Abstract
Atopic dermatitis, food allergy, allergic rhinitis, and asthma are among the most common diseases in childhood. They are heterogeneous diseases, can co-exist in their development, and manifest complex associations with other disorders and environmental and hereditary factors. Elucidating these intricacies by identifying clinically distinguishable groups and actionable risk factors will allow for better understanding of the diseases, which will enhance clinical management and benefit society and affected individuals and families. Artificial intelligence (AI) is a promising tool in this context, enabling discovery of meaningful patterns in complex data. Numerous studies within pediatric allergy have and continue to use AI, primarily to characterize disease endotypes/phenotypes and to develop models to predict future disease outcomes. However, most implementations have used relatively simplistic data from one source, such as questionnaires. In addition, methodological approaches and reporting are lacking. This review provides a practical hands-on guide for conducting AI-based studies in pediatric allergy, including (1) an introduction to essential AI concepts and techniques, (2) a blueprint for structuring analysis pipelines (from selection of variables to interpretation of results), and (3) an overview of common pitfalls and remedies. Furthermore, the state-of-the art in the implementation of AI in pediatric allergy research, as well as implications and future perspectives are discussed.
Conclusion: AI-based solutions will undoubtedly transform pediatric allergy research, as showcased by promising findings and innovative technical solutions, but to fully harness the potential, methodologically robust implementation of more advanced techniques on richer data will be needed.
|
What is Known: • Pediatric allergies are heterogeneous and common, inflicting substantial morbidity and societal costs. • The field of artificial intelligence is undergoing rapid development, with increasing implementation in various fields of medicine and research. |
|
What is New: • Promising applications of AI in pediatric allergy have been reported, but implementation largely lags behind other fields, particularly in regard to use of advanced algorithms and non-tabular data. Furthermore, lacking reporting on computational approaches hampers evidence synthesis and critical appraisal. • Multi-center collaborations with multi-omics and rich unstructured data as well as utilization of deep learning algorithms are lacking and will likely provide the most impactful discoveries. |
Supplementary Information
The online version contains supplementary material available at 10.1007/s00431-024-05925-5.
Keywords: Allergic rhinitis, Allergy, Asthma, Artificial intelligence, Atopic dermatitis, Childhood, Children, Eczema, Infants, Machine learning, Pediatrics, Teenagers, Wheezing
Introduction
Artificial intelligence (AI) is defined as computer systems that are capable of performing tasks that typically require human intelligence. AI is arguably the most transformative technology of the modern age, and has revolutionized science in waves over the past decades [1, 2], from hard-coded rule-based systems, to machine learning (ML) algorithms that learn from data, and most recently generative AI that enable interactive synthesis and generation of text and images/audio [3, 4]. AI is fundamental to automate and optimize patient management [3], accelerate drug discovery [5], and democratize healthcare-related knowledge [6]. AI even holds potential as a digital assistant in the conceptualization of research and its communication [7].
Pediatric allergic diseases are heterogeneous and intricately interrelated [8]. In combination with increased availability of large-scale (bio)medical data, pediatric allergy research is a suitable context for AI-driven research [9, 10]. The high prevalence of pediatric allergies [11–14] also promises societal benefits from AI applications in clinical practice (e.g., decision support systems) and targeted preventive measures. However, challenges need to be met, including patient privacy, validity, generalizability, specificity, contextualization, bias, and explainability [10, 15, 16]. Furthermore, given the rapid development in AI, up-to-date methodological insight is vital, but most clinicians lack such training and many research groups do not include/co-operate with specialists [17]. This review is intended as a guide and reference for conducting AI-based research in pediatric allergy. It is composed of three sections: (1) introduction of relevant AI terminologies/concepts, techniques, common pitfalls and remedies, and a blueprint for structuring and reporting AI-based studies; (2) overview of studies implementing AI in pediatric allergy in unique and impactful ways; (3) a discussion of limitations and possibilities of AI in pediatric allergy.
Brief background into the field of AI
Machine learning
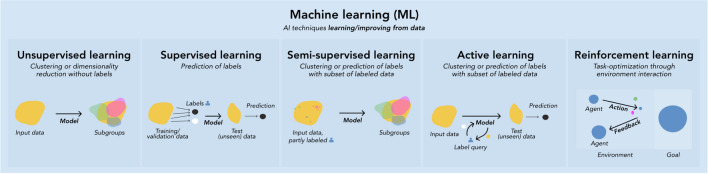
Machine learning (ML) is a subset of AI. As with other AI techniques, ML algorithms (models) mimic human intelligence by solving problems and performing tasks “intelligently”. In ML, however, there are no preprogrammed rules for how to perform tasks or solve problems; instead, these are derived from patterns that the models learn from data based on mathematical principles. The underlying objective is to measure and enhance specific tasks, e.g., identifying subgroups in a patient sample [18]. ML is commonly categorized by the mechanism by which it learns from data. Briefly, in supervised learning, the model learns patterns associated with provided labels, aiming to predict labels in new data (e.g., unseen patients with certain symptoms having a disease or not). Supervised learning can be divided into classification (categorical labels, e.g. asthma subtypes) and regression (continuous labels, e.g. lung function results). In unsupervised learning, the model does not involve labels, and instead “independently” explores distinct patterns (subgroups) within data (e.g. trajectories of eczema) onto which it provides label suggestions. Semi-supervised and active learning are useful on large data in which a small subset is labeled and for which labeling is difficult/time-consuming, and differ mainly by the mechanisms by which they assign labels to unlabeled data [19]. Finally, in reinforcement learning, a virtual artificial environment is created, within which a task (e.g. drug dosing to reduce complications while maximizing survival/recovery rates) is simulated by the model and optimized based on negative/positive feedback from the virtual artificial environment [20, 21] (Fig. 1).
Fig. 1.
Subdomains of machine learning divided by learning mechanism
Artificial neural networks (ANNs) are a subtype of ML conceptually inspired by the neuron structures in the brain. The basic unit consists of neurons, receiving input from and transmitting output to other neurons based on specific functions/criteria. ANNs are composed of at least one hidden (processes therein cannot be directly observed) neuron layer and an input and output layer. Deep neural networks (DNN) are ANNs with multiple hidden layers [22], applications of which are commonly referred to as deep learning (DL). Numerous neural network types have been developed for specific tasks, such as convolutional neural networks, suitable for image analyses [23], and recurrent neural networks, applicable in sequential data/trajectory analyses [24]. The model used must be thoughtfully selected, due to model assumptions regarding data, type(s) of patterns learned, and output interpretation. Summaries of common models are presented in Table 1. Many methods used today have a long history, originally stemming from fields such as statistics [25]. Due to the development of computing power, they can nowadays be used on large datasets and constitute standard components of the ML toolbox.
Table 1.
Commonly used machine learning models
| Model namea | Appropriate use-cases / assumptions of data | Advantages/strengths | Disadvantages/limitations | Examples in pediatric allergy / further reading |
|---|---|---|---|---|
| Unsupervised learning | ||||
| k-means / k-means + + [26], (kernel) fuzzy c-means [27] etc |
▪ Cross-sectional continuous data ▪ Assumes clusters to be homogeneous with similar within-cluster variation along all variables |
▪ Implementations widely available, with variants for various types of data and clustering objectives, e.g., fuzzy c-means (probabilistic labeling) [28] ▪ Computationally efficient |
▪ Unsuitable for categorical/mixed data ▪ Unsuitable if clusters have strongly different within-cluster variation ▪ Prioritizes within-cluster homogeneity over between-cluster separation |
▪ Endotypes of seasonal AR based on cytokine patterns [29] ▪ Immunologic endotypes of AD in infants [30] ▪ Phenotypes of asthma based on demographics, comorbidities, and medication [31] ▪ Biomarker-based phenotypes of AD [32] ▪ Transcriptomic clusters of asthma [33] |
| k-medoids / partitioning around medoids (PAM) / FastPAM / CLARA / CLARANS [34–37] / fuzzy k-medoids [38] etc |
▪ Cross-sectional continuous or mixed data ▪ More flexible than k-means but still focuses on within-cluster homogeneity |
▪ Relatively non-sensitive to outliers ▪ Can use any distance metric [39] ▪ Somewhat computationally costly, but high-performance variants are available, e.g., [34, 35, 40]) ▪ Implementations widely available, with various iterations for specific clustering objectives, e.g., with soft labeling [38] |
▪ Spherical/convex clusters are more likely to be identified [41] ▪ DBSCAN and other methods may be more appropriate in case of very different cluster sizes, and if between-cluster separation is more important than within-cluster homogeneity [42] |
▪ Longitudinal phenotypes of AR based on mHealth symptoms/medication data [43] ▪ Phenotypes of AR based on demographic, heredity, and clinical data [44] ▪ Phenotypes of eczema based on longitudinal disease patterns [45] ▪ Longitudinal phenotypes of wheezing [46, 47] ▪ Endotypes of asthma based on exhaled breath condensate [48] |
| Hierarchical clustering (HCA) / HCA on principal components (HCPC) [49] etc | ▪ Cross-sectional continuous, categorical, or mixed data |
▪ Accommodates any distance metric and a variety of linkage functions [50] ▪ Collinear/high-dimensional data can be managed automatically in HCPC [49], which combines feature extraction with HCA on the principal components ▪ Implementation widely available, as are various hierarchical methods with different focus, e.g., on homogeneity or separation, outliers etc. [51] |
