Abstract
Background
The diversity-generating retroelements (DGRs) are a family of genetic elements that can produce mutations in target genes often related to ligand-binding functions, which possess a C-type lectin (CLec) domain that tolerates massive variations. They were first identified in viruses, then in bacteria and archaea from human-associated and environmental genomes. This DGR mechanism represents a fast adaptation of organisms to ever- changing environments. However, their existence, phylogenetic and structural diversity, and functions in a wide range of environments are largely unknown.
Results
Here we present a study of DGR systems based on metagenome-assembled genomes (MAGs) from host-associated, aquatic, terrestrial and engineered environments. In total, we identified 861 non-redundant DGR-RTs and ~ 5.7% are new. We found that microbes associated with human hosts harbor the highest number of DGRs and also exhibit a higher prevalence of DGRs. After normalizing with genome size and including more genome data, we found that DGRs occur more frequently in organisms with smaller genomes. Overall, we identified nine main clades in the phylogenetic tree of reverse transcriptases (RTs), some comprising specific phyla and cassette architectures. We identified 38 different cassette patterns and 6 of them were shown in at least 10 DGRs, showing differences in terms of the numbers, arrangements, and orientations of their components. Finally, most of the target genes were related to ligand-binding and signaling functions, but we discovered a few cases in which the VRs were situated in domains different from the CLec.
Conclusions
Our research sheds light on the widespread prevalence of DGRs within environments and taxa, and supports the DGR phylogenetic divergence in different organisms. These variations might also occur in their structures since some cassette architectures were common in specific underrepresented phyla. In addition, we suggest that VRs could be found in domains different to the CLec, which should be further explored for organisms in scarcely studied environments.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11124-1.
Keywords: Diversity-generating retroelement (DGR), Metagenome assembled genome (MAG), Cassette structure, Target gene, Domain annotation, C-type lectin (CLec) fold
Background
The diversity-generating retroelements (DGRs) are discovered genetic elements capable of producing mutations in specific genes, thus favoring their diversification. The first DGR system was characterized in the bacteriophage BPP-1 that infects the genus Bordetella, the bacteria that cause whooping cough. The membrane of this bacteria is very variable to avoid being infected; however, the BPP-1 phage uses a mechanism to produce many variants of the Major tropism determinant (Mtd) gene, a tail fiber protein that binds to adhesion receptors on the surfaces of the bacteria. In this way, the phage is able to infect more bacteria despite the variations on their surfaces [1–3].
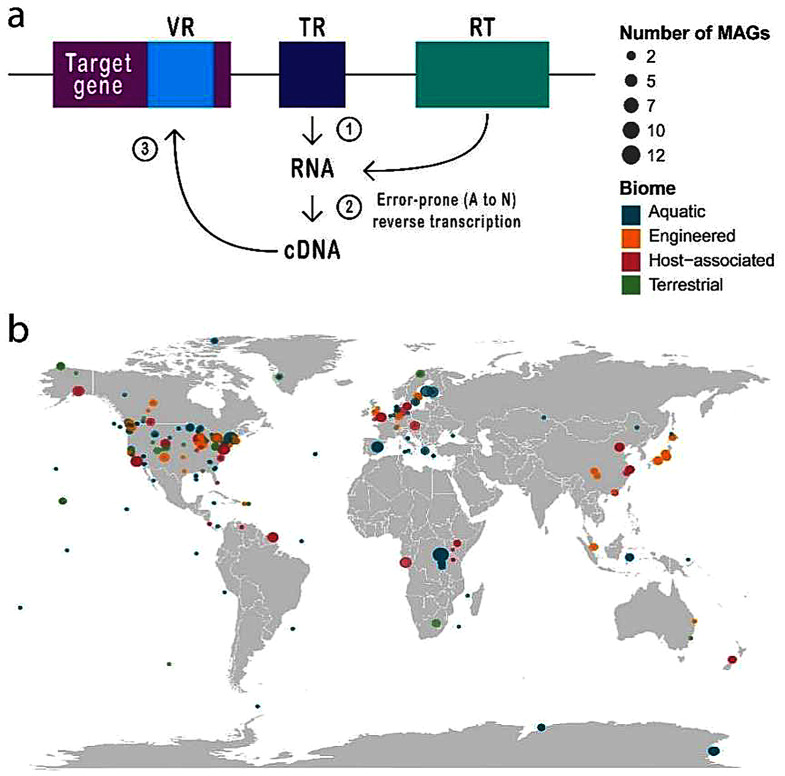
A DGR system has a cassette structure consisting of three main components: a reverse transcriptase (RT), a template region (TR), and a variable region (VR); these last two are similar in size and nucleotide composition, but the variable region is located within the target gene [2]. The typical conformation is VR-TR-RT; however, the components can be found in different arrangements and numbers in some genomes. The DGR mechanism is known as “mutagenic retrohoming”, during which the TR is transcribed into RNA, and then the reverse transcriptase converts it to complementary DNA (cDNA), but with some mutations that mainly affect adenines (A → N). Finally, this mutant cDNA is inserted into the target gene, thus causing hypervariation of the encoded protein [4] (Fig. 1a).
Fig. 1.
Schematic overview of a DGR and the geographical distribution of DGR-containing MAGs. (a) DGR components and mechanism. (b) Geographical distribution of DGR-containing MAGs. Coordinates were obtained from the GEM catalog metadata. Dot sizes indicate the number of MAGs per site
In the prototypical BPP-1 DGR, the lengths of the components are 328 amino acids (RT) and 134 base pairs (TR and VR), and the average number of potential mutation adenine sites is 23. Thus, theoretically, DGRs can produce more than 1014 different VR nucleotide sequences, corresponding to approximately 1013 different polypeptides [1]. Generally, the VR is found in the C-type lectin fold, a domain capable of tolerating a large number of mutations, which is located at the C terminus of the target protein [5, 6]. As in the BPP-1 DGR, most of the target proteins are associated with ligand-binding functions; however, recent studies have found that the ligand-binding domain that contains the variable region could be associated with another domain that has regulatory functions [7]. In fact, we could suggest that DGRs are modifying other functions that allow organisms to cope with changes in their respective habitats.
Due to the importance of this system, several tools have been developed for its detection, mainly based on the alignment with reference sequences [8–11]. Recently, a tool called Metagenomic Complex Sequence Scanning Tool (MetaCSST) was developed, which is based on the Generalized Hidden Markov Model (GHMM) for the search of remarkable sequence patterns of the RT and TR components, and it has the advantage of being faster and slightly more flexible than other alignment-based tools [12]. Overall, all these methods have allowed the detection of DGRs in genomes of viruses, bacteria and archaea, as well as in metagenomes obtained from the human microbiome and environmental samples [12–18]. A recent comprehensive study identified 13,415 non-redundant DGR-RTs at 95% AAI [18], and some clades based on the RT sequences were established, which are mainly determined by the taxonomy of the organisms. Furthermore, research by Paul BG et al. on metagenomes such as groundwater suggests that DGR systems may be associated with organisms with smaller genomes (about 0.5–1 Mb) [15]. However, studies related to the identification of DGRs in environmental genomes are still very scarce [18], although these DGRs are likely playing important roles for organisms, depending on the environment in which they live.
