Abstract
Methanol, as a non-edible feedstock, offers a promising sustainable alternative to sugar-based substrates in biochemical production. Despite progress in engineering methanol assimilation in nonmethylotrophs, the full transformation into methanol-dependent synthetic methylotrophs remains a formidable challenge. Here, moving beyond the conventional rational design principle, we engineered a synthetic methylotrophic Saccharomyces cerevisiae through genome rearrangement and adaptive laboratory evolution. This evolutionarily advanced strain unexpectedly shed the heterologous methanol assimilation pathway and demonstrated the robust growth on sole methanol. We discovered that the evolved strain likely realized methanol assimilation through a previously unidentified Adh2-Sfa1-rGly (ASrG) pathway, facilitating the concurrent assimilation of formate and CO2. Furthermore, the incorporation of electron transfer material C3N4 quantum dots obviously enhanced methanol-dependent growth, emphasizing the role of energy availability in the ASrG pathway. This breakthrough introduces a previously unidentified C1 utilization pathway and highlights the exceptional adaptability and self-evolving capacity of the S. cerevisiae metabolic network.
Evolutionary engineering transforms S. cerevisiae to grow on sole methanol with a previously unidentified C1 assimilation pathway.
INTRODUCTION
Renewable one-carbon (C1) substrates, particularly methanol and formate, present an appealing alternative to traditional sugar-based substances for biochemical production (1, 2). As methanol and formate can be efficiently derived from CO2 through advanced electrocatalysis methods, their utilization in microbial fermentation heralds a future of sustainable and carbon-neutral biochemical production (3, 4). However, the inherent complexity of methanol and formate metabolism in native microbes often limits the genetic malleability, thereby restricting their broader application in industrial processes. In recent years, there has been a burgeoning interest in endowing model microorganisms with methanol assimilation capabilities (5–8). Although great strides have been made, these advancements typically necessitate intricate and labor-intensive metabolic engineering based on a rational design. Moreover, even after the implementation of rational design, adaptive laboratory evolution (ALE) is frequently used as a critical strategy to achieve methanol-dependent growth (6–8). While this evolutionary approach is time-consuming (9, 10), it offers the possibility of converting microorganisms into efficient methylotrophs solely through evolutionary processes.
Synthetic chromosome rearrangement and modification by LoxP-mediated evolution (SCRaMbLE) is a revolutionary genome recombination technology originated from the Synthetic Yeast Project (Sc2.0) (11, 12). By enabling the rearrangement of synthetic chromosomes through Cre recombinase, SCRaMbLE can introduce a spectrum of mutations—deletions, duplications, inversions, and translocations—that can markedly alter microbe’s metabolic capabilities (13). This technology has not only facilitated the enhancement of chemical production (14–16) but also increased microbial resistance against various stress factors (17, 18). Particularly in heterozygous diploids, SCRaMbLE has demonstrated a heightened survival rate, making it a valuable tool in the development of robust synthetic organisms (16, 18).
Saccharomyces cerevisiae, known for its robustness and versatility, has proven to be an excellent chassis for constructing synthetic methylotrophs. Its ability to compartmentalize toxic intermediates and metabolic reactions provides a distinct advantage over prokaryotic systems (2, 19). A recent study has demonstrated the transformation of S. cerevisiae into a synthetic methylotroph, yet it relied on the intervention of complexed modular metabolic pathways, as well as long-term ALE (6). Actually, S. cerevisiae has been suggested to have methanol assimilation capabilities, primarily through its alcohol dehydrogenase 2 (Adh2) for methanol oxidation (20, 21). Also, endogenous components of the reductive glycine (rGly) pathway have been identified, allowing successful formate assimilation in S. cerevisiae (22, 23).
In this study, we departed from the traditional rational design to develop a synthetic methylotrophic S. cerevisiae through SCRaMbLE-mediated genome rearrangements combined with ALE. This unconventional approach allowed us to harness the inherent metabolic flexibility of S. cerevisiae, leading to the evolution of mutant strains capable of effectively oxidizing methanol without relying on any heterologous C1 compound metabolic pathway. Notably, the evolved strain primarily uses an Adh2-Sfa1-rGly (ASrG) pathway for methanol assimilation, which also enables the utilization of formate and NaHCO3. These findings substantially advance our understanding of the S. cerevisiae metabolic network’s potential, highlighting its adaptability and capacity for synthetic methylotrophy engineering.
RESULTS
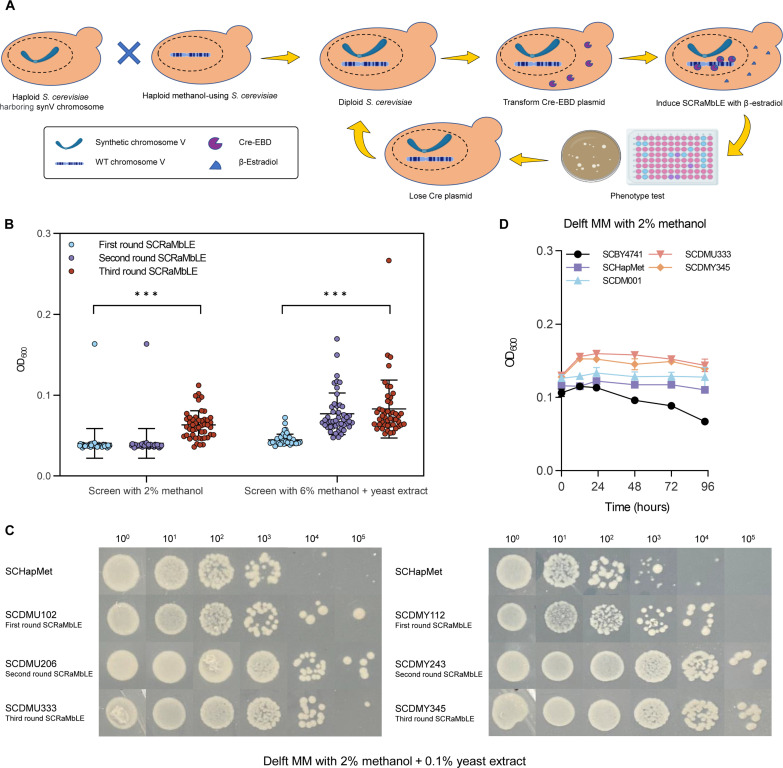
Enhancing methanol utilization in diploid methanol-using S. cerevisiae SCDM001 via SCRaMbLE
To construct methylotrophs through SCRaMbLE, we first engineered a diploid methanol-using S. cerevisiae strain, SCDM001. This diploid was created by mating a haploid strain embedded with a synthetic chromosome V (synV) with another haploid strain of SCHapMet carrying the methanol assimilation pathway derived from Pichia pastoris (24). The resulting heterozygous diploid of SCDM001 carrying distinct syn-PCRTags along with the methanol assimilation genes (fig. S1) displayed a marked increase in cell size compared to its parental haploid of SCHapMet (fig. S2). Similar to SCHapMet, SCDM001 was able to survive in 2% methanol for 96 hours, although no real growth was observed (fig. S3).
To enhance the methanol utilization capability of SCDM001, strains were subjected to successive rounds of the SCRaMbLE process. Each iteration involved cultivating the yeast in Delft minimal medium (MM) containing 2% methanol and an alternative medium composed of 6% methanol and 0.1% yeast extract to select strains with superior phenotypes (Fig. 1A). Across three iterative SCRaMbLE rounds, strains consistently demonstrated apparent improvements in growth on methanol medium (Fig. 1B). By screening the best-performing strains in each SCRaMbLE round (namely SCDMU102, SCDMY112, SCDMU206, SCDMY243, SCDMU333, and SCDMY345), we illustrated a consistent trend of enhanced methanol utilization capacity, as evidenced by serial dilution assays (Fig. 1C). Despite these advancements, even the most robust SCRaMbLEd strains, SCDMU333 and SCDMY345, showed only transient growth during the initial 12 hours (with a μ of 0.01 hour−1) and were ultimately unable to sustain prolonged growth exclusively on methanol over a prolonged period (Fig. 1D). Serial dilution assays further confirmed that none of the diploids could maintain apparent growth solely on methanol. An elevated methanol concentration of 4% did not inhibit microbial growth, suggesting an increased resilience of diploid strains to methanol toxicity (fig. S4).
Fig. 1. Screening of synthetic methylotrophic S. cerevisiae by SCRaMbLE.
(A) Workflow for screening synthetic methylotrophs by SCRaMbLE. (B) Screening statistics of three rounds of SCRaMbLE. Error bars shown are mean and SD from the 47 samples in each round SCRaMbLE. Statistical analysis was performed using a two-tailed student’s t test (***P < 0.001). (C) Serial dilution assay comparing the growth of an independent SCRaMbLEd isolate from each SCRaMbLE round with the original haploid methanol-using strain SCHapMet. SCDMU102, SCDMU206, and SCDMU333 are isolates screened on 2% methanol after one, two, and three rounds of SCRaMbLE, respectively. SCDMY112, SCDMY243, and SCDMY345 are isolates screened on 6% methanol + yeast extract after one, two, and three rounds of SCRaMbLE, respectively. (D) Growth curves of the wild-type (WT) S. cerevisiae (SCBY4741), haploid methanol-using strain (SCHapMet), diploid methanol-using strain (SCDM001), and SCRaMbLEd strain (SCDMU333 and SCDMY345) in Delft MM with 2% methanol. All data are presented as mean ± SD. Triplicate independent samples were adopted for each panel. For strains except SCBY4741, uracil (20 mg/liter) was supplemented in Delft MM to compensate for uracil auxotroph. For strain SCBY4741, uracil (20 mg/liter), histidine (20 mg/liter), leucine (100 mg/liter), and methionine (20 mg/liter) were supplemented in Delft MM.
