Abstract
Background
Sheep coccidiosis is an infectious parasitic disease that primarily causes diarrhea and growth retardation in young animals, significantly hindering the development of the sheep breeding industry. Cereal grains and animal feeds are frequently contaminated with mycotoxins worldwide, with aflatoxin B1 (AFB1) being the most common form. AFB1 poses a serious threat to gastrointestinal health upon ingestion and affects the function of parenteral organs, thus endangering livestock health. However, the impact of the combined effects of coccidia and AFB1 on the reproductive system of sheep has not been reported. Therefore, this study utilized sheep as an animal model to investigate the mechanisms underlying the reproductive toxicity induced by the individual or combined effects of AFB1 and Eimeria ovinoidalis (E. ovinoidalis) on the gut–blood–reproductive axis.
Results
The results showed that AFB1 and coccidia adversely affect the reproductive system defense of sheep by altering uterine histopathology and hormone levels and triggering inflammation, which is associated with changes in the gut microbiota and metabolites. Moreover, co-exposure to AFB1 and coccidia disrupted the intestinal structure of the colon, resulting in reduced crypt depth. The impaired barrier function of the colon manifests primarily through the suppression of barrier protein expression, changes in the gut microbiome composition, and disruptions in gut metabolism. Importantly, the levels of blood inflammatory factors (IL-6, IL-10, TNF-α, and LPS) increased, suggesting that exposure to AFB1 and coccidia compromises the function of uterine organs in sheep by perturbing the gut–blood–reproductive axis. Blood metabolomics analysis further revealed that the differential metabolites predominantly concentrate in the amino acid pathway, particularly N-acetyl-L-phenylalanine. This metabolite is significantly correlated with IL-6, TNF-α, LPS, ERα, and ERβ, and it influences hormone levels while inducing uterine damage through the regulation of the downstream genes PI3K, AKT, and eNOS in the relaxin signaling pathway, as demonstrated by RNA sequencing.
Conclusions
These findings reveal for the first time that the combined effects of AFB1 and E. ovinoidalis on sheep uterine function operate at the level of the gut–blood–reproductive axis. This suggests that regulating gut microbiota and its metabolites may represent a potential therapeutic strategy for addressing mycotoxins and coccidia-co-induced female reproductive toxicity.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01966-y.
Keywords: Mycotoxin, Aflatoxin B1, Coccidia, E. ovinoidalis, Reproduction toxicity, Gut microbiota
Introduction
Sheep coccidiosis, caused by the genus Eimeria, is an infectious disease that primarily affects young animals, developing in the intestines [1]. The clinical infections, characterized by diarrhea and subclinical infections, which result in stunted growth and development, caused by this parasite lead to a deterioration in the quality of meat, skin, wool, and other derived products. Under intensive breeding conditions, accompanied by high animal density and productivity, the prevention and control of sheep coccidiosis hold significant economic importance [2]. Eimeria ovinoidalis (E. ovinoidalis), a type of coccidian parasite, has been identified as the most pathogenic species among sheep worldwide [3]. In sheep, the primary clinical symptoms include diarrhea and catarrhal enteritis, along with specific pathological findings [4, 5]. The colon, ileum, and cecum are the organs primarily affected by the pathological changes induced by E. ovinoidalis.
The contamination of mycotoxins is a significant challenge faced by global agricultural production. Research indicates that approximately 75% of feed worldwide is contaminated with mycotoxins, presenting serious health risks to livestock [6, 7]. Aflatoxin B1 (AFB1), a mycotoxin produced by Aspergillus flavus and Aspergillus parasiticus, is a member of the aflatoxin family and is commonly found in cereals and animal feed all over the world, together with other fungal toxins [8, 9]. The toxicity and dangers of aflatoxins have gained considerable attention, making their detection and management a crucial aspect in the field of food safety. The Food and Drug Administration (FDA) of the USA has established the maximum limits for total aflatoxins (B1, B2, G1, and G2) in all foods at 20 µg/kg, with a specific restriction of 100 µg/kg for peanut and corn feed products [10, 11]. Meanwhile, the European Commission has established lower thresholds of 2 µg/kg for AFB1 and 4 µg/kg for total aflatoxins [12]. Notably, excessive exposure to AFB1 has been linked to some serious health issues in animals, including immunotoxicity, hepatotoxicity, nephrotoxicity, carcinogenicity, and reproductive and developmental toxicity [13–15]. Furthermore, AFB1 and its metabolites can persist in major body tissues, posing significant health risks to people consuming contaminated meat [9], while most studies on the effects of AFB1 on animal tissues and organs have primarily concentrated on the liver and kidney [16, 17]. It is essential to note that high doses of AFB1 can also lead to reproductive toxicity in female animals, resulting in reduced fertility and potential miscarriages [18, 19]. Given the widespread prevalence of aflatoxins in grains and feed, investigating their effects on reproductive toxicity is of paramount importance.
The gastrointestinal tract is the initial organ in the body that interacts with harmful substances. Research has shown that AFB1 and coccidia invasion can alter the abundance of the gut microbiota and its metabolites, leading to the proliferation of pathogenic or opportunistic pathogens and compromising intestinal barrier function [20, 21]. As the first line of defense against harmful substances, the intestine plays a crucial role in regulating the immune system, the endocrine system, and the digestive process [13]. Following intestinal intake and absorption, mycotoxins can cause direct damage through biotransformation and metabolism of target organs. They can also induce changes in blood-related substances via the intestinal mucosa or red blood cell metastasis [22, 23]. To date, it remains unclear whether AFB1-induced reproductive toxicity is associated with changes in gut microbiota-derived metabolites or blood metabolism influenced by the synergistic effects of coccidia. Therefore, gaining a deeper understanding of the interactions among the host, microbiota-derived metabolites, and blood metabolism could help elucidate potential mechanisms underlying the reproductive toxicity induced by the combined effects of AFB1 and coccidia.
Sheep are a significant source of meat, leather, wool, and dairy products worldwide, making them vital for global food security. However, the sheep farming industry faces considerable challenges due to the presence of fungal toxins in feed. Moreover, the susceptibility and resistance to sheep coccidiosis urgently require the attention of researchers [24]. While numerous studies have focused on animals with compromised AFB1 toxicity and coccidia infection alone, the pervasive presence of mycotoxins and the high infection rate of coccidia mean that sheep often experience co-exposure involving multiple toxins or various coccidia [25–27]. Therefore, it is of great economic significance to investigate cases of AFB1 exposure in conjunction with coccidia infection. Furthermore, enhancing our understanding of the interactions between these two pathogenic factors within the gut–blood–reproductive axis may improve feed efficiency and overall health in sheep farming, thereby promoting the development of sustainable sheep production practices based on scientific knowledge. Consequently, this study aims to explore the main effects of the combined action of AFB1 and coccidia on the uterus and intestines of sheep, while also investigating the interactions between these two pathogenic factors in the context of the gut–blood–reproductive axis. This study will elucidate the underlying mechanisms and provide novel insights into the combined toxic effects of AFB1 and coccidia.
Materials and methods
Chemical reagents
AFB1 was purchased from Qingdao Pribolab Chemical Inc. (Shandong, China) and was dissolved in dimethyl sulfoxide (DMSO) solution before use. DMSO was sourced from Tianjin Fuyu Fine Chemical Co., Ltd. (Tianjin, China). The unsporulated oocysts of E. ovinoidalis were obtained from the Laboratory of Parasites at Henan Agricultural University. The oocysts were treated with a 2.5% potassium dichromate solution facilitate sporulation. Enzyme-linked immunosorbent assay (ELISA) kits for the following assays were supplied by Shanghai Enzyme-linked Biotechnology Co., Ltd. (Shanghai, China): interleukin-6 (IL-6, #YJ622840), interleukin-10 (IL-10, #YJ507611), tumor necrosis factor-α (TNF-α, #YJ220635), lipopolysaccharides (LPS, #YJ100325), estrogen receptor α (ERα, #YJ155970), and estrogen receptor β (ERβ, #YJ155971).
Animal ethics
All animal experimental procedures were conducted following the guidelines of the Animal Welfare and Ethics Research Committee of the College of Veterinary Medicine at Henan Agricultural University (ethical permission code: HNND2022030818) in Zhengzhou, China. There were no signs of pain or suffering exhibited by the animals prior to their euthanasia during the experiment. Furthermore, all the possible measures were taken to ensure the welfare of the sheep throughout the study.
Experimental procedure
The experiment was conducted at Luoyang Xinning Animal Husbandry Technology Co., Ltd. in Luoyang City, Henan Province. One hundred healthy weaned ewes, 35 days old, were selected and 1 mg/kg of Diclazuril was administered for two consecutive days to eliminate potential coccidia infections. The ewes were housed in newly constructed, flame-sterilized pens. On the 11th day after medication, each ewe underwent rectal sampling and microscopic examination for 3 consecutive days. Sixteen healthy female Hu sheep weighing approximately 8–12 kg and free from coccidia, cestode, and trematode infections were selected and assigned to four groups: control (Con) (n = 4), AFB1 group (n = 4), E. ovinoidalis (E.o) group (n = 4), and AFB1 + E.o group (n = 4). Each group was housed in separate new enclosures, with one enclosure left empty between two experimental enclosures to prevent mingling among the lambs.
The experimental design is illustrated in Fig. 1a. The Con group received daily oral administration of PBS solution and DMSO. The AFB1 group received daily oral administration of AFB1 at a dosage of 60 μg/kg. The E.o group received a single oral administration of 2 × 104 sporulated oocysts on day 0 only. The AFB1 + E.o group received a single oral administration of 2 × 104 sporulated oocysts on day 0 along with daily oral administration of AFB1 at a dosage of 60 μg/kg. The experimental sheep were fed lamb starter pellets and roughage, consisting of peanut vine and corn stalk, sourced from Liantai Biotechnology Co., Ltd. (Hebi City, Henan Province). The lambs were raised in a free-range environment with unlimited access to water and were fed a fixed amount of pellets feed and peanut vine daily at 8 am and 4 pm. The unconsumed feed was collected and weighed daily. Lamb body weights were recorded throughout the 14-day experimental period.
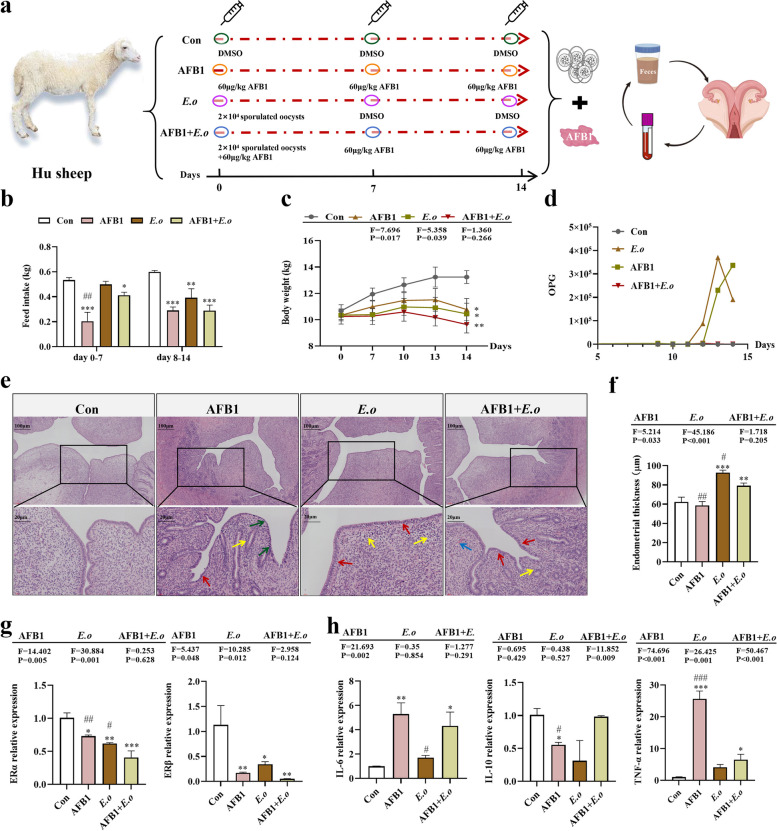
Fig. 1.
