Abstract
Despite their pivotal role, the evolutionary origins of vertebrate digestive systems remain enigmatic. We explored the cellular characteristics of the amphioxus (Branchiostoma floridae) digestive tract, a model for the presumed primitive chordate digestive system, using bulk tissue companioned with single-cell RNA sequencing. Our findings reveal segmentation and a rich diversity of cell clusters, and we highlight the presence of epithelial-like, ciliated cells in the amphioxus midgut and describe three types of endocrine-like cells that secrete insulin-like, glucagon-like, and somatostatin-like peptides. Furthermore, Pdx, Ilp1, Ilp2, and Ilpr knockout amphioxus lines revealed that, in amphioxus, Pdx does not influence Ilp expression. We also unravel similarity between amphioxus Ilp1 and vertebrate insulin-like growth factor 1 (Igf1) in terms of predicted structure, effects on body growth and amino acid metabolism, and interactions with Igf-binding proteins. These findings indicate that the evolutionary alterations involving the regulatory influence of Pdx over insulin gene expression could have been instrumental in the development of the vertebrate digestive system.
Single-cell profiling of the amphioxus digestive tract reveals scattered distribution of three endocrine-like cell clusters.
INTRODUCTION
Vertebrate animals share a unique digestive accessory organ, the pancreas, consisting of a multiplex of exocrine and endocrine cell types with debated evolutionary origin (1). Specifically, the vertebrate pancreas contains multiple endocrine cell types that secrete different peptide hormones including insulin, glucagon, and somatostatin, which allow fine-tuning of glucose metabolism, appetite, and body growth (2). Among the many vertebrate pancreatic markers, the ParaHox gene Pdx [also referred to as Xlox (3), XlHbox 8 (4), IPF1 (5), IDX-1 (6), STF-1 (7), and IUF1 (8)] is highlighted as a master regulator of pancreatic development (9, 10). This highly conserved transcription factor (TF) defines a highly restricted region in the gut endoderm of bilaterian animals (3), and gene knockout experiments in vertebrates have established that Pdx is essential for formation of the pancreas (11, 12). In addition, Pdx expression in adult vertebrates is essential for maintaining pancreatic β cell identity and is required for normal regulation of insulin gene expression (13).
Outside the vertebrate clade, less is known about Pdx function and much less about potential interaction between Pdx and the pancreatic hormone insulin. In Ecdysozoa, the Pdx gene is secondarily lost in almost all investigated animals, including the two most popular invertebrate model animals, the fruit fly (Drosophila melanogaster) and roundworm (Caenorhabditis elegans) (14). However, both animals encode a number of different insulin-like peptides (ILPs), among which some serve functions similar to vertebrate insulin. Three of eight ILPs in the fruit fly play an essential role in glucose level control and fat storage, but are secreted by specialized cells in the brain and not the digestive tract (15, 16). For the roundworm, 40 putative ILPs have been identified in sensory neurons and interneurons, and some are crucial for metabolism control and entrance/exit from different life stages under starved conditions (17, 18). In Lophotrochozoa, a study on the Pacific oyster (Crassostrea gigas) suggests a potential regulatory role of the oyster Pdx protein over Ilp gene expression (19). In contrast, little is known about Pdx and Ilp interaction in invertebrate deuterostomes, although both Pdx and Ilp expression patterns in the sea urchin (Strongylocentrotus purpuratus) and tunicate (Ciona intestinalis) have been characterized in detail using in situ hybridization (20–23). Specifically for tunicates, expression of Pdx, Ilp, and multiple pancreas-related digestive enzyme genes have been detected in different regions along the adult and juvenile intestinal chamber, suggesting that multiple regions rather than one digestive region may serve functions similar to the vertebrate pancreas (21, 23).
The cephalochordate Branchiostoma floridae (referred to as amphioxus) has been a popular model animal used for research into the evolution of vertebrates given its close phylogenetic relationship to the vertebrate lineage (24–26). Dissecting amphioxus embryo development and adult tissue profiles has shed light on the evolution of vertebrate organs such as the brain (27, 28), kidney (29, 30), and pituitary (31, 32). For the pancreas, many have debated the existence of pancreatic “glands” and cell types in the amphioxus gut (33–35). Four amphioxus gut cell types have been repeatedly mentioned in the existing literature, including “nondigestive” ciliated cells, endocrine-like cells, “digestive” cells that carry out intracellular digestion, and zymogen-rich enzyme-secreting cells (35, 36). Among pancreatic endocrine-like cells, ILP-secreting cells have been detected using antisera targeting mammal insulin peptides; however, they have not been described in detail (36, 37). In addition, cells secreting glucagon-like peptides have been detected in both the amphioxus gut and Hatschek’s pit (31), a structure connected to the ventral cerebral vesicle and proposed to be functionally similar to the vertebrate adenohypophysis (38, 39). Aside from these cell types, whether a pancreatic-like structure exists in amphioxus remains unclear as recent studies have characterized amphioxus whole gut tissue without making distinction between segments (40) or have only characterized single gene profiles along the amphioxus gut (41). Even in existing amphioxus single-cell datasets, only a preliminary profile of gut tissue is provided due to a relatively low number of genes detected in each cell (42). To the extent of our knowledge, there is only one report of a midgut-specific gene knockout amphioxus line, in which the authors describe lack of green fluorescence in a highly restricted area of the midgut in Pdx mutant 1- to 7-week-old larvae (43). No significant change in Ilp1 gene expression was found in these larvae; however, the authors were not able to test this in postmetamorphosis individuals (43).
In this study, we performed in-depth bulk tissue and single-cell level transcriptomics of wild-type and Pdx mutant amphioxus, and we also used Ilp1, Ilp2, and Ilpr knockout amphioxus to further probe the possible interaction of the Pdx protein and Ilp genes, along with Ilp function in this invertebrate. These data show that Ilp gene expression is not controlled by Pdx, and Ilp mainly regulates body growth in amphioxus. We argue that Pdx-mediated regulation of insulin gene expression and subsequent clustering of insulin-secreting endocrine cells in the gut is likely an acquired trait in the last vertebrate ancestor.
RESULTS
Pronounced segmentation of the amphioxus gut
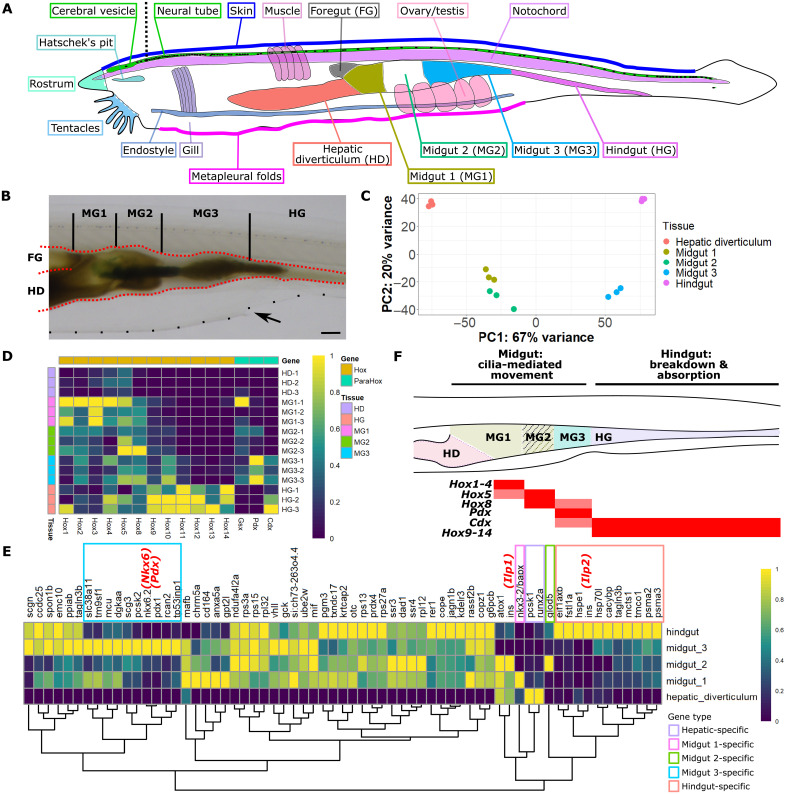
To explore whether the amphioxus gut shows segmentation along the anterior-posterior (A-P) axis, we separated the amphioxus gut into four sections based on anatomical characteristics. We adopt terminology such as “midgut (MG)” and “hindgut (HG)” as opposed to “stomach” or “esophagus” when describing sections in the amphioxus digestive system. The only exception is the “hepatic diverticulum (HD),” a structure forming a protruding sac connected to the amphioxus gut (Fig. 1A). To describe the gut, we use “foregut (FG)” to refer to the short section of gut tissue located anterior to the connection point of the hepatic diverticulum to the gut (Fig. 1, A and B). This section is very short and is difficult to remove from gill and pharyngeal tissue; hence, we focus on the “midgut,” a gut section located posterior to the “foregut” and that is considerably wider in circumference. The “hindgut” refers to the region located posterior to the atriopore, an opening approximately located at the two-thirds of the animal. Within the midgut region, we further separated this region into three sections, including the “midgut 1 (MG1)” section, which is located between the point where the hepatic diverticulum joins the gut and the “midgut 2 (MG2)” section (Fig. 1B). MG2 is visually distinguishable from the other midgut sections as it has a deeper color (Fig. 1B), and we use this section as an anatomical landmark. Subsequently, we defined a “midgut 3 (MG3)” section, which is located posterior to MG2 and anterior to the hindgut.
Fig. 1. Transcriptomic profile of the amphioxus gut.
(A) Adult amphioxus with the location and name of dissected tissues indicated using different colors. A black dotted line indicates the approximate location of the incision made to separate the “cerebral vesicle” and “neural tube.” “Foregut (FG)” is labeled to indicate the location of this section, which was removed during sampling. (B) Gut tissue is highlighted in red, whereas the metapleural folds at the ventral side of body are indicated by a dotted black line. A black arrow indicates the atriopore. Scale bar, 1 mm. (C) PCA plot showing the distribution of all 15 digestive tissue samples. Each type of tissue has three biological repeats obtained from two amphioxus individuals. Normalized expression of (D) amphioxus Hox and ParaHox genes and (E) amphioxus orthologs to zebrafish pancreatic β cell–specific genes. Genes with less than five TPM expression in the five digestive tissues are omitted. All gene IDs are in table S1. In (E), gene names shown are their zebrafish orthologs, with amphioxus Ilp1 and Ilp2 genes highlighted in red, and genes enriched in different gut sections marked in different colored boxes. (F) Summary of gut section profiles in adult amphioxus. Expression pattern of all Hox genes included in (D), and two ParaHox genes, Pdx and Cdx, are indicated below a simplified depiction of the amphioxus gut. Each of the five gut sections are shown in a different color, with the shaded region indicating the section of the gut that is deeper in appearance to the naked eye. Midgut and hindgut function, as predicted by transcriptomic analysis, is indicated above this simplified diagram. HD, hepatic diverticulum; FG, foregut; MG, midgut; HG, hindgut.
We sampled gut sections, the hepatic diverticulum, and other nondigestive tissues from at least two wild-type adult amphioxus individuals (only exceptions are ovary and testis) and compared the transcriptome of these tissues (see Materials and Methods and the Supplementary Materials for details on dissection and sample size; fig. S1A and table S1). All individuals were well fed and maintained under standard conditions unless otherwise specified. Digestive tissue can be separated into four groups, with MG1 and MG2 clustering together whereas the hepatic diverticulum, MG3, and hindgut are relatively separate (Fig. 1C and fig. S1B). Transcriptome data from different sections of the amphioxus gut provide the opportunity to probe detailed molecular profiles and answer whether a pancreas-like structure is present in this invertebrate chordate.
Expression of the midgut marker Pdx is strongly enriched in only MG3 samples, whereas the hindgut marker Cdx is highly enriched in hindgut samples (Fig. 1D). Hox genes show linear colinearity along the A-P axis of the gut, with genes Hox3-7 enriched in MG1, Hox8 in MG2, and Hox9-12 in the hindgut, whereas both Hox and ParaHox expression are barely detectable in the hepatic diverticulum [maximum expression is Hox4, with average transcripts per million (TPM) of 7.59; table S1]. Aside from these genes, we also assessed the expression of amphioxus orthologs to zebrafish pancreas β cell–specific genes (see Materials and Methods). Among these 65 amphioxus genes, only two show strong enrichment in the hepatic diverticulum (orthologs to pcsk1 and runx2a), whereas 10 are strongly enriched in MG3 (Fig. 1E). MG3-enriched genes include amphioxus orthologs of two vertebrate pancreatic TF genes, Pdx and Nkx6 (Fig. 1, D and E). However, expression of the amphioxus orthologs to the zebrafish insulin gene, Ilp1 and Ilp2, was not enriched in MG3. Ilp1 expression is strongest in MG2 and present in the hepatic diverticulum and MG1, whereas Ilp2 expression is enriched in the hindgut.
To look beyond Hox, ParaHox, and known pancreatic marker genes, we profiled the expression pattern of all genes along the gut A-P axis. To do this, we set hepatic diverticulum samples as “baseline,” and we defined 10 clusters of genes with enriched expression in different sections of the gut (fig. S1C). Gene ontology (GO) analysis of midgut-enriched genes returned terms related to “actin binding” and “lipid binding,” whereas genes only enriched in MG3 returned terms related to cilia, including “cilium” and “microtubule.” In addition, genes enriched in both MG3 and hindgut suggested dynamic transcription processes going on in this region of the gut, with more than 40 of these genes related to “mRNA metabolic process.” For hindgut-enriched genes, top GO terms include “proteolysis,” “carbohydrate-derived biosynthesis,” and “intracellular protein transport.”
Together, these results support functional segmentation in the amphioxus gut, and in this invertebrate chordate, specific regions are likely defined by ParaHox genes Pdx and Cdx, which mark the midgut (pancreas and duodenum) and hindgut (small and large intestines), respectively, in vertebrates. We propose that most processes related to the breakdown and absorption of macromolecules likely take place in the hindgut region, whereas the midgut region is mainly responsible for transporting and moving food particles along the gut (Fig. 1F). Specifically, MG3 is a special region in the amphioxus gut that may include the cilia-rich “ilio-colon (ring)” described by previous anatomical studies. We also find strong enrichment of several vertebrate pancreatic marker TFs in MG3, although amphioxus orthologs of pancreatic hormones are not enriched here.
Pdx-positive midgut cells are epithelial-like ciliated cells and do not secrete ILPs
Having established that the amphioxus gut is sectioned into different segments with drastically different gene expression patterns, we next wondered whether amphioxus had cell types similar to vertebrate pancreas cells.
Control adult amphioxus digestive tissue was dissected from three individuals and separated into different sections for dissociation into single-cell suspensions, of which one biological repeat was included for each tissue section (fig. S2A). We did not separate the midgut sample further into MG1, MG2, and MG3 segments, and we relied on section-specific marker genes identified by bulk RNA sequencing (RNA-seq) of these segments to determine cell clusters enriched in MG1, MG2, and MG3 segments in the midgut sample. For each sample, between 30,000 and 40,000 cells were loaded onto one cartridge and processed as an individual sequencing library. On average, 72.5% of all clean reads were uniquely aligned to the reference genome, and only these reads were used for further analysis (table S2). This resulted in a total of 131,016 high-quality cells, among which 36.2% cells were obtained from the hepatic diverticulum, 27.2% from midgut, and 36.6% from hindgut tissue, with even distribution from the two parallel repeats for the hepatic diverticulum (fig. S2A and table S2). A slightly higher number of cells were obtained from midgut sample 1 compared to sample 2, and more cells were obtained from hindgut sample 2 compared to sample 1; however, the overall distribution of cells projected into a two-dimensional (2D) t-distributed stochastic neighbor embedding (TSNE) plot remained similar (fig. S2A). A median of 1044 genes were detected in each cell, with a median of 707 genes detected in cells dissociated from the hepatic diverticulum, 1195 genes in cells from midgut tissue, and 1281 genes in cells from the hindgut. This is much higher than a previously published dataset that generated single-nuclei transcriptome data for the amphioxus “anterior intestine” (median gene number 144) and “posterior intestine” (median gene number 148) (42). We were not able to directly integrate and compare this previous dataset with our new data due to discrepancies in the reference genome used; however, we predict that this strong difference in gene number should lead to more cell types observed in our new dataset.
For downstream analysis unless otherwise mentioned, cells from repeat samples were treated as one sample. Cells from control and mutant samples (described later) were initially projected into the same 2D space without integration, but they did not cluster well (fig. S2B). Therefore, we compared two integration methods: the canonical correlation analysis method provided by the Seurat package (referred to here as “CCA-Seurat”) (44) and Harmony (45). Both methods were able to integrate cells from different genotypes (fig. S2B). Cells integrated with CCA-Seurat were split into 74 cell clusters, whereas cells integrated with Harmony were split into 78 cell clusters (fig. S2C). Disagreement between the two methods was found in some relatively larger cell clusters. For example, cluster 3 in the Harmony integrated dataset contains cells not integrated by CCA-Seurat (split into non-neighboring clusters 0 and 6), whereas cluster 1 in the CCA-Seurat integrated dataset contains cells not integrated by Harmony (split into non-neighboring clusters 0 and 4) (fig. S2C and table S2). However, both integration methods were able to identify the same small cell clusters and assign them far from other cells. Given the strong similarity in both integrated datasets, we have selected the CCA-Seurat dataset to proceed with analysis, and we accounted for the discrepancies in integration of some of the larger cell clusters by manually merging those with similar transcriptome profiles, resulting in 26 cell clusters that are referred to using alphabetical letters throughout this paper (fig. S2D). We also assessed the reproducibility of our data by comparing parallel biological samples for each tissue type using the Milo algorithm (46). For all three tissue types, most cell clusters showing significant differences in abundance were relatively small (fig. S2E). This applies to clusters Q, G, P, N, and S2 in the two midgut samples (each making up less than 5% in each sample), clusters AS, P, V, and J in the two hepatic diverticulum samples, and clusters M, R, Z, and P in the two hindgut samples (table S2). The exceptions are cluster L cells, which take up 32.5% (4581 of 14,093) of Ctrl-MG-2 whereas only 8.4% (1804 of 21,590) of cells from Ctrl-MG-1 belong to cluster L, and cluster J cells, which take up 16.1% (3192 of 19,835) of Ctrl-HG-1 whereas only 2.1% (591 of 28,056) of cells from sample Ctrl-HG-2 belong to cluster J.
