Abstract
Evaluation of the parameters such as tumor microenvironment (TME) and tumor budding (TB) is one of the most important steps in colorectal cancer (CRC) diagnosis and cancer development prognosis. In recent years, artificial intelligence (AI) has been successfully used to solve such problems. In this paper, we summarize the latest data on the use of artificial intelligence to predict tumor microenvironment and tumor budding in histological scans of patients with colorectal cancer. We performed a systematic literature search using 2 databases (Medline and Scopus) with the following search terms: ("tumor microenvironment" OR "tumor budding") AND ("colorectal cancer" OR CRC) AND ("artificial intelligence" OR "machine learning " OR "deep learning"). During the analysis, we gathered from the articles performance scores such as sensitivity, specificity, and accuracy of identifying TME and TB using artificial intelligence. The systematic review showed that machine learning and deep learning successfully cope with the prediction of these parameters. The highest accuracy values in TB and TME prediction were 97.7% and 97.3%, respectively. This review led us to the conclusion that AI platforms can already be used as diagnostic aids, which will greatly facilitate the work of pathologists in detection and estimation of TB and TME as instruments and second-opinion services. A key limitation in writing this systematic review was the heterogeneous use of performance metrics for machine learning models by different authors, as well as relatively small datasets used in some studies.
Keywords: Colorectal cancer, Systematic review, Tumor microenvironment, Tumor budding, Artificial intelligence
Introduction
One of the most important problems in oncopathology is the accuracy of diagnosis. A correct diagnosis determines the prognosis and the most effective patient treatment trajectory (PTT). TNM classification system is used as a standard for stratification of colorectal cancer patients into prognostic groups. However, survival rate is heterogeneous within the same tumor stages. This compels us to use additional predictive markers.1,2 Among the diagnostic methods, biopsy is a widely used and recommended procedure for cancer diagnosis. Histopathological evaluation serves as the basis for the definitive diagnosis of CRC, and routine tissue sections stained with hematoxylin and eosin (H&E) are indispensable for predicting patient life expectancy and choosing treatment tactics. There are several significant prognostic factors that a pathologist should pay attention to when studying tissue specimens. This causes considerable difficulty among pathologists and results in a lack of consensus on interpretations of these parameters. One of these factors is a detailed assessment of the tumor microenvironment (TME) and the presence of tumor buds. Tumor budding (TB) is a single tumor cell or a cell cluster consisting of 4 tumor cells or less. TB is counted on H&E and is assessed in 1 hotspot (in a field measuring 0.785 mm2) at the invasive front. There are peritumoral budding (PTB, tumor buds at the tumor front) and intratumoral budding (ITB, tumor buds in the tumor center). PTB could only be evaluated by intestinal resection specimens, and ITB could also be evaluated by biopsy. ITB assessment in pre-operative biopsies could help to select patients for neo-adjuvant therapy and could potentially predict tumor regression.2 TB is an independent predictor of rapid local spread, predictor of lymph node metastases, predictor of survival in stage II CRC, and worsening patient prognosis.2,3,4 Tumor budding is a phenomenon resulting from the loss of differentiation, uncoupling, and acquisition of more aggressive features of cells in the invasive tumor margin.2,5 In this regard, the International Tumor Budding Consensus Conference decided to include TB in guidelines and protocols for colorectal cancer reporting.2
TME is characterized by a complex interaction between tumor cells, immune cells as well as stromal cells. This interaction, among other things, determines the malignant potential, the rate of progression, and the ability to control the cancer development in each patient.6,7 The TME includes parameters such as lipid microenvironment,8 tumor cells, blood vessels, the extracellular matrix, fibroblasts, lymphocytes, bone marrow-derived suppressor cells, signaling molecules,9 myxoid stroma, and evaluating the tumor–stroma ratio (TSR). Adipocytes as the main component of the lipid microenvironment can act as energy providers and metabolic regulators. Thus, lipid microenvironment is able to promote the proliferation, invasion, and resistance to therapy of CRC.8 Accumulation of fibroblasts and myofibroblasts in tumor tissue leads to excessive production of collagenous extracellular matrix—desmoplasia. Desmoplasia is also connected with poor prognosis and resistance to therapy. Rapid proliferation of tumor cells leads to ischemia and hypoxia. Ischemic and hypoxic cancer cells secrete vascular endothelial growth factor (VEGF). Angiogenesis promotes the migration of tumor cells into blood vessels, metastasis, and poor prognosis. Tumor cells can induce normal fibroblasts to differentiate into cancer-associated fibroblasts (CAF). CAF secrets variety of growth factors, chemokines, cytokines, and metalloproteinases. It stimulates tumor growth, angiogenesis, invasion, and metastasis. Lymphocytes are the main immune cells of tumors—T- and B cells inhibit tumor growth.9 On the contrary, tumors with extensive stromal component have poor prognosis. TSR is an independent poor prognostic factor in stage I–III CRC.10 High myxoid stromal ratio connected with worse prognostic outcomes in CRC.11 Currently, therapeutic targets for CRC are inhibition VEGF (antiangiogenic therapy), inhibition immune checkpoint (restore immune function lymphocytes), adoptive cell therapy (utilizing the immune cells to achieve anti-tumor effects). There are therapeutic targets such as tumor-derived exosomes therapy, cancer vaccine, and oncolytic virus therapy. However, the listed issues require further study.9 Additionally, it is worth saying that a salient feature of mesenchymal CRC is the accumulation of hyaluronan and it predicts poor survival. Several parameters can be used to assess TME. In reviewed studies, we found that peritumoral stroma (PTS) score,12 deep stroma score,13 adipocytes,8 CD8+ T cells and tumoral spatial heterogeneity,14 tumor infiltration phenotypes (TIPs),15 tumor-infiltrating lymphocytes (TILs),10,17 and tumor–stroma ratios (TSR)17,18 were used. AI technologies have enormous potential for use in assessing and evaluating TME. Computer vision can automate or semi-automate this process and also provide a second opinion for physicians.
Since a reliable objective morphological conclusion is crucial for the following stratification of patients and optimal treatment trajectory, an automated method that increases the precision and objectivity of a morphological study would successfully fill this niche. The advent of validated gold-standard for CRC diagnosis would significantly increase the detection rate and standardization, making pathomorphological conclusions more objective. In recent years, artificial intelligence capabilities have been successfully used to solve such problems.19,20 AI analysis of a digital image can reduce the time spent by pathologists on CRC diagnosis and eliminate or at least reduce the differences in TB scores occurring among pathologists.21
In this study, we analyzed how artificial intelligence technologies are used to identify TME and TB and how well is this topic covered in modern literature.
Materials and methods
Search strategy, inclusion criteria
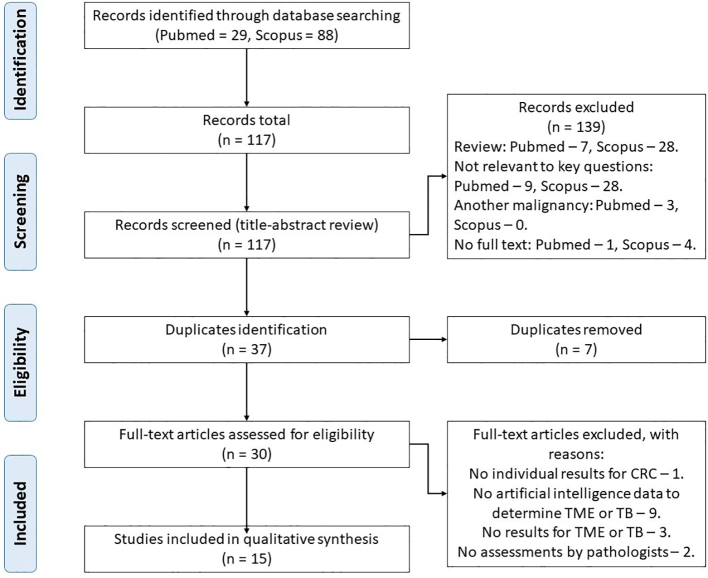
This systematic review followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (see PRISMA statements, Fig. 1). The detailed search strategy and review protocol has been published in Prospero (ID CRD42022337237). The scope of the review according to PICO process (Patient, Intervention, Comparison, Outcomes) is as follows:
Fig. 1.
