Abstract
Background
Functional mitral regurgitation (MR) is a common form of mitral valve dysfunction that often persists even after surgical intervention, requiring reoperation in some cases. To advance our understanding of the pathogenesis of functional MR, it is crucial to characterize the cellular composition of the mitral valve leaflet and identify molecular changes in each cell subtype within the mitral valves of MR patients. Therefore, we aimed to comprehensively examine the cellular and molecular components of mitral valves in patients with MR.
Methods
We conducted a single-cell RNA sequencing (scRNA-seq) analysis of mitral valve leaflets extracted from six patients who underwent heart transplantation. The cohort comprised three individuals with moderate-to-severe functional MR (MR group) and three non-diseased controls (NC group). Bioinformatics was applied to identify cell types, delineate cell functions, and explore cellular developmental trajectories and interactions. Key findings from the scRNA-seq analysis were validated using pathological staining to visualize key markers in the mitral valve leaflets. Additionally, in vitro experiments with human primary valvular endothelial cells were conducted to further support our results.
Results
Our study revealed that valve interstitial cells are critical for adaptive valve remodelling, as they secrete extracellular matrix proteins and promote fibrosis. We discovered an abnormal decrease in a subpopulation of FABP4 (fatty acid binding protein 4)-positive proliferating valvular endothelial cells. The trajectory analysis identifies this subcluster as the origin of VECs. Immunohistochemistry on the expanded cohort showed a reduction of FABP4-positive VECs in patients with functional MR. Intervention experiments with primary cells indicated that FABP4 promotes proliferation and migration in mitral valve VECs and enhances TGFβ-induced differentiation.
Conclusions
Our study presented a comprehensive assessment of the mitral valve cellular landscape of patients with MR and sheds light on the molecular changes occurring in human mitral valves during functional MR. We found a notable reduction in the proliferating endothelial cell subpopulation of valve leaflets, and FABP4 was identified as one of their markers. Therefore, FABP4 positive VECs served as proliferating endothelial cells relates to functional mitral regurgitation. These VECs exhibited high proliferative and differentiative properties. Their reduction was associated with the occurrence of functional MR.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-024-03791-4.
Keywords: ScRNA-seq, Functional mitral regurgitation, Valvular endothelial cell, FABP4
Highlights
This comprehensive single-cell atlas of the mitral valve from patients with functional mitral regurgitation provided insights into the involvement of specific endothelial cell subtypes in the pathogenesis of regurgitation.
Our results revealed a decrease in proliferating subpopulation of valvular endothelial cells in functional mitral regurgitation.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-024-03791-4.
Background
Mitral regurgitation (MR), a commonly occurring valvular disorder, is estimated to affect around 3% of the general population, impacting more than 176 million individuals on a global scale [1]. Functional MR resulting from left ventricular dysfunction, left atrial dilation, and pulmonary hypertension accounts for 65% of MR cases [2]. In structurally normal mitral valves, functional MR can occur when mitral leaflet coaptation fails due to dysfunction in the left atrium (LA) or left ventricle (LV). Even mild functional MR can result in adverse cardiovascular outcomes [3]. A randomized controlled trial showed that despite the temporary elimination of regurgitation by mitral valvuloplasty, the rates of heart failure-related adverse events and cardiovascular rehospitalization remained high during follow-up. Up to 58.8% of patients experienced recurrent moderate-to-severe mitral valve regurgitation, suggesting that valve function may continue to deteriorate even after regurgitation is corrected [4]. A small retrospective study has shown that the midterm results for isolated surgical edge-to-edge mitral repair are not inferior to those of conventional repair techniques with annuloplasty [5]. This suggests that the mitral valve leaflets undergo alterations and that regurgitation cannot be compensated by solely repairing the annulus or subvalvular structures. Chaput et al. found that the leaflet area increases with the dilatation of the left ventricle. But in patients with functional MR, such adaptation is insufficient to meet the needs for increased leaflet area imposed by the tethered leaflet configuration [6]. To accommodate hemodynamic changes caused by LV or LA dysfunction, mitral valve leaflets undergo a process called mitral remodelling [7]. Inadequate restructuring of the leaflets plays an essential role in the development of MR [8]. However, there is limited knowledge about the changes in valvular cells during this process. During this process, the loss or increase of certain components in valve cells may result in inadequate leaflet adaptation.
Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool for the reliable identification of closely related cell subtypes and the assessment of gene expression heterogeneity in individual cell subtypes in physiological and pathological conditions [9]. To evaluate cell type variations during functional MR, we performed scRNA-seq of mitral valve leaflets from patients with moderate-to-severe functional MR and nondiseased mitral valve tissues collected from patients who underwent heart transplantation [10].
Our first objective was to determine whether unbiased single-cell clustering could identify valve cell types, discover new cell phenotypes using established markers, and improve existing classifications. Our second objective was to investigate the potential of single-cell analysis for elucidating the underlying mechanisms of leaflet adaptation. This study provides a foundation for investigating the mechanisms of valve remodelling and the potential therapeutic benefits of leaflet adaptation and inhibition of fibrosis for functional MR, a topic currently under ongoing debate in the field [11]. Despite the lack of research evidence, individuals exhibiting inadequate leaflet adaptation may gain increased benefits from leaflet enlargement. Previous clinical research has demonstrated that patients with a less adequate augmented mitral valve are likely to benefit more from mitral valve replacement surgery than patients with an adequate augmented mitral valve. This phenomenon is known as proportionate functional mitral regurgitation [12]. Understanding the changes in valvular cells can help us better explain how the mitral valve adapts to hemodynamic changes. Hopefully, this comprehensive single-cell map of the mitral valve will not only contribute to the treatment of functional MR but also potentially identify new therapeutic targets.
Methods
Collection of human mitral valve specimens and histology
The cohort comprised three individuals with moderate-to-severe functional MR (MR group) and three non-diseased controls (NC group). Regurgitant human mitral valve specimens were obtained from explanted hearts, while nondiseased control (NC) specimens were collected from transplanted hearts with normal mitral structure and function [10]. The selection of these samples was based on the patients’ medical history and ultrasound results [13]. All 6 specimens were donated by patients who underwent cardiac surgery at Fuwai Hospital, Beijing (Human ethics approval number: FW-2022–1658). Detailed information on the patients was provided in Tables S1 and S2 (Additional file 1: Table S1, S2). The functional MR diagnosis was validated through a comprehensive evaluation of echocardiographic, clinical, and pathological findings. The pathology specimens were evaluated by two pathologists at Fuwai Hospital. (Additional file 1:FigureS1). The use of human mitral valve specimens was approved by the Human Ethics Committee of Fuwai Hospital, Chinese Academy of Medical Sciences. Mitral valve specimens were collected in Dulbecco’s modified Eagle’s medium (Gibco, 11,885,084) and kept on ice immediately after separation. Specimens were removed from each sample and fixed in 4% paraformaldehyde overnight. Then, the specimens were embedded in paraffin and cut into sections. The remainder of each sample was made into a single-cell suspension.
Sequencing data processing
Raw gene expression matrices were generated for each sample by the Cell Ranger (version 3.1.0). Pipeline coupled with human reference version GRCh38-2020-A. The output filtered gene expression matrices were analysed by R software (version 4.1.2) with the Seurat package (version 4.1.1). A custom R script was used to combine the expression data and metadata from all libraries corresponding to a single batch. The expression data matrix was loaded into a Seurat object along with the library metadata for downstream processing. The percentage of mitochondrial transcripts for each cell (percent.mt) was calculated and added as metadata to the Seurat object. Cells were further filtered before dimensionality reduction (nFeature_RNA-min. 200; nFeature_RNA-max. 4000; percent.mt-max. 10%). Low-quality libraries identified were removed from the dataset. Split the dataset into a list of six seurat objects (NC1, NC2, NC3, MR1, MR2, MR3), normalize (expression values were scaled to 10,000 transcripts per cells and Log-transformed), and identify variable features for each dataset independently. We calculated variable features by the FindVariableFeatures function with selection.method = “vst” and nfeatures = 2000.
Statistical analyses
Statistical analysis was conducted using R (version 4.1.2). The Mann–Whitney U test was performed with Seurat (version 4.1.1). FindAllMarkers was used to identify differentially expressed genes (DEGs) between the cell clusters. The cell ratios of each group were compared using logit transformation, followed by Student’s t test (with or without Welch’s correction) or the Mann–Whitney U test according to the results of the normality test and variance homogeneity test. For differential expression and cellular transcriptomic signatures, p values were adjusted for multiple hypothesis testing using the Benjamini‒Hochberg method. A p value < 0.05 was considered to indicate statistical significance.
Multiple dataset integration
First, we selected features that were consistently variable across datasets for integration. Then we identified anchors using the FindIntegrationAnchors function, which takes a list of Seurat objects as input. We used these anchors to integrate the datasets together with IntegrateData. The integrated data were then used for subsequent analysis.
Dimensionality reduction
Effects of the variable (percent.mito) were estimated and regressed out using a GLM (ScaleData function, model.use = 'linear'), and the scaled and centred residuals were used for dimensionality reduction and clustering. To reduce the dimensionality of the datasets, the RunPCA function was conducted with default parameters on linear-transformation scaled data generated by the ScaleData function. Next, the ElbowPlot and DimHeatmap functions were used to identify proper dimensions of each dataset.
Cell clustering and cluster identification
After non-linear dimensional reduction and projection of all cells into two dimensional space by UMAP, Clustering was run using the FindClusters function using the original Louvain algorithm and 10 interactions. Clustering was performed at varying resolution values, and we chose a value of 0.5 for the resolution parameter for the initial stage of clustering. Clusters were preliminarily assigned identities based on the expression of combinations of known marker genes for major cell classes and types.
Differential proportion analysis of cell populations or subpopulations was performed using central logit transformation of cell ratios, and then t-tests between each group.
Differential expression genes (DEGs) identification and functional enrichment
First, we removed lowly expressed genes that had fewer than 10 cells with any counts. Then, we aggregated counts to the sample level by summing counts of each sample within each cell type. Pseudobulk differential expression analysis was performed by DESeq2 (v1.34.0). A DESeq2 object was created by the DESeqDataSetFromMatrix function to prepare for running the DE analysis. DE analysis was performed by running the DESeq function with default parameters on the DESeq2 object created earlier. Enrichment analysis for the functions of the DEGs was conducted using the clusterProfiler (version 4.2.2) R package [14].
