Abstract
It has been established that essential amino acids (EAA) regulate protein synthesis in mammary epithelial cells by rapidly altering the phosphorylation state of translation factors. However, the long-term transcriptional response to EAA supply has been investigated much less. Eight transcription factors were selected as candidate mediators of EAA effects on mammary cell function via the amino acid response (ATF4, ATF6), mitogen-activated protein kinase (JUN, FOS, EGR1), and mechanistic target of rapamycin complex 1 (MYC, HIF1A, SREBF1). The objective was to determine if and when expression of these candidate genes was affected in primary cultures of bovine mammary epithelial cells more than 24 h after imposing an EAA deficiency, and to evaluate effects of EAA deficiency on protein synthesis, endoplasmic reticulum size, cell proliferation, and lipogenesis. Differentiated cells were cultured in 1 of 3 treatment media representing normal physiological concentrations of all amino acids (CTL), low lysine (LK), or low methionine (LM) for 24, 40, 48, or 60 h. Both LK and LM suppressed protein synthesis and activated ATF4 expression, indicating the classic amino acid response pathway had been triggered. However, there was no effect of LK or LM on endoplasmic reticulum size, possibly related to elevated ATF6 expression on LM. Expression of early response genes JUN, FOS, EGR1 and MYC was not elevated by EAA deficiency but LM decreased EGR1 expression. LM also increased expression of HIF1A. The EGR1 and HIF1A expression results are consistent with the decrease in cell proliferation rate observed. Variable responses in SREBF1 expression to LK and LM at different timepoints may have contributed to a lack of effect on lipogenesis rates. These findings indicate that EAA deficiency may inhibit mammary protein synthesis and cell proliferation through transcription factors.
Introduction
Lysine (Lys) and methionine (Met) are essential amino acids (EAA) often found to be in deficient supply for maximal milk protein synthesis in the mammary glands of lactating dairy cows [1]. It is well-documented that EAA can regulate milk protein yield by altering the phosphorylation state of translation factors through mechanistic target of rapamycin complex 1 (mTORC1), integrated stress response (ISR), and glycogen synthase kinase-3 signaling [2–5]. This translational response does not appear to persist for long periods (days and weeks) over which dietary effects on milk protein yields are sustained and may just be part of the initial response [4,6]. In addition to rapid effects on mRNA translation, there may be changes in gene transcription that contribute to the long-term effects of EAA on activities of the mammary epithelial cell. However, the transcriptional response has been investigated much less.
The ISR is the name given to the group of reactions induced by phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α). Four different eIF2α kinases sense a diverse array of nutritional, endoplasmic reticulum (ER), viral, and redox stresses and cause a decrease in global mRNA translation but an increase in translation of the mRNA for activating transcription factor 4 (ATF4) [7]. ATF4 belongs to the basic leucine-zipper family of transcription factors, including ATF6, FOS, and JUN, with which it homo- or hetero-dimerizes to induce expression of amino acid transporters, mRNA translation factors, and ER-resident proteins in what has been called the amino acid response pathway (AARP) [8]. ATF6 is itself an ATF4 target [9] whose gene product can be activated by the ISR [10]. An alternative to the ATF4 pathway for transcription factor activation by EAA deficiency involves RAS/RAF/MEK/ERK signaling that stimulates the expression of FOS, JUN, and EGR1 [11–13]. Furthermore, mTORC1 inactivation by low EAA concentrations can decrease the expression of several genes encoding transcription factors such as ATF4, MYC, HIF1A, and SREBF1 [14–17].
The transcriptional response to EAA deficiency has been studied little in the mammary glands. It has been demonstrated that the AARP is stimulated in mammary epithelial cells subjected to EAA deficiency through the eIF2α kinase GCN2 [18]. Supplementing all EAA to lactating cows [19] or depleting arginine [20] produced gene expression patterns in the mammary glands consistent with the operation of the AARP. Addition of valine or methionine to cultures of mammary epithelial cells enhanced expression of the SREBF1 product, SREBP-1c, and fat accumulation, likely through mTORC1 activation [21,22]. Of the suite of potentially EAA-responsive transcription factors described above, ATF4, ATF6 and JUN regulate ER biogenesis [23–26], FOS, JUN, EGR1, MYC and HIF1A regulate cell proliferation [27–32], and SREBP-1c regulates lipogenesis [33,34]. All four cellular processes are important determinants of milk yield and protein and fat content.
For this study, we hypothesized that the deprivation of Lys or Met can inhibit protein synthesis, ER biogenesis, cell proliferation and lipogenesis through a chronic, long-term (i.e. after 24 h) transcriptional response in bovine mammary epithelial cells (BMEC). The transcription factor genes of interest were ATF4, ATF6, JUN, FOS, EGR1, MYC, HIF1A, and SREBF1. Objectives were to evaluate if and when their expression in BMEC is affected after imposing an EAA deficiency in vitro, and to evaluate effects of EAA deficiency on protein synthesis, ER size, cell proliferation and lipogenesis.
Materials and methods
Materials
All reagents and chemicals for cell culture were procured from Sigma-Aldrich (Oakville, ON, Canada) or Thermo Fisher Scientific (Waltham, MA, USA), except where noted. Kits for sample preparation and analysis were purchased from Thermo Fisher Scientific. Collagenase Type III was obtained from Worthington Biochemical Corporation (Lakewood, NJ, USA). Medium 170 was acquired from U.S. Biological (Salem, MA, USA). Radioisotopes were sourced from PerkinElmer (Waltham, MA, USA).
