Abstract
The covalently attached cofactor biotin plays pivotal roles in central metabolism. The top-priority ESKAPE-type pathogens, Acinetobacter baumannii and Klebsiella pneumoniae, constitute a public health challenge of global concern. Despite the fact that the late step of biotin synthesis is a validated anti-ESKAPE drug target, the primary stage remains fragmentarily understood. We report the functional definition of two BioC isoenzymes (AbBioC for A. baumannii and KpBioC for K. pneumoniae) that act as malonyl-ACP methyltransferase and initiate biotin synthesis. The physiological requirement of biotin is diverse within ESKAPE pathogens. CRISPR-Cas9–based inactivation of bioC rendered A. baumannii and K. pneumoniae biotin auxotrophic. The availability of soluble AbBioC enabled the in vitro reconstitution of DTB/biotin synthesis. We solved two crystal structures of AbBioC bound to SAM cofactor (2.54 angstroms) and sinefungin (SIN) inhibitor (1.72 angstroms). Structural and functional study provided molecular basis for SIN inhibition of BioC. We demonstrated that BioC methyltransferase plays dual roles in K. pneumoniae infection and A. baumannii colistin resistance.
Bacterial BioC methyltransferase has multifaceted roles in biotin nutritional immunity, representing an attractive drug target.
INTRODUCTION
The water-soluble vitamin B7, biotin, is an indispensable micronutrient throughout the three domains of life (1). Unlike plants and most of bacterial species that program biotin synthesis pathways (1), humans cannot make this coenzyme. In general, gut commensals behave as a major group of biotin suppliers for mammalian biotin-dependent carboxylases/decarboxylases (2, 3). Granted that its dietary supplement benefits the notorious inflammatory bowel disease (4), the manipulation of biotin constitutes a promising option to ameliorate the kind of metabolic disorders caused by inherited biotin deficiency.
The de novo synthesis of an organic sulfur-containing biotin cofactor consists of a primary step and a late stage. The long-settled late segment proceeds via four conserved enzymes BioF/A/D/B to assemble the fused heterocyclic rings of biotin. A variety of early steps is committed to generate pimeloyl moiety as a fatty acid–like “arm” of biotin (i.e., an α,ω-dicarboxylic acid with seven carbons). Notably, the primary stage varies distinctly in diverse bacterial lineages. So far, no less than three routes have been assigned to pimelate production, namely, (i) a prototypical “BioC-BioH” strategy (5–7), (ii) the “BioI-BioW” machinery (8–10), and (iii) a noncanonical BioZ pathway (11, 12). The prevalent BioC-BioH form exploits a type II fatty acid synthesis (FAS II) to elongate a temporarily disguised primer of methyl malonate, giving the methyl-pimeloyl moiety product (5–7). In contrast to BioH esterase removing methyl moiety from methyl-pimelate, BioC methyltransferase introduces a methyl disguise to produce a surrogate methyl-malonate. In Bacillus subtilis, the P450 enzyme BioI oxidatively cleaves long-chain fatty acyl-ACP esters to give pimeloyl-ACP, a cognate biotin precursor (10), and then BioW ligase converts pimelic acid to pimeloyl–coenzyme A (CoA) ester (8, 9), an alternative substrate for BioF (8-amino-7-oxononanoate synthase) (13). As an atypical β-ketoacyl-ACP synthase III (FabH)-like enzyme, α-proteobacterial BioZ condenses a malonyl–acyl carrier protein (Mal-ACP) with a glutaryl-CoA donated from lysine degradation, producing pimeloyl-ACP, a cognate precursor for biotin synthesis (11, 12). The growing arsenal of BioH isoenzymes largely extends our understanding of complexity in the paradigm BioC-BioH early step (14), whereas the remaining cousin BioC appears as a conundrum in that it is biochemically unamenable and lacks structural insights (7, 15). Structure and catalysis of the recalcitrant BioC enzyme represent the last piece of “golden fleece” in structural biology of bacterial biotin synthesis.
Antimicrobial resistance (AMR) is a devastating “one health” threat (16). Of around 5 million AMR-related deaths worldwide, ~1.27 million is attributable to AMR in 2019 (17). The AMR dissemination stimulates the World Health Organization to recognize top priority “ESKAPE” pathogens (18), which namely include (i) Enterococcus faecalis, (ii) S. aureus, (iii) K. pneumoniae, (iv) A. baumannii, (v) Pseudomonas aeruginosa, and (vi) Enterobacter species. Moreover, biotin deficiency–causing intestinal dysbiosis engenders the dynamic role of ESKAPE-type microbes in the transition from microbiota compositions to opportunistic agents (19, 20). To tackle the ESKAPE-causing crisis, it is required to develop certain pathogen-specific, narrow-spectrum antimicrobials with unique drug targets. The maintenance of bacterial biotin homeostasis is postulated to be the case. This is because mycobacterial biotin synthesis and its biotinylation is a validated druggable antituberculosis (anti-TB) pathway (21, 22). Recently, Carfrae and colleagues (23) reported that targeting the late stage of biotin synthesis impairs pathogenicity of two ESKAPE members (K. pneumoniae and A. baumannii) during mice infection mimicking human environment. Similarly, virulence attenuation was seen for an additional ESKAPE agent, P. aeruginosa, upon the removal of BioH, a gatekeeper of biotin synthesis (24). However, the primary step of biotin generation in K. pneumoniae and A. baumannii awaits experimental demonstration. Because it is the coenzyme for AccB, a core component of acetyl-CoA carboxylase (ACC) complex that initiates FAS II pathway (25), biotin determines cell envelope lipid remodeling that is implicated in successful lung infections with Mycobacterium abscessus (26). Central to biotin, FAS II route was recently found to be required for bacterial insusceptibility to colistin, a “last-resort” defense against AMR-producing superbugs (27). Collectively, we favored that the “100-year-old” vitamin biotin is a multifaceted player.
Here, we proposed that a BioC-BioH paradigm is shared by the two distantly related ESKAPE microbes, A. baumannii and K. pneumoniae. Using SUMO fusion expression strategy, our continued efforts allowed the success in obtaining the soluble form of the recalcitrant BioC enzyme. Apart from the genetic and biochemical definition, we presented two high-resolution x-ray structures of A. baumannii BioC (AbBioC) bound to S-adenosyl-l-methionine (SAM) cofactor (2.54 Å) and sinefungin (SIN) inhibitor (1.72 Å). Because it functions as a pivotal player in the primary step of biotin synthesis, we explored clinical roles of BioC in K. pneumoniae infection and A. baumannii colistin resistance. In summary, this work solves a “long-standing” puzzle in structural exploration of bacterial biotin synthesis (1) and underlines BioC as a potential anti-ESKAPE drug target.
RESULTS
Genetic organization of ESKAPE biotin synthesis
Most of current knowledge on the early step of biotin synthesis arises from studies with the Gram-negative Escherichia and Gram-positive Bacillus (1). Unlike the paradigm Escherichia coli that relies on BioC-BioH pair to make biotin precursor, B. subtilis exploits a distinct pimeloyl-CoA BioW to compensate the genetic loss of bioC-bioH, whereas its relative Bacillus cereus retains the canonical BioC-BioH route because it contains a bifunctional type II biotin protein ligase (BirA) (BC1537)–regulated operon of bioADFHCB (fig. S1). The component of gut microbiota community, E. faecalis, seemed to be the only biotin auxotrophic member of ESKAPE microbes, and this is because only a locus of bioY (encoding biotin transporter) rather than a bio cluster that is encoded on its chromosome (fig. S1). Similar to B. subtilis that is characterized by a bioWAFDBI operon, S. aureus also adopts BioW machinery for biotin production, except with a different arrangement of bioDABFWX. It was noted that the small membrane protein BioX is assumed to be involved in biotin metabolism but awaits functional assignment (fig. S1). As for the E. coli bioC-bioH pair, bioC is integrated into the bioBFCD operon adjacent to bioA, whereas bioH is free-standing (fig. S1). An identical genetic context was noticed in K. pneumoniae, an opportunistic ESKAPE pathogen with an origin of intestinal microbiota (fig. S1). Similar scenarios were observed for the other two Enterobacter species, Enterobacter cloacae, and Enterobacter aerogenes (fig. S1). In particular, P. aeruginosa is an unusual ESKAPE pathogen in which a complete pathway of biotin synthesis is domesticated into a single “bioA/BFHCD” cluster (24, 28). Except for bioB that is scattered on chromosome, a similar operon “bioHAFCD” appears in A. baumannii (fig. S1). The arrangement (bioA/BFHCD and bioHAFCD) is postulated to confer physiological advantage in assuring parallel expression of BioH and BioC, two important players for biotin precursor pimeloyl-ACP generation (28). Thus, genetic organization for biotin primary step varies markedly among ESKAPE-type pathogens (Fig. 1A and fig. S1).
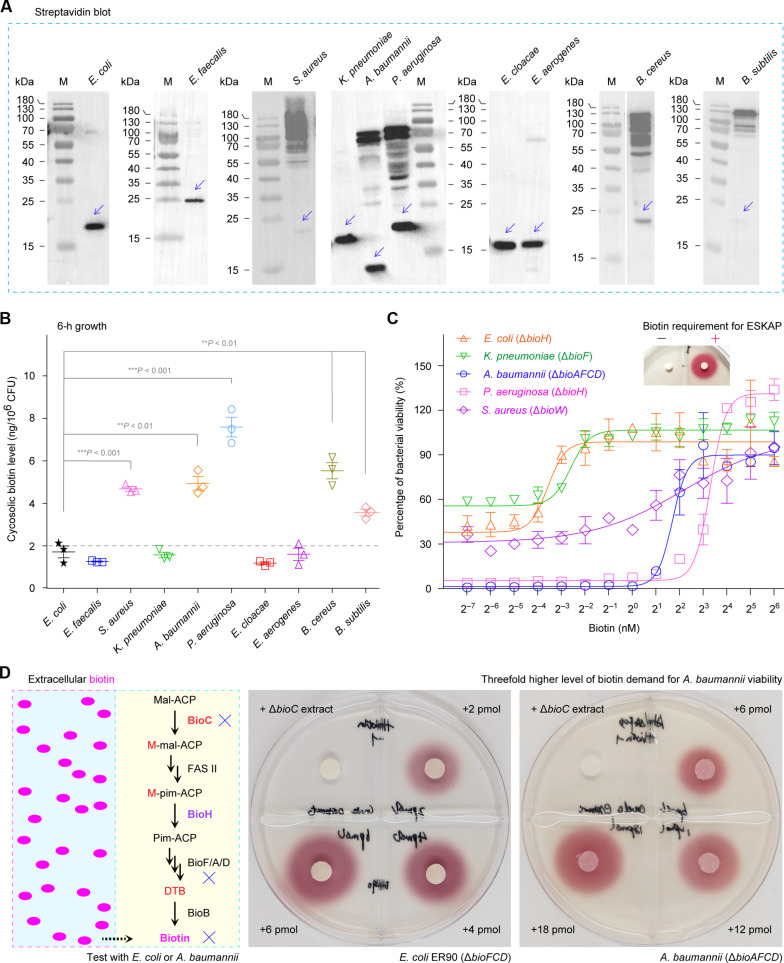
Fig. 1. Physiological requirement of biotin in ESKAPE-including microbes.
(A) Diversity in protein biotinylation of ESKAPE microbes. To detect biotin modification, Western blot was performed with crude extracts of diverse ESKAPE species, in which anti-streptavidin primary antibody was included. A representative photograph from three biological replicates was displayed. The biotinylated forms of BCCP (biotin carboxy carrier protein; AccB) were highlighted with blue arrows. (B) Varied level of cytosolic biotin in ESKAPE microbes. Data were presented as an average ± SD of three independent experiments. This was assayed by two-tailed analysis of variance (B). (C) Determination of biotin demand in certain ESKAPE-type pathogens. On the basis of the nonlinear regression model provided by GraphPad Prism, bacterial viability curves were plotted here, of which each data point is given as means ± SD (n = 3). Four biotin auxotrophic microbes were tested here, namely, E. coli (ΔbioH), K. pneumoniae (ΔbioF), A. baumannii (ΔbioAFCD), and P. aeruginosa (ΔbioH). The inside graph of biotin bioassay represents the viability of the biotin auxotroph ER90 (ΔbioF/C/D) engendered by the addition of exogenous biotin. The symbols of minus (−) and plus (+) denotes the absence or presence of 4-pmol biotin. (D) Biotin bioassays suggested that the supply of exogenous biotin at around a threefold higher level render A. baumannii viable at the comparable level to that of E. coli. The magenta oval dots on the left hand of (D) denote a pool of biotin molecules. Of three independent DTB/biotin bioassays, a representative result was given here. M, protein standards; BioH, methyl-pimeloyl ACP ester demethylase; BioC, Mal-ACP O-methyltransferase; BioD, dethiobiotin synthetase. h, hour.
Diverse demands for biotin by ESKAPE
In total, 10 diverse microbes including ESKAPE were systematically analyzed to seek for the alteration on physiological demand of biotin. Apart from the profile of biotinylated protein (Fig. 1A), cytosolic biotin level was measured (Fig. 1B), as well as the minimum biotin requirement for bacterial viability (Fig. 1C). In terms of streptavidin blotting, all the 10 species were roughly divided into two groups (Fig. 1A). Unlike a single AccB protein that is biotinylated in the five species (E. coli, E. faecalis, K. pneumoniae, E. cloacae, and E. aerogenes), biotin modification of multiple proteins were found in the remaining five species (S. aureus, A. baumannii, P. aeruginosa, B. cereus, and B. subtilis). The enzyme-linked immunosorbent assay (ELISA) measurement unveiled that (i) five bacterial species with a single AccB biotinylated (such as E. coli and E. faecalis) consistently contain a pool of intracellular biotin at a comparable level of almost 2 ng/106 colony-forming units (CFU) and (ii) the other five species characterized by multiple protein biotinylation (e.g., S. aureus and A. baumannii) display the relatively high level (4 to 8 ng/106 CFU) of cytosolic biotin (Fig. 1B). Apart from an assignment to biotin transporters having altered activities, a varied level of cytosolic biotin in diverse ESKAPE microbes largely accounts for the distinct profile of protein biotinylation among different species (Fig. 1, A and B). In addition, this observation is correlated with the fact that the minimal requirement of biotin in E. coli and K. pneumoniae is substantially less than those in A. baumannii, P. aeruginosa, and S. aureus (Fig. 1C). Biotin bioassays also validated that the minimal demand of biotin is around threefold higher in A. baumannii compared to E. coli (Fig. 1D).
Next, we focused on the recalcitrant ESKAPE member, A. baumannii, and performed in silico search for potential biotin-requiring proteins. As a result, seven candidates were returned (fig. S2A), which invariantly feature a biotinylated lysine (K) residue at the conserved motif EAMK (i.e., four continuous residues: glutamate, alanine, methione, and lysine) (fig. S2B). Consistent with the bioinformatic analysis, multiple proteins are biotinylated (rather than one single AccB band) in our assay of streptavidin blot (fig. S2, C and D). To verify this observation, four of seven candidates [HKO16_10520 (AccB), HKO16_06425, HKO16_06925, and HKO16_15815] were selected for further analyses. All the four recombinant forms (AccB and HKO16_06425 in full length and HKO16_06925 plus HKO16_15815 in an EAMK-covering truncated version) were overexpressed and purified to homogeneity (fig. S2E). Furthermore, streptavidin blot demonstrated that they are indeed biotin-modified forms (fig. S2F), which is quite similar to those of the plant causative agent, Agrobacterium tumefaciens (29), and the human pathogen surrogate, Mycobacterium smegmatis (30, 31). This constitutes an explanation for the relatively high biotin requirement by A. baumannii (Fig. 1, B and C), highlighting the complexity in biotin demand by diverse ESKAPE pathogens.
Genetic definition of two bioC homologs
The prior study led by Cronan’s laboratory implicated that E. coli BioC (EcBioC) functions as an initiator for the prototypic BioC-BioH primary pathway of biotin synthesis (Fig. 2, A and B) and is genetically replaced by the distantly related paralog of B. cereus (BcBioC) with only 26.94% identity (fig. S3) (5, 32). Phylogeny of BioC suggested its origin of methyltransferase (Fig. 2, C and D). Different from K. pneumoniae that arranges bioH (KPSH_49340) isolated from bioC (KPSH_16290), A. baumannii recruits bioH (HKO16_03840) into the same “bioHAFCD” operon as bioC (HKO16_03860) does (Fig. 2A). In addition, KpBioC seems alike EcBioC rather than BcBioC (63.75% versus 25.71% identity), whereas AbBioC requires several gaps for the alignment with both EcBioC and BcBioC (25 to 26.61% identity; fig. S3). To evaluate in vivo roles of distinct bioC homologs, we combined two genetic approaches, namely, (i) functional complementation of the E. coli ΔbioC mutant and (ii) knockout of bioC from A. baumannii (or K. pneumoniae). By using the CRISPR-Cas9 system (fig. S4, A and B) (33), we inactivated the bioC locus from A. baumannii. The mutant of A. baumannii devoid of bioC was validated by multiplex polymerase chain reaction (PCR) and Sanger DNA sequencing (fig. S4C). As expected from bacterial viability, the removal of bioC rendered A. baumannii biotin auxotrophic (fig. S4D). We noticed that the plasmid-borne expression of AbBioC restores robust growth of the E. coli ΔbioC mutant on the nonpermissive condition (Fig. 2, E and F). The complementation of AbBioC corrected the inability of A. baumannii ΔbioC strain to appear on the biotin-lacking condition (Fig. 2, G and H). In addition, KpBioC genetically replaces EcBioC as the positive control BcBioC does (fig. S5A). As expected, the ΔbioC isogenic mutant of K. pneumoniae that we created here is also characterized by a biotin auxotroph and can be restored upon an introduction of either AbBioC or KpBioC (fig. S5B).
