Abstract
We developed a new sodium magnetic resonance fingerprinting (23Na MRF) method for the simultaneous mapping of and sodium density with built-in (radiofrequency transmission inhomogeneities) and corrections (frequency offsets). We based our 23Na MRF implementation on a 3D FLORET sequence with 23 radiofrequency pulses. To capture the complex spin dynamics of the 23Na nucleus, the fingerprint dictionary was simulated using the irreducible spherical tensor operators formalism. The dictionary contained 831,512 entries covering a wide range of factor and parameters. Fingerprint matching was performed using the Pearson correlation and the resulting relaxation maps were weighted with a subset of the highest correlation coefficients corresponding to signal matches for each voxel. Our 23Na MRF method was compared against reference methods in a 7-compartment phantom, and applied in brain in five healthy volunteers at 7 T. In phantoms, 23Na MRF produced values comparable to those obtained with reference methods. Average sodium relaxation time values in cerebrospinal fluid, gray matter and white matter across five healthy volunteers were in good agreement with values previously reported in the literature.
1. Introduction
Sodium ions (Na+) plays a critical role in the human body and are invariably linked to the maintenance of ionic homeostasis as well as many physiological and electrochemical processes of metabolism (1, 2). For this reason, the non-invasive detection of these ions from the nuclear magnetic resonance (NMR) signal of the sodium isotope 23Na (spin ) has become an important modality for the study of metabolic regulation and biochemical status throughout the body (1, 2). In brain, 23Na MRI has been used to study neurodegenerative disease (1–7), tumor pathology (1, 2, 8, 9) and neurological events, such as stroke (2, 10) and traumatic brain injury (11). The intracellular and extracellular spaces in brain tissues both represent motion-restricted environments that give rise to signal contributions coming from the central and satellite transitions, which are strongly influenced by the quadrupolar interactions of the 23Na nuclear spin system with its surroundings (2). These dynamics result in a biexponential transverse relaxation decay, i.e. a long component () and a short component (), in both the intracellular and extracellular spaces. As a consequence, both gray matter (GM) and white matter (WM) in brain will also exhibit an overall 23Na biexponential transverse relaxation, as a weighted average of the relaxation processes from the intracellular and extracellular spaces. Similarly, 23Na longitudinal relaxation follows the same biexponential pattern from the intracellular and extracellular spaces. However, in soft biological tissues, both the short and long components are often very close to each other and longitudinal relaxation is usually measured as a monoexponential process () in GM and WM. In fluids such as cerebrospinal fluid (CSF), the quadrupolar interaction averages to zero as a result of rapid motion allowing the signal dynamics to be often modeled as a monoexponential relaxation for both transverse and longitudinal magnetization components. Collectively, the spin nature of the 23Na nucleus and the low concentration of Na+ ions in brain tissue (on the order of 40–50 mM on average) combined with the inhomogeneous structure of the brain, makes it difficult to simultaneously quantify 23Na relaxation times and density (1, 2, 12).
Proton magnetic resonance fingerprinting (1H MRF) has become a popular technique for the simultaneous quantification of physical properties within a system (13–17). Recently, sodium MRF (23Na MRF) studies in the brain have demonstrated promising initial results. Kratzer et al. (18, 19) implemented a version of 23Na MRF capable of quantifying relaxation parameters in CSF and brain tissue (combined GM and WM) that utilized a 3D radial sequence with variable repetition times (TR), echo times (TE) and flip angles (FA). Our group previously introduced a multi-pulse approach to multicompartmental Na+ concentration quantification (20), which we now expanded for quantifying 23Na relaxation in the brain.
In this work, we present a 23Na MRF technique that is sensitive enough to quantify differences in average relaxation times over whole GM, WM, and CSF. Our method simultaneously maps 23Na , sodium density (SD), and experimental imperfections arising from radiofrequency (RF) transmission inhomogeneities ( factor) and frequency offsets from B0 inhomogeneities . The 23Na MRF pulse train with variable FAs and phase angles (PA) was designed by incorporating the irreducible spherical tensor operator (ISTO) formalism into a genetic algorithm (GA) that minimizes signal correlation between GM and WM, assuming average relaxation times from the literature for these two tissues (21–23) during this optimization phase. A 3D Fermat looped orthogonally encoded trajectory (FLORET) (24) was used to fully sample k-space with constant TE. The proposed 23Na MRF sequence can acquire data over the full brain with 5-mm isotropic resolution in about 30 min at 7 T. We tested our 23Na MRF approach in a 7-compartment phantom and in five healthy volunteers.
2. Material and Methods
23Na spin dynamics simulation
The dynamics of the 23Na spin were modeled using the ISTO framework under the conventions described in Madelin et al. (2), Lee et al. (12), and Gilles et al. (20). Within this formalism, the evolution of the 23Na spin system is described by the Liouville-von Neumann (master) equation (with convention ):
| (1) |
where is the density operator of the spin system at thermal equilibrium, is the total spin Hamiltonian and is the Redfield relaxation superoperator. The total Hamiltonian is the sum of the main Hamiltonians acting on the density operator, such as the Zeeman Hamiltonian (interaction of the spins with the constant field), the residual quadrupolar coupling Hamiltonian (interaction of the quadrupole moment of the nuclei with the residual average electric field gradient in anisotropic environments), and the RF field Hamiltonian (interaction of the spins with time-varying transmit RF field ). In soft quasi-isotropic brain tissues, can be considered negligible. Since we are also operating in the rotating frame for the spin dynamics simulation, the main Hamiltonians acting on the spin system are related to inhomogeneities only, and .
The Redfield relaxation superoperator acting on the density operator of the spin system is described by:
| (2) |
where is a constant that depends on the convention used to describe spectral densities, is the ISTO of rank 2 and order is the spectral density function, and is the imaginary term associated with dynamic frequency shift, which in our case can be omitted due to its negligible observable effect in soft tissues (2). The matrix formulation of and relaxation rates with , are given by (12):
| (3) |
| (4) |
| (5) |
| (6) |
When simulating the spin dynamics for 23Na MRF pulse train optimization or for generating the final fingerprint dictionary, the algorithm takes the relaxation times as input, then calculates the spectral density functions according to Equations 3–6 and uses the results to construct the Redfield relaxation superoperator in Equation 2, which is then added to the Liouville equation. After each time step of the simulation (100 μs), the simulated 23Na MR signal, which corresponds to the MR-observable transverse magnetization, is calculated as the average rank-1 single quantum coherence using the standard formula , where is the trace of matrix .
Pulse sequence design for 3D 23Na MRF
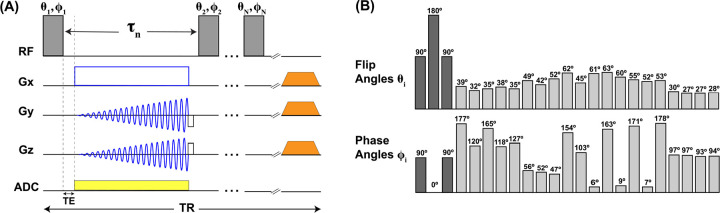
Figure 1(A) presents the 23Na MRF sequence. System excitation was driven by a series of non-selective rectangular RF pulses with and followed by a delay , forming the MRF pulse train shown in Figure 1(B). An initial magnetization inversion using a 90°–180°–90° (dark gray) composite 3-pulse block (25) was used to increase sensitivity and improve RF homogeneity for the inversion before initiating the 20-pulse variable FA/PA train (light gray).
Figure 1. Pulse sequence diagram for 3D 23Na MRF.
The overall pulse scheme is shown in (A). The diagram in (B) represents the variable flip angle (FA) and phase angle (PA) MRF train. There were a total of non-selective rectangular RF pulses of duration . Each pulse was followed by a time period . On the RF channel in (A), the gray rectangles correspond to a single and combination in train (B). In (B), the dark gray rectangles represent a 3-pulse inversion composite block (25) used to increase sensitivity of the sequence and improve RF homogeneity for the magnetization inversion. Within the composite block, . The light gray rectangles indicate the 20-pulse variable FA and PA train. All and are listed at the top of each rectangle and for . During each delay , a time period TE = 0.2 ms was followed by the ADC event, indicated by a yellow block on the ADC channel. A 3D spiral encoding scheme using the FLORET trajectory (20, 24, 26) (3 hubs at 45°, 100 interleaves/hub), indicated in blue, was played over the gradient channels. Immediately after the ADC, a rewind gradient was also played out and then the next RF pulse in the MRF train was initiated. After pulses and delays have played out, a 5-ms spoiling gradient at 70% maximum gradient strength, indicated by an orange trapezoid, was applied in all directions. The entirety of this scheme represented one TR of the sequence.
