Abstract
Voltage-gated sodium channels (VGSCs) are transmembrane protein complexes that are vital to the generation and propagation of action potentials in nerve and muscle fibers. The canonical VGSC is generally conceived as a heterotrimeric complex formed by 2 classes of membrane-spanning subunit: an α-subunit (pore forming) and 2 β-subunits (non–pore forming). NaV1.5 is the main sodium channel α-subunit of mammalian ventricle, with lower amounts of other α-subunits, including NaV1.6, being present. There are 4 β-subunits (β1–β4) encoded by 4 genes (SCN1B–SCN4B), each of which is expressed in cardiac tissues. Recent studies suggest that in addition to assignments in channel gating and trafficking, products of Scn1b may have novel roles in conduction of action potential in the heart and intracellular signaling. This includes evidence that the β-subunit extracellular amino-terminal domain facilitates adhesive interactions in intercalated discs and that its carboxyl-terminal region is a substrate for a regulated intramembrane proteolysis (RIP) signaling pathway, with a carboxyl-terminal peptide generated by β1 RIP trafficked to the nucleus and altering transcription of various genes, including NaV1.5. In addition to β1, the Scn1b gene encodes for an alternative splice variant, β1B, which contains an identical extracellular adhesion domain to β1 but has a unique carboxyl-terminus. Although β1B is generally understood to be a secreted variant, evidence indicates that when co-expressed with NaV1.5, it is maintained at the cell membrane, suggesting potential unique roles for this understudied protein. In this review, we focus on what is known of the 2 β-subunit variants encoded by Scn1b in heart, with particular focus on recent findings and the questions raised by this new information. We also explore data that indicate β1 and β1B may be attractive targets for novel antiarrhythmic therapeutics.
Keywords: SCN1B (β1/β1B), Voltage-gated sodium channel, Arrhythmia, Brugada syndrome, Ephaptic coupling, Perinexus
Introduction
Voltage-gated sodium channels (VGSCs) are transmembrane protein complexes that are vital to the propagation of action potential and cell-to-cell communication in nerves and muscle.1,2 As the name suggests, VGSCs form pores in the cell membrane and respond to changes in membrane voltage, triggering the channel to open or close to selectively regulate the passage of sodium ions. The present consensus is that the canonical VGSC is heterotrimeric, being formed by 2 classes of membrane-spanning subunits: an α-subunit (pore forming) and one or more β-subunits (non–pore forming).3,4 Overall membrane topology for VGSCs is shown in Figure 1A. Both the α- and β-subunits contain multiple isoforms that are expressed differentially in skeletal, cardiac, and nervous system tissues, including 9 α-subunit isoforms (NaV1.1–NaV1.9) and 4 β-subunit isoforms (β1–β4), each encoded by distinct genes.5–7 The β1–β4 subunits share a consistent modular organization, being composed of an extracellular V-type immunoglobulin (Ig) domain, a transmembrane alpha-helical domain, and a relatively disordered intracellular carboxyl (C)-terminal domain (Figure 1B).4,8–10 An alternately spliced isoform of the Scn1b gene encoding the β1 protein, β1B, incorporates the same amino (N)-terminal extracellular regions as β1, followed by a unique C-terminus (Figure 1C).11–13
Figure 1.
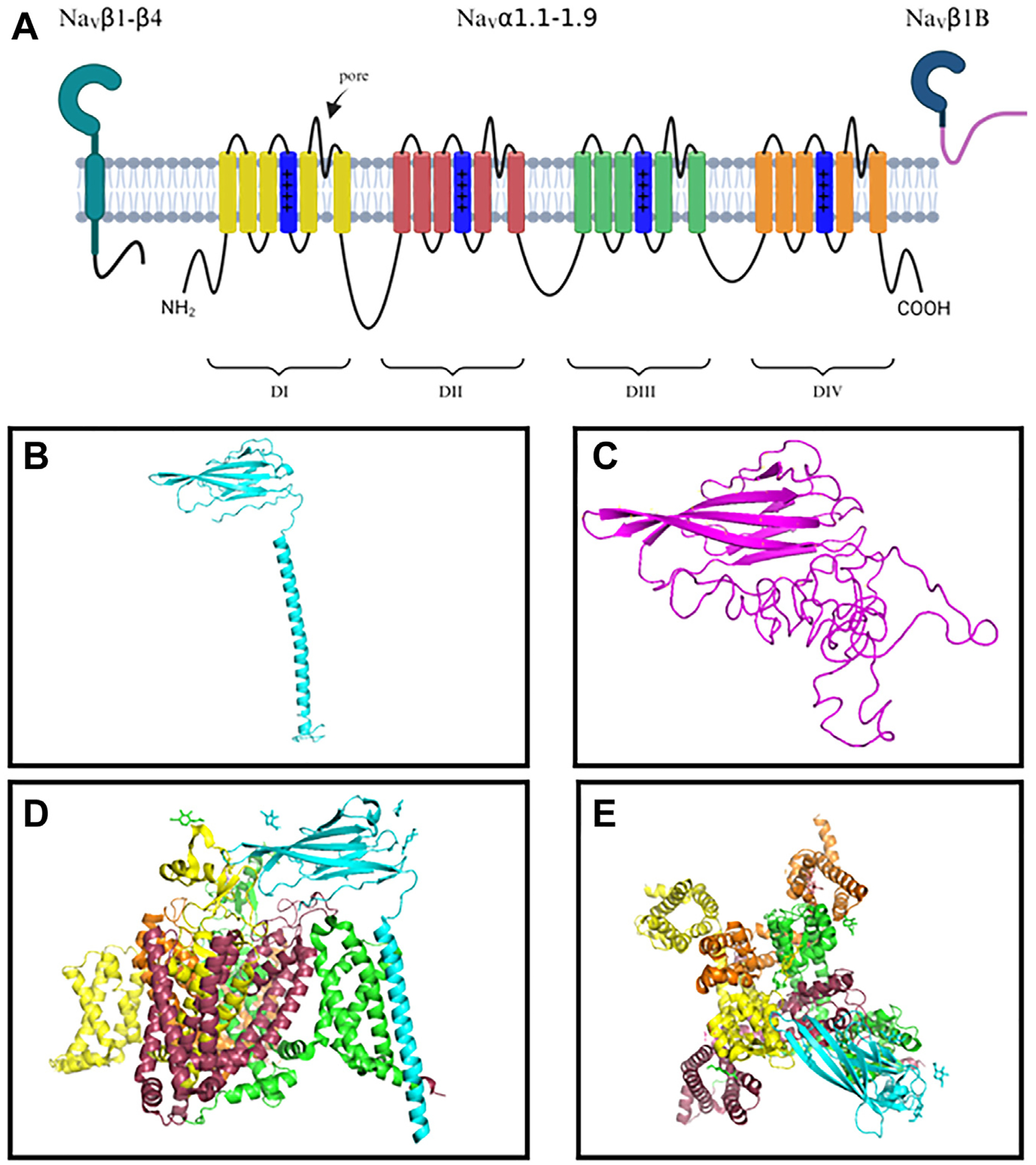
Voltage-gated sodium channel (VGSC) subunit structure. A: Membrane topology of VGSC α- and β-subunits. The α-subunit is composed of 4 homologous domains (DI–DIV), each composed of 6 transmembrane regions. B: VGSC β1-subunit predicted folding. β1 comprises an extracellular Ig domain, an alpha-helical transmembrane region, and a small intracellular domain. C: SCN1B splice-variant β1B predicted folding. β1B contains an identical immunoglobulin domain to that of β1 but differs in the C-terminal domain. D: Resolved structure for β1 (blue) in complex with NaV1.7. β1 interacts with domain 3 of NaV1.7 (green) as viewed from a cross-section of the membrane.26 Glycosylation sites for β1 and NaV1.7 are indicated by attached sugars, in the same color as the domain they are associated with. E: Structure for β1 (blue) in complex with NaV1.7. β1 interacts with domain 3 of NaV1.7 (green) as viewed down the α-subunit pore. Models were created using The PyMOL Molecular Graphics System, Version 1.2r3pre (Schrödinger, LLC), and predicted folding was performed using I-TASSER.27,28
The α-subunit polypeptide forms a functional VGSC channel via 4 loop-connected transmembrane domains, which confer voltage-sensing, ion selectivity, and gate inactivation.14–18 Type 1 transmembrane β-subunits associate with the α-subunit covalently or noncovalently, depending on isoform.19 Noncovalent interaction of β1 with NaV1.7 is shown in Figures 1D and 1E. In 1980, Scn1b/β1 became the first VGSC β-subunit to be reported.20–25 Initially described as a modulator of the α-subunit, β-subunits are now known to have a wide variety of functions, including trafficking of α-subunits to the membrane,29,30 modulating channel kinetics and gating,10,31 and participating in adhesion interactions.32–39
As has been characterized for other membrane proteins such as Notch,40 β-subunits can also modulate transcription, undergoing a process of regulated intramembrane proteolysis (RIP) generating an intracellular cleavage product (ICD) that translocates to the nucleus.41–43 The β1B isoform (also known as β1A in rat) was originally identified by Kazen-Gillespie et al11 in the embryonic rat brain and adult adrenal glands and heart, and was reported to increase VGSC density and function. This study also provided evidence for participation of β1B in cell adhesion. Importantly, because of its unique C-terminus, β1B does not undergo RIP like β1 and thus is incapable of modulating gene expression in the same manner.