Analysis of TF regulatory network
TF regulatory network analysis was performed using pySCENIC [15] (version 0.11.2). First, coexpression modules were inferred using a regression per-target approach (pyscenic grn function). Next, the modules with indirect targets were filtered using cis-regulatory motif discovery (pyscenic ctx function). Lastly, the activity of each regulon in each cell was scored using the AUCell algorithm (pyscenic aucell function). The AUC scores for each cell were attached to the Seurat object. We identified regulons that are specific to clusters using the regulon specificity score (RSS), and then selected the top 5 and top 20 regulons of each cluster. The top 20 regulons were visualized using the Seurat VlnPlot function, and the top 5 regulons were visualized using the pheatmap R package (version 1.0.12).
Assign cell-cycle scores
We assigned each cell a score based on its expression of G2/M and S phase markers using the Seurat CellCycleScoring function. Then we visualized the co-expression of S.Score and G2M.Score simultaneously using the Seurat FeaturePlot function (blend = TRUE). The phase scores for each cell were calculated based on gene expression data using the cyclone function of the scran (version 1.22.1) R package [16]. We visualized the phase scores using pheatmap (version 1.0.12) R package.
Gene set enrichment analysis (GSEA)
DEGs were analysed using the Seurat FindAllMarkers function (test.use = 't', min.cells.group = 0, min.pct = 0, logfc.threshold = -Inf). Only significant genes (P < 0.05) and genes with an avg_logFC higher than log(1.5) or lower than − log(1.5) were used to perform GSEA. We performed GO gene set enrichment analysis with the gseGO function provided by the clusterProfiler (version 4.2.2) package, Reactome pathway gene set enrichment analysis using the gsePathway function of the ReactomePA [17] (version 1.38.0) package, and hallmark gene set enrichment analysis using the GSEA function of the clusterProfiler (version 4.2.2) package.
Cell–cell communication analysis
We used CellChat [18] (v1.4.0) to infer cell–cell communication based on a prior ligand-receptor interaction database. We loaded the normalized counts data of each group (NC/MR) separately that were extracted from the Seurat object into CellChat and followed the workflow recommended in CellChat to infer and visualize the cell–cell communication network. Then, we merged the two CellChat objects together, compared the total number of interactions and interaction strength using the compareInteractions function, and compared the number of interactions and interaction strength among different cell clusters with the netVisual diffInteraction function and the netVisual_ heatmap function. Finally, we compared the overall information flow of each signalling pathway using the rankNet function, and used the netVisual_bubble function to compare the communication probabilities mediated by ligand-receptor pairs from some cell clusters to others.
RNA velocity analysis
We used velocyto [19] (v0.17.17) to calculate RNA velocity based on the ratio of spliced and unspliced reads. Loom files were generated using the ‘velocyto run’ function and were used for downstream RNA velocity analysis using the scVelo [20] (v0.2.5) package. We followed the scVelo analysis pipeline on the website (https://scvelo.readthedocs.io/VelocityBasics/) to estimate RNA velocity and visualize the results.
Trajectory inference
Trajectory inference was performed using the slingshot [21] (version 1.8.0) R package with default settings for the slingshot function, and using the UMAP or PCA embeddings from the subclustering for each cell type. Results were visualized with the ggplot2 R package (version 3.3.6). We used the expression count matrix and the trajectory information as input for the fitGAM function (nknots = 7) of the tradeSeq [22] (version 1.8.0) package to fit the NB-GAM for each gene to smooth each gene’s expression in each lineage. Then, we visualized the expression pattern of a gene over pseudotime using the plotSmoothers function.
Isolation and treatments of valvular endothelial cells
Isolation of valvular endothelial cells (VECs) was performed using a modified method described by Songia et al. [23] and Gould et al. [24]. Samples for primary cell isolation were obtained from patients undergoing cardiac transplantation at the cardiac surgery department, Fuwai Hospital (Beijing, China). After obtaining the mitral valve samples, we proceeded to dissect the chordae tendineae and papillary muscles. The posterior leaflet of the mitral valve was selected for experimental purposes. Mitral leaflets were placed in 2 mg/mL type II collagenase (LS004176, Worthington Biochemical Corp.) in Advanced Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% foetal bovine serum (FBS, Gibco), 1% Penicillin, and 1% Streptomycin solution (Gibco), and incubated for 20 min at 37 °C. Sterile cotton swabs were used to remove the endothelial layer. Following this, the cell suspension was centrifuged and resuspended, after which it was seeded into T25 culture flasks. After reaching confluence, the cells were washed and isolated using Dynabeads conjugated with CD31 (130-091-935, Miltenyi). All experiments were performed on cultured cells between their third and fourth passages. The recombinant proteins used for cell treatment were FABP4 (RD172036100, Biovendor) and TGFβ1 (P01137, MedChemExpress).
For additional methodological details regarding molecular biology experiments and pathological analyses, please refer to Supplementary Material 1.
Results
Human functional MR mitral valve cell atlas
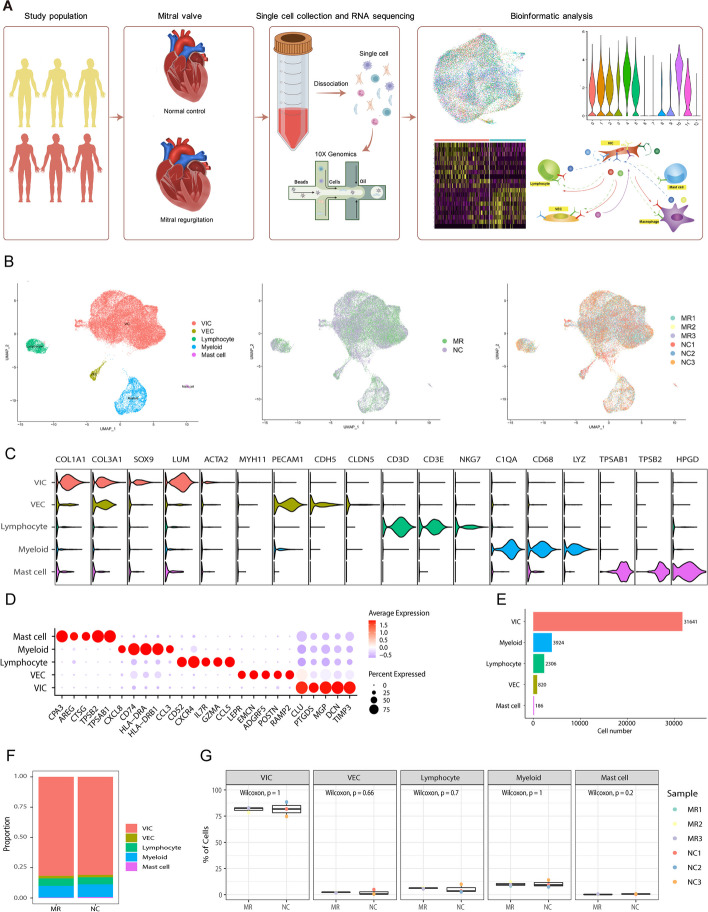
To clarify the cell composition within the functional MR mitral valve leaflets, we obtained high-quality data from three nondiseased control (NC) mitral valves and three functional MR samples obtained from six individuals using the 10X Genomics platform (Additional file 1: Table S1, Fig. 1A). The data included 43,381 cells and 2011 genes were detected per cell (Additional file 1: Figure S2, Table S2). Then, we performed graph-based clustering of the dataset and annotated the clusters using established marker genes (Fig. 1B, C) [25–27]. Cluster 3, identified as of low quality, was excluded from further analysis. (Additional file 1: Figure S2C, D). Subsequent analysis revealed several distinct clusters, which could be assigned to valvular interstitial cells (VICs), valvular endothelial cells (VECs), lymphocytes (Lym), myeloid cells (Mye), and mast cells (Fig. 1C). Clusters 0, 1, 2, 3, 4, 5, 10, and 11 contained VICs, which highly expressed COL1A1, COL3A3, SOX9, and LUM (Fig. 1C, D). Cluster 8 contained VECs, which highly express PECAM1 and CDH5. Cluster 7 was composed of lymphocytes (CD3D, CD3E, and NKG7). Myeloid cells, characterized by high expression of C1QA, CD68, and LYZ, were found in Clusters 6 and 9. Cluster 12 was composed of mast cells, which highly express TPSAB1, TPSB2, and HOGD (Additional file 1: Figure S3A, B). The marker genes for each cluster are available in the supplementary table (Additional file 2, Major cluster DEGs). VICs accounted for the absolute majority of cells in both the diseased and normal specimens (Fig. 1E, F). However, when comparing the proportion of each type between normal mitral valves and their MR counterparts, the Wilcoxon rank sum test showed that there were no significant differences in the proportions of each cell type between the two groups (Fig. 1G). Therefore, we investigated the differences in cell subpopulations between the NC and MR groups.
Fig. 1.
Clustering and identification of cell types in nondiseased and functional regurgitated human mitral valves with scRNA-seq data. A Six mitral valves from three normal controls and three patients with MR were harvested, and scRNA-seq was separately performed. B The UMAP plot of the combined six samples shows clusters, sample types, individual patients, and cell types. C The gene markers and each cell type identified are shown in the violin plot. D Dot plot of the top 5 marker genes in VIC clusters. The dot size corresponds to the proportion of cells within the group expressing each gene, and the dot color corresponds to the expression level. E The number of cells measured in six specimens. F The composition of each cell type in the NC and MR groups. G Box plots comparing the proportions of each cell type between the two groups. MR, mitral regurgitation; scRNA-seq, single-cell RNA sequencing; UMAP, Uniform Manifold Approximation and Projection; VIC, valvular interstitial cell; VEC, valvular endothelial cell
As there was one female patient and two male patients in the MR group, we conducted a sex-related correlation analysis on the 3 patients in the MR group. The results showed that sex differences did not significantly impact gene expression within the MR group (Additional file 1: Figure S4). Considering the age differences, we also performed age-related correlation tests on the children (< 18 years) and adult (> 18 years) subgroups within the MR group. Similarly, age did not appear to have a pronounced effect on gene expression (Additional file 1: Figure S5).
