Abstract
Background
The NOD-like receptor protein (NLRP)3 inflammasome is at the signaling hub center to instigate inflammation in response to pathogen infection or oxidative stress, and its tight control is pivotal for immune defense against infection while avoiding parallel intensive inflammatory tissue injury. Acetylation of NLRP3 is critical for the full activation of NLRP3 inflammasome, while the precise regulation of the acetylation and deacetylation circuit of NLRP3 protein remained to be fully understood.
Methods
The interaction between histone deacetylase 10 (HDAC10) and NLRP3 was detected by immunoprecipitation and western blot in the HDAC10 and NLRP3 overexpressing cells. The role of HDAC10 in NLRP3 inflammasome activation was measured by immunofluorescence, real-time PCR and immunoblotting assay in peritoneal macrophages and bone marrow-derived macrophages after the stimulation with LPS and ATP. To investigate the role of HDAC10 in NLRP3-involved inflammatory diseases, the Hdac10 knockout (Hdac10−/−) mice were used to construct the LPS-induced acute endotoxemia model and folic acid-induced acute tubular necrosis model. Tissue injury level was analyzed by hematoxylin and eosin staining, and the serum level of IL-1β was measured by enzyme-linked immunosorbent assay (ELISA). The conservative analysis and immunoprecipitation assay were performed to screen the precise catalytic site regulated by HDAC10 responsible for the switching from the acetylation to ubiquitination of NLRP3.
Results
Here we demonstrated that HDAC10 directly interacted with NLRP3 and induced the deacetylation of NLRP3, thus leading to the inhibition of NLRP3 inflammasome and alleviation of NLRP3 inflammasome-mediated acute inflammatory injury. Further investigation demonstrated that HDAC10 directly induced the deacetylation of NLRP3 at K496 residue, thus switching NLRP3 acetylation to the ubiquitination modification, resulting in the proteasomal degradation of NLRP3 protein. Thus, this study identified HDAC10 as a new eraser for NLRP3 acetylation, and HDAC10 attenuated NLRP3 inflammasome involved acute inflammation via directly deacetylating NLRP3.
Conclusions
This study indicated that HDAC10 switched NLRP3 modification from acetylation to ubiquitination and attenuated acute inflammatory diseases, thus it provided a potential therapeutic strategy for NLRP3 inflammasome-associated diseases by targeting HDAC10.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01992-1.
Keywords: HDAC10, NLRP3 inflammasome, Deacetylation, Ubiquitination, Acute inflammation
Plain language summary
The NOD-like receptor protein (NLRP)3 inflammasome is the core signaling hub and key instigator of acute inflammation in response to infection and oxidative stress to promote inflammatory cascade responses. Although inflammasome activation protects against infection and injury, excessive NLRP3 inflammasome activity yields excessive cytokine secretion and a pathological hyperinflammatory state. Of increasing interest is the critical contribution of NLRP3 inflammasome to acute inflammatory injury. Thus, the activation of NLRP3 inflammasome must be tightly controlled to ensure its efficient biological function without excessive side effect. NLRP3 protein is rich in lysine and susceptible to both acetylation and ubiquitination modification, while these two modifications usually had opposing effects on the protein degradation and stability. Various post-translational modifications in inflammasome signaling can be cross-regulated by each other, whereas the machinery involved in their spatial coordination during the process of NLRP3 inflammasome activation remains to be fully understood. Here we demonstrated that HDAC10 negatively regulated NLRP3 inflammasome by switching from acetylation to ubiquitination modification, leading to the proteasome-mediated degradation of the NLRP3 protein and attenuation of NLRP3 inflammasome activation. The further ubiquitination and acetylation analysis revealed that the Lys496 residue of NLRP3 protein is the pivotal convergence point involved in this PTM switch, indicating the critical switching node effect of this lysine residue. Altogether, our work suggested a novel fine-tuned regulatory mechanism of NLRP3 inflammasome activation by HDAC10, and provided a potential therapeutic target for NLRP3 inflammasome-associated diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01992-1.
Introduction
Acute inflammation is a defense mechanism characterized by a cascade of signaling in response to pathogens or oxidative stress. Whereas proper inflammation is protective, intensive inflammatory cascade signaling may induce severe tissue damage, leading to fatal tissue injuries even death. Despite the tremendous development of therapeutic strategies, acute inflammatory injury diseases constitute a major health burden across the world. As the central instigators of the inflammatory response, NOD-like receptor protein 3 (NLRP3) and the inflammasome assembled from it, have emerged as a key regulator in diverse acute inflammatory injury.
The NLRP3 inflammasome is formed by a cytosolic sensor NLRP3 protein, an adaptor ASC protein, and the cysteine protease procaspase-1 [1, 2]. NLRP3 comprises an N-terminal pyrin domain (PYD), a central nucleotide-binding and oligomerization domain (NOD or NACHT), and a C-terminal leucine-rich repeat (LRR) domain. NLRP3 is a crucial component of the cytosolic immunosurveillance system of mammals and it could detect signature components of pathogens and consequently trigger immune response. Assembly and activation of NLRP3 inflammasome triggers proteolytic cleavage of procaspase-1 into active caspase-1, leading to the production of pro-inflammatory cytokines including interleukin (IL)-1β and IL-18. In addition, caspase-1 also cleaves gasdermin D and promotes a rapid pro-inflammatory cell death designated as pyroptosis [3, 4]. NLRP3 inflammasome activation protects against infection and injury, whereas uncontrolled activity yields excessive cytokine secretion and a pathological hyper inflammatory state. Thus, the activation of NLRP3 inflammasome must be tightly controlled to ensure its efficient biological function without excessive side effect. However, the current understanding of the NLRP3 inflammasome is still limited and some unresolved questions remain to be answered.
NLRP3 is a prototypical sensor protein connecting cellular stress to proinflammatory signaling, and accumulating evidence suggests that posttranslational modifications (PTMs) are critical for NLRP3 to shape distinct inflammatory reactions in response to different stimuli [5]. Protein acetylation is a type of PTM involved in a varieties of biological processes, including protein stability, protein–protein interactions, and subcellular localization, et al. [6]. The most recent study demonstrated that acetylation is required for the full activation of the NLRP3 inflammasome [7], indicating the importance of the precise regulation of acetylation and deacetylation circuit for NLRP3. Histone deacetylase (HDAC) family is a group of enzymes that remove acetyl groups from the lysine residues of its target proteins [8]. HDACs are grouped into four classes, class I (HDAC1, 2, 3 and 8), class II (HDAC 4, 5, 6, 7, 9, and 10), class III (SIRTs family) and class IV (HDAC11) according to their characteristics [9, 10]. Distinct from other class II members, HDAC10 has unique leucine-rich domain and is enriched in the cytoplasm [11], which indicated its unique role aside from the conventional histone modification effect. In recent years, HDAC10 is also recognized as a polyamine deacetylase [12], and both the protein and polyamine deacetylase activities are reported in several cellular and animal models [13, 14]. The amazing diversity of its biological activity also indicates a potent complex regulatory role in multiple physiological and pathological processes. Though the recent emerging literatures of HDACs in innate immunity indicate these enzymes as important control points in host immune defense [15], the role of HDAC10 in the regulation of NLRP3 inflammasome activation remains to be clarified.
In this study, we demonstrated that HDAC10 played a critical role in attenuating NLRP3 inflammasome involved inflammation via direct deacetylating NLRP3 protein, thereafter leading to the NLRP3 ubiquitination and proteasome-mediated degradation. Thus, our data suggested that HDAC10 acted as a pivotal PTM switch of NLRP3 to shift from acetylation to ubiquitination, thereby negatively regulating the NLRP3 inflammasome activation and alleviated acute inflammatory injury.
