Abstract
Background
The typical pathological feature of pancreatic ductal adenocarcinoma (PDAC) is a significant increase in stromal reaction, leading to a hypoxic and poorly vascularized tumor microenvironment. Tumor cells undergo metabolic reprogramming, such as the Warburg effect, yet the underlying mechanisms are not fully understood.
Methods
Interference and overexpression experiments were conducted to analyze the in vivo and in vitro effects of USP7 on the growth and glycolysis of tumor cells. Small-molecule inhibitors of USP7 and transgenic mouse models of PDAC were employed to assess the consequences of targeting USP7 in PDAC. The molecular mechanism underlying USP7-induced c-Myc stabilization was determined by RNA sequencing, co-IP and western blot analyses.
Results
USP7 is abnormally overexpressed in PDAC and predicts a poor prognosis. Hypoxia and extracellular matrix stiffness can induce USP7 expression in PDAC cells. Genetic silencing of USP7 inhibits the glycolytic phenotypes in PDAC cells, while its overexpression has the opposite effect, as demonstrated by glucose uptake, lactate production, and extracellular acidification rate. Importantly, USP7 promotes PDAC tumor growth in a glycolysis-dependent manner. The small-molecule inhibitor P5091 targeting USP7 effectively suppresses the Warburg effect and cell growth in PDAC. In a transgenic mouse model of PDAC, named KPC, P5091 effectively blocks tumor progression. Mechanistically, USP7 interacts with c-Myc, enhancing its stability and expression, which in turn upregulates expression of glycolysis-related genes.
Conclusions
This study sheds light on the molecular mechanisms underlying the Warburg effect in PDAC and unveils USP7 as a potential therapeutic target for improving PDAC treatment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05962-6.
Keywords: Deubiquitinating enzymes, HAUSP, Aerobic glycolysis, Glucose metabolism
Introduction
Energy metabolism reprogramming, exemplified by the Warburg effect, is a crucial characteristic of cancer cells [1]. The Warburg effect describes the metabolic phenomenon where tumor cells generate energy predominantly through glycolysis, leading to increased lactic acid production even in the presence of adequate oxygen levels. Unlike normal cells that primarily rely on oxidative phosphorylation for energy production, cancer cells exhibit a preference for glycolysis. The Warburg effect can drive tumor advancement through various mechanisms, including but not limited to providing cellular buildings to sustain the rapid growth of tumor cells, generating an acidic microenvironment, facilitating immune evasion, and strengthening drug resistance [2, 3]. Pancreatic ductal adenocarcinoma (PDAC) is distinguished by its rich stromal components and the absence of oxygen and blood supply in the tumor microenvironment [4–6], leading to increased reliance of tumor cells on the glycolytic pathway. Previously, accumulated studies have reported several key regulators in PDAC, such as the KRAS mutation [7], p38gamma MAPK [8], and the transcriptional factor ONECUT3 [5].
Deubiquitinases (DUBs) are enzymes that reverse the process of protein ubiquitination within the ubiquitin-proteasome system [9, 10]. DUBs play a crucial role in maintaining cellular homeostasis by removing ubiquitin molecules from proteins, thereby regulating their stability, localization, and activity [11, 12]. DUBs are categorized into are categorized into several families based on their structural and functional similarities, including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph disease protein domain proteases (MJDs), and The JAMM (JAB1/MPN/MOV34) family. Among the USPs, ubiquitin-specific protease 7 (USP7) stands out as a well-studied cysteine protease with the ability to modulate multiple cellular proteins involved in various pathways, including viral replication, immune response, apoptosis, DNA damage and repair, and epigenetic regulation [13–18]. The dysregulation of USP7 in many cancers positions it as a promising target for cancer therapy, as it plays a significant role in tumor initiation and progression through interacting with oncogenic proteins and tumor suppressors [19]. Additionally, USP7 contributes to tumor evasion from the immune system by affecting the functions of regulatory T cells and effector T cells [20, 21]. Previously, we revealed that USP7 interacts with WDR1 to prevent ubiquitination-mediated degradation of β-Catenin in PDAC [22]. Recently, accumulated studies documented diverse oncogenic roles of USP7 in human cancers, such as lung cancer [23, 24], gastric cancer [25], glioma [26], and breast cancer [27]. At present, a variety of small molecular inhibitors, natural compounds and small molecular peptides against USP7 have shown specific USP7 inhibition and certain anti-tumor activity [20], such as USP7 inhibitor P5091 [28], which can reduce Wnt signal pathway activity by enhancing ubiquitin-mediated degradation of β-catenin.
c-Myc plays a pivotal role in the Warburg effect. By directly regulating several key genes in the glycolytic pathway, c-Myc promotes the occurrence and maintenance of the Warburg effect. One of the genes activated by c-Myc is glucose transporter 1 (GLUT1), which enhances glucose uptake into cancer cells [29]. Additionally, c-Myc stimulates the expression of HK2 and pyruvate kinase isoform M2 (PKM2) [29, 30], two key enzymes crucial for glycolysis. Moreover, c-Myc upregulates lactate dehydrogenase A (LDHA) [31], which converts pyruvate to lactate, leading to increased production of lactic acid.
In this study, through bioinformatics analysis and experimental validation, we identified USP7 as a key regulator of the Warburg effect in PDAC. USP7 is induced by extracellular matrix stiffness, a mechanical cues of the tumor microenvironment. Inhibition of USP7 resulted in decreased glycolytic phenotype and tumor growth in PDAC cells, highlighting its potential as a therapeutic target. Moreover, USP7 controls c-Myc protein stability through ubiquitination, leading to increased c-Myc expression and subsequent activation of glycolysis-related genes, enhancing the Warburg effect in PDAC cells.
Methods and materials
Cell culture and reagents
Human PDAC cells (AsPC1, BxPC3, CFPAC-1, MiaPaca-2, and PANC1) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The non-malignant HPDE cells were a gift from Shanghai Cancer Institute. All the cells were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2, using Dulbecco’s Modified Eagle Medium (DMEM) or RPMI-1640 medium supplemented with 100 U/mL penicillin G and 100 mg/mL streptomycin and 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, USA). All cells were authenticated by STR profiling. The collagen I-coated soft (0.5 kPa) and stiff (12 kPa) 6-well culture plates were sourced from Matrigen (Irvine, CA, USA). Cycloheximide (CHX) was purchased from (Cat. 239765, St. Louis, MO, USA). 2-Deoxy-D-glucose (2-DG; Cat. S4701) was acquired from Selleck (Shanghai, China). USP7 inhibitor P5091 (Cat. HY-15667), HIF1α inhibitor HY-13,671 (Cat. HY-13671), and MG132 (Cat. HY-13259) were obtained from MedChemExpress. The specific shRNAs against USP7 and pcDNA3.1-USP7 plasmid were synthesized by Genepharma (Shanghai, China), as reported previously [22].
PDAC samples
In this study, two cohorts of PDAC samples were employed. A tissue microarray containing paired PDAC and nontumor pancreas tissues was used to determine the expression of USP7 in PDAC. The second cohort including PDAC cases with 18 F-FDG-PET/CT scanning was used to investigate the association between USP7 expression and the maximum standardized uptake value (SUVmax). The investigational protocol was approved by the Research Ethics Committee of Huashan Hospital, Fudan University, and all the patients were provided with written informed consent before enrollment.
Western blotting analysis
Whole cell lysates from PDAC cells were prepared using IP lysis buffer (Beyotime, Shanghai, China). The concentration of the total protein in each sample was accurately determined using the BCA kit. Subsequently, 20 µg of protein samples was loaded and subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for separation, after which proteins were transferred to polyvinylidene fluoride membranes. After blocking, the membranes were incubated with following primary antibodies: anti-USP7 antibody (1:1,000; ab4080; Abcam, Shanghai, China), anti-c-Myc antibody (1:1,000; #18583; Cell Signaling Technology, Shanghai, China), anti-β-actin antibody (1:1,000; ab8226; Abcam, Shanghai, China). The next day, the membranes underwent incubation with a horseradish peroxidase-conjugated secondary antibody (1:1000; GE Healthcare Biosciences, based in NY, USA) for 1 h at room temperature. The protein signal was detected using enhanced ECL reagents (GE Healthcare Biosciences, NY, USA), and the protein bands were quantitatively analyzed utilizing ImageJ software developed by NIH Image. For protein half-life experiment, PDAC cells were treated with CHX (20 µg/ml) for durations as indicated, and proteins were extracted for Western blotting analysis.