Here we present our study aiming to detect DGRs in genomes from a wide range of environments and organisms, and characterize their prevalence, diversity and roles. It will advance DGR study by using the most comprehensive global microbial genome catalog published to date, the Genomes from Earth’s Microbiomes catalog (GEM), consisting of 52,515 MAGs obtained from over 10,000 samples collected from diverse microbial environments (aquatic, host-associated, terrestrial, engineered, and outdoor air) [19].
Results and discussion
Prevalence of DGRs across different environments and taxa
The Genomes from Earth’s Microbiomes (GEM) catalog contains 52,515 MAGs obtained from 10,450 metagenomes. We identified DGRs in 2,014 MAGs (3.84%) from 1,270 metagenomes representing a wide range of environments (280 aquatic, 164 engineered, 756 host-associated, 70 terrestrial) from different geographic locations. Among the DGRs identified from MAGs, 66.5% are located on viral contigs, 29.8% on cellular contigs, and the remaining 3.7% are unknown. This ratio is different in different biomes. Among the host-associated microorganisms, the proportion of viruses is the highest. Among the microorganisms in the aquatic environment, the proportion of cellular is the highest. This result is similar to previous studies, indicating that the living environment of microorganisms has a large impact on the distribution of DGRs in viruses and bacteria, among others (Additional file: Fig. S1). The number of DGR-containing MAGs per metagenome ranges from 1 to 13 (Fig. 1b) (Additional file: Table S1, S2).
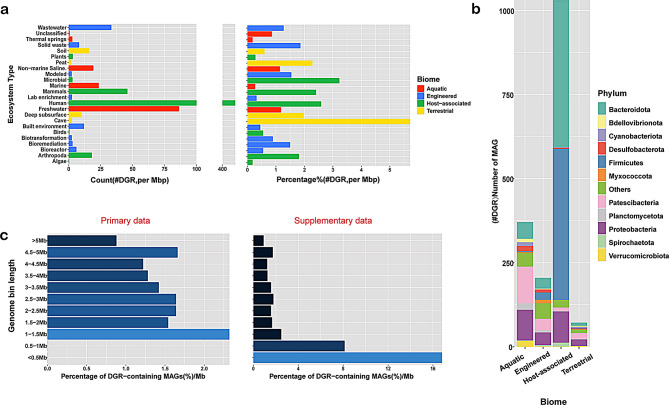
The 2,014 DGR-containing MAGs mainly belong to host-associated environments, specifically to the human microbiomes (1,033 MAGs); however, a considerable number of them belong to aquatic, engineered, or terrestrial environments. Among human-associated microbiomes, the digestive system, especially gut flora-associated microbiom, contains the majority of DGRs. A recent study of epsilon crAss-like phages has shown that a significant fraction of temperate phages in the gut microbiome contain DGRs [20]. When we calculated the percentage of DGR-containing MAGs compared to the total number of MAGs for each environmental category, we found that cave- and microbial-associated organisms showed a higher prevalence of DGRs than human microbiomes (Fig. 2a) (Additional file: Table S4 – S6). Furthermore, our initial dataset consisted of 3,038 archaeal MAGs, from which only 20 were found to have DGRs. We found that 1,994 bacterial MAGs contain DGRs, of which the phyla Bacteroidota, Firmicutes, Proteobacteria, and Patescibacteria were the most abundant across environments(Fig. 2b). Overall, DGRs were found in 4 archaeal and 45 bacterial phyla (Additional file: Table S7).
Fig. 2.
Prevalence of DGRs across environments, taxa and different genome sizes. (a) After being corrected by the amount of Mbp, the number of DGR-containing MAGs per ecosystem category (left) and percentage of DGR-containing MAGs compared to the total number of MAGs in each ecosystem category (right), (b) Number of DGR-containing MAGs per biome colored by phylum, (c) The frequency of DGRs in genomes of different sizes after normalizing for genome length, primary data(left) and Supplementary data (right)
While previous studies have reported the presence of DGRs in genomes from the human microbiome [12] and groundwater [13], our study leverages the most comprehensive catalog of environmental genomes built to date in order to expand the number of known DGRs. Recently, Roux et al.. (2021) [18] published their findings on DGRs from public metagenomes they collected from the Integrated Microbial Genomes and Microbiomes (IMG/M) database, which was also used to build the GEM catalog. They identified 13,415 non-redundant DGR-RTs at 95% AAI from 9,467 metagenomes. Our study uses MAGs obtained from 10,450 metagenomes also from the IMG/M database, from which 3,532 were analyzed for the first time. In 682 new metagenomes (of 3,532), a total of 1,037 DGR-containing MAGs (out of 2,014) were identified. (Additional file: Table S3).
Prevalence of DGRs across different genome sizes
We also analyzed the presence of DGRs in organisms with different genome sizes. For this purpose, we only considered the high-quality MAGs (9,143 out of 52,515), which have completeness over 90%. From the 2,014 DGR-containing MAGs, only 408 were in high quality, and most of them have genome lengths over 2 Mbps. When checking the number of MAGs per ecosystem category, we could see that for host-associated environments, 268 MAGs belong to human hosts and, to a less extent, to mammals and arthropods. For the aquatic, engineered, and terrestrial biomes, the amounts of MAGs per ecosystem category were more evenly distributed (Additional file: Fig. S2) (Additional file: Table S8). In these biomes, the DGRs were more frequent in genomes longer than 2 Mb from freshwater, marine, wastewater, bioreactor, deep subsurface, and built environments.
However, this result does not indicate that DGR is more frequent in larger genomes. After normalizing with genome length, we obtained different results. After correlation analysis of the percentage of MAGs containing DGR (per Mbp) and genome lengths, the p-value was 0.71, indicating that there was no significant association between the two. However, the results are still not accurate enough considering the lack of data with genome sizes between 0 and 1 Mbps. In order to avoid the bias of the genome size, we performed additional analysis of DGR data from a different dataset, derived from 31,007 DGRs previously detected in 3,123 MAGs [18]. In addition to the original 408 high-quality MAGs, we analyzed the genome sizes of a total of 3,531 MAGs, and the results indicated that DGRs tended to occur more frequently in organisms with smaller genomes (Fig. 2c, Spearman correlation, r=-0.8, p-value = 0.003). This result is consistent with previous results that DGRs tend to be found in organisms with smaller genomes [15]. However, Bourguignon et al. (2020) noted that smaller genomes in prokaryotes are linked to faster evolutionary rates [21]. This may also contribute to this trend.
