Abstract
Backgrounds
Diagnostic prediction is a central application that spans various medical specialties and scenarios, sequential diagnosis prediction is the process of predicting future diagnoses based on patients' historical visits. Prior research has underexplored the impact of irregular intervals between patient visits on predictive models, despite its significance.
Method
We developed the Multi-task Fusion Visit Interval for Sequential Diagnosis Prediction (MISDP) framework to address this research gap. The MISDP framework integrated sequential diagnosis prediction with visit interval prediction within a multi-task learning paradigm. It uses positional encoding and interval encoding to handle irregular patient visit intervals. Furthermore, it incorporates historical attention residue to enhance the multi-head self-attention mechanism, focusing on extracting long-term dependencies from clinical historical visits.
Results
The MISDP model exhibited superior performance across real-world healthcare dataset, irrespective of the training data scarcity or abundance. With only 20% training data, MISDP achieved a 4. 2% improvement over KAME; when training data ranged from 60 to 80%, MISDP surpassed SETOR, the top baseline, by 0. 8% in accuracy, underscoring its robustness and efficacy in sequential diagnosis prediction task.
Conclusions
The MISDP model significantly improves the accuracy of Sequential Diagnosis Prediction. The result highlights the advantage of multi-task learning in synergistically enhancing the performance of individual sub-task. Notably, irregular visit interval factors and historical attention residue has been particularly instrumental in refining the precision of sequential diagnosis prediction, suggesting a promising avenue for advancing clinical decision-making through data-driven modeling approaches.
Keywords: Sequential diagnosis prediction, Multi-task learning, Irregular visit intervals, Historical attention residue
Background
Clinical decision support systems are critical in the healthcare sector [1, 2], with diagnostic prediction being a central application [3–5] that spans various medical specialties and scenarios [4, 6, 7], and sequential diagnosis prediction is one kind of prediction based on patients' historical visits. Research in diagnostic prediction evolves and iterates with technological advancements. Early diagnostic prediction models relied on heuristics and expert systems, such as the MYCIN system [8], which was limited by high maintenance costs due to manual rule curation. Advances in traditional machine learning approaches [9] have since automated the training of classifiers from electronic medical records, treating diagnosis prediction as a multi-class classification task. Deep learning, particularly with recurrent neural networks (RNNs) and attention mechanisms, has shown significant promise in capturing temporal dynamics within medical records [10]. Models like Med2Vec [11] and MiME [12] utilize multi-level representation learning to integrate visit sequence and medical code co-occurrence data. Others, including Dipole [13] and RETAIN [14], directly apply RNNs to model temporal relationships in patient histories. Attention-based models such as MMORE [15], MusaNet [16], and HiTANet [17] focus on capturing multi-scale temporal features. Although these models considered the temporal information of patient visits, they fail to effectively capture deeper information, such as the irregularity of visit intervals. SETOR [18] employs a neural ordinary differential equation addressing visit intervals and length of stay in an end-to-end learning manner, which significantly enhances the prediction effect, while the irregularity of visit intervals has not been fully explored. This paper mainly focused on handle this problem.
Electronic medical records often present diagnostic imbalance. To enhance medical code embedding [19] and predictive performance, some studies have incorporated external medical ontologies [20]. Choi et al. proposed the Graph-based Attention Model for Representation Learning (GRAM) [21], which integrates medical ontologies with attention mechanisms and RNNs for representation learning. Ma et al. additionally considered auxiliary information for learning embeddings from non-leaf nodes in medical ontologies [22] to extend GRAM. SETOR [18] utilized Ontological Representation and transformer. Hongyi Zhang et al. addressed the imbalance issue through the active balancing mechanism for imbalanced medical data [23], and Hsu-Hsiang Chang et al. proposed a meta-learning approach for electronic health records with a high imbalanced ratio [24]. This paper employed the widely-adopted methodology of integrating ontologies to effectively addressing the diagnostic imbalance encountered in medical data.
Multi-task learning enhances model generalization across various tasks [25]. Caruana et al. proposed the hard parameter sharing paradigm [26], in which hidden layers are shared among all tasks in neural network, and each task has its own independent output layer, reducing the risk of overfitting on individual tasks. It has been applied in medicine for joint diagnosis and prognosis [27, 28]. Haque et al. developed MULTIMIX [29], which jointly learns disease classification and lesion segmentation in a cautiously supervised manner. Harutyunyan et al. employed LSTM for multi-task joint training [30], demonstrating excellent performance on multiple patient outcome prediction tasks. Mulyar et al. proposed the MT-Clinical BERT model [31], leveraging the multi-task BERT architecture for text encoding and simultaneously learning features for multiple clinical task prediction heads, achieving higher F1 scores on the i2b2-2012 dataset. This paper utilized Multi-task learning for modeling.
