ABSTRACT
The wood frog (Rana sylvatica) endures whole‐body freezing over the winter, with extensive extracellular ice formation and halted physiological activities. Epigenetic mechanisms, including reversible histone lysine methylation, enable quick alterations in gene expression, helping to maintain viability during freeze‐thaw cycles. The present study evaluated eight histone lysine methyltransferases (KMTs), 10 histone lysine demethylases (KDMs), and 11 histone marks in wood frog kidneys. Using immunoblotting, significant changes in relative protein levels of multiple KMTs and KDMs were observed in response to freezing, with variable alterations during thawing. Specifically, the repressive methyl marks H3K27me1 and H4K20me3 significantly decreased during freezing, whereas H3K9me3, H3K27me3, and H3K36me2 decreased during thawing. These results demonstrate that the regulation of histone methylation and demethylation play crucial roles in controlling gene expression over the freeze‐thaw cycle and the maintenance of normal renal physiology.
Keywords: freeze tolerance, gene expression, hypometabolism, kidney, Rana sylvatica
Summary
This research explores how histone methylation and demethylation regulate gene expression in the kidneys of freeze‐tolerant frogs, providing insights into how these animals survive prolonged extreme cold.
Key findings reveal specific epigenetic modifications that enable frogs to endure freezing temperatures, offering a model for understanding similar survival mechanisms in other species.
This work is significant because it further expands avenues for cryobiology research and may inspire innovative strategies for tissue preservation and organ transplantation, with potential impacts on both conservation biology and medical science.
1. Introduction
Freeze tolerance is a unique adaptation that allows animals to endure prolonged subzero temperatures over the winter. The wood frog (Rana sylvatica) is the best‐studied freeze‐tolerant species and can withstand prolonged exposure to subzero temperatures with over half of its body water frozen as extracellular ice [1, 2]. During freezing, multiple physiological processes are halted including heartbeat, breathing, and muscle movement [1, 2]. Given the energetically costly nature of gene expression and the fact that many biochemical pathways are tightly regulated in their first steps, stress‐tolerant species rely on wide‐scale epigenetic regulation to endure prolonged and challenging conditions such as freezing, anoxia, hibernation, and so forth [3, 4, 5].
Histone modifications occur on different amino acid residues, with lysine being an important residue that is modified by lysine methyltransferase (KMT) enzymes (Figure 1). Histone lysine methylation and the KMTs that catalyze their addition play important roles in transcriptional control, cell cycle, genome stability, and nuclear structure [6]. Recently, progress has been made in illustrating the roles of methyl modifications in controlling gene expression and the KMTs and lysine demethylases (KDMs) that regulate them [6]. Various posttranslational modifications, including methyl marks added by KMTs on specific amino acids of histone proteins lead to alterations of chromatin structure and recruitment of enzyme modifying complexes, activating or repressing transcription [7, 8]. KDMs are also important in gene expression regulation by removing methyl marks, and this demethylation can either activate or repress gene expression, depending on the specific lysine residue and the degree of methylation [6]. Since KDMs are responsible for removing methyl groups from histones and histones are significant components of DNA regulation, the addition or removal of methyl groups on histones has a large impact on gene expression.
Figure 1.
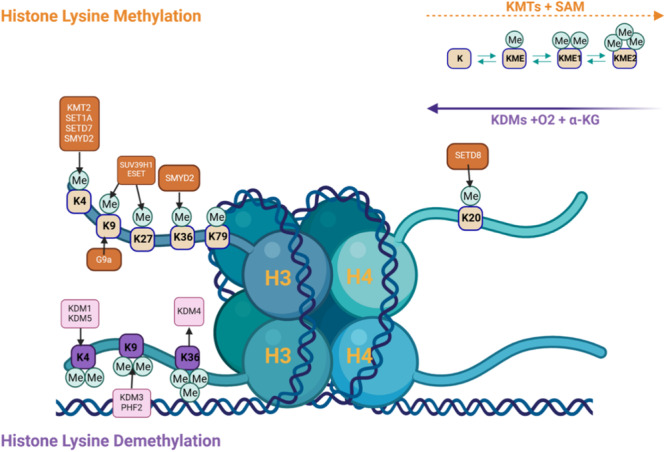
Histone lysine methylation and demethylation, involving the addition or removal of methyl groups (Me) on specific lysine residues of histones H3 and H4, which affects gene expression. Important enzymes involved in methylation are histone methyltransferases (HMTs or KMTs), such as KMT2, SET1A, and SUV39H1, using S‐adenosylmethionine (SAM) as the methyl donor. Demethylation is performed by histone demethylases (KDMs), in the presence of oxygen (O2) and alpha‐ketoglutarate (α‐KG). Methylation states (mono‐, di‐, tri‐methylation) and specific lysine targets (e.g., K4, K9, K27) determine whether gene expression is activated or repressed indicating dynamic regulation of gene activity. Created with BioRender.com.