Features of the DGR components: length and mutation bias
Overall, we identified 2,263 reverse transcriptases (RTs). The number of RTs per MAG was up to 4. We initially considered the number of DGRs based on the number of RT sequences; however, we found that 31 DGR systems have up to 2 copies of RTs in some genomes. It is worth noting that the DGRs with 2 RTs were found in all four biomes; they were not restricted to a specific environment. Overall, the number of DGRs was 2,232 (1,384 in Host-associated, 485 in Aquatic, 268 in Engineered, and 95 in Terrestrial environments) (Table 1).
Table 1.
Number of MAGs analysed, DGR-containing MAGs and DGR-RT sequences per biome
| Biome | # of MAGs | # of MAGs with DGRs | # RTs | # nr-RTs (≥ 100aa) | # nr-RTs (with reverse transcriptase domains) |
|---|---|---|---|---|---|
| Air | 21 | 0 | 0 | 0 | 0 |
| Aquatic | 19,300 | 445 | 492 | 290 | 287 |
| Engineered | 8,265 | 246 | 275 | 171 | 163 |
| Host-associated | 21,518 | 1,237 | 1,400 | 369 | 355 |
| Terrestrial | 3,411 | 86 | 96 | 59 | 56 |
| Total | 52,515 | 2,014 | 2,263 | 889 | 861 |
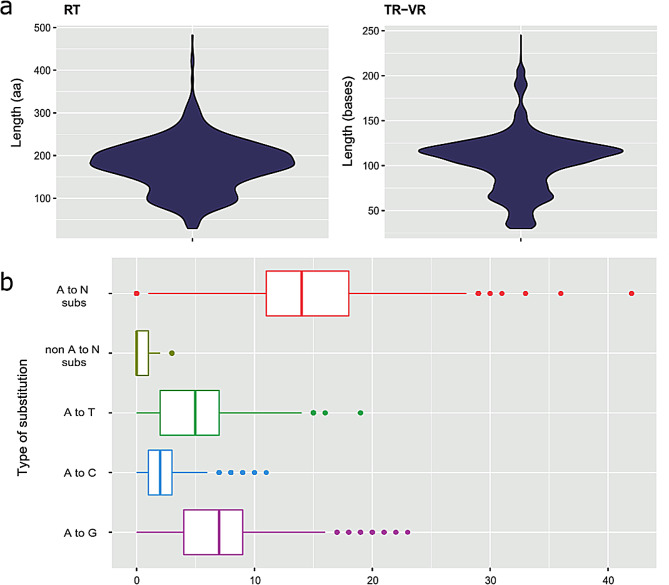
The median RT length is 177 amino acids, with maximum and minimum values of 482 and 29, respectively. Only MetaCSST, a motif-based method, has detected small RT sequences [12], and other studies have reported RTs longer than 100 aa [18]. For the subsequent phylogenetic analyses, we used 889 RTs with lengths > 100aa. The median length of the TR/VR region was 110 base pairs, with the maximum and minimum values of 245 and 30 base pairs, respectively (Fig. 3a). However, the tool also detected three TR/VR regions with atypical lengths of 3,832, 3,837, and 5,002 base pairs. It should be pointed out that MetaCSST only detects TR/VR regions longer than 30 base pairs [12]. In addition, in some cases, the tool identified very large TR/VR regions that may overlap or completely include other shorter TR/VR regions detected in the same contig.
Fig. 3.
Features of DGR components. (a) Length distribution of RT sequences (in amino acids) (median: 177, min: 29, max: 482) and length distribution of TR/VR regions (in nucleotides) (median: 110, min: 30, max: 245), (b) Mutation bias in the TR/VR pairs. As reported, it exists a mutation bias towards adenines, with an average of 14 substitutions per sequence and a maximum value of 42
For the three special TR/VR regions (with atypical length) we previously identified, their atypical lengths were 3,832, 3,837, and 5,002 bps, respectively. We analyzed these three DGRs, and found that their structural patterns are unusual(Additional file: Fig. S3). The TR, VR, and RT regions of the first two DGRs overlap, while the TR and RT of the third DGR overlap. This is inconsistent with the previous definition of DGRs. In addition, studies have reported that lectin-like proteins have many repeats that are prone to misassembly [22]. After examining the repetitive regions, we found that they correspond to C-type lectins. Therefore, we filtered out these three atypical DGRs and excluded them from subsequent analysis. Additionally, we have added the function to detect and filter overlaps into the MetaCSST detection tool.
We also analyzed the mutation bias of the TR/VR pairs. As expected, most of the mutations correspond to the Adenines (A → N), with an average of 14 mutations per sequence and a maximum value of 42. Among all possible substitutions, A → G was the most frequent with a median of 7 and a maximum value of 23 (Table 2; Fig. 3b).
Table 2.
Mutation bias of TR/VR pairs
| A-to-N-subs | Non-A-to-N-subs | A-T-subs | A-C-subs | A-G-subs | |
|---|---|---|---|---|---|
| Median | 14 | 0 | 5 | 2 | 7 |
| Min | 0 | 0 | 0 | 0 | 0 |
| Max | 42 | 3 | 19 | 11 | 23 |
In the prototypical DGR system (the one characterized from the bacteriophage BPP-1), the TR-VR pair length is 134 bps with 23 mutation sites (A ◊ N), and the RT length is 328 aa [1, 2]. Still, other studies have reported DGRs with RT lengths ranging from 149 to 648 aa [18]. In addition to those results supported by the previous studies, we found smaller RT sequences and up to 3 non-A-to-N substitutions in some TR/VR regions, due to the slight flexibility of the MetaCSST tool.
Phylogenetic analysis of DGR-RTs
In order to explore the DGR diversity, we considered the 2,263 RT sequences. First, we removed redundancy with CD-HIT v4.8.1 [23], which resulted in 1,012 unique RT sequences. The protein sequences were obtained using the EMBOSS Transeq online tool [24]. We further removed the RT sequences shorter than 100 aa, resulting in 889 nr-RTs. Using InterProScan, we identified known catalytic domains in 889 RT sequences. Out of the 889 RT sequences, 861 were found to contain complete reverse transcriptase domains, confirming their ability to perform reverse transcription (Additional file: Table S13). We have used these 861 RT sequences for the consequent analysis to ensure the quality of the corresponding DGRs. Their multiple sequence alignment and trimming were done using MAFFT v7.222 [25] and TrimAl v1.2 [26], respectively. Finally, the phylogenetic tree was built using FastTree v2.1.11 [27]. We annotated the environments and taxonomy taken from the GEM catalog metadata file.