Isolation and culture of bovine mammary epithelial cells
To produce cultures of BMEC, organoids prepared from mammary tissue were incubated in growth media for approximately 2 weeks to allow new cells to proliferate and occupy the plate. Multiple passages and trypsin treatment were used to remove fibroblasts from the cultures and the remaining BMEC were differentiated with lactogenic hormones in preparation for experimental treatments. Lactating Holstein cows were obtained from the Ontario Dairy Research Centre (Elora, ON) and were slaughtered humanely with a captive bolt followed by exsanguination. The protocol of this study was approved by the University of Guelph Animal Care Committee (Animal Utilization Protocol #4307). To obtain organoids, mammary tissue was harvested from a rear quarter at slaughter and promptly placed in 1:1 (vol/vol) Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) supplemented with 1× antibiotics/antimycotics (penicillin at 100 units/ml, streptomycin at 100 μg/ml, and amphotericin B at 0.25 μg/ml). The tissue was transported on ice to the laboratory and transferred into 100-mm cell plates. Visible fat, blood, milk, and extraneous connective tissue were removed with scalpel blades. The remaining mammary tissue was minced with scalpels into approximately 1 mm3 fragments in a sterile environment in 5 ml of Hanks’ balanced salt solution containing 1× antibiotics/antimycotics (HBSS+). Minced tissue was transferred into a 50-ml centrifuge tube and resuspended in ice-cold HBSS+. After hand shaking the contents, the tube was placed on ice for 5 min. This procedure was repeated 4 to 5 times until the medium was clear. The washed tissue fragments were then transferred for enzymatic dissociation into sterile DMEM/F12 supplemented with 300 U/ml collagenase Type III, 1 mg/ml DNase I and 1× antibiotics/antimycotics. Digestion was carried out at 37°C with continuous shaking at 120 rpm for 4 h. Post-digestion, the preparation was filtered through a sterile sieve with 200-μm mesh and transferred into a 50-ml centrifuge tube for centrifugation at 80 × g for 30 s at 4°C. The supernatant was discarded, and the pellet of highly enriched epithelial organoids was either cryopreserved in a -80 °C freezer or seeded onto 100 mm collagen-coated dishes to allow BMEC to proliferate. If previously frozen, organoids were thawed at room temperature and then seeded onto collagen. The growth medium was composed of 1:1 (vol/vol) DMEM/F12:MCDB170, 10% fetal bovine serum, 0.1% (wt/vol) albumax II, 7.5 μg/mL bovine insulin, 0.3 μg/mL hydrocortisone, 5 ng/mL recombinant human epidermal growth factor, 2.5 μg/mL bovine apo-transferrin, 5 μM isoproterenol, 5 pM 3,3′,5-triiodo-l-thyronine, 0.5 pM β-estradiol, 0.1 nM oxytocin, and 1× antibiotics/antimycotics. To allow growth of new BMEC from the mammary organoids, 5 ml media were added every 3 d in the first week without discarding the medium in the plates. Once most of the organoids were attached to the dish, then medium was replaced every 2 d. Dishes were incubated at 37°C in a humidified atmosphere of 5% CO2. When cells reached 50–60% confluency, they were passaged onto 100-mm cell culture plates with fresh growth medium containing 0.25% fetal bovine serum. To remove fibroblasts, cultures were incubated with 0.15% trypsin plus 0.02% EDTA for 2 minutes at 37°C. During the incubation, fibroblasts detached more rapidly than mammary epithelial cells, as observed under a microscope. The trypsin-EDTA solution containing detached fibroblasts was carefully aspirated, ensuring minimal disruption to the adherent mammary epithelial cells. Complete growth medium was then added to neutralize the trypsin and preserve the epithelial cell population. All experiments were performed in third passage BMEC which were randomly assigned and seeded onto 6-well plates and grown to confluency. When all wells reached confluency, cells were differentiated to a lactational phenotype by incubating for 5 d in DMEM containing 3.5 mM D-glucose, 2 mM sodium acetate, 200 μM L-glutamine, and supplemented with lactogenic hormones (5 μg/mL each of bovine insulin, prolactin, and hydrocortisone), 5 μg/mL apo-transferrin, 0.5 mg/mL BSA, and 1× antibiotics/antimycotics. Media were replaced every 2 d.
Differentiated BMEC were cultured in triplicate in each of the three treatment media representing normal physiological concentrations of all amino acids (CTL), low Lys (LK), or low Met (LM) for 24, 40, 48, or 60 h. The base medium was DMEM containing 3.5 mM D-glucose, 2 mM sodium acetate, and 200 μM L-glutamine supplemented with lactogenic hormones (5 μg/mL each of bovine insulin, prolactin, and hydrocortisone), 5 μg/mL apo-transferrin, 0.5 mg/mL BSA, and 1× antibiotics/antimycotics. All amino acids except Met and Lys were dissolved in the base medium at their normal physiological concentrations [35,36] of (in μM) 180 L-alanine, 80 L-arginine, 50 L-Asparagine, 10 L-Aspartic acid, 12 L-Cysteine; 60 L-Glutamic acid, 170 L-Glutamine, 200 L-Glycine, 40 L-Histidine, 120 L-Isoleucine, 180 L-Leucine, 50 L-Phenylalanine, 70 L-Proline, 70 L-Serine, 110 L-Threonine, 30 L-Tryptophan, 50 L-Tyrosine, and 240 L-Valine. Treatments were base medium with 1) 80 μM L-Lys and 20 μM L-Met (CTL), 2) 20 μM L-Lys and 20 μM L-Met (LK), and 3) 80 μM L-Lys and 5 μM L-Met (LM). At each termination time, wells were rinsed twice and then cells were harvested into either lysis buffer from the RNA isolation kit for gene expression analysis, or RIPA buffer with protease inhibitor cocktail for protein analysis. All samples were stored at -80 °C until RNA isolation and protein analysis were performed. Experiments were performed on 6 different occasions and the cells were from a different cow for each occasion.
Gene expression
Total RNA was extracted from cells with the Invitrogen PureLink RNA mini-Kit according to the manufacturer’s protocol. The RNA concentration and purity were quantified by measuring absorbance at 260 and 260/280 nm, respectively, on a NanoDrop OneC microvolume spectrophotometer (Thermo Fisher Scientific). All the 260/280 ratios were above 1.9. All RNA samples were tested for RNA integrity by using Agilent TapeStation 4150 at Advanced Analysis Centre in University of Guelph (Guelph, Canada), and the RNA integrity number were above 8. Isolated RNA was treated with DNase to avoid DNA contamination following the Invitrogen DNase I kit protocol. cDNA was synthesized from 1,000 ng DNase-treated RNA using the Applied Biosystems high-capacity RNA-to-cDNA kit. Primers of candidate genes were designed with the NCBI Primer-BLAST tool to yield qPCR amplification products of 80 to 150 bp (Table 1). Target genes were ATF4, ATF6, FOS, JUN, EGR1, MYC, HIF1A, and SREBF1, and reference genes were PPIA, RPS6KB1, and UXT. Primer oligos were purchased from Integrated DNA Technologies (Iowa, USA). Real-Time PCR was performed using PerfeCta SYBR Green FastMix with a StepOnePlus Real-Time PCR system (Applied Biosystems, Waltham, MA, USA). Technical duplicates were assayed for each sample. Relative gene expression was obtained by normalizing cycle threshold values for target genes to the geometric means of those for the three reference genes.