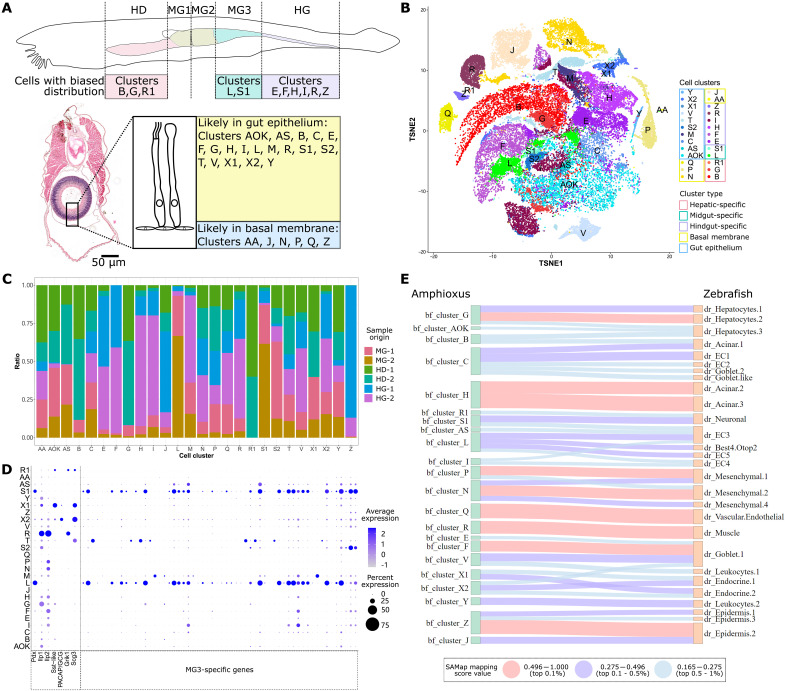
To profile these cell clusters (Fig. 2, A and B), we analyzed the tissue of origin and compared the transcriptomic profile of these cells to vertebrate digestive cells. Among these 26 cell clusters, 11 consist of cells enriched in one gut section (over 66.7% sampled from one tissue). Six hindgut-enriched cell clusters include clusters E (87.5% from hindgut tissue), F (97.1%), I (81.5%), H (85.1%), R (77.9%), and Z (98.9%) (Fig. 2, A and C). Three cell clusters are hepatic diverticulum–enriched, including clusters B (88.3% from the hepatic diverticulum), G (91.7%), and a rare cell cluster R1 (100%). The only two cell clusters that are enriched in midgut tissue are clusters L (93% from midgut tissue) and S1 (87%). Among parallel biological repeats, only cluster Z was strongly contributed to by one sample (86.8% from sample Ctrl-HG-1) (Fig. 2C and table S2). To further infer which cell clusters are specific to different sections of the gut, we compared marker genes for each cell cluster against a list of digestive tissue marker genes calculated from bulk transcriptome data. For the two hepatic diverticulum–enriched clusters (B and G), 43.5% cluster B marker genes are also hepatic diverticulum–specific genes, whereas 39.3% cluster G marker genes are also hepatic diverticulum–specific (fig. S3A). Four of the six hindgut-enriched clusters (H, E, I, and F) are marked by hindgut-specific genes, with genes sharing both identities taking up 47.7, 30.2, 17.9, and 16.2% of all marker genes for each cluster, respectively. Although most cluster R cells were isolated from hindgut tissue, only 3.6% of cluster R marker genes are also hindgut-specific genes. Even when expanding the definition of “tissue-specific” beyond just digestive tissue, 95.9% cluster R marker genes are not enriched in a single type of tissue (fig. S3B). For hindgut-enriched cluster Z, 42.6% cluster Z marker genes are skin-specific genes, and 32.1% are not expressed at high levels in digestive tissue. For midgut-enriched cell clusters L and S1, 15.5 and 10.4% of cluster marker genes are also MG3-specific genes, respectively. A total of 6.5% of the cluster AS marker genes are also MG1-specific genes, and the only marker genes that are also MG2-specific genes include one cluster T marker gene and one cluster Z marker gene.
Fig. 2. Single-cell profiling of wild-type amphioxus digestive tissue.
(A) Predicted location of amphioxus cells digestive tissue. Different regions along the gut A-P axis are shown in different colors, with clusters enriched in these regions listed below. A cross section sampled in the MG3 region of a juvenile individual is shown with a cartoon indication of gut epithelia and cells making up the gut basal membrane. Cluster IDs of cell predicted to be located in the gut epithelia and basal membrane are listed. Scale bar, 50 μm. (B) TSNE plot of adult amphioxus cell types from the hepatic diverticulum, midgut, and hindgut tissue. Clusters predicted in (A) to be enriched in different gut locations are marked in different colored boxes. (C) Summary of cell cluster identity and their sample of origin. (D) Dot plot of marker genes used to characterize cell clusters L, S1, S2, R, X1, and X2. Gene ID for each amphioxus gene is shown below, with gene names provided for genes discussed in the main text. (E) Sankey plot for amphioxus gut cell clusters and zebrafish gut cell types. Cell types in the two species with strong similarity (ranking top 1%) are connected by a red line, those with medium similarity (ranking 1 to 5%) are connected by a purple line, and those showing weak similarity (ranking 5 to 10%) are connected by a blue line. Predicted similarities with arbitrary mapping scores not in the top 10% tier are not shown. HD, hepatic diverticulum; MG, midgut; HG, hindgut.
We further deduced cell identity based on genes of interest such as Pdx, a gene only expressed in the two midgut-enriched cell clusters (L and S1) along with a number of MG3-specific genes (Fig. 2D). Clusters L and S1 have highly similar gene expression profiles, with cluster S1 showing enrichment of cilia-related tektin genes (Tekt1, Tekt2, and Tekt3). GO enrichment analysis of marker genes for clusters L and S1 both return top terms including microtubule, and differential expression analysis of genes enriched in cluster S1 compared to cluster L are mainly cilium-related genes (fig. S3C). Amphioxus orthologs of genes encoding vertebrate pancreatic hormones [insulin/insulin-like growth factor (IGF) Ilp1 and Ilp2 (37, 47, 48), somatostatin/cortistatin-like Sst-like (49), and glucagon-like PACAP/GCG (31)] were not detected in Pdx-expressing clusters L and S1 (Fig. 2D). Rather, Ilp1 and Ilp2 are strongly coexpressed in cluster R cells along with glutamate receptor gene Grik1, whereas Ilp1 is weakly present in clusters G, AOK, S2, X1, and Y, and Ilp2 is weakly present in clusters F, E, I, H, N, and P. Most cluster R cells are in hindgut tissue (77.9%), whereas cluster G is mostly located in the hepatic diverticulum (91.7%), and clusters F, E, I, and H are mostly found in hindgut tissue (97.1, 87.5, 81.5, and 85.1%) (Fig. 2C and table S2). Sst-like and secretory vesicle-related gene Scg3 are strongly coexpressed in cluster X1 cells, with trace Sst-like expression detected in clusters R1 and X2, and both PACAP/GCG and Scg3 are enriched in cluster X2, with trace PACAP/GCG expression detected in cluster X1 (Fig. 2D). Cluster X1 cells are mainly distributed in the anterior gut, with 60.2% cluster X1 cells sampled from the hepatic diverticulum and 39.8% sampled from midgut tissue (Fig. 2C and table S2). In contrast, cluster X2 cells are mainly found in the hindgut (64.3%) and midgut (29.9%), with rare presence in the hepatic diverticulum (5.8%).
Cell clusters L, S1, R, X1, and X2 make up a small portion of this dataset (9.5%) (table S2). Most (74.9%) of the amphioxus gut cells express Fabp (fatty acid–binding protein) (fig. S4, A and B), and these cells (clusters AOK, F, E, I, B, G, T, H, M, S2, Y, V, and AS) are likely the main components of the amphioxus gut epithelium (Fig. 2A). We have assigned cell identity based on their location and marker genes, and we briefly highlight some clusters here. The largest cluster is cluster AOK (30.1%), and most are located in the hepatic diverticulum (50.5%) and midgut (45.8%) (Fig. 2C and table S2). Cluster T cells express Chit1 (chitinase), and cluster H cells express Ctrb and Ctrl (chymotrypsin), whereas cluster M cells express Pnlip (phospholipase) and Amy1 (amylase) (fig. S4, A and B). Cluster S2 express tektin (related to microtubule stabilization) (50) but do not express Pdx, and cluster Y are cells undergoing cell division as marker genes such as Ccnb1 (51), Kif11 (52), and Kif20a (53) encode mitosis proteins. The remaining clusters (R, R1, Z, J, P, N, Q, and AA, 15.6% of all cells) express Fabp at low levels (fig. S4, A and B). Clusters R and R1 both express glutamate transporter gene VGLUT, but they are different in location (cluster R mainly in the hindgut and cluster R1 restricted to the hepatic diverticulum) and have different marker genes (cluster R express Ilp1 and Ilp2 and cluster R1 express dihydropyrimidinase gene Dpys and neuron-related synaptotagmin gene Syt) (fig. S4, A and B). Cluster Q likely represents vascular endothelial–like cells marked by AngX (also referred to as Angptl7), a gene involved in hematopoietic cell development (54), and Pdvegfr, a hematopoietic marker (55). The remaining clusters are marked by gene encoding structural proteins such as collagen (cluster P), adhesion molecule endo16 and perlecan (cluster N), intermediate filaments (IFs) (cluster Z), and contactin (cluster J). Cluster AA is marked by TF gene Scx, a gene involved in tendon and ligament development (56).
We compared amphioxus digestive cells to zebrafish gut cell types using the SAMap algorithm (57, 58). Pdx-positive amphioxus cluster L shows similarity to three epithelial cell (EC) types (EC3, EC5, and EC4) and Best4/Otop2 cells, a special EC type enriched for expression of genes involved in pH sensing and electrolyte balance (59), whereas cluster S1 (Pdx-positive ciliated) shows similarity to zebrafish neuronal cells and EC3 cells (Fig. 2E). Cluster R (Ilp1 and Ilp2) shows similarity to zebrafish muscle cells, whereas both cluster X1 (Sst-like) and cluster X2 (PACAP/GCG) are similar to zebrafish endocrine cells. We also identified amphioxus cell clusters with transcriptomic profiles highly similar to zebrafish cell types including hepatocytes (amphioxus cluster G), goblet cells (cluster F), acinar cells (cluster H), mesenchymal cells (clusters P and N), epidermis cells (cluster Z), and vascular endothelial cells (cluster Q), and all marker genes for these clusters in both species are listed (table S2).
The amphioxus gut has endocrine-like cell clusters (X1 and X2) that secrete peptide hormones Sst-like and PACAP/GCG. Cluster X1 is absent from the hindgut, whereas cluster X2 cells are mainly located in the hindgut. Cells coexpressing Ilp1 and Ilp2 (cluster R) show more similarity to vertebrate muscle cells than endocrine cells, and Pdx is not expressed in this hindgut-enriched cell cluster. In summary, amphioxus has gut endocrine-like cells (clusters X1 and X2) that not only express peptide hormone mRNA but also show strong overall transcriptomic similarity to zebrafish endocrine cell types. However, these amphioxus endocrine-like cells are enriched in different regions of the gut, and they do not express vertebrate pancreatic marker Pdx.
A Pdx mutant line reveals Pdx-independent development of endocrine-like cells in amphioxus
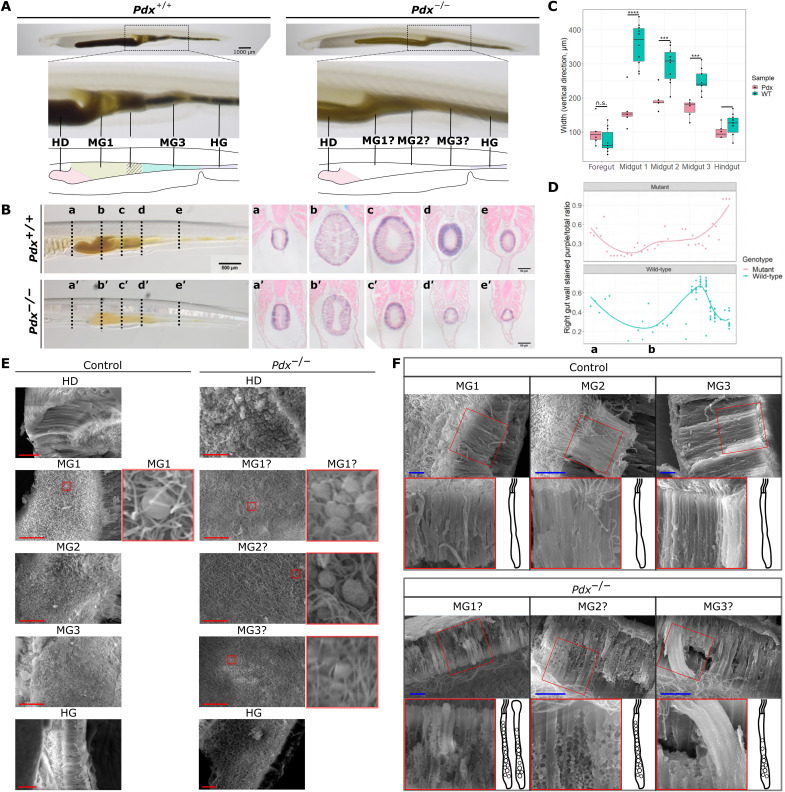
To explore the role of Pdx in cell clusters L and S1, we used a Pdx heterozygous mutant amphioxus line that carries a frameshift deletion upstream of the Pdx gene homeobox sequence (fig. S5A) (43). We found that homozygous Pdx mutant amphioxus lack visible midgut patterning and cannot properly compact food particles (Fig. 3A; see movies S1 and S2 for comparison of food particle movement). Comparison of hematoxylin and eosin (H&E) stained cross sections of fixed individuals show that all midgut regions (MG1, MG2, and MG3) are significantly thinner in Pdx mutant animals, whereas no difference was found between the pharynx (foregut) and hindgut of control and mutant animals (Fig. 3, B and C). In addition, the control MG3 region stands out from other sections as gut cells in this region have a higher portion that is stained purple, indicative of a different distribution of cell nuclei in this region (Fig. 3B). This is trend is not observable in Pdx mutants (Fig. 3, B and D).
Fig. 3. Pdx mutant amphioxus.
(A) Representative control and Pdx mutant juveniles. Magnification of the midgut region shows lack of midgut patterning in mutants, summarized by a simplified cartoon below. Scale bar, 1 mm. (B) H&E-stained sections of control and Pdx mutants. The approximate locations where sections were sampled from are indicated using lowercase letters. Scale bar, 500 μm for fixed individuals. Scale bars, 50 μm for stained sections. (C) Foregut, midgut, and hindgut width in control and Pdx mutants. Each dot represents one slide section sampled from one individual. Statistical analysis was performed using two-sided t tests; n.s., no significance, ***P = between 0.001 and 0.0001, ****P = below 0.0001. (D) Ratio of tissue in the gut wall that is stained purple in control and Pdx mutant samples. (E) Digestive tissue under a scanning electron microscope. One representative photo from the HD, MG1, MG2, MG3, and HG regions was selected for the control and Pdx mutant groups. Midgut regions with rounded cells are highlighted in red, and these regions are magnified and shown on the right side of the original photograph. Scale bars, 20 μm. (F) Scan of MG1, MG2, and MG3 gut cross sections in control and Pdx mutant samples. One region indicated using a red box is magnified and rotated to depict the gut cell with its apical end pointing upward. A cartoon of a representative gut cell in each photo is provided to assist interpretation. Scale bars, 20 μm. All enlarged regions are magnified by a scale of 2. HD, hepatic diverticulum; MG, midgut; HG, hindgut.
Fixed gut sections were scanned using a scanning electron microscope. Scanning electron microscopy (SEM) results echo paraffin sections as we observe a clear increase in the length of gut cells from section MG1 to MG3 in control samples, whereas this is not obvious in Pdx mutants (Fig. 3, E and F). Furthermore, this increased resolution shows that all sections of the midgut are lined by one layer of tall, column-like cells with cilia at their apical end (side-facing gut lumen) in control animals whereas cells with a rounded apical tip are largely present in the MG1 section of mutants. Scan of the gut lumen surface reveals the rare presence of these rounded cells “buried” under a carpet of cilia in the control MG1 section, whereas this not obvious in the MG2 and MG3 sections (Fig. 3E). However, clusters of rounded cells are highly prominent in Pdx mutants throughout the entire midgut. This trend is also very severe in the Pdx mutant hepatic diverticulum, where cilia are barely observable in sections of this tissue (Fig. 3E). In addition, mutant gut samples have wider, shorter cells with large granular inclusions distributed throughout the entire length of the cell body (Fig. 3F). The tufts of cilia at the apical tip of these cells also appear to be shorter than that observed in control animals. In summary, Pdx mutants do not form proper midgut morphological structure and show dysfunctional movement of food particles inside the gut. In addition, the mutant midgut appears thinner and lacks ciliated cell types enriched in the MG3 section of the wild-type midgut.