To this end, this paper proposed the Multi-task Fusion Visit Interval for Sequential Diagnosis Prediction (MISDP) framework. It featured private layers for diagnostic prediction and visit interval prediction, with the former leveraging ontologies to enhance diagnosis representation and the latter using positional encoding and interval encoding to capture irregular visit intervals. Shared layers integrated these features, followed by an optimization step using historical attention residue within a multi-head self-attention mechanism. This refined the representation of historical disease diagnoses. The decoding layer then generated the final sequential diagnosis predictions through a classifier, yielding disease prediction outcomes.
In summary, our primary contributions are:
The proposal of a multi-task framework named "MISDP", which employed a visit interval prediction sub-task to enhance Sequential Diagnosis Prediction and bolster its generalization capability.
The utilization of positional encoding and interval encoding in the visit interval prediction sub-task to fully captured the intervals and sequence of visits.
The introduction of historical attention residue to optimize the transformer's multi-head self-attention module, enhancing the model's capacity to learn from long-term dependencies.
Methods
Dataset and processing
Our analysis was conducted using data extracted from the Medical Information Mart for Intensive Care III (MIMIC-III) database, a publicly accessible and comprehensive resource for critical care research. The MIMIC-III database, which encompasses detailed clinical information from patients admitted to intensive care units at a major tertiary care hospital in Boston, spans the period from June 1, 2001, to October 10, 2012 [32]. For this study, we utilized the MIMIC-III v1. 4 dataset, released on September 2, 2016.
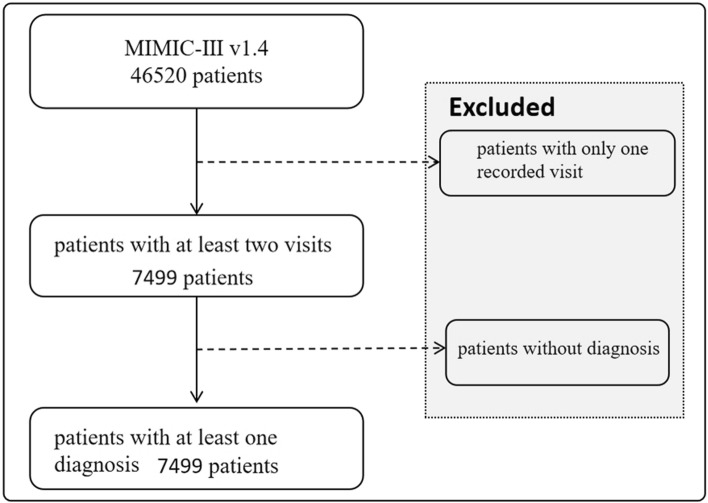
To ensure a robust analysis, we focused on patients with a minimum of two recorded visits and one diagnosis within the database, as shown in Fig. 1. This criterion resulted in a final cohort comprising 7, 499 unique patients. The average number of visits per patient was 2. 66, with an average of 13. 1 ICD-9 codes documented per visit, ranging up to a maximum of 39 ICD-9 codes per visit.
Fig. 1.
Flowchart of enrolment
Model framework
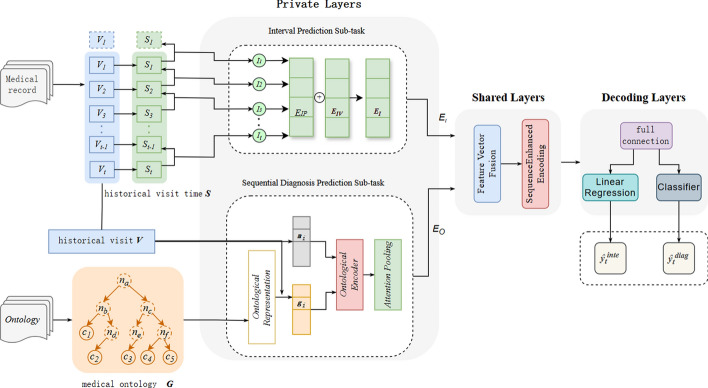
In this section, we propose the Multi-task Fusion Visit Interval for Sequential Diagnosis Prediction (MISDP) model, the model architecture, as depicted in Fig. 2, is composed of private layers for two sub-tasks, shared layers for feature integration, and decoding layers for generating the final output.
