Abstract
Importance:
Persons with substance use disorders (SUD) often suffer from additional comorbidities, including psychiatric conditions and physical health problems. Researchers have explored this overlap in electronic health records (EHR) using phenome wide association studies (PheWAS) to characterize how different indicators are related to all conditions in an individual’s EHR. However, analyses have been largely cross-sectional in nature.
Objective:
To characterize whether various social and genetic risk factors are associated with time to comorbid diagnoses in electronic health records (EHR) after the first diagnosis of SUD.
Design:
Leveraging those with EHR and whole-genome sequencing data in All of Us (N = 287,012), we explored whether social determinants of health are associated with lifetime risk of SUD. Next, within those with a diagnosed SUD (N = 17,460), we examined whether polygenic scores (PGS) were associated with time to comorbid diagnoses performing a phenome-wide survival analysis.
Setting:
Participating health care organizations across the United States.
Participants:
Participants in the All of Us Research Program with available EHR and genomic data,
Exposures:
Social determinants of health and polygenic scores (PGS) for psychiatric and substance use disorders,
Main Outcomes and Measures:
Phecodes for diagnoses derived from International Statistical Classification of Diseases, Ninth and Tenth Revisions, Clinical Modification, codes from EHR.
Results:
Multiple social and demographic risk factors were associated with lifetime SUD diagnosis. Most strikingly, those reporting an annual income <$10K had 4.5 times the odds of having an SUD diagnosis compared to those reporting $100-$150K annually (OR = 4.48, 95% CI = 4.01, 5.01). PGSs for alcohol use disorders, schizophrenia, and post-traumatic stress disorder were associated with time to their respective diagnoses (HRAUD = 1.10, 95% CI = 1.06, 1.14; HRSCZ = 1.13, 95% CI = 1.06, 1.20; HRPTSD = 1.15, 95% CI = 1.08, 1.22). A PGS for ever-smoking was associated with time to subsequent smoking related comorbidities and additional SUD diagnoses HRSMOK = 1.6 to 1.16).
Conclusions and Relevance:
Social determinants, especially those related to income have profound associations with lifetime SUD risk. Additionally, PGS may include information related to outcomes above and beyond lifetime risk, including timing and severity.
INTRODUCTION
Psychiatric disorders have far-reaching consequences for affected individuals, their families, communities, and the broader society1–4. Substance use disorders (SUD) in particular are extremely detrimental. An estimated 107,000 Americans died as the result of an overdose in 20215. In 2016, alcohol use contributed 4.2% to the global disease burden and other drug use contributed 1.3%4. Excessive alcohol use and illicit drug use cost the United States an annual $250 billion6 and $190 billion7 respectively. Given the substantial human and economic costs of substance use disorders, understanding the ways these can contribute to risk for comorbidity has important public health implications.
The proliferation of large-scale biobanks – such as The Million Veteran Program8, FinnGen9, Biobank Japan10, UK Biobank11, and more recently, All of Us12 – linking individual-level genomic data with electronic health records (EHRs) presents opportunities to further explore the relationships between SUD, or any contributory indicators (e.g., clinical diagnoses, polygenic scores), and a wide range of outcomes. This hypothesis-free approach, referred to as a phenome-wide association study (PheWAS)10, can aid us in better understanding patterns of comorbidity. Recent PheWAS using polygenic scores (PGS) have identified widespread associations between PGSs and a host of psychiatric and other medical diagnoses11–13 spanning all bodily systems (e.g., suicidal thoughts and behaviors, drug induced psychosis, viral hepatitis, etc.) and have significant potential implications for public health strategies.
However, previous PheWAS have primarily used cross-sectional data among populations most similar to European reference panels. In the current study, we move beyond the focus of lifetime diagnosis in PheWAS to characterize whether different aspects of genetic risk, in the form of PGS, are associated with time to appearance of comorbid diagnoses in the EHR after the first indication of SUD. We do so by utilizing longitudinal EHR data from the All of Us Research Program, which is more demographically inclusive, including Black/African-American (34%) and Hispanic or Latino/a/x (15%) participants, than previous work in this area. Harnessing the longitudinal quality of the All of Us Research Programs’ EHR data, we explored whether: 1) social determinants of health were associated with lifetime risk of SUD and 2) PGSs for psychiatric and substance use disorders were associated with the onset of comorbidities post-SUD diagnosis, by performing a phenome-wide survival analysis. Our analyses inform the degree to which genetic and environmental risk factors are important in the course of SUD related medical problems (e.g., suicidal thoughts and behaviors, drug induced psychosis, viral hepatitis, etc.).