Materials and methods
Cell culture and transfection
HEK293T cells were cultured in DMEM (R10-013-CV, Corning, USA) supplemented with 10% FBS (S711-001 S, LONSERA, Uruguay), penicillin (100 U/ml) and streptomycin (100 µg/ml). All the cells were cytogenetically tested and authenticated prior to use. To obtain mouse primary peritoneal macrophages, C57BL/6J mice (male, 6–8 weeks old) were intraperitoneally injected with sterile 6% starch broth. Peritoneal exudate cells (PECs) were harvested 72 h after the injection, and further cultured in DMEM supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 µg/ml). Adherent monolayer cells were harvested as peritoneal macrophages 4 h after the attachment. For the generation of BMDMs, bone marrow cells were isolated from the mice (BALB/c, 6 to 8 weeks) tibia and femur, and further plated into dishes at the density of 1 × 106 /ml. The isolated cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM) containing 20% (v/v) fetal bovine serum (FBS) and 30% (v/v) L929 supernatants for 4 days before replacement of the medium. The cells were further cultured for another 3 days before the adherent BMDMs were harvested. HDAC1-HDAC10, KAT5, P300 and CBP plasmids were cloned into pcDNA3.1 vector. The NLRP3 plasmid was got as previously described [16]. The CBL-B plasmids were synthesized by Miaoling Biology (Wuhan, China). NLRP1 and AIM2 plasmids were from Dr Dapeng Ma. HDAC10 mutants and NLRP3 mutants were constructed by using a site-directed mutagenesis kit (TOYOBO Life Science, Osaka, Japan) according to the manufacture’s instruction. Small interference RNA(SiRNA) was synthesized from Sigma (St. Louis, USA), and the cells were transfected with SiRNA by RNAiMax transfection reagent (catalog no. 13778150, Invitrogen, USA) according to the manufacture’s instruction.
Mice
C57BL/6J mice were purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd (Beijing, China). Hdac10−/− mice on C57BL/6J background were generated by Cyagen Biosciences (Guangzhou, China) using CRISPR-Pro technology. Hdac10fl/flLyz2-cre mice were generated by crossing Lyz-cre+/− and Hdac10fl/fl mice, which were both purchased from Cyagen (Guangzhou, China). All mice were housed in a specific pathogen-free facility in the Laboratory Animal Center of Shandong University in accordance with the National Institute of Health Guide for Care and Use of Laboratory Animals. All of the mice experiments were approved by the Scientific Investigation Board of Shandong University.
Animal models
C57BL/6J wild type and Hdac10−/− male mice (6–8 weeks old) were used for the construction of folic acid-induced acute tubular necrosis mice model and LPS-induced endotoxemia mouse model in this study. Hdac10fl/fl mice and Hdac10fl/flLyz2-cre mice (6–8 weeks old) were used for the construction of LPS induced endotoxemia model. The mice were randomly assigned, and the background of these mice was blinded to the major investigator. Folic acid-induced acute tubular necrosis mice model and LPS induced endotoxemia model were performed and evaluated as previously described [17]. All mice were bred in specific pathogen-free conditions at the Laboratory Animal Center of Shandong University. All of the mice experiments were performed in accordance with the general guidelines of Institutional Animal Care and Use Committee.
Reagents and antibodies
LPS (Escherichia coli, 0111: B4, L4130), folic acid (F7876), ATP (A6419), and anti-Flag antibody (F1804) were bought from Sigma-Aldrich (St. Louis, USA). The antibodies including anti-NLRP3 (AG-20B-0014), anti-Caspase1 p20 (AG-20B-0042), anti-ASC (AG-25B-0006) antibodies were purchased from Adipogen (San Diego, USA). The anti-GAPDH (60004-1-Ig), anti-β-actin (66009-1-Ig), anti-HA (51064-2-AP), anti-GFP (66002-1-Ig), anti-His (66005-1-Ig), anti-TOM20 (66777-1-Ig), anti-NLRP3 (19771-1-AP) and anti-HDAC10 (24913-1-AP) antibodies were from Proteintech (Chicago, USA). The anti-IL-1β (12242 S) antibody was from Cell Signaling Technology (Danfoss, USA). The anti-Myc (TA150054) antibody was from OriGene (Maryland, USA). The protein A/G plus agarose (sc-2003) for IP and the anti-ubiquitin (sc-8017) antibody were from Santa Cruz Biotechnology (California, USA). Chlorhexidine (CHX) was bought from Selleck (Houston, USA).
Western blot, immuno-precipitation (IP) and immunofluorescence assay
Western blot, immuno-precipitation (IP) and immunofluorescence microscopy assays were performed and evaluated as previously described [18, 19]. Cell culture supernatants were harvested and concentrated for immunoblotting with Amicon Ultra10 K (UFC5010) from Millipore (Massachusetts, USA). Full length western blots were provided as a single supplemental material file.
Real-time quantitative PCR and ELISA
Total RNA was prepared with Trizol reagent from Tiangen (Beijing, China) according to the manufacturer’s instructions. Total RNA was reversely transcribed using a PrimeScript™ RT reagent Kit with gDNA Eraser (RR047A) from TAKARA (Dajin, Japan). Real-time PCR was performed using FastSYBR Mixture (CW0659s) from CWBio (Jiangsu, China). Specific primers for PCR amplification will be provided upon request. The ELISA assay for the detection of mouse IL-1β, TNF-α and IL-6 was performed by ELISA kit (Biolegend, USA) according to the manufacturer’s instructions.
Immunohistochemistry
Immunohistochemistry was carried out to explore the expression of NLRP3 and Caspase-1 in lung tissues from LPS-induced acute endotoxemia mice models and kidney tissues from FA-induced kidney injury models. Briefly, tissue sections were soaked in xylene for dewaxing and graded alcohol for rehydrating. Antigen retrieval was performed by placing in citrate buffer and high-pressure heating to expose epitopes, and 5% BSA was added to block the nonspecific epitopes. Sections were incubated with primary antibodies against NLRP3 (1:200) and Caspase-1 (1:300) at 4 °C overnight. Sections were cultured with biotin-labeled secondary antibody for 1 h according to the manufacturer’s instructions. The images were photographed by the panoramic scanning and image analysis system (VS120, USA) for defining the pathological section, and the calculated mean optical density (MOD) values were analyzed by Image Pro-plus software.
In vitro binding assay
Myc-NLRP3, Flag-HDAC10 and Flag-HDAC3 proteins were expressed by a TNT Quick Coupled Transcription/Translation System (Promega) according to the instructions of the manufacturer, and the in vitro protein interaction assay were further performed by immunoprecipitation assay.
Statistical analysis
Data were statistically analyzed using GraphPad Prism software (GraphPad 9, CA, USA). Statistical significance among groups was evaluated with two-tailed student’s t test or two-way analysis of variance. Difference in the survival was statistically analyzed by using a Log-Rank test. P value < 0.05 (two-tailed) was considered statistically significant, and the data were presented as mean ± SD.
Results
HDAC10 directly interacted with NLRP3
Acetylation modification is an important class of protein post-translational modifications, which actively participated in many biological processes [20]. To define the role of acetylation in NLRP3 inflammasome activation, we first co-transfected HEK293T cells with NLRP3 plasmid and the constructs encoding three most common acetyltransferases (CBP, P300 and KAT5) for further acetylation analysis. Both the western blot and immunoprecipitation assay showed that NLRP3 protein could be acetylated, and KAT5 is the most potent acetyltransferase for NLRP3 (Fig. S1A, S1B). We treated the NLRP3 overexpressing cells with HDAC family specific inhibitor TSA and the data showed that the acetylation level of NLRP3 was also significantly upregulated after TSA treatment (Fig. S1C, S1D), which indicated that the deacetylation of NLRP3 was regulated by HDAC family members. We further isolated primary mouse peritoneal macrophages and detected the effect of TSA on NLRP3 inflammasome activation. The data showed that the endogenous NLRP3 protein level was significantly increased, and the cleavage of caspase-1 and IL-1β was also significantly increased in the TSA treated macrophages (Fig. S1E), which indicated that deacetylation of NLRP3 was regulated by HDAC proteins and the inhibition of NLRP3 deacetylation by TSA promoted the NLRP3 inflammasome activation.