Fig. 2.
Schematic representation of the MISDP model architecture, which includes private layers for interval prediction and sequential diagnosis prediction, shared layers for feature integration, and decoding layers for final output generation
Private layers
The MISDP model's private layers are designed to handle two distinct sub-tasks: visit interval prediction and sequential diagnosis prediction.
Interval prediction sub-task
The interval prediction sub-task focused on modeling the intervals between patient visits, which could indicate the stability or acute changes in a patient's health condition. The visit intervals are calculated by extracting the time intervals between consecutive visits. The time interval between visits and is denoted as , with the first visit interval being zero by definition. The embedding representation of these intervals is represented by the vector . The order of visit intervals is encoded using the positional embedding approach from the Transformer model, as defined in Eq. (1).
| 1 |
where represents the absolute position of the visit intervals, start from one, then get the order of visit intervals .
By connecting and , we obtain the integrated expression for the visit intervals and sequence, as shown in Eq. (2),
| 2 |
Sequential diagnosis prediction sub-task
The sequential diagnosis prediction sub-task involves encoding both visit records and medical ontologies, fusing their heterogeneous features, and applying attention pooling to derive a single context-aware vector representation. This process employed the same method as the Ontological Encoder and Attention Pooling used in SETOR [18].
Shared layers
The shared layers facilitated information sharing between tasks. Initially, feature vectors from the interval prediction sub-task and the sequential diagnosis prediction sub-task were integrated through concatenation, as shown in Eq. (3).
| 3 |
where represented the diagnostic code for the patient's t-th visit, represented the encoding of the t-th intervals and sequence, denoted the patient's t-th visit, represented the visit vector for the t-th visit.
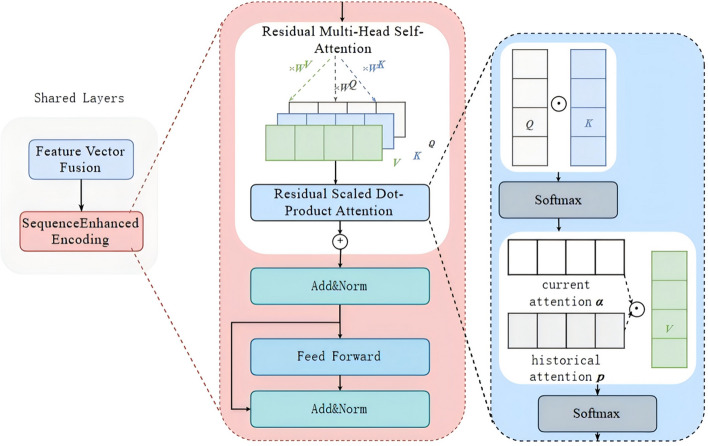
Subsequently, the model employed a multi-layer bidirectional Transformer structure for encoding, generating hidden representations for each visit within the patient's visit history, as detailed in Eq. (4). The Transformer Encoder utilized multi-head attention, feedforward neural networks, and residual connections to effectively learning the dependencies between visits.
| 4 |
Here we innovatively introduced historical attention into Transformer, implementing residual multi-head self-attention to better capture the long-term dependencies between visit sequences, as shown in Fig. 3. This module initially employed a 1 × 1 convolutional layer to process the historical attention probability matrix, linearly combining the information of each attention head through convolution. Subsequently, a learnable rate parameter was used to weight and sum the current attention probability matrix with the historical attention matrix. Finally, a softmax operation was applied to normalize the weighted sum of the attention probability matrix into a probability distribution.
Fig. 3.
The schematic diagram of the shared layer, illustrating the integration of diagnostic and interval features
Decoding layers
The sequential diagnosis prediction sub-task was formulated as a multi-class classification problem, targeting various disease diagnostic categories. In the decoding layer, the feature vector output from the shared layer was first subjected to a fully connected layer for dimensionality reduction, yielding a one-dimensional vector . The softmax function was then applied to obtain the predictive results. Assuming all parameters during the model training process are denoted by , the classifier transforms into the conditional probability distribution of the diagnostic outcome , was the ICD-9 codes set, as illustrated in Eq. (5).
| 5 |
After obtaining the probabilities for each diagnosis, the top k diagnoses with the highest probabilities were selected as the predictive results .