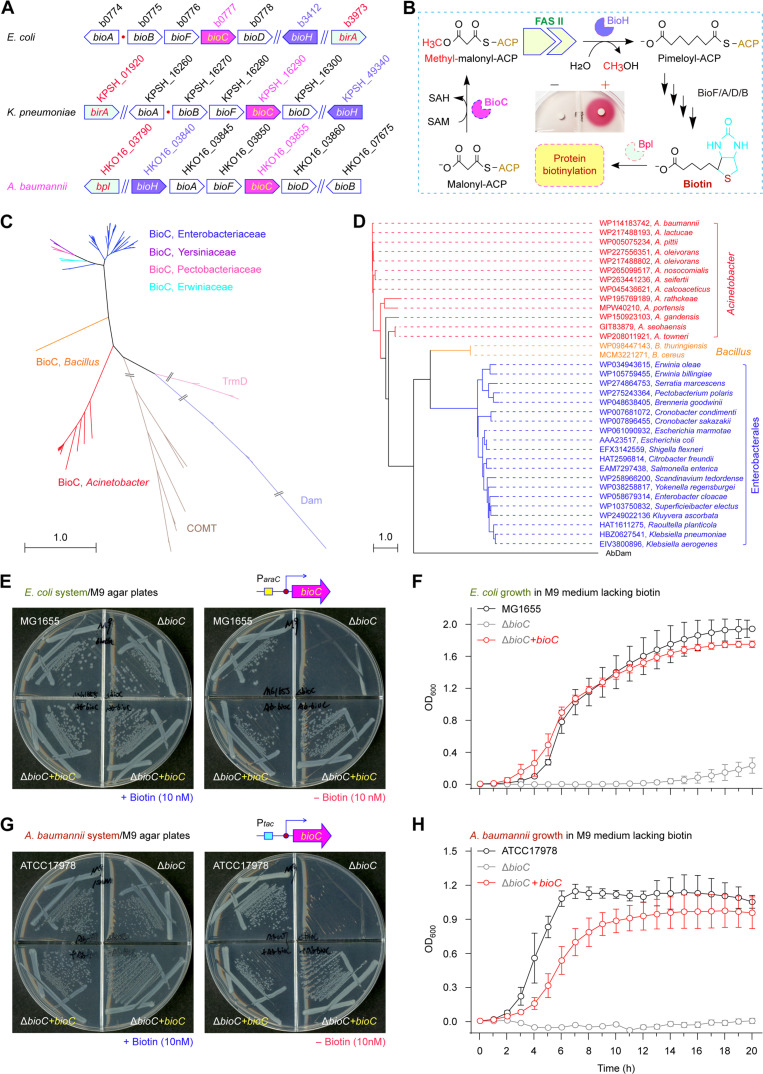
Fig. 2. Functional identification of A. baumannii bioC.
(A) The cluster of biotin synthesis operon. (B) The BioC-BioH path for biotin synthesis. As described in Fig. 1C, the inside graph indicates the viability of the biotin auxotroph ER90 empowered by the supplementation of exogenous biotin. (C) Phylogeny for the family of methyltransferases. The maximum likelihood (ML)–based phylogeny was inferred from 1000 bootstrap replicates and exhibited in a radial form. Apart from three distantly related enzymes TrmD (light pink), Dam (light blue), and COMT (brown), certain BioC enzymes of six different origins were included. Namely, they refer to Enterobacteriaceae (blue), Yersiniaceae (purple), Pectobacteriaceae (pink), Erwiniaceae (cyan), Bacillus (orange), and Acinetobacter (red). (D) An unrooted tree of bacterial BioC orthologs. The evolutionary history of BioC was also produced in terms of the ML method with 1000 bootstrap replicates. The percentages of replicate trees in which the associated taxa are clustered in the bootstrap test were labeled next to the branches. Accession numbers of individual BioC members were indicated accordingly. A. baumannii Dam (AbDam) acted as an internal reference for phylogeny. (E) The A. baumannii bioC paralog genetically restores the inability of biotin auxotrophic E. coli ΔbioC mutant on the nonpermissive condition. (F) Growth curves of the biotin auxotrophic E. coli ΔbioC mutants with or without a plasmid-borne A. baumannii bioC paralog. (G and H) The CRISPR-Cas9–based inactivation of A. baumannii bioC renders it biotin auxotrophic, and this is restored by in trans expression of the plasmid-borne bioC. Of three independent viability experiments, a representative photograph was given [(E) and (G)]. Growth curves were plotted [(F) and (H)], and each data point was shown as mean ± SD (n = 3). BioA, 7,8-diaminononanoate synthase; BioB, biotin synthase; Bpl, biotin protein ligase; OD600, optical density at wavelength of 600 nm.
The BioC toxicity characterized by bacterial growth inhibition is partially attributable to the depletion of Mal-ACP, a building block for the canonical FAS II pathway (7). Therefore, overproduction of BioC resembles the inactivation of malonyl-CoA-ACP transacylase (FabD) that catalyzes the conversion of malonyl-CoA (Mal-CoA) to Mal-ACP (7, 34). We adopted an arabinose-inducible promoter (ParaC)–driven construct for the SUMO-BioC fusion enzyme, along with its native form (fig. S6A). Consistent with the prior observation with BcBioC by Lin and Cronan (7), basal expression of AbBioC can support the viability of the biotin auxotroph E. coli ΔbioC on nonpermissive condition, while its arabinose-induced overexpression poisons the recipient E. coli (fig. S6, B and D). In contrast, the SUMO-BioC form of A. baumannii displayed its activity in vivo only when expressed at a high level (fig. S6, C and D). Because glucose is a repressor for ParaC promoter, we sought for altered activities of AbBioC in both native and chimeric versions (fig. S7, A and C). Unlike the native AbBioC form that virtually remains active in E. coli on the condition containing 0.2% glucose, regardless of the inducer arabinose addition (fig. S7B), the activity of SUMO-BioC hybrid form is arabinose dose dependent, independently of either 0.2% glucose (fig. S7B) or 0.2% glycerol (fig. S7C) as sole carbon source. The combined data genetically demonstrate the presence of BioC-like activity in certain ESKAPE members (A. baumannii and K. pneumoniae) and offer a means to produce the toxic protein BioC via the SUMO tag–based fusion expression strategy.
Characterization of AbBioC
The BioC enzyme belongs to a class I SAM-dependent O-methyltransferase. The phylogenetic analyses suggested that the class I methyltransferase is composed of diverse members placed into distinct subclades (Fig. 2C). This is largely attributed to the variation in the enzymatic substrate specificity (nucleic acids, small molecules, and lipids). In brief, (i) both TrmD [tRNA (guanosine(37)-N1)-methyltransferase] and Dam (adenine-specific DNA methyltransferase) recognize the substrate of nucleic acids; (ii) COMT (catecholamine O-methyltransferase) is specific to small molecule, and (iii) BioC prefers the malonyl group linked to ACP, a short-chain fatty acid. The unrooted tree illustrated that AbBioC-including clade restricted to Acinetobacter is appreciably closer to the Bacillus cousins (e.g., BcBioC) than EcBioC and KpBioC (Fig. 2D). It was assumed that the solubility of AbBioC is similar to that of the cousin BcBioC. Next, we engineered KpBioC and AbBioC by using the strategy of SUMO fusion expression. The native BioC enzymes were obtained upon the removal the N-terminal 6x His-tagged SUMO subunit (fig. S8). Similar to EcBioC in inclusion body, KpBioC without its SUMO tag steadily precipitates. It was noted that soluble AbBioC (~25 kDa) was liberated from its fusion version (~40 kDa) upon the removal of a SUMO tag (16 kDa) by the ubiquitin-like protease 1 (ULP1) (Fig. 3A and fig. S8). Size exclusion chromatography (SEC) revealed that AbBioC exists as a monomer (Fig. 3B), whose identity was verified with quadrupole orthogonal acceleration–time-of-flight mass spectrometry (fig. S9). The availability of soluble AbBioC enabled its structure-to-function study.
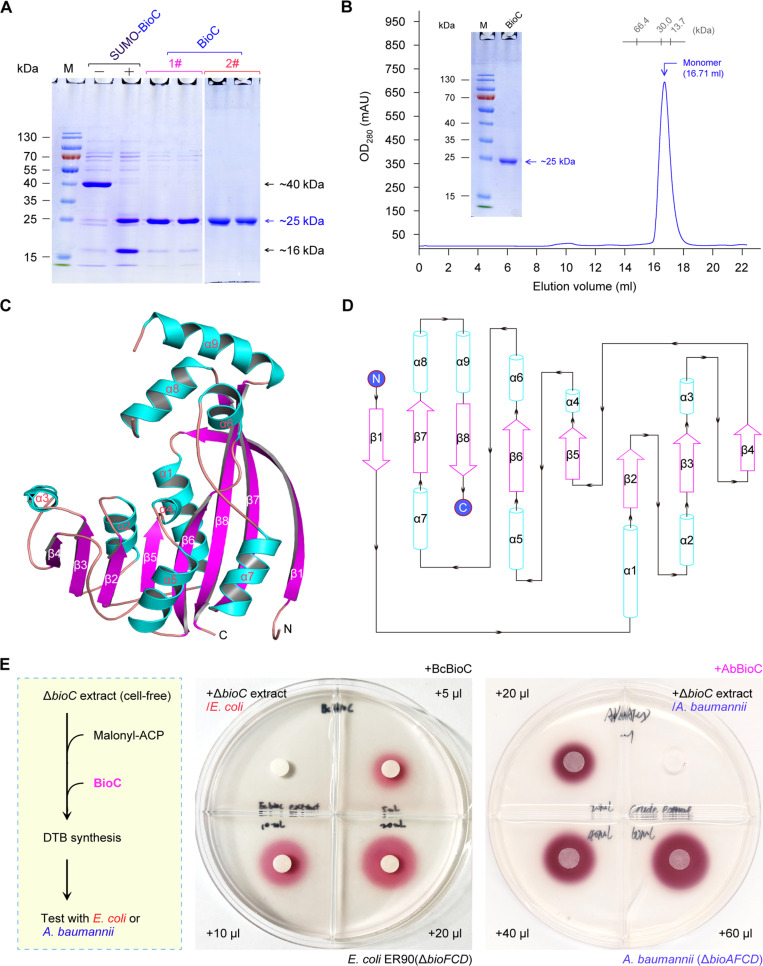
Fig. 3. Structure and function of AbBioC methyltransferase.
(A) Preparation of soluble AbBioC by expressing the N-terminal 6x His-tagged SUMO-fused BioC protein. The AbBioC is ~25 kDa; SUMO is ~16 kDa, and the resultant SUMO-BioC is estimated to be ~40 kDa. The symbols of plus (and/or minus) separately denote the presence (and/or absence) of ULP1 enzyme, which is designed to specifically remove the N-terminal 6x His-tagged SUMO. The numbers (1# and 2#) represent two rounds of nickel column–based protein purification. It was generated by the combination of two individual gels. (B) Gel filtration profile of Acinetobacter BioC protein. SEC of BioC protein was conducted using Superdex 75 column. The purity of the recombinant BioC (~25 kDa) is illustrated in the inside gel of 15% SDS–polyacrylamide gel electrophoresis (SDS-PAGE). (C) Overall architecture of AbBioC. The α helices (α1 to α9) and β strands (β1 to β8) are shown as cylinders and fancy sheets, respectively. (D) Topological characterization of AbBioC. (E) Biotin bioassay demonstrated that BioC protein begins biotin synthesis in vitro. A representative result of both SDS-PAGE [(A) and (B)] and biotin/DTB bioassays (E) was given here (n = 3). A scheme on the left hand of (E) was formulated, illustrating the path of BioC-initiated biotin synthesis reconstituted in vitro. Here, the biotin-free ΔbioC crude extract (20 μl) is the negative control. Two types of BioC enzymes (BcBioC and AbBioC) were examined. As for the BcBioC control, it was assayed with the well-studied reporter strain of E. coli ER90(ΔbioFCD) (6, 63). Notably, the A. baumannii ΔbioAFCD mutant that we developed here was used as an indicator strain for assaying AbBioC function in vitro. N, N terminus; C, C terminus; BcBioC, Bacillus cereus BioC.
To examine the production of Mal-ACP methyl ester (M-Mal-ACP) by AbBioC (fig. S10A), we prepared its cognate substrate Mal-ACP in vitro, via the reaction catalyzed by Sfp (surfactin production) of Bacillus (fig. S10B). The canonical substrate for Sfp (apo-ACP) was given by the cleavage of holo-ACP by acyl carrier protein phosphodiesterase (AcpH) to eliminate the 4′-phosphopanthetheine (Ppan) prosthetic group (fig. S10C). Sfp converted apo-ACP to (i) holo-ACP by addition of CoA (fig. S10D) and (ii) Mal-ACP via ligation of Mal-CoA (fig. S10E). As expected from our AbBioC reaction with Mal-ACP substrate (measured mass versus theoretical value: 8933.789 versus 8933.39; fig. S10E), the final product of M-Mal-ACP was readily given, of which the actual mass is 8948.037 (fig. S10F). Next, we reconstituted the dethiobiotin (DTB) synthesis system in vitro using the biotin-starved ΔbioC crude extract (Fig. 3E). As observed for the reaction catalyzed by BcBioC as the positive control, the mixture of AbBioC catalysis allowed robust growth of the reporter strain ER90(ΔbioF/C/D) on the nonpermissive condition. This was judged by the reduction of the redox indicator 0.01% TTC (2,3,5-triphenyl tetrazolium chloride) in the agar to a bright-red, insoluble formazan deposited that is proportional to the level of DTB intermediate (Fig. 3E). In conclusion, AbBioC is a functional member of Mal-ACP methyltransferases, beginning bacterial biotin synthesis (Figs. 2B and 3E).
Structure of BioC methyltransferase
To elucidate molecular basis for BioC catalysis, we performed x-ray crystallography studies. Of four BioC enzymes, two amenable cousins (BcBioC and AbBioC) were prepared in large scale and subjected to extensive crystallization trials. As a result, we solved one AbBioC/SAM complex structure at 2.54 Å resolution (Table 1). This crystal structure belongs to the P21 space group (Table 1) and contains four protomers per asymmetric unit (fig. S11). Each AbBioC protomer is assembled into two domain-swapped dimers (fig. S11A). However, the SEC profile demonstrated that AbBioC appears monomeric (Fig. 3B). As indicated by the low root mean square deviation values (0.29 to 0.35 Å2), overall α/β-foldings of the four AbBioC protomers are virtually identical (fig. S11, B and C). As described with each AbBioC protomer (Fig. 3, C and D), its architecture is composed of nine α helices (α1 to α9) and eight β strands (β1 to β8). The central stands (β2 to β8) form one flat sheet. Except for β8, the rest of β strands are parallel to each other. It was noted that the domain-swapped β1 (residues 1 to 20) replaces an α helix motif incorrectly predicted by AlphaFold (fig. S11, D to F). Helices α1 to α3 reside on one side of the β sheet, whereas α4, α5, and α7 are located on the opposite side (Fig. 3C). Notably, the remaining helices α6, α8, and α9 sit on this β sheet layer, and their orientations are roughly vertical to each other (Fig. 3C).
Table 1. Crystal data collection and refinement statistics of AbBioC in complex with its SAM cofactor or SIN inhibitor.
RMSDs, root mean square deviations.
| Structures | AbBioC-SAM complex | AbBioC-SIN complex |
|---|---|---|
| PDB ID | 8X8I | 8X8J |
| Data collection* | ||
| Space group | P21 | C2 |
| Cell parameter | ||
| a, b, c (Å) | 71.06, 57.23, 143.67 | 116.70, 56.55, 70.68 |
| α, β, γ (°) | 90.00, 101.72, 90.00 | 90.00, 101.13, 90.00 |
| Wavelength (Å) | 0.9792 | 0.9792 |
| Resolution (Å) | 70.34–2.54 | 57.25–1.72 |
| High-resolution shell (Å) | 2.67–2.54 | 1.81–1.72 |
| Completeness (%) | 99.3(99.9) | 98.9(99.9) |
| Redundancy | 3.2(3.3) | 5.8(5.8) |
| Rmerge (%) | 10.0(64.0) | 10.5(140.2) |
| I/σ(I) | 9.4(2.4) | 8.4(1.9) |
| Refinement | ||
| Resolution (Å) | 70.34–2.54 | 40.48–1.72 |
| No. of reflections | 37621 | 47247 |
| Rwork (%)/Rfree (%) | 23.61/28.75 | 22.02/25.03 |
| No. of atoms | ||
| Protein | 7478 | 3662 |
| SAM/SIN | 4 | 2 |
| Water | 30 | 94 |
| RMSDs | ||
| Bond length (Å) | 0.003 | 0.005 |
| Bond angle (°) | 0.587 | 0.733 |
| Ramachandran plot (%) | ||
| Most favorable | 97.71 | 98.69 |
| Additional allowed | 2.29 | 1.31 |
| Outlier | 0.00 | 0.00 |
*Values in parentheses are for the high-resolution shell.
Compared to the two putative methyltransferases [A. tumefaciens Tam (AtTam) and Anabaena variabilis Ava_0823], AbBioC gave the limited identity (19.07% for AtTam and 18.34% for Ava_0823; fig. S12A). The DALI search revealed that the architecture of AbBioC is most similar to that of AtTam [Protein Data Bank (PDB): 2P35] and Ava_0823 (PDB: 3CCF) (35). The N-terminal orientation differed markedly, albeit the conservation in their core structures (fig. S12, B and C). In our AbBioC structure (PDB: 8X8I), the SAM cofactor is occupied into a small cavity (figs. S11 to S13). Instead of SAM, one S-adenosyl-l-homocysteine (SAH) molecule is present in the AtTam structure (PDB: 2P35; fig. S12D). Probably, lacking the methyl group leads to different orientations of the amino motifs. In contrast, the conformations of the sugar puckers and the nucleobases are quite similar. The three SAM-interacting residues are extremely conserved and adopt similar conformations in the AbBioC/SAM complex and the AtTam/SAH structure (fig. S12D). To the best of our knowledge, this provides a previously unidentified glimpse of BioC architecture in the past 50 years (36, 37).
