Abstract
Cytomorphology evaluation of bone marrow cell is the initial step to diagnose different hematological diseases. This assessment is still manually performed by trained specialists, who may be a bottleneck within the clinical process. Deep learning algorithms are a promising approach to automate this bone marrow cell evaluation. These artificial intelligence models have focused on limited cell subtypes, mainly associated to a particular disease, and are frequently presented as black boxes. The herein introduced strategy presents an engineered feature representation, the region-attention embedding, which improves the deep learning classification performance of a cytomorphology with 21 bone marrow cell subtypes. This embedding is built upon a specific organization of cytology features within a squared matrix by distributing them after pre-segmented cell regions, i.e., cytoplasm, nucleus, and whole-cell. This novel cell image representation, aimed to preserve spatial/regional relations, is used as input of the network. Combination of region-attention embedding and deep learning networks (Xception and ResNet50) provides local relevance associated to image regions, adding up interpretable information to the prediction. Additionally, this approach is evaluated in a public database with the largest number of cell subtypes (21) by a thorough evaluation scheme with three iterations of a 3-fold cross-validation, performed in 80% of the images (n = 89,484), and a testing process in an unseen set of images composed by the remaining 20% of the images (n = 22,371). This evaluation process demonstrates the introduced strategy outperforms previously published approaches in an equivalent validation set, with a f1-score of 0.82, and presented competitive results on the unseen data partition with a f1-score of 0.56.
Keywords: Deep learning, Bone marrow cell subtypes, Cytomorphology, Interpretability, Biomedical image processing
Introduction
Diagnosis of hematopoietic diseases such as leukemia, lymphoma, or anemia, relies on differential bone marrow (BM) cell counting.1 This procedure is carried out by trained pathologists who evaluate cellular characteristics of BM samples, after standardized protocols2 including visual location, identification, and counting of cells, tedious and hardly reproducible tasks, even by trained technicians.3 BM samples are commonly acquired by aspiration or biopsy, both minimally invasive procedures4 and usually obtained from the posterior iliac crest because this site is superficial and rarely complicated.5 The aspiration procedure extracts a sample of BM fluid by a customized needle, whereas biopsy withdraws a sample from the BM solid portion. BM aspiration specimens are important for differential cell counting, cell morphology characterization, and more complex evaluation schemes (cytogenetic analysis, molecular diagnostics, flow cytometry evaluation),4 whereas BM histology biopsy is used to assess BM architecture, i.e., the tissue structure relative to the cellular content, in addition to hematopoietic cellular characterization.6 Recent hyperspectral/multispectral imaging advances provide further reproducible and automatable advantages with respect to BM cell evaluation and differentiation. Such improvements are reached by combining conventional digital imaging and spectroscopy tools,7 with device-independent and reproducible results, hardly obtained in microscopy image analysis.3 By means of Full Spectral Flow Cytometry, InfraRed spectroscopy, and Spectral karyotyping, hematopoietic cell differentiation and characterization become a more relevant/frequent procedures to quantify intrinsic relations between cells and more complex tasks, e.g., leukemia cell detection, treatment response prediction, survival prediction, and minimal residual disease quantification.8 With these advances, spectral analysis has become the gold-standard for cell-based patient management. However, despite such technological improvements, morphology-based cellular classification systems are still the primal diagnosis reference9, mainly due to the high technology dependency of spectral alternatives. Typically, classical microscopy image analysis allows morphological cell subtype characterization in terms of whole-cell/nucleus/cytoplasm characteristics, maturation levels, and cell differentiation stage.10 Nevertheless, as explained before, both the description quality and the specialist experience end up by being bottlenecks because of the large number of possible cell subtypes, the necessary time for the analysis, and the associated inner inter- and intra-observer variability. The most important risk of manually counting cells lies on the possibility of a wrong or missed diagnosis.11
These aforementioned drawbacks have been approached by several artificial intelligence (AI) strategies. These methodologies range from traditional machine learning (ML) algorithms to deep learning (DL) approaches. However, a successful application of these approaches in clinical scenario not only relies in highly accurate classification of particular BM cells, but also in building a model which generates confidence among medical specialists. To this end, model generalizability and interpretability of AI approaches are commonly referenced as complementary conditions to foster prediction trustworthiness.12 Importantly, such conditions are still unmet by training data limitations that closely approximate clinical scenario.
Related work and contribution
In case of classical ML strategies, segmentation, feature extraction, and classification, are the usual pipeline. Segmentation has been approached by thresholding different color spaces like RGB13 or HSV combined with Otsu10 to delineate nucleus and cytoplasm, followed by a refining step with watershed,14 or clustering15 or fuzzy16 algorithms. Segmentation has been also improved by enhancing the object using pixel-gradient magnitude and orientation in combination with a common graph-cut algorithm.17 Likewise, myeloblast has been segmented by a conditional generative adversarial network that generates pix2pix-based cell masks.18 Once potential cells are segmented, features are extracted from them. Most works have used mean intensity values of the segmented region, either red or green channels14 from Lab color space.19 Other representations also add texture descriptors such as gray-level co-occurrence matrix (GLCM),20 Gabor or Fourier features,13 in combination with shape-related cell signatures approximated by morphology features.21 Recently, Hilbert-Huang transform was applied to quantify acute lymphocytic leukemia with an empirical mode decomposition, which reduces feature space dimensionality reduction.22
These features feed classical classifiers to assign samples to one of the available labels, including support vector machines (SVMs),23 particle swarm optimisation SVM,24 random forest (RF),11,25 bagging ensemble,26 and multilayer perceptron.27 These approaches are highly interpretable and have shown competitive results at classifying a limited number of blood cell subtypes, in both public and private databases. Nevertheless, these efforts have focused in discriminating leukemia-associated cells from healthy ones,28 a simplification, quite far from the required population characterization. Lately, these classical handcrafted and ML strategies have been outperformed by developed DL methodologies.
These DL architectures have been adapted for the task of differentiating multiple cellular classes, either a feature extractor along with SVM,29 eXtreme gradient boosting (XGB), RF,30 or a classifier like the one proposed by Thomas et al., with a Visual Geometry Group (VGG) network,31 Residual Network (ResNet) presented by Fan et al., to identify leukocytes in blood smear images,32 or detection transformer models.33 Other DL applications use customized architectures to detect anemia,34 or combinations of deep neural networks like VGG16 and mobileNet,35 Alexnet-GoogleNet-SVM,36 to classify up to five subtypes of cells. Likewise, fusion of randomly generated convolutional neural networks (CNNs)37 classify white blood cells, whereas more compact CNNs have been used to identify particular leukemia types,38 and white blood cells in peripheral blood.39,40 However, these DL approaches and previous ones have been all evaluated with small numbers of BM cell subtypes and few large databases.
Currently, after publication of larger databases, new DL architectures have improved performance results in multiple subtypes, a closer scenario to the herein presented investigation. Particularly, You Only Look Once architecture was introduced to differentiate 15 classes in peripheral blood images,41 training with 18,365 images and reporting a precision of 0.944 and an accuracy of 0.992. Moreover, Multiple Instance Learning for Leukocyte Identification,42 an annotation-free DL strategy assigned labels from weakly supervised models that detected different types of acute leukemia with an AUC of 0.94. These binary classification approaches required few white blood cell subtypes, as well as optimized convolutional networks43 with similar performance (f1-score = 0.94), or by duplet-CNNs44 with better results, i.e., 0.97 in accuracy by separately training gradient boosting algorithm CatBoost (Categorical Boost) and XGBoost (Extreme Gradient Boost).
Other DL methodologies have been applied to differentiate multiple BM cell subtypes. Specifically, Residual Next (ResNeXt) network45 reported an overall f1-score of 0.5946 in the largest BM database. Afterward, HematoNet applied a convolution/attention model to classify 17 cell subtypes out of the available 21 and informed a f1-score of 0.86,47 by using a combination of ConvNet and Transformer (CoAtNet). In the same database, Ananthakrishnan et al.48 explored different strategies to classify the 21 subtypes, including a CNN in combination with SVM and Xgboost, both outperformed by a siamese network which reported an accuracy of 0.84 and f1-score of 0.81 for the validation set. A more recent work explored the InceptionResNetV2 model as backbone of a CNN with ImageNet-based weights over the same cell classes, obtaining a validation accuracy of 0.96 in a single partition test, which also presents relative low precision and recall values, 0.6 and 0.5968, respectively.49 In contrast, a subset of seven cell subtypes were differentiated by a DenseNet121 model with an attention mechanism, obtaining an accuracy rate of 0.97. Other works, in private image collections, inform competitive performances by discriminating a fewer number of classes. Specifically, EfficientNetV2L, as backbone of a CNN with ImageNet-weights,50 reported a mean AUC of 0.78 for 11 cell subtypes, whereas the SqueezeNet architecture reduced the number of parameters by estimating each pixel entropy to classify five white blood cell types with an accuracy of 0.99.51
Although these results look promising, their interpretability is still limited, even after showing activation maps.47,52 While these maps identify which image areas are more relevant, since they do not provide any explanation about how the model uses them, this strategy is still far from being integrated to a clinic line of reasoning or decision.53 Furthermore, the operation of DL prediction is not only a legal and ethical requirement, but this is also crucial towards boosting real DL clinical applications.12
Unlike previously described approaches, the herein introduced methodology improves the performance of DL models when classifying a large population of 21 cell BM subtypes, while it provides extra local/regional interpretable information. These performance and interpretability improvements are achieved by integrating DL networks and pre-extracted cell image features associated with stain response, chromatin and morphology changes, per each predicted class. To this end, engineered shape, color, and texture features are extracted from segmented cell regions (nucleus, cytoplasm, and whole-cell), and arranged in square feature maps named region-attention embedding. Afterwards, this feature arrangement trains a DL network that outperforms state-of-the-art results when differentiating 21 BM cell subtypes, in the largest publicly available database.46 Both ML and DL approaches demonstrate the advantages of engineered features and region-attention embedding when discriminating 21 BM cell subtypes, either by training classical ML algorithms with the feature vector, or feeding ResNet-5054 and XCeption55,56 architectures with the region-attention embedding. Additionally, the most relevant cell features/regions are highlighted by masking the embedding with thresholded DL activation maps, granting interpretability of DL cell subtype prediction. This process increases the level of detail of post-hoc interpretations, and facilitate biological associations to the spatial attention maps.
In summary, the main contributions of the present methodology rely on:
-
1.
A method that classifies BM cell subtypes (21 classes) with state-of-the-art competitive results, by using previously designed DL architectures as backbone in combination with a new region-attention embedding image representation.
-
2.
A thorough experimental comparison of different classification strategies, that demonstrates the aptness of the mixed feature/DL approach to discriminate blood cell subtypes.
-
3.
