. 1A). When comparing DMG subgroups and astrocytes, we identified genes exhibiting differential expression specifically in each cell cycle phase and genes with differential expression across several phases (Fig. 3A–C and Additional file 1: Fig. S3A–C). Although most of the genes with differential expression were shared among all cell cycle phases (34% for H3.1K27M versus astrocytes (Fig. 3A and Additional file 1: Fig. S3A), 39% for H3.3K27M versus astrocytes (Fig. 3B and Additional file 1: Fig. S3B), and 29% for H3.1K27M versus H3.3K27M (Fig. 3C and Additional file 1: Fig. S3C)), many genes presented differential gene expression uniquely in S phase (21% for H3.1K27M versus astrocytes (Fig. 3A and Additional file 1: Fig. S3A), 19% for H3.3K27M versus astrocytes (Fig. 3B and Additional file 1: Fig. S3B), and 21% for H3.1K27M versus H3.3K27M (Fig. 3C and Additional file 1: Fig. S3C)).
Fig. 3.
H3K27-altered DMG differentially express DNA replication-related pathways in S phase. A-C Pie charts representing the percentages of genes differentially expressed uniquely in G1, S, or G2, shared between 2 and 3 cell cycle phases, or shared across all phases between H3.1K27M DMG cell lines and astrocytes (A), between H3.3K27M DMG cell lines and astrocytes (B), and between H3.1K27M and H3.3K27M cell lines (C). D–E Gene Ontology analysis of the biological processes for differentially expressed genes in S phase in H3.1K27M compared to astrocytes (D) and in H3.3K27M compared to astrocytes (E). Fold enrichment = over-representation over background
Gene ontology analyses [21–23] of genes differentially expressed in S phase between DMG cell lines and astrocytes revealed enrichment in DNA replication-related pathways in both H3.1K27M and H3.3K27M (Fig. 3D–E). While the number of differentially expressed genes in S phase between H3.1K27M and H3.3K27M subgroups were too small for statistically significant results after gene ontology analyses (420 genes, Additional file 1: Fig. S3C), the top two PANTHER pathways were threonine biosynthesis and salvage pyrimidine deoxyribonucleotides (data not shown). Of note, thymidine kinase 1, an enzyme regulating intracellular thymidine pool and tightly regulated across the cell cycle [24, 25], presented significantly higher expression in H3.1K27M specifically in the early S phase.
H3.1K27M and H3.3K27M DMG exhibit altered cell cycle regulation of DNA replication genes
Because genes involved in DNA replication-related pathways were differentially expressed between DMG cell lines and normal astrocytes in S phase, we took the list of 39 genes under the DNA replication category of PANTHER [23] to assess their expression across the cell cycle in our cell lines. For this, we computed the ratio of expression between each consequent phase in the cell cycle (G1 to ES, ES to LS, LS to G2, G2 to G1) and applied hierarchical clustering to the gene expression ratios [26]. From this analysis, we observed six major clusters across 38 DNA replication genes (one gene was removed from the results because of its lack of expression in all cell lines used, Fig. 4A).
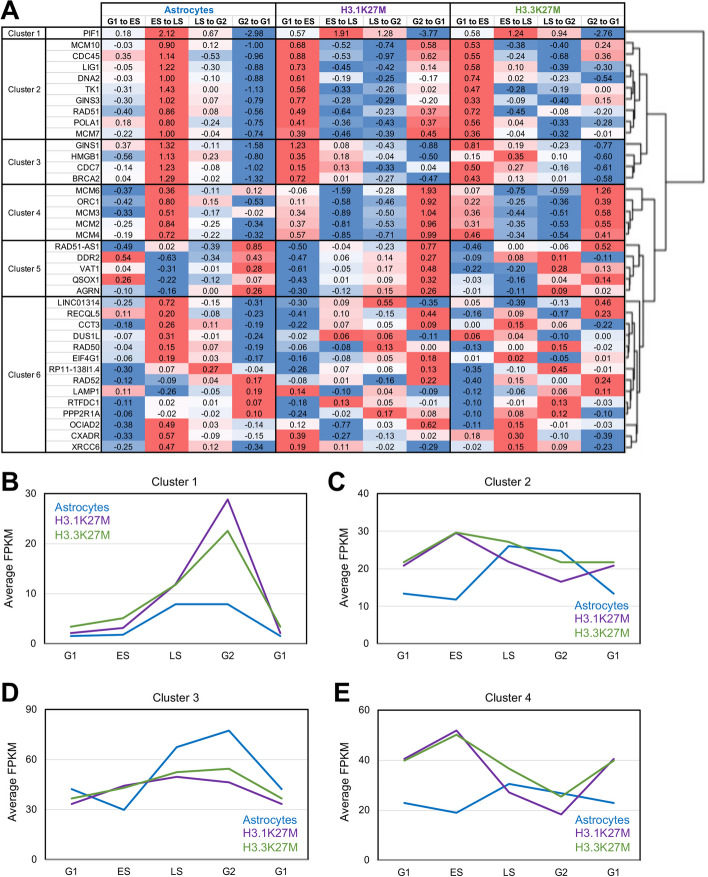
Fig. 4.
H3.1K27M and H3.3K27M DMG exhibit altered cell cycle regulation of DNA replication genes. A Heatmap representing the clustering of DNA replication genes (from PANTHER) according to their differential expression in subsequent phases of the cell cycle in astrocytes, H3.1K27M, and H3.3K27M DMG cell lines. Numbers represent the ratio of expression in one phase compared to the previous one (G1 to ES: expression in ES divided by expression in G1, etc.). Color: scale of blue to red with blue = downregulated from one phase to the next phase and red = upregulated from one phase to the next phase. B–E Graphs representing the average of FPKM (fragments per kilobase per million mapped fragments) of genes from cluster 1 (B), cluster 2 (C), cluster 3 (D), and cluster 4 (E) for each phase of the cell cycle and each subgroup (astrocytes in blue, H3.1K27M in purple, H3.3K27M in green)
Clusters one, two, three, and four contained genes directly involved in DNA replication such as the replication complex itself and the homologous recombination DNA repair machinery involved in replication fork preservation [27]. These clusters presented the most variation of expression across the cell cycle. The only gene in cluster one, PIF1, encoding an RNA helicase at replication forks [28], started to be upregulated between G1 and ES in DMG cells and continued to be until G2. In contrast, it was only upregulated between ES and LS in normal astrocytes (Fig. 4A–B). While genes in clusters two and three were upregulated between ES and LS in astrocytes and downregulated between G2 and G1, these genes were upregulated earlier in DMG cells, between G1 and ES (Fig. 4A, C–D). Of note, some of these genes even started to be upregulated between G2 and G1 in H3.1K27M DMG specifically (cluster two, Fig. 4A). In astrocytes, genes in cluster four were upregulated at the same time as the ones in clusters two and three (between ES and LS), but they were upregulated much earlier in DMG cells, between G2 and G1 (Fig. 4A, E).
