Abstract
NF-Y is a class of heterotrimeric transcription factor composed of three subunits; NF-YA, NF-YB, and NF-YC. This complex binds to the CCAAT box found in eukaryotic promoters and is involved in the plant development and proliferation at various stages. Although many studies were conducted on NF-Y gene family in various species, but no study has been reported yet in switchgrass (Panicum virgatum L.). In this study, 47 PvNF-Y genes (17 PvNF-YA, 18 PvNF-YB, and 12 PvNF-YC) have been identified and named according to their subfamily. Chromosome location analysis revealed that all 47 PvNF-Y genes are randomly distributed across nine chromosomes. Moreover, multiple sequence alignment showed the DNA-binding domain and NF-YA/NFYB interacting domains flanking with non-conserved domains. In addition, prediction of functional similarities among PvNF-Ys genes phylogenetic tree was constructed corresponding to Arabidopsis. The gene structure, conserved domains and motifs analysis of PvNF-Ys genes demonstrated their specificity and functional conservation. Cis-regulatory elements analysis identified numerous key CREs that are significantly associated with light, hormone, stress and plant development responses. Expression profiling indicated higher expression levels of many PvNF-YA genes during drought and heat stress. Additionally, qRT-PCR analysis showed that some PvNF-Ys genes have high expression level in root. In conclusion, the findings of this study could provide a foundation for further cloning and functional analysis of NF-Y genes in switchgrass.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11092-6.
Keywords: Switchgrass, NF-Y gene family, Expression analysis, Genome-wide analysis
Introduction
Transcription factors (TFs) can bind to cis-acting sites in eukaryotic gene promoter regions, thereby regulating the activation or inhibition of transcription and gene expression at particular growth and development stages [1]. Nuclear Factor Y (NF-Y), also referred to as heme activator protein (HAP) or CCAAT-binding factor (CBF), is a heterotrimeric transcription factor found across eukaryotes. In plants, NF-Y consists of three conserved subunits: NF-YA (HAP2), NF-YB (HAP3/CBF-A), and NF-YC (HAP5/CBF-C). These subunits work together to regulate various biological processes by modulating gene expression at critical developmental stages and in response to environmental cues [2]. Initially, the NF-YB and NF-YC subunits form a dimer in the cytoplasm. Then, this dimer translocates to the nucleus, where it associates with the NF-YA subunits to create a heterotrimeric complex. Finally, NF-Y heterotrimer subsequently binds to the CCAAT-box in the promoter regions to modulate the transcription of specific genes [2, 3]. In plants, subfamilies of NF-Y are essential for a wide array of developmental processes, including flowering time regulation [4], bud and root differentiation [5], embryogenesis [6], seed germination [6, 7] and chloroplast biogenesis [8]. Beyond development, these subfamilies are also essential for conferring tolerance to various abiotic stresses, such as drought, temperature extremes and salinity [9–12].
Recent studies have investigated the NF-Y gene family in various species, such as Arabidopsis thaliana [13], Oryza sativa [14], Brassica napus [15], Glycine max [16], Brassica rapa [17], Cucumis melo [18], Prunus persica [19], Solanum lycopersicum [20], Sorghum [21], foxtail millet [22], Vitis vinifera [23], and Hordeum vulgare [24], indicating the significance of the NF-Y gene family in physiological ecology and abiotic stress responses across these diverse species. Previous studies have reported that AtNF-YA1 is linked to post-germinative growth under salt stress in Arabidopsis thaliana [10]. Over-expression of AtNF-YA5 leads to enhanced resistance to drought by triggering stress-responsive genes in Arabidopsis thaliana [8]. AtNF-YB1 functions through a previously undescribed mechanism to confer improved drought performance in Arabidopsis [25]. OsNF-YC5 negatively affects salt tolerance in Oryza sativa under abiotic stress [26]. Over-expression of Ginkgo biloba substantially enhances heat shock factor expression (GbHSF) in callus tissue under heat stress, indicating that GbNF-YA6 plays a critical role in improving plant heat tolerance [27].
Switchgrass (Panicum virgatum L.) is a perennial warm-season C4 (NAD-malic enzyme) type grass that has a bunchgrass-like appearance and native to the tallgrass prairies of North America [28, 29]. Switchgrass being a C4 plant is a member of poaceae family and is considered a promising biofuel crop because of its high productivity rate, substantial genetic diversity, and broad native geographic distribution [30, 31]. Compared to traditional crops, switchgrass needs minimal management and utilizes resources more efficiently, particularly water, making it ideal for sustainable bioenergy production [32]. Switchgrass can confer resistance to salinity, drought, and inadequate nutrition [33]. Recent advancements in genetic linkage mapping, gene expression, genome high-throughput sequencing, and assembly have established switchgrass as a model species for energy crops [34, 35]. Additionally, microRNAs and long non-coding RNAs were sequenced and analyzed to understand how switchgrass regulates its response to drought stress [36, 37].
Although NF-Y gene family has been widely reported in various plant species. However, its roles and functions in switchgrass remain poorly understood till now. In this study, many bioinformatics tools were used to analyze the NF-Y gene family in switchgrass genome. To identify members of NF-Y genes, a deterministic approach was used including multiple sequence alignment, gene structure, conserved domains, motifs analysis, chromosome distribution, comparative phylogenetic analysis, cis-regulatory analysis, enrichment and expression pattern analysis. This study could provide valuable insights for identifying candidate NF-Y genes involved in regulating growth, development, and response to abiotic stresses in switchgrass.
Materials and methods
Identification of NF-Y gene family in the switchgrass
NF-Y protein sequences of Arabidopsis thaliana were downloaded from TAIR (https://www.arabidopsis.org/browse/genefamily/index.jsp/ accessed on 03 April 2024), and the genome file of switchgrass (Panicum virgatum L.) was downloaded from Phytozome v13.0 (https://phytozome-next.jgi.doe.gov/ was accessed on 03 April 2024). To identify NF-Y proteins in switchgrass, BLAST-P (https://blast.ncbi.nlm.nih.gov/Blast.cgi/ accessed on 05 April 2024) was performed with E-value ≤ 1e-5 as default parameter, and PvNF-Ys proteins were successfully matched to corresponding AtNF-Ys proteins (File S1). The physicochemical properties were calculated through ExPASy program (https://web.expasy.org/protparam/ accessed on 08 April 2024), to predict the length of protein, theoretical isoelectric point (pI), and molecular weight (MW) [38]. In silico subcellular localization was conducted by WOLF PSORT (https://wolfpsort.hgc.jp/ accessed on 09 April 2024) [39].
Functional domain and conserved motif analysis
Additionally, all candidate PvNF-Ys proteins were further analyzed to confirm the presence of conserved domains using the NCBI-Batch CD Search (accessed on 07 April 2024). Finally, only the genes containing conserved patterns of domains were selected for subsequent analysis. Further, the conserved motifs of the identified PvNF-Ys genes were predicted by using MEME-suite tool (https://meme-suite.org/tools/meme, accessed on 12 April 2024) to understand the structure and peptide sequence of PvNF-Ys genes with default values and maximum 10 number of motifs set or other variables [40].
Multiple sequence alignment and phylogenetic analysis
To further identify the evolutionary relationship between NF-Y proteins in switchgrass. Multiple sequence alignment was performed by using ClustalW program with default parameters. Then, phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA 11 software. The tree was then annotated and beautified by using the iTOL tool (accessed on 10 April 2024) [41].
Gene structure, and chromosomal distribution analysis
To predict the gene structure and chromosomal distribution of all the predicted PvNF-Ys proteins, we used TBtool for identification and visualization [42]. Each PvNF-Ys gene was mapped to switchgrass chromosomes according to the gene number and location, where duplicated gene pairs were determined on the bases of higher sequence identity (> 70–80) and stronger evolutionary relationships and linked with a colour line.