We obtained bulk tissue RNA from the postmetamorphic juvenile (1 cm) control and Pdx mutant hepatic diverticulum, midgut, and nondigestive tissue. Here, we use the term “adult” to refer to amphioxus that have visible gonads, whereas “juvenile” describes postmetamorphic animals below 2 cm in body size and do not have visible gonads. The term “larvae” refers to animals that have formed an open mouth and have not begun to metamorphose (60). For hindgut tissue, we did not manage to obtain high-quality RNA samples, possibly due to the strong presence of digestive enzyme secreting acinar-like cells (clusters H and M) in this region (Fig. 2C). Compared to a previously published five-gill slit larvae dataset (43), more differentially expressed genes were detected in the postmetamorphic juvenile (1 cm) samples taken in this study (fig. S5B). Specifically, the highest number of genes was differentially regulated in mutant juvenile midgut tissue (1187 in total) (fig. S5B and table S3). In mutant midgut tissue, 10.5% of significantly down-regulated genes are MG3-specific, 0.9% are MG1-specific, and 1.3% are MG2-specific (fig. S5C). Among significantly up-regulated genes, 17.2% are hepatic diverticulum–specific whereas 6.2% are hindgut-specific. Similar to five-gill slit larvae samples (43), both Pdx and Ilp1 do not show a significant change in expression in mutant juvenile midgut tissue (fig. S5D). However, Nkx6 is down-regulated in mutant juvenile midgut tissue and Ilp2 is up-regulated (fig. S5D). None of the Hox genes or Cdx gene showed a significant change in expression in any of the juvenile Pdx mutant tissues analyzed (table S3).
Results from bulk tissue RNA-seq suggest lack of midgut-specific cells and possible invasion of hepatic-specific and hindgut-specific cells in the midgut of Pdx mutants. We also performed single-cell RNA-seq (scRNA-seq) on live cells from the Pdx mutant hepatic diverticulum, midgut, and hindgut tissue. Similar to control samples, 73.6% of clean reads obtained from three adult Pdx mutant individuals, with two parallel samples for each tissue type, aligned uniquely to the reference genome and were used for downstream processing (fig. S6A and table S4). Of the 157,964 high-quality cells, 56,136 (35.5%) were obtained from two hepatic diverticulum samples, 53,371 (33.8%) from midgut samples, and 48,457 (30.7%) from hindgut samples, with a similar number of cells obtained from the two parallel samples for each tissue (fig. S6B and table S4). Most parallel samples contributed an equal number of cells to each cell cluster, with the exception being cluster R1 (82.9% from sample KO-HD-1) and cluster Y (50% from hepatic diverticulum sample KO-HD-2 and 37.1% from midgut sample KO-MG-1) (fig. S6B and table S4).
To assess the reproducibility of Pdx mutant data, we compared parallel samples for each tissue type using the Milo algorithm (46). For all samples, most differences in cell abundance are observed in rare cell clusters, including AA, G, L, Q, and N in midgut tissue; G, J, and L in the hepatic diverticulum; and B, M, S2, and AOK in the hindgut (each taking up less than 6% of all cells from the respective sample) (fig. S6C and table S4). In Pdx mutant midgut tissue, only two larger cell clusters, clusters H and J, showed a weak increase in cell abundance in sample KO-MG-2 [2763 (9.4%) cluster H cells and 2304 (7.8%) cluster J cells] versus sample KO-MG-1 [382 (1.6%) cluster H cells and 886 (3.7%) cluster J cells]. In the hepatic diverticulum, cluster AOK was more abundant in sample KO-HD-2 (9532 cells, 29.1%) versus KO-HD-1 (2834 cells, 12.1%) whereas cluster N was more abundant in sample KO-HD-1 (3093 cells, 13.2%) compared to KO-HD-2 (815 cells, 3.5%). In Pdx mutant hindgut tissue, a large difference was observed in cluster I abundance, with 7517 cells (30%) obtained from sample KO-HG-2 whereas only 1449 (6.2%) cells were from sample KO-HG-1.
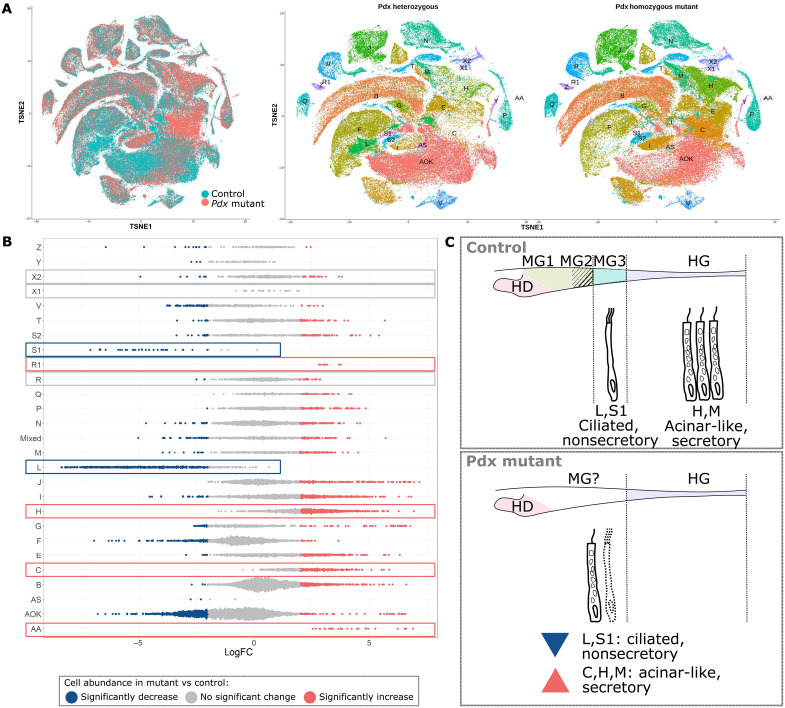
For all analysis aside from cell cluster abundance, cells from the two biological repeats for each tissue type were integrated and treated as one sample. Control and mutant cells were integrated into the same 2D mapping space for comparison and assigned the same cluster IDs (Fig. 4A). For Pdx mutants, a lower percentage of clusters L and S1 cells were dissociated from midgut tissue compared to control (fig. S7A). Specifically, for control cluster L cells, 93% are from the two midgut samples (26.3% from Ctrl-MG-1 and 66.7% from sample Ctrl-MG-2) whereas, for Pdx mutant cluster L cells, only 53.4% are from the two midgut samples (40.7% from KO-MG-2 and 12.8% from KO-MG-1) (fig. S7A). Similarly, 87% of all control S1 cells are from the two midgut samples (61.2% from Ctrl-MG-2 and 25.4% from Ctrl-MG-1) whereas only 39.2% of Pdx mutant S1 cells are from midgut samples (21.6% from KO-MG-2 and 17.5% from KO-MG-1). Using the Milo algorithm (46), we show Pdx mutants have fewer midgut-enriched clusters L and S1 cells whereas more clusters AA, R1, C, and H cells are present (Fig. 4B). Ilp-expressing cluster R cells showed similar abundance levels between control and mutant samples (Fig. 4B), but 77.9% control cluster R cells are from the hindgut (52.2% from Ctrl-HG-1 and 25.7% from Ctrl-HG-2), whereas 53% Pdx mutant cluster R cells are from the midgut (8.3% KO-MG-1 and 44.7% KO-MG-2) (fig. S7A). For the two endocrine-like cell clusters, neither X1 nor X2 showed strong variation in cell abundance between the control and mutant (Fig. 4B), and tissue origin remained similar, with 60.2% of control cluster X1 cells obtained from the hepatic diverticulum (30.3% Ctrl-HD-1 and 29.9% Ctrl-HD-2) and 63.6% mutant X1 cells obtained from the hepatic diverticulum (33.9% KO-HD-2 and 29.8% KO-HD-1) (fig. S7A). A total of 64.3% of the control cluster X2 cells are from the hindgut (35.1% Ctrl-HG-2 and 29.2% Ctrl-HG-1), and 72.1% mutant cluster X2 cells are also from the hindgut (47.3% KO-HG-1 and 24.9% KO-HG-2) (fig. S7A).
Fig. 4. Single-cell profiling of Pdx mutant amphioxus digestive tissue.
(A) TSNE plot of control and Pdx mutant gut cells. Cells are colored by genotype in the left panel and by cell cluster in the right panel. (B) Difference in cell abundance between control and Pdx mutant. miloR assigns cells into neighborhoods that may not necessarily belong to the same manually defined cluster (46). Hence, the y axis includes an additional category named “Mixed” that correspond to all miloR-defined neighborhoods that have less than 50% of cells assigned to the same manually defined cluster. Dots in red are neighborhoods that are significantly increased in cell abundance in Pdx mutants, and those in blue are significantly decreased. Neighborhoods in gray do not have significant difference in abundance. (C) Pdx mutants lack ciliated, nonsecretory clusters L and S1 cells in the MG3 region of their midgut, whereas acinar-like secretory clusters C, H, and M cells show a significant increase in abundance. HD, hepatic diverticulum; MG, midgut; HG, hindgut.
We compared gene expression in control and Pdx mutant clusters using two methods: first taking a cell-level view by treating replicates as one sample using the Wilcoxon rank sum test within the Seurat package (61), which we refer to as the “sc test,” and the second taking a sample-level view by calculating the average expression of all genes in each cluster of one replicate and performing differential expression analysis using the bulk RNA analysis DESeq2 algorithm (62), which we refer to as the “pseudobulk test.” The sc test predicts an average of 958.6 differentially expressed genes between all cell clusters in mutant samples compared to control, whereas only 36.3 are predicted using the pseudobulk test (fig. S7B and table S4). Between both tests, an average of 14.7 differentially expressed genes is shared in each cell cluster between the two methods (fig. S7B). According to the sc test, cluster L has the largest number of differentially expressed genes, with 2117 down-regulated and 412 up-regulated genes, whereas the pseudobulk test returns 15 down-regulated and 19 up-regulated genes. Twenty-three differentially expressed genes are shared between these two tests, including down-regulation of the TF gene Nkx6 and a midgut marker mucin gene bf1_1088, along with up-regulation of pancreatic lipase gene Pnlip, and hindgut markers bf4-314 and lipoxygenase gene Alox (fig. S7C and table S4). Both Ilp1 and Ilp2 genes were not differentially regulated in cluster L.
To explore whether these differentially expressed genes may be regulated directly by Pdx, we tested for enrichment of vertebrate Pdx1-specific motifs in the promoter region of strongly differentially expressed genes (defined as absolute average log2foldchange value above one) using the HOMER algorithm (63). First, among all genes nominated by the sc test, only down-regulated genes in clusters C, L, S1, S2, and X2 were enriched for Pdx-target motifs, among which only clusters L and S1 showed strong Pdx expression in control samples (fig. S7D and dataset S1). Specifically, in cluster L, 27.8% of the 245 down-regulated genes have a Pdx-binding motif present in the promoter region [defined as 300 base pairs (bp) upstream to 50 bp downstream of the transcription start site] (table S4). Even when the only 12 down-regulated genes shared between the two differential gene expression analysis methods were selected for motif analysis, the third ranking motif still contained a “TAAT” binding core, although the automatically generated best TF prediction for this motif was not Pdx but another homeodomain-containing TF, Hoxb4 (fig. S7D). For up-regulated genes nominated by both methods, none of the top enriched motifs are predicted to be Pdx binding sites.
In contrast to the strong depletion of cluster L cells in mutant Pdx tissue, cluster C cells are more abundant in mutant tissue compared to control (813, 0.6% cells in control and 10,515, 6.7% cells in mutant) (tables S2 and S4). Cluster C is rare in control samples and evenly distributed (30.4% from the hepatic diverticulum, 33.5% midgut, and 36.2% hindgut) (Fig. 2C). In contrast, cluster C cells take up a larger portion in mutant samples, and most are sampled from midgut tissue (70%) (fig. S7A). Both integration methods (CCA-Seurat and Harmony) support identification of this cell cluster as a group of cells different from cluster L, although some cells assigned cluster L are placed in very close proximity to cluster C cells in a 2D plot (fig. S7E). Although we do not detect Pdx expression in cluster C cells in control or mutant samples (fig. S7F), down-regulated genes (sc test) in mutant cluster C cells have enriched motifs that may be recognized by vertebrate Pdx1 (fig. S7D), and most differentially regulated genes in mutant cluster C cells are also differentially regulated in mutant cluster L cells (69.4% down and 79.1% up) (table S4). In general, mutant cluster C cells are marked by genes that are ubiquitously up-regulated in several cell clusters, most of which are expressed at very low levels in control cells (fig. S7G). These genes include chitinase genes, a number of hindgut marker genes, and the amphioxus ortholog to vertebrate Olfm2, a regulator of smooth muscle cell differentiation (fig. S7F).
Pdx mutant amphioxus midgut lacks epithelial-like cluster L cells but have an abundance of secretory cluster C cells (Fig. 4C). In mutant cluster L cells, some down-regulated genes are predicted to be regulated directly by Pdx whereas we do not find support for Pdx regulation of up-regulated genes, suggesting that Pdx likely promotes the expression of cluster L marker genes but does not actively suppress genes leading to a different cell fate. Furthermore, across all mutant cell clusters, we observe up-regulation of hindgut-specific genes, especially genes encoding proteins predicted to be secreted out of the cell. This may echo the abundance of secretory vesicle-like structures observed in mutant amphioxus midgut sections observed using SEM (Fig. 3F).
In conclusion, amphioxus Pdx may regulate the expression of some conserved TFs involved in vertebrate pancreatic specification, such as Nkx6 (64); however, Pdx-positive cells (clusters L and S1) do not exhibit endocrine cell-like characteristics. Rather, these amphioxus midgut cells mainly serve to propel food particle movement along the gut.
Pdx control over insulin gene expression is likely an acquired function in vertebrates
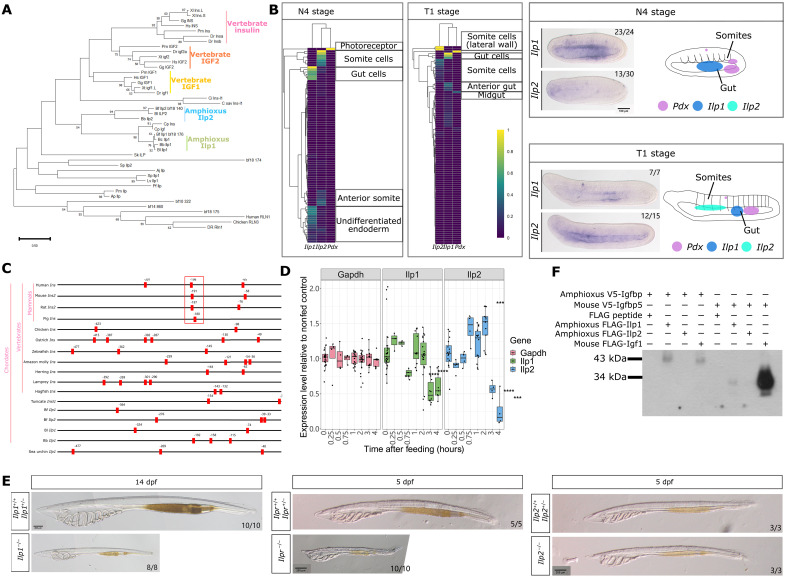
Within vertebrate pancreatic β cells, Pdx is a direct transcriptional activator of insulin and is essential for maintaining β cell identity (13). Amphioxus has two homologs to vertebrate insulin and Igf genes, Ilp1 and Ilp2, and we verified the full coding sequence of both genes using cDNA synthesized from amphioxus embryo samples (table S5). Phylogenetic tree reconstruction using the full coding sequence of deuterostome insulin, Igf, Ilp, and relaxin genes suggest a close relationship between two tunicate genes (Chelyosoma productum; gene names Ins and Igf assigned by a previous paper) (65) with amphioxus Ilp1 genes, whereas amphioxus Ilp1 and Ilp2 genes are placed as a sister group to both vertebrate insulin and IGF genes (Fig. 5A). However, amphioxus Pdx and Ilp gene transcripts are not detected in the same cell in adult digestive tissue (Fig. 2D) or in embryos (Fig. 5B). Single-cell data from amphioxus mid-neurula (referred to as N4 stage) and late neurula (T1 stage) embryos (66) do not support coexpression of Pdx, Ilp1, or Ilp2 in the same cluster (Fig. 5B), and Pdx expression patterns reported previously (43, 67) support expression in the future photoreceptor cells forming the first pigment spot and a dorsal and ventral area in the posterior gut at N4 stage, whereas at T1 stage, signal remains detectable near the first pigment spot, and expression in the gut shifts to a slightly more central position (Fig. 5B). We probed Ilp1 and Ilp2 expression patterns in N4 and T1 stage embryos, and we show that Ilp1 is consistently expressed in a gut area that is anterior to the region of Pdx expression, whereas Ilp2 expression does not overlap with Pdx or Ilp1 (Fig. 5B). Furthermore, in Pdx mutant midgut tissue, Ilp1 expression does not show significant change, although Ilp2 shows a weak increase in expression (fig. S5C).
Fig. 5. Amphioxus Ilp1, Ilp2, and Ilpr function.