Various VIC subtypes play different roles in valve adaptation to haemodynamic changes
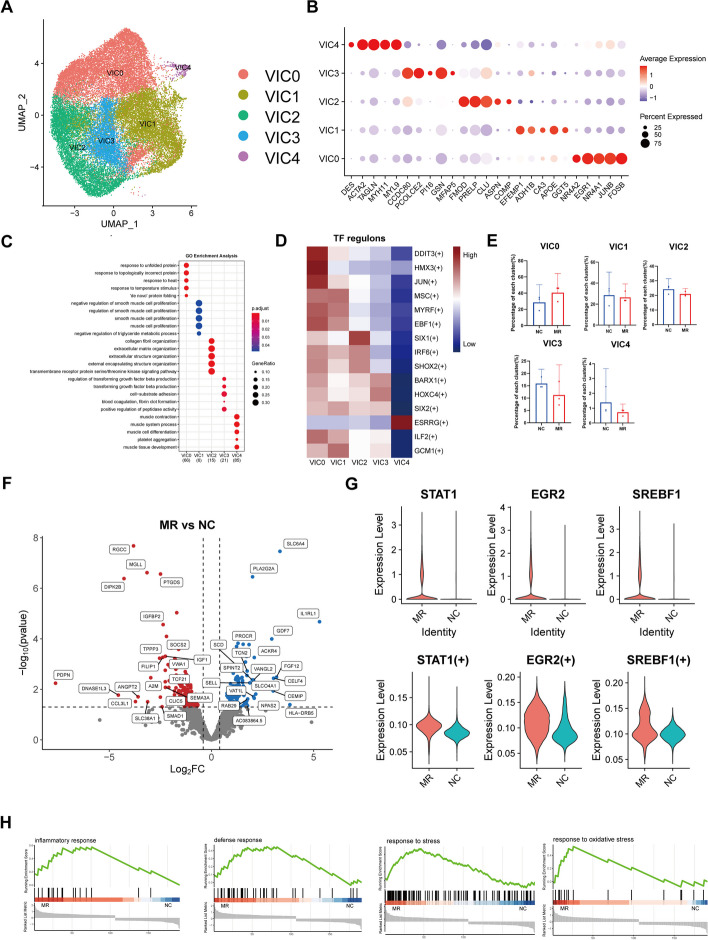
VICs comprised the vast majority of cells in the mitral valve leaflets of the NC and MR groups, with percentages of 81.0% and 81.8%, respectively. A total of 31,170 VIC cells from all 6 samples were combined, and then integrative unsupervised clustering was performed, resulting in the identification of five distinct clusters (Fig. 2A). The marker genes were identified in these VIC clusters (Fig. 2B, Additional file 1: Figure S6A).
Fig. 2.
Dynamic VIC cluster in human mitral valve leaflet. A A UMAP plot of all VIC cells coloured according to cluster. B Dot plot of the top 5 marker genes in VIC clusters. The dot size corresponds to the proportion of cells within the group expressing each gene, and the dot color corresponds to the expression level. C The enriched GO terms for each VIC cluster. D Heatmap of TF regulons in each VIC cluster. E The proportion of each VIC cluster in six samples. F Volcano plot showing the DEGs between the NC group and MR group in VIC. G The expression level and regulon activity of the TFs STAT1, EGR2, and SREBF1 in each VIC cluster. H Enrichment plots for representative pathways dysregulated in VIC in the MR. The vertical lines in the enrichment plot indicate the members of the gene set appear in the ranked list of genes. VIC, valvular interstitial cell; UMAP, uniform manifold approximation and projection; GO, Gene Ontology; MR, mitral regurgitation; TF, transcription factor; DEGs, differentially expressed genes
As the largest VIC subcluster, VIC0 was identified as stress response-related VICs expressing NR4A2 [28], JUNB [29], and FOSB [30]. The enrichment analysis of DEGs revealed that VIC0 was related to different external stimuli (Fig. 2C,Additional file 1: Figure S6B). VIC1 expressed high levels of IGFBP3 and GUCY1A1, which are involved in nitric oxide (NO) signalling [31, 32]. Thus, VIC1 can be identified as NO-related VICs. VIC1 also expressed high levels of the antioxidative stress gene CA3, along with NO, which has antifibrotic and anti-inflammatory effects within the mitral valve, indicating the protective role of VIC1 [33, 34]. VIC2 highly expressed PRELP and FMOD, which are associated with proteoglycan generation [35, 36]. Other ECM-related genes, such as COL6A2,COL1A1, and COL1A2, were also the top markers of VIC2 (Additional file 1: Figure S6A), indicating a structural VIC phenotype. VIC3 demonstrated high levels of GSN and PCOLCE2, which are implicated in fibrosis processes [37, 38]. Continual findings from the DEG enrichment analysis consistently highlighted the role of VIC3 in TGFβ signal transduction, a pathway associated with mitral valve fibrosis [39]. Thus, VIC3 may play a profibrotic role in the mitral valve. VIC4 was characterized as an active VIC phenotype (aVIC, ACTA2high VIC) with relatively high levels of ACTA2 and MYH11. These cells could be myofibroblasts, as previously reported. Their activation indicates increased fibrosis activity [40]. The supplementary table contains the marker genes related to each subcluster (Additional file 3, VIC subcluster DEGs).
Each cell cluster exhibited different functions in the heart valve. Transcription factors (TFs) play important roles in the control of genes that cause crucial changes in valvular disease [41]. Consequently, we utilized single-cell regulatory network inference and clustering (SCENIC) analysis to evaluate the TFs responsible for variations in gene expression among distinct cell clusters [42] (Fig. 2D). For example, EGR3, a differentiation regulator that mediates fibrotic gene expression [43], is a marker TF of VIC0. KLF7 is involved in the expression of proinflammatory factors and matrix metalloproteinases and is a marker TF of VIC3, demonstrating that VIC3 promotes valvular disease [44] (Additional file 1: Figure S6C). We analysed the high expression of TFs and downstream genes in various subgroups of VIC. The network diagram shows two high expression TFs of VIC1, MYRF, and EBF1, jointly regulate the expression of NR2F2 (Additional file 1: Figure S6D). This gene is thought to mitigate fibrosis by inhibiting endothelial cell senescence [45]. These align with the association between VIC1 and nitric oxide (NO). Information on the highly expressed TFs in each VIC subpopulation and their downstream regulated genes is displayed in a supplementary table (Additional file 4,VIC_cluster_regulon) and network diagrams (Additional file 5,VIC_TFandTarget_network). We also analysed the highly expressed genes in each VIC subpopulation (Additional file 6,VIC_top5DEGs_regulon).
Contrasting the cluster compositions between the NC and MR groups highlighted a distinct increase in the relative representation of VIC0 (Fig. 2E), indicating a cellular stress response when regurgitation occurs. A large number of DEGs between the NC and MR groups were identified via pseudobulk DEG analysis (Fig. 2F, G). In the MR group, we identified a series of fibrosis-related genes, such as CCL26 [46], CXCL9 [47], S100A8, and S100A9 [48]. We conducted an analysis of differential gene counts among various subpopulations of VIC (Additional file 1: Figure S6E). We observed that VIC2 exhibited the greatest number of DEGs associated with ECM components, such as PLA2G2A [49], ENPP2 [50], and SEMA5A [51] (Additional file 1: Figure S6F). The DEGs between the NC and MR groups are available in a supplementary table (Additional file 7, VIC DESeq2 DEG). We next sought to identify VIC-specific dysregulated pathways during the development of MR via gene set enrichment analysis (GSEA) (Fig. 2H). Consistent with the activation of VICs in MR, the inflammatory response pathway, as well as the stress and oxidative stress pathways, was significantly enriched in the VICs. On the other hand, the activation of the defence response pathway also indicated the protective function of VIC during regurgitation. Reactome pathway analysis found that the expression of defence reaction-related pathways in VIC was increased (Additional file 1: Figure S6G). HE staining revealed thickening and fibrosis of the mitral valve in the MR group compared to the NC group (Additional file 1: Figure S1), which aligns with the results of previous studies [52]. Together, these findings demonstrate that VIC plays an important role in valve adaptation in response to haemodynamic changes.
Dynamic VEC cluster in human mitral valve leaflet
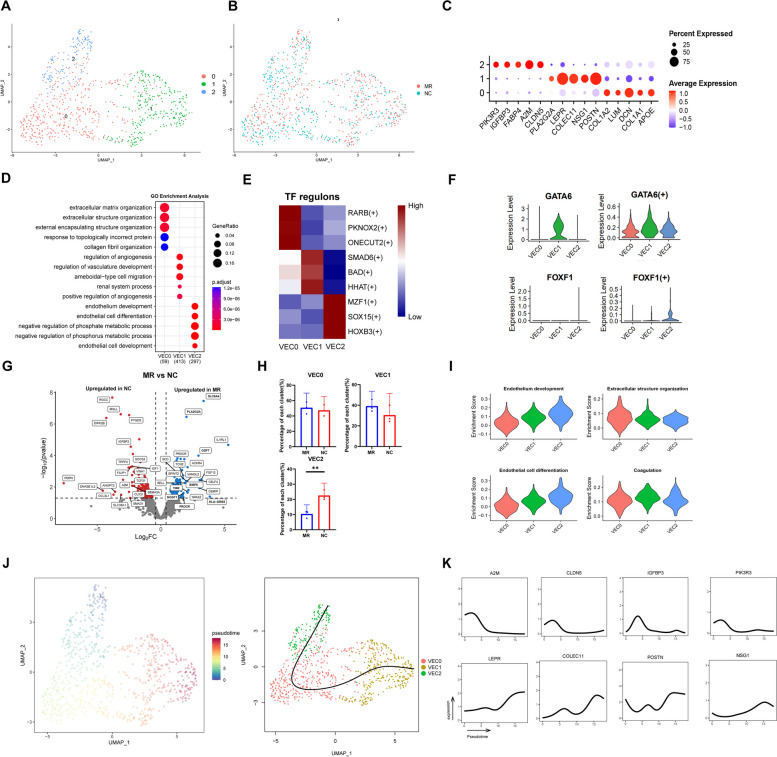
A total of 820 VECs from the NC and MR groups were grouped into three VEC subclusters (Fig. 3A, B, Additional file 1:Figure S7A). VEC0 made up more than half of the VEC subpopulation in both groups. Despite the expression of the same classical VEC markers, there were distinct gene expression profiles among the three subclusters, indicating their different roles in MR development (Fig. 3C). Based on the GO enrichment analysis, we defined three VEC subclusters (Fig. 3C and D). VEC0 was identified as structural VECs because they express DCN, LUM, and APOE, which act as crucial regulators of valve integrity and participate in collagen fibrosis [36, 53]. VEC1 was identified as protective VECs. High levels of the protective genes POSTN (which protects VIC from apoptosis [54]) and COLEC11 (which has anti-autoimmune effects [55]) were detected in VEC1 cells. KEGG enrichment analysis revealed that VEC1 was also enriched in antigen processing and presentation (Additional file 1: Figure S7B). VEC2 was identified as an IGFBP3Chigh VEC (expressing A2M, IGFBP3C, FABP4 and CLDN5). IGFBP3C (insulin-like growth factor binding protein 3) is closely related to the proliferation of adult cardiac progenitor cells [56]. FABP4 has a promotive effect on the proliferation of endothelial cells [57]. GO enrichment analysis (Fig. 3D, I) revealed that VEC2 can be identified as endothelium development-related VECs. Therefore, VEC2 is interpreted as proliferating endothelial cells. The supplementary table contains the marker genes related to each subcluster (Additional file 8, VEC subcluster DEGs).