Cis-regulatory elements and synteny analysis
Cis-regulatory elements (CREs) within the promoter region of PvNF-Ys genes were analyzed using sequences 1000 bp upstream of the start codon from Phytozome. The CREs of the PvNF-Ys genes were identified by PlantPAN 4.0 database (http://PlantPAN.itps.ncku.edu.tw/, accessed on 18 April 2024) [43]. For synteny analysis, genomes of Panicum virgatum L., Arabidopsis thaliana, Oryza sativa, and Zea mays were downloaded from the Phytozome. MCScanX (Multiple Collinearity Scan Toolbox) was utilized with default settings to assess gene duplication events [44]. Dual syntenic maps were created by using TBtools to identify synteny relationships between paralogous Panicum virgatum L. genes and orthologous PvNF-Ys genes in Arabidopsis, Oryza sativa, and Zea mays.
Functional annotation and transcriptome analysis of PvNF-Ys genes
To explore the functional annotation of the predicted PvNF-Ys genes, GO ontology analysis was carried out by using Eggnog (http://eggnog5.embl.de/ accessed on 22 April 2024) and visualized by WEGO 2.0 (https://wego.genomics.cn/ accessed on 22 April 2024). To investigate the expression level of all the identified PvNF-Ys genes, we obtained previously reported RNA-seq data from NCBI-GEO (https://www.ncbi.nlm.nih.gov/geo/, accessed on 2 May 2024) (accession no. GSE132772, and GSE174278) [45–47] for switchgrass subjected to drought and combination of drought and heat stress treatment.
Plant growth, expression profiling under tissue/organ development and qRT-PCR analysis
To investigate expression profiling under tissue/organ development, data was retrieved from JGI database (https://phytozome-next.jgi.doe.gov/geneatlas/, 3 May 2024) and to validate the organ specific expression, switchgrass seedlings were grown at room temperature of 25 ± 1 °C, 16 h light/20 ± 1 °C, a total RNA was extracted from various part of six weeks old switchgrass plants including root, node, leaf blade, leaf sheath, shoot, and seeds using EasyPure Plant RNA Kit (Transgene Biotech, Beijing, China), and according to the manufacturer’s instructions, cDNA was synthesized through the EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). qRT-PCR was conducted by using SYBR Green Mix (Magic-bio, Hangzhou, China). The gene specific primers were designed by PrimerQuest™ Tool, and eEF1α was used as a reference gene. are shown in Table S4. The quantitave data was analyzed by 2−(ΔΔCt) method and excel was used for gene expression map.
Results
Identification of PvNF-Y genes in Switchgrass
Based on amino acid residues of NF-Y gene family of Arabidopsis thaliana, a total of 47 non-redundant protein sequences (17 PvNF-YA, 18 PvNF-YB, 12 PvNF-YC) representing primary transcripts were identified in switchgrass by using BLAST-P, after removing duplicates and incomplete sequences. According to their sub-family, structure and ID, genes were named as PvNF-YA01 to PvNF-YA17, PvNF-YB01 to PvNF-YB18, and PvNF-YC01 to PvNF-YC12 (Table S1). Further, we have conducted physiochemical properties analysis of all the predicted PvNF-Ys proteins. Results revealed that the protein length of all the 47 PvNF-Ys proteins ranged from 128 (PvNF-YC04, PvNF-YC12) to 376 (PvNF-YC08) residues (Table 1). The molecular weight ranged from 13550.4 (PvNF-YC12) to 40334.4 (PvNF-YC08) kDa, and the isoelectric point varies from 4.76 (PvNF-YC02) to 10.86 (PvNF-YA12). Further, the GRAVY index ranged from − 1.358 (PvNF-YC08) to -0.135 (PvNF-YC03), indicating that PvNF-Ys proteins were mainly hydrophilic, though the extent of their hydrophilicity varied (Table 1). A comparison of the three subfamilies of PvNF-Y exhibit distinct physiochemical properties. We found that PvNF-YA gene family is characterized by larger proteins with higher pI, which are generally associated with DNA binding and transcriptional activation, as NF-YA directly interacts with DNA in the NF-Y complex [16, 17]. In contrast, PvNF-YB and PvNF-YC subfamilies have lower molecular weight often act in specialized regulatory roles, interacting with other proteins to modulate gene expression [16].
Table 1.
Physiochemical properties of all the predicted PvNF-Ys genes
| Sub-family | Genes name | Gene ID | Protein length (AA) | Molecular weight (MV) | Theoretical (pI) |
|---|---|---|---|---|---|
| PvNF-YA01 | Pavir.2NG042700.1 | 256 | 27451.8 | 9.96 | |
| PvNF-YA02 | Pavir.2NG583300.1 | 292 | 31,777 | 8.44 | |
| PvNF-YA03 | Pavir.1KG112061.1 | 268 | 29206.9 | 10.76 | |
| PvNF-YA04 | Pavir.3KG543700.1 | 214 | 23366.6 | 7.98 | |
| PvNF-YA05 | Pavir.3KG551100.2 | 328 | 35,332 | 9.19 | |
| PvNF-YA06 | Pavir.9KG612800.1 | 240 | 26501.4 | 8.68 | |
| PvNF-YA07 | Pavir.9KG392700.5 | 314 | 34434.5 | 9.24 | |
| NF-YA | PvNF-YA08 | Pavir.9KG040100.5 | 269 | 28460.6 | 9.39 |
| PvNF-YA09 | Pavir.9KG069000.2 | 346 | 36702.2 | 9.51 | |
