Abstract
Cytosolic thiouridylase is a conserved cytoplasmic tRNA thiolase composed of two different subunits, CTU1 and CTU2. CTU2 serves as a scaffold protein, while CTU1 catalyzes the 2-thiolation at the 34th wobble uridine of the anticodon loop. tRNAGlnUUG, tRNAGluUUC, and tRNALysUUU are the tRNA substrates that are modified with a thiol group at the C2 positions (s2) by CTU1, and also with a methoxycarbonylmethyl group at the C5 positions (mcm5) by Elongator and ALKBH8. mcm5s2U34 modification of the three tRNAs, and their modifying enzymes are involved in human disease and development. Elongator mutant animals exhibit severe phenotypes, while the biological function of Ctu1 in vertebrate animal models remains poorly characterized. Here, we applied antisense morpholino oligonucleotides targeting cytosolic thiouridylase subunit1 (ctu1) transcripts in a zebrafish model and small interfereing RNA against CTU1 transcript in human endothelial cells to define the phenotypes. We found that deficiency of ctu1 causes impaired angiogenesis and development in zebrafish embryos, and CTU1 is involved in proliferation, migration, and tube formation of human endothelial cells. We employed single-cell RNA sequencing to acquire the transcriptomic atlas from ctu1 and control morphant zebrafish. Comprehensive bioinformatics analysis, including pseudo-time, RNA velocity, cell-cell communication, and gene regulatory network inference revealed that ctu1 deficiency leads to the arrest of cell cycle, and the defects of nerve development and erythrocyte differentiation and the attenuation of several pro-angiogenic signaling pathways, e.g., angpt-tek and dll4-notch. Our findings show for the first time that CTU1 is essential for angiogenesis and embryonic development in vertebrates.
Introduction
tRNA undergoes extensive post-transcriptional chemical modifications in the nucleotides of the anticodon loop [1]. In eukaryotes, the cytosolic tRNAGlnUUG, tRNAGluUUC, and tRNALysUUU are the three types of tRNA that have uridine at the wobble position 34 (U34). The U34 of these tRNAs is chemically modified with a thiol group at the C2 positions (s2), and (commonly) a methoxycarbonylmethyl group at the C5 positions (mcm5), which ultimately results in a mcm5s2U34 [2, 3]. The mcm5s2U34 modification occurs through two pathways. In one pathway, urm1 acts as a sulfur donor for cytosolic tRNA thiouridylase subunit 1 (CTU1), which catalyzes the thiolation of cytosolic tRNAGlnUUG, tRNAGluUUC, and tRNALysUUU [4]; while the other one consists of ALKBH8 (alkB homolog 8, tRNA methyltransferase) and the Elongator complex (ELP1-ELP6), which participate in the formation of the mcm5 side chain [5].
mcm5s2U34 modification and its modifying enzymes have been regarded as critical regulators of gene expression and protein homeostasis [6]. mcm5s2U34 modifying enzymes play crucial roles in many important physiological and pathological processes [7, 8]. Several clinical cases indicate that mutations in the mcm5s2U34 modifying enzymes, like Elongator subunits (ELP2, ELP4, and ELP6), ALKBH8 and CTU2, are associated with human neurological diseases [9–11]. Elp2 mutant mice exhibit a severe neuro developmental phenotype [12], and Elp3 knock-out mice show embryonic lethality [13]. Double deletion of the partner enzymes (CTU1 and ELP3) for mcm5s2U34 modification is lethal to the cell in yeast [14]. Single deletion of the s2U34 modification enzyme (Ctu1 or Ctu2) in the nematode and yeast causes development abnormalities, thermosensitivity [15], and sensitivity to various exogenous stresses [16, 17]; however, the in vivo role of Ctu1 in vertebrates, and the underlying mechanisms remain to be clarified.
Single-cell RNA sequencing (scRNA-seq) technology enables the unraveling of genetic and transcriptional heterogeneity among cells and provides novel insights into gene function [18]. In this study, we employed morpholino (MO) antisense oligonucleotides targeting ctu1, and scRNA-seq in zebrafish embryos to define the phenotypes and the single-cell transcriptomic atlas in response to ctu1 deficiency. We also applied lentivirus particles in human endothelial cells to establish CTU1 overexpression (CTU1-OE) and CTU1 knockdown (CTU1-KD) endothelial cells, and examined their angiogenesis-related behaviors. Our data demonstrated that CTU1 plays a vital role in proliferation and development, and also in nerve and erythrocyte differentiation. Several pro-angiogenic signaling cascades, e.g., angpt-tek and dll4-notch, are the potential molecular mechanisms underlying the ctu1-related angiogenesis defects.
Materials and methods
Maintenance and genetic manipulation of zebrafish (Danio rerio)
All zebrafish experiments were carried out with the approval of the Animal Care and Use Committee of Shanghai University of Medicine and Health Sciences, and followed the regulations specified in EU Directive 2010/63/EU. Tg(fli1a-EGFP;casper) lines of zebrafish were maintained in the facility of the Shanghai Research Center for Model Organisms. Antisense MO was designed by Gene Tools (http://www.gene-tools.com/), and microinjected into fertilized one-cell stage embryos at 4 ng using MN-151 micromanipulator (Narishige, Japan) according to standard protocols [19]. Antisense MO targeting the intron 2-exon 3 splice of ctu1 was 5’-GCATTGTGACCTGCTCATCAAACAA-3’. Antisense MO targeting the exon 2-intron 2 of urm1 was 5’-GAATACTTTTCCATACTCACAGGGA-3’. The standard control MO sequence was 5’-CCTCTTACCTCAGTTACAATTTATA-3’. All zebrafish embryos employed in this study were without sexual differentiation.
Zebrafish embryonic angiogenesis
After MO microinjection, embryos were dechorionated at 2 days post fertilization (2-dpf), and anesthetized with 0.016% MS-222 (tricaine methanesulfonate, Sigma-Aldrich, St. Louis, MO). Zebrafish larvae were then oriented on lateral side (anterior, left; posterior, right; dorsal, top), and mounted with 3% methylcellulose in a depression slide for observation and photography by fluorescence microscopy with digital cameras (Nikon SMZ 1500). A subset of images was adjusted for levels, brightness, contrast, hue and saturation with Adobe Photoshop 7.0 software (Adobe, San Jose, California) to optimally visualize the expression patterns. Quantitative image analyses processed using image based morphometric analysis (NIS-Elements D3.1, Japan) and ImageJ software (U.S. National Institutes of Health, Bethesda, MD, USA; http://rsbweb.nih.gov/ij/).
Cell culture and lentivirus infection
Human microvascular endothelial cells (HMEC-1) (SUNNCELL, Shanghai, China) were cultured in MCDB131 medium supplemented with 10 ng/mL epidermal growth factor (Thermo, USA), 1 μg/mL hydrocortisone (Sigma, Germany), 10 mM L-Glutamine (Gibco, New York, USA), 10% FBS (Gibco, USA), and 1% penicillin-streptomycin solution (Gibco, USA), and kept in an incubator (Thermo, USA) at 37°C, with 5% CO2. The lentiviral vectors pSLenti-U6-sh(CTU1)-CMV-EGFP-F2A-Puro-WPRE, pSLenti-U6-CMV-EGFP-F2A-Puro-WPRE, and pSLenti-CMV-Ctu1-3xELAG-PGK-Puro-WPRE were constructed, and packaged into lentivirus particles by OBiO Technology (OBiO, Shanghai, China). Cells were infected with the lentivirus and 10 μg/ml Polybrene at 37°C. Then the cells were seeded into a 96-well plate (7000–8000 cells per well) and the cell viability was determined by Cell Counting Kit-8 (CCK-8, Beyotime, China) after 24, 48, and 72 hours’ culture.
RNA isolation and PCR analysis
Total RNA was extracted using the RNA-easy Isolation Kit (Vazyme Biotech, China) following the manufacturer’s guidelines. Subsequently, 1 μg of the isolated RNA was reverse transcribed using the HiScript® III All-in-one RT SuperMix Perfect for qPCR kit (Vazyme Biotech, China). Using the SYBR green reaction mix (Vazyme Biotech, China, #Q711-02), a QuantStudio™ 5 Real-Time PCR System (Cell Signaling Technology, MA) was utilized to conduct a quantitative PCR analysis. For the RT-PCR analysis of the gene expression after MO injection, primers spanning ctu1 exon 2 and exon 3 were used as 5’-GCAGGTGGGCTTGAAGAATAACT-3’ (forward), and 5’-GTGCAGCGGCGGAGACGAG-3’ (reverse) for the ctu1 I2E3-MO; primers spanning urm1 exon 1 and exon 3 were used as 5’-TGAGATTACTTCCGGGTTTTACAA-3’ (forward) and 5’-GTGCAGCGGCGGAGACGAG-3’ (reverse) for urm1 e2i2-MO. The primer ef1α sequences used as the internal control were 5’-GCTCTGGGCGCTCCTTTAG-3’ (forward) and 5’-GATACCAGCCTCAAACTCACC-3’ (reverse). The primer sequences for human CTU1 were 5’-GGTCGTGGCCTACGAAGA-3’ (forward) and 5’-AGTTCATGAGCACGGTCTCC-3’ (reverse). The primer sequences for the internal control human gene gene ACTB were 5’-TGGCACCACACCTTCTACAA-3’ (forward) and 5’-CCAGAGGCGTACAGGGATAG-3’ (reverse).