The resulted tree has three main branches. Even though none of them exclusively belong to a specific phylum or environment type, some of the sub-branches did. There are small clusters belonging to either host-associated or environmental prokaryotic genomes; however, we noticed that DGR-RT clusters were determined mainly by the taxonomy of the organism. For instance, even though the number of archaeal DGRs was low, most of them clustered together or were in an isolated branch. This indicates that DGRs from archaea have differentiated from those in other organisms, as previously reported [13, 15]. The most abundant phyla in the tree were Firmicutes, Bacteroidota, Patescibacteria, and Proteobacteria, which were scattered throughout the tree, except for Patescibacteria (also known as the Candidate Phyla Radiation group). The lineages in the CPR group are characterized for having small genomes and lacking several biosynthetic pathways. The DGR-RTs belonging to this group were clustered together on the tree, and they predominantly belong to aquatic environments.
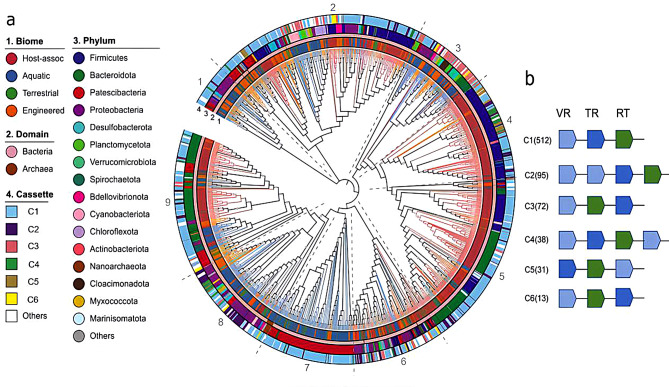
Overall, we could identify nine major clades in our phylogenetic tree (Fig. 4a). DGRs in clades 4 and 5 belong almost entirely to host-associated environments and Firmicutes and Bacteroidota phyla. The other clades were constituted by DGRs from multiple phyla and environments, particularly clades 2 and 6. However, it is worth noting that DGRs from some underrepresented phyla such as Cyanobacteria, Chloroflexota, and Cloacimonadota phyla clustered together (Fig. 4a). Past research has shown that cyanobacterial DGR-RTs represent a monophyletic clade, different from other bacterial DGRs. Hence, it would not be surprising that DGRs from other phyla have also diverged in different clades [7]. Similar patterns were shown in the individual trees we built for host-associated and environmental genomes, respectively, showing the ecosystem categories. DGR-RT clusters were mainly determined by the phyla, and there was no clear distinction based on the ecosystem categories (Additional file: Fig. S4). For comparison, we performed similarity analyses using RTs from other systems including phage defense (retrons), retrotransposons, group II introns, and DGR-RTs (Additional file: Fig. S5). The identity percentages between the non-DGR-RTs and DGR-RTs are all less than 30%, indicating that the non-DGR-RTs are very different from the DGR-RTs, which also supports previous findings that the DGR-RTs have a different evolutionary origin from the other RTs.
Fig. 4.
(a) Phylogenetic tree of DGR reverse transcriptases (amino acid sequences). The inner ring represents the environment (biome), middle rings show the taxonomic annotation at domain and phylum level, and the outer ring represents the cassette architecture. The branches are colored according to their environments(biomes). We identified 9 main clades that are delimited by the dashed lines. (b) Representation of the 6 most common DGR cassette architectures
In addition, we analyzed the novelty of the 861 non-redundant DGR-RTs found in our study. We compared our sequences against a database of 12,283 nr-RTs (at 90% AAI) [12, 16, 18] using BLAST + v2.10.1 [28], and we found that 49 RT sequences (~ 5.7%) are novel. Among these, 22 RTs are associated with human host-related genomes, while the remaining RTs are found in various other environments (see detailed results in supplementary file).
Though our study identified a much smaller number of RTs compared to the dataset of 12,283 nr-RTs, we found some new sequences and DGRs were identified in genomes from all four biomes and 24 ecosystem categories. Especially, we used complete or almost complete genomes (MAGs) for the detection of DGRs, which made it possible to obtain complete information about the frequency of DGRs and their target genes in each genome, considering their length and taxonomic and environmental affiliation. Furthermore, we could determine the cassette architectures based on the coordinates of the DGR components for each genome.
Identification of the most common cassette architectures
We characterized the cassette architectures for the DGRs containing the 861 nr-RTs with a custom Python script. However, we manually checked the DGRs with two or more TR sequences and filtered some TR/VR pairs detected by the MetaCSST tool. Overall, we identified 35 different cassette architectures with variations in the arrangement, orientation, and number of components (RT, TR, VR), and we determined 6 main groups (i.e., the most common groups shown in ≥ 10 MAGs) (Fig. 4b) (See Additional file: Table S9 for the complete list of 38 cassette architectures). The most common cassette was, as expected, the prototypical VR-TR-RT (512 DGRs have this conformation), followed by a VR-VR-TR-RT arrangement (95 DGRs). Overall, these two conformations represent 60.2% of all DGRs, and some features of other frequent cassette architectures were: multiple VR sequences and components situated in different strands. Furthermore, we identified 12 DGRs with two RT sequences and 53 DGRs with two or more TRs, whose coordinates were manually checked to remove the overlapping TR/VR pairs. In general, the cassette patterns we detected were consistent with previous studies that have reported variations in the DGR components organization, although few DGRs showed 2 RT sequences [3, 8, 12, 16].
In an attempt to check the evolution of the DGR structures, we included the cassette annotation in the phylogenetic tree. As expected, the prototypical DGR cassette VR-TR-RT is widespread among various taxa and environments. Interestingly, clade 3 has many DGRs with cassettes from groups 5 and 6, which can be found in multiple phyla. Moreover, we noticed that some small clusters had a specific cassette type, like the ones within clades 2, 4 and 8. In general, most DGRs from host-associated environments possess the prototypical DGR conformation. We noticed that DGRs from aquatic, terrestrial, and engineered environments are more diverse in structures since most of the other cassettes are found in organisms from these environments (Fig. 4a). A previous study reported the most common cassette architectures in DGRs from the human microbiome. They found 122 different cassettes, some of them similar to the ones presented in this study. However, the conformation VR-TR-RT in the negative strand was not very common; instead, they found that DGRs with two or three VRs were abundant and were associated with Proteobacteria [12]. In our phylogenetic tree, cassettes from groups 3, 4, 7, and 8 were the ones with two VRs, and most of them clustered together in clade 8, which has many representatives from phylum Proteobacteria.