(A) Phylogenetic relationship of vertebrate insulin, Igf, and invertebrate Ilp constructed using maximum likelihood analysis. Vertebrate relaxin is used as an outgroup. (B) Expression of Ilp1 and Ilp2 in mid-neurula (N4 stage) and late neurula (T1 stage) amphioxus embryos. Normalized expression at the single-cell level is summarized in heatmaps, with Pdx expression shown for reference. In situ hybridization results for Ilp1 show expression in the gut endoderm in N4 embryos, which becomes localized to a small region in the gut in T1 embryos. Ilp2 expression is not detected in N4 embryos whereas expression in the anterior pharyngeal region is detected in T1 embryos. A rough summary of Pdx, Ilp1, and Ilp2 expression in N4 and T1 stage embryos, with Pdx expression patterns obtained from published in situ results (43, 67), is provided with areas of gene expression indicated using different colors. Scale bar, 100 μm. (C) Location of the Pdx-core binding motif TAAT in the 500-bp upstream region of vertebrate insulin genes and invertebrate Ilp genes. The highly conserved Pdx-binding A3 box region in mammals is highlighted using a red box. (D) Relative expression level of Gapdh, Ilp1, and Ilp2 genes in starved wild-type larvae stimulated with food compared to the seawater control. Statistical analysis was performed using two-sided t tests; **P = between 0.01 and 0.001, ***P = between 0.001 and 0.0001, ****P = below 0.0001. (E) Phenotype of Ilp1 mutant larvae 14 days after fertilization. Scale bar, 200 μm. (F) Interaction between amphioxus Ilp1, Ilp2, Igfbp, and mouse Igf1 and Igfbp5 proteins.
The homeodomain region of the amphioxus Pdx protein is highly conserved compared to other chordates, with only 4 amino acids of the 60 in this region different from vertebrates (fig. S8A). Comparison of these amino acids with previously reported Pdx protein binding results (68) shows that they are not key for DNA binding. On the basis of the vertebrate Pdx protein binding sequence, we assessed the promoter of amphioxus Ilp1 and Ilp2 genes. Although we found a potential Pdx binding site upstream of both amphioxus Ilp1 and Ilp2 transcription start sites, the location of these potential binding sites is not conserved between these two paralogs, and we do not find evidence for conservation of potential Pdx binding sites in any of the nonmammal species assessed (Fig. 5C and dataset S2). Using HOMER, we carried out a genome-wide search for Pdx binding motifs within the core promoter region of all amphioxus genes, and Ilp1 and Ilp2 were not identified as potential Pdx-target genes (table S4).
One important feature of vertebrate insulin is rapid release of stored peptides and up-regulation of mRNA transcription following food intake (69). Because of lack of amphioxus-specific antibodies, we were not able to directly measure Ilp protein levels. We focused on testing whether a change in amphioxus Ilp1 and Ilp2 transcription could be observed postfeeding. Total mRNA was extracted from three batches of starved three-gill slit larvae stimulated with food, with each batch consisting of ~2000 wild-type individuals, with around 200 individuals sampled at each time point (see Materials and Methods for detailed experimental setup). Ilp1 expression showed weak increase within 15 to 30 min of food stimulation, whereas Ilp2 expression showed the strongest increase 2 hours after food stimulation (Fig. 5D). Both Ilp1 and Ilp2 transcript levels show a drastic drop 3 to 4 hours postfeeding. These results show that Ilp transcription has a very limited response to food ingestion, and postfeeding expression may be suppressed rather than stimulated.
To further assess Ilp1 and Ilp2 genes and their function in amphioxus, we generated Ilp1 and Ilp2 knockout lines and an Ilpr (predicted receptor of both Ilp1 and Ilp2 in amphioxus) knockout line. Because of the close location of amphioxus Ilp1 and Ilp2 genes on chromosome 18, we were not able to directly obtain Ilp1 and Ilp2 double knockout mutants by crossing the two lines. Both Ilp1 and Ilpr knockout larvae show abnormalities by development of the first gill slit, with knockout larvae failing to develop the third gill slit and showing no body growth beyond this stage, although the mutant animals are able to feed and swim (Fig. 5E). In contrast, Ilp2 knockout animals do not display any obvious phenotypic differences compared to control animals. Thus, we focused on three-gill slit stage Ilp1 mutant larvae, and transcriptomic comparison of mutant against control larvae showed no change in Pdx expression, whereas Ilp2 expression in mutant larvae is significantly up-regulated by ~2.55-fold. (fig. S8B). To interpret these transcriptome results, we also defined marker genes for different cell types in the late neurula (T1 stage) using published embryo scRNA-seq results (66) by calculating the average expression of all genes in each cell type and then using the same criteria for adult tissue to define marker genes. A total of 94.7% of the differentially expressed genes in Ilp1 mutant larvae are not embryo cell type–specific, and only one significantly up-regulated gene (bf11_692) is a gut (Pdx-negative region) cell type marker (table S5). When comparing differentially expressed genes in Ilp1 mutants against adult tissue-specific markers, 77.7% are not tissue-specific, with 10 up-regulated genes specific to the digestive tract (gut and hepatic diverticulum), whereas 7 down-regulated genes are also digestive-specific. Overall, transcriptome results suggest that the main function of Ilp1 may not be restricted to gut development or digestion-related functions.
Although we did not find a strong link between Ilp1 and gut-related gene expression, we wondered whether this peptide played a part in sugar metabolism, similar to vertebrate insulin and its regulation over glucose metabolism (70). We measured the concentration of different amino acids, nucleotides, and sugars in three-gill slit control and Ilp1 knockout larvae using gas chromatography–mass spectrometry (GC-MS). In brief, we snap froze samples consisting of ~200 individuals, with a total of three parallel samples for each group. Ilp1 knockout larvae have a strong decrease in amino acid concentration, with 19 amino acids all significantly decreased in mutants (fig. S8, C and D). Enrichment of metabolite sets returns top terms relevant to amino acid synthesis and tRNA biosynthesis, suggesting a strong decrease in protein synthesis speed in knockout larvae (fig. S8C). We did not observe a significant change in glucose content between control and mutant samples. Only two saccharides showed significant differences in content between control and Ilp1 mutant larvae, with the monosaccharide mannose showing a significant decrease in mutants and the disaccharide maltose showing a significant increase.
The amphioxus Ilp peptide structure was also compared to vertebrate IGF and insulin. We made use of the fact that vertebrate Igf proteins interact with members of the IGF binding protein (IGFBP) family whereas the insulin protein does not (21). By setting up a protein-protein interaction system, we were able to detect V5-labeled mouse Igfbp5 protein in a mixture of Igfbp5-V5–tagged and FLAG-Igf1–tagged proteins immunoprecipitated with anti-FLAG antibodies (Fig. 5F and fig. S8E). In contrast, the very weak presence of mouse Igfbp5 is detectable in samples overexpressing the amphioxus Ilp1 peptide, whereas no mouse Igfbp5 protein is detected in samples overexpressing only the FLAG tag and amphioxus Ilp2. When amphioxus Igfbp, the only member of the Igfbp family, was incubated with amphioxus Ilp peptides and mouse Igf1 protein, amphioxus Igfbp was weakly detectable in samples overexpressing amphioxus Ilp1 and mouse Igf1. This suggests that interaction between members of the insulin/Igf and Igfbp protein family is likely a conserved trait in chordates, and amphioxus Ilp1 shows more structural similarity to vertebrate Igf1 than amphioxus Ilp2.
We conclude that, in amphioxus, we did not find evidence for Pdx control over Ilp1 and Ilp2 expression. Pdx expression is restricted to a specific region of the adult amphioxus midgut, whereas amphioxus Ilp1 and Ilp2 are expressed in cells distributed throughout the digestive system. Ilp1 likely serves an essential function in controlling larvae body growth, a function that is similar to vertebrate IGF, whereas both Ilp1 and Ilp2 expression show weak response to feeding after a period of starvation. Furthermore, amphioxus Ilp1 peptide is capable of interacting with amphioxus Igfbp protein, a trait that is typical of vertebrate Igf1 and Igfbp proteins.
DISCUSSION
Digestive systems with different levels of complexity have been proposed to evolve through multiple mechanisms. Evolution of traits in the vertebrate gut, including differences in intestinal motility and transit (71), gut cell protection by secreted mucosa (72), and loss of structures such as the stomach in different lineages (73), have been correlated with cell type gain or loss, which subsequently may be linked to change in gene regulatory or coding regions.
Unique cell types in the amphioxus digestive tract
Anatomical studies describe a strongly ciliated ilio-colon ring in the amphioxus gut that is responsible for formation of the “food cord,” packed food particles observable by the naked eye (33, 35). In this study, transcriptome profiling of amphioxus gut sections reveals enrichment of Pdx-regulated gut epithelial-like cells (cluster L) and ciliated cells (cluster S1) in this region (named MG3 here). A previous work had linked Pdx expression with a fluorescence-rich area in the larvae, but it was unclear whether this area was correlated to the MG3 region in adults (43). In this study, we link this region to Pdx, shedding light on the function of this midgut TF in the adult amphioxus gut.
We show that the only two Pdx-positive cell clusters (clusters L and S1) in amphioxus do not express peptide hormones Ilp or Sst-like and are not enriched for genes relevant to endocrine function. Rather, cluster S1 is a group of ciliated cells that are likely restricted to the MG3 section and propels food particles along the gut. The digestive tract of both amphioxus and tunicates are lined by only one layer of gut epithelium cells and do not have a muscle layer akin to that in the vertebrate digestive system (74). Instead, movement of food particles along the invertebrate chordate digestive tract relies on mucus secretion and cilia movement (74). We propose that cluster S1 represents a midgut-restricted, invertebrate chordate-specific group of ciliated cells.
Aside from these special cell clusters (L and S1) in the ilio-colon ring, amphioxus is also proposed to have “digestive cells,” which have large granular secretory vesicles observable under light microscopy and are capable of uptake and intracellular digestion of food particles (35, 36). Recent advances in electron microscopy support the presence of these digestive cells in the hepatic diverticulum and the hindgut (75). In this study, genes relevant to intracellular digestion, especially genes encoding lysosome protease cathepsin, are highly expressed in hindgut-specific clusters E and F. Both clusters show transcriptomic similarity to vertebrate goblet cells, a cell type dedicated to secreting mucin (1, 76). However, for the three major hepatic diverticulum enriched cell clusters AOK, B, and G, all three show transcriptomic similarities to vertebrate hepatocytes, and we did not find evidence of strong expression of genes relevant to intracellular digestion. Thus, additional evidence is needed to support the presence of cells capable of performing intracellular digestion in the amphioxus hepatic diverticulum.
Endocrine-like amphioxus cells are scattered throughout the digestive tract
In mammals, pancreatic endocrine cell types include five different hormone-secreting cell types, glucagon-secreting α cells, insulin-secreting β cells, somatostatin-secreting δ cells, ghrelin-producing ε cells, and pancreatic polypeptide (PP)–secreting PP cells (77). These pancreatic endocrine cells form islet structures, concentrated clusters of mixed pancreatic endocrine cell types forming highly vascularized micro-organs scattered throughout the pancreas (78). Immunostaining results in mice show lack of ε cells in adult pancreas tissue, whereas this cell type is prominent in the embryonic pancreas (79). PP cells are only found in tetrapods (80), whereas α, β, and δ cells are found in adult pancreas tissue of jawed vertebrates (81). In lampreys and hagfish, only β cells and δ cells have been detected in pancreatic tissue (82). Genome-wide searches for somatostatin and insulin peptide and receptor genes have suggested that somatostatin may have a chordate origin (49), whereas ILPs are present in multiple invertebrate lineages (83, 84). Therefore, it is thought that, among pancreatic endocrine cell types, insulin-secreting β cells are the most ancient (85).
We identify amphioxus gut cells (cluster R) that coexpress both Ilp paralogs, Ilp1 and Ilp2. Although most cluster R cells were dissociated from hindgut tissue, a portion of these cells are also found from the hepatic diverticulum and midgut tissue. In contrast to insulin, other pancreatic peptide hormones are less conserved, with somatostatin proposed to be chordate-specific and glucagon vertebrate-specific (49). Amphioxus has one gene encoding a somatostatin-like peptide (49), but most attempts to characterize expression of this peptide have not been focused on the digestive system (38). For glucagon, no orthologs of the vertebrate glucagon gene have been found in invertebrates, although one member of the glucagon-like peptide superfamily is present in amphioxus (31). We identify both peptide genes in the amphioxus gut, with Sst-like expressed in cluster X1 and PACAP/GCG expressed in cluster X2 along with very low levels of calcitonin (CT)/CT gene-related peptide (CGRP) gene Ctfp2, another peptide hormone expressed in amphioxus gut tissue (86). Sst-positive X1 cells are mainly located in the hepatic diverticulum, whereas PACAP/GCG-positive X2 cells are mainly located in the hindgut. This supports the presence of two gut cell clusters dedicated to producing somatostatin-like peptide and glucagon-like peptide, respectively, in the last chordate ancestor.
Published immunofluorescence results in lampreys and hagfish show close clustering of insulin and somatostatin-producing endocrine cells in the gut, whereas glucagon is not detectable (87, 88). A previous study also shows that no PACAP expression was detectable in lamprey intestine tissue, and both the ligand and its predicted receptors were mainly expressed in the brain (89). In amphioxus, using antisera raised against mammal peptide hormones can lead to false-positive results caused by antibody cross-reactivity and protein sequence divergence (90). For example, previous searches for PP cells in bony and cartilaginous fishes have identified fish gut peptides with equal levels of similarity to neuropeptide Y (NPY) and peptide YY (PYY) and can return a positive signal when tested against antimammalian PYY or PP antibodies (80, 81, 91). An unambiguous conclusion was reached only when peptides from the dogfish central nervous system (CNS) and gut tissue were sequenced (91). Gene sequencing results predict limited structural similarities between amphioxus peptide hormones and their vertebrate orthologs, with some such as Sst-like only sharing some key amino acids with vertebrate somatostain (49). Therefore, using antisera against mammal peptide hormones may fail to pick up true Ilp and Sst-like presence in amphioxus cells. We could not obtain reliable antibodies specific to amphioxus peptide hormones, and thus the exact location of amphioxus endocrine-like cells was not verified in this study. However, scRNA-seq results strongly suggest scattered distribution of endocrine cells along the entire A-P axis of the amphioxus digestive tract.
Although we do not find support for colocalization of ILP, somatostatin-like peptide, and glucagon-like peptide secreting cell clusters in amphioxus, the likely coappearance of these three cell types in the last chordate ancestor falls in line with shared TF profiles in mice and human α, β, and δ cells. Pancreatic endocrine cells share a common progenitor cell type, and α, β, and δ cells have a mutually repressive relationship that is controlled by TFs, which down-regulate and up-regulate different genes leading to cell maturation, and by cell intercommunication (77). First, for TFs, mice knockout experiments show that Pax4 promotes β cell development while suppressing α cell development, and Arx is essential for α cell development whereas loss of Arx promotes β and δ cell development (92). Pdx1 also promotes β cell development while actively suppressing α cell marker genes including the glucagon gene (93). Second, targeted deletion of Pdx1 function in β cells leads to loss of β cells whereas α and δ cells show significant overgrowth although the Pdx1 function has not been affected in these two cell types (94). β Cells may be capable of inhibiting proliferation of other endocrine cell types through cell intercommunication, although the exact factors responsible for this remains unclear (94). Here, the amphioxus digestive system scRNA-seq dataset was obtained from adult tissue; hence, we can only speculate on whether ILP, somatostatin-like peptide, and glucagon-like peptide secreting cell clusters shared a common TF program during differentiation and whether they shared a common progenitor cell type that was distributed throughout the developing digestive tract. scRNA-seq data from amphioxus embryos show that ILP secreting cells are present in the gut as early as mid-neurula (N4) stage, but somatostatin-like, and glucagon-like peptide secreting cells are only present in CNS cells (66). We do not know how somatostatin-like and glucagon-like peptide secreting cells form in the amphioxus digestive tract and when these cells appear.
Cross-species comparison of single-cell transcriptomic profiles
We compared the transcriptomic profile of amphioxus digestive tract cells with a published zebrafish digestive system dataset (57) at the single-cell level using a popular algorithm, SAMap (58). SAMap ranks cell-cell similarities based on expression of both orthologous and paralogous genes and is commonly used to assign cell identity in new cell atlases or for finding ancestral or conserved cell types across different lineages (58, 95). In most cases, we assume similarities in gene expression profiles is equal to homology between cell types; however, when this assumption falls through, we can come to very confusing conclusions. For example, in our results, there is high similarity between amphioxus cluster S1 cells (MG3-specific ciliated Pdx+ cells) and zebrafish gut neuronal cells (Fig. 2E), both of which are ciliated cell types but serve completely different functions and are likely of different developmental origins. Enteric neurons provide intrinsic innervation to the intestine and are specific to vertebrates as they originate from neural crest cells (96), which do not have a clear homolog in amphioxus (97). For adult amphioxus, although peripheral neurons are prominent around the gut (98), and it is possible that some neurons have been dissected along with gut tissue, it is highly unlikely that cluster S1 cells have neuronal identity as they do not express any known amphioxus peripheral neuron markers. Even if cluster S1 cells were peripheral neurons, it would be a very far stretch to claim that amphioxus peripheral neurons are similar to vertebrate enteric neurons given that both have been concluded to be unique to the respective species (96, 98). In this case, genes involved in forming cilia, tektin genes, are highly conserved at sequence level and strongly expressed in ciliated cell types (99, 100), which has skewed the SAMap results in concluding that amphioxus cluster S1 cells are most similar to zebrafish gut neuronal cells. This, along with published benchmarking assays comparing cross-species analysis methods (95), suggests that biological properties of the cell should be taken into consideration when making conclusions based on algorithms such as SAMap, and similarity does not always translate into homology or shared ancestry.