The clinical utility of PGSs is an ongoing area of debate13–18. In the current analysis, we add to the discussion by exploring the potential use of polygenic scores within individuals who are already diagnosed. We move beyond a focus of lifetime associations in PheWAS to characterize whether different elements of genetic risk, in the form of polygenic scores, are associated with onset of comorbid diagnoses appearing in the EHR after the first record entry of substance use disorder (SUD). Exploring the influence of polygenic scores for SUD and other co-occurring psychiatric and medical diagnoses or traits can help characterize the vast pleiotropic impact of genetic risk for SUD, which could reflect shared genetic effects (biological pleiotropy, e.g., a common liability contributes to risk for both SUD and PTSD, explaining the comorbidity) or a causal chain of events (mediated pleiotropy, e.g., liability for AUD contributes to increased alcohol use, resulting in liver disease). Knowledge gained regarding the potential medical complications likely to arise in individuals with SUD with and without genetic risk for SUD can lead to preventative actions or interventions that may reduce the onset of these additional comorbidities. Lastly, we integrated social and genomic data in our analyses given the importance of social and clinical determinants of health in SUD19.
METHODS
The All of Us (AoU) Research Program
All of Us is a prospective, nationwide cohort study aiming to study the effects of environment, lifestyle, and genomics on health outcomes. Participant recruitment is predominantly done through participating health care provider organizations and in partnership with Federally Qualified Health Centers. Interested participants can enroll as direct volunteers, visiting community-based enrollment sites. Enrollment, informed consent, and baseline health surveys are administrated digitally through the All of Us program website (https://joinallofus.org)20. Participants are then invited to undergo a basic physical exam and biospecimen collection at an affiliated healthcare site. There are two types of participant follow-up: passive via linkage with EHR and active by periodic follow-up surveys. We included the full sample for associations between social risk factors and lifetime SUD diagnosis (N = 287,079). For the genomic analyses, we included all participants who had electronic health record (EHR) and short-read whole genome sequence (WGS) data from release 7 (May 6, 2018 to February 23, 2023). Additionally, we excluded those with self-reported military service to remove potential overlap in current genome wide association studies (GWAS) such as the Million Veteran Program (MVP), leaving the final sample with a lifetime SUD and genomic data at N = 12,831.
Genotyping
Approximately 245,000 individuals in the current release of AoU have available short-read sequencing data. To ensure consistency across sites for DNA extraction and sequencing, AoU developed a standardized process and QC metrics across sites. Sequencing was performed on the Illumina NovaSeq 6000 instrument. A full description of the collection, harmonization, sequencing, and population assignment pipeline has been published elsewhere21. To adjust for population structure in genetic analyses, participants were assigned to genetic similarity clusters of those most similar to European reference panels (EUR-like) and those most similar to African reference panels (AFR-like)22. Genetic principal components (PCs) were generated using the hwe_normalized_pca in Hail23.
Electronic health records (EHRs)
We used phecodes, which are clusters of ICD-9/10-CM codes in the EHR24,25, validated previously26,27, to create lifetime diagnoses for SUD as well as the main outcomes for the PheWAS. For lifetime SUD diagnosis, we considered individuals as meeting criteria for an SUD if they had two or more outpatient or one inpatient occurrence of phecodes 316 (Substance addiction and disorders) or 317 (Alcohol-related disorders) in their EHR. Prior analyses have shown that 2 or more phecodes is a good predictor of diagnosis28,29. We focus exclusively on those with diagnoses of alcohol and drug-related disorders, excluding tobacco use disorders. Alcohol and other drug use disorders have the strongest evidence of a common etiology30,31, and this shared risk only overlaps minimally with risk for tobacco related disorders32. For PheWAS outcomes, we used the presence of any phecode following the initial diagnosis of SUD. We excluded phecodes for which there were fewer than 50 individuals that met criteria for a diagnosis.
Social and demographic risk factors
We included a variety of social and demographic measures available from the baseline AoU survey. For the current analysis we included age, gender (man, woman, transgender/non-binary), race-ethnicity (Non-Hispanic White, Black/African-American, Hispanic or Latino/a/x, Asian American/Pacific Islander, Multi-racial, and other race-ethnicity), education (less than high school, HS diploma or GED, some college, college graduate, and advanced degree), household income (less than $10k to more than $200k), marital status (never married, married, cohabitating, divorced, separated, and widowed), health insurance (have vs not), and place of birth (US born vs foreign born). We focused on the association between social risk factors and lifetime SUD diagnosis, only. Because these items are measured at baseline entry into AoU and not necessarily time of SUD diagnoses, we lack the ability to establish whether potentially time-variant measures were the same before their first SUD diagnosis.