In summary, our results suggest that the DGR cassettes are not restricted to a unique organization. Some organisms might have developed DGRs with different arrangements and numbers of components, which might play a significant role in their adaptation to specific environments. Therefore, future research could deeply explore these DGR conformations and determine if they are associated with particular organisms or environments.
Functional annotation of target genes
We next sought to determine the target genes. For this purpose, we first searched the open reading frames (ORFs) on the 2,014 DGR-containing MAGs and carried out the functional annotation using the command-line tool PROKKA v1.14.6 [29]. We obtained the coordinates of the ORFs per MAG and then used a custom Python script to check the overlaps between the ORF and VR sequences.
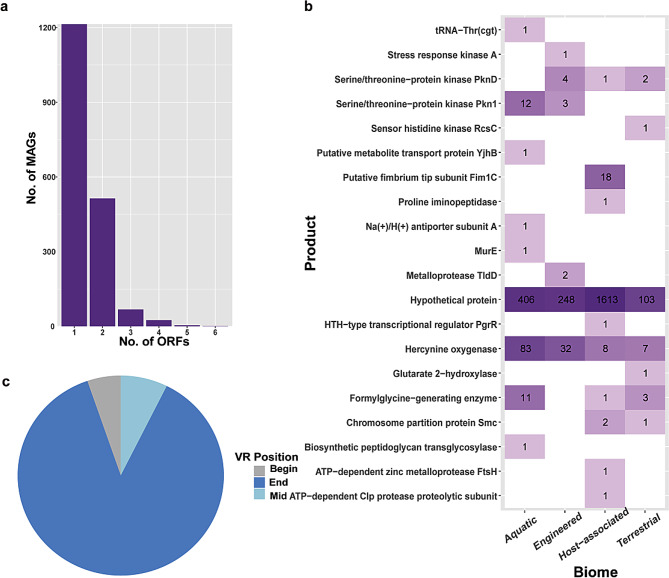
MetaCSST identified 3,180 VRs. However, some of them entirely overlap since they were hit when searched by two or more different TRs. The putative target genes were determined based on the coordinates of the VRs and ORFs. Overall, 1,826 (out of 2,014) MAGs were found to have VRs overlapped with ORFs, i.e., putative target genes, and some MAGs had up to 6 target genes (Fig. 5a). The total number of target genes identified was 2,572. Most of them (2,370) are hypothetical proteins, and only 202 predicted proteins are proteins that have now been found to have some functions, representing 19 different products (Fig. 5b). The most frequent product was Hercynine oxygenase (etgB), found in all four biomes and predominant in Aquatic and Engineered environments. It was detected in 10 ecosystem categories but with a greater presence in freshwater and wastewater samples (Additional file: Table S10). This protein catalyzes the oxidative sulfurization of Hercynine, a step in the biosynthesis pathway of Ergothioneine, which is an antioxidant that protects mycobacteria (phylum Actinobacteria) from oxidative stress; however, it was found in various phyla, such as Proteobacteria, Bacteroidota, Planctomycetota, among others. Furthermore, homologues to the etgB include the formylglycine-generating sulfatase enzyme (FGE), which has been shown to accommodate variable residues in the FGE subtype of the C-type lectin (CLec) - fold. The CLec fold is a general ligand-binding domain, but can also have enzymatic activity as seen in FGE [6, 9, 16, 30].
Fig. 5.
(a) Number of MAGs with multiple target genes, (b) Functional annotation of target genes, count per biome. The data was scaled by the sum of each row, (c) Percentage of VRs located at the beginning (containing the start codon), middle or end (closest to the stop codon) of the ORF
Various studies have reported the role of DGRs in the modification of binding proteins. For instance, in the Bordetella phage BPP-1 DGR, the target gene is the mtd (major tropism determinant), a tail fiber protein located at the distal ends of the fibers that bind to the adhesion receptors on the host surface. The phage can alter its tropism by modifying this protein, thereby determining the host range. DGR systems can generate many ligand-binding protein variants, thus changing the host range of BPP-1 [2]. Many other DGR target proteins have been associated with ligand-binding functions [14, 16, 18], and interestingly, prior reports have shown the existence of multi-domain target proteins, for example, in cyanobacteria, in which ligand-binding domains were often paired with a second domain linked to signal transduction [7]. Non-phage DGR systems can generate highly variable protein sequences, increasing binding diversity of the target proteins. For example, in Treponema denticola, the DGR system produces hyper-variable proteins containing a C-type lectin fold, such as Treponema variable protein A (TvpA) [14]. This structural framework supports extensive sequence variation without compromising function. Furthermore, DGR systems in different organisms modulate surface protein diversity, enhancing interactions between hosts and pathogens. In addition, recent studies revealed that DGR-diversified targets act as antigen sensors that confer a form of adaptive immunity to multicellular bacteria, implying that the DGR system has both phage-promoting and antiviral defense roles [31]. Most of these functions are related to mechanisms associated with phage/host attachment.
Our findings shed light on the possibility that DGRs are diversifying genes with different functions, depending on the environment type, that might help organisms to cope with the changes in environmental conditions. However, the accurately annotated proteins in our dataset represent only 7.9%, and there were many more hypothetical proteins in each environment category. Furthermore, few products had a remarkable frequency, such as Hercynine oxygenase, Putative fimbrium tip subunit Fim1C (only found in human environments), Serine/threonine-protein kinase Pkn1, Formylglycine-generating enzyme, and Serine/threonine-protein kinase PknD. The rest of the products were found only once or twice and only in one environmental category (Fig. 5b) (Additional file: Table S10).
Nevertheless, it is worth mentioning that, despite the advantages that MAGs have for the study of non-culturable organisms and their genes, we should be aware of the reliability of MAGs in capturing all population core and variable genes compared to isolate genomes. For instance, in a recent study comparing gene variability of Escherichia coli isolates against MAGs, it was shown that MAGs missed approximately 25% of the population core genes and 50% of the variable genes, showing the limitations that even high-quality MAGs might have [32]. Therefore, there could be other target proteins diversified by DGRs that have not been detected in this study since MAGs cannot fully capture the genes present in an organism.
Analysis of the target genes and identification of the target protein-domains
A hallmark of DGR target proteins is the terminal position of the VRs, which are known to be located at the C-terminal tail of the protein sequence [5]. Congruently, in our data, most of the VRs were situated at the end section of the target gene (87%) (Fig. 5c). In some cases, we found multiple VRs on the same target gene. The majority has only one VR, but we found 4 and 3 target genes with 2 VRs and 3 VRs, respectively (Additional file: Fig. S6).
DGR targets are highly variable in their sequences (approximately 17% sequence identity), but it has been shown that they share a C-type lectin (CLec) domain, where the VR is situated. This CLec fold is able to tolerate massive sequence variation, and it has been reported as a general ligand-binding site in many structurally characterized variable proteins, such as Mtd, TvpA, LdtA, and AvpA [5, 6, 14, 30, 33]. Thus, we sought to determine the internal structure of the target proteins found in our study by using the InterProScan tool [34] to search the conserved domains and sites.