The RF pulse durations were fixed at , and interpulse delay periods were fixed at within the composite block and for the next pulses . The interpulse delays were set according to our prior work on simultaneous 1H/23Na MRI (27, 28) and in anticipation of integrating this method into simultaneous 1H/23Na MRF.
The FLORET trajectory (20, 24, 26) was used to readout the signal followed by a rewinder to balance the gradient moment. After pulses and delays, a spoiling gradient (duration = 5 ms, 70% of maximum gradient strength) was applied simultaneously in all directions to ensure complete dephasing of residual transverse magnetization prior to beginning the next TR. A delay of ~300 ms was inserted between RF pulse trains to allow recovery of the longitudinal magnetization and reduce specific absorption rate (SAR). A constant TE = 0.2 ms was used throughout the sequence.
The variable 20-pulse FA/PA train was optimized using a genetic algorithm (GA) implemented in MATLAB R2020b (The MathWorks Inc., Natick, Massachusetts, USA) on an Apple MacBook Pro (16-inch, 2019) laptop with a 2.4 GHz 8-Core Intel Core i9 processor. The 3-pulse block used in the initial inversion was included in the optimization with FA and PA kept constant, resulting in a final 23-pulse train. The GA minimized an objective function estimating the Pearson correlation (PC) coefficient (29) between 23Na MR signals arising from GM and WM. Input tissue relaxation times for the simulation were based on average values reported in the literature (1, 2, 30): for GM, , , ; for WM, , and . Initial values for the -pulse train (where and ) were and with period . The composite block, corresponding to with , was also included in the simulation as a non-variable parameter. Limitations were imposed on FA () and PA () in consideration of SAR limits. The GA optimized the system for 100 generations in 8 h. The algorithm was applied a total of three times, first using the initial input values for the variable 20-pulse part of the pulse train, followed by two iterations in which the solution of the previous computation was taken as input for the next one. This way, the optimized pulse train could be inspected after every 100th generation cycle.
Fingerprint dictionary simulation
Simulation of the fingerprint dictionary was performed in MATLAB R2020b on a Cray CS500–1211 cluster with Intel Xeon Gold 6148 high memory CPUs at the NYU Langone High Performance Computing Core facility (New York City, NY, USA). The simulation code is freely available in Matlab File Exchange (see Data Availability section for the link). Signals were simulated starting from thermal equilibrium and propagated under the optimized 23-pulse 23Na MRF train (Figure 1(B)). Parameter ranges ([begin:step:end]) to build the dictionary were , , , factor = [0.7:0.1:1.3] (applied as a multiplying factor to the RF amplitude FA) and . For quantification, we originally assumed that . To insure that spectral densities where were included in the dictionary, a constant (±ΔT1) that added 1 ms to every entry for and subtracted 1 ms to every entry for was included in the simulations.
Parameter combinations where and were omitted from the computation. Due to the memory expense incurred by the simulation, the dictionary was generated in two parts and concatenated. In total, 831,512 entries were generated. The total simulation time was 6.4 days.
Experiments
All experiments were performed at 7 T (MAGNETOM, Siemens, Erlangen, Germany) using a 16-channel transmit/receive 1H/23Na RF brain coil constructed in-house (8 1H channels, 8 23Na channels) (31).
Phantom
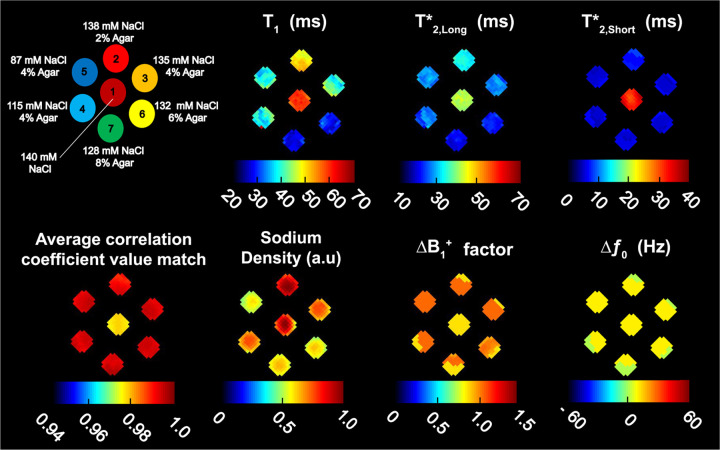
Our test phantom was constructed using a 2.3-L cylinder (outer diameter OD = 20 cm, length = 35 cm) filled with a solution of 70 mM NaCl and which contained seven 50-mL polypropylene cylinders (OD = 30 mm, length = 115 mm) arranged as 6 outer samples each containing a different concentration of NaCl and agar circling the 7th sample placed in the middle and containing a solution of 140 mM NaCl. A diagram of the phantom with sodium/agar concentrations in the axial orientation is shown in Figure 2. The regions-of-interest (ROI) in the 7 samples were generated from a 3D mask of equal diameter to each sample such that all ROI volumes would be the same.
Figure 2. 23Na MRF maps of the 7-compartment phantom.
Fingerprint matching was performed over an average of 2 center axial slices with 20 correlation coefficients included in the weighted average. A diagram of the 7-compartment phantom is shown in the top left corner. Maps for , and are shown on the top row. A map showing the average correlation coefficient values matched for 20 correlations is shown at bottom left, followed by maps for normalized SD in arbitrary units (a.u), factor (unitless) and in Hz.
For 23Na MRF, one scan consisting of 16 averages with TR = 511 ms, isotropic resolution = 5 mm and isotropic FOV = 320 mm, was acquired with the FLORET parameters: 3 hubs/45° with 100 interleaves/hub, TE = 0.2 ms, total scan time = 40:52 min.
We measured the reference using a saturation recovery (SR) reference experiment that consisted of a series of eight scans. Each scan consisted of a FLORET sequence (3 hubs/45° with 100 interleaves per hub) with different TRs = [60, 100, 140, 180, 260, 300, 360, 420] ms and fixed TE = 0.1 ms, 4 averages, isotropic resolution = 5 mm, isotropic FOV = 320 mm, rectangular RF excitation pulses with FA = 90° and . The total scan time for the SR experiments running sequentially was 1:49 h.
Similarly, we measured the reference mono- and biexponential using a multi-TE experiment that consisted of a series of 13 scans. Each scan consisted of a FLORET sequence (3 hubs/45° with 100 interleaves per hub) with TEs = [0.1, 0.5, 1.0, 1.5, 2, 5, 7.5, 10, 15, 25, 35, 50, 65] ms and fixed TR = 150 ms, 4 averages, isotropic resolution = 5 mm, isotropic FOV = 320 mm, rectangular RF excitation pulses with FA = 90° and . The total scan time for the multi-TE experiments running sequentially was 1:57 h.
Brain
Five healthy volunteers (1 female, 4 males, mean age 36 ± 8.5 years) were recruited under a protocol approved by the New York University Grossman School of Medicine institutional review board. All parts of the study were performed in accordance with the relevant guidelines and regulations set forth by the Human Research Protections Program. Informed consent was obtained before each scanning session. For 23Na MRF, we acquired 4 separate scans consisting of 2 averages per scan. These 4 scans were acquired consecutively within 29.8 ± 1.3 min. We chose to divide the 23Na MRF scanning portion of the session into separate 2-average scans in order to communicate with the volunteers during the acquisition. The average TR over all volunteer scans was TR = 704 ± 4 ms, which varied between subjects due to head size and coil loading. The minimum TR was calculated by the scanner to keep SAR within the maximum limit of 100%. Setting TR >700 ms allowed for full recovery for about 380 ms after the last RF pulse, which was more than 5 times the longest expected 23Na in brain from CSF (T1 ~ 50–60 ms (2, 19)). All brain scans were acquired with isotropic resolution = 5 mm and FOV = 320 mm.
For reference, a 1H MPRAGE was acquired with 1 average, TR = 2300 ms, TE = 2.84 ms, FOV = 256×216 mm2, slice thickness = 1 mm, 176 slices/slab and GRAPPA acceleration factor 2, for a total acquisition time of 4:32 min. The average total session time for calibration, shimming and scanning across all volunteers was 55 ± 3 min.