Converting S. cerevisiae into a synthetic methylotroph capable of growing with sole methanol via ALE
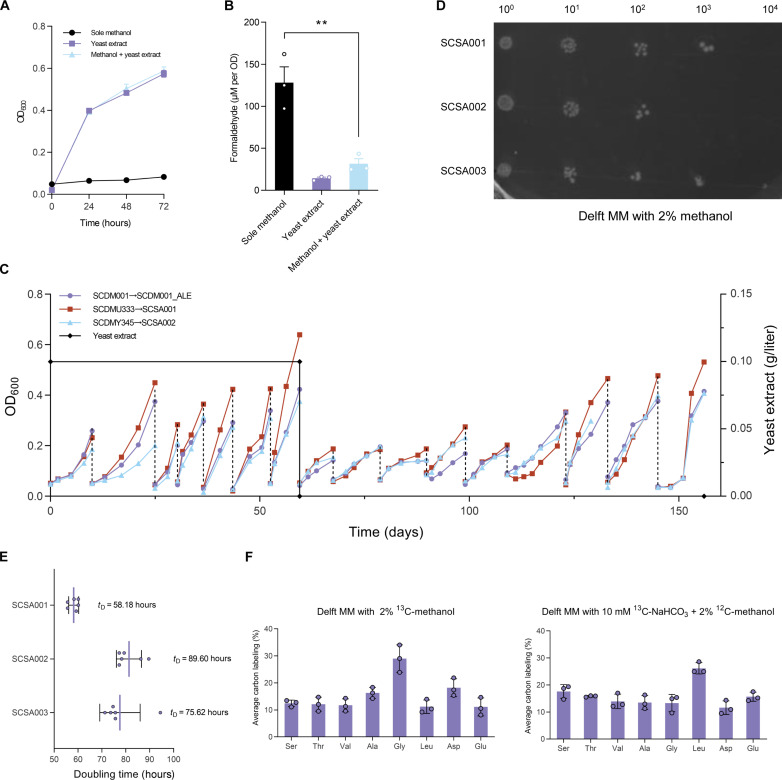
In terms of methanol assimilation, formaldehyde detoxification stands out as a critical bottleneck for both engineered and natural methylotrophs (1, 25). A prior study has demonstrated that formaldehyde-induced DNA-protein cross-linking can prevent Escherichia coli from growing on sole methanol (7). In line with these findings, we hypothesized that the accumulation of formaldehyde would similarly hinder the growth of our diploid strains on methanol. In the diploid strain SCDMU333, we observed an apparent increase in formaldehyde concentration, reaching up to 128 μM per optical density (OD) when grown on sole methanol (Fig. 2, A and B). However, the addition of mere yeast extract (0.1 g/liter) to the medium remarkably reduced formaldehyde accumulation (Fig. 2B), suggesting an ameliorative effect on methanol toxicity and potential implications for the methanol boost effect of yeast extract (26, 27). Besides, no notable differences in growth were observed between yeast extract alone and yeast extract + methanol, indicating the limited contribution of methanol to strain growth.
Fig. 2. Isolation and characterization of a of synthetic methylotrophic S. cerevisiae SCSA001 capable of using methanol as the sole carbon source via ALE.
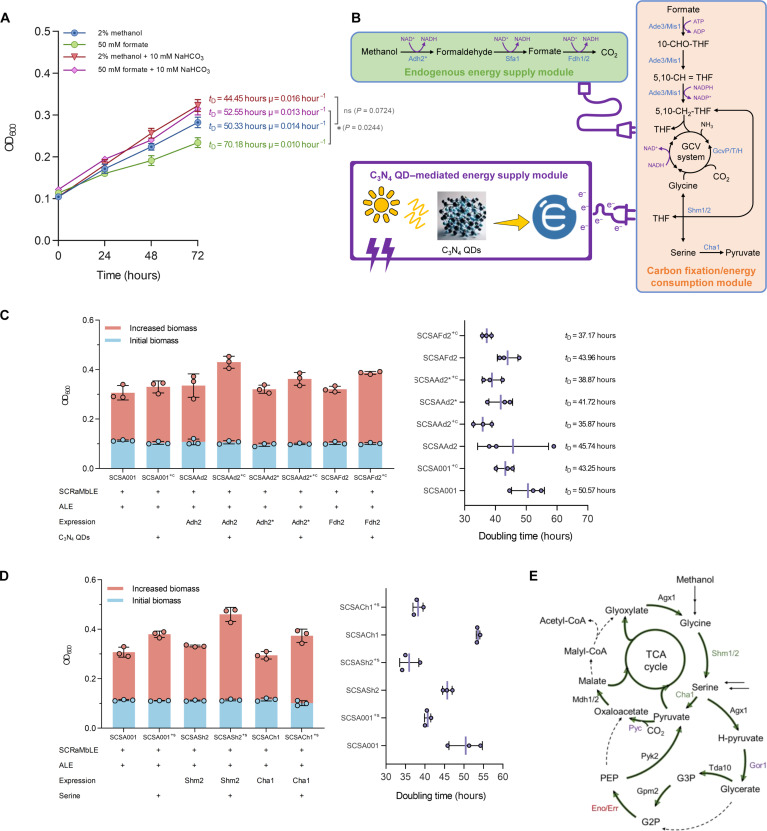
(A and B) Growth curves and formaldehyde accumulation of SCDMU333 in Delft MM added with sole methanol, yeast extract, and methanol + yeast extract [uracil (20 mg/liter) was supplemented to compensate for uracil auxotroph]. (C) The evolution trajectory of SCDM001_ALE, SCDMU333_ALE, and SCDMY345_ALE. The black solid line represents the yeast extract percentage in the medium. During ALE process, uracil (20 mg/liter) was supplemented to compensate for uracil auxotroph. (D) Serial dilution assay comparing the growth of SCSA001, SCSA002, and SCSA003 in Delft MM with 2% methanol. (E) Calculated doubling time of the three evolved strain. Error bars shown are mean and SD from six technical replicates. (F) The average carbon labeling of intracellular amino acids of SCSA001 in Delft MM supplemented with 13C-labeled methanol or 10 mM 13C-labeled NaHCO3 + 2% 12C-methanol. Strain SCSA001 was precultured three times in Delft MM with 13C-labeled methanol before being cultured for in vivo 13C-methanol metabolic tracer assays. All data are presented as mean ± SD. Triplicate independent samples were adopted for each panel except for the doubling time calculations.
To obtain strains capable of growing on sole methanol, we initiated a yeast extract–assisted ALE journey with strains SCDMU333, SCDMY345, and SCDM001. The evolution process began in Delft MM [uracil (20 mg/liter) added] supplemented with 2% methanol and yeast extract (0.1 g/liter). After seven generations, yeast extract was removed to increase selective pressure (Fig. 2C). Following ALE, the evolved strains SCDMU333_ALE, SCDMY345_ALE, and SCDM001_ALE were transformed with an empty pRS426 plasmid to compensate for uracil auxotrophy, generating strains SCSA001, SCSA002, and SCSA003, respectively (fig. S5). Fortunately, all evolved strains exhibited apparent growth improvements and were able to use methanol as the sole carbon source within a relatively short ALE period of less than 160 days (Fig. 2D). To verify the stability and robustness of these evolved phenotypes, six independent replications were conducted for each strain in Delft MM with 2% methanol. The best-performing strain, SCSA001, demonstrated a remarkable reduction in doubling time to 58.18 hours (Fig. 2E). Even the non-SCRaMbLEd diploid SCSA003 displayed apparent growth on sole methanol, with a doubling time of 75.62 hours, surpassing that of SCSA002 (89.60 hours). This finding demonstrated the potential for methanol assimilation in diploids, even in the absence of extensive chromosomal rearrangements induced by SCRaMbLE. To investigate the detailed growth performance, strain SCSA001 was cultivated in Delft MM containing 2% methanol over 240 hours. With continuous methanol supplementation, strain SCSA001 maintained continuous growth for 120 hours following a 40-hour lag phase, reaching a final OD600 of 0.547 (fig. S6). Notably, the specific growth rate (μ) remained stable at a high level between 48 and 72 hours, reaching a peak (μmax) of 0.0153 hour−1 at 56 hours.
To further evaluate the methanol uptake and incorporation, we performed in vivo 13C-methanol metabolic tracer assays on the evolved strain SCSA001. While all amino acids were notably 13C-labeled, the labeled fraction was unexpectedly lower than expected (fig. S7A), with glycine showing the highest 13C enrichment at 29.00% and glutamic acid the lowest at 11.11% (Fig. 2F). Given that methanol was the sole added carbon source, we hypothesized that CO2 might also infiltrate the central metabolism, contributing to the microbial growth. To test this, reverse labeling using 13C-NaHCO3 and 2% 12C-methanol as cosubstrates was used. As anticipated, the addition of 13C-NaHCO3 enhanced methanol-dependent growth and led to apparent 13C enrichment in all amino acids (fig. S7B). This compellingly indicates that CO2 was fixed and incorporated into the cells, playing an even more crucial role in supporting methanol-based growth than methanol itself.
Demonstrating an endogenous methanol assimilation pathway via genomic sequencing and physiological characterization
To unravel the genetic underpinnings of methanol assimilation, the whole-genome sequencing (WGS) was carried out for strains SCDM001, SCSA001, and SCSA003. Reference comparisons with diploid SCDM001 illuminated distinct recombination events and mutations in SCSA001 and SCSA003. Notably, the average sequencing depth of chromosome V in SCSA001 highlighted two deletions in the intergenic regions between 400 and 500 kb compared to SCDM001 (fig. S8). Both SCSA001 and SCSA003 exhibited an extensive array of more than 10,000 single-nucleotide polymorphisms (SNPs) and more than 2000 small insertions/deletions. SCSA001, in particular, showed a higher prevalence of copy number variations (CNVs) and structural variations, likely attributed to the SCRaMbLE-mediated genome recombination (fig. S9). These mutations are detailed in data S1 and categorized by Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (figs. S10 to S13), with major enrichments in pathways related to cell growth and death, carbohydrate metabolism, amino acid metabolism, and the metabolism of cofactors and vitamins.
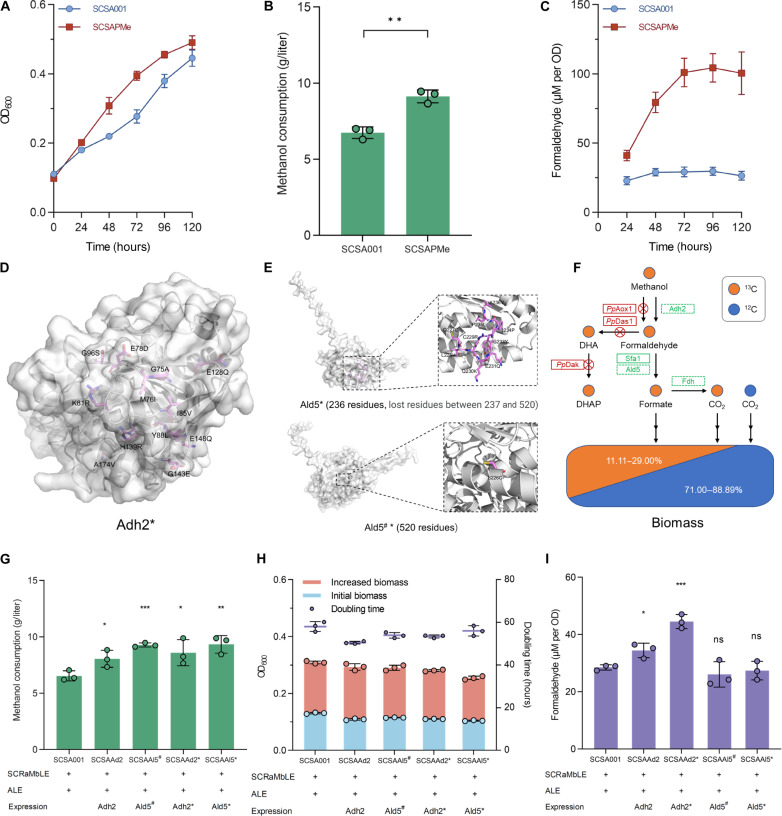
A pivotal revelation from WGS was the complete eradication of the P. pastoris–derived methanol assimilation pathway in ALE-adapted strains SCSA001 and SCSA003 (fig. S14). To evaluate the functionality of this heterologous pathway, the missing pathway was reintroduced into the evolved strain SCDMU333_ALE, generating strain SCSAPMe. Compared to the control strain SCSA001, SCSAPMe exhibited a 10.15% increase in final biomass and a 35.48% increase in methanol consumption (Fig. 3, A and B). This improvement is primarily attributed to the reduced lag phase, which resulted in a faster μ during the first 48 hours (0.0240 hour−1 versus 0.0144 hour−1). In addition, formaldehyde accumulation in SCSAPMe was more than double that observed in SCSA001 (Fig. 3C), suggesting enhanced methanol oxidation due to the heterologous pathway introduction.