BioC binding of SAM cofactor
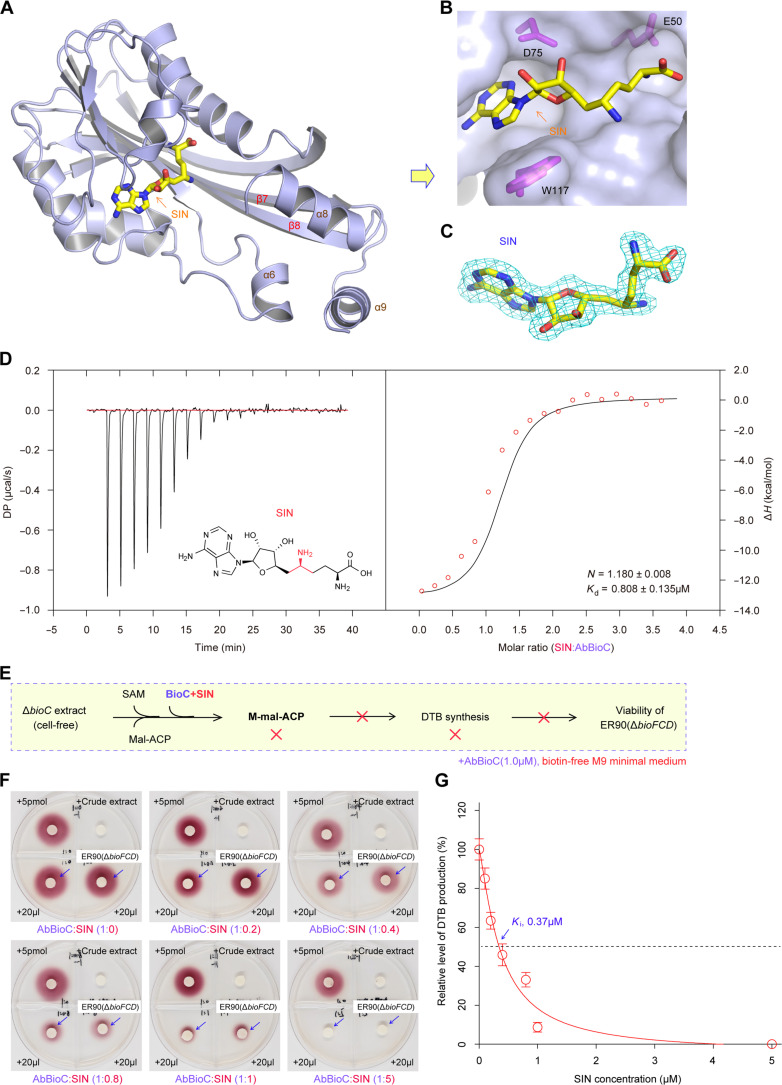
Compared to Ava_0823 (PDB: 3CCF) and AtTam/SAH (PDB: 2P35) structures, our SAM-liganded AbBioC structure offered more direct evidence for SAM as a cofactor to donate methyl group (figs. S12D and S13, A and B). Accordingly, each AbBioC protomer contains a SAM molecule (Fig. 4A and fig. S13A). As supported by the 2Fo-Fc electron density maps, the SAM molecule is well defined and adopts one extended conformation (Fig. 4B). The sugar pucker of SAM forms two hydrogen bond (H-bond) interactions with the side chain of aspartate-75 (D75), via its two hydroxyl groups (2.4 Å for 2′-OH and 3.0 Å for 3′-OH; fig. S13C). The nucleobase of SAM lies in a narrow cavity, flanked by leucine-76 (L76) on one side and tryptophan-117 (W117) on the other side. The N6 atom of the nucleobase interacts with the side-chain O atom of aspartate-96 (D96; fig. S13C). The amino motif of SAM inserts into AbBioC via a deep pocket (Fig. 4A and fig. S13C). Apart from its direct H-bond interaction with glycine-52 (G52), the amino motif also forms an extensive water-mediated H-bonding network across glutamic acid–50 (E50), cysteine-53 (C53), and leucine-58 (L58; fig. S13D). Both G52 and C53 locate at the “β2-α2” linker that connects β2 strand and α2 helix. The E50 of β2 strand and threonine-59 (T59) of α2 helix forms one stable H-bond interaction, favoring the formation of “β2-α2” linker (fig. S13D).
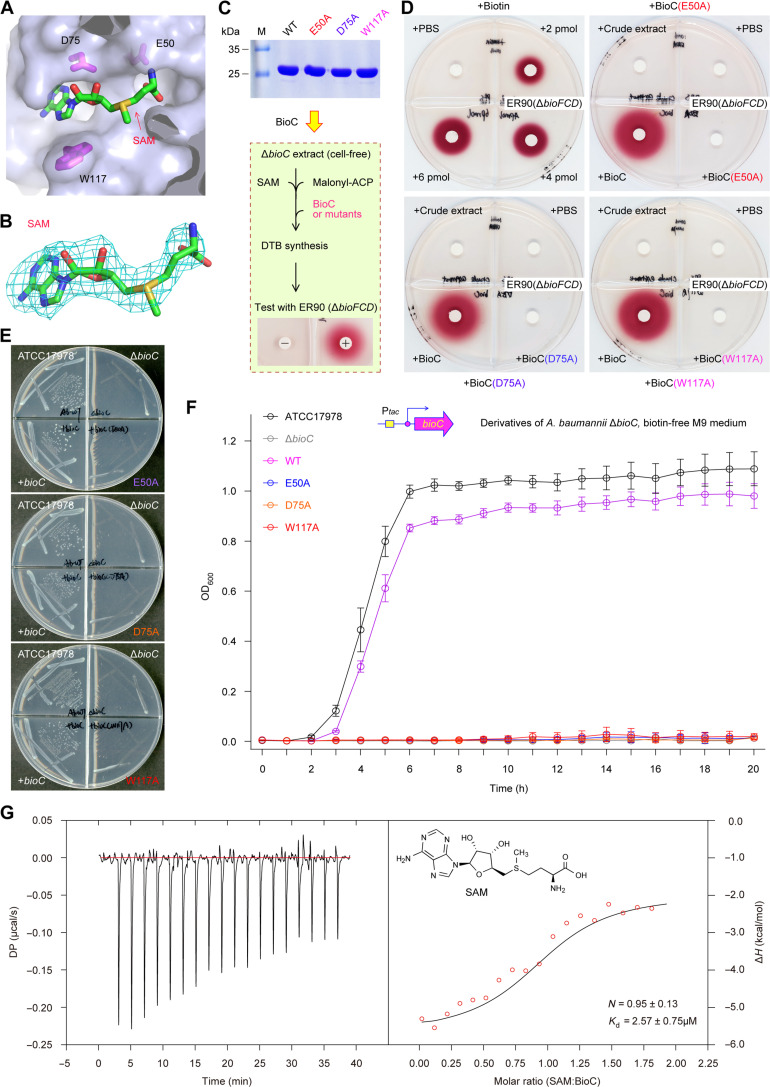
Fig. 4. Structural and functional definition of the SAM substrate recognition by AbBioC.
(A) The SAM-binding cavity of AbBioC enzyme. SAM molecule and AbBioC are shown as sticks and surface, respectively. Namely, three critical residues for SAM binding (indicated with magenta sticks) included E50, D75, and W117. (B) Fo-Fc omit the electron density map of SAM molecule. It was contoured at 2.5 sigma level. (C) SDS-PAGE (15%) profile for BioC mutants (E50A, D75A, and W117A) and BioC-based scheme for the system of biotin synthesis reconstituted in vitro. (D) Biotin/DTB bioassay revealed that the three SAM-binding residues are critical for enzymatic activity of BioC in the initiation of biotin synthesis. As described in Fig. 3E, the biotin/DTB synthesis system was reconstituted in vitro. The WT BioC added on paper discs acted as the positive control. Namely, the three AbBioC mutants with the defection in SAM binding here denote E50A, D75A, and W117A. (E) Structure-guided, site-directed mutagenesis suggested that three SAM-binding sites are essential for AbBioC activity. (F) The analyses of growth curves showed that none of the three single AbBioC mutants (E50A, D75A, and W117A) retains enzymatic activity. Growth curves (F) were produced, in which each data point was given as an average ± SD (n = 3). The ΔbioC strain of A. baumannii that is biotin auxotrophic acted as a recipient for plasmid-borne AbBioC mutants. (G) ITC analysis for stoichiometry of AbBioC binding to SAM cofactor. As for each of the bellowed experiments ranging from biotin bioassays (D), bacterial viabilities (E), to ITC assays (G), a representative result was shown here (n = 3). Of note, the values of both N and Kd were expressed as means ± SD (n = 3). N, stoichiometry; DP, differential power; ΔH, enthalpy.
Next, we applied structure-guided, site-directed mutagenesis to investigate the importance of SAM recognition in AbBioC function. In total, three single mutants of AtBioC (i.e., E50A, D75A, and W117A) were constructed and purified to homogeneity (Fig. 4C). Similar to the wild-type (WT) AbBioC protein, we tested whether these mutants remain active in vitro (Fig. 4D). In our DTB/biotin bioassay, the reporter strain ER90 displayed obvious growth that is evidenced by the deposition of insoluble red pigment in a biotin dose–dependent manner (Fig. 4D). Unlike the situation arising from both the blank PBS control and the negative control (A. baumannii ΔbioC crude extract), the WT AbBioC enzyme enabled the in vitro reconstitution of crude extract–based DTB/biotin synthesis (Fig. 4D), whereas none of the three AbBioC mutants can do this job in vitro (Fig. 4D). The similar scenarios were seen in the three plasmid-borne bioC mutants in the context of bacterial viability in vivo (Fig. 4, E and F). Moreover, our isothermal titration calorimetry (ITC) experiment determined that a SAM cofactor displays appreciably tight binding to AbBioC enzyme with the stoichiometry of N = 0.95 ± 0.13 and dissociation constant (Kd) = 2.57 ± 0.75 μM (Fig. 4G). Collectively, these data pinpointed that SAM ligand recognition by AbBioC plays an important role in the initiation of biotin synthesis.
Molecular basis for inhibition of BioC by SIN
The natural product SIN is long recognized as a potent inhibitor against an arsenal of SAM-dependent methyltransferases (38). Lin and Cronan (7) reported that SIN can efficiently inactivate BcBioC enzyme in vitro; however, the structural mechanism remained unexplored. To close this gap, we also dissected the AbBioC/SIN complex structure (Fig. 5, A to C, and fig. S13, E to H). This crystal belongs to the C2 space group and contains two AbBioC/SIN complexes per asymmetric unit (PDB: 8X8J; Table 1). Structural comparison revealed that SIN is bound at the SAM-binding pocket of AbBioC (Fig. 5, A to C, and fig. S13, E to H). Similar to that of SAM (fig. S13, C and D), the nucleobase of SIN is sandwiched by L76 and W117 and forms one H-bond interaction with D96 (Fig. 5, A and B, and fig. S13E). The sugar pucker of SIN forms two H-bonds with the side chain of D75 (fig. S13E). However, the conformation of the amino motif of SIN (Fig. 5, A and B, and fig. S13, G and H) is different from that of SAM (Fig. 4, A and B, and fig. S13, A to D). As depicted in fig. S13F, unlike the nitrogen atom at the epsilon position (NE) of SIN forming only one H-bond with the main chain oxygen atom of S113, the N atom of SIN forms two direct H-bonds. The conformation of SIN amino motif is further stabilized by water-mediated H-bond interactions (fig. S13F). The ITC analyses determined that the SIN inhibitor can competitively bind to AbBioC enzyme (N = 1.180 ± 0.008 and Kd = 0.808 ± 0.135 μM; Fig. 5D). Compared with the physiological ligand SAM, the binding affinity of SIN is around threefold tighter (Fig. 4G). Subsequent DTB/biotin bioassay confirmed that SIN efficiently interferes with the ability of AbBioC in initiating biotin synthesis (Fig. 5, E and F). Consistent with its potent binding (Fig. 5D), the dose-dependent activity of SIN against AbBioC methyltransferase was featured by its inhibition constant (Ki) value of ~0.37 μM (Fig. 5G). In summary, the structural and biochemical characterization of AbBioC/SIN interplay provided insights into an inhibitory mechanism for BioC methyltransferase.
Fig. 5. Structural and functional evidence that the SIN inhibitor inactivates AbBioC activity.
(A) Overall structure of AbBioC complexed with SIN inhibitor. (B) The SIN inhibitor–binding cavity displayed on the surface of the AbBioC enzyme. The SIN molecule and AbBioC are shown as sticks and surface, respectively. Namely, three critical residues for SIN binding (indicated with magenta sticks) include E50, D75, and W117. (C) 2Fo-Fc electron density map of SIN molecule, contoured at 1.0 sigma level. (D) Use of ITC assay to probe binding stoichiometry between AbBioC and its SIN inhibitor. A representative ITC profile was given here (n = 3), and the N (and/or Kd) value was calculated as mean ± SD (n = 3). The ITC experiments with a SIN inhibitor were performed as described for SAM cofactor in Fig. 4G. The SIN molecule was shown inside the left-hand graph of (D). In comparison with the stoichiometry of SAM binding (N = 0.95 ± 0.13; Kd = 2.57 ± 0.75 μM; Fig. 4G), SIN affinity with AbBioC is appreciably stronger because of the threefold lower value of Kd (0.808 ± 0.135 μM). (E) Scheme for an impairment of the BioC-BioH pathway by the SIN inhibitor. The cross symbol denoted an inactivated step/pathway. (F) DTB/biotin bioassay revealed that the SIN inhibitor effectively kills AbBioC in the in vitro reconstituted system of DTB/biotin synthesis. Of three independent bioassays for SIN-stressed AbBioC, a representative photograph was shown here. (G) Determination of an inhibition constant Ki assigned to the SIN inhibitor on the basis of cell viability. To illustrate the Ki curve for the SIN inhibitor, three independent experiments were performed. The resultant output was given via nonlinear regression fitting curve (provided by GraphPad Prism), of which each point was expressed as mean ± SD (n = 3). Ki value was measured to be ~0.37 μM.
Importance of β1 strand and R60 in BioC action
The β1 strand (residues 8 to 20) is unusual in that it enables AbBioC protein to form a domain-swapped dimer in the crystal structure (fig. S11A). It contained a conserved tyrosine-19 (Y19) residue (fig. S3), positioned at the C-end (fig. S14, A and B). Apart from the interaction between the two β1 strands, each β1 strand forms an extensive H-bond network with the β7 strand from the partner molecule. The β1 strand is distant from the SAM-binding site (fig. S14B), whereas two positively charged residues [lysine-27 (K27) and arginine-60 (R60)] are quite close to the binding pocket of the amino motif of SAM (fig. S14, B and C). The side chain of K27 points toward the outer surface of AbBioC, but the side chain of R60 folds back and gives H-bond interactions with two residues [serine-55 (S55) and glycine-56 (G56); fig. S14C]. To clarify their functional roles, we generated three single mutants (Y19A, K27A, and R60A), as well as the β1-deleted mutant, designated bioC(ΔN, 1-20). As expected from genetic complementation, functional loss of AbBioC was consistently observed upon either the Y19A substitution or β1 deletion (fig. S14, D and E). This largely agreed with the BcBioC(Y19F) mutant described by Lin and Cronan (7). Although K27 had no detective role, the R60A mutant of AbBioC failed to restore the growth of the biotin auxotroph, A. baumannii ΔbioC, on the nonpermissive condition (fig. S14, F and G). Similar to the E50 residue (fig. S13D), R60 also interacts with residues from the “β2-α2” linker. The essentiality of both E50 and R60 indicated that proper folding of the “β2-α2” linker is critical for the ligand SAM binding and action of AbBioC. To further ascertain the biochemical roles of β1 strand, we performed DTB/biotin bioassays. Unlike the WT AbBioC that initiates biotin synthesis in vitro, none of the BioC(Y19A), BioC(ΔN, 1-20), and BioC(R60A) mutants retained enzymatic activity in our trials (fig. S14H). Together, we favored that the β1-mediated conformational maintenance and the R60 residue–originated H-bond network both are implicated in AbBioC catalysis.
BioC recognition of primer Mal-ACP
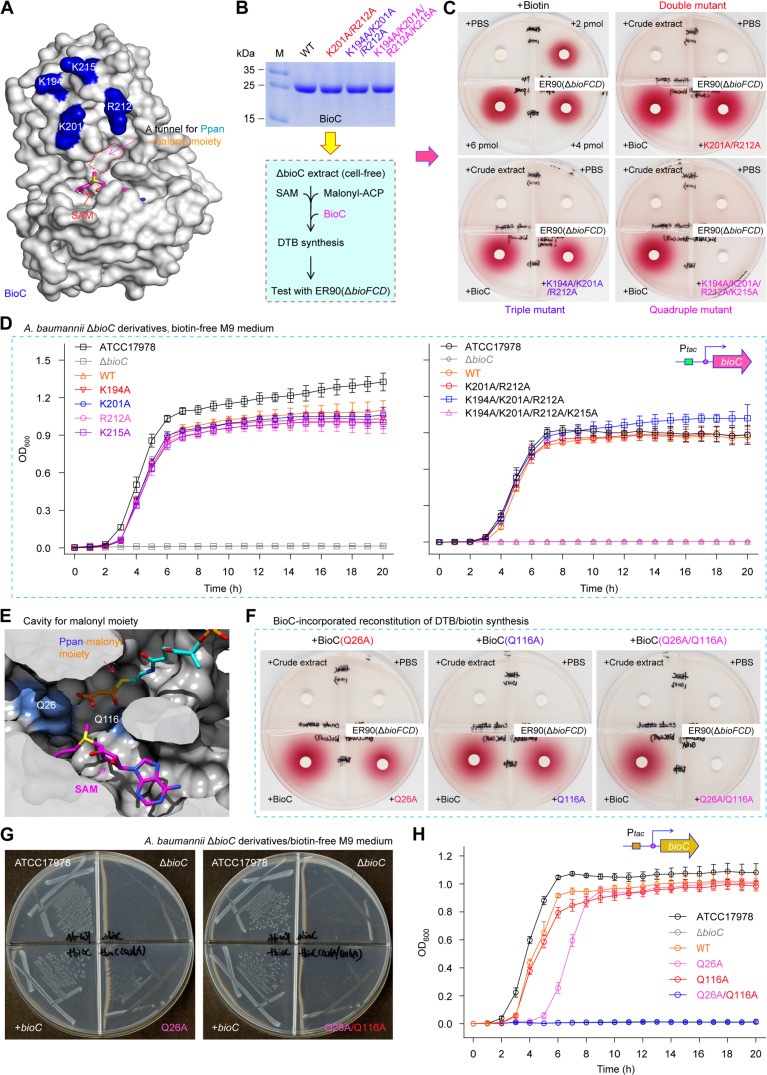
Our continued efforts were unsuccessful in obtaining complex structure of AbBioC with its primer substrate Mal-ACP. This is, in part, if not all, due to its transient interaction with the primer. In addition, no substrate-bound complex structures were reported for the two distantly related methyltransferase (AtTam and Ava_0823). To investigate the dynamic binding, we built one AbBioC/Mal-ACP complex model using the autodocking program (fig. S15). Presumably, the malonyl moiety adopts one extended conformation, placing the carboxyl group next to the SAM for methylation (fig. S15A). The ACP carrier for malonyl moiety sits near the α8 and α9 helices (fig. S15A), and the overall shape of ACP and AbBioC are roughly complementary with each other (fig. S15B). Because that ACP is a negatively charged protein rich in certain acidic amino acids (i.e., Asp and Glu), it was postulated to interact with BioC through extensive electrostatic interactions. Surface structural analyses revealed that (i) the α8 and α9 helices of AbBioC contain three lysine residues (i.e., K194, K201, and K215), and (ii) the α8-α9 connecting linker has one arginine residue, R212. Moreover, all the four positively charged residues are relatively close in space (Fig. 6A) and located on the interfaces between AbBioC and the docked ACP molecule (fig. S15, A and B). To confirm functional roles of the K/R residues, an arsenal of mutants were assayed in vitro and in vivo (Fig. 6, A to D, and fig. S16). As expected from bacterial viability tests, (i) none of the four single AbBioC mutants (K194A, K201A, R212A, and K215A) loses the activity of binding cognate ACP carrier, and (ii) both the double mutant (K201A/R212A) and the triple mutant (K194A/K201A/R212A) are functionally indistinguishable from its parental version (Fig. 6D and fig. S16). In contrast, the quadruple mutation (K194A/K201A/R212A/K215A) impaired the function of BioC in restoring the biotin auxotrophic ΔbioC strains of E. coli and A. baumannii (Fig. 6D). We then prepared the mutated AbBioC proteins for enzymatic analysis (Fig. 6B). Consistent with the observation for genetic complementation (Fig. 6D and fig. S16), our DTB/biotin bioassay showed that only the quadruple derivative of AbBioC(K194A/K201A/R212A/K215A) is nonfunctional (Fig. 6C). Similar to those of BioH [and/or BioJ (39)] demethylase with the cognate ACP moiety (6, 14), the four positively charged AbBioC residues are assumed to play a synergistic role in recognition of the canonical ACP carrier.