Transwell assays
Transwell assays were conducted with the 24-well plate (Corning, USA) according to manufacturer’s instructions. In brief, after 12 hours of starvation, 1.5 × 104 HMEC-1 cells were seeded in the upper chambers of the inserts, with the culture medium supplemented with 20% FBS used in the lower chambers. After 24 h of incubation at 37°C and removal of those non-invasive cells, the remaining cells were then fixed and stained with 1% crystal violet for 10 minutes at room temperature. Finally, those migrated cells were visualized and quantified through microscopic examination at a magnification of ×200 (Leica, Germany).
Tube formation assays
The matrigel-based tube formation assay were conducted following a previously established protocol [20]. In brief, a 50 μL layer of Matrigel (Corning, New York, USA) was evenly coated in a 96-well plate, then 1.5 × 104 HMEC-1 cells were seeded and incubated at 37°C with 5% CO2 for 16 h. Then, the formed tubes were observed and photographed under a microscope (Eclipse Ts2R; Nikon, Japan). ImageJ software (National Institutes of Health, USA) was used for the quantification of the branch points, tube branch length, and total covered area.
Single-cell library preparation
Single cell preparation, library construction, and sequencing were performed by Sinotech Genomics Co., Ltd. Shanghai, China. In brief, about 20 zebrafish fertilized eggs at 2-dpf were collected from control and ctu1 morphant. Following the digestion process as before [21], cell pellets were collected, and finally loaded as a single-cell suspension into the BD Rhapsody system (BD Biosciences, San Jose, CA). The cDNA library was generated from double strand full length cDNA by random priming amplification with the BD Rhapsody cDNA Kit (#633773, BD Biosciences) and the BD Rhapsody Targeted mRNA & AbSeq Amplification Kit (#633774, BD Biosciences). All the libraries were sequenced in a PE150 mode (Pair-End for 150bp read) in the X Ten instrument (Illumina, San Diego, CA).
scRNA-seq data processing and quality control
The BD Rhapsody Whole Transcriptome Assay Analysis Pipeline was used. The FASTQ documents were filtered to generate a single cell expression profile matrix. The R software (version 4.2.2) and Seurat (version 4.3.0) were utilized for downstream clustering and visualization. After filtering cells with unique features over 2,000 or less than 200 and mitochondrial counts exceeding 20%, we normalized the data with the “LogNormalize” method. Subsequently, the “RunPCA” function was used for Principal Component Analysis (PCA). Clustering analysis was then conducted utilizing the “FindClusters” function. For visualization, Uniform Manifold Approximation and Projection (UMAP) coordinates were determined through the “RunUMAP” function. The marker genes of each cluster were calculated by the “FindAllMarkers” function with log2FC.threshold = 0.25; min.pct = 0.25.
Cell cycle, differential expression, and gene set enrichment analysis (GSEA)
Initially, zebrafish genes were transformed into human genes using the “homologene” function. Subsequently, the cell cycle state was characterized using the cell cycle-related signature sets incorporated in the Seurat package and the “CellCycleScoring” function. Ultimately, the cell cycle phase was labeled. The differentially expressed genes (DEGs) were identified by calculating the “FindMarkers” function with min.pct = 0.25, log2FC.pct = 0.25. Genes with an absolute log2FC greater than 0.5 and an adjusted p-value less than 0.05 were considered as DEGs with significant difference between the two groups. GSEA of the DEGs were also performed by R package clusterProfiler (version 4.2.2). The “gseKEGG” and “gseGO” functions for analysis were used with OrgDb = "org.Dr.eg.db".
Pseudo-time analysis
Monocle 2, version 2.22.0 was used to determine the pseudotemporal ordering of erythroid cells. Briefly, erythroid cells of wild type (WT) and Ctu1 morphant were subclustered and a cellDataSet object was created in Monocle2 with the function “newCellDataSet” with standard arguments enabled. Genes with an average expression value greater than 0.1 were used to sort the cells in the quasi-time trajectory. Dimensionality reduction was conducted using discriminative dimensionality reduction with trees (DDRTree). Erythroid cells were ordered in pseudotime using the “orderCells” function. The dynamic expression of genes was visualized using the “plot_genes_in_pseudotime” function.
RNA velocity analysis
The ratio between unspliced and spliced mRNA levels can predict the subsequent concentration of mature mRNA, which subsequently forecasts the future cellular state [22]. The spliced and unspliced reads of scRNA-seq data were calculated based on the bam file and saved the results as loom files. Loom files for individual libraries from control and Ctu1 morphant, were combined using loompy, version 2.0.16. The scVelo, version 0.2.2 was employed to estimate the velocity vector based on the RNA velocity dynamic model with default parameters, assess the transition probability matrix in a dynamic system, infer alterations in cell state over time, and map the RNA velocity results onto UMAP images.
Cell-cell communication analysis
CellChat version 1.6.1 was used to analyze intercellular communication networks [23]. We performed sub-clustering and normalization of mesoderm cells as input data for CellChat to infer the cell-cell communication networks. The “homologene” function was used for cross-species homologous gene conversion. The significant ligand-receptor interactions were identified using the “identifyOverExpressedInteractions” (p = 0.05) function. The function “computeCommunProb” was utilized to ascertain the probability of intercellular communication, with the parameters set to raw.use = FALSE, nboot = 100, Hill function parameter kn = 0.5. The function“computeCommunProbPathway” was employed to infer potential communication pathways between cells, using a threshold of 0.05. The cell-cell communication networks were calculated via the aggregateNet function, also with a threshold of 0.05.
Gene regulatory network (GRN) inference
To infer the GRN between control and Ctu1 morphant, the SCENIC analysis was conducted utilizing the pySCENIC software [24]. After quality control and homologous gene conversion in Seurat, the normalized data was exported to a matrix and then converted into a loom file. A list of human transcription factors (TFs) (https://github.com/aertslab/pySCENIC/blob/master/resources/hs_hgnc_tfs.txt) was utilized and regulatory interactions between them and their potential target genes were inferred using GRNBoost2. CisTarget motif enrichment was conducted using SCENIC’s RcisTarget and ranking databases (hg19-tss-centered-10kb-7species.mc9nr.genes_vs_motifs.rankings.feather). The activity of the regulons was computed using SCENIC’s “AUCell” function. The activity of regulons in cells under different conditions is determined by Regulon Specificity Scores (RSS) and Z-scores [25].
Statistical analysis
Statistical analysis were performed using R software and GraphPad Prism software. The difference between the two groups was analyzed using the Wilcoxon signed-rank test or t-test. For comparisons involving three or more groups, the Kruskal-Wallis test was employed. Unless otherwise specified, statistical significance was defined as p < 0.05.
Results
Phenotype resulting from ctu1 deficiency in zebrafish embryo
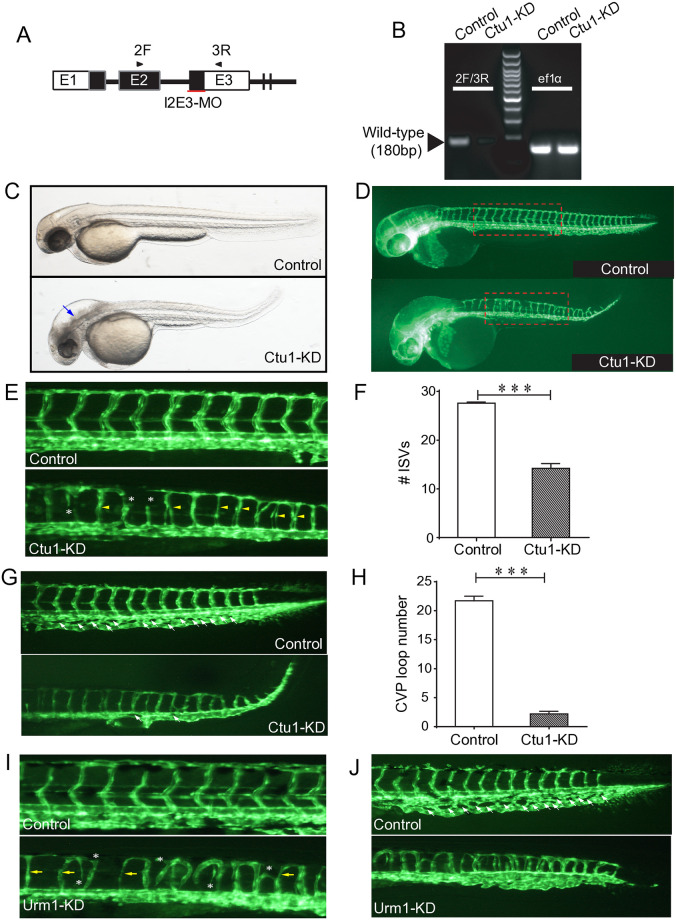
The workflow of our study was illustrated as Fig 1. Tg(fli1a:EGFP)y1 zebrafish line is an ideal model for visualizing the embryonic vasculature formation [26]. Its embryo is transparent, and its endothelial cells were green fluorescent labeled. We applied Tg(fli1a:EGFP)y1 zebrafish line, and targeted the expression of ctu1 by specific MO antisense strategies (Fig 2A). Two days after the MO injection, nucleic acids were extracted from the embryos for RT-PCR analysis. The PCR data revealed that the antisense Ctu1-i2e3-MO effectively knocked down ctu1 expression (Fig 2B). Representative bright field and fluorescence image analysis revealed that ctu1 morphant embryo showed a developmental abnormalities and defective angiogenesis. The ctu1 morphant embryos display significant morphological abnormalities including enlarged brain ventricles, pronounced hindbrain edema, pericardial edema, a misshapen spine, and a curled-up tail (Fig 2C and 2D, S1A and S1B Fig). Meanwhile, the ctu1 morphant exhibit a reduced number of incomplete and thinner intersegmental vessels (ISVs), dorsal longitudinal anastomotic vessels (DLAV), caudal vein plexus (CVPs), ectopic sprouts of dorsal aorta (DA) and posterior cardinal vein (Fig 2E–2H). Meanwhile, as Urm1 acts as a sulfur donor for Ctu1 in the thiolation of cytosolic tRNAGlnUUG, tRNAGluUUC, and tRNALysUUU, we also examined the effects of MO-mediated down-regulation of urm1 (S1C and S1D Fig). The phenotypes in urm1 morphant are quite similar as the ones in ctu1 morphant (Fig 2I and 2J, S1E–S1H Fig). The result further strengthens the evidence supporting that the Ctu1 is required for vascular networks and embryonic development.