The competing roles of KMTs and KDMs are crucial for gene regulation due to their ability to dynamically regulate chromatin structure and function. Several lysine residues on histone H3 (e.g., H3K4, H3K36, and H3K79) are markers of active transcription [9], whereas repressive marks including H3K9me3 and H3K27me3 are mainly associated with silencing gene expression and condensing chromatin [6, 10, 11]. Altering these methyl marks in response to stress enables either activation or repression of gene expression, ultimately altering chromatin accessibility and facilitating or hindering transcriptional machinery binding [9, 12].
Recent research on R. sylvatica showed tissue‐specific alterations in KMTs and their targets in response to freeze/thaw [13, 14]. Moreover, research on mammalian models demonstrated the importance of histone modifications in kidney development, physiology, and pathology [15, 16]. However, information regarding epigenetic controls on wood frog kidney function during freeze/thaw has not been investigated to date. Hence, the present study investigated protein levels of various KMTs and KDMs, and the distribution of lysine methylation in wood frog kidney tissue over the freeze‐thaw cycle, with the hypothesis that epigenetic factors associated with transcriptional activation would be downregulated during freezing.
2. Materials and Methods
2.1. Ethics Statement
All animals were cared for in accordance with the guidelines of the Canadian Council on Animal Care and experimental procedures had the prior approval of the Carleton University Animal Care Committee (protocol #106935).
2.2. Animal Collection
Male wood frogs were collected during the spring breeding season from meltwater ponds near Ottawa, Ontario, Canada, and all animal procedures were carried out as previously described [14]. Frogs were bathed in tetracycline and placed in plastic boxes containing damp sphagnum moss to acclimate at 5°C for at least 1 week. Control frogs were sampled from this condition. The remaining frogs were placed in plastic containers lined with damp paper towels in a −4°C incubator for 45 min to drop their body temperatures to below zero and trigger ice nucleation. The temperature was then raised to −2.5°C for 24 h, after which, half of the frozen frogs were randomly selected for sampling. The remaining frozen frogs were returned to the 5°C incubator for 8 h until fully thawed. For sampling, control, 24 h frozen, and 8 h thawed frogs were euthanized by pithing, and kidney tissue was excised and frozen in liquid nitrogen. Tissues were stored at −80°C until use.
2.3. Total Protein Isolation
Total protein was isolated from samples of frozen kidney tissue (~50 mg each) (n = 4 separate individuals from each condition) as previously described [14]. Briefly, tissues were crushed with a mortar and pestle under liquid nitrogen, then homogenized using a Polytron PT10 homogenizer in pre‐chilled mixture of 1× lysis buffer (1:5 w:v; Cat#43‐045; MilliporeSigma, Burlington, MA), 1 mm Na3VO4, 10 mm NaF, 10 mm β‐glycerophosphate, and 10 μL·mL−1 protease inhibitor cocktail (Cat#PIC002; Bioshop Canada Inc., Burlington, ON). Homogenates were left on ice for 30 min with occasional vortexing, then were centrifuged (14,000 × g; 20 min; 4°C) and supernatants containing soluble proteins were collected. Sample protein concentrations were determined using the BioRad protein assay (Cat#500‐0002; BioRad Laboratories, Hercules, CA), and samples were standardized to 10 μg/μL with lysis buffer and mixed 1:1 v:v with Tris buffer containing sodium dodecyl sulfate (SDS) to a final concentration of 5 μg/μL. Samples were then boiled for 10 min and stored at −40°C until use.
2.4. Histone Isolation
Histone protein samples were prepared from frozen kidney samples (~100−200 mg) from control, 24 h frozen, and 8 h thawed frogs (n = 4 biological replicates for each condition) as previously described [17]. Briefly, frozen tissues were homogenized with a Dounce homogenizer in Triton Extraction Buffer (TEB), vortexed, and then incubated on ice for 30 min. Homogenates were centrifuged (10,000 × g; 10 min; 4°C) and supernatants were discarded. Pellets were washed with TEB and centrifuged (10,000 × g; 10 min; 4°C), then supernatants were discarded and the pellet containing nuclei was resuspended in 0.2 M H2SO4. Samples were incubated (4°C; 30 min), then centrifuged (12,000 × g; 10 min; 4°C), and supernatants containing histones were transferred into fresh tubes. Next, 100% trichloroacetic acid was added to the histone solution to make a final concentration of 33%, followed by incubation on ice for 30 min. Samples were centrifuged (12,000 × g; 10 min; 4°C) and supernatants were removed. Histone pellets were washed with ice‐cold acetone and centrifuged (12,000 × g; 5 min; 4°C). Supernatants were removed and pellets air‐dried for 20 min at room temperature (RT). Pellets were resuspended in ddH2O and sonicated, and samples were quantified for histone concentrations and separation of histone proteins.