Co-immunoprecipitation experiments
PDAC cells (CFPAC-1 or PANC1) and 293T cells were collected cells until they reached approximately 80% confluence. Then cells were washed twice with cold 1× PBS to remove any residual serum and media. Following this, the cells were harvested using a cell scraper and lysed with IP lysis buffer supplemented with protease/phosphatase inhibitors (Roche, 04693132001). The cell suspension was incubated on ice for 30 min with occasional gentle mixing to ensure complete lysis. After incubation, the lysate was subjected to sonication for 10 s at a low amplitude to shear DNA and reduce viscosity, followed by clarification through centrifugation at 14,000 rpm for 15 min at 4 °C. A total of 200 µl of the clarified cell lysates were mixed with 50 µl of Protein G-agarose suspension (Millipore, USA) to facilitate the binding of target proteins to the beads. This mixture was incubated for 2 h at 4 °C with gentle agitation to allow efficient binding. After this initial incubation, the beads were collected by centrifugation, and the supernatant was discarded. The beads were then incubated with primary antibodies against USP7 or control IgG for an additional 2 h at 4 °C. Following this, 100 µl of fresh Protein G-agarose was added, and the incubation continued overnight at 4 °C to maximize antibody-antigen interactions. The immunoprecipitates were collected the next day and washed three times with 1× PBS to remove unbound proteins. Finally, loading buffer was added to the beads, and the mixture was boiled to elute the immunoprecipitated proteins, which were then prepared for analysis via Western blotting.
Immunohistochemical (IHC) analysis
IHC was performed as reported elsewhere [32]. Briefly, Paraffin-embedded slides are routinely deparaffinized and rehydrated. Antigen retrieval is performed by steaming for 15 min in Tris-ethylenediaminetetraacetic acid (EDTA) buffer. To block endogenous peroxidase, the slides were then incubated in a solution of 0.3% H2O2-methanol. After blocking with 10% normal horse serum, the slides are then incubated overnight at 4°C with the primary antibodies, including anti-USP7 antibody (1:400; ab4080; Abcam, Shanghai, China), anti-c-Myc antibody (1:200; #18583; Cell Signaling Technology, Shanghai, China), anti-Ki67 antibody (1:2,000; 27309-1-AP; Proteintech), anti-GLUT1 antibody (1:1,000; 21829-1-AP; Proteintech), anti-HK2 antibody (1:300; 66974-1-Ig; Proteintech), and anti-LDHA antibody (1:200; 19987-1-AP; Proteintech). Subsequently, a biotin-conjugated secondary antibody is applied, followed by incubation and rinsing with cold TBS before adding streptavidin-peroxidase and 3,3’-Diaminobenzidine for visualization.
Plate colony formation experiment
To assay cell proliferation, 500 cells/well were seeded into a 6-well plate or 12-well plate. The culture medium was replaced every 2–3 days. After culture for 10–12 days, the colonies were fixed with 4% paraformaldehyde and stained with crystal violet dye.
Animal experiments
To establish a subcutaneous xenograft model, male BALB/c nude mice at 8 weeks of age were used. These mice were injected with 1 × 106 PANC1 cells in the lower back region. Once the tumors reached a size of approximately 100 mm3, P5091 was administered. Throughout the study, the mice were monitored and their tumor volume and weight were recorded. Tumor volumes were calculated using the formula length × width2/2. Upon completion of the study, the mice were euthanized, and the weight of the isolated tumors was recorded for further analysis. The procedure of KPC mice experiments was performed as reportedly previously [33]. In brief, the tumor size of KPC mice was assessed through palpation. KPC mice with tumors ranging from approximately 5–8 mm in size were randomly assigned to different treatment groups. These groups were then subjected to treatment with P5091 over a period of about 4 weeks. Following the treatment duration, the mice were euthanized, and their tissues were collected for further analysis. All mice were housed in a controlled facility with a 12-h dark/light cycle to mimic natural day-night patterns and were provided with free access to food and water to meet their nutritional and hydration needs throughout the study period. All procedures and manipulations conducted in the study were approved by the Research Ethics Committee of Huashan Hospital, Fudan University. All mice were randomly assigned to experimental groups. The investigator conducting the experiment remained blinded to treatments, and a second blinded investigator handled the subsequent data analysis.
RNA isolation and real-time qPCR
Total RNA from the PDAC cell or xenograft tumor tissues was extracted utilizing TRIzol reagent (Takara, Japan). Subsequently, cDNA synthesis was carried out following standard protocols with the primeScript RT Master kit (Takara, Japan). To quantify the mRNA levels of specific genes, a SYBR green-based real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay was employed with the ViiA7 System (Applied Biosystems, Foster City, CA). The following primers were used in this study:
USP7 forward, 5’-GGAAGCGGGAGATACAGATGA-3’;
USP7 reverse, 5’-AAGGACCGACTCACTCAGTCT-3’;
c-Myc forward, 5’-GGCTCCTGGCAAAAGGTCA-3’;
c-Myc reverse, 5’-CTGCGTAGTTGTGCTGATGT-3’;
SLC2A1 forward, 5’-ATTGGCTCCGGTATCGTCAAC-3’;
SLC2A1 reverse, 5’-GCTCAGATAGGACATCCAGGGTA-3’;
HK2 forward, 5’-TTGACCAGGAGATTGACATGGG-3’;
HK2 reverse, 5’-CAACCGCATCAGGACCTCA-3’;
LDHA forward, 5’-ATGGCAACTCTAAAGGATCAGC-3’;
LDHA reverse, 5’-CCAACCCCAACAACTGTAATCT-3’;
ACTB forward, 5’-CATGTACGTTGCTATCCAGGC-3’;
ACTB reverse, 5’-CTCCTTAATGTCACGCACGAT-3’.
The relative mRNA expression levels were determined using the 2^(−ΔΔCt) method, which allows for the comparison of gene expression levels between different samples by normalizing to an internal control gene ACTB ( encoding β-actin).
RNA sequencing analysis
sh-Ctrl or sh-USP7−1 PANC1 cells, or with wild-type PANC1 cells treated with 5 µM P5091 or DMSO for 12 h, were used for this experiment. RNA isolation was performed followed by RNA sequencing using the Illumina NovaSeq 6000 platform, with the sequencing process managed by Illumina’s data collection software. For enrichment analysis Gene Set Enrichment Analysis (GSEA) was conducted using hallmark gene sets from the GSEA database (https://www.gsea-msigdb.org/gsea/index.jsp). A false discovery rate (FDR) score of less than 0.05 was considered indicative of a significant difference in gene expression.
HIF1α transcriptional activity assay
The HIF-1α DNA binding activity assay was conducted using the HIF-1α Transcription Factor Assay Kit (Abcam, ab133104). In brief, nuclear extracts from the indicated PDAC cell samples were added to the wells of the HIF-1α transcription factor plate and then allowed to incubate overnight at 4 °C without agitation. Subsequently, a diluted HIF-1α primary antibody was added to each well and incubated for 1 h at room temperature. Following this, the goat anti-rabbit HRP conjugate was added to each well and incubated for 1 h at room temperature. Finally, the levels of HIF-1α DNA binding activity were assessed at 450 nm using a microplate reader after the addition of the stop solution.
Glucose and lactate detection
For detection of glucose uptake, PDAC cells were seeded at a density of 5,000 cells per well in a 96-well culture plate. Cells were first incubated in 90 µl of Glucose Uptake Buffer (AAT Bioquest) per well for 1 h. Subsequently, 10 µl 2-DG of (AAT Bioquest) was added to each well and cells were further incubated for 30 min. Finally, 50 µl of the Uptake Assay Mixture (AAT Bioquest) was added to each well. The samples were finally analyzed using a Microplate Reader (TECAN) with excitation/emission wavelengths set at 570/610 nm. For measurement of lactate level in the culture medium, 2 × 105 PDAC cells with indicated genetic modification cell were seeded into six-well plates. After culture for 24 h, the cell supernatant was harvested and subjected for lactate detection with Lactate Assay Kit (BioVision, K607-100).
Seahorse analysis of ECAR
The glycolytic stress test, named extracellular acidification rate (ECAR), was conducted using the glycolytic stress test kit from Seahorse Bioscience and analyzed with an XFe96 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA, USA). Briefly, PDAC cells were plated at a density of 1 × 105 cells per well in the Seahorse XF cell culture microplate. After temperature and pH equilibration, sequential compound injections were performed. The compounds included glucose, oligomycin, and 2-DG at final concentrations of 10 mM, 0.5 µM, and 50 mM, respectively. Following the assay, the wells were washed, cells were lysed, and the Bradford assay was carried out to determine the protein content in the samples, as a normalization to the final data presentation.
TCGA data analysis
The TCGA pancreatic cancer dataset was acquired from the TCGA database (https://portal.gdc.cancer.gov/). The hypoxia-associated gene signature (ACOT7, ADM, ALDOA, CDKN3, ENO1, LDHA, MIF, MRPS17, NDRG1, P4HA1, PGAM1, SLC2A1, TPI1, TUBB6 and VEGFA) was adopted from a previous study [34]. The correlation analysis of USP7 with glycolytic genes and hypoxia-associated gene signature was performed with the GEPIA2 online database (http://gepia2.cancer-pku.cn/#index).