Polygenic scores (PGS)
We estimated PGSs, which are aggregate measures of the number of risk alleles individuals carry weighted by effect sizes from GWAS summary statistics, from multiple large-scale GWASs33–38 (Supplemental Table S1). We included these specific PGSs because: 1) there is strong genetic overlap between psychiatric and substance use disorders34–36,38–40, and 2) each of these GWAS included results for AFR-like participants, allowing us to move beyond EUR-like only analyses. We created PGSs using PRS-CSx41, a Bayesian regression and continuous shrinkage method that estimates the posterior effect sizes for each SNP in a given set of GWAS summary statistics. PGS accuracy decays continuously as target samples differ in ancestral background from the discovery GWAS, even within relatively homogenous genetic clusters42. PRS-CSx uses inputs from multiple genomic similarity clusters to improve power of PGS in underpowered samples, typically those who are not in the EUR-like groups. We standardized all PGSs to Z-scores.
We note that our paper includes language related to both race-ethnicity, which reflects socially-constructed categories, and genetic similarity, which uses empirical assignment based on available reference panels, because both are relevant for the current analyses. First, prior work has established race-ethnicity (and with it, racism and discrimination) are relevant for disparities in SUDs43–47. Second, informed by best practices for current approaches to handling genetic data from diverse populations48, we stratified analyses by genetic similarity and included genetic principal components, which limit the possibility of false positives due to population stratification. The inclusion of both concepts is in no way endorsing the notion that these reflect discrete biological categories.
Analytic plan
First, we examined the association between social, demographic, and genetic measures and lifetime SUD diagnosis using logistic regression. Next, we expanded on the traditional PheWAS approach by performing a phenome wide survival analysis. Rather than focusing on lifetime diagnosis, as is the case in the typical PheWAS approach, we utilized Cox proportional hazards models to estimate the association (in the form of hazard ratios, or HR) between time from SUD diagnosis to first documented phecode diagnosis. In survival analysis, the hazard ratio (HR) is a measure used to compare the risk of an event occurring at any given point in time. A HR greater than 1 indicates an increased risk of the event, while a HR less than 1 suggests a reduced risk. For example, if the HR for a PGS is 1.5, it suggests that they are 1.5 times more likely to experience the comorbidity earlier for every one unit increase in PGS. We included the earliest diagnosis for each EHR code. In the PGS models, we stratified by genetic similarity grouping (AFR-like and EUR-like) and included age at SUD diagnosis, gender, and the first 10 genetic principal components as covariates. We performed all survival models using the survival package (version 3.7.0) in R. Finally, we meta-analyzed results via fixed-effects meta-analysis in the meta package (version 7.0.0) in R. To correct for multiple testing, we applied a false discovery rate (FDR)49 of 5%.
RESULTS
Defining persons with lifetime SUD in EHR
Using the 2+ outpatient/1+ inpatient definition of substance use disorder diagnosis, we identified N = 17,460 individuals who met our criteria for a lifetime SUD (excluding nicotine/tobacco use disorders and those who reported active duty military service). Demographic characteristics for the full sample with available EHR data and those who met our criteria for SUD are presented in Table 1. Relative to the full sample, the SUD group was more likely to be younger, men, born in the United States, and identify as Black/African-American. This demographic breakdown of the SUD sample largely reflects the patterns seen in nationally representative samples43,44.
Table 1:
Sample Demographics in All of Us participants with available EHR data
| Full sample (N = 287, 012) | SUD (N = 17,460) | ||||
|---|---|---|---|---|---|