PRISMA statement.
P − patients with colorectal cancer, in whom artificial intelligence was used for the identification of tumor microenvironment and tumor budding.
I − identification of tumor microenvironment and tumor budding.
C − tissue histology.
O − identification of tumor microenvironment and tumor budding using artificial intelligence.
We performed a systematic literature search using 2 databases (Medline (PubMed) and Scopus) with the following search terms: ("tumor microenvironment" OR "tumor budding") AND ("colorectal cancer" OR CRC) AND ("artificial intelligence" OR "machine learning" OR "deep learning"). At first, 2 authors (OL and AK) independently reviewed headings and abstracts to exclude irrelevant publications such as reviews, comments, papers in languages other than English, and articles which dealt with other malignancy. In case of disagreement between the authors, articles were retained for the following step of selection. Then, the same 2 authors excluded articles that were focused on artificial intelligence data without identifying TME or TB. In case of disagreement, OL and AK sought to justify their decision to resolve the disagreement. In case of an unsuccessful attempt, a senior researcher (TD) made the final decision. Our systematic review includes all original articles from 2018 to August 2022 containing data on artificial intelligence for TME and TB identification in colorectal cancer. The following raw data were extracted manually from the articles: number of patients, methodology of TME and TB identification, sensitivity, and specificity of a method. The experts of the Laboratory for Medical Decision Support Based on Artificial Intelligence analyzed AI technologies described in the articles based on the presented metric values including accuracy, sensitivity, and specificity, if they were stated in the studies. As a result, key features of AI technologies, the most effective methods and approaches to their development, as well as their areas of application for TB and TME identification were defined.
Risk of bias and applicability concerns of the included studies were assessed independently by 2 authors (OL and AK) using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool. In case of disagreement between the authors, a senior researcher (TD) made the final decision.
The primary outcome was evaluation of the artificial intelligence applicability and metrics for tumor microenvironment and tumor budding identification in CRC.
The secondary outcome was the analysis of used AI models’ architectures in tumor microenvironment and tumor budding identification in CRC.
Results
After applying all selection criteria, the final sample included 15 articles related to the CRC. 7 of them investigated tumor budding16,21, 22, 23, 24, 25, 26 and 9 tumor microenvironment.8,10,12, 13, 14, 15, 16, 17, 18 All studies were retrospective. The levels of evidence articles were assessed using the Oxford Center for Evidence-Based Medicine scale: 1b13,23; 2b.8,10,12,14, 15, 16, 17, 18,21,22,24, 25, 26 The median number of CRC cases across all studies was 307. Most frequently, the study was performed with H&E stain.8,10,12,13,16,18,22,25 However, some studies have also included immunohistochemical (IHC) methods. TB was assessed by antibodies to pan-cytokeratin (PanCK),23,26 CK8-18,21 and cytokeratin.24 TME was assessed by antibodies to Ki67, CD3,15,17 CD4,15 and CD8.14,15,17 According to the QUADAS-2 scale, the risk of bias in all studies was assessed as low.
There was no uniformity in the reported results of predicting TB and TME using AI.
This article provides a systematic review of publications on the use of artificial intelligence technologies to identify the tumor microenvironment and tumor budding in colorectal cancer. Selected studies were published between 2018 and 2022 and reflect current trends in AI application, machine learning, and deep learning. All work focuses on the development of machine learning models to identify and evaluate certain biomarkers for diagnosing and predicting the development of colorectal cancer. Information about the models described in the analyzed articles is presented in Tables 1 (TB) and 2 (TME).
Table 1.
Applications and performance of AI algorithms in tumor budding diagnosis of colorectal cancer.
| Article | Year | Aim | Neural network type | Neural network model | Data | Biopsies/resection specimens | CRC stage | Sample size | AUC | Sensitivity | Specificity | Accuracy | Precision | Mean IoU |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Banaeeyan R. et al. Tumor Budding Detection in H&E-Stained Images Using Deep Semantic Learning | 2020 | Tumor bud detection and segmentation | CNN | SegNet | CRC tissue slides stained with H&E | – | – | Training (4 slides) Testing (1 slides) |
– | – | – | – | – | 0.49 |
| Bergler M. et al. Automatic detection of tumor buds in pan-cytokeratin stained colorectal cancer sections by a hybrid image analysis approach | 2019 | Tumor bud detection | CNN | AlexNet | CRC tissue slides stained with PanCK | – | – | Training (51 slides, 27 980 tumor buds), Validation (18 slides, 4362 tumor buds), Testing (18 slides, 8169 tumor buds) | – | 0.934 | – | – | 0.977 | – |
| Bokhorst J. M. et al. Automatic detection of tumor budding in colorectal carcinoma with deep learning | 2018 | Tumor bud detection | CNN | VGG16 | CRC tissue slides stained with H&E and CK8-18 | – | – | Training (36 images, 194 buds), Validation (14 images, 73 buds), Testing (10 images, 38 buds) | – | – | – | – | – | – |
| Lu J. et al. Development and application of a detection platform for colorectal cancer tumor sprouting pathological characteristics based on artificial intelligence | 2022 | Tumor bud detection | Faster R-CNN | VGG16 | CRC tissue slides stained with H&E | – | – | Training (600 slides), Validation (400 slides), Testing (400 slides) | 0.96 | 0.94 | 0.83 | 0.89 | 0.855 | – |
| Fisher N. C. et al. Development of a semi-automated method for tumour budding assessment in colorectal cancer and comparison with manual methods | 2022 | Tumor bud detection | – | Binary threshold classifier built within open software QuPath | CRC tissue slides stained with H&E and CK | – | II, III | 186 TMA cases | – | – | – | – | – | – |
| Pai R. K. et al. Development and initial validation of a deep learning algorithm to quantify histological features in colorectal carcinoma including tumour budding/poorly differentiated clusters | 2021 | Evaluation of tumour budding and poorly differentiated clusters | CNN | – | CRC tissue slides stained with H&E | – | I–IV | H&E slides of 230 carcinomas | – | – | – | – | – | – |
| Weis C. A. et al. Automatic evaluation of tumor budding in immunohistochemically stained colorectal carcinomas and correlation to clinical outcome | 2018 | Tumor bud detection | CNN | MatConvNet (CNN-toolbox in MATLAB) | CRC tissue slides stained with H&E and PanCK | – | – | 20 whole tissue slides | – | – | – | – | 0.88 | – |
Abbreviation: TME – tumor microenvironment, TB – tumor budding, AUC – area under the curve, CNN – convolutional neural network, CRC – colorectal cancer, H&E – hematoxylin and eosin, CK – cytokeratin.
Table 2.
Applications and performance of AI algorithms in tumor microenvironment diagnosis of colorectal cancer.