Statistical analysis
Data were presented as mean ± standard deviation (SD), and statistical analyses were performed using GraphPad Prism software version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t-test or one-way ANOVA was used to assess significant differences between different groups. Cumulative survival time was calculated using the Kaplan-Meier method and analyzed with the log-rank test. The χ2 test was utilized for determining differences in the groups during IHC analysis. Correlation analysis was conducted using Spearman’s test.
A significance level of P < 0.05 was considered statistically significant.
Results
USP7 is identified as a glycolysis-related DUB in PDAC
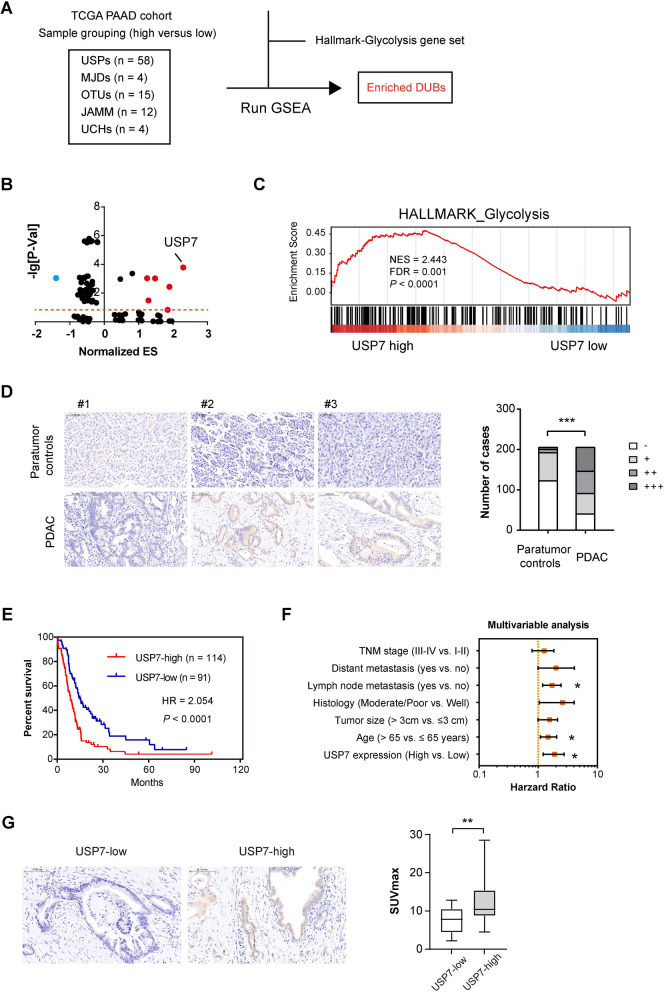
To identify the key DUBs implicated PDAC glycolysis, we used RNAseq data of PDAC samples in the TCGA cohort for Gene Set Enrichment analysis (GSEA). Sample grouping was made based on the median expression of each DUB, and the Hallmark glycolysis gene set was employed for running GSEA (Fig. 1A). The results showed that USP7 had the highest enrichment score among DUBs whose Normalized p-value is less than 0.5 and FDR value is less than 0.25 (Fig. 1B–C). Therefore, USP7 was selected for further study. To investigate the expression pattern of USP7 in PDAC, we conducted immunohistochemical analysis in human PDAC tissues and found the cytoplasmic distribution of USP7. Compared to normal control pancreatic tissue, higher USP7 expression can be found in tumor tissues (Fig. 1D). Importantly, high USP7 expression predicted a poor patient prognosis in PDAC (Fig. 1E). Multivariate analysis identified USP7 as an independent prognostic factor for PDAC (Fig. 1F). To confirm the link between USP7 and the glycolytic phenotype in PDAC, we analyzed a PDAC cohort with 18 F-FDG PET-CT data. Interestingly, the SUVmax value in the USP7 high expression group was notably higher than that of the USP7 low expression group (Fig. 1G), suggesting a potential association between USP7 expression and tumor glucose uptake.
Fig. 1.
USP7 is identified as a glycolysis-related DUB in PDAC. A Flow chart of the screen setup for glycolysis-related DUBs. USPs, ubiquitin-specific proteases; UCHs, ubiquitin C-terminal hydrolases; OTUs, ovarian tumor proteases; MJDs, Machado-Joseph disease protein domain proteases; JAMM, JAB1/MPN/MOV34 family. B Volcano plot of glycolysis-related DUBs, as identified by gene set enrichment analysis. ES, enrichment score. C GSEA plot of Hallmark_glycolysis, sample grouping was made based on the median expression level of USP7. D IHC analysis showed the expression of USP7 in human PDAC and corresponding nontumor tissues (n = 205); scale bar: 100 μm. E Kaplan-Meier analysis showed the overall survival (OS) of PDAC patients based on the protein expression of USP7; n = 114 for USP7-high and n = 91 for USP7-low. F Multivariate analysis of independent prognostic factors of PDAC. G Analysis of the relationship between USP7 expression and SUVmax value; scale bar: 100 μm; n = 14 for USP7-high and n = 8 for USP7-low. Values were compared by Fisher’s exact test (D), the log-rank test (E), multivariate Cox regression analyses (F), and the Student’s t test (G). *P < 0.05, **P < 0.01, ***P < 0.001
USP7 enhances the glycolytic metabolism of PDAC cells
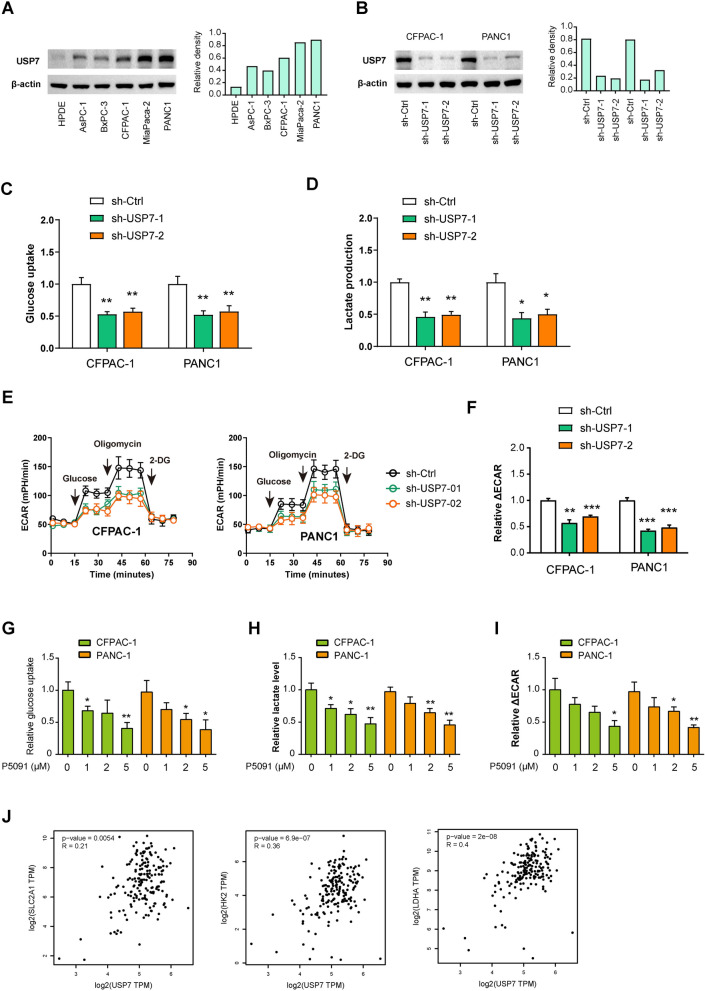
To investigate the potential regulatory role of USP7 in PDAC cell glycolysis, we conducted loss-of-function experiments in two PDAC cells, CFPAC-1 and PANC1, which exhibit higher endogenous USP7 expression (Fig. 2A). Western blotting analysis showed that two shRNAs targeting USP7 significantly reduced USP7 levels at protein levels (Fig. 2B). Compared to control cells, PDAC cells expressing sh-USP7 showed decreased glucose uptake and reduced lactate production (Fig. 2C, D). Seahorse analysis revealed a significant decrease in the extracellular acidification rate of CFPAC-1 and PANC1 cells following USP7 knockdown (Fig. 2E, F). To further validate these results, we pharmacologically inhibited USP7 activity using a specific inhibitor, P5091. Treatment with P5091 led to a dose-dependent inhibition of glucose uptake, lactate production, and extracellular acidification rate (Fig. 2G–I). Consistently, real-time qPCR analysis showed that the glucose transporter GLUT1 and glycolytic genes (HK2 and LDHA) were all reduced by either USP7 knockdown or pharmacological inhibition (Figure S1). In the TCGA cohort, USP7 expression was also closely associated with the expression of these glycolytic components (Fig. 2J). Taken together, these findings strongly support the notion that USP7 acts as a regulator of glycolysis in PDAC cells.