Clusters five and six contained genes more indirectly involved in DNA replication and presented a more stable expression across the cell cycle in astrocytes and DMG subgroups (Fig. 4A and Additional file 1: Fig. S4A–B). Of note, genes in cluster five were overall less expressed in H3.3K27M DMG in comparison to astrocytes and H3.1K27M DMG (Additional file 1: Fig. S4A), while genes in cluster six were less expressed in both DMG subgroups in comparison to astrocytes (Additional file 1: Fig. S4B).
Together, DNA replication-related genes were differentially regulated across the cell cycle in DMG H3 K27-altered compared to astrocytes, with commonalities and dissimilarities between DMG subgroups.
DMG cell lines present shorter inter-origin distance and slower replication forks than normal astrocytes
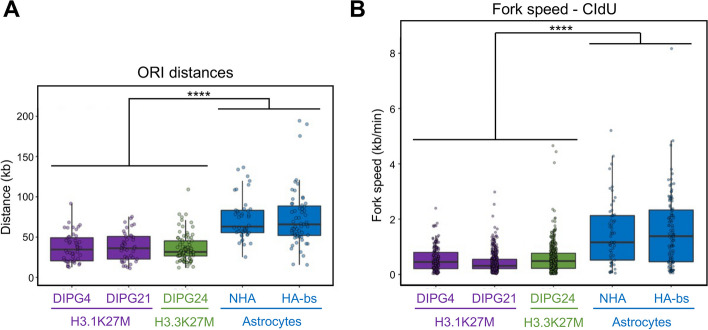
Because alteration of replication timing can be linked to changes in DNA replication forks [29], we then performed DNA fiber assays to determine potential alterations in replication fork dynamics genome wide. These assays revealed that DMG cells from both subgroups presented a significantly shorter inter-origin distance (ORI) than normal astrocytes (median of ~ 35 kb compared to ~ 65 kb, Fig. 5A), suggesting that more origins are fired in these cells during S phase. Furthermore, fork speed was significantly slower in DMG cells in comparison to normal astrocytes (median of ~ 0.5 kb/min compared to ~ 1 kb/min, Fig. 5B and Additional file 1: Fig. S5A).
Fig. 5.
H3K27-altered DMG have more densely active origins of replication and slower replication fork speed. A Box plot representing the distance in kilobases between putative active origins of replication in H3.1K27M (purple), H3.3K27M (green), and astrocytes (blue). B Box plot representing DNA replication fork speed in kilobase per minute analyzed by chlorodeoxyuridine (CldU) labeling in H3.1K27M (purple), H3.3K27M (green), and astrocytes (blue). Box plots show the median and the interquartile range (IQR), and whiskers depict the smallest and largest values within 1.5 × IQR. Statistics: Wilcoxon’s t test: **** p < 0.0001
Because slow replication fork speed is a hallmark/feature of replication stress [7], we anticipated that these cells would have increased DNA damage. Consistent with this notion, DMG cell lines from both subgroups presented phosphorylation of the histone variant H2A.X at Serine 139 when compared to normal astrocytes (gH2AX, Additional file 1: Fig. S5B).
Collectively, these data suggest that H3K27M promotes replication-associated cell cycle gene misregulation as well as replication timing alterations that are associated with replication stress.
DMG tumors feature a replication stress signature
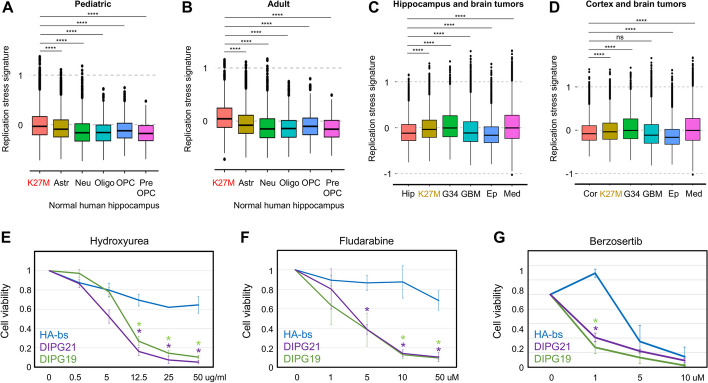
We then aimed to determine whether DMG tumors presented replication stress. We analyzed single-cell RNA sequencing data from DMG tumors and normal human hippocampus and assessed the presence of a replication stress response signature defined by the overlap between three key characteristics associated with replication stress (oncogene amplification, phospho-CHK1 protein expression, cell cycle checkpoint inhibitor response) [30]. Pediatric and adult tumors presented a higher replication stress signature than other cell types within the normal brain hippocampus (astrocytes, neurons, oligodendrocytes, oligodendrocyte precursor cells, pre-oligodendrocyte precursor cells; Fig. 6A–B).
Fig. 6.