Fig. 3. Functional assays of Adh2 and Ald5.
(A to C) Growth curves, methanol consumption, and formaldehyde accumulation of strain SCSA001 and SCSAPMe in Delft MM with 2% methanol. (D and E) Simulated structure of the evolved Adh2 and Ald5 based on AlphaFold2. Mutated residues are additionally annotated. (F) Schematic illustration of the methanol utilization pattern in strain SCSA001. The orange and blue circles represent 13C- and 12C-labeled carbon atoms, respectively. Endogenous enzymes are represented by green boxes, and heterologous enzymes derived from P. pastoris are represented by red boxes. (G to I) Methanol consumption, biomass (OD600) formation, doubling time, and formaldehyde accumulation after 72 hours of cultivation in Delft MM with 2% methanol (growth curves are shown in fig. S15). All strains were precultured in yeast extract, peptone, and dextrose (YPD) medium and washed twice using phosphate-buffered saline (PBS) (pH 7.2) before cultivated in Delft MM. All data represent the mean ± SD. Triplicate independent samples were adopted for each panel. Statistical analysis was performed using a two-tailed Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001). ns, not significant.
Anyhow, reexpression of the heterologous pathway did enhance methanol utilization and cell growth, yet its absence in the evolved strains implies that the heterologous pathway is not essential for methanol-dependent growth. Instead, the evolved strains likely assimilated methanol through adjustments in the endogenous metabolic network. This transition is supported by genome sequencing data, particularly by specific mutations in Adh2 and acetaldehyde dehydrogenase 5 (Ald5). As shown in Fig. 3D, SCSA003 retained the wild-type Adh2, while SCSA001 harbored a mutated Adh2 (Adh2*) with 12 residue changes (G75A, M76I, E78D, K81R, I85V, Y88L, G96S, E128Q, H139R, G143E, E148Q, and A174V). Concurrently, Ald5 in SCSA003 (Ald5S226C, referred to as Ald5#) incurred a single mutation, whereas Ald5 in SCSA001 (Ald5*) underwent extensive changes, including a complete alteration of residues 227 to 236 and a total loss from 237 to 520 (Q227C, L228V, C229R, Q230K, E231Q, G233Y, I234P, P235L, A236V, and G237X) (Fig. 3E).
Given the reported methanol oxidation capacity of Adh2 (20), we propose a plausible endogenous pathway involving Adh2, formaldehyde/acetaldehyde dehydrogenase (Sfa1/Ald5), and formate dehydrogenase (Fdh1/2) to support methanol utilization (Fig. 3F). Functional assays were further performed to clarify the roles of Adh2 and Ald5 in methanol and formaldehyde oxidation. Both mutated and wild-type variants of these enzymes, including Adh2, Adh2*, Ald5#, and Ald5*, were expressed in SCDMU333_ALE. Despite increased methanol consumption (Fig. 3G), none of the strains exhibited a corresponding improvement in methanol-dependent growth (Fig. 3H and fig. S15). This suggests that although the mutant proteins may have enhanced catalytic activity, it is insufficient to facilitate strain growth if formaldehyde cannot be rapidly assimilated into central metabolism. Conversely, heightened methanol oxidation activity could lead to formaldehyde accumulation, potentially hindering growth. Although minimal formaldehyde accumulation was observed in all strains (less than 50 μM per OD600), which had negligible impacts on the microbial growth, the expressions of both Adh2 and Adh2* resulted in elevated formaldehyde production by 20.81 and 56.19%, respectively (Fig. 3I). In vitro enzymatic assays comparing the catalytic efficiency of wild-type Adh2 and Adh2* against P. pastoris alcohol oxidase 1 (PpAox1) revealed that Adh2 and Adh2* generated 0.22 and 0.37 M formaldehyde from 1 M methanol in 2 hours, corresponding to 29.02 and 48.10% of PpAox1’s catalytic efficiency, respectively (fig. S16).
Moreover, in vivo 13C-methanol metabolic tracer assays were performed on SCSAAd2 and SCSAAd2* to delineate methanol assimilation routes. Although no microbial growth advantage was evident, both strains exhibited an apparent 30% increase in 13C enrichment in serine, indicating substantial methanol incorporation into central metabolism (fig. S17).
Unveiling methanol utilization via the rGly pathway in evolved strains with transcriptome analysis
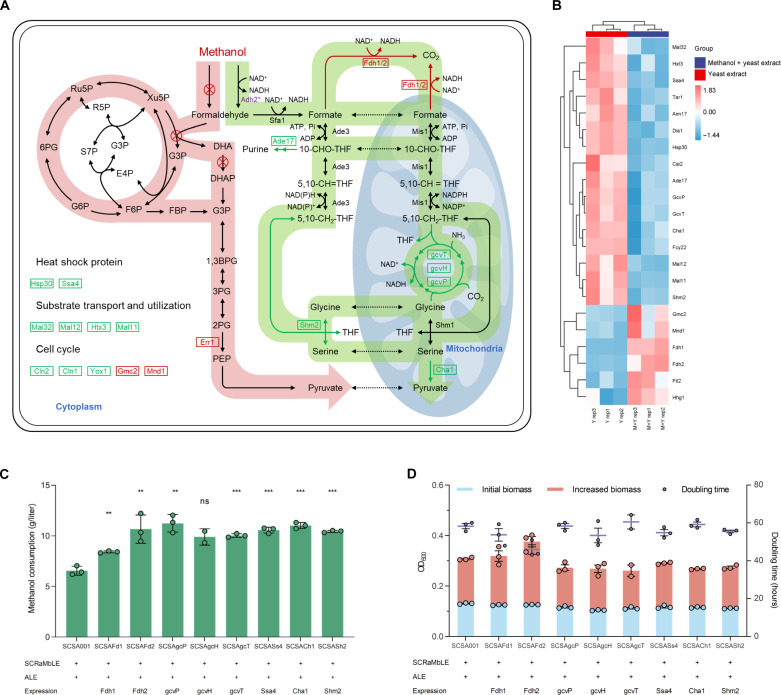
To elucidate the genetic and metabolic adaptations facilitating methanol metabolism, transcriptome analysis of strain SCSA001 grown in Delft MM containing yeast extract (1 g/liter), with or without 2% methanol, was carried out. Functional category analysis indicated substantial differential expression in genes associated with glycine, serine, and threonine metabolism, as well as C1 metabolism pathways (fig. S18). A cluster of genes integral to the rGly pathway predominantly showed down-regulation, with nearly a threefold decrease in expression levels (Fig. 4, A and B). In S. cerevisiae, the native rGly pathway typically serves to generate energy and reducing equivalents, driving the net flux in the reverse direction toward the oxidation of serine and glycine to formate and CO2 (23). The marked down-regulation likely indicates an adaptive redirection of metabolic flux, favoring the incorporation of formate into serine and glycine biosynthesis, possibly as a response to methanol. Conversely, an apparent up-regulation was observed in formate dehydrogenases of Fdh1 and Fdh2, with 22.57-fold and 6.94-fold increases, respectively. This pronounced up-regulation suggests an amplified cellular capacity for oxidizing formate to CO2, likely serving dual roles in formate detoxification and meeting heightened energy demands under methanol stress. Furthermore, this upsurge in CO2 production is a vital step in synthesizing glycine from CH2-THF, NH3, and CO2, underscoring a multifaceted adaptive response to methanol.
Fig. 4. Transcriptomic analysis and the validation of reverse engineering of the rGly pathway.
(A) Differentially expressed enzymes related to central metabolism and their effect on methanol utilization. The pathway with red background represents the heterologous methanol assimilation pathway, which was eradicated in SCSA001 and did not exhibit significant differences in expression. The pathway with green background represents the possible methanol utilization pathway in SCSA001, in which Fdh1/2 is significantly up-regulated and the rGly pathway is significantly down-regulated. Pi, inorganic phosphate; 10-CHO-THF, 10-Formyltetrahydrofolate. (B) Cluster analysis related to differentially expressed enzymes. Yeast cells for transcriptional analysis were cultivated and collected in Delft MM with 0.1% yeast extract or 0.1% yeast extract and 2% methanol for 48 hours. (C and D) Methanol consumption, biomass (OD600) formation, and doubling time after 72 hours of cultivation in Delft MM with 2% methanol (growth curves are shown in fig. S20). All strains were precultured in YPD medium and washed twice using PBS (pH 7.2) before cultivated in Delft MM. All data represent the mean ± SD. Triplicate independent samples were adopted for each panel. Statistical analysis was performed using a two-tailed Student’s t test (**P < 0.01, and ***P < 0.001). ADP, adenosine diphosphate.