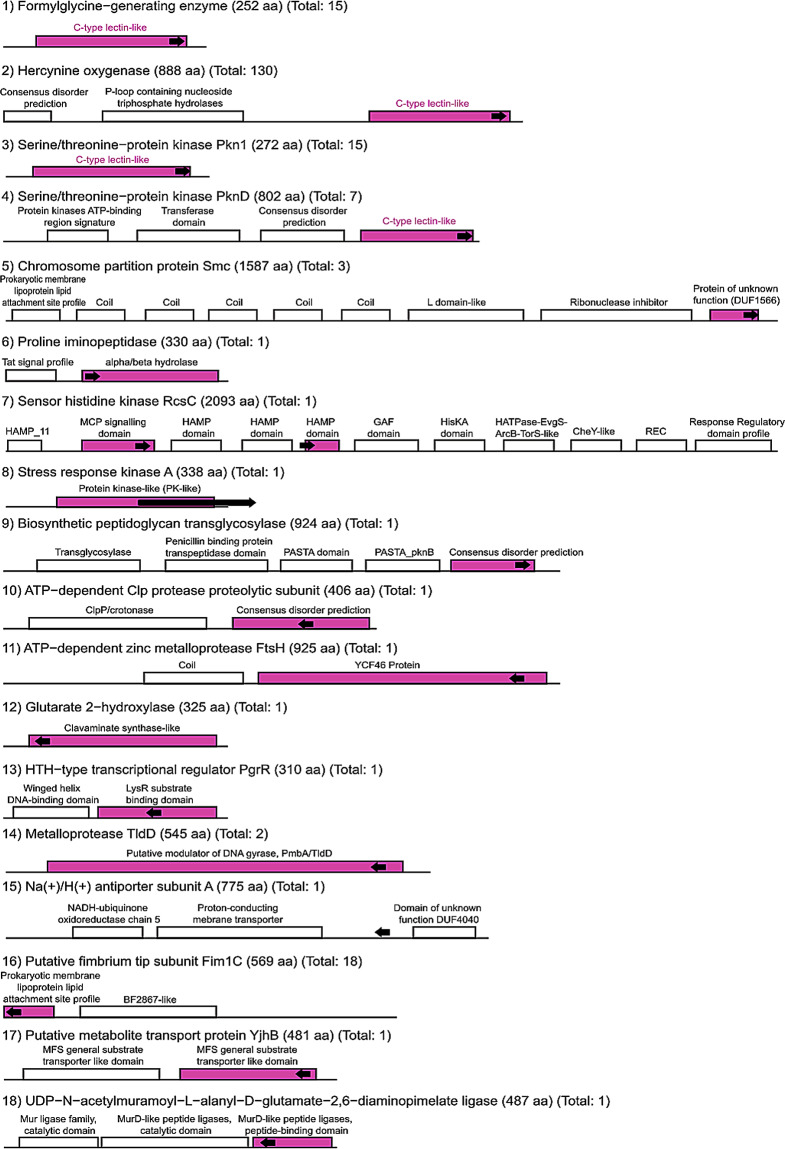
We analyzed the protein structure for each product separately. While products such as Hercynine oxygenase, Formylglycine−generating enzyme, Serine/threonine−protein kinase Pkn1, and Serine/threonine−protein kinase PknD had a CLec domain where the VR was situated, the other 14 products did not possess this domain in their internal structures (Fig. 6). Interestingly, Putative fimbrium tip subunit Fim1C, found only in the human digestive system, has the VR within a prokaryotic membrane lipoprotein lipid attachment site. This is similar to a variable lipoprotein found in Legionella pneumophila, codified by the target gene LdtA, and the C-terminus region that contains the VR was predicted to adopt a CLec fold [30]. Other target proteins that were less frequent also showed domains different from the CLec (Fig. 6), some of which have not been reported in past studies that analyzed the domain compositions [16]. Indeed, we found that many of these domains were related to binding and signaling functions, such as the LysR substrate-binding domain, alpha/beta hydrolase fold, PmbA/TldD, MFS general substrate transporter-like domain, HAMP domain, MCP signaling domain, MurD-like peptide ligase domain.
Fig. 6.
Target protein domains. The domains are shown for one representative sequence of each product; the sequence length in amino acids (aa) and the total number of sequences found per product are in parentheses. The regions highlighted in pink represent the domain that contains the VR, which is symbolized by the black arrow. The direction of the arrows indicates if it was found on the positive (right) or negative (left) DNA strand. Only four products have a C-type Lectin domain that contains the variable region
Furthermore, we analyzed the hypothetical proteins found in our study, representing 92% of all putative target genes. However, only 1,286 out of 2,370 obtained a domain annotation. Not surprisingly, the majority showed a multidomain conformation with the VR located at a C-type Lectin or a DUF1566 domain, which was reported to have a CLec-fold too [15]. Nonetheless, we also found other domains containing the VR sequence, such as Fibronectin type III, which has an Immunoglobulin-like fold (Ig) and was found only in human-associated MAGs. Interestingly, while the C-type Lectin domain was detected in proteins from almost all environments, mainly from human-associated, we noticed that the DUF1566 domains were more frequent in freshwater and arthropod-associated environments (Additional file: Table S11).
Another interesting finding was the Fibrobacter succinogenes major paralogous domain, annotated in 43 target proteins belonging to mostly freshwater and wastewater MAGs. This domain has an apparent lipoprotein signal sequence and has been found in Fibrobacter succinogenes, a bacterium essential for degrading cellulose components in ruminant animals [35]. However, it can also be present in proteins from organisms belonging to the FCB bacteria superphylum, which comprises the phyla Fibrobacteres, Chlorobiota, and Bacteroidota. Accordingly, we found this domain in Bacteroidota organisms from our dataset. Still, its roles in freshwater and wastewater environments remain unclear. However, we hypothesize that it may act as a signal peptide to translocate cellulases to the outer membrane of the cell, as previously suggested in F. succinogenes [35].
Although scarcely represented, other peculiar domains holding a VR were SaV-like, CalX-like domain, Bacterial general secretion pathway protein G-type pilin, Bacterial TonB-dependent receptor, RmlC-like cupin domain, among others (Additional file: Table S11). To date, DGRs obtained from different environments and organisms have been associated with signal peptide and ligand-binding functions [18]. Our results are another example that supports this recurring DGR role. Nevertheless, only 1,286 out of 2,370 hypothetical proteins obtained a domain annotation by InterProScan, and just 669 showed domains overlapped with a VR. Even so, we were able to identify new domains from environments different from human-associated, which deserve further attention to explore novel roles of the DGRs and interactions with other mechanisms.