Although β1 and its counterparts β1B and β1-ICD have been described in excitable tissues such as the brain and skeletal muscle and in nonexcitable tissue,3,31 recent evidence has highlighted the potential for unique assignments in the heart, such as potentially maintaining conditions required for ephaptic coupling and altering transcription of VGSC subunits. Recent reviews address the role of cardiac β1 in the contexts of α-subunit modification10 and adhesion.4,44 In the current review, we consider data that point to new roles for β1, β1B, and β1-ICD in the heart and novel therapeutic possibilities that may emerge from such insights.
Cardiac β1 and β1B: Spatial and temporal expression
VGSC β-subunits are expressed differentially in atrial and ventricular working myocardial tissues.9 β-subunit studies have focused on mammalian isoforms, although there is literature on expression in birds, reptiles, amphibians, and fish.45 Cardiac VGSCs are composed of one or more β-subunits that interact with 1 α-subunit, NaV1.5. In ventricular myocardium, NaV1.5 is principally localized in the intercalated disc, with lesser amounts in lateral sarcolemma, where it associates with the dystrophin complex.46–48 This is in contrast to other α-subunit isoforms in cardiac muscle, including tetrodotoxin-sensitive neuronal sodium channels NaV1.1, NaV1.3, and NaV1.6, which have been mainly reported to be in transverse (t)-tubules.49–51 Although there is differential spatial and temporal expression of β isoforms in heart, data suggest that Scn1b gene products are more highly expressed in atria compared to ventricles, with preferential localization in intercalated discs (Figure 2) and possibly in the t-tubule system.8,38,49,52,53 In contrast to neuronal β1 and neuronal/cardiac β3 expression, which diminishes throughout embryonic development, cardiac Scn1b/β1 expression increases during development and does not decrease in adulthood.11,54–56
Figure 2.
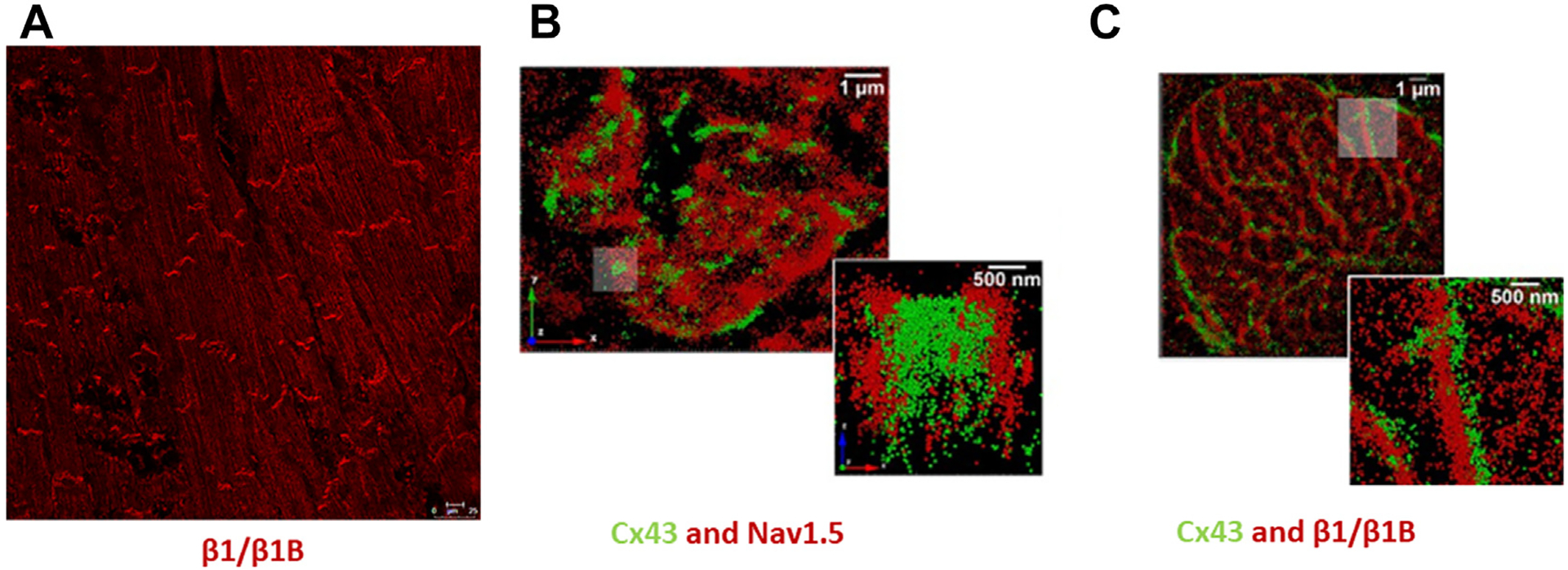
Cardiac voltage-gated sodium channel (VGSC) expression. A: β1/β1B labeling showing intercalated disc localization in adult rat ventricle using an N-terminal antibody described previously.38 B, C: Stochastic optical reconstruction microscopic images demonstrating localization of NaV1.5 and β1/β1B with connexin43 in en face intercalated discs from adult guinea pig ventricle. (Reproduced and modified under the CC BY 4.0 license.38) Scale bar in A = 10 μm; in B and C = 1 μm; in inset = 500 nm.
Similar to β1, β1B is expressed throughout heart development and adulthood11 and is thought to be present in a similar range of cell types. However, alternatively spliced soluble proteins often are differentially expressed compared to full-length counterparts.57 Thus, although widely expressed in heart, β1 and β1B likely are present at varying levels in different cardiac tissues, including the specialized conduction system, which displays unique gap junctional (GJ) proteins58–60 and altered conduction properties compared to atrial and ventricular tissues.61 It also is possible that β1 and β1B manifest differing subcellular localizations. Further elucidating patterns of Scn1b gene product distribution and function likely will advance understanding of the multifaceted roles that this gene is likely to play in the heart.
β1, β1B, and β1-ICD: Synthesis, biogenesis, and function
The β1–β4 subunits are encoded by 4 genes, SCN1B, SCN2B, SCN3B, and SCN4B, encoding full-length proteins that include 3 primary functional domains: a large extracellular N-terminal domain with a single Ig loop, a single transmembrane alpha-helical region, and a disordered intracellular C-terminal domain.4,9,10,22,62 SCN1B pre-mRNA encodes the β1 protein and is generated from 6 exons (human-NM_001037.5; rat-NM_001271045.2 and NM_017288.3). Alternative splicing of SCN1B pre-mRNA yields a truncated variant that encodes β1B, the “so-called” soluble counterpart of β1, due to intron retention following exon 3, just before sequence encoding the transmembrane domain of β1 (human-NM_199037; rat-AF182949.1) (Figure 3). Consequently, β1B lacks the intracellular and transmembrane domains but retains a conserved extracellular region including the Ig domain, which is fully homologous with β1, although followed by a C-terminal sequence that shows variability between mammalian species.11 The splicing out of the β1 transmembrane domain is thought to result in nonretention of β1B in membranes and secretion of the protein under most circumstances.63 Details of the mechanisms that regulate SCN1B pre-mRNA splicing remain largely unexplored. In addition, the β1 protein can be modified post-translationally by a mechanism that include phosphorylation, S-palmitoylation, RIP, and glycosylation.64 Sialylation of β1 modifies gating properties of multiple NaV α-subunits, including NaV1.5.65
Figure 3.