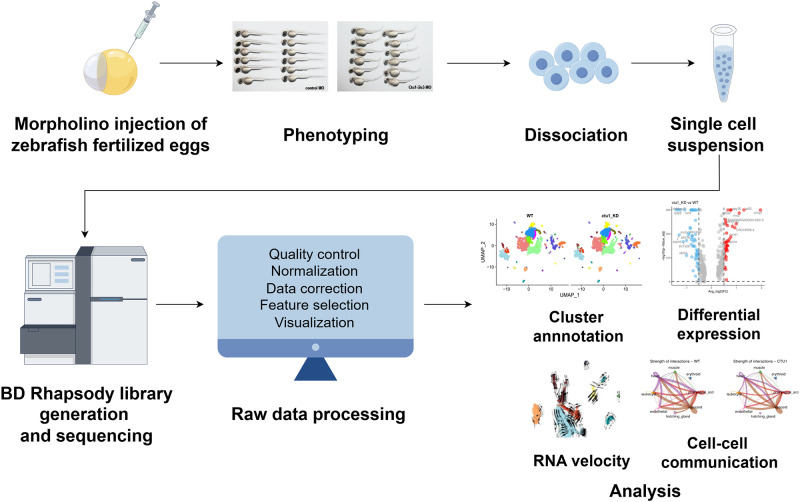
Fig 1. Schematic representation of workflow, figured (by Figdraw https://www.figdraw.com/).
Antisense MO oligonucleotides were microinjected into fertilized one-cell stage embryos. At 2-dpf, zebrafish larvae were either photographed for phentying, or collected for single-cell preparation and sequencing using BD Rhapsody system. After filtering the raw data, further single-cell data analysis was performed such as cluster annotation, DEGs analysis, RNA velocity, cell-cell communication.
Fig 2. ctu1 morphant zebrafish larvae exhibits developmental defects.
(A) Ctu1-targeted MO design strategy. (B) PCR analysis of control and Ctu1 morphant. (C and D) Bright-fieldand EGFP fluorescentimages depict the overall morphology of control and Ctu1 morphant at 2-dpf. Blue arrows indicate expanded brain ventricle and hindbrain edema in ctu1 morphant compared with control. The dotted square regions are shown at higher magnification in E. (E and G) Image of trunk regions. Compared with control MO, embryos injected with ctu1-i2e3-MO present a lower number of incomplete and thinner intersegmental vessels (ISVs, yellow arrows), and ectopic sprouts (asterisk) of dorsal aorta (E, lower panel). In control embryos, caudal vein plexus (CVP, white arrows) were formed honeycomb-like structures at the tail around 2-dpf (G, upper panel, arrowheads). In contrast, ctu1 deficency resulted in specific defects in CVP formation (G, lower panel, arrowheads). Quantification of the number of complete ISVs (F) and CVP (H). Columns, mean; bars, SEM (n = 10; unpaired student’s t-test; ***, p < 0.001). (I and J) Image of trunk regions. Compared with control MO, embryos injected with urm1-i2e2-MO present a lower number of incomplete and thinner ISVs (yellow arrows), and ectopic sprouts (asterisk) of dorsal aorta (I, lower panel). In control embryos, CVP (white arrows) were formed honeycomb-like structures at the tail around 2-dpf (J, upper panel, arrowheads). In contrast, urm1 deficency resulted in specific defects in CVP formation (J, lower panel, arrowheads).
Single-cell transcriptome atlas of the ctu1 morphant and control zebrafish embryos
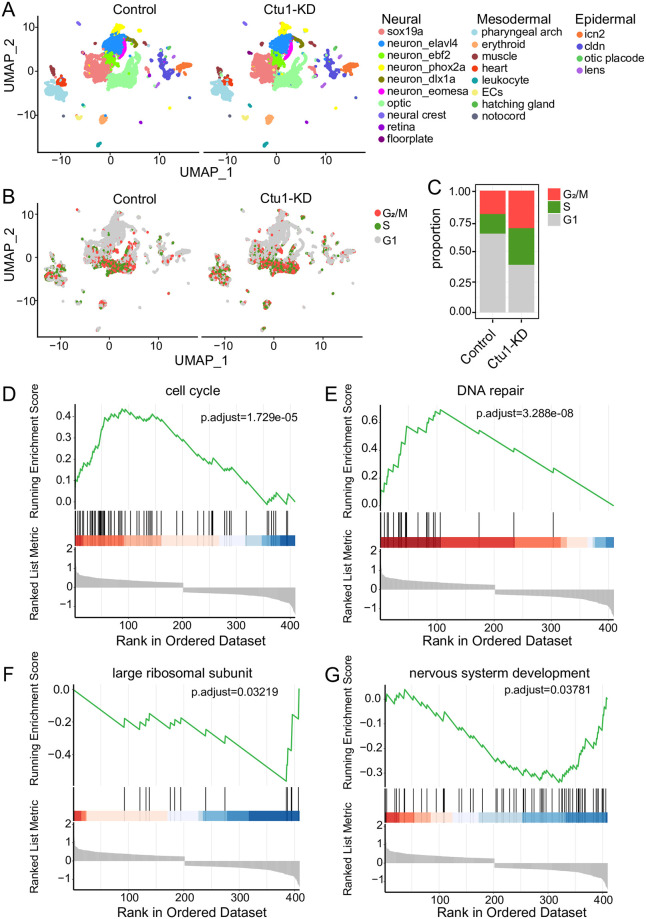
In order to create a transcriptome map of the zebrafish with ctu1 KD, we used 20 zebrafish larvae at 2-dpf after MO microinjection from the control and ctu1 morphant group for tissue digestion and scRNA-seq. In total, 20,788 cells were retained for subsequent analysis. Cells were clustered based on the gene expression profiles, and cell types were annotated based on the published literature and the CellMarker database [27, 28] (S2A and S2B Fig). UMAP visualization of a cluster analysis identifies 22 different cell types, which were both observed in the control and ctu1 morphant group (Fig 3A). The ctu1 morphant demonstrated a higher proportion of cells in the G2/M phase and S phase (Fig 3B and BC). We found 57 downregulated genes and 41 upregulated genes in the ctu1 morphant samples. The upregulated genes are mainly enriched in cell cycle (Fig 3D) and DNA replication (Fig 3E), while the downregulated genes are predominantly enriched in the large ribosomal subunit (Fig 3F) and neuron system development (Fig 3G). These results suggest that ctu1 deficiency induces DNA damage, thereby causing cell-cycle arrest at G2/M phase and activating DNA repair response.
Fig 3. Single-cell transcriptome profiles of the ctu1 morphant and control zebrafish embryos.
UMAP visualization of all zebrafish cells, which are color-coded by cell type (A), and cell cycle phase (B). (C) The relative proportion of each cell cycle phase in the control and Ctu1 morphant. Gene sets of GSEA analysis shown are cell cycle (D), DNA repair (F), large ribosomal subunit (F) and nervous system development (G).