The reproductive toxicity of sheep induced by AFB1 and coccidia exposure is associated with alterations in uterine hormones and inflammation. a Experimental design flowchart. b Feed intake of sheep over 0–14 days (n = 4). c Body weight changes of sheep over 0–14 days (n = 4). d Number of oocysts per gram (OPG) in each group (n = 4). e H&E staining of uterine tissue (100×, 400×; n = 4). Red arrows indicate endometrial epithelial cells, yellow arrows indicate inflammatory cells, green arrows denote nuclear shrinkage and dense staining, and blue arrows indicate bleeding spots. f Measurement of endometrial thickness (n = 4). g Expression levels of ERα and ERβ mRNA in uterine tissue (n = 3). h mRNA expression levels of IL-6, IL-10, and TNF-α in uterine tissue (n = 3). Statistical significance is denoted by *p < 0.05, **p < 0.01, and ***p < 0.001 (AFB1, E.o, and AFB1+E.o vs Con); or #p < 0.05, ##p < 0.01, and ###p < 0.001 (AFB1+E.o vs AFB1 and E.o)
Sample collection
Starting from day 7, rectal sampling of each lamb was conducted daily, with 5 g of feces collected in CPE gloves. Lamb identification and fecal consistency were recorded and scored according to the fecal scoring criteria outlined in Table S1. On both day 7 and day 14 of the experiment, 7 mL of blood was collected from the jugular vein of each lamb before morning feeding. Of the 7 mL collected, 2 mL of whole blood sample was placed in blood collection tubes, gently inverted to mix, and kept on dry ice within an insulated box before being immediately sent for hematology analysis. The remaining 5 mL of blood was centrifuged at 4 °C and 3000 rpm for 15 min, with the supernatant stored at − 80 °C for further analysis. Additionally, fresh feces were collected on day 14 and stored at 4 °C and − 80 °C for subsequent analysis. After 14 days, the sheep were fasted for 12 h and euthanized via exsanguination through the jugular vein. The uterus of each sheep was collected, washed with physiological saline, and weighed for record-keeping. The midsection of the colon and uterine tissues was fixed in 4% paraformaldehyde for histopathological examination. The remaining colon and uterine tissues were rapidly frozen in liquid nitrogen and stored at − 80℃ for subsequent molecular experiments.
Histopathological analysis and coccidia oocysts detection
The tissues from the uterus and colon were preserved in 4% paraformaldehyde. All specimens were deparaffinized in xylene, dehydrated in graded ethanol solutions, and embedded in paraffin using a fully enclosed tissue processor (ASP300S, Leica Biosystems, Buffalo Grove, IL, USA). The sections were stained with hematoxylin and eosin (HE) and then visualized using a Mito More Tan microscope. Additionally, the crypt depth of the intestines and the thickness of the uterine endometrium were evaluated using Motic Images Advanced 3.2 software (Motic China Group, Co., China).
Hematology analysis
The complete blood count parameters were analyzed by the Zhengzhou Furunde Pet Hospital in Henan Province, using the Maestro BC-5000Vet analyzer. An ELISA method was employed to measure the levels of inflammatory factors IL-6, IL-10, TNF-α, and LPS, as well as the expression of ERα and ERβ in plasma.
RNA extraction and RT-qPCR
Trizol reagent and liquid nitrogen were utilized to extract total mRNA from the colon and uterus. The quality of the mRNA was assessed using a Nanodrop spectrophotometer, followed by cDNA synthesis through reverse transcription according to the manufacturer’s protocol. After cDNA synthesis was completed, real-time fluorescence quantitative PCR (qPCR) was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd., China). The 2−ΔΔCT calculation method was employed to determine mRNA expression levels. Specific primers for GAPDH mRNA and target gene sequences (including ERα, ERβ, IL-6, IL-10, TNF-α, PI3K, AKT, eNOS, COL4A4, FOSB, COL11A1, GNG2, Claudin1, Claudin4, Occludin, and ZO-1) were designed using Primerbank from Shanghai Biotech Co., Ltd. and synthesized by Tsingke Biological Technology Co., Ltd. The primers used in this study are listed in Table S2. The GAPDH gene served as an internal reference.
16S rRNA sequence analysis
Microbial genomic DNA samples were extracted from feces using the QIAamp DNA kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions and stored at − 20 °C prior to further analysis. The quantity and quality of the extracted DNA were measured using a spectrophotometer and assessed via agarose gel electrophoresis, respectively. PCR amplification of the bacterial 16S rRNA gene V3–V4 region was conducted using the forward primer 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and the reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’). DNA libraries were sequenced on the Illumina MiSeq 250 platform. Sequence data analysis was primarily performed using QIIME2 with the dada2 plugin and the R software package (v3.2.0). Microbial community diversity and abundance were subsequently evaluated through α-diversity and β-diversity analysis. The Gene Cloud tool (https://www.genescloud.cn) was utilized for analyzing the 16S sequencing-related data.
Metabolomic analysis of feces and blood
Fecal and blood samples were collected and added to a pre-cooled methanol/acetonitrile/water solution (2:2:1, v/v) after being slowly thawed at 4℃. Following vortexing and 30 min of low-temperature ultrasonication, the mixture was allowed to stand at − 20℃ for 10 min. After centrifugation at 14,000 g for 20 min at 4℃, the supernatant was vacuum dried. For mass spectrometry analysis, 100 μL of an acetonitrile–water solution (1:1, v/v) was added for re-dissolution. The mixture was then vortexed and centrifuged at 14,000 g for 15 min at 4℃. After filtration through a 0.22-μm membrane, 300 μL of the supernatant was transferred to a detection vial for untargeted metabolomics liquid chromatography-mass spectrometry (LC–MS) analysis.
The raw data were converted to mzXML format using ProteoWizard, followed by peak alignment, retention time correction, and peak area extraction which were performed using XCMS software. The data extracted by XCMS underwent metabolite structure identification and preprocessing, followed by an assessment of experimental data quality, and ultimately data analysis. All analyses were based on an in-house database (Shanghai Applied Protein Technology). Metabolites with a log fold change (log2FC) ≥ 1 and p-value < 0.05 were considered significantly regulated metabolites. The identified metabolites were annotated, mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database, and further subjected to metabolite set enrichment analysis. Principal component analysis (PCA), orthogonal partial least squares discriminant analysis (OPLS-DA), and volcano plots were conducted using the online tools available at https://www.omicstudio.cn/tool. And KEGG enrichment analysis was performed using https://www.omicshare.com/tools/. The tool available at (https://www.genescloud.cn) was utilized to analyze network diagrams and network heat maps. The chord and heat maps were generated using https://hiplot.com.cn/home/index.html.
Uterine transcriptomics analysis
Total RNA was extracted from the uterus using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). Following the manufacturer’s protocol, RNA-seq libraries were prepared at BGI Genomics (Shenzhen, China) using the Illumina TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA). The raw sequencing data underwent quality control using the Fastp software for Q20 and Q30, resulting in clean reads that were subsequently visualized using the FastQC. The quality-controlled clean reads were aligned to the corresponding species’ reference genome and gene structure annotation using the HISAT2 software. HTSeq v0.6.1 software was employed to count the number of reads, and then the number of fragments per kilobase of transcript and per million mapped fragments for each gene was calculated. To accurately reflect the expression level of transcripts, normalization was performed based on the number of mapped reads in the sample and the length of the transcripts. Fragments per kilobase of transcript per million fragments mapped (FPKM) were used as the metric for gene expression levels. Differential gene expression analysis was performed using DESeq2 (with biological replicates)/edgeR (without biological replicates), with the filtering criteria for differentially expressed genes set to |log2FC|≥ 1 and p-value < 0.05. Differential gene annotation information and enrichment pathways were analyzed using the KEGG database. The volcano plot was generated using the online tools from https://www.omicstudio.cn/tool. The trend heat map and Sankey bubble map were created using the online tool http://bioinformatics.com.cn/.
Statistical analysis
All data were analyzed using SPSS software (version 26.0, SPSS, Inc., Chicago, IL, USA). A one-way analysis of variance (ANOVA) was performed to assess the significance of differences among treatment groups, and a two-way ANOVA was used to evaluate potential synergy between the two factors. The data were subjected to the Shapiro–Wilk normality test, and non-parametric analysis (Kruskal–Wallis test) was employed if the data did not follow a normal distribution. GraphPad Prism 8.0.2 (GraphPad Software, La Jolla, CA, USA) was used for bar or line graph analysis. Pearson correlation test was conducted to analyze the relationships between metabolites, core microorganisms, and related genes. All data are presented as mean ± standard error of the mean (SEM). p-value < 0.05 was considered statistically significant.
Results
The establishment and validation of the AFB1 and E.o group
To verify whether the model of the AFB1 and E.o treatment groups was successfully established, we conducted statistical analyses on the clinical performance, feed intake, weight changes, and fecal score of the experimental sheep. The results indicated that, compared to the Con group, the feed intake of the AFB1, E.o, and AFB1 + E.o treatment groups significantly decreased, particularly on days 8–14 (p < 0.001, p = 0.003, and p < 0.001, respectively). Furthermore, the AFB1 group exhibited significantly lower feed intake compared to the AFB1 + E.o group (p = 0.002) (Fig. 1b). Similarly, the trend in weight showed a comparable decline; all treatment groups (AFB1, E.o, and AFB1 + E.o) significantly decreased in weight compared to the Con group (p = 0.016, p = 0.03, and p = 0.004, respectively), with no significant difference observed between the AFB1 and AFB1 + E.o group (Fig. 1c). Additionally, it was observed that the number of oocysts per gram (OPG) in the E.o and AFB1 + E.o groups significantly increased after 12 days (Fig. 1d). Moreover, the fecal scores of the E.o and AFB1 + E.o treatment groups were significantly higher than that of the Con group (p < 0.001 and p < 0.001, respectively) (Fig. S1a-b), while no difference in the AFB1 treatment group, indicating the successful establishment of the E. ovinoidalis infection model. Unfortunately, we did not observe a synergistic effect between AFB1 and coccidia.
The combined effects of AFB1 and coccidia induce damage to the uterus
This study investigates the impact of consuming AFB1-contaminated daily feed on the reproductive system of sheep, with a focus on uterine damage under the coccidia challenge. Observations revealed that the AFB1 treatment group displayed mild congestion and yellow staining during uterine morphology assessments. The uteri in the AFB1 + E.o group exhibited obvious bleeding and yellow staining, with uterine weight was significantly lower than that of the Con group (p = 0.023) (Fig. S2a-b). In contrast, there were no significant differences in uterine morphology between the E.o and Con groups. Additionally, histopathological findings indicated that endometrial cells in the Con group were arranged consistently, whereas the endometrial epithelial cells in the uteri of the AFB1, E.o, and AFB1 + E.o groups were disorganized and exhibited severe inflammatory lesions. Moreover, the AFB1-treated group showed increased chromatin density and nuclear shrinkage, whereas bleeding lesions were evident in the AFB1 + E.o group (Fig. 1e). Statistical analysis of uterine endometrial thickness revealed significant increases in the E.o and AFB1 + E.o groups compared to the Con group, attributed to the disordered arrangement of endometrial epithelial cells (p < 0.001 and p = 0.005, respectively). However, no significant changes were observed in the AFB1 group (p = 0.499). Compared to the AFB1 + E.o group, endometrial thickness was significantly reduced in the AFB1 group (p = 0.001) and significantly increased in the E.o group (p = 0.019) (Fig. 1f). These results indicate that exposure to AFB1 and infection with coccidia can lead to degeneration and disorganization of endometrial epithelial cells, which in turn affects endometrial thickness. Nonetheless, the uneven arrangement of cells may explain the inconsistent observations regarding endometrial thickness across different treatment groups. Overall, exposure to AFB1 and infection with coccidia have caused significant pathological damage to the uterus.