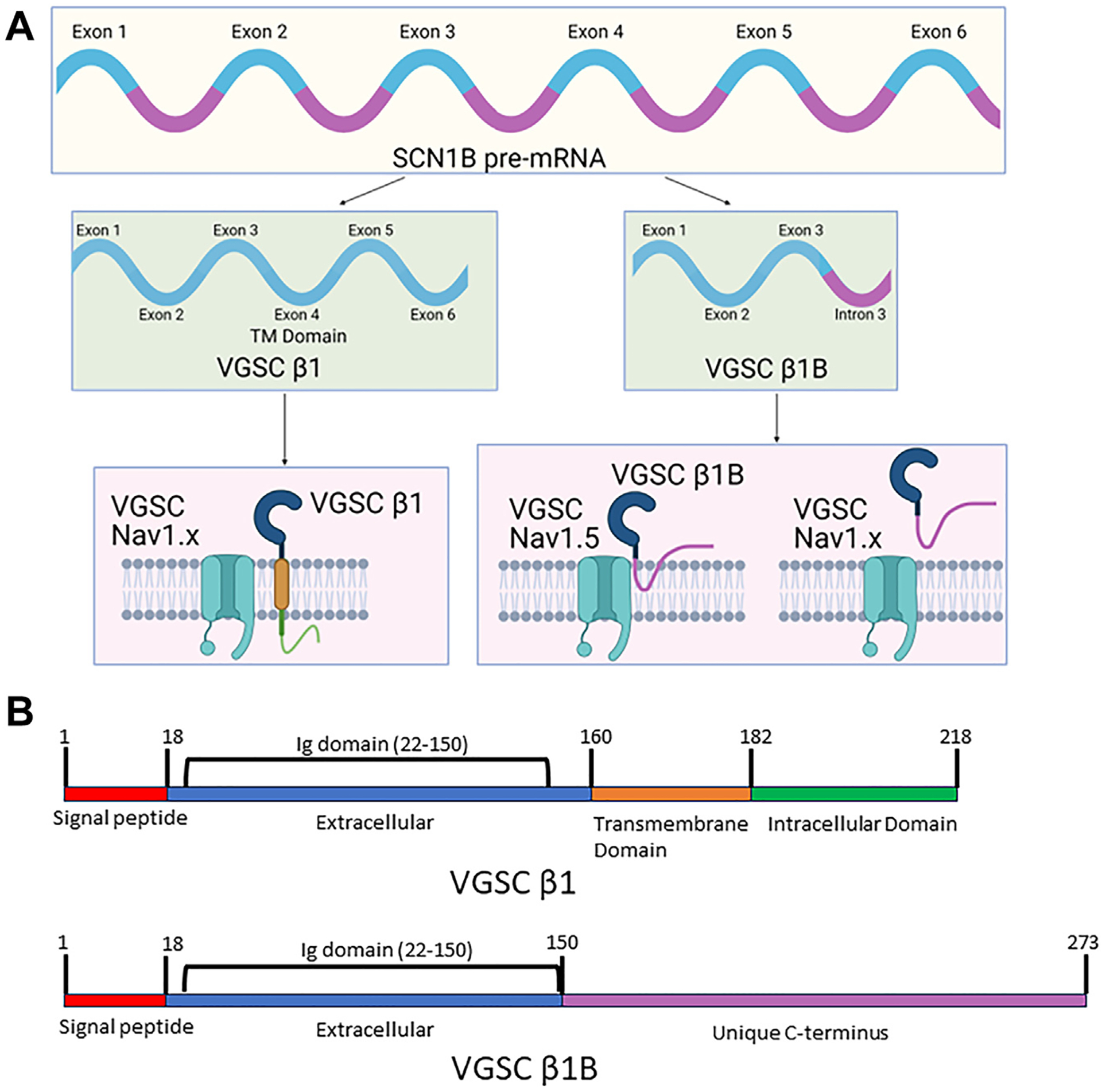
Scn1b gene products β1 and β1B. A: Schematic illustrating formation of splice variants β1 and β1B from the Scn1b gene, resulting in canonical β1, which has similar tertiary structure to other β-subunits β2–β4, and the soluble β1B variant formed by intron 3 retention. β1 is transmembrane regardless of associated α-subunit. While β1B is thought secreted, it is retained at the membrane via an unknown mechanism when coexpressed with NaV1.5. B: Schematic showing the different domains of the β1 and β1B proteins. They contain identical immunoglobulin domains from residues 22–150. After residue 150 the C-terminuses of the 2 variants differ. VGSC = voltage-gated sodium channel.
The distinctions between β1 isoforms give rise to domain-related function, including association with extracellular proteins and NaV1.5. The β1 extracellular and transmembrane domains interact with α-subunits noncovalently, in contrast to covalent bonding by β2 and β4.66–71 Crystal structures of β1 with NaV1.4 and NaV1.7 reveal ionic and hydrogen bonding between the Ig domain and the extracellular loops of α-subunits.4,26,72 However, interaction of NaV1.5 with β-subunits, including β1, likely is unique because of structural differences within NaV1.5. For example, NaV1.5 does not present accessible cysteine residues for disulfide bonds with β2 and β4 Ig domains,26,73 thus shifting their characteristic covalent bonding to noncovalent. In addition, a unique N-linked glycosylation site (Asn319) on NaV1.5 may block noncovalent interactions with the β1 Ig domain,4,26 which then will be less constrained and free to interact with other extracellular proteins. Crystal structures of β1 with NaV1.5 have been elusive, likely because of the relatively loose interaction.74–76 However, coexpression of NaV1.5 and β1B in HEK cells leads to β1B membrane retention and increases in sodium current,63 indicating potential for a unique function for β1B in the cardiac context where high levels of NaV1.5 occur. It has been proposed that β1B may regulate β1 function, particularly in trans interactions between β1 molecules in GJ-adjacent perinexal nanodomains located within intercalated discs.4
The C-terminus of β1 does not seem to be required for channel function, although it likely is important for channel assembly.68 The sequential proteolysis of the C-terminal domain that constitutes β1 RIP was first reported in 2005 and is a process that gives rise to soluble extracellular and intracellular fragments.43 During RIP, the extracellular domain of β1 is proteolysed by the β-site amyloid precursor protein cleaving enzyme-1, leaving behind a carboxy-terminal fragment composed of transmembrane domains and ICDs.42,43,64 The second cleavage is performed by γ-secretase, which releases the β1 ICD from the transmembrane domain (Figure 4), then translocating to the nucleus where it participates in transcriptional regulation of numerous genes, including many encoding ion channels.41–43 Genes affected by the β1-ICD include those related to cell adhesion, proliferation, calcium binding, immune function, and NaV α-subunit expression. Chen et al77 recently demonstrated that the Scn1b variant R89C, associated with Dravet syndrome, undergoes normal RIP but results in increased expression of Scn2a, Scn3a, Scn5a, and Scn1b. The RIP process is further regulated by β1 S-palmitoylation, which localizes β1 to the plasma membrane, where RIP likely takes place,78 although it also has been reported that endosomal β1 is cleaved by γ-secretase.79
Figure 4.
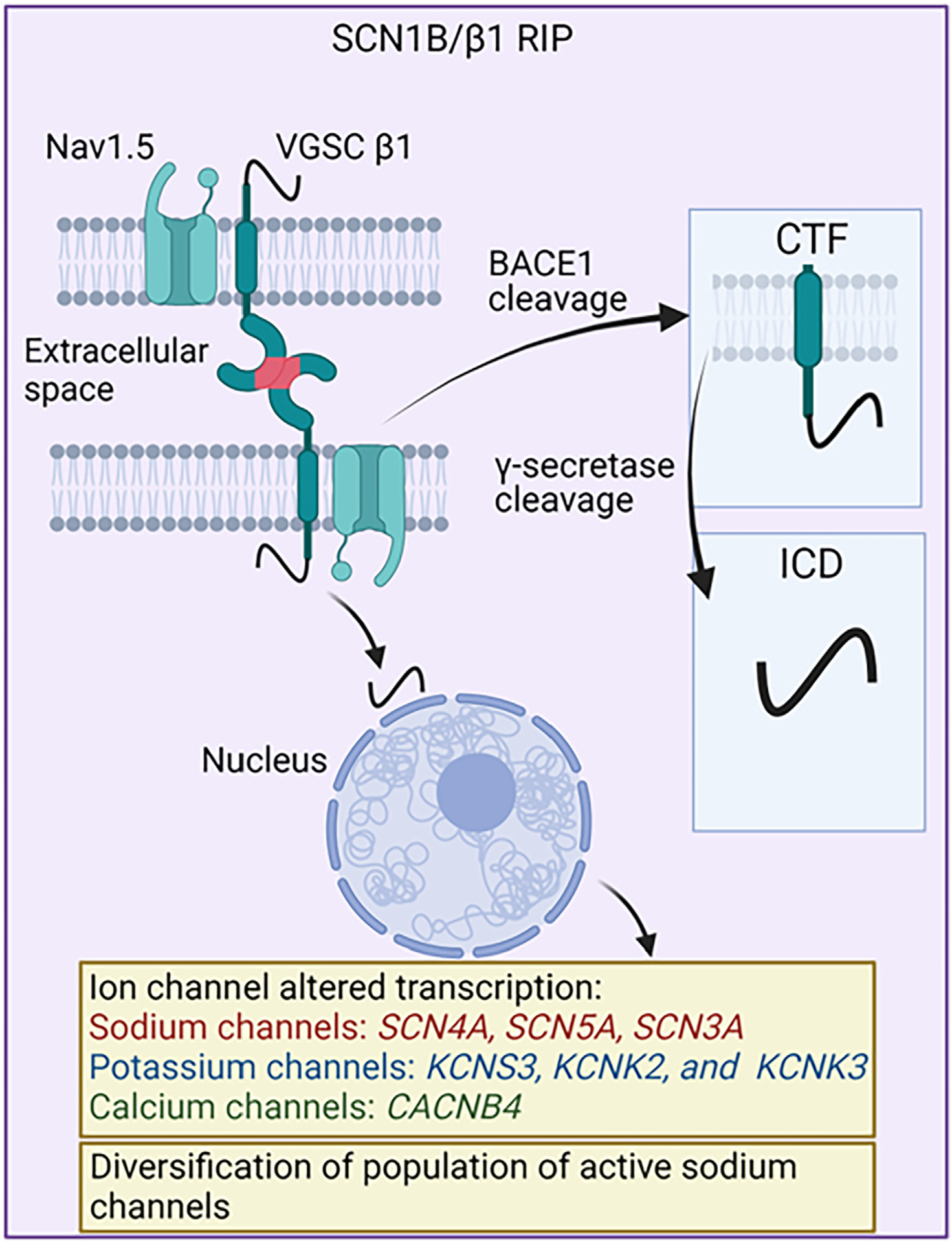
β1-regulated intramembrane proteolysis. Two-step cleavage of β1 by β-site amyloid precursor protein cleaving enzyme-1 (BACE1) and γ-secretase is shown. The resulting product of the cleavage, the intracellular domain, translocates to the nucleus and alters transcription of various ion channels and diversifies the population of active sodium channels. Cx43 = connexin43.
