Abstract
Background
Planetary plastic pollution poses a major threat to ecosystems and human health in the Anthropocene, yet its impact on biogeochemical cycling remains poorly understood. Waterlogged rice paddies are globally important sources of CH4. Given the widespread use of plastic mulching in soils, it is urgent to unravel whether low-density polyethylene (LDPE) will affect the methanogenic community in flooded paddy soils. Here, we employed a combination of process measurements, short-chain and long-chain fatty acid (SCFAs and LCFAs) profiling, Fourier-transform ion cyclotron resonance mass spectrometry, quantitative PCR, metagenomics, and mRNA profiling to investigate the impact of LDPE nanoplastics (NPs) on dissolved organic carbon (DOC) and CH4 production in both black and red paddy soils under anoxic incubation over a 160-day period.
Results
Despite significant differences in microbiome composition between the two soil types, both exhibited similar results to NPs exposure. NPs induced a change in DOC content and CH4 production up to 1.8-fold and 10.1-fold, respectively. The proportion of labile dissolved organic matter decreased, while its recalcitrance increased. Genes associated with the degradation of complex carbohydrates and aromatic carbon were significantly enriched. The elevated CH4 production was significantly correlated to increases in both the PCR-quantified mcrA gene copy numbers and the metagenomic methanogen-to-bacteria abundance ratio. Notably, the latter was linked to an enrichment of the hydrogenotrophic methanogenesis pathway. Among 391 metagenome-assembled genomes (MAGs), the abundance of several Syntrophomonas and Methanocella MAGs increased concomitantly, suggesting that the NPs treatments stimulated the syntrophic oxidation of fatty acids. mRNA profiling further identified Methanosarcinaceae and Methanocellaceae to be the key players in the NPs-induced CH4 production.
Conclusions
The specific enrichment of Syntrophomonas and Methanocella indicates that LDPE NPs stimulate the syntrophic oxidation of LCFAs and SCFAs, with Methanocella acting as the hydrogenotrophic methanogen partner. Our findings enhance the understanding of how LDPE NPs affect the methanogenic community in waterlogged paddy soils. Given the importance of this ecosystem, our results are crucial for elucidating the mechanisms that govern carbon fluxes, which are highly relevant to global climate change.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01974-y.
Keywords: Plastics; Carbon flow; Methane; Fatty acids; FT-ICR-MS, MAGs; Metatranscriptomics
Background
Plastic pollution is of growing global concern for its potential to alter the carbon cycling in terrestrial ecosystems [1]. Indeed, a large amount of plastic debris accumulates in soils and terrestrial ecosystems, primarily due to agricultural plastic film degradation, atmospheric fallout, and the use of sewage sludge as fertilizer [2–4]. For instance, film mulching has become an essential agricultural practice in rice paddies, due to the seasonal arid climate and low precipitation [5]. The accumulation of microplastics in long-term film-mulched paddy soil is estimated to reach 18.1 million particles ha−1 annually in China [5]. The degradation of plastic materials results in the formation of microplastics (MPs < 5 mm) and nanoplastics (NPs < 1 μm). Both MPs and NPs have a significant potential to adversely affect soil ecology [6, 7]. Their adverse effects on the terrestrial carbon and nitrogen cycles may impact soil microbial activities, plant growth, and litter decomposition, and these particles can be toxic for microorganisms in the soil environment [1, 8, 9]. NPs have a larger specific surface area and higher adsorption capacity and mobility than MPs, making them more easily absorbed or ingested by various organisms.
The effect of NPs on microbial communities may vary depending on the plastic type investigated and the environmental conditions. On the one hand, NPs have been shown to significantly inhibit CH4 production in different anaerobic wastewater and sludge digestion systems [10–12]. On the other hand, there is growing evidence that dissolved organic carbon (DOC) leaching from plastics stimulates microbial activity and directly or indirectly affects carbon sequestration capacity in marine and soil ecosystems [13–16]. DOC can be directly utilized by bacteria capable of breaking down complex carbon polymers into simpler organic compounds, which involves the expression and activity of carbohydrate-active enzymes (CAZymes) [17, 18]. However, the response of methanogens to the accumulation of NPs in agricultural soils is not yet known.
Rice field soils are one of the most important agricultural sources of atmospheric CH4. They thus represent an excellent model system for investigating the microbial mechanisms of CH4 production [17]. The methanogenic degradation pathway of organic matter in submerged rice paddies and anoxic wetlands follows common principles and involves a microbial food web composed of different functional guilds of the domains Bacteria and Archaea [19]. These microbial guilds participate in a cascade of anaerobic degradation steps that involve polymer hydrolysis, fermentation, syntrophic conversion of fatty acids, homoacetogenesis, and methanogenesis [20]. Three different methanogenic pathways, including acetoclastic, hydrogenotrophic, and methylotrophic methanogenesis, are typically active in paddy soils [21, 22]. Acetoclastic methanogens, such as Methanosarcina and Methanothrix, utilize acetate to form CO2 and CH4 [23]. Hydrogenotrophic methanogens, including Methanocellales, Methanobacteriales, and Methanomicrobiales, use H2 and CO2 to produce CH4 [24]. Members of the Methanosarcinales and Methanomassiliicoccales are known for their methylotrophic capabilities, able to utilize various methylated compounds, such as methanol, methylamine, and dimethylamine, as carbon and energy sources for CH4 production [25, 26].
Here, we combined process measurements and Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) with functional genome-centric metagenomics and quantitative PCR (qPCR) to disentangle the impact of low-density polyethylene nanoplastics (LDPE NPs) on the methanogenic communities in flooded rice field soil. More specifically, we aimed to elucidate whether LDPE NPs affect the DOC content in anoxic paddy soils and thus CH4 production, and if so, what is their effect on the metagenomic potential for polymer breakdown and major methanogenesis pathways. LDPE is one of the most produced and discarded synthetic plastics globally and is known to accumulate in various ecosystems, including rice paddies [27, 28]. Despite this environmental threat, the impact of LDPE on the anaerobic microbiota in flooded rice paddies remains largely unexplored. We applied our research strategy to two major soil types widely used for rice farming in China: black and red soil. The organic carbon content in red soil is significantly higher than in black soil (Table S1). Our study sites are known to represent areas of the highest CH4 production rates among Chinese black and red soils [29]. As a corroborative approach, we also obtained functional gene expression profiles from particular samples. Our research fills a knowledge gap concerning the effects of NPs on DOC and CH4 metabolism in rice field soils with potentially broad implications for ecosystem fluxes and global climate change.
Materials and methods
Microcosms setup and soil sampling
Soil samples were collected from two typical rice-growing areas that differ in their soil types, defined as black soil (BS) and red soil (RS) in China. BS was collected at the Jiansanjiang Agricultural Experimental Station in Heilongjiang (47°14′N, 132°37′E), while RS was sampled at the Changsha China National Rice Institute in Changsha in Hunan (28°11′N, 112°58′E). Major physicochemical properties of BS and RS are shown in supplementary materials (Additional File 3: Table S1). LDPE (density: 0.91 g/cm3) NPs with a size of 50 nm were purchased from Zhongxin Plastic (Guangdong, China). Nanoplastics were sterilized with methanol, dried at 40 °C, and stored at 4 °C for further use [16].
Four different concentrations of 50-nm LPDE NPs were applied in our BS and RS microcosm experiments. The concentration of nanoplastics refers to the dried paddy soil and is calculated based on its dry weight. The NPs concentrations were 0% (CK), 0.5% (0.1-g NPs), 1% (0.2-g NPs), and 5% (1-g NPs). Although there is not yet a reliable method available to quantify the accumulation of NPs in paddy soils, the amounts are likely in the range of 0 ~ 6.7% microplastics as detected in other soils [30]. Each treatment had four replicates. The microcosms were prepared by adding 20 g of dry soil, 40 mL of autoclaved water, and the appropriate amounts of NPs to sterile 100-mL bottles, followed by thorough mixing (all experimental units experienced the same physical disturbance by mixing, including the controls). The bottles were then sealed with butyl rubber stoppers and aluminum caps and flushed with N2 for 10 min to establish anoxic conditions. All bottles were incubated in a climate chamber at 25 °C in the dark for up to 160 days. In total, 64 microcosms were set up (2 soils × 4 treatments × 2 time points × 4 replicates). These microcosms were first used for gas measurements (CH4, CO2) and then destructively sampled for measurement of metabolites (liquid) and, in addition to dissolved organic carbon (DOC) and matter (DOM), for molecular analysis (soil). The sampling time points reflect our knowledge that (i) hydrolytic decomposition of easily degradable polymer substances occurs over the first 30 days of anaerobic incubation [17], and (ii) microplastic degradation has long-lasting ecological effects [31]. The detailed experimental setup is shown in Fig. S1 (Additional File 2). Soil samples were taken after 30 and 160 days of incubation. To test whether carbon is abiotically released from LDPE NPs and to what extent, sterilized (60Co γ-radiation at a dose of 25 kGy to kill the soil indigenous microbes) control and NPs-amended (0.5%, 5%) microcosms were incubated in triplicates for 30 days. The carbon released from the LDPE NPs was neglectable. The exact values were 1.83 × 10−3 mg g−1 and 2.43 × 10−3 mg g−1 dry paddy soil in the 0.5% and 5% NPs treatments, respectively. LDPE is about 80% carbon [1]. Thus, the total amount of LDPE carbon added per gram dry paddy soil in the 0.5% NPs and 5% NPs treatments was 4 mg and 40 mg, respectively.