Fig. 3.
Dynamic VECclusters in human mitral valve leaflet. A A UMAP plot of all VECs coloured according to cluster. B A UMAP plot of all VECs according to phase. C Dot plot of the top 5 marker genes in VEC clusters. The dot size corresponds to the proportion of cells within the group expressing each gene, and the dot colour corresponds to the expression level. D The enriched GO terms of each VEC cluster. E SCENIC analysis of the expression of different TF regulons between each cluster. The data are coloured according to their expression levels. F The expression level and regulon activity of the TFs GATA6 and FOXF in each VEC cluster. G Differential gene expression of VECs in the NC group and MR group is shown in a volcano plot. H Comparison of the percentage of each cell type among the three clusters of VECs. I The enrichment scores of four different biological processes in each VEC subcluster. J Pseudotime single-cell trajectory reconstructed by slingshot for VEC. K Plot of marker and functional genes along the VEC2 trajectories. VEC, valvular endothelial cell; UMAP, uniform manifold approximation and projection; GO, Gene Ontology; MR, mitral regurgitation; TF, transcription factor; DEGs, differentially expressed genes
As revealed by SCENIC analysis, each subcluster had specific TFs. Sox15, an important marker of mesoderm progenitor cells [58], was highly expressed in VEC2. VEC1 was identified as a Smad6+ cluster, and the loss of Smad6 can be linked to disrupted endothelial cell junctions [59] (Fig. 3E). GATA6, known to be involved in the protection of endothelial cells from undergoing apoptosis [60], showed greater expression and activity in Cluster 1 than in the other clusters (Fig. 3F). This finding also proves the protective effect of VEC1 on other factors. Additionally, the activity and expression of FOXF1 were unique to VEC2. This finding was linked to the crucial role of VEC2 in the strong proliferating abilities of endothelial cells [61], suggesting that VEC2 may closely associate with the proliferation of endothelial cells. (Fig. 3F). We also analysed the highly expressed TFs and their downstream genes. HOXB3 is highly expressed in VEC2 and can regulate the expression of downstream KDM4A, which is closely related to the differentiation of embryonic stem cells into endothelial cells (Additional file 1: Figure S7C) [62]. Information on the highly expressed TFs in each VEC subpopulation and their downstream regulated genes is displayed in a supplementary table (Additional file 9,VEC_cluster_regulon) and network diagrams (Additional file 10,VEC_TFandTarget_network). We also analysed the highly expressed genes in each VEC subpopulation. We found that FABP4, a marker of VEC2, is associated with the upstream TF SMAD, which is closely involved in EndMT (Additional file 11,VEC_top5DEGs_regulon) [63]. This may indicate a potential relationship between FABP4 and EndMT.
By comparing the expression differences between the NC group and MR group, genes related to inflammation (HLA-DRB5) and fibrosis (PLA2G2A) were found to be increased in the MR group. SLC6A4 is a 5-HT (5-hydroxytryptamine) transporter protein that was also upregulated in the MR group. 5-HT plays an important role in the remodelling of mitral valves following ischaemic mitral regurgitation [64]. Moreover, differential expression of genes closely associated with EndMT was observed in the MR group (Fig. 3G), including TGM2 [65], PROCR [66], and MGST1 [67]. The TGF-β signalling pathway is a key regulator of EndMT [68]. Two genes in this pathway, BMP6 and GDF7, were also differentially expressed in the MR group. The decrease in the proportion of proliferating endothelial cells (VEC2) indicated that the EndMT process was related to the occurrence of MR. During this process, specific endothelial cell subclusters detach from cell junctions, leading to delamination of the valve surface. These cells exhibit invasiveness and migratory capacity, allowing them to traverse the basement membrane into the valve interstitial tissue. Subsequently, the thickness and density of the mitral valve are increased to accommodate regurgitation [69]. The analysis of DEG counts revealed that VEC2 had the greatest number of DEGs (Additional file 1: Figure S7D). This indicates that changes in VEC2 are most strongly associated with MR. The DEGs between NC and MR are available in a supplementary table (Additional file 12, VEC DESeq2 DEG).
For a more thorough examination of the pseudotime trajectories of the VEC subclusters, we utilized Slingshot to conduct trajectory analysis [21]. Considering the high level of IGFBP3C expression [56], Cluster 2 was identified as the origin of VECs. After mapping the subcluster to the pseudotime single-cell trajectory, there was a tendency for transformation from VEC2 to VEC0 and finally to VEC1 (Fig. 3J,Additional file 1: S7E). Along the VEC lineage, protective genes (COLEC11 [55], POSTN [70]) and immune cell infiltration-related genes (NSG1 [71], LEPR [72]) were upregulated, while genes associated with fibrosis (A2M, IGFBP3 [73], CLDN5 and PIK3R3 [74]) were downregulated (Fig. 3K). This finding was consistent with the protective function of VEC1.
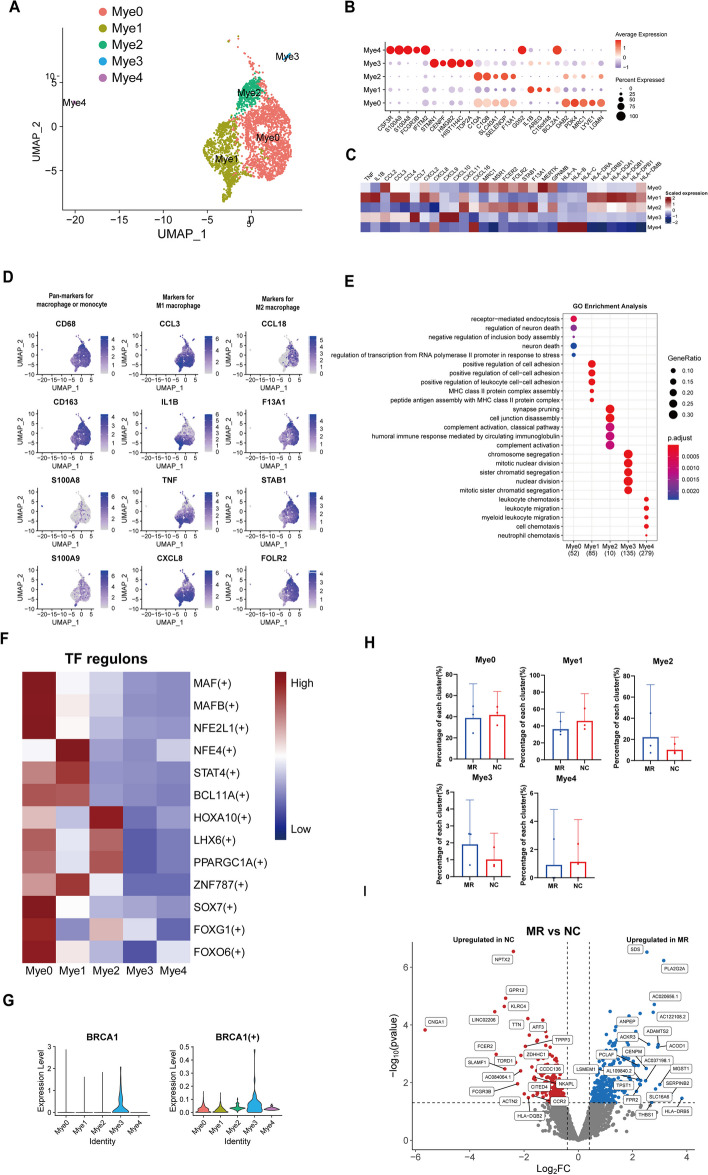
Dynamic myeloid clusters in the mitral valve
A total of 2922 myeloid cells from the NC and MR groups were divided into five subgroups (Fig. 4A, Additional file 1:Figure S8A). Myeloid0 highly expressed M2 macrophage biomarkers such as DAB2 [75], MAF [76], MERTK [77], MRC1 [78], and STAB1 [79] (Fig. 4B, C). Additionally, Myeloid0 highly expresses LGMN, a gene specifically expressed by cardiac resident macrophages that contributes to acute inflammation resolution and organ function [80]. AREG, CCL3, TNF (tumour necrosis factor), and IL1B, which are biomarkers of M1 macrophages, were highly expressed in Myeloid1 [81]. The proteins transcribed by the DEGs in Myeloid1 included inflammatory cytokines (IL-1B, TNF, C15orf48 [82], and BCL2A1 [83]), chemokines (CCL3, CCL4, CXCL2, and CXCL8), and HLA (Fig. 4B, C). Thus, Myeloid1 and Myeloid0 can be identified as M1-like and M2-like macrophages, respectively. Panmarkers for monocytes (S100A8 and S100A9) were highly expressed in Myeloid4, indicating it is a monocyte-origin cluster (Fig. 4D). Myeloid3 showed high expression of STMN1 (cellular microtubule dynamics) [84], TOP2A (DNA replication) [85], HIST1H4C (nucleosome assembly and DNA packaging) [86], and HMGB2 (DNA repair and gene transcription) [87]. In addition, the GO terms about cell mitosis were enriched in this cluster (Fig. 4E,Additional file 1: S8B). To validate this result, we derived phase S-scores and phase G2/M-scores for all Mφs based on the expression of corresponding marker genes using Seurat CellCycleScoring. Our observation revealed that most Myeloid3 cells exhibited either high S-scores or high G2/M-scores (Additional file 1: Figure S8C). Taken together, these results confirm that Myeloid3 plays a role in macrophage proliferation. MRC1 and FOLR2 were highly expressed in Myeloid2, which was annotated as FOLR2high tissue-resident macrophages. These cells have low expression of inflammatory cytokines but are closely related to the complement system (Fig. 4B, C, Additional file 1: S8D). Li et al. [88] reported a subcluster in alveolar macrophages that has the same biomarkers (FOLR2 and SELENOP). This subcluster had a close relationship with exogenous antigen presentation, which was similar to the results of the GO analysis of Myeloid2 (Fig. 4E). The complement system plays an essential role in macrophage polarization [89]. Therefore, we identified Myeloid2 as complement-related macrophages. S100A8 and S100A9 are associated with the infiltration of various immune cells in the progression of CAVD [90]. Based on the GO analysis, we named Myeloid 4 as S100A8high macrophage. The marker genes for each subcluster are available in a supplementary table (Additional file 13, Myeloid subcluster DEGs).