A fully repeatable evaluation process applied to an open access dataset with the largest published number of labeled BM cell subtypes.46
-
4.
A strategy that enhances DL interpretability and improves DL-based classification of BM cells by engineering feature maps.
Materials and methods
Database
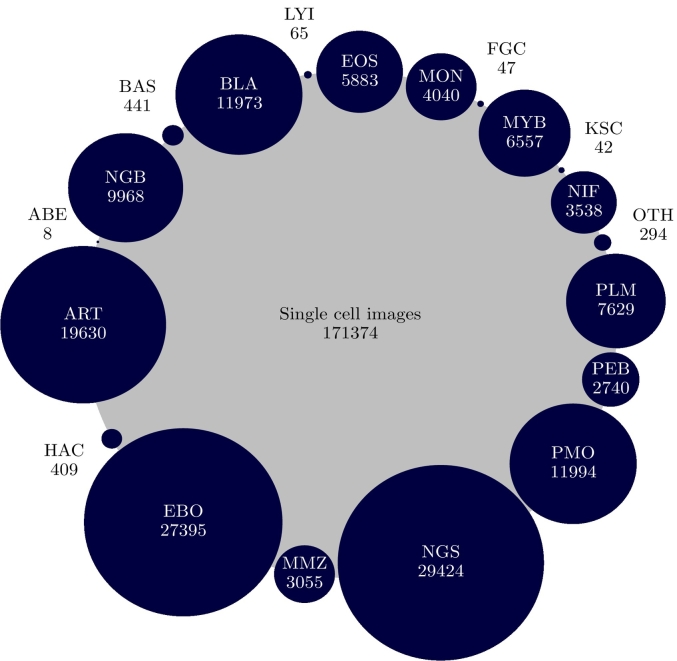
Experiments were conducted with different partitions of the public image database “An Expert-Annotated Dataset of Bone Marrow Cytology in Hematologic Malignancies (Bone-Marrow-Cytomorphology MLL Helmholtz Fraunhofer)”,46 a collection containing 171,374 single-cell images coming from BM smear samples of 945 patients with ages from 18 to 92 years, stained with Grünwald-Giemsa/Pappenheim and diagnosed with different hematological diseases. Single cell images of 250×250 pixels were acquired with a brightfield microscope at ×40 magnification and oil immersion. Each image was annotated by morphologists, providing labels for 21 classes, namely abnormal eosinophil (ABE), artifact (ART), band neutrophil (NGB), basophil (BAS), blast (BLA), eosinophil (EOS), erythroblast (EBO), faggott cell (FGC), hairy cell (HAC), inmature lymphocyte (LYI), lymphocyte (LYT), metamyelocyte (MMZ), monocyte (MON), myelocyte (MYB), not identifiable element (NIF), other cell (OTH), plasma cell (PLM), proerythroblast (PEB), promyelocyte (PMO), segmented neutrophil (NGS), and smudge cell (KSC). As inferred from Fig. 1, the class distribution of this database was highly unbalanced and therefore, a more challenging context for the multi-class differentiation task: whereas only eight images were available for the ABE class, 29,424 images were labeled as NGS.
Fig. 1.
BM cell subtype distribution along the 171,374 images within the used dataset (“An Expert-Annotated Dataset of Bone Marrow Cytology in Hematologic Malignancies”46). The blue/gray circle ratio corresponds to the proportion of each class in the dataset. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Image pre-processing
A first step of this methodology involved identification of the whole-cell boundaries and segmentation of the two main cellular components, i.e., nucleus and cytoplasm. Initially, nucleus segmentation was performed as proposed by Tavakoli et al.,57 who applied different operations to several color spaces, i.e., RGB, HLS, and CMYK. In summary, the whole procedure was:
-
•
Obtaining a color-balanced RGB image.58
-
•
Transforming the color-balanced RGB image to CMYK and HLS color spaces.
-
•
Increasing nucleus contrast by operating K and M channels from the CMYK color space as KM = (Kchannel −Mchannel).
-
•
Reducing the intensities of other image components but the nucleus by keeping minimum intensity M and S channels, from CMYK and HLS, respectively.
-
•
Operating nucleus contrast enhanced matrix KM and non-nucleus element reduction matrix MS according to soft map = MS-KM.
-
•
Applying Otsu thresholding algorithm to the soft map output to set the final segmented nucleus.
Then, the cell was segmented as the difference between the S [saturation] channel from the HLS color space and the Y [yellow] channel from the CMYK color space, from the color-balanced version of each image. Subsequently, the Otsu algorithm was applied to this difference to obtain the segmentation of the whole cell. Finally, the cytoplasm mask was obtained as a XOR operation between the nucleus and whole cell segmentations.
Considering the presented approach is based on supervised steps and color channel operations performed image to image, no further training is required. Nevertheless, to evaluate the performance of this segmentation approach, this was tested with a subset of 20 randomly selected images per cell subtype (except for ABE with only eight images), whose nucleus and cytoplasm were manually segmented. The Dice coefficient and Jaccard index, between manual and automatic segmentations, were computed for the 407 test images. The obtained results showed a reliable nucleus segmentation performance, a Dice coefficient of 0.86 and a Jaccard index of 0.78, while for the segmentation of the cytoplasm the Dice coefficient was 0.71 and the Jaccard index 0.60.
Finally, considering that for feature extraction it is necessary to have the entire region of interest (nucleus, cytoplasm and whole-cell), all images with partial cell presentation were excluded from the experimental subset, thus reducing from the available 171,374 to 111,855 cell images.
Feature extraction
Once cell, nucleus, and cytoplasm were automatically segmented, three different types of features were extracted for each cell region of interest (whole-cell, nucleus, and cytoplasm), including: (i) shape characteristics aiming to capture morphological signatures, (ii) color-based features to describe differences in terms of stain, and (iii) texture features which capture high-frequency changes commonly associated to nuclear chromatin patterns. These feature types were chosen by their known discrimination power, broadly explored in BM cell segmentation,59 classification,25 or both tasks applied in peripheral blood images.57,60
In summary, for each region of interest, eight shape features were extracted, namely convexity, compactness, elongation, eccentricity, roundness, solidity, area, and perimeter. Regarding color quantification, the first five statistic moments were estimated for each channel of the RGB color space, i.e., intensity mean, intensity variance, intensity kurtosis, skewness, and entropy. Furthermore, five texture features (contrast, dissimilarity, homogeneity, energy, and correlation) were also calculated using a GLCM with a neighborhood size of one pixel and orientations varying at 0°, 45°, 90°, and 135°. Additionally, Minkowski–Bouligand dimension features were computed, looking for capturing fractal nature variations of each cell component.61
Finally, all above-described features were concatenated as a feature vector of 144 elements, while a min-max normalization strategy was applied to reduce the range effect of each feature space in the classification process.
Region-attention embedding
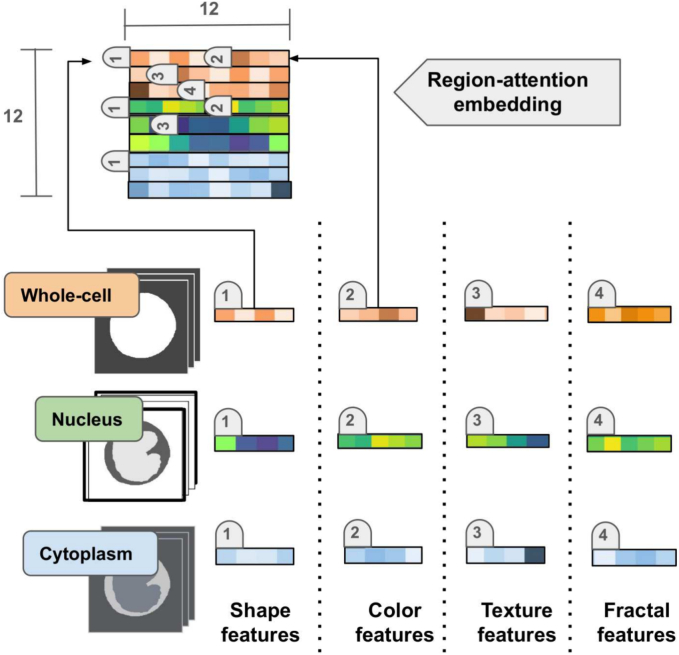
As mentioned in the introduction section, BM cell subtypes are clinically commonly described in terms of whole-cell/nucleus/cytoplasm characteristics, maturation levels, and cell differentiation stage. Likewise, spatial relations defined by the elements within these regions are fundamental to find out conceptual links. Attempting to capture these underlying concepts, the presented methodology introduces a representation herein named the region-attention embedding. The basic idea of this approach was to code spatial cell relations at ordering engineered features within the feature vector after the particular cell region, namely cytoplasm, nucleus, and whole-cell. Specifically, as show in Fig. 2, features were sorted out by cell region within the feature vector (1×144) and then presented to any DL network as a squared matrix (12×12). This particular organization facilitates a convolutional network learns ordinal relations between the different cell components, and makes the network to concentrate on particular relevant features. In contrast to visual transformer, the attention here was not exhaustively learned from millions of connected small patches, but it consists of a much smaller set of units of information with a reduced number of connections, coming from the extracted features and the source regions. The result was then a soft attention learning with much less dependency on large databases. Finally, the region-attention embedding enhances interpretability of DL-based predictions, given the possibility of identifying relevant associations between features and cell regions.
Fig. 2.
Region-attention embedding organization scheme. Numeric labels present the order of each feature vector type (shape, color, texture, fractal) after a particular cell component (whole-cell, nucleus, cytoplasm), within the region-attention embedding.
Classification
After feature extraction, the obtained characteristics were used to differentiate the proposed 21 classes. Such task was carried out by two different classification approaches which used the above-mentioned features in different organizations in combination with ML- and DL-based classifiers.
Regarding the classical ML approaches, different models were obtained with RF and SVM (linear, radial basis function - RBF, and polynomial kernel) classifiers, which were previously compared by Krappe et al.59 for cell subtype differentiation, also explored by Dincic et al.61 for acute myeloid leukemia detection, even in peripheral blood smear,11 and as baseline to be compared with DL approaches.26 Specifically, these models were trained by the above-described features, in a common vectorwise representation. All classifiers were optimized under a grid search scheme to reach the best possible performance under the described conditions.
With respect to the DL strategies, these classifiers were combined with the previously extracted features in a matrix-like presentation. To this end, ResNet-50 and Xception architectures were modified by replacing the input layer with a repeated version of the presented region-attention embedding (12×12×3). These two networks have shown outstanding results in image-related tasks, including white blood cell classification using pre-trained ResNet,62 and pre-trained Xception as feature extractor,63 followed by logistic regression.64 However, in contrast to such applications of these networks, here both of them were used to improve feature filtering given by region-attention embedding.