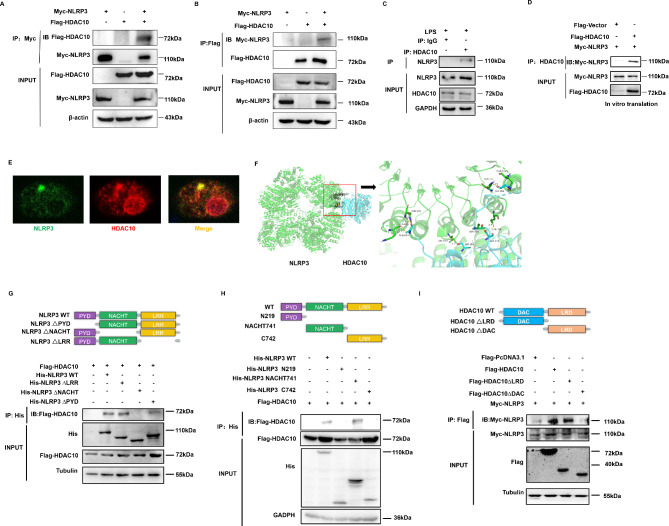
To define the precise HDAC protein responsible for the regulation of NLRP3 inflammasome activity, we screened the possible interacting proteins with NLRP3 by immunoprecipitation (IP) assay, and HDAC10 was screened out as the candidate protein (Fig. S2A-2B). Reciprocal IP assay verified that HDAC10 interacted with NLRP3 (Fig. 1A and B). The endogenous IP in mouse peritoneal macrophages also indicated that HDAC10 could bind with NLRP3 (Fig. 1C), and the interaction between NLRP3 and HDAC10 was abrogated in the Hdac10−/− macrophages (Fig. S2C). Though HDAC3 was also screened out as a candidate interacting protein with NLRP3 (Fig. S2A), both the literature [11] and our IP data (Fig. S2D-E) demonstrated that HDAC10 interacted with HDAC3. Thus, we further tried to define whether HDAC3, HDAC10, or both could directly interact with NLRP3. We expressed the HDAC3, HDAC10 and NLRP3 proteins by an in vitro transcription and translation system, and the IP assay revealed that only HDAC10 protein could directly interact with NLRP3 (Fig. 1D, S2F), indicating that the interaction between HDAC3 and NLRP3 was an indirect binding via HDAC10. Confocal microscopy assay showed that HDAC10 and NLRP3 co-localized in the mouse peritoneal macrophages (Fig. 1E), which further verified their intracellular interaction.
Fig. 1.
HDAC10 directly interacted with NLRP3. (A) Exogenous IP assay of the interaction between HDAC10 and NLRP3. (B) Reciprocal exogenous IP assay of the interaction between NLRP3 and HDAC10. (C) Endogenous immunoprecipitation assay of the interaction between HDAC10 and NLRP3 in mouse peritoneal macrophages primed with LPS for 6 h. (D) HDAC10 and NLRP3 proteins were expressed and obtained from the in vitro translation system and their interactions were further analyzed. (E) Confocal microscopy assay of the interaction between HDAC10 and NLRP3 in mouse peritoneal macrophages. NLRP3 was stained by green fluorescence and HDAC10 was stained by red fluorescence. Scale bars, 10 μm. (F) Protein-protein docking analysis of the interactions between NLRP3 (green cartoon) and HDAC10 (blue cartoon). (G, H) Schematic diagram of human NLRP3 (WT) and its truncation mutants was presented (top), and the interaction of NLRP3 (NLRP3 truncation mutants) with HDAC10 was detected by IP assay. Both the domain deleted mutant (G) and the domain overexpression mutant (H) of NLRP3 were presented. (I) Schematic diagram of human HDAC10 (WT) and its truncation mutants was presented (top), and the interaction of HDAC10 truncation mutants with NLRP3 was detected by IP assay. Data are representative of three independent experiments with similar results in (A)-(D) and (G)-(I)
Protein-protein docking is the prediction of the complex structure, based on the physicochemical complementarity at the protein-protein interface, thus simulating the proteins that recognize and interact with each other in a crowded environment [21]. To further verify the interaction between NLRP3 and HDAC10, we simulated the spatial binding between NLRP3 and HDAC10 by analyzing their interaction with the ClusPro platform (https://cluspro.bu.edu/), which verified their spatial physicochemical interaction (Fig. 1F). To further clarify the interacting domain of HDAC10 and NLRP3, we analyzed the protein structure and constructed a series of domain-deleted mutants of these two proteins. NLRP3 protein includes a PYD domain, a NACHT domain and a LRR domain, while HDAC10 contains an N-terminal DAC domain and a C-terminal LRD domain. IP assay revealed that the NACHT domain deleted NLRP3 mutant abrogated the interaction between these two proteins (Fig. 1G), and the domain overexpression constructs verified that the NACHT domain of NLRP3 was the interacting domain with HDAC10 (Fig. 1H). Further investigation showed that the DAC domain deleted HDAC10 mutant abolished their interactions (Fig. 1I), indicating that DAC domain is the interacting domain with NLRP3. Altogether these data indicated that HDAC10 directly interacted with NLRP3, and the NACHT domain of NLRP3 and the N-terminal DAC domain of HDAC10 were the molecular basis for their direct interaction.
Deletion of HDAC10 aggravated the NLRP3 inflammasome-mediated acute inflammatory injury
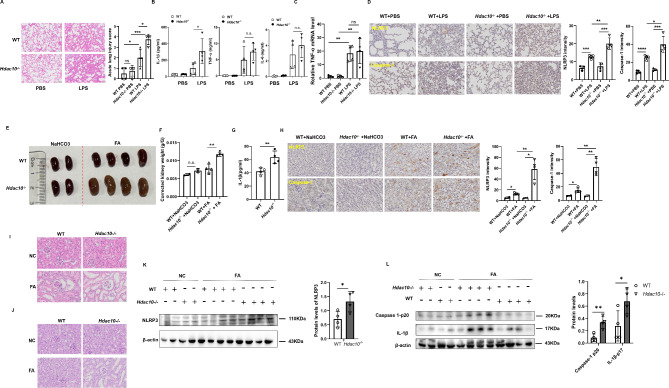
We have defined the direct interaction between HDAC10 and NLRP3, we are further interested in defining whether HDAC10 has any regulatory effect on the NLRP3 involved inflammatory diseases. We constructed a LPS induced acute endotoxemia model by intraperitoneal injection of LPS in Hdac10−/− mice and wild type mice, and further analyzed the tissue injury in these acute endotoxic mice. The data showed that the extent of lung injury was significantly increased in LPS challenged Hdac10−/− mice compared with their wild type counterparts (Fig. 2A), and the serum level of IL-1β in the Hdac10−/− mice was significantly increased while the serum level of lipopolysaccharide (LPS)-induced proinflammatory cytokines, including IL-6 and TNF-α and the mRNA level of TNF-α were not changed (Fig. 2B and C), indicating that HDAC10 specifically inhibited NLRP3 inflammasome activation.
Fig. 2.