Process measurements
Gas samples (100 µl) were taken from the headspace of the bottles using a Pressure-Lock syringe (VICI). Cumulative gas emissions were measured continuously using a gas chromatography system equipped with a Porapak Q stainless-steel column as previously described (Additional File 2: Fig. S2) [32, 33]. However, due to the highly similar gas measurement results obtained for the 0.5% and 1.0% NPs treatments, our further study was limited to the following three NPs treatments: 0% NPs (CK), 0.5% NPs (0.1 g), and 5% NPs (1 g). The contents of DOC were determined using a total organic carbon analyzer (Elementar, Langenselbold, Germany). The total amount of microbially accessible carbon released by the NPs treatments in the microcosms was estimated by summing up the difference in DOC content between the NPs treatments and the control treatments, as well as the carbon converted into CH4 and CO2 (Additional File 3: Table S2). In addition, the calculated CO2 values may slightly underestimate the released carbon because carbon fixed into cell biomass could not be considered. The solid-phase extraction of dissolved organic matter (DOM) from the CK and 0.5% NPs treatments were conducted using Fourier transform ion cyclotron resonance (FT-ICR-MS) as previously described [16]. The extraction procedure is further detailed in Additional File 1.
Targeted metabolomics
A 2-mL liquid sample of each microcosm was centrifuged for 15 min at 17,949 × g at 4 °C according to a previous study [17]. Short-chain fatty acids (SCFA) were analyzed using an HP 6890 gas chromatograph (Agilent Technologies). Another 2-mL liquid sample of each microcosm was homogenized with 300 μL of isopropanol/acetonitrile (1:1) and subsequently centrifuged at 17,949 × g for 10 min. Then the supernatant was subjected, in relation to a mixed internal standard, to long-chain fatty acids (LCFAs) analysis using UHPLC-MS (ExionLC™ AD UHPLC-QTRAP® 6500 +).
DNA/RNA extraction and quantification of mcrA genes
Extraction, purification, and quantification of total soil DNA and RNA were carried out as previously reported (see Additional File 1 for detailed methods description). The numbers of genes encoding methyl coenzyme-M reductase (mcrA) were determined using quantitative real-time PCR (qPCR) as described previously [17]. All qPCR reactions were performed in three biological replicates with three technical replicates.
Metagenomics and metatranscriptomics
Thirty-six metagenomic libraries were generated and sequenced on an Illumina HiSeq 2000 instrument at Novogene Bioinformatics Technology in a 2 × 150 bp paired-end mode. The sequences were quality-checked using Trimmomatic (version 0.35) [34]. The high-quality paired-end reads from each sample were individually assembled into contigs using MEGAHIT (version 1.1.3) [35] and evaluated using QUAST 2.3. Metagenomic contigs were queried in Prokka and BLAST against the NCBI nonredundant (nr) protein database and the clusters of orthologous groups (COG) database using MEGAN6 Ultimate Edition (version 6.20.5) [36, 37]. Functions related to carbon metabolic categories were classified into aromatic and carbohydrate carbon classes (Additional File 3: Table S3), as described in a previous study [38]. Taxonomic assignment of the H2 evolving hydrogenases genes was achieved by extracting their sequences from the metagenomic contigs. The extracted sequences were then blasted against NCBI’s nonredundant protein database using Diamond with default settings. Genome binning of the assembled contigs was carried out using metaWRAP [39]. The completeness and contamination of the metagenome-assembled genomes (MAGs) were evaluated using CheckM (Version 1.1.2) [40]. MAGs were annotated using Prokka (version 1.14.6) and searched against NCBI-nr and KEGG databases [41, 42]. The maximum-likelihood phylogenomic trees were constructed from the multiple sequence alignments (MSAs) generated by GTDB-Tk software and visualized in the online iTol platform (https://itol.embl.de/) [43]. CoverM and the average nucleotide identity (ANI) of MAGs were calculated to determine their abundances and similarities [44]. The detailed procedure of carbon annotation, genome binning, and annotation is described in Additional File 1.
Furthermore, total RNA extracted from the three replicate microcosms of a given treatment after 160 days of incubation was mixed to produce composite samples for cDNA library preparation. Metatranscriptomic libraries could be created for the following three experimental treatments: red soil with 0% NPs (RS-CK), red soil with 0.5% NPs (RS-0.5% NPs), and black soil with 0.5% NPs (BS-0.5% NPs). Total RNA extraction from the BS control treatment was attempted but failed. Sequencing was done using Illumina MiSeq in a 2 × 250 bp paired-end mode as described previously [17]. The detailed procedure of cDNA library preparation and metatranscriptomic data analysis is described in Additional File 1.
Statistical analyses
All statistical analyses were conducted in R (Version 4.0.1). Principal coordinate analysis (PCoA) based on Bray‒Curtis distances of carbon functional gene profiles was carried out to compare the variance in beta diversity across samples. Analysis of similarities (ANOSIM) based on Bray‒Curtis distances was performed to estimate the effect of NPs on carbon functional gene diversity. In addition, we applied one-way ANOVA followed by post hoc multiple comparisons using the Tukey HSD test to assess the significant difference in DOC levels, mcrA gene copy numbers, NPs-induced metagenomic abundance changes of the methanogen-bacteria ratio, and the genes encoding the degradation of aromatics and complex carbohydrates. The relative metagenomic abundance refers to the annotated counts per million reads. Statistical significance was established at a FDR-corrected P-value < 0.05. LEfse (linear discriminant analysis effect size) analysis was used to investigate the metagenomic abundance of MAGs that exhibited significant enrichment in specific treatment groups.
Results
Impact of the NPs treatments on DOC and DOM
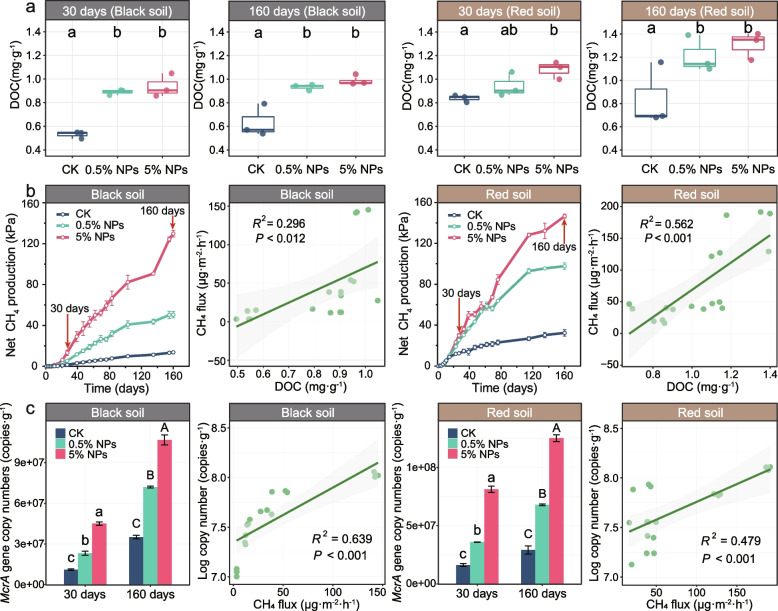
Scanning electron micrographs revealed that NPs had a strong capacity to form rod-shaped aggregates (Additional File 2: Fig. S3). The addition of NPs to the microcosms significantly increased the DOC content in the black soil and the red soil by 80% and 60%, respectively. No significant difference in DOC increase was observed between the 30-day and 160-day incubation periods (Fig. 1a). Concomitantly to the NPs-induced increase in DOC after 30 days of incubation, the relative proportion of recalcitrant DOM molecules in both the black soil [molecular lability boundary (MLBL)ck = 0.144, (MLBL)NPs = 0.115)] and the red soil [(MLBL)ck = 0.134, (MLBL)NPs = 0.121)] increased due to a decrease in labile DOM (Additional File 2: Fig. S4; Additional File 3: Table S4).
