Abstract
Background
Acute myeloid leukemia (AML) is a common hematological tumor, but it is difficult to treat. DNMT1 is a DNA methyltransferase whose main function is to maintain stable DNA methylation during the DNA replication process. DNMT1 also plays an important role in AML, but its function in cytokine-induced memory-like natural killer (CIML NK) cell activity remains unclear.
Methods and results
In this study, we isolated primary NK cells from the peripheral blood of healthy volunteers and AML patients and treated them with 10 ng/mL IL-12, 50 ng/mL IL-15 and 50 ng/mL IL-18 to promote their differentiation into CIML NK cells. The activity of CIML NK cells was evaluated by RT‒qPCR, western blotting, ELISAs, and flow cytometry. DNMT1 was highly expressed in NK cells from AML patients. Knocking down DNMT1 significantly increased the expression of CD25, CD137, CD107a, IFN-γ, and TNF-α and increased the activity of CIML NK cells. Mechanistically, knocking down DNMT1 promoted autophagy by activating the AMPK/mTOR signaling pathway, thereby enhancing the activity of CIML NK cells and alleviating the progression of AML.
Conclusions
Our study revealed that the downregulation of DNMT expression may be a new target for the treatment of AML.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11033-024-10181-9.
Keywords: Acute myeloid leukemia, Memory-like NK cells, DNMT1, Autophagy, AMPK/mTOR signaling pathway, DNA methylation
Introduction
Acute myeloid leukemia (AML) is a type of hematological malignancy with substantial heterogeneity and poses a significant challenge in clinical practice [1]. The current standard treatment for AML achieves remission in less than 30% of patients. Those with refractory or relapsed AML who cannot undergo hematopoietic stem cell transplantation (HCT) have limited treatment options and a poor prognosis [2]. On the other hand, allogeneic HCT offers a potential cure through immune-based therapy utilizing allogeneic T cells and NK cells. However, the high rates of treatment-related complications and mortality make this approach unsuitable for most AML patients [3, 4]. Therefore, an urgent need is to develop less toxic and more effective targeted cell therapies.
The main immune mechanism of allogeneic HCT, known as graft-versus-leukemia (GVL), is conventionally attributed to T cells. Nevertheless, a recent study indicated that natural killer (NK) cells also play a vital role in the GVL process [5, 6]. NK cells, a type of innate lymphocyte, are capable of eliminating virus-infected and malignant cells. These cells possess several critical features essential for successful cancer therapy, such as their innate ability to kill target cells while carrying minimal risks of graft-versus-host disease, cytokine release syndrome, or neurotoxicity [7]. Moreover, their inherent tendency to induce myeloid cell death makes them particularly attractive for AML treatment [8]. Studies have shown that NK cells can be induced to exhibit memory-like properties [9, 10]. By subjecting NK cells to brief in vitro preactivation utilizing IL-12, IL-15, and IL-18 (lasting 12 to 16 h), memory-like NK cells with potent antileukemic properties can be generated [11, 12]. Furthermore, these memory-like NK cells have produced favorable responses in more than 50% of individuals with relapsed/refractory AML, with no significant adverse effects [2]. Therefore, exploring the molecular mechanism by which cytokine-induced memory-like (CIML) NK cell activity is enhanced is highly important for the clinical treatment of AML.
Abnormal methylation of DNA is commonly observed in tumors and contributes to genome instability and controlling the expression of important genes, thus facilitating the malignant progression of tumors [13]. DNA methylation is primarily managed by DNA methyltransferases (DNMTs). DNMT1, a crucial member of the DNMT family, aids in retaining methylation during DNA replication [14]. DNMT1 has been shown to impact tumor progression. For example, in pancreatic ductal adenocarcinoma, DNMT1 can promote cell cycle progression and growth while hindering the differentiation of pancreatic ductal adenocarcinoma cells [15]. Furthermore, DNMT1 also plays a vital role in the tolerance and effectiveness of AML treatment [16]. Nonetheless, the influence of DNMT1 on the function of CIML NK cells in AML remains unexplored.
Furthermore, DNMT1 can regulate the expression of the Beclin 1 and LC3 proteins, which performs dual functions in the regulation of autophagy [17]. Autophagy is a conserved eukaryotic process that upholds intracellular protein quality control and cellular equilibrium in response to environmental stressors [18]. Importantly, autophagy also governs the enrichment and localization of NK cells [19]. Moreover, a growing body of research indicates that the AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signaling pathway is crucial for regulating autophagy [20]. AMPK acts as an energy sensor and an autophagy inducer. Once activated, AMPK reduces mTOR phosphorylation levels, thereby increasing autophagy in cells [21]. Therefore, this study explored the regulatory role of the AMPK/mTOR pathway in NK cell autophagy.
In summary, this study aimed to explore the effect of DNMT1 on CIML NK cell activity in AML through the modulation of the AMPK/mTOR signaling pathway. This study sought to elucidate its mechanism of action and provide a theoretical foundation for developing new treatments for AML.
Materials and methods
Clinical sample collection
Blood samples were obtained from 16 healthy volunteers and 16 AML patients, with an equal distribution of males and females between the ages of 30 and 60 years at the First Affiliated Hospital of Kunming Medical University. All participants provided informed consent before any study procedures were performed.
Isolation, culture and treatment of NK cells
NK cells were initially isolated from the peripheral blood of healthy volunteers and patients with AML via CD56-positive selection (Miltenyi Biotec, Germany). For CIML and control NK cells, primary NK cells were preactivated using a previously documented protocol [22]. The NK cells from AML patients were cultured at a density of 4 × 106 cells/mL and exposed to a mixture of 10 ng/mL IL-12, 50 ng/mL IL-15, and 50 ng/mL IL-18, or control conditions involving 3 ng/mL IL-15, and were preactivated for 16 h on Day 1. After this preliminary 16-hour activation, the NK cell preparations were washed three times with PBS from HyClone to remove cytokines before being placed into complete AIM-V medium supplemented with 10% fetal bovine serum and 3 ng/mL IL-15 to increase survival. The medium was changed every 2 or 3 days, and the medium was supplemented with IL-15. On Day 7, the cells were collected for subsequent experiments. Azacitidine (AZA, Sigma‒Aldrich), chloroquine (CQ, Sigma‒Aldrich), and the AMPK/mTOR signaling pathway inhibitor dorsomorphin (Comp C; Selleck Chemicals) were added to the CIML NK cells at concentrations of 40 µM, 10 µM, and 10 µM, respectively, and incubated for a period of 48 h.