Recent studies by Bouza et al provide important new information on β1 RIP, including characterization of genes modulated by β1-ICD when human SCN1B is heterologously expressed in Chinese hamster lung (CHL) cells.42 In CHL cells, β1-ICD alters transcription of sodium channels (SCN4A, SCN5A, SCN3A), potassium channels (KCNS3, KCNK2, and KCNK3), and calcium channels (CACNB4).42 Furthermore, the population of active sodium channels may be significantly altered by β1-ICD, as demonstrated by (1) overexpression of the β1-ICD in MDA-MB-231 cells, a breast cancer cell line, which resulted in greater sodium current that was less tetrodotoxin-resistant79; and (2) by transient transfection of HEK-hNaV1.5 cells with β1-ICD-V5–2A-eGFP that showed no change in sodium current density.42 Together, these results indicate that β1-ICD transcriptional regulation depends on sodium channel populations and cell type. This also suggests that targeting β-subunit adhesion domains may have downstream effects on transcription through RIP, with implications for modulating sodium currents and cellular electrical interactions. Further supporting this conclusion is the finding that genes identified as downregulated by β1-ICD overexpression in heterologous cells were upregulated in Scn1b-null heart, which lack β1-ICD signaling.42 This being said, it should be noted that β1 RIP has yet to be directly shown to occur in myocardial tissues, providing an important open question for the field.
Evidence for β1 in ephaptic coupling
β-subunits possess an extracellular Ig domain and are members of the cell adhesion molecule (CAM) superfamily.41 Via this ectodomain, β-subunits mediate interactions with other CAMs and extracellular matrix proteins, such as contactin, N-cadherin, NCAM, neurofascin-155, neurofascin-186, VGSC β2, and tenascin-R.9,80 In addition to transheterophilic interactions, β1/β1b participates in transhomophilic cell adhesion,33,81 which may play a unique role within a specialized nanodomain of the intercalated disc, termed the perinexus.82,83
The intercalated disc is a zone of electromechanical interaction between cardiomyocytes responsible for maintaining conduction and coordinated muscle contraction.84–87 Within the intercalated disc and adjacent to GJs, the perinexus comprises a narrow (<30 nm in width), pocketlike cleft of extracellular space.88,89 Numerous GJs are located within intercalated discs, with large GJs ringing disc edges in many species, including humans.90 Consequently, there are large numbers of perinexuses found between GJ-adjoined cells.38,44,91,92 Scn1b/β1 is enriched at intercalated discs (Figure 2), with antibodies to the β1 N-terminus, as well as NaV1.5, showing particular associations with GJ perinexuses.38,93,94 It is in this perinexal region that ephaptic coupling of cardiomyocytes has been proposed to take place.88,95,96
Ephaptic coupling is a mechanism of connecting neighboring cells that allows for the intercellular propagation of electrical signals (eg, action potentials). In the ventricle, ephaptic coupling has been hypothesized to occur in parallel with GJ-based coupling and has been proposed to be mediated by transients in sodium ion (Na+) concentrations within the perinexus.46,88,97–99 The theory of cardiac ephaptic coupling has been gaining in prominence over the past 2 decades and is supported by a growing number of modeling and experimental studies.38,46,91,100–121 With its focus on VGSC β-subunits, it is beyond the scope of this review to describe this work in depth. Readers are directed in particular to 2 recent papers from the groups of Seth Weinberg122 and Jan Kucera,97 who each have created sophisticated mathematical models that uncover unexpected subtleties in ephaptic mechanisms.
For ephaptic coupling between cardiomyocytes to function, cell-to-cell alignment of VGSC clusters across the perinexus is thought to be required,46 in addition to intermembrane distances <30 nm.91 Recent evidence suggests that the β1-subunit, or perhaps membrane-retained β1B,63 facilitates both these distance and alignment parameters through its Ig domain.33,38,123 Moreover, these interactions are disrupted by the β1/β1B Ig domain mimetic peptide, βadp1, resulting in loss of GJ-associated VGSCs in cultured neonatal cardiomyocytes and a widened perinexus and increased incidence of arrhythmia in a Langendorff-perfused guinea pig model.38 β1 physically interacts with the intercalated disc–bound transmembrane protein-65 (Tmem65) at the perinexus. Disruption of this interaction with Tmem65 shRNA results in aberrant perinexuses and disrupted localization of NaV1.5 and connexin43 at the intercalated disc.124 Building on these findings, we modeled β1-subunit extension from the plasma membrane to between 5 and 10 nm,44 which is consistent with a total perinexus width between 2 apposed and transadherent subunits of 10–20 nm. This is consistent with the modeling predictions of Mori et al,91 who posited that an intermembrane spacing ≤30 nm was required for operation of an ephaptic nanodomain/ephapse in the heart.
Schematics often show a single NaV1.5 with a single β1-subunit on one cardiomyocyte interacting with a similar configured complex on the opposing cardiomyocyte. However, because of the location of the purported transhomophilic β1 binding site (aa 66–86), it remains unclear how this arrangement may account for a requirement for cell-to-cell alignment of channel pores across the perinexus (Figure 5), as has been proposed.46 This being said, VGSCs do not seem to exist as single pores; rather, they assemble and gate as dimers.125 VGSCs have also been shown to form spontaneous supramolecular clusters in heterologous expression systems, facilitated by β3 modification of NaV1.5 that may alter single α-subunit alignment.126 The β3-subunit is closely homologous to β1 and assembles as a trimer via the Ig domains.127 It is possible that β1 may also trimerize at the plasma membrane, as the structural components required for trimeric assembly, including a Cys2-24 disulfide bond and a membrane-buried glutamic acid, are conserved between β1 and β3.4,39 This offers a hypothetical paradigm for framing β1 transhomophilic adhesion from a single Ig domain interaction across the perinexus to a multimeric interface, in which 3 NaV1.5 α-subunits are clustered together to align the channel pores. Although we pose possibilities for addressing pore alignment here, it seems likely that over time other hypotheses for the pore alignment problem will emerge.
Figure 5.
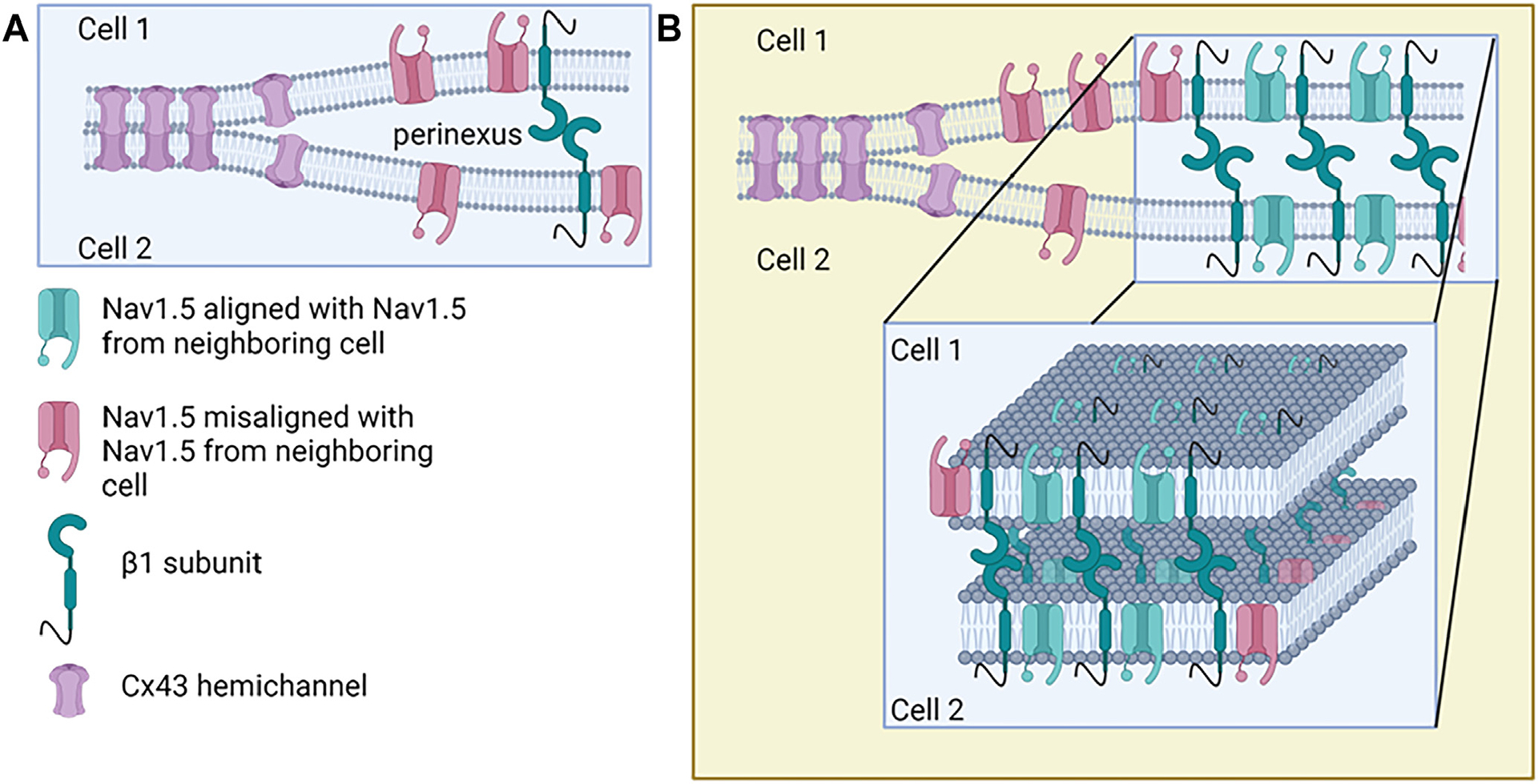
Sodium channel alignment by β1 transadhesion in the perinexus. A: Standard representation of β1-subunits interacting in transadhesion, while associated with NaV1.5 at the perinexus. Although the model only indicates a few key players, multiple other proteins are involved in maintaining perinexal width and nanostructure. The red sodium channels represent misalignment of pores across the perinexus, indicating a fundamental issue that needs to be addressed by the field. B: Extension of A indicating β1 association with NaV1.5 allows for alignment of pores across the perinexus in the center of the pool of voltage-gated sodium channels (VGSCs) found at the perinexus but still leaves orphaned sodium channels at the rim of the cluster. BACE1 = β-site amyloid precursor protein cleaving enzyme-1; CTF = carboxy-terminal fragment; ICD = intracellular domain.