Heterogeneity analysis of the three main embryonic germ layers
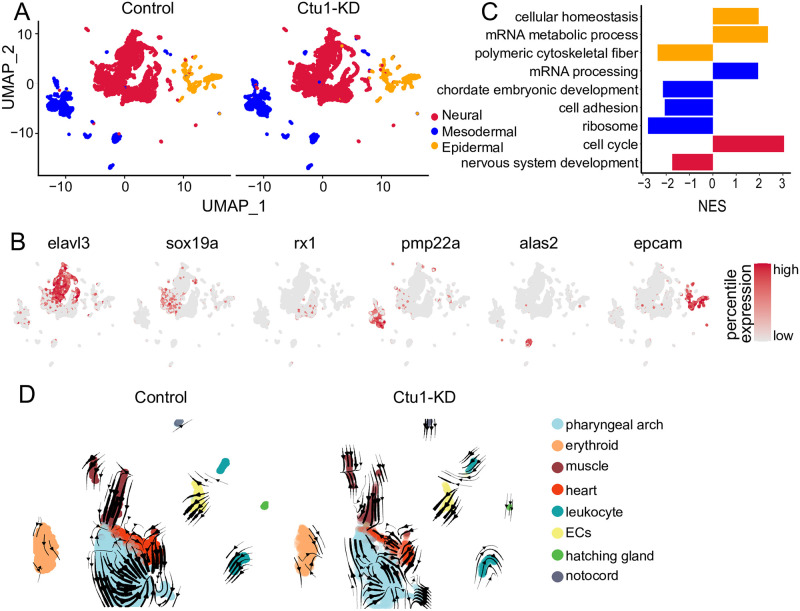
Three germ layers were isolated and DEGs analysis was conducted separately in the three germ layers-cells [27] (Fig 4A and 4B). For the epidermal cells, we identified four types, including the epidermal otic placode and three other cell subgroups marked by icn2, cldn, and lens, respectively (S3A Fig). GSEA analysis of the epidermal cells revealed that the absence of ctu1 significantly upregulation of genes associated with cellular homeostasis and mRNA metabolic process pathways, while downregulation of genes related to polymeric cytoskeletal fibers (Fig 4C, yellow). The mesoderm cells were also sub-grouped clusters including pharyngeal arches, erythrocytes, muscles, heart, leukocytes, endothelial cells, hatching glands, and notochord (S3B Fig). GSEA results of the mesoderm cells indicated that genes involved in the mRNA processing pathway were significantly upregulated. In contrast, genes associated with chordate embryonic development, cell adhesion, and ribosome pathways were significantly downregulated in ctu1 morphant (Fig 4C, blue). The neural cells were classified into the subgroups including the optic nerve, neural crest, retinal nerve, floorplate, and subpopulations marked by sox19a, elavl4, ebf2, phox2a, dlx1a and eomesa (S3C Fig). The GSEA analysis of neural cells demonstrated that a deficiency in ctu1 resulted in a significant increase in genes associated with the cell cycle pathway and a significant decrease in genes related to the neural development pathway (Fig 4C, red). In summary, DEGs analysis in the three germ layers cells implied that ctu1 plays a vital role in the mesodermal cells, while its impact on epidermal cells is not significant. We then conducted RNA velocity analysis on the mesodermal cells and observed that muscle differentiation primarily advances towards the pharyngeal arch and heart. Notably, the differentiation ability of the heart and pharyngeal arch was reduced in ctu1 morphant (Fig 4D). Consequently, ctu1 is essential for the normal growth and functional maintenance of the pharyngeal arch, heart and muscles.
Fig 4. Comparative analysis of the control and ctu1 morphant across three main embryonic germ layers.
(A) UMAP visualization of three germ layers. (B) UMAP visualization of key marker genes expression. Color scale represents log-normalized expression. The marker genes for neural cells are elavl3, sox19a, rx1; for mesodermal cells, the markers are pmp22a and alas2; and for e pidermal cells, the marker gene is epcam. (C) The bar plot shows signal pathways affected by ctu1 deficiency in epidermal, mesodermal, and neural cells as identified by GSEA. All terms demonstrate significant enriched (adj.p < 0.05) and normalized enrichment scores (NES) are shown. (D) RNA velocity plot of control and ctu1 morphant mesodermal cells.
ctu1 deficiency suppresses erythrocyte differentiation
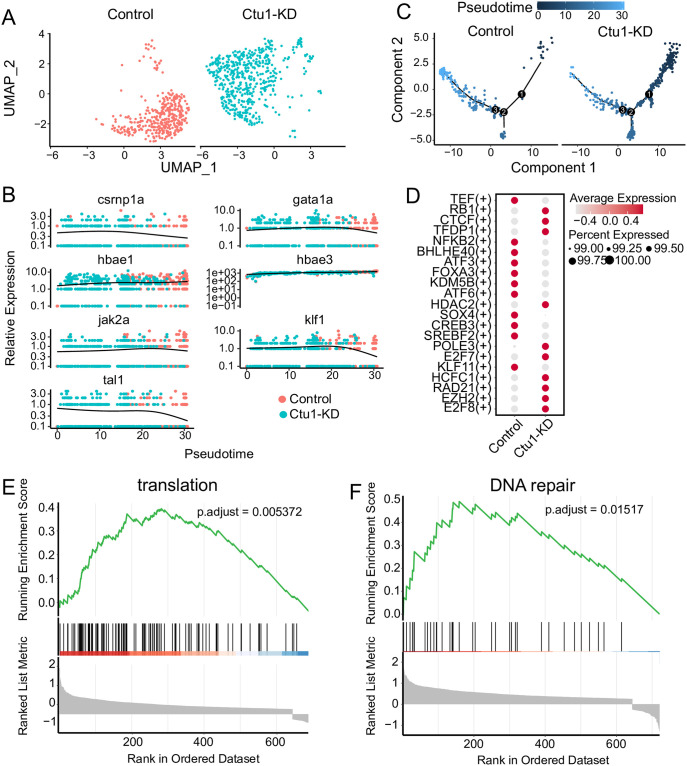
We extracted the erythrocyte populations of interest in our study for in-depth study. The transcriptome profiles of erythroid cells in the two groups were significantly different (Fig 5A). Previous research has established that crnp1a, gata1a, jak2a, klf1 and tal1 are necessary for the development of primitive hematopoiesis progenitors in zebrafish [29–33]. Primitive erythrocytes express embryonic globin genes (hbae3 and hbbe1.1) [34]. We calculated the relative expression levels of these hematopoietic marker genes as they changed over time in two samples (Fig 5B). The results showed that crnp1a, gata1a, jak2a, klf1, and tal1 were highly expressed in the ctu1 morphant, with their expression decreasing in the control group as pseudotime changes. On the other hand, the expressions of hbae1 and hbae3 are ubiquitous, with the highest expression levels in control group. The results of the pseudotime analysis showed that ctu1 morphant cells were distributed in the early stages of the differentiation trajectory (Fig 5C). Compared to the control, we identified regulators with high activity (Fig 5D), including e2f7, e2f8, hcfc1, and hcfc2, which play important roles in hematopoiesis [35, 36].rb1, tedp1, and ezh2 affect the development of erythroid cells [37–40]. In contrast, the activity-regulating factor klf11 [41], which is expressed in mature red blood cells, and the factors srebf2, creb3, and atf6 involved in endoplasmic reticulum stress [42–44], as well as the nfkb2 factor that plays a role in early differentiation of the bone marrow lineage [45], all show decreased activity. These results suggest that those specific regulons which are active in erythroid cells, exhibit difference between the control and ctu1 morphants. GSEA results indicated that significant upregulated genes associated with translation (Fig 5E), and DNA repair (Fig 5F) pathways following ctu1 gene silencing. These findings reinforce the heterogeneity of the two samples of erythroid cells and highlight that ctu1 knockdown significantly inhibits erythroid differentiation.
Fig 5. ctu1 deficiency suppresses erythrocyte differentiation.