Fig. 1.
Changes in DOC concentration, CH4 flux, and mcrA gene copy numbers in response to the addition of 0.5% and 5% LDPE NPs to black and red soils. a The increased DOC levels were determined after an incubation period of 30 and 160 days. Different letters (a, b, c) indicate significant differences between CK, 0.5% NPs, and 5% NPs treatments (P < 0.05). Error bars denote standard deviation (n = 3). b CH4 production and the relationship between CH4 flux and DOC concentration in black and red soils. The arrows indicate the time points of soil sampling for downstream analysis. Error bars denote standard deviation (n = 4). c mcrA gene copy numbers and the relationship between CH4 flux and mcrA gene copy numbers in black and red soils. The measurements were made over an incubation period of 160 days. Different letters (a, b, c [30 d]; A, B, C [160 d]) in c indicate significant differences between CK (control), 0.5% NPs, and 5% NPs treatments (P < 0.05). Error bars denote standard deviation (n = 3)
Fatty acid profiles and CH4 production
In the black soil, all the metabolites, including acetate, propionate, valerate, and lactate, showed a transient peak concentration after 30-day incubation. The greatest transient concentrations were observed for acetate (2 mM [5% NPs]), valerate (0.4 mM [0.5%, 5% NPs], and lactate (0.8 mM [CK]). Compared to the black soil, the metabolite turnover patterns markedly differed in the red soil. In particular, acetate exhibited its transient peak concentration already after 9-day incubation. The concentration of all metabolites had decreased to low levels in both soils after 160-day incubation but was still detectable in some cases (e.g., acetate and propionate in the 5% NPs treatment in the black soil) (Additional File 2: Fig. S5). The concentration of LCFAs in the black soil had increased with increasing LDPE NPs concentration after 30-day incubation but was decreased after 160-day incubation. This was most obviously for the monounsaturated C18 fatty acids oleic acid and cis-vaccenic acid (P < 0.05) (Additional File 2: Fig. S6).
Relative to the control, the addition of LDPE NPs induced a change in the CH4 production rate of up to 10.1-fold in the black soil and 4.5-fold in the red soil (Fig. 1b; Additional File 2: Figs. S2, S7). However, relative to the red soil, the black soil had an extended lag phase of approximately 20 days until CH4 production was detectable. Consequently, the amount of CH4 produced was significantly lower in the black soil after 30-day incubation than in the red soil. Although similar amounts of CH4 were measured in the 5% NPs treatments of both soils after 160 days of incubation, CH4 production in the black soil remained significantly lower over the complete 160-day incubation period than in the red soil (Fig. 1b; Additional File 2: Fig. S2). In the black soil, the total excess amounts of microbially accessible carbon released in the 5% NPs treatments relative to the control treatments were 18.69 mg (30 days) and 39.3 mg (160 days). The corresponding values for the red soil were 12.76 mg (30 days) and 43.84 mg (160 days). The values calculated for the 5% NPs treatments were significantly higher than those calculated for the 0.5% NPs treatments, primarily due to the increased amount of carbon converted into CH4 (Additional File 3: Table S2).
The CH4 production rate was significantly and positively correlated with the DOC content across both NPs treatments (0.5%, 5%) relative to CK for both sampling time points (30 days, 160 days). This correlation was highly significant for both the black soil (R2 = 0.296, P < 0.012) and the red soil (R2 = 0.562, P < 0.001) (Fig. 1b). Furthermore, the copy numbers of the mcrA gene significantly increased (P < 0.001) after the addition of NPs (Fig. 1c; Additional File 2: Fig. S8), indicating a positive correlation between methanogen abundance and CH4 production in both the black soil (R2 = 0.639, P < 0.001) and the red soil (R2 = 0.479, P < 0.001) (Fig. 1c).
Metagenomics and enriched MAGs
Shotgun metagenomic sequencing was performed for 36 soil samples, producing more than 593 GB of Illumina sequence data. A total of 1190 metagenome-assembled genomes (MAGs) were recovered. Among them, 391 MAGs were identified to be of high quality (completeness > 70% and contamination < 10%) (Additional File 2: Fig. S9). Among the bacterial MAGs, the relative abundance of 45 MAGs was significantly and positively correlated with DOC content. Most of these MAGs belonged to Syntrophomonadia (21), Lentimicrobiaceae (6), and Ignavibacteriaceae (4). In addition, a total of 38 high-quality methanogen MAGs were obtained (Additional File 2: Fig. S10; Additional File 3: Table S5), which belonged to the Methanocellaceae (17), Methanobacteriaceae (11), Methanotrichaceae (6), Methanosarcinaceae (3), and Methanomassiliicoccaceae (1). Among these MAGs, the relative abundance of 12 methanogen MAGs was significantly and positively correlated with CH4 production. These were affiliated with Methanocella (9) and Methanobacterium (3).
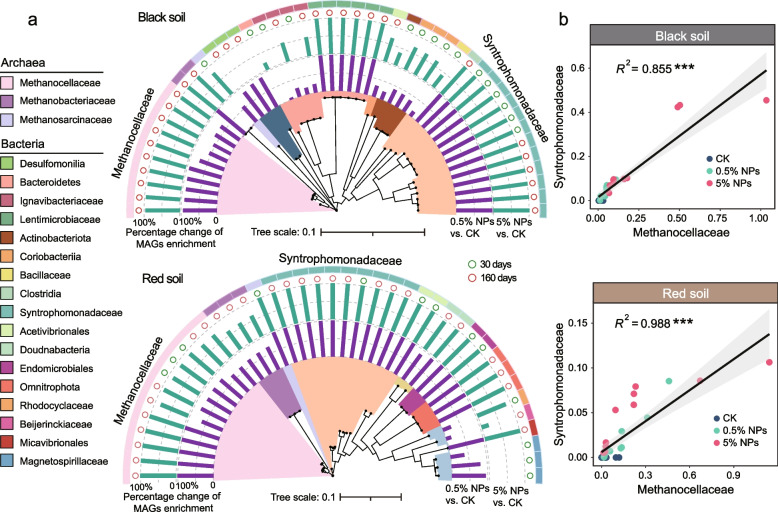
Most of the MAGs affiliated with Syntrophomonadaceae and Methanocellaceae were found in both the black and red soils to be significantly enriched in the NPs treatments relative to the control (Fig. 2a; Additional File 2: Fig. S10). In both soils, we indeed observed a significant correlation between the NPs-induced changes in the metagenomic abundance of Syntrophomondaceae and Methanocellaceae MAGs (P < 0.001) (Fig. 2b). The NPs-induced enrichment of the Syntrophomonadaceae was also highly evident in the family-level profiles obtained for the total metagenomic data (from 0.46% up to 2.42%) (P < 0.001) and the community-wide composition of hydrogenase genes (from undetectable levels to a range from 8.55% to 20.13%) (P < 0.001) (Additional File 2: Figs. S11, S12), with the latter being significantly related to an increase in the total hydrogenase gene abundance (Additional File 2: Fig. S12).
Fig. 2.
Phylogenomic trees of MAGs affiliated with the Syntrophomodaceae and Methanocellaceae and linear correlation graphs showing their NPs-induced metagenomic abundance increases in the black soil and the red soil. a Phylogenomic trees of MAGs that were recovered from black and red soils and exhibited significant changes in their metagenomic abundance between the different treatments (CK, 0.5% NPs, and 5% NPs). Multiple sequence alignments of 120 bacterial and 122 archaeal marker genes by GTDB-Tk were used to construct the phylogenomic trees of bacteria and methanogens, respectively. The maximum-likelihood phylogenies were calculated for the multiple sequence alignments (MSAs) in the format of an IQ-TREE using the LG + F + R10 model. b Graphs showing the significant correlation between the NPs-induced metagenomic abundance increases of MAGs affiliated with the Syntrophomodaceae (y-axis) and Methanocellaceae (x-axis). The correlation analysis of Syntrophomonadaceae and Methanocellaceae MAGs did not involve dereplication, because our focus was on the genomic variations among the taxa of interest. “***” means P < 0.001
In the black soil, the NPs-induced abundance increases were observed for various MAGs belonging to Syntrophomondaceae (12), Lentimicrobiaceae (6), Ignavibacteriaceae (4), Desulfomonilia (3), Coriobacteriia (3), Bacteroidetes (1), Acetivibrionales (1), Actinobacteriota (1), Bacillaceae (1), Clostridia (1), Methanosarcinaceae (1), Methanobacteriaceae (2), and Methanocellaceae (10). In the red soil, 10 Syntrophomondaceae MAGs were significantly enriched in the NPs treatments. In addition, MAGs belonging to Magnetospirillaceae were detected with increased abundance but only in the 0.5% NPs treatment. Among methanogens, MAGs affiliated with Methanosarcinaceae (B15_bin_34), Methanobacteriaceae, and Methanocellaceae were significantly enriched in the NPs treatments after 30-day incubation.