Xception network
This variant of the Inception architecture replaced classical convolution modules by depthwise separable ones, reducing the number of parameters in the network and improving its efficiency. This DL-network is composed by 36 convolutional layers structured into 14 modules, which are divided into three main stages, as follows55:
-
•
Entry flow: It normally took an input of 299×299×3, but here it was modified to receive an embedding of 12×12×3, which is filtered out by eight convolutional layers, ReLU activation, and max-pooling, which provide reduced equivalent feature maps.
-
•
Middle flow: Three depthwise separable convolution and ReLU activations were applied to the entry flow output. This process was repeated eight times to generate 19×19×728 feature maps.
-
•
Exit flow: Consisted in different units of ReLU activation, depthwise separable convolution, and max-pooling applied four times to the middle flow output, followed by the global average pooling layer.
Finally, an optional fully connected layer with logistic regression was applied to the 2048-dimensional vector to generate the output.
ResNet-50 network
A convolutional network based on a residual blocks, designed to incorporate connections from the first block input to the second block output. These residual blocks resemble subnets, featuring Conv2D and GlobalMaxPooling2D convolution layers along with an activation function. Additionally, this architecture introduces the possibility of linking direct connections between layers. This network comprises 50 layers of residual blocks, consisting of a convolutional layer, 48 residual blocks, and a classifier layer with a small filter of 1×1 and 3×3, all of which use ReLU activation functions.54
Improving DL-interpretability
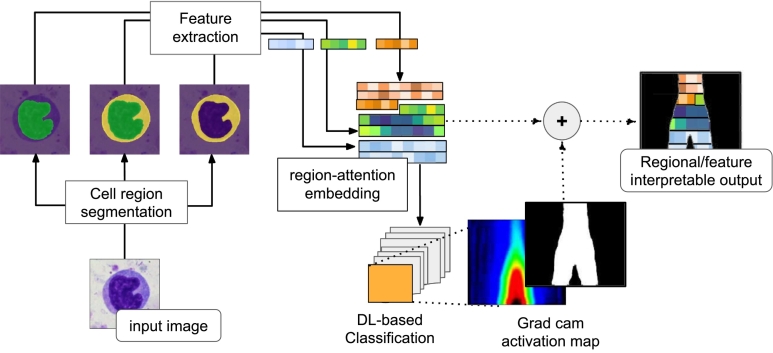
Interpretability mainly refers to the possibility a human operator infers a connection between automatic decisions and identifiable input patterns.65 These connections have become crucial in DL-based medical image analysis, because it is a common requirement for successfully plugging AI models into the clinic.66 However, DL outcome is usually interpreted by highlighting scales of the outcome relevance with activation maps67 which are not conceptual but rather they establish a per patch level of importance.12 In contrast, the analysis herein introduced not only returned the cell region that mostly contributed to the subtype discrimination, but it also quantified the stain response and morphology/chromatin changes within these regions, in terms of the extracted features. As illustrated in Fig. 3, features were selected by thresholding the class activation maps coming from the Grad cam algorithm,68 and superimposing the resulting mask to the correspondent region-attention embedding. Such binary masks were obtained by setting to one (1) all values above 90% of each activation map maximum and zero otherwise. Once the above-described process was applied, the retrieved features were linked to each cell subtype to associate them with stain response variabilities and morphology/chromatin changes. Likewise, in terms of region interpretability, this methodology used the location index of relevant features, which established a connection between a feature and the cell region.
Fig. 3.
DL-prediction interpretability improvement process. This methodology extracted features associated to each cell-region: whole-cell, nucleus, and cytoplasm, and mixed them in a squared matrix with a predefined organization (region-attention embedding). Finally, the strategy identified relevant features per class, and the feature-source cell-region, by using the activation maps provided by Grad cam algorithm.
Overall, these two contributions in terms of local feature relevance and cell region contribution, were aligned with what clinical protocols use to report and characterize each cell subtype, taking into account the content of cellular regions.
Experimental setup and evaluation
Performance of the proposed methodology approach was assessed by two experiments:
Experiment 1: Evaluating the selected engineered features to differentiate the 21 BM cell subtypes by ML strategies
This experiment applied different ML classifiers (RF and SVM with linear, polynomial, and RBF kernels) to evaluate the utility of applying the proposed feature space to discriminate the 21 cell subtypes. Furthermore, this experiment provided a baseline to compare classic classification approaches with DL-based ones.
All classifiers were trained with the whole image set and using the parameters obtained by applying a grid search scheme. In case of RF, parameters were set as follows: 700 trees, gini information criteria, and a maximum of 30 features considered during the search for the optimal split. For the SVM with RBF kernel, the regularization parameter was set to 100, and the stopping tolerance to 0.001. On the other hand, for SVM with a linear kernel, the regularization parameter was chosen to be 10, and the stopping tolerance to 0.001. Likewise, for SVM with a polynomial kernel, the degree was 2, while the regularization parameter and the kernel coefficient were 500 and 0.01, respectively.
Experiment 2: Evaluating DL performance with the region-attention embedding to differentiate the 21 BM cell subtypes
In this experiment the introduced region-attention embedding was combined with ResNet-50 and XCeption architectures to classify the available 21 cell subtypes. Improvement evaluation was performed by using the same data distribution to train/validate both selected network models but using original images and fine tuning on imagenet-based transfer learning. For ResNet-50 and Xception architectures, the batch size was 32 and the stochastic gradient descent, the optimizer. The chosen loss function was the categorical cross-entropy, together with a learning rate of 0.001. Training process, from scratch, consisted of 1865 iterations over 40 epochs for both architectures.
Statistical analysis and evaluation metrics
All classification experiments herein performed for both, ML and DL strategies, were trained and tested by using different data subsets of the above-described database. Particularly, the whole set of images was split in two groups, 80% of the images was used to train and validate the models, and the remaining independent set of 20% to test. This partition scheme follows a stratified strategy in which partitions are set to ensure the same class distribution on both, training/validation and test data groups, i.e., each cell subtype is equally represented in training/validation and testing partitions. Here, training/validation was performed under three iterations of 3-fold cross-validation scheme. This validation strategy was implemented to reduce possible batch effect in the obtained results, and stood for a more thorough evaluation of the presented approach.
The performance of all classifiers was assessed using five evaluation metrics, namely accuracy, precision, recall, f1-score, and specificity. These metrics are reported in terms of macro and weighted performance averaging schemes, for all models obtained in the validation, as well as in the test. These averaging strategies are presented considering that classification performance may be biased by the class distribution differences, so macro averaging is presented assuming all classes equally contributed to the reported averaged metric. Nevertheless, due to the high number of classes, and the large unbalance between them, weighted-averaged is here used as the main reference value in overall classification, because the contribution of each class to the average is weighted by the proportion between the number of images and the whole dataset size.
Results
All herein presented classification experiments were conducted using Python 3.9, along with Scikit-learn for ML and Keras packages with Tensorflow as backend for DL. These experiments were performed on Tesla T4 GPU, where the whole validation scheme took 4 h for ML algorithms, and 9 h for running each DL model.
Experiment 1: Evaluating the selected engineered features to differentiate the 21 BM cell subtypes
As shown in Table 1, the best BM cell subtype classification results were obtained by a SVM with a polynomial kernel, presenting an overall mean weighted f1-score of 0.60. This clearly represented a major advance because these results are comparable with the ones reported in the original database publication, with an f1-score of 0.59.45 Interestingly, other ML classifiers out of SVM-polynomial presented similar results, including SVM-RBF and SVM-linear kernel, and RF, respectively with a weighted f1-scores of 0.60, 0.57, and 0.54, all of which with a low standard deviation (Table 1), demonstrated the robustness of the extracted features for the proposed task. More detailed information, including micro- and macro-validation results, were also included in Table 1, where lower performance is presented due to the class unbalance and limitation of the implemented ML classifiers. Nevertheless, it is important to highlight that all results presented in Table 1, show only training and validation values according to the herein proposed evaluation scheme. Hence, independent testing results by using the best validation model show that SVM-polynomial achieves weighted and macro f1-score of 0.60 and 0.36, respectively, with corresponding weighted accuracy of 0.61 and macro accuracy of 0.35.
Table 1.
Comparison of training/validation performance for different machine learning classifier, while differentiation 21 BM cell subtypes. All classification results are presented in terms of mean and standard deviation (SD), of accuracy, precision, recall, f1-score, and specificity.
| Averaging | Metric | Classification approach |
|||
|---|---|---|---|---|---|
| Random forest | SVM linear kernel | SVM RBF kernel | SVM polynomial | ||
| Macro | Accuracy | 0.27 ± 0.01 | 0.30 ± 0.01 | 0.33 ± 0.01 | 0.33 ± 0.01 |
| Precision | 0.35 ± 0.02 | 0.36 ± 0.02 | 0.37 ± 0.01 | 0.37 ± 0.01 | |
| Recall | 0.27 ± 0.01 | 0.30 ± 0.01 | 0.33 ± 0.01 | 0.33 ± 0.01 | |
| F1-score | 0.28 ± 0.01 | 0.31 ± 0.01 | 0.34 ± 0.01 | 0.35 ± 0.01 | |
| Specificity | 0.98 ± 0.01 | 0.98 ± 0.01 | 0.98 ± 0.01 | 0.98 ± 0.01 | |
| Weighted | Accuracy | 0.56 ± 0.01 | 0.59 ± 0.01 | 0.61 ± 0.01 | 0.61 ± 0.01 |
| Precision | 0.55 ± 0.01 | 0.57 ± 0.01 | 0.59 ± 0.01 | 0.60 ± 0.01 | |
| Recall | 0.56 ± 0.01 | 0.59 ± 0.01 | 0.61 ± 0.01 | 0.61 ± 0.01 | |
| F1-score | 0.54 ± 0.01 | 0.57 ± 0.01 | 0.60 ± 0.01 | 0.60 ± 0.01 | |
| Specificity | 0.95 ± 0.01 | 0.95 ± 0.01 | 0.96 ± 0.01 | 0.96 ± 0.01 | |
Experiment 2: Evaluating DL performance with the region-attention embedding to differentiate the 21 BM cell subtypes
For the sake of clarity, results of this particular experiment were split, on the one hand, a classic classification problem is assessed, while on the other results show the added interpretability of the presented strategy.
Regarding the classification performance at differentiating 21 BM cell subtypes, the obtained validation results of ResNet-50 and Xception networks, in combination with region-attention embedding, were presented in Table 2. In this case, the best performance was achieved by using the region-attention embedding with Xception architecture, which obtained weighted f1-score of 0.82, while ResNet-50 presented lower performance, with weighted f1-score of 0.67. The above-mentioned results outperformed ML classifiers of experiment 1 (weighted f1-score of 0.6 and macro f1-score of 0.33), and previously published DL approaches validated using exactly the same database, like ResNext architecture with reported f1-score of 0.5945, Siamese Network which presented a f1-score of 0.81,48 and Inception-ResNetV2 with f1-score of 0.57.49 Additionally, as shown in Table 2, there is a clear improvement of DL-based classification performance, by using the proposed architectures and region-attention embedding representation together, against the same networks trained with original images, where the best obtained f1-score was of 0.69, with an Xception architecture.