▪ Computationally costly with large data [50, 52] ▪ Meaningful clusters may only occur at low levels of the hierarchy, requiring potentially many clusters (some of which may just be outlier clusters) ▪ Clusters merged once cannot be separated again, which can result in suboptimal solutions |
▪ Endotypes of rhinitis [53] ▪ Endotypes of seasonal AR [29] ▪ Phenotypes of AD based on allergic sensitization patterns [54] ▪ Phenotypes of asthma based on comorbidity, demographics, and asthma symptoms/lung function [55] ▪ Clusters of family adaptation to child’s FA [56] ▪ Clusters of asthma treatment outcome [57] |
| Latent class analysis (LCA) / longitudinal latent class analysis (LLCA) | ▪ Longitudinal or cross-sectional categorical data |
▪ Computationally efficient ▪ Informative by providing probability of assignment (soft clustering) ▪ Statistically principled approach to estimate number of clusters available ▪ Interpretability of original variables |
▪ Inter-class heterogeneity is possible ▪ Compared to other methods used for trajectory analysis, performance may be lower [58] |
▪ Trajectories of wheezing, rhinoconjunctivitis, and eczema symptoms [59] ▪ Grass/mite sensitization trajectoriess[60] ▪ Longitudinal phenotypes of FA and AD [61] ▪ Subtypes of AR based on comorbidity, heredity, and sensitization [62] ▪ Sensitization patterns [63] |
| Growth mixture modeling (GMM) | ▪ Longitudinal data (continuous, but some implementations can handle categorical variables) |
▪ Implementations widely available ▪ Allows for within-class variation ▪ Statistically principled approach to estimate number of clusters available |
▪ More computationally demanding than e.g., LCGA [64] |
▪ Trajectories of wheezing and allergic sensitization [65] ▪ Trajectories of allergic sensitization [66] |
| Latent class growth analysis (LCGA) / group-based trajectory modelling (GBTM) |
▪ Longitudinal data (continuous, but some implementations can handle categorical data) ▪ Small sample or complex model with convergence issue [67] |
▪ Implementations widely available ▪ High homogeneity due to no within-class variation allowed |
▪ May necessitate larger number of classes due to no within-class variation [68], which may prove problematic if sample size is small |
▪ Trajectories of asthma/wheezing based on dispensing data and hospital admissions [69] ▪ Trajectories of wheezing [70] ▪ Trajectories of early-onset rhinitis [71] |
| Supervised learning | ||||
| k-nearest neighbors (k-NN) |
▪ Primarily for classification but may also be used for regression ▪ Varying degrees of noise, data size, and label numbers [74] |
▪ One of the most widely used methods, with easy implementation, available in most statistical software ▪ Robust to outliers/noise [75] |
▪ High computational demand on large datasets [76] |
▪ Prediction of persistent asthma [77] ▪ Prediction of asthma diagnosis [78] ▪ Multi-omics based prediction of asthma [79] ▪ Prediction of asthma exacerbations based on blood markers, FeNO, and clinical characteristics [80] |
| Support vector machine (SVM) | ▪ High-dimensional data (continuous, but categorical variables can be supported [81]) | ▪ Performs well on high-dimensional and complex predictor data |
▪ Prone to overfitting, more so than many other supervised learning methods [77] ▪ Low explainability |
▪ Prediction of asthma diagnosis [78, 82] ▪ Prediction of symptomatic peanut allergy based on microarray immunoassay [83] ▪ Prediction of AD based on transcriptome/microbiota data [84] |
| Decision trees (DT) |
▪ Classification or regression ▪ Complex large data [85] of both continuous, categorical, or mixed nature |
▪ Easily interpretable output |
▪ Prone to overfitting ▪ Low accuracy compared to ensembles of DTs [86] |
▪ Prediction of asthma diagnosis [78, 82] ▪ Prediction of hospitalization need for asthma exacerbation [87] ▪ Prediction of symptomatic peanut allergy based on microarray immunoassay [83] |
| Random forests (RF) |
▪ Classification or regression ▪ Continuous, categorical or mixed data |
▪ Relatively low risk of overfitting [88] | ▪ Limited interpretability [88] |
▪ Prediction of persistent asthma [77] ▪ Prediction of asthma diagnosis [78, 82] ▪ Prediction of hospitalization need for asthma exacerbation [87] ▪ Prediction of AD based on transcriptome/microbiota data [84] ▪ Prediction of asthma exacerbations based on AI stethoscope, parental reporting [89] |
| Bayesian network | ▪ Continuous (although often discretized), categorical, or mixed data based on a probabilistic graphical model (directed acyclic graph) |
▪ Adaptive by possibility to refine the network with new information [90] ▪ Capability of displaying and analyzing complex relationships [91] ▪ Accommodating missingness by utilization of all variables in model [90] ▪ Intuitive interpretation due to probabilistic labeling [92] |
▪ Loss of information in discretization [90] ▪ High computational demand on large datasets [93] |
▪ Prediction of asthma exacerbation [91] ▪ Prediction of response to short-acting bronchodilator medication [92] ▪ Metabolomic prediction of asthma [94] ▪ Prediction of eczema and asthma, respectively, based on SNP signatures [95] |
| Naïve Bayes | ▪ Primarily for classification |
▪ Needs relatively little training data [75] ▪ Computationally efficient [96] |
▪ Strong assumptions of independence between the features [97] ▪ May need discretization of continuous variables [98] |
▪ Prediction of OFC outcome [99] ▪ Prediction of asthma [78] ▪ Prediction of persistent asthma [77] |
| Multilayer perceptron (MLP) | ▪ Complex high-dimensional continuous data | ▪ Ability to learn complex patterns from high-dimensional big data | ▪ May require large data for training, particularly if deep/complex network architecture [100] |
▪ Prediction of asthma diagnosis [78] ▪ Multi-omics-based prediction of asthma [79] |
| Extreme gradient boosting (XGBoost) |
▪ Classification or regression ▪ Continuous and categorical data [75] ▪ High-dimensional and/or sparse data [101] |
▪ Improving accuracy through ensemble of weak prediction models ▪ Computationally efficacious [10] |
▪ Performance issues may arise in imbalanced data [101] ▪ High memory usage |
▪ Prediction of persistent asthma [77] ▪ Prediction of AD based on transcriptome/microbiota data [84] |
| Adaptive boosting (Adaboost)/ |
▪ Classification, but can be adapted for regression ▪ If crucial to boost performance of simple models through ensemble methods |
▪ Combines multiple weak classifiers to create a strong classifier, improving accuracy ▪ Often achieves better performance with less tweaking of parameters compared to other complex models |
▪ Sensitive to noisy data and outliers, which can lead to decreased performance if not handled properly ▪ Can overfit if the number and size of weak classifiers is not controlled |
▪ Prediction of allergy [102] ▪ Allergy diagnosis framework [103] ▪ Asthma treatment outcome prediction [104] |
| Logistic Regression (LR) |
▪ Primarily for binary (but can be extended to multiclass, e.g., with one-vs-rest strategy) classification ▪ Suitable for models where the outcome is a probability 0–1 |
▪ Simple implementation ▪ Efficient to train ▪ Well-understood and widely used ▪ Comparable performance to more advanced models in specific contexts of binary classification problems [105] |
▪ Prone to underfitting when relationship between features and target is non-linear, unless feature engineering is applied ▪ Susceptible to overfitting with high-dimensional data if not regularized appropriately ▪ Assumes linear connection between dependent and independent variables, which can be limiting in complex scenarios [106] |
▪ Prediction of house dust mite-induced allergy [107] ▪ Prediction of asthma persistence [77] ▪ Prediction of FA [108] ▪ Prediction of AD based on transcriptome and microbiota data [84] |
The list is not intended to be comprehensive or cover all relevant/possible use-cases, but rather to provide an overview of common and promising algorithms. Abbreviations. AD, atopic dermatitis; AI, artificial intelligence; AR, allergic rhinitis; FA, food allergy; FeNO, fraction of exhaled nitric oxide; N/A, not available; OFC, oral food challenge; SNP, single nucleotide polymorphism
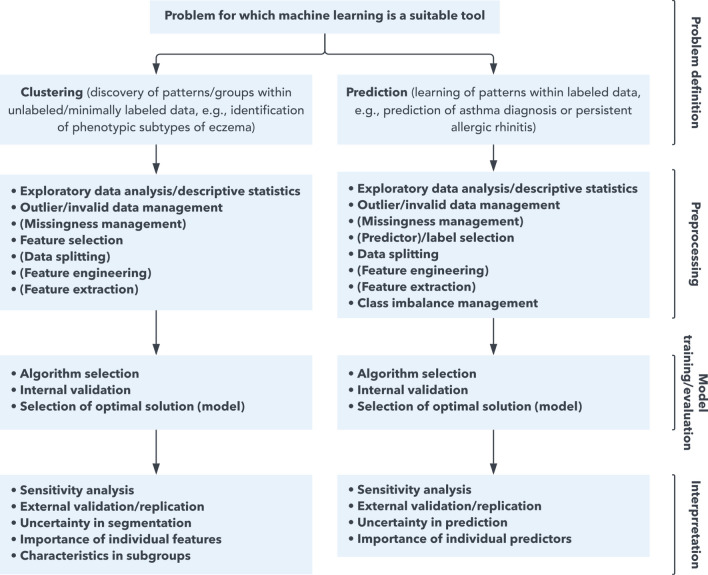
The following subsections describe terminology, techniques, and pitfalls (and remedies) for each step of an ML pipeline — from preprocessing to interpretation of results. A flowchart aimed to guide structuring of such pipelines is presented in Fig. 2.
Fig. 2.
Recommended flowchart for building a machine learning pipeline
A non-comprehensive general workflow with recommended steps to include in a study utilizing machine learning. In specific contexts, certain steps may not be needed or appropriate. Likewise, some studies may warrant specific steps not mentioned here. Items fully encapsulated in parentheses indicate that some machine learning models manage said issue “automatically” or performance evaluation may not be clear.
Main steps of machine learning pipelines
Preprocessing
Preprocessing is often the most time-consuming step and heavily influences model performance/output. Exploratory data analysis typically precedes, to understand data distribution, patterns of missingness, and differences between the analyzed subsample and excluded subjects. Commonly, data need to be transformed to suitable forms (feature engineering), e.g., one-hot encoding categorical variables, log-transforming skewed data, and standardization/normalization of variables on different scales (e.g., height and income, where the latter could otherwise dominate by magnitude). In many datasets, redundant/non-informative/ “noisy” variables are present, which may increase run-time and hamper performance and interpretation, necessitating feature selection. Beyond manually selecting sensible variables, a simple example of feature selection is based on correlation matrices, in which correlated variables are dropped. A related technique (feature extraction) also reduces data dimensionality, by transforming variables into a lower-dimensional subspace while minimizing information loss. Principal component analysis is a simple feature extraction technique, also managing correlated variables. Some preprocessing techniques may be counterproductive, e.g., categorizing continuous variables, which should be avoided if possible to retain maximal differentiating information. See Table 2 for a summary of common preprocessing techniques.
Table 2.