Fig. 4.
Dynamicmyeloid cluster in the human mitral valve leaflet. A A UMAP plot of all myeloid cells coloured according to cluster. B Dot plot of the top 5 marker genes in myeloid clusters. The dot size corresponds to the proportion of cells within the group expressing each gene, and the dot colour corresponds to the expression level. C Heatmap of inflammatory cytokines, chemokines, and HLA in each cluster. D UMAP of myeloid canonical markers. E The enriched GO terms of each myeloid cluster. F SCENIC analysis of the expression of different TF regulons between each cluster. The data are coloured according to their expression levels. G The expression level and regulon activity of the TF BRCA1 in each myeloid cluster. H Comparison of the percentage of each cell type among the five clusters of myeloid cells. I Differential gene expression of myeloid cells in the NC group and MR group is shown in a volcano plot
According to the SCENIC analysis, each subcluster exhibited specific TFs. Myeloid2 was found to be a HOXA10+ subcluster, and the upregulation of HOXA10 can be linked to the promotion of M2 polarization [91] (Fig. 4F). These findings further demonstrated that Myeloid2 plays an important role in macrophage polarization. BRCA1 is a DNA repair enzyme. The expression and activity of BRCA1 in Myeloid3 was higher than those in other clusters (Fig. 4G), providing further evidence of the proliferative effect of Myeloid3 [92]. Information on the highly expressed TFs in each Myeloid subpopulation and their downstream regulated genes is displayed in a supplementary table (Additional file 14,Mye_cluster_regulon) and network diagrams (Additional file 15,Mye_TFandTarget_network). The proportion of various myeloid subclusters between the two groups does not differ significantly; however, the expression of DEG still remains noteworthy (Fig. 4H). Myeloid cells in the MR group highly expressed SERPINB2 (Fig. 4I), which could be activated by GPNMB, a glycoprotein secreted by macrophages, which ultimately leading to the activation of fibroblasts in idiopathic pulmonary fibrosis [93]. The DEGs between the NC and MR groups are available in a supplementary table (Additional file 16, Myeloid DESeq2 DEGs).
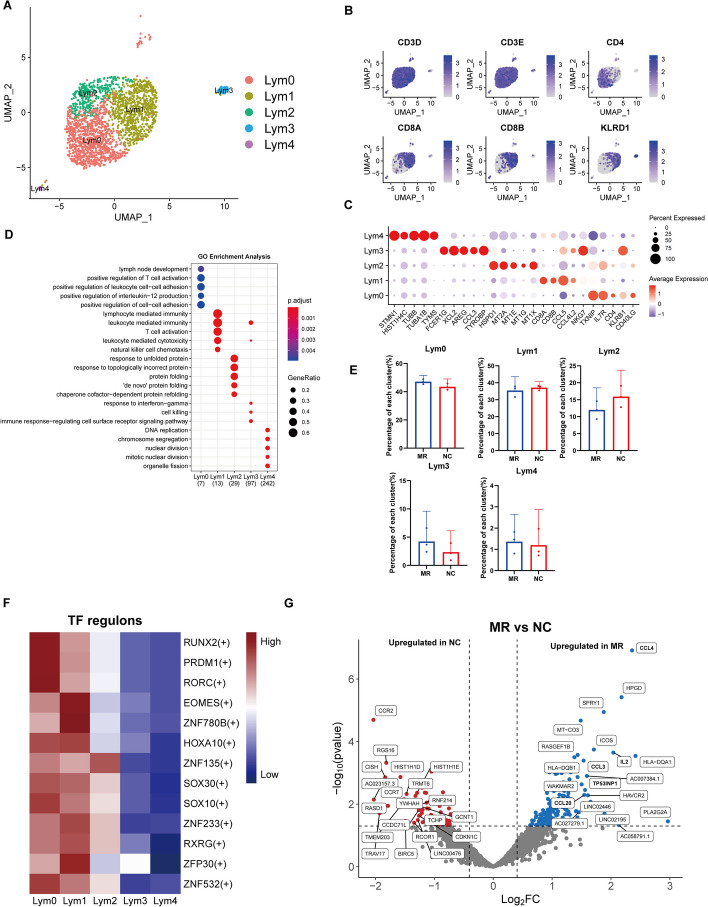
Dynamic lymphocyte clusters in the mitral valve
A total of 2106 lymphocytes from six distinct samples were stratified into five subclusters (Fig. 5A). We first examined the expression of highly conserved lymphocyte genes within these cells (Fig. 5B). Lym0 cells were found to belong to the CD4+ T-cell subset, while Lym1 cells were identified as CD8+ T cells. Interestingly, Lym2 and Lym4 were coexpressed in both CD4+ and CD8+ T cells. Furthermore, Lym3 was specifically classified as a natural killer cell based on its distinctive expression of KLRD1. Finally, we noted that pan-T-cell markers (CD3D and CD3E) were expressed across all remaining subclusters.
Fig. 5.
Dynamiclymphocyte clusters in the human mitral valve leaflet. A A UMAP plot of all lymphocyte cells coloured according to cluster. B UMAP of canonical lymphocyte cell markers. C Dot plot of the top 5 marker genes in the lymphocyte clusters. The dot size corresponds to the proportion of cells within the group expressing each gene, and the dot colour corresponds to the expression level. D The enriched GO terms of each lymphocyte cluster. E Comparison of the percentage of each cell type among the five clusters of lymphocytes. F SCENIC analysis of the expression of different TF regulons between each cluster. The data are coloured according to their expression levels. G Differential gene expression of myeloid cells in the NC group and MR group is shown in a volcano plot
To further characterize these subclusters, we explored the DEGs in each subcluster (Fig. 5C, Additional file 1: Figure S9A). Lym0 was characterized by high expression of IL7R and KLRB1 (CD161), which are known markers of CD4+ resident cells [94, 95]. Additionally, CD40LG, a protein that promotes the secretion of IgG by B cells, was highly expressed [96]. TXNIP, a scavenger of oxidative enzymes involved in regulating the activity of Th2 cells and the generation of CD4+ memory cells, was also significantly highly expressed in Lym0 [97]. Lym1 expressed a range of cytotoxicity-associated genes, including NKG7 [98], KLRD1, and the granzyme family (GZMB, GZMK, and GZMN) [99]. Inflammatory chemokines such as CCL4L2 and CCL5 were also highly expressed in this subcluster, indicating that these cells are proinflammatory CD8 + T cells. Lym2 expressed genes encoding Metallothionein (MT) family proteins, a group of molecules closely related to T-cell activation and cellular oxidative stress [100, 101], as well as heat shock protein (HSP) family proteins, confirming its role as a stress-related T cell. Lym3 expressed high levels of NK cell markers, including XCL2, XCL1, TYROBP, FCER1G, and KLRD1. Lym4 expressed microtubule-related genes such as STMN1, TUBA1B, and TUBB, which indicates that these cells are proliferating T cells. GO analysis also revealed that DNA replication and nuclear division were enriched in Lym4 (Fig. 5D, Additional file 1: Figure S9B). The proportion differences between the subpopulations of the two groups of cells were minimal, but the proportion of Lym4 in the MR group was slightly greater than that in the NC group, indicating increased cellular proliferation in the context of regurgitation (Fig. 5E). The marker genes for each subcluster are available in a supplementary table (Additional file 17, Lymphocyte subcluster DEGs).
SCENIC and clustering analyses were subsequently performed to identify the TFs that could regulate the functions of different subpopulations (Fig. 5F). Among these TFs, RORC exhibited high expression levels in Lym0. In the context of infective endocarditis, RORC can be highly activated in VICs to stimulate the development of Th17 cells, which has been linked to autoimmunity and inflammation [102]. Furthermore, RORC has been found to be closely associated with the secretion of interleukin-17 (IL-17), a cytokine that has been implicated in valve damage observed in rheumatic mitral valve disease [103]. IL-17A, a cytokine from the IL-17 family, is also highly expressed in Lym0 (Additional file 1: Figure S9C). Additionally, Lym0 exhibited high expression levels of the gene encoding ZNF532, a zinc finger protein involved in the regulation of the extracellular matrix, as well as in the reduction of EndMT in laryngeal squamous cell carcinoma cells [104]. Information on the highly expressed TFs in each Lym subpopulation and their downstream regulated genes is displayed in a supplementary table (Additional file 18,Lym_cluster_regulon) and network diagrams (Additional file 19,Lym_TFandTarget_network). Despite the highly similar cell proportions between the two groups, differential analysis revealed significant differences in gene expression profiles (Fig. 5E, G). The expression of inflammatory factors such as CCL3, CCL4, CCL20, and IL2 was increased in the MR group. This suggests that proinflammatory lymphocytes are present in the mitral valves of patients with MR. TP53INP1 inhibits the activation of cardiac fibroblasts and protects cardiac cells from damage [105, 106]. Thus, in MR, TP53INP1 may have a protective effect by preventing fibrosis. The DEGs between the NC and MR groups are available in a supplementary table (Additional file 20, Lymphocyte DESeq2 DEGs).