| PvNF-YA10 | Pavir.2KG064700.1 | 262 | 27864.6 | 10 | |
| PvNF-YA11 | Pavir.2KG529900.14 | 290 | 31,641 | 9.24 | |
| PvNF-YA12 | Pavir.1NG526800.1 | 267 | 29006.7 | 10.86 | |
| PvNF-YA13 | Pavir.9NG726300.1 | 239 | 26406.3 | 8.03 | |
| PvNF-YA14 | Pavir.9NG184100.1 | 346 | 36937.4 | 9.34 | |
| PvNF-YA15 | Pavir.9NG146600.2 | 267 | 28302.5 | 9.44 | |
| PvNF-YA16 | Pavir.3NG276300.1 | 331 | 35534.2 | 9.05 | |
| PvNF-YA17 | Pavir.3NG282900.1 | 214 | 23449.7 | 6.9 | |
| PvNF-YB01 | Pavir.2NG582200.1 | 217 | 22826.6 | 6.21 | |
| PvNF-YB02 | Pavir.6KG075900.1 | 252 | 25471.1 | 5.89 | |
| PvNF-YB03 | Pavir.4KG139640.1 | 245 | 26,541 | 9.09 | |
| PvNF-YB04 | Pavir.3KG485155.1 | 180 | 18951.1 | 6.31 | |
| PvNF-YB05 | Pavir.3KG314300.3 | 141 | 15493.2 | 5.17 | |
| PvNF-YB06 | Pavir.9KG391200.2 | 214 | 22,612 | 6.3 | |
| PvNF-YB07 | Pavir.2KG531100.1 | 217 | 22670.3 | 6.3 | |
| PvNF-YB08 | Pavir.5NG619400.1 | 159 | 17933.9 | 6.43 | |
| NF-YB | PvNF-YB09 | Pavir.5NG619500.1 | 164 | 17466.4 | 6.2 |
| PvNF-YB10 | Pavir.5NG576500.1 | 161 | 17649.8 | 5.96 | |
| PvNF-YB11 | Pavir.1NG467100.1 | 269 | 28294.3 | 6.59 | |
| PvNF-YB12 | Pavir.1NG409819.1 | 252 | 26847.7 | 6.35 | |
| PvNF-YB13 | Pavir.9NG528800.1 | 213 | 22601.9 | 6.3 | |
| PvNF-YB14 | Pavir.5KG753500.1 | 154 | 17250.2 | 6.43 | |
| PvNF-YB15 | Pavir.5KG607100.3 | 164 | 17846.9 | 5.82 | |
| PvNF-YB16 | Pavir.3NG079767.2 | 180 | 18,879 | 6.31 | |
| PvNF-YB17 | Pavir.3NG180531.2 | 138 | 15,105 | 5.3 | |
| PvNF-YB18 | Pavir.4NG231400.1 | 249 | 26821.2 | 9.57 | |
| PvNF-YC01 | Pavir.6NG263700.1 | 208 | 22254.5 | 5.48 | |
| PvNF-YC02 | Pavir.6NG097100.1 | 330 | 35403.6 | 4.76 | |
| PvNF-YC03 | Pavir.2NG380900.4 | 140 | 15427.6 | 5.36 | |
| PvNF-YC04 | Pavir.7NG445700.1 | 128 | 13568.4 | 6.83 | |
| PvNF-YC05 | Pavir.6KG104000.1 | 332 | 35965.2 | 5.2 | |
| NF-YC | PvNF-YC06 | Pavir.6KG339000.1 | 208 | 22171.4 | 5.66 |
| PvNF-YC07 | Pavir.1KG073400.2 | 259 | 28857.6 | 5.04 | |
| PvNF-YC08 | Pavir.9KG018900.1 | 376 | 40334.4 | 10.53 | |
| PvNF-YC09 | Pavir.1NG065700.5 | 260 | 29,147 | 5.05 | |
| PvNF-YC10 | Pavir.9NG568300.2 | 249 | 26153.4 | 5.2 | |
| PvNF-YC11 | Pavir.4NG275600.1 | 246 | 27417.1 | 5.13 | |
| PvNF-YC12 | Pavir.7KG370900.1 | 128 | 13550.4 | 6.83 |
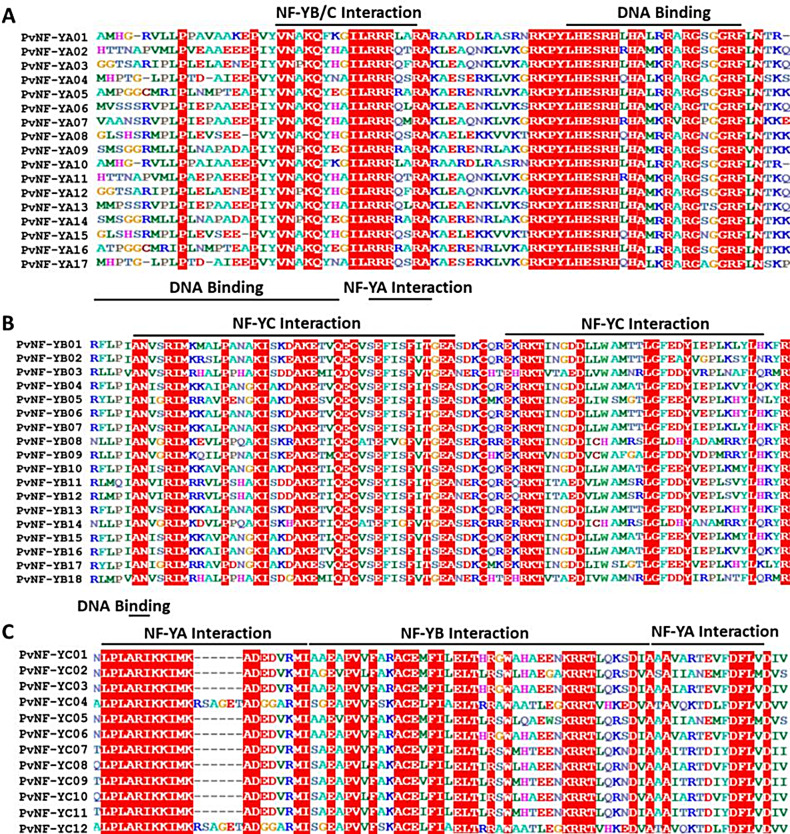
Multiple sequence alignment of PvNF-Ys proteins
To identify and analyze the conserved domains, the sequences of PvNF-YA, PvNF-YB and PvNF-YC proteins were subjected to conduct multiple sequence alignment through ClustalW (Fig. 1). A highly conserved interaction and the DNA-binding domains were found in each member of these three subunit PvNF-Ys proteins. Each member of PvNF-YA contains DNA binding and NF-YB/C-interaction domain (Fig. 1A). Similarly, the PvNF-YB members have two NF-YC interaction domains separated by DNA binding and NF-YA domain (Fig. 1B). However, PvNF-YC members have two NF-YA interactions, one NF-YB interaction, and a two amino acid DNA-binding domain (Fig. 1C).
Fig. 1.
Multiple sequence alignment analysis of PvNF-Ys proteins. (A) PvNF-YA (B) PvNF-YB (C) PvNF-YC alignment of highly conserved domains in switchgrass
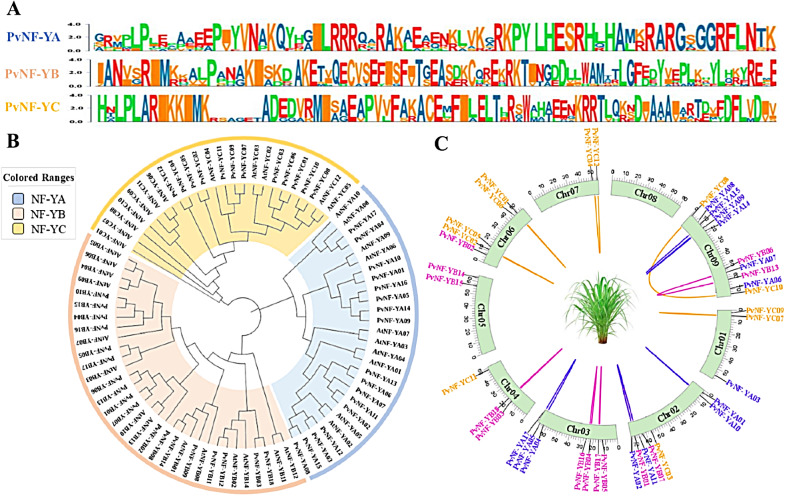
Conserved motifs, phylogenetic and chromosome distribution analysis
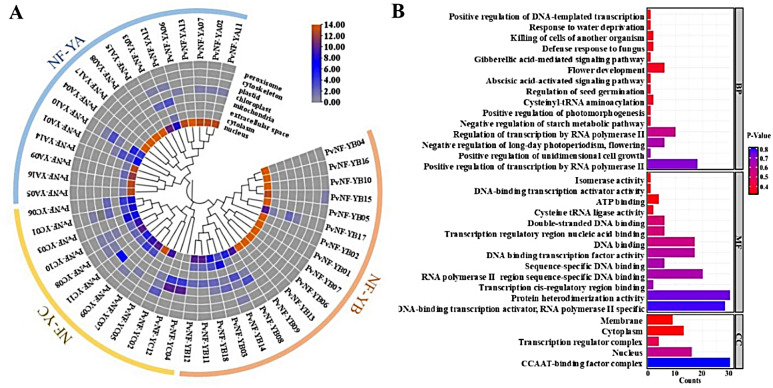
A conserved motifs analysis was carried out through an online MEME program by using protein sequences of PvNF-Ys, and their highly conserved sequence logos are presented in (Fig. 2A). To further identify the evolutionary relationship between switchgrass and Arabidopsis, a phylogenetic tree through the neighbor-joining method was constructed by using 35 AtNF-Ys and 47 PvNF-Ys protein sequences. A total of 47 PvNF-Ys was divided into three subfamilies such as PvNF-YA, PvNF-YB and PvNF-YC based on the classification of Arabidopsis NF-Y gene families and the composition of conserved domains in the PvNF-Ys proteins. Among these subfamilies, PvNF-YC was smallest group with 12 genes. Whereas PvNF-YB had largest group with 18 genes. The PvNF-YA subfamily had 17 gene members (Fig. 2B).
Fig. 2.
Conserved motif logos, phylogenetic tree, and chromosome distribution analysis in the PvNF-Ys genes. (A) A highly conserved sequence logos in the three subunits of PvNF-Ys proteins. (B) phylogenetic tree analysis of PvNF-Ys genes. (C) Chromosome distribution of PvNF-Ys genes are marked with different colors (NF-YA, blue; NF-YB, pink; NF-YC, orange)
We analyzed the genome of switchgrass to study the chromosomal distribution of PvNF-Ys genes. Our analysis showed that all the 47 PvNF-Y genes were unevenly distributed across 9 chromosomes. The maximum number of PvNF-Ys genes were found on chr9 (10), chr2 (7), chr3 (7) and chr6 (5). Chromosome 1 and 4 contained three PvNF-Y genes, while chr7 and chr5 each had only two PvNF-Y genes. however, no gene was found on chr8 (Fig. 2C).