To target the core enzymes affecting methanol metabolism at the transcriptional level, we further constructed a protein-protein interaction network using the STRING database (28, 29) for the 22 differentially expressed enzymes, along with Adh2 and Ald5. The network revealed close connections among Adh2, Ald5, methanol oxidation, the rGly pathway, and heat shock proteins (fig. S19). On the basis of these connections, we selected eight enzymes for functional verification in SCDMU333_ALE via reverse engineering. These included two formate dehydrogenases (Fdh1 and Fdh2), five enzymes from the rGly pathway (GcvP, GcvH, GcvT, Shm2, and Cha1), and the heat shock protein Ssa4 (also known as Hsp70). The uniform increase in methanol consumption across all overexpressed enzymes (Fig. 4C) points to a metabolic shift to accommodate the alternative carbon source. However, only the overexpression of Fdh2 markedly enhanced the methanol-dependent growth, reducing the doubling time from 58.33 to 46.03 hours (Fig. 4D and fig. S20). This finding highlights the pivotal role of Fdh2 in facilitating the energy production by promoting the formate oxidation to CO2—potentially alongside the generation of the reduced form of nicotinamide adenine dinucleotide (oxidized form) (NAD+) (NADH) necessary for cellular growth. This strategic adaptation allows the cell to harness formate as an energy source rather than merely assimilating it (30, 31). In addition, the overexpression of Fdh2 increased CO2 concentration, promoting the condensation of formate and CO2 into glycine in the rGly pathway. An intriguing observation is that despite the down-regulation of the rGly pathway during methanol stress, their overexpression enhances methanol consumption. This phenomenon is consistent with prior studies (22, 23), showing that the catalytic direction of the rGly pathway is regulated by both expression intensity and substrate concentration. When the rGly pathway is stably expressed under the control of a strong constitutive promoter, high substrate concentrations could drive the pathway to reverse the net flux toward the assimilation of formate and CO2. Anyhow, these results indicate that SCSA001 assimilates formate together with CO2 through the rGly pathway, indirectly achieving methanol utilization.
Elucidating the ASrG pathway for methanol assimilation
Since the rGly pathway is the sole intrinsic formate assimilation pathway in S. cerevisiae (22), we suspected that strain SCSA001 may incorporate methanol via a previously unidentified ASrG pathway. To verify this hypothesis, we cultured SCSA001 in Delft MM supplemented with various substrates. As predicted, the ASrG pathway facilitated formate-dependent growth in SCSA001, albeit at a modest μ of 0.010 hour−1. Cofeeding 10 mM NaHCO3 with formate obviously activated the ASrG pathway, improving μ to 0.013 hour−1 (Fig. 5A). Higher NaHCO3 concentrations were inhibitory, while lower concentrations (2.5 to 10 mM) enhanced the methanol-dependent growth (fig. S21). These findings underscore the indirect methanol assimilation in SCSA001, mediated through the formate and CO2 assimilation.
Fig. 5. Identification and characterization of the ASrG pathway.
(A) Growth curves of strain SCSA001 on Delft MM with different carbon sources. (B) Schematic illustration of the ASrG pathway and C3N4 QD–mediated energy supply module. (C) Biomass (OD600) formation and doubling time of strain SCSA001, SCSAAd2, SCSAAd2*, and SCSAFd2 after 72 hours of cultivation in Delft MM containing 2% methanol with or without C3N4 QD addition (growth curves are shown in fig. S22). (D) Biomass (OD600) formation and doubling time of strain SCSA001, SCSASh2, and SCSACh1 after 72 hours of cultivation in Delft MM containing 2% methanol with or without serine addition (growth curves are shown in fig. S25). (E) Schematic illustration of the potential serine cycle in S. cerevisiae. Green solid arrows represent endogenously encoded pathways; black dashed arrows represent uncoded pathways; green enzymes represent down-regulated enzymes; red enzymes represent up-regulated enzymes; purple enzymes represent mutated enzymes. All strains were precultured in YPD medium and washed twice using PBS (pH 7.2) before cultivated in Delft MM. All data represent the mean ± SD. Triplicate independent samples were adopted for each panel.
The ASrG pathway could be diverted into two branches: (i) the energy-providing branch for methanol oxidation involving Adh2, Sfa1, and Fdh1/2 and (ii) the energy-consuming branch for formate and CO2 assimilation through the rGly pathway (Fig. 5B). Despite the high energy content of methanol, its assimilation through the ASrG pathway is energetically demanding, requiring two adenosine triphosphates (ATPs) for each pyruvate produced (table S1). To maintain the energy homeostasis (32), the oxidation branch exhibited apparent up-regulation, while the assimilation branch exhibited down-regulation. This adjustment was evident in the improved methanol-dependent growth of strains overexpressing Fdh2, likely benefiting from an augmented NADH pool that facilitates formate and CO2 assimilation.
To illustrate the impact of energy availability on the ASrG pathway, we integrated an external energy supply module using C3N4 quantum dots (QDs) (Fig. 5B). These semiconductor materials are adept at converting light into electrons, which can then be transferred to microorganisms. Leveraging these materials, our earlier research demonstrated efficient hydrogen generation in E. coli, driven by the electrons generated from C3N4 QDs (33). Building upon this success, C3N4 QDs were used to increase the energy available for methanol-dependent growth in strain SCSA001 and its derivatives, including SCSAAd2, SCSAAd2*, and SCSAFd2. As expected, the addition of C3N4 QDs significantly improved the methanol-dependent growth across all strains, reducing doubling times by 14.5 to 21.6% (Fig. 5C and fig. S22). This indicates a successful reallocation of metabolic flux toward the assimilation pathway, culminating in enhanced microbial growth.
We further detected formate accumulation on methanol to characterize the role of formate detoxification in the ASrG pathway. Although expression of Adh2 and Fdh1/2 led to either increased or decreased formate accumulation, respectively, all strains exhibited minimal formate levels, remaining below 0.25 mM (fig. S23). Toxicity assays conducted in Delft MM with glucose (20 g/liter) and varying concentrations of formate revealed that while SCSA001 can use formate, concentrations above 0.1 M apparently inhibited microbial growth (fig. S24A). Strains SCSASs4 and SCSASh2 demonstrated enhanced growth on formate (fig. S24, B and C). In contrast, strain SCSACh1 that carries the serine-degrading Cha1 exhibited poor growth, emphasizing the role of serine in formate utilization. Further culturing in Delft MM with 2% methanol plus serine decreased doubling times for SCSA001, SCSASh2, and SCSACh1 by 19.3, 21.4, and 28.7%, respectively (Fig. 5D and fig. S25). This finding aligns with a recent study suggesting that S. cerevisiae can incorporate formaldehyde via a partial serine cycle and the glyoxylate cycle (6). Despite the absence of certain enzymes, such as malate–coenzyme A (CoA) ligase and phosphoenolpyruvate carboxylase, our findings indicate a variant or incomplete serine cycle in S. cerevisiae for C1 substrate assimilation (Fig. 5E).
Although the ASrG pathway catalyzes the assimilation of C1 feedstocks, its expression in wild-type S. cerevisiae BY4741 cannot support cell growth on sole methanol (fig. S26). In general, while the strain carrying the ASrG pathway demonstrated improvements in growth compared to the original strain—such as maintaining biomass on methanol for a limited period or achieving short-term biomass growth with yeast extract addition—it performed similarly to, or even less effectively than, the haploid strain SCHapMet. This result is expected, as the current consensus holds that synthetic methylotrophs cannot be achieved by simply introducing a functional methanol assimilation pathway (6, 7). Alternatively, nonrational strategies such as ALE are typically indispensable for achieving the global redistribution of metabolic networks necessary for the construction of synthetic methylotrophs (6, 7, 34). Together, these results substantiate the capability of SCSA001 to grow on methanol or formate as a sole carbon source, leveraging a reprogramming of its endogenous metabolic pathways.
DISCUSSION
The evolution of synthetic biology has substantially expanded the grasp of metabolic networks, enabling the assembly and optimization of intricate pathways. Although metabolic engineering guided by the rational design has heralded numerous achievements, it often relies on precise, targeted modifications that favor linear, simple, and traditional pathways (35–39). Yet, natural methanol utilization pathways, such as the ribulose monophosphate (RuMP) pathway, xylulose monophosphate (XuMP) pathway, and serine cycle, usually adopt complex, cyclic, multistep and nonorthogonal strategies, posing challenges in the construction of synthetic methylotrophs. Moreover, the rational design frequently grapples with reconciling various factors—such as enhancing enzyme activity, recycling intermediaries, balancing cofactors, generating energy, regulating genes, synthesizing amino acids, and reducing intermediate toxicity—all of which are recognized bottlenecks in C1 compound assimilation (6, 7, 40–47). Addressing these multifaceted challenges through artificial design often falls short due to the nonorthogonality of a metabolic network, as microorganisms spontaneously regulate their metabolic networks to counteract local metabolic changes. Alternatively, strategies like SCRaMbLE and ALE ingeniously harness the innate adaptive capacities of microorganisms to construct methylotrophs under methanol stress (6, 8, 20, 31). In this study, ALE facilitated the un-SCRaMbLEd diploid SCSA003 to use methanol solely at a μ of 0.009 hour−1, showcasing the extraordinary adaptability of microbial metabolic networks and the potential of diploids to harbor beneficial mutations.
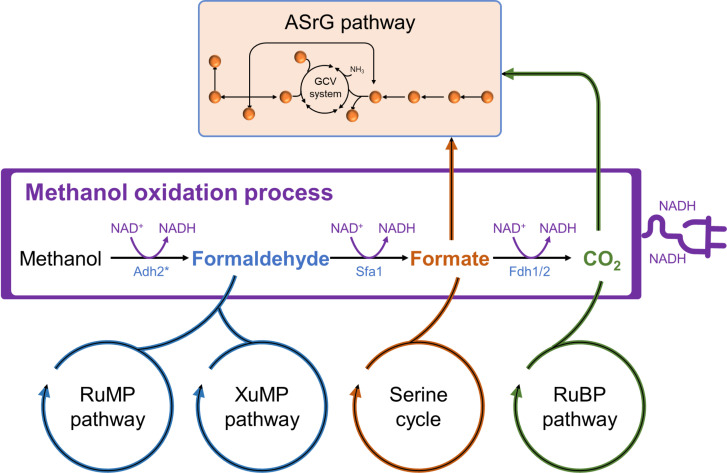
Four primary methanol assimilation pathways have been identified in native methylotrophs: the RuMP pathway (48), the XuMP pathway (35, 49, 50), the serine cycle (37, 51), and the ribulose bisphosphate (RuBP) pathway (52, 53). These pathways fundamentally assimilate the oxidation products of methanol (formaldehyde, formate, and CO2) through methanol oxidation process to generate energy (Fig. 6). Notably, the RuBP pathway, used by bacteria such as Paracoccus denitrificans, involves the complete oxidation of methanol to CO2, followed by refixation via the Calvin-Benson-Bassham cycle. Our study unveiled the unique ASrG pathway in synthetic methylotroph SCSA001, demonstrating its capability to grow on sole methanol or formate. Since energy is provided by the stepwise oxidation of methanol including Adh2, Sfa1, and Fdh1/2, the ASrG pathway’s formate-dependent growth (μ = 0.010 hour−1) is inferior to its methanol-dependent growth (μ = 0.014 hour−1) and cannot support the microbial growth solely on CO2. Admittedly, the growth rate of the evolved strains using the ASrG pathway is still relatively slow. Although the doubling time can be shortened to less than 40 hours (corresponding to μ = 0.017 hour−1) by adding C3N4 QDs or serine, it is still far below the level of natural methylotrophic yeast P. pastoris (μ = 0.085 hour−1). Besides, by introducing the P. pastoris–derived XuMP pathway combined with ALE, another synthetic methylotroph, S. cerevisiae CX01F, achieved a μmax of 0.051 hour−1 in Delft MM (6). This difference in growth performance is at least partially attributable to the energy inefficiency of the ASrG pathway. Theoretically, the ASrG pathway is less energetically efficient than the natural RuMP and XuMP pathways, as it consumes two moles of ATP for every three moles of methanol to produce one mole of pyruvate. Correspondingly, additional energy supply can apparently enhance the methanol assimilation of the ASrG pathway, as exemplified by the increased μ observed with the addition of electron donor C3N4 QDs. The energy demand of this pathway can be compensated by tactically regulating metabolic fluxes, as also demonstrated by the formate-dependent growth. In general, microorganisms cannot assimilate formate without an additional energy source (5, 23, 54–56), as the reduction degree of formate (2) is lower than that of average biomass CH1.76N0.17O0.56 (4.13) (57). Consistent with artificially designed cases (54–56), the ASrG pathway primarily relies on Fdh1/2-catalyzed formate oxidation for energy, resulting in the notable up-regulation of Fdh1 and Fdh2 (30, 31, 58).