2.5. Western Immunoblotting
Western immunoblotting of each protein target was performed as previously described [17]. Briefly, equal amounts of protein (15−40 μg, depending on target protein) from the kidney of control, 24 h frozen, and 8 h thawed frogs were loaded into SDS‐polyacrylamide gels (6%−15%, depending on the protein target molecular weight). BLUeye prestained protein ladder (Cat#PM007‐0500; FroggaBio, Toronto, ON) was also loaded to determine target protein size. Gels were run (30−180 min; 180 V) in a BioRad Mini Protean III system to separate proteins, which were then transferred onto 0.45 μm pore PVDF membranes by electroblotting (RT; 45−180 min; 160 mA). To prevent nonspecific antibody binding, membranes were incubated on a rocker (RT; 30 min) in 1X TBST buffer with 1%−10% nonfat skim milk. Membranes were washed 3 × 5 min with TBST and probed with primary antibodies (File S1; 1:1000 v:v in TBST) at 4°C overnight. Following incubation, blots were washed 3 × 5 min with TBST and probed with HRP‐conjugated anti‐rabbit or ‐mouse secondary antibodies (1:8000 v:v in TBST; Cat#APA002P, BioShop Canada Inc.) at RT for 30 min. Membranes were washed for 3 × 5 min in TBST and then visualized by chemiluminescence (1:1 v:v H2O2 and Luminol) using a ChemiGenius Bio Imaging System (Syngene, Frederick, MD). Membranes were then stained with Coomassie brilliant blue dye to visualize all protein bands for loading standardization.
2.6. Statistical Analysis
Band densities were standardized against the combined intensity of Coomassie‐stained bands in the same lane that did not show differential expression between conditions and were well separated from the band of interest. Data for each experimental condition are expressed as mean ± SEM with n = 4 samples from different animals. Statistical analysis was performed using a one‐way ANOVA and Tukey's post hoc test, with p < 0.05 accepted as a significant difference via RBioPlot [18].
3. Results
3.1. KMT Expression
KMT expression was evaluated in wood frog control, frozen, and thawed kidney (Figure 2; File S2). Expression of ASH2L, RBBP5, SETD7, SETD8, and EHMT2/G9a all followed a pattern of reduced levels in tissues from frozen, as compared with controls; however, only changes in ASH2L and RBBP levels reached statistical significance, falling to about 70% and 60% of the control values, respectively. By contrast, there were no significant changes between control and thawed, suggesting that the effects of freezing were quickly reversed after thawing. Relative protein expression of SUV39H1 in frozen frogs showed significant upregulation compared to controls.
Figure 2.
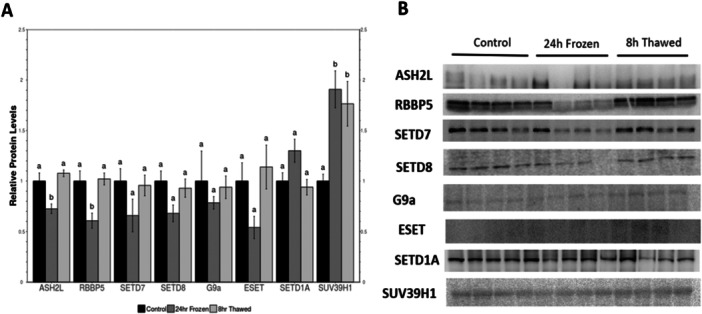
Relative protein levels of lysine methyltransferases (KMTs) in Rana sylvatica kidney via western immunoblotting. (A) Histogram showing mean ± SEM (n = 4) standardized expression levels under control, 24 h freezing, and 8 h thawed conditions. For each protein target, values that share the same letter designations are not significantly different from one another, while values with different letter designations are significantly different (p < 0.05). (B) Representative western blots for individual targets under each experimental condition. Data were analyzed using a one‐way ANOVA with a Tukey's post hoc test.