Loss function
The sequential diagnosis prediction task was addressed as a multi-class classification problem using Focal Loss (FL), introduced by Lin et al. at ICCV 2017 [37], as depicted in Eq. (6).
| 6 |
Here represents the estimated probability of the model for the recommended diagnosis, denotes the adjustable focusing parameter.
The visit interval prediction task, predicting continuous numerical values, was treated as a regression task, employing Mean Squared Error Loss (MSE Loss, MSE), as shown in Eq. (7).
| 7 |
To balance the demands and optimization objectives of multi-task learning more effectively, this study designs a joint utilization of Focal Loss (FL) for multi-class classification problem and Mean Squared Error (MSE) for regression task, as shown in Eq. (8).
| 8 |
where and denoted hyperparameters used to adjust the weight factors of the respective loss functions.
Effect evaluation
This study employed the Accuracy@k metric, following baseline model KAME [22], MMORE [15] and SETOR [18], to evaluate the model's performance. This metric quantified the proportion of correct diagnoses within the top k predictions for each test case, as delineated in Eq. (9).
| 9 |
Specifically, we counted the number of positive labels fall into top-k and computed the ratio over the number of all positive labels in this sample. averaged the ratios on the samples from the entire test set.
Results
Model configuration and training parameters
Ontological Representation and Encoder Settings The parameters for the ontological representation and encoder in our model are aligned with those utilized in SETOR [18].
Transformer encoder setting
Hidden Layer Dimension: The model employed a hidden layer dimension of 512.
Feedforward Neural Network Layer Dimension: A dimension of 2084 was used for the feedforward neural network layers.
Number of Encoding Layers: The Transformer encoder consisted of 6 encoding layers.
Attention Head Dimension: Each attention head operated with a dimension of 64.
Non-linear Activation Function: ReLU was employed as the non-linear activation function.
Training setting
Pre-training Learning Rate: An initial learning rate of 0. 002 was used for pre-training.
Maximum Total Input Sequence Length: The model was configured to handle a maximum sequence length of 64.
Total Training Batch Size: A batch size of 32 was used for training.
Learning Rate: The learning rate for the main training phase was set to 0. 00005.
Multi-task Ratio: A ratio of 0. 7 was applied.
Pre-training Proportion: The pre-training phase constitutes 0. 1 of the overall training.
Historical Attention to Current Attention Ratio: The model integrated a ratio of 0. 01 for historical attention relative to current attention, emphasizing the importance of past consultations.
Optimizer: Adadelta was selected as the optimizer.
Training Epochs: The model undergoed 20 training epochs.
Dataset Proportions: The dataset was divided into training, validation, and test sets in a ratio of 14:3:3.
These meticulously calibrated parameters and training configurations ware designed to optimize the model's predictive accuracy and generalization capabilities across a range of clinical prediction tasks.
Feature evaluation
Loss trend analysis To assess the model's learning dynamics, we charted the progression of training and validation loss across 20 epochs, as illustrated in Fig. 4. Each epoch encompassed a batch size of 32. The trajectories of both losses are congruent, evidencing a downward trend that plateaus, indicative of model convergence.
Fig. 4.
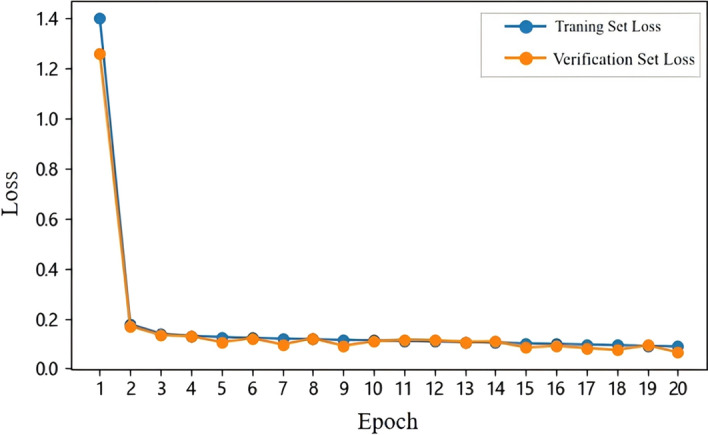
Illustration of the training and validation loss trend
Performance metric assessment Further analysis involved monitoring the accuracy and precision metric (k = 20) for both the validation and test dataset throughout the training process, as presented in Fig. 5. The accuracy and precision metrics for both sets paralleled each other closely, exhibiting a sharp ascent from baseline values and attaining a stable, elevated level by the 5th epoch. This pattern underscored the model's robust learning capacity and its ability to generalize effectively, as evidenced by the comparable performance on both dataset post-convergence.