H3K27-altered DMG present replication stress and are vulnerable to replication stress therapy. A Replication stress signature score (as defined in [30]) in pediatric DMG tumors (n = 14) and normal human hippocampus (n = 6) from single-cell RNA sequencing. K27M = DMG, Astr = astrocytes, Neu = neurons, Oligo = oligodendrocytes, OPC = oligodendrocyte precursor cells, Pre-OPC = pre- oligodendrocyte precursor cells. B Replication stress signature in adult DMG tumors (n = 4) and normal hippocampus (n = 6) from single-cell RNA sequencing. K27M = DMG, Astr = astrocytes, Neu = neurons, Oligo = oligodendrocytes, OPC = oligodendrocyte precursor cells, Pre-OPC = pre- oligodendrocyte precursor cells. C Replication stress signature in normal hippocampus (Hip, n = 6), DMG tumors (K27M, n = 18), diffuse hemispheric glioma G34-mutant (G34, n = 9), glioblastoma (GBM, n = 35), ependymoma (Ep, n = 18), and medulloblastoma (Med, n = 36) from single-cell RNA sequencing. D Replication stress signature in normal cortex (Cor, n = 48), DMG tumors (K27M, n = 18), diffuse hemispheric glioma G34-mutant (G34, n = 9), glioblastoma (GBM, n = 35), ependymoma (Ep, n = 18), and medulloblastoma (Med, n = 36) from single-cell RNA sequencing. Statistics: adjusted p-value: ns p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. E–G Cell viability assays in the presence of the replication stress therapeutic hydroxyurea (E), fludarabine (F), and berzosertib (G) for 72 h (CellTiter-Glo 2.0 Cell Viability Assay, Promega). HA-bs (astrocytes), DIPG21 (H3.1K27M), and DIPG19 (H3.3K27M) are represented. Cell viability was calculated as a fraction of the control (water for HU and DMSO for fludarabine and berzosertib) viability. Statistics: Student’s t test: * p < 0.05 related to HA-bs
We further explored whether this replication stress signature would be shared with additional brain tumors. Our comparison of normal hippocampus (Fig. 6C) or normal cortex (Fig. 6D) to DMG, diffuse hemispheric glioma G34-mutant, glioblastoma, ependymoma, and medulloblastoma demonstrates the replication stress signature to be present in oncohistone-driven brain tumors (DMG, G34) and medulloblastoma (Fig. 6C–D).
DMG cells are susceptible to replication stress therapeutics in comparison to normal astrocytes
Based on the multi-omics data we generated in the cell models and linked to tumors, we assessed whether DMG cell lines could be sensitive to replication stress therapy. Specifically, we tested three clinically used compounds: the anti-metabolite hydroxyurea, which reduces the production of dNTPs, the purine analog fludarabine, which inhibits DNA synthesis through multiple mechanisms, and the ATR inhibitor berzosertib, which prevents DNA damage checkpoint activation. DMG cell lines were much more sensitive to all three compounds than astrocytes (Fig. 6E–G and Additional file 1: Fig. S6). Of note, the only DMG cell line that did not present increased sensitivity to replication stress therapeutics in comparison to normal astrocytes (Additional file 1: Fig. S6) presented very low levels of gH2AX in comparison to other DMG cell lines (Additional file 1: Fig. S5B). Together, these data established a pharmacologic vulnerability in DNA replication, which resolved a potentially novel therapeutic strategy for this non-curable disease.
Discussion
Despite the recent advances in the understanding of the biology of DMG H3 K27-altered, these brain cancers are still not curable [31]. The K27M mutation is found in two histone genes: HIST1H3B/C, coding for the replicative histone H3.1, and H3F3A, coding for the histone variant H3.3 [11, 12]. These mutations define two DMG subgroups presenting differences in phenotypes, response to therapy, and prognosis [13].
In contrast to histone variants that are expressed throughout the cell cycle, replicative histones are expressed only during the S phase, so that newly synthesized DNA becomes properly chromatinized. H3.1 and H3.3 are enriched at specific places of the genome and have independent and complementary roles [14]. During DNA replication, old histones are recycled, and new histones are deposited. In the context of histone H3, the newly deposited histones are the replicative H3.1/2 [14]. Later during the S phase, H3.3 is repopulated at the regions it was enriched before replication [14]. This highly regulated deposition was recently shown to be important for proper DNA replication [14]. However, to the best of our knowledge, the consequences of the K27M oncogenic mutations in respective histones H3 variants on the timing of DNA replication had still to be determined.
Here, we demonstrate that DMG cell lines harboring the H3.1K27M or H3.3K27M mutations presented differential replication timing in comparison to astrocytes and between themselves. We further demonstrate that regions presenting differential replication timing also presented differential gene expression, with regions replicating earlier during the S phase increasing gene expression, and regions replicating later during the S phase decreasing gene expression. These results are consistent with active genomic compartments replicating early and inactive genomic compartments replicating late [2]. During development, different cell types harbor differential replication timing and gene expression [3]. The observation of differential replication timing for 10–12% of the genome in DMG cell lines compared to astrocytes may not be surprising, considering these to be different cell types [DMG being stalled in a cancer stem-cell-like state [17]]. However, a causal relationship between replication timing and gene expression has not been firmly established, and, in some instances, such relationships do not exist. For example, the differential replication timing due to the overexpression of a lysine demethylase in diploid cells did not correlate with changes in gene expression [1]. While further studies are necessary to determine potential causalities between replication timing and gene expression, a possibility would be for such relationships to exist when considering different cell types or when cell states are altered. Therefore, the fact that differential timing between H3.1K27M and H3.3K27M DMG cell lines correlated with differential gene expression could suggest these two DMG subgroups to be in different cell states and/or stalled in distinct differentiation stages. This would be consistent with recent findings highlighting the possibility of different cells of origin for these two subgroups [32].
Expression of genes involved in DNA replication, such as ORC1 and MCMs, is tightly regulated across the cell cycle [33, 34]. However, a limitation of these previous studies is the use of drugs altering DNA replication to synchronize the cells, which could itself alter gene expression. Using cell sorting based on DNA content, we demonstrate that, in normal astrocytes, genes encoding the origin recognition complex, DNA replication licensing factors, DNA polymerase, and DNA repair machinery associated with DNA replication, presented increased expression between early S and late S phase. In DMG cell lines, these genes presented increased expression much earlier during the cell cycle, between G2 and G1 or between G1 and ES. Furthermore, these genes, especially cluster four (ORC1, MCM2-3–4–6), presented higher expression in DMG cells compared to astrocytes. ORC1 acts as a nucleating center for origin recognition complex assembly and then pre-replication complex assembly [35], having more ORC1 expressed earlier during the cell cycle could result in more origins firing, as observed in our assays, and in alteration in the timing of replication. The specific relationship between gene expression of DNA replication genes and alterations of DNA replication will need to be determined in future studies.