Cell transfection
NK cells and CIML NK cells were seeded into 24-well plates at a density of 1 × 105 cells per well one day before the infection. Following an overnight culture, a lentivirus supernatant (Genepharma, Shanghai, China) containing either sh-NC or sh-DNMT1 was utilized to infect the cells at a multiplicity of infection (MOI) of 20. The transfection efficiency was evaluated at 48 h postinfection, and the cells were subsequently used in further experiments.
DNA methylation analysis
Genome-wide DNA methylation (5-mC) levels in NK cells were measured using a MethylFlash DNA quantification (coloration) kit (Epigentek). Genomic DNA (100 mg) was isolated from NK cells using a Genomic DNA Isolation Kit (Qiagen). For the quantification of the 5-mC content, specific monoclonal antibodies were applied in combination with both detection and secondary antibodies. Standard curves were generated to quantify the 5-mC content, utilizing the negative and positive DNA controls provided in the kit. The colorimetric estimation of the 5-mC content was conducted using a microplate reader according to the manufacturer’s protocols. First, the genomic DNA from each sample was added to the wells of the assay plate. Upon color development, the absorbance was measured at 450 nm with a microplate reader according to the manufacturer’s guidelines.
RT‒qPCR
Total RNA was extracted from NK cells with TRIzol reagent (Invitrogen, 15596026). The RNA was then reverse-transcribed to generate cDNA using a first-strand cDNA synthesis kit (Genenode, China). The synthesized cDNA was subsequently subjected to real-time fluorescence quantitative PCR with a SYBR Green kit (Solarbio, China). The PCR analysis utilized GAPDH as an internal reference, and the data were analyzed using the 2−ΔΔCt method. The specific sequences of the primers used can be found in Table 1.
Table 1.
Primer sequences
| Target | Sequence (F: forward primer; R: reverse primer) (5´-3´) |
|---|---|
| DNMT1 |
F: TACCAGGGAGAAGGACGGG R: ACCACCAGACGCCACAT |
| GAPDH | F: GGGAAACTGTGGCGTGAT |
| R: AAUGGTGGAGGAGTGGGT |
Western blot analysis
Proteins were obtained from NK cells and CIML NK cells using RIPA buffer (Sigma‒Aldrich, USA) containing 1% protease inhibitors and phosphatase inhibitors. The protein concentration was determined with a bicinchoninic acid (BCA) assay kit (Thermo Scientific, USA) according to the manufacturer’s protocol. The total proteins were separated on SDS‒PAGE gels and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA). The membranes were subsequently blocked with 5% skim milk for 1.5 h at room temperature. Primary antibodies against DNMT1 (1:1000, ab188453, Abcam, UK), LC3B (1:1000, ab192890, Abcam, UK), Beclin1 (1:1000, ab302669, Abcam, UK), p62 (1:1000, ab109012, Abcam, UK), AMPK (1:1000, #2532, CST, USA), p-AMPK (1:1000, #2535, CST, USA), mTOR (1:1000, ab32028, Abcam, UK), p-mTOR (1:2000, ab109268, Abcam, UK), and GAPDH (1:2000, ab8245, Abcam, UK) were applied. The membranes and antibodies were then incubated overnight at 4 °C. A secondary antibody (1:4000, ab97051, Abcam, UK) was subsequently added and incubated with the membrane at room temperature for 1 h before being developed using an enhanced chemiluminescence (ECL) kit (Millipore, USA). Finally, a semiquantitative analysis of the bands was performed using ImageJ software.
ELISA
In accordance with the instructions of the TNF-α (SEKH-0047, Solarbio) and IFN-γ (SEKH-0046, Solarbio) kits, NK cell or CIML NK cell culture media were transferred to sterile centrifuge tubes. After centrifugation at 1,000×g for 10 min, the supernatant was evenly distributed into small EP tubes for analysis. After each working solution was added according to the manufacturer’s instructions, the levels of cytokines (TNF-α and IFN-γ) were measured by detecting the optical density at 450 nm using a microplate reader.
Flow cytometry
NK cells and CIML NK cells were resuspended in FACS buffer composed of 0.5% (w/v) BSA and 2 mM EDTA in a Ca2+− and Mg2+-free PBS solution. Next, the cells were transferred to 1.5 mL EP tubes, ensuring that the samples from each group contained at least 1 × 10⁶ cells. The supernatant was removed via centrifugation. The antibodies were then diluted at a 1:50 ratio in PBS with 1% BSA and incubated in the dark for 45 min. After washes with FACS buffer, the samples were analyzed using a BD FACSVerse flow cytometer. Antibodies, including PE-conjugated CD56, CD25, and CD137 antibodies (Abcam, UK), were used for the flow cytometry analysis. Isotype-matched antibodies were used as controls to ensure proper gating. The detailed steps of the gating strategy are provided in Supplementary Figure S1.
CD107a degranulation assay
NK cells and CIML NK cells were suspended in 200 µL of cell culture medium, and 5 µL of the PE-conjugated CD107a antibody (Abcam, UK) or isotype control antibody was added to the cells. After an incubation for 1 h, GolgiStop (monensin), a protein transport inhibitor (BD Biosciences), was added. Cells were incubated for an additional 3 h. The cells were washed with PBS and analyzed by flow cytometry.