Table 1. Primer sequences used in qPCR.
| Transcript | Accession number | Primer Sequence |
|---|---|---|
| ATF4 | NM_001034342 | 5’-CATCATGGGTTCTCCTGCGA |
| 3’-GGAGAAAGCATCCTCCTTGC | ||
| ATF6 | XM_024989876 | 5’-GAACGTGTGTTTCGGGGGAA |
| 3’-GCAAACAGGGCAGAATCCCA | ||
| FOS | NM_182786 | 5’-GCGAATCCGAAGGGAAAGGA |
| 3’-GTTGGTCTGTCTCCGCTTGG | ||
| JUN | NM_001077827 | 5’-CGAAGTGACGGACTGTTCTATG |
| 3’-CCGTTGCTGGACTGTATGATTA | ||
| EGR1 | NM_001045875 | 5’-CACCTGACCGCAGAGTCCTTT |
| 3’-GGTGGTTTGGCTGGGGTAA | ||
| MYC | NM_001046074 | 5’-CAGAGAAGCTCTTCTGCCTTTT |
| 3’-CATCGCTGCAAGCCCGTATT | ||
| HIF1A | NM_174339 | 5’-CGGGCACCGATTCACCAT |
| 3’-TTCGACGTTCAGAACTTATCTTTTT | ||
| SREBF1 | NM_001113302 | 5’-GCTGACCGACATAGAAGACATGC |
| 3’-TCAGGACTGGCAGGGTCTG | ||
| UXT | NM_001037471 | 5’-TTGACACAGTGGTCCCAGAC |
| 3’-CTTGGTGAGGTTGTCGCTGA | ||
| PPIA | NM_178320 | 5’-CGCGTCTCTTTTGAGCTGTT |
| 3’-TCGCCATAGATGGACTTGCC | ||
| RPS6 | NM_001015548 | 5’-ATGGCAGGGGTGTTTGACAT |
| 3’-ACAATGTTCCATGCCAAGTTCA |
Cell activity
Eighteen wells of confluent differentiated BMEC were cultured with each of CTL, LK and LM treatment media for 48 and 60 h. Rates of protein synthesis, cell proliferation and lipogenesis were determined by quantifying, respectively, the incorporation of label from L-[2,3,4,5,6-3H]phenylalanine and [methyl-3H]thymidine into an acid precipitate, and [1-14C]acetate into a hexane:isopropanol extract, over the last 2 h of the treatment period as follows. After 46 and 58 h of treatment, [3H]phenylalanine, [3H]thymidine and Na-[1-14C]acetate were each added to a final concentration of 1 μCi/ml into 3 different wells, for a total of 9 wells per treatment and timepoint. Two hours after isotope addition, wells were washed 3 times with PBS to remove extracellular tracer and cells were harvested into 0.5 ml RIPA lysis buffer and homogenized. Cells for protein and DNA analysis were harvested at 48 and 60 h from 9 additional wells per treatment into 0.2 ml RIPA buffer containing protease inhibitor cocktail and stored immediately at -80 °C.
DNA concentrations in 10 μl cell lysate were measured with the DNA Qubit Assay. Protein and DNA in homogenates from [3H]phenylalanine and [3H]thymidine incubations were precipitated with 0.5 ml ice-cold 20% TCA on ice for 30 min and then centrifuged at 15,000 × g for 5 min. Pellets were washed 3 times by resuspension and centrifugation with 5% TCA. After washing, pellets were dissolved in 0.5 M NaOH and 0.5 ml were transferred to 5 ml scintillation cocktail for counting in a 3H window. Rates of protein synthesis and cell proliferation were expressed as disintegrations per minute (DPM) per nanogram of DNA.
Lipid in lysates from [14C]acetate incubations was extracted into 200 ml hexane: isopropanol (3:2 vol/vol). 1 ml of the solvent layer was transferred into 5 ml scintillation cocktail and counted on a Beckman liquid scintillation counter for 10 min per sample. Lipid synthesis rate was expressed as DPM per nanogram of DNA.
Western blot
Cell proteins were extracted in RIPA buffer containing protease inhibitor cocktail and thawed on ice for 15 min. A 200-μl aliquot of the cell lysate was taken to measure protein concentration with a BCA protein assay kit using BSA as standard. Another aliquot of cell lysate was mixed with 2X Laemmli sample buffer and heated at 70°C for 10 min. 30 μg protein in cell lysates were resolved by SDS-PAGE and then transferred onto polyvinylidene difluoride membranes. Membranes were blocked in 5% (wt/vol) non-fat milk in Tris-buffered saline containing 0.1% (vol/vol) Tween 20 (TBST) at room temperature for 1 h and then incubated for 16 h with primary antibodies against the ER tracker protein disulfide isomerase (PDI; rabbit mAb #3501, RRID: AB_2156433, Cell Signaling Technologies, Danvers, MA, USA) and a loading control GAPDH (rabbit mAb #2118, RRID:AB_561053) diluted in 5% non-fat milk in TBS-T at 4°C. After washing 3 times in TBS-T for 5 min, membranes were incubated with secondary antibodies diluted 1:10,000 in 5% non-fat milk in TBS-T at room temperature for 1 h with constant shaking. After washing 6 times in TBS-T, bound horseradish peroxidase-linked secondary antibodies were visualized by chemiluminescence (Bio-Rad Laboratories, Mississauga, ON, Canada). Signal intensities were quantified using Image Lab Software (Bio-Rad Laboratories) and PDI abundance was expressed relative to GAPDH.
Statistical analysis
Observations were subjected to analysis of variance by the GLIMMIX procedure of SAS Studio (SAS Institute Inc., Cary, NC, USA). Cells were cultured on 6 different occasions from 6 different cows with sampling timepoints ranging from 24 to 60 h. Not all timepoints were sampled at every occasion so a meta-analysis approach was used for ANOVA with study as a fixed effect, according to the following model:
where μ is the overall mean, studyi is the random effect of study (i = 1 to 6), timej is the fixed effect of time (j = 24, 40, 48, 60, or j = 48, 60), trtk is the fixed effect of treatment (k = 1 to 3), and εijk is random variation. Normality of residuals was confirmed visually with density histograms and normal probability plots. Effects of treatments within and across timepoints were estimated as linear contrasts between CTL, LK, and LM. Contrasts were considered statistically significant when P ≤ 0.05 and trends when 0.05 < P ≤ 0.15.
Results
Gene expression
To evaluate if and when the selected transcription factors ATF4, ATF6, JUN, FOS, EGR1, MYC, HIF1A and SREBF1 were affected by Lys or Met deprivation, their mRNA expression was measured at 24, 40, 48 and 60 h after Lys or Met subtraction to ¼ the normal plasma concentration. All data supporting this study are available on Borealis and can be accessed at https://doi.org/10.5683/SP3/CEHUCP.