Taxonomic and functional profiles of carbohydrate and aromatic C utilization
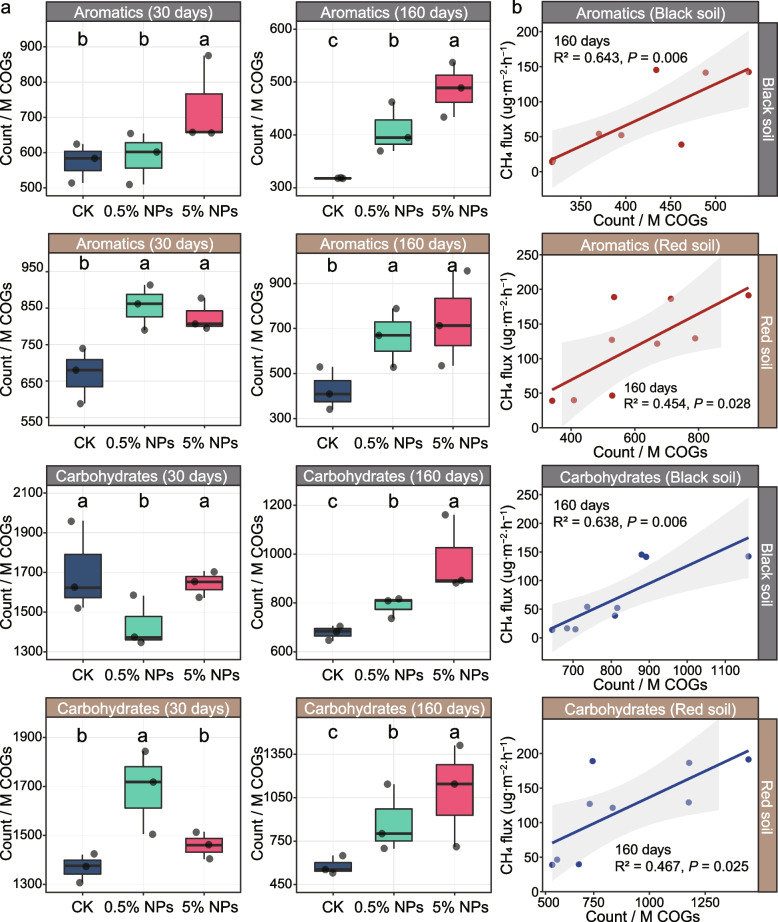
The addition of LDPE NPs significantly affected the taxonomic and functional profiles of genes involved in polymer breakdown. PCoA indicated that in both the black soil (P < 0.001) and the red soil (P < 0.01), the incubation time had a more significant impact on the beta diversity of genes encoding the degradation of complex carbohydrates and aromatic C than the NPs treatments (Additional File 2: Fig. S13). However, the NPs treatments had in both soil types a significant effect on the functional composition of genes encoding the degradation of aromatic C but not on those involved in utilizing complex carbohydrates (BS: RTreatment = 0.192, P < 0.05; RS: RTreatment = 0.169, P < 0.05) (Additional File 2: Fig. S13). In addition, the relative metagenomic abundance of genes encoding aromatics degradation was significantly correlated with DOC content in both soils after 160-day incubation, but not after 30-day incubation (Additional File 2: Fig. S14). The genes encoding the hydrolysis of complex carbohydrates exhibited a higher metagenomic abundance than those encoding the degradation of aromatic C in both the black soil and the red soil (Fig. 3a). Relative to CK, their metagenomic abundance had consecutively and significantly increased with the concentration of NPs after the 160-day incubation period, but their abundance was significantly lower than after 30 days of incubation (Fig. 3a; Additional File 2: Fig. S15). Furthermore, in black and red soils, the relative metagenomic abundance of genes encoding the degradation of carbohydrates and aromatic C was significantly correlated with both the log copy number of the mcrA genes and the CH4 production rate on day 160, but not on day 30 (Fig. 3b; Additional File 2: Figs. S16, S17).
Fig. 3.
Effect of the different treatments (CK, 0.5% NPs, and 5% NPs) in black and red soils on the metagenomic abundance and phylum-level composition of genes encoding the decomposition of aromatics and complex carbohydrates. a Changes in the relative abundance of genes involved in the degradation of aromatics and complex carbohydrates between control (CK), 0.5% NPs, and 5% NPs treatments after 30-day and 160-day incubations. The metagenomic abundance changes are indicated for both black and red soils in counts per million reads. Error bars denote standard deviation (n = 3). Different letters (a, b, c) indicate significant differences between the treatments (P < 0.05). b The treatment-specific relationships between CH4 production and the abundance of genes encoding the degradation of aromatics (red lines) and complex carbohydrates (blue lines) after 160-day incubation. The metagenomic abundances are indicated as counts per million reads
The taxonomic assignment of genes encoding the degradation of carbohydrates and aromatic C showed that the bacterial communities were dominated by species affiliated to the Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria (Additional File 2: Fig. S18). Among the genes encoding carbohydrate degradation, Syntrophomonadaceae, Peptococcaceae, Marinilabiliaceae, and Paenibacillaceae were significantly enriched by the NPs treatments in the black soil, while Mycobacteriaceae, Oxalobacteraceae, and Comamonadaceae showed a significant enrichment by the NPs treatments in the red soil (Additional File 2: Fig. S19). Among the genes encoding aromatic C decomposition, Clostridiaceae, Peptococcaceae, and Paenibacillaceae were significantly enriched by the NPs treatment in the black soil, while unclassified Proteobacteria showed an increased relative abundance in NPs-treated red soil (Additional File 2: Fig. S20).
The methanogen community
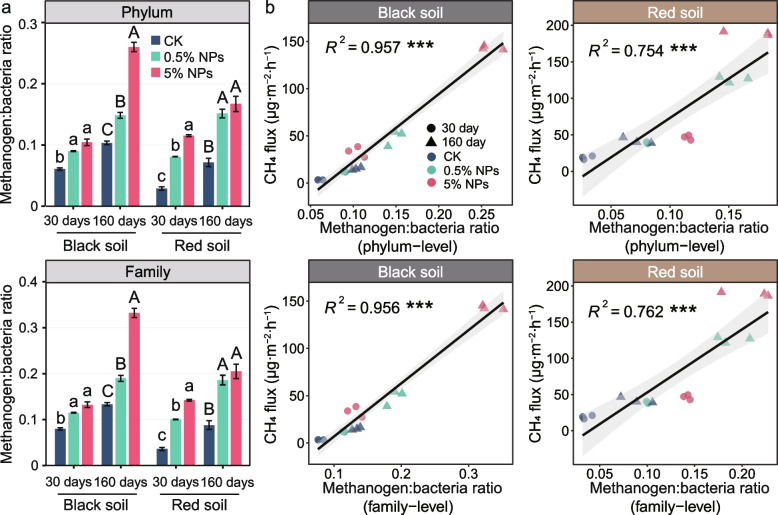
The metagenomic abundance of the methanogens relative to bacteria increased with both the concentration of NPs and the incubation time (Fig. 4a). In addition, the increase in the metagenomic methanogen-to-bacteria abundance ratio showed a highly significant correlation (P < 0.001) with CH4 production in both the black soil and the red soil (Fig. 4b). Likewise, the methanogen-to-bacteria transcript ratio and the expression level of mRNA affiliated to the KEGG level 3 category “methane metabolism” had, relative to the control, markedly increased in the red soil 0.5% NPs treatment after the 160-day incubation period. Both the methanogen-to-bacteria abundance ratio and the expression level of the methane metabolism-affiliated mRNA were comparable between the black soil and the red soil (Additional File 3: Table S6).
Fig. 4.