Next, we investigated whether AFB1 and coccidia treatment are accompanied by changes in uterine hormone levels and trigger inflammation. Therefore, we detected the estrogen receptors (ER) α and β that are present in the uterus and play a crucial role in reproduction. As shown in Fig. 1g, both AFB1 and E.o treatments significantly downregulated the expression of the ERα (p = 0.016 and p = 0.003, respectively) and ERβ genes (p = 0.008 and p = 0.021, respectively). The combination of AFB1 and E.o treatment resulted in an even more pronounced decrease in ERα (p = 0.005) and ERβ (p = 0.004) mRNA expression levels compared to the individual AFB1 or E.o treatment groups. Additionally, the inflammatory response induced by AFB1 or E.o treatment led to an upregulation of uterine inflammatory factors, including IL-6 and TNF-α mRNA, accompanied by a reduction in IL-10 expression. This inflammatory response was primarily driven by AFB1 exposure (IL-6, p = 0.003; IL-10, p = 0.016; TNF-α, p < 0.001). However, in the AFB1 + E.o group, it was observed that E.o could inhibit the overexpression of inflammatory factors rapidly induced by AFB1 exposure (Fig. 1h). Taken together, these data suggest that the toxic effects of AFB1 and coccidia treatment on the reproductive system are associated with alterations in uterine hormones and the induction of inflammation.
The combined effects of AFB1 and coccidia compromise the integrity of the intestinal barrier
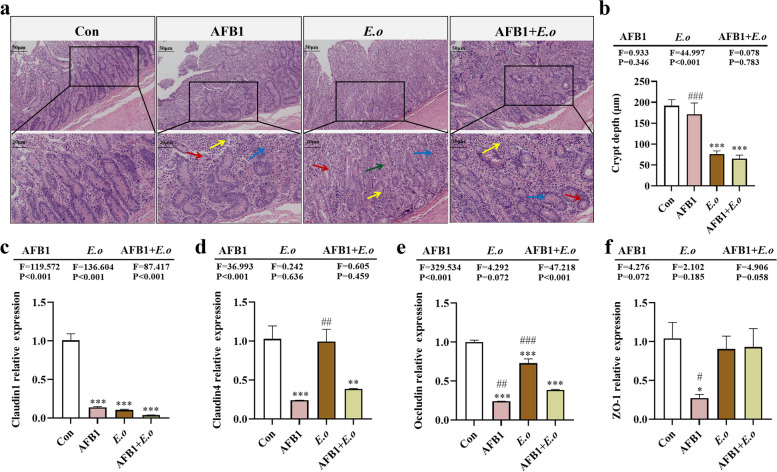
Given that coccidia infection mainly affects the intestine, we observed pathological changes in the colon. Histopathological examination of tissue sections revealed various stages of coccidia in both the E.o and AFB1 + E.o groups (Fig. S3). The intestinal structure in the E.o and AFB1 + E.o groups was severely disrupted, characterized by significant inflammatory cell infiltration and congestion. Similar pathological changes were also observed in the AFB1 group, although to a lesser extent (Fig. 2a). Moreover, coccidia infection significantly reduced the depth of colonic crypts in both the E.o and AFB1 + E.o groups compared to the Con group (p < 0.001 for both), and there was a significant difference in crypt depth between the AFB1 and AFB1 + E.o groups (p < 0.001). Although the difference was not statistically significant, the reduction in crypt depth attributed to coccidia was notably more pronounced with the addition of AFB1 (Fig. 2b). These results indicate that both AFB1 and coccidia treatment can disrupt intestinal structure, leading to impaired intestinal barrier function, with a more pronounced effect in the E.o group. Therefore, the expression levels of genes and proteins associated with intestinal barrier function were examined next. The results demonstrated that AFB1 treatment significantly downregulated the mRNA expression levels of Claudin1 (p < 0.001), Claudin4 (p = 0.001), Occludin (p < 0.001), and ZO-1 (p = 0.016) (Fig. 2c, d). Coccidia infection significantly downregulated the mRNA expression levels of Claudin1 (p < 0.001) and Occludin (p < 0.001). Furthermore, the combination of both treatments led to significant downregulation of mRNA expression levels of Claudin1 (p < 0.001), Claudin4 (p = 0.004), and Occludin (p < 0.001). Notably, the expression of Claudin1 mRNA was influenced not only by AFB1 and coccidia treatments individually but also showed a synergistic effect between the two factors (F = 87.417, p < 0.001).
Fig. 2.
The intestinal barrier of sheep is compromised by AFB1 and coccidia treatment. a Histological examination of colon tissue via H&E staining (200×, 400×; n = 4). Red arrows indicate intestinal epithelial cells, yellow arrows denote inflammatory cells, blue arrows represent bleeding points, and green arrows represent coccidia oocysts. b Statistics on crypt depth (n = 6). c–f Expression levels of intestinal barrier-related genes Claudin1, Claudin4, Occludin, and ZO-1 (n = 3). Statistical significance is indicated by *p < 0.05, **p < 0.01, and ***p < 0.001 (AFB1, E.o, and AFB1+E.o vs Con); or #p < 0.05, ##p < 0.01, and ###p < 0.001 (AFB1+E.o vs AFB1 and E.o)
The combined effects of AFB1 and coccidia induce alterations in the composition and metabolites of the intestinal microbiota
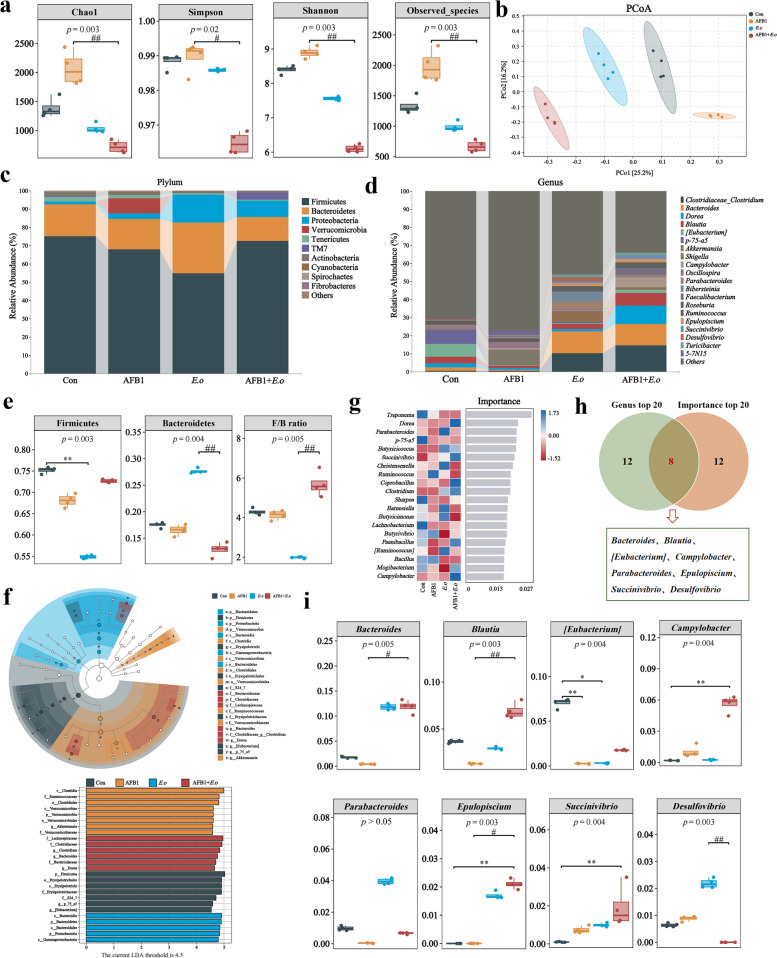
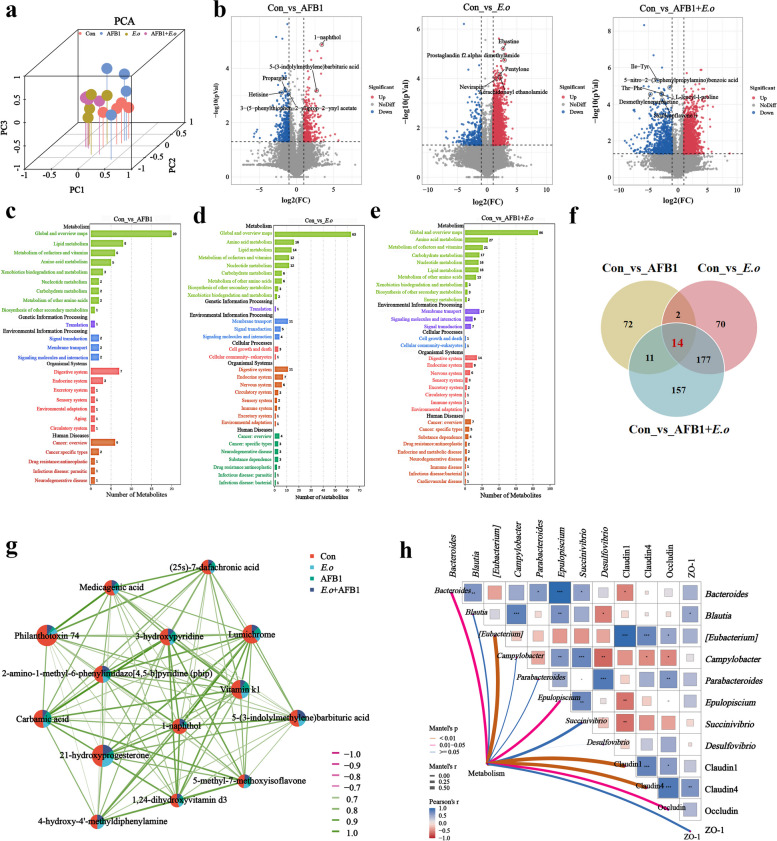
To elucidate the mechanism of gut damage caused by the combined effect of AFB1 and coccidia, we investigated alterations in the gut microbiota and metabolites, as well as screening for potential biomarkers. We analyzed the microbial composition in fecal samples from sheep treated with AFB1 and coccidia using 16S rRNA gene sequencing technology. Firstly, alpha diversity indicators including Chao1, Shannon index, Simpson index, and observed species were employed to assess the richness and diversity of individual taxa (Fig. 3a). The exposure to both AFB1 and coccidia significantly affected the diversity of the gut microbiota, as evidenced by lower microbial community abundances and diversities shown by the E.o and AFB1 + E.o groups compared to the Con group and the considerably larger trend in alpha diversity indices shown by the AFB1 group compared to the AFB1 + E.o group. Principal co-ordinates analysis (PCoA) revealed differences in microbial structure among the groups (Fig. 3b). Additionally, the composition of species at the phylum level was analyzed and high-abundance phyla including Firmicutes, Bacteroidetes, and Proteobacteria were found (Fig. 3c). Differential analysis of Firmicutes, Bacteroidetes, and the F/B ratio between groups showed a significant decrease in the relative abundance of Firmicutes in the E.o group (p = 0.003) and an increase in the relative abundance of Bacteroidetes. Moreover, the E.o group exhibited a significant decrease in the F/B ratio compared to the AFB1 + E.o group (p = 0.005) (Fig. 3e). The results indicated that Firmicutes, Erysipelotrichales, Erysipelotrichi in the Con group, Clostridia, Ruminococcaceae, Clostridiales in the AFB1 group, Bacteroidetes, Bacteroidia, Bacteroidales in the E.o group, and Lachnospiraceae, Clostridiaceae, Clostridium in the AFB1 + E.o group possessed relatively high abundance (Fig. 3f). Moreover, we overlapped the top 20 genera with high abundance observed at the genus level and the top 20 genera identified by random forest analysis using a Venn diagram, which revealed 8 genera shared among the groups (Fig. 3d, g-h). We analyzed the expression patterns of these shared genera among the groups. Among these, the relative abundances of Bacteroides, Parabacteroides, Epulopiscium, Succinivibrio, and Desulfovibrio increased in the E.o and AFB1 + E.o groups and dropped in the AFB1 group. Conversely, the relative abundances of Blautia and [Eubacterium] dropped in the AFB1 and E.o groups, but increased in the AFB1 + E.o group (Fig. 3i).