| Article | Year | Aim | Neural network type | Neural network model | Data | Biopsies/resection specimens | CRC stage | Sample size | Sensitivity | Specificity | Accuracy | Precision | Recall |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kather J. N. et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study | 2019 | Segmentation for tissue classes: adipose tissue, background, debris, lymphocytes, mucus, smooth muscle, normal colon mucosa, cancer-associated stroma, and CRC epithelium | CNN | VGG19, AlexNet, SqueezeNet v. 1.1, GoogLeNet, Resnet50 | CRC tissue slides stained with H&E | – | – | 100 000 image patches from 86 H&E slides, Training (70%), Validation (15%), Testing (15%) | – | – | >0.94 | – | – |
| Kwak M. S. et al. Deep convolutional neural network-based lymph node metastasis prediction for colon cancer using histopathological images | 2021 | Segmentation for tissue classes: normal colon mucosa, stroma, lymphocytes, mucus, adipose tissue, smooth muscle, and colon cancer epithelium | CNN | U-Net | CRC tissue slides stained with H&E | – | I–III | 100 000 image patches, Training (80%), Validation (10%), Testing (10%) | 0.677 | – | ≈0.85 | – | – |
| Lin A. et al. Deep learning analysis of the adipose tissue and the prediction of prognosis in colorectal cancer | 2022 | Segmentation for tissue classes: tumor and non-tumor sections | CNN | VGG19 | CRC tissue slides stained with H&E | – | – | >100 000 image patches | – | – | 0.973 | – | – |
| Gong C. et al. Quantitative characterization of CD8+ T cell clustering and spatial heterogeneity in solid tumors | 2019 | Detection CD8+ T cells and quantify the spatial heterogeneity | – | – | CRC tissue slides stained with CD8 | – | – | 29 slides | – | – | – | 0.881 | 0.742 |
| Jakab A., Patai Á. V., Micsik T. Digital image analysis provides robust tissue microenvironment-based prognosticators in patients with stage I-IV colorectal cancer | 2022 | Evaluation of tumor-stroma ratio | Machine learning–based algorithm | SlideViewer software 2.4 version and its QuantCenter module | CRC tissue slides stained with H&E | Resection specimens | I–IV | 185 slides | 0.671 (tumor epithelium) 0.646 (stroma) |
0.865 (tumor epithelium) 0.783 (stroma) |
0.80 (tumor epithelium) 0.724 (stroma) |
– | – |
| Failmezger H. et al. Computational tumor infiltration phenotypes enable the spatial and genomic analysis of immune infiltration in colorectal cancer | 2021 | Identification of spatial tumor infiltration phenotypes | Lasso regression model | – | CRC tissue slides stained with Ki67, CD3/CD4, CD3/CD8 | Resection specimens | – | 80 slides | – | – | – | – | – |
| Pai R. K. et al. Development and initial validation of a deep learning algorithm to quantify histological features in colorectal carcinoma including tumour budding/poorly differentiated clusters | 2021 | Segmentation for tissue classes: carcinoma, stroma, mucin, necrosis, fat, and smooth muscle | CNN | – | CRC tissue slides stained with H&E | – | I–IV | – | 0.88 | 0.73 | – | – | – |
| Yoo S. Y. et al. Whole-Slide Image Analysis Reveals Quantitative Landscape of Tumor–Immune Microenvironment in Colorectal CancersTIME Analysis via Whole-Slide Histopathologic Images | 2022 | Evaluation of tumor-infiltrating lymphocytes and tumor-stroma ratio | Random forest classifier | – | CRC tissue slides stained with CD3, CD8 | Resection specimens | II, III | Training cohort (260 slides), Discovery cohort (590 patients), Validation cohort (293 patients). | – | – | – | – | – |
| Zhao K. et al. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer | 2020 | Evaluation of tumor-stroma ratio | CNN | VGG19 | CRC tissue slides stained with H&E | Resection specimens | – | Training set (283100 slides), Test set 1 (6300 slides), Test set 2 (22500 slides) | 0.728 | – | 0.9746 | – | – |
Abbreviation: TME – tumor microenvironment, TB – tumor budding, AUC – area under the curve, CNN – convolutional neural network, CRC – colorectal cancer, H&E – hematoxylin and eosin, CK – cytokeratin.
TB assessment
In this study, we summarized the data on the existence prediction of TB and predictive value of TB scores (if these data were provided to the authors).
TB assessment using convolutional neural networks (CNN) in the analyzed articles was performed by various methods. Banaeeyan et al. in their study report an average intersection over union of 0.11 for TB, with mean and weighted IoU 0.49 and 0.83 respectively ("the IoU metric measures the number of pixels common between the target and prediction masks divided by the total number of pixels present across both masks").22 Bergler et al. report 0.068 and 0.967 for precision and sensitivity of TB estimated by AI respectively. At the same time, the minimum number of false-negative and false-positive results during data validation were 144 and 58 131, respectively.23 In their study, Bokhorst et al. achieved a maximum score of F1-score of 0.36, with a recall of 0.72 when compared to the hand-annotated TB.21 Performance measures for TB assessment in Lu et al. study were the following: accuracy rate—0.89, precision—0.855, sensitivity—0.94, specificity—0.83, and negative-predictive value—0.933. The sensitivity of AI model in assessing TB was 0.94, which was slightly lower than that of pathologists (0.9765), however, this correlation was not statistically significant (P=.031).25 When comparing automatic TB assessment with the manual one, Weis et al. achieved an R2-value of 0.86.26
TME assessment
In this study, we consider such indicators of TME as the lymphocytes,8,12, 13, 14, 15 tumor-infiltrating lymphocytes (TILs),16,17 lipid microenvironment,8 cancer-associated stroma,8,13 tumor–stroma ratio (TSR),10,17,18 peritumoral stroma (PTS),12 and tumor infiltration phenotypes (TIPs).15
TME assessment using neural networks was also performed by various methods. According to Kather et al., neural networks precisely define tissue classes, such as adipose tissue, background, debris, lymphocytes, mucus, smooth muscle, normal colon mucosa, cancer-associated stroma, and CRC epithelium. The overall identification accuracy was close to 99% on an internal testing set and 94.3% on an external testing set.13 Lin et al. obtained an accuracy of 97.3% when identifying the same classes.8 Kwak et al. also report high quality segmentation of their model, with test mean dice similarity coefficient (DSC) of 0.892. DSC for adipose tissue, lymphocytes, mucus, smooth muscle, normal colon mucosa, stroma, and colon cancer epithelium were 0.938, 0.968, 0.841, 0.732, 0.928, 0.815, and 0.930, respectively.12 Jakab et al. report sensitivity, specificity, and accuracy for software-based tumor assessment of 67.1%, 86.5%, and 80%; stroma recognition—64.6%, 78.3%, and 72.4%, respectively. Moreover, the intraclass correlation coefficient (ICC) of prediction of the tumor–stroma ratio (TSR) between the 2 observers was 0.945; between visual measurements and software it was 0.759. The researchers also note that the ICC values between all areas were comparable for both software and visual measurements. This indicates similar performance of both estimation methods.10 Zhao et al. also trained AI to evaluate TSR and achieved high efficiency. At the same time, the researchers note that a high correlation was achieved between CNN model prediction and the pathologist annotation.18 Pai et al. compared mismatch repair-proficient (MMRP) and mismatch repair-deficient (MMRD) tumors. As a result, differences were found between MMRD and MMRP tumors in percentage of inflammatory stroma (P=.02), mucin (P=.04), and tumor-infiltrating lymphocytes (TILs) per mm2 carcinoma (P<.001).16 Moreover, all parameters were higher in MMRD tumors, which is consistent with our knowledge about the features of this type of tumor.27 As a result of cut-off of >44.4 TILs per mm2, the authors achieved sensitivity of 88% and specificity of 73% in identifying MMRD tumors.16
Failmezger et al. in their study report an accuracy of 85% for computation of tumor infiltration phenotypes.15 CD8+ T cell point estimate and their standard error using AI to be Recall, R (%)=74.2±0.7 and Precision, P (%)=88.1±0.6. Moreover, the level of correspondence between the segmentation outcome and that by manual counts was also assessed. Spearman's rank correlation coefficient was 0.985, which indicates a high degree of consistency of the results.14
Kwak et al. set a new stromal microenvironment parameter—peritumoral stroma (PTS)—to evaluate lymph node metastasis (P<.001). PTS score is calculated as the sum of pixels of stromal tissue within the tumor region boundaries/the sum of pixels of the tumor.12
Yoo et al. in their study proposed an analytic pipeline for quantifying TILs and TSR from whole-slide images obtained after CD3 and CD8 IHC staining. According to these and other TME parameters, the authors divided all cases of CRC into 5 clinicopathologically relevant subgroups, that in the future may allow determining the patient's prognosis without performing a molecular tumor analysis.17
Applications of AI models in CRC diagnosis
In the reviewed studies, the models were aimed at object detection,21, 22, 23,25 segmentation,12,14,22 classification,8,13 and quantification16, 17, 18,26 of various types of tissues and areas of TB. In addition, a number of works resort to the use of AI technologies not only for automated detection of standard biomarkers, but also for identifying their predictive capabilities: it has been shown that a deep neural network can decompose a complex tissue into its constituent parts and give a predictive estimate based on their ratios, which can improve the prognosis of patient survival compared to the staging system of the Union for International Cancer Control (UICC).13 The study of prognostic capabilities in colon cancer was also carried out in relation to lymph node metastases,12 tumor infiltrate phenotypes,15 and TSR.18 IHC stained14,15,17,23,24 or, in most cases, H&E-stained medical images,8,13,16,18,21,22,25 were used as input data. Weis et al. used images obtained by both methods.26 The choice is due to the accumulated volumes of such data: these images are most often used in medical practice for manual cancer diagnosis during the analysis of biopsy and surgical material.