Statistical analysis
All the experimental data in this study are presented as the means ± standard deviations (means ± SDs). GraphPad Prism 8.2 was used for data analysis and visualization. A t test was conducted to compare two groups, whereas one-way ANOVA was used for comparisons of multiple groups. Pairwise comparisons between groups were performed with two-way ANOVA. Statistical significance was considered at P < 0.05.
Results
The methylation level is increased in NK cells from AML patients, and the expression of DNMT1 is upregulated
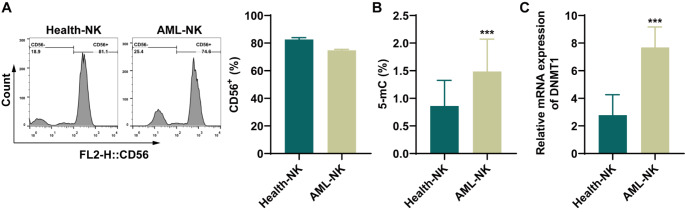
Initially, NK cells were isolated from the peripheral blood of both healthy volunteers and patients diagnosed with AML. The expression of the marker CD56 was examined to confirm the isolation of NK cells, and the results confirmed successful isolation (Fig. 1A). Next, the DNA methylation levels in the NK cells were measured. The methylation level in NK cells from AML patients was significantly higher than that in cells from healthy volunteers (Fig. 1B). Furthermore, DNMT1 mRNA levels in NK cells from AML patients were significantly elevated compared with those in NK cells from healthy volunteers (Fig. 1C). These findings suggest that both DNA methylation levels and DNMT1 gene expression are increased in NK cells from patients with AML.
Fig. 1.
The methylation level was increased in NK cells from leukemia patients, and the expression of DNMT1 was upregulated. A: The expression of the NK cell marker CD56 in healthy volunteers and AML patients was detected by flow cytometry. B: The DNA methylation level in NK cells from healthy volunteers and AML patients was detected using a kit. C: RT‒qPCR was used to detect DNMT1 mRNA levels in NK cells from healthy volunteers and AML patients. ***P < 0.001 compared with Health-NK cells
Knocking down DNMT1 increases the activity of memory-like NK cells
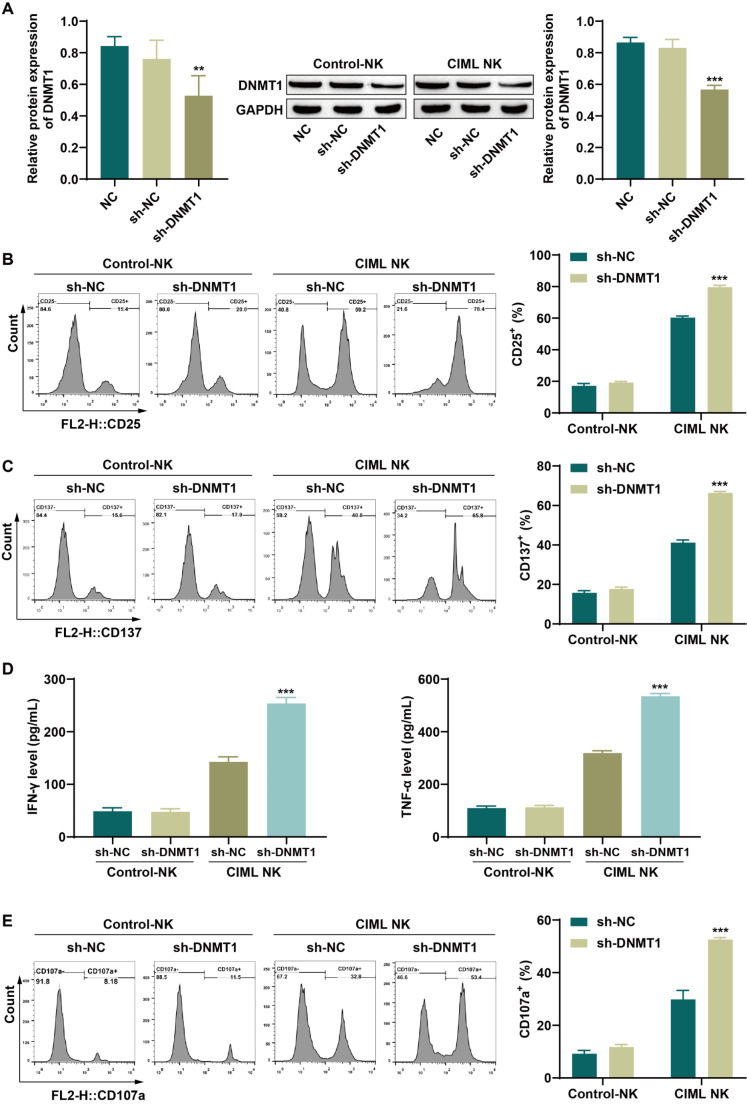
We investigated the effect of DNMT1 on the activity of CIML NK cells in AML patients, and the expression of DNMT1 was noticeably reduced following DNMT1 knockdown in both NK cells and CIML NK cells (Fig. 2A). A subsequent analysis focused on detecting the expression of NK cell activation markers. The levels of CD25 and CD137 in NK cells remained unchanged after DNMT1 knockdown, whereas a marked increase in CD25 and CD137 expression was observed in CIML NK cells (Fig. 2B-C). The ELISA results revealed that DNMT1 knockdown did not significantly alter the secretion of IFN-γ and TNF-α by NK cells, but a significant increase in the levels of these cytokines was noted in CIML NK cells (Fig. 2D). Degranulation assays are commonly utilized to determine NK cell cytotoxicity levels, with CD107a serving as a marker of the extent of degranulation. While DNMT1 knockdown did not impact CD107a expression in NK cells, CIML NK cells presented a significant increase in CD107a levels (Fig. 2E), indicating a significant increase in CIML NK cell cytotoxicity upon DNMT1 knockdown. These findings suggest that DNMT1 suppression can effectively increase the activity of CIML NK cells without significantly affecting NK cell activity.