Fig. 6. Probing interplay between AbBioC and its physiological substrate Mal-ACP.
(A) Surface structure of AbBioC enabled the visualization of four positively charged residues (K194, K201, R212, and K215). It was generated from structural presentation via the rotation by 90° counterclockwise (fig. S15B). (B) BioC-based scheme for the reconstitution of DTB/biotin synthesis system in vitro. (C) Biotin/DTB bioassay suggested that the quadruple mutant (K194A/K201A/R212A/K215A) of AbBioC losses its ability to trigger the DTB/biotin synthesis. (D) Combined site-directed mutagenesis and growth curves allowed us to determine functional loss of the quadruple mutant BioC (K194A/K201A/R212A/K215A). (E) Structural analysis of the malonyl moiety–loading tunnel within AbBioC methyltransferase. The phosphopantetheine (Ppan)–malonyl group arises from the cognate substrate Mal-ACP thioester. It seemed likely that the malonyl-recognizable channel converges with the SAM cofactor. The SAM cofactor and Ppan-malonyl moiety appear as sticks. The carbon atoms of SAM, PPan, and malonyl are shown with magenta, cyan, and orange, respectively. The sectioned view of AbBioC is displayed as surface. The other atoms are shown with red, blue, and yellow for oxygen, nitrogen, and sulfur atoms, respectively. Green dashed lines denote H-bonds. Two putative neutralizing residues of AbBioC are colored light blue, namely, Q26 and Q116. AbBioC is shown as cartoon colored gray. (F) The double mutations of Q26 and Q116A inactivated the role of AbBioC in the system of biotin/DTB synthesis in vitro. (G and H) In vivo evidence that relative to the residue Q116, Q26 plays a major role in neutralizing the malonyl group of substrate Mal-ACP. As for both biotin bioassays [(C) and (F)] and bacterial viabilities (G), a representative result was presented (n = 3). Growth curves were generated on the basis of three independent measurements [(D) and (H)], and each data point was expressed as mean ± SD (n = 3).
A malonyl moiety-loading tunnel was mapped to converge with the SAM cofactor–occupied cavity. Presumably, two glutamines (Q26 and Q116) localized in the cavity bottom appear to neutralize the free carboxyl group of Mal-ACP substrate (Fig. 6E and fig. S17A). The neutralization pattern might represent a common mechanism for FAS-relevant enzymes. This is because similar scenarios have been observed for the Mal-CoA:ACP transacylase FabD (40, 41), the pimeloyl-CoA synthetase BioW (8, 9), and an atypical biotin synthesis enzyme BioZ (11). To ascertain this postulate, we generated three AbBioC derivatives that include two single mutants (Q26A and Q116A) and a double mutant (Q26A/Q116A). Among them, only the double-mutant AbBioC (Q26A/Q116A) enzyme (fig. S17B) was disabled to reconstitute the DTB/biotin synthesis in vitro (Fig. 6F). This was further supported by the fact that only the single mutant (i.e., Q26A and Q116A) is permitted to render robust viability of the biotin auxotrophic ΔbioC strain of A. baumannii (and/or E. coli) on the biotin-free M9 medium (Fig. 6, G and H, and fig. S18, A and B). Unlike the two known channels that rely on the basic amino acid, arginine [R117 for FabD (40, 41) and R159/R201 for BioW (8)], the AbBioC counterpart, exploits two polar residues Q26/Q116 to neutralize the free carboxyl group of malonyl substrate (fig. S17A). Therefore, the combined data in vitro and in vivo illustrated the landscape of AbBioC interplay with its Mal-ACP substrate.
The involvement of BioC in mobile colistin resistance
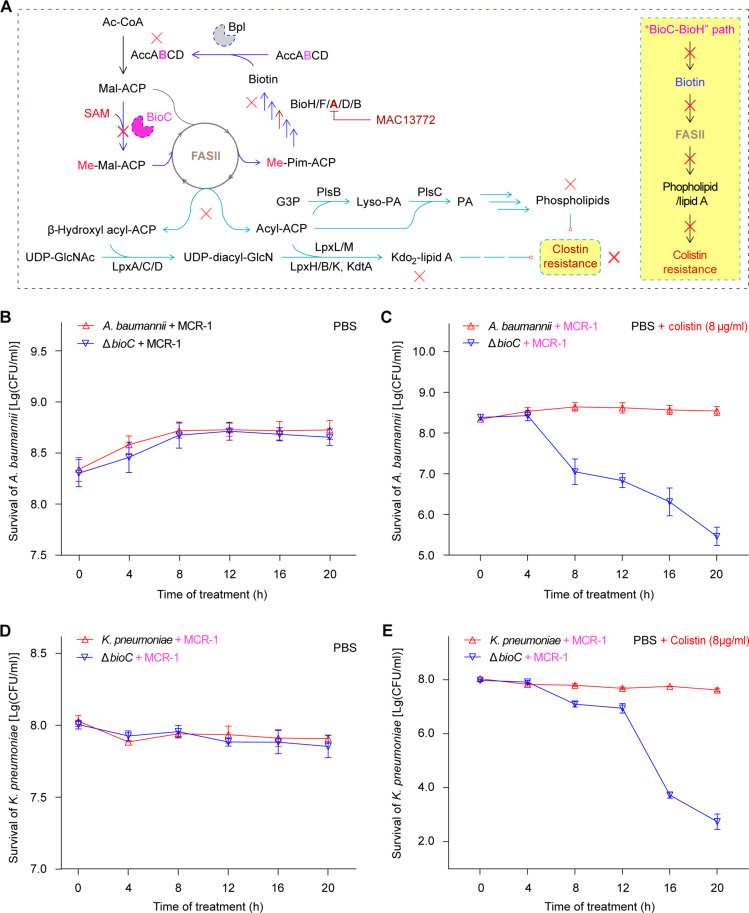
Not only does biotin synthesis rely on FAS II pathway (5), but it also provides biotin ligand for the biotinylated acetyl-CoA carboxylase dedicated to a first-committed step of FAS II cycle (Fig. 7A). Perturbation of FAS II route by certain inhibitors (e.g., cerulenin for FabB/F and triclosan for FabI) abolished phospholipid/lipid A synthesis via restricting the pool of free fatty acids (Fig. 7A). The depletion of lipid A and phosphatidylethanolamine (PE), two substrates for mobile colistin resistance (MCR) modifying enzyme, explained, in part (if not all), the resensitivity of mcr-1–positive ESKAPE pathogens to polymyxin, a last-resort defense antibiotic (Fig. 7A) (27). We therefore posited that the BioC primary step of biotin synthesis is implicated in colistin-induced lysis resistance provided by MCR-catalyzed lipid A modification in certain clinically relevant ESKAPE pathogens (Fig. 7).
Fig. 7. A role of BioC methyltransferase in Acinetobacter MCR-1 colistin resistance.
(A) Scheme for BioC-BioH pathway of biotin synthesis that connects FAS II–centering phospholipid/lipid A synthesis with the phenotypic colistin resistance route. It seemed likely that targeting biotin synthesis can bypass bacterial colistin resistance. (B) The bioC deletion fails to markedly alter bacterial survival of mcr-1–containing A. baumannii in PBS buffer. (C) Removal of bioC can resensitize MCR-1–producing, polymyxin-resistant A. baumannii to colistin at a level of up to 8 μg/ml. (D) The viability of K. pneumoniae ΔbioC mutant is indistinguishable from its parental strain in PBS buffer. (E) Expression of MCR-1 fails to render the K. pneumoniae ΔbioC mutant insusceptible to colistin. The colistin killing curves [(B) and (E)] were plotted with the Excel software on the basis of three independent trials, and the value of each data point was displayed as mean ± SD (n = 3). Ac-CoA, acetyl-CoA; Acc, acetyl-CoA carboxylase; AccABCD, four subunits of the Acc enzyme complex (AccA, α subunit of carboxyltransferase; AccB, BCCP; AccC, a biotin carboxylase; and AccD, β subunit of carboxyltransferase); ACP, acyl carrier protein; Mal-ACP, malonyl-ACP; Me-M-ACP, monomethyl Mal-ACP; Me-Pim-ACP, monomethyl pimeloyl-ACP; BioC, Mal-ACP methyltransferase; BioH, methyl-pimeloyl ACP ester demethylase; BioF, 7-keto-8-aminopelargonic acid synthase, the pyridoxal 5′-phosphate (PLP)–dependent enzyme (AON synthase); BioA, 7,8-diaminononanoate synthase, the PLP-dependent transaminase; FAS II, type II fatty acid synthase system; G3P, glycerol-3-phosphate; PlsB, G3P acyltransferase; PA, phosphatidic acid; Lyso-PA, lysophosphatidic acid; PlsC, Lyso-PA acyltransferase; UDP-GlcNAc, uridine diphosphate N-acetylglucosamine; UDP-diacyl-GlcN, uridine diphosphate glucosamine; Kdo2-lipid A, 3-deoxy-d-manno-octulosonic acid-lipid A; LpxA, the first acyltransferase of the Raetz pathway for lipid A synthesis; LpxC, UDP-3-O-(R-3-hydroxyacyl)-N-acetylglucosamine deacetylase; LpxD, acyl-ACP–dependent N-acyltransferase; LpxL, lauroyltransferase; LpxM, myristoyltransferase; LpxH, UDP-2,3-diacylglucosamine hydrolase; LpxB, a membrane-associated glycosyltransferase; LpxK, tetraacyldisaccharide 4′-kinase; KdtA, Kdo transferase of Raetz pathway.
To examine this hypothesis, we analyzed bacterial lysis as recommended by Carfrae et al. (27). Similar to the ΔbioC mutant of A. baumannii ATCC 17978 devoid of an early step of biotin synthesis (Fig. 2B and fig. S4), the strain ATCC 43816 of classical hypervirulent K. pneumoniae (hvKP) was also subjected to CRISPR-Cas9 manipulation, giving its ΔbioC derivative. All the four strains were engineered to express mcr-1 carried by the corresponding recombinant plasmids. As expected, the subinhibitory levels of neither MAC13772 inhibitor (4 μg/ml) nor colistin (8 μg/ml) lysed bacterial cells (fig. S19). In contrast, the combination of MAC13772 with colistin was synergistically active against mcr-1–expressing A. baumannii (fig. S19). Consistent with the observation for E. coli by Carfrae and coworkers (27), this largely verified our A. baumannii–based lysis assays. Similar to those of the negative control, A. baumannii (Fig. 7B), the treatment of PBS buffer had no impacts on the viability of K. pneumoniae and its ΔbioC mutant cells, regardless of MCR-1 (Fig. 7D). The removal of bioC elicited the vulnerability of mcr-1–expressing A. baumannii cells to bactericidal colistin even at the subinhibitory level of 8 μg/ml (Fig. 7C). As for mcr-1–harboring K. pneumoniae cells (Fig. 7E), CRISPR knockout of bioC phenocopied the inactivation of E. coli BioA (27) and mimicked the phenotypes observed with synergistic perturbation of MAC13772/colistin on E. coli (27) and A. baumannii (fig. S19). In sum, we concluded that (i) BioC-initiating biotin synthesis is a metabolic prerequisite for the formation of MCR-mediated polymyxin resistance and (ii) biotin synthesis inhibitors synergize colistin against infections with mcr-1–expressing ESKAPE agents.
Targeting biotin synthesis to bypass colistin resistance
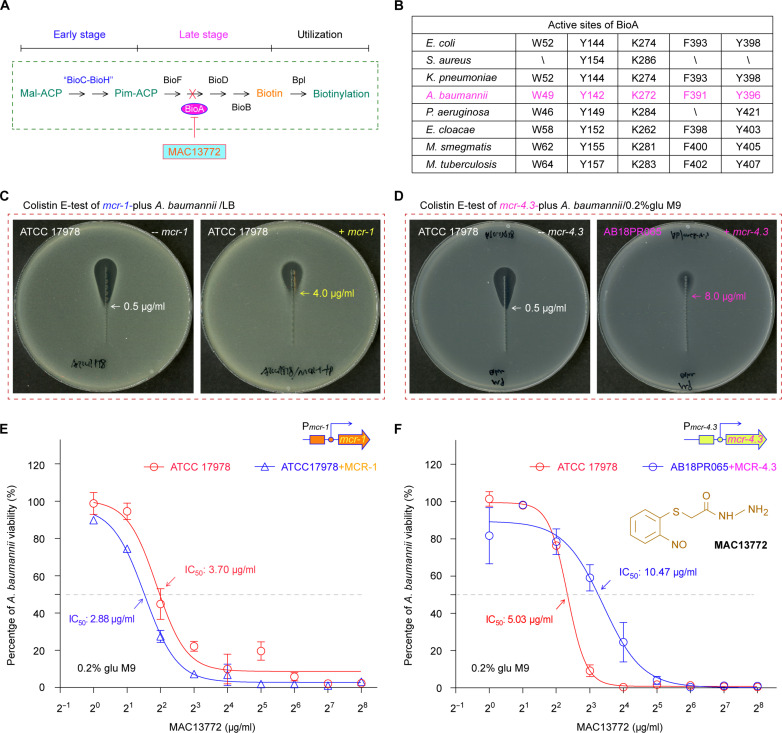
The acquisition of mcr-1 determinant facilitated A. baumannii to become a top priority ESKAPE pathogen causing untreatable nosocomial infections (42). This was because (i) MCR-1, the mcr-1 gene product, acts as a member of the lipid A–phosphoethanolamine (PEA) transferase family and modifies lipid A moiety linked to lipopolysaccharide (LPS) on a bacterial surface (43) and (ii) the production of MCR-1 in clinical isolates compromises the renewed interest of the last-resort antibiotic colistin in a health care setting (44, 45). Consistent with mycobacterial biotin synthesis as a validated druggable pathway (21, 22), certain ESKAPE members can be eliminated (23), following the treatment with MAC13772, the BioA inhibitor (Fig. 8, A and B) (46). It is well known that the MAC13772 inhibitor covalently binds pyridoxal-5′-phosphate (PLP) and gives an adduct of SAM cofactor/MAC13772 (fig. S20), tightly fixed at the catalytic center via an extensive H-bond network (K274 and Y398) along with hydrophobic interaction (W52, Y144, and F393; Fig. 8B) (23). However, it is an unanswered question of whether interfering biotin synthesis remains active in combating the recalcitrant mcr-harboring A. baumannii with colistin resistance.
Fig. 8. Acinetobacter biotin synthesis is a druggable pathway.
(A) Schematic diagram for biotin synthesis blocked by MAC13771 inhibitor. (B) Analyses for five active sites in BioA dedicated to the late stage of biotin synthesis. Top: A scheme for BioA having a role in the late step of biotin synthesis. Bottom: Conservation analyses of BioA active sites among a panel of ESKAPE-including bacteria. E-test measurement reveals that expression of plasmid-borne mcr-1 (C) and/or mcr-4.3 (D) enable the A. baumannii strain to be insusceptible to the last-resort antibiotic colistin. A representative photograph for colistin E-test (n = 3) was given here (C and D). (E) Measurement of MAC113772 half-maximum inhibitory concentration (IC50) for A. baumannii ATCC 17978, regardless of mcr-1. (F) Comparable level of MAC113772 IC50 for the A. baumannii strain with/without mcr-4.3. To determine the IC50 value of MAC13772, the BioA inhibitor, three independent evaluations were conducted [(E) and (F)]. The nonlinear regression curve fit (provided by GraphPad Prism) was applied, and data points were expressed as means ± SD (n = 3). Unlike the engineered strain of A. baumannii ATCC 17978 that harbors a plasmid-borne mcr-1 [(C) and (E)], AB18PR065 is a clinical isolate of A. baumannii bearing mcr-4.3, an additional MCR subtype [(D) and (F)]. Pim-ACP, pimeloyl-ACP; mcr-1, a prototype of mobile colistin resistance determinant; mcr-4.3, the fourth new subtype of MCR resistance elements.
We then constructed a colistin-resistant A. baumannii strain that harbors a plasmid-borne mcr-1 (47, 48), in addition to the swine isolate of A. baumannii AB18PR065 containing mcr-4.3 (49), a distinct subtype of the MCR family (50, 51). As expected from our colistin E-test, an introduction of mcr-1 into the ATCC 17978 strain of A. baumannii led to an increment of colistin minimum inhibitory concentration (MIC) from 0.5 to 4.0 μg/ml (Fig. 8C). The mcr-4.3–carrying strain of A. baumannii AB18PR065 displayed a colistin MIC of 8.0 μg/ml, twofold higher than that of the mcr-1–bearing strain of A. baumannii (Fig. 8D). The analyses of growth curves revealed that the MAC13772 inhibitor is functional in interfering viability of the reference strain of A. baumannii ATCC 17978 with or without mcr-1 (fig. S21, A and B). The half-maximum inhibitory concentration (IC50) of MAC13772 was determined at the almost identical level for the strain ATCC 17978, regardless of mcr-1 (Fig. 8E). A similar scenario was observed for the mcr-4.3–positive strain of A. baumannii (fig. S21, C and D), of which the MAC13772 IC50 is labeled with 10.47 μg/ml, only around twofold higher than that of the colistin-susceptible control strain (Fig. 8F). The results demonstrated that the MAC13772 inhibitor is active in dampening the survival of A. baumannii albeit of MCR-1/4 colistin resistance.