Common/recommended preprocessing steps/techniques
| Preprocessing step | Substep or rationale/use-case | Techniques |
|---|---|---|
| Exploratory data analysis (EDA) / descriptive statistics | Provide an overview of patterns and potential imbalance in data. In case of stratified analyses (e.g., by sex) or subset analysis (e.g., those completing follow-up), comparison with table of characteristics and statistical tests for difference is useful |
▪ Plots (ordinal/continuous data: histograms, nominal data: bar plots) ▪ Tables (for nearly any type of data; informative but should preferably be combined with visual presentations for ease) |
| Outlier/invalid data management | Outliers may be naturally occurring, but may also be due to measurement error, erroneous transfer of data etc. Removing outlier/invalid data is sensible (to improve representativeness and decrease impact of unusual measures) depending on the data and model/task at hand. Exploratory data analysis is needed to evaluate this. Importantly, over-removing outliers may reduce generalizability and increase the risk of overfitting [109]. If negative impact is suspected, sensitivity analysis can be performed to compare robustness/trends in the output. The ML models used also influence the need for removing outliers, as some are (more) robust to outliers. Invalid data management requires manual assessment with exploratory data analysis and domain-expertise |
▪ Manual removal, e.g., of measures below/above a certain factor (commonly 1.5; 3 may be considered as extreme outliers) of interquartile ranges [IQR]. If assuming Gaussian distribution, removing measures outside of the 95th confidence interval may also be sensible ▪ Data-driven approaches (e.g., local outlier factor [LOF] [110]) ▪ For unsupervised learning, DBSCAN, HDBSCAN, and trimming approaches can facilitate identifying and handling noise in the data ▪ Model diagnostics can identify outliers after fitting a model; sometimes this is needed to see that an observation is outlying with respect to the modelled general pattern |
| Missingness management | The (likely) mechanism of missingness [111] must be evaluated to decide a suitable approach to substitute missing value or exclude subjects with (certain degrees of) missingness. Exploratory data analysis is key in this evaluation. Multiple imputation is generally preferred, to reduce the uncertainty in individual substitutions by producing several imputed datasets. The minimum number of imputed datasets is debated, with various rules of thumb [112]. There are also data-driven methods to decide the number of datasets needed for a specified precision of standard error [112]. Selecting an imputation method requires attention to data type (e.g., cross-sectional or longitudinal [113]) and association between variables [114]. In many cases, the analysis should be performed in each imputed dataset and pooled [115] |
▪ Complete case analysis (either as main analysis or sensitivity analysis) ▪ Available case analysis (similar to the above, but dynamically select subjects with sufficient data for each individual analysis) [111] ▪ Imputation (e.g., multiple imputation) [113, 114, 116, 117] |
| Feature/predictor/label selection | In some datasets, particularly high-dimensional data, there may be irrelevant/non-informative/ “noisy” variables. While it is important to consider all variables, the inclusion of e.g., variables that are overlapping to a high degree clinically may decrease model performance, cause overfitting, and complicating interpretation |
▪ Literature/expert-based selection, based on experience or literature ▪ Data-driven methods [118]: - Recursive feature elimination (RFE) [78] - Sequential feature algorithms [119, 120] - LASSO (combines variable selection and regularization to improve prediction accuracy and model interpretability [121]) - Tree-based approaches (e.g., decision trees, random forests) [118], and wrapper around said approaches, e.g., Boruta’s algorithm |
| Data splitting | To reduce overfitting, data typically needs to be split into a training set (for learning patterns), and a validation set (for performance assessment, to guide hyperparameter fine-tuning). Ideally, a third test set, agnostic of the preceding steps, is useful to assess the model performance on unseen data. In supervised learning, results from several approaches can be compared on a data subset not used for tuning; in unsupervised learning performance measurement is less obvious |
▪ Cross-validation (k-fold validation, leave-one-out [LOO] etc.) [122] ▪ Bootstrapping ▪ Stability exploration in unsupervised learning |
| Feature engineering |
Feature scaling Useful in continuous data and varying scale between features (e.g., height and annual income, the latter often on scales many times larger). Models (e.g., k-nearest neighbors) relying on distance metrics [123] perform better following feature scaling, as sheer magnitude of certain variable otherwise overinfluences the algorithm. Feature scaling should be performed after data splitting, as the sets otherwise introduce information to each other |
▪ Normalization (rescales data to [typically] range 0–1; sensitive to outliers, useful when scale of variables are of higher importance, e.g., neural networks) [124] ▪ Standardization (rescales data to mean 0 and standard deviation 1; more robust to outliers and generally recommended for linear models, e.g., support vector machines) [124] |
|
Discretizing/continuizing continuous/discrete variables Discretizing converts continuous data into categorical bins, simplifying modeling, handling non-linear relationships, and improving interpretability by mitigating the effects of outliers. However, it causes information loss, and this is often a reason to avoid it. In contrast, continuizing converts categorical or ordinal data into continuous formats, capturing ordinal relationships and enhancing performance for algorithms that prefer numerical inputs, although this may not be feasible at times |
▪ Entropy-MDL discretization ▪ Equal frequency discretization ▪ Equal width discretization ▪ One-hot encoding ▪ Frequency or mean encoding |
|
| Feature extraction | In addition to feature selection, dimensionality can also be reduced by creating new features that aggregate information in the original features. Many algorithms perform suboptimally with high-dimensional data (“curse of dimensionality”) [125]. In such cases, dimensionality reduction can be applied to reduce runtime and improve performance |
▪ Linear techniques (e.g., principal component analysis [PCA; continuous data], multiple correspondence analysis [MCA; categorical data], MFA/FAMD [mixed data]; perform feature scaling prior to this step [126]) ▪ Non-linear techniques (e.g., autoencoders, t-SNE, UMAP) ▪ Subject-matter knowledge to define informative indexes summarizing several features |
| Class imbalance management | In supervised learning, there may be substantial imbalance of labels among the subjects. This imbalance can reduce the performance of the model | ▪ Oversampling of minority class subjects (e.g., adaptive synthetic sampling approach [ADASYN] [127]) and/or undersampling of majority class subjects (e.g., by random exclusion) |
The table describe common steps taken during preprocessing. The list is intended as a guide with steps presented in consecutive order as they should be performed, but it is not meant to be comprehensive or universally applicable. It is recommended, particularly for unique applications, to evaluate previous similar implementations or relevant technical literature.
Model training/evaluation
As per the “no free lunch” theorem [128], no model is ideal for all contexts. Thus, multiple models should be evaluated, the choice of which depends on the (1) nature of the data (density/distribution; tabular/non-tabular; numerical, categorical, or mixed; presence of outliers/noise etc.), and (2) task (relevant learning mechanism, desired utility/interpretability of output etc.). See Table 1 for a summary of common methods (additional methods are described in Table S1). Furthermore, it is important to accommodate differing and clinically influential factors (such as sex and age) of allergies and their presentation and outcomes. An AI application may otherwise produce biased and non-generalizable output when such factors are not accounted for [129]. Such unfairness in AI can be addressed using various approaches, e.g., by so-called “active fairness”, which essentially involves incorporating important “sensitive” attributes into the algorithm [130].
An important issue in cluster analysis is that there is no unique definition of an “optimal”/ “true” clustering. Most datasets allow for different reasonable clustering according to different criteria, e.g., requiring clusters to be homogeneous (low within-cluster distances), or making cluster separation the dominating criterion (which may lead to heterogeneous clusters if no gap separates dissimilar observations/subjects). It may also be reasonable to make model assumptions (e.g. Gaussian). These choices cannot be made from data alone; rather, they need to be made with the clustering requirements in mind [51, 131]. Ensemble clustering provides an alternative approach, in which clustering output from several algorithms are run through a consensus function to derive a final solution [132], and can be attractive in difficulty of deciding what kind of clusters are sought after. It must be noted, however, that ensemble clustering does not necessarily provide a more “correct” solution, as clustering algorithms are based on different concepts of what an optimal cluster is; this discrepancy is similar to meta-analysis, in which studies are often too heterogeneous for pooling. The corresponding subdomain in supervised learning (ensemble learning) [88, 133–135] is more often appropriate, as the different learners/models aim to solve the same underlying problem.
In unsupervised clustering, selecting a distance measure (i.e., measure of similarity/dissimilarity between subjects) is of importance, as it heavily influences evaluation metrics [50, 51, 136–138]. Another crucial issue in clustering is the choice of the number of clusters. In many situations the data do not determine an unambiguous number of clusters; rather, this will depend on the “granularity” of the clustering required for the application. Lower numbers of clusters are often easier to handle, but often larger numbers of clusters can improve the model fit. Several indices can be used for determining the number of clusters, but they commonly give conflicting results [51, 139]. Clinical interpretation is central in weighing between different alternatives.
Overfitting occurs when a model is overly specific to the training data, thus not generalizing well to unseen data. The risk of overfitting increases with model complexity and the number of variables in relation to the sample size [140]. Overfitting may also result from improperly separating training and validation data [10]. A common approach to combat overfitting is splitting data into: (1) training set (for training the model); (2) validation set (for fine-tuning); (3) test set (for performance evaluation) [122, 141, 142]. In neural networks, dropout regularization is a common technique to reduce the risk of overfitting. Underfitting is the opposite issue: the complexity of the data is not reflected in the model, e.g., too few variables or insufficient/non-representative training sample, and the performance is suboptimal on both training and unseen data [140, 143, 144].
Fine-tuning of hyperparameters (model settings) is generally necessary to optimize performance, typically done by comparing performance using different hyperparameters. A common technique is grid search (brute force search of all possible hyperparameter settings) [145], the practicality of which is limited in contexts with many possible settings. Random search is a compromise in such cases. Optuna [146], a hyperparameter optimization framework, and Bayesian optimization [147] are additional alternatives. The metrics assessed in training vary depending on the analysis aim(s), learning mechanism, and data, but should include multiple [148] metrics, e.g., accuracy, recall etc. (supervised learning) or intra-cluster separation, inter-cluster homogeneity etc. (unsupervised learning). In cluster analysis, some choices (e.g., dissimilarity measure) should aim at valid representation of the data within the context rather than optimizing cluster validity measures, as the meaning of criteria such as between-cluster separation and ultimately the clustering itself relies on the dissimilarity measure. Beyond optimizing a model on specific data, it is also valuable to stress-test the application under different conditions, such as assessing the stability of the algorithm with introduction of noise to the data, or by comparing the output of multiple algorithms. A summary of common evaluation approaches is presented in Table 3.
Table 3.
Common/recommended model evaluation techniques
| Technique | Rationale/use-case | Examples of implementation/approaches |
|---|---|---|
| Unsupervised learning | ||
| Robustness | Unsupervised learning solutions robust to noise are reasonably (more) generalizable and less overfit to the trained-on data |
▪ Bootstrapping ▪ Sensitivity analysis |
| Clinical interpretability | In unsupervised learning, algorithms may derive clusters/trajectories that differ by statistical measures, but are so similar that they are not clinically distinguishable. Assessing clinically the characteristics of subgroups is highly relevant. In supervised learning, this objective is less obvious, but clinically sensible predictor variables driving predictions most strongly could indicate a sensible/practically useful solution |
▪ Tables and other textual representation ▪ Figures, e.g., heatmaps, line plots etc |
| Homogeneity within clusters and dissimilarity across clusters | The classical definition of well-defined clusters/trajectories, i.e,. that the subjects within the derived subgroups are similar to each other and dissimilar to subjects in the other subgroups). From a clinical perspective, this may also be done with e.g., heatmaps |
▪ Silhouette score ▪ Calinski-Harabasz score ▪ Davies-Bouldin score |
| Model fit / complexity | Statistical measures of how well a model fits the observed data (with varying penalty for complexity of the solution) provides an easily interpretable metric for selecting the optimal model in a data-driven manner |
▪ Akaike information criterion (AIC; commonly suggests more complex solutions and larger number of subgroups) ▪ Bayesian information criterion (BIC; in contrast to AIC emphasizes simpler solutions [149]) |
| Supervised learning | ||
| Performance metrics | Various measures are available to assess how accurately the model predicts based on new input data, varying widely depending on the learning task and data at hand. Ideally, multiple metrics need to be evaluated to assess model performance, some being more suitable in specific types of prediction models [148, 150, 151] |
▪ R2, adjusted R2, MAE, RMSE, MAPE etc. (regression) ▪ MCC, F1 score, Cohen’s kappa, Brier score, AUROC, AUPRC, log-loss, accuracy, recall, specificity, NPV/PPV etc. (classification) |
| Validation curve(s) | Diagnostic tool to assess the performance of a model in relation to hyperparameter settings (for example number of trees in a random forest model). More specifically, this can be used to e.g., evaluate hyperparameters that influences the model’s tendency to overfit/underfit | ▪ Plot of performance metric on y-axis and hyperparameter on x-axis |
The table describe common techniques used in model evaluation. The list is not meant to be comprehensive or universally applicable. It is recommended, particularly for unique applications, to evaluate previous similar implementations or relevant technical literature.