Fig. 2.
USP7 enhances the glycolytic metabolism of PDAC cells.A Western blotting analysis showed USP7 protein expression in PDAC cells and the nonmalignant HPDE cells. B Western blotting showed USP7 knockdown efficiency in CFPAC-1 and PANC1 cells. C, D The effects of USP7 knockdown on the glucose uptake and lactate production in CFPAC-1 and PANC1 cells (n = 3 per group). E, F Seahorse experiment showed the effect of USP7 knockdown on extracellular acidification rate (ECAR) of CFPAC-1 and PANC1 cells. ΔECAR represents the difference between the ECAR values induced by oligomycin and 2-DG (n = 3 per group). G–I The effects of USP7 inhibition by different concentrations of P5091 on the glucose uptake, lactate production, and ECAR in CFPAC-1 and PANC1 cells (n = 3 per group). J Correlation analysis of USP7 with glucose transporter (GLUT1, encoded by SLC2A1) and glycolytic genes (HK2 and LDHA) in pancreatic cancer (n = 178) Values were compared by the one-way ANOVA multiple comparisons with Tukey’s method among groups (C, D, F, G) and Spearman’s rank correlation methods (J). Assays for glycolysis were independently repeated three times with similar results. *P < 0.05, **P < 0.01, ***P < 0.001
Hypoxia and extracellular matrix stiffness induces the expression of USP7 in PDAC cells
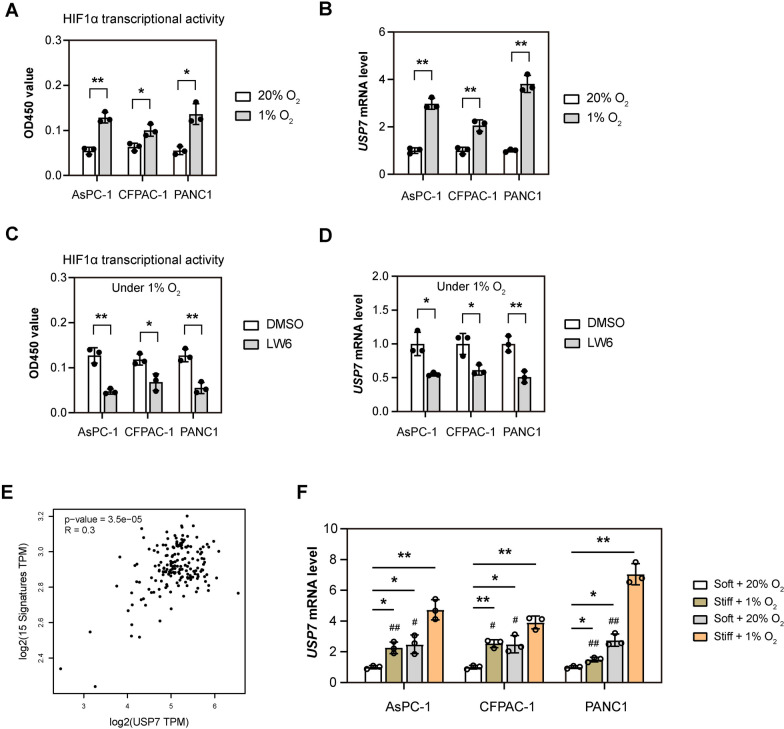
Hypoxia and abundant extracellular matrix components are the two most typical pathological features of PDAC [35–37]. So next, we explored whether hypoxia and extracellular matrix stiffness can induce USP7 expression. Firstly, we cultured PDAC cells (AsPC-1, CFPAC-1 and PANC1) under normoxic (5% O2) and hypoxic (1% O2) conditions, and detected the expression of USP7 after 24 h. As a validation of hypoxia, the HIF1α transcriptional activity was monitored (Fig. 3B). It was found that the mRNA level of USP7 was up-regulated by 2–4 times under hypoxic conditions (Fig. 3B). To demonstrate whether this effect is mediated by HIF1α, we employed an inhibitor for HIF1α, LW6, and found that hypoxia-induced USP7 expression was downregulated by LW6, suggesting that HIF1α-dependent regulation of USP7 expression (Fig. 3C, D). Using a well-documented hypoxia-related gene signature [34], we also observed a close link betweenUSP7 and the HIF signature in PDAC from the TCGA cohort (Fig. 3E).
Fig. 3.
Hypoxia and extracellular matrix stiffness induces the expression of USP7 in PDAC cells. A PDAC cells (AsPC-1, CFPAC-1 and PANC1) were cultured under normoxic (5% O2) and hypoxic (1% O2) conditions for 24 h. Then, HIF1α transcriptional activity in PDAC cells was measured (n = 3). B The mRNA level of USP7 in groups as indicated in (A) was determined by real-time qPCR analysis (n = 3). C PDAC cells (AsPC-1, CFPAC-1 and PANC1) were cultured under hypoxic (1% O2) conditions for 24 h, with or without treatment of an HIF1α inhibitor LW6. Then, HIF1α transcriptional activity in PDAC cells was measured (n = 3). D The mRNA level of USP7 in groups as indicated in (B) was determined by real-time qPCR analysis (n = 3). E USP7 mRNA levels in PDAC samples from TCGA database were compared with the HIF1α signature using correlation analysis (n = 178). F CFPAC-1 and PANC1 seeded on soft (0.5 kPa) and stiff matrices (12 kPa) 6-well plates and cultured under normoxic (5% O2) or hypoxic (1% O2) conditions for 24 h. The mRNA level of USP7 was determined by real-time qPCR analysis (n = 3) Values were compared by the Student’s t test (A–D), Spearman’s rank correlation methods (E), and one-way ANOVA multiple comparisons with Tukey’s method among groups (F). Experiments were independently repeated three times (A–D, F) with similar results. *P < 0.05 and **P < 0.01; # indicates comparison with Stiff + 1% O2 group, #P < 0.05 and ##P < 0.01
To further investigate the contribution of extracellular matrix stiffness, PDAC cells were seeded on soft (0.5 kPa) and stiff matrices (12 kPa) and cultured under either normoxic or hypoxic conditions. Compared to soft culture condition, USP7 mRNA level was upregulated under stiff culture condition (Fig. 3F). Importantly, USP7 expression can be further boosted under hypoxic condition, indicating a synergistic effect between extracellular matrix stiffness and hypoxia in inducing USP7 expression (Fig. 3F). Together, both hypoxia and extracellular matrix stiffness contribute to the expression of USP7 in PDAC.
USP7 promotes PDAC tumor growth in a glycolysis-dependent manner
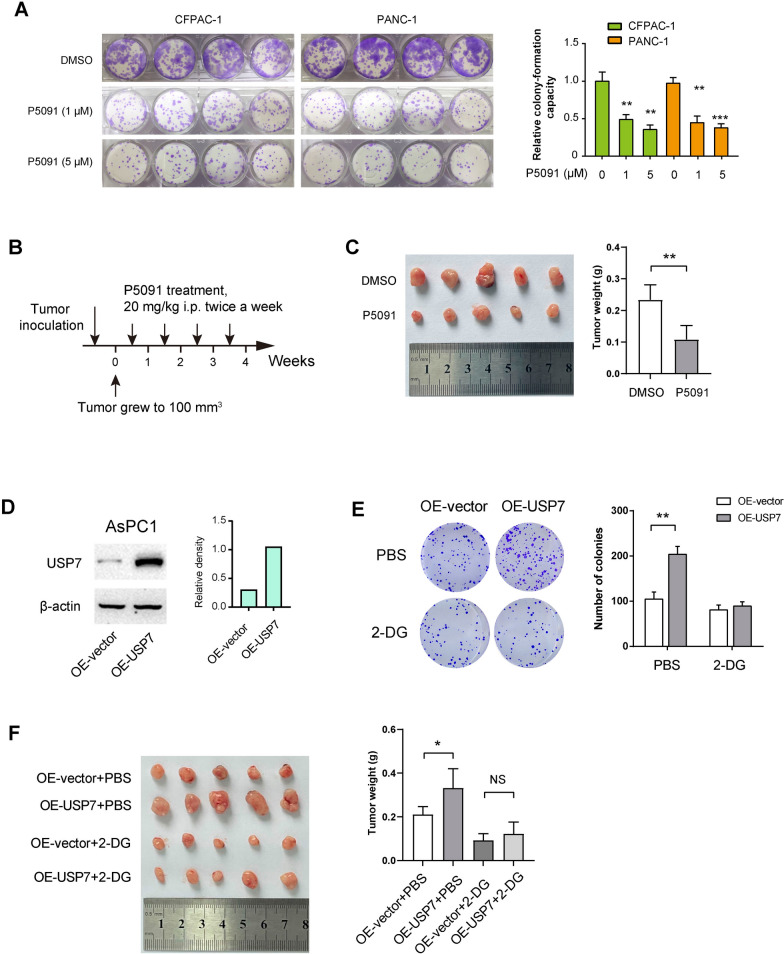
To examine the oncogenic roles of USP7 in PDAC, we conducted a colony formation assay upon USP7 inhibition. The results demonstrated that P5091 inhibited the colony-forming ability of CAFAC-1 and PANC1 cells in a dose-dependent manner (Fig. 4A). Additionally, we established a subcutaneous xenograft model in Balb/c nude mice by injecting PANC1 cells. Once the tumors reached a size of 100 mm3, P5091 was administered via intraperitoneal injection (Fig. 4B). After four weeks, we observed a significant reduction in tumor burden following P5091 treatment (Fig. 4C).