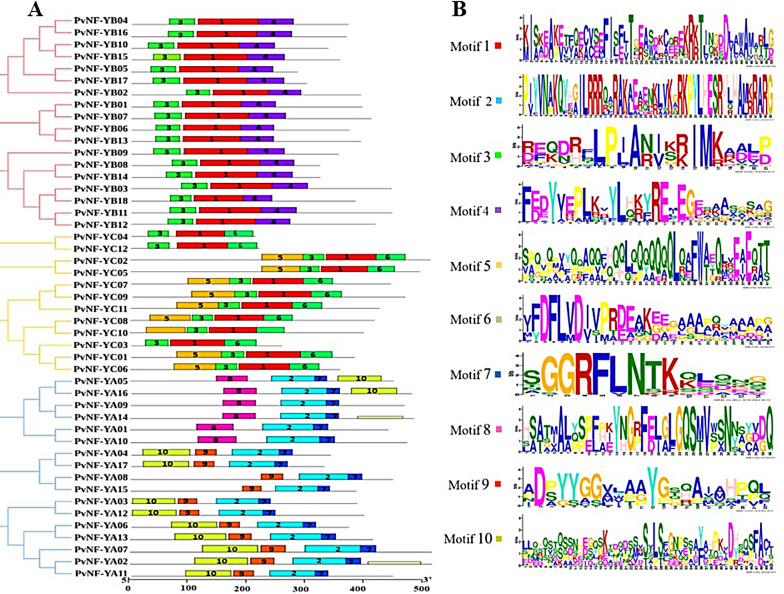
Across the three subunit PvNF-Ys proteins, a total of ten motif logos with distinct amino acid sequences were identified. In the PvNF-YA subunit members, motifs 2,7,8,9 and 10 were detected, with motifs 2 and 7 found consistently across all members of the PvNF-YA subfamily. In the PvNF-YB subfamily, all the members shared similar distribution of motifs including 1,3 and 4 motifs. However, motifs 1 and 3 were also detected in the PvNF-YC subfamily members along with motif 5 and 6 (Fig. 3A). All the ten-motif logos were displayed in (Fig. 3B).
Fig. 3.
The distribution of conserved motifs in the PvNF-Ys genes. (A) Analysis of conserved motifs along with rectangular phylogenetic tree. Different color bars represented motif types in each subunit of PvNF-Ys proteins. The length of proteins can be estimated using the scale at the bottom. (B) Sequence information of 10 conserved motifs
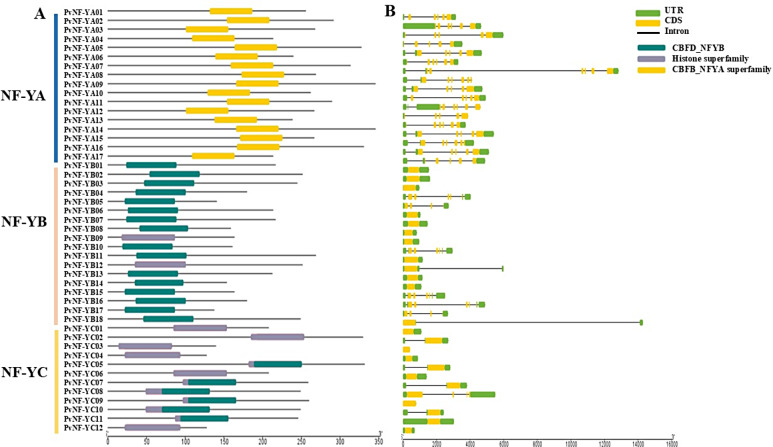
Functional domain and gene structure analysis of PvNF-Ys genes
The conserved domain distribution was identified by NCBI conserved domain database. Analysis results revealed that members of PvNF-YA family contain conserved domain CBFB-NFYA (CCAAT-binding transcription factor B-NFYA), or the CBFB-NFYA superfamily (Fig. 4A). PvNF-YB and PvNF-YC families possess conserved regions within the CBFD-NFYB-HMF (CCAAT-binding transcription factor D-NFYB histone-like transcription factor) family or Histone superfamily (Fig. 4A). Additionally, to improve our understanding of gene architecture, the genomic DNA sequences of the PvNF-Y genes were analyzed to assess and compare the structures and counts of exons, introns, and UTRs. The results showed that majority of PvNF-YB and PvNF-YC genes had either no or only a few introns. Moreover, each PvNF-YA gene have maximum number of introns and exons. The number of UTRs were found in PvNF-Ys ranging from 1 to 4. However, there was no UTR found in PvNF-YC03 and PvNF-YC09 genes (Fig. 4B).
Fig. 4.
Distribution of domains and gene structure prediction of PvNF-Ys genes in switchgrass. (A) Conserved domains of PvNF-Ys genes. Different colour boxes represent the conserved domains. (B) The number of introns, CDS and UTR were represented in black lines, yellow bars, and green bars in each gene
Orthologous gene pairs among switchgrass and various plant species
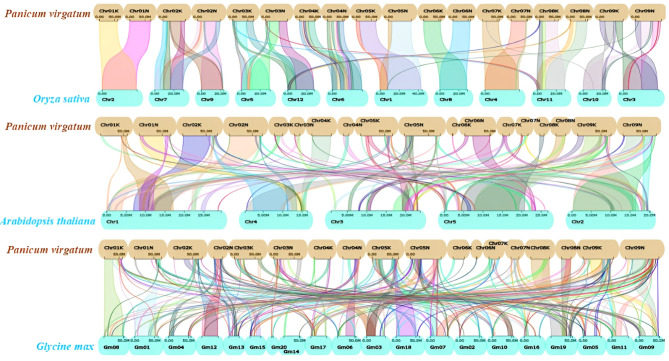
The orthologous gene pairs offer insights into the evolutionary relationships among different plant species. To gain a deeper understanding of the evolutionary connections of PvNF-Ys genes in dicot and monocot plants, a dual synteny analysis was performed between switchgrass (Panicum virgatum L.), Arabidopsis, Oryza sativa and Glycine max. These results indicate that collinear relationship between switchgrass, Glycine max and Arabidopsis thaliana was most significant while between Oryza sativa was less prominent (Fig. 5). These results provide clues for examining the relationship between functional genes. Beyond that, these findings can help to demonstrate the homologous feature in the genome organization of different plants and further explore the way of functional evolution of NF-Y gene pairs in the process of plant evolution.
Fig. 5.
Dual synteny analysis of switchgrass PvNF-Ys genes with dicot and monocot plant. Yellow bars represent chromosomes of switchgrass, while blue bars represent chromosomes of Arabidopsis, Oryza sativa and Glycine max. Colour lines emphasize syntenic PvNF-Ys gene pairs within the genomes of switchgrass and other plants. Dispute lines denote collinear blocks, while fine lines in the background emphasize syntenic NF-Y gene pairs within the genomes of switchgrass and other plants
Subcellular localization, functional annotation and cis-regulatory elements analysis
To determine the subcellular location of PvNF-Ys proteins, we performed an in silico sub-cellular localization analysis using WOLF PSORT. The results showed that most of the predicted PvNF-Ys proteins were predominantly located in the nucleus except the PvNF-YC04 and PvNF-YC12, were found in the mitochondria (Fig. 6A). To further analyze functional annotation, all the identified 47 PvNF-Ys genes were successfully annotated for their potential functions using Eggnog. The Gene ontology (GO) analysis results showed that PvNF-Ys genes were characterized into three categories; molecular function, biological process and cellular components. The PvNF-Ys genes were significantly involved in many biological processes including positive regulation of transcription by RNA polymerase II, negative regulation of long-day photoperiodism, flower development and positive regulation of unidimensional cell growth. Further, PvNF-Ys genes were predicted to be more frequently involved in many molecular functions such as RNA polymerase II-specific, protein heterodimerization activity, cis-regulatory region sequence-specific DNA binding, and DNA-binding transcription factor activity. Furthermore, PvNF-Ys were involved significantly in the CCAAT-binding factor complex, nucleus, and cytoplasm as illustrated in cellular component category (Fig. 6B).