Interpretation
Models differ by assumptions and statistical methods and thus necessitate appropriate interpretation of results. As an example, in hard clustering algorithms (e.g., k-means), each subject is assigned to one cluster; however, it is possible that some subjects are not similar to subjects within their cluster, necessitating cautious conclusions about cluster homogeneity. Multiple techniques can be used to assess outliers, e.g. heatmaps [47]. A substantial part of interpreting the performance of a model is to elucidate determinant factors. SHapley Additive exPlanations (SHAP) [152] is a common tool to visualize the contribution/importance of each variable for prediction or clustering. Finally, externally validating the results in a comparable independent cohort is useful to evaluate model robustness and generalizability. A list of common interpretation approaches are shown in Table 4.
Table 4.
Common/recommended approaches for interpreting model results
| Approach | Rationale/use-case | Examples |
|---|---|---|
| Replication with independent data (external validation) | Deriving comparable results from a model in an external (independent) cohort/sample of individuals indicates that the patterns learned by the model are (relatively) generalizable. Caution is needed in interpretating such results, however, as it may be that the underlying characteristics are very similar in the external data, or conversely, very dissimilar, which complicates interpretation | ▪ Re-running analysis in independent external cohort/population |
| Relation with external information | Mostly applicable in unsupervised learning. If derived subgroups indeed represent clinically distinct entities, it may be that these differ meaningfully in aspects not characterized in the model. For example, if clusters based solely on skin prick test results are composed of children with very similar sociodemographic background factors, it increases the probability that there may be non-random and clinically meaningful pathophysiological differences between the clusters |
▪ Tables/plots of characteristics (with appropriate statistical significance tests) ▪ Figures, e.g., spider plots, bar plots etc |
| Explanation of feature importance | Machine learning models, particularly with higher degree of complexity, are not directly interpretable based on the output. To get a sense of the attributes and patterns that influenced the model the most, multiple techniques are available to visually/numerically present the most important parts of the data according to the model. Individual methods may or may not be applicable in specific models/contexts |
▪ Class Activation Maps (CAM) [158] ▪ Local Interpretable Model-agnostic Explanations (LIME) [159] ▪ SHapley Additive exPlanations (SHAP) [152] ▪ Mean decreased Gini value [160] |
| Subgroup homogeneity | In unsupervised learning, the derived subgroups may have within-subgroup heterogeneity impacting interpretation of recognizability in the clinical setting, subsequent association analyses etc. For this reason, it is useful to present (indirect) measures of homogeneity/heterogeneity within derived classes. For trajectory analyses, the 95% confidence interval (95%CI) around the predictor prevalences/probabilities may indicate the degree of variation within trajectories. Tabular data may provide similar information as well |
▪ Tables/plots of characteristics (with 95%CIs) |
The table describes common techniques used in model evaluation. The list is not meant to be comprehensive or universally applicable. It is recommended, particularly for unique applications, to evaluate previous similar implementations or relevant technical literature.
Practical aspects of implementation
Software necessary for implementing AI-based applications are widely available, often through free open-source packages for Python and R statistical software. However, the major hurdle often lies in hardware and technical know-how. In terms of hardware, most “shallow” algorithms, as well as DL with small/moderate-sized data, can be performed on regular modern laptops/computers. For advanced modelling or analysis of large data, the use of dedicated hardware is often needed, in which case it is crucial to ascertain that sensitive data are not accessible by unauthorized parties. Although computational skills are much more time-consuming to develop, there are many resources aimed at lowering the barrier to implementing AI, e.g., low-code/no-code platforms/tools for performing AI-based analyses, such as MLpronto [153], Orange Data Mining, KNIME, as well as various online courses and certifications in AI for clinicians [154]. While everyone in a healthcare team does not necessarily need to learn how to program or develop in-depth knowledge about machine learning, it is important that colleagues have at least a basic understanding of fundamental concepts, to efficiently discuss AI implementations.
Ethical considerations and reporting
Beyond understanding of technical intricacies, AI-based research in pediatric allergy necessitates particular attention to ethical aspects, e.g., pertaining to data protection, as AI algorithms are often fed vast amounts of sensitive information. Frameworks guiding safe inclusion of pediatric data in AI-based research include ACCEPT-AI [129].
Detailed reporting from all steps of the pipeline is crucial for critical appraisal and reproducibility/synthesis. For example, as allergic diseases demonstrate heterogeneous developmental patterns across age, it is essential to clearly report on age (and variation thereof) in the study population. Multiple checklists have been developed, e.g., Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD + AI) [155] for prediction models and Guidelines for Reporting on Latent Trajectory Studies (GRoLTS) [156] for trajectory analyses, although well-established equivalents are lacking in many areas, e.g. cluster analysis. While items essential to report vary depending on analysis and data, a general guideline is provided in Table 5. Reporting recommendations of AI-based clinical studies have also been proposed [157]. Additional guidelines can be found at the EQUATOR (Enhancing the QUAlity and Transparency Of health Research) Network (https://www.equator-network.org/).
Table 5.
Recommended elements to report in machine learning-based studies
| Step | Rationale/general description |
|---|---|
| Preprocessing |
▪ Exploratory data analysis/descriptive statistics - Cohort characteristics: Describe in detail the used cohort/sample. Present the initial participation rate and relevant background factors for evaluation of generalizability and representability - Analysis subset characteristics and comparison: If a subset of the cohort/sample was used for the analysis, compare background factors between the full cohort/sample and the subset - Correlation analysis: Report correlations between variables (e.g., through a correlation matrix). Understanding these relationships can guide feature engineering and model selection, as well as reveal potential (multi)collinearity issues - Missingness: Visualization of the degree of missingness (across subjects and variables, preferably with a measure of patterns across missingness between variables) provides useful information as well ▪ Outlier/invalid data management: Describe the presence/degree of outliers and/or data deemed invalid, and if any processing of these was performed. Visualization is particularly useful, e.g., with simple box plots. If possible, provide code/syntax (in a repository) ▪ Management of missingness: Visualize/tabulate missingness and patterns thereof. Provide the rationale for using a particular imputation algorithm or other approach, including details (preferably including plots) on evaluation/validation of the imputation. If possible, provide code/syntax (in a repository) ▪ Feature selection: Provide explicit rationale for the used variables. Ideally, add a table (in the supplementary material) where the reason(s) for inclusion/exclusion for each variable that could potentially have been of relevance are listed. Importantly, feature selection processes should be described in detail, including narrative summary and output of data-driven approaches and tables. If data-driven methods were used, provide code/syntax (in a repository) ▪ Feature scaling: Report if the variables were inputted as-is or if any scaling was performed (preferably providing code/syntax) ▪ Dimensionality reduction: Report the tools and hyperparameters/settings used (preferably providing the actual code/syntax), together with details on the percentage of variance explained in the reduced subspace, loss, or other relevant information to assess the performance/representability of the new data |
| Model training / evaluation |
▪ Algorithm selection: Describe the rationale for the selection of models. Preferably, select at least two models to assess the robustness of the chosen solution ▪ Model implementation: Explain in detail how the model(s) were implemented, which hyperparameter settings were tested and the underlying rationale. Preferably, provide the actual implementation code/syntax (in a repository) ▪ Model evaluation: Provide a detailed log of the model(s) with different hyperparameters, so as to make the selection of the optimal solution transparent and clear for the reader. For example, if a cluster analysis was performed and 3–5 cluster-solutions were assessed as the top 3 models, provide clinical characteristics and evaluation metrics for at least these (but preferably all tested solutions) |
| Interpretation |
▪ Characteristics: Most relevant for unsupervised analyses. Provide rich details on the subgroups, including on parameters included and not included in the model (e.g., background factors, comorbidities, sociodemographic factors etc.) ▪ Influencing factors/explanation of the model: Provide as much detail as possible on how the model derived its output, e.g., feature importance ▪ Uncertainty in findings: Describe the uncertainty in the model (e.g., 95% confidence intervals of the predictions or subject characteristics) ▪ External validation: Provide an analysis of the generalizability of the results, preferably by externally validating the model in a different cohort/sample ▪ Limitations: Could the analyses have been done differently in an optimal setting? Transparently describe challenges and drawbacks/compromises |
Implementation in clinical practice and research
Implementation of AI across disciplines varies massively. For example, across United States Food and Drug Administration-approved medical AI devices, a majority have been developed for radiology, while none are listed under allergy [161]. Despite this, the need is paramount and the possibilities evident, given the availability of rich data and computational power, as well as rapid development of algorithms. While applications in clinical practice are largely absent, the utilization of AI within research is more established. In the following subsections, promising/impactful AI-based studies in pediatric allergy are summarized.
Diagnosis of disease
In a study by Kothalawala et al., models were evaluated for predicting asthma at 10 years based on hospital records, clinical assessments, and questionnaire responses from multiple time points. Notably, just a few predictors (cough, atopy, and wheeze) contributed substantially [78]. He et al., conversely, focused on predicting asthma at 5 years, reporting limited predictive power of markers from infancy, with earliest reliable model being based on data at age 3 years. The authors also noted a clear progression of feature importance at different ages [162]. Overall, substantial heterogeneity is present across prediction models of asthma, and generalizability is moderate, rendering clinical implications unclear [77, 163]. This uncertainty is furthered by reports indicating comparable performance to regression models [164]. Sophisticated AI methods are demanding to implement; thus, they are rare and may not live up to their potential. Although prediction models using multi-omics data are scanty, there are indications that specific omics combination provide superior performance [79, 165]. Prediction models have also been applied in diagnosis of atopic dermatitis (AD) [84], allergic rhinitis (AR) [166], and more specific diagnostic labels, e.g. oral food challenge positivity to specific foods [99, 167]. Overall, these studies demonstrate applicability of various predictors and meaningful results, but also substantial heterogeneity and limited clinical utility due to lacking methodological reporting and impracticality of synthesis/comparisons.
Subtyping of disease
Subtyping can be performed on cross-sectional (cluster analysis) or repeated measures (trajectory analysis). In a study by Havstad et al. [63], a cluster analysis of sensitization patterns, found four clusters, which were more strongly associated to AD and wheeze/asthma compared to any atopy. Studies focused on endotypic markers have also exemplified the heterogeneity of allergy, e.g., a study by Malizia et al. [29], in which three inflammatory patterns of seasonal AR were identified based on cytokine patterns. Longitudinal disease patterns have commonly been derived from binary (presence/absence) markers, and although clinically meaningful patterns have been identified, e.g. wheezing trajectories associated with (actionable) risk factors [168] and outcomes, such as allergic disease/sensitization in adulthood [169], important disease characteristics are omitted. Some studies have included severity measures, e.g., by Mulick et al. on eczema trajectories [170]. Given the reported uncertainty/within-class heterogeneity with latent class analysis [46, 171], a common algorithm in trajectory analysis, alternative methods, based on cluster analysis of longitudinal data condensed into characteristics such as onset-age, number of episodes with symptoms etc., as implemented by Haider et al., have contributed with homogeneous trajectories of eczema [45] and wheezing [46, 47]. Studies incorporating data from multiple sources, such as parental report and prescribed medication/physician assessment [172–174], have further contributed with distinct subgroups and highlighted the complexity underlying pediatric allergies and utility of multi-dimensional characterization.