In addition, we extended an assay of MAC13772 inhibition to the colistin-resistant, virulent K. pneumoniae, a critically prioritized ESKAPE member. Colistin E-test showed that the strain ATCC 43816 of K. pneumoniae exhibits colistin MIC of 32 μg/ml upon MCR-1 expression, 64-fold higher than that of the recipient strain alone (fig. S22A). The kind of insusceptibility to colistin was also observed for the strain ZZW20 of wastewater origin (52), an ST11 K. pneumoniae isolate producing MCR-8, a distinct variant of the MCR family (53). As a result, the mcr-1–bearing K. pneumoniae gave the MAC13772 IC50 of 59.96 μg/ml, at a comparable level to that of ATCC 43816 strain alone (i.e., 78.39 μg/ml). Meanwhile, MAC13772 IC50 of 156 μg/ml was assigned to the strain ZZW20 having MCR-8 colistin resistance (fig. S22B). Consistent with the observation for K. pneumoniae by Carfrae et al. (23), approximately 20- to 30-fold elevation of MAC13772 IC50 was noted for K. pneumoniae relative to A. baumannii (fig. S22B). We favored that the limited efficacy of MAC13772 inhibition is mainly due to the poor membrane permeability of K. pneumoniae caused by its capsular hypermucoviscosity (54).
A role of BioC in K. pneumoniae hypervirulence
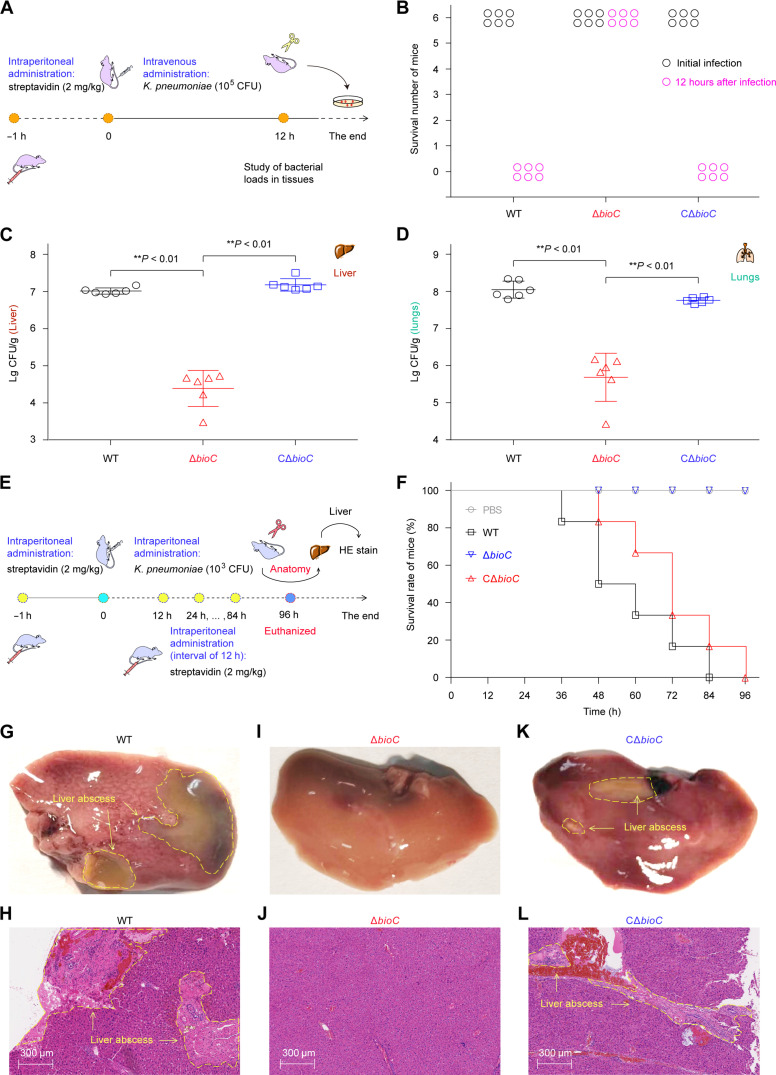
The majority of canonical hvKP isolates that are positive in string tests accounts for certain deadly community-acquired infections featuring pyogenic liver abscess (55–58). The phenotypic hypervirulence is largely due to the acquisition of some pLVPK-like virulence plasmids (59–61), which generally contain, but is not limited to, the rmpA/A2 regulatory gene controlling mucoid phenotype A (61, 62). Of the two dominant K1/K2 capsule types, the ATCC 43816 strain is a classical K2-type hvKP strain (55–58). Because the M-pim-ACP demethylase BioH/BioJ is implicated in bacterial infection (24, 63), we asked the question of whether its pairing enzyme, BioC methyltransferase, has a similar role (Fig. 9). To test this postulate, we evaluated the effects of bioC inactivation on the hvKP strain ATCC 43816 by using CD-1 mice models, namely, (i) intravenous administration (Fig. 9A) and (ii) intraperitoneal infection (Fig. 9E). Given that an average plasma biotin level of ~13.5 ng/ml (~55.4 nM) in CD-1 mice is around 5.5-fold higher than that (2.4 ng/ml, ~9.8 nM) of human plasma (fig. S23) (24), we leveraged intraperitoneal preadministration with streptavidin (2 mg/kg) at 1 hour before the mouse challenge with K. pneumoniae, resembling human plasma environment (23). First, we performed intravenous challenge of CD-1 mice with 1 × 105 CFU of K. pneumoniae and categorized them into three groups (six mice each) that were separately inoculated with WT, ΔbioC, and CΔbioC. Unlike that the entire WT-inoculating group was dead at 12 hours after infections; all the six challenged mice invariantly survived upon the removal of bioC (Fig. 9B). In contrast, the reintroduction of plasmid-borne bioC restored full virulence of the recipient ΔbioC (Fig. 9B). Next, we sought to evaluate varied levels of bacterial loads in four organs consisting of the liver (Fig. 9C), lungs (Fig. 9D), spleen (fig. S24A), and kidneys (fig. S24B). As expected, bacterial loads of ΔbioC recovered from all the examined organs declined markedly, in that it is over 2-log lower than its parental type. Replication defects of ΔbioC in different tissues were entirely reversed upon genetic complementation with a plasmid-borne bioC (Fig. 9, C and D, and fig. S24).
Fig. 9. The BioC-aided early step of biotin synthesis is required for full virulence of K. pneumoniae, an important ESKAPE member.
(A) Scheme for a model of CD-1 mice intravenously challenged with K. pneumoniae ATCC 43816. (B) Survival of CD-1 mice at 12 hours after infection. (C) The reduced survival of the K. pneumoniae ΔbioC mutant in the mouse liver. (D) The removal of bioC compromised survival of K. pneumoniae in the mouse lungs. As for intravenous infections [(B) to (D)], each symbol (circle/triangle/square) represents one mouse per group (n = 6). The data are expressed as means ± SD, and the significance of difference was checked with one-way ANOVA provided by GraphPad Prism. **P < 0.01. (E) Schematic diagram for the survival and pathological alteration of CD-1 mice infected with 103 CFU of K. pneumoniae. (F) Comparative analysis for survival curves of CD-1 mice challenged with different K. pneumoniae strains (103 CFU). (G) Anatomical presentation of pyogenic abscess from CD-1 mouse infected with K. pneumoniae. (H) Hematoxylin and eosin (HE) staining displayed the pyogenic abscess–related lesions from the infected CD-1 mice. (I) No lesions on the mouse liver after infection with the K. pneumoniae ΔbioC mutant. (J) Use of HE staining to visualize liver tissues from the CD-1 mouse infected with the ΔbioC mutant. (K) The plasmid-borne expression of bioC restores the ΔbioC mutant’s pathogenesis in the CD-1 mice model, leading to the clinical symptom of liver abscess. (L) HE staining demonstrated that pyogenic abscess is reproduced in the liver section from the CD-1 mouse infected with the CΔbioC strain. As for both anatomy [(G), (I), and (K)] and HE stains [(H), (J), and (L)], a representative result was presented from one mouse per group (n = 6). The symptoms of liver abscess were circled with yellow dashed lines white arrows.
To analyze putative lesions of liver abscess, we used the systemic/intraperitoneal model of CD-1 mice, which were challenged with 1 × 103 CFU of K. pneumoniae at 1 hour after administration of streptavidin (2 mg/kg). Notably, an administration with streptavidin (2 mg/kg) was intraperitoneally introduced at regular intervals of 12 hours within the whole monitoring period of 96 hours (Fig. 9E). In total, 24 CD-1 mice included here were divided into four groups (six mice each). As shown in survival curves, none of the six ΔbioC-infecting mice died within 96 hours, which is almost identical to that of the negative control (Fig. 9F). As expected, all the mice were gradually killed by K. pneumoniae expressing bioC, regardless of its carriage on chromosome or plasmid (Fig. 9F). Then, we conducted histopathological analyses for mouse livers obtained from experimental groups. The combinations of anatomical assays with hematoxylin and eosin (HE) staining clearly unveiled that (i) pyogenic abscess occurs on the mice liver from the WT-infecting group (Fig. 9, G and H) but not for the negative control (fig. S25, A and B) and the ΔbioC-infecting group (Fig. 9, I and J) and (ii) typical lesions of liver abscess invariantly recur in the group challenged with the CΔbioC strain (Fig. 9, K and L). Therefore, these findings provided functional proof that BioC methyltransferase is a virulence factor in K. pneumoniae, representing a promising anti-ESKAPE drug target.
DISCUSSION
Biotin acts as a nutritional virulence factor (24, 64). Although biotin synthesis is a validated druggable pathway (21, 22), most of lead compounds [e.g., MAC13772 (46)] are confined to the late-stage enzyme BioA (22, 65). The delayed discovery of inhibitors against the primary step is largely due to the incomplete understanding of the dominant BioC-BioH pathway (66, 67). The local BLASTp search against the National Center for Biotechnology Information database allowed us to track the carriage of AbBioC homolog (19.64 to 100% identity; 200-311aa long) from 118,209 of 286,825 bacterial genomes as of May 2023 (~41.2%). Apart from the promiscuous activity of BioH that renders it challengeable to obtain a specific inhibitor (5), the nonavailability of the BioC structure hampers in silico screen and design of lead compounds (7, 22). The prototypic version, EcBioC, is poorly active in the preparation in vitro (5, 7). In addition to its close relative of the soil bacterium Pseudomonas putida (PpBioC, 37.45% identity), EcBioC is genetically replaced with the distantly related paralogs from two Gram-positive bacteria, a foodborne pathogen B. cereus and the food spoilage–causing agent Kurthia (7, 32). Despite the fact that BcBioC and its counterpart of Kurthia (~50% identity) are metabolically engineered to produce odd-carbon dicarboxylic acids of industrial importance in E. coli (32), the solubility of BcBioC is intrinsically unsuitable for protein crystallization. To solve this puzzle, we concentrated on the top priority ESKAPE pathogens. Fortunately, we had a success in solving the high-resolution crystal structures of AbBioC liganded with SAM cofactor (Figs. 2E and 4A) and SIN inhibitor (fig. S13, E to I). As a SAM analog naturally produced by Streptomycetes (38), SIN is a promiscuous pan-MTase inhibitor against diverse pathogens ranging from viral domains (68) to bacterial lineages (7, 69). Given the criteria (i) functional equivalence of AbBioC to BcBioC (fig. S5) and (ii) SIN, a mimicry of SAM cofactor, the AbBioC/SAM and AbBioC/SIN complex structures functionally define inhibitory mechanism of BioC methyltransferase (figs. S13 and S14). Moreover, it laid a foundation for structural modification central to SAM/SIN scaffold. Considering its in vivo release and clinical efficacy, SIN-loaded nanoparticle is expected to be coupled with an appropriate delivery system (70). This explains partially (if not all) the inability of free SIN in diminishing viability of bioC-expressing bacterium.
Recently, Tam (MSMEG_0629) of M. smegmatis was proposed as MsBioC O-methyltransferase that retains limited activity of replacing EcBioC, instead of detoxifying trans-aconitate (15). However, the poor yield of MsBioC/Tam hampers our structural/biochemical investigation (15). A similar scenario was observed for the refolded EcBioC form (5). AbBioC, as we determined here, is also annotated as a Tam-like methyltransferase, albeit its limited identity to MsBioC/Tam (18.67%). In contrast to MsBioC/Tam that has low BioC-like activity (15), AbBioC is capable of enabling robust growth of the ΔbioC mutants of both E. coli and A. baumannii on nonpermissive condition (figs. S5 and S6). Genomic context revealed that bioC is situated either in the biotin gene cluster or as a chimeric bioHC (or bioCD) version (fig. S26), suggesting some evolutionary advantage. Our results indicated that the disguise of AbBioC with an N-terminal SUMO tag efficiently benefits large-scale preparation of soluble AbBioC proteins engaged in structural and functional exploration. It was noted that we have captured four functional modules. Namely, they included (i) a SAM cofactor–binding cavity (Fig. 4), (ii) the ACP-interacting surface rich in positive residues (Fig. 6, A to D), (iii) the tunnel for Ppan-linked malonyl moiety loading (Fig. 6, E to H), and (iv) an essential “Y19-containing β1 sheet” motif (fig. S14). Therefore, the data provided the previously unelucidated architecture and functional insights into BioC methyltransferase.
The transferability of polymyxin resistance is a global challenge of health care concern (71). This is largely due to MCR determinants (mcr-1 to mcr-10) carried by diverse plasmids (47, 72). In principle, the kind of integral membrane protein MCR catalyzes the transfer of PEA moiety from the lipid PE donor to the lipid A group tethered to surface LPS of Gram-negative bacterium (43). The resultant neutralization of the net negatively charged surface is desensitized to killing with cationic antimicrobial polypeptides (such as colistin), a “last-line” defense against multidrug-resistant ESKAPE-type pathogens. Carfrae et al. (27) reported that the disruption of bacterial FAS II pathway circumvents polymyxin resistance. This is because FAS II machinery provided building blocks for PE and lipid, two essential substrates of MCR catalysis (Fig. 7A) (43). Because the AccB subunit of ACC enzyme requires biotin modification to start bacterial FAS II pathway (25), biotin synthesis and its utility is implicated in the formation of bacterial insusceptibility to colistin (Fig. 7A). This assumption is validated by the fact that (i) Acinetobacter bioF (A1S_0807) encoding BioF, participates in formation of natural colistin resistance (73) and (ii) MAC13772, an effective BioA inhibitor, restores polymyxin sensitivity in mcr-1–expressing bacterial species (27). Unlike BioF and BioA that are two enzymes involved in the biotin late step (Fig. 2B), we observed that CRISPR-Cas9–aided elimination of the biotin primary stage enzyme BioC resensitizes A. baumannii (Fig. 7, B and C) and K. pneumoniae (Fig. 7, D and E) to colistin. We thereafter reasoned that the other BioH player of BioC-BioH path is also involved in phenotypic polymyxin resistance. Along with a recent finding of Carfrae et al. (27), our genetic and microbial investigation pinpointed that the biotin/fatty acid synthetic pathway functions as a metabolic basis for MCR polymyxin resistance. Notably, our work reported a previously unidentified role of BioC in impairing bacterial hypervirulence of K. pneumoniae with a hallmark of liver abscess (Fig. 9). Retrospectively, a number of BioH/J isoforms have been implicated in bacterial pathogenesis, which extends from the two intracellular agents, Francisella (63, 74) and M. tuberculosis (21), to an extracellular ESKAPE pathogen, P. aeruginosa (24). Thus, we believed that targeting BioC-directed primary step is beneficial to elucidate some lead compounds against untreatable infections with recalcitrant ESKAPE pathogens. Because the single mycobacterial Bpl seemed as an alternative anti-TB target (21), further evaluation of BplA as a potential target is also deserved in the anti-ESKAPE therapy.
In summary, structural and biochemical characterization in this study illustrates a comprehensive landscape of BioC methyltransferase. Apart from its initiating biotin synthesis, BioC seems moonlighted into two unrelated functions, polymyxin resistance and nutritional virulence. Therefore, targeting the dominant BioC-BioH primary pathway is prioritized as an approach of “killing two birds with one stone,” expanding the current repertoire of limited lead inhibitors against the nosocomial ESKAPE threats.
MATERIALS AND METHODS
Ethics statement
The maintenance and manipulation of all the ESKAPE agents and their isogenic mutants plus complemented strains were performed in the biosafety level 2 (BSL-2) laboratories at the Zhejiang University School of Medicine. As for all the experiments of mice infections, they were carried out under the guidelines and regulations of the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China (11-14-1988). The protocol for studies on mice was licensed by the Laboratory Animal Welfare and Ethics Committee, Zhejiang University (ZJU20230459). Under appropriate regulation, plasma samples of healthy individuals were kindly provided by General ICU of the Second Affiliated Hospital, Zhejiang University School of Medicine.
Bacterial strains and growth conditions
In total, 10 kinds of different bacterial species were cultivated here. Apart from three reference strains (E. coli, B. subtilis, and B. cereus), the rest of the bacteria corresponded to ESKAPE members (table S1). Namely, they included (i) E. faecalis, (ii) S. aureus, (iii) K. pneumoniae, (iv) A. baumannii, (v) P. aeruginosa, (vi) E. cloacae, and (vii) E. aerogenes. Except for E. faecalis grown on Tryptic soy agar, all the remaining bacteria were kept on Luria-Bertani (LB) broth at 37°C. All the E. coli strains were derived from the model strain MG1655 with appropriate genetic modification (75). The two strains of E. coli, ER90(ΔbioF/ΔbioC/ΔbioD) (5, 76) and STL24 (ΔbioC) (5, 7), were biotin auxotroph (36) and required biotin for viability. Using CRISPR-Cas9 technology, two types of ΔbioC deletion mutants (i.e., FYJ6108 and FYJ6184) were separately created from A. baumannii ATCC 17978 (33) and K. pneumoniae ATCC 43816 (table S1) (77). Before functional cloning and protein expression of BioC enzymes, varied bioC orthologs were amplified with PCR accordingly. Namely, they corresponded to (i) EcBioC/E. coli MG1655, (ii) BcBioC from B. cereus ATCC 10987 (78), (iii) AbBioC for A. baumannii ATCC 17978 (33), and (iv) KpBioC arising from K. pneumoniae ATCC 43816 (77). Unlike DH5α that was a cloning host, BL21(DE3) acted as an expression host for heterogeneous protein production. The assays of bacterial viability and growth curves were conducted with M9 minimal medium to evaluate biotin requirement. When necessary, either kanamycin (50 μg/ml) or apramycin (50 μg/ml) was supplemented for colony selection.