Fig. 3.
AFB1 and coccidia exposure altered the intestinal microorganisms of sheep. a Alpha diversity index. Chao1 and Observed species indices characterize richness, while diversity is assessed using Shannon and Simpson indices. b Principal coordinates analysis (PCoA) was performed to calculate beta diversity on a distance matrix of Bray–Curtis indices. c Changes of intestinal microbial composition at the phylum level (TOP 10). d Abundance analysis of the gut microbiota at the genus level (TOP 20). e Relative abundance of Bacteroidetes, Firmicutes, and F/B ratio. f Linear discriminant analysis effect size (LEfSe) comparison analysis between the groups. g Random Forests analysis (top 20 at the genus level). h Venn diagram analysis of differential genera. The differential genera screened by the top 20 genera and the importance top 20 genera from Random Forest analysis are presented in a Venn diagram, with the coincidence part indicating the potential biomarkers. i Relative abundance analysis of common differential genera. The groups were compared by the Kruskal–Wallis nonparametric test, and the two groups were compared by Dunn’s test. *p < 0.05, **p < 0.01, and ***p < 0.001 (AFB1, E.o, and AFB1+E.o vs Con); or #p < 0.05, ##p < 0.01, and ###p < 0.001 (AFB1+E.o vs AFB1 and E.o)(n = 4)
Fecal metabolomics was employed to further investigate the potential mechanisms underlying intestinal barrier damage induced by the interaction between AFB1 and coccidia. A total of 19,492 metabolites were identified using both combined positive and negative ion modes. These metabolites exhibited a distinct distribution across different groups, as illustrated in Fig. 4a. Additionally, differential metabolites were screened based on |log2FC|≥ 1 and p-value < 0.05, with the five most significantly different metabolites were highlighted in Fig. 4b. Compared to the Con group, the AFB1 group identified a total of 786 differential metabolites, comprising 366 upregulated and 420 downregulated metabolites (Table S3). These differential metabolites were mainly enriched in pathways such as lipid metabolism, metabolism of cofactors and vitamins, and amino acid metabolism (Fig. 4c). Compared to the Con group, the E.o group identified a total of 3204 differential metabolites, including 2886 upregulated and 318 downregulated metabolites (Table S4), which were mainly enriched in pathways such as amino acid metabolism, lipid metabolism, and cofactor and vitamin metabolism (Fig. 4d). The AFB1 + E.o group identified a total of 3974 differential metabolites, including 3154 upregulated metabolites and 820 downregulated metabolites (Table S5). These metabolites were predominantly enriched in pathways such as amino acid metabolism, cofactor and vitamin metabolism, and carbohydrate metabolism (Fig. 4e).
Fig. 4.
AFB1 and coccidia exposure altered intestinal metabolites in sheep. a The distribution of the metabolites was analyzed using principal component analysis (PCA). b Differentially expressed metabolites (DEMs) selected based on |log2FC| ≥ 1, p-value < 0.05 are represented by volcanic plots. c–e The KEGG enrichment pathways of the DEMs. f Overlapped differential metabolites between groups (VIP > 1, p-value< 0.05). g Network correlation diagram between overlapping DEMs. Nodes represent the proportion of each metabolite in each group; line thickness indicates the strength of correlation, while color denotes the nature of the correlation: green for positive and red for negative. h Correlation heatmap of overlapping DEMs with differential bacteria and intestinal barrier indices (Claudin-1, Claudin-4, Occludin, and ZO-1). Pearson correlation analysis and Mantel test were utilized to assess the correlation coefficients (r-values) and significance (p-values). The heatmap reflects correlation values, with deeper colors and larger lattice sizes indicating greater absolute correlation values. A closer affinity to blue signifies a stronger positive correlation, while a shift towards red indicates a stronger negative correlation. Asterisk denote significance levels. The network diagram illustrates the relationships between overlapping DEMs, differential genera, and intestinal barrier indices. Line thickness represents correlation strength; thicker lines indicate stronger correlations. Line color reflects significance, with red indicating p-values< 0.01 (n = 4)
According to the criteria of VIP > 1 and p-value < 0.05, we further screened for metabolites with significant differences. Among them, 263, 99, and 359 differential expression metabolites (DEMs) were identified in the E.o, AFB1, and AFB1 + E.o groups, respectively. A Venn diagram analysis identified 14 common DEMs (Fig. 4f). Additionally, the network graph demonstrated the interactions among these 14 DEMs (Fig. 4g). We then considered these 14 common DEMs as potential biomarkers and performed a network heatmap analysis to correlate them with eight gut microbiota markers and intestinal barrier-related indicators. As shown in Fig. 4h, the common DEMs were significantly positively correlated with Bacterioides, [Eubacterium], Epulopiscium, Claudin1, Claudin4, and Occludin, while they exhibited negative correlations with Blautia, Campylobacter, Parabacterioides, Succinivibrio, Desulfovibrio, and ZO-1. In conclusion, these results indicate that exposure to AFB1 and coccidia induces intestinal barrier dysfunction by altering gut microbiota composition and interfering with intestinal metabolism.
The combined effects of AFB1 and coccidia induce blood inflammation and hormonal alterations, concurrently affecting blood metabolism
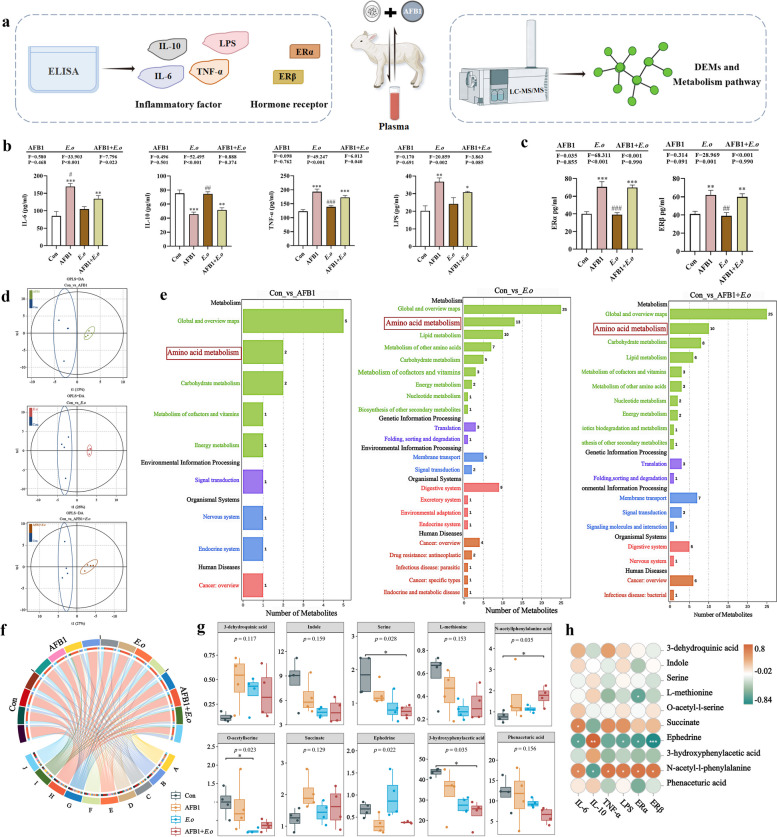
We have concluded that AFB1 and coccidia disrupt the intestinal barrier by perturbing the gut microbiota and metabolites. However, the relationship between uterine damage caused by AFB1 and coccidia and intestinal impairment remains unclear. Thus, we hypothesized that the destruction of the intestinal barrier may permit enterotoxins to enter the bloodstream and cause circulatory inflammation [22, 23]. The expression levels of inflammatory factors and hormones in the blood were first assessed. Routine blood analysis revealed an increase in inflammatory cells in the blood induced by AFB1 and E.o treatment (Fig. S4). Then, plasma was subjected to ELISA and metabolomic assays (Fig. 5a). The levels of inflammatory factors IL-6, IL-10, TNF-α, and LPS in plasma were measured using ELISA (Fig. 5b). The levels of pro-inflammatory factors IL-6 (p < 0.001 and p = 0.007, respectively), TNF-α (p < 0.001 and p = 0.001, respectively), and LPS (p = 0.002 and p = 0.019, respectively) were significantly elevated in the AFB1 and AFB1 + E.o treatment groups, while the expression level of the anti-inflammatory factor IL-10 exhibited an opposite trend (p < 0.001 and p = 0.002, respectively). Furthermore, we also measured the levels of estrogen receptors (ER) in plasma. Compared to the Con group, the expression levels of ERα (p < 0.001 and p < 0.001, respectively) and ERβ (p = 0.005 and p = 0.009, respectively) were significantly increased in the AFB1 and AFB1 + E.o groups (Fig. 5c). Therefore, we speculate that uterine damage may result from altered intestinal function, allowing endotoxins to enter the bloodstream and leading to a series of metabolic abnormalities.
Fig. 5.