Management of disease
Successful management can be defined in various ways, including survival, treatment adherence, and remission. Typically, the same models as for diagnosis prediction are applicable. Tailored sequential models have shown promising accuracy in predicting adherence to subcutaneous immunotherapy in AR treatment [175]. Disease progress can also be modelled as trajectories. In a study by Belhassen et al. [176], trajectories based on use of inhaled corticosteroids before and after asthma-related hospitalization were derived and related to, among others, sociodemographic factors, which may provide guidance in individualized treatment. DL image analysis of eczema lesions [177] coupled with a mobile app has shown promising results in e.g., patient-oriented eczema measures [178], although large-scale studies are needed to validate such findings, and engagement rates need to improve. Similarly, an AI-assisted clinical support was evaluated in asthma management, showing comparable asthma exacerbation rates as with regular care, albeit reducing administrative burden [179], indicating limitations in clinical decision-making and potential in streamlining time-consuming tasks.
Outcome prediction
Models for outcome prediction, typically architecturally equivalent to diagnosis prediction models, have been used in various settings, from relatively simplistic contexts and predictor sets, such as symptom and treatment recordings of eczema, aiming to predict future severity [180], to large-scale medical record data to predict asthma persistence [77]. In general, despite incremental improvement, even models incorporating > 100 predictors and large study samples are not yet fully refined for broad clinical implementation [181]. Associated health outcomes, such as suicidality in adolescents with AR, have also been investigated, adding important contribution to our understanding of the complex multi-dimensional nature and implication of pediatric allergy [182]. DL implementations are scanty [183].
Limitations and future perspectives
Advanced AI is data hungry [184], and data are limited in many contexts. Possible solutions include data augmentation, active learning, and transfer learning [185]. This challenge is exacerbated by data protection laws limiting data sharing, particularly within the European Union. Non-disclosive distributed learning frameworks may facilitate collaboration, while large language models may aid in tedious standardization/harmonization. As an example, DataSHIELD is a freely available open-source framework for co-analysis of individual-level data, which allows original data to remain unseen by other entities [186]. Broad multi-disciplinary collaboration may also facilitate incorporation of multi-omics data, and ultimately aid in streamlining AI training through increased concentration of relevant expertise. National and international consortia require substantial administrative efforts, particularly at the early stage, but are increasingly needed to meet the demands of data and context-specific expertise.
Explainability is another pertinent issue, which has largely persisted, particularly for advanced algorithms [187–189], and which may introduce elusive biases. Increased involvement of patient-representative groups is key to continuously safe-guard and appropriately adjust models. Decision-makers are also often uninformed of technical specifics, potentials, and limitations of AI applications, resulting in unequal distribution of funding. Increased familiarity with AI is needed, as are well-established guidelines and easy-to-use frameworks for conducting analyses in a reproducible and appropriate fashion. These characteristics are largely lacking in the extant AI-based studies in pediatric allergy. Furthermore, there are important gaps in the literature. For example, few studies are focused on food allergy and allergic rhinitis in comparison to eczema and particularly asthma. It is likely that the most impactful discoveries will be made using DL approaches with large-scale multi-omics data (including wearables and other accessible/non-invasive gadgets) as well as nationwide/multi-national register data with cross-linkage, in which input is as unfiltered and comprehensive as possible. This will maximally harness the computational power and data-driven analyses enabled by such technologies.
Conclusion
AI has immense potential in pediatric allergy, although implementation in research and clinical practice is still at the foundational stage. Improved methodology, increased detail in reporting of computational aspects in studies, and increased collaboration are central to accelerate impactful discoveries. Advanced models are also underutilized, as are multi-omics and rich unstructured data. Finally, decision-makers and the general public (not least patient-representative groups) should be involved in development of AI-based technologies in order to increase awareness of utility, instill trust, and reduce bias of such applications.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- AD
Atopic dermatitis
- AI
Artificial intelligence
- ANN
Artificial neural network
- AR
Allergic rhinitis
- DL
Deep learning
- DNN
Deep neural network
- ML
Machine learning
Authors contribution
DL and BIN conceptualized the article. DL performed the literature review and drafted (with consultation from RB, TD, CH, and SAS) and revised (with critical input and consultation from RB, TD, CH, SAS, GW, EG, and BIN) the manuscript. All authors read and approved of the final version of the article.
Funding
Open access funding provided by University of Gothenburg. The present work was supported by funding from Herman Krefting Foundation, Swedish Research Council, Swedish government under the ALF agreement between the Swedish government and the county councils (Västra Götaland), and the Swedish Heart and Lung Foundation. The funders had no role in the design of this work, the literature review, the preparation of the manuscript, or the decision to submit.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval
This is a review work; therefore, ethical approval is not warranted.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Xu Y, Liu X, Cao X, Huang C, Liu E, Qian S et al (2021) Artificial intelligence: a powerful paradigm for scientific research. Innovation (Camb) 2:100179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulikowski CA (2019) Beginnings of Artificial Intelligence in Medicine (AIM): computational artifice assisting scientific inquiry and clinical art - with reflections on present AIM challenges. Yearb Med Inform 28:249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alowais SA, Alghamdi SS, Alsuhebany N, Alqahtani T, Alshaya AI, Almohareb SN et al (2023) Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med Educ 23:689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy S (2024) Generative AI in healthcare: an implementation science informed translational path on application, integration and governance. Implement Sci 19:27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A et al (2024) Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630:493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen A, Liu L, Zhu T (2024) Advancing the democratization of generative artificial intelligence in healthcare: a narrative review. J Hospital Manag Health Policy 8
- 7.Macdonald C, Adeloye D, Sheikh A, Rudan I (2023) Can ChatGPT draft a research article? An example of population-level vaccine effectiveness analysis. J Glob Health 13:01003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Custovic A, Custovic D, Fontanella S (2024) Understanding the heterogeneity of childhood allergic sensitization and its relationship with asthma. Curr Opin Allergy Clin Immunol 24:79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahuja AS (2019) The impact of artificial intelligence in medicine on the future role of the physician. PeerJ 7:e7702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Breugel M, Fehrmann RSN, Bügel M, Rezwan FI, Holloway JW, Nawijn MC et al (2023) Current state and prospects of artificial intelligence in allergy. Allergy 78:2623–2643 [DOI] [PubMed] [Google Scholar]
- 11.Serebrisky D, Wiznia A (2019) Pediatric asthma: a global epidemic. Ann Glob Health 85(1):6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D, Zheng J (2022) The burden of childhood asthma by age group, 1990–2019: a systematic analysis of global burden of disease 2019 data. Front Pediatr 10:823399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licari A, Magri P, De Silvestri A, Giannetti A, Indolfi C, Mori F et al (2023) Epidemiology of allergic rhinitis in children: a systematic review and meta-analysis. J Allergy Clin Immunol Pract 11:2547–2556 [DOI] [PubMed] [Google Scholar]
- 14.Mallol J, Crane J, von Mutius E, Odhiambo J, Keil U, Stewart A (2013) The International Study of Asthma and Allergies in Childhood (ISAAC) phase three: a global synthesis. Allergol Immunopathol 41:73–85 [DOI] [PubMed] [Google Scholar]
- 15.Hoque F, Poowanawittayakom N (2023) Future of AI in medicine: new opportunities & challenges. Mo Med 120:349 [PMC free article] [PubMed] [Google Scholar]
- 16.Eigenmann P, Akenroye A, Atanaskovic Markovic M, Candotti F, Ebisawa M, Genuneit J et al (2023) Pediatric Allergy and Immunology (PAI) is for polishing with artificial intelligence, but careful use. Pediatr Allergy Immunol 34:e14023 [DOI] [PubMed] [Google Scholar]
- 17.Ferrante G, Licari A, Fasola S, Marseglia GL, La Grutta S (2021) Artificial intelligence in the diagnosis of pediatric allergic diseases. Pediatr Allergy Immunol 32:405–413 [DOI] [PubMed] [Google Scholar]
- 18.Razavian N, Knoll F, Geras KJ (2020) Artificial intelligence explained for nonexperts. Semin Musculoskelet Radiol 24:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazakis N, Kanas VG, Aridas CK, Karlos S, Kotsiantis S (2019) Combination of active learning and semi-supervised learning under a self-training scheme. Entropy (Basel) 21(10):988. 10.3390/e21100988
- 20.Khezeli K, Siegel S, Shickel B, Ozrazgat-Baslanti T, Bihorac A, Rashidi P (2023) Reinforcement learning for clinical applications. Clin J Am Soc Nephrol 18:521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutton RS, Barto AG (2018) Reinforcement learning: an introduction. A Bradford Book, Cambridge, MA, USA, chapter 1, pp 1–13
- 22.Kufel J, Bargieł-Łączek K, Kocot S, Koźlik M, Bartnikowska W, Janik M et al (2023) What is machine learning, artificial neural networks and deep learning?-Examples of practical applications in medicine. Diagnostics (Basel). 13(15):2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights Imaging 9:611–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montesinos López OA, Montesinos López A, Crossa J (2022) Fundamentals of artificial neural networks and deep learning. multivariate statistical machine learning methods for genomic prediction: Springer, p 379–425 [PubMed]
- 25.Murtagh F (2015) A brief history of cluster analysis. CRC Press, Handbook of Cluster Analysis, pp 21–30 [Google Scholar]
- 26.Arthur D, Vassilvitskii S (2007) k-means++: the advantages of careful seeding. In: Proceedings of the eighteenth annual ACM-SIAM symposium on Discrete algorithms (SODA '07). Society for Industrial and Applied Mathematics, USA, 1027–1035. 10.1145/1283383.1283494
- 27.Suganya R, Shanthi R (2012) Fuzzy c-means algorithm-a review. Int J Sci Res Publ 2:1 [Google Scholar]
- 28.Bezdek JC (2013) Objective function clustering. In: Pattern recognition with fuzzy objective function algorithms. Advanced Applications in Pattern Recognition. Springer, Boston, MA. 10.1007/978-1-4757-0450-1_3
- 29.Malizia V, Ferrante G, Cilluffo G, Gagliardo R, Landi M, Montalbano L et al (2021) Endotyping seasonal allergic rhinitis in children: a cluster analysis. Front Med (Lausanne) 8:806911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauffer F, Baghin V, Standl M, Stark SP, Jargosch M, Wehrle J et al (2021) Predicting persistence of atopic dermatitis in children using clinical attributes and serum proteins. Allergy 76:1158–1172 [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Bian J, Fishe JN (2023) Pediatric and adult asthma clinical phenotypes: a real world, big data study based on acute exacerbations. J Asthma 60:1000–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakker DS, de Graaf M, Nierkens S, Delemarre EM, Knol E, van Wijk F et al (2022) Unraveling heterogeneity in pediatric atopic dermatitis: identification of serum biomarker based patient clusters. J Allergy Clin Immunol 149:125–134 [DOI] [PubMed] [Google Scholar]