(A) UMAP visualization of erythroid cells, colored according to samples. (B) The relative expression of the marker genes associated with erythroid differentiation in the pseudotime trajectories of control and ctu1 morphant. (C) Pseudotime trajectories of erythroid cells. (D) Dot plots show changes in the expression of transcription factors across different samples. The color and size of circles indicate the average expression level and percentage of cells. GSEA analysis of control and ctu1 morphant. Gene sets shown are translation (E), and DNA repair (F).
ctu1 deficiency reduces the activity of the angpt and notch signaling pathways originated from the endothelial cells in the mesoderm
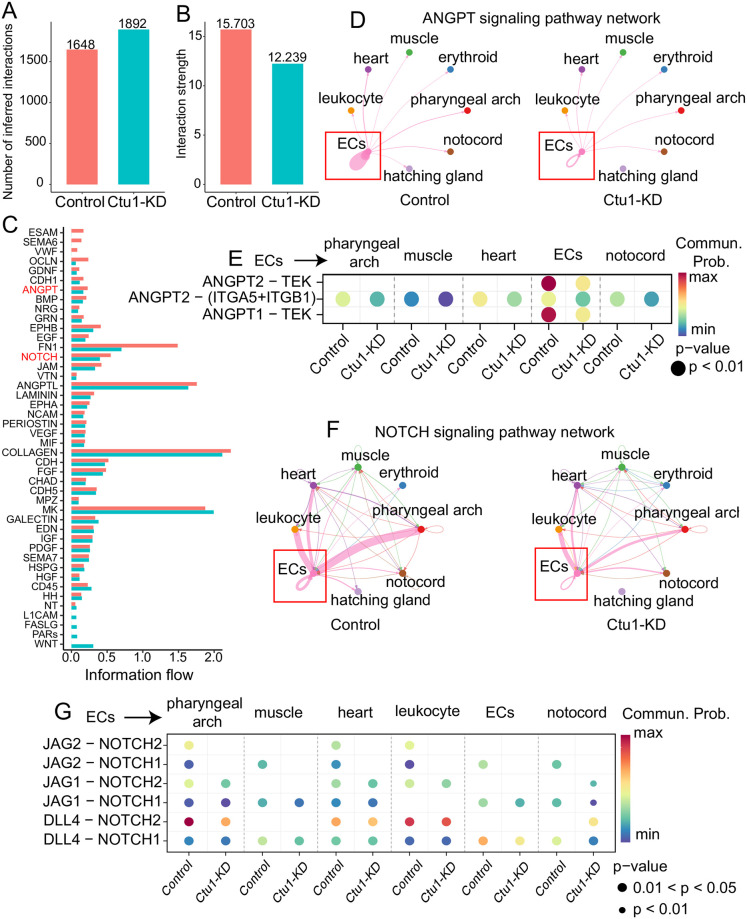
To explore further insights into the communication network among mesoderm cells in ctu1 morphant, we utilized the CellChat to infer the ligand-receptor pairs involved in interactions between different cell types. Initially, a comprehensive assessment of both the volume and intensity of cellular communication was conducted. The total communication volume with control and ctu1 morphant increases, yet the overall communication intensity diminishes (Fig 6A and 6B). In particular, the intensity of several pro-angiogenesis signaling cascades, like endothelial cell-selective adhesion molecule (esam), semaphorin 6A (sema6), Von Willebrand factor (vwf), occludin (ocln), and bone morphogenetic protein (bmp), were significantly attenuated when Ctu1 was comprised (Fig 6C). In particularly, cell-cell communication showed that the strength of angiopoietin (angpt) signaling pathway network which originated from endothelial cells was significantly attenuated in ctu1 morphant compared to the control (Fig 6D). The probability of angpt-tek communication was significantly attenuated in the ctu1 morphant (Fig 6E). The dll4-mediated notch signaling pathway plays an important role in the occurrence and formation of embryonic blood vessels [46, 47]. Reduced levels of dll4-notch expression in zebrafish lead to abnormal endothelial cell numbers and exhibit ISV vascular defects [48, 49]. Here our data indicated that dll4-notch signaling which was predominantly derived from endothelial cells was significantly attenuated in pharyngeal arch, muscle, and endothelial cells (Fig 6F and 6G). In conclusion, the decreased autocrine angpt-tek and dll4-notch signalings in endothelial cells are probably responsible for the defective angiogenesis after ctu1 suppression in zebrafish.
Fig 6. ctu1 deficiency reduces the activity of the angpt and notch signaling pathways originated from the endothelial cells in mesoderm.
Bar plot shows overview number (A) and strength (B) in control and ctu1 morphant. (C) Ranking of active signaling pathways in control and ctu1 morphant based on their overall information flow within the inferred cellular networks. Signaling pathways are colored according to condition where they are enriched. (D) The chord plot shows angpt signaling in sending and receiving cells. Nodes are colored by celltypes. The thickness of the line represents the strength of the signal. (E) Dot plots show communication probability of angpt signaling between endothelial cells (senders) and each celltypes (receivers). Blue, low communication probability; red, high communication probability. Size of circle represents the pvalue of cells with communication probability. Chord plots (F) and Dot plots (G) shows notch signaling pathway network.
Effects of CTU1 upon endothelial cells
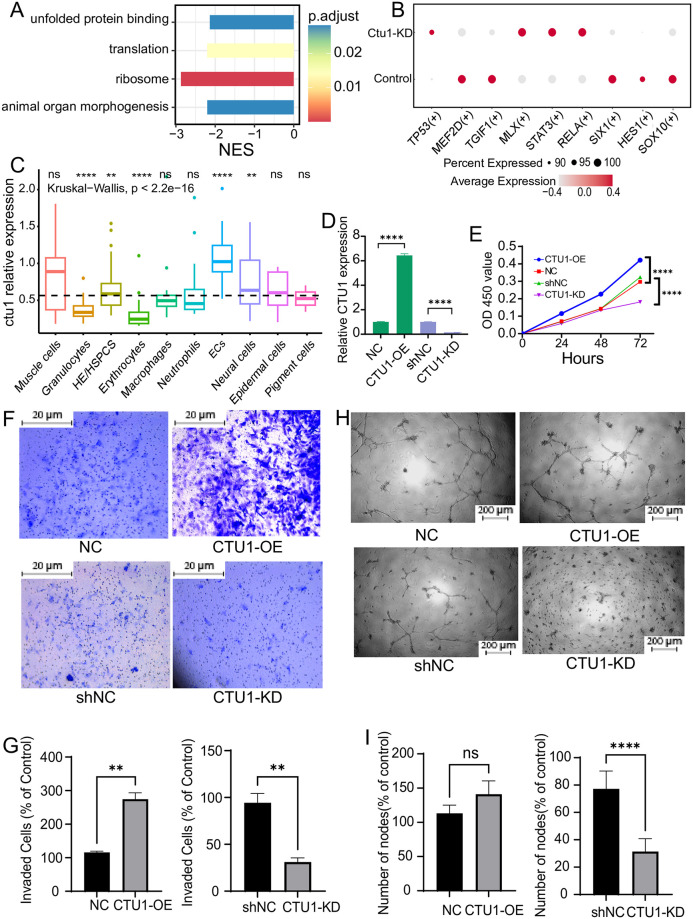
Endothelial cells play critical roles in angiogenesis. We performed DEGs analysis with the single-cell transcriptome data of the zebrafish endothelial cells (S3D Fig), and found that the decreased genes in the ctu1 morphant was enriched in unfolded protein binding, translation, ribosome, and animal organ morphogenesis (Fig 7A). Furthermore, we conducted GRN analysis by utilizing pySCENIC to elucidate the transcriptional regulatory changes of ctu1. The results revealed that ctu1-deficient endothelial cells exhibited elevated activity levels of tp53, mlx, stat3, rela and decreased activity of mef2d, tgif1, six1, hes1 and sox10 (Fig 7B). The myocyte enhancement factor mef2d promotes tumor angiogenesis in vitro and in vivo and induces the expression of pro-antigenic cytokines in colorectal cancer cells [50]. sox10 stem cells have the differentiate into perivascular cells, which play a crucial role in stabilizing newly formed microvessels [51].
Fig 7. Single-cell data analysis of the endothelial cells in zebrafish, and the angiogenesis-related behaviors of human endothelial cells with differential expression of CTU1.
(A) The bar plot shows signal pathways affected by ctu1 deficiency in endothelial cells as identified by GSEA. (B) The dot plot illustrates the variation in transcription factor expression in control and ctu1 morphant, with the color and size of the circles representing the average expression level and the proportion of cells, respectively. (C) The box plot illustrates the relative expression levels of ctu1 of each cell type in 36-hpf zebrafish. ****, p < 0.0001. (D) The CTU1 expression levels of CTU1-KD and CTU1-OE HMEC-1 cells after exposing to the corresponding lentivirus. (E) The growth curve of CTU1-KD, CTU1-OE, and vehicle control HMEC-1 cells. Representative images (F) and quantifications of the migrated cells (G, n = 3 independent experiments) of HMEC-1 cells with different expression levels of CTU1 in the transwell assay. Scale bars represent 20 μm. Comparisons between each group were analyzed using Student’s t-test. **, p < 0.01. Representative images (H) and quantifications of the branch points (I, n = 3 independent experiments) of HMEC-1 cells with different expression levels of CTU1 in the tube formation assay. Scale bar, 200 μm. Comparisons between each group were analyzed using Student’s t-test. ****, p < 0.0001.
By analyzing the GSE186423 dataset, which encompasses a total of 4,583 endothelial and hematopoietic progenitor cells from 36-hpf zebrafish embryos, we discovered that ctu1 exhibits the highest expression levels in zebrafish endothelial cells (Fig 7C). To confirm the effect of CTU1 on angiogenesis, we also applied human endothelial cells HMEC-1 in vitro, and transfected them with CTU1 over-expressing or silencing lentivirus to get CTU1-OE and CTU1-KD HMEC-1 cells (Fig 7D). We evaluated the cell growth, migration and tube formation ability of those HMEC-1 cells. Our results indicated that CTU1-OE significantly increased cell number, while CTU1-KD notably reduced cell viability after 72 hours’ culture (Fig 7E). In addition, the overexpression of CTU1 significantly enhanced the migratory capacity of HMEC-1 cells, and reducing CTU1 expression modestly decreased their migratory capacity (Fig 7F and 7G). In the tube formation assay, our experiments clearly showed that at the 16-hour time point, there were significant differences in tube branch length and total area covered among the CTU1-KD, CTU1-OE, and control HMEC-1 cells. In particular, CTU1-KD HMEC-1 cells failed to form vasculature branches (Fig 7H and 7I).