Increased replication initiation and/or origin firing can lead to the depletion of nucleotide pools, and, in turn, to slower fork speed and replication stress [7]. We noted the increased expression of thymidine kinase 1 (TK1) specifically in the early S phase of H3.1K27M DMG cells. TK1 is a cell cycle-regulated cytosolic enzyme from the DNA salvage pathway involved in regenerating thymidine for DNA synthesis [24, 25]. Future studies will need to determine whether this enzyme is a dependency for this subgroup of DMG. Of note, a genome-wide CRISPR screen identified de novo pyrimidine biosynthesis as a dependency in DMG cells. Inhibiting an enzyme from this pathway arrested the cell cycle by stalling replication forks, leading to replication stress and apoptosis in DMG cells but not in astrocytes [36]. Replication defects due to the overexpression of H3.3K27M were also reported in another study, revealing increased genomic instability upon replication stress [37]. Moreover, expression of H3.1K27M in fibroblasts decreased double-strand break repair induced by ionizing radiation and increased genome instability [38].
Standard of care therapy in DMG is radiation therapy, with a short-lived response. Replication stress therapy has the potential to increase sensitivity to radiotherapy. Hydroxyurea has been used as a radiosensitizer in combination therapies against cancers such as head and neck and cervix [39]. Furthermore, hydroxyurea, fludarabine, and berzosertib have the capacity to cross the blood–brain barrier, a limitation of therapeutic strategies in brain cancers. Future studies will need to assess whether such a combination could be beneficial in mice models of DMG.
Conclusions
In this study, we demonstrated that, in comparison to normally dividing brain cells, DMG cell lines harboring the H3.1K27M or H3.3K27M mutations presented differential replication timing that correlated with differential gene expression. We further demonstrated that DNA replication-related genes were differentially regulated across the cell cycle and that DMG cell lines presented a smaller distance between the origin of replication and slower replication forks. Confirming findings in patient samples, DMG tumors presented a higher replication stress signature than the normal hippocampus. Finally, we uncovered a specific susceptibility of DMG cells to replication stress therapy. In summary, a comprehensive characterization of DNA replication timing in DMG and associated gene expression revealed targeting DNA replication as a potentially novel therapeutic strategy against DMG.
Methods
Cell culture
DMG cell lines were obtained from Michelle Monje (Stanford University). DMG cell lines were cultured in a base medium consisting of equal parts Neurobasal-A Medium (Thermofisher, 10,888,022) and DMEM/F-12 (Gibco, 11,330,032) to which were added 1% of the following: 1 M HEPES (Gibco, 15,630,106), GlutaMAX Supplement (Gibco, 35,050,061), 100 × MEM Non-Essential Amino Acids (Gibco, 11,140,050), 100 mM Sodium Pyruvate (Gibco, 11,360,070), and 100 × Antibiotic–Antimycotic (Gibco, 15,240,096). This base medium was supplemented with the following growth factors: 20 ng/ml EGF (STEMCELL, 78,006), 20 ng/ml FGFb (STEMCELL, 78,003), 10 ng/ml PDGF-AA (Shenandoah Biotechnology, 100-16AF-100UG), and 10 ng/ml PDGF-BB (Shenandoah Biotechnology, 100–18-100UG); and with 0.2% of B-27 Supplement Minus Vitamin A (50 ×) (Gibco, 1,287,010) and Heparin Solution (STEMCELL, 07980). Cells were passaged using the cell separator TrypLE Express (Gibco, 12,604,039) and Hanks’ Balanced Salt Solution (Corning, 21–022-CV) and resuspended in media as a single cell suspension. Cells were allowed to form neurospheres in preparation for experiments.
Normal human astrocytes (NHA) (Lonza, CC-2565) and Human Astrocytes-brain stem (HA-bs) (Sciencell, 1840) were purchased directly from the suppliers and cultured following the manufacturer’s instructions.
Cell lines were not authenticated after obtention. Cell lines were used at low passages and were free of mycoplasma contamination.
Cell viability assays
Cells were seeded at 1 × 103 density in triplicate in 96 well plates. After 24 h, media containing 2 × drug concentration was added in equal parts to media and cells in the plate. Hydroxyurea (Selleck Chemicals, S1896), fludarabine (MedChem Express, HY-B0069), and berzosertib (Selleck Chemicals, S7102) were incubated with cell lines for 72 h. Then, cell viability was measured using CellTiter-Glo 2.0 Cell Viability Assay (Promega, G9243) following the manufacturer’s instructions.
DNA fiber assays
DNA fibers were prepared as described [29] based on [40]. Cells were sequentially pulse-labeled with 25 mM IdU (Sigma, I7125) and 50 mM CldU (Sigma, C6891) for 30 min each and harvested. Labeled cells were lysed by the lysis buffer (0.5% SDS in 200 mM Tris–HCl, pH7.4, 50 mM EDTA) and extracted DNA fibers were stretched onto the slide glass by tilting. The fibers were fixed in methanol/acetic acid (3:1), then denatured with 2.5 M HCl for 1 h, neutralized with PBS, and blocked with blocking buffer (1% BSA, 0.1% Tween 20 in PBS). CldU and IdU tracks were detected with anti-BrdU antibodies recognizing CldU (Abcam, Ab6326) and IdU (BD Biosciences, 347,580), respectively, and appropriate secondary antibodies (Thermo Fisher SCIENTIFIC, A11001 and A11077). Images were acquired on a Leica SP8 confocal microscope using a 40 × Plan/Apo NA1.3 oil immersion objective (Leica) at 2048 × 2048 pixels at an effective pixel size of 142 nm. Images were converted from Leica image format to TIFF files and fibers were annotated manually with a custom Fiji macro. From the saved fiber regions of interest, fiber patterns were automatically found in both CldU and IdU channels by a custom-build software written in Python (https://github.com/IES-HelmholtzZentrumMunchen/dna-fibers-analysis) followed by filtering for ongoing forks. Statistical analysis and preparation of plots was carried out in “R” version 4.1.2 with ggplot2 version 3.3.6. To calculate fork speed, we used the established conversion 1 mm = 2 kb [41]. To obtain the inter-origin distances, label boundaries were selected manually from annotated fiber regions aided by a custom Fiji macro. Origins were defined as the middle between two consecutive label boundaries along each fiber. All distances between neighboring origins were then used for statistical evaluation and plotting in “R,” using the same conversion factor as above.