Ilp does not overlap with Pdx expression
Vertebrate Pdx1 directly regulates expression of insulin (101), a proposed ancient interaction evidenced by possible interaction between the mollusk Pdx protein and Ilp gene regulatory region (19). However, although the Pdx homeodomain in amphioxus and other vertebrate species is highly conserved, no sites within the core promoter region (−300 to +50 bp) of amphioxus Ilp1 and Ilp2 genes were supported by HOMER analysis to be potential Pdx1 binding sites. Furthermore, coexpression of Ilp and Pdx genes were not detected in the same cell in scRNA-seq data from amphioxus embryos and adult digestive tissue. Lack of direct regulatory relationship between amphioxus Pdx protein and the Ilp1 gene is also supported by bulk RNA transcriptome data from a published dataset (43) and our dataset, where both datasets show no change in Ilp1 expression in Pdx mutant five-gill slit larvae and in postmetamorphic juveniles.
In jawless vertebrates, hagfish lack a functional Pdx gene (102) but still have insulin-secreting cells clustered at the base of the bile duct and scattered throughout the digestive tract (88), whereas lampreys have a Pdxa gene that is specifically expressed in pancreas-like tissue (103) and have insulin-secreting cells clustered in this region (82) whereas somatostatin-secreting cells are distributed throughout the intestine (104). In the tunicate, an invertebrate chordate, expression of Insl-2 (one of the three tunicate Ilp orthologs) and Pdx are both observable in the anterior palps in mid-tailbud larva (21), and coexpression of both genes in the same cell is supported by embryo scRNA-seq data (105). Expression of Insl-3 and Pdx are both observable in the anterior endoderm (21); however, coexpression in the same cell is not supported by scRNA-seq data (105). In 1-week-old juveniles, Pdx expression is found in multiple regions along the A-P axis of the digestive tract (23) whereas, in 2- to 3-week-old juveniles, Pdx expression becomes restricted to the posterior stomach (106). Tunicate Insl-2 expression in 3-day-old juveniles is distributed throughout the esophagus and entire stomach, whereas in adults, expression of Insl-1 is only weakly detectable in stomach tissue, Insl-2 is expressed in the stomach and strongly expressed in different sections of the intestine, and Insl-3 is strongly expressed in the stomach and weakly present in some sections of the intestine (21). Because of lack of single-cell level data for juvenile and adult tunicate digestive tissue, we cannot pinpoint tunicate Ilp expression to a specific cell type. The relationship between Pdx and Ilp is also unclear in nonchordate deuterostomes, with Ilp-secreting cells distributed throughout the gut of sea urchin larva whereas Pdx (designated SpLox) is confined to a specific region of the gut endoderm in both sea urchins and sea stars (20, 22, 107). Early vertebrate genome duplications do seem to correlate with major innovations in the vertebrate pancreas; however, more evidence is required before we can conclude whether Pdx-regulation of Ilp genes was present in the last chordate ancestor and whether Pdx-regulated expression of the insulin gene is a necessary requirement for formation of clustered endocrine cells.
Ilp1 is structurally and functionally similar to Igf1
Vertebrate insulin and Igf proteins share a common peptide structure consisting of three domains (B, C, and A), with two additional domains (D and E) located at the C terminus of Igf proteins (84). The full amphioxus Ilp1 gene coding sequence predicts the presence of all five domains (B, C, A, D, and E) in the protein product, whereas Ilp2 consists of only three domains (B, C, and A) and has stronger structural similarity to vertebrate insulin. In addition, protein interaction experiments show that Ilp1 has weak binding capability to amphioxus Igfbp, a protein containing the characteristic vertebrate Igf binding domain (108).
Vertebrate Igf1 and Igf2 regulate cell proliferation and body growth, whereas insulin is a highly specific metabolic hormone that mainly regulates glucose levels in the body (84). For invertebrate species, many studies have linked ILPs to body growth (109), whereas studies conducted in protostomes such as flies (110), silkworms (111), and prawns (112) have also unraveled mechanisms allowing control of metabolism, specifically circulating glucose levels, through ILPs. However, for invertebrate deuterostomes, “circulating” and “blood glucose” are rarely discussed concepts as these animals are assumed to lack a complex vascular system and “blood” containing highly specialized blood cells (113). Although both Ilp1 and Ilpr mutant amphioxus are viable, they maintain the form of two- to three-gill slit larva throughout their entire lives. This inability to grow in body size strongly resembles the greatly impaired body size observed in Igf1 and Igf2 knockout mice (114). In addition, ablation of amphioxus Ilp1 does not affect body glucose concentration, and for Ilp2 mutant larvae, we did not observe any obvious change in phenotype. Furthermore, we do not find strong support for food-induced up-regulation of both Ilp1 and Ilp2 expression in wild-type amphioxus larva. We conclude that both Ilp genes are likely not key regulators of glucose metabolism in amphioxus.
Together, distribution of endocrine-like cells (clusters R, X1, and X2) throughout the amphioxus gut provides evidence against clustering of these cells in the same location, suggesting that clustering of endocrine cells and subsequent formation of a pancreas-like structure very likely occurred in the last vertebrate ancestor. Expression of midgut marker Pdx and Ilp in different amphioxus gut cells points to lack of Pdx regulation over Ilp expression in amphioxus, and we propose that evolution of new regulatory elements allowing Pdx control over insulin and the emergence of the new insulin gene were both crucial for formation of the pancreas in vertebrates.
MATERIALS AND METHODS
Amphioxus tissue dissection and RNA extraction
Amphioxus (B. floridae) obtained from a stock maintained by J.-K. Yu originating from Tampa, Florida, and all juvenile or adult animals regardless of genotype were housed in a temperature-controlled (19°C) facility at Xiamen University (115). Before dissection, adult amphioxus larger than 2 cm in body length was transferred to 19°C filtered seawater containing 20 mM MgCl2 (Sigma-Aldrich, 208337) for 10 min to sedate the animals, as inspired by an observation in cephalopods (116). Juvenile amphioxus smaller in size was directly transferred to 19°C filtered seawater without extra MgCl2. All animals were dissected under a stereoscope (Siontae, SZM45B2), and tissue was quickly rinsed in filtered seawater prior to transfer to TRIzol reagent (Invitrogen, 15596026). Samples were immediately processed using a glass grinder pretreated at 150°C overnight to remove ribonuclease (RNAse). All samples were stored on ice prior to RNA extraction performed following the standard TRIzol RNA extraction protocol.
Library preparation and sequencing
mRNA libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, E7530) and sequenced on an Illumina NovaSeq 6000 platform (Illumina) as 150-bp paired-end reads. All raw sequencing and processed files are openly accessible in the Open Archive for Miscellaneous Data hosted by the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (117, 118) and can be downloaded at https://ngdc.cncb.ac.cn/omix under accession number OMIX006304.
Bulk tissue transcriptome analysis
Raw reads containing adapter or poly-N sequences and low-quality reads were filtered prior to mapping to an updated B. floridae reference genome (a polished version of the Huang et al. annotated genome) (119). This reference genome is available at https://github.com/daiyc-zoo/amphioxus_singlecell, and an interactive BLAST platform is available at http://bio-add.org/InTranslg/. Reads were aligned using STAR v2.7.7a with --quantMode GeneCounts, --twopassMode Basic, --alignIntronMax 50000, --alignMatesGapMax 1000000, and --outFilterMultimapNmax 40 (https://github.com/alexdobin/STAR) (120). Gene counts were summarized using featureCounts v2.0.1 with -p and -O (https://subread.sourceforge.net/) (121). The DESeq2 v1.36.0 package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) (62) in R v4.2.0 (R Core, https://r-project.org/) was used to generate principal components analysis (PCA) plots.
Gene expression heatmaps were generated by first calculating the average TPM of all genes in all tissue types. TPM values were calculated for each sample by taking the gene count for each gene generated by featureCounts and dividing this value by gene length in kilobase. This value was then divided by the scale factor, a number equivalent to the sum of mapped reads to transcript normalized by transcript length for the entire sample divided by 1 million. For tissue samples with biological repeats (only exceptions being ovary and testis), the average TPM for each gene across all repeats was used for downstream analysis. To filter out genes with low expression, those with maximum expression less than five TPM in all tissues of interest were excluded. Normalized gene expression values for remaining genes were minimum-maximum normalized within each gene. This was then plotted using the pheatmap v1.0.12 package (https://CRAN.R-project.org/package=pheatmap).
Identification of tissue-specific amphioxus genes
First, we filtered out “not expressed” genes that have less than five TPM in all tissues. Tissue-specific genes were defined by ranking the TPM of each gene in all tissues from highest to lowest. If the top TPM value was at least five times higher than the second top value, then this gene was tissue-specific. If not, this gene was labeled “not specific.” We defined tissue-specific genes in all adult tissue types and tissue-specific genes in only the five digestive tissue types (hepatic diverticulum, MG1, MG2, MG3, and hindgut).
Identification of orthologous genes across species
Zebrafish pancreatic β cell–specific genes were identified from published adult zebrafish transcriptome data (table S1). Specifically, raw reads were downloaded and processed using the amphioxus transcriptome analysis pipeline described above. The reference zebrafish genome used was assembly GRCz11 (GenBank GCA_000002035.4). After average TPM values were summarized for all zebrafish genes and tissues, we filtered out genes with maximum TPM below five and identified 76 zebrafish pancreatic β cell–specific genes (table S1) using the standards described above for amphioxus.
OrthoFinder v2.5.4 (https://github.com/davidemms/OrthoFinder) (122, 123) was used to find zebrafish and amphioxus orthologs. Reference protein sequences for both species were extracted from the reference genomes described above. All amphioxus orthologs of zebrafish pancreatic β cell–specific genes were selected, regardless of whether they were one-to-one, one-to-many, or many-to-many. Selected amphioxus genes were filtered, normalized, and displayed as described above.
Amphioxus genes with similar trends in expression throughout the digestive tract were identified using the expression clustering algorithm STEM v1.3.13 (https://cs.cmu.edu/~jernst/stem/) (124). The hepatic diverticulum was set to “time point zero,” and MG1, MG2, MG3, and hindgut tissue expressions were assigned artificial “time points” of one, two, three, and four, respectively. STEM assigned genes into different expression profile clusters, among which 10 clusters showed statistical significance (fig. S1C). All heatmaps for genes in each cluster were plotted using the method described above. For GO enrichment, all amphioxus genes in each cluster were first matched to their zebrafish orthologs (obtained using OrthoFinder as described above). The Entrez ID of these zebrafish orthologs were used as an input for GO enrichment analysis using the org.Dr.eg.db v3.18.0 package (https://bioconductor.org/packages/release/data/annotation/html/org.Dr.eg.db.html) (125).
Single-cell sample collection and library construction
Tissues used for single-cell sequencing were dissected as described above. In brief, live cells sampled from the hepatic diverticulum, midgut, and hindgut were used for library preparation separately, and two biological samples for each section were taken from different individuals and sequenced separately. Considering the total cell number required for each sample to ensure a sufficient amount survive the dissociation and sample preparation process, we did not separate the midgut sample further into MG1, MG2, and MG3 segments.
To obtain single cells, we optimized existing protocols (105, 126) for adult amphioxus tissue. In brief, dissected tissue was transferred to ice-cold calcium-magnesium–free artificial seawater (CMF-ASW) (127). After the tissue slowly descended to the bottom of the tube, most CMF-ASW was removed, leaving behind ~25 μl to avoid exposing the tissue to air. Five hundred microliters of an ice-cold enzyme mix [5% trypsin, collagenase (2 mg ml−1), 0.5% pronase, and cellulase (1 mg ml−1) in CMF-ASW] was added, and the tissue was pipetted up and down rapidly in the mix for 1 min to speed up dissociation and then transferred to a 37°C incubator with a nutating shaker. The tissue was incubated for a total of 30 min, aided with manual pipetting every 5 min. Cell dissociation was monitored under an inverted microscope. Before termination of digestion, Calcein AM (BD Biosciences, 564061) at a final concentration of 10 μM was added to the tissue mix and incubated at 37°C with shaking for 5 min. Digestion was terminated by adding 1 ml of an ice-cold quenching solution [20% fetal bovine serum (FBS) and glycine (2 mg ml−1) in CMF-ASW]. Cells were passed through a 40-μm cell strainer and centrifuged at 500g, 4°C for 5 min. The supernatant was removed while ensuring the cell pellet was not exposed to air, and 500 μl of an RNase-free 3X phosphate-buffered solution (PBS) was added to resuspend the cells.
Cell viability and concentration were assessed again on a BD Rhapsody Scanner (BD Biosciences, 633701) to guarantee that the same number of cells was loaded for different samples. Following protocol guidelines, ~30,000 to 40,000 cells were loaded for each sample, and they were captured using the BD Rhapsody Express System and BD Rhapsody Cartridge Kit (BD Biosciences, 633733). Sequencing libraries were generated using the BD Rhapsody cDNA Kit (BD Biosciences, 633773) and BD Rhapsody WTA Amplification Kit (BD Biosciences, 633801) according to the manufacturer’s protocol. Concentration and quality of the libraries were assessed using a Qubit 3.0 Fluoromenter and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, #Q32851) and Agilent 2100 Bioanalyzer. All libraries were sequenced on an Illumina NovaSeq 6000 platform (Illumina) with aim to reach 80% sequencing saturation for each sample.
Processing of single-cell sequencing reads
Raw reads were processed with the BD Rhapsody WTA Analysis Pipeline v1.12.1 (https://bitbucket.org/CRSwDev/cwl/src/master/) on the Seven Bridges platform (https://sevenbridges.com/). All R1 and R2 reads were first tested for overlap and then trimmed and filtered for identification of cell label sequences (CLS), common linker sequences (L), and unique molecular identifier (UMI) sequence using custom code included in the Analysis Pipeline. Subsequently, R2 reads were aligned to the same amphioxus reference genome used for bulk tissue transcriptome analysis using default parameters for STAR (120). Specifically, only alignments beginning within the first five nucleotides of a R2 read, with length of the alignment match (can be a match or mismatch) in the Compact Idiosyncratic Gapped Alignment Report (CIGAR) string scoring above or equal to 37, and not aligning to phiX174 were retained for downstream analysis. Expression matrices for each sample were processed using the Seurat v4.1.1 package (https://satijalab.org/seurat/) (128) in R v4.1.1 (R Core, https://r-project.org/). Cells with less than 200 detected genes and genes detected in less than three cells were filtered out. To filter out doublet cells, all samples went through the scrublet v0.2.3 pipeline (https://github.com/swolock/scrublet) (129) in Python (Python Software Foundation, https://python.org/) using the parameters expected_doublet_rate = 0.06, sim_doublet_ratio = 2, n_neighbors = 30, min_count = 3, min_cells = 3, vscore_percentile = 85, and n_prin_comps = 30. Cells with doublet scores over 0.2 were labeled as doublets and removed from the sample prior to further analysis.
Only singlet cells were used for downstream analysis. We also filtered out cells with more than 20% RNA mapping to the same gene. Each sample was normalized independently by a default scale factor, and the top 2000 genes with highest variance were identified. To correct for batch effect, all 12 samples (six from control group and six from Pdx mutant group, as described below) were integrated using canonical correlation analysis (44). Specifically, features repeatedly identified as variable features across all 12 samples (referred to as “anchors”) were used for integration by the FindIntegrationAnchors function, with parameter dims = 1:50. Then, all samples were integrated (128) using the IntegrateData function with parameters anchor.features = anchors, k.filter = 200, and dims = 1:50. This one integrated dataset was used to carry out standard linear transformation followed by PCA and uniform manifold approximation and projection (UMAP) reduction, and we performed Louvain clustering on the 50 PCs with default parameters. We tested a range of resolution parameter settings ranging from one to four, and we settled for resolution = 3 as the best balance between identifying rare cell types and resolving larger, less differentiated cell types. For cell cluster identification, we manually merged adjacent clusters in samples based on expression of top marker genes nominated using the FindAllMarkers function in the Seurat package. For the manually merged clusters, we also identified cluster-specific marker genes using this function. To increase the reliability of nominated genes, we filtered out all genes with pct1/pct2 lower than 2.
To compare single-cell integration methods, we also analyzed all control and mutant cells without integration. Specifically, all cells were normalized as one sample by a default scale factor and the top 2000 genes with highest variance were identified, followed directly by PCA analysis and TSNE plotting. For Harmony integration (45), the same Seurat object and PCA used for CCA-Seurat analysis was imported and analyzed using Harmony v1.2.0 (https://github.com/immunogenomics/harmony) using all 50 PCA dimensions.
To compare amphioxus cell clusters with zebrafish gut cell types, we downloaded control group cells from a zebrafish dataset (57). SAMap v1.0.15 (https://github.com/atarashansky/SAMap) (58) was used for cross-species single-cell comparison. We used the get_mapping_scores function to obtain a statistical value for the degree of similarity between all amphioxus and zebrafish cell clusters. By ranking all values from highest to lowest, we identified the top 1, 5, and 10% cell-cell similarities. The top 10% amphioxus cell cluster to zebrafish cell cluster predictions were used to plot a Sankey plot using the networkD3 v0.4 package (https://CRAN.R-project.org/package=networkD3).
Cell abundance and gene differential expression between control and Pdx mutants
Using cell cluster identities defined above and taking into consideration biological repeat identity, we tested for differences in cell cluster abundance using Milo v1.2.0 (https://github.com/MarioniLab/miloR) (46). A Milo object was created from a merged dataset containing all control and mutant samples, and a k-nearest neighbor (KNN) graph was constructed using the parameters k = 30, d = 30, and reduced.dim = “PCA”. Representative neighborhoods on the KNN graph were defined using prop = 0.1, k = 30, d = 30, refined = TRUE, and reduced_dims = “PCA”. Cell identity was transferred from Seurat clustering results, and cell neighborhoods identified by Milo that consisted of a mix of cell identities with less than 50% cells sharing the same identity were labeled as “mixed.” When we used Milo to compare cell abundance between two biological repeats, we first randomly split one biological repeat into two artificial repeats of the same size using the sample function provided by the Seurat package, with set.seed set to 13, and then took artificial replicate identity along with batch identity into account when defining experimental design. When Milo was used to compare control and mutant samples, sample genotype, tissue, and batch identity were all taken into account.