Discussion
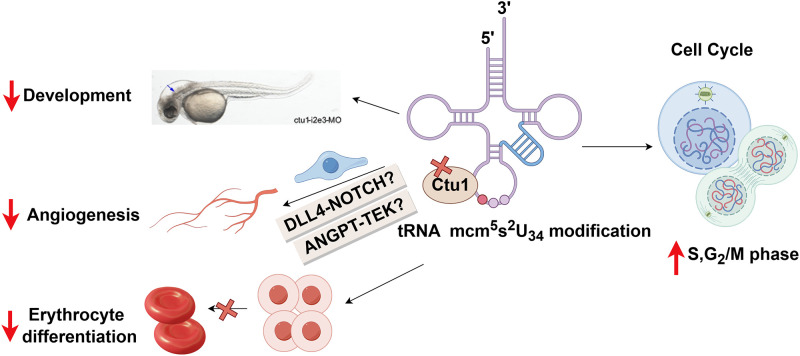
In our study, we demonstrate that the conserved wobble uridine tRNA modifying enzyme CTU1 is critical for zebrafish embryo angiogenesis, development and differentiation. We show that ctu1 deficiency leads to defects in cell cycle progression and nerve development, as well as the disturbed differentiation of nerve cell and erythrocyte. Endothelial cells are the most affected cell types when comprising Ctu1, with the decreased cell to cell communication of several pro-angiogentic signaling cascades, e.g. esam5, bmp, ocld, dll4-notch and angpt-tek (Fig 8). Our work is of great significance in revealing the vital role and the underlying mechanisms of CTU1 in angiogenesis, development and differentiation in vertebrates.
Fig 8. Model of the consequences of Ctu1 deficiency in zebrafish, figured by Figdraw.
Recently, the essential role of mcm5s2U34 tRNA modification and their modifying enzymes in proliferation and development has been demonstrated from yeast to mammals [7]. For example, double deletion of the partner enzymes (Ctu1 and Elp3) is lethal to the cell in yeast [14]. Inactivation of the CTU complex leads to a thermosensitive decrease with aberrant development in the nematode and fission yeast [15]. In mice, a deficiency in Elp1 causes male infertility [52], while Elp3-KO embryos exhibits significant growth retardation and fails to develop beyond E12.5 [13]. Here our finding provides the first experimental evidence that s2U34 tRNA modifying enzyme CTU1 is essential in cellular proliferation and development in vertebrates.
mcm5s2U34 tRNA modifications and their modifying enzymes are involved in human neurological diseases, and normal neurons development and generation of neurons as well [53]. Mutations in the six subunits of Elongator, and Alkbh8 is associated with human neurological diseases [9, 10, 54, 55]. Comprehensive transcriptome analysis in mouse embryos indicates that Elp1 is essential for the expression of genes responsible for nervous system development [56]. Elp1 and Elp2 mutation also causes a severe neurodevelopmental phenotype in mice models [12, 57]. Meanwhile, homozygous Ctu2 mutation was identified in patients diagnosed with a novel multiple congenital anomalies syndrome called DREAM-PL which is characterized by dysmorphic facies, renal agenesis, ambiguous genitalia, microcephaly, and lissencephaly [11, 58]. Here our analysis demonstrated that ctu1 deficiency causes downregulation of genes in neuron system development, which provides further evidence and also the underlying molecular mechanisms that ctu1 regulate neurological development in zebrafish.
Additionally, our analysis shows that the expression of the marker genes for early hematopoietic and mature erythrocyte in the control and ctu1 deficient erythrocytes were separately distributed by pseudotime, and the ctu1 deficient ones were mainly distributed in the early stages along the differentiation trajectory. The erythropoisis defects in ctu1 deficient zebrafish embryo, together with the fact that loss of Elp3 causes bone marrow failure and compromises the grafting activity of hematopoietic stem cells [59], highlights the vital role of mcm5s2U34 tRNA modifications in hematopoisis.
We observed defective angiogenesis in ctu1 morphant zebrafish larvae. During vertebrate embryogenesis, the pharyngeal arch artery connects the heart to the dorsal aorta, and defects in pharyngeal bursa development are commonly associated with abnormal vascular development [60]. RNA velocity analysis reveals reduced flow flux of muscle-pharyngeal arches in the ctu1 deficient embryo, which maybe responsible for the observed vascular defects. Furthermore, cell communication analysis indicates that several pro-angiogenesis genes, like esam [61], sema6a [62], ocln, bmp, fn1, angpt [63], and notch [46, 47] are all reduced in the mesodermal layer of the ctu1 morphant zebrafish. Their critical roles in neovascularization and angiogenesis have been well documented. angpt and tek serve as major regulators of angiogenesis in both physiological and pathologic conditions [64]. ocln is a functional marker of vascular endothelial cells on tube-forming activity plays, and plays a critical role in tube formation, sprouting, and proliferation [65]. fn1 and bmp signaling has been shown to be important for angiogenesis [66]. Reduced levels of dll4-notch expression in zebrafish lead to abnormal endothelial cell numbers and exhibit ISV vascular defects [48, 49], which was similar with our observation in the transparent zebrafish emybro after ctu1 deficiency. In summary, the reduced communication of those pro-angiogenic genes and signaling cascades, and the defective angiogenesis phenotype, identifies ctu1 as a novel regulator for angiogenesis and neuovasculogenesis in zebrafish.
Furthermore, as ctu1 is highly expressed in endothelial cells, and endothelial cells are hot target after ctu1 deficiency by bioinformatics analysis. We further analyzed the transcriptome data of those endothelial cells, and found that ctu1 knockdown significantly inhibited the expression of ribosomal genes in endothelial cells. Ribosomes are the main effector of the translational machinery to synthesize proteins. Decrease of the ribosomal subunit-selective homeostasis and inhibition of the translation process has been documented to be decreased with reduced tRNA biosynthetic activity [67]. The existence of a connection between tRNA modification biology and proteins in the ribosome has been shown in S. pombe and S. cerevisiae [68]. Here our analysis data also links mcm5s2U34 tRNA modifying enzyme Ctu1 to ribosomal gene expression, and the biological function of endothelial cells.
In conclusion, this is the first study which implies that cytosolic thiouridylase CTU1 is essential for angiogenesis and embryonic development. Our discovery indicates that the epigenetic modification of tRNA-U34, especially s2U34 modification and its modifying enzymes, are key regulators of angiogenesis and differentiation in vertebrates.
Supporting information
(A and B) Bright-fieldand EGFP fluorescentimages depict the overall morphology of control and ctu1 morphant at 2-dpf. (C) Urm1-targeted MO design strategy. (D) PCR analysis of control and urm1 morphant. (E and F) Bright-fieldand EGFP fluorescentimages depict the overall morphology of control and urm1 morphant at 2-dpf. Quantification of the number of complete ISVs (G) and CVP (H). Columns, mean; bars, SEM (n = 10; unpaired student’s t-test; ***, p < 0.001).
(TIF)
(A) UMAP visualization of zebrafish cells, which are colored by clusters. (B) The violin plot shows the expression of top marker genes in each cluster.
(TIF)
(A) UMAP visualization of epidermal cells, which are colored by cell types. (B) UMAP visualization of mesoderm cells, which are colored by cell types. (C) UMAP visualization of neural cells, which are colored by cell types. (D) UMAP visualization of endothelial cells, which are colored by clusters.
(TIF)
(TIF)
Data Availability
All data are accessible in NODE (https://www.biosino.org/node) with the accession number OEX00002144 or through the URL: https://www.biosino.org/node/experiment/detail/OEX00002144.
Funding Statement
This study was funded by the Science and Technology Commission of Shanghai Municipality (22010502500).