Fig. 4.
USP7 promotes PDAC tumor growth in a glycolysis-dependent manner.A Plate colony formation experiment showed that different doses of P5091 treatment on the proliferation of CFPAC-1 and PANC1 cells (n = 4 per group). B Treatment schedule of P5091 in a subcutaneous xenograft model (n = 5 per group). Once the tumors grew to an approximate size of 100 mm³, P5091 was administered. C The tumor weight of PANC1-derived subcutaneous xenograft tumors upon P5091 treatment. D Western blotting analysis showed the overexpression efficiency of USP7 in AsPC1 cells. E Plate colony formation experiment showed that the effect of USP7 overexpression on the proliferation of AsPC1 cells, with or without 2-DG treatment (n = 3 per group). F The tumor weight of subcutaneous xenograft tumors formed from OE-Control and OE-USP7 AsPC1 cells (n = 5 per group). Mice were given intraperitoneal injections of 500 mg/kg of 2-DG every two days Values were compared by one-way ANOVA multiple comparisons with Tukey’s method among groups (A) and the Student’s t test (C, E, F). Experiments were independently repeated three times (C–E) with similar results. Animal experiments (C, F) were not repeated. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant
To further elucidate the relationship between the oncogenic functions of USP7 and glycolysis, we conducted experiments where USP7 was overexpressed in a PDAC cell line, AsPC1, which has lower endogenous USP7 expression (Fig. 4D). The overexpression of USP7 not only enhanced the colony formation ability of AsPC1 cells but also increased their glycolysis (Fig. 4E and S2). Moreover, the inhibition of glycolysis with 2-DG effectively counteracted the growth-promoting effect induced by USP7 overexpression (Fig. 4E). In an in vivo setting, AsPC1 cells with USP7 overexpression developed larger tumors compared to control cells. To evaluate the impact of glycolysis inhibition, mice were administered intraperitoneal injections of 500 mg/kg of 2-DG every other day. Notably, treatment with 2-DG reversed the growth advantage conferred by USP7 overexpression (Fig. 4F). These findings collectively suggest that USP7 plays a crucial role in linking glycolysis to promote PDAC growth.
USP7 regulates the stability of the c-Myc protein through ubiquitination modification
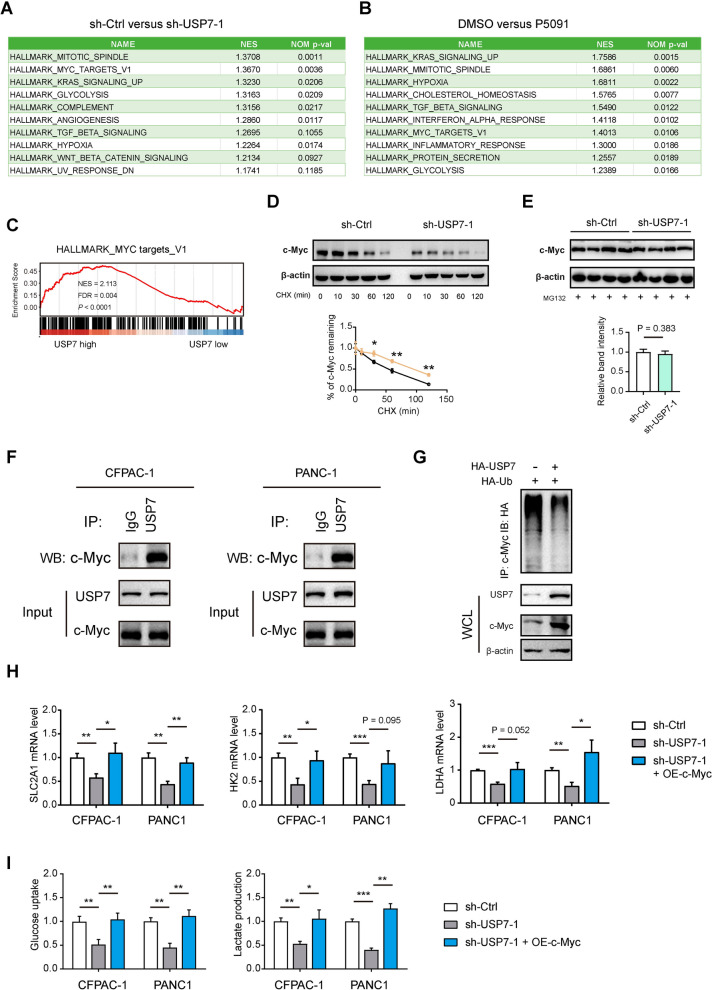
To investigate the molecular mechanism of USP7 in PDAC, RNA sequencing analysis was conducted to analyze the extensive transcriptional changes in PANC1 cells following USP7 knockdown and treatment with P5091 (Fig. 5A, B). Using Hallmark gene sets, we performed GSEA analysis and showed the top 10 pathways. The results indicated that both genetic silencing and pharmacological inhibition of USP7 effectively downregulated several pathways, including MITOTIC SPINDLE, KRAS SIGNALING_UP, GLYCOLYSIS, MYC TARGETS_V1, and HYPOXIA. Given that c-Myc is a key regulator of cell proliferation, growth, and glycolytic metabolism, we focused on c-Myc for further investigation. Using RNA-seq data in the TCGA cohort, we also revealed a close association between USP7 and the expression of c-Myc target genes (Fig. 5C). Consistently, interference with USP7 led to accelerated degradation of the c-Myc protein, suggesting that USP7 influences the stability of c-Myc protein (Fig. 5D). Furthermore, we treated sh-Ctrl and sh-USP7-1 PANC1 cells with MG132, which inhibits proteasomal degradation, and the result showed that the protein levels of c-Myc remain comparable after treatment (Fig. 5E). Co-immunoprecipitation experiments demonstrated the interaction between USP7 and c-Myc (Fig. 5F), while ubiquitin experiments in 293T cells revealed a significant decrease in the ubiquitination level of c-Myc protein upon USP7 overexpression (Fig. 5G). These findings suggest that USP7 may modulate the ubiquitination of c-Myc protein, thereby impacting its stability.
Fig. 5.
USP7 regulates the stability of the c-Myc protein through ubiquitination modification.A, B GSEA analysis showed the top 10 pathways affected by USP7 knockdown or P5091 in PANC1 cells. GSEA was performed with RNA sequencing data of indicated PANC1 cells and the Hallmark gene sets. C GSEA plot showed the link between USP7 expression and c-Myc target genes. Data was generated in the TCGA cohort. D The sh-Ctrl and sh-USP7-1 PANC1 cells were treated with 100 µg/ml CHX for the indicated times (0, 10, 30, 60, 90, 120 min); then, the cell extracts were harvested, and subjected to immunoblotting with c-Myc antibodies. E Western blotting showing the protein levels of c-Myc in sh-Ctrl and sh-USP7-1 PANC1 cells treated with 10 µM MG132 for 6 h (n = 4 per group). F Co-IP analysis of the interaction among USP7 and c-Myc in CFPAC-1 and PANC1 cells. G Cell lysates from OE-Control and OE-USP7 AsPC1 cells were immunoprecipitated with anti-c-Myc, and the immunocomplexes were immunoblotted with antibodies against HA. H Real-time qPCR analysis showed SLC2A1, HK2, and LDHA expression in sh-Ctrl and sh-USP7-1 PDAC cells after ectopic expression of c-Myc (n = 3 per group). I Measurement of glucose uptake and lactate production in sh-Ctrl and sh-USP7-1 PDAC cells after ectopic expression of c-Myc (n = 3 per group) Values were compared by the Student’s t test (D, E) and one-way ANOVA multiple comparisons with Tukey’s method among groups (H, I). Experiments were independently repeated two (F, G) or three times (D, E, H, I) with similar results. RNA sequencing analysis (A, B) was not repeated. *P < 0.05, **P < 0.01, ***P < 0.001
c-Myc is a transcription factor known to stimulate the expression of important glycolytic components such as SLC2A1, HK2, and LDHA [29, 38]. Real-time qPCR revealed that the decreased the mRNA expression of these glycolytic genes induced by USP7 knockdown was significantly reversed by introducing exogenous c-Myc (Fig. 5H). Consistently, glucose uptake and lactate production in sh-USP7-1 CFPAC-1 and PANC1 cells were also restored by overexpression of c-Myc (Fig. 5I), highlighting the critical involvement of the USP7-c-Myc axis in regulating PDAC cell glycolysis.