Differential gene expression was calculated between specific cell clusters in control and mutant samples using two methods: one being the Seurat Wilcoxon rank sum test (sc test) and the second being the pseudobulk method. For the sc test, parallel biological repeats for each tissue type were considered as one sample and differentially expressed genes were defined as genes with padj < 0.05 and absolute log2foldchange > 0.25. For the pseudobulk test, cells were first split by cluster and sample identity, and then the average expression levels of all cells assigned the same sample and cluster identity were calculated. These values were used for analysis using the DESeq2 v1.36.0 package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) (62), with each biological repeat considered as different samples, and differentially expressed genes were defined as genes with padj < 0.05 and absolute log2foldchange > 0.25.
Embedding and paraffin sectioning
Postmetamorphosis juvenile amphioxi were fixed in 4% paraformaldehyde (PFA)-PBS (pH 7.4) (Sigma-Aldrich, 158127) for 1 hour at room temperature and then embedded in 1.5% low melting agarose (Sigma-Aldrich, A2790). Agarose blocks were prepared for paraffin embedding via an ethanol and xylene series: 30, 50, 60, 70, 80, 95, and 100% ethanol, xylene:ethanol (1:1), xylene, xylene:paraffin (1:1), xylene:paraffin (1:2), xylene:paraffin (1:3), and lastly paraffin (SCR, 8002-74-2). The prepared embryos were transferred to a paraffin block mold (Citotest, 80203-0009) and mounted on their ventral side. Paraffin blocks were sectioned using a Leica RM2016 microtome at a thickness of 5 μm. The sections were left to float on a 42°C dry bath (Coyote, H2O3-PRO) with water manually added to the metal surface and collected on microscope slides (Sail Brand, 7107) precoated with 18 μl of poly-l-lysine (2.5 mg/ml; Macklin, P875130). After the sections flattened at 42°C, water was manually dried away using water absorbent paper and then further fixed to the slides overnight at 37°C. Slides were stained with H&E and then photographed under a Leica DM4B microscope. Tissue width was measured using the ImageJ software (130).
Scanning electron microscopy
Extra incisions were made to the side of amphioxus gut tissue to expose the gut lumen. Tissues from different regions of the gut were fixed overnight at 4°C in 2% glutaraldehyde-seawater in separate tubes. Fixed tissue was then rinsed for 30 s in distilled water, dehydrated in ethanol and acetone, and processed by critical point drying in a Leica EM CPD300. Dried samples were mounted with the side-facing gut lumen upward on double-sided adhesive tape fixed to SEM stubs, coated with gold, and viewed in an FEI Quanta 650 FEG scanning electron microscope.
Generation of stable mutant Ilp1, Ilp2, and Ilpr amphioxus lines
Ilp1 mutants were established using a published transcription activator-like effector nuclease (TALEN)–guided method (131). Two TALEN mRNAs were coinjected into unfertilized amphioxus eggs, and gene editing success was assessed using a published protocol (table S5) (132). Ilp2 and Ilpr mutants were generated using CRISPR-Cas9 editing targeting the Ilp2 and Ilpr genes, respectively. We designed four guide RNA (gRNA) sequences targeting the Ilp2 first and second exon and five gRNA sequences targeting the Ilpr second exon (table S5). To test gRNA efficiency, we followed an established protocol (132). In brief, selected gRNA sequences were synthesized as DNA oligos and ligated into a pT7-gRNA plasmid (133). Template DNA fragments were polymerase chain reaction (PCR) synthesized from ligated pT7 vectors, and purified DNA containing a T7 promoter sequence and the correct gRNA sequence verified by Sanger sequencing was purified using the QIAquick PCR Purification Kit (Qiagen). gRNA was synthesized using the MEGAshortscript T7 Transcription Kit (Invitrogen) and purified using LiCl precipitation, concentrated to 1 μg/μl, and then mixed with Cas9 protein (Takara) in 100% glycerol and fluorescein isothiocyanate–labeled dextran (FITC-dextran, 10,000 molecular weight, Thermo Fisher Scientific) for microinjection. Each gRNA microinjection mix contained 0.25 μl of 100% glycerol, 0.125 μl of FITC-dextran (5 mg/ml), 0.875 μl of purified gRNA, and 0.25 μl of Cas9 protein (3 μg/μl).
To obtain mutants, we injected wild-type eggs with Cas9-gRNA and fertilized injected eggs after microinjection. Embryos were maintained in a 30°C incubator, and larvae were used for assessment of editing efficiency. We designed PCR primer pairs to amplify one region near the Ilp2 or Ilpr target sites (table S5) and assessed editing efficiency using restriction enzymes specific to the gRNA target site following a published protocol (132). We selected one gRNA with the highest editing efficiency for each gene and larvae injected with these gRNAs were maintained in ambient seawater maintained at 30°C until they metamorphosed. Gamete gene editing efficiency in each F0 individual was assessed using the same protocol described for gRNA efficiency assessment. Ripe F0 individuals carrying the Ilp2 or Ilpr mutant allele were then mated to wild-type individuals to produce F1 individuals with a heterozygous mutant genotype. Ripe F1 individuals were then crossed to produce F2 embryos, among which 25% were homozygous mutants.
Phylogenetic analysis and Pdx-binding prediction
Vertebrate insulin, Igf1, Igf2, relaxin, and invertebrate Ilp cDNA were obtained from NCBI (https://ncbi.nlm.nih.gov/), UniProt (https://uniprot.org/), and Ensembl Metazoa (https://metazoa.ensembl.org/index.html) where possible (table S5). Ilp sequences for a tunicate, C. productum, were obtained from the original paper (65). Ilp sequences for an acorn worm, Ptychodera flava, were obtained from genome data hosted by the Satoh lab, Okinawa Institute of Science and Technology (https://marinegenomics.oist.jp/acornworm/gallery) (134), and sea cucumber, Apostichopus japonicus, Ilp sequences were obtained from genome data hosted by the Institute of Oceanology, Chinese Academy of Sciences (http://www.genedatabase.cn/aja_genome_20161129.html) (135). BLASTN was used to identify potential amphioxus Ilp and relaxin-like genes using vertebrate insulin, Igf1, and relaxin protein sequences as a query. All proteins were aligned using ClustalW in MEGA11 (https://megasoftware.net/) (136). Phylogeny in Fig. 5A was generated using the maximum likelihood method in MEGA11 with bootstrap set to 500.
A 500-bp upstream region of the transcription start site for selected vertebrate insulin genes and invertebrate Ilp genes was extracted, and TAAT, the core Pdx-binding sequence, was manually identified. To predict potential Pdx-regulated amphioxus genes, we also used HOMER v4.11 (http://homer.ucsd.edu/homer/index.html) (63) to search for Pdx protein binding motifs in the core promoter region (−300 to +50 bp of the annotated transcription start site) of all amphioxus genes.
Plasmid construct preparation and whole-mount in situ hybridization
Full-length coding sequences of Ilp1 and Ilp2 were cloned from a neurula cDNA library (table S5), ligated into a pGEM-T-Easy vector (Promega, A1360), and verified by Sanger sequencing. Plasmids containing Ilp1 or Ilp2 were used to amplify a DNA template for antisense riboprobe synthesis using digoxigenin (DIG) (Roche, 11209256910) with T7 RNA polymerase (Promega, P2075). Whole-mount in situ hybridization with single probes was performed as previously described (137). Embryos were hybridized with one probe labeled with DIG and detected with an anti-DIG-alkaline phosphatase (AP) antibody (Roche, 11093274910). After staining, embryos were mounted in 80% glycerol and photographed using an inverted microscope (Olympus, IX73).
Larvae feeding and quantitative PCR
Larvae were starved for 5 days in filtered seawater maintained at 19°C and were maintained in two large glass petri dishes. Before the start of the feeding experiment, larvae were visually inspected under an inverted microscope to ensure that the gut of most animals were empty. Control group animals were stimulated with 1 ml of filtered seawater, and treatment group animals were stimulated with 1 ml of algae. Approximately 100 larvae were removed from each group at each designated time point in Fig. 5D and immediately transferred to TRIzol reagent. Total RNA was extracted and purified following the standard TRIzol RNA extraction protocol. cDNA was prepared from 1 μl of total RNA for each sample using the PrimeScript One Step RT-PCR Kit Ver.2 (Takara, RR055A).
Primers targeting amphioxus Ilp1 and Ilp2 were designed using the NCBI Primer-BLAST tool (https://ncbi.nlm.nih.gov/tools/primer-blast/) (table S5). Primers targeting amphioxus housekeeping gene Gapdh were obtained from a previous study (138). Primer specificity was verified prior to quantitative PCR (qPCR) using standard PCR with wild-type larvae cDNA as the template. qPCR was performed using 1X GoTaq SYBR Mix (Promega, M7132) with 20 ng of cDNA in a final volume of 20 μl. Each run consisted of one cycle of 5 min at 95°C followed by 45 cycles of 15 s at 95°C and 45 s at 60°C. Specific amplification of the target sequences was verified by constructing melting curves (80 cycles of 10 s, temperature increased by 0.5°C step from 55° to 95°C).
Larva metabolomics
For each sample used for GC-MS analysis, control or Ilp1 mutant individuals were snap frozen in liquid nitrogen. Larvae were fed normally (twice a day) and raised in the same petri dish prior to sample collection. Before GC-MS, 0.1 mg of larvae was directly homogenized in ice-cold methanol/water (4/1, v/v) by pipetting up and down rapidly. After vortexing for 30 s, the mixture was centrifuged (13,000 rpm, 4°C, 15 min) to precipitate proteins and tissue residues. Then, 800 μl of the supernatant was transferred into a fresh tube and lyophilized in a vacuum centrifuge. The residue was dissolved in 50 μl of the methoxyamine solution (20 mg/ml in pyridine) and incubated in a 37°C water bath for 1.5 hours, followed by a silylation reaction by adding 40 μl of N-methyl-N-(trimethylsilyl)trifluoroacetamide and incubating at 37°C for 1 hour. After centrifugation, the supernatant was aspirated and used for GC-MS analysis on a QP 2010Plus GC-MS system equipped with an AOC-20i autosampler (Shimadzu, Kyoto, Japan). Peak intensity data were normalized to tissue wet weight and analyzed using MetaboAnalyst v5.0 (https://metaboanalyst.ca/) (139).
Protein-protein interactions and immunoblotting analysis
The full-length coding sequence of amphioxus Ilp1, Ilp2, and Igfbp genes were cloned from embryo cDNA and ligated into a pcDNA3.3 expression vector (Invitrogen, K830001). Mouse Igf1 and Igfbp5 genes were cloned from a mouse cDNA library maintained by the School of Life Sciences, Xiamen University. For amphioxus Igfbp and mouse Igfbp5 genes, the full-length coding sequence excluding the stop codon was subcloned with a V5 tag at the C terminus into a pcDNA3.3 vector (fig. S6E). For amphioxus Ilp1 and Ilp2, and mouse Igf1, a FLAG tag was inserted between the signal peptide and start of the B peptide as described by a published study (140). Subcloned pcDNA3.3 vectors containing tagged amphioxus or mouse genes were transformed into Escherichia coli strain DH5α [American Type Culture Collection (ATCC), PTA-1977], followed by culturing in LB medium in a shaker at 200 rpm at 37°C. Plasmid DNA was purified using an in-house protocol and transfected into human embryonic kidney (HEK) 293T cells (ATCC, CRL-3216) maintained in Dulbecco’s modified Eagle’s medium (high glucose) supplemented with NaHCO3 (3.7 g/liter), 10% FBS, 100 IU of penicillin, and streptomycin (100 mg/ml) at 37°C in a humidified incubator containing 5% CO2. Polyethylenimine (PEI) transfection reagent (Polysciences, 23966) at a final concentration of 10 μM was used to transfect HEK293T cells. The total DNA to be transfected for each plate was adjusted to the same amount by using the relevant empty vector, which served as a negative control. Cells transfected with different plasmids were seeded into separate wells on a 6-well plate and harvested at 24 hours after transfection.
Cells transfected with different plasmids were lysed separately using 250 μl of an ice-cold lysis buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% (v/w) Triton X-100, 2.5 mM sodium pyrophosphate, and 1 mM β-glycerophosphate, with protease inhibitor cocktail]. Cells underwent sonication and centrifugation at 4°C for 15 min, and the supernatant containing FLAG-tagged proteins were mixed with the supernatant containing V5-tagged proteins, and then incubated with rabbit anti-V5 antibodies (Sigma-Aldrich, V8137) bound to protein A/G agarose beads overnight at 4°C. Protein A/G agarose beads were prepared in advance by mixing Protein A Sepharose Fast Flow beads (Cytiva, 17127904) and Protein G Sepharose 4 Fast Flow beads (Cytiva, 17061806) at a ratio of 1:1 and then balanced twice with 100 times volume of the lysis buffer. After incubation, the beads were washed three times with the lysis buffer and, at the final step, beads were directly mixed with an equal volume of 2× SDS sample buffer and boiled for 10 min before immunoblotting.
For immunoblotting, 1.0-mm-thick SDS-polyacrylamide gels (8% resolving gel) were prepared in-house following a published protocol (141). Fifteen microliters of each sample was loaded into wells, and electrophoresis was run at 100 V by a Mini-PROTEAN Tetra Electrophoresis Cell (Bio-Rad). Following previously described conditions, the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (0.45 μm, Merck, IPVH00010) (141). The blotted PVDF membrane was then blocked by 5% (w/v) nonfat milk dissolved in TBST [40 mM Tris, 275 μM NaCl, and 0.2% (v/v) Tween 20 (pH 7.6)] for 2 hours on an orbital shaker at 60 rpm at room temperature, followed by rinsing with TBST for twice, 5 min each. The PVDF membrane was incubated with rabbit anti-FLAG antibodies (Sigma-Aldrich, F7425) diluted at a ratio of 1:1000 overnight at 4°C on an orbital shaker at 60 rpm, followed by rinsing three times with TBST, 5 min each at room temperature. Goat anti-rabbit immunoglobulin G horseradish peroxidase conjugate antibody (Thermo Fisher Scientific, 31460) was added at a ratio of 1:2000 and incubated for 3 hours at room temperature with gentle shaking. Antibodies were then collected for reuse, and the membrane was washed three times using TBST, 5 min each at room temperature. PVDF membranes were incubated in an enhanced chemiluminescence (ECL) mixture (by mixing equal volumes of ECL solution and peroxide solution for 5 min) and then placed into a cassette with Medical X-Ray Film (FUJIFILM). The films were then developed with X-OMAT MX Developer and Replenisher and X-OMAT MX Fixer and Replenisher solutions (Carestream) on a Medical X-Ray Processor (Carestream) using Developer (Model 002, Carestream). The developed films were scanned using a Perfection V850 Pro scanner (Epson) with the Epson Scan software (v.3.9.3.4) and were cropped using the Inkscape software.
Sample sizes, statistics, and reproducibility
For all procedures, amphioxus of the same genetic background was randomized for dissection, photography, and downstream processing. No statistical method was used to predetermine the sample size. The investigators were not blinded to allocation during experiments and outcome assessment. For replicate samples taken on the same day, animals that could not be dissected at the same time were supplied with algae constantly to ensure that they remained under a well-fed condition (unless they were in the starved group). For replicate samples taken on different days, we made an effort to align the time of dissection to account for day-night cycles and time postfeeding.
For bulk RNA-seq of wild-type adult amphioxus tissues, each sample consisted of the same tissue sections dissected from at least two different individuals (table S1). The only exceptions are ovary and testis, which were rich in RNA content, and the Hatschek’s pit, which is difficult to dissect. At least three samples were processed in parallel for each tissue type, with the only exceptions being ovary, testis, and tentacles. For transcriptome comparison between the control and Pdx mutant tissue, the tissue dissected from five juvenile postmetamorphosis (1 cm) individuals were pooled into one tube for total RNA extraction and treated as one biological sample. A total of two biological samples for each group and tissue type were used for analysis (i.e., two control midgut samples versus two Pdx mutant midgut samples). Transcriptome comparison between control and Ilp1 three-gill slit stage larva was carried out by separating individuals displaying different phenotypes by visual inspection under a stereoscope (Olympus, szx16). Approximately 150 individuals were pooled into one tube for total RNA extraction as one biological repeat, and a total of three biological repeats for each group were used for analysis. Similarly, for metabolome comparison between the control and Ilp1 three-gill slit stage larva, ~200 individuals were pooled into the same tube as one biological repeat and snap frozen in liquid nitrogen. A total of three biological repeats for each group were used for analysis.
For scRNA-seq of the control and Pdx mutant digestive tissue, we display the total number of adult individuals and tissues used in two figures (figs. S2A and S6A). In brief, three control individuals and three Pdx mutant individuals were used for tissue sampling, with one individual used for midgut dissection, one for both hepatic diverticulum and hindgut dissection, and one for dissection of all three tissue sections. Therefore, for each of the three tissue types used for scRNA-seq, two biological replicates from different individuals were used for analysis.
For paraffin sectioning, five juvenile postmetamorphosis (1 cm) control and eight juvenile postmetamorphosis (1 cm) Pdx mutant amphioxus individuals were used. For SEM scanning, two adult control and two adult Pdx mutant amphioxus individuals were used. For in situ analysis and photography, the total number of individuals in each sample is indicated in the relevant figure. For qPCR analysis, each batch of three-gill slit wild-type larva were starved for 5 days in filtered seawater and separated into two groups, each consisting of around 1000 individuals. For each time point, ~100 to 150 individuals were randomly sampled and used for total RNA extraction. Data from three batches of larva were used for analysis.