Fig. 6.
In silico subcellular localization and functional annotation analysis of all the predicted PvNF-Ys proteins. (A) Subcellular localization analysis in the PvNF-Y proteins, side bar shows highest count present in each category. (B) Gene ontology analysis of the predicted PvNF-Ys genes significantly enriches < 1 P-value
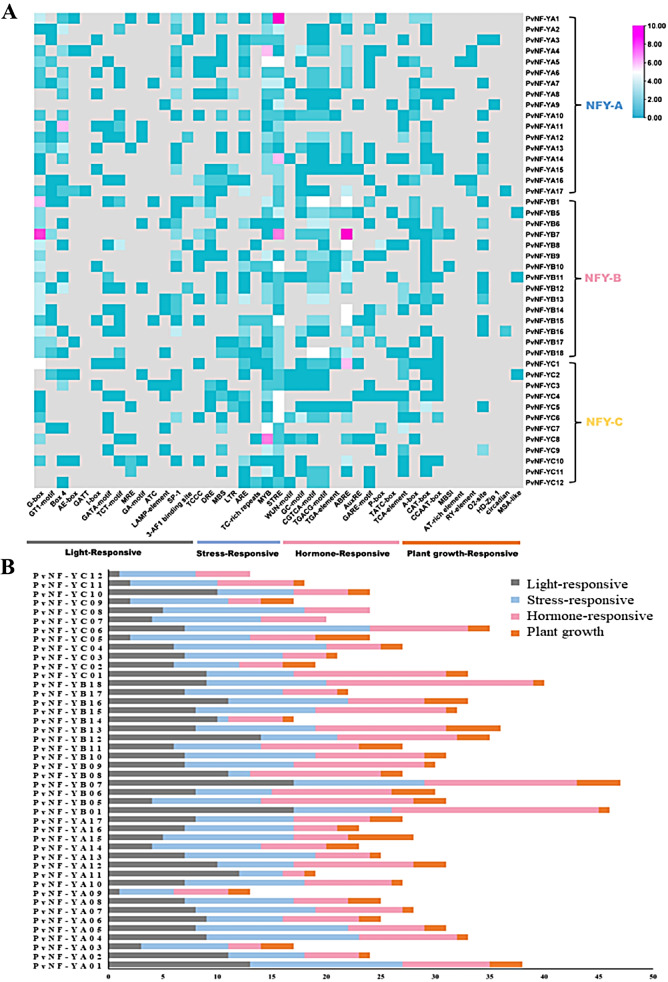
Cis-regulatory elements are DNA-binding motifs located in the promoter regions of genes that regulate transcription. In silico analysis of cis-regulatory elements can be performed to evaluate the potential functions of various genes. In PvNF-Ys genes many cis-regulatory elements were detected at the promoter region. Several phytohormone and stress-related motifs including DRE, MBS, LTR, ARE, TC-rich repeaters, MYB, STRE, WUN-motif, GC-motif, CGTCA-motif, TGACG-motif, ABRE (Abscisic acid-responsive element), GARE-motif, P-box, TATC-box (Gibberellin-responsive element), AuxRE, TGA-element (Auxin-responsive element) and TCA-element (Salicylic acid-responsive element) were detected in the promoter region, suggesting that the expression of PvNF-Ys genes may be regulated by multiple phytohormone. Moreover, stress-related cis-regulatory elements motifs findings indicated that PvNF-Ys genes may be closely associated with multiple biotic and abiotic stress responses. Furthermore, many motifs related to plant growth, development and other elements such as A-box, CAT-box, CCAAT-box, MBSI, AT-rich element, RY-element, O2-site, HD-Zip 1, Circadian and MSA-like were found in PvNF-Ys genes. Several light responsive such as G-box, GT1-motif, Box 4, AE-box, GATT, I-box, GATA-motif, TCT-motif, MRE, GA-motif, ATC, LAMP-element, SP-1, 3-AF1 binding site, and TCCC were detected in the PvNF-Ys genes (Fig. 7A, Table S2). Overall, statistics of cis-regulatory elements in the PvNF-Y gene family were calculated. All the predicted cis-regulatory elements are categorized by different colors based on their association with light, stress, hormone and plant growth (Fig. 7B). The results indicated that stress-responsive and hormone-related motifs were significantly dominant among the predicted PvNF-Ys genes (Fig. 7B, Table S3).
Fig. 7.
In-silico cis-regulatory elements analysis of the predicted PvNF-Ys genes. (A) Cis-regulatory elements motifs analysis in their respective groups were associated with different phytohormones, responses to stress and light, growth and developmental processes. (B) Different color bars represent the highest count based on the number of times a particular cis-element occurs within the promoter in each member of PvNF-Y gene family
Expression patterns of PvNF-Ys genes under drought and heat stress
To explore the potential role of PvNF-Ys genes under drought stress and plant development. Here, we compiled the expression pattern of PvNF-Ys genes by using data from NCBI-GEO. The experiment (accession no; GSE132772) was conducted on gene expression and physiological responses in switchgrass under drought stress [47]. This study led to understanding the effects of whole genome duplication and water stress on growth, physiology, and gene expression. Tetraploid liberty and its neo-octoploid cultivars were subjected to drought and recovery conditions and mRNA sequencing of collected samples was performed using Illumina. The expression pattern of all PvNF-Ys genes was retrieved from this dataset. Among the 47 PvNF-Ys genes, most of the PvNF-YA genes exhibited high expression patterns compared to PvNF-YB and PvNF-YC gene families (Fig. 8A).
Fig. 8.
The expression profile of PvNF-Ys genes under drought stress treatment. (A) The colors bar represents level of expression based on FPKM values: orange/high expression, yellow/low expression, blue/no expression. (B) The expression profile of PvNF-Ys genes under drought and combined drought and heat stress depend on FPKM values: green/high expression and Yellow/low expression
Another RNA-Seq study (accession no; GSE174278) investigated the transcriptomic response of switchgrass (Panicum virgatum L.), genotype Alamo AP13, to drought and combined drought and heat stress [45, 46]. The experiment involved drought treatment alone and a combination of drought with heat stress (35 °C/25°Cday/night). Samples were collected at multiple time points (0, 72, 96, 120, 144, and 168 h). The expression profile of all PvNF-Ys genes was re-analyzed from this dataset, we found that most of the PvNF-YA genes showed high expression level compared to PvNF-YB and PvNF-YC family’s members under different drought stress levels (Fig. 8B).
qRT-PCR analysis of specific tissue/organ development for the PvNF-Ys gene family
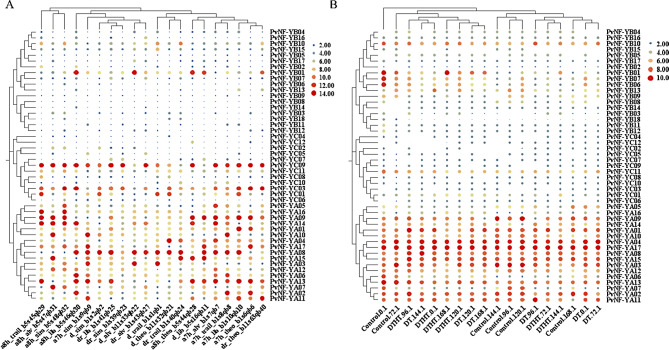
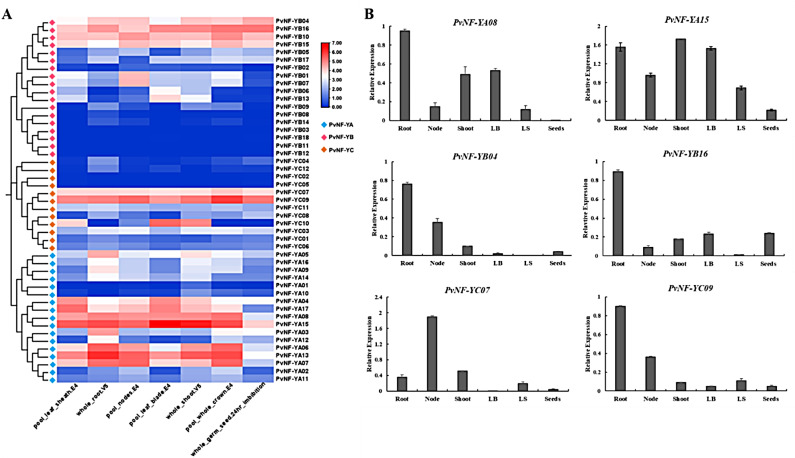
To further investigate the potential role of PvNF-Ys genes in plant growth and development stages, we conducted expression profiling of PvNF-Ys genes across various tissue/organ using data from the JGI database. The results showed that most of the PvNF-Ys genes were expressed in most tissue/organ, but the level of expression varied (Fig. 9A). To validate the organ/tissue specific expression profile of PvNF-Ys genes, we carried out qRT-PCR experiment utilizing gene-specific primers. Several PvNF-Ys genes were selected based on evolutionary relationship between them, we found that almost all the selected PvNF-Ys genes except PvNF-YC07 shown high expression level in root compared to other organs (Fig. 9B).