The metagenomic abundance ratios of methanogens to bacteria across the experimental treatments (CK, 0.5% NPs, 5% NPs) and incubation times (30 days, 160 days) at the phylum and family levels (a) and their correlations with methane production in black and red soils (b). Error bars in the column plots denote standard deviation (n = 3). Letters represent significant differences between treatments (a, b, c for 30 days; A, B, C for 160 days). Significance levels were as follows: *P < 0.05, **P < 0.01, and ***P < 0.001
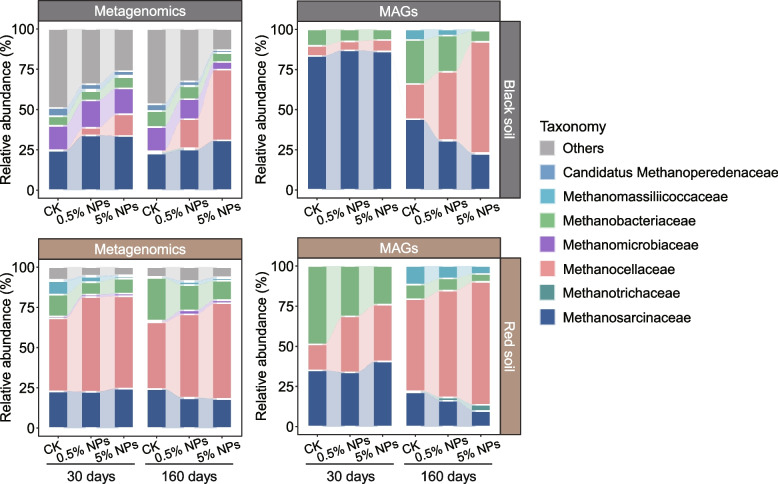
Metagenomic analysis and taxonomic classification of the methanogen MAGs collectively confirmed that Methanosarcinaceae, Methanotrichaceae, Methanocellaceae, and Methanobacteriaceae were the dominant methanogenic families (Fig. 5; Additional File 2: Fig. S21; Additional File 3: Table S7). In black soil, Methanosarcinaceae was the most abundant methanogen group, but the addition of NPs induced a shift towards an increase in the relative abundance of Methanocellaceae. In particular, the relative abundance of Methanocellaceae exceeded that of Methanosarcinaceae in the 5% NPs treatment after 160-day incubation, but not after 30-day incubation. A strong NPs-induced increase in Methanocellaceae abundance after 160 days of incubation was also well evidenced by the assembled MAGs (Fig. 5). In red soil, Methanocellaceae was the predominant family-level group, while both Methanosarcinaceae and Methanotrichaceae were of lower abundance. The addition of 0.5% and 5% NPs induced, relative to the control, a significant increase in the relative metagenomic abundance of the Methanocellaceae after an incubation period of 30 and 160 days, while no treatment effect was observed for Methanosarcinaceae and Methanotrichaceae. The strong increase in Methanocellaceae abundance at both incubation times was further corroborated by the assembled MAGs (Fig. 5). Transcript analysis of methanogenic mRNA showed that the addition of NPs induced predominant activity of the Methanosarcinaceae and Methanocellaceae, accompanied by a relative abundance shift in the expression of genes involved in methylotrophic methanogenesis from Methanomassiliicoccaceae towards Methanosarcinaceae in the red soil (Additional File 2: Figs. S22, S23).
Fig. 5.
Family-level changes in the metagenomic abundance of the methanogen community and the methanogen MAGs across the different treatments. Data represents means of three replicates
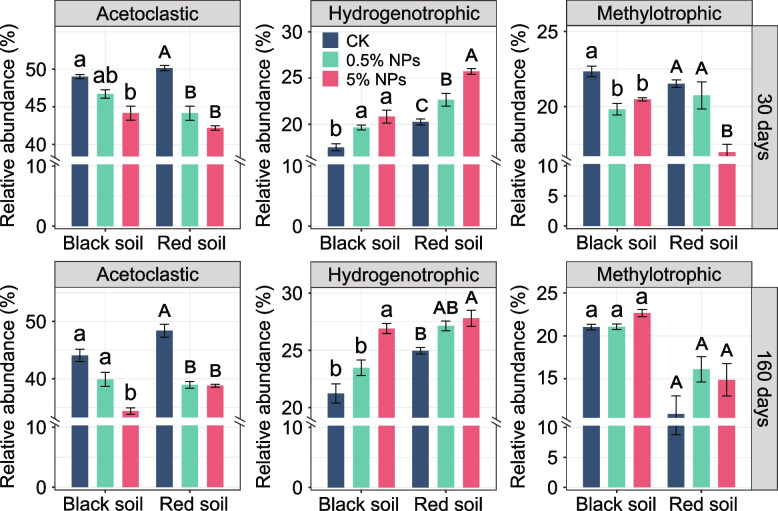
Analysis of the functional potential revealed that among the three methanogenic pathways, genes encoding acetoclastic methanogenesis were most abundant. However, their relative proportion decreased with increasing NPs concentration in both black and red soils (Fig. 6; Additional File 2: Figs. S24, S25). Concomitantly, the relative metagenomic abundance of genes encoding hydrogenotrophic methanogenesis significantly increased (Fig. 6; Additional File 2: Fig. S24, S25). Notably, the relative metagenomic abundance of genes encoding hydrogenotrophic methanogenesis was greater in the red soil across all the experimental treatments, which agrees well with the predominance of Methanocellaceae in this soil type (compare Figs. 5 and 6). The genetic potential for methylotrophic methanogenesis was increased in both soil types only after 160 days of incubation, and this increase was more pronounced in the red soil (Fig. 6). The NPs-induced increase in the relative metagenomic abundance of genes encoding hydrogenotrophic (R2BS = 0.591, P < 0.001; R2RS = 0.45, P < 0.05) and methylotrophic (R2BS = 0.458, P < 0.05; R2RS = 0.577, P < 0.05) methanogenesis showed a significant and positive correlation with DOC after 160 days of incubation, whereas the genes encoding acetoclastic methanogenesis did not display a significant correlation (Additional File 2: Fig. S27).
Fig. 6.
Changes in the relative abundance of the three methanogenesis pathways in response to the addition of 0.5% and 5% LDPE NPs to the black and red soils after 30-day and 160-day incubations. The calculation of the pathway-specific abundances is based on the data shown in the heat plot in Fig. S24. General biomarkers for methanogenesis pathways (mcrA, mtr) were not considered for calculating the pathway-specific abundances. The changes in metagenomic abundance are based on the cumulative abundances of those genes highly specific for each particular methanogenesis pathway. The cumulative relative abundance of these three methanogenesis pathways, along with the relative abundance of mcr and mtr genes, as shown in Fig. S26, collectively accounts for 100%. Letters indicate significant differences between the treatments (a, b, c for black soil; A, B, C for red soil). Error bars denote standard deviation (n = 3)
Further, functional KEEG-based annotation of the methane metabolism-affiliated mRNA agreed well with the metagenomic results. The mRNA transcripts highly specific for one of the three methanogenesis pathways revealed prevalent expression of acetoclastic methanogenesis. However, the expression level of hydrogenotrophic and methylotrophic methanogenesis increased relative to acetoclastic methanogenesis in the red soil 0.5% NPs treatment compared to the control treatment (Additional File 3: Table S6). Transcript mapping of methanogenic mRNA onto the pooled 38 methanogen MAGs showed that in both the control treatment and the 0.5% NPs treatment, the methanogen populations expressed all three methanogenic (acetoclastic, hydrogenotrophic, methylotrophic) pathways after 160 days of incubation (Additional File 2: Figs. S28, S29). The methylotrophic pathway was characterized by an exclusive expression of mtaBC (Additional File 2: Fig. S29). These two genes encode the methyltransferase/methanol corrinoid protein, a specific biomarker for methanol-dependent methanogenesis.
Discussion
Here, we investigated the microbiome-driven effects of LDPE NPs on the DOC content and CH4 production in two distinct paddy soil types, the black soil and the red soil. Through simulating the accumulation of LDPE NPs in soil microcosms, we particularly aimed to assess whether paddy soils majorly differing in their microbiome composition (see Additional File 1 for further details) show a common response pattern to the accumulation of LDPE NPs (Additional File 2: Fig. S11). Although the methanogenic communities in both soil types showed a marked difference in the duration of their lag phase for CH4 production (Fig. 1b; Additional File 2: Fig. S2) and related activities (Fig. 3a; Additional File 2: Figs. S5, S6), we observed a common response pattern primarily characterized by a significant NPs-induced increase in soil DOC. The increased soil DOC subsequently triggered a correlated increase in the methanogen community (determined via qPCR of mcrA gene copy numbers) and CH4 production (Fig. 1; Additional File 2: Figs. S7, S8). Correspondingly, the metagenomic methanogen-to-bacteria abundance ratio shifted in both soils towards a significant increase in the methanogen community (Fig. 4), which is further corroborated by the mRNA patterns obtained for the red soil after 160 days of incubation (Additional File 3: Table S6).