Data preparation methods for AI models
An important stage of research work on the use of AI technologies is to prepare data for analysis by the model. Most often, researchers had several dozens of whole-slide images of tumor tissue, which were subsequently divided into several thousands of smaller images (patches) of a standardized size, usually with an aspect ratio of 1×1.8,12,13,18,21,26 As a result of splitting images into patches, the calculations performed by the model during analysis is simplified, and its speed is increased.8 In order to unify patches with a height and width less than the target size, Kwak et al. supplemented them by mirror reflection.12 To increase the number of training images and, as a result, increase the stability of the network to variations in data sets and improve the results, data augmentation was used in several works. For this, horizontal and vertical image reflection, rotation and scaling,13,21,22 deformation and blurring,21 brightness change,21 and histogram equalization (an image processing method for adjusting contrast)12 were used. Another important step in working with data was the exclusion of inappropriate and distorted images: for example, with tissue folds, torn tissues, or without tumor tissue.8,13
The specific issue in studies was often to determine if an object belongs to the area of tumor budding. In Bergler et al.,23 TB was considered to be an association of 4 or fewer tumor cells, while cells were considered connected if the distance between them was less than the average diameter of a tumor cell. In Bokhorst et al., and Weis et al.,21,26 TB was defined as isolated epithelial cells or their clusters, including not more than 5 such cells, located near the invasive front of the tumor, but not touching the main tumor. In some studies, in addition to the areas of TB, poorly differentiated clusters (PDCs) were also used as detected biomarkers—according to the researchers,16 TB/PDCs measure outperformed both automated counting of TB areas and its manual detection. In Kather et al.,13 the problem of misclassifications between muscle and stroma classes, as well as between lymphocytes and detritus/necrosis was also described. Thus, to improve the quality of the work of AI models, it is necessary to clearly define the criteria for assigning identified objects to each group, and also consider the possibility of creating a larger number of output classes of areas and objects, which will be discussed more specifically in the next subsection.
Types of models and architectural solutions used
The data show that CNNs are most often used for the analysis of medical images8,12,13,16,18,21, 22, 23,26 in different variations. This choice on the part of researchers is due to the fact that CNNs are designed specifically to work with data with a spatial structure which is common to all images. The main advantage is in the ability of CNNs to automatically extract hierarchical features from images of different types on different network layers. The following CNN models were used: AlexNet,23 SegNet,22 U-Net,12 MatConvNet,26 and VGGNet (VGG1621,25 and VGG198,13,18). In the analyzed articles, the architecture of the models used from 826 to 47 convolutional layers.13 Model training did not always occur from scratch: to save resources and time, many studies used models pre-trained on the ImageNet dataset.8,13,18,23 Optimization of the "upper" layers of the network for research purposes was carried out using the transfer learning methods, most frequently used one—stochastic gradient descent with momentum (SGDM),13,18,22 which ensures faster convergence of the model to the optimal value. In Bokhorst et al.,21 stochastic gradient descent with the Nesterov moment was used, which theoretically has somewhat better characteristics, but it is impossible to compare these approaches in practice due to the inconsistency of the model metrics chosen by different researchers.
In Bergler et al.,23 at the first stage, potential tumor growths were detected using classical image processing methods: threshold processing, filtering (median filter), and morphological operations, that were also used in Weis et al.26 This approach allows reducing image noise, removing staining residues, and obtaining a large number of candidate objects. Thus, the classical image processing methods used in the work demonstrated high sensitivity, but poor accuracy. The AlexNet CNN was applied to the obtained results to distinguish between “true-positive” and “false-positive” tumor bud candidates, which significantly increased the accuracy of detecting TB cases, although it led to a slight decrease in the sensitivity. In this study, the AlexNet model’s weights were retrained on all convolutional and fully connected layers, while the pre-trained weights of the ImageNet dataset were used as initialization. This architecture was chosen by Bergler et al. as it has already been successfully aplied for similar tasks.23 The AlexNet model also showed the best accuracy value (97.7%) among all 6 works using this metric and also showed the second-best sensitivity value (93.4%) among the 6 works where this metric was considered.
SegNet model adaptation was applied by Banaeeyan et al.22 to detect and segment tumor buds. The proposed architecture is an encoder–decoder network with a pixel-level classification layer embedded in last layer. It consists of 18 convolutional layers, each one followed by a batch normalization and rectified linear unit layer. Binary TB class probabilities are obtained by feeding the output of the latest decoder layer into a softmax classifier. Dilated convolution was also applied.22 Since standard convolutional networks were mainly intended for image classification problems and not for dense prediction, extended convolution was proposed to fill this gap by systematically aggregating large-scale context information. This solution made it possible to provide an exponential expansion of receptive fields and at the same time avoid losses in image resolution or pixel coverage. Though, this study has significant limitations such as very small sample size (only 58 images from 5 WSI) and assumption that the main tumor front was known for the model.
Model based on the U-Net architecture was described by Kwak et al. for the segmentation of TME-related features, as this architecture was initially proposed to improve the performance of segmentation particularly for biomedical images.12 The image segmentation process was conducted by multi-threshold technique where threshold values were empirically selected. Adam (Adaptive Moment estimation) algorithm was used for cross-entropy loss minimization and the adaptive momentum algorithm was used for smooth convergence. Model training was terminated by researchers when the mean Dice similarity coefficient (DSC) for the validation dataset did not increase by at least 0.1% after 10 additional epochs from its epoch with the best performance. In result, Kwak et al, achieved relatively balanced classification performance for all of 7 observed TME features, as DSC values for those features varies from 0.732 to 0.968.12 The training and test accuracy curves converged on approaching 56 epochs, where training met criterion for termination.
Weis et al. used MatConvNet which is CNN-toolbox in MATLAB to decide if a tumor bud candidate contained a single tumor bud or not.26 They constructed an 8-layer CNN consisting of 2 blocks of a combination of convolutional, rectifier, and pooling layers and a fully connected layer. The resulting CNN was trained for 10 000 epochs with a constant learning rate of 10–5. The overall process of core analysis includes also color deconvolution and k-means clustering, which were also implemented with MATLAB built-in tools.
VGGNet models are most often used in observed studies. Bokhorst et al. developed VGG-like network with 2 configurations: with 2 output classes (background and peritumoral budding) and with 3 output classes (background, peri-, and intratumoral budding).21 L2 regularization was applied because of the relatively small amount of data (total of 60 images) and related risk of overfitting. For training, an adaptive learning rate scheme was used, where the learning rate was initially set to 0.00001 and then multiplied by a factor of 0.7 after every 5 epoch if no increase in performance was observed. Procession of hard-negative mining also was performed, which contain objects of negative classes and, as they were more demonstrative for the model the model training process became more efficient. The use of this technique has significantly reduced the number of false-positive cases carried out by the network. As a result, the best results were shown by the model with 3 output classes and hard-negative mining. Even with any other modifications of the models, the one that had 3 rather than 2 output classes showed more effectiveness.
Lu et al.25 used one of the modifications of CNNs, Faster Regional Convolutional Neural Network (Faster R-CNN) on the basis of VGG-16 model. This architecture is formed by the fusion of Region Proposal Network (RPN) and Fast R-CNN technologies, where the first allows determining hypotheses, and the second is used to process them and identify class boundaries. This enables to use less data to extract high-precision information about the image, their effective recognition and further segmentation. By selecting the appropriate parameters and increasing negative examples in the samples, the retraining of Faster R-CNN models can be reduced, and the accuracy of object detection can be improved.25 For the same tasks, the artificial addition of negative cases was used by Lu et al., where tumor budding areas containing non-cancerous tissue cells were deliberately selected and labeled as negative cases. Even when using smaller amounts of training data, training results were better for samples with a 6:4 positive-to-negative feature ratio.
This problem can be also solved by increasing the output classes of the network, according to which the identified objects are distributed. Lin et al.8 where Lin et al. achieved the best value of accuracy (97.3%) among the reviewed articles, the sections of histological images were completely divided into 9 output categories. Thus, we can conclude that a moderate increase in the number of output classes has a beneficial effect on the efficiency of the model since it allows the algorithms to more accurately classify the identified objects according to their classes. For that purpose, Lin et al. constructed a classifier on the basis of VGG19 model through transfer learning. Kather et al.13 Zhao et al. also used VGG19 model for which final classification layer was replaced by a 9-category layer corresponding to 9 tissue classes.