Fig. 2.
Knockdown of DNMT1 increased the activity of memory-like NK cells. A: DNMT1 knockdown efficiency in NK cells and CIML NK cells was detected by western blotting. B: The expression of the activation marker CD25 in NK cells and CIML NK cells was detected by flow cytometry. C: The levels of the activation marker CD137 in NK cells and CIML NK cells were detected by flow cytometry. D: ELISA was used to detect the levels of the cytokines IFN-γ and TNF-α secreted by NK cells and CIML NK cells. E: CD107a expression in NK cells and CIML NK cells was detected by flow cytometry. **P < 0.01 and ***P < 0.001 compared with the NC or sh-NC group
The effect of the hypomethylation-inducing drug azacitidine on the activity of memory-like NK cells is consistent with that of DNMT1 knockdown
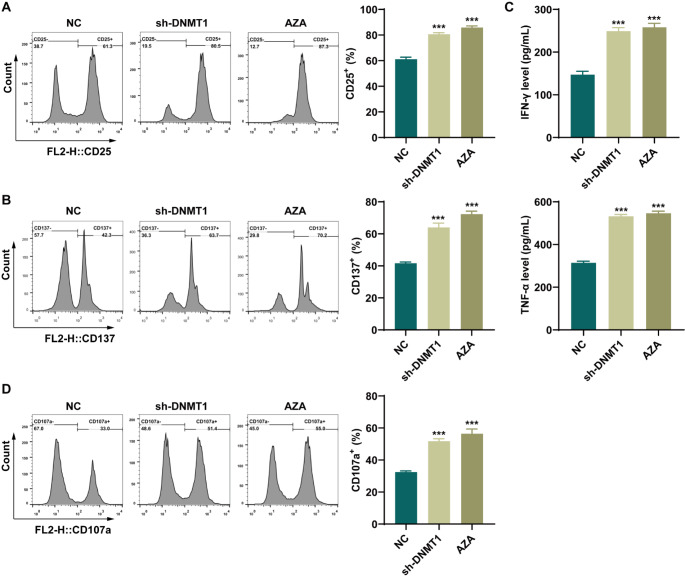
The impact of DNA methylation on AML-CIML NK cell activity was subsequently investigated through the introduction of AZA to CIML NK cells. The flow cytometry data indicated that the expression of the activation markers CD25 and CD137 in CIML NK cells was significantly increased after DNMT1 knockdown or AZA treatment compared with that in the NC group (Fig. 3A-B). ELISAs revealed significant increases in IFN-γ and TNF-α levels in CIML NK cells following DNMT1 knockdown or AZA treatment compared with those in the NC group (Fig. 3C). Moreover, the level of CD107a was noticeably increased in CIML NK cells upon DNMT1 knockdown or AZA treatment (Fig. 3D). These findings suggest that the suppression of DNA methylation can increase the activity of CIML NK cells, with results similar to those of the knockdown of DNMT1.
Fig. 3.
Effect of a hypomethylation-inducing drug (azacitidine) on the activity of memory-like NK cells, which was consistent with the effect of DNMT1 knockdown. A: The expression of the activation marker CD25 in CIML NK cells was detected by flow cytometry. B: The expression of the activation marker CD137 in CIML NK cells was detected by flow cytometry. C: The levels of IFN-γ and TNF-α secreted from CIML NK cells were detected via ELISAs. D: CD107a expression in CIML NK cells was detected by flow cytometry. ***P < 0.001 compared with the NC group
Knockdown of DNMT1 increases the level of autophagy in memory-like NK cells
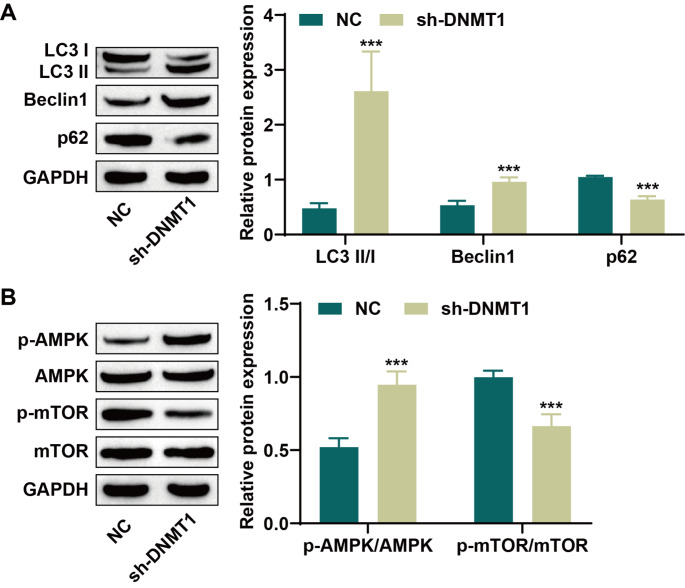
Studies have shown that autophagy is involved in the development and functional regulation of NK cells [23]. Therefore, we further investigated whether DNMT1 affects the activity of AML-CIML NK cells through the regulation of autophagy. Western blot analysis revealed that DNMT1 inhibition noticeably increased Beclin1 and LC3 II/I expression levels while decreasing p62 expression (Fig. 4A). The AMPK/mTOR pathway is a crucial regulator of autophagy [21]; therefore, we also examined the levels of proteins related to this pathway. Knockdown of DNMT1 significantly increased AMPK phosphorylation and decreased mTOR phosphorylation (Fig. 4B). These results suggest that DNMT1 suppression may induce autophagy in CIML NK cells by activating the AMPK/mTOR pathway.
Fig. 4.