- 33.Yeh YL, Su MW, Chiang BL, Yang YH, Tsai CH, Lee YL (2018) Genetic profiles of transcriptomic clusters of childhood asthma determine specific severe subtype. Clin Exp Allergy 48:1164–1172 [DOI] [PubMed] [Google Scholar]
- 34.Schubert E, Rousseeuw PJ (2021) Fast and eager k-medoids clustering: O(k) runtime improvement of the PAM, CLARA, and CLARANS algorithms. Inf Syst 101:101804 [Google Scholar]
- 35.Schubert E, Rousseeuw PJ (2019) Faster k-medoids clustering: improving the PAM, CLARA, and CLARANS algorithms. In: Similarity search and applications: 12th International Conference, SISAP 2019, Newark, NJ, USA, Proceedings 12. Springer International Publishing, pp 171–187. https://arxiv.org/abs/1810.05691
- 36.Ng RT, Han J (2002) CLARANS: a method for clustering objects for spatial data mining. IEEE Trans Knowl Data Eng 14:1003–1016 [Google Scholar]
- 37.Kaufman L, Rousseeuw PJ (2009) [Chapter 2] Partitioning around medoids (Program PAM). In: Kaufman L, Rousseeuw PJ (eds) Finding groups in data, pp 68–125. 10.1002/9780470316801.ch2, [Chapter 3] Clustering large applications (Program CLARA). In: Kaufman L, Rousseeuw PJ (eds) Finding groups in data, pp 126–163. 10.1002/9780470316801.ch3
- 38.Krishnapuram R, Joshi A, Nasraoui O, Yi L (2001) Low-complexity fuzzy relational clustering algorithms for web mining. IEEE Trans Fuzzy Syst 9:595–607 [Google Scholar]
- 39.Preud’homme G, Duarte K, Dalleau K, Lacomblez C, Bresso E, Smaïl-Tabbone M et al (2021) Head-to-head comparison of clustering methods for heterogeneous data: a simulation-driven benchmark. Sci Rep 11:4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufman L, Rousseeuw PJ (1986) Clustering large data sets. In: Gelsema ES, Kanal LN (eds) Pattern Recognition in Practice. Elsevier, Amsterdam, pp 425–437 [Google Scholar]
- 41.Pina AF, Meneses MJ, Sousa-Lima I, Henriques R, Raposo JF, Macedo MP (2023) Big data and machine learning to tackle diabetes management. Eur J Clin Invest 53:e13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esnault C, Rollot M, Guilmin P, Zucker JD (2022) Qluster: an easy-to-implement generic workflow for robust clustering of health data. Front Artif Intell 5:1055294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giordani P, Perna S, Bianchi A, Pizzulli A, Tripodi S, Matricardi PM (2020) A study of longitudinal mobile health data through fuzzy clustering methods for functional data: the case of allergic rhinoconjunctivitis in childhood. PLoS ONE 15:e0242197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berna R, Mitra N, Hoffstad O, Wan J, Margolis DJ (2020) Identifying phenotypes of atopic dermatitis in a longitudinal United States cohort using unbiased statistical clustering. J Invest Dermatol 140:477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haider S, Granell R, Curtin JA, Holloway JW, Fontanella S, Hasan Arshad S et al (2023) Identification of eczema clusters and their association with filaggrin and atopic comorbidities: analysis of five birth cohorts. Br J Dermatol 190:45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haider S, Granell R, Curtin J, Fontanella S, Cucco A, Turner S et al (2022) Modeling wheezing spells identifies phenotypes with different outcomes and genetic associates. Am J Respir Crit Care Med 205:883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCready C, Haider S, Little F, Nicol MP, Workman L, Gray DM et al (2023) Early childhood wheezing phenotypes and determinants in a South African birth cohort: longitudinal analysis of the Drakenstein Child Health Study. The Lancet Child Adolesc Health 7:127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinha A, Desiraju K, Aggarwal K, Kutum R, Roy S, Lodha R et al (2017) Exhaled breath condensate metabolome clusters for endotype discovery in asthma. J Transl Med 15:262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Husson F, Josse J, Pages J (2010) Principal component methods-hierarchical clustering-partitional clustering: why would we need to choose for visualizing data. Applied Mathematics Department 17. https://www.sthda.com/english/upload/hcpchussonjosse.pdf
- 50.Coombes CE, Liu X, Abrams ZB, Coombes KR, Brock G (2021) Simulation-derived best practices for clustering clinical data. J Biomed Inform 118:103788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hennig C (2015) Clustering strategy and method selection. Handb Cluster Anal 9:703–730 [Google Scholar]
- 52.Zhang X, Lauber L, Liu H, Shi J, Wu J, Pan Y (2021) Research on the method of travel area clustering of urban public transport based on Sage-Husa adaptive filter and improved DBSCAN algorithm. PLoS ONE 16:e0259472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JY, Lee S, Suh DI, Kim DW, Yoon HJ, Park SK et al (2022) Distinct endotypes of pediatric rhinitis based on cluster analysis. Allergy Asthma Immunol Res 14:730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yum HY, Lee JS, Bae JM, Lee S, Kim YH, Sung M et al (2022) Classification of atopic dermatitis phenotypes according to allergic sensitization by cluster analysis. World Allergy Organ J 15 [DOI] [PMC free article] [PubMed]
- 55.Yoon J, Eom EJ, Kim JT, Lim DH, Kim WK, Song DJ et al (2021) Heterogeneity of childhood asthma in Korea: cluster analysis of the Korean childhood asthma study cohort. Allergy Asthma Immunol Res 13:42–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fedele DA, McQuaid EL, Faino A, Strand M, Cohen S, Robinson J et al (2016) Patterns of adaptation to children’s food allergies. Allergy 71:505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banić I, Lovrić M, Cuder G, Kern R, Rijavec M, Korošec P, Turkalj M (2021) Treatment outcome clustering patterns correspond to discrete asthma phenotypes in children. Asthma Res Pract 7:11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sijbrandij JJ, Hoekstra T, Almansa J, Reijneveld SA, Bültmann U (2019) Identification of developmental trajectory classes: comparing three latent class methods using simulated and real data. Advances in Life Course Research 42:100288 [DOI] [PubMed] [Google Scholar]
- 59.Forster F, Ege MJ, Gerlich J, Weinmann T, Kreißl S, Weinmayr G et al (2022) Trajectories of asthma and allergy symptoms from childhood to adulthood. Allergy 77:1192–1203 [DOI] [PubMed] [Google Scholar]
- 60.Custovic A, Sonntag H-J, Buchan IE, Belgrave D, Simpson A, Prosperi MCF (2015) Evolution pathways of IgE responses to grass and mite allergens throughout childhood. J Allergy Clin Immunol 136:1645–52.e8 [DOI] [PubMed] [Google Scholar]
- 61.Peng Z, Kurz D, Weiss JM, Brenner H, Rothenbacher D, Genuneit J (2022) Latent classes of atopic dermatitis and food allergy development in childhood. Pediatr Allergy Immunol 33:e13881 [DOI] [PubMed] [Google Scholar]
- 62.Yavuz ST, Oksel Karakus C, Custovic A, Kalayci Ö (2021) Four subtypes of childhood allergic rhinitis identified by latent class analysis. Pediatr Allergy Immunol 32:1691–1699 [DOI] [PubMed] [Google Scholar]
- 63.Havstad S, Johnson CC, Kim H, Levin AM, Zoratti EM, Joseph CL et al (2014) Atopic phenotypes identified with latent class analyses at age 2 years. J Allergy Clin Immunol 134:722–7.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguena Nguefack HL, Pagé MG, Katz J, Choinière M, Vanasse A, Dorais M et al (2020) Trajectory modelling techniques useful to epidemiological research: a comparative narrative review of approaches. Clin Epidemiol 12:1205–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altman MC, Calatroni A, Ramratnam S, Jackson DJ, Presnell S, Rosasco MG et al (2021) Endotype of allergic asthma with airway obstruction in urban children. J Allergy Clin Immunol 148:1198–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lau HX, Chen Z, Chan YH, Tham EH, Goh AEN, Van Bever H et al (2022) Allergic sensitization trajectories to age 8 years in the Singapore GUSTO cohort. World Allergy Organ J 15:100667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Nest G, Lima Passos V, Candel MJJM, van Breukelen GJP (2020) An overview of mixture modelling for latent evolutions in longitudinal data: modelling approaches, fit statistics and software. Adv Life Course Res 43:100323 [DOI] [PubMed] [Google Scholar]
- 68.Feldman BJ, Masyn KE, Conger RD (2009) New approaches to studying problem behaviors: a comparison of methods for modeling longitudinal, categorical adolescent drinking data. Dev Psychol 45:652–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lewis KM, De Stavola BL, Cunningham S, Hardelid P (2023) Socioeconomic position, bronchiolitis and asthma in children: counterfactual disparity measures from a national birth cohort study. Int J Epidemiol 52:476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai R, Miliku K, Gaddipati S, Choi J, Ambalavanan A, Tran MM et al (2022) Wheeze trajectories: determinants and outcomes in the CHILD Cohort Study. J Allergy Clin Immunol 149:2153–2165 [DOI] [PubMed] [Google Scholar]
- 71.Loo EXL, Liew TM, Yap GC, Wong LSY, Shek LP, Goh A et al (2021) Trajectories of early-onset rhinitis in the Singapore GUSTO mother-offspring cohort. Clin Exp Allergy 51:419–429 [DOI] [PubMed] [Google Scholar]
- 72.Hu C, Duijts L, Erler NS, Elbert NJ, Piketty C, Bourdès V et al (2019) Most associations of early-life environmental exposures and genetic risk factors poorly differentiate between eczema phenotypes: the Generation R Study. Br J Dermatol 181:1190–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziyab AH, Mukherjee N, Zhang H, Arshad SH, Karmaus W (2022) Sex-specific developmental trajectories of eczema from infancy to age 26 years: a birth cohort study. Clin Exp Allergy 52:416–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uddin S, Haque I, Lu H, Moni MA, Gide E (2022) Comparative performance analysis of K-nearest neighbour (KNN) algorithm and its different variants for disease prediction. Sci Rep 12:6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarker IH (2021) Machine Learning: algorithms, real-world applications and research directions. SN Comput Sci 2:160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salvador-Meneses J, Ruiz-Chavez Z, Garcia-Rodriguez J (2019) Compressed kNN: K-nearest neighbors with data compression. Entropy 21(3):234. 10.3390/e21030234 [DOI] [PMC free article] [PubMed]
- 77.Bose S, Kenyon CC, Masino AJ (2021) Personalized prediction of early childhood asthma persistence: a machine learning approach. PLoS ONE 16:e0247784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kothalawala DM, Murray CS, Simpson A, Custovic A, Tapper WJ, Arshad SH et al (2021) Development of childhood asthma prediction models using machine learning approaches. Clin Transl Allergy 11:e12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang XW, Wang T, Schaub DP, Chen C, Sun Z, Ke S et al (2023) Benchmarking omics-based prediction of asthma development in children. Respir Res 24:63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Vliet D, Alonso A, Rijkers G, Heynens J, Rosias P, Muris J et al (2015) Prediction of asthma exacerbations in children by innovative exhaled inflammatory markers: results of a longitudinal study. PLoS ONE 10:e0119434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jung T, Kim J (2023) A new support vector machine for categorical features. Expert Syst Appl 229:120449 [Google Scholar]
- 82.Jeddi Z, Gryech I, Ghogho M, El Hammoumi M, Mahraoui C (2021) Machine learning for predicting the risk for childhood asthma using prenatal, perinatal, postnatal and environmental factors. Healthcare (Basel) 99(11):1464. 10.3390/healthcare9111464 [DOI] [PMC free article] [PubMed]
- 83.Lin J, Bruni FM, Fu Z, Maloney J, Bardina L, Boner AL et al (2012) A bioinformatics approach to identify patients with symptomatic peanut allergy using peptide microarray immunoassay. J Allergy Clin Immunol 129:1321–8.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang Z, Li J, Kong N, Kim JH, Kim BS, Lee MJ et al (2022) Accurate diagnosis of atopic dermatitis by combining transcriptome and microbiota data with supervised machine learning. Sci Rep 12:290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Song YY, Lu Y (2015) Decision tree methods: applications for classification and prediction. Shanghai Arch Psychiatry 27:130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kingsford C, Salzberg SL (2008) What are decision trees? Nat Biotechnol 26:1011–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patel SJ, Chamberlain DB, Chamberlain JM (2018) A machine learning approach to predicting need for hospitalization for pediatric asthma exacerbation at the time of emergency department triage. Acad Emerg Med 25:1463–1470 [DOI] [PubMed] [Google Scholar]