Amino acid metabolism plays a role in blood inflammation and hormonal changes during the AFB1 and coccidia exposure. a Schematic representation of the experimental design and program diagram. b Expression levels of inflammatory factors IL-6, IL-10, TNF-α and LPS. c Expression levels of estrogen receptors ERα and ERβ (n = 3). d OPLS-DA score plots for the Con and AFB1, E.o and AFB1+E.o groups. e KEGG enrichment pathway analysis of DEMs. f Expression of 10 metabolites enriched in the amino acid metabolic pathways in different groups. A, 3-dehydroquinic acid. B, indole. C, serine. D, L-methionine. E, O-acetyl-l-serine. F, succinate. G, ephedrine. H, 3-hydroxyphenylacetic acid. I, N-acetyl-l-phenylalanine. J, phenaceturic acid. g Differential analysis of the ten metabolites enriched in amino acid metabolic pathways. Groups were compared using the Kruskal–Wallis nonparametric test, and the two groups were compared using Dunn’s test. h Association analysis of the ten metabolites with inflammatory factors and blood hormones. Statistical significance was indicated by *p < 0.05, **p < 0.01, and ***p < 0.001 (AFB1, E.o, and AFB1+E.o vs Con); or #p < 0.05, ##p < 0.01, and ###p < 0.001 (AFB1+E.o vs AFB1 and E.o)(n = 4)
Subsequently, changes in plasma metabolites were detected through comprehensive non-targeted metabolomics. The differences in metabolites between groups were illustrated in the OPLS-DA score plot (Fig. 5d). DEMs were selected based on |log2FC|≥ 1 and p-value < 0.05, with the top five significantly different metabolites were labeled (Fig. S5). Compared to the Con group, the AFB1 group exhibited a total of 402 DEMs, with 235 upregulated and 167 downregulated (Table S6). The E.o group showed a total of 978 DEMs, with 325 upregulated and 653 downregulated (Table S7). The AFB1 + E.o group had a total of 1,211 DEMs, with 874 upregulated and 337 downregulated (Table S8). Additionally, KEGG enrichment analysis revealed that the three comparison groups were mainly enriched in the amino acid metabolism pathway (Fig. 5e). Therefore, we selected metabolites enriched in this pathway and demonstrated their distribution among the groups (Fig. 5f). Furthermore, the differential expression analysis of these metabolites between groups is shown in Fig. 5g. Compared to the Con group, the levels of 3-dehydroquinic acid, succinate, and N-acetyl-L-phenylalanine showed an increasing trend in the AFB1, E.o, and AFB1 + E.o groups, while the levels of indole, serine, L-methionine, O-acetyl-L-serine, 3-hydroxyphenylacetic acid, and phenaceturic acid were significantly decreased. Next, we conducted a correlation analysis between these shared DEMs and blood inflammatory factors as well as hormone-related indicators (Fig. 5h). N-acetyl-L-phenylalanine showed a strong positive correlation with IL-6 (r = 0.580, p = 0.050), TNF-α (r = 0.699, p = 0.014), LPS (r = 0.664, p = 0.022), ERα (r = 0.706, p = 0.013), and ERβ (r = 0.685, p = 0.017), while there was a significant negative correlation with IL-10 expression (r = − 0.650, p = 0.025). In summary, disrupted gut microbiota and metabolites are responsible for the blood inflammation and hormonal alterations induced by AFB1 and coccidia, with amino acid metabolism potentially being a major contributing factor.
The combined effects of AFB1 and coccidia damage the uterus by regulating the PI3K/AKT/eNOS pathway
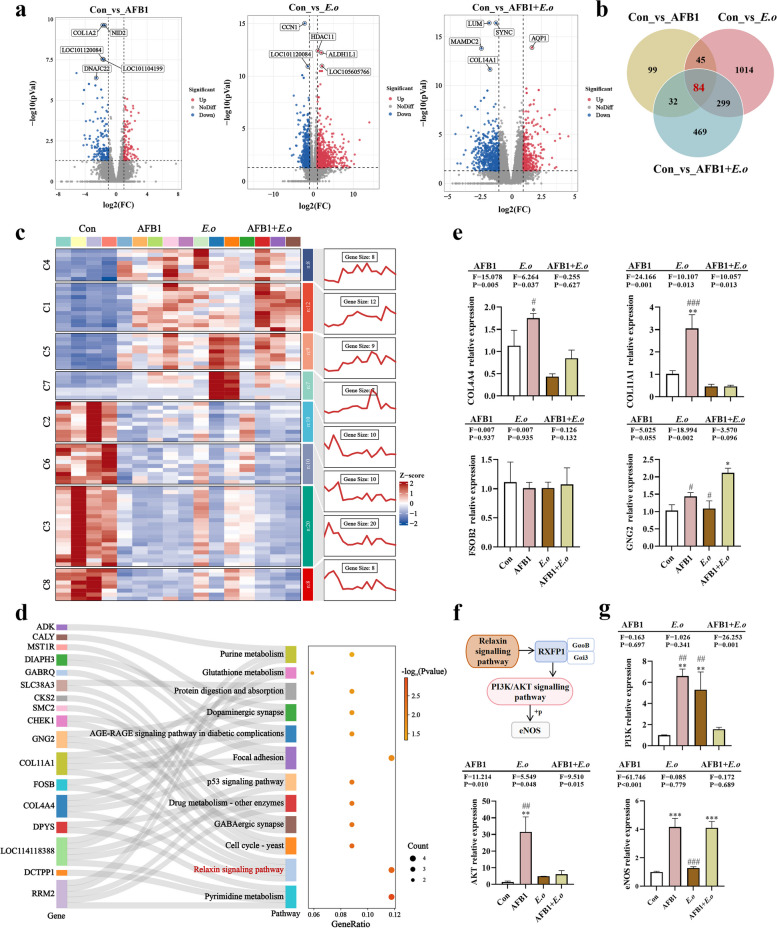
Next, we explored the mechanisms of direct damage caused by AFB1 and coccidia in the uterus through uterine transcriptome analysis. Differentially expressed genes (DEGs) were selected based on |log2FC|≥ 1 and p-value < 0.05, and these were illustrated using volcano plots, with the top five significantly altered genes labeled (Fig. 6a). Compared to the Con group, the AFB1 group exhibited a total of 260 DEGs, with 97 upregulated and 163 downregulated (Table S9). The top five DEGs in this group were COL1A2, NID2, LOC101120084, DNAJC22, and LOC101104199. Similarly, there were 1442 DEGs, with 888 upregulated and 554 downregulated in the E.o group, and the top 5 DEGs were CCN1, HDAC11, LOC101120084, ALDHIL1, and LOC105605766 (Table S10). There were 884 DEGs, with 321 upregulated and 563 downregulated in the AFB1 + E.o group, and the top 5 DEGs were LUM, SYNC, AQP1, MAMDC2, and COL14A1 (Table S11). A Venn diagram was then used to illustrate the overlap of 84 common DEGs (Fig. 6b), which were further categorized into different clusters and visually presented through a heatmap (Table S12; Fig. 6c). Additionally, a Sankey bubble chart was used to display the significant enrichment of 12 pathways along with the corresponding DEGs enriched in each pathway (Fig. 6d). In this study, we specifically focused on the relaxin signaling pathway and validated the expression levels of four enriched DEGs, COL11A1, COL4A4, FOSB, and GNG2 (Fig. 6e). Compared to the Con group and the AFB1 + E.o group, exposure to AFB1 showed a significantly upregulated in the expressions of COL11A1 (p = 0.002 and p < 0.001, respectively) and COL4A4 (p = 0.044 and p = 0.015, respectively). Furthermore, exposure to both AFB1 and coccidia significantly increased the mRNA level of GNG2 compared to the Con group (p = 0.002). However, the mRNA level of GNG2 was significantly downregulated in the AFB1 and coccidia single exposure compared to the AFB1 + E.o group (p = 0.019 and p = 0.002, respectively). No significant change in the expression of FOSB was observed following AFB1 and coccidia exposure. The KEGG map illustrates the important downstream pathways regulated by the relaxin signaling pathway, including the PI3K/AKT/eNOS pathway, which is known to regulate cell proliferation and differentiation and is involved in the development of various diseases (Fig S6; Fig. 6f). The results indicated that exposure to AFB1 and coccidia activated the PI3K (Con vs AFB1, p = 0.010; Con vs E.o, p = 0.002), AKT (Con vs AFB1, p = 0.002), and eNOS (Con vs AFB1, p < 0.001; Con vs AFB1 + E.o, p < 0.001) genes in the uterus (Fig. 6g). These findings suggest that the occurrence of uterine pathological inflammation is regulated by the PI3K/AKT/eNOS pathway, and changes in hormone levels (ERα and ERβ) also contribute to uterine damage.
Fig. 6.
AFB1 and coccidia exposure induced uterine damage through the relaxin signaling pathway. a Differentially expressed genes (DEGs) were identified using criteria of |log2FC| ≥ 1 and p-value < 0.05, and are represented in volcano plots. b Overlapped DEGs between groups (VIP > 1, p-value < 0.05). c A trend heatmap illustrates the expression status and trend of DEGs. The mfuzz algorithm was utilized to classify 84 DEGs into different clusters, and then the heat map+each cluster line map was drawn to visually display the trend. d The Sankey bubble map displays 12 significantly different pathways and their enriched DEGs. The left side is the Sankey diagram, indicating the genes within each pathway. The right side features the bubble diagram. The bubble size represents the number of genes that the pathway belongs to, and the bubble color represents the p-value. e Expression levels of COL11A1, COL4A4, FOSB, and GNG2 genes (n = 4). f The activation of the PI3K/AKT/eNOS pathway by relaxin signaling pathway. Relaxin signal binds to the receptor relaxin/insulin-like family peptide receptor 1 (RXFP1), which first interacts with Gαs to activate adenylate cyclase (AC), resulting in increased cyclic adenosine monophosphate (cAMP) production, while also negatively regulating through interaction with GαoB. Subsequently, RXFP1 associates with Gαi3 to activate the PI3K/AKT signaling pathway, leading to their phosphorylation and subsequent activation of eNOS. g Gene expression levels of PI3K, AKT and eNOS (n = 3). Statistical significance is indicated by *p < 0.05, **p< 0.01, and ***p < 0.001 (AFB1, E.o, and AFB1+E.o vs Con); or #p< 0.05, ##p < 0.01, and ###p < 0.001 (AFB1+E.o vs AFB1 and E.o)
Discussion
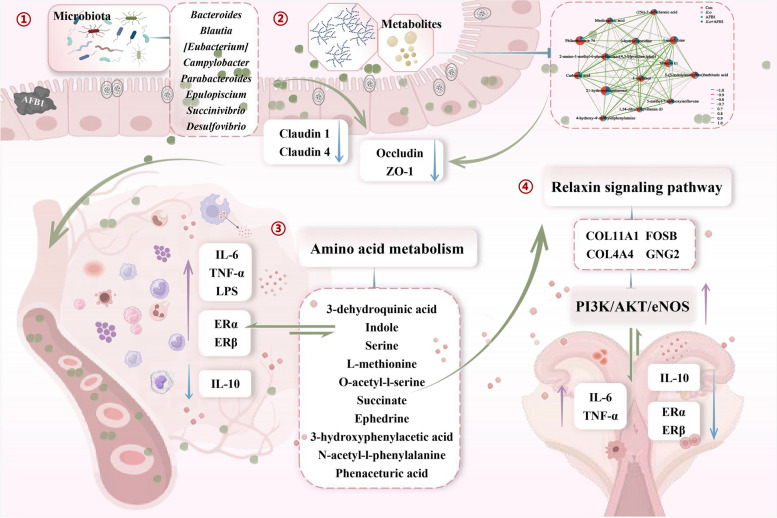
In this study, we utilized a livestock infection model to investigate the mechanisms of uterine injury caused by individual and mixed infections of AFB1 and coccidia (see Fig. 7). The results showed that the toxicity induced by AFB1 and coccidia initially disrupts the gut microbiota and metabolites, leading to increased production of LPS due to the predominance of Gram-negative bacteria, including Bacterioides, Parabacterioides, Epulopiscium, Succinivibrio, Desulfovibrio, Blautia, and Campylobacter. This gut dysbiosis and altered metabolism compromise intestinal barrier function, which exacerbates systemic inflammation in conjunction with elevated plasma LPS levels and hormonal changes. Moreover, the systemic inflammation and hormonal alterations induced by AFB1 and coccidia contribute to the development of inflammatory lesions in the reproductive system, while also triggering the relaxin signaling pathway and activating the PI3K/AKT/eNOS pathway, resulting in the upregulation of relevant gene expression levels. In summary, AFB1 is initially absorbed and metabolized in the intestine (a common route of exposure to this toxin), which is also a target organ for coccidia parasitism. Damage to gut function and alterations in microorganisms and metabolites (such as short-chain fatty acids, bile acids, etc.) are directly translocated to reproductive organs through systemic circulation (intestinal mucosal cells or red blood cells). Additionally, the metabolites themselves exert toxic effects on the reproductive system through their biotransformation reactions [22, 23]. Therefore, we conclude that co-exposure to AFB1 and coccidia compromises intestinal barrier function and triggers systemic inflammation through alterations in gut microbiota-derived metabolites, thereby directly and/or indirectly affecting reproductive system function.