Table 2.
Comparison of training/validation performance for Xception and ResNet50 networks, at differentiating 21 BM cell subtypes. All classification results are presented in terms of mean and standard deviation (SD), of accuracy, precision, recall, f1-score, and specificity.
| Averaging | Metric | Classification approach |
|
|---|---|---|---|
| Xception + Region-attention embedding | ResNet50 + Region-attention embedding | ||
| Macro | Accuracy | 0.66 ± 0.25 | 0.46 ± 0.19 |
| Precision | 0.74 ± 0.25 | 0.55 ± 0.19 | |
| Recall | 0.66 ± 0.25 | 0.46 ± 0.19 | |
| F1-score | 0.69 ± 0.25 | 0.48 ± 0.19 | |
| Specificity | 0.99 ± 0.01 | 0.98 ± 0.01 | |
| Weighted | Accuracy | 0.82 ± 0.15 | 0.67 ± 0.14 |
| Precision | 0.82 ± 0.15 | 0.67 ± 0.14 | |
| Recall | 0.82 ± 0.15 | 0.67 ± 0.14 | |
| F1-score | 0.82 ± 0.15 | 0.67 ± 0.14 | |
| Specificity | 0.99 ± 0.01 | 0.98 ± 0.14 | |
In terms of macro-averaged performance, the obtained results with the combination of region-attention embedding and Xception network also presented overall competitive values when compared to the state of the art (f1-score of 0.69), regardless the highly unbalanced distribution of the dataset, as shown in bold values within Table 2.
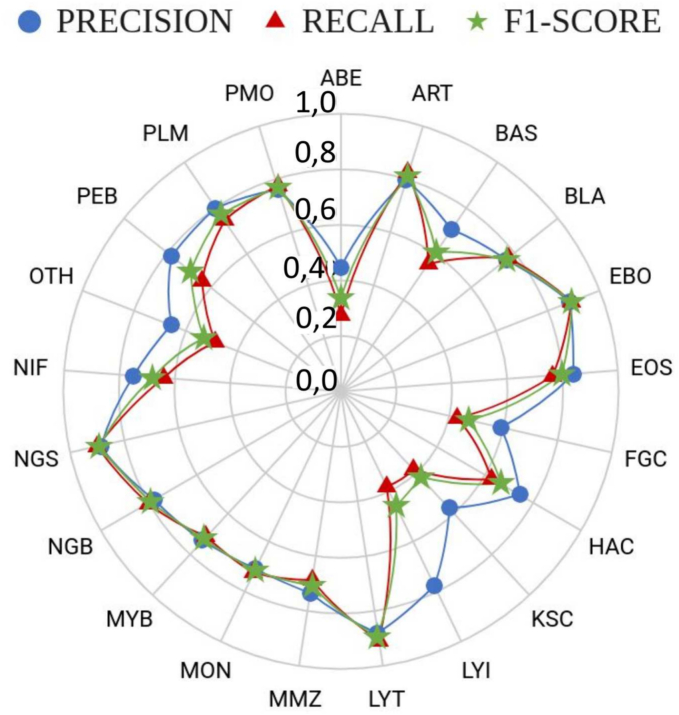
Regarding the testing performance for the two implemented networks in an independent partition group, the inclusion of the proposed region-attention embedding led to a weighted f1-score of 0.56, in combination with Xception network, as presented in Table 3. This outstanding result was obtained with an unseen set of test images, and this was still comparable to what was previously reported in the original database publication but in a training-validation scheme. Additionally, the high overall specificity for validation and testing, shown in Table 2, Table 3, corresponded to the pairwise classification process performed between a particular cell subtype vs. the remained ones, which is highly unbalanced. Furthermore, detailed performance metrics per cell subtype are presented in Fig. 4, where the best testing and validation results stood for the segmented neutrophils, with mean f1-score = 0.89. This is an expected result considering that such cell subtype had the higher number of images in the database (n = 14,649). In contrast, the cell subtype with the lowest performance corresponded to abnormal eosinophil with f1-score = 0.33, which is the class with seven images from a total of 111,855 images in the database.
Table 3.
Comparison of test classification performance in an unseen data partition, for previously obtained Xception and ResNet50 models at differentiating 21 BM cell subtypes, in a unseen set partition. All classification results are presented in terms of mean and standard deviation (SD), of accuracy, precision, recall, f1-score, and specificity.
| Averaging | Metric | Classification approach |
|
|---|---|---|---|
| Xception + Region-attention embedding | ResNet50 + Region-attention embedding | ||
| Macro | Accuracy | 0.32 | 0.30 |
| Precision | 0.34 | 0.34 | |
| Recall | 0.32 | 0.30 | |
| F1-score | 0.33 | 0.31 | |
| Specificity | 0.98 | 0.98 | |
| Weighted | Accuracy | 0.56 | 0.54 |
| Precision | 0.56 | 0.54 | |
| Recall | 0.56 | 0.54 | |
| F1-score | 0.56 | 0.54 | |
| Specificity | 0.95 | 0.95 | |
Fig. 4.
Per-class validation performance of region-attention embedding in combination with Xception network, in terms of precision, recall, and f1-score (more detailed results available in Table A1 within the supplementary material).
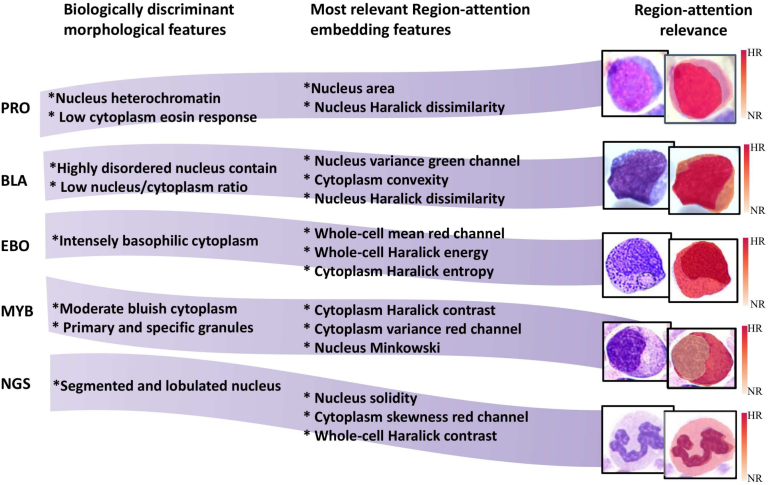
In terms of interpretability of the presented method, the obtained model found out patterns commonly associated with cytological information at a local level, and it also associates them to a defined cell region (nucleus, cytoplasm, and whole-cell). As an evidence of such findings, shown in Fig. 5, the most relevant region-attention embedding features were coupled with biological patterns, reported as clinically discriminant for a subset of five cell subtypes, namely promyelocites, blasts, erythroblast, myelocytes, and segmented neutrophils. Here, interpretability consisted in associating information of a cell subtype prediction with particular stain patterns or chromatine/morphology-related features. In fact, both local changes and the quantified biological characteristics worked as complement of the activation maps (region-based interpretations), which led to region relevance maps as presented in the final column of Fig. 5. Particularly, as shown in the first row of the table in Fig. 5, the nucleus Haralick dissimilarity, nucleus area, and cytoplasm blue channel skewness were more relevant for the promyelocyte class than any other region-attention embedding characteristics (column 2). These features quantified higher nucleus size and local textural heterogeneities associated with hyper-granularity within the nucleus, both reported as a common signature of this cell subtype.69 A similar association for the other cell subtypes will be presented in the discussion section for extending the interpretability advantages of the presented method.
Fig. 5.
Interpretable output of the presented combination of region-attention embedding and Xception network. This table presents a match between the most relevant features within the region-attention embedding for five different BM cell subtypes (column 2), and clinically discriminant morphology features (column 3). Images in the right panel graphically show the cell region where the most relevant features are found, in a heat map scale going from highly relevant (HR) region to a non-relevant (NR) region.
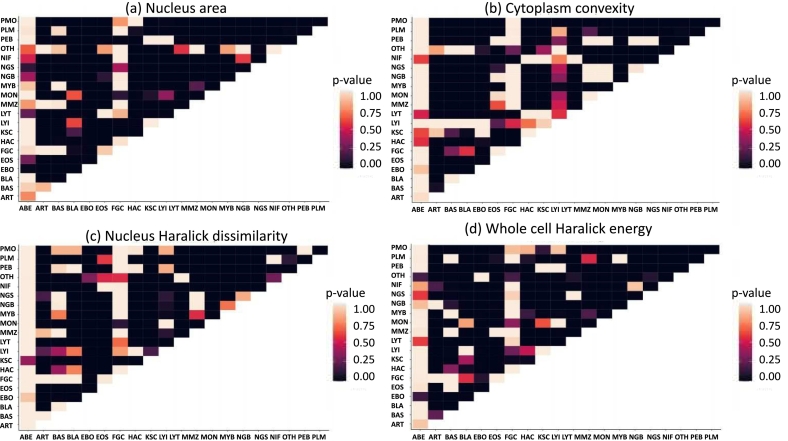
In addition, as shown in Fig. 6, the per class representativity of the obtained relevant features was independently supported by a Tuckey statistic test, which revealed the pairwise cell subtype discriminancy of all available features (n = 144). Specifically, by setting the significance level , the relevance of certain features was demonstrated according to the frequency of feature occurrence for a particular cell class. This occurrence was exemplified in Fig. 6(b), where for the blast class (BLA column), the single feature turned out to be crucial to differentiate this cell subtype against the others, because a large part of the column shows low p-values. This is an important result considering that there is an overlap between them and the features that region-attention-based approach found as relevant, previously presented in Fig. 5 (nucleus Haralick dissimilarity, cytoplasm convexity and mean intensity of blue channel).
Fig. 6.