Data processing
All images were reconstructed offline in MATLAB. For each channel, raw k-space data acquired during the 23Na MRF sequence was filtered with a Hamming kernel then reconstructed using gridding (32, 33) to produce a set of 23 complex images with a final nominal resolution 2.5×2.5×2.5 mm3 (128×128×128 matrix). The images from the 8 channels were combined using coil sensitivity profiles as described by Bydder et al. (34). The average time for image reconstruction in the brain across all five volunteers was 42 s, and 38 s in the phantom. For the brain images, an additional denoising step was performed on the complex images using the Marchenko-Pastur method (35–37). This added another 15 s to the brain image reconstruction time. Supplementary Figure S1 shows 23 axial images for a center slice in the phantom. Supplementary Figures S2 and S3 show 23 reconstructed axial images for a center slice in the brain of volunteer 5.
Phantom images from FLORET for and reference experiments were reconstructed in the same way as the 23Na MRF data. Curve fitting was applied voxelwise over the axial plane in the central slice of the phantom using the Levenberg-Marquardt algorithm applied using lsqcurvefit in MATLAB. A monoexponential kernel was assumed for (38) according to Equation 7:
| (7) |
where coefficient and noise floor were variable over the fit optimization. values were restricted to a lower bound of 20 ms and upper bound of 80 ms, and TR was the repetition time from the FLORET acquisitions. Time required for this process was 33 s.
For , we applied the biexponential kernel shown in Equation 8 as descibed by Ridley et. al (38, 39).
| (8) |
The amplitude scaling factor was initialized as 1 and allowed to vary during the optimization with the lower bound set at 0 and the upper bound set at infinity. The Rician noise parameter was initialized as 0.1 based on the average noise measured in a background region outside of the phantom on the magnitude image, as described in Qian et al. (40). In this case, the upper bound was set at 1 with a lower bound of 0.01. The sodium signal fraction was initialized at 0.6 and allowed to vary between a lower bound of 0.4 and an upper bound of 0.8. The TE variable corresponded to TEs from the FLORET acquisitions. was bound between 0.5 ms and 60 ms and was bound between 10 ms and 80 ms.
In both fits, the upper and lower bounds for relaxation times were chosen to coincide with those parameter ranges simulated in the 23Na MRF dictionary. Finally, the average relaxation values from the 7 individual phantom samples were calculated from the resultant 2D relaxation maps after masking. The time required for fitting was 27 s. Collectively, this process was referred to as the reference method (RM).
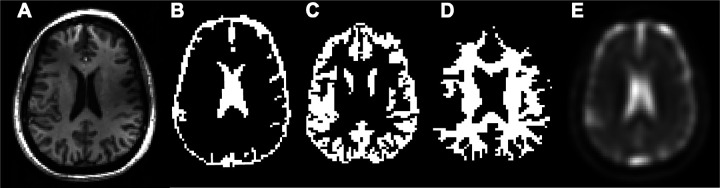
For each volunteer, images from the 1H MPRAGE DICOM datasets were co-registered to the 23Na MRF data using SPM12 (UCL, London, UK) (28, 41). The 23Na MRF image used for co-registration was the one acquired just after the first pulse in the 23Na MRF train. Tissue probability maps were generated from the normalized and co-registered MPRAGE images and segmented into CSF, GM and WM tissue classes using SPM12 (28, 41). The segmentated regions for CSF, GM and WM were then binarized with a 90% threshold to generate tissue ROI masks in MATLAB. These images are presented in Figure 3. The high threshold was chosen to reduce the likelihood of contamination between different tissues.
Figure 3. Images of a center slice of the brain in the axial position from volunteer 5.
(A) 1H MPRAGE. (B) Binary mask for CSF. (C) Binary mask for GM. (D) Binary mask for WM. (E) 23Na image acquired after first RF pulse of the MRF pulse train.
Fingerprint dictionary matching
The fingerprint dictionary (size = 831,512 entries) was matched voxelwise to the complex 23Na image data using Pearson correlation. In the phantom, matching was performed on the average of two center slices in the axial plane. Matching required 10:34 min per slice followed by reconstruction of the correlation-weighted maps which added another 10 s. In brain, matching was performed over one center slice in each of the axial, coronal and sagittal positions for all five volunteers. Additionally, a slab of 20 axial slices were matched for volunteer 5. Matching required an average of 9:42 min per slice plus 10 s for reconstruction of the correlation-weighted maps. All these processes were also performed on an Apple MacBook Pro (2019, 2.4 GHz 8-Core Intel Core i9).
Because of the dictionary size, and due to the low SNR of the sodium images, it was therefore possible that more than one match could generate a high correlation for a single voxel. To account for this, we included matches for a subset of the top correlations for each voxel , and generated the final maps by calculating the correlation-weighted parameter from the dictionary of values corresponding to the matching correlation coefficient according to:
| (9) |
where was the maximum number of correlation coefficients used for weighting and and were the unweighted and weighted parameters , , , factor or , respectively.
SD was calculated as the mean absolute value of signal per correlation for each voxel , normalized by the highest voxel intensity in the image. Weighting was then done according to Equation 9, where was the unweighted SD and was the weighted SD.
Correlation coefficient weighting
We investigated the effect of the correlation weighting on the maps by directly evaluating both phantom and brain 23Na MRF maps weighted with the maximum correlation coefficient only (k = 1) though the k = 1000 highest correlation coefficients.
To better choose the number of correlation coefficients to apply as a weighting factor in the final maps, we devised a method of selection using limits based on our RM and a priori information. First, for each relaxation parameter, a 2D map was generated using 23Na MRF for every level of unweighted correlation (). For the phantom experiments, a range of , and determined by the RM were used as limits. For the brain data range, values from the literature for in CSF, GM and WM, and for GM and WM were used (19, 21–23, 39, 42, 43). We chose to omit in brain from this analysis due to the lack of 23Na values reported at 7 T in the literature.
These limits were then applied to the maps to generate a set of indices providing the location of pixels where the value of the masked 23Na MRF mapped parameter fell within the range of the reference. This index set was then used to create a binary mask which was applied back to the original 23Na MRF 2D maps. These results were plotted as the maximum number of matches made to the subset of pixels within the reference range versus the number k included in matching.
Statistical Analysis
We used the two-sided Wilcoxon rank-sum test (WRST) (44) in MATLAB to compare the values in the ROIs of the 7-compartment phantom, mapped using the RM against 23Na MRF. Similarly, we used the same test to examine the sensitivity of 23Na MRF for distinguishing between CSF, GM and WM in brain.
3. Results
Phantom
Figure 2 shows a diagram of the 7-compartment phantom and the maps from 23Na MRF: , , , SD, factor, , as well as a map of the average correlation coefficient associated with the signals matched in each ROI (average from 20 correlation coefficients). The ROIs are numbered in correspondence with the data given in Table 1, which lists the mean relaxation times calculated for each individual ROI, with their respective standard deviations (STDV), measured using 23Na MRF and with RM.
Table 1. 23Na relaxation times measured in the 7-compartment phantom.
Measurements are shown as mean value ± standard deviation, from our 23Na MRF method and from the average of two repetitions of the reference method.
| Concentration |
23NaMRF |
Reference Method |
||||||
|---|---|---|---|---|---|---|---|---|
| ROI | Agar (%) | NaCl (mM) | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) |
|
| ||||||||
| 1 | 0 | 140 | 58.9 ± 1.2 | 40.6 ± 0.9 | 32.9 ± 1.9 | 57.4 ± 2.0 | 52.1 ± 3.5 | 37.2 ± 2.7 |
| 2 | 2 | 138 | 49.8 ± 2.0 | 32.3 ± 1.3 | 7.6 ± 0.8 | 49.4 ± 1.9 | 30.5 ± 2.6 | 10.4 ± 0.8 |
| 3 | 4 | 135 | 41.2 ± 2.9 | 26.5 ± 1.5 | 6.2 ± 0.6 | 42.8 ± 2.3 | 26.6 ± 1.4 | 6.1 ± 0.4 |
| 4 | 4 | 115 | 40.0 ± 4.7 | 25.5 ± 2.2 | 6.2 ± 0.6 | 43.0 ± 1.1 | 26.3 ± 1.1 | 6.5 ± 0.7 |
| 5 | 4 | 87 | 39.4 ± 2.5 | 27.2 ± 1.2 | 6.1 ± 0.4 | 44.7 ± 0.6 | 25.9 ± 0.8 | 6.4 ± 0.2 |
| 6 | 6 | 132 | 29.7 ± 1.9 | 21.9 ± 1.1 | 6.2 ± 0.5 | 38.3 ± 1.6 | 23.6 ± 0.8 | 4.5 ± 0.3 |
| 7 | 8 | 128 | 28.5 ± 1.3 | 20.2 ± 1.2 | 6.3 ± 0.4 | 36.8 ± 0.7 | 23.0 ± 0.7 | 5.7 ± 0.3 |
The average relaxation times listed in Table 1 for 23Na MRF measurements can be easily estimated by visual inspection of the relaxation maps in Figure 2.