Loss of proliferating endothelial cells in the endothelium associated with the inadequate adaptation to regurgitation
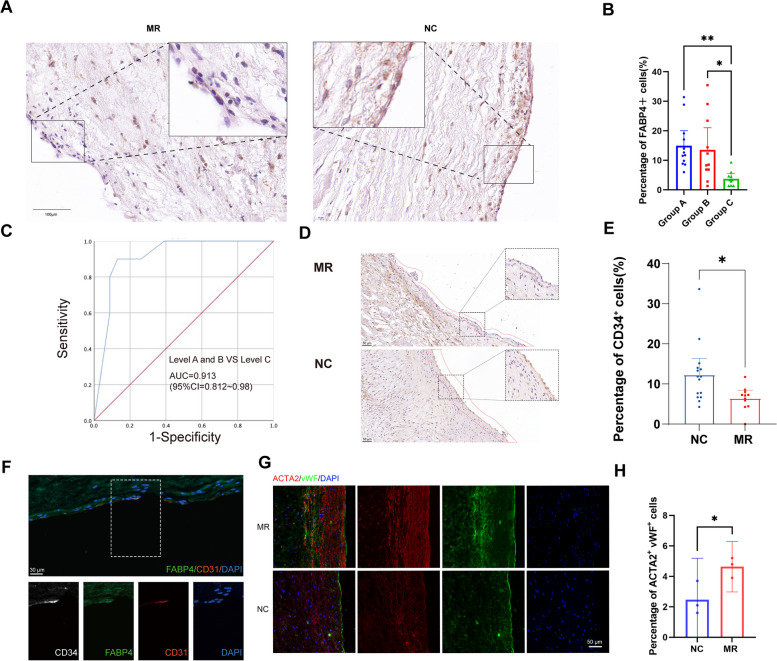
In MR, the surface area of the mitral valve increases to accommodate the increased effective regurgitant orifice area (EROA) resulting from left ventricular dysfunction. Our results revealed a reduction in the population of proliferating endothelial cells in the regurgitant group (Fig. 3H). To confirm this observation, we selected FABP4 as a marker for identifying the proliferating endothelial cell population, as shown in Fig. 3C. We performed immunohistochemical staining on mitral valves from an additional 21 patients with MR and 12 patients with normal mitral valve function (Fig. 6A). Since severe MR is associated with a poor prognosis, intervention is often needed. Therefore, we chose to categorize these patients into three groups. Group A (n = 12) included patients with no or mild MR who typically do not require specific interventions for MR. Patients with moderate MR, whose treatment may vary depending on their clinical situation, were classified into Group B (n = 11). Group C (n = 10) consisted of patients with severe MR. We observed the most significant decrease in FABP4+ cells in the C group (Fig. 6B). We further validated whether the number of FABP4+ cells could predict the degree of mitral valve regurgitation. We found that a proportion of FABP4+ VECs less than 6.5% was often associated with severe regurgitation, with an area under the curve (AUC) of 0.913 (Fig. 6C).
Fig. 6.
Loss of proliferating endothelial cells in the endothelium associated with inadequate adaptation to regurgitation. A Immunohistochemical staining of FABP4 in NC and MR specimens. B Quantification of the FABP4+ cell ratio in the endothelium per image (n = 12 in Group A, 11 in Group B, and 10 in Group C). C ROC curve for the predictive ability of FABP4+ VECs on the severity of mitral valve regurgitation. D Immunohistochemistry results for CD34 in the NC and MR groups. E Differential expression of CD34 between the NC and MR groups in IHC. F Immunostaining of FABP4, CD34 and CD31. G Immunostaining of ACTA2 (αSMA) and vWF in the NC and MR groups. H Quantification of the percentage of positive cells per image (n = 3 per condition). Mean ± SEM, two-tailed t test. Group A indicates patients with no or mild MR; Group B indicates patients with moderate MR; Group C indicates patients with severe MR. ROC, receiver operating characteristic; NC, non-diseased control; MR, mitral regurgitation
In order to investigate the relationship between proliferating endothelial cells and endothelial progenitor cells, we performed immunohistochemical staining using the classic endothelial progenitor cell marker CD34. The proportion of CD34+ cells was also reduced in the MR group (Fig. 6D, E). Multiple fluorescence immunohistochemistry (mIHC) demonstrated colocalization of CD34 and FABP4 in the tissue (Fig. 6F). This suggests that VEC2 is associated with the endothelial progenitor cells, which might explain its strong proliferative and EndMT capabilities.
To investigate the potential involvement of EndMT in MR, we performed our study by employing multicolour immunohistochemical staining and used von Willebrand factor (vWF) as a marker for VECs and α-smooth muscle actin (αSMA) as a marker for VICs (Fig. 6G, H). We found that EndMT was enhanced in the MR group.
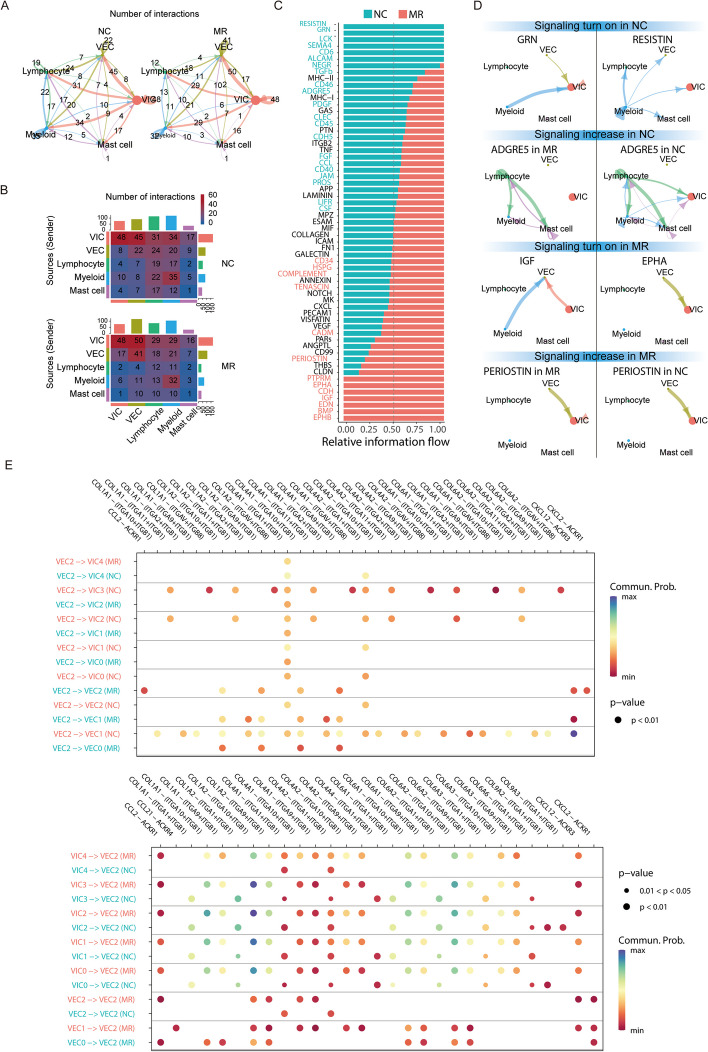
Extensive cell–cell interactions in the mitral valve
Using CellChat, we investigated the cell–cell interaction network among the cell types identified in our present work. The number of cell–cell interactions between different cell types was decreased in most MR specimens (Fig. 7A). VEC released the most number of ligands and expressed the most number of receptors. Compared with those in NC specimens, VEC-VEC interactions were increased in MR specimens, followed by the VEC-VIC interactions, while myeloid-lymphocyte interactions were decreased (Fig. 7B).
Fig. 7.
Extensive cell–cell interactions in the mitral valve. A Overall intercellular communication between each pair of cell populations in the comparison of NC and MR. The width of bands corresponding to the number of ligand‒receptor pairs. B Heatmaps of the number of interactions between NC and MR, showing the outgoing and incoming signalling in each cell group in greater detail (the top coloured bar plot represents the sum of each column of values displayed in the heatmap (incoming signalling). The right coloured bar plot represents the sum of each row of values (outgoing signalling). C Significant signalling pathways were ranked based on differences in the overall information flow within the inferred networks between NC and MR. The overall information flow of a signalling network was calculated by summarizing all communication probabilities in that network. D Circos plots showing the inferred intercellular communication network among VEC2 and other cell subtypes in the NC and MR groups. E Comparison of the specific significant ligand‒receptor pairs between the NC and MR groups, which contribute to signalling among VEC2 and the subclusters of VEC and VIC types. The dot colour reflects the communication probability, and the dot size represents the computed p value. An empty space means that the communication probability is zero. p values were computed from a one-sided permutation test
In our subsequent analysis, we compared the flow of information within each signalling pathway between NC and MR specimens. Notably, pathways such as those involving PTN, MK, NOTCH, and VEGF (black in Fig. 7C) exhibited consistent patterns across both groups. This coherence suggests their similar importance in the mitral valve under both conditions, indicating a limited role in regurgitation. In contrast, other pathways prominently changed their information flow in MR specimens compared with NC specimens: (i) deactivation of pathways involving the mitigating valve fibrosis and osteoblastic differentiation-GRN [107], and tissue-reparative gene RESISTIN [108]), (ii) decrease (migration inhibitor ADGRE5 [109]) (red in Fig. 7C, D), (iii) turn on (cell proliferation, cell survival, cell differentiation, and cell transformation positive regulator IGF [110], and apoptosis inhibitor EPHA [111, 112]), or (iv) increase (pro-fibrosis pathway PERIOSTIN [113]) (green in Fig. 7C, D). As VEC and VIC interacted frequently, the subcluster VEC2 mainly play a role in MR valves; therefore, we compared the interaction probabilities mediated by ligand–receptor pairs among VEC2 and the subclusters of VEC/VIC and identified specific ligand–receptor pairs that were only enriched in NC or MR specimens, including as significant signalling pathways (Fig. 7E). For example, in NC, VEC2 especially secreted a variety of ECM proteins to interact with integrin in VIC3, VIC2, and VEC1. Previous studies have indicated that integrin [114] interact with ECM components to enhance cellular adhesion. Thus, such interactions were necessary for valve cells in mitral valves to resist continuous hemodynamic stress. In the MR group, VEC2 cells showed high expression of ACKR1 [115], and they received the pro-inflammatory cytokine CCL2 from other subclusters of VEC and VIC cells, indicating that inflammation was induced in both VEC and VIC cells.