Fig. 7.
A schematic diagram of the effects of AFB1 and coccidia exposure on the reproductive system of sheep through the gut–blood–reproductive axis
Due to the high prevalence of mycotoxins in grain feed and animal feed, investigating the reproductive toxicity caused by these toxins in animals is essential. Studies have shown that high doses of mycotoxins consumed by female animals can cause reproductive toxicity, leading to reduced fertility and even miscarriage [18, 19]. In addition, under intensive breeding conditions characterized by high animal density and productivity, coccidiosis can become a significant economic concern in small ruminants. Therefore, we utilized female sheep as an animal model to examine the effects of AFB1 and coccidia individually or in combination on the reproductive system. Our clinical characterization data, changes in feed intake, body weight, and uterine morphology confirmed the detrimental effects of AFB1 and coccidia on reproductive health. Subsequently, the mechanisms underlying the reproductive damage caused by AFB1 and coccidia were further elucidated through the application of fecal 16S rRNA sequencing, fecal metabolomics, blood metabolomics, and uterine transcriptomics techniques, during which several potential biomarkers were identified.
First, the results of the 16S rRNA sequencing revealed that under the influence of AFB1 and coccidia factors, the relative abundance of Firmicutes, which is mainly influenced by coccidia factor, decreased under the combined effects of AFB1 and coccidia. In contrast, the relative abundance of Bacteroides was increased. Firmicutes are considered important metabolic players in the gut, as they can break down complex sugars, polysaccharides, and fatty acids from feed, producing energy and nutrients for absorption and utilization by the host [28]. Additionally, they play a crucial role in restoring gut homeostasis as well as inhibiting or eradicating gas-producing clostridia [29]. Coccidian infection considerably increases the abundance of the phylum Bacterioides, which can further damage intestinal epithelial cells and enhance the invasion of other pathogens, thereby initiating or exacerbating enteritis [30]. Therefore, we speculate that the decrease in Firmicutes and the increase in Bacterioides may be due to secondary infections caused by coccidia. The microbiome, which includes bacteria, viruses, fungi, protozoa, and parasites, plays a critical role in host health through symbiotic, pathogenic, or parasitic relationships [31, 32]. Numerous studies have demonstrated complex interactions between coccidia and other intestinal parasites, fungi, or bacteria [33–36], which may result in more severe clinical manifestations in the host. Furthermore, fungal toxins have been proposed to negatively affect the cellular immune response to parasites [34], and the reduced microbial diversity observed under the combined action of AFB1 and coccidia may contribute to this possibility. Furthermore, we further screened out eight common genera, Bacteroides, Parabacteroides, Epulopiscium, Succinivibrio, Desulfovibrio, [Eubacterium], Blautia, and Campylobact, most of them are gram-negative bacteria, which likely create favorable conditions for the production of LPS. These findings indicate that AFB1 and coccidia, either alone or in combination, significantly alter the gut microbiota, resulting in the enrichment of some opportunistic pathogens, which can induce or exacerbate intestinal inflammation, and impair intestinal barrier function. This is supported by histopathological observations of intestinal tissue and the expression of intestinal barrier-related genes. Additionally, both gut metabolites and the microbiota play an important role in maintaining gut homeostasis, and disruption of intestinal barrier function is inevitably correlated with these metabolites. Through metabolomic screening, 14 common DEMs were identified under the individual or combined action of AFB1 and coccidia. Correlation analysis with microbiota markers and intestinal barrier-related indicators revealed significant positive correlations with Bacteroides, [Eubacterium], Epulopiscium, Claudin1, Claudin4, and Occludin. These findings suggest that the disruption of the intestinal barrier caused by AFB1 and coccidia is mediated through the interference of microbial communities and metabolites.
Previous research has shown that gut microbial metabolites also affect various physiological functions of other visceral organs and participate in mediating the pathogenesis of different diseases in these organs [37–41]. After ingestion and absorption in the intestine, mycotoxins mainly undergo biotransformation and metabolism via bile, urine, plasma, liver, and uterus and may be transferred through the intestinal mucosa or red blood cells [22, 23]. Therefore, we speculate that in the case of a compromised intestinal barrier, microbial-derived LPS and their related molecules can enter the bloodstream and transfer to the reproductive system, potentially leading to uterine damage and inflammation. Increased levels of white blood cells, IL-6, TNF-α, and LPS expression in plasma indicate the presence of inflammation in the body. Estrogens are known to be regulators of immune function in the monocyte–macrophage system [42], playing a critical role in suppressing cytokine production [43–46]. Transcripts of estrogen receptors ERα and ERβ have been identified in human macrophages and monocytes derived from original primitive hematopoietic progenitor cells [47]. IL-6 is one of the main cytokines involved in monocyte function related to chronic inflammation. It is established that 17β-estradiol (E2) can inhibit the expression of TNF-α, IL-1, and IL-6 [48]. Additionally, in macrophages with ERα deficiency, the release of LPS-induced TNF-α is significantly increased, suggesting that ERα plays a prominent role in mediating the anti-inflammatory effects of estrogen [49]. Our results demonstrate that the expression of ERα and ERβ in plasma is significantly increased under the dominant influence of AFB1, indicating a positive role of these receptors ERα and ERβ in suppressing AFB1 and coccidia-induced blood inflammation. Additionally, through blood metabolomics, we enriched the amino acid metabolism pathway and identified N-acetyl-L-phenylalanine as a highly correlated metabolite in this pathway. Amino acid metabolism is closely associated with inflammation [50]. On the one hand, amino acid metabolites are involved in regulating the body’s inflammatory response, such as arginine can inhibit the activity of key enzymes in inflammatory cells, such as eNOS and cyclooxygenase, thereby suppressing the synthesis and release of inflammatory factors [51]. On the other hand, amino acid metabolism significantly impacts the function of immune cells. During inflammation, immune cells such as macrophages and T cells have increased metabolic demands, requiring more amino acids for energy and synthesis. An insufficient supply of some essential amino acids during the inflammatory state may impair various immune cell functions, including cell proliferation, cell signaling, and cytotoxicity [52, 53]. Moreover, it is interesting to note that the supplementation of dietary amino acids can activate unliganded ERα in the liver, with this activation pathway relying on mTOR signaling to regulate uterine growth [54, 55]. This may explain the correlation between amino acid metabolism and estrogen receptors. However, the relationship between enhanced amino acid metabolism in the blood and exposure to AFB1 and coccidia remains unclear. Consistent with our findings, previous studies have demonstrated that exposure to AFB1 or co-exposure with other toxins can disrupt amino acid metabolism and affect cellular redox homeostasis [56–58]. Additionally, parasitic infections have been demonstrated to induce the production of amino acid metabolites with antimicrobial and antiviral properties, inhibiting the growth and reproduction of parasites [59]. Furthermore, amino acid metabolites can regulate the function of host immune cells and inflammatory responses. For instance, L-arginine is a key amino acid required for nitric oxide (NO)-mediated parasite killing and polyamine-mediated parasite reproduction within host target cells [60, 61]. It can activate phagocytes to produce NO and oxidative intermediates, exhibiting cytotoxic activity against intracellular pathogens. In combination with our findings, the alterations in blood metabolism provide a fundamental mechanistic explanation for the impairment of intestinal barrier function induced by AFB1 and coccidia, with amino acid metabolism being involved in systemic inflammation and disruption of the reproductive system.
We have elucidated the direct mechanisms underlying uterine injury induced by the separate or combined actions of AFB1 and coccidia through uterine transcriptomics. KEGG enrichment analysis highlighted the relaxin signaling pathway and identified four DEGs enriched in this pathway, namely COL11A1, COL4A4, FOSB, and GNG2. COL11A1 and COL4A4 play supportive and connective roles in the extracellular matrix. Due to their low expression in normal tissues, changes in COL11A1 expression are considered excellent prognostic markers for disease progression [62–64]. FOSB is involved in regulating gene transcription and is associated with the regulation of cell proliferation, differentiation, apoptosis, and transformation [65]. GNG2 participates in multiple cell signaling pathways, playing a crucial role in cell proliferation and angiogenesis, and is a potential molecular target for the treatment of various diseases [66–68]. Our results demonstrated that AFB1 exposure significantly upregulated the expression of the COL11A1 and COL4A4 genes in the uterus, while the combined action of AFB1 and E.o significantly upregulated the expression of the GNG2 gene, explaining the activation of uterine relaxin signaling pathway primarily driven by AFB1 exposure.
Furthermore, the downstream genes regulated by the PI3K/AKT/eNOS signaling pathway can regulate various biological processes, including cell survival, proliferation, and differentiation. NO is an important signaling molecule produced by endothelial cells, which can induce smooth muscle relaxation. NO activates eNOS to convert L-arginine to L-cyclic guanosine monophosphate (cGMP), promoting vasodilation and relaxation of blood vessels [69]. In addition, the PI3K/AKT pathway, when activated, can enhance the production of NO in endothelial cells, leading to the phosphorylation and activation of eNOS, regulating vasodilation and hemodynamics [70, 71]. Specifically, PI3K activates the AKT signaling molecule, which phosphorylates serine1177 of eNOS, enhancing its enzymatic activity and maintaining the anti-adhesive and anti-inflammatory properties of endothelial cells [72]. Moreover, the PI3K/AKT pathway is implicated in AFB1-induced animal toxicity, as AFB1 activates the PI3K/AKT/mTOR signaling pathway to induce testicular oxidative stress in mice [73, 74]. Furthermore, the activation of the PI3K/AKT signaling pathway has been demonstrated to inhibit the increased apoptosis of cecal epithelial cells caused by E. tenella infection [75]. Additionally, proteomic analysis has revealed that differential protein expression in cecal epithelial cells infected with E. tenella is also associated with the PI3K/AKT pathway [76]. Our data are consistent with the aforementioned findings, as the PI3K/AKT pathway is activated in response to AFB1 and coccidia treatments, enhancing the enzymatic activity of eNOS and regulating the proliferation of endometrial epithelial cells. The inflammatory damage to the uterus caused by AFB1 and coccidia, as demonstrated in pathological sections of uterine tissue and the expression of inflammatory factors IL-6, IL-10, and TNF-α, is regulated by the PI3K/AKT/eNOS pathway, as currently discovered. Considering the reproductive toxicity of AFB1 and coccidia, one potential endocrine-disrupting effect of their metabolites may be related to the disruption of pituitary hormone production [77]. AFB1 can induce oxidative stress, apoptosis, and interfere with testicular function in sheep, damaging the hypothalamic–pituitary–gonadal axis and causing hormonal imbalance in rats [13, 78–80]. ERα and ERβ are two subtypes of estrogen receptors that regulate reproductive activities in female animals. ERα is mainly involved in the regulation of endometrial proliferation and angiogenesis, while ERβ is responsible for endometrial differentiation and regeneration [81]. In this study, the downregulation of ERα and ERβ expression under the combined influence of AFB1 and E.o demonstrates the role of estrogen receptors in mediating uterine damage. Furthermore, supplementation with L-arginine has been shown to increase ERα expression in the endometrium of non-pregnant sheep, highlighting the crucial role of amino acid metabolism in the regulation of steroid hormone receptor expression in the endometrium [82].