Knockdown of DNMT1 increased the level of autophagy in memory-like NK cells. A: The levels of the autophagy-related proteins LC3 II/I, Beclin1 and p62 were detected via western blotting. B: Western blot analysis of the expression of proteins related to the AMPK/mTOR signaling pathway. ***P < 0.001 compared with the NC group
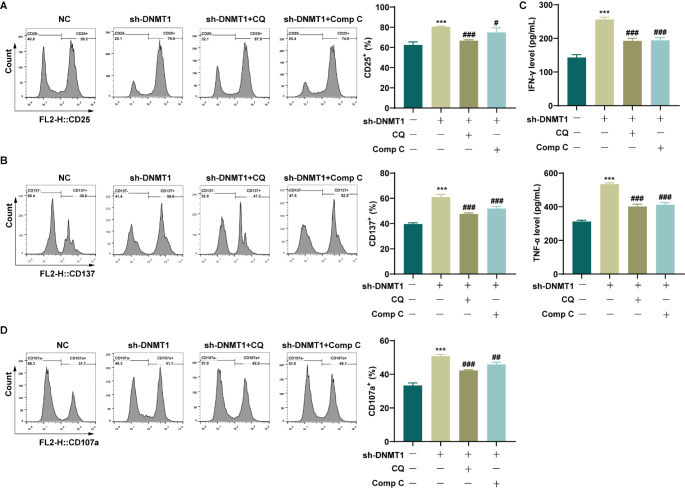
Autophagy inhibitors and AMPK inhibitors weaken the effect of DNMT1 knockdown on memory-like NK cells
AML-CIML NK cells were exposed to the autophagy inhibitor CQ or the AMPK pathway inhibitor Comp C to confirm the role of DNMT1 knockdown in regulating autophagy in CIML NK cells and its impact on CIML NK cell activity via the activation of the AMPK/mTOR signaling pathway. The results from the flow cytometry analysis revealed a significant decrease in the expression of the activation markers CD25 and CD137 in CIML NK cells when treated with either CQ or Comp C compared with the sh-DNMT1 group (Fig. 5A-B). Additionally, the ELISA results revealed notable reductions in the levels of IFN-γ and TNF-α in the CQ and Comp C groups compared with those in the sh-DNMT1 group (Fig. 5C). Moreover, the addition of CQ or Comp C led to a substantial decrease in CD107a expression in CIML NK cells (Fig. 5D). These findings suggest that autophagy inhibitors or AMPK pathway inhibitors can weaken the effect of DNMT1 knockdown on increasing CIML NK cell activity.
Fig. 5.
Autophagy inhibitors and signaling pathway inhibitors weakened the effect of DNMT1 knockdown on memory-like NK cells. A: The expression of the activation marker CD25 in CIML NK cells was detected by flow cytometry. B: The expression of the activation marker CD137 in CIML NK cells was detected by flow cytometry. C: The levels of IFN-γ and TNF-α secreted from CIML NK cells were detected via ELISAs. D: CD107a expression in CIML NK cells was detected by flow cytometry. ***P < 0.001 compared with the NC group; #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with the sh-DNMT1 group
Discussion
AML is a form of leukemia characterized by the abnormal proliferation of immature myeloid cells within the bone marrow, posing substantial treatment challenges. Our study focused on evaluating the potential antileukemic effects of CIML NK cells to investigate novel therapeutic strategies for AML [24]. First, we isolated NK cells from healthy volunteers and AML patients for our experiments. Importantly, abnormal DNA methylation is a key factor contributing to the progression of various tumors, including AML [25]. This epigenetic alteration is known to drive disease progression and resistance to therapies. DNMT1, a DNA methyltransferase, is responsible for maintaining DNA methylation patterns and neonatal methylation [17]. DNMT1 is often upregulated in many cancer types, leading to poor outcomes, and AML is no different [26, 27]. Mizuno et al. [28] investigated DNMT levels in 33 AML patients using a competitive PCR analysis, revealing significantly elevated DNMT1 levels in most patients. The overexpression of DNMT1 is believed to contribute to AML development and recurrence by causing the hypermethylation of tumor suppressor genes [28]. In this study, we found that the DNA methylation level in the NK cells of AML patients was significantly increased, and the expression of DNMT1 in the NK cells of AML patients was also significantly higher than that in the NK cells of healthy volunteers. Compared with previous studies, which only reported that DNMT1 was highly expressed in AML, our study further revealed that DNMT1 was highly expressed in the NK cells of AML patients. These findings suggest that high expression of DNMT1 may be an important factor affecting the activity of CIML NK cells.
In recent years, the focus on distinct NK cell subsets exhibiting CIML characteristics has grown. Due to their enhanced effector functions and extended half-lives, CIML NK cells have emerged as valuable tools in immunotherapy for a variety of tumors [29]. CIML NK cells were initially identified in mice that were costimulated with IL-12, IL-15, and IL-18 [30], a finding that was subsequently validated in human NK cells [31]. Both preclinical and clinical investigations indicate that CIML NK cells offer benefits within the tumor microenvironment and may play a role in preventing the relapse of hematopoietic malignancies [2, 32]. Consequently, understanding the molecular mechanisms that enhance the activity of CIML NK cells is essential for AML treatment. In our study, upon NK cell differentiation, CIML NK cells exhibited increased activity and toxicity. Furthermore, silencing DNMT1 was found to augment CIML NK cell activity. Other reports also indicate that other signals can contribute to the induction of CIML NK cell activity, including the activation of the receptor CD16, which binds to cytokines to enhance the function of CIML NK cells [33]. Upon activation by the high-affinity anti-CD16a/CD30 bispecific protein AFM13, CD16 functionality and subsequent ITAM signaling were shown to increase CIML NK cell reactions [34]. Moreover, our findings revealed that AZA had similar effects to the knockdown of DNMT1, both of which increased the activity of CIML NK cells. Based on previous studies, we showed for the first time that the knockdown of DNMT1 could increase the activity of CIML NK cells, suggesting that it may be an important target for the treatment of AML.
