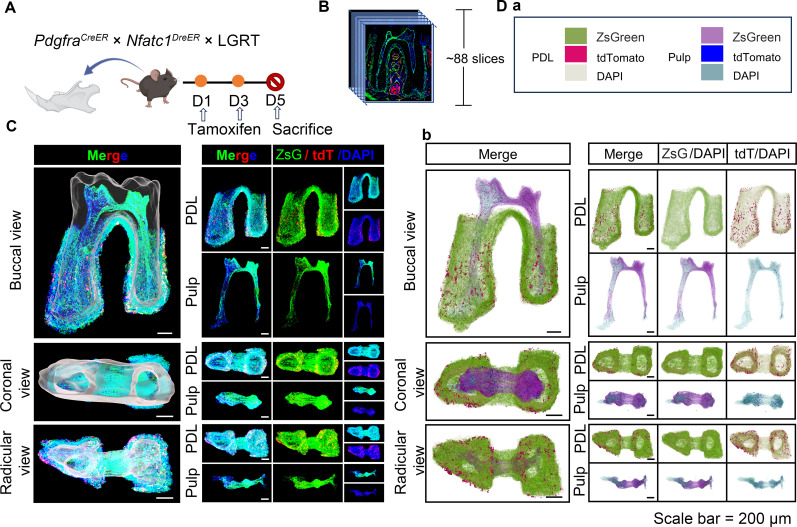
Figure 4. Using traditional serial section for imaging of mandible of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse).
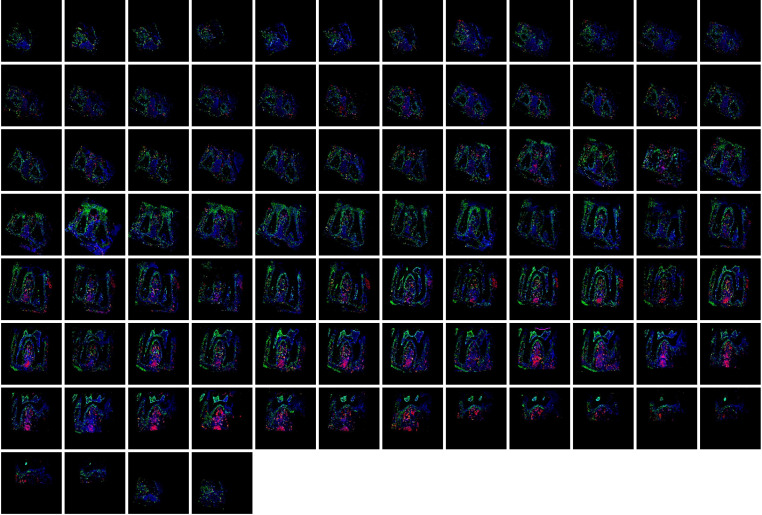
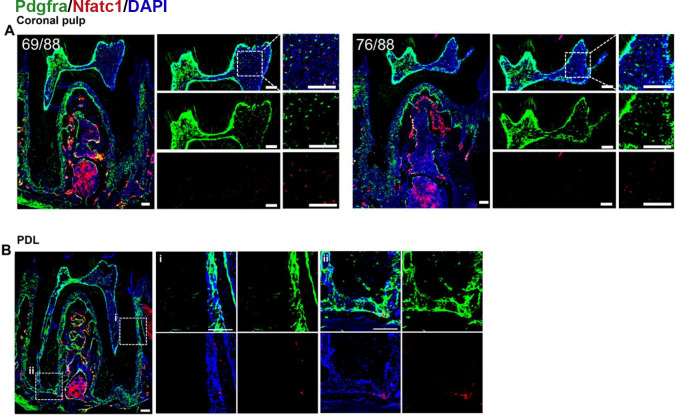
(A) Schematic illustration of lineaging tracing in PdgfraCreER×Nfatc1DreER× LGRT mice. (B) Operation process of frozen section. Using this procedure, a total of 88 slices were collected in this sample. (C) Mandible M1 after 3D reconstruction by Imaris, including pulp and PDL with virtual dentin shell (white) in buccal view, coronal view and radicular view (scale bar = 200 μm). (D) 3D reconstruction of mandible M1 by DICOM-3D; in PDL, ZsGreen+ cells in green, tdTomato+ cells in rose red; in pulp, ZsGreen+ cells in purple, tdTomato+ cells in blue. The image stack was also displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively (D-b), scale bar = 200 μm. (D-a): The legend of (D-b).