Deletion of HDAC10 aggravated the NLRP3 inflammasome-mediated acute inflammatory injury. (A) Histopathological analysis of the lung tissues by hematoxylin and eosin (H&E) staining. The quantified lung injury was depicted by defined clinical parameters in ALI score (n = 4 mice/group). Scale bar: 100 μm. (B) ELISA analysis of IL-1β, TNF-α and IL-6 in the serum of WT or Hdac10−/− mice after intraperitoneal injection with LPS (n = 4/group). (C), The mRNA level of TNF-α in the lung tissues of WT or Hdac10−/− mice after intraperitoneal injection with LPS(n = 4/group). (D), Immunohistochemical assay of NLRP3 and Caspase-1 in the lung tissues of WT or Hdac10−/− mice after intraperitoneal injection with LPS(n = 4/group). Scale bar: 50 μm. (E, F) Folic acid-induced acute tubular necrosis mice model was constructed. Image (E) and weight (F) of spleen injury of WT and Hdac10-/- mice after intraperitoneal (i.p.) injection with folic acid were presented. (G) ELISA analysis of IL-1β in the serum of WT or Hdac10−/− mice after intraperitoneal injection with folic acid. n = 4. (H), Immunohistochemical assay of NLRP3 and Caspase-1 in kidney tissue after intraperitoneal injection with FA. n = 2 (NaHCO3 group), n = 4 (FA group). Scale bar: 20 μm. (I) H&E staining of the kidney tissue sections. Scale bars 20 μm. (J) Periodic acid-Schiff (PAS) staining of the kidney tissue sections. Scale bars 20 μm. (K, L) WT or Hdac10−/− mice were i.p. injected with FA, followed by immunoblot analysis of NLRP3(K), and the activation of its downstream signaling Caspase1 p20 and IL-1β p17 (L) in the kidney tissue samples. The expression level of NLRP3, Caspase1-p20 and IL-1β p17 were quantified by Image J software and normalized to β-actin. Data and error bars are mean ± SD in (A)-(D), (F), (G), (H), (K) and (L). * P < 0.05, ** P < 0.01, *** P<0.001 and **** P<0.0001. Data are representative of three independent experiments with similar results. Statistical analysis was done with student’s t test
In addition, immunohistochemical assay was used to test the inflammation markers, and the data showed that the levels of NLRP3 and Caspase-1 were significantly increased in Hdac10−/− mice (Fig. 2D). Because the pro-inflammatory cytokine IL-1β is also possibly released by activation of other inflammasomes including the AIM2 inflammasome and the NLRP1 inflammasome, we further tested the interaction of HDAC10 with NLRP1 and AIM2. The data showed that HDAC10 could not interact with NLRP1 or AIM2 (Fig S2G), indicating the unique regulation of NLRP3 inflammasome by HDAC10. Thus, these data indicated that HDAC10 attenuated LPS induced endotoxemia possibly via its specifical regulatory effect on NLRP3 inflammasome.
We further investigated the role of HDAC10 in a folic acid (FA)-induced acute tubular necrosis (ATN) model by intraperitoneal injection of FA in mice. Thirty-six hours after the injection of FA, the mice were sacrificed and further assay showed that the extent of kidney injury and kidney weight in the Hdac10−/− mice was significantly increased compared with the wild-type mice (Fig. 2E and F). The serum level of IL-1β in the Hdac10−/− mice was also significantly increased compared with that of their wild-type counterparts (Fig. 2G). Immunohistochemical assay was used to test the inflammation markers, and the data showed that the level of NLRP3 and Caspase-1 were significantly increased in Hdac10−/− mice (Fig. 2H). H&E staining and periodic acid-Schiff (PAS) staining showed that the renal inflammation and edema were both significantly exacerbated in Hdac10−/− mice compared with their wild-type counterparts (Fig. 2I and J). In addition, the expression level of NLRP3 in the kidney was significantly upregulated in the Hdac10−/− mice, and the cleavage of pro-Caspase 1 and pro-IL-1β were also significantly increased in Hdac10−/− mice compared with the wild type mice (Fig. 2K and L). These data indicated that HDAC10 significantly attenuated the NLRP3 inflammasome involved acute inflammatory tissue injury, probably via its negative regulatory effect on NLRP3 inflammasome.
HDAC10 inhibited NLRP3 by its deacetylation activity
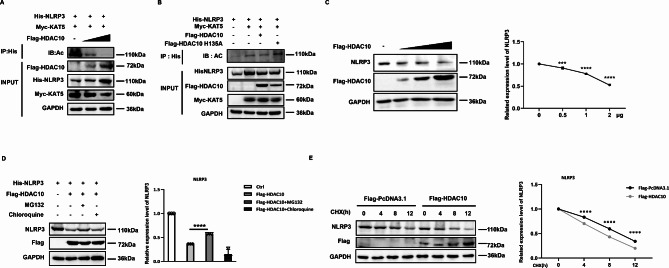
In consideration of the protective effect of HDAC10 on acute inflammation, we are interested in defining the exact regulatory mechanism of HDAC10 in NLRP3 inflammasome activation. HDAC10 is a member of the class II histone deacetylases [22], thus we further try to define whether HDAC10 exerts its effect via deacetylating NLRP3. The acetylation assay showed that exogenous overexpression of HDAC10 induced the deacetylation of NLRP3 in a dose-dependent manner (Fig. 3A). To verify whether the effect of HDAC10 was mediated by its protein deacetylation activity, we constructed an enzyme dead mutant of HDAC10 (H135A) by replacing histidine with alanine at the position of 135. Further investigation showed that the HDAC10 (H135A) mutant significantly abolished HDAC10-mediated deacetylation of NLRP3 (Fig. 3B), which verified the involvement of HDAC10 deacetylase activity in the regulation of NLRP3.
Fig. 3.
HDAC10 inhibited NLRP3 by deacetylating NLRP3. (A) Immunoprecipitation analysis of the exogenous NLRP3 acetylation in HEK293T cells co-transfected with NLRP3 plasmid, HDAC10 plasmid and KAT5 plasmid. (B) Immunoprecipitation analysis of the exogenous NLRP3 acetylation in HEK293T cells co-transfected with NLRP3 plasmid, HDAC10 or HDAC10 H135A plasmid and KAT5 plasmid. (C) Immunoblot analysis of endogenous NLRP3 protein expression in HEK293T cells transfected with different doses of HDAC10 plasmids. NLRP3 expression levels were quantified by ‘Image J’ software and normalized to GAPDH (right panel). (D) Immunoblot analysis of NLRP3 in NLRP3 and HDAC10 transfected HEK293T with the treatment of MG132 or Chloroquine for 4 h before being harvested. NLRP3 expression levels were quantified by ‘Image J’ software and normalized to GAPDH (right panel). (E) Immunoblot analysis of NLRP3 in HEK293T cells transfected with NLRP3 and HDAC10 plasmids, followed with the cycloheximide (CHX) treatment. NLRP3 expression levels were quantified by ‘Image J’ software and normalized to GAPDH (right panel). Data and error bars are mean ± SD in (C)-(E). * P < 0.05, ** P < 0.01, *** P<0.001 and **** P<0.0001. Data are representative of three independent experiments with similar results. The western blot experiments were repeated three times independently with similar results in (A)-(E). Statistical analysis was done with student’s t test and two-way ANOVA
To further clarify the regulatory effect of HDAC10 on NLRP3 protein, we transfected HEK293T cells with HDAC10 construct and further analyzed the NLRP3 protein. The data showed that the protein level of NLRP3 was significantly down-regulated in a dose-dependent manner in the HDAC10 overexpressing cells (Fig. 3C). We further treated the cells with the proteasome inhibitor MG132 or the lysosomal inhibitor chloroquine. The data showed that HDAC10-induced NLRP3 degradation was significantly rescued by MG132 (Fig. 3D), which indicated that HDAC10 reduced the stability of NLRP3 via the proteasome-mediated degradation. When the de novo protein synthesis was blocked by cycloheximide (CHX), the exogenous overexpression of HDAC10 significantly increased the protein degradation of NLRP3 (Fig. 3E), which indicated that HDAC10 reduced the protein stability of NLRP3. Altogether, these data indicated that HDAC10 negatively regulated the level of NLRP3 protein by inducing deacetylation of NLRP3 protein.