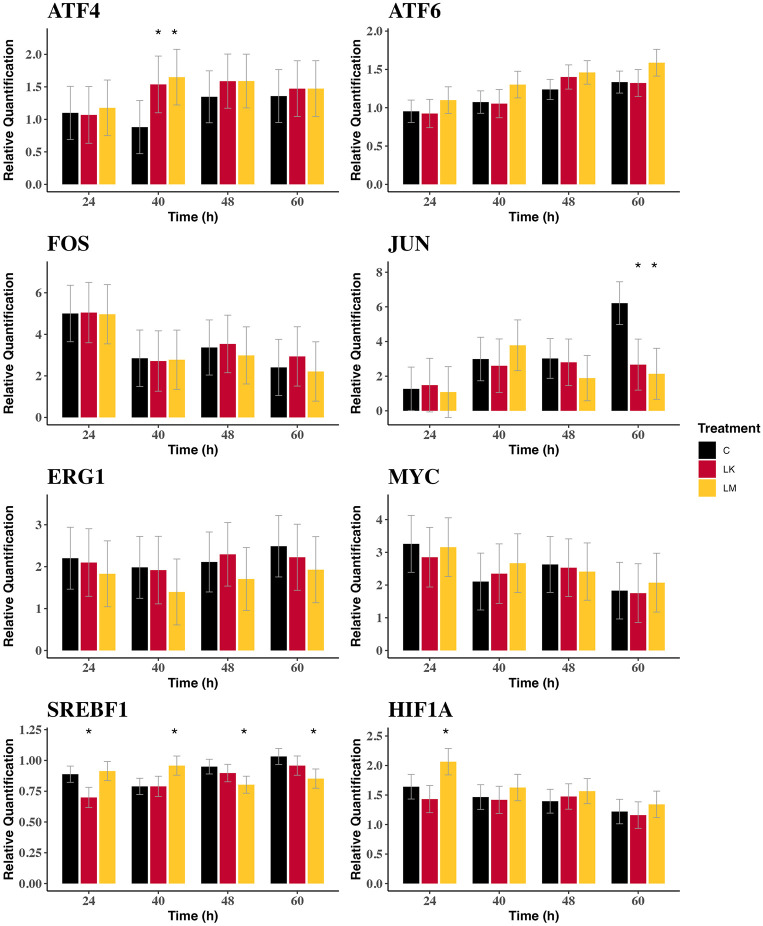
Compared to CTL, LK and LM both increased ATF4 expression (P < 0.05; Table 2), particularly at 40 h (P < 0.01; Fig 1). LM increased ATF6 expression across all timepoints (P = 0.012; Table 2) while LK did not (P = 0.78). LM tended to increase ATF6 expression at 40 (P = 0.08) and 60 h (P = 0.11) compared to CTL (Fig 1). The expression of JUN tended to increase with LM (P = 0.08) but was not affected by LK (P = 0.17). At 60 h, both LK and LM exhibited lower expression of JUN (P < 0.01) compared to CTL (Fig 1) which appears to have been due to an increase in JUN expression in CTL incubations (Ptime < 0.01) that did not occur on LK or LM (Ptime > 0.05). The expression of JUN was significantly affected by time (P = 0.03). There was no effect of either LK or LM on the expression of FOS or MYC (Table 2; Fig 1). The expression of EGR1 was decreased by LM (P = 0.04; Table 2) but there was no significant effect at any single timepoint for either of the EAA deficiency treatments (Fig 1). LM increased the expression of HIF1A across all timepoints (P < 0.01; Table 2), and at 24 h (P < 0.01; Fig 1), and tended to increase HIF1A expression at 48 h (P = 0.12). SREBF1 expression was decreased by LK (P = 0.04) but not by LM. LK decreased SREBF1 expression at 24 h (P = 0.01; Fig 1) while LM increased SREBF1 expression at 40 h (P = 0.02) and decreased its expression at 48 and 60 h (P = 0.01).
Table 2. Gene expression of candidate transcription factors in bovine mammary epithelial cells across all timepoints of 24, 40, 48 and 60 h after lysine or methionine subtraction to ¼ the normal concentration1.
| Gene | Treatment2 | SEM | P-value3 | |||
|---|---|---|---|---|---|---|
| CTL | LK | LM | LK | LM | ||
| ATF4 | 1.17 | 1.42 | 1.47 | 0.40 | 0.04 | 0.01 |
| ATF6 | 1.15 | 1.18 | 1.36 | 0.13 | 0.78 | 0.01 |
| FOS | 3.41 | 3.56 | 3.24 | 1.30 | 0.71 | 0.65 |
| JUN | 3.37 | 2.39 | 2.22 | 1.10 | 0.17 | 0.09 |
| EGR1 | 2.20 | 2.13 | 1.72 | 0.71 | 0.81 | 0.05 |
| MYC | 2.46 | 2.37 | 2.58 | 0.85 | 0.69 | 0.54 |
| HIF1A | 1.43 | 1.37 | 1.65 | 0.20 | 0.42 | <0.01 |
| SREBF1 | 0.91 | 0.84 | 0.88 | 0.06 | 0.04 | 0.33 |
1Values are least squares means ± SE (n = 12) in arbitrary units.
2CTL = control; LK = low lysine; LM = low methionine.
3Linear contrast of LK or LM vs. CTL.
Fig 1. Gene expression of candidate transcription factors in bovine mammary epithelial cells.
(A) Gene expressions were measured at 24, 40, 48, 60 h after lysine or methionine subtraction to ¼ the normal concentration. (B) Values are least square means ± SE (n = 3) in arbitrary units. (C) CTL = control; LK = low lysine; LM = low methionine. (D) *P ≤ 0.05, †0.05 < P ≤ 0.15 compared to CTL.