Fig. 5.
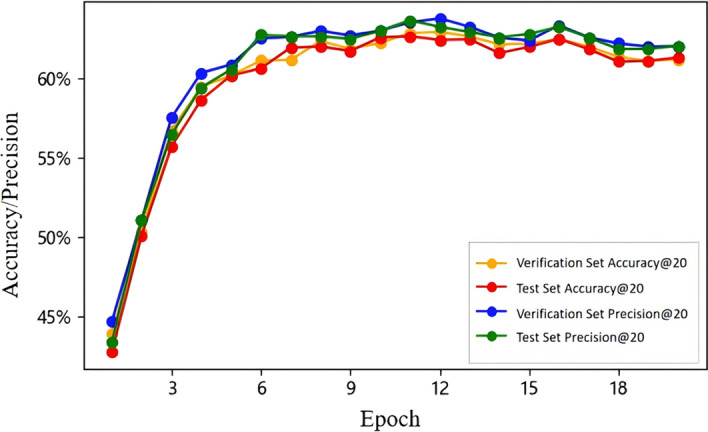
Visualization of the performance metric trends for validation and test dataset
Comparison with state-of-the-art algorithms
This section outlined the state-of-the-art approaches for sequential diagnosis prediction task in healthcare, and detailed their implementation. Our proposed model's performance was benchmarked against the following leading models, as summarized in Table 1.
Table 1.
Introduction to baseline models
| Baseline models | Introduction |
|---|---|
| KAME [22] | Utilizes medical ontologies to learn representations of medical codes and their hierarchies, which are then input into a neural network to predict sequential diagnoses |
| MMORE [15] | Generates multiple representations for each disease diagnosis via attention mechanisms, offering clinically enriched sub-classifications |
| SETOR [18] | Employs neural ordinary differential equations to manage irregular intervals between patient visits and captures dependencies through multi-layer transformer blocks, integrating medical ontologies to enhance data scarcity challenges |
We segmented the training dataset into proportions of 20%, 40%, 60%, and 80% to evaluate model performance variability. With an average of 13. 1 diagnostic codes per visit, we set k = 20 for evaluating Accuracy@20, as detailed in Table 2.
Table 2.
Accuracy@20 (%) comparison among models at different training data proportions
| Model | 20% | 40% | 60% | 80% |
|---|---|---|---|---|
| KAME | 53.9 | 55.8 | 57.9 | 60.0 |
| MMORE | 55.0 | 57.4 | 60.0 | 61.9 |
| SETOR | 57.4 | 59.2 | 61.9 | 62.4 |
| MISDP | 58.1 | 59.4 | 62.7 | 63.2 |
The bold font is utilized to emphasize the results of the MISDP method
The MISDP model notably outperformed other models, especially under data scarcity conditions. Specifically, with only 20% training data, MISDP achieved a 4. 2% improvement over KAME, highlighting its robustness. When training data ranged from 60 to 80%, MISDP surpassed SETOR, the top baseline, by 0. 8%. The results indicated that MISDP predicted better outcomes than other baselines both when the training data is insufficient and sufficient.
Through iterative experimentation, we identified an optimal training set proportion of 0. 7 for peak model performance. A comparison of Accuracy@k, parameter count, and floating-point operations (FLOPs) under these conditions was presented in Table 3.
Table 3.
Comparative results at optimal k values
| Model | Accuracy@5 | Accuracy@10 | Accuracy@20 | Accuracy@30 | Parameters(M) | FLOPs (G) |
|---|---|---|---|---|---|---|
| KAME | 27.98 | 41.81 | 57.31 | 68.02 | – | – |
| MMORE | 28.97 | 43.74 | 56.18 | 71.61 | – | – |
| SETOR | 31.18 | 45.80 | 62.36 | 72.46 | 9.39 | 9.01 |
| MISDP | 31.61 | 46.56 | 63.17 | 72.97 | 9.48 | 9.06 |
The bold font is utilized to emphasize the results of the MISDP method
The finding indicated that the MISDP model achieved superior accuracy across various k-values when compared to established baseline models. Specifically, MISDP's accuracy exceeded that of the benchmark SETOR model by up to 0. 81%, highlighting its enhanced predictive capabilities. This improvement was attained with a minimal increase in computational resources: a mere 0. 09 rise in parameter count and a 0. 05 augmentation in floating-point operations (FLOPs). In the context of medical diagnostics, where precision was paramount, even marginal gained in accuracy can significantly impact patient outcomes. The MISDP model's ability to deliver these improvements without a substantial increase in computational expense underscores its efficiency and practicality for real-world clinical applications.