HDAC10 significantly inhibited NLRP3 inflammasome activation
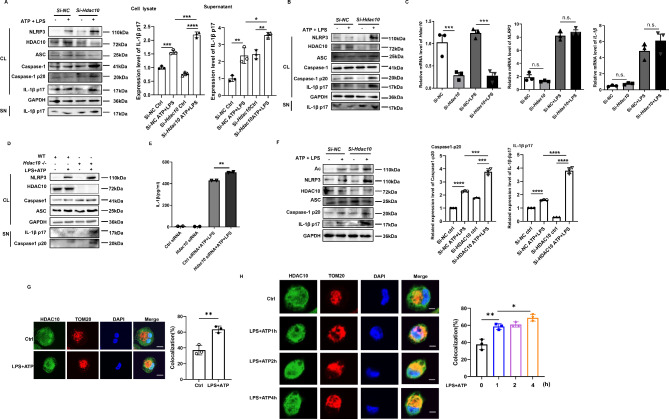
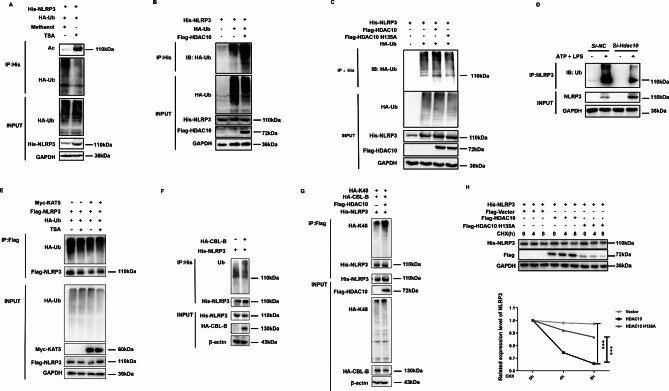
To further define the effect of HDAC10 on NLRP3 inflammasome activation, we knocked down Hdac10 in mouse peritoneal macrophages and BMDM cells by Si-Hdac10. The data showed that inhibition of Hdac10 significantly increased the protein level of NLRP3, and the cleavage of Caspase1 and IL-1β under the stimulation with LPS/ATP was also significantly enhanced in mouse peritoneal macrophages and BMDM cells (Fig. 4A and B). The qRT-PCR data revealed that inhibition of HDAC10 did not affect the mRNA level of NLRP3 and IL-1β (Fig. 4C), indicating that HDAC10 did not exert its negative regulatory effect on the transcription level of NLRP3. We further isolated the peritoneal macrophages from Hdac10-/- mice, and the data showed that the NLRP3 inflammasome activation was significantly increased under the stimulation of LPS/ATP (Fig. 4D). Moreover, ELISA assay showed that inhibition of HDAC10 significantly enhanced the IL-1β secretion in the LPS/ATP stimulated macrophages (Fig. 4E). In addition, we further knocked down Hdac10 in mouse peritoneal macrophages, and the data verified that inhibition of HDAC10 blocked the deacetylation of NLRP3 and further promoted the NLRP3 inflammasome activation (Fig. 4F), which indicated that HDAC10 induced NLRP3 deacetylation and further attenuated the NLRP3 inflammasome activation.
Fig. 4.
HDAC10 significantly inhibited NLRP3 inflammasome activation. (A) Immunoblot analysis of NLRP3 inflammasome activation of the Si-Hdac10 transfected mouse peritoneal macrophages followed with LPS priming for 6 h and ATP treatment for 30 min. (B) Immunoblot analysis of NLRP3 inflammasome activation of the Si-Hdac10 transfected BMDM cells followed with LPS priming for 6 h and ATP treatment for 30 min. (C) qRT-PCR analysis of NLRP3 and IL-1β mRNA levels in in the LPS-primed Si-Hdac10 transfected mouse peritoneal macrophages. (D) Immunoblot analysis of NLRP3 inflammasome activation in the LPS and ATP treated WT and Hdac10-/- mouse peritoneal macrophages. (E) ELISA analysis of IL-1β in the supernatant of Si-Hdac10 transfected mouse peritoneal macrophages. (F) Immunoblot analysis of NLRP3 acetylation in mouse peritoneal macrophages transfected with Si-Hdac10 and further treated with LPS and ATP. Caspase1-p20 expression levels (left) and IL-1β p17 (right) were quantified by ‘Image J’ software and normalized to GAPDH. (G) Confocal microscopy assay of the interaction between HDAC10 and TOM20 in THP-1 cells. HDAC10 was stained by green fluorescence and TOM20 was stained by red fluorescence, and the colocalization of HDAC10 (green) and TOM20 (red) was visualized as yellow fluorescence in the merged panel. Scale bars, 10 μm. (H) Confocal microscopy assay of the interaction between HDAC10 and TOM20 in THP-1 cells treated with LPS for 1 h, 2 h, 4 h and ATP for 30 min. HDAC10 was stained by green fluorescence and TOM20 was stained by red fluorescence, and the colocalization of HDAC10 (green) and TOM20 (red) was visualized as yellow fluorescence in the merged panel. Scale bars, 10 μm. Data and error bars are mean ± SD in (C) and (E)-(H). * P < 0.05, ** P < 0.01, *** P<0.001 and **** P<0.0001. Data are representative of three independent experiments with similar results in C, E and G-H. The western blot experiments were repeated three times independently with similar results in (A), (B), (D) and (F). Statistical analysis was done with student’s t test
During the activation of NLRP3 inflammasome, NLRP3 transferred from endoplasmic reticulum to mitochondria, and further induced the formation of NLRP3 multi-protein platform and NLRP3 inflammasome activation [23]. Thus, we further tried to define whether the interaction between HDAC10 and NLRP3 occurred at the mitochondria. The confocal microscopy assay showed that HDAC10 co-localized with mitochondria marker TOM20 in the THP-1 cells, and NLRP3 inflammasome activation increased the colocalization (Fig. 4G). Further investigation revealed that during the activation process of NLRP3 inflammasome, the amount of HDAC10 protein localized at the mitochondria was dramatically increased (Fig. 4H), indicating the active participation of HDAC10 in the NLRP3 inflammasome activation. Altogether, these data suggested that HDAC10 significantly inhibited NLRP3 inflammasome activation by negatively regulating NLRP3 protein.
Deacetylation of NLRP3 by HDAC10 promoted NLRP3 ubiquitination
We have demonstrated that HDAC10 deacetylates NLRP3 and further inhibits NLRP3 through the proteasomal degradation pathway, which usually mediated degradation of the ubiquitinated protein; thus we are further interested in defining whether HDAC10 promotes NLRP3 ubiquitination. The acetylation and ubiquitination assay showed that after treatment with the HDAC inhibitor TSA, the acetylation of NLRP3 was significantly increased, while the protein ubiquitination was significantly inhibited (Fig. 5A), indicating the possible involvement of a PTM switch in the regulation of NLRP3 protein. Further investigation showed that exogenous overexpression of HDAC10 significantly increased the ubiquitination of NLRP3 (Fig. 5B), and the HDAC10 enzyme dead mutant (H135A) dramatically rescued the ubiquitination of NLRP3 (Fig. 5C), which further validated that ubiquitination of NLRP3 was dependent on the deacetylation activity of HDAC10. When Hdac10 was inhibited by its specific SiRNA (Si-Hdac10) in mouse peritoneal macrophages, the ubiquitination of NLRP3 was also significantly inhibited (Fig. 5D). In addition, when KAT5-induced NLRP3 acetylation was maintained after the treatment with TSA, the ubiquitination of NLRP3 was also significantly inhibited (Fig. 5E). Altogether, these data indicated that the ubiquitination of NLRP3 was a subsequent consequence of HDAC10 activation. It is recently recognized that proteasomal degradation of NLRP3 was induced by casitas B cell lymphoma-B (CBL-B), an E3 ubiquitination ligase mediated K48-linked ubiquitination [24], thus we further try to define the effect of HDAC10 on CBL-B induced ubiquitination. The data revealed that exogenous overexpression of HDAC10 significantly increased the CBL-B-mediated K48-linked ubiquitination of NLRP3 (Fig. 5F and G), indicating that the ubiquitination of NLRP3 was a subsequent event following its deacetylation. The CHX assay further revealed that the enzyme dead mutant of HDAC10 (H135A) significantly rescued the HDAC10-mediated degradation of NLRP3(Fig. 5H), indicating that HDAC10 mediated NLRP3 degradation is a subsequent effect of its deacetylation activity.