Fig. 9.
Expression analysis of specific tissue/organs of the PvNF-Ys genes. (A) Expression profiling of PvNF-Ys in tissue and organ development using data through JGI database (B) Quantitative-real-time PCR showed expression of genes in different organs
Discussion
NF-Ys factor is essential for plant development and proliferation at different stages. Abiotic stressors like salt, drought, and temperature significantly impact plant development, leading to adaptation mechanisms for root growth, plant-microbe interactions, and water stress response [48]. NF-Y genes have garnered a lot of interest in various areas of agricultural research. In this study, we found 47 PvNF-Ys genes, with 17 PvNF-YA, 18 PvNF-YB and 12 PvNF-YC (Table 1). However, number of genes varies over different species. A total of 24 PpNF-Ys were found in Prunus persica [19] with 6 PpNF-YAs, 12 PpNF-YBs, and 6 PpNF-YCs, while in Petunia hybrida have 27 PhNF-Ys with 10 PhNF-YAs, 13 PhNF-YBs and 12 PvNF-YCs [49].
A phylogenetic study revealed a strong relationship between switchgrass (Panicum virgatum L.) PvNF-Y genes and Arabidopsis AtNF-Y genes. All the 47 identified PvNF-Ys genes were categorized into three subgroups corresponding to AtNF-Y genes (Fig. 2B). That is similar to the NF-Ys discovered in Arachis hypogaea [19], Prunus persica L [49], and Solanum tuberosum L [50]. The roles of PvNF-Y genes can be deduced from the phylogenetic tree’s Arabidopsis genes with known functions. The PvNF-YB2, B11, B17, and B05 share lineage with AtNF-YB2 and B3 specifying their role in the regulation of flowering time [23]. Additionally, AtNF-YC4 makes clade with PvNF-YC7, C09 and C11 plays significant role in blooming and photomorphogenesis [4]. Moreover, PvNF-YB03, B11, B12, and B18 contained aspartic acid residue at position 55 indicating that they may be LEC1 type genes while others are non-LEC type [52]. In Arabidopsis thaliana AtNF-YA2 and A10 genes were seen to be highly expressed in roots. In the present case, according to qRT-PCR screening expression levels of selected PvNF-Ys genes are elevated in root tissue. Sideways, PvNF-Ys genes have a greater number of stress-responsive elements in promoter sequences. This leads to the conformation of their stress-resilient capabilities [5].
Multiple sequence alignments (Fig. 1) have shown that the PvNF-YA, PvNF-YB, and PvNF-YC subfamilies are characterized by DNA-binding domains that bind to specific CCAAT binding sites [51]. A sequence of 21 amino acids is displayed in the conserved region at the C-terminal of PvNF-YA: Y-L-H-E-S-R-H-x-H-A-x-x-R-x-R-G-x-G-G-R-F. This sequence, which comes from the DNA binding domain of PvNF-YA present in plants, mammals, and yeasts, is assumed to be linked to DNA at CCAAT locations. PvNF YA’s conserved area’s N-terminal region has the sequence Y-V-N-A-K-Q-x-x-x-I-L-R-R-R-x-x-R-A-K-L-E. The structure and amino acid composition of the conserved areas of NF-YB were comparable to those of H2B motifs. Based on the equivalent portions of NF-YB members in Arabidopsis, the 31-amino acid sequence R-L-P-x-I-A-N-x-x-R-I-M-x-x-x-x-P-x-x-x-K-I-x-x-x-A-K-E-T-x-Q was identified as the DNA-binding domain of PvNF-YB [52]. During the alignment examination of 12 PvNF-YC members, the interaction domain of Arabidopsis revealed similarities with a conserved 74 amino-acid sequence L-P-L-A-R-I-K-K-I-M-K-x-x-x-x-A-D-x-x-V. This led to the conclusion that PvNF-YC related to PvNF-YA/PvNF-YB through this fragment, which served as its centre area. Furthermore, just two critical residues from series “A” and “R” were present in PvNF-YC’s DNA-binding domain, which was necessary for heterotrimeric NF-Y and DNA to form a complete structure as reported in previous study [53].
In addition, motifs analysis (Fig. 3A) revealed only three (motifs 1, 3, and 4) were substantially shared by all PvNF-YA, PvNF-YBs have motifs 1, 3, 5, and 6, except for PvNF-Y04 and PvNF-YB12, which do not have motif 5. PvNF-YCs have a lot of motifs 2, 5, 8, 9, and 10. Similar to the previous research on peach [19]. Gene structure analysis, 27 PvNF-Y sequences had introns, whereas one-third of PvNF-Y sequences had single exons and zero introns (Fig. 4B). The results aligned with earlier studies that demonstrated that short intron length or absence increased plant gene expression [53, 54]. According to earlier research, the NF-Y members’ gene structure was strongly correlated with their evolutionary relationships [55–57]. Research on other plants has revealed that whereas NF-YBs and NF-YCs are more varied, the majority of NF-YAs have three to six introns in their genome sequence [22, 58, 59]. Additionally, a number of NF-YB and NF-YC members have been discovered without any intron like in chickpea [60], S. bicolor [55], Ricinus cummunis [61].
The location of genes are very important to determine their biological activity in the cells [62]. The majority of genes are located in the nucleus indicating its role in gene expression, with a smaller proportion being found in the cytoplasm and other organelles. Numerous cis-acting elements linked to plant growth and development as well as stress response were also found during the examination of the panicum virgatum NF-Y gene families. The CREs results indicated that stress-responsive and hormone-related motifs were significantly dominant among the predicted 47 PvNF-Ys. Phytohormones play a significant role in plant growth and development [63]. Light and plant growth responses were also found (Fig. 7B).
The expression profiles of every PvNF-Ys gene under drought stress [47] were examined in this work. PvNF-YAs exhibited high resilience to circumstances of drought and recovery (Fig. 8A). Further examination of drought and heat stress was conducted [45, 46]. In contrast to the PvNF-YB and PvNF-YC gene families, the majority of the PvNF-YA genes showed strong expression patterns (Fig. 8AB). The PvNF-YA members show elevated expression level during stress maybe because of its critical involvement in the transcriptional regulation associated with stress tolerance and having high number of stress-responsive elements in promoter regions. Recent studies have demonstrated that NF-YA subunits can activate or modulate genes related to water retention, osmotic adjustment, and efficient nutrient mobilization, which are essential under drought stress [64].
Gene ontology results (Fig. 6B) showed that these genes were significantly involved in regulating transcription by RNA polymerase II, regulation of cell growth, and unidimensional cell growth processes, while protein heterodimerization activity and DNA-binding transcription activator activities were most observed molecular functions of NFY transcription factor which are characteristic of NF-Y members and are found in a wide range of organisms, from plants to mammals [65]. In results, CAAT-binding factor complex found as a most significant cellular function [59]. Tissue/organ specific expression profiling results revealed that most of the PvNF-Ys genes were expressed in different organs, but the level of expression differed (Fig. 9A). Furthermore, we conduct qRT-PCR experiment (Fig. 9B) to verify the expression levels of particular PvNF-Ys genes in different organs by using gene-specific primers [66]. These findings are crucial for identifying potential genes that increase molecular breeding’s efficiency. Also, our results help choose Panicum virgatum L. cultivars with strong resilience to adversity and can be a vital resource for understanding how NF-Y transcription factors function in stressful conditions.