Ablation study
We performed a detailed ablation study to assess the impact of two critical components in MISDP model for this study: the visit interval prediction sub-task and the attention residue. The evaluation metric employed was Accuracy@K. The results of the ablation study were presented in Table 4.
w/o Interval: remove the visit interval prediction sub-task from the private layer.
w/o Residue: remove the historical attention residue from the shared layer.
Table 4.
Ablation Performance Comparison
| Ablation Module | Accuracy@5 | Accuracy@20 |
|---|---|---|
| MISDP | 31.61 | 63.17 |
| w/o Interval | 31.18 | 62.36 |
| w/o Residue | 31.34 | 62.91 |
The bold font is utilized to emphasize the results of the MISDP method
Ablated interval The visit interval prediction sub-task, mainly analyzing the intervals and sequence of patient visits, was identified as a significant factor in the model's predictive accuracy. It played a crucial role in capturing the fluctuations in a patient's health status. Following the removal of this component, the model exhibited a decrease in accuracy by 0. 43% at k = 5 and by 0. 81% at k = 20, underscoring the importance of interval prediction in enhancing the model's performance.
Ablated residue The historical attention residue accounts for the patient's visit history modeling long-term dependencies. The MISDP model's complete architecture showed superior results compared to the variant without this mechanism, with a 0. 27% decrease in Accuracy@5 and a 0. 26% decrease in Accuracy@20. This indicated that the residue mechanism can improve the model's accuracy to certain extent.
Discussion
The present study introduced the Multi-task Fusion Visit Interval for Sequential Diagnosis Prediction (MISDP) model, designed to address the shortcomings of current predictive models in handling the irregularity of patient visit intervals, a factor that significantly impacts the accuracy of sequential diagnosis predictions. The model incorporated two novel features to capitalize on sequential information extracted from visit histories. Firstly, multi-task learning is employed to fuse visit interval prediction and sequential diagnosis prediction, capturing both the sequence and intervals of visits. Positional encoding represented the sequence of visits, while interval encoding reflected the stability or acute changes in a patient's health condition. Secondly, the integration of historical attention information into current attention calculations enables the model to maintain long-term dependencies.
In comparison with baseline models such as KAME [22], which relied on neural networks, and MMORE [15], which employed attention mechanisms, MISDP demonstrated an enhanced ability to learn sequence information by integrating intervals and sequences of visits into a transformer architecture. Furthermore, when compared to SETOR [18], MISDP leverages multi-task learning to fuse visit interval prediction and sequential diagnosis prediction. Multi-task learning enhanced model generalization across various tasks [25]. Notably, with only 20% training data, MISDP achieved a 4. 2% improvement over KAME [22] and a 0. 7% improvement over best baseline model SETOR [18].
Computational efficiency and parameter setting were important for model training and clinical application. In terms of computational efficiency, The system was powered by an Intel Xeon Platinum 8350C CPU, enhanced with hardware acceleration from an NVIDIA GeForce RTX 3090 GPU, and was equipped with 42 GB of memory and 12 GB of video memory. In the training process, multiple epochs were employed and Iterative experimentation identified optimal training parameters like the learning rate, data partition ratio of 14:3:3, ReLU activation function etc., which improved predictive performance and stability. For clinical deployment, GPU is recommended to facilitate predictive capabilities. The parameter K is optimally set to 5 in outpatient settings, where the focus is on immediate visit diagnoses and the number of diagnoses is limited, and 20 in inpatient settings, where a comprehensive view of the patient's diagnoses is required and the number of diagnoses is higher.
This study also has some limitations. Firstly, the dataset only contained records in intensive care units, it is hard to fully represent patients' medical condition change. Furthermore we will use hospital records combining outpatient and inpatient data for model validation. Secondly, the model mainly used diagnosis for prediction, laboratory and imaging results are crucial for disease prediction, while these indicators are specific to certain specialties and are more suitable for predicting specific diseases. Thirdly, the interval sequence was mainly represented by interval encoding and positional encoding, furthermore we can explore relative positional encoding, complex embedding or large language model to capture visit records better. Moreover, the study primarily focused on the accuracy of sequential diagnosis prediction, with less emphasis on the prediction of visit intervals, we will continue to explore visit interval prediction to enhance the model's understanding of patient visitation patterns.