Replication timing sequencing
We adapted our replication timing sequencing protocol from two published protocols [18, 19]. Cells were incubated with 100 mM BrdU for 2 h, trypsinized, and filtered through a 100 mm Nylon mesh before being fixed with 100% EtOH. Cells were then resuspended in PBS/1%FBS with 1% Propidium Iodide and 0.25 mg/ml RNAse A and incubated for 1 h prior sorting. Cells were sorted in four equal fractions from S phase as described in Fig. 1A and in [18]. Two hundred thousand cells were collected per phase. Cells were then treated with proteinase K and DNA was extracted following the Zymo Quick-DNA Microprep Kit extraction instructions (Zymo Research, D3020). DNA was sonicated using a QSonica Q700 (10 min at 20% amplitude). Libraries were constructed using the NEBNext Ultra DNA Library Prep Kit (New England Biolabs, E7370L) as described in [19]. BrdU-labeled DNA was then immunoprecipitated and purified, followed by PCR amplification and purification as described in [19].
Replication timing analysis
Repli-seq reads in each of the surveyed cell cycle phases (S1, S2, S3, S4) were mapped to the hg19 reference genome, followed by the removal of duplicates and counting reads over 50-kb bins across the genome. These counts were then quantile normalized and LOESS smoothed as described in [19].
Replication timing index (RTI) and differential RT
The traditional metric of RT, early-to-late (E/L) ratio, is based on two time points (early and late S-phase). We generalized this metric to a more sensitive numerical value to quantify RT based on four time points in our experiments, or any other number of profiled timepoints N3 2. The RT index (RTI) is based on a weighted sum of normalized replication signals (Repli-seq read densities Dn) from each time point n:
where n is the time point of the cell cycle (1 to 4, corresponding to time points S1 to S4) and Dn is the density of BrdU reads (per bp) within the given region at this time point. Regions of differential RT between cell types were identified using the difference of RTI in a given genomic bin, based on the cutoff of two standard deviations in pairwise comparisons of biological replicates, similar to [42, 43].
Classification of local replication patterns
To identify types of local replication patterns, we analyzed RTI values at 50 kb genomic bins and surveyed RTI patterns over a window of 10 adjacent bins sliding across the genome. To represent the local shape of the RT pattern in each window, the RTI value in each of the 10 bins was normalized by the average RTI across window. The local patterns of constant RT, local RTI minimum, local RTI maximum, and RTI slope corresponded to constant timing regions (CTR, defined as a window with RTI variance < 0.006 among 10 bins), initiation zones (IZ, a local minimum of RTI), termination sites (TS, a local maximum of RTI) and the remaining genomic windows classified as timing transitioning regions (TTR), similar to [9]. We further subdivided the constant timing regions (CTRs) into initiation constant replication regions (ICTR) and termination constant replication regions (TCTR) based on RTI profiles in the flanking 500 kb windows.
Cell cycle RNA sequencing
Cell cycle RNA-sequencing experiments were performed as described in [1]. Cells were incubated with Hoechst 33,342 (Invitrogen, H3570) at 1:3000 dilution for 1 h. Cells were then trypsinized, resuspended in Hoechst-containing media, filtered using a 100-mM filter, and sorted in four phases (250,000 cells per phase) as described in Fig. 1A in 1 mL of lysis buffer containing 10 mL of beta-mercapto-ethanol. RNA was then extracted following the PureLink RNA Mini kit (Invitrogen, 12,183,025) instructions with DNAse treatment (Invitrogen, 12,185,010). Libraries were prepped following the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina instructions with rRNA depletion (NEB #E6310), followed by paired-end sequencing.
RNA sequencing analysis
STAR aligner was used to map sequencing reads to transcripts in the hg19 reference genome [44]. Read counts for individual transcripts were produced with HTSeq-count [45], followed by the estimation of expression values using EdgeR [46]. Differential expression analysis was performed using EdgeR after normalizing read counts and including only genes with count per million reads (CPM) > 1 for one or more samples. Differentially expressed genes were defined based on the criteria of > twofold change in expression value and false discovery rate (FDR) < 0.05.
Single-cell RNA sequencing
Single-cell RNA sequencing from DMG were performed in [47], from hippocampus in [48], from cortex in [49], from G34 in [50], from GBM in [51], from ependymoma in [52], and from medulloblastoma in [53].
Western blot
Cell lines were lysed in RIPA. Lysates were sonicated in a QSonica Q800R for 10 min with 30 s “on” 30 s “off” intervals at 95% amplitude. Twenty-five milligrams of lysates were analyzed by western blot with antibodies against phospho-H2A.X(Ser139) (Sigma-Aldrich clone JBW301, #05–636) and b-actin (Abcam #8226).
Supplementary Information
Additional file 1: this file contains supplementary figures S1 to S6, supplementary figures legends, and supplementary tables legends.
Additional file 2: this file contains supplementary tables S1 to S3.
Additional file 3: this file contains the uncropped blots of Figure S5B.
Acknowledgements
We would like to thank Dr. Michelle Monje for providing all DIPG cell lines, and Drs. Johnathan Whetstine and Boning Chen for helpful discussions.
Peer review information
Tatiana Kutateladze and Wenjing She were the primary editors of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. The peer-review history is available in the online version of this article.