References
- 1.Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol. 2021;22(6):375–92. Epub 2021/03/05. doi: 10.1038/s41580-021-00342-0 [DOI] [PubMed] [Google Scholar]
- 2.Nakai Y, Nakai M, Yano T. Sulfur Modifications of the Wobble U34 in tRNAs and their Intracellular Localization in Eukaryotic Cells. Biomolecules. 2017;7(1). doi: 10.3390/biom7010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaffrath R, Leidel SA. Wobble uridine modifications-a reason to live, a reason to die?! RNA biology. 2017;14(9):1209–22. Epub 2017/03/10. doi: 10.1080/15476286.2017.1295204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leidel S, Pedrioli PGA, Bucher T, Brost R, Costanzo M, Schmidt A, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458(7235):228–32. doi: 10.1038/nature07643 [DOI] [PubMed] [Google Scholar]
- 5.Rapino F, Zhou Z, Roncero Sanchez AM, Joiret M, Seca C, El Hachem N, et al. Wobble tRNA modification and hydrophilic amino acid patterns dictate protein fate. Nat Commun. 2021;12(1):2170. Epub 2021/04/17. doi: 10.1038/s41467-021-22254-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuorto F, Lyko F. Genome recoding by tRNA modifications. Open Biol. 2016;6(12). Epub 2016/12/16. doi: 10.1098/rsob.160287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. 2018;361(6409):1346–9. doi: 10.1126/science.aau1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Tao E-W, Tan J, Gao Q-Y, Chen Y-X, Fang J-Y. tRNA modifications: insights into their role in human cancers. Trends in Cell Biology. 2023;33(12):1035–48. doi: 10.1016/j.tcb.2023.04.002 [DOI] [PubMed] [Google Scholar]
- 9.Gaik M, Kojic M, Wainwright BJ, Glatt S. Elongator and the role of its subcomplexes in human diseases. EMBO Mol Med. 2023;15(2):e16418. Epub 2022/12/01. doi: 10.15252/emmm.202216418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monies D, Vågbø CB, Al-Owain M, Alhomaidi S, Alkuraya FS. Recessive Truncating Mutations in ALKBH8 Cause Intellectual Disability and Severe Impairment of Wobble Uridine Modification. The American Journal of Human Genetics. 2019;104(6):1202–9. doi: 10.1016/j.ajhg.2019.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaheen R, Mark P, Prevost CT, AlKindi A, Alhag A, Estwani F, et al. Biallelic variants in CTU2 cause DREAM-PL syndrome and impair thiolation of tRNA wobble U34. Hum Mutat. 2019;40(11):2108–20. Epub 2019/07/14. doi: 10.1002/humu.23870 [DOI] [PubMed] [Google Scholar]
- 12.Kojic M, Gawda T, Gaik M, Begg A, Salerno-Kochan A, Kurniawan ND, et al. Elp2 mutations perturb the epitranscriptome and lead to a complex neurodevelopmental phenotype. Nat Commun. 2021;12(1):2678. Epub 2021/05/13. doi: 10.1038/s41467-021-22888-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo H, Son D, Jang Y-J, Hong K. Indispensable role for mouse ELP3 in embryonic stem cell maintenance and early development. Biochemical and Biophysical Research Communications. 2016;478(2):631–6. doi: 10.1016/j.bbrc.2016.07.120 [DOI] [PubMed] [Google Scholar]
- 14.Bjork GR, Huang B, Persson OP, Bystrom AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13(8):1245–55. Epub 2007/06/27. doi: 10.1261/rna.558707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci U S A. 2008;105(14):5459–64. doi: 10.1073/pnas.0709404105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Vazquez J, Vargas-Perez I, Sanso M, Buhne K, Carmona M, Paulo E, et al. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013;9(7):e1003647. doi: 10.1371/journal.pgen.1003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damon JR, Pincus D, Ploegh HL. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Molecular biology of the cell. 2015;26(2):270–82. doi: 10.1091/mbc.E14-06-1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm L, Mason E, Yu H, Dudczig S, Panara V, Chen T, et al. Single-cell analysis of lymphatic endothelial cell fate specification and differentiation during zebrafish development. EMBO J. 2023:e112590. Epub 2023/03/14. doi: 10.15252/embj.2022112590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasevicius A, Ekker SC. Effective targeted gene ’knockdown’ in zebrafish. Nat Genet. 2000;26(2):216–20. doi: 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- 20.Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nature Protocols. 2010;5(9):1518–34. doi: 10.1038/nprot.2010.113 [DOI] [PubMed] [Google Scholar]
- 21.Bresciani E, Broadbridge E, Liu PP. An efficient dissociation protocol for generation of single cell suspension from zebrafish embryos and larvae. MethodsX. 2018;5:1287–90. Epub 2018/10/27. doi: 10.1016/j.mex.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018;560(7719):494–8. Epub 2018/08/10. doi: 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. Epub 2021/02/19. doi: 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15(7):2247–76. Epub 2020/06/21. doi: 10.1038/s41596-020-0336-2 [DOI] [PubMed] [Google Scholar]
- 25.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–6. Epub 2017/10/11. doi: 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S. Transgenic Zebrafish. In: Tuan RS, Lo CW, editors. Developmental Biology Protocols: Volume II. Totowa, NJ: Humana Press; 2000. p. 375–83. [Google Scholar]
- 27.Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. 2018;360(6392):981–7. doi: 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu C, Li T, Xu Y, Zhang X, Li F, Bai J, et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023;51(D1):D870–D6. Epub 2022/10/28. doi: 10.1093/nar/gkac947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klymkowsky M, Espina J, Feijóo CG, Solís C, Glavic A. csrnp1a Is Necessary for the Development of Primitive Hematopoiesis Progenitors in Zebrafish. PLoS ONE. 2013;8(1). doi: 10.1371/journal.pone.0053858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma ACH, Ward AC, Liang R, Leung AYH. The role of jak2a in zebrafish hematopoiesis. Blood. 2007;110(6):1824–30. doi: 10.1182/blood-2007-03-078287 [DOI] [PubMed] [Google Scholar]
- 31.Tyrkalska SD, Pérez-Oliva AB, Rodríguez-Ruiz L, Martínez-Morcillo FJ, Alcaraz-Pérez F, Martínez-Navarro FJ, et al. Inflammasome Regulates Hematopoiesis through Cleavage of the Master Erythroid Transcription Factor GATA1. Immunity. 2019;51(1):50–63.e5. doi: 10.1016/j.immuni.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki H, Ogawa T, Fujita S, Sone R, Kawahara A. Cooperative contributions of the klf1 and klf17 genes in zebrafish primitive erythropoiesis. Sci Rep. 2023;13(1):12279. Epub 2023/08/11. doi: 10.1038/s41598-023-39196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodell MA, Qian F, Zhen F, Xu J, Huang M, Li W, et al. Distinct Functions for Different scl Isoforms in Zebrafish Primitive and Definitive Hematopoiesis. PLoS Biology. 2007;5(5). doi: 10.1371/journal.pbio.0050132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, et al. Characterization of embryonic globin genes of the zebrafish. Developmental Biology. 2003;255(1):48–61. doi: 10.1016/s0012-1606(02)00041-6 [DOI] [PubMed] [Google Scholar]
- 35.Thurlings I, Martinez-Lopez LM, Westendorp B, Zijp M, Kuiper R, Tooten P, et al. Synergistic functions of E2F7 and E2F8 are critical to suppress stress-induced skin cancer. Oncogene. 2017;36(6):829–39. Epub 2016/07/28. doi: 10.1038/onc.2016.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Yu M, Yang C, Li Q. Upregulation of HCFC1 expression promoted hepatocellular carcinoma progression through inhibiting cell cycle arrest and correlated with immune infiltration. J Cancer. 2023;14(8):1381–97. Epub 2023/06/07. doi: 10.7150/jca.84579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Liu D, Xue F, Zhang H, Yang Q, Sun L, et al. Stage-specific dual function: EZH2 regulates human erythropoiesis by eliciting histone and non-histone methylation. Haematologica. 2023;108(9):2487–502. Epub 2023/04/07. doi: 10.3324/haematol.2022.282016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Loyd MR, Randall MS, Morris JJ, Shah JG, Ney PA. Repression by RB1 characterizes genes involved in the penultimate stage of erythroid development. Cell Cycle. 2015;14(21):3441–53. Epub 2015/09/24. doi: 10.1080/15384101.2015.1090067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taimatsu K, Takubo K, Maruyama K, Suda T, Kudo A. Proliferation following tetraploidization regulates the size and number of erythrocytes in the blood flow during medaka development, as revealed by the abnormal karyotype of erythrocytes in the medaka TFDP1 mutant. Dev Dyn. 2015;244(5):651–68. Epub 2015/02/05. doi: 10.1002/dvdy.24259 [DOI] [PubMed] [Google Scholar]
- 40.Nakajima R, Deguchi R, Komori H, Zhao L, Zhou Y, Shirasawa M, et al. The TFDP1 gene coding for DP1, the heterodimeric partner of the transcription factor E2F, is a target of deregulated E2F. Biochem Biophys Res Commun. 2023;663:154–62. Epub 2023/05/04. doi: 10.1016/j.bbrc.2023.04.092 [DOI] [PubMed] [Google Scholar]