Conclusions
The present study leverages the most comprehensive catalog of environmental genomes built to date, which allowed us to assess the prevalence of the DGR mechanism in various organisms from different taxa and environments and with different genome lengths. Our research findings support the widespread prevalence of DGRs, a preference for the distribution of viral and cellular DGRs in different environments, as well as a higher-frequency distribution of DGRs in organisms with smaller genomes.
In addition to results from a previous study that used metagenomes from a wide range of environments, we found 49 new DGR-RT sequences. Furthermore, our phylogenetic analyses revealed that DGR systems have diverged based on the host taxonomy and environments, resulting in nine main clades. Some taxonomic categories have developed unique clades of DGRs, which also seem to have their own conformational diversity, i.e., variations in the number, arrangement, and orientation of their components. We identified 35 different cassette patterns and 6 were the most frequent. The differentiation of clades based on the environment was less evident, but we could still observe certain differences between DGRs from host-associated and environmental MAGs.
To date, few DGRs and their target genes have been fully explored. This study showed that DGRs modify various enzyme proteins and binding proteins, accommodating variations in a CLec-fold. However, our domain annotations showed that domains other than the CLec may also harbor a VR. Taken together, these findings highlight the need for characterizing DGR systems in organisms from understudied environments since the DGR mechanism may play essential roles in the adaptation of organisms to harsh conditions or potential threats to their survival, which deserve further attention.
Methods
Data collection
The public medium- and high-quality metagenome-assembled genomes (MAGs) from the Genomes from Earth’s Microbiomes (GEM) catalog and its associated environmental metadata were manually checked and downloaded from DOE-JGI (IMG/M + GOLD) (published in December 2019) [19] (Additional file 1: Table S1).
This catalog comprises 52,515 bacterial and archaeal MAGs from 10,450 metagenomes from the IMG/M database, corresponding to 527 studies and 10,331 samples collected from diverse microbial environments (aquatic, host-associated, terrestrial, engineered, and outdoor air). They all meet the medium-quality level of the MIMAG standard (mean completeness = 83%, mean contamination = 1.3%). The clustering of these MAGs based on 95% whole-genome ANI revealed 18,028 species-level OTUs and, based on taxonomic annotations, they cover 137 known phyla, 305 known classes, and 787 known orders. Overall, from the 52,515 MAGs in the GEM catalog, only 3,038 represent archaea, and 49,477 represent bacteria.
Identification of diversity-generating retroelements (DGRs)
We used the Metagenomic Complex Sequence Scanning Tool (MetaCSST) [12] for the detection of DGRs. The tool was run in all 52,525 MAGs using default parameters, and it detected DGRs in 2,014 MAGs. The number of DGRs found per environment was based on the number of RT sequences. Besides, we used geNomad [36] to identify whether the DGRs are located in viral contigs. This is a tool designed for the classification and detection of viral sequences in metagenomic data. It helps in distinguishing viral contigs from host sequences and is useful for identifying viral components, including integrated proviruses and metagenomic viral contigs. By using geNomad, we aimed to determine whether the DGR RTs were encoded in viral sequences or not.
In order to check whether it exists a pattern for the presence of DGRs, several bar charts were drawn, considering the number of DGR-containing MAGs per environment, taxa, and genome length ranges. To analyze the presence of DGRs in organisms with different genome sizes, we only considered the 9,143 high-quality MAGs (HQ), which have completeness > 90% and contamination < 5%.
Phylogenetic analyses
The total number of DGRs was based on the number of RT sequences. Therefore, these sequences were further analyzed. First, to obtain the unique RT sequences, we removed redundancy using CD-HIT v4.8.1 based on a sequence identity threshold of 90% [23]. The protein sequences were obtained using the EMBOSS Transeq online tool [24]. We further removed the RT sequences shorter than 100 aa. Then, the multiple sequence alignment of the RT protein sequences was done using MAFFT v7.222 [25], using the default parameters and automatic selection of accuracy-oriented and speed-oriented methods. This alignment was trimmed using TrimAl v1.2 with the –gappyout option [26]. Finally, the phylogenetic tree was built using FastTree v2.1.11 [27] with the GTR + CAT model of nucleotide evolution, and the tree was then visualized with iTOL (Interactive Tree of Life) v6 [37]. Taxonomic annotations were taken from the GEM catalog metadata file.
Furthermore, we included RT sequences of Bordetella phage BPP-1 and Group II introns for comparison with the DGR-RT sequences found in prokaryotes.
Identification of DGR cassette structures
For the unique DGRs, we characterized their cassette architectures with a custom Python script that can be found at Script_cassettes.py. This program can read the MetaCSST output file and order the DGR components (TR, VR, and RT) based on their coordinates. However, if a genome sequence had ≥ 2 TRs, the program would mark them. We then manually checked these cases (9 in total) and removed them in the subsequent analysis.
Identification and functional annotation of target genes
The putative target genes were identified by the ORF positions containing the VR sequences. First, the functional annotation for the DGR-containing MAGs was done with PROKKA v1.14.6 [29], using default parameters. This tool makes use of Prodigal for the identification and translation of ORFs and RNA regions. It then uses BLAST and HMMER to compare the translated sequences against public databases, such as CDD, PFAM, and TIGRFAM, to identify their products. The start and end positions of these sequences on the genomes were also given. Next, we used a custom Python script to check if the start and end positions of the VR sequences – which were provided by the MetaCSST tool – were localized within the ORF or RNA sequences. The script is at Script_functional_annotation.py.
Analysis of the target protein domains
We run InterProScan v5.54-87.0 [34] with default parameters to annotate all domains and sites on the target proteins. The output file contained the detected domains with their coordinates, which we used to check the position of the VR within them, using a custom Python script that can be accessed through Script_overlap_domains.py.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the High Performance Computing (HPC) Centre of Shanghai Jiao Tong University for the computation.
Abbreviations
- DGR
Diversity-Generating Retroelements
- MAG
Metagenome Assembled Genome
- ORF
Open Reading Frame
- RT
Reverse Transcriptase Gene
- TR
Template region
- VR
Variable Region
- CLec
C-type Lectin
Author contributions
CCW conceived the study. MCV and CXW implemented the analysis pipeline. MCV and CXW collected the metagenome-assembled genomes. MCV and CXW analyzed the data. MCV, CXW and CCW wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by grants from National key R&D program (2023YFF1001600), National Natural Science Foundation of China (32170643, 61472246), Natural Science Foundation of Shanghai (20ZR1428200, 22ZR1433600), Shanghai Key Program of Computational Biology (23JS1400800), and Joint International Research Laboratory of Metabolic & Developmental Sciences Joint Research Fund (MDS-JF-2019A07). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
All scripts for this study can be found under https://github.com/mariela-ecv/DGRs_MAGs. The resulted MAGs containing DGRs, the annotation of DGRs, and 6 additional datasets can be found at https://cgm.sjtu.edu.cn/DGRs-MAGs/index.html.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mariela Carrasco-Villanueva and Chaoxian Wang contributed equally to this work and should be considered co-first authors.