3.2. Histone Methylation Levels
Histone lysine methylation analysis (Figure 3; File S2) revealed that some histone marks expression levels changed, ranging from slight to significant repression, under frozen, and thawed conditions compared to control. For example, significant repression was observed for H3K9me3, H3K27me1/3, H3K36me2, and H4K20me3. Specifically, relative levels of H3K9me3 remained unchanged between control and freezing conditions, but decreased significantly more than 50% of control values in thawed. Expression levels of H3K27me1 showed a significant decrease of approximately 55% during freezing and a further decrease of approximately 70% after thawing. For H3K27me3, there was a slight reduction of around 90% during freezing and a significant decrease of 60% after thawing. Relative levels of H3K36me2 remained unchanged in freezing, but there was a significant decrease of at least 70% in thawed compared to control. Relative expression levels of H4K20me3 also showed a significant decrease of at least 60% in both the freezing and thawed compared to the control. No significant changes were observed in H3K4me1/2/3, H3K36me3, H3K79me3, and H4K20me1 across all conditions.
Figure 3.
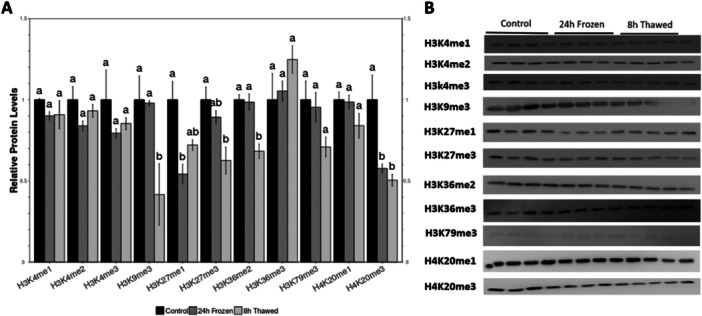
Relative protein levels of methylated histone residues in Rana sylvatica kidneys. All other information as in Figure 2.
3.3. KDM Expression
KDM relative expression levels were also analyzed in the wood frog kidney (Figure 4; File S2). KDM4A, KDM4C, and KDM5B protein levels fell significantly during freezing to about 60%, 25%, and 70% of the control values, respectively. Relative protein expression of KDM1A, KDM4B, KDM5C, and KDM7C showed no difference in frozen frogs, with an upregulation during thawing, however not to a statistically significance level.
Figure 4.
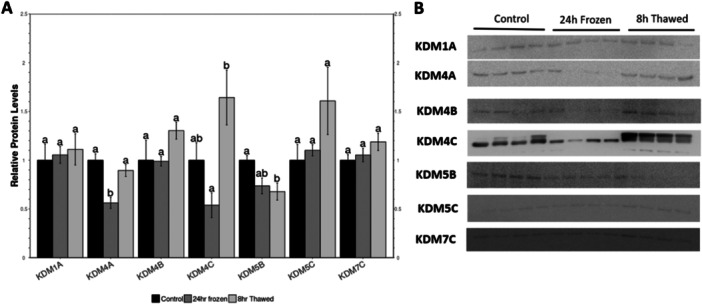
Relative protein levels of lysine demethylases (KDMs) in Rana sylvatica kidneys. All other information as in Figure 2.
4. Discussion
The hypometabolic state characteristic of freeze‐induced MRD requires dramatic suppression of energy expenditure while still maintaining crucial pro‐survival processes [19, 20]. Various physiological and biochemical adaptations interconnect to enable R. sylvatica to survive whole‐body freezing, including transcriptional, posttranscriptional, translational, and posttranslational regulatory mechanisms [13, 17, 21, 22, 23, 24, 25, 26]. Histone modifications play an important role in regulating gene expression, with histone methylation recently being studied in R. sylvatica over the freeze‐thaw cycle [14, 24]. Herein, histone lysine methylation and demethylation in kidney of R. sylvatica was examined over the freeze‐thaw cycle.
SET enzymes are important in gene expression regulation for their ability to add methyl groups to histones. Methylation alters chromatin packaging, leading to activation or repression of gene expression [27, 28]. For example, SET enzymes are responsible for adding methyl groups onto specific lysine residues on histone H3, with H3K9 methylation associated with gene silencing and the formation of heterochromatin [28]. The SETs examined presently achieve selective regulation of gene expression via chromatin structure modifications, with their dysregulation being associated with pathologies including cancer [29]. Alternatively, differential regulation is seen in response to environmental stress. Herein, ASH2L and RBBP enzymes showed significant downregulation in kidneys from frozen frogs compared to the control and thawed conditions; however, there was no difference between control and thawed conditions, indicating a rapid return to the control physiological state upon thawing. Furthermore, SUV39H1 was upregulated in freezing compared to control, which has been seen previously in a study reporting that KMTs associated with active transcription were mostly downregulated in frozen wood frogs, whereas those linked with gene silencing were sustained [14]. The KMTs involved in the methylation of H3K4, including ASH2L, RBBP5, SETD7, and SETD8 where significantly downregulated during freezing or showed no changes in the relative protein levels. Such downregulation of these enzymes that methylate H3K4 provides further confirmation of their transcriptional activation activities, and, as such, their downregulation suggests that they are transcriptional repressors in the frozen wood frogs.