In some studies, several CNN architectures have been tested simultaneously. Kather et al.,13 evaluated the effectiveness of 5 models for classification problems at the same time: AlexNet, SqueezeNet, GoogLeNet, Resnet50, and VGG19. The latter architecture showed the best result (98.7%) in this study and was used in further experiments, while all other models (except for SqueezeNet) showed a proportion of correctly classified objects above 97%. It should also be noted that the VGGNet models were used in studies with best values achieved for the following metrics: area under the curve and sensitivity in detection problems,25 as well as the proportion of correctly classified objects (accuracy) in classification problems.8 However, due to the uneven availability of metrics for the cited works, it is far too early to claim the absolute advantage of this model.
Even though several studies have suggested that better results can be obtained using advanced neural network models,13 like deep CNN architectures as VGGNet, AlexNet, GoogLeNet, and ResNet, some of observed studies used machine learning algorithms built within medical software like QuPath. Fischer et al. built a semi-automated method for tumor budding assessment based on a (immunopositive/immunonegative) threshold pixel classifier built within QuPath by combining image downsampling, stain separation using colour deconvolution, Gaussian smoothing, and global thresholding within a single step.24 This classifier was used for identification of connective discrete areas of immunopositivity as tumor buds were defined by the area of immunopositivity. Yoo et al. also used QuPath for identification of 2 types of objects by their quantitative features related to shape, intensity, and texture. After manual segmentation, the random forest method developed with R was used to construct classifiers to identify lymphocytes, tumor, and stroma from IHC images.17 Jakab et al. used SlideViewer, digital pathology software, and its QuantCenter module without developing any CNN architectures.10 Though it was still effective enough for tumor tissue and stroma recognition.
In some studies, as in Pai et al.,16 several CNNs (though no information about used CNN model were stated) were used simultaneously to analyze 4 subgroups of tumor tissue characteristics. A total of 3 neural networks segmented the image into 13 regions and 1 more neural network detected objects. The first neural network isolated layers of carcinoma (excluding TB/PDCs), TB/PDCs, stroma, mucins, necrosis, fat, and smooth muscle. The second neural network inside the identified stroma determined its state: immature, mature, or inflammatory. The third neural network segments carcinoma into the categories of low-, high-grade, and cricoid. The fourth neural network detects TILs as objects in the carcinoma layer. Theoretically, TB/PDCs could also be considered as objects in this algorithm; however, due to noticeable differences in size between objects of such a group, they were classified as a tissue region. The more detailed results of the segmentation algorithm had strong associations with adverse pathological features and metastases, demonstrating the potential of deep learning to provide an accurate assessment of CRC histological features. As a result, the estimates of the identified characteristics either contained minor errors or did not contain them at all. All pathologist-evaluators did not reveal consistent errors in the work of the model.16
As an another tactic for the model, Weis et al.26 tested the method of manual counting of TB areas, which involves concentration on the hotspots and absolute counting of TB. However, the researchers did not reveal any association between the obtained results and the actual clinical data. Meanwhile, a significant correlation was shown by the detection of TB areas using spatial clustering. In Failmezger et al.,15 the authors used the Ripley L-function to identify spatial heterogeneity, which identifies the nearest cell neighbors at a certain distance; this made it possible to classify various phenotypes of tumor infiltration. In Gong et al.,14 the HDBSCAN clustering algorithm was used to extract clusters of CD8+ T-lymphocytes, which can identify cluster hierarchies based on the density and distances between objects.
Algorithms and data augmentation methods used in studies also presented in Tables 3 (TB) and 4 (TME).
Table 3.
Used algorithms and methods of data augmentation in tumor budding diagnosis of colorectal cancer.
| Article | Data augmentation methods | Algorithms |
|---|---|---|
| Banaeeyan R. et al. Tumor Budding Detection in H&E-Stained Images Using Deep Semantic Learning | Horizontal reflection, vertical reflection, rotation, and scaling | - Encoder–decoder network with a pixel-level classification layer embedded in its last layer (an adaptation of the original SegNet model); - A total of 18 convolutional layers, each one followed by a batch normalization and rectified linear unit layer. The fully connected layer is not embedded in the architecture; - Every encoder has its own corresponding decoder and therefore the decoder block contains 18 layers. Binary TB class probabilities are obtained by feeding the output of the latest decoder layer into a softmax classifier. |
| Bergler M. et al. Automatic detection of tumor buds in pan-cytokeratin stained colorectal cancer sections by a hybrid image analysis approach | Median-filter to smoothen the image by reducing noise | - Classical methods like thresholding, filtering, and morphological operations for the detection of candidates. - AlexNet model for distinguishment between “true-positive” and “false-positive” tumor bud candidates. |
| Bokhorst J. M. et al. Automatic detection of tumor budding in colorectal carcinoma with deep learning | Random flipping, rotating, elastic deformation, blurring, brightness (random gamma), and contrast changes. | - VGGlike network with 2 configurations: one with 2 output classes (TB versus Background) and one with 3 output classes (TB, TG, Background); - L2 regularization and dropout layers added after the 2nd and 4th max-pool layer; - Multinomial logistic regression objective optimization (softmax), using stochastic gradient descent with Nesterov momentum. |
| Lu J. et al. Development and application of a detection platform for colorectal cancer tumor sprouting pathological characteristics based on artificial intelligence | – | - Faster RCNN developed by fusing the region proposal network (RPN) on the basis of Fast R-CNN; - VGG16 framework for feature extraction, RPN for feature region proposal and Fast RCNN network for boundary box classification and regression; |
| Fisher N. C. et al. Development of a semi-automated method for tumour budding assessment in colorectal cancer and comparison with manual methods | – | - The semi-automated method based on a binary threshold classifier built within QuPath (v0.2.3); - A pixel classifier created in QuPath to identify connective discrete areas of immunopositivity by combining image downsampling, stain separation using colour deconvolution, Gaussian smoothing, and global thresholding within a single step. |
| Pai R. K. et al. Development and initial validation of a deep learning algorithm to quantify histological features in colorectal carcinoma including tumour budding/poorly differentiated clusters | – | - CNNs were trained to segment each of 4 layers; - The first CNN segmented tissue into carcinoma (exclusive of TB/PDCs), TB/PDCs, stroma, mucin, necrosis, fat, and smooth muscle; - The second CNN segmented stroma into immature, mature, and inflammatory; - The third CNN segmented carcinoma into low-, high-grade, and signet ring cell carcinoma; - The fourth CNN identified TILs within the carcinoma layer. |
| Weis C. A. et al. Automatic evaluation of tumor budding in immunohistochemically stained colorectal carcinomas and correlation to clinical outcome | – | - 8-layer MatConvNet CNN model; - Color deconvolution to separate the background and foreground staining; - k-means clustering for thresholding. |
Table 4.
Used algorithms and methods of data augmentation in tumor microenvironment diagnosis of colorectal cancer.