Plasmids and molecular manipulations
In total, four types of plasmids were applied here, including (i) pBAD24H constructs (50), (ii) pET/pET-SUMO series (79), (iii) pWSK129 derivatives (80), and (iv) pCas9-borne versions (table S2) (33). First, an arabinose-induced expression vector pBAD24H was exploited for a functional assay of putative bioC in the biotin auxotroph STL24 (ΔbioC) as a recipient strain. The resultant plasmids included (but are not limited to) the major three constructs of pBAD24::AbbioC, pBAD24::KpbioC, and pBAD24::BcbioC. It was noted that the version of pBAD24::SUMO-AbBioC was also created, which was designed to neutralize the potential toxicity of AbBioC overexpression (table S2). Second, unlike BcbioC that was dependent on pET28a to give solubility, AbbioC (and a number of its mutated variants) plus KpbioC were cloned into pET-SUMO that is a fusion expression vector with a SUMO tag at N terminus, giving a panel of recombinant plasmids, such as pET-SUMO::AbbioC and pET-SUMO::KpbioC (table S2). To prepare the apo-form of ACP (81), an additional enzyme of E. coli ACP phosphodiesterase (AcpH; b0404) was required because it essentially liberates Ppan from the residue serine-36 of ACP (82). Before the tag-free ACP production with pBAD24::acpP(b1094), pET28a::acpH(b0404) was created to give the N-terminal hexahistidine (6x His)–fused AcpH version (table S2) (82). B. subtilis Sfp (Ppan transferase)–encoding gene sfp was amplified with PCR and cloned into pET28a via homologous recombination, resulting in pET28a::sfp (83). The purified Sfp enzyme could convert Mal-ACP from apo-ACP in the presence of Mal-CoA (7). As for AbBioC reaction that ligates Mal-ACP with SAM to give M-Mal-ACP, the byproduct SAH was believed to interfere with the activity of BioC methyltransferase. The gene mtn/pfs(b0159) of E. coli was cloned to give a recombinant plasmid pET28a::mtn(b0159), of which protein product is 5′-methylthioadenosine(MTA)/SAH nucleosidase and eliminates the feedback inhibition by SAH (table S2) (76, 84). Using the method of homologous recombination, the four A. baumannii genes encoding putative biotinylated proteins [HKO16_10520 (AccB), HKO16_06425, HKO16_06925, and HKO16_15815] were obtained by PCR and introduced into pET28a via two restriction sites of Bam HI and Hind III (table S2). Third, the mobile colistin resistance determinant (mcr-1) was introduced into the constitutive expression vector pWSK129 (11, 80), producing the recombinant form of pWSK129::mcr-1 that was dedicated to resistance evaluation in K. pneumoniae (table S2). Fourth, the expression vector for A. baumannii, designated pΔCas9, was specifically developed through removing the Cas9 from the parental vector pCasAb-apr. The resultant pΔCas9 contained the unique multiple cloning sites, Eco RI at 5′-end and Hind III followed by a 6x His tag. This allowed us to generate the recombinant plasmids of pΔCas9::AbbioC, pΔCas9::KpbioC, and pΔCas9::mcr-1 (table S2).
To define extensive interaction of AbBioC with (i) its SAM cofactor (or SIN inhibitor) and (ii) Mal-ACP substrate, a collection of AbbioC mutants were produced using the site-directed mutagenesis with various pairs of specific primers like AbbioC(E50A)-F plus AbbioC(E50A)-R (table S3). As for these trials, all the three sets of recombinant plasmids (pBAD24::AbbioC, pΔCas9::AbbioC, and pET-SUMO::AbbioC) functioned as PCR templates accordingly. As a result, in total, 16 AbbioC mutants on each plasmid backbone were generated (table S2), including 12 single mutants [e.g., pBAD24::AbbioC(E50A), pΔCas9::AbbioC(E50A), and pET-SUMO::AbbioC(E50A)], 2 double mutants [i.e., pBAD24:: AbbioC (Q26A/Q116A) and pBAD24::AbbioC(K201A/R212A)], a triple mutant [such as pBAD24::AbbioC(K194A/K201A/R212A)], and a quadruple mutant [such as pBAD24::AbbioC(K194A/K201A/R212A/K215A)]. In addition, a deletion mutant of AbbioC(ΔN, 1-20) was engineered to evaluate a functional role of β1 sheet in AbBioC activity. Thus, it involved three constructs (table S2), namely, (i) pBAD24::AbbioC(ΔN,1-20), (ii) pΔCas9::AbbioC(ΔN,1-20), and (iii) pET-SUMO::AbbioC(ΔN,1-20). Along with routine PCR detection, all the acquired plasmids were validated by Sanger sequencing.
CRISPR-Cas9 knockout of A. baumannii bioC
As Wang and coworkers (33) established, the CRISPR-Cas9 system consisting of pCasAb-apr and pSGAb-km (table S2), was adopted to knockout the putative bioC gene from the chromosome of A. baumannii ATCC 17978. In principle, pCasAb-apr plasmid expresses both Cas9 nuclease and RecAb recombination system, and pSGAb-km produces sgRNA against the target gene (33). Of several sgRNA oligos for AbbioC that we designed, the optimal one that was annealed with the two complementary primers (AbbioC-sgRNA-F/R; table S3) was ligated into the pSGAb-km vector to obtain pSGBioC-sgRNA. Following the expression of Cas9 and RecAb induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the A. baumannii ATCC 17978 that carries pCasAb-apr was prepared as competent cells, which were ready to receive pSGBioC-sgRNA plasmid along with the AbbioC repair template via an electroporation. Using two pairs of specific primers (AbbioC-Up400-F/R plus AbbioC-Dn400-F/R; table S3), the fusion PCR was performed to gain an AbbioC-targeting repair template [800 base pairs (bp)] that cover the two neighboring regions of AbbioC (400 bp each). The candidate ΔbioC mutants were screened with multiplex PCR assays and verified with direct DNA sequencing of PCR products. Next, the two plasmids of pCasAb-apr and pSGBioC-sgRNA were cured from the interested ΔbioC mutant by growth on LBA plates containing 10% sucrose (Shanghai Hushi Environmental Protection Reagent Technology Co. Ltd., Shanghai, China). The biotin auxotrophic phenotype of the resultant A. baumannii ΔbioC mutant, termed as FYJ6108 (table S1), was analyzed on biotin-lacking M9 minimal medium. Similarly, CRISPR-Cas9 manipulation enabled producing strain FYJ6184, i.e., the ΔbioC derivative of K. pneumoniae ATCC 43816 (table S1). A collection of genetically complemented strains of A. baumannii ΔbioC were obtained via an electroporation with appropriate pΔCas9-borne derivatives such as pΔCas9-AbBioC and pΔCas9-KpBioC (table S2). Similar to the recipient strain STL24 (ΔbioC) that carried a variety of pBAD24.8x His-borne AbbioC mutants, the complemented strains of A. baumannii ΔbioC with diverse pΔCas9-borne bioC derivatives were analyzed using growth curves and bacterial viability on the basis of the nonpermissive, biotin-deficient condition of M9 defined medium as described for the biotin requirement of E. coli (5, 29, 63) and P. aeruginosa PAO1 (24).
Genetic analyses for bioC homologs
The functions of AbbioC and KpBioC were genetically assayed with three sets of bacterial mutants devoid of BioC methyltransferase. In addition to the paradigm ΔbioC mutant of E. coli STL96 that was developed by Lin and Cronan (7), we also created two ΔbioC mutants (FYJ6108 and FYJ6184) that separately arose from A. baumannii ATCC 17978 and K. pneumoniae ATCC 43816 (table S1). In comparison to the positive control, BcBioC-expressing E. coli ΔbioC strain, bacterial viabilities of STL96 derivatives that produced AbBioC (or KpBioC) carried by pBAD24.8x His, were determined on the nonpermissive, biotin-free condition of M9 minimal medium. The SUMO tag was expected to disguise AbBioC, thereby circumventing the potential toxicity of its overexpression. The expression profile of pBAD-borne SUMO-AbbioC fusion induced with arabinose was systematically evaluated according to the growth of the biotin auxotroph FYJ6113 on the nonpermissive condition of M9 defined medium with 0.2% glycerol (or glucose) as the sole carbon source (table S1). An expression system of pΔCas9 carried by FYJ6184, the bioC-defective K. pneumoniae allowed us to probe a functional replacement of AbBioC with its paralog KpBioC. To finely characterize determinants of AbBioC activity, in total, 17 mutants were engineered, consisting of 1 deletion mutant of AbBioC(ΔN, 1-20) and 16 point mutants covering substrate/cofactor-binding cavities. Unlike the system of MG1655 ΔbioC mutant that carried pBAD24.8x His-borne AbbioC derivatives, the biotin-requiring strain FYJ6108 (i.e., A. baumannii ΔbioC) acted as a recipient host for all the 17 pΔCas9-based AbbioC variants (FYJ6115 to FYJ6127 and FYJ6190 to FYJ6193). All the derivatives of STL96 and FYJ6108 were subjected to the analyses of bacterial viabilities and growth curves on the nonpermissive condition of M9 defined medium with biotin restricted. As described with BioZ (11) and BioH (14) [or its isoform BioJ (39, 63)], all the recombinant strains were passaged and subcultured (1:200) into 96-well plate containing biotin-free M9 liquid medium (200 μl per well) that was kept in a spectrophotometer (Spectrum Lab S32A) set at 200 rpm. The bacterial optical density at wavelength of 600 nm (OD600) values at 37°C were regularly recorded at hourly intervals for the whole monitoring period of 20 hours. Three independent trials were developed, of which each point was in triplicate.
Measurement of cytosolic and plasma biotin levels
The ELISA assay (E-IR-R501, Elabscience) was used to determine the abundance of biotin in different organisms (24). Apart from CD-1 mice blood samples, the plasma of healthy individuals were gifts obtained from hospitals. Because a list of biotin-requiring proteins markedly varied among ESKAPE-including 10 species, the profiles of intracellular biotin pools were concerned. To minimize biotin contamination, the log-phase cultures of 10 diverse bacterial species (such as E. coli and A. baumannii) were washed three times, resuspended in 1× PBS of appropriate volume, and then lysed by French pressure cell. After the removal of debris, the cleaned lysates (50 μl each well) in appropriate dilution were supplemented into the 96-well ELISA plate that was precoated with biotin antigen. After a couple rounds of routine procedures, all the samples were subjected to the determination of optical density at 450-nm wavelength (OD450). This led to the calculation of biotin levels through the comparison of OD450 values with the standard curve that we plotted. Similarly, plasma biotin levels were also detected. Unlike plasma sample that is expressed in ng/ml, biotin level in bacterial samples was converted to ng/106 CFU.
Protein expression, purification, and identification
In total, 28 recombinant proteins were prepared here. The majority of them (20 in total) involved 3 BioC isoforms of different origins (BcBioC, AbBioC, and KpBioC) and 17 mutated versions of AbBioC. The remaining eight proteins consisted of four putative biotinylated enzymes (AccB, HKO16_06425, HKO16_06925, and HKO16_15815), two components for apo-ACP generation [AcpH (82) and AcpP (81, 85)], and two elements [Sfp (83, 86) and Pfs/Mtn (76, 84)] engaged in converting from Mal-ACP to M-Mal-ACP ester. Strain FYJ5039 was used to give the soluble form of BcBioC; FYJ6169 and FYJ6170 were separately applied in SUMO-fused versions of AbBioC and KpBioC (table S1). Unfortunately, KpBioC invariantly appeared as precipitates regardless of the SUMO tag. It was noted that the deletion mutant of AbBioC(ΔN,1-20) was successfully obtained using the two engineered strains as follows: (i) FYJ6136 carrying pBAD24::AbbioC(ΔN,1-20) and (ii) FYJ6220 bearing pET-SUMO::AbbioC(ΔN,1-20). All the rest of AbBioC mutants that we produced arose from the BL21/pET-SUMO expression system, involving 16 strains (FYJ6153 to FYJ6159 and FYJ6196 to FYJ6204; table S1).
To harvest AbBioC protein, 1 liter of bacterial culture of FYJ6169 in mid-logarithmic phase (OD600: ~0.8) was induced with 0.3 mM IPTG for 12 hours at 16°C. Bacterial pellets collected by centrifugation were suspended in lysis buffer [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 10% glycerol, 20 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride] and then subjected to the lysis by French pressure cell (JNBIO, Guangzhou Juneng Nano & Biotech Co. Ltd.). After the removal of cell debris by 1 hour of centrifugation (16,800 rpm/min, at 4°C), the resultant lysate was incubated with Ni-NTA agarose beads (QIAGEN) for 1 hour. Before an elution of the 6x His-tagged version of SUMO-AbBioC with the elution buffer [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 5% glycerol, and 300 mM imidazole], the protein-bound beads received three rounds of treatment with wash buffer [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 5% glycerol, and 50 mM imidazole] to eliminate contaminated proteins. Before an overnight incubation in the dialysis buffer [20 mM tris-HCl (pH 8.0) and 300 mM NaCl], the SUMO-AbBioC protein was concentrated to a total volume of ~10 ml (~3 mg/ml) and then mixed with the yeast ULP1 enzyme (0.02 mg/ml) for the cleavage the SUMO tag (87, 88). Next, the mixture of protein digestion was loaded on a Ni-NTA column for two to three rounds, thereafter giving SUMO tag–free AbBioC enzyme in the effluent. This was because both an intact SUMO-AbBioC version and ULP1 enzyme are nickel beads-bound, whereas AbBioC is not. Subsequently, AbBioC sample was concentrated to ~0.6-ml volume (~10 mg/ml) with the ultrafiltration tube of 30-kDa cut-off (Millipore), which was followed by the analysis of SEC using a Superdex 200 Increase 10/300 column (GE Healthcare). The AbBioC protein collected from the interested peak was verified by the separation with 15% SDS–polyacrylamide gel electrophoresis (SDS-PAGE). After trypsin digestion of AbBioC cut from the gel, the identity of resultant polypeptides was further validated by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Using a similar protocol, all the other AbBioC mutants were overexpressed and purified to homogeneity.
Western blot
Streptavidin blot was applied to address the varied repertoire of protein biotinylation in diverse ESKAPE members. Apart from ESKAPE, three additional microbes (i.e., E. coli, B. cereus, and B. subtilis) were cultivated to prepare bacterial lysates. As described by Shi and coworkers (24), streptavidin AP conjugate (Roche) functioned as the primary antibody of 1000-fold dilution in these blot assays. Using CDP-Star substrate, the signals of chemiluminescence were captured (89). To further validate the profile of multiple proteins biotinylated in A. baumannii, the four candidates (AccB, HKO_06425, HKO_06925, and HKO_15815) that were consistently overexpressed in the 6x His–fused recombinant forms (table S2) were subjected to both streptavidin blot and anti–6x His tag Western blot. For the latter blot, mouse anti–6x His tag monoclonal antibody (EarthOx Life Science) acted as the primary antibody (1:8000), and HRP-goat anti-mouse IgG (EarthOx Life Science) was the secondary antibody (1:5000) (29, 31).
Preparation of apo-ACP
The apo-form of ACP was prepared as described by Yin and coauthors (85) with minor alteration. Along with FYJ6171, an AcpP-expressing strain, FYJ6174 that encodes AcpH (table S1) was cultivated individually (1 liter each), and the resultant two bacterial pellets were suspended in the same tube containing 50 ml of lysis buffer [50 mM tris-HCl (pH 8.8), 25 mM MgCl2, and 1 mM dithiothreitol (DTT)]. After cell disruption by French pressure cell, bacterial lysates were spun to obtain cleaned supernatants, which then were maintained at 37°C for around 4 hours, before the precipitation with 2× volumes of cold isopropanol. Following the removal of the precipitated contaminants by 1 hour of centrifugation (13,000 rpm) at 4°C, the supernatants were dialyzed against the dialysis buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT] overnight. Last, the dialyzed apo-ACP protein was purified by an anion-exchange chromatography with HiTrap Q HP column (GE Healthcare), which was validated by separation with conformationally sensitive gel of 0.5 M urea/PAGE (pH 9.5 and 17.5%; fig. S10B).
Assays for BioC methyltransferase in vitro
AbBioC was assumed as an O-methyltransferase (fig. S3) that catalyzes transferring a methyl group from SAM cofactor to Mal-ACP, producing M-Mal-ACP (7). Because the substrate Mal-ACP is not commercially available, the in vitro synthesis system was set up. This was based on the principle that B. subtilis Sfp ligates mal-CoA (Sigma-Aldrich) with its recipient apo-ACP to give Mal-ACP (fig. S10A). In general, the Sfp-catalyzed reaction system (100 μl in total) consisted of 100 mM MES (pH 6.0), 10 mM MgCl2, 10 μM Sfp, 0.5 mM apo-ACP, and 0.75 mM Mal-CoA. As expected from conformationally sensitive PAGE (pH 9.5 and 17.5%) containing 0.5 M urea (fig. S10B), the substrate Mal-ACP we made was distinguishable from its apo-form of ACP. As Lin and Cronan described (7), the enzymatic reaction by BioC (100 μl in total) was built to proceed to 1 hour of incubation at 37°C, which composed of 100 mM tris-HCl (pH 7.5), 10 mM MgCl2, 10% glycerol, 200 mM NaCl, 0.3 μM BioC, 3 μM Mtn, 0.5 mM Mal-ACP, and 1 mM SAM. Notably, the presence of Mtn restricted the inhibition of BioC activity by SAH, the by-product of SAM hydrolysis (76, 84). Last, MALDI-TOF/electrospray ionization mass spectrometry was applied to determine the product of M-Mal-ACP arising from its reactant Mal-ACP, as well as apo-ACP and holo-ACP. In principle, the theoretical mass values [mass/charge ratio (m/z)] were assigned as follows: (i) 8508.3 for holo-ACP (fig. S10C), (ii) 8847.3 for apo-ACP (fig. S10D), (iii) 8933.39 for Mal-ACP (fig. S10E), and (iv) 8947.39 for M-mal-ACP (fig. S10F).
Crystallization and data collection
The AbBioC protein was concentrated to 68 mg/ml for crystallization screens. For the complex formation, the protein and SAM ligand (or SIN inhibitor) were mixed and incubated at room temperature for 30 min. The final protein AbBioC, SAM, and SIN concentrations were 55 mg/ml, 18 mM, and 20 mM, respectively. The crystallization conditions were identified at 18°C using the Gryphon crystallization robot system and commercial crystallization kits. The sitting-drop vapor diffusion method was applied during screening process of both complexes. The drop contains equal volumes of protein sample and reservoir solution. As a result, the AbBioC/SAM crystals were obtained under the A5 condition of the Wizard Classic 1/2 kit, composed of 30% (v/v) polyethylene glycol, molecular weight 400 and 0.1 M CAPS/sodium hydroxide (pH 10.5), whereas the AbBIoC-SIN crystals were harvested under F11 condition of the Crystal Screen HT kit, consisted of 1.6 M ammonium sulfate, 10% (v/v) 1,4-dioxane, and 0.1 M MES monohydrate (pH 6.5).