These findings indicate that SET enzymes play a role in freeze‐tolerance likely by regulating chromatin structure and supporting the expression of genes involved in cryoprotection or other pro‐survival processes. Additionally, the KMTs involved in the methylation of H3K9 were either significantly upregulated during freezing or remained unchanged. SUV39HI, a known transcription suppressor, was significantly upregulated during freeze/thaw, indicating that there is a reduction in energy‐consuming processes like transcription. Further supporting this finding, H3K9me3, while unchanged during freezing, was significantly downregulated in thawed kidneys. Moreover, H3K27 was significantly downregulated during freezing, and H3K36me2, which is known for its recruitment of the HDAC complex Rpd3c to act on histone H4 [30], was downregulated during thawing. H4K20me3 was also significantly downregulated in both frozen and thawed states. The suppression of KDM4, a known transcriptional activator that is involved in demethylating repressive marks (H3K9 and H3K36), during freezing further highlights the transcriptional repression occurring in kidneys under frozen conditions.
H3K9me3/me2 and H3K36me3/me2 are considered to be transcription repressive marks and removal of their methyl groups increase gene expression [31, 32]. Decreased gene expression is expected during freezing to support MRD until normal conditions are reestablished. Herein, expression levels of histone marks exhibited a range of changes over the freeze‐thaw cycle. H3K9me3, H3K27me1/3, H3K36me2, and H4K20me3 showed strong and significant decreases under both frozen and thawed conditions, compared with controls. H3K9me3 is a well‐known histone modification associated with transcriptional repression [10], and its significant decrease after thawing, but not during freezing, suggests a dynamic regulation of gene expression in response to thawing. This observation aligns with the need for rapid transcriptional reactivation of genes that are crucial for post‐thaw cellular repair and recovery. The unchanged levels of H3K9me3 during freezing may also help maintain MRD, potentially conserving energy by keeping non‐essential genes silenced.
H3K27me1, which is associated with active gene transcription, showed a significant decrease during both freezing and thawing, highlighting a role for this histone mark in suppressing gene activity. By contrast, H3K27me3, a mark known for gene silencing, exhibited a slight reduction during freezing and a significant decrease after thawing. These differential changes suggest that H3K27me1 and H3K27me3 are crucial for modifying the transcriptional landscape over the freeze‐thaw cycle. The repression of H3K27me1 may reflect a strategic downregulation of metabolic activity, whereas the reduction of H3K27me3 upon thawing likely facilitates the reactivation of genes essential for recovery. The significant decrease in H3K36me2 levels during thawing, but not during freezing, indicates that transcription elongation processes are downregulated during recovery from freezing. Unchanged H3K36me2 levels during freezing suggest that transcription elongation is sustained during this period, possibly to ensure essential renal functions. The substantial reduction upon thawing points to shifting cellular priorities, with transcriptional resources potentially redirected toward immediate repair and survival mechanisms rather than elongation of existing transcripts. The significant reduction in H4K20me3 during both freezing and thawing also highlights its role in chromatin relaxation and DNA damage response. H4K20me3 has been implicated in genome stability and heterochromatin organization [33], with the significant decrease seen presently suggesting that H4K20me3 may be downregulated to allow for a more accessible chromatin structure, facilitating repair processes during freezing and promoting genomic stability during thawing. This aligns with the need to repair potentially freeze‐damaged DNA and reestablish chromatin organization post‐thaw.
It is important to note that while the aforementioned histone marks showed significant changes, other histone protein residues (H3K4me1/2/3, H3K36me3, H3K79me3, and H4K20me1) did not exhibit significant alterations across all conditions. This suggests that these marks are less regulated by the freeze‐thaw cycle in R. sylvatica kidney. Increased H3K36me2 may be associated with activation of genes involved in energy metabolism and the stress response, whereas reduced H3K27me3 and H4K20me3 may allow for the expression of genes involved in tissue repair and regeneration. Overall, these changes in lysine methylation during freeze/thaw suggest a dynamic epigenetic response to environmental and physiological stresses that contribute to freezing survival. Previous studies further support these data, with levels of H3K9me3 and H3K36me2 found to be maintained in skeletal muscle of R. sylvatica [14].