Conclusions
This study presented the Multi-task Fusion Visit Interval for Sequential Diagnosis Prediction (MISDP) model, a novel approach that integrated visit interval factors and historical attention residue within a multi-task learning framework to enhance the accuracy of sequential diagnosis predictions. Our experiments on the MIMIC III dataset, including comparative and ablation studies, had validated the effectiveness of these innovations in improving predictive accuracy. The MISDP model exhibited superior performance even with limited training data, outperforming existing baseline models. The result highlighted the advantage of multi-task learning in synergistically enhancing the performance of individual sub-task, Notably, irregular visit interval factors and historical attention residue had been particularly instrumental in refining the precision of sequential diagnosis prediction. Future research will concentrate on optimizing visit encoding, visit interval prediction and Model validation based on other data, with the goal of refining the model's capabilities and providing a more robust tool for clinical decision-making.
Acknowledgements
Not applicable.
Author contributions
S.Z. wrote the main manuscript text, S.Z. and R.Y. conceived, designed and implemented this study, Z.P. prepared data and revised the manuscript, X.T. and H.J. revised the manuscript. All authors reviewed and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang S, Liang Y, Li J, Li X. Applications of clinical decision support systems in diabetes care: scoping review. J Med Internet Res. 2023;8(25):e51024. 10.2196/51024.PMID:38064249;PMCID:PMC10746969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Standiford TC, Farlow JL, Brenner MJ, Conte ML, Terrell JE. Clinical decision support systems in otolaryngology-head and neck surgery: a state of the art review. Otolaryngol Head Neck Surg. 2022;166(1):35–47. 10.1177/01945998211004529. [DOI] [PubMed] [Google Scholar]
- 3.Toh ZA, Berg B, Han QYC, Hey HWD, Pikkarainen M, Grotle M, He HG. Clinical decision support system used in spinal disorders: scoping review. J Med Internet Res. 2024;19(26):e53951. 10.2196/53951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harada T, Miyagami T, Kunitomo K, Shimizu T. Clinical decision support systems for diagnosis in primary care: a scoping review. Int J Environ Res Public Health. 2021;18(16):8435. 10.3390/ijerph18168435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao L, Zhang C, Zeng L, et al. Accuracy and effects of clinical decision support systems integrated with BMJ best practice-aided diagnosis: interrupted time series study[J]. JMIR Med Inf. 2020. 10.2196/16912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyachi Y, Ishii O, Torigoe K. Design, implementation, and evaluation of the computer-aided clinical decision support system based on learning-to-rank: collaboration between physicians and machine learning in the differential diagnosis process. BMC Med Inform Decis Mak. 2023;23(1):26. 10.1186/s12911-023-02123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groh M, Badri O, Daneshjou R, Koochek A, Harris C, Soenksen LR, Doraiswamy PM, Picard R. Deep learning-aided decision support for diagnosis of skin disease across skin tones. Nat Med. 2024;30(2):573–83. 10.1038/s41591-023-02728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sotos JG. MYCIN and NEOMYCIN: two approaches to generating explanations in rule-based expert systems[J]. Aviat Space Environ Med. 1990;61(10):950–4. [PubMed] [Google Scholar]
- 9.Lin D, Vasilakos AV, Tang Y, et al. Neural networks for computer-aided diagnosis in medicine: a review[J]. Neurocomputing. 2016;216:700–8. [Google Scholar]
- 10.Lipton ZC, Kale DC, Elkan CP, and Wetzel RC. Learning to diagnose with LSTM recurrent neural networks. CoRR abs/1511. 03677 (2015): n. pag.
- 11.Choi E, Bahadori MT, Searles E et al. Multi-layer representation learning for medical concepts [C]. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, 2016; pp 1495–1504.
- 12.Choi E, Xiao C, Stewart WF et al. MiME: multilevel medical embedding of electronic health records for predictive healthcare[C]. In: Proceedings of the 32nd international conference on neural information processing systems, 2018; pp 4552–4562.
- 13.Ma F, Chitta R, Zhou J et al. Dipole: diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks[C]. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, 2017; pp 1903–1911.