This study investigated the individual and combined effects of AFB1 and coccidia on the reproductive system using a two-factor analysis of variance. The results demonstrated that the effects were not solely driven by individual pathogenic factors, and our objective was to observe the impact of both pathogenic factors on the reproductive system. We observed a synergistic effect of AFB1 and coccidia on the expression levels of uterine IL-10, TNF-α, AKT, and COL11A1, as well as intestinal Claudin1 and Occludin gene levels, along with IL-6 and TNF-α levels in plasma. Moreover, it is undeniable that AFB1 and coccidia as two pathogenic factors can cause significant damage to animal health. Exploring the interaction between these two factors was a focus of our study before its initiation. Unfortunately, the findings indicated that the interaction between AFB1 and coccidia was not optimal, possibly due to substantial individual differences among large animals. In addition, our research results cannot conclusively determine the causal contributions of the gut microbiota and their corresponding metabolites in relation to AFB1 and coccidia. Therefore, potential biomarkers identified require further validation through more direct experimental evidence, such as gut microbiota transplantation studies and intervention studies on differential metabolites. In summary, AFB1 and coccidia exhibit toxic effects on the reproductive system in sheep, mediated by the disruption of intestinal barrier function, alterations in gut microbiota-derived metabolites, and the induction of systemic inflammation, all of which directly and/or indirectly interfere with reproductive function.
Conclusions
In summary, our research findings highlight a potential mechanism linking gut microbiota-derived metabolites, such as LPS, with compromised intestinal–reproductive axis defense induced by AFB1 and coccidia through systemic inflammation. Thus, this comprehensive study reveals a novel toxic mechanism of action for AFB1 and coccidia in the gut–blood–reproductive axis and suggests that regulating gut microbiota and its metabolites could be a promising strategy to mitigate potential threats to farm animals and humans through dietary modifications.
Supplementary Information
Authors’ contributions
S.C.H: Funding acquisition, Supervision, Resources, Writing – review & editing. K.L.L: Data curation, Formal analysis, Visualization, Writing an original draft. P.C: Methodology, Data curation, Investigation. B.W.X: Data curation, Methodology, Investigation. W.L.D: Data curation, Methodology. T.J.Y and Y.N.L: Samples collected. S.Y.L: Data analysis, Investigation. J.K.L: Sample prepare, Writing – review & editing. F.C.J: Funding acquisition, supervision, Resources. All authors reviewed the manuscript.
Funding
This study was partly supported by the National Key R&D Program (no. 2023YFD1801205), the China Agriculture (Sheep and Goats) Research System (no. CARS-38), the National Natural Science Foundation of China (no. 32202876), the China Postdoctoral Science Foundation (no. 2023T160198), and the Key Scientific and Technological Project of the Henan Province Department of China (no. 232102111046).
Data availability
The bacterial 16S rRNA sequencing data obtained from the fecal have been deposited in the NCBI Sequence Read Archive (SRA) database under the accession number (PRJNA1067344).
Declarations
Ethics approval and consent to participate
All sheep experiments adhered to the animal welfare guidelines of Henan Agricultural University (ethical permission code: HNND2022030818) in Zhengzhou, China.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shu-cheng Huang and Kai-li Liu contributed equally to this work.
Contributor Information
Jia-kui Li, Email: lijk210@mail.hzau.edu.cn.
Fu-chun Jian, Email: jfchun2008@163.com.
References
- 1.Mohamaden WI, Sallam NH, Abouelhassan EM. Prevalence of Eimeria species among sheep and goats in Suez Governorate. Int J Vet Sci Med. 2018;6(1):65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chartier C, Paraud C. Coccidiosis due to Eimeria in sheep and goats, a review. Small Ruminant Res. 2012;103(1):84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrau T, Silva LMR, Pérez D, Failing K, Martínez-Carrasco C, Macías J, et al. de Ybáñez RR. Associated risk factors influencing ovine Eimeria infections in southern Spain. Vet Parasitol. 2018;263:54–58. [DOI] [PubMed] [Google Scholar]
- 4.Bangoura B, Bardsley KD. Ruminant coccidiosis. Vet Clin North Am Food Anim Pract. 2020;36(1):187–203. [DOI] [PubMed] [Google Scholar]
- 5.Khodakaram Tafti A, Maryam M. Pathologic lesions of naturally occurring coccidiosis in sheep and goats. Comp Clin Pathol. 2008;17:87–91. [Google Scholar]
- 6.Streit E, Schatzmayr G, Tassis P, Tzika E, Marin D, Taranu I, Oswald IP, et al. Current situation of mycotoxin contamination and co-occurrence in animal feed–focus on Europe. Toxins (Basel). 2012;4(10):788–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng WX, Marchal JLM, van der Poel AFB. Strategies to prevent and reduce mycotoxins for compound feed manufacturing. Anim Feed Sci Tech. 2018;237:129–53. [Google Scholar]
- 8.Frisvad JC, Hubka V, Ezekiel CN, Hong SB, Nováková A, Chen AJ, et al. Houbraken J. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud Mycol. 2019;93:1–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rushing BR, Selim MI. Aflatoxin B1: a review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem Toxicol. 2019;124:81–100. [DOI] [PubMed] [Google Scholar]
- 10.Office of Regulatory Affairs, Center for Food Safety and Applied Nutrition. Compliance Policy Guides - CPG Sec. 555.400 Aflatoxins in Human Food. 2021. https://www.fda.gov/media/149666/download. Accessed June 2021.
- 11.Office of Regulatory Affairs, Center for Food Safety and Applied Nutrition. Compliance Policy Guides - CPG Sec. 570.375 Aflatoxins in Peanuts and Peanut Products. 2021. https://www.fda.gov/media/72073/download. Accessed June 2021.
- 12.Regulation A. Setting maximum levels for certain contaminants in foodstuffs as regards aflatoxins. Off J Eur Union. 2010;165:8–12. [Google Scholar]
- 13.Lin LX, Cao QQ, Zhang CD, Xu TT, Yue K, Li Q, et al. Jian FC. Aflatoxin B1 causes oxidative stress and apoptosis in sheep testes associated with disrupting rumen microbiota. Ecotoxicol Environ Saf. 2022;232:113225. [DOI] [PubMed] [Google Scholar]
- 14.Cao QQ, Lin LX, Xu TT, Lu Y, Zhang CD, Yue K, et al. Jian FC. Aflatoxin B1 alters meat quality associated with oxidative stress, inflammation, and gut-microbiota in sheep. Ecotoxicol Environ Saf. 2021;225:112754. [DOI] [PubMed] [Google Scholar]
- 15.Sui Y, Lu Y, Zuo S, Wang H, Bian X, Chen G, et al. Dong H. Aflatoxin B1 exposure in sheep: insights into hepatotoxicity based on oxidative stress, inflammatory injury, apoptosis, and gut microbiota analysis. Toxins. 2022;14(12):840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Kang W, Mao X, Ge L, Du H, Li J, Huang K, et al. Melatonin mitigates aflatoxin B1-induced liver injury via modulation of gut microbiota/intestinal FXR/liver TLR4 signaling axis in mice. J Pineal Res. 2022;73(2):e12812. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Liu F, Zhou X, Liu M, Zang H, Liu X, Feng X, et al. Alleviation of oral exposure to aflatoxin B1-induced renal dysfunction, oxidative stress, and cell apoptosis in mice kidney by curcumin. Antioxidants (Basel). 2022;11(6):1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu LL, Chen S, Shen MY, Huang QY, Li HG, Sun SC, Luo XQ, et al. Aflatoxin B1 impairs porcine oocyte quality via disturbing intracellular membrane system and ATP production. Ecotoxicol Environ Saf. 2023;263:115213. [DOI] [PubMed] [Google Scholar]
- 19.Yaacobi-Artzi S, Kalo D, Roth Z. Effect of the aflatoxins B1 and M1 on bovine oocyte developmental competence and embryo morphokinetics. Reprod Toxicol. 2023;120:108437. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Y, Zeng D, Zhang Y, Ni XQ, Wang J, Jian P, Jing B, et al. Lactobacillus plantarum BS22 promotes gut microbial homeostasis in broiler chickens exposed to aflatoxin B1. J Anim Physiol Anim Nutr. 2018;102(1):e449–59. [DOI] [PubMed] [Google Scholar]
- 21.Liew WP, Mohd-Redzwan S, Than LTL. Gut microbiota profiling of aflatoxin B1-induced rats treated with Lactobacillus casei shirota. Toxins. 2019;11(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalska K, Habrowska-Górczyńska DE, Piastowska-Ciesielska AW. Zearalenone as an endocrine disruptor in humans. Environ Toxicol Pharmacol. 2016;48:141–9. [DOI] [PubMed] [Google Scholar]
- 23.Metzler M, Pfeiffer E, Hildebrand A. Zearalenone and its metabolites as endocrine disrupting chemicals. World Mycotoxin J. 2010;3(4):385–401. [Google Scholar]
- 24.Odden A, Enemark HL, Robertson LJ, Ruiz A, Hektoen L, Stuen S. Treatment against coccidiosis in Norwegian lambs and potential risk factors for development of anticoccidial resistance-a questionnaire-based study. Parasitol Res. 2017;116(4):1237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan X, Liu M, He S, Tong T, Liu Y, Ding K, et al. An epidemiological study of gastrointestinal nematode and Eimeria coccidia infections in different populations of Kazakh sheep. PLoS One. 2021;16(5):e0251307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bangoura B, Bhuiya MAI, Kilpatrick M. Eimeria infections in domestic and wild ruminants with reference to control options in domestic ruminants. Parasitol Res. 2022;121(8):2207–32. [DOI] [PubMed] [Google Scholar]
- 27.Klarić MS, Cvetnić Z, Pepeljnjak S, Kosalec I. Co-occurrence of aflatoxins, ochratoxin A, fumonisins, and zearalenone in cereals and feed, determined by competitive direct enzyme-linked immunosorbent assay and thin-layer chromatography. Arh Hig Rada Toksikol. 2009;60(4):427–34. [DOI] [PubMed] [Google Scholar]
- 28.Megrian D, Taib N, Witwinowski J, Beloin C, Gribaldo S. One or two membranes? Diderm Firmicutes challenge the Gram-positive/Gram-negative divide. Mol Microbiol. 2020;113(3):659–71. [DOI] [PubMed] [Google Scholar]
- 29.Fasina YO, Newman MM, Stough JM, Liles MR. Effect of Clostridium perfringens infection and antibiotic administration on microbiota in the small intestine of broiler chickens. Poult Sci. 2016;95(2):247–60. [DOI] [PubMed] [Google Scholar]
- 30.Lu C, Yan Y, Jian F, Ning C. Coccidia-microbiota interactions and their effects on the host. Front Cell Infect Microbiol. 2021;11:751481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desselberger U. The mammalian intestinal microbiome: composition, interaction with the immune system, significance for vaccine efficacy, and potential for disease therapy. Pathogens. 2018;7(3):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baines D, Sumarah M, Kuldau G, Juba J, Mazza A, Masson L. Aflatoxin, fumonisin and Shiga toxin-producing Escherichia coli infections in calves and the effectiveness of Celmanax®/Dairyman’s Choice™ applications to eliminate morbidity and mortality losses. Toxins. 2013;5(10):1872–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girgis GN, Barta JR, Brash M, Smith TK. Morphologic changes in the intestine of broiler breeder pullets fed diets naturally contaminated with Fusarium mycotoxins with or without coccidial challenge. Avian Dis. 2010;54(1):67–73. [DOI] [PubMed] [Google Scholar]