Targeting USP7 in a transgenic mouse model of PDAC inhibits tumor progression and glycolytic metabolism
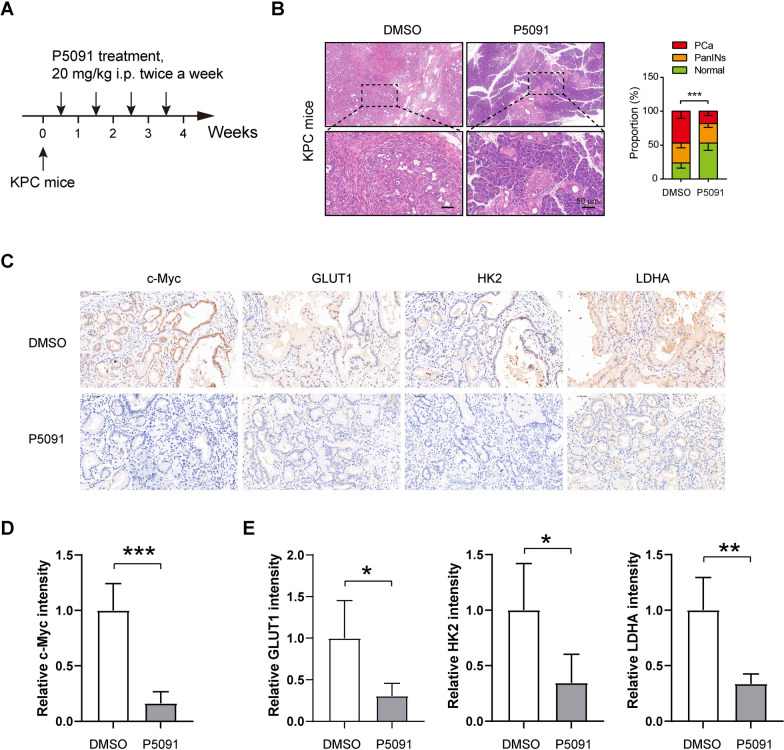
To investigate the potential targeting benefits of USP7, we utilized the established transgenic mouse model for spontaneous pancreatic cancer (LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx1-Cre; KPC) [33, 39], and employed the inhibition of USP7 using P5091 (Fig. 6A). Mice treated with 20 mg/kg of P5091 for 4 weeks exhibited a significant inhibition in the progression of precancerous lesions and tumors in KPC mice. Specifically, there was an increase in the area of normal pancreas, while the tumor area decreased, as displayed in Fig. 6B. Furthermore, immunohistochemical analysis revealed that c-Myc protein level in ductal cancer cells was markedly decreased by P5091 treatment (Fig. 6C, D). Additionally, the expression levels of glycolysis-related proteins, such as GLUT1, HK2, and LDHA, were also reduced following P5091 treatment (Fig. 6C and E). These findings provide strong evidence supporting the role of USP7 in promoting tumor glycolysis and cell growth.
Fig. 6.
Targeting USP7 in a transgenic mouse model of PDAC inhibits tumor progression and glycolytic metabolism.A Treatment schedule of P5091 in the KPC mice (n = 5 per group). B Histological analysis of pancreas lesions upon P5091 treatment in KPC mice. PanINs: pancreatic intraepithelial neoplasia. C IHC analysis of c-Myc, GLUT1, HK2, and LDHA in KPC pancreas lesions upon P5091 treatment. Scale bar: 50 μm. D Quantification data of c-Myc staining in C (n = 5 per group). E Quantification data of GLUT1, HK2, and LDHA staining in C (n = 5 per group) Values were compared by Fisher’s exact test (B) and the Student’s t test (C, E). Animal experiment in this figure was not repeated. *P < 0.05, **P < 0.01, ***P < 0.001
USP7 expression correlates c-Myc and glycolytic proteins in clinical samples
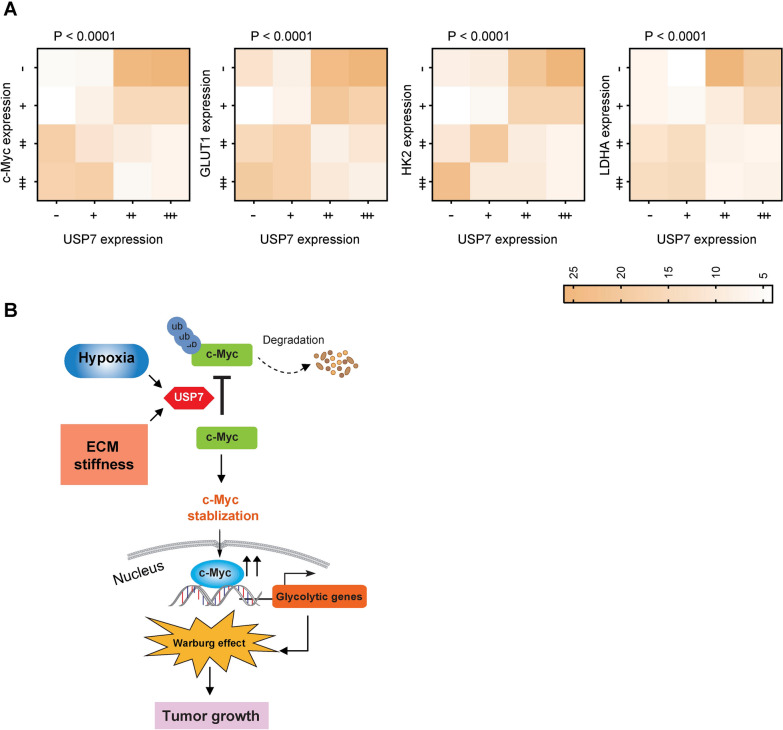
To further elucidate the relationship between USP7 and the c-Myc-glycolysis axis, we conducted IHC analysis on a tissue microarray comprising 205 samples. Consecutive sections were employed for this purpose. The results revealed a significant association between USP7 and c-Myc expression in PDAC tissues (Fig. 7A). Additionally, a similar correlation was observed between USP7 and key glycolytic proteins, including GLUT1, HK2, and LDHA (Fig. 7A). This finding supports the close link among the USP7-c-Myc-glycolysis axis within a clinical context.
Fig. 7.
USP7 expression correlates c-Myc and glycolytic proteins in clinical samples.A Analysis of the correlation between USP7, c-Myc, and glycolytic proteins (GLUT1, HK2, and LDHA) in a PDAC tissue microarray. P values were compared by the Spearman’s rank correlation methods. B The schematic diagram illustrates that the stiffness of the extracellular matrix and a hypoxic microenvironment trigger the expression of USP7. USP7, in turn, boosts the stability of c-Myc by facilitating its ubiquitination. Consequently, c-Myc promotes the transcription and regulation of glycolytic genes, leading to elevated glycolysis levels and fostering tumor growth
Discussion
USP7 plays multifaceted roles in cancer biology, including its impact on cell proliferation, invasiveness, immune evasion, and stem cell maintenance [15, 23, 26, 40], underscoring its significance as a potential therapeutic target for various cancer types. In this study, we further broaden the knowledge of USP7 in the Warburg effect of PDAC cells and decipher c-Myc as a direct target of USP7 to regulate the glycolytic metabolism (Fig. 7B).