Conclusions
The complete analysis screened 47 PvNF-Ys genes in switchgrass (Panicum virgatum L.), and is divided into three subfamilies (NF-YA, NF-YB, NF-YC). This study revealed the maximum number of NF-Ys residues on the ninth chromosome. Multiple sequence alignment exhibits DNA binding along with interacting domains. Phylogenetic analysis revealed the evolutionary relation of PvNF-Ys and AtNF-Ys. Gene structure showed one-third of PvNF-Ys lack intron. Cis-regulatory elements associated with stress-responsive and hormone-responsive elements motifs in the promoter region. Previously reported RNA seq data suggested that PvNF-YAs gene family showed high expression towards drought, combination of drought and heat stress compare to PvNF-YBs and PvNF-YCs gene family. qRT-PCR analysis showed that PvNF-YA08, PvNF-YB04, PvNF-YB16, and PvNF-YC09 highly expressed in roots, (except PvNF-YC07 gene). These findings deepen our understanding of how PvNF-Y genes play a role in cellular processes and environmental responses, with implications for crop resilience and biotechnology applications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP-2024R369), King Saud University, Riyadh, Saudi Arabia.
Author contributions
HH and TAS project administration, methodology, data analyze; HH, NF, MS, IM, MN, YMH, KA, KAA data curation, experiment conduct, writing–original draft preparation; TAS funding acquisition. The authors read and approved the final manuscript.
Funding
The work was supported by the Researchers Supporting Project number (RSP-2024R369), King Saud University, Riyadh, Saudi Arabia.
Data availability
All the data generated or analyzed during this study are included in this published article and its supplementary data files. The genome file of switchgrass (Panicum virgatum L.) was downloaded from Phytozome v13.0 (https://phytozome-next.jgi.doe.gov/). The Protein sequences of NFYs transcription factor gene families of Arabidopsis thaliana were obtained from TAIR (https://www.arabidopsis.org/browse/genefamily/index.jsp).
Declarations
Informed consent
Not applicable.
Institutional review board statement
Not applicable.
Disclaimer
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hadia Hussain and Noor Fatima contributed equally to this work.
References
- 1.Asensi-Fabado MA, Amtmann A, Perrella G. Plant responses to abiotic stress: the chromatin context of transcriptional regulation. Biochim Biophys Acta Gene Regul Mech. 2017;1860(1):106–22. [DOI] [PubMed] [Google Scholar]
- 2.Kim IS, Sinha S, de Crombrugghe B, Maity SN. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol. 1996;16(8):4003–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gusmaroli G, Tonelli C, Mantovani R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene. 2002;283(1–2):41–8. [DOI] [PubMed] [Google Scholar]
- 4.Ke-Lin H, Yang L, Huan W, Jing T, Yi-Fan F, Yong Z, Xue-Bao L. NF–YC3, NF–YC4 and NF–YC9 are required for CONSTANS-mediated, photoperiod‐dependent flowering in Arabidopsis thaliana. Plant J. 2010;63(3):379–91. [DOI] [PubMed] [Google Scholar]
- 5.Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais-Brière C, Njo MF, Beeckman T, Crespi MD, Hartmann C. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014;202(4):1197–211. [DOI] [PubMed] [Google Scholar]
- 6.Jinye M, Helin T, Sulei H, Yan L, Jianru Z. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant. 2013;6(1):188–201. [DOI] [PubMed] [Google Scholar]
- 7.Jenny B, Saori E, Mitsuru K, Jennifer M, Yajun W. Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol Biochem. 2011;49(6):579–83. [DOI] [PubMed] [Google Scholar]
- 8.Katherine MW, Snehali U, Jennifer Y, Julia A, Samuel IH, Yevgeniya RL, Mary BA, Lon SK. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143(4):1590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donald EN, Peter PR, Tom RA, Robert AC, Jingrui W, David CW, Don CA, Robert JB, Paolo PC, Meghan GD, Brendan SH, Roderick WK, Don RM, Roger DC, Katherine AK, Stanton BD, Neal G, Oliver JR, Jacqueline EH. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. PNAS.2007;104(42):16450–16455. [DOI] [PMC free article] [PubMed]
- 10.Li Y, Fang Y, Fu Y, Huang J, Wu C, Zheng C. NFYA1 is involved in regulation of post germination growth arrest under salt stress in Arabidopsis. PLoS ONE. 2013;8(4):e61289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Ye T, Zhong B, Liu X, Jin R, Chan Z. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014;203(2):554–67. [DOI] [PubMed] [Google Scholar]
- 12.Hikaru S, Junya M, Hidenori T, Kyonosin M, Feng Q, Yuriko O, Kyoko M, Teppei O, Kazuya K, Maika N, Kazuo S, Kazuko YS. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell. 2014;26(12):4954–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149(2):625–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenjie Y, Zhanhua L, Yufei X, Jialing Y. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J. 2017;5(1):21–31. [Google Scholar]
- 15.Liang M, Yin X, Lin Z, Zheng Q, Liu G, Zhao G. Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L). Planta. 2014;239:107–26. [DOI] [PubMed] [Google Scholar]
- 16.Quach TN, Nguyen HT, Valliyodan B, Joshi T, Xu D, Nguyen HT. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol Genet Genom. 2015;290:1095–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Z, Wang Y, Li W, Wang Y, Liu X, Ou X, Su W, Song S, Chen R. Genome-wide identification of the NF-Y gene family and their involvement in bolting and flowering in flowering Chinese cabbage. Int J Mol Sci. 2023;24(15):11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Du Q, Li J, Wang H, Xiao H, Wang J. Genome-wide identification and chilling stress analysis of the NF-Y Gene Family in Melon. Int J Mol Sci. 2023;24(8):6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao L, Guixiang L, Wei L, Xiaomin D, Anning Z. Genome-wide analysis of the NF-Y gene family in peach (Prunus persica L). BMC Genom. 2019;20:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan L, Ka L, Zheng J, Dongyan C, Daqi F, Hongliang Z, Benzhong Z, Yunbo L. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016;17:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maheshwari P, Kummari D, Palakolanu SR, Nagasai TU, Nagaraju M, Rajasheker G, Jawahar G, Jalaja N, Rathnagiri P, Kavi KPB. Genome-wide identification and expression profile analysis of nuclear factor Y family genes in Sorghum bicolor L.(Moench). PLoS ONE. 2019;14(9):e0222203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng ZJ, He GH, Zheng WJ, Lu PP, Chen M, Gong YM, Ma YZ, Xu ZS. Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front Plant Sci. 2015;6:1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong R, Zhan Z, Yi W, Shaohua L, Zhenchang L. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L). BMC Genom. 2016;17:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahman P, Seyyed AM, Kamil R, Hossein AH, Mohammad ZM. Genome-wide identification and co-expression network analysis of nuclear factor-Y in barley revealed potential functions in salt stress. Physiol Mol Biol Plants. 2019;25:485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni Z, Hu Z, Jiang Q, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol biol. 2013;82:113–29. [DOI] [PubMed] [Google Scholar]
- 26.Xin Y, Mengtian H, Shuai L, Zhiyan L, Jiexiu O, Xin W, Pengfei L. A member of NF-Y family, OsNF-YC5 negatively regulates salt tolerance in rice. Gene. 2024;892:147869. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Yuepeng R, Yifan J, Shasha H, Jiayi W, Guangdong W. Characterization of NF–Y gene family and their expression and interaction analysis in Phalaenopsis orchid. Plant Physiol Biochem. 2023;204:108143. [DOI] [PubMed] [Google Scholar]
- 28.Michael DC, Christian MT, Shawn MK, Buell CR, Wang ZY, Peijian C, Jeremy S, Pamela R. The switchgrass genome: tools and strategies. Plant Genom.2011;4(3).