Feature relevance maps by applying pairwise subtype differentiation based on a Tuckey test, for: (a) nucleus area, (b) cytoplasm convexity, (c) nucleus Haralick dissimilarity, and (d) whole cell Haralick energy. Here, the most significant differences are represented by lower p-values (<0.05), which means the darkest purple matrix points. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Data ablation
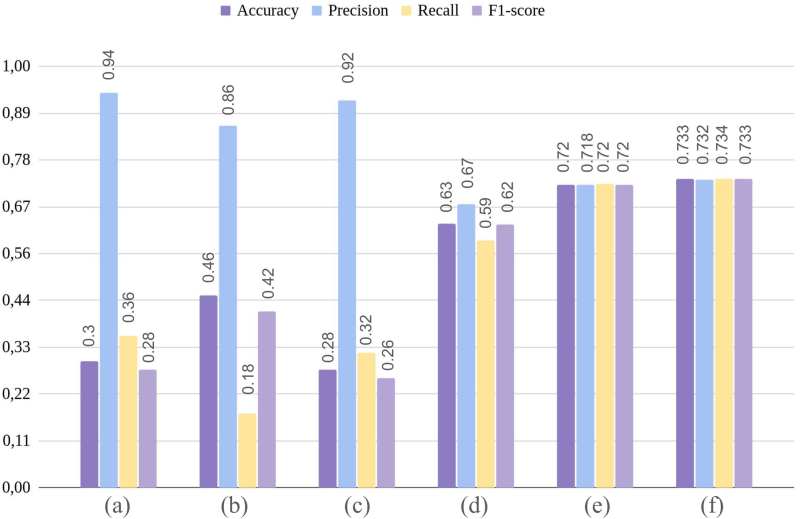
Considering that one of the current challenges in AI is to deal with reduced number of images for training/validating DL based strategies, a data ablation experiment was additionally performed. In this test, the number of training images per cell subtype was consecutively reduced to 1000 and 2000, and a new validation of region-attention embedding was executed when combined with Xception network, in differentiating the 21 classes. This experiment tests the robustness of the introduced approach under variable data size conditions, and it is then compared against the commonly used image-based input for DL classification. To this end, the Xception network was trained/validated by separately using both, original RGB images and their corresponding region-attention embeddings. Specifically, for image-based network feeding, data augmentation was applied by image rotation and flipping, to meet a balanced version of the cell subtypes in terms of the proposed number of examples per-class, n:{1000, 2000}. Additionally, two different training strategies were applied, i.e., using RGB images to train the network from scratch, and using transfer learning with imageNet weights, both under a 40 epocs and fine-tuned training scheme. In contrast, the region-attention embedding approach did not use any data augmentation, because applying rotation and flipping image transformations may led to a biased model given by the rotation invariance of all implemented features within the embedding. As presented in Fig. 7(e,f), Xception network in combination with region-attention embeddings, outperformed the corresponding network version trained/validated with RGB images, when classifying 21 BM cell subtypes. Particularly, by using region-attention embedding with Xception, trained with 1000 and 2000 images per-class respectively, the classification performance achieved weighted f1-score of 0.72 and 0.73, respectively. Under the same data conditions, but using RGB images and ImageNet-based transfer learning, Xception network obtained a f1-score of 0.42 with 1000 images (see Fig. 7(b)), and 0.62 with 2000 images (see Fig. 7(d)), which evidenced no stability when working with such limited data, and supported the advantage of using region-attention embeddings against RGB images. An extended version of the obtained results in these complementary experiments is shown in Table A3 within supplementary material.
Fig. 7.
Performance of Xception network by using images and the presented region-attention embedding, with 1000 and 2000 images per-class. Particularly, the bars present accuracy, precision, recall, and f1-score, for Xception network trained with: (a) 1000 RGB images and imageNet weights, (b) 1000 RGB images from scratch, (c) 2000 RGB images and imageNet weights, (d) 2000 RGB images from scratch, (e)1000 image equivalent region-attention embeddings, and (f) 2000 image equivalent region-attention embeddings.
Discussion
This work has introduced a new BM cell subtype classification strategy built upon engineered features which fed two different DL networks (ResNet-50 and Xception), and outperformed state-of-the art published methodologies at classifying 21 classes from a public database with the largest number of BM cell subtypes. Specifically, the particular organization of the feature assembly, herein called region-attention embedding, set the patterns that contributes the most to discriminate the 21 different classes. This region-attention embedding is assembled in a matrix-like arrangement of the extracted features, setting those from the same cell component are located nearby to preserve regional coherence and to provide explainability.
As presented in Table 4, the introduced region-attention embedding outperforms state-of-the-art approaches at classifying 21 BM cell subtypes. Particularly, these results evidenced a performance improvement when using Xception and ResNet50 in combination with region-attention embedding, rather than a transfer learning approach with original cell images, because it presents an increment of more than 8% points in all metrics. Regarding other published classification strategies, the original database publication reported an overall f1-score of 0.57 by using a tuned ResNext architecture for all the 21 cells, under a simple held-out experimental setup, which is largely outperformed by the presented strategy with f1-score of 0.82. Furthermore, independent validation results of this ResNext were graphically summarized with no specifies, but with overall lower performance values than obtained with the region-attention embedding. In comparison with a more recent strategy by Ananthakrishnan et al.,48 who addressed discrimination of all classes in the same database, the herein introduced approach not only obtain a slightly higher validation accuracy (0.82 vs 0.81), but it also provides a more thorough evaluation setup. Specifically, in that work, the implemented Siamese network was tested in a single run over two partitions of the dataset (training and testing), while with the region-attention embedding evaluation included three iterations of a 3-fold stratified cross-validation, and a test in an unseen data partition, thereby reducing the batch effect possibility. Actually, in similar held-out experimental conditions, the Inception-ResNetV2 by Meem et al.,49 presented accuracy of 0.96, precision of 0.60, and recall of 0.5968, which stand for unbalanced classification performance in 21 cell subtype differentiation. In contrast, the region-attention embedding show highly stable classification metrics, with 0.82 for both, precision and recall. Finally, a more recent work presented by Glüge et al.,70 a thorough evaluation of DL-based classification was introduced by using a large list of networks, all trained/validated with k-fold evaluation scheme. This validation strategy provided similar evaluation advantages to the one herein reported, but with lower overall f1-score of 0.762 ± 0.05 when compared to the region attention embedding that achieves a f1-score of 0.82 ± 0.15.
Table 4.
Comparison of state-of-the-art techniques challenged by classifying 21 bone marrow cell subtypes in the same image database, i.e., “An Expert-Annotated Dataset of Bone Marrow Cytology in Hematologic Malignancies (Bone-Marrow-Cytomorphology MLL Helmholtz Fraunhofer)”.46 The performance of each method is presented in terms of the number of classes, implemented network and experimental strategy for validation, as reported in the correspondent publication. In bold, the validation results obtained by the method presented in this work.
| DL approach | Experimetal configuration | Precision | Accuracy | Recall | f1-score |
|---|---|---|---|---|---|
| Xception (images) Transfer learning | Repeated 3-fold cross-validation | 0.74 ± 0.11 | 0.74 ± 0.18 | 0.66 ± 0.16 | 0.69 ± 0.15 |
| ResNet50 (images) Transfer learning | Repeated cross-validation | 0.55 ± 0.17 | 0.56 ± 0.14 | 0.46 ± 0.22 | 0.48 ± 0.2 |
| ResNext45 Transfer learning | Hold-out | 0.51 | 0.69 | 0.69 | 0.57 |
| Regnet_y_32gf70 pretrained on ImageNet | 5-fold cross-validation | 0.79 ± 0.06 | – | 0.75 ± 0.06 | 0.76 ± 0.05 |
| Siamese Network48 | Hold-out | 0.84 | – | – | 0.81 |
| Inception-ResNetV249 | Hold-out | 0.6 | 0.962 | 0.59 | 0.57 |
| Region -attention embedding + Xception | Repeated cross-validation | 0.82 ± 0.15 | 0.83 ± 0.15 | 0.82 ± 0.15 | 0.82 ± 0.15 |
Other strategies have presented higher BM cell subtype classification performance, but overall tested with lower number of cell classes, or evaluated in private databases. Particularly, Thipathi et al.,47 published HematoNet as a network designed to classify 17 cell subtypes, which was evaluated using the same database that the presented region-attention embedding, but using single held-out partition scheme. HematoNet achieved a f1-score of 0.86, by introducing the CoAtNet, a combination of both ConvNet network and transformer attention layers. However, as informed in the original publication, CoAtNet validation was performed with a single training/testing partition, which stands for an oversimplified representation of the sample space. In contrast, region-attention embedding evaluation avoid biased training/testing process by applying three iterations of a 3-fold cross-validation, and by using the available dataset images, without any data augmentation. In fact, data augmentation, as the most common strategy to increase training images, is also recognized as a process that reduces model generalizability and increases overfitting possibility. In a similar evaluation scheme, Ahmad et al.,71 introduced a white blood cell classification method that uses entropy-controlled Deep Feature optimization to extract the best latent space representation from multiple DL networks. This approach presented an accuracy of 99.6, at differentiating five cell subtypes, which represented a reduced version of the herein addressed task. In terms of large number of classes, the works presented by Hazra et al.56 and Wang et al.72 have shown to succeed in multiple subtype classification, but still using private databases, and fewer cell subtypes. The former with 12 cell subtypes reported a f1-score of 0.89 by using generative adversarial networks and sequential CNN, and the second reported a f1-score of 0.86 at differentiating 19 classes in non-open access image set, and both evaluated using single hold-out evaluation. In contrast, region-attention embedding evaluation includes higher number of classes 21, coming from a large open acces dataset, which stands for a reproducible method and evaluation. Additionally, the presented evaluation scheme minimize bias possibility in both training and testing processes, by applying three iterations of a 3-fold cross-validation, and by using the available dataset images, without any data augmentation. In fact, data augmentation, as the most common strategy to increase training images, is also recognized as a process that reduces model generalizability and increases overfitting possibility.
Although the presented results point out this technology is potentially applicable, a crucial part of this discussion is that classification performance is not enough to ensure translation to the clinical practice. Even though image-based BM cell characterization reaches human-level performance, spectral analysis and morphology evaluation remain the more interpretable techniques at identifying such hematopoietic elements. Importantly, in comparison with those strategies, the herein presented approach is not limited by technology equipment and test panels, and also provides reproducible and interpretable results. These last advantages are fundamental because AI model trust is a condition that depends on the evaluation thoroughness and interpretability of the algorithm outcome.12 In this regard, the herein introduced work provides a more thorough evaluation of the classification task, demonstrating competitive state-of-the-art validation results at classifying the 21 BM cell subtypes, under 3-fold validation scheme and further test partition evaluation. As explained in the experimental setup and evaluation section, validation results were obtained by a cross-validation scheme, in contrast with the hold-out scheme of other publications which may be biased by particular data partitions (batch effect). In addition, as shown in Table 3, another evaluation of this method, which was much less-biased, consisted in classifying an unseen partition, i.e., to set aside the test set from the beginning, resulting in a weighted f1-score of 0.56, which is similar to the f1-score = 0.57 published by the database owners but using a unique validation partition.