Fig. 5.
Deacetylation of NLRP3 by HDAC10 leads to NLRP3 ubiquitination. (A) Ubiquitination analysis of NLRP3 in NLRP3 and HA-UB transfected HEK293T cells followed with TSA treatment for 24 h. (B) Ubiquitination analysis of NLRP3 in HEK293T cells transfected with NLRP3, HDAC10 and HA-UB plasmids. (C) Ubiquitination analysis of NLRP3 in the HEK293T cells transfected with NLRP3, HDAC10 (or the H135A mutant) and HA-UB plasmids. (D) IP analysis of NLRP3 ubiquitination in Si- Hdac10 transfected mouse peritoneal macrophages with the LPS and ATP treatment. (E) Ubiquitination analysis of NLRP3 in NLRP3, HA-UB and KAT5 transfected HEK293T cells with TSA treatment for 24 h. (F) Ubiquitination analysis of NLRP3 in HEK293T cells transfected with NLRP3 and HA-CBL-B plasmids. (G) K48 ubiquitination analysis of NLRP3 in HEK293T cells transfected with NLRP3, HDAC10, CBL-B and HA-UB-K48 plasmids. (H) Immunoblot analysis of NLRP3 in NLRP3 plasmid and HDAC10 (H135A) mutant transfected HEK293T cells under the treatment with CHX for 0 h, 4 h and 8 h. NLRP3 expression levels were quantified by ‘Image J’ software and normalized to GAPDH (right panel). Data and error bars are mean ± SD in (H). * P < 0.05, ** P < 0.01, *** P<0.001 and **** P<0.0001. Data are representative of three independent experiments with similar results. The western blot experiments were repeated three times independently with similar results in (A)-(H). Statistical analysis was done with two-way ANOVA
HDAC10 induced deacetylation and ubiquitination of NLRP3 converging at lys496 residue
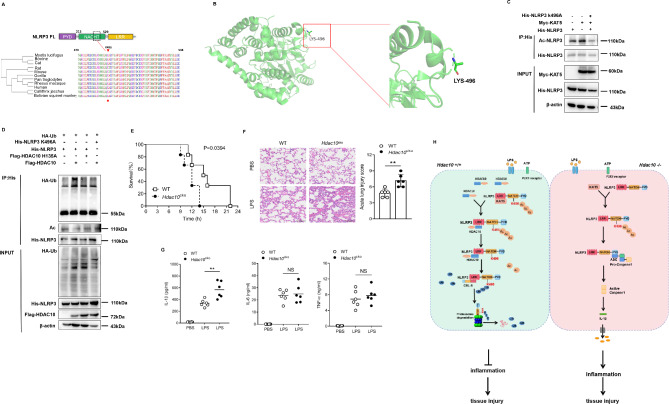
To define the precise residue involved in the modification switch, we screened the possible acetylation sites of NLRP3 (http://pail.biocuckoo.org) and further analyzed their modification potential of ubiquitination (Fig. S3). According to the analysis of the possible modification residues, the K496, K567, K689 and K927 residues of NLRP3 were screened out as the potential amino acid residues acting as the modification switch induced by HDAC10 (Fig. S4A). We constructed a series of NLRP3 mutants(K496A, K567A, K689A and K927A) by replacing lysine with alanine at these residues and analyzed the modification of these residues under the effect of HDAC10. Both the ubiquitination and acetylation analysis revealed that K496 residue was the potential switching site from acetylation to ubiquitination induced by HDAC10 (Fig. 6A and D, S4B). Thus, these data indicated that HDAC10 promoted the ubiquitination degradation of NLRP3 by deacetylating NLRP3, and K496 residue of NLRP3 is possibly the pivotal convergence point involved in this PTM switch.
Fig. 6.
HDAC10 deacetylated NLRP3 and further induced its ubiquitination converging at lys496. (A) Conservation analysis of NLRP3 Lys496 among species by the Clustal Omega program. (B) Protein structure analysis of the NLRP3 NACHT domain (left) and K496 site (right). (C) Immunoprecipitation analysis of the exogenous NLRP3 acetylation in HEK293T cells co-transfected with NLRP3 plasmid, NLRP3 K496A plasmid and KAT5 plasmid. (D) Ubiquitination and acetylation analysis of NLRP3 in the HEK293T cells transfected with NLRP3 (or the K496A mutant), HDAC10 (or the H135A mutant) and HA-UB plasmids. (E) Survival status of WT and Lyz-cre; Hdac10f/f mice after intraperitoneal injection with LPS (n = 6 mice/group). (F) Histopathological analysis of the lung tissues by hematoxylin and eosin (H&E) staining. The quantified lung injury was depicted by defined clinical parameters in ALI score (n = 5 mice/group). Scale bar: 100 μm. (G) ELISA analysis of IL-1β, TNF-α and IL-6 in the serum of WT or Lyz-cre; Hdac10f/fmice after intraperitoneal injection with LPS. (H) Schematic diagram of the regulatory role of HDAC10 in NLRP3 inflammation activation. HDAC10 directly induced the deacetylation of NLRP3 at K496 residue, thus switching NLRP3 acetylation to the ubiquitination modification, leading to the proteasomal degradation of NLRP3 protein. Data and error bars are mean ± SD in (F) and (G). * P < 0.05, ** P < 0.01, *** P<0.001 and **** P<0.0001. Data are representative of three independent experiments with similar results in (C)-(D). Statistical analysis was done with student’s t test
To verify the role of macrophage expressed HDAC10 in acute inflammation, we constructed another LPS-induced endotoxemia animal model and compared the tissue injury between the Hdac10fl/fl Lyz2-cre mice and the Hdac10fl/fl mice, with the intension to further validate the role of macrophage derived HDAC10 in acute inflammation. Our data showed that the overall survival time of LPS-challenged Hdac10fl/fl Lyz2-cre mice was significantly decreased compared with the Hdac10fl/fl mice (Fig. 6E). Twelve hours after the injection, the mice were sacrificed and further assay showed that the extent of lung injury in Hdac10fl/fl Lyz2-cre mice was significantly increased compared with the Hdac10fl/fl mice (Fig. 6F). The serum level of IL-1β in Hdac10fl/fl Lyz2-cre mice was also significantly increased, while the levels of TNF-α and IL-6 were not significantly altered compared with that of their Hdac10fl/fl counterparts (Fig. 6G). Thus, in the macrophages conditional Hdac10 knockout mice, we further validated the macrophages derived HDAC10 played a pivotal protective effect on the NLRP3 mediated acute inflammatory injury.
Altogether, this study demonstrated that HDAC10 inhibited NLRP3 inflammasome activation by directly targeting NLRP3 and inducing acetylation to ubiquitination switch of NLRP3 protein, which suggested a potential therapeutic strategy for these NLRP3 inflammasome involved inflammatory diseases by targeting HDAC10 (Fig. 6H).
Discussion
NLRP3 inflammasome is the core signaling hub and key instigator of acute inflammation in response to infection and oxidative stress to promote inflammatory cascade responses. Though inflammasome activation protects against infection and injury, excessive NLRP3 inflammasome activity yields excessive cytokine secretion and a pathological hyperinflammatory state. Of increasing interest is the critical contribution of NLRP3 inflammasome to acute inflammatory injury. Therefore, precise regulation of inflammasome activities is pivotal for adequate immune protection while avoiding collateral side effect to maintain the global tissue homeostasis. In this study, we identified HDAC10 as a novel negative regulator of NLRP3 inflammasome and deletion of HDAC10 aggravated acute inflammatory injury.