All crystals were cryoprotected in reservoir solution supplemented with 25% (v/v) glycerol and snap-frozen in liquid nitrogen. The x-ray diffraction data were collected on beamline BL18U1 of National Facility for Protein Science Shanghai (NFPS). Data were automatically processed by the auto PROC program developed by the beamline staff. The data collection and processing statistics were listed in Table 1.
Structural determination and refinement
The AbBioC/SAM complex structure was solved by molecular replacement (MR) method using the Phaser program (90) of the CCP4i suite (91); the AbBioC structure predicted by the AlphaFold2 program (92) was used as the search model. The resulting models were refined against the diffraction data using the Refmac5 program. The 2Fo-Fc and Fo-Fc electron density maps were regularly calculated and used as guide for the building of SAM and water molecules in COOT (93). About 5% randomly selected data were set aside for free R-factor cross-validation calculations during the refinement. The AbBioC/SIN complex structure was dissected by MR method using the AbBioC/SAM structure as the search model. The final refinement of the structure was performed using the phenix.refine program. The structural refinement statistics were also summarized in Table 1.
Isothermal titration calorimetry
To reveal the stoichiometry of AbBioC binding to SAM cofactor (or SIN inhibitor), ITC experiments were conducted using a microcalorimeter (MicroCal PEAQ-ITC) at room temperature (25°C) as described for BioZ (11) with little alteration. SAM was purchased from Sigma-Aldrich, and SIN was available in MedChemExpress. Titration reaction buffer consisted of 20 mM tris (pH 8.0) and 300 mM NaCl. The concentration of AbBioC used here is 20 mM, and the ligand SAM (or SIN) was 200 μM in the same buffer. The cuvette contained 300 μl of AbBioC protein (20 μM), and the syringe included 70 μl of SAM/SIN (200 μM). During titration, SAM (or SIN) was syringed into the cuvette in 20 drops. Except for an initial droplet of 0.4 μl, the rest of the 19 injections proceeded in a volume of 2.0 μl at an interval of 120 s. Three independent replicates were performed, and the integral titration data were fitted with a single binding site model in Origin 7.0 software. The resultant binding stoichiometry (N) and Kd were expressed as mean ± SD.
Reconstitution of BioC-initiated DTB synthesis
To ascertain altered roles of diverse AbBioC mutants in biotin synthesis, the biotin/DTB synthetic system was reconstituted in vitro as earlier described for Ehrlichia chaffeensis BioC ortholog with appropriate changes (94). To visualize whether biotin or its precursor DTB was produced in this reaction system, the biotin bioassay was used, which relies on the indicator strain of A. baumannii ΔbioAFCD that we created here (table S1), as well as a well-studied report strain, the E. coli ER90 (ΔbioFCD) mutant (5). Apart from the WT AbBioC and the positive control BcBioC, in total, nine mutated AbBioC versions were examined here. Namely, they included five single mutants (E50A, D75A, W117A, Q26A, and Q116A), two double mutants (K201A/R212A and Q26A/Q116A), a triple mutant of K194A/K201A/R212A, and a quadruple mutant of K194A/K201A/R212A/K215A (table S1).
As earlier recommended by Lin and colleagues (5) with little change, the cell-free crude extract of Strain STL96 (MG1655, ΔbioC) was prepared, providing the kit of enzymes engaged in the FAS II–coupled biotin synthesis pathway. In brief, the strain STL96 was grown at 37°C to mid-log phase (OD600, 0.8) in a flask containing 200 ml of M9 defined medium supplemented with 1 nM biotin. Following the removal of residual biotin through two rounds of wash with M9 media (20 ml each), bacterial cells were subcultured into 1 liter of biotin-free M9 minimal medium at 37°C for over 5 hours, which led to an intracellular biotin starvation or depletion. Subsequently, the pelleted STL96 cells were lysed by two rounds of passages through a French pressure cell, which was followed by 0.5 hours of centrifugation (16,800 rpm) at 4°C. The resultant supernatants were further subjected to ammonium sulfate precipitation (85% saturation) and then centrifuged at 12,800 rpm for 1 hour. It was noted that ACP species plus small molecules are assumed to remain soluble in the above solution. The solubilized precipitates were dialyzed against 2 liters of PBS buffer at 4°C overnight, benefiting the elimination of side effects by ammonium sulfate and residual small molecules. The concentration of STL96 crude extract was obtained by the NanoDrop-based measurement (Thermo Fisher Scientific). As a result, the final output was around 175 mg/ml for stock solution.
Next, the BioC-directed DTB/biotin system (300 μl in total) included the following components: (i) 3 mg of STL96 crude extract, (ii) 6 μg of AbBioC enzyme, (iii) 150 μg of substrate of Mal-ACP, (iv) 0.1 μmol of SAM cofactor, (v) 0.1-μmol l-alanine, (vi) 0.01-μmol PLP, (vii) 0.1-μmol glucose-6-phosphate, (viii) 0.1-μmol NADPH, (ix) 0.1-μmol ATP, (x) 1.0-μmol MgCl2, (xi) 0.1-μmol KHCO3, and (xii) 0.5-μmol DTT. In general, the reaction was initiated upon an addition of BioC enzyme, maintained at 37°C for ~3 hours, and quenched by immersion into the metal bath at 100°C for 10 min (29, 63). The mixture of AbBioC reaction was subjected to 10 min of centrifugation at 12,000 rpm, and the resultant supernatant that was supposed to contain DTB/biotin products was transferred to a fresh EP tube and stocked at −20°C until the use in the bioassay.
To prepare biotin indicative M9 agar plates, the biotin-requiring strain ER90 (ΔbioFCD) was used as a reporter sensing the availability of extracellular biotin/DTB pool. In brief, overnight culture of ER90 in 5 ml of M9 chemically defined medium containing 2 nM biotin was pelleted by the 6 min of centrifugation at 6000 rpm, washed twice with M9 medium, and subcultured to 100 ml of biotin-free M9 minimal medium. Presumably, the 5 hours of bacterial starvation caused the depletion of cytosolic biotin/DTB metabolites out of the ER90 cells. The pelleted cells were washed twice, suspended in 1 ml of PBS buffer without any biotin, and stocked at 4°C until use. In general, the ER90 cells that we prepared (100 to 150 μl) were mixed with 100 ml of melted M9 agar medium (~55°C), containing 0.01% (w/v) of TTC, an indicator for bacterial respiration. As a result, quadruple-sectored petri dishes were produced, each sector of which harbored 4- to 5-ml M9 agar medium covered with a paper disc. The supply of biotin/DTB spotted on the paper disc rendered ER90 to be of viability, which, in turn, reduced TTC to give insoluble red formazan deposited around colonial growth, whereas the indicator strain cannot display any growth signal.
Antibacterial activity of MAC13772
Because MAC13772 is a validated anti-TB inhibitor specifically targeting the late step BioA enzyme (46), we attempted to extend such antibacterial activity to the recalcitrant agents of A. baumannii (or K. pneumoniae) with transferable colistin resistance, as initially investigated by Carfrae and colleagues (23) with little change. In total, six strains were examined here, consisting of three A. baumannii and three K. pneumoniae. In brief, the strain AB18PR065 was a natural isolate of A. baumannii expressing MCR-4.3 (49). A. baumannii ATCC 17978 was introduced with a recombinant plasmid pΔCas9::mcr-1, giving the strain FYJ6179 with MCR-1 colistin resistance (table S1). Similarly, the plasmid pWSK129::mcr-1 was transformed into K. pneumoniae ATCC 43816, which resulted in colistin-resistant strain FYJ6185. The strain ZZW20 was a natural ST11 isolate of K. pneumoniae expressing MCR-8 (52). Relative to the two negative controls that are mcr-negative, all the four MCR-producing strains were subjected to determination of minimal inhibitory concentration (MIC) based on colistin susceptibility test strip (E-test) on LB (or 0.2% glucose M9) agar plates. Next, a single colony from the aforementioned strains was suspended in 20 μl of M9 buffer and subcultured (1: 200) into 200 μl of biotin-free M9 chemically defined medium supplemented with the MAC13772 compound in a series of twofold dilution (1, 2, 4,…, 256 μg/ml). Notably, the avidin (0.1 μg/ml) that can chelate biotin was added into M9 buffer to make biotin-free minimal medium. Bacterial growth at 37°C proceeded in a spectrophotometer (Spectrum Lab S32A) with a shaking speed of 200 rpm. The growth curves were plotted within a 20-hour period, in which the OD600 value was recorded at an interval of 1 hour. The relative ratio of bacterial viability at the end of monitoring period allowed to determine the half inhibitory concentration (IC50) of MAC13772 (24). The final output was presented in an average ± SD of three replicates.
Bacterial lysis assay
Very recently, Carfrae et al. (27) proposed that inhibiting a FAS II pathway restores bacterial susceptibility to colistin. To address the contribution of BioC to polymyxin resistance, extensive assays for bacterial killing by colistin were performed by integrating the ΔbioC mutant of A. baumannii (and K. pneumoniae) with certain mcr-1–carrying plasmid. In brief, four strains were engineered here (table S1), namely, (i) FYJ6179 (A. baumannii ATCC 17978 carrying pΔCas9::mcr-1), (ii) FYJ6181 (ΔbioC of A. baumannii carrying pΔCas9::mcr-1), (iii) FYJ6185 (K. pneumoniae ATCC 43816 carrying pWSK129::mcr-1), and (iv) FYJ6186 (ΔbioC mutant of K. pneumoniae carrying pWSK129::mcr-1). The bacterial lysis experiments were divided into five groups. Apart from the positive control group [FYJ6179 with synergistic effect by colistin (8 μg/ml) and MAC13772 (4 μg/ml)], they included (i) FYJ6179 (or its ΔbioC mutant FYJ6181) in PBS buffer, (ii) FYJ6179 (or its ΔbioC mutant FYJ6181) stressed with colistin (8 μg/ml), (iii) FYJ6185 (or its ΔbioC mutant FYJ6186) in PBS buffer, and (iv) FYJ6185 (or its ΔbioC mutant FYJ6186) treated with colistin (8 μg/ml).
In brief, 2 ml of overnight cultures of A. baumannii (or K. pneumoniae) were prepared with M9 defined medium retaining minimal biotin level (10 nm for A. baumannii and 4 nm for K. pneumoniae) and then transferred to 50 ml of M9 minimal medium containing 5-nm biotin, until the mid-logarithmic phase (OD600, ~0.6). Following three rounds of wash with 1 ml of biotin-free PBS buffer, the pelleted cells were resuspended and then subseeded at the dose of 2 × 108 CFU/ml into 2 ml of PBS buffer with or without the aforementioned colistin treatment. As for the positive control, the combination of treatments was described as follows: (i) MAC13772 (4 μg/ml) alone, (ii) colistin (8 μg/ml) alone, and (iii) MAC13772 combined with colistin (8 μg/ml). They were kept in a shaker at 37°C for 20 hours, and bacterial samples were taken at 0, 4, 8, 12, 16 and 20 hours to check cell viability by plate counting in serial 10-fold dilutions.
Virulence assays with mouse infections
To ascertain the importance of BioC in hypervirulence of K. pneumoniae, two sets of experimental infection models were applied with 5-week-old CD-1 mice. Namely, they denoted (i) an intravenous infection and (ii) an intraperitoneal infection. Before bacterial challenge, the K. pneumoniae strains were washed three times to eliminate excess biotin and suspended in biotin-free PBS buffer. In addition to WT (i.e., the reference strain ATCC 43816), both the bioC-inactivated mutant ΔbioC of K. pneumoniae (FYJ6184) and its genetically complemented CΔbioC strain FYJ6188 that carried by a plasmid pΔCas9::AbbioC were included here. It was noted that cell density for K. pneumoniae (OD600, 0.1) was calculated to be 107 CFU.
In total, 18 CD-1 mice (5 weeks old) were used in the intravenous infections and classified into three groups (6 mice each). The mouse was inoculated via the tail vein with 105 CFU of ATCC 43816 strain, ΔbioC (FYJ6184) and its complemented strain FYJ6188 expressing AbBioC. Because that mouse plasma contained biotin at over sixfold higher level than that of human plasma, the CD-1 mice were required to mimic the human environment via the intraperitoneal inoculation of streptavidin (2 mg/ml) at 1 hour before infection with K. pneumoniae strains (23). All the mice were dead 12 hours after challenge with WT or CΔbioC, except for the ΔbioC-infected group with humanitarian execution. Next, the four organs extracted (i.e., liver, lungs, spleen, and kidneys) were weighed, homogenized in 1 ml of PBS buffer, diluted appropriately, and inoculated on LB agar to calculate the bacterial loads accordingly. As a result, bacterial localization of different K. pneumoniae strains in various mouse organs was given by calculating bacterial load index log10 (CFU/g).
As for the systemic/intraperitoneal infection, 24 CD-1 mice were divided into four groups (six mice each). Apart from the negative control group inoculated with PBS buffer, the rest of the experimental groups were challenged with different strains of K. pneumoniae (103 CFU/mouse). Unlike an intravenous challenge of mice that only required an intraperitoneal injection of streptavidin (2 mg/ml) 1 hour before the K. pneumoniae administration (23), the systemic infection demanded regular injection of streptavidin (2 mg/ml) every 12 hours to simulate human plasma biotin level until the end of animal experiment. The survival of infected mice was recorded within the entire monitoring period. All the CD-1 mice were euthanized at the end of 96 hours, allowing the visualization of liver abscess, a hallmark of the infection with hypervirulent K. pneumoniae. In addition, the mouse liver sections were subjected to analysis of HE staining (54).
Phylogenetic analysis
The amino acid sequences of BioC orthologs and other methyltransferases were obtained by protein BLAST. Sixty-five unique amino acid sequences were used for the phylogenetic analysis. Multiple sequence alignment was conducted by MUSCLE v5.1 (https://github.com/rcedgar/muscle) using default parameters (95). ModelFinder (96) was used to identify the best-fit protein substitution model, and the best model was used to generate a maximum likelihood (ML) tree with 1000 bootstrap replicates. The ML phylogenic trees were constructed by using the LG amino acid substitution model with extended model selection followed by tree inference using IQ-TREE v2.0.3 (https://github.com/iqtree/iqtree2) (97). The results are presented as a radial phylogram. A subset of the 35 sequences was reanalyzed using the same method to obtain a detailed phylogenetic tree, which was visualized with FigTree v1.4.4 and iTOL v6 (98).
Acknowledgments
Funding: This work was supported by National Key Research & Development Program of China (2023YFC2307100 and 2023YFC2308403, Y.F.; 2022YFC2303900 and 2022YFC3704700, Y.X.), National Science Fund for Distinguished Young Scholar (32125003, Y.F.), National Natural Science Foundation of China (32141001 and 31830001, Y.F.), and Natural Science Foundation of Shanxi Province (2023-JC-YB-159, T.C.). We thank Y. Xu (Analysis Center for Agrobiology and Environmental Sciences, Zhejiang University) for technical assistance in MALDI-TOF mass spectrometry.
Author contributions: Conceptualization: Y.F., J.G., and Z.-G.H. Methodology: Y.F., J.G., R.Y., and T.C. Investigation: Y.F., Z.S., W.Z., Y.S., T.C., Y.X., R.Y., M.H., C.Z., H.Z., T.L., J.Q., and J.G. Formal analysis: Y.F., Z.S., W.Z., Y.S., T.C., H.Z., and J.G. Software: Y.F., Z.S., W.Z., T.C., R.Y., C.Z., and J.G. Data curation: Y. F., Z.S., W.Z., Y.S., T.C., R.Y., and J.G. Visualization: Y.F., M.H., C.Z., T.L., J.G., and Z.-G.H. Validation: Y.F., J.G., and Z.-G.H. Supervision: Y.F. Resources: Y.F., M.H., J.G., and Z.-G.H. Project administration: Y.F. Funding acquisition: Y.F., T.C., and Y.X. Writing (original draft): Y.F., J.G., H.Z., Z.S., T.L., and Z.-G.H. Writing (review and editing): Y.F., J.G., and Z.-G.H.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The coordinates and structure factors of AbBioC were deposited in the RCSB Protein Data Bank under accession numbers 8X8I and 8X8J. All data needed to evaluate the conclusions in this paper are present in the paper and/or the Supplementary Materials. All the bacterial strains (E. coli, A. baumannii, and K. pneumoniae) are provided by Zhejiang University pending scientific review and a completed material transfer agreement (MTA). Requests for the strains and plasmids should be submitted to Y.F. (fengyj@zju.edu.cn).