Furthermore, several KDMs showed significant alterations during freezing with KDM4A, KDM4C, and KDM5C all significantly reduced. KDM4A and KDM4C are associated with downregulation of H3K9me3 and H3K36me2/me3 [34], with KDM4C demethylating overexpressed H3K9 [35]. Frozen frogs showed suppressed KDM4C protein compared to control, but rebounding during thawing. This suggests that another energy‐consuming process that is highly linked to aggressive cancers is significantly repressed in frozen animals [35, 36]. Additionally, regulation of KDMs can lead to altered transcriptional activation of specific genes. For instance, KDM4A and KDM4C are both involved in the regulation of genes associated with cell differentiation and development, whereas KDM5B has been linked to the regulation of genes involved in metabolism, immune response, and cell cycle control [34, 37]. All of these cellular processes are energy‐consuming, and, therefore, it is not surprising that these lysine demethylases are suppressed to preserve pro‐survival activities during freezing when intracellular energy supply is low.
5. Conclusion
Overall, this study reports significant changes in KMTs, KDMs, and associated histone methyl marks in R. sylvatica kidneys in response to the freeze‐thaw cycle. The redistribution of depleted energy resources before freezing is crucial for survival, achieved through transcriptional control during hypometabolism. The relative expression of histone methylation and demethylation provides a framework for downregulating non‐essential pathways while sustaining key pathways. Adequate knowledge of the molecular/biochemical mechanisms underlying kidney adaptation to freezing in wood frogs holds great significance not only for basic biological research but also for conservation biology and human medicine. While our results reveal general histone modification trends, future studies focusing on the specific genes and pathways regulated by these histone marks are needed.
Ethics Statement
All animals were cared for in accordance with the guidelines of the Canadian Council on Animal Care and experimental procedures had the prior approval of the Carleton University Animal Care Committee (protocol #106935).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Supplementary File S1. Western immunoblot antibody information.
Supplementary File S2. Relative expression data for all western immunoblot targets.
Acknowledgments
We thank J.M. Storey for the editorial review and T. Bloskie for assistance in data collection/analysis. This study was funded by an NSERC Discovery Grant to KBS (RGPIN‐2020‐04733). S.A.B. holds an NSERC CGS‐D.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Lee R. E., Costanzo J. P., Davidson E. C., and Layne J. R., “Dynamics of Body Water During Freezing and Thawing in a Freeze‐Tolerant Frog (Rana sylvatica),” Journal of Thermal Biology 17, no. 4–5 (1992): 263–266, 10.1016/0306-4565(92)90064-M. [DOI] [Google Scholar]
- 2. Storey K. B. and Storey J. M., “Freeze‐Tolerance in Animals,” Physiological Reviews 68, no. 1 (1988): 27–84, 10.1152/PHYSREV.1988.68.1.27. [DOI] [PubMed] [Google Scholar]
- 3. Morin P. and Storey K. B., “Mammalian Hibernation—Differential Gene Expression and Novel Application of Epigenetic Controls,” International Journal of Developmental Biology 53, no. 2–3 (2009): 433–442, 10.1387/IJDB.082643PM. [DOI] [PubMed] [Google Scholar]
- 4. Storey K. B. and Storey J. M., “Metabolic Rate Depression and Biochemical Adaptation in Anaerobiosis, Hibernation and Estivation,” Quarterly Review of Biology 65, no. 2 (1990): 145–174, 10.1086/416717. [DOI] [PubMed] [Google Scholar]