Fig. 6. Schematic illustration of the ASrG pathway and natural methanol utilization pathways.
The RuMP and XuMP pathways assimilate formaldehyde; the serine cycle assimilates formate; the RuBP pathway assimilates CO2; the ASrG pathway simultaneously assimilates formate and CO2. The methanol oxidation process is an energy supply branch common to all pathways. GCV system, glycine cleavage system.
We also identified serine as a pivotal node in the ASrG pathway. Enhanced serine levels, either through direct supplementation or overexpression of the enzyme Shm2, apparently promoted the methanol-dependent growth, indicating the role of a variant or incomplete serine cycle in formate utilization. This hypothesis was supported by comparative genomic (data S1) and transcriptomic analyses (data S2), which revealed SNP mutations in enzymes such as pyruvate carboxylase (Pyc) and glyoxylate reductase (Gor1), as well as a substantial up-regulation of enolase-related protein 1 (59) . These findings suggest adaptations in fixing HCO3−/ CO2 and generating intermediates in the serine cycle. The introduction of NaHCO3 notably enhanced the microbial growth, likely due to enhanced HCO3−/ CO2 capture and utilization. This phenomenon aligns with the observed diversity in carboxylase and decarboxylase variants in WGS, particularly Pyc and acetyl-CoA carboxylase (Acc). Pyc (catalyzing the conversion of pyruvate to oxaloacetate) plays a key role in the anaplerotic reaction, which is also crucial for incorporating carbon into the tricarboxylic acid (TCA) cycle (60). Given that almost half of the amino acids are directly synthesized from TCA cycle metabolites, an efficient Pyc is vital for the continuous replenishment of metabolites through replenishment reactions (61). Acc, typically involved in assimilation and biosynthesis in bacteria and yeasts, is a key enzyme in the CO2 fixation pathways of many archaea (62).
The SCRaMbLE system facilitated extensive phenotype variations through deletions, duplications, inversions, and translocations, which offers a genome-wide exploration and characterization (14–18). Implementing SCRaMbLE in diploids provided a unique advantage, allowing genotypic diversity to be stably inherited (16, 18, 63). The massive genomic variations revealed by WGS underscore the potential of diploidy to harbor various mutations, including the deletion of essential genes. While this diversity increases the likelihood of obtaining desired phenotypes, implementing SCRaMbLE alone failed to obtain a true synthetic methylotroph. In addition, because SCRaMbLE generally results in large-scale genomic rearrangements, identifying the key mutations responsible for phenotypic changes amid extensive genomic alterations is challenging. Therefore, we did not conduct genome sequencing on strains that underwent SCRaMbLE alone. Instead, we sequenced the ALE-adapted strains and focused on reverse engineering and characterizing genes implicated in methanol oxidation and those differentially expressed in the transcriptome. Although 13C metabolite tracer analysis and reverse engineering supported the ASrG pathway, the complexity of methanol/formaldehyde/formate toxicity indicates that basic metabolic processes might also influence the C1 compound utilization (25). In addition, the diversity of methanol utilization pathways in different ALE-derived strains poses challenges for comprehensive analysis. Here, computational biology tools such as deep learning might offer avenues to integrate, predict, and model the system network of C1 feedstock assimilation (64–66).
In conclusion, this study used a nonrational strategy to convert S. cerevisiae to grow on sole methanol or formate, using genome-scale chromosomal rearrangements and ALE. While the methanol-dependent growth performance is yet to surpass that achieved by rational design combined with ALE, this research posits a more versatile and general approach. The metabolic flexibility of S. cerevisiae diploids allows them to evolve into desired phenotypes, expanding substrate spectrum, increasing tolerance to toxic substances, and enhancing product accumulation. The elucidation of the methanol metabolic pathway in strain SCSA001 will not only inform future rational design of synthetic methylotrophs but also broaden our understanding of microbial adaptability and metabolic engineering strategies.
MATERIALS AND METHODS
Plasmids, strains, and genes
All strains and plasmids used in this study are listed in tables S2 and S3, respectively. The flowchart of S. cerevisiae construction in this study is shown in fig. S27. S. cerevisiae strain TAM-AOX-CAT-DAS2-DAK [AOX(PPGK)-DAS2(PTDH)-CAT(PPDC)-DAK(PFBA) MATα Δpdc1,5,6 mutx ura3-52] (24) was used as the initial host and renamed as SCHapMet. The diploid methanol-using S. cerevisiae SCDM001 was obtained by mating synV (11, 15) with SCHapMet. Strain SCDM001 was used as the background strain for SCRaMbLE. Strains SCDM001, SCDMU333, and SCDMY345 were used as the background strain for ALE. Strain SCDMU333_ALE was used as the background strain for all genetic manipulations. Genes fdh1, fdh2, gcvP, gcvH, gcvT, ssa4, cha1, and shm2 and the mutated adh2* and ald5* were from strain SCSA001 genome. Unmutated adh2 and mutated ald5# were from strain SCSA003 genome. Genes aox1, das2, cta, dak, and their corresponding promoters and terminators were cloned from strain SCHapMet genome. Sequence of enhanced peroxisome signaling peptide (LGRGRRSKL) was obtained according to (67) and synthesized directly by attaching to the primer. Sequences of mutant genes used in this study are listed in table S5. Promoter TEF1 and terminator ENO2tt used in the pRS426 plasmid were amplified from the genome of strain SCBY4741.
DNA manipulation and transformation
The different elements of expression cassettes, including promoters, genes, and terminators, were amplified using high-fidelity DNA polymerase PrimeSTAR (Thermo Fisher Scientific). Each individual gene expression plasmid was constructed by Gibson assembly. All primers used in this study are listed in table S4. Exceptionally, strain SCBYArG was constructed according to the method we described previously (24), where each individual gene expression cassette (e.g., PTEF1-adh2*-TPGI) was first assembled by overlap extension polymerase chain reaction and then integrated into the δ site of S. cerevisiae BY4741 by a DNA assembler (68) in one step. Yeast transformation was performed by electroporation as previously described (24). Yeast extract, peptone, and dextrose (YPD) medium (50 ml) was inoculated with a 0.5-ml S. cerevisiae seed culture and shaken at 30°C and 250 rpm for 4 to 5 hours until OD600 reached 0.8. Yeast cells were harvested by centrifugation at 4°C and 4000 rpm for 5 min. The supernatant was discarded, and cell pellets were washed with 50 ml of ice-cold deionized water, followed by another wash step with cold 1 M sorbitol and lastly resuspended in 250 ml of cold sorbitol. A mixture of 50 ml of yeast cells and 4 ml of DNA was electroporated in a 0.2-cm cuvette at 1.5 kV for 5 ms. The transformed cells were immediately mixed with 1-ml YPD medium and shaken at 30°C for 1 hour. In the following step, the cells were harvested by centrifugation, washed with 1 M sorbitol twice to remove the remaining medium, and lastly resuspended in 1 ml of sorbitol. Aliquots of 30 to 50 ml were spread on synthetic dropout medium [yeast nitrogen base (6.7 g/liter) without amino acids and glucose (20 g/liter)] with uracil (20 mg/liter) or YPD with hygromycin (200 mg/liter).
Media and growth conditions
E. coli strains were grown at 37°C on LB medium containing ampicillin (100 mg/liter). If not otherwise stated, Delft MM (6, 57) {(NH4)2SO4 [7.5 g/liter], KH2PO4 [11.93 g/liter], K2HPO4 [2.14 g/liter], MgSO4·7H2O [0.5 g/liter], vitamin solution [1 ml/liter] [biotin (0.05 g/liter), p-amino benzoic acid (0.2 g/liter), nicotinic acid (1 g/liter), Ca-pantothenaat (1 g/liter), pyridoxine-HCl (1 g/liter), thiamine-HCl (1 g/liter), and myo-inositol (25 g/liter) (pH 6.5)] and trace metal solution [2 ml/liter] [EDTA (15.0 g/liter; sodium sa1t), ZnSO4·7H2O (4.5 g/liter), MnCl2·2H2O (0.84 g/liter), CoC12·6H2O (0.3 g/liter), CuSO4·5H2O (0.3 g/liter), Na2MoO4·2H2O (0.4 g/liter), CaC12·2H2O (4.5 g/liter), FeSO4·7H2O (3 g/liter), H3BO3 (1 g/liter), and KI (0.1 g/liter) (pH 4.0)]} containing 2% methanol was used for strain characterization and metabolite quantification. For auxotrophic strains, uracil (20 mg/liter), histidine (20 mg/liter), leucine (100 mg/liter), and methionine (20 mg/liter) were supplemented in Delft MM if necessary.
For all experiments for methanol utilization, strains were precultured in YPD [yeast extract (10 g/liter), peptone (20 g/liter), and glucose (20 g/liter)] for 16 to 18 hours and then washed twice with MM, which was subsequently transferred into MM with the initial OD600 of 0.1. In particular, to verify the role of the C3N4 QD–mediated energy supply module, C3N4 QDs (40 mg/liter) was supplemented into Delft MM with 2% methanol. To verify the role of serine in formate utilization, 5 mM serine was supplemented into Delft MM with 2% methanol.