Supplementary Materials
This PDF file includes:
Tables S1 to S3
Figs. S1 to S26
References
REFERENCES AND NOTES
- 1.Sirithanakorn C., Cronan J. E., Biotin, a universal and essential cofactor: Synthesis, ligation and regulation. FEMS Microbiol. Rev. 45, fuab003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belda E., Voland L., Tremaroli V., Falony G., Adriouch S., Assmann K. E., Prifti E., Aron-Wisnewsky J., Debedat J., Le Roy T., Nielsen T., Amouyal C., Andre S., Andreelli F., Bluher M., Chakaroun R., Chilloux J., Coelho L. P., Dao M. C., Das P., Fellahi S., Forslund S., Galleron N., Hansen T. H., Holmes B., Ji B., Krogh Pedersen H., Le P., Le Chatelier E., Lewinter C., Manneras-Holm L., Marquet F., Myridakis A., Pelloux V., Pons N., Quinquis B., Rouault C., Roume H., Salem J. E., Sokolovska N., Sondertoft N. B., Touch S., Vieira-Silva S., MetaCardis C., Galan P., Holst J., Gotze J. P., Kober L., Vestergaard H., Hansen T., Hercberg S., Oppert J. M., Nielsen J., Letunic I., Dumas M. E., Stumvoll M., Pedersen O. B., Bork P., Ehrlich S. D., Zucker J. D., Backhed F., Raes J., Clement K., Impairment of gut microbial biotin metabolism and host biotin status in severe obesity: Effect of biotin and prebiotic supplementation on improved metabolism. Gut 71, 2463–2480 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neophytou C., Pitsouli C., Biotin controls intestinal stem cell mitosis and host-microbiome interactions. Cell Rep. 38, 110505 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Erbach J., Bonn F., Diesner M., Arnold A., Stein J., Schroder O., Aksan A., Relevance of biotin deficiency in patients with inflammatory bowel disease and utility of serum 3-hydroxyisovaleryl carnitine as a practical everyday marker. J. Clin. Med. 11, 1118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin S., Hanson R. E., Cronan J. E., Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat. Chem. Biol. 6, 682–688 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal V., Lin S., Lukk T., Nair S. K., Cronan J. E., Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis. Proc. Natl. Acad. Sci. U.S.A. 109, 17406–17411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S., Cronan J. E., The BioC O-methyltransferase catalyzes methyl esterification of malonyl-acyl carrier protein, an essential step in biotin synthesis. J. Biol. Chem. 287, 37010–37020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estrada P., Manandhar M., Dong S. H., Deveryshetty J., Agarwal V., Cronan J. E., Nair S. K., The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes. Nat. Chem. Biol. 13, 668–674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M., Moynié L., Harrison P. J., Kelly V., Piper A., Naismith J. H., Campopiano D. J., Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters. Nat. Chem. Biol. 13, 660–667 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Cryle M. J., Schlichting I., Structural insights from a P450 carrier protein complex reveal how specificity is achieved in the P450(BioI) ACP complex. Proc. Natl. Acad. Sci. U.S.A. 105, 15696–156701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S., Xu Y., Guan H., Cui T., Liao Y., Wei W., Li J., Hassan B. H., Zhang H., Jia X., Ouyang S., Feng Y., Biochemical and structural characterization of the BioZ enzyme engaged in bacterial biotin synthesis pathway. Nat. Commun. 12, 2056 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y., Cronan J. E., α-Proteobacteria synthesize biotin precursor pimeloyl-ACP using BioZ 3-ketoacyl-ACP synthase and lysine catabolism. Nat. Commun. 11, 5598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manandhar M., Cronan J. E., A canonical biotin synthesis enzyme, 8-amino-7-oxononanoate synthase (BioF), utilizes different acyl chain donors in B. subtilis and E. coli. Appl. Environ. Microbiol. 84, e02084–e02017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y., Yang J., Li W., Song S., Shi Y., Wu L., Sun J., Hou M., Wang J., Jia X., Zhang H., Huang M., Lu T., Gan J., Feng Y., Three enigmatic BioH isoenzymes are programmed in the early stage of mycobacterial biotin synthesis, an attractive anti-TB drug target. PLOS Pathog. 18, e1010615 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji K., Xu Y., Sun J., Huang M., Jia X., Jiang C., Feng Y., Harnessing efficient multiplex PCR methods to detect the expanding Tet(X) family of tigecycline resistance genes. Virulence 11, 49–56 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Neill J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. London: Review on Antimicrobial Resistance (2014). https://amr-review.org/sites/default/files/AMR Review Paper - Tackling a crisis for the health and wealth of nations_1.p.
- 17.Antimicrobial Resistance Collaborators , Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang M., Liu L., Li X., Shi Y., Zhang H., Lu T., Feng Y., Heterogeneity and clinical genomics of blaKPC-2-producing, carbapenem-resistant Pseudomonas aeruginosa. hLife 2, 314–319 (2024). [Google Scholar]
- 19.Hayashi A., Mikami Y., Miyamoto K., Kamada N., Sato T., Mizuno S., Naganuma M., Teratani T., Aoki R., Fukuda S., Suda W., Hattori M., Amagai M., Ohyama M., Kanai T., Intestinal dysbiosis and biotin peprivation induce alopecia through overgrowth of Lactobacillus murinus in mice. Cell Rep. 20, 1513–1524 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Hanachi M., Maghrebi O., Bichiou H., Trabelsi F., Bouyahia N. M., Zhioua F., Belghith M., Harigua-Souiai E., Baouendi M., Guizani-Tabbane L., Benkahla A., Souiai O., Longitudinal and comparative analysis of gut microbiota of Tunisian newborns according to delivery mode. Front. Microbiol. 13, 780568 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiwari D., Park S. W., Essawy M. M., Dawadi S., Mason A., Nandakumar M., Zimmerman M., Mina M., Ho H. P., Engelhart C. A., Ioerger T., Sacchettini J. C., Rhee K., Ehrt S., Aldrich C. C., Dartois V., Schnappinger D., Targeting protein biotinylation enhances tuberculosis chemotherapy. Sci. Transl. Med. 10, eaal1803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bockman M. R., Mishra N., Aldrich C. C., The biotin biosynthetic pathway in Mycobacterium tuberculosis is a validated target for the development of antibacterial agents. Curr. Med. Chem. 27, 4194–4232 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Carfrae L. A., MacNair C. R., Brown C. M., Tsai C. N., Weber B. S., Zlitni S., Rao V. N., Chun J., Junop M. S., Coombes B. K., Brown E. D., Mimicking the human environment in mice reveals that inhibiting biotin biosynthesis is effective against antibiotic-resistant pathogens. Nat. Microbiol. 5, 93–101 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Shi Y., Cao Q., Sun J., Hu X., Su Z., Xu Y., Zhang H., Lan L., Feng Y., The opportunistic pathogen Pseudomonas aeruginosa exploits bacterial biotin synthesis pathway to benefit its infectivity. PLOS Pathog. 19, e1011110 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cronan J. E., The classical, yet controversial, first enzyme of lipid synthesis: E. coli acetyl-CoA carboxylase. Microbiol. Mol. Biol. Rev. 85, e0003221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan M., McGowen K., Liu Q., Akusobi C., Young D., Mayfield J., Raman S., Wolf I. D., Moody D. B., Aldrich C. C., Muir A., Rubin E. J., Biotin-dependent cell envelope remodelling is required for Mycobacterium abscessus survival in lung infection. Nat. Microbiol. 8, 481–497 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carfrae L. A., Rachwalski K., French S., Gordzevich R., Seidel L., Tsai C. N., Tu M. M., MacNair C. R., Ovchinnikova O. G., Clarke B. R., Whitfield C., Brown E. D., Inhibiting fatty acid synthesis overcomes colistin resistance. Nat. Microbiol. 8, 1026–1038 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Cao X., Zhu L., Hu Z., Cronan J. E., Expression and activity of the BioH esterase of biotin synthesis is independent of genome context. Sci. Rep. 7, 2141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y., Zhang H., Cronan J. E., Profligate biotin synthesis in α-proteobacteria – A developing or degenerating regulatory system? Mol. Microbiol. 88, 77–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Q., Li X., Zou T., Zhang H., Wang Y., Gao R., Li Z., He J., Feng Y., Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol. Microbiol. 94, 1006–1023 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Wei W., Zhang Y., Gao R., Li J., Xu Y., Wang S., Ji Q., Feng Y., Crystal structure and acetylation of BioQ suggests a novel regulatory switch for biotin biosynthesis in Mycobacterium smegmatis. Mol. Microbiol. 109, 642–662 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Haushalter R. W., Phelan R. M., Hoh K. M., Su C., Wang G., Baidoo E. E., Keasling J. D., Production of odd-carbon dicarboxylic acids in E. coli using an engineered biotin-fatty acid biosynthetic pathway. J. Am. Chem. Soc. 139, 4615–4618 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Wang Z., Chen Y., Hua X., Yu Y., Ji Q., A highly efficient CRISPR-Cas9-based genome engineering platform in A. baumannii to understand the H2O2-sensing mechanism of OxyR. Cell Chem. Biol. 26, 1732–1742 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Harder M. E., Ladenson R. C., Schimmel S. D., Silbert D. F., Mutants of E. coli with temperature-sensitive malonyl coenzyme A-acyl carrier protein transacylase. J. Biol. Chem. 249, 7468–7475 (1974). [PubMed] [Google Scholar]
- 35.Holm L., Dali server: Structural unification of protein families. Nucleic Acids Res. 50, 210–215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleary P. P., Campbell A., Deletion and complementation analysis of biotin gene cluster of E. coli. J. Bacteriol. 112, 830–839 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleary P. P., Campbell A., Chang R., Location of promoter and operator sites in the biotin gene cluster of E. coli. Proc. Natl. Acad. Sci. U.S.A. 69, 2219–2223 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borchardt R. T., Eiden L. E., Wu B., Rutledge C. O., Sinefungin, a potent inhibitor or S-adenosylmethionine: Protein O-methyltransferase. Biochem. Biophys. Res. Commun. 89, 919–924 (1979). [DOI] [PubMed] [Google Scholar]
- 39.Wei W., Guan H., Zhu T., Zhang S., Fan C., Ouyang S., Feng Y., Molecular basis of BioJ, a unique gatekeeper in bacterial biotin synthesis. iScience 19, 796–808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oefner C., Schulz H., D’Arcy A., Dale G. E., Mapping the active site of E. coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 62, 613–618 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Serre L., Verbree E. C., Dauter Z., Stuitje A. R., Derewenda Z. S., The E. coli malonyl-CoA:acyl carrier protein transacylase at 1.5-A resolution. Crystal structure of a fatty acid synthase component. J. Biol. Chem. 270, 12961–12964 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Novović K., Jovčić B., Colistin resistance in A. baumannii: Molecular mechanisms and epidemiology. Antibiotics 12, 516 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Srinivas S., Xu Y., Wei W., Feng Y., Genetic and biochemical mechanisms for bacterial lipid A modifiers associated with polymyxin resistance. Trends Biochem. Sci. 44, 973–988 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Xu Y., Lin J., Cui T., Srinivas S., Feng Y., Mechanistic insights into transferable polymyxin resistance among gut bacteria. J. Biol. Chem. 293, 4350–4365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y., Wei W., Lei S., Lin J., Srinivas S., Feng Y., An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio 9, e02317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zlitni S., Ferruccio L. F., Brown E. D., Metabolic suppression identifies new antibacterial inhibitors under nutrient limitation. Nat. Chem. Biol. 9, 796–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye H., Li Y., Li Z., Gao R., Zhang H., Wen R., Gao G. F., Hu Q., Feng Y., Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio 7, e00177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao R., Hu Y., Li Z., Sun J., Wang Q., Lin J., Ye H., Liu F., Srinivas S., Li D., Zhu B., Liu Y. H., Tian G. B., Feng Y., Dissemination and mechanism for the MCR-1 colistin resistance. PLOS Pathog. 12, e1005957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma F., Shen C., Zheng X., Liu Y., Chen H., Zhong L., Liang Y., Liao K., Xia Y., Tian G. B., Yang Y., Identification of a novel plasmid carrying mcr-4.3 in an A. baumannii strain in China. Antimicrob. Agents Chemother. 63, e00133-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., Hou M., Xu Y., Srinivas S., Huang M., Liu L., Feng Y., Action and mechanism of the colistin resistance enzyme MCR-4. Commun. Biol. 2, 36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H., Wei W., Huang M., Umar Z., Feng Y., Definition of a family of nonmobile colistin resistance (NMCR-1) determinants suggests aquatic reservoirs for MCR-4. Adv. Sci. 6, 1900038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bai S., Li R., Yu Y., Liao X. P., Sporadic dissemination of mcr-8-ST11 Klebsiella pneumoniae isolates in China. Enferm. Infecc. Microbiol. Clin. 40, 95–97 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Ullah S., Ji K., Li J., Xu Y., Jiang C., Zhang H., Huang M., Feng Y., Characterization of NMCR-2, a new non-mobile colistin resistance enzyme: Implications for an MCR-8 ancestor. Environ. Microbiol. 23, 844–860 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Liu L., Lou N., Liang Q., Xiao W., Teng G., Ma J., Zhang H., Huang M., Feng Y., Chasing the landscape for intrahospital transmission and evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae. Sci. Bull. 68, 3027–3047 (2023). [DOI] [PubMed] [Google Scholar]
- 55.Nadasy K. A., Domiati-Saad R., Tribble M. A., Invasive Klebsiella pneumoniae syndrome in North America. Clin. Infect. Dis. 45, e25–e28 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Li W., Sun G., Yu Y., Li N., Chen M., Jin R., Jiao Y., Wu H., Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 58, 225–232 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Catalan-Najera J. C., Garza-Ramos U., Barrios-Camacho H., Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence 8, 1111–1123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qu T. T., Zhou J. C., Jiang Y., Shi K. R., Li B., Shen P., Wei Z. Q., Yu Y. S., Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 15, 161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y. T., Chang H. Y., Lai Y. C., Pan C. C., Tsai S. F., Peng H. L., Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337, 189–198 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Yang X., Xie M., Xu Q., Ye L., Yang C., Dong N., Chan E. W., Zhang R., Chen S., Transmission of pLVPK-like virulence plasmid in Klebsiella pneumoniae mediated by an Incl1 conjugative helper plasmid. iScience 25, 104428 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang H. L., Chiang M. K., Liou W. J., Chen Y. T., Peng H. L., Chiou C. S., Liu K. S., Lu M. C., Tung K. C., Lai Y. C., Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur. J. Clin. Microbiol. Infect. Dis. 29, 689–698 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Cheng H. Y., Chen Y. S., Wu C. Y., Chang H. Y., Lai Y. C., Peng H. L., RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192, 3144–3158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng Y., Napier B. A., Manandhar M., Henke S. K., Weiss D. S., Cronan J. E., A Francisella virulence factor catalyses an essential reaction of biotin synthesis. Mol. Microbiol. 91, 300–314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woong Park S., Klotzsche M., Wilson D. J., Boshoff H. I., Eoh H., Manjunatha U., Blumenthal A., Rhee K., Barry C. E., Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLOS Pathog. 7, e1002264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai R., Wilson D. J., Geders T. W., Aldrich C. C., Finzel B. C., Inhibition of Mycobacterium tuberculosis transaminase BioA by aryl hydrazines and hydrazides. ChemBioChem 15, 575–586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin S., Cronan J. E., Closing in on complete pathways of biotin biosynthesis. Mol. Biosyst. 7, 1811–1821 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Cronan J. E., Lin S., Synthesis of the α,ω-dicarboxylic acid precursor of biotin by the canonical fatty acid biosynthetic pathway. Curr. Opin. Chem. Biol. 15, 407–413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pugh C. S., Borchardt R. T., Stone H. O., Sinefungin, a potent inhibitor of virion mRNA(guanine-7-)-methyltransferase, mRNA(nucleoside-2′-)-methyltransferase, and viral multiplication. J. Biol. Chem. 253, 4075–4077 (1978). [PubMed] [Google Scholar]
- 69.Yadav M. K., Park S. W., Chae S. W., Song J., Sinefungin, a natural nucleoside analogue of S-adenosylmethionine, inhibits Streptococcus pneumoniae biofilm growth. Biomed. Res. Int. 156987, 10.1155/2014/156987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalimouttou S., Skiba M., Bon P., Pierre D., Arnaud P., Lahiani-Skiba M., Sinefungin-PLGA nanoparticles: Drug loading, characterization, in vitro drug release and in vivo studies. J. Nanosci. Nanotechnol. 9, 150–158 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Feng Y., Transferability of MCR-1/2 polymyxin resistance: Complex dissemination and genetic mechanism. ACS Infect. Dis. 4, 291–300 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Wang Q., Sun J., Li J., Ding Y., Li X. P., Lin J., Hassan B., Feng Y., Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 5, 70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hood M. I., Becker K. W., Roux C. M., Dunman P. M., Skaar E. P., Genetic determinants of intrinsic colistin tolerance in A. baumannii. Infect. Immun. 81, 542–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Napier B. A., Meyer L., Bina J. E., Miller M. A., Sjostedt A., Weiss D. S., Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc. Natl. Acad. Sci. U.S.A. 109, 18084–18089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Y., Cronan J. E., A new member of the E. coli fad regulon: Transcriptional regulation of fadM (ybaW). J. Bacteriol. 191, 6320–6328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi-Rhee E., Cronan J. E., A nucleosidase required for in vivo function of the S-adenosyl-L-methionine radical enzyme, biotin synthase. Chem. Biol. 12, 589–593 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Budnick J. A., Bina X. R., Bina J. E., Complete genome sequence of Klebsiella pneumoniae strain ATCC 43816. Microbiol. Resour. Announc. 10, e01441-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Økstad O., Hegna I., Lindbäck T., Rishovd A., Kolstø A., Genome organization is not conserved between B. cereus and B. subtilis. Microbiology 145, 621–631 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Guerrero F., Ciragan A., Iwai H., Tandem SUMO fusion vectors for improving soluble protein expression and purification. Protein Expr. Purif. 116, 42–49 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Wang R. F., Kushner S. R., Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in E. coli. Gene 100, 195–199 (1991). [PubMed] [Google Scholar]
- 81.Cronan J. E., The acyl carrier proteins of lipid synthesis are busy having other affairs. Biochem. J. 480, 855–873 (2023). [DOI] [PubMed] [Google Scholar]
- 82.Thomas J., Rigden D. J., Cronan J. E., Acyl carrier protein phosphodiesterase (AcpH) of E. coli is a non-canonical member of the HD phosphatase/phosphodiesterase family. Biochemistry 46, 129–136 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Quadri L. E., Weinreb P. H., Lei M., Nakano M. M., Zuber P., Walsh C. T., Characterization of Sfp, a B. subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595 (1998). [DOI] [PubMed] [Google Scholar]
- 84.Cornell K. A., Riscoe M. K., Cloning and expression of E. coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: Identification of the pfs gene product. Biochim. Biophys. Acta 1396, 8–14 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Yin Y., Li R., Liang W. T., Zhang W. B., Hu Z., Ma J. C., Wang H. H., Of its five acyl carrier proteins, only AcpP1 functions in Ralstonia solanacearum fatty acid synthesis. Front. Microbiol. 13, 1014971 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakano M. M., Marahiel M. A., Zuber P., Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in B. subtilis. J. Bacteriol. 170, 5662–5668 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takahashi Y., Mizoi J., Toh E. A., Kikuchi Y., Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J. Biochem. 128, 723–725 (2000). [DOI] [PubMed] [Google Scholar]
- 88.Mossessova E., Lima C. D., Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Choi-Rhee E., Schulman H., Cronan J. E., Promiscuous protein biotinylation by E. coli biotin protein ligase. Protein Sci. 13, 3043–3050 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bunkoczi G., Echols N., McCoy A. J., Oeffner R. D., Adams P. D., Read R. J., Phaser.MRage: Automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 69, 2276–2286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Potterton E., Briggs P., Turkenburg M., Dodson E., A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 (2003). [DOI] [PubMed] [Google Scholar]
- 92.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 94.Hang X., Zeng Q., Zeng L., Jia J., Bi H., Functional replacement of the BioC and BioH proteins of E. coli biotin precursor biosynthesis by E. chaffeensis novel proteins. Curr. Microbiol. 76, 626–636 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakano M. M., Corbell N., Besson J., Zuber P., Isolation and characterization of sfp: A gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 232, 313–321 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3
Figs. S1 to S26
References