In addition to a potential direct role of β1 Ig domains in coupling, we recently reported findings suggesting that modulation of β-subunit adhesion may also have indirect impacts on electrical interactions between myocytes via β1 RIP.128 We found that treating β1-CHL–expressing cells, with the β1 mimetic peptide βadp1, results in increased RIP and increased levels of β1 immunolabeling after treatment for 48 hours.128 These results suggest a connection between β1-subunit adhesion and β1 RIP. They further indicate the prospect that RIP may modulate the ephaptic mechanism via its effects on β1 levels. However, in fairness it should be noted that although indirect evidence for cardiac ephaptic conduction has been growing in the last decade, direct evidence for the phenomenon has yet to be provided. As with the case β1 RIP, important questions on cardiac ephaptic conduction, as well as the role of β1 in this putative mechanism, remain open.
β1- and β1B-related cardiac pathologies
β-subunit pathology has been studied extensively in the cardiac context, and because of the important functions and new insights detailed earlier, β-subunits are emerging as prospects for therapeutic targets. β1/β1B variants are associated with atrial fibrillation,129,130 long QT syndrome,7 and Brugada syndrome.131–134 Recently, Angsutararux et al76 investigated variants in β1 linked to either atrial fibrillation or Brugada syndrome to learn more about the mechanisms underlying arrhythmia. Of the variants studied, the majority were in the extracellular domains, including R85H, E87Q, and D153N in β1. Some important known mutations in β1 associated with arrhythmia or Brugada syndrome are shown in Figure 6. Interestingly, none of these mutations are associated with α-subunit interaction, but rather integrity of the Ig domain or, in the case of D153N, located in the linker region between the Ig domain and the transmembrane domain. The β1 mutations were associated with a variety of impacts on sodium channel function and expression, including altering levels of cell surface NaV regulation of NaV channel activation and inactivation gating, and direct effects on VSD-III activation, which has been shown to affect the response of the channel to antiarrhythmic drugs.8,76 This study reiterates the importance of the extracellular domain of the β1-subunit, especially the putative adhesion domain shared by β1 and β1B (ie, amino acids 66–86). Interestingly, a region in β1B has been shown by multiple groups to be associated with either Brugada syndrome130,135 or long QT syndrome7 within a 4-amino-acid range (residues 210–214), which could represent a future target for drug development. Selected mutations reported in β1 and β1B associated with cardiac pathologies are indicated in Figure 6.
Figure 6.
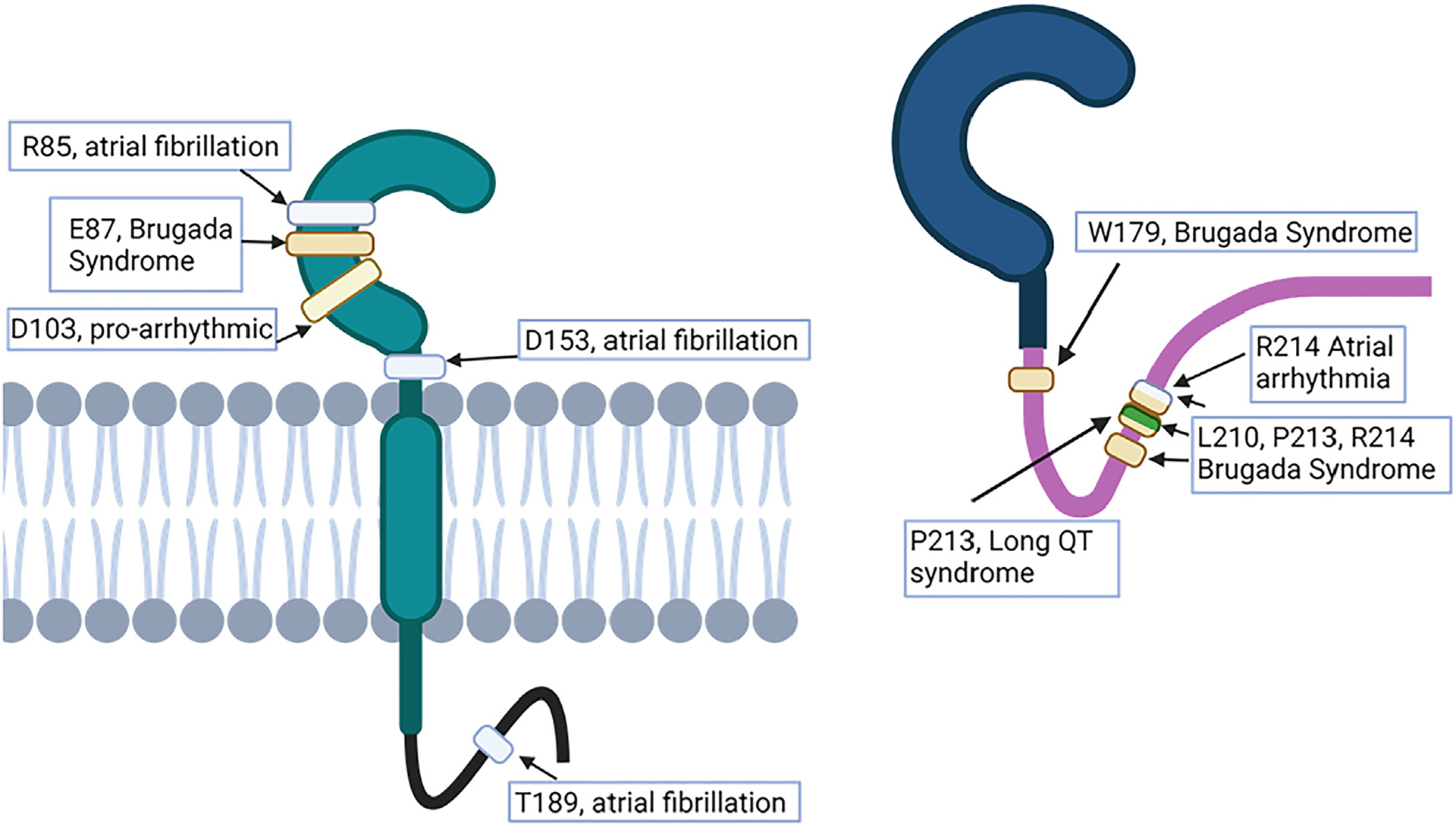
Residues associated with natural mutations in β1/β1B that result in conduction pathologies. Indication of location and pathology associated with selected mutations mentioned in the text resulting in conduction abnormalities. Mutations in the immunoglobulin domain are shown only for β1. Of particular interest in β1 are the mutations around the putative transadhesion binding region (residues 66–86) that are associated with atrial fibrillation and Brugada syndrome. A region of interest unique to β1B is residues 210–214, which has been shown by multiple groups to contain naturally occurring mutations that result in conduction pathologies.
Strong evidence supports that subunits encoded by SCN1B are vital to normal cardiac electrical function, and that their disruption may result in conduction abnormalities. For example, mice from a Scn1b null mouse line, which lacks both β1 and β1B, generally do not survive past 3 weeks post-natal.136 Their isolated null myocytes show slowed repolarization, and the mice exhibit prolonged RR (ventricular rhythm) and QT (ventricular activation/recovery) intervals, suggesting association with a long QT syndrome and the importance of Scn1b to normal cardiac electrophysiology.137 In addition, Scn1b null mice demonstrate atrial dysfunction, including sinoatrial node dysfunction, increased atrial collagen, and atrial fibrillation.138 Scn1b null mice also have widened perinexuses, supporting the hypothesis that β1/β1B is vital to maintaining perinexal width.38 Cardiac-specific Scn1b null mice also show increased susceptibility to arrhythmias, although these mice do survive past 3 weeks of age.139 In humans with persistent atrial fibrillation, perinexal width in atrial appendages has been shown to be approximately 3 nm wider than controls (without atrial fibrillation), and SCN1B (β1/β1B) was confirmed to be located in human perinexuses, providing further support for a vital role of β1/β1B in normal cardiac conduction.110
Potential for β1/β1B as antiarrhythmic drug targets
The history of antiarrhythmic therapeutics is replete with studies that reveal the promise, but also the caveats, associated with arrhythmia drug use and development. One such example is the Cardiac Arrhythmia Suppression Trial (CAST), undertaken in 1989, which tested the ability of 2 sodium channel blockers in treating ventricular arrhythmia after myocardial infarction.140 Although the treatment decreased the total number of arrhythmias experienced by participants, the number of sudden cardiac deaths increased over the study.140 The investigators concluded that encainide and flecainide should not be used to treat patients with minor or asymptomatic ventricular arrhythmias postmyocardial infarction. However, flecainide has been in use in the clinic consistently since the CAST results were released, with effectiveness demonstrated in atrial fibrillation, atrioventricular nodal reentrant tachycardia, and ventricular arrhythmias in patients without cardiac structural disease.141,142 Overall, the number of new antiarrhythmic drugs has decreased in the last decades, and current options have significant limitations, although there is a push for repurposing existing drugs.143 This illustrates the need for better understanding of current antiarrhythmic drugs and the need for development of new antiarrhythmic drugs.