Additionally, it should be strengthen out that a crucial factor which improved DL performance in the presented region-attention strategy relied in the important dimensionality reduction executed by the introduced strategy, where a 250×250×3 image input was transformed into 12×12 region-attention embedding. In fact, this low-dimensional space distilled out discriminative image patterns, increasing cell subtype separability before DL-based classification. Furthermore, the embedded spatial information in the input helped DL network to capture high level feature/spatial relations that defined inter-class differences. Particularly, an additional experiment in which feature organization within the proposed embedding was randomly performed, showed a decrease of the 21 class performance, i.e., a f1-score of 0.77. This interesting result suggests it is crucial to keep the spatial feature dependence before the convolutional layers, because this order captures not only local relations between engineered characteristics, but also integrates region/spatial cell information to improve subtype separability. Additionally, a second supplementary experiment that differentiated the 21 BM cell subtypes, by using region-attention embedding and a linear neural network (no convolution layers), also pointed out the importance of DL convolutional layers for the proposed task. Specifically, as presented in the validation results of experiment 2, Xception network with a set of convolutional layers obtained a better validation performance (f1-score = 0.82) than the presented with a linear neural network (f1-score = 0.48). The hypothesis behind these performance differences is that convolutions capture complex spatial relations between pre-extracted engineered features, as a result of the organization of the region-attention embedding. Detailed results of these additional experiments were summarized in Table A2 in the supplementary material.
BM cell subtype prediction interpretability
Presently, DL models have become a real actor in many medical applications, including BM cell classification,73 and leukemia cell detection.74 However, the role of these models as part of the clinical pipeline is still limited by the little-to-none understanding about how a particular prediction is made, because DL strategies have been commonly presented as black boxes.75 In fact, transparency has appeared as an important concept because interpretability is now a law requirement for approval of any DL decision medical system, after the European Union's General Data Protection Regulation law.76 In this context, DL model transparency has been 2-fold tackled, either modifying the architecture of the networks, or providing post-hoc explanation about how prediction is achieved,12 which are commonly based in tools that highlight specific image regions as a measure of relevance.77 Nevertheless, very few of these interpretable approaches have been applied to BM cell subtype differentiation, and currently research is centered on classification rather than explainability of model decision. In contrast, the presented region-attention embedding offered both, highly accurate classification and detailed information regarding most discriminative features/cell-location per subtype. Specifically, as a post-prediction interpretation strategy, the introduced approach use prior biological knowledge as complement for most popular alternatives which used both, latent space explanations and attribution maps. However, rather than projecting high-dimensional latent spaces to two dimensions to give prediction interpretations,78,79 the region-attention embedding allow a direct feature/subtype association, similar to presented by Krappe et al.,25 who used knowledge-based hierarchical tree classifier to differentiate 16 leukemia-related cells from regular ones, in peripheral blood images. But in addition, region-attention embedding enhances outcome explanation in terms of the feature location, which is a further information source working on top of prior knowledge used to constrain the tree. These feature-type/cell–region–source combination presented a complementary advantage, particularly in comparison with filter-relation interpretations for Resnext50 proposed in 2019 by Prelberg et al., that only retrieve low-level relevant features when classifying white blood cells into normal B-lymphoid and malignant B-lymphoblasts.80
As complement, attribution maps offered interpretation by highlighting regions of the input which were relevant for the outcome.81 However, these spatial representation of relevance by itself, usually provided few information about which pattern within salient regions helped the most to the prediction.53 Importantly, this lack of detail within these methodologies is covered by the presented work by using the spatial attribution maps to locate pattern descriptors and locations in a single processing step, without requiring major modifications to backbone network architecture. Particularly, by modifying the DL structure, some methods predicted high-level image concepts (semantic related features) to perform automatic tasks.82 These models, also known as concept learning models, have proved to be more competitive when mixing hidden neurons and freely trained ones extracting features from scratch,83 or by capsuling complex diagnostic concepts in vectored structures instead of using scalar feature maps used in CNN,84 but again missing the detail that herein introduced embedding is providing.
Other works, also explored handcrafted feature explainability to improve DL model transparency by indicating the most relevant patterns defining cell differences, but focused on leukemia cell lineage differentiation, like acute myeloid leukemia by Dincic et al.61 In detail, these authors studied different morphological, fractal, and textural descriptors to indicate the most relevant patterns defining cell differences, but lack on detailed position for such discriminant patterns. For peripheral blood images, a more balanced approach found accurate classification and spatial-location explanation with compact classification model based on a combination of attention layer and CNN blocks, that distinguished promyelocytes from normal leukocytes, in a binary classification that uses attention maps to facilitate model prediction interpretability.38 However, herein implemented approach provides similar spatial awareness for multi-class classification of BM cell population, enhancing subtype discrimination interpretability, far from unspecific DL activation maps presented by Matek et al.,45 and heavy attention maps.47
Unlike the previously published works, the introduced region-attention embedding naturally offers both, local feature and cell region information, identifying whether a DL model prediction is using nucleus/cytoplasm, chromatin changes or cell shape, and discriminating among all possible cell subtypes. To this end, the region-attention engineered features are particularly selected to quantify the above-described biological characteristics. In addition, the feature location within the matrix-like structure allows to track not only the most relevant characteristic, but also the correspondent region of origin. As shown in Fig. 5, the proposed method output includes a list of paired features/cell-regions, on top of the cell subtype prediction, which enables a direct comparison against clinical discriminant features. This option reinforces AI trustworthiness, in contrast with previously works in BM cell subtype classification, which are limited to show gradient based attribution maps. Specifically, Gradcam and SmoothGrad algorithms used by Matek et al.46 and Tripathi,47 show spatial relevance maps that highlight the regions that mostly contribute to label prediction, but nothing about the local patterns within such cell regions. Other approaches have used transformer-based attention to identify relevant regions within the cell, introducing B-cos Vision Transformer and B-cos Swin Transformer to reduce attention map fragmentation and enhance model explainability.85 However, even when these approximations show more detailed relevance maps, they have no description about the particular patterns/features that attention modules are using from the highlighted regions, to reach the cell subtype prediction.
Overall, interpretable information provided by the presented strategy facilitates integration to the clinical workflow, because obtained features are easily associated to biological concepts which medical experts are familiar with. For instance, as shown in Fig. 5, the erythroblast prediction mainly uses the cytoplasm Haralick entropy and whole cell characteristics like mean intensity value of the red channel or Haralick energy. These characteristics describe texture heterogeneity, a concept close to cytoplasm hypergranularity, and low eosin stain response presented in basophilic cytoplasm. Regarding myelocytes, the most discriminative biological features are related with cytoplasm, a condition captured by the method because the frequency of cytoplasm features is higher than the one from nucleus characteristics. This finding is illustrated by the spatial heat map in the myelocyte row within Fig. 5, where the nucleus is marked with a lower relevant value than cytoplasm. In contrast, segmented neutrophil relevance map shows the model uses nucleus features rather than cytoplasm ones. Particularly, nucleus solidity appeared as the most frequent region-attention embedding feature, which describes differences of segmented and lobulated nucleus patterns. Furthermore, the relevance of the region-attention embedding features was also validated by a Tuckey test (see Fig. 6), to determine whether a feature defines statistical differences between pairwise cell subtypes. Specifically, the relevance for some particular features is presented in heat map matrices, where a lower intensity value (black) corresponds to statistical significant differences between two defined classes. For example, as depicted in Fig. 6(a), nucleus area appeared as a feature with statistically significant differences which likely facilitated promyelocyte identification, because PMO row/column showed low p-values when this subtype was compared against the other cell classes. In the case of whole cell Haralick energy as a determinant characteristic, Fig. 6(d) shows that the EBO row/column was one of the very few composed almost entirely by low p-values, i.e., whole cell Haralick energy presented a feature space with promyelocytes separated from other cell subtypes. These quantitative results provide an independent support to the feature relevance obtained with the presented method, because the most frequent characteristics in this Tuckey tables matches the selection by the region-attention embedding based strategy.
Limitations of the proposed strategy come from its dependency to cell region segmentation and the lack an independent dataset within the evaluation process. Regarding segmentation, an accurate delimitation of pre-defined cell regions (cytoplasm, nucleus, and whole cell) is a major requirement for constructing region-attention embeddings, which may affect the performance of the introduced approach. However, as presented in Image pre-processing, the evaluation of the herein implemented segmentation strategy demonstrated a high performance in recognizing such image components, reducing possible effects of a wrong segmentation within the whole classification process. A second limitation of the presented strategy, no independent dataset was used for a multi-center validation, which is a common mistake in previously published BM cell subtype classification works. However, herein presented evaluation scheme included a test in an unseen partition of the dataset, which represented a more exhaustive evaluation in comparison with previously published works, and also stands a further step to demonstrate the generalizability of the presented approach.
Conclusions
The herein introduced strategy improved BM cell subtype classification by mixing DL architectures with engineered features, arranged in a region-attention embedding. This method outperforms previously published approaches by classifying 21 different classes, while a thorough evaluation is implemented on a large publicly available database. This approach provided new levels of information in terms of local features and cell regions, that allows for medical users to relate a model output with clinical features they use for discriminating BM cells. Future efforts will be focused on model interpretability assessment, because herein presented evaluation includes only qualitative results based on matching region-attention embedding features with published clinical features. This explainability analysis may be enriched by including domain-expert perception of the provided relevant cell subtype feature/region, which is a commonly used strategy to boost model trustworthy. Nevertheless, interpretability and explainability, both with different meanings, are still subjective by their dependency on the particular application, and propagation of the DL model uncertainty to their interpretability validation. Furthermore, the lack of ground truth interpretable patterns within the BM cell image context makes evaluation and quantification of interpretability improvement a challenge which remains open.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Eduardo Romero reports financial support was provided by Colombia Ministry of Science Technology and Innovation. Jonathan Tarquino reports a relationship with Colombia Ministry of Science Technology and Innovation that includes: funding grants. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
This work was supported in part by the project with code 110192092354 and entitle “Program for the Early Detection of Premalignant Lesions and Gastric Cancer in urban, rural and dispersed areas in the Department of Nariño” of call No. 920 of 2022 of MinCiencias.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2024.100390.
Appendix A. Supplementary data
Supplemetary material includes different tables with detailed performance information for multiclass classification task, region-attention embedding organization changes, and data ablation experiment, including all metrics presented within the main manuscript.