The center ROI 1 contained a solution of 140 mM NaCl and showed slight variations in uniformity for , , and SD maps, but appeared uniform in maps of factor and . The mean for 23Na MRF was 58.9 ± 1.2 ms versus 57.4 ± 2.0 ms for RM. The mean for 23Na MRF was shorter than for 23Na MRF by 18.2 ± 2.4 ms. For RM, the mean (53.1 ± 3.5) was similar to the mean (57.4 ± 2.0 ms). The difference between mean for 23Na MRF and RM was 11.5 ± 3.6 ms, where RM was higher. Mean values for both 23Na MRF and RM were lower than and . The difference between and in 23Na MRF was 26 ± 2.2 ms while the difference between and was 7.7 ± 2.1 ms. For RM, the difference between and was 20.2 ± 3.4 ms and the difference between and was 14.9 ± 4.4 ms.
Loss of uniformity in the phantom compartments was seen in the maps of ROI 3 (41.2 ± 2.9 ms), ROI 4 (40.0 ± 4.7 ms) and ROI 5 (39.4 ± 2.5 ms). Out of these ROIs, the most notable artifact appeared in the map for ROI 4. In this case, we can align the artifact in the map with perturbations in the maps of factor and . The variation noted for the map in ROI 4 could also be outlined in the maps for and . Mean in ROI 4 was 25.5 ± 2.2 ms, which represented the highest STDV for out of all ROIs. Mean in ROI 4 was 6.2 ± 0.6 ms.
Relaxation time data is also presented as boxplots in Figure 4 for comparing 23Na MRF with RM. SD data is shown in the same figure as a comparison between 23Na MRF measurements and ground truth (GT), presented as mean value ±5% STDV.
Figure 4. Boxplots of , , and SD in the phantom compartments: 23Na MRF vs. reference method (RM).
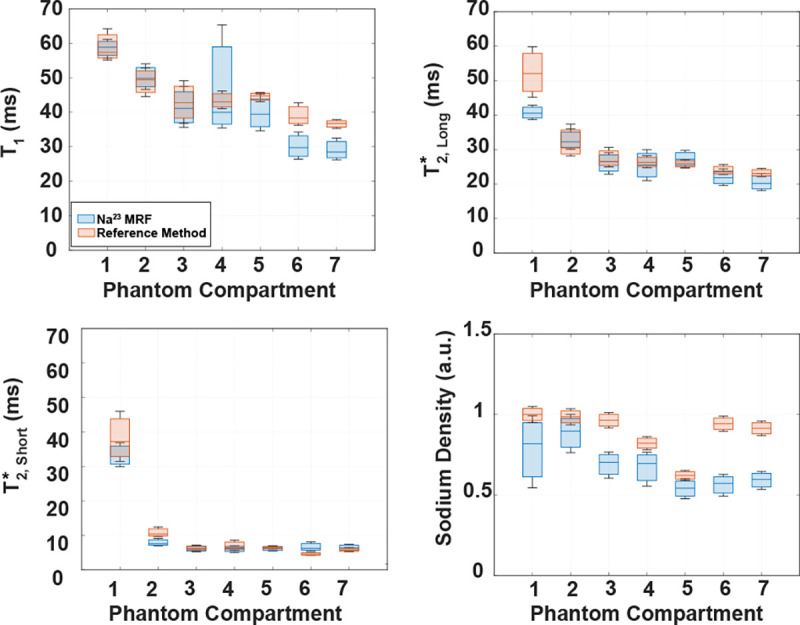
Relaxation time data corresponds to data listed in Table 1. SD calculated using 23Na MRF was compared to the ground truth (mean value ±5%) (see Figure 2). Blue boxes represent data from our 23Na MRF method. Red boxes represent data from RM for relaxation times, and from ground truth for SD.
The boxplots for overlap within the interquartile range for ROIs 1 through 5 of the 7-compartment phantom. For ROI 6, the maximum for 23Na MRF falls 1.93 ms below the minimum for the RM . In ROI 7 the for 23Na MRF was 2.82 ms below the minimum for RM . A non-statistically significant difference between RM and 23Na MRF was found only in ROI 2 with WRST (p = 0.0526).
The boxplot for for ROI 1 shows that the maximum 23Na MRF is 2.31 ms below the minimum RM . For ROIs 2 through 7, there is overlap within the interquartile ranges between RM and 23Na MRF. However, a non-statistically significant difference between RM and 23Na MRF was only found for ROI 4 with WRST (p = 0.0930).
Interquartile overlap was noted between 23Na MRF and RM for in ROIs 1, 3, 4, 5 and 7. The WRST analysis for these ROIs supported non-statistically significant difference between 23Na MRF and RM in ROIs 4, 5 and 7 with p = 0.7919, 0.0802 and 0.9054, respectively. In ROI 2, the RM minimum was ~0.6 ms greater than the maximum from 23Na MRF. Finally, for ROI 6, the minimum for 23Na MRF was ~0.6 ms greater than the maximum for RM.
SD of the GT was consistently higher than SD from 23Na MRF. Overlap in the distributions occured only between the 2nd quartile of the GT and 4th quartile of 23Na MRF in ROI 1, between the 2nd quartile of GT and 3rd quartile of 23Na MRF in ROI 2, and between the 1st quartile of GT and 4th quartile of 23Na MRF in ROI 5.
Supplementary Figure S4 shows scatter plots with linear fits between 23Na MRF and RM for , , , and between 23Na MRF and GT (mean value ±5% STDV) for SD, using data from Table 1.
Brain
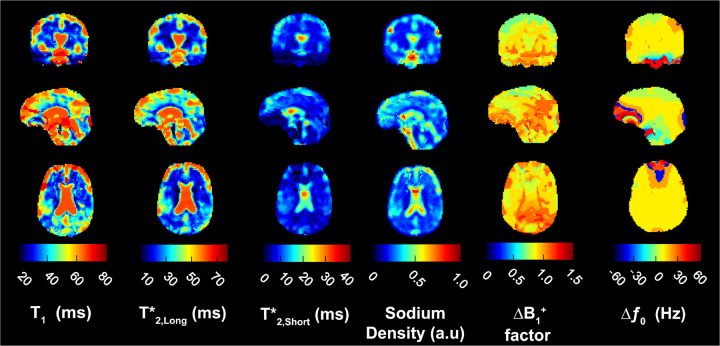
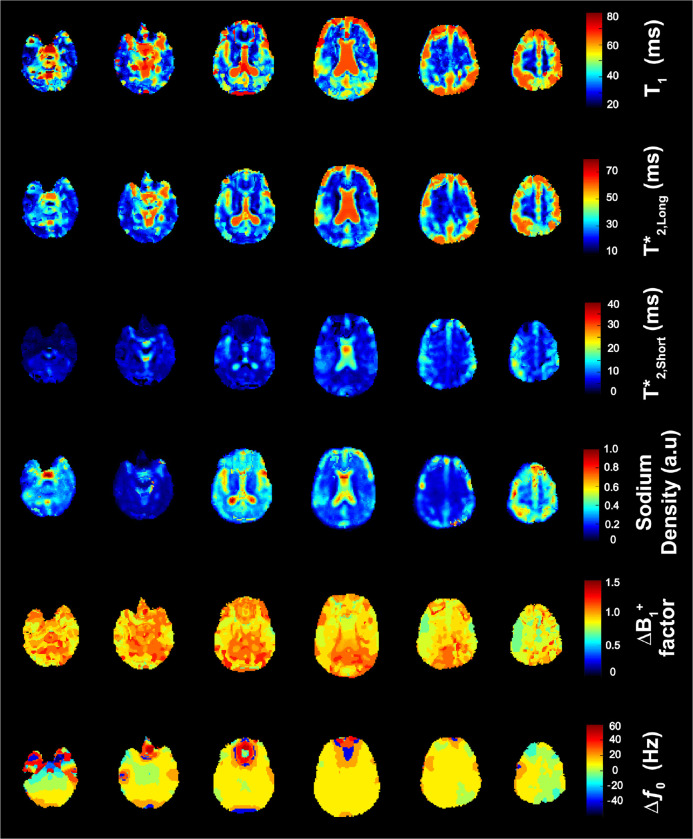
Figure 5 shows the maps for , , , , and from volunteer 5 in coronal, sagittal and axial slices. We used a weighting factor of k = 20 correlation coefficients during the PC matching process for all volunteers. The most apparent feature in the relaxation times and SD maps was the CSF-filled central ventricles. Contrast from the long and values in CSF dominate the central ventricular structure in all three planes. Contrast from CSF can also be seen within subarachnoid spaces and the cavity along the gyri of the cerebral cortex overall cross sections. In the sagittal slice, we identified the third and fourth ventricles and the occipital horn of the lateral ventricle in maps for , , and SD. The overall contrast for in CSF was reduced compared to and . Here the highest values for were concentrated in the center of the central ventricle in each cross-section. Mean values reported in Table 2 were the result of this unequal distribution of over the collective CSF ROI. Mean in the voxels at the center of the central ventricle across all 5 volunteers was 40.1 ± 0.1 ms.