- 29.Bragg LH, McMillan C. Ecotypic differentiation within four north American prairie grasses. II. Behavioral variation within transplanted community fractions. Am J Bot. 1965;52(1):55–65. [Google Scholar]
- 30.Parrish DJ, Fike JH. The biology and agronomy of switchgrass for biofuels. Volume 24. BPTS; 2005. pp. 423–59. 5–6.
- 31.Wright L. Historical perspective on how and why Switchgrass was selected as a model High-Potential Energy Crop. ORNL; 2007.
- 32.Sanderson MA, Reed RL, McLaughlin SB, Wullschleger SD, Conger BV, Parrish DJ, Wolf DD, Taliaferro C, Hopkins AA, Ocumpaugh WR, Hussey MA, Read JC, Tischler CR. Switchgrass as a sustainable bioenergy crop. Bioresour Technol. 1996;56(1):83–93. [Google Scholar]
- 33.McLaughlin SB, Kszos LA. Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy. 2005;28(6):515–35. [Google Scholar]
- 34.Yajun X, Chunxiang F, Yaxin G, Rangaraj N, Hiroshi H, Joseph B, Zeng-Yu W. Agrobacterium-mediated Transformation of Switchgrass and inheritance of the transgenes. Bioenergy Res. 2009;2(4):275–83. [Google Scholar]
- 35.Ji-Yi Z, Panicum virgatum L. Plant J. 2013;74(1). [DOI] [PubMed]
- 36.Zhang C, Tang G, Peng X, Sun F, Liu S, Xi Y. Long non-coding RNAs of switchgrass (Panicum virgatum L.) in multiple dehydration stresses. BMC Plant Biol. 2018;18(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandana H, Yun Z, Chandra O, Reddy P, Guru J, Kanchana G, Vijaya GK, Abdelali B, Ramanjulu S. Characterization of drought- and heat-responsive microRNAs in switchgrass. Plant Sci. 2016;242:214–23. [DOI] [PubMed] [Google Scholar]
- 38.Elisabeth G, Christine H, Alexandre G, S’everine D, Marc RW, Ron DA, Amos B. Protein Identification and Analysis Tools on the ExPASy Server, in The Proteomics Protocols Handbook, J.M. Walker, Editor. Humana Press.2005.571-607.
- 39.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(2):585–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43(1):39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y, Xia R. TBtools-II: a one for all, all for one bioinformatics platform for biological big-data mining. Mol Plant. 2023;16(11):1733–42. [DOI] [PubMed] [Google Scholar]
- 43.Chow CN, Yang CW, Wu NY, Wang HT, Tseng KC, Chiu YH, Lee TY, Chang WC. PlantPAN 4.0: updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 2024;52(D1):D1569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Li J, Paterson AH. MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinform. 2013;29(11):1458–60. [DOI] [PubMed] [Google Scholar]
- 45.Hayford RK, Serba DD, Xie S, Ayyappan V, Thimmapuram J, Saha MC, Wu CH, Kalavacharla VK. Global analysis of switchgrass (Panicum virgatum L.) transcriptomes in response to interactive effects of drought and heat stresses. BMC Plant Biol. 2022;22(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayyappan V, Sripathi VR, Xie S, Saha MC, Hayford R, Serba DD, Subramani M, Thimmapuram J, Todd A, Kalavacharla VK. Genome-wide profiling of histone (H3) lysine 4 (K4) tri-methylation (me3) under drought, heat, and combined stresses in switchgrass. BMC Genom. 2024;25(1):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lisa C, Jennifer B, Prisca C, Sheyla A, Gautam S, Nathan P, Serge E, Christian MT. Gene expression and physiological differences in neo-octoploid Switchgrass Subjected to Drought stress. Bioenergy Res. 2020;13(1):63–78. [Google Scholar]
- 48.Zanetti ME, Rípodas C, Niebel A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim Biophys Acta. 2017;1860(5):645–54. [DOI] [PubMed] [Google Scholar]
- 49.Qian W, Lu L, Xiurong Z, Yuying L, Suqing Z, Lingrang K, Yongshan W, Fengzhen L, Kun Z. Genome-wide identification and abiotic stress response pattern analysis of NF-y gene family in peanut (Arachis hypogaea l). Trop Plant Biol. 2021;14:329–44. [Google Scholar]
- 50.Li S, Zhang N, Zhu X, Ma R, Liu S, Wang X, Yang J, Si H. Genome-wide analysis of NF-Y genes in potato and functional identification of StNF-YC9 in drought tolerance. Front Plant Sci. 2021;12:749688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Myers ZA, Holt BF. Nuclear factor-Y: still complex after all these years? Curr Opin Plant Biol. 2018;45:96–102. [DOI] [PubMed]
- 52.Nerina G, Roderick WK, Swadhin S, Matteo C, Chamindika S, David SH, Ben FH, Roberto M. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell. 2017;29(6):1516–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Betty YWC, Cas S, Andrew EF, Chris MB, Roger PH. Effect of 5’UTR introns on gene expression in Arabidopsis thaliana. BMC Genom. 2006;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren XY, Vorst O, Fiers MW, Stiekema WJ, Nap JP. In plants, highly expressed genes are the least compact. Trends Genet. 2006;22(10):528–32. [DOI] [PubMed] [Google Scholar]
- 55.Malviya N, Jaiswal P, Yadav D. Genome-wide characterization of nuclear factor Y (NF-Y) gene family of sorghum [Sorghum bicolor (L.) Moench]: a bioinformatics approach. Physiol Mol Biol Plants. 2016;22:33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Q, Wen S, Lan C, Yu Y, Chen G. Genome-wide identification and expression Profile Analysis of the NF-Y Transcription Factor Gene Family in Petunia hybrida. Volume 9. Plants; 2020. p. 336. 3. [DOI] [PMC free article] [PubMed]
- 57.Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant. 2012;5(4):876–88. [DOI] [PubMed] [Google Scholar]
- 58.Le HH, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem sci. 2003;28(4):215–20. [DOI] [PubMed] [Google Scholar]
- 59.Wang P, Zheng Y, Guo Y, Chen X, Sun Y, Yang J, Ye N. Identification, expression, and putative target gene analysis of nuclear factor-Y (NF-Y) transcription factors in tea plant (Camellia sinensis). Planta. 2019;250(5):1671–86. [DOI] [PubMed] [Google Scholar]
- 60.Chu HD, Nguyen KH, Watanabe Y, Le DT, Pham TLT, Mochida K, Tran LP. Identification, structural characterization and gene expression analysis of members of the nuclear factor-Y family in chickpea (Cicer arietinum L.) under dehydration and abscisic acid treatments. Int J Mol Sci. 2018;19(11):3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Xu W, Chen Z, Han B, Haque ME, Liu A. Gene structure, expression pattern and interaction of Nuclear Factor-Y family in castor bean (Ricinus communis). Planta. 2018;247:559–72. [DOI] [PubMed] [Google Scholar]
- 62.Silhavy TJ, Benson SA, Emr SD. Mechanisms of protein localization. Microbiol rev. 1983;47(3):313–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maheen N, Shafiq M, Sadiq S, Farooq M, Ali Q, Habib U, Shahid MA, Ali A, Ali F. Genome Identification and Characterization of WRKY Transcription Factor Gene Family in Mandarin (Citrus reticulata). Agriculture.2023;13(6):1182.
- 64.Lv M, Cao H, Wang X, Zhang K, Si H, Zang J, Xing J, Dong J. Identification and expression analysis of maize NF-YA subunit genes. PeerJ. 2022;7(10):e14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mantovani R. The molecular biology of the CCAAT-binding factor. NF-Y Gene. 1999;239(1):15–27. [DOI] [PubMed] [Google Scholar]
- 66.Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF. Tissue specific expression patterns of Arabidopsis thaliana NF-Y transcription factors suggest. Mol Biol. 2008;215:403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated or analyzed during this study are included in this published article and its supplementary data files. The genome file of switchgrass (Panicum virgatum L.) was downloaded from Phytozome v13.0 (https://phytozome-next.jgi.doe.gov/). The Protein sequences of NFYs transcription factor gene families of Arabidopsis thaliana were obtained from TAIR (https://www.arabidopsis.org/browse/genefamily/index.jsp).