Stimulative effect of LDPE NPs on the bacterial activity
Light (UV)-induced aging and deterioration of LDPE after extended exposure time has been repeatedly reported [13, 45], but the occurrence of such effects can be excluded from our study due to the incubation of the soil microcosms under anoxic conditions in the dark. The lack of a significant carbon release from the pristine NPs in the sterile microcosms after 30-day incubation already provides unambiguous evidence that the significant increase in soil DOC is due to a LDPE NPs-induced increase in microbial activity [16]. While neglectable amounts of carbon were released abiotically from the LDPE NPs, soil DOC had — relative to the control — significantly increased in the LDPE NPs treatments in both the black soil and the red soil after the 30-day incubation period (Fig. 1a). Various bacteria and fungi have been demonstrated to be able to degrade and utilize polyethylene [46], such as Rhodococcus spp., Cladosporium spp., and Fusarium spp. [46–48]. In addition, soil protists, as major consumers of bacteria and fungi, may play a critical role in mitigating the impacts of microplastics pollution [49, 50]. However, all of them degrade polyethylene only under aerobic conditions [46]. The introduction of oxygen into the alkane structure and depolymerization are the two key limiting steps for its biodegradation [46, 47]. Indeed, previous studies have shown that polyethylene is not biodegradable under anaerobic conditions. For example, no degradation of polyethylene was observed in a liquid waste disposal bioreactor operated under anaerobic conditions for over 500 days, even at temperatures as high as 50 °C [51]. Furthermore, we observed no significant change in the metagenomic abundance of genes described to be involved in the aerobic degradation of the polyethylene, such as the flavin-binding monooxygenase (almA) and alkane 1-monooxygenase (alkB) [52] (Additional File 2: Fig. S30). Nonetheless, given the complexity and emerging nature of the research on nanoplastics, we admit that the anaerobic degradation of LDPE NPs by a yet unknown mechanism cannot be completely ruled out. This, however, would have to be a highly efficient mechanism, given that, for example, the observed NPs-induced carbon flux observed already after 30 days of incubation in the black soil corresponds to an equivalent of 16.2% of the total carbon added via nanoplastics to the 0.5% LDPE treatments (Additional File 3: Table S2).
The NPs-induced increase in DOC is most likely a stimulative effect on the hydrolytic microbiota activity, leading to an increased transformation of complex polymeric carbon into DOC that the soil microbiota could utilize. This view is strongly supported by the significant NPs-induced increase in the microbially accessible carbon that with incubation time was detectable as DOC or had already been converted into CH4 and CO2 (Additional File 3: Table S2). In particular, the strong and positive correlation between the metagenomic abundance of genes involved in degrading aromatic C and the DOC contents suggests that complex aromatic compounds were a major source for the NPs-induced increase in DOC levels (Additional File 2: Fig. S14), a conclusion further supported by the observed increase in the DOM recalcitrance after the 30-day incubation period (Additional File 2: Fig. S4; Additional File 3: Table S4). Furthermore, both the NPs treatments and the incubation time had a significant effect on the functional composition of the aromatic genes in the black soil and the red soil but only the incubation time on the genes encoding the decomposition of complex carbohydrates. Thus, in addition to soil lipids, the most likely sources for the NPs-induced increase in soil DOC are plant-derived humic substances and lignin. The three polymers share similar functional groups, such as carboxyl, phenolic/aliphatic hydroxyl, and methoxyl groups but, most importantly, aromatic moieties [53].
Concomitantly, the metagenomic abundance of genes encoding the decomposition of complex carbohydrates and aromatics was positively and significantly correlated with both the mcrA gene copy numbers and the CH4 production (Fig. 3b; Additional File 2: Figs. S16, S17). This finding suggests that the NPs-induced increase in the genetic potential for polymer hydrolysis significantly contributed to CH4 production. Specifically, the tenfold increase in the NPs concentration, from 0.5 to 5%, resulted in a 1.5-fold increase in CH4 production in red soil and a threefold increase in black soil, following the 160-day incubation period (Fig. 1b; Additional File 3: Table S2).
LDPE NPs significantly enriched for Syntrophomonadaceae and Methanocellaceae
Despite significant differences in the bacterial taxa responding to the NPs (Additional File 2: Fig. S11), the methanogen community showed a highly similar response pattern between the two paddy soil types. Both the black soil and the red soil shared a significant shift in the metagenomic potential from acetoclastic methanogenesis towards hydrogenotrophic methanogenesis (Fig. 6) and, at the taxonomic level, a specific abundance increase in Methanocella spp. (Fig. 5). While this common community response was highly significant at the DNA level, the mRNA profiles obtained for the control and 0.5% NPs treatments of the red soil further corroborated the metagenomic results (Additional File 2: Fig. S29; Additional File 3: Table S6). Concomitantly, the LDPE NPs induced a significant metagenomic abundance increase in Syntrophomonas-affiliated MAGs (Additional File 2: Fig. S10) and, even more evident, their hydrogenase genes (Additional File 2: Fig. S12). The correlated abundance increases in Syntrophomonadaceae and Methanocellaceae MAGs in response to the NPs treatments were highly significant for both black and red soils (Fig. 2; Additional File 2: Fig. S10).
Syntrophomonas is known to be a keystone taxon for the anaerobic oxidation of fatty acids of four (butyrate) or more carbons by β-oxidation. Indeed, Syntrophomonas spp. are able to degrade saturated and unsaturated monocarboxylic fatty acids of up to 18 carbons [54] by syntrophic association with hydrogen- or formate-utilizing partner organisms and depend on this association for thermodynamic reasons. To become thermodynamically feasible in our study, the β-oxidation of butyrate, valerate, or longer fatty acids, such as oleic acid and cis-vaccenic acid (Additional File 2: Figs. S5, S6), had to be syntrophically coupled to the activity of a hydrogenotrophic methanogen partner. While these two fatty acids are typical molecular marker for plant-derived SOC [55, 56], the high valerate concentrations may result from the ongoing syntrophic oxidation of LCFAs, which is most evident for the black soil (Additional File 2: Fig. S31). Methanocella has been shown to be intrinsically adaptive to low H2 concentrations. Their higher affinity to hydrogen allows these methanogens to outcompete Methanobacterium under substrate-limiting conditions [57]. By contrast, Methanobacterium is adapted to survive in high hydrogen conditions [58]. Indeed, the metagenomic abundance of Methanobacteriaceae was depleted in our NPs treatments. In particular, Methanocellaceae outcompeted Methanobacteriaceae during the later incubation period, likely because the partial H2 pressure fell below the level thatMethanobacteriaceae can effectively utilize [17, 59]. Thus, it is reasonable to conclude that in both soil types, the syntrophy between Syntrophomonas and Methanocella played a major role in the conversion of SCFAs (Fig. S5) and LCFAs (Additional File 2: Fig. S6) to acetate. Notably, the genomic composition of the Syntrophomonas assemblages clearly differed between the black and red soils. Their differences in soil characteristics and microbiome composition, but also the varying length range of fatty acids degraded by different Syntrophomonas spp., may well explain the differing genomic composition [60]. By contrast, the genomic composition of Methanocella spp. was similar, with the same MAGs being specifically enriched in both soil types (e.g., A28_bin_31 and A13_bin_23) (Additional File 2: Fig. S10).
Although the metagenomic abundance of genes encoding methylotrophic methanogenesis showed no significant difference between control and NPs treatments, their relative abundance in the NPs treatments was significantly and positively correlated to soil DOC after the extended 160-day incubation period (Additional File 2: Fig. S27). This correlation was accompanied in the red soil by a shift in the pathway expression from hydrogen-dependent methylotrophy operated by Methanomassiliicoccaceae to hydrogen-independent methylotrophy operated by Methanosarcinaceae (Additional File 2: Figs. S22, S23). Given this shift, LDPE NPs induced a relative decrease in the expression of the Methanosarcina-affiliated acetoclastic pathway but relative increase in the expression of Methanosarcina-affiliated methylotrophy. This was related to an exclusive expression of the methanol-specific mtaBC genes (Additional File 2: Fig. S29) [25]. Certain amounts of methanol will be released during the decomposition of lignin [61]. The view of lignin as the possible source of methanol agrees well with the fact that the metagenomic abundance of genes involved in degrading aromatic C showed a strong and positive correlation with DOC (P < 0.05) and CH4 production (P < 0.05) (Fig. 3b; Additional File 2: Figs. S14, S17). The mRNA profiles also confirmed that Methanocella were the dominant player in hydrogenotrophic methanogenesis (Additional File 2: Figs. S22, S23).