| Article | Data augmentation methods | Algorithms |
|---|---|---|
| Kather J. N. et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study | Image color normalization with the Macenko method, random horizontal and vertical flips of the training images | - The classification layer was replaced and the whole network was trained with stochastic gradient descent with momentum (SGDM); - Evaluated the performance of five different CNN models: a VGG19 model, AlexNet, SqueezeNet version 1.1, GoogLeNet, and Resnet50. VGG19 was the best performance model with suitable learning time. |
| Kwak M. S. et al. Deep convolutional neural network-based lymph node metastasis prediction for colon cancer using histopathological images | Image color normalization with the Macenko method, histogram normalization. | - U-Net architecture; - Adam for minimizing the cross-entropy loss during stochastic optimization; - Adaptive momentum algorithm for smooth convergence. |
| Lin A. et al. Deep learning analysis of the adipose tissue and the prediction of prognosis in colorectal cancer | – | - VGG19 model trained by migration learning; - Constructed a classifier called VGG19CRC to classify tumor and non-tumor sections of colorectal tissue. |
| Gong C. et al. Quantitative characterization of CD8+ T cell clustering and spatial heterogeneity in solid tumors | – | - Software HALO (v2.2.1870.31) from Indica Labs (Corrales, NM) to perform segmentation of digitized pathological images, using the module “Indica Labs–CytoNuclear v1.6.”; - Clustering algorithm Hierarchical DBSCAN (HDBSCAN). R package “largeVis” was used for cluster analysis, in which a variation of the HDBSCAN algorithm is implemented. |
| Jakab A., Patai Á. V., Micsik T. Digital image analysis provides robust tissue microenvironment-based prognosticators in patients with stage I-IV colorectal cancer | – | - Slides were assessed and annotated using SlideViewer software 2.4 version and its 116 QuantCenter module (3DHistech, Budapest, Hungary). |
| Failmezger H. et al. Computational tumor infiltration phenotypes enable the spatial and genomic analysis of immune infiltration in colorectal cancer | – | - A proprietary machine learning algorithm for detection and classification based on color, intensity, texture, object shape; - 1d logistic fused lasso regression model based on each tile’s feature vector; - Ripley’s L function for determination the number of neighbors of another type of cell within a certain distance. |
| Pai R. K. et al. Development and initial validation of a deep learning algorithm to quantify histological features in colorectal carcinoma including tumour budding/poorly differentiated clusters | – | - CNNs were trained to segment each of 4 layers; - The first CNN segmented tissue into carcinoma (exclusive of TB/PDCs), TB/PDCs, stroma, mucin, necrosis, fat, and smooth muscle; - The second CNN segmented stroma into immature, mature, and inflammatory; - The third CNN segmented carcinoma into low-, high-grade, and signet ring cell carcinoma; - The fourth CNN identified TILs within the carcinoma layer. |
| Yoo S. Y. et al. Whole-Slide Image Analysis Reveals Quantitative Landscape of Tumor–Immune Microenvironment in Colorectal Cancers TIME Analysis via Whole-Slide Histopathologic Images | – | - QuPath for analyzing digital pathology images; - Quantitative features related to shape, intensity, and texture were subsequently computed, and exported to R (www.r-project.org) along with manually assigned labels; - K-means–based consensus clustering using the R/Bioconductor ConsensusClusterPlus package; - Support vector machine classifier with Gaussian kernel was constructed using the scikit-learn library of Python. |
| Zhao K. et al. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer | – | - VGG-19 model; - The final classification layer was replaced by a 9-category layer (corresponding to 9 tissue classes). - Training with stochastic gradient descent with momentum (SGDM). |
Discussion
The use of AI methods significantly reduces the time spent by pathologists. According to Lu et al., it took the pathologists who used the AI model significantly less time (0.03±0.01 s) to make a diagnosis comparing to those using traditional methods (13±5 s) (P<.01). However, there were no statistical differences in the accuracy of detecting benign and malignant lesions between these 2 groups.25
TB assessment
Detection of all TB on H&E-stained whole-slide imaging is a rather difficult task, since tumor buds have a different shape and are very similar to other cells, which greatly complicates their detection and segmentation.22 Peritumoral inflammatory infiltrate, reactive stromal cells on H&E, and apoptotic corpuscles and cellular debris on pankeratin IHC can complicate the TB diagnosis.2 Bokhorst et al. reported that only 20% of the total number of manually detected tumor buds was diagnosed by both pathologists in H&E, and 27% of tumor buds were detected by both pathologists on IHC (CK8-18). For the reference standard, the authors advise choosing tumor buds that are chosen by the majority of votes in a group of specialists.21 These data show the urgent need for reliable mechanisms of assessing TB.
Fisher et al. state that 4 times more total bud numbers were detected with CK staining than with the H&E method. The semi-automated method detected 10 times more TB than the H&E method and 3 times more than the CK method. The result is statistically significant (P < .0001). However, researchers note the problem of false-positive results with the semi-automated method, which is associated, firstly, with the designation of any discrete CK-positive area as a bud, regardless of the signs taken into account during its manual assessment. The second reason is the designation of tumor cells accumulated in the gland lumen as a bud. To solve this problem, the authors tried to fill the gland lumen in QuPath; however, when staining was not circumferential, such glands were mistaken TB by the semi-automated method.24 Lu et al. note a similar problem: the high sensitivity of the AI model leads to a decrease in specificity. The authors name the main reasons for false-positive results, when other structures were mistaken for buds, and a low overlap rate due to the large area of malignant lesions obtained by the AI model segmentation, including some benign areas.25
The number of spatial clusters of tumor buds (budding hotspots) was highly dependent on the presence or absence of lymph node metastases (P = .003 for N0 vs. N1-2). In this regard, the authors conclude that it is the spatial TB clustering in hotspots (and especially the hotspots number) and not the absolute TB number, suitable for assessing TB.26 Pai et al. suggest using TB/PDCs as a tool for assessing TB since the algorithm measures of this parameter showed stronger correlations with lymph node metastasis and distant metastasis (P=.004) than with TB (P=.04) and TB/PDC counts (P=.06).16
Lu et al. argue that it is possible to sacrifice some specificity to increase the sensitivity of cancer detection.25
TME assessment
Anti-tumor immunotherapy treatment has been successfully used to treat patients with CRC in recent years. However, the number of positive responses to therapy fluctuates, and for a more accurate selection of therapy, it is necessary to study TME in detail.15 Gong et al. found that the number of high-density CD8+ T-cells clusters, especially round and elongated ones is significantly higher in patients who respond well to treatment. In such high-density clusters, most false-negative results occur due to the limitations of segmentation algorithm. However, in these areas cell density is much higher compared to others, despite the underestimation.14 Kather et al. report that misclassifications of structures occurred mostly between lymphocytes and debris/necrosis, since necrosis is often infiltrated by immune cells.13
In their study, Failmezger et al. propose to investigate tumor infiltration phenotypes (TIPs) along with established classifications for CRC, e.g., by consensus molecular subtypes or the MSI status. There is 2 TIPs: TIP inflamed represent patterns where tumor cells and immune cells are co-clustered and TIP excluded represent patterns where immune cells are segregated from tumor cells. However, the authors note that the role of TIP as a prognostic or predictive biomarker has yet to be explored.15 Kwak et al. in turn suggest using the peritumoral stroma score ([stromal tissue within the tumor region boundaries]/[tumor area]) as a prognostic biomarker to assess lymph node metastases.12
Jakab et al. suggest evaluating invasive front and whole tumor areas along with hotspot areas because these patterns are independent prognostic factors for overall survival (OS). Moreover, carcinoma percentage and carcinoma–stroma percentage are independent predictors of local recurrence and distant metastases.10 Zhao et al. in their study also note the significance of the TSR score for OS prognosis.18 However, Kather et al. report that most frequent structure classification errors occurred between the muscle and stroma classes as was expected since muscle and stroma have a fibrous architecture. This may cause further difficulties in determining TSR.13
Lin and colleagues studied the effect of present adipose tissue (ADI) on CRC prognosis. They found negative feedback between ADI and immune infiltration scores, such as lymphocyte infiltration signature score and TILs regional fraction.8
Analysis of the use of CNNs and deep learning
As it can be seen from the reviewed articles, the conclusion can be made that CNNs and deep learning are relatively accessible tools for diagnosing and predicting the CRC course in patients on the basis of histological images.10,13 Due to the adaptation to the spatial structure of the data, CNNs can solve tissue classification problems better than specialized equipment for histological tissue classification13 and traditional machine learning methods.18 The quality of AI model predictions is comparable to the quality of manual assessment or even higher: implementation of a semi-automated method surpassed manual assessment approximately 2.5 times in tumor clusters detection, and when implementing deep learning algorithms, the estimated potential was even higher.24 Even with a comparable quality of diagnosis by a model and a clinical specialist, the average time required for a model to make a diagnosis based on a single image is far less compared to those for a specialist. In Lu et al.,25 it was determined that detecting a tumor-sprouting model takes an average of 0.03 s, while it takes 13 s for a pathologist to perform the same task. It is worth mentioning that this time does not include time spent for scanning specimens and supposes a comparison of 2 already scanned slides. In Weis et al.,26 the convolution layer accuracy was evaluated by 4 pathologists specializing in the gastrointestinal tract and not involved in algorithm training: all 4 evaluators did not reveal consistent errors in the model.