The regulatory role of FABP4+ VECs in VEC proliferation, migration, and EndMT
To explore the function of FABP4 in VEC cells, we isolated primary VECs from patients who underwent heart transplantation. Immunofluorescence staining for FABP4 and CD34 was performed on VECs cultured to the third generation (Additional file 1:Figure S10A). A total of 1427 cells were counted, resulting in 525 FABP4+ cells and 542 CD34+ cells. There were 436 cells co-expressing both markers, representing 80.4% of the FABP4+ cells and 83.0% of the CD34+ cells (Additional file 1: Figure S10B). This demonstrates that the VEC2 subpopulation largely overlaps with CD34-positive endothelial cells, further indicating the association between VEC2 and endothelial progenitor cells. FABP4-positive VECs account for nearly 40% of the total VECs, which is higher than the 10–20% proportion of VEC2 observed in bioinformatics and pathological results (Figs. 2H, and 6B), indicating that after a period of culture (3–4 generations), the proportion of this subpopulation increases by more than 50% compared to the initial level, demonstrating the strong proliferative capacity of this subpopulation.
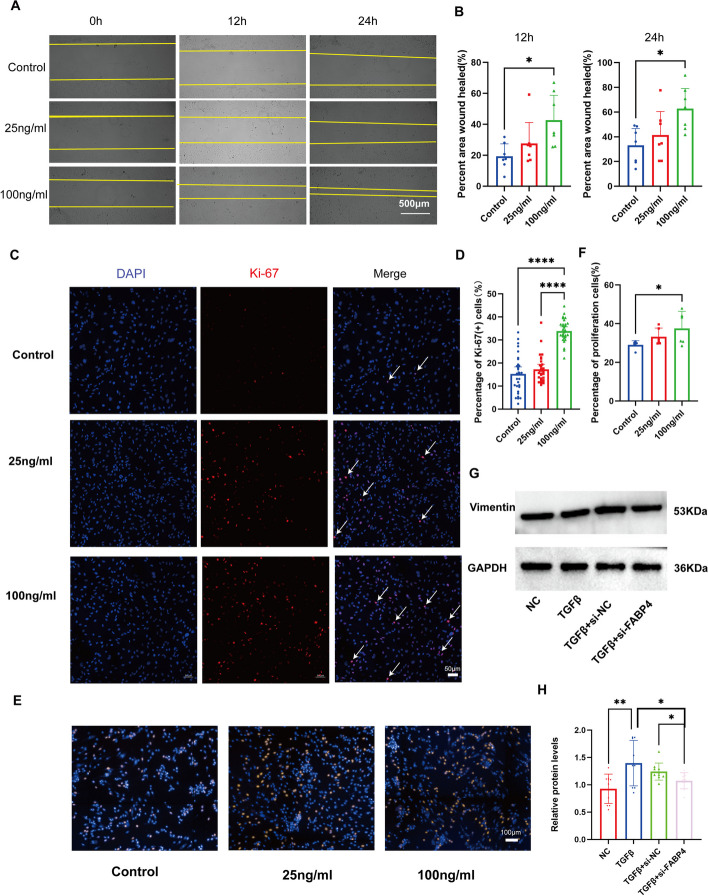
Subsequently, we treated VECs with FABP4 at concentrations of 25 and 100 ng/ml [116, 117]. We conducted a cell scratch assay and observed that FABP4 promoted VEC migration at 12 and 24 h after treatment in a concentration-dependent manner (Fig. 8B, C). Next, we assessed the impact of FABP4 on VEC proliferation using Ki-67 immunofluorescence and EdU (5-Ethynyl-2’-deoxyuridine) staining. Our results demonstrated that FABP4 also enhances VEC proliferation, and this effect is more pronounced at higher FABP4 concentrations (Fig. 8D, E, F, G). Then we used small interfering RNA (siRNA) to reduce FABP4 expression (Additional file 1: Figure S10C, D). We found that si-FABP4-2 reduced the expression of FABP4 by more than 70% after 48 h of transfection. We observed that the cell migration ability was significantly reduced after the expression of FABP4 was decreased (Fig. 8H, I). Similarly, Ki-67 immunofluorescence staining and Edu staining experiments revealed a marked reduction in the proportion of proliferating cells (Fig. 8J, K,Additional file 1: S10D, E). These indicate that the FABP4 may play a positive regulatory role in the overall migration and proliferation of VECs.
Fig. 8.
The regulatory role of FABP4+ VECs in VEC proliferation, migration, and EndMT. A Representative images of human VECs from the cell scratch assay. B Quantitative analysis of the results of the cell scratch assay. C Representative images of human VECs from Ki-67 proliferation assay. D Quantitative analysis of the results of Ki-67 proliferation assay. E Representative images of EdU staining. F Quantitative analysis of the results of EdU staining. G Representative images of Western blotting. H Quantitative analysis of the results of Western blotting. A Scale bar, 500 μm. C Scale bar, 50 μm. E Scale bar, 100 μm. Points in each group in B, D, F, H represent 7, 30, 6, and 10 biological replicates, respectively. The data were normally distributed and had equal variance. One-way ANOVA followed by Tukey’s multiple comparison test was used for multiple groups. A value of P<0.05 was considered statistically significant
Finally, we investigated the impact of FABP4 on TGFβ-induced EndMT. We divided cells into four groups: a normal control group, a group treated with 100 ng/ml FABP4, a group treated with 10 ng/ml TGFβ1, and a combined intervention group in which both substances were applied simultaneously. We observed that although FABP4 at a concentration of 100 ng/ml appeared to have a limited effect on cell differentiation, there was a noticeable trend towards enhanced promotion of EndMT when it was coadministered with TGFβ1 (Additional file 1: Figure S10F). Next, we investigated whether inhibition of FABP4 could attenuate TGF-β-induced EndMT. VEC was transfected by siRNA, and then the cells were treated with 10 ng/ml of TGF-β for 48 h. It was observed that the marker of EndMT, vimentin, increased after TGF-β treatment. However, in cells transfected with si-FABP4, vimentin expression decreased after TGF-β treatment (Fig. 8L, M). These indicated that the inhibition of FABP4 can reduce TGF-β-induced EndMT of VEC. These findings demonstrated the pro-migratory and pro-proliferative function of FABP4 on VECs. In addition, FABP4 also plays a positive regulatory role in EndMT.
Discussion
In this study, we employed scRNA-seq to profile the transcriptome of the mitral valve in both NC and MR specimens at a single-cell resolution. This investigation establishes a single-cell atlas of mitral valve, contributing to a better understanding of the changes at the single-cell level in functional mitral regurgitation. Our cellular composition data revealed one VEC subpopulation in the NC specimens. VEC2 cells exhibited high expression of FABP4 and were identified as proliferating endothelial cells. A previous study showed that as the left ventricle underwent remodelling with cavity enlargement, the MV leaflet area increased to limit and reduce MR [118]. This increase in area was not only achieved by passive stretching but also the result of VEC activation through EndMT [119]. The trajectory analysis of valve endothelial cells revealed that VEC2 can differentiate into two other subtypes of VEC. Pathological and in vitro experiments revealed that FABP4 and CD34 are co-expressed in such VEC subpopulation. These suggest that the proliferative capacity of VEC2 may be associated with endothelial progenitor cells. These findings suggested that the loss of valvular proliferating endothelial cells may be an important factor contributing to the impaired adaptability of regurgitant mitral valves.
Using cell–cell interaction analysis, we found that several pathways were disrupted or maintained in MR specimens. Then, we ascertained the central role of VECs in the pathophysiology of MV. VECs secrete and receive several factors that interact with other cell populations. Such VEC-based intercellular communication might be critical for sustaining haemostasis of the valvular microenvironment in the NC and contributing to the pathology of MR.
We isolated and cultured primary human mitral VECs. By using recombinant protein intervention and si-RNA intervention, we revealed that FABP4 promotes the migration and proliferation of VECs, similar to its function in vascular endothelial cells [116, 120]. Additionally, inhibiting the expression of FABP4 is able to reduce the EndMT of VECs in vitro. Jin et al. have reported that FABP4, in cooperation with TGFβ, will promote EndMT in cervical cancer cells [117]. Further in-depth analysis is needed to explore the mechanisms by which FABP4 regulates VECs.
There are several limitations in this study. First, we integrated scRNA-seq datasets from three MR samples and three NC samples. These samples were obtained from heart transplant recipients. Thus, they may have exhibited molecular or cellular changes in the mitral valve related to cardiomyopathy. Second, the age and sex of the patients from whom the MR and NC samples were obtained were not well matched (Table S1). Although there was no perfect sex match between the two groups, as there was one female patient in the MR group, we conducted a sex-related correlation analysis on the 3 patients in the MR group. The results showed that sex differences did not significantly impact gene expression within the MR group. Considering the age differences, we also performed age-related correlation tests on the paediatric (< 18 years) and adult (> 18 years) subgroups within the MR group. Similarly, age did not appear to have a pronounced effect on gene expression. This is consistent with previous research findings [10]. However, considering the absence of samples from elderly individuals in this study, future research may include samples from older adults (> 50 years old) to further investigate the relationship between aging and mitral valve regurgitation. To address these limitations, future studies should include specimens from diverse sources. Furthermore, discrepancies in recovery rates among cell types may arise from their distinct characteristics and distribution patterns. It is also plausible that certain cell types are more vulnerable to digestive processes, resulting in increased cell loss. Therefore, our dataset might not accurately portray the precise cellular proportions within the human mitral valve. A potential solution to overcome this recovery bias is to digest the fibre layer, intermediate layer, and atrium/ventricle separately and adjust the processing time based on the sensitivity of each layer to the digestive solution. Finally, although clinical pathological validation and cell experiments can support the bioinformatics results, these experiments do not fully reveal the specific regulatory processes of this subcluster in MR. Further cell experiments, such as separating this cell subpopulation using techniques such as flow cytometry, have not been conducted. This is limited by the difficulty in obtaining and culturing primary VECs from the human mitral valve. Despite these limitations and technical challenges, our study yielded several interesting and novel findings. In subsequent studies, we anticipate continuing to collect relevant samples for RNA sequencing or single-cell RNA sequencing of the chordae tendineae or papillary muscles of regurgitant mitral valves. This could further enhance our understanding of the functional aspects of mitral valve regurgitation. Moreover, we will further optimize the isolation and culture methods for primary mitral valve endothelial cells. We will also use specific markers identified in this study (e.g. FABP4) for sorting and further exploring the functions of this specific subset. Moreover, given the advanced stage of development in large animal models for functional MR, we will conduct further large animal experiments to validate our findings [121].