- 34.Antonissen G, Martel A, Pasmans F, Ducatelle R, Verbrugghe E, Vandenbroucke V, Croubels S, et al. The impact of Fusarium mycotoxins on human and animal host susceptibility to infectious diseases. Toxins. 2014;6(2):430–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukata T, Baba E, Arakawa A. Growth of Salmonella typhimurium in the caecum of gnotobiotic chickens with Eimeria tenella. Res Vet Sci. 1984;37(2):230–3. [PubMed] [Google Scholar]
- 36.Ruff MD, Rosenberger JK. Interaction of low-pathogenicity reoviruses and low levels of infection with several coccidial species. Avian Dis. 1985;29(4):1057–65. [PubMed] [Google Scholar]
- 37.Wang W, Zhai S, Xia Y, Wang H, Ruan D, Zhou T, et al. Ochratoxin A induces liver inflammation: involvement of intestinal microbiota. Microbiome. 2019;7(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146(6):1513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20(2):145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan S, Ge W, Wang J, Liu W, Zhao Y, Shen W, et al. Zearalenone-induced aberration in the composition of the gut microbiome and function impacts the ovary reserve. Chemosphere. 2020;244:125493. [DOI] [PubMed] [Google Scholar]
- 41.Huang S, Lin L, Wang S, Ding W, Zhang C, Shaukat A, et al. Total flavonoids of Rhizoma Drynariae mitigates aflatoxin B1-induced liver toxicity in chickens via microbiota-gut-liver axis interaction mechanisms. Antioxidants. 2023;12(4):819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dama A, Baggio C, Boscaro C, Albiero M, Cignarella A. Estrogen receptor functions and pathways at the vascular immune interface. Int J Mol Sci. 2021;22(8):4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell L, Emmerson E, Williams H, Saville CR, Krust A, Chambon P, et al. Estrogen receptor-alpha promotes alternative macrophage activation during cutaneous repair. J Invest Dermatol. 2014;134(9):2447–57. [DOI] [PubMed] [Google Scholar]
- 44.Trenti A, Tedesco S, Boscaro C, Trevisi L, Bolego C, Cignarella A. Estrogen, angiogenesis, immunity and cell metabolism: solving the puzzle. Int J Mol Sci. 2018;19(3):859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Straub RH. The complex role of estrogens in inflammation. Endocr Rev. 2007;28(5):521–74. [DOI] [PubMed] [Google Scholar]
- 46.Miller AP, Feng W, Xing D, Weathington NM, Blalock JE, Chen YF, et al. Estrogen modulates inflammatory mediator expression and neutrophil chemotaxis in injured arteries. Circulation. 2004;110(12):1664–9. [DOI] [PubMed] [Google Scholar]
- 47.Pelekanou V, Kampa M, Kiagiadaki F, Deli A, Theodoropoulos P, Agrogiannis G, et al. Estrogen anti-inflammatory activity on human monocytes is mediated through cross-talk between estrogen receptor ERα36 and GPR30/GPER1. J Leukoc Biol. 2016;99(2):333–47. [DOI] [PubMed] [Google Scholar]
- 48.Calippe B, Douin-Echinard V, Delpy L, Laffargue M, Lélu K, Krust A, et al. 17Beta-estradiol promotes TLR4-triggered proinflammatory mediator production through direct estrogen receptor alpha signaling in macrophages in vivo. J Immunol. 2010;185(2):1169–76. [DOI] [PubMed] [Google Scholar]
- 49.Lambert KC, Curran EM, Judy BM, Lubahn DB, Estes DM. Estrogen receptor-alpha deficiency promotes increased TNF-alpha secretion and bacterial killing by murine macrophages in response to microbial stimuli in vitro. J Leukoc Biol. 2004;75(6):1166–72. [DOI] [PubMed] [Google Scholar]
- 50.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16(9):553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy C, Newsholme P. Importance of glutamine metabolism in murine macrophages and human monocytes to L-arginine biosynthesis and rates of nitrite or urea production. Clin Sci. 1998;95(4):397–407. [PubMed] [Google Scholar]
- 52.Rath M, Müller I, Kropf P, Closs EI, Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front Immunol. 2014;5:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109(4):1568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Della Torre S, Rando G, Meda C, Stell A, Chambon P, Krust A, et al. Amino acid-dependent activation of liver estrogen receptor alpha integrates metabolic and reproductive functions via IGF-1. Cell Metab. 2011;13(2):205–14. [DOI] [PubMed] [Google Scholar]
- 55.Della Torre S, Benedusi V, Pepe G, Meda C, Rizzi N, Uhlenhaut NH, et al. Dietary essential amino acids restore liver metabolism in ovariectomized mice via hepatic estrogen receptor α. Nat Commun. 2021;12(1):6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi H, Peng J, Hao J, Wang X, Xu M, Li S. Growth performance, digestibility, and plasma metabolomic profiles of Saanen goats exposed to different doses of aflatoxin B1. J Dairy Sci. 2022;105(12):9552–63. [DOI] [PubMed] [Google Scholar]
- 57.Cheng J, Huang S, Fan C, Zheng N, Zhang Y, Li S, et al. Metabolomic analysis of alterations in lipid oxidation, carbohydrate and amino acid metabolism in dairy goats caused by exposure to aflotoxin B1. J Dairy Res. 2017;84(4):401–6. [DOI] [PubMed] [Google Scholar]
- 58.Ma X, Ye Y, Sun J, Ji J, Wang JS, Sun X. Coexposure of cyclopiazonic acid with aflatoxin b1 involved in disrupting amino acid metabolism and redox homeostasis causing synergistic toxic effects in hepatocyte spheroids. J Agric Food Chem. 2022;70(16):5166–76. [DOI] [PubMed] [Google Scholar]
- 59.Kropf P, Fuentes JM, Fähnrich E, Arpa L, Herath S, Weber V, et al. Arginase and polyamine synthesis are key factors in the regulation of experimental leishmaniasis in vivo. FASEB J. 2005;19(8):1000–2. [DOI] [PubMed] [Google Scholar]
- 60.Iniesta V, Gómez-Nieto LC, Molano I, Mohedano A, Carcelén J, Mirón C, Corraliza I, et al. Arginase I induction in macrophages, triggered by Th2-type cytokines, supports the growth of intracellular Leishmania parasites. Parasite Immunol. 2002;24(3):113–8. [DOI] [PubMed] [Google Scholar]
- 61.Iniesta V, Gómez-Nieto LC, Corraliza I. The inhibition of arginase by N(omega)-hydroxy-l-arginine controls the growth of Leishmania inside macrophages. J Exp Med. 2001;193(6):777–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer H, Stenling R, Rubio C, Lindblom A. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis. 2001;22(6):875–8. [DOI] [PubMed] [Google Scholar]
- 63.Fischer H, Salahshor S, Stenling R, Björk J, Lindmark G, Iselius L, et al. COL11A1 in FAP polyps and in sporadic colorectal tumors. BMC Cancer. 2001;1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raglow Z, Thomas SM. Tumor matrix protein collagen XIα1 in cancer. Cancer Lett. 2015;357(2):448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qian X, Lin G, Wang J, Zhang S, Ma J, Yu B, et al. CircRNA_01477 influences axonal growth via regulating miR-3075/FosB/Stat3 axis. Exp Neurol. 2022;347:113905. [DOI] [PubMed] [Google Scholar]
- 66.Lai WS, Ding YL. GNG7 silencing promotes the proliferation and differentiation of placental cytotrophoblasts in preeclampsia rats through activation of the mTOR signaling pathway. Int J Mol Med. 2019;43(5):1939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leung T, Chen H, Stauffer AM, Giger KE, Sinha S, Horstick EJ, et al. Zebrafish G protein gamma2 is required for VEGF signaling during angiogenesis. Blood. 2006;108(1):160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao A, Li D, Mao X, Yang M, Deng W, Hu W, et al. GNG2 acts as a tumor suppressor in breast cancer through stimulating MRAS signaling. Cell Death Dis. 2022;13(3):260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyatt AW, Steinert JR, Mann GE. Modulation of the L-arginine/nitric oxide signalling pathway in vascular endothelial cells. Biochem Soc Symp. 2004;71:143–56. [DOI] [PubMed] [Google Scholar]
- 70.Shi F, Wang YC, Zhao TZ, Zhang S, Du TY, Yang CB, et al. Effects of simulated microgravity on human umbilical vein endothelial cell angiogenesis and role of the PI3K-Akt-eNOS signal pathway. PLoS One. 2012;7(7):e40365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei X, Guan L, Fan P, Liu X, Liu R, Liu Y, et al. Direct current electric field stimulates nitric oxide production and promotes no-dependent angiogenesis: Involvement of the PI3K/AKT signaling pathway. J Vasc Res. 2020;57(4):195–205. [DOI] [PubMed] [Google Scholar]
- 72.Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y, Walsh K, et al. Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature. 1999;399(6736):597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai C, Tian E, Hao Z, Tang S, Wang Z, Sharma G, et al. Aflatoxin B1 toxicity and protective effects of curcumin: molecular mechanisms and clinical implications. Antioxidants. 2022;11(10):2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang W, Cao Z, Zhang J, Ji Q, Li Y. Aflatoxin B1 promotes autophagy associated with oxidative stress-related PI3K/AKT/mTOR signaling pathway in mice testis. Environ Pollut. 2019;255(Pt 2):113317. [DOI] [PubMed] [Google Scholar]
- 75.Zhang X, Li S, Zheng M, Zhang L, Bai R, Li R, et al. Effects of the PI3K/AKT signaling pathway on the apoptosis of early host cells infected with Eimeria tenella. Parasitol Res. 2020;119(8):2549–61. [DOI] [PubMed] [Google Scholar]
- 76.Zhao Z, Zhao Q, Zhu S, Huang B, Lv L, Chen T, et al. iTRAQ-based comparative proteomic analysis of cells infected with Eimeria tenella sporozoites. Parasite. 2019;26:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Storvik M, Huuskonen P, Kyllönen T, Lehtonen S, El-Nezami H, Auriola S, et al. Aflatoxin B1–a potential endocrine disruptor–up-regulates CYP19A1 in JEG-3 cells. Toxicol Lett. 2011;202(3):161–7. [DOI] [PubMed] [Google Scholar]
- 78.Owumi SE, Otunla MT, Arunsi UO, Oyelere AK. Apigeninidin-enriched Sorghum bicolor (L. Moench) extracts alleviate aflatoxin B1-induced dysregulation of male rat hypothalamic-reproductive axis. Exp Biol Med. 2022;247(15):1301–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rotimi OA, Onuzulu CD, Dewald AL, Ehlinger J, Adelani IB, Olasehinde OE, et al. Early life exposure to aflatoxin B1 in rats: alterations in lipids, hormones, and DNA methylation among the offspring. Int J Environ Res Public Health. 2021;18(2):589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu K, Liu M, Wang H, Rajput SA, Al Zoubi OM, Wang S, et al. Effect of zearalenone on aflatoxin B1-induced intestinal and ovarian toxicity in pregnant and lactating rats. Ecotoxicol Environ Saf. 2023;258:114976. [DOI] [PubMed] [Google Scholar]
- 81.Drew BG, Hamidi H, Zhou Z, Villanueva CJ, Krum SA, Calkin AC, et al. Estrogen receptor (ER)α-regulated lipocalin 2 expression in adipose tissue links obesity with breast cancer progression. J Biol Chem. 2015;290(9):5566–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao XX, Zhang QF, Zhu M, Li XH, Guo YX, Pang J, et al. Effects of l-arginine on endometrial estrogen receptor α/β and progesterone receptor expression in nutrient-restricted sheep. Theriogenology. 2019;138:137–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacterial 16S rRNA sequencing data obtained from the fecal have been deposited in the NCBI Sequence Read Archive (SRA) database under the accession number (PRJNA1067344).