We summarize here the potential for β1/β1B as new targets for antiarrhythmic drugs as outlined earlier. Several studies that we have focused on here highlighted potential advantages of targeting β-subunits to treat or prevent arrhythmias: (1) directly, via targeting Ig adhesion, which may be responsible for maintaining normal cardiac conduction and is disrupted at the perinexus in patients with atrial fibrillation; (2) indirectly, by targeting the interaction between β- and α-subunits (to impact drug efficacy); or (3) by targeting the RIP process, with potential to alter sodium, potassium, and calcium channels and diversify and regulate sodium currents.8,9,38,41,144 Currently, there are no known β-subunit–targeting drugs. However, β1-specific mimetic peptides show promise as prodrugs.38,128 For example, βadp1 seems to acutely promote β1 RIP, and, in the longer term, increases levels of β1 in cells heterologously expressing Scn1b.128 On the other hand, peptides containing dimeric repeats based on the β1 Ig domain seem to promote intercellular adhesion. Whether the gain-of-function effects mediated by these peptidic constructs show antiarrhythmic benefits in vivo awaits further study.
Conclusion
Recent investigations have revealed new insights on the roles of β1 and its related proteins β1B and β1-ICD, suggesting these proteins play a more critical role in cardiac electrophysiology than previously understood. Specifically, these findings highlight the likely importance of β1 RIP signaling and assignments in ephaptic coupling. This review discussed emerging opportunities and posed several key questions based on these new insights. Significant gaps remain in the field, notably the need for direct evidence of (1) the occurrence and role of β1 RIP in cardiac tissues and (2) ephaptic conduction in the heart. Other critical areas of interest include understanding the distinct roles of β1 vs β1B in cardiac function. The unique interactions of the β1 and β1B extracellular domains with NaV1.5 require further exploration. Additionally, there is interest in the broader Scn1b interactome beyond its association with α-subunits like NaV1.5. Proteins such as Tmem65, which directly interacts with β1/β1B, and the Coxsackie and adenovirus receptor (CAR), which coprecipitates with NaV1.5, represent promising starting points for further study. Moreover, mutations in the conserved β1/β1B extracellular Ig domain are linked to arrhythmogenic pathologic conditions. This underscores the critical role of the cardiac β1/β1B adhesion domain in cardiac physiology and likely presents valuable targets for both understanding and developing therapeutic strategies to address cardiac arrhythmias.
Funding Sources:
This research was supported by National Institutes of Health/National Heart, Lung, and Blood Institute grants to Dr Gourdie (1R35HL161237-01) and Dr Williams (1F31HL164088-01).
Abbreviations
- CAM
cell adhesion molecule
- GJ
gap junction
- ICD
intracellular domain
- Ig
immunoglobulin
- RIP
regulated intramembrane-proteolysis
- VGSC
voltage-gated sodium channel
Footnotes
Disclosures:
Dr Gourdie and Dr Williams have submitted patent PCT/US2023/063525 on peptides mentioned in the text. All other authors have no conflicts of interest to disclose.
References
- 1.Wang J, et al. Distribution and function of voltage-gated-sodium-channels in the nervous system. Channels (Austin) 2017;11:534–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, et al. Ventricular voltage-gated ion channels: detection, characteristics, mechanisms, and drug safety evaluation. Clin Transl Med 2021;11:e530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hull JM, Isom LL. Voltage-gated sodium channel β-subunits: the power outside the pore in brain development and disease. Neuropharmacology 2018;132:4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salvage SC, et al. Cell-adhesion properties of β-subunits in the regulation of cardiomyocyte sodium channels. Biomolecules 2020;10:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.deLera Ruiz M, Kraus RL. Voltage-gated sodium channels: structure, function, pharmacology, and clinical indications. J Med Chem 2015;58:7093–7118. [DOI] [PubMed] [Google Scholar]
- 6.Dehghani-Samani A, et al. Mutations of voltage-gated ionic channels and risk of severe cardiac arrhythmias. Acta Cardiol Sin 2019;35:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riuró H, et al. A missense mutation in the sodium channel β1b subunit reveals SCN1B as a susceptibility gene underlying long QT syndrome. Heart Rhythm 2014;11:1202–1209. [DOI] [PubMed] [Google Scholar]
- 8.Zhu W, et al. Modulation of the effects of class Ib antiarrhythmics on cardiac NaV1.5-encoded channels by accessory NaVβ-subunits. JCI Insight 2021; 6:e143092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouza AA, Isom LL. Voltage-gated sodium channel β-subunits and their related diseases. Handb Exp Pharmacol 2018;246:423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edokobi N, Isom LL. Voltage-gated sodium channel β1/β1b subunits regulate cardiac physiology and pathophysiology. Front Physiol 2018;9:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazen-Gillespie KA, et al. Cloning, localization, and functional expression of sodium channel beta1A subunits. J Biol Chem 2000;275:1079–1088. [DOI] [PubMed] [Google Scholar]
- 12.Patel F, Brackenbury WJ. Dual roles of voltage-gated sodium channels in development and cancer. Int J Dev Biol 2015;59:357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, et al. SCN1B genetic variants: a review of the spectrum of clinical pheno-types and a report of early myoclonic encephalopathy. Children 2022;9:1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auld VJ, et al. A rat brain Na+ channel alpha subunit with novel gating properties. Neuron 1988;1:449–461. [DOI] [PubMed] [Google Scholar]
- 15.Kruger LC, Isom LL. Voltage-gated Na+ channels: not just for conduction. Cold Spring Harb Perspect Biol 2016;8:a029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catterall WA. Voltage gated sodium and calcium channels: discovery, structure, function, and pharmacology. Channels (Austin) 2023;17:2281714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catterall WA, et al. The conformational cycle of a prototypical voltage-gated sodium channel. Nat Chem Biol 2020;16:1314–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen-Izu Y, et al. Na+ channel function, regulation, structure, trafficking and sequestration. J Physiol 2015;593:1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baroni D, Moran O. On the multiple roles of the voltage gated sodium channel β1 subunit in genetic diseases. Front Pharmacol 2015;6:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beneski DA, Catterall WA. Covalent labeling of protein components of the sodium channel with a photoactivable derivative of scorpion toxin. Proc Natl Acad Sci U S A 1980;77:639643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isom LL, et al. Auxiliary subunits of voltage-gated ion channels. Neuron 1994; 12:1183–1194. [DOI] [PubMed] [Google Scholar]
- 22.Isom LL, et al. Primary structure and functional expression of the beta-1 subunit of the rat brain sodium channel. Science 1992;256:839–842. [DOI] [PubMed] [Google Scholar]
- 23.Isom LL, et al. Structure and function of the β2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell 1995;83:433–442. [DOI] [PubMed] [Google Scholar]
- 24.Morgan K, et al. β3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci U S A 2000;97:2308–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu FH, et al. Sodium channel beta4, a new disulfide-linked auxiliary subunit with similarity to beta2. J Neurosci 2003;23:7577–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen H, et al. Structures of human Na(v)1.7 channel in complex with auxiliary subunits and animal toxins. Science 2019;363:1303–1308. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy A, et al. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010;5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortada E, et al. Trafficking and function of the voltage-gated sodium channel β2 subunit. Biomolecules 2019;9:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cusdin FS, et al. Trafficking and cellular distribution of voltage-gated sodium channels. Traffic 2008;9:17–26. [DOI] [PubMed] [Google Scholar]
- 31.Brackenbury W, Isom L. Na+ channel β-subunits: overachievers of the ion channel family. Front Pharmacol 2011;2:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Z-C, et al. Tenascin-R is a functional modulator of sodium channel β-subunits. J Biol Chem 1999;274:26511–26517. [DOI] [PubMed] [Google Scholar]
- 33.Malhotra JD, et al. Sodium channel β-subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell-cell contact. J Biol Chem 2000; 275:11383–11388. [DOI] [PubMed] [Google Scholar]
- 34.McEwen DP, et al. The voltage-gated Na+ channel β3 subunit does not mediate trans homophilic cell adhesion or associate with the cell adhesion molecule contactin. Neurosci Lett 2009;462:272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yereddi NR, et al. The immunoglobulin domain of the sodium channel β3 subunit contains a surface-localized disulfide bond that is required for homophilic binding. FASEB J 2013;27:568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ratcliffe CF, et al. Sodium channel β1 and β3 subunits associate with neurofascin through their extracellular immunoglobulin-like domain. J Cell Biol 2001; 154:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazarinova-Noyes K, et al. Contactin associates with Na+ channels and increases their functional expression. J Neurosci 2001;21:7517–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veeraraghavan R, et al. The adhesion function of the sodium channel beta subunit (β1) contributes to cardiac action potential propagation. Elife 2018; 7:e37610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Namadurai S, et al. A new look at sodium channel β-subunits. Open Biology 2015;5:140192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sprinzak D, Blacklow SC. Biophysics of notch signaling. Annu Rev Biophys 2021; 50:157–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Malley HA, Isom LL. Sodium channel β-subunits: emerging targets in channelopathies. Annu Rev Physiol 2015;77:481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouza AA, et al. Sodium channel β1 subunits participate in regulated intramembrane proteolysis-excitation coupling. JCI Insight 2021;6:e141776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong H-K, et al. β-subunits of voltage-gated sodium channels are novel substrates of β-site amyloid precursor protein-cleaving enzyme (BACE1) and γ-secretase. J Biol Chem 2005;280:23009–23017. [DOI] [PubMed] [Google Scholar]