Furthermore, abnormal demethylation and methylation of DNA can lead to irregular autophagy in states of prolonged inactivity or excessive activation [35]. Autophagy plays a crucial role in diverse physiological processes, including the immune response, pathological aging, cell reprogramming, tumor prevention, and elimination of foreign microorganisms [36, 37]. Recent studies suggest that autophagy is highly important for the differentiation and function of NK cells. Wang et al. [38] have shown that autophagy is vital for the formation of NK cells and promotes their survival by removing damaged mitochondria and intracellular ROS. O’Sullivan [39] also underscored the importance of autophagy in the creation and survival of CIML NK cells. Our study demonstrated that decreasing the levels of DNMT1 can increase autophagy in CIML NK cells, leading to increased NK cell activity. These results are consistent with those of prior studies suggesting that autophagy regulation impacts CIML NK cell activity. By investigating the relationships among DNMT1, autophagy, and CIML NK cell activity, we improved our comprehension of this connection. Notably, within the autophagy regulatory pathways, activated mTOR signaling inhibits autophagy, whereas AMPK activation can promote autophagy by suppressing mTOR signaling [40]. For instance, Shen et al. [41] documented that activating the AMPK/mTOR pathway can reduce kidney damage by inducing autophagy. Our findings indicate that inhibiting DNMT1 can induce autophagy by activating the AMPK/mTOR pathway, thereby enhancing CIML NK cell activity. Based on previous studies, we elucidated a new mechanism by which the DNMT1/AMPK/mTOR signaling axis regulates CIML NK cell activity through autophagy, and our study also identified a new target for the treatment of AML.
Nonetheless, this research has several limitations. First, intracellular signaling pathways are extremely complex and often cross-regulated. The involvement of DNMT1 may not be limited to AMPK/mTOR regulation. In the future, our research group will explore whether DNMT1 regulates other pathways and affects the activity of memory-like NK cells. Second, this study only examined the basal levels of IFN-γ, TNF-α and CD107a of NK cells, which is insufficient to fully understand their function in a dynamic environment. Further functional analysis with specific target cells is needed in the future to evaluate the cytolytic capacity of CIML NK cells. Furthermore, our study explored this mechanism only at the cellular level, and lacks animal and clinical studies. The utilization of DNMT1 in AML therapy requires extensive exploration in future research.
Conclusions
In conclusion, our findings suggest that downregulating DNMT1 promotes autophagy through activation of the AMPK/mTOR pathway, thereby enhancing CIML NK cell activity and alleviating AML progression. These findings suggest that inhibiting DNMT1 may be a potentially effective strategy for the treatment of AML.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Conceptualization, Yixun Li, Chong Guo and Kun Wu; methodology, Yixun Li and Fujia Zhang; software, Shenju Cheng and Yanhong Li; validation, Chong Guo and Yaling Zhao; formal analysis, Yixun Li, Fujia Zhang and Shan Luo; investigation, Chong Guo, Shenju Cheng, Yun Zeng and Kun Wu; resources, Yaling Zhao and Kun Wu; writing—original draft preparation, Yixun Li and Chong Guo; writing—review and editing, Yaling Zhao and Kun Wu; visualization, Shan Luo and Yun Zeng; supervision, Yixun Li, Yanhong Li and Kun Wu; and funding acquisition, Kun Wu. All the authors have read and agreed to the published version of the manuscript.
Funding
The present study was supported by the Basic Research Project of Yunnan Science and Technology Department (202201AY070001-058), the “Xingdian Talents” Project of Yunnan Province (RLMY20200020), and the Medical Discipline Leader Training Program of Yunnan Provincial Health Commission (D-2019003).
Data availability
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethical approval and consent to participate
All the authors confirm that all the procedures were conducted in accordance with ethical standards and that informed consent was obtained from all the participants. The study involving humans was approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University (Approval No. L-10) and was conducted in accordance with the Declaration of Helsinki.
Consent for publication
All the authors agreed with the publication of this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yixun Li and Chong Guo contributed equally to this work.
Contributor Information
Yaling Zhao, Email: imzhaoyaling@163.com.
Kun Wu, Email: wukun@ydyy.cn.
References
- 1.Wang Y, Tang X, Zhu Y, Yang XX, Liu B (2023) Role of interleukins in acute myeloid leukemia. Leuk Lymphoma 64:1400–1413. 10.1080/10428194.2023.2218508 [DOI] [PubMed] [Google Scholar]
- 2.Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, Leong JW, Abdel-Latif S, Schneider SE, Willey S, Neal CC, Yu L, Oh ST, Lee YS, Mulder A, Claas F, Cooper MA, Fehniger TA (2016) Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med 8:357ra123. 10.1126/scitranslmed.aaf2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz MA, Sierra J, Tallman MS, Löwenberg B, Bloomfield CD (2010) Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115:453–474. 10.1182/blood-2009-07-235358 [DOI] [PubMed] [Google Scholar]