Cell activity
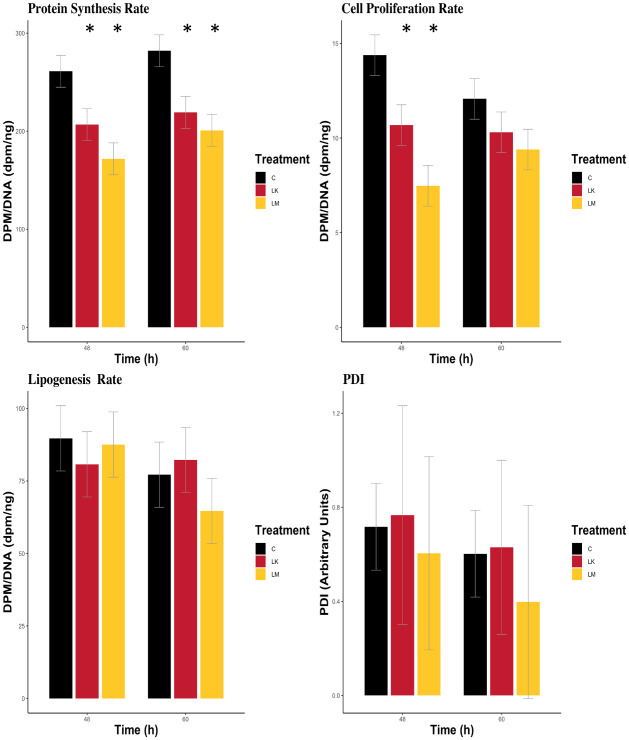
As presented in Fig 2 and Table 3, LK decreased protein synthesis 21% at 48 h (P = 0.04) and 22% at 60 h (P = 0.02), while LM decreased protein synthesis 34% at 48 h (P < 0.01) and 29% at 60 h (P < 0.01). Neither LK nor LM had any effects on fat synthesis rates. LK decreased cell proliferation 26% at 48 h (P = 0.03) but showed no effect at 60 h. LM dropped cell proliferation 48% at 48 h (P < 0.01) and 22.2% at 60 h (P = 0.01). PDI expression was measured as an ER abundance marker. There were no effects of LK or LM on PDI expression at either 48 h or 60 h.
Fig 2. Cell activities in bovine mammary epithelial cells at 48 and 60 h after lysine or methionine subtraction to ¼ the normal concentration.
(A) Values are least square means ± SE (n = 3). (B) CTL = control; LK = low lysine; LM = low methionine. (C) *P ≤ 0.05 compared to CTL.
Table 3. Cell activities in bovine mammary epithelial cells across both timepoints of 48 and 60 h after lysine or methionine subtraction to ¼ the normal concentration1.
| Item | Treatment2 | SEM | P-value3 | |||
|---|---|---|---|---|---|---|
| CTL | LK | LM | LK | LM | ||
| Protein synthesis rate | 272 | 213 | 186 | 11.5 | <0.01 | <0.01 |
| Fat synthesis rate | 83.4 | 81.5 | 76.1 | 8.0 | 0.87 | 0.53 |
| Cell proliferation rate | 13.2 | 10.5 | 8.4 | 0.8 | 0.03 | <0.01 |
| PDI (arbitrary units) | 0.66 | 0.70 | 0.50 | 0.03 | 0.93 | 0.72 |
1Values are least squares means ± SE (n = 6) in dpm/ng DNA unless otherwise stated.
2CTL = control; LK = low lysine; LM = low methionine; PDI = endoplasmic reticulum tracker protein disulfide isomerase.
3Linear contrast of LK or LM vs. CTL.
Discussion
To elucidate the effects of reduced concentrations of a single EAA on BMEC activity and expression of genes for 8 transcription factors hypothesized to be regulated by EAA following different signaling mechanisms and affecting different downstream activities related to milk synthesis, experiments were performed with BMEC in culture. Treatments were designed to provide Lys and Met concentrations at ¼ of normal physiological concentrations in plasma of lactating dairy cows while maintaining all other amino acids at normal levels. Measurements were made after 24 h to focus on longer term responses to chronic EAA deficiency. While changes in HIF1A and SREBF1 expression were detected at 24 h, changes in expression of ATF4, ATF6, EGR1, HIF1A and SREBF1 occurred after 40 h. Similarly, others have reported that expression of ATF4 and its downstream targets is not upregulated in response to deprivation of a single amino acid until more than 12 h of incubation [18,37] and can remain elevated for 72 h [38,39]. FOS and JUN are considered immediate-early genes and have been found to peak in expression around 6 h into an EAA deficiency and return to normal by 24 h [11], which is consistent with our finding of no effect after 24 h. EGR1 expression can also constitute part of the early response to stress [12] but Papež et al. [39] found no effect of glutamine deprivation on EGR1 expression in CHO cells until 48 h, when it declined. HIF1A expression was also downregulated in response to glutamine deprivation at 48 h of incubation [39]. Because most of the transcription factor changes occurred after 40 h, we chose 48 and 60 h as timepoints for cell activity measurements.
Deficiencies of both Lys and Met suppressed protein synthesis and activated ATF4 expression, indicating the classic AARP had been triggered, likely through free tRNA-mediated activation of GCN2 [18]. ATF4 homo- and hetero-dimerizes with ATF6, FOS and JUN, among other basic leucine-zipper transcription factors, to induce expression of genes involved in resolving and adapting to the EAA deficiency, such as amino acid transporters and ER-resident proteins [8]. In addition to being up-regulated when global protein synthesis is inhibited by eIF2α phosphorylation, ATF4 and its partners can also induce expression of activators and inhibitors of protein synthesis, possibly to affect how the system responds to other regulatory influences via mTORC1, such as growth factors and ATP concentration [17]. Interestingly, muscle-specific knockout of ATF4 promoted expression of anabolic genes involved in protein synthesis and prevented the age-related decline in muscle mass [40]. Thus, the upregulation of ATF4 expression we observed during EAA deficiency may have contributed to the decreased rate of protein synthesis.
Despite a decrease in global protein synthesis, there was no effect of Lys or Met deficiency on ER size indicated by PDI abundance. Compared to the measurements of cell activity by isotope dilution, the PDI abundance assay by immunoblot was highly variable and may have obscured treatment differences. However, the lack of an EAA effect may indicate that proteostasis was achieved by ATF4 up-regulation. ATF6 is itself an ATF4 target [9] whose gene product can be activated by the ISR to selectively increase the expression of genes related to ER biogenesis and mRNA translation [41]. A mix of branched-chain amino acids dose-dependently increased ATF6 expression in mouse cardiac myocytes [42], and EAA activated the ATF6 target XBP1 in the mammary glands of cows to allow for greater milk protein yields [19,43]. The increase in ATF6 expression we detected during Met deficiency may have contributed to maintenance of the capacity for synthesis of the secretory proteins of milk.
Expression of JUN and FOS to produce hetero-dimerization partners of ATF4 is considered part of an early stress response through RAS/RAF/MEK/ERK signaling that promotes cell survival by stimulating proliferation and suppressing apoptosis [44]. Exposing cells to an absence of a single EAA, or to histidinol to block histidyl-tRNA synthesis, caused increases in JUN and FOS expression within 12 h [11,12,45,46]. In our experiments, after 24 h, there were no differences in JUN or FOS expression besides a lack of increase in JUN at 72 h. The findings suggest JUN and FOS do not participate in the long-term signaling responses in BMEC.