| N/Mean | %/SD | N/Mean | %/SD | X 2 | |
| Age (mean) | 55.10 | 0.02 | 51.40 | 0.29 | * |
| Woman | 171,431 | 59.73 | 8,310 | 47.59 | * |
| Man | 108,209 | 37.70 | 8,585 | 49.17 | * |
| Transgender/Non-Binary | 1,893 | 0.66 | 142 | 0.81 | |
| Gay/Lesbian/Bisexual | 24,706 | 8.61 | 2,259 | 12.94 | * |
| Straight/Heterosexual | 252,386 | 87.94 | 14,406 | 82.51 | * |
| Asian | 8,038 | 2.80 | 81 | 0.46 | * |
| Black/African American | 57,177 | 19.92 | 5,883 | 33.69 | * |
| Hispanic or Latino/a/x | 47,699 | 16.62 | 2,668 | 15.28 | * |
| MENA | 1,605 | 0.56 | 50 | 0.29 | * |
| Non-Hispanic White | 150,402 | 52.40 | 7,216 | 41.33 | * |
| Native Hawaiian/Pacific Islander | 297 | 0.10 | 23 | 0.13 | * |
| Multiple race-ethnicities | 10,771 | 3.75 | 668 | 3.83 | * |
| Other race-ethnicity not listed | 3,017 | 1.05 | 261 | 1.49 | * |
| Born outside the US | 43,635 | 15.20 | 1,095 | 6.27 | * |
| US-born | 238,162 | 82.98 | 15,969 | 91.46 | * |
| Less than high school | 27,867 | 9.71 | 3,417 | 19.57 | * |
| High school diploma or GED | 56,778 | 19.78 | 5,822 | 33.34 | * |
| Some college | 72,938 | 25.41 | 4,771 | 27.33 | * |
| College graduate | 62,550 | 21.79 | 1,664 | 9.53 | * |
| Advanced degree | 57,696 | 20.10 | 922 | 5.28 | * |
p < .05, MENA = Middle East and Nort Africa
Social and Demographic results
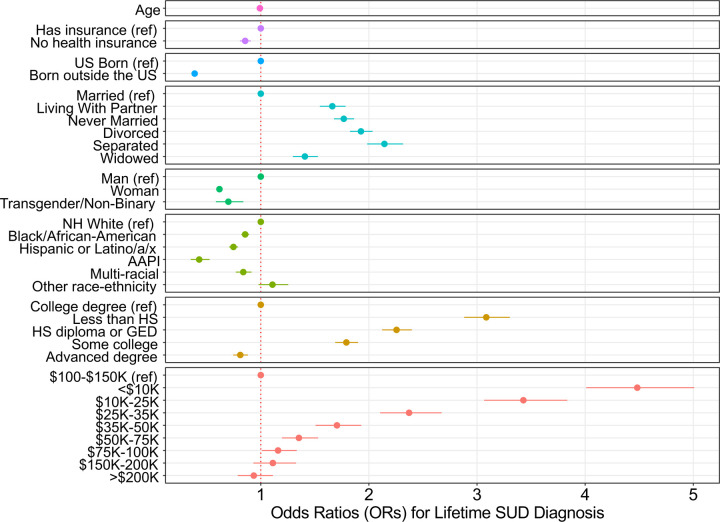
Figure 1 presents the results from the multivariable logistic regression model examining the association between social and demographic risk factors and lifetime SUD diagnosis in the full phenotypic sample (N = 287,079). These results largely mimicked prior analyses in earlier versions of the AoU data50, whereby there was a stark gradient in lifetime SUD diagnosis across education and income, and those with the lowest income (OR = 4.482; CI = 4.013, 5.006) and education (OR = 3.085; CI = 2.883, 3.302) were at greatest risk for diagnosis. Additionally, women (OR = 0.618; CI = 0.598, 0.639), transgender and non-binary participants (OR = 0.700; CI = 0.585, 0.838), and those born outside of the US (OR = 0.388; CI = 0.361, 0.418) all had lower odds of a lifetime diagnosis. Relative to non-Hispanic White participants, all other racial-ethnic groups were at lower risk. Interestingly, though the base rates of lifetime SUD diagnosis for Black/African-American participants were higher than others, this relationship was reversed after accounting for the other social factors (OR = 0.856; CI = 0.821, 0.892). The full results are available in Supplemental Table S2.
Figure 1: Association results for social risk factors and lifetime SUD diagnosis.
Adjusted odds ratios (OR) and corresponding 95% confidence intervals presented on the x-axis. Reference categories indicated by (ref). Models included each of the above measures, simultaneously. NH = Non-Hispanic, AAPI = Asian American or Pacific Islander, HS = high school, GED = general educational development.
PGS Survival PheWAS
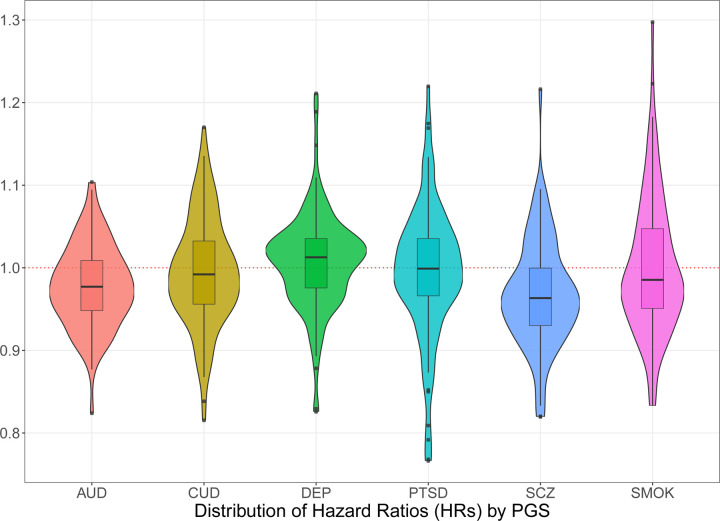
Moving from the full sample to those with an SUD diagnosis only, we further constrained to those of AFR-like and EUR-like genetic similarity for subsequent within-SUD survival models using PGSs (N = 12,831, see Supplemental Table S3 for stratified results). Figure 2 present the distribution of HRs for each of the PGSs, highlighting the varying magnitude of effects sizes. Associations between each PGS and phecodes were enriched for reduced time to corresponding diagnoses. After adjusting for multiple testing, only 19 of the associations remained significant (Table 2, see Supplemental Table S4 for full results). Of all the PGSs, the PGS for lifetime smoking initiation (SMOK) showed the greatest number of trait associations, the strongest being with chronic airway obstruction (HR =1.163; CI = 1.108, 1.220), emphysema (HR = 1.140; CI = 1.061, 1.226), congestive heart failure NOS (HR = 1.140; CI = 1.070, 1.214), and viral hepatitis C (HR = 1.124; CI = 1.065, 1.186). Importantly, PGSs for SCZ, PTSD, and AUD were associated with time to SCZ (HR = 1.126; CI = 1.060, 1.195), PTSD (HR = 1.147; CI = 1.082, 1.217), and AUD (HR = 1.103; CI = 1.064, 1.142), respectively.