NLRP3 protein is subjected to multi-layered regulation at the transcriptional and posttranslational levels to ensure an efficient and appropriate effect for its powerful inflammatory machinery [25]. The NLRP3 protein is reported to be modified by acetylation at the lysine residues in the PYD domain, and its deacetylation by sirtuin 2 (SIRT2), a cytosolic deacetylase, further prevents and reverses aging-associated inflammation and insulin resistance [26]. Histone deacetylases (HDACs) are a group of enzymes responsible for the removal of acetyl groups from the acetylated histone and non-histone proteins [27]. Though several HDAC family members including HDAC3, HDAC6 and HDAC11 are reported to regulate the NLRP3 inflammasome activity, these regulatory effects are all independent of their deacetylation regulation [28–31]. HDAC10 is a member of the class II histone deacetylases and its role in many biological processes, including embryonic development, genome stability, and RNA transcription has been identified in recent years [32, 33]. In this study, we revealed that HDAC10 directly interacted with NLRP3 via the DAC domain of HDAC10 and the NACHT domain of NLRP3, and the protein-protein interaction simulation further verified the spatial docking between HDAC10 and NLRP3. The acetylation assay revealed that KAT5 catalyzed NLRP3 acetylation, while HDAC10 directly induced NLRP3 deacetylation and attenuated NLRP3 inflammasome activation. The most recent study demonstrated that NLRP3 acetylation is critical for the full activation of NLRP3 inflammasome [7], while here in this study we identified KAT5-HDAC10 as a new writer-eraser couple in the acetylation and deacetylation circuit during NLRP3 inflammasome activation.
We demonstrated that HDAC10 induced the deacetylation of NLRP3 and further mediated the proteasomal degradation of NLRP3, leading to the reduction of its protein abundance. Further investigation demonstrated that the negative regulatory effect of HDAC10 was abolished by its enzyme-dead mutant (H135A), indicating the critical involvement of the catalytic activity of HDAC10 in the regulation of NLRP3 protein. We further tried to define the potential link between the HDAC10-mediated deacetylation and proteasomal degradation of NLRP3, and the data revealed that HDAC10 acted as a PTM switch to reconstitute the NLRP3 protein modification by replacing the acetylation with ubiquitination, further leading to the ubiquitination mediated proteasomal degradation.
NLRP3 protein is rich in lysine and susceptible to both acetylation and ubiquitination modification, while these two modifications usually had opposing effects on the protein degradation and stability [34]. Various post-translational modifications in inflammasome signaling can be cross-regulated by each other [2], whereas the machinery involved in their spatial coordination during the process of NLRP3 inflammasome activation remains to be fully understood. Here we demonstrated that HDAC10 negatively regulated NLRP3 inflammasome by switching from acetylation to ubiquitination modification, leading to the proteasome-mediated degradation of the NLRP3 protein and attenuation of NLRP3 inflammasome activation. The further ubiquitination and acetylation analysis revealed that the Lys496 residue of NLRP3 protein is the pivotal convergence point involved in this PTM switch, indicating the critical switching node effect of this lysine residue. There are 68 lysine residues in the human NLRP3 protein, however only K689 and K496 residues have been verified as ubiquitination acceptor sites [35]. In these only two ubiquitin residues, both ubiquitination and sumoylation of K689 residue was reported [36, 37], whereas for the other K496 residue, the related studies are quite limited and no other PTM except ubiquitination has been reported. Here in this study, we identified a novel acetylation to ubiquitination switch at the K496 site induced by HDAC10, which may provide clues for further molecular and pharmacological approaches in controlling NLRP3 involved inflammatory injury. Though the regulatory tuning effect between acetylation and ubiquitination was recognized in recent studies [38], the negative regulatory role of HDAC10 in NLRP3 inflammasome activation was reported here for the first time. The present study provided new insights into the negative regulatory mechanisms of NLRP3 inflammasome by HDAC10 and provided potential therapeutic strategy for NLRP3 inflammasome-associated diseases by targeting HDAC10.
Conclusion
In summary, we demonstrated here that HDAC10 attenuated NLRP3 inflammasome mediated acute inflammatory injury by direct deacetylating NLRP3 and further leading to the ubiquitination modification and proteasomal degradation of NLRP3. The present study provided deeper insight into the negative regulatory mechanism of NLRP3 inflammasome and further offered the potential therapeutic target for NLRP3 inflammasome-associated inflammatory diseases. This work contributes further to our fundamental knowledge and understanding of the dynamic and complex molecular interactions between NLRP3 inflammasome and its regulation protein. It also paved new avenues of immunomodulatory therapy for inflammatory diseases based on the regulatory mechanisms of NLRP3 inflammasome by HDAC10.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work. All relevant data that support the findings of this study are available from the corresponding author upon request.
Author contributions
LH designed the research, conceived funding and wrote manuscript, MY, ZQ, and YL performed most experiments and contributed to the writing of this manuscript, XC, CS participated in the cellular experiments, XZ, HX participated in the plasmid construction, XC and WM participated in the macrophage isolation experiments. MY, ZQ, XZ, and DM generated and prepared the data of Figs. 1, 2, 3 and 5. MY and YL generated and prepared the data of Fig. 4. MY generated and prepared the data of Fig. 6. ZQ generated and prepared the data of figure S1. MY generated and prepared the data of figure S2 -S4. XZ and HX constructed the mutant plasmids in Fig. 4 and figure S3. XC and WM prepared the macrophage isolation in Figs. 1 and 5. XC and CS prepared the cells in Figs. 2 and 4 and figure S1. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82171748, No. 82472725 and 32200709), Distinguished Professor of Taishan Scholars (No. tstp20221109), Shandong Provincial Natural Science Foundation Joint Fund (ZR2023LZL010), the Postdoctoral Innovation Fund Project of Shandong Province (SDCX-ZG-202203044), the Major Innovation Project of Shandong Province (No. 2021GXGC011305), and Natural Science Foundation of Shandong Province (ZR2022QC061).
Data availability
The molecular docking was operated on the ClusPro 2.0 platform (https://cluspro.bu.edu/). The possible acetylation sites of NLRP3 were predicted from (http://pail.biocuckoo.org). All the data supporting the conclusions of this article are available from the corresponding author upon reasonable request. All data are available in the main text or the supplementary materials
Declarations
Ethics approval and consent to participate
All mouse experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee, and were approved by the Medical Ethics Committee of Shandong University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Min Yang and Zhenzhi Qin contributed equally to this work and share first authorship.