A common gap in the reviewed articles is the insufficient justification for the choice of model—most often the choice of CNN model is presented as a given. Speaking about specific models, the best results of the metrics given in the studies were shown by the VGGNet model, namely its variations—VGG16, which was used for feature extraction,25 and VGG19, used for the problem of tissue classification8 and tissue–stroma ratio evaluation.18 The number in these model names indicates the number of convolutional layers used which is the only difference between variants of VGGNet models. The main difference of VGGNet from other CNN architectures is smaller receptive field (3×3), which allows the network to focus on more local image details, get more generalizable features and reduce over-fitting. VGGNet models also known for their relative simplicity and uniform architecture which allows them to be widely used in many areas, including digital pathology. Though VGGNet models have huge number of parameters, about 138 million, what can make the calculations quite expensive. Used in Bergler et al.,23 AlexNet is one of the first CNN architectures and a prototype of VGGNet, which inherits the concept of using ReLU (Rectified Linear Unit) activation functions from AlexNet. In comparison to VGGNet, it has a larger receptive field size in the earlier layers (11×11 and 5×5) and smaller receptive field size (3×3) in the later layers. In the same time, AlexNet also contains a large number of parameters (about 62 million); it lacks scalability and is not optimized for parallel calculations.
In Bergler et al.,23 authors also suggest that using a more advanced model, such as Google's Inception V3, can further improve the classification of tumor clusters, thereby reducing the number of false-positive cases. There was no Inception models used in observed studies, though Kather et al. evaluated the effectiveness of several CNN models for classification including GoogLeNet, which is actually the first version of Inception model with a total of 22 layers. The main innovation of this architecture is the use of inception modules, which use multiple filter sizes in parallel to capture different levels of image information. However, despite the higher computational efficiency of GoogLeNet, which analyzed a dataset of 100 000 images approximately 2 times faster, the VGGNet architecture still showed slightly better accuracy metrics in Kather et al. study.13 Thus, GoogLeNet model was not properly explored later in the article and this gap should be addressed in future studies, as well as exploring the capabilities of Inception models in digital pathology. Later versions of Inception, such as V2, V3, and V4, introduce additional optimizations like factorized convolutions to further reduce the number of parameters significantly, what will also lead to reductions in time and financial costs for CNN calculations. Kather et al.,13 also put forward plans to use a model of the Mask R-CNN type; in Lu et al.,25 the Faster R-CNN network proved to be effective as it shows assessment results similar to manual evaluation by senior pathologists in much shorter time period.
Currently, pathologists are often required to pre-contour areas of TB and confirm diagnostic results with systems despite Yoo et al. study, where fully automated method that requires no manual input on HE-stained WSI was developed, though it does not contain CNN architectures.17 Therefore, AI platforms can be used as diagnostic aids, but, so far, they cannot completely replace pathologists and fully automated workflow for TB detection is still to be developed. In the future, elimination of the limitations and establishment of uniform standards for image coloring methods and metrics for evaluating the effectiveness of AI models will enable to adapt such technologies for its more effective use in practical diagnostics. Nevertheless, even at the current stage of development, AI models may be of value in helping pathologists to diagnose and predict the course of CRC in patients.16,18,25,26
Limitations
The following limiting factors of research were mentioned in the analyzed articles: the insufficient volume of training data sets (medical images with samples of tumor budding areas),13,16,22 labor-intensive work on image labeling, and the human factor in its implementation, the limitations of models to predict only one indicator,8,14 high sensitivity of the models and, as a result, a large number of false-positive cases.14,24,25 As part of the work, some assumptions were also made due to limitations in model operation and the high complexity of their training. These assumptions include the designation of the main tumor front as known,22 and the preferential selection of images with a well-defined disease morphology.12 The global lack of medical laboratories digitalization also makes it difficult to further implement AI technologies due to the incomplete digitization of medical data flows in the laboratories.
A key limitation in writing this systematic review was the heterogeneous use of performance metrics for machine learning models by different authors. More often than others, the traditional concept of object proportion classified correctly by the model (its accuracy) was used—it was encountered in 5 works out of 15 selected.8,10,12,13,25 The sensitivity and precision metrics were used in studies10,16,23,25 and studies14,23,25,26 respectively. The specificity,10,16,25 recall,14 area under the curve,25 and intersection-over-union (IoU)22 metrics were used less frequently. Thus, no unambiguous conclusion can be drawn about the effectiveness of approaches in developing models for identifying TB areas. Also, we had deficient basis to recommend architectures for different future research settings, as very few studies presented information about specimen type and CRC stage and they often differ in the form of description of the data used for models training.
Due to the high heterogeneity in the studies with regards to methodology (heterogeneous use of performance metrics for machine learning models), it was not possible to perform a meta-analysis. Thus, the authors conducted a qualitative narrative synthesis of the obtained data.
Conclusion
To date, AI models may be of value in assisting pathologists in making a diagnosis of CRC when evaluating parameters such as TME and TB. However, modern methods are yet not enough for self-diagnosis without medical supervision. One way to improve the results of AI models can be to increase the amount of input data and the number of output classes. This issue is currently more relevant than ever and requires further investigation, especially in implementing more advanced CNN architectures such as Google Inception versions, which has not yet been done and described in public domain. The other key prospect is a development of fully automated workflows, which will not require manual input to significantly relief pathologists and will help to avoid human factor. A key limitation in writing this systematic review was the heterogeneous use of performance metrics for machine learning models by different authors, as well as a relatively small number of samples used in some studies.
Funding
This work was financed by the Ministry of Science and Higher Education of the Russian Federation within the framework of state support for the creation and development of World-Class Research Centers "Digital biodesign and personalized healthcare" № 075-15-2022-304 and was conducted within the framework of the strategic project “Translational Research in Medicine and Pharmaceutics” of Sechenov University financed by the strategic academic leadership program “Priority 2030”.
Authors
1. Olga A. Lobanova – Corresponding author, Junior Researcher, World-Class Research Center "Digital biodesign and personalized healthcare", Assistant, Institute of Clinical Morphology and Digital Pathology, Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-6813-3374
Scopus ID: 57484405500
WOS Researcher ID: GYU-0505-2022
lobanova_o_a@staff.sechenov.ru
2. Anastasia O. Kolesnikova – student, Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-6893-2109
3. Valeria A. Ponomareva – IT-analyst, LLC “Intelligent analytics”, Moscow, Russia
https://orcid.org/0009-0003-2280-2442
lera.7608@gmail.com
4. Ksenia A. Vekhova – student, Sechenov University, Moscow, Russia
https://orcid.org/0000-0003-0900-4721
Scopus ID: 57810355000
WOS Researcher ID: ABI-5681-2022
vka2002@bk.ru
5. Anaida L. Shaginyan – Lead Data Scientist, Laboratory for Medical Decision Support Based on Artificial Intelligence, Sechenov University, Moscow, Russia
https://orcid.org/0009-0006-1670-7176
WOS Researcher ID: GYU-0480-2022
shaginyan_a_l@staff.sechenov.ru
6. Alisa B. Semenova – Junior Researcher, Laboratory for Medical Decision Support Based on Artificial Intelligence Sechenov University, Moscow, Russia
https://orcid.org/0009-0009-9827-8089
alisa7009@yandex.ru
7. Dmitry P. Nekhoroshkov – IT-analyst, LLC “Intelligent analytics”, Moscow, Russia
https://orcid.org/0009-0004-5746-2887
dm.nekhoroshkov@gmail.com