Authors’ contributions
AEH was involved in conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing-original draft, writing-review and editing. KC was involved in formal analysis and visualization. TN was involved in investigation, data curation, and writing-review and editing. JGM was involved in investigation, formal analysis and visualization. AE was involved in formal analysis, visualization, and writing-review and editing. CAOB was involved in formal analysis and visualization. AGS was involved in investigation and data curation. RP was involved in investigation and data curation. MGF was involved in resources and supervision. METP was involved in resources, supervision, and writing-review and editing. RIS was involved in methodology and supervision. CVR was involved in conceptualization, resources, supervision, funding acquisition, writing-original draft, writing-review and editing, and project administration.
Authors’ X handles
X handles: @VanRechemLab (Capucine Van Rechem)
Funding
C.V.R. was supported by the Distinguished Scientist Award from the Sontag Foundation, the DIPG DMG Research Funding Alliance, and the Super Sam Foundation. A.E.H. was supported by the Stanford Maternal and Child Health Research Institute. A.G.S. was supported by a Walter V. and Idun Berry fellowship. M.G.F. was supported by the Distinguished Scientist Award from the Sontag Foundation and an NIH Director New Innovator Award (1DP2NS127705-01). R.I.S. was supported by an NIH P30 grant (DK040561).
Sorting was performed on instruments in the Stanford Shared FACS Facility obtained using NIH S10 Shared Instruments Grant S10RR025518-01 (Falstaff); PICI was purchased by the Parker Institute for Cancer Immunotherapy.
Data availability
The data generated in this study are publicly available in GEO under accession numbers GSE268775 and GSE268776 [54, 55]. Single cell RNA sequencing from DMG were previously published and are publicly available [47, 56], from hippocampus in [48, 57], from cortex in [49], from G34 in [50, 58 ], from GBM in [51], [59], from ependymoma in [52, 60], and from medulloblastoma in [53, 61]. DNA fiber assays analyses were performed using a previously published and publicly available custom-build software written in Python (https://github.com/IES-HelmholtzZentrumMunchen/dna-fibers-analysis) [29, 62]. No other scripts and software were used other than those mentioned in the Methods section.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
MGF is a consultant for Blueprint Medicines; other authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Van Rechem C, Ji F, Chakraborty D, Black JC, Sadreyev RI, Whetstine JR. Collective regulation of chromatin modifications predicts replication timing during cell cycle. Cell Rep. 2021;37(1):109799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20(6):761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakatani T, Schauer T, Altamirano-Pacheco L, Klein KN, Ettinger A, Pal M, et al. Emergence of replication timing during early mammalian development. Nature. 2024;625(7994):401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, Pope BD, et al. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res. 2012;22(10):1833–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Q, Bert SA, Armstrong NJ, Caldon CE, Song JZ, Nair SS, et al. Replication timing and epigenome remodelling are associated with the nature of chromosomal rearrangements in cancer. Nat Commun. 2019;10(1):416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin YL, Pasero P. Replication stress: from chromatin to immunity and beyond. Curr Opin Genet Dev. 2021;71:136–42. [DOI] [PubMed] [Google Scholar]
- 7.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16(1):2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Costa A, Chowdhury D, Shapiro GI, D’Andrea AD, Konstantinopoulos PA. Targeting replication stress in cancer therapy. Nat Rev Drug Discov. 2023;22(1):38–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao PA, Sasaki T, Gilbert DM. High-resolution Repli-Seq defines the temporal choreography of initiation, elongation and termination of replication in mammalian cells. Genome Biol. 2020;21(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Rechem C, Whetstine JR. Examining the impact of gene variants on histone lysine methylation. Biochim Biophys Acta. 2014;1839(12):1463–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–31. [DOI] [PubMed] [Google Scholar]
- 12.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130(6):815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaney K, Weiss N, Almouzni G. The cell-cycle choreography of H3 variants shapes the genome. Mol Cell. 2023;83(21):3773–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarthy JF, Meers MP, Janssens DH, Henikoff JG, Feldman H, Paddison PJ, et al. Histone deposition pathways determine the chromatin landscapes of H3.1 and H3.3 K27M oncohistones. Elife. 2020;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaney K, Strobino M, Wenda JM, Pankowski A, Steiner FA. H3.3K27M-induced chromatin changes drive ectopic replication through misregulation of the JNK pathway in C. elegans. Nat Commun. 2019;10(1):2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whetstine JR, Van Rechem C. Protocol to isolate cells in four stages of S phase for high-resolution replication-timing sequencing. STAR Protoc. 2022;3(1):101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchal C, Sasaki T, Vera D, Wilson K, Sima J, Rivera-Mulia JC, et al. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat Protoc. 2018;13(5):819–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji F, Van Rechem C, Whetstine JR, Sadreyev RI. Computational workflow for integrative analyses of DNA replication timing, epigenomic, and transcriptomic data. STAR Protoc. 2022;3(4):101827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gene Ontology C, Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, et al. The gene ontology knowledgebase in 2023. Genetics. 2023;224(1):iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou LP, Mi H. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31(1):8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang ZF, Huang DY, Chi LM. Serine 13 is the site of mitotic phosphorylation of human thymidine kinase. J Biol Chem. 1998;273(20):12095–100. [DOI] [PubMed] [Google Scholar]
- 25.Skovgaard T, Uhlin U, Munch-Petersen B. Comparative active-site mutation study of human and Caenorhabditis elegans thymidine kinase 1. FEBS J. 2012;279(10):1777–87. [DOI] [PubMed] [Google Scholar]
- 26. Team RC. _R: a language and environment for statistical computing_. Vienna: R Foundation for Statistical Computing; 2023.