As its name suggests, EGR1 is also categorized as an early response gene and, like FOS and JUN, its expression is activated by the RAS/RAF/MEK/ERK signaling pathway [29]. Supplementation with histidinol or induction of ER stress with thapsigargin increased the expression of EGR1 in HepG2 cells from 8 to 24 h of incubation [12,13]. Contrary to these findings, our data demonstrate that Met deficiency led to a long-term suppression of EGR1 expression. Glutamine deficiency in CHO cells also led to a long-term decrease in EGR1 expression at 48 h without a prior early response [39]. EGR1 is a noted driver of cell proliferation [47] and its downregulation may have contributed to the decrease in cell proliferation we observed in BMEC exposed to LM.
Another stress-induced regulator of cell proliferation is HIF1A, which is responsible for cell cycle arrest during hypoxia [27,48]. The arrest mechanism involves antagonism by HIF1A of the proliferative effects of the transcription factor MYC [48,49], and it has been proposed that the balance between HIF1A and MYC influences the decision to progress through the cell cycle. The increased expression of HIF1A during Met deficiency in our experiment, accompanied by no change in MYC expression, may have contributed to the observed decrease in cell proliferation rate. We can only speculate as to how EAA deficiency may provoke HIF1A and MYC expression. MYC expression can be upregulated by the same RAS/RAF/MEK/ERK signaling pathway responsible for JUN, FOS and EGR1 up-regulation [50], which we did not observe. Alternatively, expression of both transcription factors can be stimulated by mTORC1 signalling [14,15] which is depressed by EAA deficiency [2,51]. However, removal of any EAA from CHO cell cultures increased the abundance of MYC mRNA within 1 to 8 h [46], while removal of glutamine increased MYC expression and decreased HIF1A expression after 48 h of incubation [39]. Further research may help clarify the relationship between EAA deficiency and the HIF1A/MYC balance.
SREBP-1c is the key lipid biosynthesis transcription factor in various tissues [33,34], and is a major regulator of milk fat synthesis in BMEC [52,53]. The expression of SREBF1 and its lipogenic targets is diminished by inhibiting mTORC1 [14,53,54], which EAA can regulate directly in mammary cells [2]. Indeed, valine addition to porcine mammary epithelial cell cultures increased nuclear SREBP-1c and intracellular triacylglycerol contents in an mTORC1-dependent manner [22], and Met addition to supraphysiological concentrations for BMEC enhanced SREBP-1c abundance and fat accumulation [21]. In our experiments, Met and Lys deficiency failed to affect lipogenesis rates at 48 and 60 h even though LM decreased the expression of SREBF1 at those timepoints. However, LM also increased SREBF1 expression at 40 h and this inconsistent effect may have clouded the lipogenic response.
Our results demonstrate the effects of Lys and Met on the mRNA expression of candidate transcription factors involved in protein synthesis and cell proliferation. However, there are limitations that should be acknowledged. Gene expression in this study was measured exclusively at the transcriptional level using qPCR, which does not account for post-transcriptional modifications, such as mRNA translation, protein phosphorylation, proteolytic cleavage and nuclear entry. To obtain a more complete understanding of the involvement of these transcription factors in the amino acid response of BMEC, future studies should explore their post-transcriptional status. Additionally, this experiment was conducted in a cell culture model of BMEC, which does not replicate the conditions found in vivo, including communication with other mammary cell types and control of blood flow. Further in vivo studies are needed to validate these findings in a more complex biological system.
Conclusions
In this study, EAA deficiency decreased the rate of protein synthesis and increased the expression of ATF4 which indicates the classic AARP was triggered in BMEC. The alternative RAS/RAF/MEK/ERK stress signaling pathway did not appear to be an important player in the long-term response to EAA deficiency as evidenced by a lack of increased expression of JUN, FOS or EGR1 between 24 and 60 h. The lower protein synthesis rate can affect cell proliferation rates but the decreased expression of EGR1 and increased expression of HIF1A during Met deficiency may have also contributed to the reduced cell proliferation rates. These results suggest that a transcriptional response to EAA deficiency contributes to effects on protein synthesis and cell proliferation in BMEC.
Acknowledgments
The authors wish to thank Julie Kim, Sara Ahmady, Jillian Wang, and Brian MacDougall at the University of Guelph for technical assistance.
Data Availability
All data supporting this study are available on Borealis and can be accessed at https://doi.org/10.5683/SP3/CEHUCP.
Funding Statement
Financial support for this study was provided in part by Trouw Nutrition Canada Inc. (https://www.trouwnutrition.ca/en-ca/) and NSERC Canada (https://www.nserc-crsng.gc.ca/index_eng.asp) under Collaborative Research and Development grant 513256-17 awarded to JPC. JD was a salaried employee of Trouw Nutrition, Amersfoort, The Netherlands. The funders had no additional role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Trouw Nutrition and NSERC CRDPJ-513256-17 NSERC Discovery RGPIN-2014-05600.