References
- 1.Huang Y, Xu W, Zhou R. NLRP3 inflammasome activation and cell death. Cell Mol Immunol. 2021;18(9):2114–27. 10.1038/s41423-021-00740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paik S, Kim JK, Silwal P, Sasakawa C, Jo EK. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell Mol Immunol. 2021;18(5):1141–60. 10.1038/s41423-021-00670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Liu X, Shi H, Yu Y, Yu Y, Li M, et al. NLRP3 inflammasome, an immune-inflammatory target in pathogenesis and treatment of cardiovascular diseases. Clin Transl Med. 2020;10(1):91–106. 10.1002/ctm2.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Hauenstein AV. The NLRP3 inflammasome: Mechanism of action, role in disease and therapies. Mol Aspects Med. 2020;76:100889. 10.1016/j.mam.2020.100889. [DOI] [PubMed] [Google Scholar]
- 5.Qin Y, Meng X, Wang M, Liang W, Xu R, Chen J, et al. Posttranslational ISGylation of NLRP3 by HERC enzymes facilitates inflammasome activation in models of inflammation. J Clin Invest. 2023;133(20). 10.1172/JCI161935. [DOI] [PMC free article] [PubMed]
- 6.Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20(3):156–74. 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Luo L, Xu X, Wu J, Wang F, Lu Y, et al. Acetylation is required for full activation of the NLRP3 inflammasome. Nat Commun. 2023;14(1):8396. 10.1038/s41467-023-44203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Li W, Schulz VP, Zhao H, Qu X, Qi Q, et al. Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency. Blood. 2021;138(17):1615–27. 10.1182/blood.2020007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt O, Nehls N, Prexler C, von Heyking K, Groll T, Pardon K, et al. Class I histone deacetylases (HDAC) critically contribute to Ewing sarcoma pathogenesis. J Exp Clin Cancer Res. 2021;40(1):322. 10.1186/s13046-021-02125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jänsch N, Meyners C, Muth M, Kopranovic A, Witt O, Oehme I, et al. The enzyme activity of histone deacetylase 8 is modulated by a redox-switch. Redox Biol. 2019;20:60–7. 10.1016/j.redox.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong JJ, Liu J, Bertos NR, Yang X-J. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 2002;30(5):1114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hai Y, Shinsky SA, Porter NJ, Christianson DW. Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat Commun. 2017;8:15368. 10.1038/ncomms15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart TM, Foley JR, Holbert CE, Klinke G, Poschet G, Steimbach RR, et al. Histone deacetylase-10 liberates spermidine to support polyamine homeostasis and tumor cell growth. J Biol Chem. 2022;298(10):102407. 10.1016/j.jbc.2022.102407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, Wang J, Wang X, Wang B, Zhao Z, Fu J, et al. Degradation of HDAC10 by autophagy promotes IRF3-mediated antiviral innate immune responses. Sci Signal. 2022;15(765):eabo4356. 10.1126/scisignal.abo4356. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Stuart JH, Talbot-Cooper C, Agrawal-Singh S, Huntly B, Smid AI, et al. Histone deacetylase 4 promotes type I interferon signaling, restricts DNA viruses, and is degraded via vaccinia virus protein C6. Proc Natl Acad Sci U S A. 2019;116(24):11997–2006. 10.1073/pnas.1816399116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Q, Guo P, Mu K, Zhang Y, Zhao W, Huai W, et al. Estrogen suppresses hepatocellular carcinoma cells through ERβ-mediated upregulation of the NLRP3 inflammasome. Lab Invest. 2015;95(7):804–16. 10.1038/labinvest.2015.63. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y, Lv X, Sun C, Sun Y, Yang M, Ma D, et al. TRIM50 promotes NLRP3 inflammasome activation by directly inducing NLRP3 oligomerization. EMBO Rep. 2022;23(11):e54569. 10.15252/embr.202154569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huai W, Zhao R, Song H, Zhao J, Zhang L, Zhang L, et al. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat Commun. 2014;5:4738. 10.1038/ncomms5738. [DOI] [PubMed] [Google Scholar]
- 19.Ma X, Ma X, Qiu Y, Zhu L, Lin Y, You Y, et al. TRIM50 suppressed hepatocarcinoma progression through directly targeting SNAIL for ubiquitous degradation. Cell Death Dis. 2018;9(6):608. 10.1038/s41419-018-0644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol. 2022;23(5):329–49. 10.1038/s41580-021-00441-y. [DOI] [PubMed] [Google Scholar]
- 21.Vakser IA. Protein-protein docking: from interaction to interactome. Biophys J. 2014;107(8):1785. 10.1016/j.bpj.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferro A, Pantazaka E, Athanassopoulos CM, Cuendet M. Histone deacetylase-based dual targeted inhibition in multiple myeloma. Med Res Rev. 2023;43(6):2177–236. 10.1002/med.21972. [DOI] [PubMed] [Google Scholar]
- 23.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–5. 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 24.Tang J, Tu S, Lin G, Guo H, Yan C, Liu Q, et al. Sequential ubiquitination of NLRP3 by RNF125 and Cbl-b limits inflammasome activation and endotoxemia. J Exp Med. 2020;217(4). 10.1084/jem.20182091. [DOI] [PMC free article] [PubMed]
- 25.Fischer FA, Mies LFM, Nizami S, Pantazi E, Danielli S, Demarco B, et al. TBK1 and IKKε act like an OFF switch to limit NLRP3 inflammasome pathway activation. Proc Natl Acad Sci U S A. 2021;118(38). 10.1073/pnas.2009309118. [DOI] [PMC free article] [PubMed]
- 26.He M, Chiang HH, Luo H, Zheng Z, Qiao Q, Wang L, et al. An Acetylation Switch of the NLRP3 Inflammasome Regulates Aging-Associated Chronic Inflammation and Insulin Resistance. Cell Metab. 2020;31(3):580–91. 10.1016/j.cmet.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Wang Z, Liu J. Role of HDACs in normal and malignant hematopoiesis. Mol Cancer. 2020;19(1):5. 10.1186/s12943-019-1127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magupalli VG, Negro R, Tian Y, Hauenstein AV, Di Caprio G, Skillern W, et al. HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science. 2020;369(6510). 10.1126/science.aas8995. [DOI] [PMC free article] [PubMed]
- 29.Yao F, Jin Z, Zheng Z, Lv X, Ren L, Yang J, et al. HDAC11 promotes both NLRP3/caspase-1/GSDMD and caspase-3/GSDME pathways causing pyroptosis via ERG in vascular endothelial cells. Cell Death Discov. 2022;8(1):112. 10.1038/s41420-022-00906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi Z, Chen S, Xu T, Zhen W, Yu W, Jiang D, et al. Histone Deacetylase 3 Couples Mitochondria to Drive IL-1beta-Dependent Inflammation by Configuring Fatty Acid Oxidation. Mol Cell. 2020;80(1):43–58. 10.1016/j.molcel.2020.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Hwang I, Lee E, Jeon SA, Yu JW. Histone deacetylase 6 negatively regulates NLRP3 inflammasome activation. Biochem Biophys Res Commun. 2015;467(4):973–8. 10.1016/j.bbrc.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Fernandez C, Lorda-Diez CI, Duarte-Olivenza C, Hurle JM, Montero JA. Histone Epigenetic Signatures in Embryonic Limb Interdigital Cells Fated to Die. Cells. 2021;10(4). 10.3390/cells10040911. [DOI] [PMC free article] [PubMed]
- 33.Wu Y, Chen H, Lu J, Zhang M, Zhang R, Duan T, et al. Acetylation-dependent function of human single-stranded DNA binding protein 1. Nucleic Acids Res. 2015;43(16):7878–87. 10.1093/nar/gkv707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Z, Damianou A, Di Daniel E, Kessler BM. Inflammasome activation controlled by the interplay between post-translational modifications: emerging drug target opportunities. Cell Commun Signal. 2021;19(1):23. 10.1186/s12964-020-00688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barry R, John SW, Liccardi G, Tenev T, Jaco I, Chen C-H, et al. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat Commun. 2018;9(1):3001. 10.1038/s41467-018-05321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan P, Zhang Q, Liu W, Jia Y, Ai S, Wang T, et al. Cullin1 binds and promotes NLRP3 ubiquitination to repress systematic inflammasome activation. FASEB J. 2019;33(4):5793–807. 10.1096/fj.201801681R. [DOI] [PubMed] [Google Scholar]
- 38.Zhao K, Zhang Y, Xu X, Liu L, Huang L, Luo R, et al. Acetylation is required for NLRP3 self-aggregation and full activation of the inflammasome. bioRxiv. 2019. 10.1101/2019.12.31.891556. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The molecular docking was operated on the ClusPro 2.0 platform (https://cluspro.bu.edu/). The possible acetylation sites of NLRP3 were predicted from (http://pail.biocuckoo.org). All the data supporting the conclusions of this article are available from the corresponding author upon reasonable request. All data are available in the main text or the supplementary materials