- 41.Emery DW, Gavriilidis G, Asano H, Stamatoyannopoulos G. The transcription factor KLF11 can induce gamma-globin gene expression in the setting of in vivo adult erythropoiesis. J Cell Biochem. 2007;100(4):1045–55. Epub 2006/11/30. doi: 10.1002/jcb.21093 [DOI] [PubMed] [Google Scholar]
- 42.Oh-Hashi K, Yamamoto A, Murase R, Hirata Y. Comparative Analysis of CREB3 and CREB3L2 Protein Expression in HEK293 Cells. Int J Mol Sci. 2021;22(5). Epub 2021/04/04. doi: 10.3390/ijms22052767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter F, O’Brien A, Concannon CG, Dussmann H, Prehn JHM. ER stress signaling has an activating transcription factor 6alpha (ATF6)-dependent "off-switch". J Biol Chem. 2018;293(47):18270–84. Epub 2018/10/06. doi: 10.1074/jbc.RA118.002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong G, Huang X, Wu L, Jiang S, Tan Q, Chen S. SREBF2 triggers endoplasmic reticulum stress and Bax dysregulation to promote lipopolysaccharide-induced endothelial cell injury. Cell Biol Toxicol. 2022;38(1):185–201. Epub 2021/03/08. doi: 10.1007/s10565-021-09593-1 [DOI] [PubMed] [Google Scholar]
- 45.De Molfetta GA, Luciola Zanette D, Alexandre Panepucci R, Dos Santos AR, da Silva WA Jr., Antonio Zago M. Role of NFKB2 on the early myeloid differentiation of CD34+ hematopoietic stem/progenitor cells. Differentiation. 2010;80(4–5):195–203. Epub 2010/08/17. doi: 10.1016/j.diff.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 46.Gale NW, Dominguez MG, Noguera I, Pan L, Hughes V, Valenzuela DM, et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. 2004;101(45):15949–54. doi: 10.1073/pnas.0407290101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson O, Novodvorsky P, Gray C, Rothman AMK, Lawrie A, Crossman DC, et al. Blood flow suppresses vascular Notch signalling via dll4 and is required for angiogenesis in response to hypoxic signalling. Cardiovascular Research. 2013;100(2):252–61. doi: 10.1093/cvr/cvt170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Therapontos C, Vargesson N. Zebrafish notch signalling pathway mutants exhibit trunk vessel patterning anomalies that are secondary to somite misregulation. 2010;239(10):2761–8. doi: 10.1002/dvdy.22410 [DOI] [PubMed] [Google Scholar]
- 49.Lin M, Dai H, Zheng L, Yu P, Chen Y, Hong R, et al. Tryptanthrin inhibits tumor angiogenesis via Notch/Dll4 signaling pathway in zebrafish. Translational Cancer Research. 2023;12(10):2660–72. doi: 10.21037/tcr-23-925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu S, Ilyas I, Little PJ, Li H, Kamato D, Zheng X, et al. Endothelial Dysfunction in Atherosclerotic Cardiovascular Diseases and Beyond: From Mechanism to Pharmacotherapies. Pharmacol Rev. 2021;73(3):924–67. Epub 2021/06/06. doi: 10.1124/pharmrev.120.000096 [DOI] [PubMed] [Google Scholar]
- 51.Wang D, Wang A, Wu F, Qiu X, Li Y, Chu J, et al. Sox10(+) adult stem cells contribute to biomaterial encapsulation and microvascularization. Sci Rep. 2017;7:40295. Epub 2017/01/11. doi: 10.1038/srep40295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schimenti JC, Lin F-J, Shen L, Jang C-W, Falnes PØ, Zhang Y. Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression. PLoS Genetics. 2013;9(5). doi: 10.1371/journal.pgen.1003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bednářová A, Hanna M, Durham I, VanCleave T, England A, Chaudhuri A, et al. Lost in Translation: Defects in Transfer RNA Modifications and Neurological Disorders. Frontiers in Molecular Neuroscience. 2017;10. doi: 10.3389/fnmol.2017.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin TY, Abbassi NEH, Zakrzewski K, Chramiec-Glabik A, Jemiola-Rzeminska M, Rozycki J, et al. The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat Commun. 2019;10(1):625. Epub 2019/02/09. doi: 10.1038/s41467-019-08579-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waqas A, Nayab A, Shaheen S, Abbas S, Latif M, Rafeeq MM, et al. Case Report: Biallelic Variant in the tRNA Methyltransferase Domain of the AlkB Homolog 8 Causes Syndromic Intellectual Disability. Front Genet. 2022;13:878274. Epub 2022/05/17. doi: 10.3389/fgene.2022.878274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morini E, Gao D, Logan EM, Salani M, Krauson AJ, Chekuri A, et al. Developmental regulation of neuronal gene expression by Elongator complex protein 1 dosage. Journal of Genetics and Genomics. 2022;49(7):654–65. doi: 10.1016/j.jgg.2021.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kojic M, Abbassi NEH, Lin T-Y, Jones A, Wakeling EL, Clement E, et al. A novel ELP1 mutation impairs the function of the Elongator complex and causes a severe neurodevelopmental phenotype. Journal of Human Genetics. 2023;68(7):445–53. doi: 10.1038/s10038-023-01135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaheen R, Patel N, Shamseldin H, Alzahrani F, Al-Yamany R, AL A, et al. Accelerating matchmaking of novel dysmorphology syndromes through clinical and genomic characterization of a large cohort. Genetics in medicine: official journal of the American College of Medical Genetics. 2016;18(7):686–95. doi: 10.1038/gim.2015.147 [DOI] [PubMed] [Google Scholar]
- 59.Rosu A, El Hachem N, Rapino F, Rouault-Pierre K, Jorssen J, Somja J, et al. Loss of tRNA-modifying enzyme Elp3 activates a p53-dependent antitumor checkpoint in hematopoiesis. Journal of Experimental Medicine. 2021;218(3). doi: 10.1084/jem.20200662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao A, Zhang M, Li L, Liu J, Ning G, Cao Y, et al. Pharyngeal pouches provide a niche microenvironment for arch artery progenitor specification. Development. 2021;148(2). Epub 2020/12/19. doi: 10.1242/dev.192658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inoue M, Ishida T, Yasuda T, Toh R, Hara T, Cangara HM, et al. Endothelial cell-selective adhesion molecule modulates atherosclerosis through plaque angiogenesis and monocyte–endothelial interaction. Microvascular Research. 2010;80(2):179–87. doi: 10.1016/j.mvr.2010.04.005 [DOI] [PubMed] [Google Scholar]
- 62.Chen L-H, Liao C-Y, Lai L-C, Tsai M-H, Chuang EY. Semaphorin 6A Attenuates the Migration Capability of Lung Cancer Cells via the NRF2/HMOX1 Axis. Scientific Reports. 2019;9(1):13302. doi: 10.1038/s41598-019-49874-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butkiewicz D, Gdowicz-Kłosok A, Krześniak M, Rutkowski T, Krzywon A, Cortez AJ, et al. Association of Genetic Variants in ANGPT/TEK and VEGF/VEGFR with Progression and Survival in Head and Neck Squamous Cell Carcinoma Treated with Radiotherapy or Radiochemotherapy. Cancers. 2020;12(6). doi: 10.3390/cancers12061506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Augustin HG, Young Koh G, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin–Tie system. Nature Reviews Molecular Cell Biology. 2009;10(3):165–77. doi: 10.1038/nrm2639 [DOI] [PubMed] [Google Scholar]
- 65.Kanayasu-Toyoda T, Ishii-Watabe A, Kikuchi Y, Kitagawa H, Suzuki H, Tamura H, et al. Occludin as a functional marker of vascular endothelial cells on tube-forming activity. 2018;233(2):1700–11. doi: 10.1002/jcp.26082 [DOI] [PubMed] [Google Scholar]
- 66.Derwall M, Malhotra R, Lai CS, Beppu Y, Aikawa E, Seehra JS, et al. Inhibition of bone morphogenetic protein signaling reduces vascular calcification and atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32(3):613–22. Epub 2012/01/10. doi: 10.1161/ATVBAHA.111.242594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Itani OA, Zhong X, Tang X, Scott BA, Yan JY, Flibotte S, et al. Coordinate Regulation of Ribosome and tRNA Biogenesis Controls Hypoxic Injury and Translation. Curr Biol. 2021;31(1):128–37.e5. Epub 2020/11/07. doi: 10.1016/j.cub.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jackman J, De Zoysa T, Hauke AC, Iyer NR, Marcus E, Ostrowski SM, et al. A connection between the ribosome and two S. pombe tRNA modification mutants subject to rapid tRNA decay. PLOS Genetics. 2024;20(1). doi: 10.1371/journal.pgen.1011146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A and B) Bright-fieldand EGFP fluorescentimages depict the overall morphology of control and ctu1 morphant at 2-dpf. (C) Urm1-targeted MO design strategy. (D) PCR analysis of control and urm1 morphant. (E and F) Bright-fieldand EGFP fluorescentimages depict the overall morphology of control and urm1 morphant at 2-dpf. Quantification of the number of complete ISVs (G) and CVP (H). Columns, mean; bars, SEM (n = 10; unpaired student’s t-test; ***, p < 0.001).
(TIF)
(A) UMAP visualization of zebrafish cells, which are colored by clusters. (B) The violin plot shows the expression of top marker genes in each cluster.
(TIF)
(A) UMAP visualization of epidermal cells, which are colored by cell types. (B) UMAP visualization of mesoderm cells, which are colored by cell types. (C) UMAP visualization of neural cells, which are colored by cell types. (D) UMAP visualization of endothelial cells, which are colored by clusters.
(TIF)
(TIF)
Data Availability Statement
All data are accessible in NODE (https://www.biosino.org/node) with the accession number OEX00002144 or through the URL: https://www.biosino.org/node/experiment/detail/OEX00002144.