Data availability statement and ethical compliance
This research is considered exempt of ethic committee evaluation and data availability because all herein presented experiments used publicly available data, with de-identified information, from the following public resource:
An Expert-Annotated Dataset of Bone Marrow Cytology in Hematologic Malignancies (Bone–Marrow–Cytomorphology_MLL_Helmholtz_Fraunhofer) link: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=101941770
References
- 1.Thachil J., Bates I. twelfth ed ed. Elsevier Ltd.; 2017. Approach to the Diagnosis and Classification of Blood Cell Disorders. [DOI] [Google Scholar]
- 2.Ladines-Castro W., Barragán-Ibañez G., Luna-Pérez M., et al. Morphology of leukaemias. Revista Médica del Hospital General de México. 2016;79(2):107–113. doi: 10.1016/j.hgmx.2015.06.007. [DOI] [Google Scholar]
- 3.Wu Q., Zeng L., Ke H., Xie W., Zheng H., Zhang Y. Medical Imaging 2005: Image Processing. Vol. 5747. SPIE; 2005. Analysis of blood and bone marrow smears using multispectral imaging analysis techniques; pp. 1872–1882. [Google Scholar]
- 4.Tomasian A., Jennings J.W. Bone marrow aspiration and biopsy: techniques and practice implications. Skeletal Radiol. 2022;51(1):81–88. doi: 10.1007/s00256-021-03882-w. [DOI] [PubMed] [Google Scholar]
- 5.Malempati S., Joshi S., Lai S., Braner D.A., Tegtmeyer K. Bone marrow aspiration and biopsy. N Engl J Med. 2009;361(15):28. doi: 10.1056/NEJMvcm0804634. [DOI] [PubMed] [Google Scholar]
- 6.Gilotra M., Gupta M., Singh S., Sen R. Comparison of bone marrow aspiration cytology with bone marrow trephine biopsy histopathology: an observational study. J Lab Physicians. 2017;9(03):182–189. doi: 10.4103/JLP.JLP_117_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuchin V. 3rd ed. 2015. Tissue Optics: Light Scattering Methods and Instruments for Medical Diagnostics. (Bellingham, WA) [Google Scholar]
- 8.Ortega S., Halicek M., Fabelo H., Callico G.M., Fei B. Hyperspectral and multispectral imaging in digital and computational pathology: a systematic review. Biomed Optics Express. 2020;11(6):3195–3233. doi: 10.1364/BOE.386338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lantos C., Kornblau S.M., Qutub A.A. Hematology: Latest Research and Clinical Advances. 2018. Quantitative-morphological and cytological analyses in leukemia; pp. 95–113. [Google Scholar]
- 10.Mohd S., Md M., Wan W. White blood cell (WBC) counting analysis in blood smear images using various color segmentation methods. Measure J Int Measure Confederat. 2018;116:543–555. doi: 10.1016/j.measurement.2017.11.002. [DOI] [Google Scholar]
- 11.Alqudah A., Al-Ta’ani O., Al-Badarneh A. Automatic segmentation and classification of white blood cells in peripheral blood samples. J Eng Sci Technol Rev. 2018;11(6):7–13. doi: 10.25103/jestr.116.02. [DOI] [Google Scholar]
- 12.Salahuddin Z., Woodruff H.C., Chatterjee A., Lambin P. Transparency of deep neural networks for medical image analysis: a review of interpretability methods. Comput Biol Med. 2022;140 doi: 10.1016/j.compbiomed.2021.105111. [DOI] [PubMed] [Google Scholar]
- 13.Rawat J., Singh A., B. HS, Virmani J., Devgun J. Computer assisted classification framework for prediction of acute lymphoblastic and acute myeloblastic leukemia. Biocybernet Biomed Eng. 2017;37(4):637–654. doi: 10.1016/j.bbe.2017.07.003. [DOI] [Google Scholar]
- 14.Benomar M., Chikh A., Descombes X., Benazzouz M. Multi features based approach for white blood cells segmentation and classification in peripheral blood and bone marrow images. Int J Biomed Eng Technol. 2019 doi: 10.1504/ijbet.2019.10030162. [DOI] [Google Scholar]
- 15.Jagadev P., Virani H. 2017 International Conference on Trends in Electronics and Informatics (ICEI) IEEE; 2017. Detection of leukemia and its types using image processing and machine learning; pp. 522–526. [Google Scholar]
- 16.Khosrosereshki M., Menhaj M. 2017 5th Iranian Joint Congress on Fuzzy and Intelligent Systems (CFIS) IEEE; 2017. A fuzzy based classifier for diagnosis of acute lymphoblastic leukemia using blood smear image processing; pp. 13–18. [Google Scholar]
- 17.Sudha K., Geetha P. A novel approach for segmentation and counting of overlapped leukocytes in microscopic blood images. Biocybernet Biomed Eng. 2020;40(2):639–648. [Google Scholar]
- 18.Zhang Z., Arabyarmohammadi S., Leo P., et al. Automatic myeloblast segmentation in acute myeloid leukemia images based on adversarial feature learning. Comput Methods Prog Biomed. 2024;243 doi: 10.1016/j.cmpb.2023.107852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghane N., Vard A., Talebi A., Nematollahy P. Classification of chronic myeloid leukemia cell subtypes based on microscopic image analysis. EXCLI J. 2019;18:382. doi: 10.17179/excli2019-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdulhay E., Mohammed M., Ibrahim D., Arunkumar N., Venkatraman V. Computer aided solution for automatic segmenting and measurements of blood leucocytes using static microscope images. J Med Syst. 2018;42(4) doi: 10.1007/s10916-018-0912-y. [DOI] [PubMed] [Google Scholar]
- 21.Ananthi V., Balasubramaniam P. A new thresholding technique based on fuzzy set as an application to leukocyte nucleus segmentation. Comput Methods Prog Biomed. 2016;134:165–177. doi: 10.1016/j.cmpb.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Elrefaie R.M., Mohamed M.A., Marzouk E.A., Ata M.M. A robust classification of acute lymphocytic leukemia-based microscopic images with supervised Hilbert-Huang transform. Microsc Res Tech. 2024;87(2):191–204. doi: 10.1002/jemt.24425. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J., Zhang M., Zhou Z., Chu J., Cao F. Automatic detection and classification of leukocytes using convolutional neural networks. Med Biol Eng Comput. 2017;55:1287–1301. doi: 10.1007/s11517-016-1590-x. [DOI] [PubMed] [Google Scholar]
- 24.Dong N., Zhai M.-D., Chang J., Wu C.-H. A self-adaptive approach for white blood cell classification towards point-of-care testing. Appl Soft Comput. 2021;111 doi: 10.1016/J.ASOC.2021.107709. [DOI] [Google Scholar]
- 25.Krappe S., Wittenberg T., Haferlach T., Münzenmayer C. Automated morphological analysis of bone marrow cells in microscopic images for diagnosis of leukemia: nucleus-plasma separation and cell classification using a hierarchical tree model of hematopoesis. Med Imag 2016 Comput Aided Diagn. 2016;9785 doi: 10.1117/12.2216037. [DOI] [Google Scholar]
- 26.Baig R., Rehman A., Almuhaimeed A., Alzahrani A., Rauf H. Detecting malignant leukemia cells using microscopic blood smear images: a deep learning approach. Appl Sci (Switzerland) 2022;12(13) doi: 10.3390/app12136317. [DOI] [Google Scholar]
- 27.Othman M.Z., Mohammed T.S., Ali A.B. Neural network classification of white blood cell using microscopic images. Int J Adv Comput Sci Appl. 2017;8(5) [Google Scholar]
- 28.Anilkumar K., Manoj V., Sagi T. A survey on image segmentation of blood and bone marrow smear images with emphasis to automated detection of leukemia. Biocybernet Biomed Eng. 2020;40(4):1406–1420. [Google Scholar]
- 29.Elhassan T., Rahim M., Swee T., Hashim S., Aljurf M. Feature extraction of white blood cells using CMYK-moment localization and deep learning in acute myeloid leukemia blood smear microscopic images. IEEE Access. 2022;10:16577–16591. doi: 10.1109/ACCESS.2022.3149637. [DOI] [Google Scholar]
- 30.Rastogi P., Khanna K., Singh V. LeuFeatx: deep learning–based feature extractor for the diagnosis of acute leukemia from microscopic images of peripheral blood smear. Comput Biol Med. 2022;142(November 2021) doi: 10.1016/j.compbiomed.2022.105236. [DOI] [PubMed] [Google Scholar]
- 31.Thomas S., Vijaylakshmi S. 2022 2nd International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE) IEEE; 2022. Image recognition, recusion cellular classification using different techniques and detecting microscopic deformities; pp. 1053–1055. [Google Scholar]
- 32.Fan H., Zhang F., Xi L., Li Z., Liu G., Xu Y. LeukocyteMask: an automated localization and segmentation method for leukocyte in blood smear images using deep neural networks. J Biophotonics. 2019;12(7):1–17. doi: 10.1002/jbio.201800488. [DOI] [PubMed] [Google Scholar]
- 33.Leng B., Wang C., Leng M., Ge M., Dong W. Deep learning detection network for peripheral blood leukocytes based on improved detection transformer. Biomed Signal Process Control. July 2022;82 doi: 10.1016/j.bspc.2022.104518. [DOI] [Google Scholar]
- 34.Alzubaidi L., Fadhel M., Al-shamma O., Zhang J., Duan Y. Deep learning models for classification of red blood cells in microscopy images to aid in sickle cell anemia diagnosis. Electronics (Switzerland) 2020;9(3) doi: 10.3390/electronics9030427. [DOI] [Google Scholar]
- 35.Kassani S., Kassani P., Wesolowski M., Schneider K., Deters R. ICTC 2019 - 10th International Conference on ICT Convergence: ICT Convergence Leading the Autonomous Future. 2019. A hybrid deep learning architecture for leukemic B-lymphoblast classification; pp. 271–276. arXiv:1909.11866. [DOI] [Google Scholar]
- 36.Çınar A., Tuncer S. Classification of lymphocytes, monocytes, eosinophils, and neutrophils on white blood cells using hybrid Alexnet-GoogleNet-SVM, SN. Appl Sci. 2021;3(4) doi: 10.1007/s42452-021-04485-9. [DOI] [Google Scholar]
- 37.Wang J., Li A., Huang M., Ibrahim A., Zhuang H., Ali A. 2018 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT) 2018. Classification of white blood cells with patternnet-fused ensemble of convolutional neural networks (PECNN) pp. 325–330. [DOI] [Google Scholar]
- 38.Qiao Y., Zhang Y., Liu N., Chen P., Liu Y. An end-to-end pipeline for early diagnosis of acute promyelocytic leukemia based on a compact CNN model. Diagnostics. 2021;11(7):1–15. doi: 10.3390/diagnostics11071237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Fu Y., Chen P. 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 2019. WBCaps: a capsule architecture-based classification model designed for white blood cells identification; pp. 7027–7030. [DOI] [PubMed] [Google Scholar]
- 40.Chola C., Muaad A.Y., Bin Heyat M.B., et al. BCNet: a deep learning computer-aided diagnosis framework for human peripheral blood cell identification. Diagnostics. 2022;12(11) doi: 10.3390/diagnostics12112815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naing K., Kittichai V., Tongloy T., Chuwongin S. biorRxiv. 2021. The evaluation of acute myeloid leukaemia (AML) blood cell detection models using different YOLO. [Google Scholar]