References
- 1.Schwab CG, Broderick GA. A 100-Year Review: Protein and amino acid nutrition in dairy cows. J Dairy Sci. 2017. Dec;100(12):10094–112. doi: 10.3168/jds.2017-13320 [DOI] [PubMed] [Google Scholar]
- 2.Appuhamy JADRN, Knoebel NA, Nayananjalie WAD, Escobar J, Hanigan MD. Isoleucine and Leucine Independently Regulate mTOR Signaling and Protein Synthesis in MAC-T Cells and Bovine Mammary Tissue Slices,. J Nutr. 2012. Mar;142(3):484–91. doi: 10.3945/jn.111.152595 [DOI] [PubMed] [Google Scholar]
- 3.Burgos SA, Dai M, Cant JP. Nutrient availability and lactogenic hormones regulate mammary protein synthesis through the mammalian target of rapamycin signaling pathway. J Dairy Sci. 2010. Jan;93(1):153–61. doi: 10.3168/jds.2009-2444 [DOI] [PubMed] [Google Scholar]
- 4.Cant JP, Kim JJM, Cieslar SRL, Doelman J. Symposium review: Amino acid uptake by the mammary glands: Where does the control lie? J Dairy Sci. 2018. Jun;101(6):5655–66. doi: 10.3168/jds.2017-13844 [DOI] [PubMed] [Google Scholar]
- 5.Pszczolkowski VL, Arriola Apelo SI. The market for amino acids: understanding supply and demand of substrate for more efficient milk protein synthesis. J Anim Sci Biotechnol. 2020. Dec;11(1):108. doi: 10.1186/s40104-020-00514-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seymour DJ, Kim JJM, Doelman J, Cant JP. Feed restriction of lactating cows triggers acute downregulation of mammary mammalian target of rapamycin signaling and chronic reduction of mammary epithelial mass. J Dairy Sci. 2024. Aug;107(8):5667–80. doi: 10.3168/jds.2023-24478 [DOI] [PubMed] [Google Scholar]
- 7.Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005. Feb;16(1):3–12. doi: 10.1016/j.semcdb.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 8.Kilberg MS, Pan YX, Chen H, Leung-Pineda V. NUTRITIONAL CONTROL OF GENE EXPRESSION: How Mammalian Cells Respond to Amino Acid Limitation. Annu Rev Nutr. 2005. Aug 21;25(1):59–85. doi: 10.1146/annurev.nutr.24.012003.132145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neill G, Masson GR. A stay of execution: ATF4 regulation and potential outcomes for the integrated stress response. Front Mol Neurosci. 2023. Feb 7;16:1112253. doi: 10.3389/fnmol.2023.1112253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015. Jul;17(7):829–38. doi: 10.1038/ncb3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu L, Balasubramanian M, Shan J, Dudenhausen EE, Kilberg MS. Auto-activation of c-JUN Gene by Amino Acid Deprivation of Hepatocellular Carcinoma Cells Reveals a Novel c-JUN-mediated Signaling Pathway. J Biol Chem. 2011. Oct;286(42):36724–38. doi: 10.1074/jbc.M111.277673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan J, Balasubramanian MN, Donelan W, Fu L, Hayner J, Lopez MC, et al. A Mitogen-activated Protein Kinase/Extracellular Signal-regulated Kinase Kinase (MEK)-dependent Transcriptional Program Controls Activation of the Early Growth Response 1 (EGR1) Gene during Amino Acid Limitation. J Biol Chem. 2014. Aug;289(35):24665–79. doi: 10.1074/jbc.M114.565028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan J, Dudenhausen E, Kilberg MS. Induction of early growth response gene 1 (EGR1) by endoplasmic reticulum stress is mediated by the extracellular regulated kinase (ERK) arm of the MAPK pathways. Biochim Biophys Acta BBA—Mol Cell Res. 2019. Mar;1866(3):371–81. doi: 10.1016/j.bbamcr.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a Metabolic Gene Regulatory Network Downstream of mTOR Complex 1. Mol Cell. 2010. Jul;39(2):171–83. doi: 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzes KM, Swamy M, Hukelmann JL, Emslie E, Sinclair LV, Cantrell DA. Control of amino acid transport coordinates metabolic reprogramming in T-cell malignancy. Leukemia. 2017. Dec;31(12):2771–9. doi: 10.1038/leu.2017.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012. Apr;149(2):274–93. doi: 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep. 2017. May;19(6):1083–90. doi: 10.1016/j.celrep.2017.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edick AM, Audette J, Burgos SA. CRISPR-Cas9-mediated knockout of GCN2 reveals a critical role in sensing amino acid deprivation in bovine mammary epithelial cells. J Dairy Sci. 2021. Jan;104(1):1123–35. doi: 10.3168/jds.2020-18700 [DOI] [PubMed] [Google Scholar]
- 19.Nichols K, Doelman J, Kim JJM, Carson M, Metcalf JA, Cant JP. Exogenous essential amino acids stimulate an adaptive unfolded protein response in the mammary glands of lactating cows. J Dairy Sci. 2017. Jul;100(7):5909–21. doi: 10.3168/jds.2016-12387 [DOI] [PubMed] [Google Scholar]
- 20.Fox MK. The Effects of Arginine on Gene Expression in Bovine Mammary and Longissimus Dorsi Tissues. 2021.
- 21.Qi H, Meng C, Jin X, Li X, Li P, Gao X. Methionine Promotes Milk Protein and Fat Synthesis and Cell Proliferation via the SNAT2-PI3K Signaling Pathway in Bovine Mammary Epithelial Cells. J Agric Food Chem. 2018. Oct 24;66(42):11027–33. doi: 10.1021/acs.jafc.8b04241 [DOI] [PubMed] [Google Scholar]
- 22.Che L, Xu M, Gao K, Zhu C, Wang L, Yang X, et al. Valine increases milk fat synthesis in mammary gland of gilts through stimulating AKT/MTOR/SREBP1 pathway†. Biol Reprod. 2019. Jul 1;101(1):126–37. doi: 10.1093/biolre/ioz065 [DOI] [PubMed] [Google Scholar]
- 23.Cao SS, Kaufman RJ. Unfolded protein response. Curr Biol. 2012. Aug;22(16):R622–6. doi: 10.1016/j.cub.2012.07.004 [DOI] [PubMed] [Google Scholar]
- 24.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016. Oct;17(10):1374–95. doi: 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schröder M. The unfolded protein response. Mol Biotechnol. 2006;34. [DOI] [PubMed] [Google Scholar]
- 26.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001. Apr 30;20(19):2390–400. doi: 10.1038/sj.onc.1204383 [DOI] [PubMed] [Google Scholar]
- 27.Druker J, Wilson JW, Child F, Shakir D, Fasanya T, Rocha S. Role of Hypoxia in the Control of the Cell Cycle. Int J Mol Sci. 2021. May 5;22(9):4874. doi: 10.3390/ijms22094874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc Regulates Cyclin D-Cdk4 and -Cdk6 Activity but Affects Cell Cycle Progression at Multiple Independent Points. Mol Cell Biol. 1999. Jul 1;19(7):4672–83. doi: 10.1128/MCB.19.7.4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagel JI, Deindl E. Early Growth Response 1—A Transcription Factor in the crossfire of Signal Transduction Cascades. INDIAN J BIOCHEM BIOPHYS. 2011;48. [PubMed] [Google Scholar]