- 5. Zhang J. and Storey K. B., “Akt Signaling and Freezing Survival in the Wood Frog, Rana sylvatica ,” Biochimica et Biophysica Acta (BBA)‐General Subjects 1830, no. 10 (2013): 4828–4837, 10.1016/J.BBAGEN.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 6. Black J. C., Van Rechem C., and Whetstine J. R., “Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact,” Molecular Cell 48, no. 4 (2012): 491–507, 10.1016/J.MOLCEL.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kouzarides T., “Chromatin Modifications and Their Function,” Cell 128, no. 4 (2007): 693–705, 10.1016/J.CELL.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 8. Holbert M. A. and Marmorstein R., “Structure and Activity of Enzymes That Remove Histone Modifications,” Current Opinion in Structural Biology 15, no. 6 (2005): 673–680, 10.1016/J.SBI.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 9. Hyun K., Jeon J., Park K., and Kim J., “Writing, Erasing and Reading Histone Lysine Methylations,” Experimental & Molecular Medicine 49, no. 4 (2017): 324, 10.1038/EMM.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Igolkina A. A., Zinkevich A., Karandasheva K. O., et al., “H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 Histone Tags Suggest Distinct Regulatory Evolution of Open and Condensed Chromatin Landmarks,” Cells 8, no. 9 (2019): 1034, 10.3390/CELLS8091034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai Y., Zhang Y., Loh Y. P., et al., “H3K27me3‐Rich Genomic Regions Can Function as Silencers to Repress Gene Expression via Chromatin Interactions,” Nature Communications 12, no. 1 (2021): 719, 10.1038/s41467-021-20940-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sterling J., Menezes S. V., Abbassi R. H., and Munoz L., “Histone Lysine Demethylases and Their Functions in Cancer,” International Journal of Cancer 148, no. 10 (2021): 2375–2388, 10.1002/IJC.33375. [DOI] [PubMed] [Google Scholar]
- 13. Bloskie T. and Storey K. B., “Epigenetics of the Frozen Brain: Roles for Lysine Methylation in Hypometabolism,” FEBS Letters 596, no. 16 (2022b): 2007–2020, 10.1002/1873-3468.14440. [DOI] [PubMed] [Google Scholar]
- 14. Hawkins L. J. and Storey K. B., “Histone Methylation in the Freeze‐Tolerant Wood Frog (Rana sylvatica),” Journal of Comparative Physiology B 188, no. 1 (2018): 113–125, 10.1007/S00360-017-1112-7. [DOI] [PubMed] [Google Scholar]
- 15. Tessier S. N., Luu B. E., Smith J. C., and Storey K. B., “The Role of Global Histone Post‐Translational Modifications During Mammalian Hibernation,” Cryobiology 75 (2017): 28–36, 10.1016/J.CRYOBIOL.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 16. Watts A. J. and Storey K. B., “Hibernation Impacts Lysine Methylation Dynamics in the 13‐Lined Ground Squirrel, Ictidomys tridecemlineatus ,” Journal of Experimental Zoology Part A: Ecological and Integrative Physiology 331, no. 4 (2019): 234–244, 10.1002/JEZ.2259. [DOI] [PubMed] [Google Scholar]
- 17. Bloskie T., Taiwo O. O., and Storey K. B., “Reversible Histone Modifications Contribute to the Frozen and Thawed Recovery States of Wood Frog Brains,” Biomolecules 14, no. 7 (2024): 839, 10.3390/BIOM14070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J. and Storey K. B., “RBioplot: An Easy‐to‐Use R Pipeline for Automated Statistical Analysis and Data Visualization in Molecular Biology and Biochemistry,” PeerJ 4, no. 9 (2016): 2436, 10.7717/PEERJ.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bansal S., Luu B. E., and Storey K. B., “MicroRNA Regulation in Heart and Skeletal Muscle Over the Freeze‐Thaw Cycle in the Freeze‐Tolerant Wood Frog,” Journal of Comparative Physiology B 186, no. 2 (2016): 229–241, 10.1007/S00360-015-0951-3. [DOI] [PubMed] [Google Scholar]
- 20. Dawson N. J., Katzenback B. A., and Storey K. B., “Free‐Radical First Responders: The Characterization of CuZnSOD and MnSOD Regulation During Freezing of the Freeze‐Tolerant North American Wood Frog, Rana sylvatica ,” Biochimica et Biophysica Acta (BBA)‐General Subjects 1850, no. 1 (2015): 97–106, 10.1016/J.BBAGEN.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 21. Li Y., Minic Z., Hüttmann N., Khraibah A., Storey K. B., and Berezovski M. V., “Proteomic Analysis of Rana sylvatica Reveals Differentially Expressed Proteins in Liver in Response to Anoxia, Dehydration or Freezing Stress,” Scientific Reports 14, no. 1 (2024): 15388, 10.1038/S41598-024-65417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rehman S., Varma A., Gupta A., and Storey K. B., “The Regulation of m6A‐Related Proteins During Whole‐Body Freezing of the Freeze‐Tolerant Wood Frog,” Biochemistry and Cell Biology 101, no. 1 (2023): 77–86, 10.1139/BCB-2022-0164. [DOI] [PubMed] [Google Scholar]