- 27.Kwon Y, Rosner H, Zhao W, Selemenakis P, He Z, Kawale AS, et al. DNA binding and RAD51 engagement by the BRCA2 C-terminus orchestrate DNA repair and replication fork preservation. Nat Commun. 2023;14(1):432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone EG, Thompson MD, Byrd AK. Role and regulation of Pif1 family helicases at the replication fork. Int J Mol Sci. 2022;23(7):3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatani T, Lin J, Ji F, Ettinger A, Pontabry J, Tokoro M, et al. DNA replication fork speed underlies cell fate changes and promotes reprogramming. Nat Genet. 2022;54(3):318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi N, Kim S, Schultz CW, Rajapakse VN, Zhang Y, Redon CE, et al. Replication stress defines distinct molecular subtypes across cancers. Cancer Res Commun. 2022;2(6):503–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koschmann C, Al-Holou WN, Alonso MM, Anastas J, Bandopadhayay P, Barron T, et al. A road map for the treatment of pediatric diffuse midline glioma. Cancer Cell. 2024;42(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jessa S, Mohammadnia A, Harutyunyan AS, Hulswit M, Varadharajan S, Lakkis H, et al. K27M in canonical and noncanonical H3 variants occurs in distinct oligodendroglial cell lineages in brain midline gliomas. Nat Genet. 2022;54(12):1865–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Meijden CM, Lapointe DS, Luong MX, Peric-Hupkes D, Cho B, Stein JL, et al. Gene profiling of cell cycle progression through S-phase reveals sequential expression of genes required for DNA replication and nucleosome assembly. Cancer Res. 2002;62(11):3233–43. [PubMed] [Google Scholar]
- 34.Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J Biol Chem. 2003;278(42):41528–34. [DOI] [PubMed] [Google Scholar]
- 35.Fragkos M, Ganier O, Coulombe P, Mechali M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 2015;16(6):360–74. [DOI] [PubMed] [Google Scholar]
- 36.Pal S, Kaplan JP, Nguyen H, Stopka SA, Savani MR, Regan MS, et al. A druggable addiction to de novo pyrimidine biosynthesis in diffuse midline glioma. Cancer Cell. 2022;40(9):957-72 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bockaj I, Martini TEI, de Camargo Magalhaes ES, Bakker PL, Meeuwsen-de Boer TGJ, Armandari I, et al. The H3.3K27M oncohistone affects replication stress outcome and provokes genomic instability in pediatric glioma. PLoS Genet. 2021;17(11): e1009868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Chang JF, Sun J, Chen L, Yang XM, Tang HY, et al. Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J Cell Sci. 2018;131(12):jcs215525. [DOI] [PubMed] [Google Scholar]
- 39.Symonds RP, Collingwood M, Kirwan J, Humber CE, Tierney JF, Green JA, et al. Concomitant hydroxyurea plus radiotherapy versus radiotherapy for carcinoma of the uterine cervix: a systematic review. Cancer Treat Rev. 2004;30(5):405–14. [DOI] [PubMed] [Google Scholar]
- 40.Techer H, Koundrioukoff S, Azar D, Wilhelm T, Carignon S, Brison O, et al. Replication dynamics: biases and robustness of DNA fiber analysis. J Mol Biol. 2013;425(23):4845–55. [DOI] [PubMed] [Google Scholar]
- 41.Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell. 2007;18(8):3059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivera-Mulia JC, Desprat R, Trevilla-Garcia C, Cornacchia D, Schwerer H, Sasaki T, et al. DNA replication timing alterations identify common markers between distinct progeroid diseases. Proc Natl Acad Sci U S A. 2017;114(51):E10972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarni D, Sasaki T, Irony Tur-Sinai M, Miron K, Rivera-Mulia JC, Magnuson B, et al. 3D genome organization contributes to genome instability at fragile sites. Nat Commun. 2020;11(1):3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu I, Jiang L, Samuelsson ER, Marco Salas S, Beck A, Hack OA, et al. The landscape of tumor cell states and spatial organization in H3–K27M mutant diffuse midline glioma across age and location. Nat Genet. 2022;54(12):1881–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong S, Ding W, Sun L, Lu Y, Dong H, Fan X, et al. Decoding the development of the human hippocampus. Nature. 2020;577(7791):531–6. [DOI] [PubMed] [Google Scholar]
- 49.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358(6368):1318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu I, Alencastro Veiga Cruzeiro G, Bjerke L, Rogers RF, Grabovska Y, Beck A, et al. GABAergic neuronal lineage development determines clinically actionable targets in diffuse hemispheric glioma, H3G34-mutant. Cancer Cell. 2024;42(9):1528–48.e17. [DOI] [PubMed]
- 51.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835-49 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gojo J, Englinger B, Jiang L, Hubner JM, Shaw ML, Hack OA, et al. Single-cell RNA-seq reveals cellular hierarchies and impaired developmental trajectories in pediatric ependymoma. Cancer Cell. 2020;38(1):44-59 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature. 2019;572(7767):74–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hains AE, Chetal K, Nakatani T, Marques JG, Ettinge A, Biagi Jr CAO, Gonzalez-Sandoval A, Pillai R, Filbin MG, Torres-Padilla ME, Sadreyev RI, Van Rechem C. Multi-omics approaches reveal that Diffuse Midline Gliomas present altered DNA replication and are susceptible to replication stress therapy. GSE268775. Gene Expression Omnibus; 2024. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE268775.
- 55.Hains AE, Chetal K, Nakatani T, Marques JG, Ettinge A, Biagi Jr CAO, Gonzalez-Sandoval A, Pillai R, Filbin MG, Torres-Padilla ME, Sadreyev RI, Van Rechem C. Multi-omics approaches reveal that diffuse midline gliomas present altered DNA replication and are susceptible to replication stress therapy. GSE268776. Gene Expression Omnibus; 2024. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE268776.