USP7 has been implicated in driving various malignant characteristics in PDAC, including facilitating cell cycle progression, promoting resistance to apoptosis, enhancing DNA damage repair mechanisms, and inducing epithelial-to-mesenchymal transition (EMT) [41–43]. Recent studies have highlighted a novel role for USP7 in non-small-cell lung cancer (NSCLC), where it interacts with c-Abl to inhibit its K48-linked polyubiquitination, leading to increased c-Abl stability [24]. This stabilization of c-Abl by USP7 further direct phosphorylating and stabilizing hexokinase-2, which ultimately contributes to NSCLC promotes glycolysis, cell proliferation, and metastasis [24]. In line with this finding, our research has demonstrated the critical involvement of USP7 in PDAC tumor growth by modulating glycolytic metabolism. We observed that overexpression of USP7 enhances PDAC cell growth, which can be effectively inhibited by blocking glycolysis using 2-DG. Notably, we identified c-Myc as a direct target of USP7 in this context. Interestingly, inhibition of USP7 activity results in c-Myc ubiquitination, degradation, reduced nuclear translocation, and suppression of EMT signaling in lung adenocarcinoma [44]. Furthermore, the regulatory axis between USP7 and c-Myc appears to be conserved across different cellular systems, as evidenced by its involvement in hepatocellular carcinoma [45, 46] and neural stem cells [47]. These findings suggest a universal mechanism by which USP7 modulates c-Myc activity to regulate cellular processes associated with tumorigenesis and metastasis. The insights gained from our findings and others shed light on the potential therapeutic implications of targeting the USP7-c-Myc axis in various cancer contexts. However, the specific ubiquitination sites on c-Myc that are targeted by the proteasome pathway and how USP7 counteracts this process warrant further investigations. Except for c-Myc, USP7 also governs the ubiquitination of various other proteins integral to glycolytic metabolism, including HIF-1α [48]. Thus, we cannot entirely dismiss the potential that USP7 may exert its influence on additional targets, thereby impacting glycolysis in a broader context. Indeed, c-Myc serves as a prominent target gene within the Wnt signaling pathway, and β-Catenin has been shown to be influenced by USP7 [49–51]. Notably, USP7 functions as a cancer-specific activator of Wnt signaling, enhancing Wnt/β-Catenin pathway activity through the deubiquitination of β-Catenin [50]. The USP7 inhibitor P5091 has been demonstrated to effectively inhibit Wnt signaling and tumor growth, underscoring the pivotal role of USP7 in this context [28]. Previously, our research has revealed that WDR1 interacts with USP7, thereby preventing the ubiquitination-mediated degradation of β-Catenin, which subsequently promotes the expression of c-Myc [22]. This intricate interplay suggests that USP7 may engage with Wnt/β-Catenin signaling to drive glycolytic processes in PDAC.
Several approaches have been explored to inhibit USP7 and disrupt its pro-tumorigenic functions in cancers [13, 20]. To aid the evidence of targeting USP7 in PDAC, we verified the therapeutic effects of P5091 in KPC mice and showed the remarkable anti-tumor effect of P5091 in this genetically engineered mouse model. P5091 can effectively block the deubiquitinating activity of USP7, leading to the destabilization of its oncogenic substrates such as FOXM1, oncoprotein SE translocation (SET), and N-myc [27, 52, 53]. By disrupting the interaction between USP7 and its target proteins, P5091 exhibits anti-tumor activity in various cancer types. Combination therapy using P5091 with standard chemotherapeutic agents or targeted inhibitors has shown synergistic effects in suppressing tumor growth and overcoming drug resistance mechanisms. For instance, P5091 is able to downregulate FGL1, a major ligand of LAG3, thus enhancing CD8+ T cell activity [54]. Moreover, P5091 induces melanoma cell senescence and sensitizes cells to HDAC/LSD1 inhibitors [55]. In multiple myeloma, combining P5091 with lenalidomide, HDAC inhibitor SAHA, or dexamethasone shows synergistic anti-tumor activities [56]. However, whether P5091 showed a synergistic effect with the first-line chemotherapy for PDAC remain unclear. Further research is needed to investigate the functional consequences of the USP7-c-Myc interaction, particularly focusing on how inhibiting USP7 affects c-Myc target gene expression and overall cellular outcomes. Moving from preclinical studies to clinical trials involves numerous challenges, including patient selection and biomarker identification. Therefore, using USP7 expression as a potential biomarker for personalized medicine in cancer therapy is a promising approach that could help tailor treatment strategies to individual patients.
In conclusion, our study offers valuable insights into USP7-dependent glycolytic metabolism in PDAC. Targeting USP7 holds promise as a novel therapeutic strategy for disrupting glycolytic metabolism and improving treatment outcomes in PDAC. Further research into the specific pathways and interactions involved in the USP7-c-Myc-mediated regulation of glycolysis may pave the way for the development of targeted therapies tailored to combat metabolic reprogramming in PDAC.
Supplementary Information
Author contributions
Jichun Gu, Xi Xiao and Hengchao Li designed the experiments. Jichun Gu, Xi Xiao, Caifeng Zou, Yishen Mao, Chen Jin and Rongkun Li generated the methods and performed the experiments. Jichun Gu, Rongkun Li, Deliang Fu and Hengchao Li performed the data analysis and interpretation. Jichun Gu, Rongkun Li and Hengchao Li wrote the manuscript.
Funding
This work was supported by grants from National Natural Science Foundation of China [Grant No. 82273426, No. 82103105 and GZ1456] and Natural Science Foundation of Jiangsu Province [Grant No. BK20210143].
Availability of data and materials
All data generated or analyzed during this study are included in this manuscript and its supplementary information files.
Declarations
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of Huashan Hospital, Fudan University and performed in accordance with the Declaration of Helsinki. All patients involved in this study signed informed consent. All the animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Fudan university.
Consent for publication
All authors agree to submit the article for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jichun Gu, Xi Xiao, and Caifeng Zou have contributed equally to this work.
Contributor Information
Rongkun Li, Email: jiqimaoakun@163.com.
Hengchao Li, Email: lihengchao@huashan.org.cn.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 2.Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Update. 2018;38:1–11. [DOI] [PubMed] [Google Scholar]
- 3.Barba I, Carrillo-Bosch L, Seoane J. Targeting the Warburg effect in cancer: where do we stand? Int J Mol Sci. 2024;25(6):3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu YH, Zheng JH, Jia QY, Duan ZH, Yao HF, Yang J, et al. Immunosuppression, immune escape, and immunotherapy in pancreatic cancer: focused on the tumor microenvironment. Cell Oncol. 2023;46(1):17–48. [DOI] [PubMed] [Google Scholar]
- 5.Chen PC, Ning Y, Li H, Su JG, Shen JB, Feng QC, et al. Targeting ONECUT3 blocks glycolytic metabolism and potentiates anti-PD-1 therapy in pancreatic cancer. Cell Oncol. 2024;47(1):81–96. [DOI] [PubMed] [Google Scholar]
- 6.Zhu LL, Li BT, Li RK, Hu LP, Zhang YL, Zhang ZG, et al. METTL3 suppresses pancreatic ductal adenocarcinoma progression through activating endogenous dsRNA-induced anti-tumor immunity. Cell Oncol. 2023;46(5):1529–41. [DOI] [PubMed] [Google Scholar]
- 7.Ying HQ, Kimmelman AC, Lyssiotis CA, Hua SJ, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras Maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F, Qi XM, Wertz R, Mortensen M, Hagen C, Evans J, et al. p38γ MAPK is essential for aerobic glycolysis and pancreatic tumorigenesis. Cancer Res. 2020;80(16):3251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange SM, Armstrong LA, Kulathu Y, Deubiquitinases. From mechanisms to their inhibition by small molecules. Mol Cell. 2022;82(1):15–29. [DOI] [PubMed] [Google Scholar]
- 10.Dewson G, Eichhorn PJA, Komander D. Deubiquitinases in cancer. Nat Rev Cancer. 2023;23(12):842–62. [DOI] [PubMed] [Google Scholar]
- 11.Snyder NA, Silva GM. Deubiquitinating enzymes (DUBs): regulation, homeostasis, and oxidative stress response. J Biol Chem. 2021;297(3):101077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Li JL, Chen ZH, Jiang K, Yang KY, Huang F, et al. VE-822 upregulates the deubiquitinase OTUD1 to stabilize FHL1 to inhibit the progression of lung adenocarcinoma. Cell Oncol. 2023;46(4):1001–14. [DOI] [PubMed] [Google Scholar]