- 44.Hoagland DT, et al. The role of the gap junction perinexus in cardiac conduction: potential as a novel anti-arrhythmic drug target. Prog Biophys Mol Biol 2019;144:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seneci L, Mikheyev AS. Sodium channel β-subunits: an additional element in animal tetrodotoxin resistance? Int J Mol Sci 2024;25:1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hichri E, et al. Distribution of cardiac sodium channels in clusters potentiates ephaptic interactions in the intercalated disc. J Physiol 2018;596:563–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rougier JS, et al. A distinct pool of Na(v)1.5 channels at the lateral membrane of murine ventricular cardiomyocytes. Front Physiol 2019;10:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillet L, et al. Cardiac-specific ablation of synapse-associated protein SAP97 in mice decreases potassium currents but not sodium current. Heart Rhythm 2015; 12:181–192. [DOI] [PubMed] [Google Scholar]
- 49.Maier SKG, et al. Distinct subcellular localization of different sodium channel α and β-subunits in single ventricular myocytes from mouse heart. Circulation 2004;109:1421–1427. [DOI] [PubMed] [Google Scholar]
- 50.Tarasov M, et al. NaV1.6 dysregulation within myocardial T-tubules by D96V calmodulin enhances proarrhythmic sodium and calcium mishandling. J Clin Invest 2023;133:e152071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Struckman HL, et al. Super-resolution imaging using a novel high-fidelity antibody reveals close association of the neuronal sodium channel Na(V)1.6 with ryanodine receptors in cardiac muscle. Microsc Microanal 2020;26:157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaborit N, et al. Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol 2007;582:675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malhotra JD, et al. Tyrosine-phosphorylated and nonphosphorylated sodium channel beta1 subunits are differentially localized in cardiac myocytes. J Biol Chem 2004;279:40748–40754. [DOI] [PubMed] [Google Scholar]
- 54.Okata S, et al. Embryonic type Na+ channel β-subunit, SCN3B masks the disease phenotype of Brugada syndrome. Sci Rep 2016;6:34198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Domínguez JN, et al. Temporal and spatial expression pattern of beta1 sodium channel subunit during heart development. Cardiovasc Res 2005; 65:842–850. [DOI] [PubMed] [Google Scholar]
- 56.Shah BS, et al. Developmental expression of the novel voltage-gated sodium channel auxiliary subunit β3, in rat CNS. J Physiol 2001;534:763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mittendorf KF, et al. Tailoring of membrane proteins by alternative splicing of pre-mRNA. Biochemistry 2012;51:5541–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanter HL, et al. Structural and molecular determinants of intercellular coupling in cardiac myocytes. Microsc Res Tech 1995;31:357–363. [DOI] [PubMed] [Google Scholar]
- 59.Mezzano V, et al. Cell junctions in the specialized conduction system of the heart. Cell Commun Adhes 2014;21:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coppen SR, et al. Connexin45 expression is preferentially associated with the ventricular conduction system in mouse and rat heart. Circ Res 1998; 82:232–243. [DOI] [PubMed] [Google Scholar]
- 61.Ideker RE, et al. Purkinje fibers and arrhythmias. Pacing Clin Electrophysiol 2009; 32:283–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodges SL, et al. Therapeutic potential of targeting regulated intramembrane proteolysis mechanisms of voltage-gated ion channel subunits and cell adhesion molecules. Pharmacol Rev 2022;74:1028–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patino GA, et al. Voltage-gated Na+ channel β1B: a secreted cell adhesion molecule involved in human epilepsy. J Neurosci 2011;31:14577–14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bouza AA, et al. Sodium channel β1 subunits are post-translationally modified by tyrosine phosphorylation, S-palmitoylation, and regulated intramembrane proteolysis. J Biol Chem 2020;295:10380–10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson D, et al. The sialic acid component of the β1 subunit modulates voltage-gated sodium channel function. J Biol Chem 2004;279:44303–44310. [DOI] [PubMed] [Google Scholar]
- 66.Zhu W, et al. Mechanisms of noncovalent β-subunit regulation of NaV channel gating. J Gen Physiol 2017;149:813–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salvage SC, et al. Gating control of the cardiac sodium channel Nav1.5 by its β3-subunit involves distinct roles for a transmembrane glutamic acid and the extracellular domain. J Biol Chem 2019;294:19752–19763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meadows L, et al. The intracellular segment of the sodium channel β1 subunit is required for its efficient association with the channel α-subunit. J Neurochem 2001;76:1871–1878. [DOI] [PubMed] [Google Scholar]
- 69.Chopra SS, et al. Molecular cloning and analysis of zebrafish voltage-gated sodium channel beta subunit genes: implications for the evolution of electrical signaling in vertebrates. BMC Evol Biol 2007;7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buffington SA, Rasband MN. Na+ channel-dependent recruitment of Navβ4 to axon initial segments and nodes of Ranvier. J Neurosci 2013;33:6191–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C, et al. Identification of the cysteine residue responsible for disulfide linkage of Na+ channel α and β2 subunits. J Biol Chem 2012;287:39061–39069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan Z, et al. Structure of the Nav1.4-β1 complex from electric eel. Cell 2017; 170:470–482.e411. [DOI] [PubMed] [Google Scholar]
- 73.Pan X, et al. Molecular basis for pore blockade of human Na(+) channel Na(v)1.2 by the m-conotoxin KIIIA. Science 2019;363:1309–1313. [DOI] [PubMed] [Google Scholar]
- 74.Cervantes DO, et al. Scn1b expression in the adult mouse heart modulates Na+ influx in myocytes and reveals a mechanistic link between Na+ entry and diastolic function. Am J Physiol Heart Circ Physiol 2022;322:H975–H993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Signore S, et al. Late Na(1) current and protracted electrical recovery are critical determinants of the aging myopathy. Nat Commun 2015;6:8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Angsutararux P, et al. Molecular pathology of sodium channel beta-subunit variants. Front Pharmacol 2021;12:761275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen C, et al. Epilepsy and sudden unexpected death in epilepsy in a mouse model of human SCN1B-linked developmental and epileptic encephalopathy. Brain Commun 2023;5:fcad283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bouza AA, et al. Sodium channel β1 subunits are post-translationally modified by tyrosine phosphorylation, S-palmitoylation, and regulated intramembrane proteolysis. J Biol Chem 2020;295:10380–10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haworth AS, et al. Subcellular dynamics and functional activity of the cleaved intracellular domain of the Na+ channel β1 subunit. J Biol Chem 2022; 298:102174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McEwen DP, Isom LL. Heterophilic interactions of sodium channel beta1 subunits with axonal and glial cell adhesion molecules. J Biol Chem 2004; 279:52744–52752. [DOI] [PubMed] [Google Scholar]
- 81.Malhotra JD, et al. Structural requirements for interaction of sodium channel beta 1 subunits with ankyrin. J Biol Chem 2002;277:26681–26688. [DOI] [PubMed] [Google Scholar]
- 82.Gourdie RG. The cardiac gap junction has discrete functions in electrotonic and ephaptic coupling. Anat Rec (Hoboken) 2019;302:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gourdie RG, et al. Gap junctional connexin43: novel insights from the new millennium and their clinical implications. In: Jalife J, Stevenson WG, eds. Cardiac Electrophysiology: From Cell to Bedside, Eighth Edition. Amsterdam: Elsevier; 2021. . 1600. [Google Scholar]
- 84.Manring HR, et al. At the heart of inter- and intracellular signaling: the intercalated disc. Biophys Rev 2018;10:961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nielsen MS, et al. The intercalated disc: a unique organelle for electromechanical synchrony in cardiomyocytes. Physiol Rev 2023;103:2271–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Struckman HL, et al. Unraveling impacts of chamber-specific differences in intercalated disc ultrastructure and molecular organization on cardiac conduction. JACC Clin Electrophysiol 2023;9:2425–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeruva S, Waschke J. Structure and regulation of desmosomes in intercalated discs: lessons from epithelia. J Anat 2023;242:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rhett JM, et al. The perinexus: sign-post on the path to a new model of cardiac conduction? Trends Cardiovasc Med 2013;23:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rhett JM, Gourdie RG. The perinexus: a new feature of Cx43 gap junction organization. Heart Rhythm 2012;9:619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gourdie RG, et al. Gap junction distribution in adult mammalian myocardium revealed by an anti-peptide antibody and laser scanning confocal microscopy. J Cell Sci 1991;99(Pt 1):41–55. [DOI] [PubMed] [Google Scholar]