- 30.Shan J, Donelan W, Hayner JN, Zhang F, Dudenhausen EE, Kilberg MS. MAPK signaling triggers transcriptional induction of cFOS during amino acid limitation of HepG2 cells. Biochim Biophys Acta BBA—Mol Cell Res. 2015. Mar;1853(3):539–48. doi: 10.1016/j.bbamcr.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang IC, Chen YJ, Hughes DE, Ackerson T, Major ML, Kalinichenko VV, et al. FoxM1 Regulates Transcription of JNK1 to Promote the G1/S Transition and Tumor Cell Invasiveness. J Biol Chem. 2008. Jul;283(30):20770–8. doi: 10.1074/jbc.M709892200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziello JE, Jovin IS, Huang Y. Hypoxia-Inducible Factor (HIF)-1 Regulatory Pathway and its Potential for Therapeutic Intervention in Malignancy and Ischemia. [PMC free article] [PubMed]
- 33.Bauman DE, Perfield JW, Harvatine KJ, Baumgard LH. Regulation of Fat Synthesis by Conjugated Linoleic Acid: Lactation and the Ruminant Model. J Nutr. 2008. Feb;138(2):403–9. doi: 10.1093/jn/138.2.403 [DOI] [PubMed] [Google Scholar]
- 34.Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res. 2001. Nov;40(6):439–52. doi: 10.1016/s0163-7827(01)00010-8 [DOI] [PubMed] [Google Scholar]
- 35.Mackle TR, Dwyer DA, Ingvartsen KL, Chouinard PY, Ross DA, Bauman DE. Evaluation of Whole Blood and Plasma in the Interorgan Supply of Free Amino Acids for the Mammary Gland of Lactating Dairy Cows. J Dairy Sci. 2000. Jun;83(6):1300–9. doi: 10.3168/jds.S0022-0302(00)74996-4 [DOI] [PubMed] [Google Scholar]
- 36.Martineau R, Ouellet DR, Patton RA, White RR, Lapierre H. Plasma essential amino acid concentrations in response to casein infusion or ration change in dairy cows: A multilevel, mixed-effects meta-analysis. J Dairy Sci. 2019. Feb;102(2):1312–29. doi: 10.3168/jds.2018-15218 [DOI] [PubMed] [Google Scholar]
- 37.Brüggenthies JB, Fiore A, Russier M, Bitsina C, Brötzmann J, Kordes S, et al. A cell-based chemical-genetic screen for amino acid stress response inhibitors reveals torins reverse stress kinase GCN2 signaling. J Biol Chem. 2022. Dec;298(12):102629. doi: 10.1016/j.jbc.2022.102629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fomina-Yadlin D, Gosink JJ, McCoy R, Follstad B, Morris A, Russell CB, et al. Cellular responses to individual amino-acid depletion in antibody-expressing and parental CHO cell lines. Biotechnol Bioeng. 2014. May;111(5):965–79. doi: 10.1002/bit.25155 [DOI] [PubMed] [Google Scholar]
- 39.Papež M, Jiménez Lancho V, Eisenhut P, Motheramgari K, Borth N. SLAM-seq reveals early transcriptomic response mechanisms upon glutamine deprivation in Chinese hamster ovary cells. Biotechnol Bioeng. 2023. Apr;120(4):970–86. doi: 10.1002/bit.28320 [DOI] [PubMed] [Google Scholar]
- 40.Miller MJ, Marcotte GR, Basisty N, Wehrfritz C, Ryan ZC, Strub MD, et al. The transcription regulator ATF4 is a mediator of skeletal muscle aging. GeroScience. 2023. Apr 4;45(4):2525–43. doi: 10.1007/s11357-023-00772-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, et al. Stress-Independent Activation of XBP1s and/or ATF6 Reveals Three Functionally Diverse ER Proteostasis Environments. Cell Rep. 2013. Apr;3(4):1279–92. doi: 10.1016/j.celrep.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Xiong Z, Yan W, Gao E, Cheng H, Wu G, et al. Branched chain amino acids exacerbate myocardial ischemia/reperfusion vulnerability via enhancing GCN2/ATF6/PPAR-α pathway-dependent fatty acid oxidation. Theranostics. 2020;10(12):5623–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma YF, Batistel F, Xu TL, Han LQ, Bucktrout R, Liang Y, et al. Phosphorylation of AKT serine/threonine kinase and abundance of milk protein synthesis gene networks in mammary tissue in response to supply of methionine in periparturient Holstein cows. J Dairy Sci. 2019. May;102(5):4264–74. doi: 10.3168/jds.2018-15451 [DOI] [PubMed] [Google Scholar]
- 44.Bahrami S, Drabløs F. Gene regulation in the immediate-early response process. Adv Biol Regul. 2016. Sep;62:37–49. doi: 10.1016/j.jbior.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 45.Chaveroux C, Jousse C, Cherasse Y, Maurin AC, Parry L, Carraro V, et al. Identification of a Novel Amino Acid Response Pathway Triggering ATF2 Phosphorylation in Mammals. Mol Cell Biol. 2009. Dec 1;29(24):6515–26. doi: 10.1128/MCB.00489-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pohjanpeltol P, Holtta E. Deprivation of a Single Amino Acid Induces Protein Synthesis- Dependent Increases in c-jun, c-myc, and Ornithine Decarboxylase mRNAs in Chinese Hamster Ovary Cells. MOL CELL BIOL. 1990;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cesana M, Tufano G, Panariello F, Zampelli N, Ambrosio S, De Cegli R, et al. EGR1 drives cell proliferation by directly stimulating TFEB transcription in response to starvation. Gattelli A, editor. PLOS Biol. 2023. Mar 8;21(3):e3002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hubbi ME, Semenza GL. Regulation of cell proliferation by hypoxia-inducible factors. Am J Physiol-Cell Physiol. 2015. Dec 15;309(12):C775–82. doi: 10.1152/ajpcell.00279.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1α induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004. May 5;23(9):1949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerkhoff E, Houben R, Löffler S, Troppmair J, Lee JE, Rapp UR. Regulation of c-myc expression by Ras/Raf signalling. Oncogene. 1998. Jan 15;16(2):211–6. doi: 10.1038/sj.onc.1201520 [DOI] [PubMed] [Google Scholar]
- 51.Moshel Y, Rhoads RE, Barash I. Role of amino acids in translational mechanisms governing milk protein synthesis in murine and ruminant mammary epithelial cells. J Cell Biochem. 2006. Jun;98(3):685–700. doi: 10.1002/jcb.20825 [DOI] [PubMed] [Google Scholar]
- 52.Ma L, Corl BA. Transcriptional regulation of lipid synthesis in bovine mammary epithelial cells by sterol regulatory element binding protein-1. J Dairy Sci. 2012. Jul;95(7):3743–55. doi: 10.3168/jds.2011-5083 [DOI] [PubMed] [Google Scholar]
- 53.Li N, Zhao F, Wei C, Liang M, Zhang N, Wang C, et al. Function of SREBP1 in the Milk Fat Synthesis of Dairy Cow Mammary Epithelial Cells. Int J Mol Sci. 2014. Sep 23;15(9):16998–7013. doi: 10.3390/ijms150916998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP Activity Is Regulated by mTORC1 and Contributes to Akt-Dependent Cell Growth. Cell Metab. 2008. Sep;8(3):224–36. doi: 10.1016/j.cmet.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting this study are available on Borealis and can be accessed at https://doi.org/10.5683/SP3/CEHUCP.