- 4.Mrózek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, Metzeler KH, Schwind S, Wu YZ, Kohlschmidt J, Pettenati MJ, Heerema NA, Block AW, Patil SR, Baer MR, Kolitz JE, Moore JO, Carroll AJ, Stone RM, Larson RA, Bloomfield CD (2012) Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol 30:4515–4523. 10.1200/jco.2012.43.4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaffer BC, Hsu KC (2020) Selection of allogeneic hematopoietic cell transplant donors to optimize natural killer cell alloreactivity. Semin Hematol 57:167–174. 10.1053/j.seminhematol.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathioudaki A, Wang X, Sedloev D, Huth R, Kamal A, Hundemer M, Liu Y, Vasileiou S, Lulla P, Müller-Tidow C, Dreger P, Luft T, Sauer T, Schmitt M, Zaugg JB, Pabst C (2024) The remission status of AML patients after allo-HCT is associated with a distinct single-cell bone marrow T-cell signature. Blood 143:1269–1281. 10.1182/blood.2023021815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bald T, Krummel MF, Smyth MJ, Barry KC (2020) The NK cell-cancer cycle: advances and new challenges in NK cell-based immunotherapies. Nat Immunol 21:835–847. 10.1038/s41590-020-0728-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivori S, Pende D, Quatrini L, Pietra G, Della Chiesa M, Vacca P, Tumino N, Moretta F, Mingari MC, Locatelli F, Moretta L (2021) NK cells and ILCs in tumor immunotherapy. Mol Aspects Med 80:100870. 10.1016/j.mam.2020.100870 [DOI] [PubMed] [Google Scholar]
- 9.Dong H, Ham JD, Hu G, Xie G, Vergara J, Liang Y, Ali A, Tarannum M, Donner H, Baginska J, Abdulhamid Y, Dinh K, Soiffer RJ, Ritz J, Glimcher LH, Chen J, Romee R (2022) Memory-like NK cells armed with a neoepitope-specific CAR exhibit potent activity against NPM1 mutated acute myeloid leukemia. Proc Natl Acad Sci U S A 119:e2122379119. 10.1073/pnas.2122379119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terrén I, Orrantia A, Astarloa-Pando G, Amarilla-Irusta A, Zenarruzabeitia O, Borrego F (2022) Cytokine-Induced Memory-Like NK cells: from the basics to clinical applications. Front Immunol 13:884648. 10.3389/fimmu.2022.884648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun JC, Lanier LL (2018) Is there natural killer cell memory and can it be harnessed by vaccination? NK Cell memory and immunization strategies against Infectious diseases and Cancer. Cold Spring Harb Perspect Biol 10. 10.1101/cshperspect.a029538 [DOI] [PMC free article] [PubMed]
- 12.Ni J, Miller M, Stojanovic A, Garbi N, Cerwenka A (2012) Sustained effector function of IL-12/15/18-preactivated NK cells against established tumors. J Exp Med 209:2351–2365. 10.1084/jem.20120944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Fan Z, Meng Y, Liu S, Zhan H (2023) Blood-based DNA methylation signatures in cancer: a systematic review. Biochim Biophys Acta Mol Basis Dis 1869:166583. 10.1016/j.bbadis.2022.166583 [DOI] [PubMed] [Google Scholar]
- 14.Ren W, Gao L, Song J (2018) Structural basis of DNMT1 and DNMT3A-Mediated DNA methylation. Genes (Basel) 9. 10.3390/genes9120620 [DOI] [PMC free article] [PubMed]
- 15.Wong KK (2020) DNMT1 as a therapeutic target in pancreatic cancer: mechanisms and clinical implications. Cell Oncol (Dordr) 43:779–792. 10.1007/s13402-020-00526-4 [DOI] [PubMed] [Google Scholar]
- 16.Pappalardi MB, Keenan K, Cockerill M, Kellner WA, Stowell A, Sherk C, Wong K, Pathuri S, Briand J, Steidel M, Chapman P, Groy A, Wiseman AK, McHugh CF, Campobasso N, Graves AP, Fairweather E, Werner T, Raoof A, Butlin RJ, Rueda L, Horton JR, Fosbenner DT, Zhang C, Handler JL, Muliaditan M, Mebrahtu M, Jaworski JP, McNulty DE, Burt C, Eberl HC, Taylor AN, Ho T, Merrihew S, Foley SW, Rutkowska A, Li M, Romeril SP, Goldberg K, Zhang X, Kershaw CS, Bantscheff M, Jurewicz AJ, Minthorn E, Grandi P, Patel M, Benowitz AB, Mohammad HP, Gilmartin AG, Prinjha RK, Ogilvie D, Carpenter C, Heerding D, Baylin SB, Jones PA, Cheng X, King BW, Luengo JI, Jordan AM, Waddell I, Kruger RG, McCabe (2021) Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat Cancer 2 MT:1002–1017 [PMC free article] [PubMed]
- 17.Yan L, Geng Q, Cao Z, Liu B, Li L, Lu P, Lin L, Wei L, Tan Y, He X, Li L, Zhao N, Lu C (2023) Insights into DNMT1 and programmed cell death in diseases. Biomed Pharmacother 168:115753. 10.1016/j.biopha.2023.115753 [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto H, Matsui T (2024) Molecular mechanisms of Macroautophagy, Microautophagy, and chaperone-mediated autophagy. J Nippon Med Sch 91:2–9. 10.1272/jnms.JNMS.2024_91-102 [DOI] [PubMed] [Google Scholar]
- 19.Lu H, Yang HL, Zhou WJ, Lai ZZ, Qiu XM, Fu Q, Zhao JY, Wang J, Li DJ, Li MQ (2021) Rapamycin prevents spontaneous abortion by triggering decidual stromal cell autophagy-mediated NK cell residence. Autophagy 17:2511–2527. 10.1080/15548627.2020.1833515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Chen Y, Lin M, Huang B, Lin J (2021) Qingjie Fuzheng Granule inhibits EMT and induces Autophagy in Colorectal Cancer via mTOR Signaling pathways. Evid Based Complement Alternat Med 2021:9950499. 10.1155/2021/9950499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Zhu Y, Zhang W, Peng X, Zhou J, Li F, Han B, Liu X, Ou Y, Yu X (2018) Delphinidin induced protective autophagy via mTOR pathway suppression and AMPK pathway activation in HER-2 positive breast cancer cells. BMC Cancer 18:342. 10.1186/s12885-018-4231-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Y, Huang Y, Zhang Z, Chen A, Li L, Tian M, Shen J, Shao J (2023) iRGD-modified memory-like NK cells exhibit potent responses to hepatocellular carcinoma. J Transl Med 21:205. 10.1186/s12967-023-04024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Soto A, Bravo-San Pedro JM, Kroemer G, Galluzzi L, Gonzalez S (2017) Involvement of autophagy in NK cell development and function. Autophagy 13:633–636. 10.1080/15548627.2016.1274486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakhtiyaridovvombaygi M, Yazdanparast S, Mikanik F, Izadpanah A, Parkhideh S, Shahbaz Ghasabeh A, Roshandel E, Hajifathali A, Gharehbaghian A (2023) Cytokine-Induced Memory-Like NK cells: emerging strategy for AML immunotherapy. Biomed Pharmacother 168:115718. 10.1016/j.biopha.2023.115718 [DOI] [PubMed] [Google Scholar]