- 91.Mori Y, et al. Ephaptic conduction in a cardiac strand model with 3D electrodiffusion. Proc Natl Acad Sci U S A 2008;105:6463–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rhett JM, et al. Cx43 associates with Na(v)1.5 in the cardiomyocyte perinexus. J Membr Biol 2012;245:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rhett JM, et al. Cx43 associates with Na(v)1.5 in the cardiomyocyte perinexus. J Membr Biol 2012;245:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Veeraraghavan R, Gourdie RG. Stochastic optical reconstruction microscopy-based relative localization analysis (STORM-RLA) for quantitative nanoscale assessment of spatial protein organization. Mol Biol Cell 2016;27:3583–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carmeliet E Conduction in cardiac tissue. Historical reflections. Physiol Rep 2019;7:e13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Veeraraghavan R, et al. Mechanisms of cardiac conduction: a history of revisions. Am J Physiol Heart Circ Physiol 2014;306:H619–H627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ivanovic E, Kucera JP. Localization of Na(1) channel clusters in narrowed perinexi of gap junctions enhances cardiac impulse transmission via ephaptic coupling: a model study. J Physiol 2021;599:4779–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Veeraraghavan R, et al. Sodium channels in the Cx43 gap junction perinexus may constitute a cardiac ephapse: an experimental and modeling study. Pflugers Arch 2015;467:2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.George SA, et al. Modulating cardiac conduction during metabolic ischemia with perfusate sodium and calcium in guinea pig hearts. Am J Physiol Heart Circ Physiol 2019;316:H849–H861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin J, Keener JP. Modeling electrical activity of myocardial cells incorporating the effects of ephaptic coupling. Proc Natl Acad Sci U S A 2010; 107:20935–20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tsumoto K, et al. Specific decreasing of Na(1) channel expression on the lateral membrane of cardiomyocytes causes fatal arrhythmias in Brugada syndrome. Sci Rep 2020;10:19964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kucera JP, et al. Localization of sodium channels in intercalated disks modulates cardiac conduction. Circ Res 2002;91:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wei N, et al. The dual effect of ephaptic coupling on cardiac conduction with heterogeneous expression of connexin 43. J Theor Biol 2016;397:103–114. [DOI] [PubMed] [Google Scholar]
- 104.Wei N, Tolkacheva EG. Interplay between ephaptic coupling and complex geometry of border zone during acute myocardial ischemia: Effect on arrhythmogeneity. Chaos 2020;30:033111. [DOI] [PubMed] [Google Scholar]
- 105.Weinberg SH. Ephaptic coupling rescues conduction failure in weakly coupled cardiac tissue with voltage-gated gap junctions. Chaos 2017;27:093908. [DOI] [PubMed] [Google Scholar]
- 106.Greer-Short A, et al. Revealing the concealed nature of long-QT type 3 syndrome. Circ Arrhythm Electrophysiol 2017;10:e004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nowak MB, et al. Intercellular sodium regulates repolarization in cardiac tissue with sodium channel gain of function. Biophys J 2020;118:2829–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nowak MB, et al. Mechanisms underlying age-associated manifestation of cardiac sodium channel gain-of-function. J Mol Cell Cardiol 2021;153:60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tveito A, et al. A cell-based framework for numerical modeling of electrical conduction in cardiac tissue. Front Phys 2017;5:48. [Google Scholar]
- 110.Raisch TB, et al. Intercalated disk extracellular nanodomain expansion in patients with atrial fibrillation. Front Physiol 2018;9:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Veeraraghavan R, et al. Potassium channels in the Cx43 gap junction perinexus modulate ephaptic coupling: an experimental and modeling study. Pflugers Arch 2016;468:1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Veeraraghavan R, et al. Sodium channels in the Cx43 gap junction perinexus may constitute a cardiac ephapse: an experimental and modeling study. Pflugers Arch 2015;467:2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.George SA, et al. Extracellular sodium dependence of the conduction velocity-calcium relationship: evidence of ephaptic self-attenuation. Am J Physiol Heart Circ Physiol 2016;310:H1129–H1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Adams WP, et al. Extracellular perinexal separation is a principal determinant of cardiac conduction. Circ Res 2023;133:658–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morris JA, et al. Nernst-Planck-Gaussian modelling of electrodiffusional recovery from ephaptic excitation between mammalian cardiomyocytes. Front Physiol 2023;14:1280151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ivanovic E, Kucera JP. Tortuous cardiac intercalated discs modulate ephaptic coupling. Cells 2022;11:3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wei N, Tolkacheva EG. Mechanisms of arrhythmia termination during acute myocardial ischemia: role of ephaptic coupling and complex geometry of border zone. PLoS One 2022;17:e0264570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jæger KH, et al. Properties of cardiac conduction in a cell-based computational model. PLoS Comput Biol 2019;15:e1007042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y, et al. Fibroblasts in heart scar tissue directly regulate cardiac excitability and arrhythmogenesis. Science 2023;381:1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poelzing S, et al. Initiation and entrainment of multicellular automaticity via diffusion limited extracellular domains. Biophys J 2021;120:5279–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu X, et al. Hypernatremia and intercalated disc edema synergistically exacerbate long-QT syndrome type 3 phenotype. Am J Physiol Heart Circ Physiol 2021;321:H1042–H1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moise N, et al. Intercalated disk nanoscale structure regulates cardiac conduction. J Gen Physiol 2021;153:e202112897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Isom LL, Catterall WA. Na+ channel subunits and Ig domains. Nature 1996; 383:307–308. [DOI] [PubMed] [Google Scholar]
- 124.Teng ACT, Gu L, Di Paola M, et al. Tmem65 is critical for the structure and function of the intercalated discs in mouse hearts. Nat Commun 2022;13:6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Clatot J, et al. Voltage-gated sodium channels assemble and gate as dimers. Nat Commun 2017;8:2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Salvage SC, et al. Supramolecular clustering of the cardiac sodium channel Nav1.5 in HEK293F cells, with and without the auxiliary β3-subunit. FASEB J 2020;34:3537–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Namadurai S, et al. Crystal structure and molecular imaging of the Nav channel β3 subunit indicates a trimeric assembly. J Biol Chem 2014; 289:10797–10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Williams ZJ, et al. Development and characterization of the mode-of-action of inhibitory and agonist peptides targeting the voltage-gated sodium channel SCN1B/β1 subunit. bioRxiv 2019;:562974. 20232023.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Watanabe H, et al. Mutations in sodium channel β1- and β2-subunits associated with atrial fibrillation. Circ Arrhythm Electrophysiol 2009;2:268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Olesen MS, et al. SCN1Bb R214Q found in 3 patients: 1 with Brugada syndrome and 2 with lone atrial fibrillation. Heart Rhythm 2012;9:770–773. [DOI] [PubMed] [Google Scholar]
- 131.Ricci MT, et al. SCN1B gene variants in Brugada syndrome: a study of 145 SCN5A-negative patients. Sci Rep 2014;4:6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Peeters U, et al. Contribution of cardiac sodium channel β-subunit variants to Brugada syndrome. Circ J 2015;79:2118–2129. [DOI] [PubMed] [Google Scholar]
- 133.Watanabe H, et al. Sodium channel β1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest 2008; 118:2260–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hu D, et al. A novel rare variant in SCN1Bb linked to Brugada syndrome and SIDS by combined modulation of Na(v)1.5 and K(v)4.3 channel currents. Heart Rhythm 2012;9:760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.El-Battrawy I, et al. Studying Brugada syndrome with an SCN1B variants in human-induced pluripotent stem cell-derived cardiomyocytes. Front Cell Dev Biol 2019;7:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen C, et al. Mice lacking sodium channel β1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J Neurosci 2004;24:4030–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lopez-Santiago LF, et al. Sodium channel Scn1b null mice exhibit prolonged QT and RR intervals. J Mol Cell Cardiol 2007;43:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ramos-Mondragon R, et al. Neonatal Scn1b-null mice have sinoatrial node dysfunction, altered atrial structure, and atrial fibrillation. JCI Insight 2022;7:e152050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin X, et al. Scn1b deletion leads to increased tetrodotoxin-sensitive sodium current, altered intracellular calcium homeostasis and arrhythmias in murine hearts. J Physiol 2015;593:1389–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cardiac Arrhythmia Suppression Trial (CAST) Investigators. Preliminary report: effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. N Engl J Med 1989;321:406–412. [DOI] [PubMed] [Google Scholar]
- 141.Echt DS, Ruskin JN. Use of flecainide for the treatment of atrial fibrillation. Am J Cardiol 2020;125:1123–1133. [DOI] [PubMed] [Google Scholar]
- 142.Basza M, et al. Flecainide in clinical practice. Cardiol J 2023;30:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saljic A, et al. Recent advances in antiarrhythmic drug therapy. Drugs 2023; 83:1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hodges SL, et al. Therapeutic potential of targeting regulated intramembrane proteolysis mechanisms of voltage-gated ion channel subunits and cell adhesion molecules. Pharmacol Rev 2022;74:1030–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]