8. Svetlana E. Kochetkova – student, Sechenov University, Moscow, Russia
https://orcid.org/0000-0003-3542-9723
Researcher ID: GXH-3068-2022
sv.k0ch@yandex.ru
9. Natalia V. Kretova – Junior Researcher, World-Class Research Center "Digital biodesign and personalized healthcare", Pathologist, Institute of Clinical Morphology and Digital Pathology Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-4825-4830
WOS Researcher ID: GXF-9891-2022
kretova_n_v@staff.sechenov.ru
10. Alexander S. Zanozin – Pathologist, Ph.D., Institute of Clinical Morphology and Digital Pathology, Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-3254-3451
zanozin_a_s@staff.sechenov.ru
11. Maria A. Peshkova – Junior Researcher, World-Class Research Center "Digital biodesign and personalized healthcare", Institute of Regenerative Medicine, Sechenov University, Moscow, Russia
https://orcid.org/0000-0001-9429-6997
Scopus ID: 57218920751
WOS Researcher ID: ABI-2496-2020
peshkova_m_a@staff.sechenov.ru
12. Natalia B. Serezhikova – Cand. Sci (Bio), Senior Researcher, Institute of Regenerative Medicine, Scientific and Technological Park of the Biomedical Park, Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-4097-1552
Scopus ID: 57217149258
WOS Researcher ID: AFX-4314-2022
serezhikova_n_b@staff.sechenov.ru
13. Nikolay V. Zharkov – BSc., MSc., Cand. Sci (Bio), Researcher, World-Class Research Center "Digital biodesign and personalized healthcare", biologist, the Institute of Clinical Morphology and Digital Pathology, Sechenov University, Moscow, Russia
https://orcid.org/0000-0001-7183-0456
Scopus ID: 14034896300
WOS Researcher ID: N-2489-2016
zharkov_n_v@staff.sechenov.ru
14. Evgeniya A. Kogan – Pathologist, D.M., Professor, Institute of Clinical Morphology and Digital Pathology, Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-1107-3753
Scopus ID: 7102620822
kogan_e_a@staff.sechenov.ru
15. Alexander A. Biryukov – Head of Laboratory, Laboratory for Medical Decision Support Based on Artificial Intelligence, Sechenov University, Moscow, Russia
https://orcid.org/0000-0003-4102-7835
WOS Researcher ID: ADZ-1723-2022
biryukov_a_a@staff.sechenov.ru
16. Ekaterina E. Rudenko – Pathologist, Ph.D., Senior Lecturer, Deputy Director for Research, Institute of Clinical Morphology and Digital Pathology, Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-0000-1439
Scopus ID: 57202234814
rudenko_e_e@staff.sechenov.ru
17. Tatiana A. Demura – Pathologist, D.M., Professor, Director, Institute of Clinical Morphology and Digital Pathology, Sechenov University, Moscow, Russia
https://orcid.org/0000-0002-6946-6146
Scopus ID: 25936132400
demura_t_a@staff.sechenov.ru
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Chu Q.D., Zhou M., Medeiros K.L., Peddi P., Kavanaugh M., Wu X.C. Poor survival in stage IIB/C (T4N0) compared to stage IIIA (T1-2 N1, T1N2a) colon cancer persists even after adjusting for adequate lymph nodes retrieved and receipt of adjuvant chemotherapy. BMC Cancer. 2016;16(1):460. doi: 10.1186/s12885-016-2446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lugli A., Kirsch R., Ajioka Y., et al. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod Pathol. 2017;30(9):1299–1311. doi: 10.1038/modpathol.2017.46. [DOI] [PubMed] [Google Scholar]
- 3.Trotsyuk I., Sparschuh H., Müller A.J., et al. Tumor budding outperforms ypT and ypN classification in predicting outcome of rectal cancer after neoadjuvant chemoradiotherapy. BMC Cancer. 2019;19(1):1033. doi: 10.1186/s12885-019-6261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swets M., Kuppen P.J.K., Blok E.J., Gelderblom H., Van De Velde C.J.H., Nagtegaal I.D. Are pathological high-risk features in locally advanced rectal cancer a useful selection tool for adjuvant chemotherapy? Eur J Cancer. 2018;89:1–8. doi: 10.1016/j.ejca.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Ueno H., Murphy J., Jass J.R., Mochizuki H., Talbot I.C. Tumour `budding’ as an index to estimate the potential of aggressiveness in rectal cancer: tumour `budding’ in rectal cancer. Histopathology. 2002;40(2):127–132. doi: 10.1046/j.1365-2559.2002.01324.x. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 8.Lin A., Qi C., Li M., et al. Deep learning analysis of the adipose tissue and the prediction of prognosis in colorectal cancer. Front Nutr. 2022;9 doi: 10.3389/fnut.2022.869263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y., Zheng X., Wu C. The role of the tumor microenvironment and treatment strategies in colorectal cancer. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.792691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakab A., Patai Á.V., Micsik T. Digital image analysis provides robust tissue microenvironment-based prognosticators in patients with stage I-IV colorectal cancer. Hum Pathol. 2022;128:141–151. doi: 10.1016/j.humpath.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Hacking S.M., Wu D., Alexis C., Nasim M. A novel superpixel approach to the tumoral microenvironment in colorectal cancer. J Pathol Inform. 2022;13 doi: 10.1016/j.jpi.2022.100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwak M.S., Lee H.H., Yang J.M., et al. Deep convolutional neural network-based lymph node metastasis prediction for colon cancer using histopathological images. Front Oncol. 2021;10 doi: 10.3389/fonc.2020.619803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kather J.N., Krisam J., Charoentong P., et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. Butte AJ, ed. PLOS Med. 2019;16(1):e1002730. doi: 10.1371/journal.pmed.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong C., Anders R.A., Zhu Q., et al. Quantitative characterization of CD8+ T cell clustering and spatial heterogeneity in solid tumors. Front Oncol. 2019;8:649. doi: 10.3389/fonc.2018.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Failmezger H., Zwing N., Tresch A., Korski K., Schmich F. Computational Tumor Infiltration Phenotypes Enable the Spatial and Genomic Analysis of Immune Infiltration in Colorectal Cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.552331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pai R.K., Hartman D., Schaeffer D.F., et al. Development and initial validation of a deep learning algorithm to quantify histological features in colorectal carcinoma including tumour budding/poorly differentiated clusters. Histopathology. 2021;79(3):391–405. doi: 10.1111/his.14353. [DOI] [PubMed] [Google Scholar]
- 17.Yoo S.Y., Park H.E., Kim J.H., et al. Whole-slide image analysis reveals quantitative landscape of tumor–immune microenvironment in colorectal cancers. Clin Cancer Res. 2020;26(4):870–881. doi: 10.1158/1078-0432.CCR-19-1159. [DOI] [PubMed] [Google Scholar]
- 18.Zhao K., Li Z., Yao S., et al. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bera K., Schalper K.A., Rimm D.L., Velcheti V., Madabhushi A. Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16(11):703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niazi M.K.K., Parwani A.V., Gurcan M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20(5):e253–e261. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bokhorst J.M., Rijstenberg L., Goudkade D., Nagtegaal I., van der Laak J., Ciompi F. In: Computational Pathology and Ophthalmic Medical Image Analysis. Stoyanov D., Taylor Z., Ciompi F., et al., editors. Vol. 11039. Springer International Publishing; 2018. Automatic detection of tumor budding in colorectal carcinoma with deep learning; pp. 130–138. (Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 22.Banaeeyan R., Fauzi M.F.A., Chen W., et al. 2020 IEEE Region 10 Conference (TENCON) IEEE; 2020. Tumor budding detection in H&E-stained images using deep semantic learning; pp. 52–56. [DOI] [Google Scholar]
- 23.Bergler M., Benz M., Rauber D., et al. In: Digital Pathology. Reyes-Aldasoro C.C., Janowczyk A., Veta M., Bankhead P., Sirinukunwattana K., editors. Vol. 11435. Springer International Publishing; 2019. Automatic detection of tumor buds in pan-cytokeratin stained colorectal cancer sections by a hybrid image analysis approach; pp. 83–90. (Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 24.Fisher N.C., Loughrey M.B., Coleman H.G., Gelbard M.D., Bankhead P., Dunne P.D. Development of a semi-automated method for tumour budding assessment in colorectal cancer and comparison with manual methods. Histopathology. 2022;80(3):485–500. doi: 10.1111/his.14574. [DOI] [PubMed] [Google Scholar]
- 25.Lu J., Liu R., Zhang Y., et al. Development and application of a detection platform for colorectal cancer tumor sprouting pathological characteristics based on artificial intelligence. Intell Med. 2022;2(2):82–87. doi: 10.1016/j.imed.2021.08.003. [DOI] [Google Scholar]
- 26.Weis C.A., Kather J.N., Melchers S., et al. Automatic evaluation of tumor budding in immunohistochemically stained colorectal carcinomas and correlation to clinical outcome. Diagn Pathol. 2018;13(1):64. doi: 10.1186/s13000-018-0739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams D.S., Mouradov D., Jorissen R.N., et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2019;68(3):465–474. doi: 10.1136/gutjnl-2017-315664. [DOI] [PubMed] [Google Scholar]