Acknowledgments
We are grateful to Y.-J. Luo, T. Lewin, and J. Mulley for advice and feedback on the preliminary dataset, and we thank S. Shimeld for feedback on the first draft of this paper. We thank Y. Wang for helping arrange test trials on the BD Rhapsody platform. T. Yang and J. Liu provided assistance while testing the SEM platform for amphioxus tissue, and we thank L. Yao and C. Wu for assisting with SEM amphioxus tissue dehydration and gold particle coating. We thank L. Zhou for assistance setting up test trials for metabolomic analysis of amphioxus larvae samples. We are grateful to S. Lin for helpful discussions on protein interaction results. For data storage and upload to the National Genomics Data Center, we thank W. Liu for assistance. We thank C. Duan for seminal discussions around generation of Ilp1 knockout amphioxus, and we thank H. Shen and H. Lin for establishing the Ilp1 knockout line.
Funding: This work was supported by the National Science Foundation of China grant 32100335 (Y.D.); Office of China Postdoc Council 2021 Postdoctoral International Exchange Program, Talent-Introduction Program Postdoc number 273565 (Y.D.); Institute of Zoology Chinese Academy of Sciences Bingzhi Postdoctoral Fellowship Program (Y.D.); Institute of Zoology, Chinese Academy of Sciences grant 2023IOZ0104 (X.Z.), and National Science Foundation of China grants 32070458, 32270439, and 32061160471 (G.Li).
Author contributions: Conceptualization: Y.D. Sample collection: Y.D., R.P., Y.F., C.S., G.Li, and Y.S. Experimental investigation: Y.D., R.P., and Z.C. Maintenance of Pdx knockout animals: X.W. Computational analysis: Y.D. Visualization: Y.D. Establishment of the long non-coding (lnc) online database: J.L. and Z.L. Supervision: G.Li, X.Z., Q.P., M.L., and P.Z. Data interpretation: Y.D., X.Z., and G.Liu. Writing—original draft: Y.D. Writing—review and editing: Y.D., G.Li, and X.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Plasmids and amphioxus lines generated in this study can be provided by Xiamen University pending scientific review and a completed material transfer agreement. Requests for relevant plasmids and amphioxus lines should be submitted to G.Li. The data generated in this study can be downloaded in a raw and processed form from the Open Archive for Miscellaneous Data hosted by the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences and are publicly accessible at https://ngdc.cncb.ac.cn/omix under accession number OMIX006304. Publicly available datasets and common, freely available data analysis software packages used in this project are listed in the Materials and Methods section. Python and R codes are freely available at https://github.com/daiyc-zoo/amphioxus_singlecell or the Science Data Bank (142) at https://scidb.cn/en/list under DOI https://doi.org/10.57760/sciencedb.08801.
Supplementary Materials
The PDF file includes:
Figs. S1 to S8
Legends for tables S1 to S5
Legends for movies S1 and S2
Legends for datasets S1 and S2
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
Movies S1 and S2
Datasets S1 and S2
REFERENCES AND NOTES
- 1.A. Verma, S. Duggal, I. Kumar, “Gastrointestinal system” in Imaging in Critical Care Medicine (CRC Press, ed. 4, 2023), pp. 116–138. [Google Scholar]
- 2.I. Chester-Jones, P. M. Ingleton, J. G. Phillips, “The gastrointestinal endocrine system” in Fundamentals of Comparative Vertebrate Endocrinology (Springer US, 1987), vol. 122, pp. 541–578. [Google Scholar]
- 3.Brooke N. M., Garcia-Fernàndez J., Holland P. W. H., The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Wright C. V. E., Schnegelsberg P., De Robertis E. M., XlHbox 8: A novel Xenopus homeo protein restricted to a narrow band of endoderm. Development 105, 787–794 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Ohlsson H., Karlsson K., Edlund T., IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12, 4251–4259 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller C. P., McGehee R. E., Habener J. F., IDX-1: A new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 13, 1145–1156 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard J., Peers B., Johnson T., Ferreri K., Lee S., Montminy M. R., Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol. Endocrinol. 7, 1275–1283 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Boam D. S. W., Docherty K., A tissue-specific nuclear factor binds to multiple sites in the human insulin-gene enhancer. Biochem. J. 264, 233–239 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Quobaili F., Montenarh M., Pancreatic duodenal homeobox factor-1 and diabetes mellitus type 2 (review). Int. J. Mol. Med. 21, 399–404 (2008). [PubMed] [Google Scholar]
- 10.Kaneto H., Miyatsuka T., Shiraiwa T., Yamamoto K., Kato K., Fujitani Y., Matsuoka T., Crucial role of PDX-1 in pancreas development, β-cell differentiation, and induction of surrogate β-cells. Curr. Med. Chem. 14, 1745–1752 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Jonsson J., Carlsson L., Edlund T., Edlund H., Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371, 606–609 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L. M., Wright C. V. E., PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122, 983–995 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Le Lay J., Stein R., Involvement of PDX-1 in activation of human insulin gene transcription. J. Endocrinol. 188, 287–294 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Ferrier D. E. K., Holland P. W. H., Sipunculan ParaHox genes. Evol. Dev. 3, 263–270 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Liguori F., Mascolo E., Vernì F., The genetics of diabetes: What we can learn from Drosophila. Int. J. Mol. Sci. 22, 11295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grönke S., Clarke D. F., Broughton S., Andrews T. D., Partridge L., Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLOS Genet. 6, e1000857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Kennedy S. G., Ruvkun G., Daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17, 844–858 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kodama E., Kuhara A., Mohri-Shiomi A., Kimura K. D., Okumura M., Tomioka M., Iino Y., Mori I., Insulin-like signaling and the neural circuit for integrative behavior in C. elegans. Genes Dev. 20, 2955–2960 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu F., Marlétaz F., Gavriouchkina D., Liu X., Sauka-Spengler T., Zhang G., Holland P. W. H., Evidence from oyster suggests an ancient role for Pdx in regulating insulin gene expression in animals. Nat. Commun. 12, 3117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perillo M., Arnone M. I., Characterization of insulin-like peptides (ILPs) in the sea urchin Strongylocentrotus purpuratus: Insights on the evolution of the insulin family. Gen. Comp. Endocrinol. 205, 68–79 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Thompson J. M., Di Gregorio A., Insulin-like genes in ascidians: Findings in Ciona and hypotheses on the evolutionary origins of the pancreas. Genesis 53, 82–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole A. G., Rizzo F., Martinez P., Fernandez-Serra M., Arnone M. I., Two ParaHox genes, SpLox and SpCdx, interact to partition the posterior endoderm in the formation of a functional gut. Development 136, 541–549 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Iguchi R., Usui K., Nakayama S., Sasakura Y., Sekiguchi T., Ogasawara M., Multi-regional expression of pancreas-related digestive enzyme genes in the intestinal chamber of the ascidian Ciona intestinalis type A. Cell Tissue Res. 394, 423–430 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Bertrand S., Escriva H., Evolutionary crossroads in developmental biology: Amphioxus. Development 138, 4819–4830 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Bourlat S. J., Juliusdottir T., Lowe C. J., Freeman R., Aronowicz J., Kirschner M., Lander E. S., Thorndyke M., Nakano H., Kohn A. B., Heyland A., Moroz L. L., Copley R. R., Telford M. J., Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature 444, 85–88 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Putnam N. H., Butts T., Ferrier D. E. K., Furlong R. F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu K., Benito-Gutiérrez È., Dubchak I., Garcia-Fernàndez J., Gibson-Brown J. J., Grigoriev I. V., Horton A. C., De Jong P. J., Jurka J., Kapitonov V. V., Kohara Y., Kuroki Y., Lindquist E., Lucas S., Osoegawa K., Pennacchio L. A., Salamov A. A., Satou Y., Sauka-Spengler T., Schmutz J., Shin-I T., Toyoda A., Bronner-Fraser M., Fujiyama A., Holland L. Z., Holland P. W. H., Satoh N., Rokhsar D. S., The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Holland L. Z., Holland N. D., Chordate origins of the vertebrate central nervous system. Curr. Opin. Neurobiol. 9, 596–602 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Benito-Gutiérrez È., Gattoni G., Stemmer M., Rohr S. D., Schuhmacher L. N., Tang J., Marconi A., Jékely G., Arendt D., The dorsoanterior brain of adult amphioxus shares similarities in expression profile and neuronal composition with the vertebrate telencephalon. BMC Biol. 19, 110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holland N. D., The long and winding path to understanding kidney structure in amphioxus—A review. Int. J. Dev. Biol. 61, 683–688 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Kozmik Z., Holland N. D., Kalousova A., Paces J., Schubert M., Holland L. Z., Characterization of an amphioxus paired box gene, AmphiPax2/5/8: Developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development 126, 1295–1304 (1999). [DOI] [PubMed] [Google Scholar]
- 31.On J. S. W., Su L., Shen H., Arokiaraj A. W. R., Cardoso J. C. R., Li G., Chow B. K. C., PACAP/GCGa is an important modulator of the amphioxus CNS-Hatschek’s pit axis, the homolog of the vertebrate hypothalamic-pituitary axis in the basal chordates. Front. Endocrinol. 13, 850040 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holland L. Z., Albalat R., Azumi K., Benito-Gutiérrez È., Blow M. J., Bronner-Fraser M., Brunet F., Butts T., Candiani S., Dishaw L. J., Ferrier D. E. K., Garcia-Fernàndez J., Gibson-Brown J. J., Gissi C., Godzik A., Hallböök F., Hirose D., Hosomichi K., Ikuta T., Inoko H., Kasahara M., Kasamatsu J., Kawashima T., Kimura A., Kobayashi M., Kozmik Z., Kubokawa K., Laudet V., Litman G. W., McHardy A. C., Meulemans D., Nonaka M., Olinski R. P., Pancer Z., Pennacchio L. A., Pestarino M., Rast J. P., Rigoutsos I., Robinson-Rechavi M., Roch G., Saiga H., Sasakura Y., Satake M., Satou Y., Schubert M., Sherwood N., Shiina T., Takatori N., Tello J., Vopalensky P., Wada S., Xu A., Ye Y., Yoshida K., Yoshizaki F., Yu J.-K., Zhang Q., Zmasek C. M., de Jong P. J., Osoegawa K., Putnam N. H., Rokhsar D. S., Satoh N., Holland P. W. H., The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100–1111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller J., Über den Bau und die Lebenserscheinungen des Branchiostoma lubricum Costa, Amphioxus lanceolatus Yarrel. Abh. K. Preuss. Akad. Wiss. 1844, 79–116 (1844). [Google Scholar]
- 34.van Wijhe J., On the anatomy of the larva of Amphioxus lanceolatus and the explanation of its asymmetry. K. Ned. Akad. van Wet. Proc. 21, 1013–1023 (1919). [Google Scholar]
- 35.Barrington E. J. W., VI- The digestive system of Amphioxus (Branchiostoma) Lanceolatus. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 228, 269–312 (1937). [Google Scholar]
- 36.Biuw L. W., Hulting G., Fine-grained secretory cells in the intestine of the lancelet, Branchiostoma (Amphioxus) lanceolatum, studied by light microscopy. Zeitschrift für Zellforsch. und Mikroskopische Anat. 120, 546–554 (1971). [DOI] [PubMed] [Google Scholar]
- 37.Chan S. J., Cao Q. P., Steiner D. F., Evolution of the insulin superfamily: Cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. Proc. Natl. Acad. Sci. U.S.A. 87, 9319–9323 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nozaki M., Gorbman A., The question of functional homology of Hatschek’s pit of amphioxus (Branchiostoma belcheri) and the vertebrate adenohypophysis. Zoolog. Sci. 9, 387–395 (1992). [Google Scholar]
- 39.Gorbman A., Brain–Hatschek’s pit relationships in amphioxus species. Acta Zool. 80, 301–305 (1999). [Google Scholar]
- 40.Marlétaz F., Firbas P. N., Maeso I., Tena J. J., Bogdanovic O., Perry M., Wyatt C. D. R., de la Calle-Mustienes E., Bertrand S., Burguera D., Acemel R. D., van Heeringen S. J., Naranjo S., Herrera-Ubeda C., Skvortsova K., Jimenez-Gancedo S., Aldea D., Marquez Y., Buono L., Kozmikova I., Permanyer J., Louis A., Albuixech-Crespo B., Le Petillon Y., Leon A., Subirana L., Balwierz P. J., Duckett P. E., Farahani E., Aury J. M., Mangenot S., Wincker P., Albalat R., Benito-Gutiérrez È., Cañestro C., Castro F., D’Aniello S., Ferrier D. E. K., Huang S., Laudet V., Marais G. A. B., Pontarotti P., Schubert M., Seitz H., Somorjai I., Takahashi T., Mirabeau O., Xu A., Yu J. K., Carninci P., Martinez-Morales J. R., Crollius H. R., Kozmik Z., Weirauch M. T., Garcia-Fernàndez J., Lister R., Lenhard B., Holland P. W. H., Escriva H., Gómez-Skarmeta J. L., Irimia M., Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564, 64–70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun T., Zhang S., Ji G., Identification and expression of an elastase homologue in Branchiostoma belcheri with implications to the origin of vertebrate pancreas. Mol. Biol. Rep. 37, 3303–3309 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Ma P., Liu X., Xu Z., Liu H., Ding X., Huang Z., Shi C., Liang L., Xu L., Li X., Li G., He Y., Ding Z., Chai C., Wang H., Qiu J., Zhu J., Wang X., Ding P., Zhou S., Yuan Y., Wu W., Wan C., Yan Y., Zhou Y., Zhou Q. J., Wang G. D., Zhang Q., Xu X., Li G., Zhang S., Mao B., Chen D., Joint profiling of gene expression and chromatin accessibility during amphioxus development at single-cell resolution. Cell Rep. 39, 110979 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Zhong Y., Herrera-Úbeda C., Garcia-Fernàndez J., Li G., Holland P. W. H., Mutation of amphioxus Pdx and Cdx demonstrates conserved roles for ParaHox genes in gut, anus and tail patterning. BMC Biol. 18, 68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P., Raychaudhuri S., Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dann E., Henderson N. C., Teichmann S. A., Morgan M. D., Marioni J. C., Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 40, 245–253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., Guo Z.-Y., Shen L., Zhang Y.-J., Feng Y.-M., Refolding of amphioxus insulin-like peptide: Implications of a bifurcating evolution of the different folding behavior of insulin and insulin-like growth factor 1. Biochemistry 42, 9687–9693 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Lecroisey C., Le Pétillon Y., Escriva H., Lammert E., Laudet V., Identification, evolution and expression of an insulin-like peptide in the cephalochordate Branchiostoma lanceolatum. PLOS ONE 10, e0119461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mirabeau O., Joly J.-S., Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. U.S.A. 110, E2028–E2037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amos L. A., The tektin family of microtubule-stabilizing proteins. Genome Biol. 9, 229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alfonso-Pérez T., Hayward D., Holder J., Gruneberg U., Barr F. A., MAD1-dependent recruitment of CDK1-CCNB1 to kinetochores promotes spindle checkpoint signaling. J. Cell Biol. 218, 1108–1117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wojcik E. J., Buckley R. S., Richard J., Liu L., Huckaba T. M., Kim S., Kinesin-5: Cross-bridging mechanism to targeted clinical therapy. Gene 531, 133–149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirokawa N., Noda Y., Okada Y., Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr. Opin. Cell Biol. 10, 60–73 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Xiao Y., Jiang Z., Li Y., Ye W., Jia B., Zhang M., Xu Y., Wu D., Lai L., Chen Y., Chang Y., Huang X., Liu H., Qing G., Liu P., Li Y., Xu B., Zhong M., Yao Y., Pei D., Li P., ANGPTL7 regulates the expansion and repopulation of human hematopoietic stem and progenitor cells. Haematologica 100, 585–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pascual-Anaya J., Albuixech-Crespo B., Somorjai I. M. L., Carmona R., Oisi Y., Álvarez S., Kuratani S., Muñoz-Chápuli R., Garcia-Fernàndez J., The evolutionary origins of chordate hematopoiesis and vertebrate endothelia. Dev. Biol. 375, 182–192 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Chen J. W., Galloway J. L., The development of zebrafish tendon and ligament progenitors. Development 141, 2035–2045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willms R. J., Jones L. O., Hocking J. C., Foley E., A cell atlas of microbe-responsive processes in the zebrafish intestine. Cell Rep. 38, 110311 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Tarashansky A. J., Musser J. M., Khariton M., Li P., Arendt D., Quake S. R., Wang B., Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 10, e66747 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smillie C. S., Biton M., Ordovas-Montanes J., Sullivan K. M., Burgin G., Graham D. B., Herbst R. H., Rogel N., Slyper M., Waldman J., Sud M., Andrews E., Velonias G., Haber A. L., Jagadeesh K., Vickovic S., Yao J., Stevens C., Dionne D., Nguyen L. T., Villani A.-C., Hofree M., Creasey E. A., Huang H., Rozenblatt-Rosen O., Garber J. J., Khalili H., Desch A. N., Daly M. J., Ananthakrishnan A. N., Shalek A. K., Xavier R. J., Regev A., Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carvalho J. E., Lahaye F., Yong L. W., Croce J. C., Escrivá H., Yu J.-K., Schubert M., An updated staging system for cephalochordate development: One table suits them all. Front. Cell Dev. Biol. 9, 668006 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hao Y., Stuart T., Kowalski M. H., Choudhary S., Hoffman P., Hartman A., Srivastava A., Molla G., Madad S., Fernandez-Granda C., Satija R., Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gittes G. K., Developmental biology of the pancreas: A comprehensive review. Dev. Biol. 326, 4–35 (2009). [DOI] [PubMed] [Google Scholar]