- 88.Jiang T, Gradus JL, Rosellini AJ (2020) Supervised machine learning: a brief primer. Behav Ther 51:675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Emeryk A, Derom E, Janeczek K, Kuźnar-Kamińska B, Zelent A, Łukaszyk M et al (2023) Home monitoring of asthma exacerbations in children and adults with use of an AI-aided stethoscope. Ann Fam Med 21:517–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karaboga HA, Gunel A, Korkut SV, Demir I, Celik R (2021) Bayesian network as a decision tool for predicting ALS disease. Brain Sci 11(2):150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spyroglou II, Spöck G, Rigas AG, Paraskakis EN (2018) Evaluation of Bayesian classifiers in asthma exacerbation prediction after medication discontinuation. BMC Res Notes 11:522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Himes BE, Wu AC, Duan QL, Klanderman B, Litonjua AA, Tantisira K et al (2009) Predicting response to short-acting bronchodilator medication using Bayesian networks. Pharmacogenomics 10:1393–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu F, Zhang SW, Guo WF, Wei ZG, Chen L (2016) Inference of gene regulatory network based on local Bayesian networks. PLoS Comput Biol 12:e1005024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kelly RS, McGeachie MJ, Lee-Sarwar KA, Kachroo P, Chu SH, Virkud YV et al (2018) Partial least squares discriminant analysis and Bayesian networks for metabolomic prediction of childhood asthma. Metabolites 8(4):68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sharma S, Poon A, Himes BE, Lasky-Su J, Sordillo JE, Belanger K et al (2012) Association of variants in innate immune genes with asthma and eczema. Pediatr Allergy Immunol 23:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yarmus JS (2003) ABN: a fast, greedy Bayesian network classifier. J Yarmus—2003. https://citeseerx.ist.psu.edu/document?repid=rep1&type=pdf&doi=30e439fe0ad747df8ae213d75d452c13a7e96fdd
- 97.John GH, Langley P (1995) Estimating continuous distributions in Bayesian classifiers. Proceedings of the Eleventh conference on uncertainty in artificial intelligence; Montréal, Qué, Canada: Morgan Kaufmann Publishers Inc. p 338–45
- 98.Maslove DM, Podchiyska T, Lowe HJ (2013) Discretization of continuous features in clinical datasets. J Am Med Inform Assoc 20:544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gryak J, Georgievska A, Zhang J, Najarian K, Ravikumar R, Sanders G, Schuler CF (2024) Prediction of pediatric peanut oral food challenge outcomes using machine learning. Journal of Allergy and Clinical Immunology: Global 3:100252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bhattacharya S, Bennet L, Davidson JO, Unsworth CP (2022) Multi-layer perceptron classification & quantification of neuronal survival in hypoxic-ischemic brain image slices using a novel gradient direction, grey level co-occurrence matrix image training. PLoS ONE 17:e0278874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ahn JM, Kim J, Kim K (2023) Ensemble machine learning of gradient boosting (XGBoost, LightGBM, CatBoost) and attention-based CNN-LSTM for harmful algal blooms forecasting. Toxins 15(10):608. 10.3390/toxins15100608 [DOI] [PMC free article] [PubMed]
- 102.Kamphorst K, Lopez-Rincon A, Vlieger AM, Garssen J, van’ t Riet E, van Elburg RM (2023) Predictive factors for allergy at 4–6 years of age based on machine learning: a pilot study. PharmaNutrition. 23:100326 [Google Scholar]
- 103.Kavya R, Christopher J, Panda S, Lazarus YB (2021) Machine learning and XAI approaches for allergy diagnosis. Biomed Signal Process Control 69:102681 [Google Scholar]
- 104.Lovrić M, Banić I, Lacić E, Pavlović K, Kern R, Turkalj M (2021) Predicting treatment outcomes using explainable machine learning in children with asthma. Children (Basel) 8(5):376. 10.3390/children8050376 [DOI] [PMC free article] [PubMed]
- 105.de Hond AAH, Kant IMJ, Honkoop PJ, Smith AD, Steyerberg EW, Sont JK (2022) Machine learning did not beat logistic regression in time series prediction for severe asthma exacerbations. Sci Rep 12:20363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hastie T, Tibshirani R, Friedman JH, Friedman JH (200) The elements of statistical learning: data mining, inference, and prediction: Springer 9
- 107.Mondal P, Dey D, Chandra Saha N, Moitra S, Saha GK, Bhattacharya S, Podder S (2019) Investigation of house dust mite induced allergy using logistic regression in West Bengal. India World Allergy Organ J 12:100088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sugiura S, Hiramitsu Y, Futamura M, Kamioka N, Yamaguchi C, Umemura H et al (2021) Development of a prediction model for infants at high risk of food allergy. Asia Pac Allergy 11:e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cofre-Martel S, Lopez Droguett E, Modarres M (2021) Big Machinery data preprocessing methodology for data-driven models in prognostics and health management. Sensors (Basel) 21(20):6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Breunig MM, Kriegel H-P, Ng RT, Sander J (eds) (2000) LOF: identifying density-based local outliers. Proceedings of the 2000 ACM SIGMOD international conference on Management of data
- 111.Kwak SK, Kim JH (2017) Statistical data preparation: management of missing values and outliers. Korean J Anesthesiol 70:407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Graham JW, Olchowski AE, Gilreath TD (2007) How many imputations are really needed? Some practical clarifications of multiple imputation theory. Prev Sci 8:206–213 [DOI] [PubMed] [Google Scholar]
- 113.Huque MH, Carlin JB, Simpson JA, Lee KJ (2018) A comparison of multiple imputation methods for missing data in longitudinal studies. BMC Med Res Methodol 18:168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shah AD, Bartlett JW, Carpenter J, Nicholas O, Hemingway H (2014) Comparison of random forest and parametric imputation models for imputing missing data using MICE: a CALIBER study. Am J Epidemiol 179:764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Austin PC, White IR, Lee DS, van Buuren S (2021) Missing data in clinical research: a tutorial on multiple imputation. Can J Cardiol 37:1322–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dettori JR, Norvell DC, Chapman JR (2018) The sin of missing data: is all forgiven by way of imputation? Global Spine J 8:892–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee JH, Huber JC Jr (2021) Evaluation of multiple imputation with large proportions of missing data: how much is too much? Iran J Public Health 50:1372–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pudjihartono N, Fadason T, Kempa-Liehr AW, O’Sullivan JM (2022) A review of feature selection methods for machine learning-based disease risk prediction. Front Bioinform 2:927312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ferri FJ, Pudil P, Hatef M, Kittler J (1994) Comparative study of techniques for large-scale feature selection. Machine intelligence and pattern recognition. 16: Elsevier p 403–13
- 120.Pudil P, Novovičová J, Kittler J (1994) Floating search methods in feature selection. Pattern Recogn Lett 15:1119–1125 [Google Scholar]
- 121.Tibshirani R (1996) Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Stat Methodol 58:267–288 [Google Scholar]
- 122.Xu Y, Goodacre R (2018) On splitting training and validation set: a comparative study of cross-validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. J Anal Test 2:249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahmed Ouameur M, Caza-Szoka M, Massicotte D (2020) Machine learning enabled tools and methods for indoor localization using low power wireless network. Internet of Things 12:100300 [Google Scholar]
- 124.Przybyś-Małaczek A, Antoniuk I, Szymanowski K, Kruk M, Kurek J (2023) Application of machine learning algorithms for tool condition monitoring in milling chipboard process. Sensors 23(13):5850. 10.3390/s23135850 [DOI] [PMC free article] [PubMed]
- 125.Berisha V, Krantsevich C, Hahn PR, Hahn S, Dasarathy G, Turaga P, Liss J (2021) Digital medicine and the curse of dimensionality. NPJ Digit Med 4:153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ibrahim S, Nazir S, Velastin SA (2021) Feature selection using correlation analysis and principal component analysis for accurate breast cancer diagnosis. J Imaging 7(11):225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He H, Yang B, Garcia E, Li SA (2008 Jun) Adaptive synthetic sampling approach for imbalanced learning Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence) Hong Kong. ChinaIEEE
- 128.Wolpert DH, Macready WG (1997) No free lunch theorems for optimization. IEEE Trans Evol Comput 1:67–82 [Google Scholar]
- 129.Muralidharan V, Burgart A, Daneshjou R, Rose S (2023) Recommendations for the use of pediatric data in artificial intelligence and machine learning ACCEPT-AI. npj Digit Med 6:166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ruf B, Detyniecki M (2020) Active fairness instead of unawareness. arXiv preprint arXiv:200906251
- 131.Van Mechelen I, Hennig C, Kiers HA (2024) Onset of a conceptual outline map to get a hold on the jungle of cluster analysis. Wiley Interdiscip Rev: Data Minin Knowl Discov 14:e1547 [Google Scholar]
- 132.Fodeh SJ, Brandt C, Luong TB, Haddad A, Schultz M, Murphy T, Krauthammer M (2013) Complementary ensemble clustering of biomedical data. J Biomed Inform 46:436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gruber S, Krakower D, Menchaca JT, Hsu K, Hawrusik R, Maro JC et al (2020) Using electronic health records to identify candidates for human immunodeficiency virus pre-exposure prophylaxis: an application of super learning to risk prediction when the outcome is rare. Stat Med 39:3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Naimi AI, Balzer LB (2018) Stacked generalization: an introduction to super learning. Eur J Epidemiol 33:459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Naderalvojoud B, Hernandez-Boussard T (2023) Improving machine learning with ensemble learning on observational healthcare data. AMIA Annu Symp Proc 2023:521–529 [PMC free article] [PubMed] [Google Scholar]
- 136.Shirkhorshidi AS, Aghabozorgi S, Wah TY (2015) A comparison study on similarity and dissimilarity measures in clustering continuous data. PLoS ONE 10:e0144059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oyewole GJ, Thopil GA (2023) Data clustering: application and trends. Artif Intell Rev 56:6439–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boriah S, Chandola V, Kumar V (200) Similarity measures for categorical data: a comparative evaluation. Proceedings of the 2008 SIAM International Conference on Data Mining (SDM). Proceedings: Society for Industrial and Applied Mathematics; 8 p 243–54
- 139.Halkidi M, Vazirgiannis M, Hennig C (2015) Method-independent indices for cluster validation and estimating the number of clusters. Handb Clust Anal 26:595–618 [Google Scholar]
- 140.Faouzi J, Colliot O (2023) Classic machine learning methods. In: Colliot O (ed) Machine learning for brain disorders. New York, NY, pp 25–75
- 141.Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, van Velzen EJJ et al (2008) Assessment of PLSDA cross validation. Metabolomics 4:81–89 [Google Scholar]
- 142.Harrington PB (2018) Multiple versus single set validation of multivariate models to avoid mistakes. Crit Rev Anal Chem 48:33–46 [DOI] [PubMed] [Google Scholar]
- 143.Nichols JA, Herbert Chan HW, Baker MAB (2019) Machine learning: applications of artificial intelligence to imaging and diagnosis. Biophys Rev 11:111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Montesinos López OA, Montesinos López A, Crossa J (2022) Overfitting, model tuning, and evaluation of prediction performance. Springer, Multivariate statistical machine learning methods for genomic prediction, pp 109–139 [PubMed] [Google Scholar]
- 145.Saad MH, Hashima S, Sayed W, El-Shazly EH, Madian AH, Fouda MM (2022) Early diagnosis of COVID-19 images using optimal CNN hyperparameters. Diagnostics (Basel) 13(1):76. 10.3390/diagnostics13010076 [DOI] [PMC free article] [PubMed]
- 146.Akiba T, Sano S, Yanase T, Ohta T, Koyama M, editors. Optuna: a next-generation hyperparameter optimization framework. Proceedings of the 25th ACM SIGKDD international conference on knowledge discovery & data mining; 2019.