- 13.Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550(7677):481–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manea T, Nelson JK, Garrone CM, Hansson K, Evans I, Behrens A, et al. USP7 controls NGN3 stability and pancreatic endocrine lineage development. Nat Commun. 2023;14(1):2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Sun LG, Tan YJ, Wang Q, Luo T, Li CL, et al. USP7 represses lineage differentiation genes in mouse embryonic stem cells by both catalytic and noncatalytic activities. Sci Adv. 2023;9(20):eade3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumagai J, Kiuchi M, Kokubo K, Yagyu H, Nemoto M, Tsuji K, et al. The USP7-STAT3-granzyme-Par-1 axis regulates allergic inflammation by promoting differentiation of IL-5-producing Th2 cells. Proc Natl Acad Sci USA. 2023;120(49):e2302903120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czech-Sioli M, Siebels S, Radau S, Zahedi RP, Schmidt C, Dobner T, et al. The Ubiquitin-specific protease Usp7, a novel Merkel cell polyomavirus large T-antigen interaction partner, modulates viral DNA replication. J Virol. 2020;94(5):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Li J, Wang YJ, Liu XX, Yang X, Liao ZY, et al. Deubiquitinase USP7 stabilizes KDM5B and promotes tumor progression and cisplatin resistance in nasopharyngeal carcinoma through the ZBTB16/TOP2A axis. Cell Death Differ. 2024;31(3):309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park HB, Baek KH. Current and future directions of USP7 interactome in cancer study. Bba-Rev Cancer. 2023;1878(6):188992. [DOI] [PubMed] [Google Scholar]
- 20.Korenev G, Yakukhnov S, Druk A, Golovina A, Chasov V, Mirgayazova R, et al. USP7 inhibitors in cancer immunotherapy: current status and perspective. Cancers. 2022;14(22):5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LQ, Kumar S, Dahiya S, Wang F, Wu J, Newick K, et al. Ubiquitin-specific protease-7 inhibition impairs Tip60-dependent Foxp3 + T-regulatory cell function and promotes antitumor immunity. EBioMedicine. 2016;13:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li HC, Liu XH, Jiang SH, Zhou XW, Yao L, Di Y, et al. WD repeat-containing protein 1 maintains β-Catenin activity to promote pancreatic cancer aggressiveness. Br J Cancer. 2020;123(6):1012–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai XM, Lu LS, Deng SK, Meng JS, Wan C, Huang J, et al. USP7 targeting modulates anti-tumor immune response by reprogramming tumor-associated macrophages in lung cancer. Theranostics. 2020;10(20):9332–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He YM, Jiang SY, Zhong YY, Wang XG, Cui YL, Liang JP, et al. USP7 promotes non-small-cell lung cancer cell glycolysis and survival by stabilizing and activating c-Abl. Clin Transl Med. 2023;13(12):e1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ZR, Kang WT, Li OW, Qi FY, Wang JW, You YH, et al. Abrogation of USP7 is an alternative strategy to downregulate PD-L1 and sensitize gastric cancer cells to T cells killing. Acta Pharm Sin B. 2021;11(3):694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha G, Sarkar S, Mohanta PS, Kumar K, Chakrabarti S, Basu M, et al. USP7 targets XIAP for cancer progression: establishment of a p53-independent therapeutic avenue for glioma. Oncogene. 2022;41(47):5061–75. [DOI] [PubMed] [Google Scholar]
- 27.Yi JJ, Li H, Chu B, Kon N, Hu XP, Hu JP, et al. Inhibition of USP7 induces p53-independent tumor growth suppression in triple-negative breast cancers by destabilizing FOXM1. Cell Death Differ. 2023;30(7):1799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An T, Gong YX, Li X, Kong LM, Ma PC, Gong L, et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem Pharmacol. 2017;131:29–39. [DOI] [PubMed] [Google Scholar]
- 29.Fang Y, Shen ZY, Zhan YZ, Feng XC, Chen KL, Li YS, et al. CD36 inhibits β-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4 to repress colorectal tumorigenesis. Nat Commun. 2019;10:3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang WW, Zheng YH, Xia Y, Ji HT, Chen XM, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang XK, Guo SQ, Wang S, Zhang YY, Chen HJ, Wang Y, et al. EIF4A3-induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 2022;82(5):831–45. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Yao HF, Li H, Su T, Jiang SH, Wang H et al. Transglutaminases are oncogenic biomarkers in human cancers and therapeutic targeting of TGM2 blocks chemoresistance and macrophage infiltration in pancreatic cancer. Cell Oncol (Dordr). 2023;46:1473. [DOI] [PubMed] [Google Scholar]
- 33.Jiang SH, Li J, Dong FY, Yang JY, Liu DJ, Yang XM, et al. Increased Serotonin signaling contributes to the warburg effect in pancreatic tumor cells under metabolic stress and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;153(1):277–91. [DOI] [PubMed] [Google Scholar]
- 34.Ye YQ, Hu QS, Chen H, Liang K, Yuan Y, Xiang Y, et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat Metabolism. 2019;1(4):431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao JX, Yang G, Zhou WC, Qiu JD, Chen GY, Luo WH, et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J Hematol Oncol. 2021;14(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat Rev Gastro Hepat. 2020;17(8):487–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng JH, Zhu YH, Yang J, Ji PX, Zhao RK, Duan ZH, et al. A CLIC1 network coordinates matrix stiffness and the Warburg effect to promote tumor growth in pancreatic cancer. Cell Rep. 2024;43(8):114633. [DOI] [PubMed] [Google Scholar]
- 38.Ala M. Target c-Myc to treat pancreatic cancer. Cancer Biol Ther. 2022;23(1):34–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang SH, Zhu LL, Zhang M, Li RK, Yang Q, Yan JY, et al. GABRP regulates chemokine signalling, macrophage recruitment and tumour progression in pancreatic cancer through tuning KCNN4-mediated Ca(2+) signalling in a GABA-independent manner. Gut. 2019;68(11):1994–2006. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Dai YB, Ge H, Guo SS, Zhang W, Wang YL et al. The deubiquitinase USP7 promotes HNSCC progression via deubiquitinating and stabilizing TAZ. Cell Death Dis. 2022;13(8):677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng X, Zhang B, Guo F, Wu HS, Jin X. Deubiquitination of FBP1 by USP7 blocks FBP1-DNMT1 interaction and decreases the sensitivity of pancreatic cancer cells to PARP inhibitors. Mol Oncol. 2022;16(7):1591–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang KQ, Sun T, Li WD, Guo YM, Li AM, Hsieh M, et al. Inhibition of USP7 upregulates USP22 and activates its downstream cancer-related signaling pathways in human cancer cells. Cell Commun Signal. 2023;21(1):319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Zhu XL, Sun R, Ma PP, Zhang EH, Wang Z, et al. Ubiquitin-specific protease 7 is a druggable target that is essential for pancreatic cancer growth and chemoresistance. Invest New Drug. 2020;38(6):1707–16. [DOI] [PubMed] [Google Scholar]
- 44.Liu JH, Yang HL, Deng ST, Hu Z, Chen WF, Yan WW, et al. The small molecule chemical compound cinobufotalin attenuates resistance to DDP by inducing ENKUR expression to suppress MYH9-mediated c-Myc deubiquitination in lung adenocarcinoma. Acta Pharmacol Sin. 2022;43(10):2687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou RT, Li YH, Luo XJ, Zhang W, Yang HL, Zhang YW, et al. ENKUR expression induced by chemically synthesized cinobufotalin suppresses malignant activities of hepatocellular carcinoma by modulating β-catenin/c-Jun/MYH9/USP7/c-Myc axis. Int J Biol Sci. 2022;18(6):2553–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun LB, He M, Li F, Wu D, Zheng P, Zhang C et al. Oxyberberine sensitizes liver cancer cells to sorafenib via inhibiting NOTCH1-USP7-c-Myc pathway. Hepatol Commun. 2024;8(4):e0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicklas S, Hillje AL, Okawa S, Rudolph IM, Collmann FM, van Wuellen T, et al. A complex of the ubiquitin ligase TRIM32 and the deubiquitinase USP7 balances the level of c-Myc ubiquitination and thereby determines neural stem cell fate specification. Cell Death Differ. 2019;26(4):728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li QG, Sun HZ, Luo DK, Gan L, Mo SB, Dai WX, et al. 2024 Lnc-RP11-536 K7.3/SOX2/HIF-1α signaling axis regulates oxaliplatin resistance in patient-derived colorectal cancer organoids. J Exp Clin Canc Res. 2021;40(1):348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basu B, Karmakar S, Basu M, Ghosh MK. USP7 imparts partial EMT state in colorectal cancer by stabilizing the RNA helicase DDX3X and augmenting Wnt/β-catenin signaling. Bba-Mol Cell Res. 2023;1870(4):119446. [DOI] [PubMed] [Google Scholar]
- 50.Novellasdemunt L, Foglizzo V, Cuadrado L, Antas P, Kucharska A, Encheva V, et al. USP7 is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating beta-catenin deubiquitination. Cell Rep. 2017;21(3):612–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harmston N, Lim JYS, Arqués O, Palmer HG, Petretto E, Virshup DM, et al. Widespread Repression of Gene Expression in Cancer by a Wnt/β-Catenin/MAPK Pathway. Cancer Res. 2021;81(2):464–75. [DOI] [PubMed] [Google Scholar]
- 52.Chen JY, Jiao ZS, Liu YJ, Zhang M, Wang DL. USP7 interacts with and destabilizes oncoprotein SET. Biochem Biophys Res Commun. 2024;709:149818. [DOI] [PubMed] [Google Scholar]
- 53.Tavana O, Li DW, Dai C, Lopez G, Banerjee D, Kon N, et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat Med. 2016;22(10):1180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun LL, Zhao LN, Sun J, Yuan HF, Wang YF, Hou CY et al. Inhibition of USP7 enhances CD8 T cell activity in liver cancer by suppressing PRDM1-mediated FGL1 upregulation. Acta Pharmacol Sin. 2024;45:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Granieri L, Marocchi F, Melixetian M, Mohammadi N, Nicoli P, Cuomo A et al. Targeting the USP7/RRM2 axis drives senescence and sensitizes melanoma cells to HDAC/LSD1 inhibitors. Cell Rep. 2022;40(12):111396. [DOI] [PubMed] [Google Scholar]
- 56.Chauhan D, Tian Z, Nicholson B, Kumar KGS, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript and its supplementary information files.