Figure 5. Brain maps from 23Na MRF.
Examples of coronal, sagittal and axial slices for 23Na relaxation times, SD, factor and from volunteer 5. All maps were calculated with a correlation coefficient weighting of k = 20.
Table 2. Mean 23Na MRF values in brain in 5 volunteers.
Data shown represents mean relaxation times and normalized sodium density (SD) ± standard deviations (STDV) measured in three brain tissue ROIs (cerebrospinal fluid [CSF], grey matter [GM], white matter [WM]) for each volunteer (VOL). Mean values were calculated over the center slices of the axial, coronal and sagittal planes of the brain. The bottom row of each column corresponds to the mean value ± STDV of the mean values measured in each ROI across all volunteers
| CSF |
GM |
WM |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VOL | (ms) | (ms) | (ms) | SD | (ms) | (ms) | (ms) | SD | (ms) | (ms) | (ms) | SD |
|
| ||||||||||||
| 1 | 50.7 ± 15.1 | 41.7 ± 16.3 | 10.0 ± 8.8 | 0.246 ± 0.134 | 44.1 ± 12.5 | 33.0 ± 11.2 | 6.6 ± 3.6 | 0.212 ± 0.072 | 43.0 ± 13.6 | 27.9 ± 12.1 | 6.6 ± 3.9 | 0.177 ± 0.054 |
| 2 | 62.1 ± 7.0 | 55.0 ± 9.6 | 17.2 ± 9.4 | 0.467 ± 0.172 | 44.1 ± 12.0 | 31.2 ± 9.2 | 7.4 ± 3.8 | 0.234 ± 0.067 | 36.5 ± 10.5 | 23.1 ± 8.5 | 5.4 ± 2.9 | 0.191 ± 0.063 |
| 3 | 61.8 ± 10.2 | 47.3 ± 13.9 | 12.4 ±7.8 | 0.42 ± 0.169 | 42.9 ± 12.5 | 30.9 ± 9.2 | 5.3 ± 2.7 | 0.262 ± 0.711 | 39.5 ± 13.3 | 26.3 ± 9.8 | 4.6 ± 3.0 | 0.213 ± 0.061 |
| 4 | 59.1 ± 10.3 | 47.4 ± 12.9 | 9.9 ± 7.2 | 0.41 ± 0.165 | 40.4 ± 12.8 | 27.3 ± 8.3 | 6.3 ± 3.1 | 0.234 ± 0.060 | 34.2 ±11.8 | 20.2 ± 9.7 | 4.9 ± 2.6 | 0.176 ± 0.062 |
| 5 | 63.1 ± 5.7 | 57.3 ± 7.6 | 13.1 ± 7.1 | 0.43 ± 0.128 | 46.3 ± 12.9 | 35.1 ± 11.1 | 7.7 ± 3.4 | 0.242 ± 0.071 | 42.2 ± 11.0 | 28.5 ± 12.2 | 5.9 ± 2.7 | 0.185 ± 0.061 |
|
| ||||||||||||
| Mean | 59.4 ± 5.1 | 49.7 ± 6.3 | 12.5 ± 3.0 | 0.395 ± 0.086 | 43.6 ± 2.1 | 31.5 ± 2.9 | 6.7 ± 1.0 | 0.237 ± 0.018 | 39.1 ± 3.7 | 25.2 ± 3.5 | 5.5 ± 0.8 | 0.188 ± 0.151 |
The normalized SD maps demonstrated high sodium concentration within CSF and lower sodium concentration in GM and WM. For , extremely negative and positive shifts were mapped around the medial frontal gyrus, shown in the axial position, over the frontal lobe above the nasal sinus cavity, shown in the sagittal plane, and at the base of the medulla near the posterior cerebellum, shown in the coronal plane.
Figure 6 presents the 23Na MRF maps in six equidistant axial slices for volunteer 5 with a weighting factor of k = 20 correlation coefficients.
Figure 6. Examples of six equally-spaced axial slices of brain maps from 23Na MRF.
Maps shown are 23Na relaxation times, SD, factor and from volunteer 5. All maps were calculated with a correlation coefficient weighting of k = 20.
Table 2 lists the mean 23Na relaxation times and SD with their respective STDVs in CSF, WM and GM for all 5 subjects. The bottom row lists the mean values with corresponding STDV for each tissue across all five volunteers. The greatest deviation from mean relaxation time across volunteers was recorded in CSF. The shortest was measured in volunteer 1 along with the highest STDV (50.7 ± 15.1 ms) while the longest and lowest STDV were recorded in volunteer 5 (63.1 ± 5.7 ms). The wide range of these values contributed to an overall 8.6% STDV amongst volunteers. This trend was also observed for in CSF, where volunteer 1 contributed 41.7 ± 16.3 ms and volunteer 5 contributed 57.3 ± 7.6 ms, toward a mean 49.7 ± 6.3 ms across all five volunteers. For in CSF, a mean value of 12.5 ± 3.0 ms was measured over the five volunteers with 23Na MRF. Finally, a good agreement between relaxation times in GM and WM was noted across volunteers, as indicated by the low corresponding STDVs.
Figure 7 summarizes the results from Table 2 into boxplots. We found that the median in CSF was 61.8 ms and that there was no overlap in ranges between either the GM or WM groups. Strong differentiation between CSF and GM/WM was corroborated statistically by WRST (p = 0.0079). The median for GM was 44.1 ms and the median for WM was 39.5 ms. The lower adjacent value in GM (40.4 ms) was positioned below the third quartile of WM with the upper adjacent value in WM (43 ms) positioned above the first quartile of GM. Despite the overlap in these regions, a statistically significant difference between in GM and WM was observed (p = 0.0476).
Figure 7. Boxplots of the mean , , and SD in CSF, GM and WM measured in the brain of 5 volunteers.
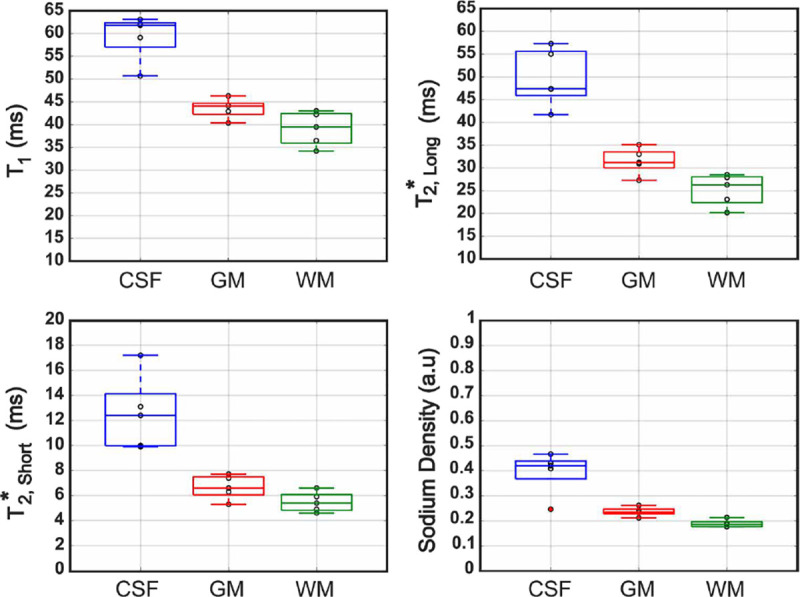
Mean values were measured on the center slice of each of the three planes (see Table 2). Data points for individual volunteers are indicated with black circles, and outliers are marked in red. The lines inside each box represent the median values. Abbreviations: GM, grey matter; WM, white matter; CSF, cerebrospinal fluid.