SCRaMbLE of heterozygous diploids
Heterozygous diploid cells were transformed with pLM050-pCLB2-CreEBD-Hph and maintained on YPD-hygromycin plates. Cells were grown overnight in liquid YPD-hygromycin media to saturation. Cultures were diluted to a starting OD600 of 0.1 in 50 ml of fresh YPD-hygromycin media. β-Estradiol was added to a final concentration of 1 μM, and cultures were incubated at 30°C with shaking at 225 rpm for 6 hours. Cultures were spun down at 3000g for 3 min and washed three times with water to wash out β-estradiol, and cells were plated onto either solid Delft MM with 2% methanol and uracil (20 mg/liter) or 6% methanol, yeast extract (0.1 g/liter), and uracil (20 mg/liter). A Cre-EBD plasmid was subsequently lost after continuous subculture in YPD for 10 days and verified by YPD-hygromycin plates.
Adaptive laboratory evolution
ALE experiments with SCDM001, SCDMU333, and SCDMY345 were performed by serial transfers in shake flasks, which involved two stages (fig. S5). In the first stage, cells were cultured in the Delft MM containing 2% methanol and yeast extract (0.1 g/liter) with an initial OD600 of ~0.02. Whenever OD600 exceeds 0.4 or a single culture reached 7 days, cells are transferred to fresh Delft MM for a new round of passage. After seven rounds of passage, the yeast extract addition was removed. The ALE was terminated upon reaching a stable growth rate in the Delft MM containing 2% sole methanol. The ALE-derived strains were transformed into the empty pRS426 plasmid, generating strain SCSA001, SCSA002, and SCSA003.
13C metabolite tracer analysis
For 13C-labeled amino acid analysis, strains were cultivated in Delft MM with 13C-labeled methanol (99% atom enrichment; Sigma-Aldrich, MO, USA) as the sole carbon source. This preculture process on 13C-labeled methanol was repeated three times, and then the cells were washed twice with phosphate-buffered saline (PBS) (pH 7.2). The cells in the exponential growth stage were transferred into a fresh Delft MM with 2% 13C-labeled methanol or 10 mM 13C-labeled NaHCO3 and 2% methanol. Gas chromatography–mass spectrometry (GC-MS) was used to analyze the amino acid profiles according to a previous method (8). Specifically, a 10-ml sample was collected after 72 hours and quickly injected into −40°C 60% (v/v) methanol for 30 s to quench the cellular metabolism. The mixture was centrifuged at 4000g for 5 min at 4°C, and the supernatant was removed. The resulting cell pellets were washed with Milli-Q water twice to remove residual media and then centrifuged at 4000g for 5 min at 4°C, and the supernatant was removed. The treated cells were ground under liquid nitrogen, and 50 mg was accurately weighed into a 1.5-ml centrifuge tube. In addition, 1 ml of −40°C 50% (v/v) methanol was added into the centrifuge tube and then frozen and thawed three times with liquid nitrogen. The mixture was centrifuged at 12,000g for 3 min at 4°C, and 200 μl of the supernatant was transferred into a new centrifuge tube. The sample was quickly frozen in liquid nitrogen and then placed in a vacuum freeze dryer to freeze-dry. For derivatization, 50 μl of methoxylamine hydrochloride (20 mg/ml) in pyridine was added to the samples, which were then vortexed occasionally and incubated at 40°C for 80 min. Then, the samples were mixed with 80 μl of N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich), vortexed occasionally, and incubated at 40°C for 80 min. After derivatization, the samples were centrifuged at 12,000g for 10 min, and the supernatant was transferred to new GC vials.
The derivatized amino acid samples were analyzed by GC/quadrupole orthogonal acceleration–time-of-flight (Q-TOF)–MS using an Agilent 7890 A GC coupled with a 7200 Accurate-Mass Q-TOF (Agilent Technologies, Germany). A DB-5MS ultra inert column (30 m by 0.25 mm, 0.25-μm film thickness) was used (Agilent Technologies, USA). The sample (1 μl) was injected into the GC in splitless mode. The oven temperature was programmed as follows: 60°C for 1 min, 8 C/min to 132°C, 2°C/min to 150°C, 5°C/min to 185°C, and 10°C/min to 325°C, 5 min hold. Mass spectra of the amino acids were in the mass range of 50 to 650 mass/charge ratio at an acquisition rate of 5 spectra/s. The temperatures of the ion source and transfer line were 250° and 290°C, respectively. Electron ionization was carried out at 70 eV. Agilent Mass Hunter Qualitative Analysis software was used for peak detection and mass spectral deconvolution. The annotation of amino acids was performed via matching their mass fragmentation patterns with those in the National Institute of Standards and Technology mass spectral library (match factor, > 80%). 13C-labeling was determined from the measured mass isotopomer data as described above. The mass spectrum data of derivatized proteinogenic amino acids were subsequently corrected for natural isotope abundance using the method described by Wahl et al. (69), and the correction was performed with an open-source MATLAB-based package (WUFlux) (70, 71).
Whole-genome resequencing and data analysis
Whole-genome DNA of SCDM001, SCSA001, and SCSA003 was extracted and sequenced using the Illumina NovaSeq and the Oxford Nanopore Technologies platform at Personalbio (Shanghai, China). For the Illumina paired-end sequencing process, a minimum of 1 μg of genomic DNA was used for the construction of sequencing libraries for each sample. A library was constructed by Illumina TruSeq Nano DNA LT following the Illumina TruSeq DNA Sample Preparation Guide. The library was quality-controlled using the Agilent High Sensitivity DNA Kit and sequenced using an Illumina NovaSeq platform (Personalbio Co. Ltd) in 2 × 150 bp paired-end mode with a 400-bp insert size of the library. High-quality data were obtained from raw data by removing adapter contamination with AdapterRemoval (version 2) (72) and collection with SOAPec (version 2.01) (https://help.rc.ufl.edu/doc/SOAPec).
The genome of SCDM001 was used as a reference to conduct difference analysis. For the other two evolved strains, resequencing was carried out. The whole genome was sequenced using Illumina NovaSeq, paired-end 300 cycles (2 × 150 bp) at Personalbio (Shanghai, China). Each sample was represented by 30.5 to 30.9 million sequence raw reads. After data processing, the clean reads had gone through reads mapping, SNP/Indel analysis [Genome Analysis Toolkit (GATK) (73) and Annotate Variation (ANNOVAR) (74)], structural variation analysis, variation map of the whole genome and CNV analysis (CNVnator, v.0.3). These results are shown in fig. S9 and data S1.
RNA extraction and transcriptome profiling
To conduct the transcriptional analysis, strain SCSA001 was cultured in Delft MM containing yeast extract (1 g/liter) with or without 2% methanol at 30°C, 220 rpm for 48 hours. Two-milliliter samples were rapidly centrifuged at 4°C and 10,000 rpm for 2 min, which was washed with cold 1‰ diethyl pyrocarbonate water and centrifuged twice. The supernatant was fully removed and then frozen in liquid nitrogen for 15 min. Then, the sample was stored at −80°C. Comparative RNA sequencing analysis was carried out by Shanghai Personalbio Biotechnology Co. Ltd. using the Illumina high-throughput sequencing platform. All treatments were conducted in batch culture in triplicates. Differential expression levels (log2 fold change) and corresponding significance levels (P values) of genes obtained are detailed in data S2.
Alcohol dehydrogenase activity assay
The Adh2, Adh2*, and PpAox1 genes were inserted into the plasmid pGAPZ(alpha) B under the control of the GAP promoter and transformed into P. pastoris GS115. Recombinant P. pastoris cells were cultured in Delft MM with 2% glucose at 30°C, 100 rpm for 3 days. After cultivation, cells were removed by centrifugation (6000g) and filtration (0.45 μm). The culture supernatant was applied to a Ni2+-affinity column. The column was washed with washing buffer containing 20 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl, and 50 mM imidazole, and purified recombinant proteins Adh2, Adh2*, and PpAox1 were eluted using elution buffer containing 20 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl, and 200 mM imidazole. Protein concentrations were measured on the basis of ultraviolet (280 nm) absorbance using a NanoDrop spectrophotometer.
The protein activity assay of Adh2 and Adh2* was performed in a total volume of 1 ml of solution containing 1 M methanol, 10 mM sodium phosphate buffer (pH 7.2), 2 mM NAD+, and a purified protein (0.05 mg/ml). Formaldehyde was measured using a previous method (75) by the addition of 1 ml of Nash reagent (2 M ammonium acetate, 0.05 M acetic acid, and 0.02 M acetylacetone) (76, 77).
Preparation of C3N4 QDs
C3N4 QDs used in this study were prepared as described in our previous study (33). In particular, the bulk C3N4 was prepared by polymerization of melamine molecules under high temperature. In detail, melamine was heated at 600°C for 2 hours under air condition with a ramp rate of about 3°C/min for both the heating and cooling processes. The obtained yellow product was the bulk C3N4 powder. Next, 1 g of bulk C3N4 was treated in the mixture of concentrated sulfuric acid (H2SO4) (20 ml) and nitric acid (HNO3) (20 ml) for about 2 hours at room temperature. The mixture was then diluted with deionized water and washed for several times, and the as-obtained white product was porous C3N4. Second, 100 mg of the porous C3N4 was dispersed in 30 ml of concentrated NH3·H2O, and then the mixed solution was transferred into a 45-ml Teflon cup and heated in a sealed autoclave at 180°C for 12 hours, during which the porous C3N4 was peeled into porous nanosheets. Upon cooling to room temperature, the precipitate was washed with water for several times to remove the adsorbed NH3 molecules. Third, 10 mg of porous C3N4 nanosheets was dispersed in 100 ml of water and then ultrasound for about 6 hours. The as-obtained aqueous suspension was then centrifuged at about 7000 rpm to remove large-sized nanoparticles.
Analytical methods
Biomass concentration was determined by measuring the OD600 using an AOE INSTRUMENTSUV1800 spectrophotometer. Methanol concentration was determined using Sieman Biosensors Analyzer S-10. Formaldehyde concentration was determined using Nash reagent as described above. Formate concentration was measured via high-performance liquid chromatography (HPLC) (UltiMate 3000 HPLC system, Dionex, USA). An ultraviolet absorption detector (UVD) 170U ultraviolet detector at a wavelength of 215 nm and an ion exchange chromatographic column (Bio-Rad Aminex HPX-87H column, USA) were used. The products were eluted at 55°C with 0.25 mM H2SO4 as the mobile phase at a flow rate of 0.5 ml/min.
Protein-protein interaction construction and analysis
STRING database (28) (https://string-db.org/) was used to construct protein-protein interaction network graphs. Twenty-four target proteins were imported into the database, and the interaction relationships among the targets were obtained on the basis of interactions with a medium combined score of >0.4.