Figure 2: Distribution of hazard ratios for PGSs from multivariable survival PheWAS.
Violin and box-and-whisker plots for conditional hazard ratios (HR) between each PGS and phecodes in the meta-analyzed results. All models included each PGS, simultaneously. Thick black line in the center of each box represents the median value. The edges of the box represent the 25th and 75th percentile. AUD = alcohol use disorder, CUD = cannabis use disorder, DEP = depression, PTSD = post-traumatic stress disorder, SMOK = ever smoker, SCZ = schizophrenia.
Table 2:
Phenome wide Significant Associations from Survival Models
| PGS | Phecode description | HR | 95% CI (lower) | 95% CI (upper) |
|---|---|---|---|---|
| SMOK | Chronic airway obstruction | 1.163 | 1.108 | 1.220 |
| PTSD | Posttraumatic stress disorder | 1.147 | 1.082 | 1.217 |
| SMOK | Emphysema | 1.140 | 1.061 | 1.226 |
| SMOK | Congestive heart failure (CHF) NOS | 1.140 | 1.070 | 1.214 |
| SMOK | Obstructive chronic bronchitis | 1.129 | 1.060 | 1.204 |
| SCZ | Schizophrenia | 1.126 | 1.060 | 1.195 |
| SMOK | Viral hepatitis C | 1.124 | 1.065 | 1.186 |
| SMOK | Tobacco use disorder | 1.116 | 1.086 | 1.148 |
| AUD | Alcohol-related disorders | 1.103 | 1.064 | 1.142 |
| SMOK | Substance addiction and disorders | 1.055 | 1.030 | 1.081 |
| AUD | GERD | 0.937 | 0.910 | 0.966 |
| AUD | Sepsis | 0.909 | 0.862 | 0.959 |
| AUD | Inflammatory and toxic neuropathy | 0.899 | 0.856 | 0.945 |
| SMOK | Hypothyroidism NOS | 0.897 | 0.846 | 0.951 |
| SCZ | Migraine | 0.896 | 0.852 | 0.942 |
| AUD | Herpes zoster | 0.795 | 0.717 | 0.881 |
| AUD | Glucocorticoid deficiency | 0.790 | 0.693 | 0.899 |
| AUD | Neutropenia | 0.747 | 0.639 | 0.872 |
| CUD | Cystic mastopathy | 0.689 | 0.576 | 0.825 |
HR = hazard ratio; PGS = polygenic score; SMOK = ever smoker; PTSD = post-traumatic stress disorder; SCZ = schizophrenia; AUD alcohol use disorder; CUD = cannabis use disorder; NOS =. Not otherwise specified; GERD = Gastroesophageal reflux disease. Full results available in Supplementary Table S4.
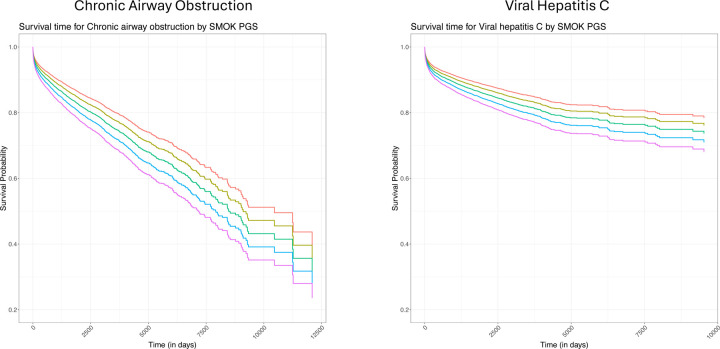
Figure 3 presents two of the top associations between the SMOK PGS and time to subsequent chronic airway obstruction and viral hepatitis C codes. Here PGSs were stratified by standard deviations (SD), ranging from −2 SD (red) to +2 SD (purple). While the overall effect of the PGSs was small, by the end of observation, the difference in probability of “survival” between these two extremes was approximately 15% lower for chronic airway obstruction and 10% lower for viral hepatitis C in the +2 SD group.
Figure 3: Survival plots for chronic airway obstruction and viral hepatis C across SMOK PGS levels.
Predicted survival curves for chronic airway obstruction and viral hepatis C as a function of SMOK PGS levels (red = −2 SD, yellow = −1 SD, green = mean, blue = +1 SD, purple = +2 SD). All models adjusted for age at diagnosis, gender, genetic similarity, and first 10 genetic principal components (PCS).