- 23. Singh G. and Storey K. B., “TXNIP Shuttling: A Key Molecular Link in Regulating Inflammation and Mitochondrial Dysfunction in Freeze‐Tolerant Wood Frogs,” Gene 857 (2023): 147184, 10.1016/J.GENE.2023.147184. [DOI] [PubMed] [Google Scholar]
- 24. Bloskie T. and Storey K. B., “DNA Hypomethylation May Contribute to Metabolic Recovery of Frozen Wood Frog Brains,” Epigenomes 6, no. 3 (2022a): 17, 10.3390/EPIGENOMES6030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Varma A. and Storey K. B., “Hepatic Citrate Synthase Suppression in the Freeze‐Tolerant Wood Frog (Rana sylvatica),” International Journal of Biological Macromolecules 242, no. pt. 1 (2023): 124718, 10.1016/J.IJBIOMAC.2023.124718. [DOI] [PubMed] [Google Scholar]
- 26. Wade S., Hadj‐Moussa H., and Storey K. B., “mRNA m6A Methylation in Wood Frog Brain Is Maintained During Freezing and Anoxia,” Journal of Experimental Zoology Part A: Ecological and Integrative Physiology 339, no. 3 (2023): 325–334, 10.1002/JEZ.2681. [DOI] [PubMed] [Google Scholar]
- 27. Kouzarides T., “Histone Methylation in Transcriptional Control,” Current Opinion in Genetics & Development 12, no. 2 (2002): 198–209, 10.1016/S0959-437X(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 28. Xiao B., Jing C., Wilson J. R., et al., “Structure and Catalytic Mechanism of the Human Histone Methyltransferase SET7/9,” Nature 421, no. 6923 (2003): 652–656, 10.1038/NATURE01378. [DOI] [PubMed] [Google Scholar]
- 29. Michalak E. M. and Visvader J. E., “Dysregulation of Histone Methyltransferases in Breast Cancer—Opportunities for New Targeted Therapies?,” Molecular Oncology 10, no. 10 (2016): 1497–1515, 10.1016/J.MOLONC.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li B., Jackson J., Simon M. D., et al., “Histone H3 Lysine 36 Dimethylation (H3K36me2) Is Sufficient to Recruit the Rpd3s Histone Deacetylase Complex and to Repress Spurious Transcription,” Journal of Biological Chemistry 284, no. 12 (2009): 7970–7976, 10.1074/JBC.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keogh M. C., Kurdistani S. K., Morris S. A., et al., “Cotranscriptional Set2 Methylation of Histone H3 Lysine 36 Recruits a Repressive Rpd3 Complex,” Cell 123, no. 4 (2005): 593–605, 10.1016/J.CELL.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 32. Peters A. H. F. M., Mermoud J. E., O'Carroll D., et al., “Histone H3 Lysine 9 Methylation Is an Epigenetic Imprint of Facultative Heterochromatin,” Nature Genetics 30, no. 1 (2002): 77–80, 10.1038/NG789. [DOI] [PubMed] [Google Scholar]
- 33. Nelson D. M., Jaber‐Hijazi F., Cole J. J., et al., “Mapping H4K20me3 Onto the Chromatin Landscape of Senescent Cells Indicates a Function in Control of Cell Senescence and Tumor Suppression Through Preservation of Genetic and Epigenetic Stability,” Genome Biology 17, no. 1 (2016): 158, 10.1186/S13059-016-1017-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hung K. H., Woo Y. H., Lin I. Y., et al., “The KDM4A/KDM4C/NF‐κB and WDR5 Epigenetic Cascade Regulates the Activation of B Cells,” Nucleic Acids Research 46, no. 11 (2018): 5547–5560, 10.1093/NAR/GKY281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee D. H., Kim G. W., Yoo J., et al., “Histone Demethylase KDM4C Controls Tumorigenesis of Glioblastoma by Epigenetically Regulating p53 and c‐Myc,” Cell Death & Disease 12, no. 1 (2021): 1–14, 10.1038/s41419-020-03380-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Young L. C. and Hendzel M. J., “The Oncogenic Potential of Jumonji D2 (JMJD2/KDM4) Histone Demethylase Overexpression,” Biochemistry and Cell Biology 91, no. 6 (2013): 369–377, 10.1139/bcb-2012-0054. [DOI] [PubMed] [Google Scholar]
- 37. Kang M., Mehrazarin S., Park N. H., and Wang C. Y., “Epigenetic Gene Regulation by Histone Demethylases: Emerging Role in Oncogenesis and Inflammation,” Oral Diseases 23, no. 6 (2017): 709–720, 10.1111/ODI.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File S1. Western immunoblot antibody information.
Supplementary File S2. Relative expression data for all western immunoblot targets.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