Thermodynamic analysis of the ASrG pathway
The physiological standard Gibbs energies of reactions (ΔrG′m in kilojoules per mole) at physiological standard reactant concentrations (1 mM), pH (6.0), ionic strength (0.1 mM), temperature (25°C), pressure (1 bar), and free magnesium ion concentration (pMg) (3) were computed using the eQuilibrator biochemical thermodynamics calculator (78).
Statistics
Statistical analysis was performed using GraphPad Prism 8.0.2 software using a two-tailed t test analysis of variance hypothesis. Significant differences are marked as *P < 0.05, **P < 0.01, and ***P < 0.001. All data are presented as mean ± SD. The number of biologically independent samples for each panel was three unless otherwise stated in the figure legends.
Acknowledgments
We thank B. Li from Tianjin University in China for providing strain SCsynV and technical support for SCRaMbLE.
Funding: This work was supported by National Key R&D Program of China (2022YFC2105900 to Y.J.), the National Natural Science Foundation of China (22078151 to W.Z., 22378199 to W.Z., and 22178169 to F.X.), Jiangsu Natural Science Fund for Distinguished Young Scholars (BK20220052 to F.X.), Open Funding Project of State Key Laboratory of Microbial Metabolism (MMLKF23-09 to W.Z.), State Key Laboratory of Materials-Oriented Chemical Engineering (KL-MCE-23A10 to F.X.), China Postdoctoral Science Foundation (no. 2023M740370 to W.Z.), and Young Elite Scientist Sponsorship Program by CAST (YESS20200174 to W.Z.).
Author contributions: Each author’s contribution to the paper is listed below: Conceptualization: F.G., Y.W., W.J., Y.J., F.X., M.J., and W.Z. Funding acquisition: Y.J., F.X., M.J., and W.Z. Investigation: F.G., K.L., Y.Q., Y.Z., W.J., Y.J., F.X., M.J., and W.Z. Methodology: F.G., C.L., Y.W., W.J., Y.J., and W.Z. Resources: Y.Q., Y.Z., Y.J., M.J., and W.Z. Supervision: Y.W., F.X., and W.Z. Writing—original draft: F.G. and W.Z. Writing—review and editing: F.G., C.L., Y.J., F.X., and M.J. Validation: F.G., K.L., W.J., Y.J., M.J., and W.Z. Formal analysis: F.G., C.L., Z.Z., W.J., and F.X. Project administration: F.G. and W.Z. Data curation: W.J. Visualization: F.G. and C.L. Software: F.X. and W.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The genome sequence data of evolved strains can be downloaded from (NCBI.SRA) (https://ncbi.nlm.nih.gov/sra): (PRJNA1085989). The RNA sequencing raw data can be downloaded from (NCBI.SRA) (https://ncbi.nlm.nih.gov/sra): (PRJNA1086020).
Supplementary Materials
The PDF file includes:
Figs. S1 to S27
Tables S1 to S5
Legends for data S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data S1 and S2
REFERENCES AND NOTES
- 1.Guo F., Qiao Y., Xin F., Zhang W., Jiang M., Bioconversion of C1 feedstocks for chemical production using Pichia pastoris. Trends Biotechnol. 41, 1066–1079 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Fabarius J. T., Wegat V., Roth A., Sieber V., Synthetic methylotrophy in yeasts: Towards a circular bioeconomy. Trends Biotechnol. 39, 348–358 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Liu Z., Wang K., Chen Y., Tan T., Nielsen J., Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 3, 274–288 (2020). [Google Scholar]
- 4.Liu Z., Shi S., Ji Y., Wang K., Tan T., Nielsen J., Opportunities of CO2-based biorefineries for production of fuels and chemicals. Green Carbon 1, 75–84 (2023). [Google Scholar]
- 5.Kim S., Lindner S. N., Aslan S., Yishai O., Wenk S., Schann K., Bar-Even A., Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat. Chem. Biol. 16, 538–545 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Zhan C., Li X., Lan G., Baidoo E. E. K., Yang Y., Liu Y., Sun Y., Wang S., Wang Y., Wang G., Nielsen J., Keasling J. D., Chen Y., Bai Z., Reprogramming methanol utilization pathways to convert Saccharomyces cerevisiae to a synthetic methylotroph. Nat. Catal. 6, 435–450 (2023). [Google Scholar]
- 7.Chen F. Y.-H., Jung H.-W., Tsuei C.-Y., Liao J. C., Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell 182, 933–946.e14 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Zhang S., Guo F., Yang Q., Jiang Y., Yang S., Ma J., Xin F., Hasunuma T., Kondo A., Zhang W., Jiang M., Improving methanol assimilation in Yarrowia lipolytica via systematic metabolic engineering combined with compartmentalization. Green Chem. 25, 183–195 (2023). [Google Scholar]
- 9.Mavrommati M., Daskalaki A., Papanikolaou S., Aggelis G., Adaptive laboratory evolution principles and applications in industrial biotechnology. Biotechnol. Adv. 54, 107795 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Sandberg T. E., Salazar M. J., Weng L. L., Palsson B. O., Feist A. M., The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 56, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Z.-X., Li B.-Z., Mitchell L. A., Wu Y., Qi X., Jin Z., Jia B., Wang X., Zeng B.-X., Liu H.-M., Wu X.-L., Feng Q., Zhang W.-Z., Liu W., Ding M.-Z., Li X., Zhao G.-R., Qiao J.-J., Cheng J.-S., Zhao M., Kuang Z., Wang X., Martin J. A., Stracquadanio G., Yang K., Bai X., Zhao J., Hu M.-L., Lin Q.-H., Zhang W.-Q., Shen M.-H., Chen S., Su W., Wang E.-X., Guo R., Zhai F., Guo X.-J., Du H.-X., Zhu J.-Q., Song T.-Q., Dai J.-J., Li F.-F., Jiang G.-Z., Han S.-L., Liu S.-Y., Yu Z.-C., Yang X.-N., Chen K., Hu C., Li D.-S., Jia N., Liu Y., Wang L.-T., Wang S., Wei X.-T., Fu M.-Q., Qu L.-M., Xin S.-Y., Liu T., Tian K.-R., Li X.-N., Zhang J.-H., Song L.-X., Liu J.-G., Lv J.-F., Xu H., Tao R., Wang Y., Zhang T.-T., Deng Y.-X., Wang Y.-R., Li T., Ye G.-X., Xu X.-R., Xia Z.-B., Zhang W., Yang S.-L., Liu Y.-L., Ding W.-Q., Liu Z.-N., Zhu J.-Q., Liu N.-Z., Walker R., Luo Y., Wang Y., Shen Y., Yang H., Cai Y., Ma P.-S., Zhang C.-T., Bader J. S., Boeke J. D., Yuan Y.-J., “Perfect” designer chromosome V and behavior of a ring derivative. Science 355, eaaf4704 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Wu Y., Li B.-Z., Zhao M., Mitchell L. A., Xie Z.-X., Lin Q.-H., Wang X., Xiao W.-H., Wang Y., Zhou X., Liu H., Li X., Ding M.-Z., Liu D., Zhang L., Liu B.-L., Wu X.-L., Li F.-F., Dong X.-T., Jia B., Zhang W.-Z., Jiang G.-Z., Liu Y., Bai X., Song T.-Q., Chen Y., Zhou S.-J., Zhu R.-Y., Gao F., Kuang Z., Wang X., Shen M., Yang K., Stracquadanio G., Richardson S. M., Lin Y., Wang L., Walker R., Luo Y., Ma P.-S., Yang H., Cai Y., Dai J., Bader J. S., Boeke J. D., Yuan Y.-J., Bug mapping and fitness testing of chemically synthesized chromosome X. Science 355, eaaf4706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dymond J. S., Richardson S. M., Coombes C. E., Babatz T., Muller H., Annaluru N., Blake W. J., Schwerzmann J. W., Dai J., Lindstrom D. L., Boeke A. C., Gottschling D. E., Chandrasegaran S., Bader J. S., Boeke J. D., Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 477, 471–476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y., Zhu R.-Y., Mitchell L. A., Ma L., Liu R., Zhao M., Jia B., Xu H., Li Y.-X., Yang Z.-M., Ma Y., Li X., Liu H., Liu D., Xiao W.-H., Zhou X., Li B.-Z., Yuan Y.-J., Boeke J. D., In vitro DNA SCRaMbLE. Nat. Commun. 9, 1935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Xie Z.-X., Ma Y., Chen X.-R., Huang Y.-Q., He B., Bin J., Li B.-Z., Yuan Y.-J., Ring synthetic chromosome V SCRaMbLE. Nat. Commun. 9, 3783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia B., Wu Y., Li B.-Z., Mitchell L. A., Liu H., Pan S., Wang J., Zhang H.-R., Jia N., Li B., Shen M., Xie Z.-X., Liu D., Cao Y.-X., Li X., Zhou X., Qi H., Boeke J. D., Yuan Y.-J., Precise control of SCRaMbLE in synthetic haploid and diploid yeast. Nat. Commun. 9, 1933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Z., Wang L., Wang Y., Zhang W., Guo Y., Shen Y., Jiang L., Wu Q., Zhang C., Cai Y., Dai J., Identifying and characterizing SCRaMbLEd synthetic yeast using ReSCuES. Nat. Commun. 9, 1930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen M. J., Wu Y., Yang K., Li Y., Xu H., Zhang H., Li B., Li X., Xiao W.-H., Zhou X., Mitchell L. A., Bader J. S., Yuan Y., Boeke J. D., Heterozygous diploid and interspecies SCRaMbLEing. Nat. Commun. 9, 1934 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao D., Liu T., Gao J., Xu J., Gou Y., Pan Y., Li D., Ye C., Pan R., Huang L., Xu Z., Lian J., De novo biosynthesis of vindoline and catharanthine in Saccharomyces cerevisiae. BioDesign Res. 2022, 0002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espinosa M. I., Gonzalez-Garcia R. A., Valgepea K., Plan M. R., Scott C., Pretorius I. S., Marcellin E., Paulsen I. T., Williams T. C., Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae. Nat. Commun. 11, 5564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinosa M. I., Williams T. C., Pretorius I. S., Paulsen I. T., Benchmarking two Saccharomyces cerevisiae laboratory strains for growth and transcriptional response to methanol. Syn. Syst. Biotechno. 4, 180–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la Cruz J. G., Machens F., Messerschmidt K., Bar-Even A., Core catalysis of the reductive glycine pathway demonstrated in yeast. ACS Synth. Biol. 8, 911–917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bysani V. R., Alam A. S., Bar-Even A., Machens F., Engineering and evolution of the complete Reductive Glycine Pathway in Saccharomyces cerevisiae for formate and CO2 assimilation. Metab. Eng. 81, 167–181 (2024). [DOI] [PubMed] [Google Scholar]
- 24.Dai Z., Gu H., Zhang S., Xin F., Zhang W., Dong W., Ma J., Jia H., Jiang M., Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae. Bioresour. Technol. 245, 1407–1412 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Gao J., Li Y., Yu W., Zhou Y., Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nat. Metab. 4, 932–943 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Guo F., Dai Z., Peng W., Zhang S., Zhou J., Ma J., Dong W., Xin F., Zhang W., Jiang M., Metabolic engineering of Pichia pastoris for malic acid production from methanol. Biotechnol. Bioeng. 118, 357–371 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Bennett R. K., Agee A., Har J. R. G., von Hagel B., Siu K.-H., Antoniewicz M. R., Papoutsakis E. T., Triggering the stringent response enhances synthetic methanol utilization in Escherichia coli. Metab. Eng. 61, 1–10 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A. L., Fang T., Doncheva N. T., Pyysalo S., Bork P., Jensen L. J., von Mering C., The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., von Mering C., STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Da Y., Bi H., Liu Y., Chen B., Wang M., Liu Z., Nielsen J., Tan T., A one-carbon chemicals conversion strategy to produce precursor of biofuels with Saccharomyces cerevisiae. Renew. Energy 208, 331–340 (2023). [Google Scholar]