- 42.Manescu P., Narayanan P., Bendkowski C., et al. Detection of acute promyelocytic leukemia in peripheral blood and bone marrow with annotation-free deep learning. Sci Rep. 2023;13(1):2562. doi: 10.1038/s41598-023-29160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talaat F.M., Gamel S.A. Machine learning in detection and classification of leukemia using c-nmc_leukemia. Multimed Tools Appl. 2023:1–14. [Google Scholar]
- 44.Devi T.G., Patil N., Rai S., Sarah C.P. Segmentation and classification of white blood cancer cells from bone marrow microscopic images using duplet-convolutional neural network design. Multimed Tools Appl. 2023:1–23. [Google Scholar]
- 45.Matek C., Krappe S., Münzenmayer C., Haferlach T., Marr C. Highly accurate differentiation of bone marrow cell morphologies using deep neural networks on a large image data set. Blood. 2021;138:1917–1927. doi: 10.1182/blood.2020010568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matek C., Krappe S., Münzenmayer C., Haferlach T., Marr C. An expert-annotated dataset of bone marrow cytology in hematologic malignancies [data set] Cancer Imaging Arch. 2021 doi: 10.7937/TCIA.AXH3-T579. [DOI] [Google Scholar]
- 47.Tripathi S., Augustin A., Sukumaran R., Dheer S., Kim E. HematoNet. Experte level classification of bone marrow cytology morphology in hemaatological malignancy with deep learning. Artif Intel Life Sci. 2022;2(100043) doi: 10.01016/j.ailsci.2022.100043. [DOI] [Google Scholar]
- 48.Ananthakrishnan B., Shaik A., Akhouri S., Garg P., Gadag V., Kavitha M.S. Automated bone marrow cell classification for haematological disease diagnosis using siamese neural network. Diagnostics. 2022;13(1):112. doi: 10.3390/diagnostics13010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meem R.F., Hasan K.T. arXiv preprint. 2023. Bone marrow cytomorphology cell detection using inceptionresnetv2. arXiv:2305.05430. [Google Scholar]
- 50.Lewis J.E., Shebelut C.W., Drumheller B.R., et al. An automated pipeline for differential cell counts on whole-slide bone marrow aspirate smears. Mod Pathol. 2023;36(2) doi: 10.1016/j.modpat.2022.100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratheesh S., Breethi A.A. Deep learning based non-local k-best renyi entropy for classification of white blood cell subtypes. Biomed Signal Process Control. 2024;90 [Google Scholar]
- 52.Wang C.-W., Huang S.-C., Lee Y.-C., Shen Y.-J., Meng S.-I., Gaol J.L. Deep learning for bone marrow cell detection and classification on whole-slide images. Med Image Anal. 2022;75 doi: 10.1016/j.media.2021.102270. [DOI] [PubMed] [Google Scholar]
- 53.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intel. 2019;1(5):206–215. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Proc IEEE Conf Comput Vis Pattern Recognit. 2016:770–778. [Google Scholar]
- 55.Chollet F. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1800–1807. arXiv:1610.02357. [DOI] [Google Scholar]
- 56.Hazra D., Byun Y.C., Kim W.J. Enhancing classification of cells procured from bone marrow aspirate smears using generative adversarial networks and sequential convolutional neural network. Comput Methods Prog Biomed. 2022;224 doi: 10.1016/j.cmpb.2022.107019. [DOI] [PubMed] [Google Scholar]
- 57.Tavakoli S., Ghaffari A., Kouzehkanan Z.M., Hosseini R. New segmentation and feature extraction algorithm for classification of white blood cells in peripheral smear images. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-98599-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hegde R.B., Prasad K., Hebbar H., Singh B.M.K. Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybernet Biomed Eng. 2019;39(2):382–392. doi: 10.1016/j.bbe.2019.01.005. [DOI] [Google Scholar]
- 59.Krappe S., Benz M., Wittenberg T., Haferlach T., Münzenmayer C. Automated classification of bone marrow cells in microscopic images for diagnosis of leukemia: a comparison of two classification schemes with respect to the segmentation quality. Med Imag 2015 Comput Aided Diagn. 2015;9414 doi: 10.1117/12.2081946. [DOI] [Google Scholar]
- 60.Prinyakupt J., Pluempitiwiriyawej C. Segmentation of white blood cells and comparison of cell morphology by linear and naïve Bayes classifiers. Biomed Eng Online. 2015;14 doi: 10.1186/s12938-015-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dinčić M., Popović T.B., Kojadinović M., Trbovich A.M., Ilić A.Ž. Morphological, fractal, and textural features for the blood cell classification: the case of acute myeloid leukemia. Eur Biophys J. 2021;50(8):1111–1127. doi: 10.1007/s00249-021-01574-w. [DOI] [PubMed] [Google Scholar]
- 62.Habibzadeh M., Jannesari M., Rezaei Z., Baharvand H., Totonchi M. Tenth International Conference on Machine Vision (ICMV 2017) Vol. 10696. SPIE; 2018. Automatic white blood cell classification using pre-trained deep learning models: Resnet and inception; pp. 274–281. [Google Scholar]
- 63.Yildirim M., Çinar A. Classification of white blood cells by deep learning methods for diagnosing disease. Revue d’Intelligence Artificielle. 2019;33(5):335–340. doi: 10.18280/ria.330502. [DOI] [Google Scholar]
- 64.Mohamed E.H., El-Behaidy W.H., Khoriba G., Li J. Improved white blood cells classification based on pre-trained deep learning models. J Commun Software Syst. 2020;16:37–45. doi: 10.24138/jcomss.v16i1.818. [DOI] [Google Scholar]
- 65.Pereira S., Meier R., McKinley R., et al. Enhancing interpretability of automatically extracted machine learning features: application to a RBM-random forest system on brain lesion segmentation. Med Image Anal. 2018;44:228–244. doi: 10.1016/j.media.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 66.Lipton Z.C. The mythos of model interpretability: in machine learning, the concept of interpretability is both important and slippery. Queue. 2018;16(3):31–57. [Google Scholar]
- 67.Ghassemi M., Oakden-Rayner L., Beam A.L. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digital Health. 2021;3(11):e745–e750. doi: 10.1016/S2589-7500(21)00208-9. [DOI] [PubMed] [Google Scholar]
- 68.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2020;128(2):336–359. doi: 10.1007/s11263-019-01228-7. arXiv:1610.02391. [DOI] [Google Scholar]
- 69.Theera-Umpon N., Dhompongsa S. Morphological granulometric features of nucleus in automatic bone marrow white blood cell classification. IEEE Trans Inf Technol Biomed. 2007;11(3):353–359. doi: 10.1109/titb.2007.892694. [DOI] [PubMed] [Google Scholar]
- 70.Glüge S., Balabanov S., Koelzer V.H., Ott T. Evaluation of deep learning training strategies for the classification of bone marrow cell images. Comput Methods Prog Biomed. 2024;243 doi: 10.1016/j.cmpb.2023.107924. [DOI] [PubMed] [Google Scholar]
- 71.Ahmad R., Awais M., Kausar N., Akram T. White blood cells classification using entropy-controlled deep features optimization. Diagnostics. 2023;13(3):352. doi: 10.3390/diagnostics13030352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang W., Luo M., Guo P., Wei Y., Tan Y., Shi H. Artificial intelligence-assisted diagnosis of hematologic diseases based on bone marrow smears using deep neural networks. Comput Methods Prog Biomed. 2023;231 doi: 10.1016/j.cmpb.2023.107343. [DOI] [PubMed] [Google Scholar]
- 73.Zolfaghari M., Sajedi H. A survey on automated detection and classification of acute leukemia and WBCs in microscopic blood cells. Multimed Tools Appl. 2022;81(5):6723–6753. [Google Scholar]
- 74.Saleem S., Amin J., Sharif M., Mallah G.A., Kadry S., Gandomi A.H. Leukemia segmentation and classification: a comprehensive survey. Comput Biol Med. 2022;106028 doi: 10.1016/j.compbiomed.2022.106028. [DOI] [PubMed] [Google Scholar]
- 75.Bi W.L., Hosny A., Schabath M.B., et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69(2):127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Temme M. Algorithms and transparency in view of the new general data protection regulation. Eur Data Prot L Rev. 2017;3:473. [Google Scholar]
- 77.Babic B., Gerke S., Evgeniou T., Cohen I.G. Beware explanations from AI in health care. Science. 2021;373(6552):284–286. doi: 10.1126/science.abg1834. [DOI] [PubMed] [Google Scholar]
- 78.Biffi C., Cerrolaza J.J., Tarroni G., et al. Explainable anatomical shape analysis through deep hierarchical generative models. IEEE Trans Med Imaging. 2020;39(6):2088–2099. doi: 10.1109/TMI.2020.2964499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Z., Bei Y., Rudin C. Concept whitening for interpretable image recognition. Nat Mach Intel. 2020;2(12):772–782. [Google Scholar]
- 80.Prellberg J., Kramer O. ISBI 2019 C-NMC Challenge: Classification in Cancer Cell Imaging: Select Proceedings. Springer; 2019. Acute lymphoblastic leukemia classification from microscopic images using convolutional neural networks; pp. 53–61. [Google Scholar]
- 81.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]
- 82.Koh P.W., Nguyen T., Tang Y.S., et al. In: Proceedings of the 37th International Conference on Machine Learning, Vol. 119 of Proceedings of Machine Learning Research. H. D. III, Singh A., editors. PMLR; 2020. Concept bottleneck models; pp. 5338–5348.https://proceedings.mlr.press/v119/koh20a.html URL. [Google Scholar]
- 83.Dai Y., Wang G., Li K.-C. Conceptual alignment deep neural networks. J Intel Fuzzy Syst. 2018;34(3):1631–1642. [Google Scholar]
- 84.Sabour S., Frosst N., Hinton G.E. Dynamic routing between capsules. Adv Neural Inf Proces Syst. 2017;30 [Google Scholar]
- 85.Tran M., Lahiani A., Dicente Cid Y., et al. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2023. B-cos aligned transformers learn human-interpretable features; pp. 514–524. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemetary material includes different tables with detailed performance information for multiclass classification task, region-attention embedding organization changes, and data ablation experiment, including all metrics presented within the main manuscript.
Data Availability Statement
This research is considered exempt of ethic committee evaluation and data availability because all herein presented experiments used publicly available data, with de-identified information, from the following public resource:
An Expert-Annotated Dataset of Bone Marrow Cytology in Hematologic Malignancies (Bone–Marrow–Cytomorphology_MLL_Helmholtz_Fraunhofer) link: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=101941770