DISCUSSION
Large-scale biobanks and genetic data are becoming increasingly prevalent in health research, offering the potential to explore multiple comorbidities simultaneously. The goal of precision medicine is to realize the potential of harnessing these sources of information from to personalize treatment and stratify risk. However, much of the work to date has focused on lifetime risk for a single diagnosis. In the current analyses, we expand upon existing research by focusing on risk longitudinally and within persons who have a SUD diagnosis to begin to understand what value genetic and social risk factors might play in helping those who already have a diagnosis.
We found multiple social and demographic risk factors associated with lifetime SUD diagnosis. These results largely mimic prior findings in AoU50 and in the population more broadly43,44, whereby those who largely occupy marginalized positions are at greater risk for having a SUD. The most striking finding is the gradient in income, whereby those reporting an annual household income less than $10k annually had 4.5 times the odds of having an SUD diagnosis, reflecting either social determinants as risk factors or the disabling impact of SUD. Importantly the increased rate of diagnosis among those who identify as Black or African-American reversed after accounting for other social determinants of health, such that they were at decreased risk relative to Non-Hispanic White participants. This finding suggests the increased rates of diagnoses in this population may reflect socioeconomic inequalities.
Focusing on genetic risk within persons who had a SUD diagnosis, polygenic scores for AUD, SCZ, and SMOK demonstrated enrichment for a longer time to corresponding diagnoses in the EHR. While this could be interpreted as some type of protective effect, the more likely explanation is those at greater risk are less likely to treatment51,52, especially as barriers to access, such as health insurance and economic resources, were key predictors in the social demographic correlates. In terms of specific PGS-phecode associations, several remained significant after correcting for multiple testing, including associations between PGSs for SCZ, PTSD, and AUD and the time to their corresponding diagnoses. These results increase confidence that associations are not merely spurious, as these PGSs demonstrated specificity for the expected outcomes. The results also highlight the potential for assessing risk in comorbid conditions among those already diagnosed with a SUD. PGS for lifetime smoking initiation was associated with risk for a variety of smoking-related conditions as well as other correlates of SUD (e.g. viral hepatitis C). Prior work on externalizing disorders suggests that “ever smoker” is a strong proxy for externalizing risk31,53, offering a glimpse into one potential mechanism linking this PGS to the comorbid diagnoses.
This work has several important limitations. First, we lacked the social and demographic risk factors measured at time of diagnosis and could not establish the time ordering between these measures. As the AoU database grows through recruitment, data generation, and passive data collection via constantly updating EHRs, we will be able to explore the prospective association between important social determinants of health and diagnoses. Second, the use of EHR data, while convenient, is also confounded by access and healthcare utilization. We cannot discern whether associations are truly reflective of the progression in disease process or whether they reflect propensity for help seeking when available in a healthcare setting. Lastly, though we have attempted to be more inclusive in the genetic analyses, we still lacked sufficient GWAS summary statistics for other genetic similarity groups to include here. As a field, we must continue to strive to ensure results from genetic discoveries can be applied to all populations, equitably48.
Harnessing the longitudinal data in the All of Us database, we have demonstrated the importance of various social conditions for the lifetime diagnosis of substance use disorders. Annual income is especially relevant in disparities of SUD. Additionally, we have also shown that in persons who have a SUD diagnosis, various measures of genetic risk were associated with time to subsequent diagnoses in their EHR, moving beyond typical cross-sectional approaches. While there is considerable work to be done for the use of social, clinical, and biological data within a healthcare setting, the current results demonstrate the potential of these approaches. Future work should endeavor to integrate across levels of analysis in a longitudinal framework.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported in part by National Institute of Mental Health (R01MH125938: Drs Peterson, Bigdeli, Chatzinakos, and Meyers); the National Institute of Drug Abuse (R01DA050721: Dr Barr; R01DA060596: Drs Meyers, Barr, Chatzinakos, and Neale), and the National Institute on Alcohol Abuse and Alcoholism (R01AA030010: Drs Meyers, Barr, Chatzinakos, and Neale). Computing costs associated with this work was supported by R01MH125938 (PI: Peterson). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276. In addition, the All of Us Research Program would not be possible without the partnership of its participants.