Conclusions
Our results clearly demonstrate that the accumulation of LDPE NPs in anoxic paddy soils leads to a significant increase in DOC and, in consequence, CH4 production. The presence of 5% NPs triggered the highest carbon flux and CH4 production, with a 1.5-fold to threefold increase in total CH4 production relative to the 0.5% NPs treatments after the 160-day incubation period. However, relative to the total amount of NPs added to the paddy soil, 0.5% NPs had a greater effect on the carbon flux and CH4 production than 5% NPs, thereby indicating that even smaller quantities of NPs can substantially influence CH4 production in rice field soils. Experimental evidence suggests that humic substances, lignin, and soil lipids are major sources for the NPs-induced increase in microbially accessible carbon. Although greatly differing in microbiome composition and the initial microbial response to NPs’ presence, both soil types exhibited a remarkably similar methanogen response. The specific enrichment of Syntrophomonas and Methanocella indicates that LDPE NPs stimulate the syntrophic oxidation of SCFAs and LCFAs, with Methanocella acting as the hydrogenotrophic methanogen partner. Methanocella has previously been shown to play a key role in H2 consumption during the syntrophic oxidation of SCFAs in Italian and Chinese rice paddy soils [17, 57]. While our research primarily focused on the effects of LDPE NPs on the methanogenic community and CH4 production in the anoxic bulk soil, the rice rhizosphere is more complex due to the oxygen diffusion facilitated by rice roots in agricultural settings. The aerenchyma in rice plants allows oxygen to diffuse into the rhizosphere, creating a mosaic of oxic and anoxic zones [62, 63]. This oxygen availability could promote the aerobic degradation of LDPE NPs, particularly through microbial processes that introduce oxygen into the alkane structure, a critical limiting step in NP depolymerization [46, 47]. Therefore, LDPE may be degraded by aerobic bacteria associated with rice roots, potentially providing “fresh carbon” that could further stimulate CH4 production in nearby micro-oxic and anoxic zones. Certain oxygen-tolerant methanogens, such as Methanocella [63, 64], may utilize the carbon derived from LDPE NPs for CH4 production, in addition to plant-derived carbon. Undoubtedly, our results have important implications for the production and release of methane in LDPE-contaminated paddy soils and, given the major contribution of this ecosystem to atmospheric methane, for global climate change. Further research is required to determine whether the common methanogenic response observed in black and red soils is similarly triggered by LDPE NPs in geographically diverse anthropogenic and natural wetlands.
Supplementary Information
Additional File 1: Supplemental materials and methods; Supplemental discussion.
Additional File 2: Supplemental Fig. S1 to S31.
Additional File 3: Supplemental Tables S1 to S7.
Acknowledgements
We thank Fusuo Zhang and Jianchao Zhang for fruitful discussions and critical comments on the paper. We are grateful to Xi Zhou, Xingjie Wu, and Ye Liu for technical support. We thank Xi Zhou for her assistance in analyzing microbial communities.
Authors’ contributions
JJP and YGZ designed the study. ZBH, YRH, and YL performed the experiments. ZBH, YRH, QCB, XL, and JJP performed the bioinformatic data analysis. ZBH, XL, YGZ, WL, MR, and JJP wrote the manuscript. All the authors revised, read, and approved the final manuscript.
Funding
This study was financially supported by the National Natural Science Foundation of China (42277307 & 41977038) and National Key Research and Development Program of China (2023YFD1900202).
Data availability
The metagenomic and metatranscriptomic raw sequencing data have been deposited in the NCBI Sequence Read Archive repository with BioProject Accession numbers PRJNA951521 and PRJNA951607. Affiliated files have been made available via FigShare; along with 391 MAGs, their taxonomic analysis, and functional annotation. The files and data are deposited under the title “Increased methane production associated with community shifts towards Methanocella in paddy soils with the presence of nanoplastics”. The online link is as follows: 10.6084/m9.figshare.25183763.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rillig MC, Leifheit E, Lehmann J. Microplastic effects on carbon cycling processes in soils. PLoS Biol. 2021;19(3):e3001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evangeliou N, Grythe H, Klimont Z, Heyes C, Eckhardt S, Lopez-Aparicio S, et al. Atmospheric transport is a major pathway of microplastics to remote regions. Nat Commun. 2020;11(1):3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, Liu Q, Jia W, Yan C, Wang J. Agricultural plastic mulching as a source of microplastics in the terrestrial environment. Environ Pollut. 2020;260:114096. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Geng S, Li Z, Song K. Effect of microplastic on anaerobic digestion of wasted activated sludge. Chemosphere. 2020;247: 125874. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Song K, Tu C, Li L, Feng Y, Li R, et al. Distribution and weathering characteristics of microplastics in paddy soils following long-term mulching: a field study in southwest China. Sci Total Environ. 2023;858(2): 159774. [DOI] [PubMed] [Google Scholar]
- 6.Hartmann NB, Huffer T, Thompson RC, Hassellov M, Verschoor A, Daugaard AE, et al. Are we speaking the same language? recommendations for a definition and categorization framework for plastic debris. Environ Sci Technol. 2019;53:1039–47. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Bank MS, Rinklebe J, Hou D. Plastic-rock complexes as hotspots for microplastic generation. Environ Sci Technol. 2023;57(17):7009–17. [DOI] [PubMed] [Google Scholar]
- 8.Kim SW, Waldman WR, Kim TY, Rillig MC. Effects of different microplastics on nematodes in the soil environment: tracking the extractable additives using an ecotoxicological approach. Environ Sci Technol. 2020;54(21):13868–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeley ME, Song B, Passie R, Hale RC. Microplastics affect sedimentary microbial communities and nitrogen cycling. Nat Commun. 2020;11(1):2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Ma D, Feng K, Lou Y, Zhou H, Liu B, et al. Polystyrene nanoplastics shape microbiome and functional metabolism in anaerobic digestion. Water Res. 2022;219: 118606. [DOI] [PubMed] [Google Scholar]
- 11.Fu S, Ding J, Zhang Y, Li Y, Zhu R, Yuan X, et al. Exposure to polystyrene nanoplastic leads to inhibition of anaerobic digestion system. Sci Total Environ. 2018;625:64–70. [DOI] [PubMed] [Google Scholar]
- 12.Wei W, Hao Q, Chen Z, Bao T, Ni B. Polystyrene nanoplastics reshape the anaerobic granular sludge for recovering methane from wastewater. Water Res. 2020;182: 116041. [DOI] [PubMed] [Google Scholar]
- 13.Romera-Castillo C, Pinto M, Langer TM, Alvarez-Salgado XA, Herndl GJ. Dissolved organic carbon leaching from plastics stimulates microbial activity in the ocean. Nat Commun. 2018;9(1):1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rillig MC, Lehmann A. Microplastic in terrestrial ecosystems. Science. 2020;368(6498):1430–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen M, Huang W, Chen M, Song B, Zhang Y. (Micro)plastic crisis: un-ignorable contribution to global greenhouse gas emissions and climate change. J Cleaner Prod. 2020;254: 120138. [Google Scholar]
- 16.Sun Y, Li X, Li X, Wang J. Deciphering the fingerprint of dissolved organic matter in the soil amended with biodegradable and conventional microplastics based on optical and molecular signatures. Environ Sci Technol. 2022;56(22):15746–59. [DOI] [PubMed] [Google Scholar]
- 17.Peng J, Wegner CE, Bei Q, Liu P, Liesack W. Metatranscriptomics reveals a differential temperature effect on the structural and functional organization of the anaerobic food web in rice field soil. Microbiome. 2018;6(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Li X, Xiao M, Feng Z, Yu Y, Yao H, et al. Effects of microplastics on soil carbon dioxide emissions and the microbial functional genes involved in organic carbon decomposition in agricultural soil. Sci Total Environ. 2022;806:150714. [DOI] [PubMed] [Google Scholar]
- 19.Schulz K, Hunger S, Brown GG, Tsai SM, Cerri CC, Conrad R, et al. Methanogenic food web in the gut contents of methane-emitting earthworm Eudrilus eugeniae from Brazil. ISME J. 2015;9(8):1778–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans PN, Boyd JA, Leu AO, Woodcroft BJ, Parks DH, Hugenholtz P, et al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol. 2019;17(4):219–32. [DOI] [PubMed] [Google Scholar]