- 147.Elshewey AM, Shams MY, El-Rashidy N, Elhady AM, Shohieb SM, Tarek Z (2023) Bayesian optimization with support vector machine model for Parkinson disease classification. Sensors (Basel). 23(4):2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chicco D, Jurman G (2022) The ABC recommendations for validation of supervised machine learning results in biomedical sciences. Front Big Data 5:979465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu Q, Charleston MA, Richards SA, Holland BR (2023) Performance of Akaike Information Criterion and Bayesian Information Criterion in selecting partition models and mixture models. Syst Biol 72:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Park SY, Park JE, Kim H, Park SH (2021) Review of statistical methods for evaluating the performance of survival or other time-to-event prediction models (from conventional to deep learning approaches). Korean J Radiol 22:1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rashidi HH, Albahra S, Robertson S, Tran NK, Hu B (2023) Common statistical concepts in the supervised Machine Learning arena. Front Oncol 13:1130229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lundberg S (2017) A unified approach to interpreting model predictions. arXiv preprint arXiv:170507874
- 153.Tjaden J, Tjaden B (2023) MLpronto: a tool for democratizing machine learning. PLoS ONE 18:e0294924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mathur P, Arshad H, Grasfield R, Khatib R, Aggarwal A, Auron M, Khare A (2024) Navigating AI: a quick start guide for healthcare professionals. Cureus 16:e72501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Collins GS, Moons KGM, Dhiman P, Riley RD, Beam AL, Van Calster B et al (2024) TRIPOD+AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 385:e078378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.van de Schoot R, Sijbrandij M, Winter SD, Depaoli S, Vermunt JK (2017) The GRoLTS-checklist: guidelines for reporting on latent trajectory studies. Struct Equ Modeling 24:451–467 [Google Scholar]
- 157.Stevens LM, Mortazavi BJ, Deo RC, Curtis L, Kao DP (2020) Recommendations for reporting machine learning analyses in clinical research. Circulation: Cardiovascular Quality and Outcomes 13:e006556 [DOI] [PMC free article] [PubMed]
- 158.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (eds) (2016) Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2921–2929
- 159.Ribeiro MT, Singh S, Guestrin C (eds) (2016) "Why should i trust you?" Explaining the predictions of any classifier. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining
- 160.Nembrini S, König IR, Wright MN (2018) The revival of the Gini importance? Bioinformatics 34:3711–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Artificial Intelligence and Machine Learning (AI/ML)-enabled medical devices: U.S. Food & Drug Administration; 2024 [Available from: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices
- 162.He P, Moraes TJ, Dai D, Reyna-Vargas ME, Dai R, Mandhane P et al (2024) Early prediction of pediatric asthma in the Canadian Healthy Infant Longitudinal Development (CHILD) birth cohort using machine learning. Pediatr Res 95:1818–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kothalawala DM, Kadalayil L, Weiss VBN, Kyyaly MA, Arshad SH, Holloway JW, Rezwan FI (2020) Prediction models for childhood asthma: a systematic review. Pediatr Allergy Immunol 31:616–627 [DOI] [PubMed] [Google Scholar]
- 164.Hamad AF, Yan L, Jafari Jozani M, Hu P, Delaney JA, Lix LM (2023) Developing a prediction model of children asthma risk using population-based family history health records. Pediatr Allergy Immunol 34:e14032 [DOI] [PubMed] [Google Scholar]
- 165.Prosperi MC, Marinho S, Simpson A, Custovic A, Buchan IE (2014) Predicting phenotypes of asthma and eczema with machine learning. BMC Med Genomics 7(Suppl 1):S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang J, Yang Y, Gong X (2024) Interpretable machine learning for allergic rhinitis prediction among preschool children in Urumqi. China Sci Rep 14:22281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang J, Lee D, Jungles K, Shaltis D, Najarian K, Ravikumar R et al (2023) Prediction of oral food challenge outcomes via ensemble learning. Inf Med Unlocked 36:101142 [Google Scholar]
- 168.Owora AH, Zhang Y (2021) Childhood wheeze trajectory-specific risk factors: a systematic review and meta-analysis. Pediatr Allergy Immunol 32:e13313 [DOI] [PubMed] [Google Scholar]
- 169.Lopez DJ, Lodge CJ, Bui DS, Waidyatillake NT, Abramson MJ, Perret JL et al (2022) Establishing subclasses of childhood eczema, their risk factors and prognosis. Clin Exp Allergy 52:1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mulick AR, Mansfield KE, Silverwood RJ, Budu-Aggrey A, Roberts A, Custovic A et al (2021) Four childhood atopic dermatitis subtypes identified from trajectory and severity of disease and internally validated in a large UK birth cohort. Br J Dermatol 185:526–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Oksel C, Granell R, Mahmoud O, Custovic A, Henderson AJ (2019) Causes of variability in latent phenotypes of childhood wheeze. J Allergy Clin Immunol 143:1783–90.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Belgrave DCM, Simpson A, Semic-Jusufagic A, Murray CS, Buchan I, Pickles A, Custovic A (2013) Joint modeling of parentally reported and physician-confirmed wheeze identifies children with persistent troublesome wheezing. J Allergy Clin Immunol 132:575–83.e12 [DOI] [PubMed] [Google Scholar]
- 173.Nakamura T, Haider S, Fontanella S, Murray CS, Simpson A, Custovic A (2022) Modelling trajectories of parentally reported and physician-confirmed atopic dermatitis in a birth cohort study*. Br J Dermatol 186:274–284 [DOI] [PubMed] [Google Scholar]
- 174.Lisik D, Wennergren G, Kankaanranta H, Basna R, Shah SA, Alm B et al (2024) Asthma and allergy trajectories in children based on combined parental report and register data. Pediatr Allergy Immunol 35:e14254 [DOI] [PubMed] [Google Scholar]
- 175.Li Y, Xiong Y, Fan W, Wang K, Yu Q, Si L et al (2024) Sequential model for predicting patient adherence in subcutaneous immunotherapy for allergic rhinitis. Front Pharmacol 15:1371504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Belhassen M, Nolin M, Jacoud F, Marant Micallef C, Van Ganse E (2023) Trajectories of controller therapy use before and after asthma-related hospitalization in children and adults: population-based retrospective cohort study. JMIR Public Health Surveill 9:e50085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Attar R, Hurault G, Wang Z, Mokhtari R, Pan K, Olabi B et al (2023) Reliable detection of eczema areas for fully automated assessment of eczema severity from digital camera images. JID Innov 3:100213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zvulunov A, Lenevich S, Migacheva N (2023) A Mobile health app for facilitating disease management in children with atopic dermatitis: feasibility and impact study. JMIR Dermatol 6:e49278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Seol HY, Shrestha P, Muth JF, Wi CI, Sohn S, Ryu E et al (2021) Artificial intelligence-assisted clinical decision support for childhood asthma management: a randomized clinical trial. PLoS ONE 16:e0255261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hurault G, Domínguez-Hüttinger E, Langan SM, Williams HC, Tanaka RJ (2020) Personalized prediction of daily eczema severity scores using a mechanistic machine learning model. Clin Exp Allergy 50:1258–1266 [DOI] [PubMed] [Google Scholar]
- 181.Luo G, He S, Stone BL, Nkoy FL, Johnson MD (2020) Developing a model to predict hospital encounters for asthma in asthmatic patients: secondary analysis. JMIR Med Inform 8:e16080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lee H, Cho JK, Park J, Lee H, Fond G, Boyer L et al (2024) Machine learning-based prediction of suicidality in adolescents with allergic rhinitis: derivation and validation in 2 independent nationwide cohorts. J Med Internet Res 26:e51473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Molfino NA, Turcatel G, Riskin D (2024) Machine learning approaches to predict asthma exacerbations: a narrative review. Adv Ther 41:534–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP et al (2018) Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface 15:20170387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22:1345–1359 [Google Scholar]
- 186.Gaye A, Marcon Y, Isaeva J, LaFlamme P, Turner A, Jones EM et al (2014) DataSHIELD: taking the analysis to the data, not the data to the analysis. Int J Epidemiol 43:1929–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Budiarto A, Tsang KCH, Wilson AM, Sheikh A, Shah SA (2023) Machine learning–based asthma attack prediction models from routinely collected electronic health records: systematic scoping review. JMIR AI 2:e46717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Miller Randolph A, Pople Harry E, Myers JD (1982) Internist-I, an Experimental computer-based diagnostic consultant for general internal medicine. N Engl J Med 307:468–476 [DOI] [PubMed] [Google Scholar]
- 189.Ojha T, Patel A, Sivapragasam K, Sharma R, Vosoughi T, Skidmore B et al (2024) Exploring machine learning applications in pediatric asthma management: scoping review. JMIR AI 3:e57983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.