For in CSF, the median value was 47.4 ms. Again, there was no overlap of for CSF with in either GM or WM. The differences were statistically significant (p = 0.0079) between GM and CSF, and between WM and CSF. The median in GM was 31.2 ms with a lower adjacent value of 27.3 ms, which fell below the third quartile of WM. The median in WM was 26.3 ms. WRST comparison between in GM and WM indicated a statistically significant difference (p = 0.0317).
Median in CSF, GM and WM were 12.4 ms, 6.6 ms and 5.4 ms, respectively. As was the case in and , the range of shown for the boxplot of CSF in Figure 7 did not align within the ranges of in GM or WM. Similarly to statistical results results for and , in CSF was statistically significantly different from in both GM and WM (p = 0.0079). There was no statistically significant difference in between GM and WM (p = 0.1111). This was corroborated by the respective boxplots in Figure 7 where we noted that the upper adjacent of WM was equal to the median of GM (6.6 ms).
For normalized SD in CSF, we found a maximum of 0.467, a median of 0.420 and a minimum of 0.0246, which was considered an outlier. The lower adjacent of CSF was 0.368 and did not overlap with any data grouped for either GM or WM. The minimum value recorded for GM was 0.212, which was nearly equivalent to the maximum of WM (0.213). A statistically significant difference was observed for SD between GM and WM (p = 0.0159).
Table 3 provides a comparison between the relaxation times obtained using our 23Na MRF method and those reported in the literature at 7 T. For in CSF measured using our proposed 23Na MRF, the mean value (59.4 ± 5.1 ms) was comparable to the mean reported by Kratzer et al. (19) (61.9 ± 2.8 ms) using their 23Na MRF method. values for CSF were reported throughout the literature as monoexponential fits with values ranging from 46.3 ± 6.3 ms (19) to 57.2 ± 6.6 ms (23). This range indicated that our measured value of 49.7 ± 6.3 ms for was in good agreement with the literature. Our mean measurement of 12.5 ± 3.0 ms for , however, was out of range compared to these same values.
Table 3. Comparison of the relaxation times measured in brain at 7 T between the correlation-weighted (CW)23Na MRF method and values from the literature.
Relaxation times values were compared in cerebrospinal fluid (CSF), grey matter (GM) and white matter (WM). “Brain Tissue” was included where a distinction was not made between GM and WM.
| CSF |
GM |
WM |
Brain Tissue |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Method | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) | (ms) |
|
| |||||||||||||
| Kratzer et al. (19) | 23Na MRF | 61.9 ± 2.8 | 46.3 ± 4.5 | 35.0 ± 3.2 | 29.3 ± 3.8 | 5.5 ± 1.3 | |||||||
| Lommen et al. (42) | DA-3DPR | 53.6 ± 6.9 | 35.7 ± 2.4 | 5.1 ± 0.8 | |||||||||
| DA-3DPR | 54.4 ± 5.7 | 34.4 ± 1.5 | 4.2 ± 0.4 | ||||||||||
| Ridley (39) | DA-3DPR | 33.9 ± 5.9 | 5.0 ± 0.9 | 34.0 ± 5.2 | 4.5 ± 0.6 | ||||||||
| Blunck et al. (23) | 3D-MERINA | 57.2 ± 6.6 | 25.9 ± 8.3 | 2.0 ± 2.1 | 22.4 ± 7.8 | 2.0 ± 2.1 | |||||||
| Niesporek (22) | DA-3DPR | 46.9 ± 2.1 | 36.4 ± 3.1 | 5.4 ± 0.2 | 23.3 ± 2.6 | 3.5 ± 0.1 | |||||||
| Nagel et al. (21) | DA-3DPR | 56.0 ± 4.0 | |||||||||||
| Fleysher et al. (43) | GRE | 54.0 ± 4.0 | 28.0 ± 2.0 | 29.0 ± 2.0 | |||||||||
|
| |||||||||||||
| Our work | CW 23Na MRF | 59.4 ± 5.1 | 49.7 ± 6.3 | 12.5 ± 3.0 | 43.6 ± 2.1 | 31.5 ± 2.9 | 6.7 ± 1.0 | 39.1 ± 3.7 | 25.2 ± 3.5 | 5.5 ± 0.8 | |||
We did not find values for specific to GM and WM during our literature search (which we limited to data recorded at 7 T), so we compared our results to the for unspecified brain tissue determined by Kratzer et al. (19). In this case, our measurement for WM was in good agreement with their value for brain tissue, but our value for GM was ~11 ms longer.
For in GM, our measured value of 31.5 ± 2.9 ms was in agreement with previous reports (22, 23, 39, 43), while our measured value of 6.7 ± 1.0 ms for was similar to that found by Ridley et al. (39), but slightly longer than other reports (22, 23).
For in WM, we calculated a mean of 25.2 ± 3.5 ms, which was within the range of 40.0 ± 5.2 ms (39) to 22.4 ± 7.8 ms (23) reported for in the literature. Our WM value of 5.5 ± 0.8 ms was within range of two sources (39, 42) and slightly longer than the other three references (22, 23, 42). Finally, both the and values that we measured in GM and WM were in good agreement with values reported for brain tissue (19).
Correlation coefficient weighting
Supplementary Figure S5 shows the maps of the 7-compartment phantom produced after matching was performed using different numbers of correlation coefficients as weighting factors, from the maximum correlation only (k = 1) through k = 1000.
Supplementary Figure S6 shows the maps for volunteer 5 representing brain results produced after matching was performed using different numbers of correlation coefficients as weighting factors, from the maximum correlation only (k = 1) through k = 1000.
Supplementary Figure S7 summarizes the results of matching in the 7-compartment phantom with different k values against the RM in a series of boxplots. When k = 20, there was more overlap between the interquartile regions of RM and as compared to the single maximum valued correlation, and the most overlap in interquartile regions for all ROIs in . Although STDV was reduced for as the number of correlation coefficients increases, there was loss of overlap between RM and 23Na MRF for in ROIs 6 and 7, respectively. Overall, did not change significantly between k = 1 and k = 200.
Supplementary Figure S8 shows a series of graphs for the relaxation times measured in each ROI of the 7-compartment phantom plotting the maximum number of matches made to a subset of pixels within a reference range limited by values from the RM. Examining the trends in the graphs for the phantom, we noted that the plot most often changed direction in from a high number of maximum correlations per pixel match to a global decrease followed by leveling off in the region between k = 10 to k = 50. Direction changes were also noted in graphs in the neighborhood of k = 100.
Supplementary Figure S9 shows similar graphs for in CSF, GM and WM and in GM and WM for each of the 5 volunteers. The maximum number of matches made to a subset of pixels within each ROI were limited by ranges defined by literature values taken from Table 3. While graphs of do not show any notable trend, there are direction changes in the graphs of in the neighborhood of k = 20 for CSF, GM and WM.
4. Discussion
In this work we demonstrated a refined approach to quantitative mapping of , , and SD using 23Na MRF with correlation coefficient weighting. We constructed a comprehensive dictionary that included combinations of , , , factor and . We implemented a 3D FLORET sequence with an optimized 23-pulse variable FA/PA MRF train capable of full brain coverage in about 30 min without varying TE or delays between the pulses in the 23Na MRF pulse train. While the ISTO spin system simulation and PC matching procedure were based on our previous work (20), this study integrated a new 23Na MRF pulse train optimization protocol and refined matching criteria. Our method was tested in a 7-compartment phantom, and successfully applied for brain mapping in five healthy volunteers at 7 T.
Because and were included as dictionary parameters, the spatial influences of transmit inhomogeneity and frequency shift on relaxation times were partially accounted for in the matching process. RF field variations were most pronounced in the phantom data (Figure 2), due to its high average relative permittivity. On the other hand, variations were more pronounced in vivo, due to air-filled structures such as the ear canal and maxillary sinus cavities (Figure 6). Some of the artifacts noted in the relaxation maps could be lined up with similar artifacts in maps of and . This indicated that broadening the ranges for and in the dictionary, or reducing the step size for these entries, might improve the appearance of these artifacts in the parameter maps.
The low resolution used to offset the sodium SNR deficits makes precise measurements in isolated tissues difficult. Even at the current resolution, low SNR remains a hurdle. Speckle noise can be seen in some of the maps in Figures 5 and 6, that overlap in regions where changes in factor or were apparent, corresponding to areas of low SNR. We first tried to overcome some of these caveats by denoising the images prior to matching. While denoising did offer subtle improvement in image quality by removing some pixels with outlying intensity, the image quality was not enhanced enough to make a difference in the matching process.