- 21.Leroy F, Gogo S, Guimbaud C, Bernard-Jannin L, Hu Z, Laggoun-Defarge F. Vegetation composition controls temperature sensitivity of CO2 and CH4 emissions and DOC concentration in peatlands. Soil Biol Biochem. 2017;107:164–7. [Google Scholar]
- 22.Zhang C, Pan J, Liu Y, Duan C, Li M. Genomic and transcriptomic insights into methanogenesis potential of novel methanogens from mangrove sediments. Microbiome. 2020;8(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferry JG. Fundamentals of methanogenic pathways that are key to the biomethanation of complex biomass. Curr Opin Biotechnol. 2011;22(3):351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berghuis BA, Yu FB, Schulz F, Blainey PC, Woyke T, Quake SR. Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc Natl Acad Sci USA. 2019;116(11):5037–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narrowe AB, Borton MA, Hoyt DW, Smith GJ, Daly RA, Angle JC. Uncovering the diversity and activity of methylotrophic methanogens in freshwater wetland soils. mSystems. 2019;4(6):e00320-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Wegener G, Williams TA, Xie R, Xiao X. A methylotrophic origin of methanogenesis and early divergence of anaerobic multicarbon alkane metabolism. Sci Adv. 2021;7(27):eabd7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burd BS, Mussagy CU, Singulani JD, Tanaka JL, Scontri M, Brasil GSP, et al. Galleria mellonella larvae as an alternative to low-density polyethylene and polystyrene biodegradation. J Polym Environ. 2023;31(3):1232–41. [Google Scholar]
- 28.Zhang Z, Kang Y, Wang W, Xu L, Liu J, Zhang Z, et al. Low-density polyethylene microplastics and biochar interactively affect greenhouse gas emissions and microbial community structure and function in paddy soil. Chemosphere. 2023;340: 139860. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Wang J, Zhang T, Li B, Yan L. Assessment of methane emissions from China’s agricultural system and low carbon measures. Environ Sci Technol. 2021;44(3):200–8. [Google Scholar]
- 30.Fuller S, Gautam A. A procedure for measuring microplastics using pressurized fluid extraction. Environ Sci Technol. 2016;50(11):5774–80. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Li Y, Lei J, Li Z, Tan Q, Xie L, et al. Time-dependent effects of microplastics on soil bacteriome. J Hazard Mater. 2023;447: 130762. [DOI] [PubMed] [Google Scholar]
- 32.Peng J, Lv Z, Rui J, Lu Y. Dynamics of the methanogenic archaeal community during plant residue decomposition in an anoxic rice field soil. Appl Environ Microbiol. 2008;74(9):2894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst L, Steinfeld B, Barayeu U, Klintzsch T, Kurth M, Grimm D, et al. Methane formation driven by reactive oxygen species across all living organisms. Nature. 2022;603(7901):482–7. [DOI] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Liu C, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–6. [DOI] [PubMed] [Google Scholar]
- 36.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43(D1):D261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huson DH, Beier S, Flade I, Gorska A, El-Hadidi M, Mitra S, et al. MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. 2016;12(6):e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartman WH, Ye R, Horwath WR, Tringe SG. A genomic perspective on stoichiometric regulation of soil carbon cycling. ISME J. 2017;11(12):2652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6(1):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. [DOI] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1(5):1–6. [DOI] [PubMed] [Google Scholar]
- 44.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79(24):7696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Royer SJ, Ferron S, Wilson ST, Karl DM. Production of methane and ethylene from plastic in the environment. PLoS ONE. 2018;13(8):e0200574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghatge S, Yang Y, Ahn JH, Hur HG. Biodegradation of polyethylene: a brief review. Appl Biol Chem. 2020;63(1):1–14. [Google Scholar]
- 47.Tao X, Ouyang H, Zhou A, Wang D, Matlock H, Morgan JS, et al. Polyethylene degradation by a Rhodococcous strain isolated from naturally weathered plastic waste enrichment. Environ Sci Technol. 2023;57(37):13901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wróbel M, Szymańska S, Kowalkowski T, Hrynkiewicz K. Selection of microorganisms capable of polyethylene (PE) and polypropylene (PP) degradation. Microbiol Res. 2023;267: 127251. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Yang R, Guo L, Gao W, Su P, Xu Z, et al. The composition, biotic network, and assembly of plastisphere protistan taxonomic and functional communities in plastic-mulching croplands. J Hazard Mater. 2022;430:128390. [DOI] [PubMed] [Google Scholar]
- 50.Liu M, Wang C, Zhu B. Drought alleviates the negative effects of microplastics on soil micro-food web complexity and stability. Environ Sci Technol. 2023;57(30):11206–17. [DOI] [PubMed] [Google Scholar]
- 51.Selke S, Auras R, Nguyen TA, Castro Aguirre E, Cheruvathur R, Liu Y. Evaluation of biodegradation-promoting additives for plastics. Environ Sci Technol. 2015;49(6):3769–77. [DOI] [PubMed] [Google Scholar]
- 52.Sun X, Chen Z, Kong T, Chen Z, Dong Y, Kolton M, et al. Mycobacteriaceae mineralizes micropolyethylene in riverine ecosystems. Environ Sci Technol. 2022;56(22):15705–17. [DOI] [PubMed] [Google Scholar]
- 53.Yin X, Cai M, Liu Y, Zhou G, Richter-Heitmann T, Aromokeye DA, et al. Subgroup level differences of physiological activities in marine Lokiarchaeota. ISME J. 2021;15(3):848–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sousa DZ, Smidt H, Alves MM, Stams AJ. Ecophysiology of syntrophic communities that degrade saturated and unsaturated long-chain fatty acids. FEMS Microbiol Ecol. 2009;68(3):257–72. [DOI] [PubMed] [Google Scholar]
- 55.Conway N, McDowell CJ. Incorporation and utilization of bacterial lipids in the Solemya velum symbiosis. Mar Biol. 1991;108:277–91. [Google Scholar]
- 56.Tsai YY, Ohashi T, Wu CC, Bataa D, Misaki R, Limtong S, et al. Delta-9 fatty acid desaturase overexpression enhanced lipid production and oleic acid content in Rhodosporidium toruloides for preferable yeast lipid production. J Biosci Bioeng. 2019;127(4):430–40. [DOI] [PubMed] [Google Scholar]
- 57.Liu P, Yang Y, Lü Z, Lu Y. Response of a rice paddy soil methanogen to syntrophic growth as revealed by transcriptional analyses. Appl Environ Microbiol. 2014;80(15):4668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Li W, Baker BJ, Zhou Y, He L, Danchin A, et al. Carbon metabolism and adaptation of hyperalkaliphilic microbes in serpentinizing spring of Manleluag, the Philippines. Environ Microbiol Rep. 2022;14(2):308–19. [DOI] [PubMed] [Google Scholar]
- 59.Li D, Ni H, Jiao S, Lu Y, Liang Y. Coexistence patterns of soil methanogens are closely tied to methane generation and community assembly in rice paddies. Microbiome. 2021;9(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61(2):262–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurth JM, Nobu MK, Tamaki H, de Jonge N, Berger S, Jetten MS, et al. Methanogenic archaea use a bacteria-like methyltransferase system to demethoxylate aromatic compounds. ISME J. 2021;15(12):3549–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, Conrad R. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science. 2005;309(5737):1088–90. [DOI] [PubMed] [Google Scholar]
- 63.Erkel C, Kube M, Reinhardt R, Liesack W. Genome of rice cluster I archaea the key methane producers in the rice rhizosphere. Science. 2006;313(5785):370–2. [DOI] [PubMed] [Google Scholar]
- 64.Lyu Z, Lu Y. Metabolic shift at the class level sheds light on adaptation of methanogens to oxidative environments. ISME J. 2018;12(2):411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional File 1: Supplemental materials and methods; Supplemental discussion.
Additional File 2: Supplemental Fig. S1 to S31.
Additional File 3: Supplemental Tables S1 to S7.
Data Availability Statement
The metagenomic and metatranscriptomic raw sequencing data have been deposited in the NCBI Sequence Read Archive repository with BioProject Accession numbers PRJNA951521 and PRJNA951607. Affiliated files have been made available via FigShare; along with 391 MAGs, their taxonomic analysis, and functional annotation. The files and data are deposited under the title “Increased methane production associated with community shifts towards Methanocella in paddy soils with the presence of nanoplastics”. The online link is as follows: 10.6084/m9.figshare.25183763.