REFERENCES
- 1.Global Ferrari A., regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry. 2022;9(2):137–150. doi: 10.1016/S2215-0366(21)00395-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray CJL, Mokdad AH, Ballestros K, et al. The state of US health, 1990–2016: Burden of diseases, injuries, and risk factors among US states. JAMA - Journal of the American Medical Association. 2018;319(14):1444–1472. doi: 10.1001/jama.2018.0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reitsma MB, Fullman N, Ng M, et al. Smoking prevalence and attributable disease burden in 195 countries and territories, 1990–2015: a systematic analysis from the Global Burden of Disease Study 2015. The Lancet. 2017;389(10082):1885–1906. doi: 10.1016/S0140-6736(17)30819-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degenhardt L, Charlson F, Ferrari A, et al. The global burden of disease attributable to alcohol and drug use in 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Psychiatry. 2018;5(12):987–1012. doi: 10.1016/S2215-0366(18)30337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.U.S. Overdose Deaths In 2021. Increased Half as Much as in 2020 - But Are Still Up 15%. Accessed May 15, 2022. https://www.cdc.gov/nchs/pressroom/nchs_press_releases/2022/202205.htm
- 6.Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. 2010 National and State Costs of Excessive Alcohol Consumption. Am J Prev Med. 2015;49(5):e73–e79. doi: 10.1016/j.amepre.2015.05.031 [DOI] [PubMed] [Google Scholar]
- 7.National Drug Intelligence Center. National Drug Threat Assessment. Vol 2019. United States Department of Justice; 2011. [Google Scholar]
- 8.Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- 9.Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–518. doi: 10.1038/s41586-022-05473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagai A, Hirata M, Kamatani Y, et al. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol. 2017;27(3):S2–S8. doi: 10.1016/j.je.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The All of Us Research Program Investigators. The “All of Us” Research Program. New England Journal of Medicine. 2019;381(7):668–676. doi: 10.1056/nejmsr1809937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis CM, Vassos E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020;12(1):1–11. doi: 10.1186/s13073-020-00742-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. doi: 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wray NR, Lin T, Austin J, et al. From Basic Science to Clinical Application of Polygenic Risk Scores: A Primer. JAMA Psychiatry. 2021;78(1):101–109. doi: 10.1001/jamapsychiatry.2020.3049 [DOI] [PubMed] [Google Scholar]
- 16.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19(9):581–590. doi: 10.1038/s41576-018-0018-x [DOI] [PubMed] [Google Scholar]
- 17.Murray GK, Lin T, Austin J, McGrath JJ, Hickie IB, Wray NR. Could Polygenic Risk Scores Be Useful in Psychiatry?: A Review. JAMA Psychiatry. 2021;78(2):210–219. doi: 10.1001/jamapsychiatry.2020.3042 [DOI] [PubMed] [Google Scholar]
- 18.Klarin D, Natarajan P. Clinical utility of polygenic risk scores for coronary artery disease. Nat Rev Cardiol. 2022;19(5):291–301. doi: 10.1038/s41569-021-00638-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barr PB, Driver MN, Kuo SIC, et al. Clinical, environmental, and genetic risk factors for substance use disorders: characterizing combined effects across multiple cohorts. Mol Psychiatry. 2022;27(11):4633–4641. doi: 10.1038/s41380-022-01801-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Institutes of Health. “All of Us” Research Program. U.S Department of Health and Human Service- National Institute of Health. 2021. https://allofus.nih.gov/ [Google Scholar]
- 21.Bick AG, Metcalf GA, Mayo KR, et al. Genomic data in the All of Us Research Program. Nature. 2024;627(8003):340–346. doi: 10.1038/s41586-023-06957-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Academies of Sciences and Medicine E. Using Population Descriptors in Genetics and Genomics Research: A New Framework for an Evolving Field. The National Academies Press; 2023. doi: 10.17226/26902 [DOI] [PubMed] [Google Scholar]
- 23.Hail Team. Hail 0.2. Accessed December 10, 2024. https://github.com/hail-is/hail
- 24.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. doi: 10.1038/nbt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: Workflow development and initial evaluation. J Med Internet Res. 2019;21(11):1–13. doi: 10.2196/14325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei WQ, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. Journal of the American Medical Informatics Association. 2016;23(e1):20–27. doi: 10.1093/jamia/ocv130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei WQ, Bastarache LA, Carroll RJ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One. 2017;12(7):e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheutlin AB, Dennis J, Linnér RK, et al. Penetrance and pleiotropy of polygenic risk scores for schizophrenia in 106,160 patients across four health care systems. American Journal of Psychiatry. 2019;176(10):846–855. doi: 10.1176/appi.ajp.2019.18091085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bigdeli TB, Voloudakis G, Barr PB, et al. Penetrance and Pleiotropy of Polygenic Risk Scores for Schizophrenia, Bipolar Disorder, and Depression among Adults in the US Veterans Affairs Health Care System. JAMA Psychiatry. 2022;79(11):1092–1101. doi: 10.1001/jamapsychiatry.2022.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barr PB, Dick DM. The Genetics of Externalizing Problems. Curr Top Behav Neurosci. 2020;47:93–112. doi: 10.1007/7854_2019_120 [DOI] [PubMed] [Google Scholar]