- 31.Wang K., Liu Y., Wu Z., Wu Y., Bi H., Liu Y., Wang M., Chen B., Nielsen J., Liu Z., Tan T., Investigating formate tolerance mechanisms in Saccharomyces cerevisiae and its application. Green Carbon 1, 65–74 (2023). [Google Scholar]
- 32.Tong T., Chen X. L., Hu G. P., Wang X. L., Liu G. Q., Liu L. M., Engineering microbial metabolic energy homeostasis for improved bioproduction. Biotechnol. Adv. 53, 107841 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Wu D., Zhang W., Fu B., Zhang Z., Living intracellular inorganic-microorganism biohybrid system for efficient solar hydrogen generation. Joule 6, 2293–2303 (2022). [Google Scholar]
- 34.Reiter M. A., Bradley T., Büchel L. A., Keller P., Hegedis E., Gassler T., Vorholt J. A., A synthetic methylotrophic Escherichia coli as a chassis for bioproduction from methanol. Nat. Catal. 7, 560–573 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Fan L., Tuyishirne P., Zheng P., Sun J., Synthetic methylotrophy: A practical solution for methanol-based biomanufacturing. Trends Biotechnol. 38, 650–666 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Schrader J., Schilling M., Holtmann D., Sell D., Filho M. V., Marx A., Vorholt J. A., Methanol-based industrial biotechnology: Current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 27, 107–115 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Yu H., Liao J. C., A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds. Nat. Commun. 9, 3992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitaker W. B., Sandoval N. R., Bennett R. K., Fast A. G., Papoutsakis E. T., Synthetic methylotrophy: Engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr. Opin. Biotechnol. 33, 165–175 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Cotton C. A. R., Claassens N. J., Benito-Vaquerizo S., Bar-Even A., Renewable methanol and formate as microbial feedstocks. Curr. Opin. Biotechnol. 62, 168–180 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Gregory G. J., Bennett R. K., Papoutsakis E. T., Recent advances toward the bioconversion of methane and methanol in synthetic methylotrophs. Metab. Eng. 71, 99–116 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez J. E., Bennett R. K., Papoutsakis E. T., Antoniewicz M. R., Methanol assimilation in Escherichia coli is improved by co-utilization of threonine and deletion of leucine-responsive regulatory protein. Metab. Eng. 45, 67–74 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Roth T. B., Woolston B. M., Stephanopoulos G., Liu D. R., Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2. ACS Synth. Biol. 8, 796–806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu T.-Y., Chen C.-T., Liu J. T.-J., Bogorad I. W., Damoiseaux R., Liao J. C., Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1. Appl. Microbiol. Biotechnol. 100, 4969–4983 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Ganesh I., Vidhya S., Eom G. T., Hong S. H., Construction of methanol-sensing Escherichia coli by the introduction of a Paracoccus denitrificans MxaY-based chimeric two-component system. J. Microbiol. Biotechnol. 27, 1106–1111 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Rohlhill J., Sandoval N. R., Papoutsakis E. T., Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol. ACS Synth. Biol. 6, 1584–1595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woolston B. M., Roth T., Kohale I., Liu D. R., Stephanopoulos G., Development of a formaldehyde biosensor with application to synthetic methylotrophy. Biotechnol. Bioeng. 115, 206–215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett R. K., Agee A., Har J. R. G., von Hagel B., Antoniewicz M. R., Papoutsakis E. T., Regulatory interventions improve the biosynthesis of limiting amino acids from methanol carbon to improve synthetic methylotrophy in Escherichia coli. Biotechnol. Bioeng. 118, 43–57 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Kato N., Yurimoto H., Thauer R. K., The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Biosci. Biotechnol. Biochem. 70, 10–21 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Kelso P. A., Chow L. K. M., Carpenter A. C., Paulsen I. T., Williams T. C., Toward methanol-based biomanufacturing: Emerging strategies for engineering synthetic methylotrophy in Saccharomyces cerevisiae. ACS Synth. Biol. 11, 2548–2563 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yurimoto H., Kato N., Sakai Y., Assimilation, dissimilation, and detoxification of formaldehyde, a central metabolic intermediate of methylotrophic metabolism. Chem Rec. 5, 367–375 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Chistoserdova L. V., Lidstrom M. E., Genetics of the serine cycle in Methylobacterium extorquens AM1: Identification of sgaA and mtdA and sequences of sgaA, hprA, and mtdA. J. Bacteriol. 176, 1957–1968 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dedysh S. N., Smirnova K. V., Khmelenina V. N., Suzina N. E., Liesack W., Trotsenko Y. A., Methylotrophic autotrophy in Beijerinckia mobilis. J. Bacteriol. 187, 3884–3888 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ochsner A. M., Sonntag F., Buchhaupt M., Schrader J., Vorholt J. A., Methylobacterium extorquens: Methylotrophy and biotechnological applications. Appl. Microbiol. Biotechnol. 99, 517–534 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Gleizer S., Ben-Nissan R., Bar-On Y. M., Antonovsky N., Noor E., Zohar Y., Jona G., Krieger E., Shamshoum M., Bar-Even A., Milo R., Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 179, 1255–1263.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antonovsky N., Gleizer S., Noor E., Zohar Y., Herz E., Barenholz U., Zelcbuch L., Amram S., Wides A., Tepper N., Davidi D., Bar-On Y., Bareia T., Wernick D., Shani I., Malitsky S., Jona G., Bar-Even A., Milo R., Sugar synthesis from CO2 in Escherichia coli. Cell 166, 115–125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gassler T., Sauer M., Gasser B., Egermeier M., Troyer C., Causon T., Hann S., Mattanovich D., Steiger M. G., The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 38, 210–216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu T., Liu Q., Wang X., Liu X., Chen Y., Nielsen J., Metabolic reconfiguration enables synthetic reductive metabolism in yeast. Nat. Metab. 4, 1551–1559 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian J., Deng W., Zhang Z., Xu J., Yang G., Zhao G., Yang S., Jiang W., Gu Y., Discovery and remodeling of Vibrio natriegens as a microbial platform for efficient formic acid biorefinery. Nat. Commun. 14, 7758 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kornblatt M. J., Albert J. R., Mattie S., Zakaib J., Dayanandan S., Hanic-Joyce P. J., Joyce P. B. M., The Saccharomyces cerevisiae enolase-related regions encode proteins that are active enolases. Yeast 30, 55–69 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Owen O. E., Kalhan S. C., Hanson R. W., The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 277, 30409–30412 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Kornberg H. L., Anaplerotic sequences in microbial metabolism. Angew. Chem. Int. Ed. 4, 558–565 (1965). [Google Scholar]
- 62.Erb T. J., Carboxylases in natural and synthetic microbial pathways. Appl. Environ. Microbiol. 77, 8466–8477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong Y., Zhang H., Zhou S., Ma L., Xiao W., Wu Y., Yuan Y.-J., Structural variations and adaptations of synthetic chromosome ends driven by SCRaMbLE in haploid and diploid yeasts. ACS Synth. Biol. 12, 689–699 (2023). [DOI] [PubMed] [Google Scholar]
- 64.Sen P., Lamichhane S., Mathema V. B., McGlinchey A., Dickens A. M., Khoomrung S., Orešič M., Deep learning meets metabolomics: A methodological perspective. Brief. Bioinform. 22, 1531–1542 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Copeland W. B., Bartley B. A., Chandran D., Galdzicki M., Kim K. H., Sleight S. C., Maranas C. D., Sauro H. M., Computational tools for metabolic engineering. Metab. Eng. 14, 270–280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L., Dash S., Ng C. Y., Maranas C. D., A review of computational tools for design and reconstruction of metabolic pathways. Syn. Syst. Biotechno. 2, 243–252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeLoache W. C., Russ Z. N., Dueber J. E., Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 7, 11152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao Z., Zhao H., Zhao H., DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 37, e16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wahl S. A., Dauner M., Wiechert W., New tools for mass isotopomer data evaluation in 13C flux analysis: Mass isotope correction, data consistency checking, and precursor relationships. Biotechnol. Bioeng. 85, 259–268 (2004). [DOI] [PubMed] [Google Scholar]
- 70.He L., Wu S. G., Zhang M., Chen Y., Tang Y. J., WUFlux: An open-source platform for 13C metabolic flux analysis of bacterial metabolism. BMC Bioinformatics 17, 444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo Y., Zhang R., Wang J., Qin R., Feng J., Chen K., Wang X., Engineering yeasts to Co-utilize methanol or formate coupled with CO2 fixation. Metab. Eng. 84, 1–12 (2024). [DOI] [PubMed] [Google Scholar]
- 72.Schubert M., Lindgreen S., Orlando L., AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo F., Wu M., Zhang S., Feng Y., Jiang Y., Jiang W., Xin F., Zhang W., Jiang M., Improved succinic acid production through the reconstruction of methanol dissimilation in Escherichia coli. Bioresour. Bioprocess. 9, 62 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Couderc R., Baratti J., Oxidation of methanol by the yeast, Pichia pastoris. Purification and properties of the alcohol oxidase. Agric. Biol. Chem. 44, 2279–2289 (1980). [Google Scholar]
- 77.Nash T., The colorimetric estimation of formaldehyde by means of the hantzsch reaction. Biochem. J. 55, 416–421 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beber M. E., Gollub M. G., Mozaffari D., Shebek K. M., Flamholz A. I., Milo R., Noor E., eQuilibrator 3.0: A database solution for thermodynamic constant estimation. Nucleic Acids Res. 50, D603–D609 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S27
Tables S1 to S5
Legends for data S1 and S2
Data S1 and S2