- 25.Cardoso C, Pestana D, Gokuladhas S, Marreiros AD, O’Sullivan JM, Binnie A, M TF and, Castelo-Branco P (2024) Identification of novel DNA methylation prognostic biomarkers for AML with Normal Cytogenetics. JCO Clin Cancer Inf 8:e2300265. 10.1200/cci.23.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Wang R, Hu X, Gao Y, Wang Z, Li J, Wong J (2019) Activated MEK/ERK Pathway drives widespread and coordinated overexpression of UHRF1 and DNMT1 in Cancer cells. Sci Rep 9:907. 10.1038/s41598-018-37258-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Zhang D (2022) DNA methyltransferase-1 in acute myeloid leukaemia: beyond the maintenance of DNA methylation. Ann Med 54:2011–2023. 10.1080/07853890.2022.2099578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H (2001) Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 97:1172–1179. 10.1182/blood.v97.5.1172 [DOI] [PubMed] [Google Scholar]
- 29.Carreira-Santos S, López-Sejas N, González-Sánchez M, Sánchez-Hernández E, Pera A, Hassouneh F, Durán E, Solana R, Casado JG, Tarazona R (2023) Enhanced expression of natural cytotoxicity receptors on cytokine-induced memory-like natural killer cells correlates with effector function. Front Immunol 14:1256404. 10.3389/fimmu.2023.1256404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM (2009) Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci U S A 106:1915–1919. 10.1073/pnas.0813192106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terrén I, Orrantia A, Mosteiro A, Vitallé J, Zenarruzabeitia O, Borrego F (2021) Metabolic changes of Interleukin-12/15/18-stimulated human NK cells. Sci Rep 11:6472. 10.1038/s41598-021-85960-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gang M, Marin ND, Wong P, Neal CC, Marsala L, Foster M, Schappe T, Meng W, Tran J, Schaettler M, Davila M, Gao F, Cashen AF, Bartlett NL, Mehta-Shah N, Kahl BS, Kim MY, Cooper ML, DiPersio JF, Berrien-Elliott MM, Fehniger TA (2020) CAR-modified memory-like NK cells exhibit potent responses to NK-resistant lymphomas. Blood 136:2308–2318. 10.1182/blood.2020006619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker-Hapak MK, Shrestha N, McClain E, Dee MJ, Chaturvedi P, Leclerc GM, Marsala LI, Foster M, Schappe T, Tran J, Desai S, Neal CC, Pence P, Wong P, Wagner JA, Russler-Germain DA, Zhu X, Spanoudis CM, Gallo VL, Echeverri CA, Ramirez LL, You L, Egan JO, Rhode PR, Jiao JA, Muniz GJ, Jeng EK, Prendes CA, Sullivan RP, Berrien-Elliott MM, Wong HC, Fehniger TA (2021) A Fusion protein complex that combines IL-12, IL-15, and IL-18 signaling to Induce Memory-Like NK cells for Cancer Immunotherapy. Cancer Immunol Res 9:1071–1087. 10.1158/2326-6066.Cir-20-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pahl JHW, Koch J, Götz JJ, Arnold A, Reusch U, Gantke T, Rajkovic E, Treder M, Cerwenka A (2018) CD16A activation of NK cells promotes NK Cell Proliferation and Memory-Like cytotoxicity against Cancer cells. Cancer Immunol Res 6:517–527. 10.1158/2326-6066.Cir-17-0550 [DOI] [PubMed] [Google Scholar]
- 35.Li R, Wei X, Jiang DS (2019) Protein methylation functions as the posttranslational modification switch to regulate autophagy. Cell Mol Life Sci 76:3711–3722. 10.1007/s00018-019-03161-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortega MA, Fraile-Martinez O, de Leon-Oliva D, Boaru DL, Lopez-Gonzalez L, García-Montero C, Alvarez-Mon MA, Guijarro LG, Torres-Carranza D, Saez MA, Diaz-Pedrero R, Albillos A, Alvarez-Mon M (2024) Autophagy in its (proper) context: molecular basis, Biological relevance, pharmacological modulation, and Lifestyle Medicine. Int J Biol Sci 20:2532–2554. 10.7150/ijbs.95122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Li X, Zhang Q, Fu C, Jiang W, Xue J, Liu S, Meng Q, Ai L, Zhi X, Deng S, Liang W (2024) Autophagy in Disease Onset and Progression. Aging Dis 15:1646–1671. 10.14336/ad.2023.0815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, Liu Y, Wang Y, Li H, Zhang RX, He MS, Chen L, Wu NN, Liao Y, Deng ZL (2016) Adenovirus-mediated siRNA targeting CXCR2 attenuates titanium particle-induced osteolysis by suppressing osteoclast formation. Med Sci Monit 22:727–735. 10.12659/msm.897243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Sullivan TE, Johnson LR, Kang HH, Sun JC (2015) BNIP3- and BNIP3L-Mediated Mitophagy promotes the generation of natural killer cell memory. Immunity 43:331–342. 10.1016/j.immuni.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H, Ouyang Y, Yin H, Cui H, Deng H, Liu H, Jian Z, Fang J, Zuo Z, Wang X, Zhao L, Zhu Y, Geng Y, Ouyang P (2022) Induction of autophagy via the ROS-dependent AMPK-mTOR pathway protects copper-induced spermatogenesis disorder. Redox Biol 49:102227. 10.1016/j.redox.2021.102227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen SH, Wang RL, Yuan Q, Jian LY, Guo HH, Li HS, Liu XP, Huang RF (2023) The roles of AMPK/mTOR autophagy pathway in the acute kidney injury-induced acute lung injury. Chin J Physiol 66:73–84. 10.4103/cjop.CJOP-D-22-00122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.