Matching with PC was performed voxelwise between the data and the dictionary. We ordered the signal matches for each individual voxel according to their respective correlation coefficients and then produced maps reflecting data that included a subset of matches for each voxel. We later refined this technique by using the correlation coefficient value per voxel signal match as a weighting factor. While the time required for matching and map reconstruction was longer than the fitting times of the RM for , and , the 23Na MRF method has the benefit of mapping not only , and but also and SD in a single acquisition. Furthermore, the combined scan time for RM based on the scan time in the phantom, would be more than 2 h versus a scan time of about 30 min for brain 23Na MRF.
Changes in the maps of correlation coefficient value averaged over increasing numbers of coefficients were minor. This was because the differences between the maximum value and an average of some subset of coefficients for any single voxel were in the thousandths. Despite such minor changes in value, each correlation represents a potential match between the dictionary and data. We acknowledge that examining correlation in non-convex space and within a noisy environment opens the possibility that some higher valued correlations were calculated for signals representing local minima as opposed to a “true” match. Our choice to include multiple matches based on correlation coeffcient weighting increased the probability that some match resulted from a “true” match as opposed to a local minimum. It may be possible to further improve matching by accounting for non-convexity in the reconstruction process (45–47).
In the phantom, we weighted the data with 20 coefficients. This reduced the slight inhomogeneity in and and provided the best agreement between 23Na MRF and the RM data. While weighting beyond 20 coefficients continued to slightly smooth artifacts and improve the agreement between 23Na MRF and RM in , the agreement to RM in began to diverge in ROIs 6 and 7 at k = 50. There was no significant change in values beyond the maximum correlation for . Similarly, for SD, an improved overlap in the boxplots between 23Na MRF and RM is seen at k = 20 with no change beyond this value.
Matching in vivo was also completed using weighting with k = 20 correlation coefficients. In this case, increasing the number of coefficients introduced signals with lower value correlation coefficients for CSF. Interestingly, this had the effect of increasing the values of within CSF while slightly decreasing and . This was combined with an overall increase in the of brain tissue and decrease in SD. Essentially, weighting signals by their correlation coefficients operated as a smoothing kernel by including a range of highly similar fingerprints. We compared the smoothing effect to that of applying a Gaussian filter prior to matching which is shown in Supplementary Figure S10. Comparison of parameter maps at the maximum correlation with and without the addition of the Gaussian filter demonstrated no effective improvement in the visual appearance for the brain data. Furthermore, this outcome did not change when additional correlations were included.
Visually, we could not differentiate between GM and WM in the brain maps of the five volunteers. However, our statistical analysis indicated that the mean and were different enough to distinguish between GM and WM. As listed in Table 3, and were reported in several studies where a wide range between the shortest and longest and was noted amongst different sources. While different fitting techniques or data acquisition schemes could be the culprit, there may also be variations in relaxation time within GM and WM. Some recent works suggest that regional differences in structural composition within GM and WM (39, 48) are contributing factors, or that local changes are induced within the cellular environment of the respective tissues (3). These considerations, combined with the inherently low SNR of sodium, make distinguishing GM and WM through relaxation mapping difficult.
The ISTO simulation that we used to model relaxation included dictionary ranges from 10 to 66 ms for and from 0.5 to 66 ms for . We hypothesized that average values for and CSF would be equal or very close to each other. However, the mean was significantly shorter than in our measurements in CSF. Similar issues with fitting in CSF have been mentioned in the literature (23, 49) where the difficulty arose from using discrete biexponential or monoexponential models. Fingerprint matching, however, operates across a pseudo-continuum and is based on the dynamics of the spin system. Theoretically, this would circumvent some of the shortcomings associated with fitting discrete models. The concentration of higher value in CSF in the center in the central ventricle indicated that partial volume effects from surrounding tissue may have contributed to the low . Eroding the tissue masks did not significantly change the distribution of in CSF, however, any contamination of the ROI could have resulted in a reduced apparent .
It is also worth noting that our 23Na MRF method, and the ISTO simulation that we used to generate the fingerprint dictionary, do not make any assumption about either the compartmental homogeneity within the voxel, nor the ratios between relaxation processes for the central and satellite transitions of the 23Na spins (i.e. the long and short relaxation components). Both 23Na MRF and the ISTO simulation simply estimate average relaxation times in each voxel, which is very likely a volume weighted average of multiple intra-voxel compartments (extracellular and intracellular spaces from multiple cell types) and local magnetic field inhomogeneities. Consequently, there is very little chance to measure the ideal ratio of 0.6:0.4 for due to quadrupolar relaxation processes. Indeed, if we were to specifically measure the ratio of with a fitting method (which MRF is not), the ratio of 0.6:0.4 would occur only in a perfectly ideal model where relaxation is purely quadrupolar. This is usually not the case in real samples, including gels, fluids and biological tissues. In reality, dipolar coupling, residual quadrupolar interaction (due to local structural anisotropies), local field inhomogeneities (chemical shift interaction) of various nature, and even spin-orbit interaction where the spin interacts with the magnetic fields generated by the rotational motion of the ion itself, can occur and influence the relaxation process of the 23Na spins. Such influences effectively act as pertubations to the main quadrupolar relaxation process, and thus generate 23Na relaxation times that deviate from the ideal case of pure quadrupolar relaxation (50–52).
5. Conclusion
In conclusion, we demonstrated an implementation of 23Na MRF from a 3D 23-pulse FLORET acquisition that enabled quantitative mapping of , , , , factor and in about 30 min over the whole brain with 5-mm isotropic resolution at 7 T. Furthermore, we introduced correlation coefficient weighting in the data reconstruction to enhance the robustness of the method and smooth the final maps.
The proposed 23Na MRF method could finally be combined with 1H MRF based on our previous work on simultaneous acquisition of 1H MRF and 23Na MRI (27, 28, 53) to generate a fully simultaneous 1H/23Na MRF technique (54).
Supplementary Material
Acknowledgements
This work was supported by the NIH/NIBIB grant R01EB026456 and performed under the rubric of the Center for Advanced Imaging Innovation and Research, a NIBIB Biomedical Technology Resource Center (P41EB017183). The authors would like to thank research coordinators Nahbila-Malikha Kumbella and Liz Aguilera, MPH, for volunteer outreach. Some computational requirements for this work were supported by the NYU Langone High Performance Computing (HPC) Core’s resources and personnel.
Footnotes
Supplementary material
Supporting information is available in the Supplementary Material file.
Data availability
Data analyzed in this study is available on GitHub: https://github.com/LaurenFODonnell/Sodium_MRF_Datasets.git. The Matlab code for sodium NMR dynamics simulation used for generating the fingerprint dictionary is available on Matlab File Exchange: https://www.mathworks.com/matlabcentral/fileexchange/67472-simulation-of-sodium-nmr.