- 65.McRory J. E., Sherwood N. M., Ancient divergence of insulin and insulin-like growth factor. DNA Cell Biol. 16, 939–949 (1997). [DOI] [PubMed] [Google Scholar]
- 66.Dai Y., Zhong Y., Pan R., Yuan L., Fu Y., Chen Y., Du J., Li M., Wang X., Liu H., Shi C., Liu G., Zhu P., Shimeld S., Zhou X., Li G., Evolutionary origin of the chordate nervous system revealed by amphioxus developmental trajectories. Nat. Ecol. Evol. 8, 1693–1710 (2024). [DOI] [PubMed] [Google Scholar]
- 67.Osborne P. W., Benoit G., Laudet V., Schubert M., Ferrier D. E. K., Differential regulation of ParaHox genes by retinoic acid in the invertebrate chordate amphioxus (Branchiostoma floridae). Dev. Biol. 327, 252–262 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Longo A., Guanga G. P., Rose R. B., Structural basis for induced fit mechanisms in DNA recognition by the Pdx1 homeodomain. Biochemistry 46, 2948–2957 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Rorsman P., Braun M., Regulation of insulin secretion in human pancreatic islets. Annu. Rev. Physiol. 75, 155–179 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Cheatham B., Kahn C. R., Insulin action and the insulin signaling network. Endocr. Rev. 16, 117–142 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Riddle M. R., Boesmans W., Caballero O., Kazwiny Y., Tabin C. J., Morphogenesis and motility of the Astyanax mexicanus gastrointestinal tract. Dev. Biol. 441, 285–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakashima K., Kimura S., Ogawa Y., Watanabe S., Soma S., Kaneko T., Yamada L., Sawada H., Tung C.-H., Lu T.-M., Yu J.-K., Villar-Briones A., Kikuchi S., Satoh N., Chitin-based barrier immunity and its loss predated mucus-colonization by indigenous gut microbiota. Nat. Commun. 9, 3402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Castro L. F. C., Gonçalves O., Mazan S., Tay B.-H., Venkatesh B., Wilson J. M., Recurrent gene loss correlates with the evolution of stomach phenotypes in gnathostome history. Proc. R. Soc. London Ser. B Biol. Sci. 281, 20132669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakayama S., Sekiguchi T., Ogasawara M., Molecular and evolutionary aspects of the protochordate digestive system. Cell Tissue Res. 377, 309–320 (2019). [DOI] [PubMed] [Google Scholar]
- 75.He C., Han T., Liao X., Zhou Y., Wang X., Guan R., Tian T., Li Y., Bi C., Lu N., He Z., Hu B., Zhou Q., Hu Y., Lu Z., Chen J.-Y., Phagocytic intracellular digestion in amphioxus (Branchiostoma). Proc. R. Soc. London Ser. B Biol. Sci. 285, 20180438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vidak E., Javoršek U., Vizovišek M., Turk B., Cysteine cathepsins and their extracellular roles: Shaping the microenvironment. Cells 8, 264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pan F. C., Wright C., Pancreas organogenesis: From bud to plexus to gland. Dev. Dyn. 240, 530–565 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Bonner-Weir S., Morphological evidence for pancreatic polarity of β-cell within islets of Langerhans. Diabetes 37, 616–621 (1988). [DOI] [PubMed] [Google Scholar]
- 79.Suissa Y., Magenheim J., Stolovich-Rain M., Hija A., Collombat P., Mansouri A., Sussel L., Sosa-Pineda B., McCracken K., Wells J. M., Heller R. S., Dor Y., Glaser B., Gastrin: A distinct fate of Neurogenin3 positive progenitor cells in the embryonic pancreas. PLOS ONE 8, e70397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conlon J. M., The origin and evolution of peptide YY (PYY) and pancreatic polypeptide (PP). Peptides 23, 269–278 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Youson J. H., Al-Mahrouki A. A., Ontogenetic and phylogenetic development of the endocrine pancreas (islet organ) in fishes. Gen. Comp. Endocrinol. 116, 303–335 (1999). [DOI] [PubMed] [Google Scholar]
- 82.Youson J. H., Cheung R., Morphogenesis of somatostatin- and insulin-secreting cells in the lamprey endocrine pancreas. Fish Physiol. Biochem. 8, 389–397 (1990). [DOI] [PubMed] [Google Scholar]
- 83.Wu Q., Brown M. R., Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 51, 1–24 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Chan S. J., Steiner D. E., Insulin through the ages: Phylogeny of a growth promoting and metabolic regulatory hormone. Am. Zool. 40, 213–222 (2000). [Google Scholar]
- 85.Madsen O. D., Pancreas phylogeny and ontogeny in relation to a ‘pancreatic stem cell’. C. R. Biol. 330, 534–537 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sekiguch T., Kuwasako K., Ogasawara M., Takahashi H., Matsubara S., Osugi T., Muramatsu I., Sasayama Y., Suzuki N., Satake H., Evidence for conservation of the calcitonin superfamily and activity-regulating mechanisms in the basal chordate Branchiostoma floridae. J. Biol. Chem. 291, 2345–2356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elliott W. M., Youson J. H., Immunocytochemical localization of insulin and somatostatin in the endocrine pancreas of the sea lamprey Petromyzon marinus L., at various stages of its life cycle. Cell Tissue Res. 243, 629–634 (1986). [Google Scholar]
- 88.Falkmer S., Emdin S., Havu N., Lundgren G., Marques M., Östberg Y., Steiner D. F., Thoma N. W., Insulin in invertebrates and cyclostomes. Am. Zool. 13, 625–638 (1973). [Google Scholar]
- 89.Ng S. Y. L., Chow B. K. C., Kasamatsu J., Kasahara M., Lee L. T. O., Agnathan VIP, PACAP and their receptors: Ancestral origins of today’s highly diversified forms. PLOS ONE 7, e44691 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reinecke M., Immunohistochemical localization of polypeptide hormones in endocrine cells of the digestive tract of Branchiostoma lanceolatum. Cell Tissue Res. 219, 445–456 (1981). [DOI] [PubMed] [Google Scholar]
- 91.Larhammar D., Evolution of neuropeptide Y, peptide YY and pancreatic polypeptide. Regul. Pept. 62, 1–11 (1996). [DOI] [PubMed] [Google Scholar]
- 92.Collombat P., Mansouri A., Hecksher-Sørensen J., Serup P., Krull J., Gradwohl G., Gruss P., Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 17, 2591–2603 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holland A. M., Góñez L. J., Naselli G., MacDonald R. J., Harrison L. C., Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of β-cells in the adult pancreas. Diabetes 54, 2586–2595 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Gannon M., Ables E. T., Crawford L., Lowe D., Offield M. F., Magnuson M. A., Wright C. V. E., Pdx-1 function is specifically required in embryonic β cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev. Biol. 314, 406–417 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song Y., Miao Z., Brazma A., Papatheodorou I., Benchmarking strategies for cross-species integration of single-cell RNA sequencing data. Nat. Commun. 14, 6495 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gershon M. D., Genes and lineages in the formation of the enteric nervous system. Curr. Opin. Neurobiol. 7, 101–109 (1997). [DOI] [PubMed] [Google Scholar]
- 97.Yu J.-K., Meulemans D., McKeown S. J., Bronner-Fraser M., Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 18, 1127–1132 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wicht H., Lacalli T. C., The nervous system of amphioxus: Structure, development, and evolutionary significance. Can. J. Zool. 83, 122–150 (2005). [Google Scholar]
- 99.Norrander J., Larsson M., Ståhl S., Höög C., Linck R., Expression of ciliary tektins in brain and sensory development. J. Neurosci. 18, 8912–8918 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas J., Morlé L., Soulavie F., Laurençon A., Sagnol S., Durand B., Transcriptional control of genes involved in ciliogenesis: A first step in making cilia. Biol. Cell 102, 499–513 (2010). [DOI] [PubMed] [Google Scholar]
- 101.Melloul D., Transcription factors in islet development and physiology: Role of PDX-1 in beta-cell function. Ann. N. Y. Acad. Sci. 1014, 28–37 (2004). [DOI] [PubMed] [Google Scholar]
- 102.Furlong R. F., Younger R., Kasahara M., Reinhardt R., Thorndyke M., Holland P. W. H., A degenerate ParaHox gene cluster in a degenerate vertebrate. Mol. Biol. Evol. 24, 2681–2686 (2007). [DOI] [PubMed] [Google Scholar]
- 103.Zhang H., Ravi V., Tay B.-H., Tohari S., Pillai N. E., Prasad A., Lin Q., Brenner S., Venkatesh B., Lampreys, the jawless vertebrates, contain only two ParaHox gene clusters. Proc. Natl. Acad. Sci. U.S.A. 114, 9146–9151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Van Noorden S., Greenberg J., Pearse A. G. E., Cytochemical and immunofluorescence investigations on polypeptide hormone localization in the pancreas and gut of the larval lamprey. Gen. Comp. Endocrinol. 19, 192–199 (1972). [DOI] [PubMed] [Google Scholar]
- 105.Cao C., Lemaire L. A., Wang W., Yoon P. H., Choi Y. A., Parsons L. R., Matese J. C., Wang W., Levine M., Chen K., Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349–354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakayama S., Satou K., Orito W., Ogasawara M., Ordered expression pattern of Hox and ParaHox genes along the alimentary canal in the ascidian juvenile. Cell Tissue Res. 365, 65–75 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Annunziata R., Andrikou C., Perillo M., Cuomo C., Arnone M. I., Development and evolution of gut structures: From molecules to function. Cell Tissue Res. 377, 445–458 (2019). [DOI] [PubMed] [Google Scholar]
- 108.Daza D. O., Sundström G., Bergqvist C. A., Duan C., Larhammar D., Evolution of the insulin-like growth factor binding protein (IGFBP) family. Endocrinology 152, 2278–2289 (2011). [DOI] [PubMed] [Google Scholar]
- 109.Leevers S. J., Growth control: Invertebrate insulin surprises! Curr. Biol. 11, 209–212 (2001). [DOI] [PubMed] [Google Scholar]
- 110.Géminard C., Arquier N., Layalle S., Bourouis M., Slaidina M., Delanoue R., Bjordal M., Ohanna M., Ma M., Colombani J., Léopold P., Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes 55, S5–S8 (2006). [Google Scholar]
- 111.Masumura M., Satake S., Saegusa H., Mizoguchi A., Glucose stimulates the release of bombyxin, an insulin-related peptide of the silkworm Bombyx mori. Gen. Comp. Endocrinol. 118, 393–399 (2000). [DOI] [PubMed] [Google Scholar]
- 112.Jiang Q., Jiang Z., Gu S., Qian L., Li X., Gao X., Zhang X., Insights into carbohydrate metabolism from an insulin-like peptide in Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 293, 113478 (2020). [DOI] [PubMed] [Google Scholar]
- 113.Ruppert E. E., Carle K. J., Morphology of metazoan circulatory systems. Zoomorphology 103, 193–208 (1983). [Google Scholar]
- 114.Accili D., Nakae J., Kim J. J., Park B.-C., Rother K. I., Targeted gene mutations define the roles of insulin and IGF-I receptors in mouse embryonic development. J. Pediatr. Endocrinol. Metab. 12, 475–485 (1999). [DOI] [PubMed] [Google Scholar]
- 115.Li G., Shu Z., Wang Y., Year-round reproduction and induced spawning of Chinese amphioxus, Branchiostoma belcheri, in laboratory. PLOS ONE 8, e75461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Messenger J. B., Nixon M., Ryan K. P., Magnesium chloride as an anaesthetic for cephalopods. Comp. Biochem. Physiol. C Comp. Pharmacol. Toxicol. 82, 203–205 (1985). [DOI] [PubMed] [Google Scholar]
- 117.Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Dong L., Zhang Z., Yu C., Sun Y., Chi L., Chen H., Zhai S., Sun Y., Lan L., Zhang X., Xiao J., Bao Y., Wang Y., Zhang Z., Zhao W., The genome sequence archive family: Toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics 19, 578–583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.National Genomics Data Center Members and Partners , Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 48, D24–D33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang Z., Xu L., Cai C., Zhou Y., Liu J., Xu Z., Zhu Z., Kang W., Cen W., Pei S., Chen D., Shi C., Wu X., Huang Y., Xu C., Yan Y., Yang Y., Xue T., He W., Hu X., Zhang Y., Chen Y., Bi C., He C., Xue L., Xiao S., Yue Z., Jiang Y., Yu J.-K., Jarvis E. D., Li G., Lin G., Zhang Q., Zhou Q., Three amphioxus reference genomes reveal gene and chromosome evolution of chordates. Proc. Natl. Acad. Sci. U.S.A. 120, e2201504120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Emms D. M., Kelly S., OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Emms D. M., Kelly S., OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ernst J., Bar-Joseph Z., STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.M. Carlson, org.Dr.eg.db: Genome wide annotation for Zebrafish (Bioconductor, 2019).
- 126.Satoh N., Tominaga H., Kiyomoto M., Hisata K., Inoue J., Nishitsuji K., A preliminary single-cell RNA-seq analysis of embryonic cells that express Brachyury in the amphioxus, Branchiostoma japonicum. Front. Cell Dev. Biol. 9, 696875 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Unson M. D., Holland N. D., Faulkner D. J., A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 119, 1–11 (1994). [Google Scholar]
- 128.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M., Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wolock S. L., Lopez R., Klein A. M., Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li G., Feng J., Lei Y., Wang J., Wang H., Shang L.-K., Liu D.-T., Zhao H., Zhu Y., Wang Y.-Q., Mutagenesis at specific genomic loci of amphioxus Branchiostoma belcheri using TALEN method. J. Genet. Genomics 41, 215–219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Su L., Shi C., Huang X., Wang Y., Li G., Application of CRISPR/Cas9 nuclease in amphioxus genome editing. Genes 11, 1311 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.-W., Xi J. J., Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Simakov O., Kawashima T., Marlétaz F., Jenkins J., Koyanagi R., Mitros T., Hisata K., Bredeson J., Shoguchi E., Gyoja F., Yue J.-X., Chen Y.-C., Freeman R. M., Sasaki A., Hikosaka-Katayama T., Sato A., Fujie M., Baughman K. W., Levine J., Gonzalez P., Cameron C., Fritzenwanker J. H., Pani A. M., Goto H., Kanda M., Arakaki N., Yamasaki S., Qu J., Cree A., Ding Y., Dinh H. H., Dugan S., Holder M., Jhangiani S. N., Kovar C. L., Lee S. L., Lewis L. R., Morton D., Nazareth L. V., Okwuonu G., Santibanez J., Chen R., Richards S., Muzny D. M., Gillis A., Peshkin L., Wu M., Humphreys T., Su Y.-H., Putnam N. H., Schmutz J., Fujiyama A., Yu J.-K., Tagawa K., Worley K. C., Gibbs R. A., Kirschner M. W., Lowe C. J., Satoh N., Rokhsar D. S., Gerhart J., Hemichordate genomes and deuterostome origins. Nature 527, 459–465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang X., Sun L., Yuan J., Sun Y., Gao Y., Zhang L., Li S., Dai H., Hamel J.-F., Liu C., Yu Y., Liu S., Lin W., Guo K., Jin S., Xu P., Storey K. B., Huan P., Zhang T., Zhou Y., Zhang J., Lin C., Li X., Xing L., Huo D., Sun M., Wang L., Mercier A., Li F., Yang H., Xiang J., The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLOS Biol. 15, e2003790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tamura K., Stecher G., Kumar S., MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu J. K. S., Holland L. Z., Amphioxus whole-mount in situ hybridization. Cold Spring Harb. Protoc. 2009, pdb.prot5286 (2009). [DOI] [PubMed] [Google Scholar]
- 138.Yuan L., Wang Y., Li G., Differential expression pattern of two Brachyury genes in amphioxus embryos. Gene Expr. Patterns 38, 119152 (2020). [DOI] [PubMed] [Google Scholar]
- 139.Pang Z., Chong J., Zhou G., de Lima Morais D. A., Chang L., Barrette M., Gauthier C., Jacques P.-É., Li S., Xia J., MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pfeffer L. A., Brisson B. K., Lei H., Barton E. R., The insulin-like growth factor (IGF)-I E-peptides modulate cell entry of the mature IGF-I protein. Mol. Biol. Cell 20, 3810–3817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang C.-S., Li M., Wang Y., Li X., Zong Y., Long S., Zhang M., Feng J.-W., Wei X., Liu Y.-H., Zhang B., Wu J., Zhang C., Lian W., Ma T., Tian X., Qu Q., Yu Y., Xiong J., Liu D.-T., Wu Z., Zhu M., Xie C., Wu Y., Xu Z., Yang C., Chen J., Huang G., He Q., Huang X., Zhang L., Sun X., Liu Q., Ghafoor A., Gui F., Zheng K., Wang W., Wang Z.-C., Yu Y., Zhao Q., Lin S.-Y., Wang Z.-X., Piao H.-L., Deng X., Lin S.-C., The aldolase inhibitor aldometanib mimics glucose starvation to activate lysosomal AMPK. Nat. Metab. 4, 1369–1401 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.L. Chengzan, H. Yanfei, L. Jianhui, Z. Lili, ScienceDB: A public multidisciplinary research data repository for eScience, in 2017 IEEE 13th International Conference on e-Science (e-Science) (IEEE, 2017), pp. 248–255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Legends for tables S1 to S5
Legends for movies S1 and S2
Legends for datasets S1 and S2
Tables S1 to S5
Movies S1 and S2
Datasets S1 and S2