- 31.Karlsson Linnér R, Mallard TT, Barr PB, et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci. Published online 2021:1–10. doi: 10.1038/s41593-021-00908-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kendler KS, Myers J. The boundaries of the internalizing and externalizing genetic spectra in men and women. Psychol Med. 2014;44(3):647–655. doi: 10.1017/S0033291713000585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders GRB, Wang X, Chen F, et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 2022 612:7941. 2022;612(7941):720–724. doi: 10.1038/s41586-022-05477-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levey DF, Stein MB, Wendt FR, et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci. Published online May 27, 2021. doi: 10.1038/s41593-021-00860-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levey DF, Galimberti M, Deak JD, et al. Multi-ancestry genome-wide association study of cannabis use disorder yields insight into disease biology and public health implications. Nat Genet. 2023;55(12):2094–2103. doi: 10.1038/s41588-023-01563-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nievergelt CM, Maihofer AX, Atkinson EG, et al. Genome-wide association analyses identify 95 risk loci and provide insights into the neurobiology of post-traumatic stress disorder. Nat Genet. 2024;56(5):792–808. doi: 10.1038/s41588-024-01707-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kranzler HR, Zhou H, Kember RL, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10(1):1499. doi: 10.1038/s41467-019-09480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou H, Kember RL, Deak JD, et al. Multi-ancestry study of the genetics of problematic alcohol use in over 1 million individuals. Nat Med. 2023;29(12):3184–3192. doi: 10.1038/s41591-023-02653-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deak JD, Zhou H, Galimberti M, et al. Genome-wide association study in individuals of European and African ancestry and multi-trait analysis of opioid use disorder identifies 19 independent genome-wide significant risk loci. Mol Psychiatry. 2022;27(10):3970–3979. doi: 10.1038/s41380-022-01709-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trubetskoy V, Pardiñas AF, Qi T, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604(7906):502–508. doi: 10.1038/s41586-022-04434-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan Y, Lin YF, Feng YCA, et al. Improving polygenic prediction in ancestrally diverse populations. Nat Genet. 2022;54(5):573–580. doi: 10.1038/s41588-022-01054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Y, Hou K, Xu Z, et al. Polygenic scoring accuracy varies across the genetic ancestry continuum. Nature. 2023;618(7966):774–781. doi: 10.1038/s41586-023-06079-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grant BF, Goldstein RB, Saha TD, et al. Epidemiology of DSM-5 alcohol use disorder results from the national epidemiologic survey on alcohol and related conditions III. JAMA Psychiatry. 2015;72(8):757–766. doi: 10.1001/jamapsychiatry.2015.0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant BF, Saha TD, June Ruan W, et al. Epidemiology of DSM-5 drug use disorder results from the national epidemiologic survey on alcohol and related conditions-III. JAMA Psychiatry. 2016;73(1):39–47. doi: 10.1001/jamapsychiatry.2015.2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams DR. Race, Socioeconomic Status, and Health The Added Effects of Racism and Discrimination. Ann N Y Acad Sci. 1999;896(1):173–188. doi: 10.1111/j.1749-6632.1999.tb08114.x [DOI] [PubMed] [Google Scholar]
- 46.Williams DR, Collins C. Racial residential segregation: a fundamental cause of racial disparities in health. Public Health Rep. 2001;116(5):404–416. http://www.ncbi.nlm.nih.gov/pubmed/12042604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams DR, Mohammed SA. Discrimination and racial disparities in health: evidence and needed research. J Behav Med. 2009;32(1):20–47. doi: 10.1007/s10865-008-9185-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterson RE, Kuchenbaecker K, Walters RK, et al. Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell. 2019;179(3):589–603. doi: 10.1016/j.cell.2019.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. http://www.jstor.org/stable/2346101 [Google Scholar]
- 50.Barr PB, Bigdeli TB, Meyers JL. Prevalence, Comorbidity, and Sociodemographic Correlates of Psychiatric Diagnoses Reported in the All of Us Research Program. JAMA Psychiatry. 2022;79(6):622–628. doi: 10.1001/jamapsychiatry.2022.0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mojtabai R, Evans-Lacko S, Schomerus G, Thornicroft G. Attitudes Toward Mental Health Help Seeking as Predictors of Future Help-Seeking Behavior and Use of Mental Health Treatments. Psychiatric Services. 2016;67(6):650–657. doi: 10.1176/appi.ps.201500164 [DOI] [PubMed] [Google Scholar]
- 52.Blanco C, Iza M, Rodríguez-Fernández JM, Baca-García E, Wang S, Olfson M. Probability and predictors of treatment-seeking for substance use disorders in the U.S. Drug Alcohol Depend. 2015;149:136–144. doi: 10.1016/j.drugalcdep.2015.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deak JD, Clark DA, Liu M, et al. Alcohol and nicotine polygenic scores are associated with the development of alcohol and nicotine use problems from adolescence to young adulthood. Addiction. 2022;117(4):1117–1127. doi: 10.1111/add.15697 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.