References
- 1.Madelin G. & Regatte R. R. Biomedical applications of sodium MRI in vivo. Journal of Magnetic Resonance Imaging 38, 511–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madelin G., Lee J.-S., Regatte R. R. & Jerschow A. Sodium MRI: Methods and applications. Progress in Nuclear Magnetic Resonance Spectroscopy 79, 14–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petracca M., Fleysher L., Oesingmann N. & Inglese M. Sodium MRI of multiple sclerosis. NMR in Biomedicine 29, 153–161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krahe J. et al. Increased brain tissue sodium concentration in Friedreich ataxia: A multimodal MR imaging study. NeuroImage: Clinical 34, 103025 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellon E. et al. Sodium MR Imaging Detection of Mild Alzheimer Disease: Preliminary Study. American Journal of Neuroradiology 30, 978–984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohamed S. et al. Evaluation of Sodium (23Na) MR-imaging as a Biomarker and Predictor for Neurodegenerative Changes in Patients With Alzheimer’s Disease. In Vivo 35, 429–435 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haeger A. et al. What can 7T sodium MRI tell us about cellular energy depletion and neurotransmission in Alzheimer’s disease? Alzheimer’s & Dementia 17, 1843–1854 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto T. et al. In vivo sodium-23 MRI in brain tumors: evaluation of preliminary clinical experience. American journal of physiologic imaging 6, 74—80 (1991). [PubMed] [Google Scholar]
- 9.Nunes Neto L. P. et al. Quantitative sodium imaging and gliomas: a feasibility study. Neuroradiology 60, 795–802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boada F. et al. Sodium MRI and the Assessment of Irreversible Tissue Damage During Hyper-Acute Stroke. Translational Stroke Research 3, 236–245 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Gerhalter T. et al. Global decrease in brain sodium concentration after mild traumatic brain injury. Brain Communications 3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.-S., Regatte R. R. & Jerschow A. Optimal excitation of 23Na nuclear spins in the presence of residual quadrupolar coupling and quadrupolar relaxation. The Journal of Chemical Physics 131, 174501 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Ma D. et al. Magnetic resonance fingerprinting. Nature 495, 187–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloos M. A. et al. Multiparametric imaging with heterogeneous radiofrequency fields. Nature Communications 7, 12445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panda A. et al. Magnetic resonance fingerprinting – An overview. Current Opinion in Biomedical Engineering 3, 56–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bipin Mehta B. et al. Magnetic resonance fingerprinting: a technical review. Magnetic Resonance in Medicine 81, 25–46 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Hsieh J. J. L. & Svalbe I. Magnetic resonance fingerprinting: from evolution to clinical applications. Journal of Medical Radiation Sciences 67, 333–344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kratzer F. J. et al. Sodium relaxometry using 23Na MR fingerprinting: A proof of concept. Magnetic Resonance in Medicine 84, 2577–2591 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Kratzer F. J. et al. 3D sodium (23Na) magnetic resonance fingerprinting for time-efficient relaxometric mapping. Magnetic Resonance in Medicine 86, 2412–2425 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Gilles A., Nagel A. M. & Madelin G. Multipulse sodium magnetic resonance imaging for multicompartment quantification: Proof-of-concept. Scientific Reports 7, 17435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagel A. M. et al. The Potential of Relaxation-Weighted Sodium Magnetic Resonance Imaging as Demonstrated on Brain Tumors. Investigative Radiology 46 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Niesporek S. C. et al. Improved determination in 23Na, 35Cl, and 17O MRI using iterative partial volume correction based on 1H MRI segmentation. Magnetic Resonance Materials in Physics, Biology and Medicine 30, 519–536 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Blunck Y. et al. 3D-multi-echo radial imaging of 23Na (3D-MERINA) for time-efficient multi-parameter tissue compartment mapping. Magnetic Resonance in Medicine 79, 1950–1961 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Pipe J. et al. A new design and rationale for 3D orthogonally oversampled k-space trajectories. Magnetic Resonance in Medicine 66, 1303–1311 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Levitt M. H. Composite pulses. Progress in Nuclear Magnetic Resonance Spectroscopy 18, 61–122 (1986). [Google Scholar]
- 26.Robison R. K., Anderson III A. G. & Pipe J. G. Three-dimensional ultrashort echo-time imaging using a FLORET trajectory. Magnetic Resonance in Medicine 78, 1038–1049 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Yu Z., Madelin G., Sodickson D. K. & Cloos M. A. Simultaneous proton magnetic resonance fingerprinting and sodium MRI. Magnetic Resonance in Medicine 83, 2232–2242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez G. G. et al. Repeatability of simultaneous 3D 1H MRF/23Na MRI in brain at 7 T. Scientific Reports 12, 14156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saidi R., Bouaguel W. & Essoussi N. Hybrid Feature Selection Method Based on the Genetic Algorithm and Pearson Correlation Coefficient, 3–24 (Springer International Publishing, Cham, 2019). [Google Scholar]
- 30.Coste A. et al. Tissue sodium concentration and sodium T1 mapping of the human brain at 3T using a Variable Flip Angle method. Magnetic Resonance Imaging 58, 116–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B. et al. A radially interleaved sodium and proton coil array for brain MRI at 7 T. NMR in Biomedicine 34, e4608 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pipe J. G. & Menon P. Sampling density compensation in MRI: Rationale and an iterative numerical solution. Magnetic Resonance in Medicine 41, 179–186 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Zwart N. R., Johnson K. O. & Pipe J. G. Efficient sample density estimation by combining gridding and an optimized kernel. Magnetic Resonance in Medicine 67, 701–710 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Bydder M., Larkman D. & Hajnal J. Combination of signals from array coils using image-based estimation of coil sensitivity profiles. Magnetic Resonance in Medicine 47, 539–548 (200). [DOI] [PubMed] [Google Scholar]
- 35.Veraart J. et al. Denoising of diffusion MRI using random matrix theory. NeuroImage 142, 394–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemberskiy G. et al. Achieving sub-mm clinical diffusion MRI resolution by removing noise during reconstruction using random matrix theory. In Proceedings of the Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM) (Montreal, Canada, 2019). [Google Scholar]
- 37.Lemberskiy G. et al. MRI below the noise floor. In Proceedings of the Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM) (Melbourne, Australia, 2020). [Google Scholar]
- 38.Gast L. V., Platt T., Nagel A. M. & Gerhalter T. Recent technical developments and clinical research applications of sodium (23Na) MRI. Progress in Nuclear Magnetic Resonance Spectroscopy 138–139, 1–51 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Ridley B. et al. Distribution of brain sodium long and short relaxation times and concentrations: a multi-echo ultra-high field 23Na MRI study. Scientific Reports 8, 4357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian Y., Zhao T., Zheng H., Weimer J. & Boada F. E. High-resolution sodium imaging of human brain at 7 T. Magnetic Resonance in Medicine 68, 227–233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.SPM12. Statistical Parametric Mapping (University College London, The Wellcome Centre for Human Neuroimaging, UCL Queen Square Institute of Neurology, London, UK, 2012–2020). [Google Scholar]
- 42.Lommen J. M. et al. Probing the microscopic environment of 23Na ions in brain tissue by MRI: On the accuracy of different sampling schemes for the determination of rapid, biexponential decay at low signal-to-noise ratio. Magnetic Resonance in Medicine 80, 571–584 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Fleysher L., Oesingmann N., Stoeckel B., Grossman R. I. & Inglese M. Sodium long-component mapping in human brain at 7 Tesla. Magnetic Resonance in Medicine 62, 1338–1341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riffenburgh R. H. Chapter 13 - Finding Probabilities of Error. In Riffenburgh R. H. (ed.) Statistics in Medicine (Second Edition), 213–226 (Academic Press, Burlington, 2006), second edition edn. [Google Scholar]
- 45.Haacke E. M., Liang Z.-P. & Boada F. E. Image reconstruction using projection onto convex sets, model constraints, and linear prediction theory for the removal of phase, motion, and Gibbs artifacts in magnetic resonance and ultrasound imaging. Optical Engineering 29, 555–566 (1990). [Google Scholar]
- 46.Zhao B. et al. Improved magnetic resonance fingerprinting reconstruction with low-rank and subspace modeling. Magnetic Resonance in Medicine 79, 933–942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duarte R., Repetti A., Gómez P. A., Davies M. & Wiaux Y. Greedy approximate projection for magnetic resonance fingerprinting with partial volumes. Inverse Problems 36, 035015 (2020). [Google Scholar]
- 48.Kolbe S. C. et al. Microstructural correlates of 23Na relaxation in human brain at 7 Tesla. NeuroImage 211, 116609 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Syeda W., Blunck Y., Kolbe S., Cleary J. O. & Johnston L. A. A continuum of components: Flexible fast fraction mapping in sodium MRI. Magnetic Resonance in Medicine 81, 3854–3864 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Madelin G. X-Nuclei Magnetic Resonance Imaging (Jenny Stanford Publishing, 2022). [Google Scholar]
- 51.Levitt M. H. Spin Dynamics: basics of nuclear magnetic resonance (John Wiley & Sons Ltd, 2008). [Google Scholar]
- 52.Kimmich R. NMR: Tomography, Diffusometry, Relaxometry (Springer Science & Business Media, 2011). [Google Scholar]
- 53.Yu Z., Madelin G., Sodickson D. K. & Cloos M. A. Simultaneous proton magnetic resonance fingerprinting and sodium MRI. Magnetic Resonance in Medicine 83, 2232–2242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez G. G., O’Donnell L., Yu Z., Cloos M. & Madelin G. Multinuclear fingerprinting (MNF): high-resolution simultaneous proton/sodium MR fingerprinting. In Proceedings of the Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM) (Singapore, 2024). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data analyzed in this study is available on GitHub: https://github.com/LaurenFODonnell/Sodium_MRF_Datasets.git. The Matlab code for sodium NMR dynamics simulation used for generating the fingerprint dictionary is available on Matlab File Exchange: https://www.mathworks.com/matlabcentral/fileexchange/67472-simulation-of-sodium-nmr.