References
- 1.Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, Miller JF. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295(5562):2091–4. [DOI] [PubMed] [Google Scholar]
- 2.Liu M, Gingery M, Doulatov SR, Liu Y, Hodes A, Baker S, Davis P, Simmonds M, Churcher C, Mungall K, Quail MA. Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J Bacteriol. 2004;186(5):1503–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10(4):388–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431(7007):476–81. [DOI] [PubMed] [Google Scholar]
- 5.McMahon SA, Miller JL, Lawton JA, Kerkow DE, Hodes A, Marti-Renom MA, Doulatov S, Narayanan E, Sali A, Miller JF, Ghosh P. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 2005;12(10):886–92. [DOI] [PubMed] [Google Scholar]
- 6.Le Coq J, Ghosh P. Conservation of the C-type lectin fold for massive sequence variation in a Treponema diversity-generating retroelement. Proc Natl Acad Sci. 2011;108(35):14649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallota-Eastman A, Arrington EC, Meeken S, Roux S, Dasari K, Rosen S, Miller JF, Valentine DL, Paul BG. Role of diversity-generating retroelements for regulatory pathway tuning in cyanobacteria. BMC Genomics. 2020;21(1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schillinger T, Lisfi M, Chi J, Cullum J, Zingler N. Analysis of a comprehensive dataset of diversity generating retroelements generated by the program DiGReF. BMC Genomics. 2012;13(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schillinger T, Zingler N. The low incidence of diversity-generating retroelements in sequenced genomes. Mob Genetic Elem. 2012;2(6):287–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye Y. Identification of diversity-generating retroelements in human microbiomes. Int J Mol Sci. 2014;15(8):14234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharifi F, Ye Y. MyDGR: a server for identification and characterization of diversity-generating retroelements. Nucleic Acids Res. 2019;47(W1):W289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan F, Yu X, Duan Z, Lu J, Jia B, Qiao Y, Sun C, Wei C. Discovery and characterization of the evolution, variation and functions of diversity-generating retroelements using thousands of genomes and metagenomes. BMC Genomics. 2019;20(1):1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul BG, Bagby SC, Czornyj E, Arambula D, Handa S, Sczyrba A, Ghosh P, Miller JF, Valentine DL. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat Commun. 2015;6(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nimkulrat S, Lee H, Doak TG, Ye Y. Genomic and metagenomic analysis of diversity-generating retroelements associated with Treponema denticola. Front Microbiol. 2016;7:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul BG, Burstein D, Castelle CJ, Handa S, Arambula D, Czornyj E, Thomas BC, Ghosh P, Miller JF, Banfield JF, Valentine DL. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat Microbiol. 2017;2(6):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Gingery M, Abebe M, Arambula D, Czornyj E, Handa S, Khan H, Liu M, Pohlschroder M, Shaw KL, Du A. Diversity-generating retroelements: natural variation, classification and evolution inferred from a large-scale genomic survey. Nucleic Acids Res. 2018;46(1):11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benler S, Cobián-Güemes AG, McNair K, Hung SH, Levi K, Edwards R, Rohwer F. A diversity-generating retroelement encoded by a globally ubiquitous Bacteroides phage. Microbiome. 2018;6(1):1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roux S, Paul BG, Bagby SC, Nayfach S, Allen MA, Attwood G, Cavicchioli R, Chistoserdova L, Gruninger RJ, Hallam SJ, Hernandez ME. Ecology and molecular targets of hypermutation in the global microb iome. Nat Commun. 2021;12(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayfach S, Roux S, Seshadri R, Udwary D, Varghese N, Schulz F, Wu D, Paez-Espino D, Chen IM, Huntemann M, Palaniappan K. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 2021;39(4):499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babkin IV, Tikunov AY, Baykov IK, Morozova VV, Tikunova NV. Genome Analysis of Epsilon CrAss-like phages. Viruses. 2024;16(4). [DOI] [PMC free article] [PubMed]
- 21.Bourguignon T, Kinjo Y, Villa-Martín P, Coleman NV, Tang Q, Arab DA, et al. Increased mutation rate is linked to genome reduction in Prokaryotes. Curr Biol. 2020;30(19):3848–e554. [DOI] [PubMed] [Google Scholar]
- 22.Liu PL, Huang Y, Shi PH, Yu M, Xie JB, Xie L. Duplication and diversification of lectin receptor-like kinases (LecRLK) genes in soybean. Sci Rep. 2018;8(1):5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–7. [DOI] [PubMed] [Google Scholar]
- 25.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. [DOI] [PubMed] [Google Scholar]
- 29.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. [DOI] [PubMed] [Google Scholar]
- 30.Handa S, Paul BG, Miller JF, Valentine DL, Ghosh P. Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements. BMC Struct Biol. 2016;16(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doré H, Eisenberg AR, Junkins EN, Leventhal GE, Ganesh A, Cordero OX, et al. Targeted hypermutation of putative antigen sensors in multicellular bacteria. Proc Natl Acad Sci U S A. 2024;121(9):e2316469121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meziti A, Rodriguez-R LM, Hatt JK, Peña-Gonzalez A, Levy K, Konstantinidis KT. The reliability of metagenome-assembled genomes (MAGs) in representing natural populations: insights from comparing MAGs against isolate genomes derived from the same fecal sample. Appl Environ Microbiol. 2021;87(6):e02593–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arambula D, Wong W, Medhekar BA, Guo H, Gingery M, Czornyj E, Liu M, Dey S, Ghosh P, Miller JF. Surface display of a massively variable lipoprotein by a Legionella diversity-generating retroelement. Proceedings of the National Academy of Sciences. 2013;110(20):8212-7. [DOI] [PMC free article] [PubMed]
- 34.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raut MP, Couto N, Karunakaran E, Biggs CA, Wright PC. Deciphering the unique cellulose degradation mechanism of the ruminal bacterium Fibrobacter succinogenes S85. Sci Rep. 2019;9(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camargo AP, Roux S, Schulz F, Babinski M, Xu Y, Hu B, et al. Identification of mobile genetic elements with geNomad. Nat Biotechnol. 2024;42(8):1303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts for this study can be found under https://github.com/mariela-ecv/DGRs_MAGs. The resulted MAGs containing DGRs, the annotation of DGRs, and 6 additional datasets can be found at https://cgm.sjtu.edu.cn/DGRs-MAGs/index.html.