- 14.Choi E, Bahadori MT, Kulas JA et al. RETAIN: an interpretable predictive model for healthcare using reverse time attention mechanism[C]. In: Proceedings of the 30th international conference on neural information processing systems, 2016; pp 3512–3520.
- 15.Song L, Cheong CW, Yin K et al. Medical concept embedding with multiple ontological representations[C]. In: Proceedings of the alternate medical concept embedding with multiple ontological representations, 2019; pp 4613–4619.
- 16.Peng X, Long G, Shen T et al. Self-attention enhanced patient journey understanding in healthcare system[C]. In: Machine learning and knowledge discovery in databases: European conference, 2021; pp 719–735.
- 17.Luo J, Ye M, Xiao C et al. Hitanet: hierarchical time-aware attention networks for risk prediction on electronic health records[C]. In: Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining, 2020; pp 647–656.
- 18.Peng X, Long G, Shen T et al. Sequential diagnosis prediction with transformer and ontological representation[C]. In: 2021 IEEE international conference on data mining (ICDM). IEEE, 2021; pp 489–498.
- 19.Liu W, Zhou P, Zhao Z et al. K-bert: enabling language representation with knowledge graph[C]. In: Proceedings of the AAAI conference on artificial intelligence. 2020; 34(03): 2901–2908.
- 20.Zhang Z, Han X, Liu Z et al. ERNIE: enhanced language representation with informative entities[C]. In: Proceedings of the 57th annual meeting of the association for computational linguistics, 2019; pp 1441–1451.
- 21.Choi E, Bahadori MT, Song L et al. GRAM: graph-based attention model for healthcare representation learning[C]. In: Proceedings of the 23rd ACM SIGKDD international conferenceon knowledge discovery and data mining, 2017; pp 787–795. [DOI] [PMC free article] [PubMed]
- 22.Ma F, You Q, Xiao H et al. Kame: knowledge-based attention model for diagnosis prediction in healthcare[C]. In: Proceedings of the 27th ACM international conference on information and knowledge management, 2018; pp 743–752.
- 23.Zhang H, Zhang H, Pirbhulal S, Wu W, De Albuquerque VHC. Active balancing mechanism for imbalanced medical data in deep learning-based classification models. ACM Trans Multimed Comput Commun Appl. 2020;16:15. 10.1145/3357253. [Google Scholar]
- 24.Chang H-H, Hsu T-C, Hsieh Y–H, and Lin C. Meta-EHR: a meta-learning approach for electronic health records with a high imbalanced ratio and missing rate. In: 2023 45th annual international conference of the IEEE engineering in medicine & biology society (EMBC), Sydney, Australia, 2023; pp 1–4. Doi: 10. 1109/EMBC40787.2023.10340634 [DOI] [PubMed]
- 25.Ruder S. An overview of multi-task learning in deep neural networks[J]. Arxiv Preprint Arxiv:170605098, 2017.
- 26.Caruna R. Multitask learning: a knowledge-based source of inductive bias[C]. In: Machine learning: Proceedings of the tenth international conference, 1993; pp 41–48.
- 27.Shao W, Wang T, Sun L, Dong T, Han Z, Huang Z, Zhang J, Zhang D, Huang K. Multi-task multi-modal learning for joint diagnosis and prognosis of human cancers. Med Image Anal. 2020;65:101795. 10.1016/j.media.2020.101795. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Ge R, Wan P, Zhu Q, Zhang D, Shao W. Multi-task multi-instance learning for jointly diagnosis and prognosis of early-stage breast invasive carcinoma from whole-slide pathological images. In: Frangi A, de Bruijne M, Wassermann D, Navab N (Eds.), Information processing in medical imaging. IPMI 2023. Lecture notes in computer science, 2023; vol 13939. Springer, Cham. 10.1007/978-3-031-34048-2_12.
- 29.Haque A, Imran A-A-Z, Wang A et al. Generalized multi-task learning from substantially unlabeled multi-source medical image data[J]. Arxiv Preprint Arxiv:211013185, 2021.
- 30.Harutyunyan H, Khachatrian H, Kale DC, et al. Multitask learning and benchmarking with clinical time series data[J]. Sci Data. 2019;6(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulyar A, Uzuner O, McInnes B. MT-clinical BERT: scaling clinical information extractionwith multitask learning[J]. J Am Med Inform Assoc. 2021;28(10):2108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016. 10.1038/sdata. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.