- 56.Liu I, Jiang L, Samuelsson ER, Marco Salas S, Beck A, Hack OA, Jeong D, Shaw ML, Englinger B, LaBelle J, Mire HM, Madlener S, Mayr L, Quezada MA, Trissal M, Panditharatna E, Ernst KJ, Vogelzang J, Gatesman TA, Halbert ME, Palova H, Pokorna P, Sterba J, Slaby O, Geyeregger R, Diaz A, Findlay IJ, Dun MD, Resnick A, Suvà ML, Jones DTW, Agnihotri S, Svedlund J, Koschmann C, Haberler C, Czech T, Slavc I, Cotter JA, Ligon KL, Alexandrescu S, Yung WKA, Arrillaga-Romany I, Gojo J, Monje M, Nilsson M, Filbin MG. The landscape of tumor cell states and spatial organization in H3–K27M mutant diffuse midline glioma across age and location. GSE184357. Gene Expression Omnibus; 2022. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE184357. [DOI] [PMC free article] [PubMed]
- 57.Zhong S, Ding W, Sun L, Lu Y, Dong H, Fan X, Liu Z, Chen R, Zhang S, Ma Q, Tang F, Wu Q, Wang X. Decoding the development of the human hippocampus. GSE119212. Gene Expression Omnibus; 2019. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119212. [DOI] [PubMed]
- 58.Liu I, Alencastro Veiga Cruzeiro G, Bjerke L, Rogers RF, Grabovska Y, Beck A, Mackay A, Barron T, Hack OA, Quezada MA, Molinari V, Shaw ML, Perez-Somarriba M, Temelso S, Raynaud F, Ruddle R, Panditharatna E, Englinger B, Mire HM, Jiang L, Nascimento A, LaBelle J, Haase R, Rozowsky J, Neyazi S, Baumgartner AC, Castellani S, Hoffman SE, Cameron A, Morrow M, Nguyen QD, Pericoli G, Madlener S, Mayr L, Dorfer C, Geyeregger R, Rota C, Ricken G, Ligon KL, Alexandrescu S, Cartaxo RT, Lau B, Uphadhyaya S, Koschmann C, Braun E, Danan-Gotthold M, Hu L, Siletti K, Sundström E, Hodge R, Lein E, Agnihotri S, Eisenstat DD, Stapleton S, King A, Bleil C, Mastronuzzi A, Cole KA, Waanders AJ, Montero Carcaboso A, Schüller U, Hargrave D, Vinci M, Carceller F, Haberler C, Slavc I, Linnarsson S, Gojo J, Monje M, Jones C, Filbin MG. GABAergic neuronal lineage development determines clinically actionable targets in diffuse hemispheric glioma, H3G34-mutant. GSE162989. Gene Expression Omnibus; 2024. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE162989. [DOI] [PubMed]
- 59.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, Gritsch S, Perez EM, Gonzalez Castro LN, Lan X, Druck N, Rodman C, Dionne D, Kaplan A, Bertalan MS, Small J, Pelton K, Becker S, Bonal D, Nguyen QD, Servis RL, Fung JM, Mylvaganam R, Mayr L, Gojo J, Haberler C, Geyeregger R, Czech T, Slavc I, Nahed BV, Curry WT, Carter BS, Wakimoto H, Brastianos PK, Batchelor TT, Stemmer-Rachamimov A, Martinez-Lage M, Frosch MP, Stamenkovic I, Riggi N, Rheinbay E, Monje M, Rozenblatt-Rosen O, Cahill DP, Patel AP, Hunter T, Verma IM, Ligon KL, Louis DN, Regev A, Bernstein BE, Tirosh I, Suvà ML. An integrative model of cellular states, plasticity, and genetics for glioblastoma. GSE131928. Gene Expression Omnibus; 2019. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131928. [DOI] [PMC free article] [PubMed]
- 60.Gojo J, Englinger B, Jiang L, Hübner JM, Shaw ML, Hack OA, Madlener S, Kirchhofer D, Liu I, Pyrdol J, Hovestadt V, Mazzola E, Mathewson ND, Trissal M, Lötsch D, Dorfer C, Haberler C, Halfmann A, Mayr L, Peyrl A, Geyeregger R, Schwalm B, Mauermann M, Pajtler KW, Milde T, Shore ME, Geduldig JE, Pelton K, Czech T, Ashenberg O, Wucherpfennig KW, Rozenblatt-Rosen O, Alexandrescu S, Ligon KL, Pfister SM, Regev A, Slavc I, Berger W, Suvà ML, Kool M, Filbin MG. Single-cell RNA-seq reveals cellular hierarchies and impaired developmental trajectories in pediatric ependymoma. GSE141460. Gene Expression Omnibus; 2020. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141460. [DOI] [PMC free article] [PubMed]
- 61.Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, DeWitt JC, Groves A, Mayr L, Weisman HR, Richman AR, Shore ME, Goumnerova L, Rosencrance C, Carter RA, Phoenix TN, Hadley JL, Tong Y, Houston J, Ashmun RA, DeCuypere M, Sharma T, Flasch D, Silkov A, Ligon KL, Pomeroy SL, Rivera MN, Rozenblatt-Rosen O, Rusert JM, Wechsler-Reya RJ, Li XN, Peyrl A, Gojo J, Kirchhofer D, Lötsch D, Czech T, Dorfer C, Haberler C, Geyeregger R, Halfmann A, Gawad C, Easton J, Pfister SM, Regev A, Gajjar A, Orr BA, Slavc I, Robinson GW, Bernstein BE, Suvà ML, Northcott PA. Resolving medulloblastoma cellular architecture by single-cell genomics. GSE119926. Gene Expression Omnibus; 2019. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119926. [DOI] [PMC free article] [PubMed]
- 62.NakataniT, Lin J, Ji F, Ettinger A, Pontabry J, Tokoro M, Altamirano-Pacheco L, Fiorentino J, Mahammadov E, Hatano Y, Van Rechem C, Chakraborty D, Ruiz-Morales ER, Arguello Pascualli PY, Scialdone A, Yamagata K, Whetstine JR, Sadreyev RI, Torres-Padilla M. DNA replication fork speed underlies cell fate changes and promotes reprogramming. Github; 2017. https://github.com/IES-HelmholtzZentrumMunchen/dna-fibers-analysis. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: this file contains supplementary figures S1 to S6, supplementary figures legends, and supplementary tables legends.
Additional file 2: this file contains supplementary tables S1 to S3.
Additional file 3: this file contains the uncropped blots of Figure S5B.
Data Availability Statement
The data generated in this study are publicly available in GEO under accession numbers GSE268775 and GSE268776 [54, 55]. Single cell RNA sequencing from DMG were previously published and are publicly available [47, 56], from hippocampus in [48, 57], from cortex in [49], from G34 in [50, 58 ], from GBM in [51], [59], from ependymoma in [52, 60], and from medulloblastoma in [53, 61]. DNA fiber assays analyses were performed using a previously published and publicly available custom-build software written in Python (https://github.com/IES-HelmholtzZentrumMunchen/dna-fibers-analysis) [29, 62]. No other scripts and software were used other than those mentioned in the Methods section.