Conclusion
In conclusion, we revealed the cellular and molecular landscape of the MR valves at the single-cell level. We found that the proportion of proliferating endothelial cells was reduced in the MR group. These cells highly express FABP4 and exhibit greater proliferation and differentiation capabilities compared to other endothelial cells. This is associated with the occurrence of MR. The implications of these findings broaden our understanding of functional MR progression and may offer potential pathways for the advancement of future treatments.
Supplementary Information
Additional file 1: Table S1. Information of patients enrolled in the study. Table S2. Quality control results. Figure S1. Histological examination (HE staining) of mitral valve corresponding to each patient. Figure S2. Quality control for scRNA-seq datasets. Figure S3. Identification of valve cells. Figure S4. Potential effects of gender on the cellular landscape of mitral regurgitation valves. Figure S5. Potential effects of age on the cellular landscape of mitral regurgitation valves. Figure S6. VIC subclusters helps the valve change its structure to accommodate hemodynamic changes. Figure S7. Supplementary figures on VEC analysis. Figure S8. Supplementary figures on myeloid analysis. Figure S9. Supplementary figures on Lymphocyte analysis. Figure S10. Supplementary figures on function of FABP4 in VEC.
Additional file 5: VIC_TF and Target_network diagram.
Additional file 6: The highly expressed genes in each VIC subpopulation (VIC_top5DEGs_regulon).
Additional file 7: DEGs between the NC and MR groups in VIC (VIC DESeq2 DEG).
Additional file 10: VEC_TF and Target_network diagram.
Additional file 11: The highly expressed genes in each VEC subpopulation (VEC_top5DEGs_regulon).
Additional file 12: DEGs between the NC and MR groups in VEC (VEC DESeq2 DEG).
Additional file 13: Myeloid cluster DEGs.
Additional file 14: Myeloid_cluster_regulon.
Additional file 15: Myeloid_cluster_TF and Target_network diagram.
Additional file 16: DEGs between the NC and MR groups in Myeloid_cluster (Myeloid DESeq2 DEG).
Additional file 17: Lymphocyte cluster DEGs.
Additional file 18: Lymphocyte cluster_regulon.
Additional file 19: Lymphocyte_cluster_TF and Target_network diagram.
Additional file 20: DEGs between the NC and MR groups in Lymphocyte cluster (Lymphocyte DESeq2 DEG).
Additional file 21: The original, uncropped gels.
Acknowledgements
We would like to express our gratitude to graduate students and teachers in the State Key Laboratory of Cardiovascular Disease, including Yu-Long Xiong, Yu Jiang, Yu-Chen Xie, Zhen-Hao Zhang, Wei-Dong Yan, Chen-Yang Qiu, Guang-Xuan Zhu, Yu-Lin Chen, Lei Han and others, for their assistance in our research, including guidance in experimental techniques and methodological discussions on research topics. We also appreciate the help provided by Zi-Rui Liu, Hang Zhang, Ren-Jie Tang, Si-Yuan Huang, Tian-Shuo Yang, Han Mo, Hao-Tian Zhang, and others in sample acquisition and the collection of case information.
Abbreviations
- DEG
Differential expressed gene
- ECM
Extracellular matrix
- EndMT
Endothelial–mesenchymal transition
- FABP4
Fatty acid binding protein 4
- GSEA
Gene set enrichment analysis
- GO
Gene Ontology
- MR
Mitral regurgitation
- NC
Nondiseased control
- NO
Nitric oxide
- SCENIC
Single-cell regulatory network inference and clustering
- scRNA-seq
Single-cell RNA sequencing
- TF
Transcription factor
- UMAP
Uniform manifold approximation and projection
- VECs
Valvular endothelial cells
- VICs
Valvular interstitial cells
Authors’ contributions
J.S. and X.X3 designed and supervised the project. X.X1, W.W and M.F. conducted the study and wrote the manuscript. M.F., X.X1 and S.S. performed data analyses and data integration. X.H. and X.C. supervised the study. R.Z., X.X2 and W.F. provided clinical samples and information. X.X3, W.W and N.Z. performed the immunostaining experiments and in vitro experiment. All authors read and approved the final manuscript.
Funding
This work was supported by the National Science Foundation for Distinguished Young Scholars of China (grant no. 82125004), Shenzhen Science and Technology Innovation Commission (grant no.JCYJ20220818103414030), the National Natural Science Foundation of China (grant no. 82300397, 81900335, 22275124), National High Level Hospital Clinical Research Funding (grant no. 2023-GSP-QN-13, 2023-GSP-ZD-1), Guangdong Provincial Enterprise Joint Foundation (grant no.2022A1515220103), and the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2019ZT08Y481).
Data availability
Sequencing data are available at Genome Sequence Archive (accession no. HRA005416), and processed gene expression data are deposited in Gene Expression Omnibus (accession no. GSE241947).
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical University, as well as by the Ministry of Science and Technology, with the approval number of FW-2022–1658. Written informed consent for tissue donation, which clearly stated the purpose of our study, was obtained from all of the patients. For participants under the age of 18, written consent was obtained from their legal guardians in accordance with ethical standards.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaohu Wang, Mengxia Fu and Weiteng Wang contributed equally to this work.
Contributor Information
Xianqiang Wang, Email: qiangxianwang@163.com.
Jiangping Song, Email: fwsongjiangping@126.com.
References
- 1.VT Nkomo JM Gardin TN Skelton JS Gottdiener CG Scott M Enriquez-Sarano 2006 Burden of valvular heart diseases: a population-based study Lancet (London, England) 368 9540 1005 1011 [DOI] [PubMed] [Google Scholar]
- 2.Chehab O, Roberts-Thomson R, Ng Yin Ling C, Marber M, Prendergast BD, Rajani R, et al. Secondary mitral regurgitation: pathophysiology, proportionality and prognosis. Heart. 2020;106(10):716–23. [DOI] [PubMed] [Google Scholar]
- 3.A Sannino RL Smith 2nd GG Schiattarella B Trimarco G Esposito PA Grayburn 2017 Survival and Cardiovascular Outcomes of Patients With Secondary Mitral Regurgitation: A Systematic Review and Meta-analysis JAMA cardiology 2 10 1130 1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D Goldstein AJ Moskowitz AC Gelijns G Ailawadi MK Parides LP Perrault 2016 Two-Year Outcomes of Surgical Treatment of Severe Ischemic Mitral Regurgitation N Engl J Med 374 4 344 353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.F Maisano G Viganò A Blasio A Colombo C Calabrese O Alfieri 2006 Surgical isolated edge-to-edge mitral valve repair without annuloplasty: clinical proof of the principle for an endovascular approach EuroIntervention : journal of EuroPCR in collaboration with the Working Group on Interventional Cardiology of the European Society of Cardiology 2 2 181 186 [PubMed] [Google Scholar]
- 6.M Chaput MD Handschumacher F Tournoux L Hua JL Guerrero GJ Vlahakes 2008 Mitral leaflet adaptation to ventricular remodeling: occurrence and adequacy in patients with functional mitral regurgitation Circulation 118 8 845 852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M Vinciguerra S Romiti E Wretschko M D'Abramo D Rose F Miraldi 2021 Mitral Plasticity: The Way to Prevent the Burden of Ischemic Mitral Regurgitation? Frontiers in cardiovascular medicine 8 794574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar M, Thompson PD, Chen K. New Perspective on Pathophysiology and Management of Functional Mitral Regurgitation. Trends Cardiovasc Med. 2023;33(6):386–392. 10.1016/j.tcm.2022.03.001. [DOI] [PubMed]
- 9.M Fu J Song 2021 Single-Cell Transcriptomics Reveals the Cellular Heterogeneity of Cardiovascular Diseases Front Cardiovasc Med 8 643519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu S, Fu M, Chen X, et al. Cellular Landscapes of Nondiseased Human Cardiac Valves From End-Stage Heart Failure-Explanted Heart. Arterioscler Thromb Vasc Biol. 2022;42(12):1429–46. 10.1161/ATVBAHA.122.318314. [DOI] [PubMed] [Google Scholar]
- 11.SH Shinn HV Schaff 2013 Evidence-based surgical management of acquired tricuspid valve disease Nat Rev Cardiol 10 4 190 203 [DOI] [PubMed] [Google Scholar]
- 12.PA Grayburn A Sannino M Packer 2019 Proportionate and Disproportionate Functional Mitral Regurgitation: A New Conceptual Framework That Reconciles the Results of the MITRA-FR and COAPT Trials JACC Cardiovasc Imaging 12 2 353 362 [DOI] [PubMed] [Google Scholar]
- 13.G Benfari C Antoine B Essayagh R Batista J Maalouf A Rossi 2021 Functional Mitral Regurgitation Outcome and Grading in Heart Failure With Reduced Ejection Fraction JACC Cardiovasc Imaging 14 12 2303 2315 [DOI] [PubMed] [Google Scholar]
- 14.G Yu LG Wang Y Han QY He 2012 clusterProfiler: an R package for comparing biological themes among gene clusters OMICS 16 5 284 287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.B Sande Van de C Flerin K Davie M Waegeneer De G Hulselmans S Aibar 2020 A scalable SCENIC workflow for single-cell gene regulatory network analysis Nat Protoc 15 7 2247 2276 [DOI] [PubMed] [Google Scholar]
- 16.A Scialdone KN Natarajan LR Saraiva V Proserpio SA Teichmann O Stegle 2015 Computational assignment of cell-cycle stage from single-cell transcriptome data Methods 85 54 61 [DOI] [PubMed] [Google Scholar]
- 17.G Yu QY He 2016 ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization Mol Biosyst 12 2 477 479 [DOI] [PubMed] [Google Scholar]
- 18.S Jin CF Guerrero-Juarez L Zhang I Chang R Ramos CH Kuan 2021 Inference and analysis of cell-cell communication using Cell Chat Nat Commun 12 1 1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.G Manno La R Soldatov A Zeisel E Braun H Hochgerner V Petukhov 2018 RNA velocity of single cells Nature 560 7719 494 498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.V Bergen M Lange S Peidli FA Wolf FJ Theis 2020 Generalizing RNA velocity to transient cell states through dynamical modeling Nat Biotechnol 38 12 1408 1414 [DOI] [PubMed] [Google Scholar]
- 21.K Street D Risso RB Fletcher D Das J Ngai N Yosef 2018 Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics BMC Genomics 19 1 477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Berge K, Roux de Bézieux H, Street K, Saelens W, Cannoodt R, Saeys Y, et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun. 2020;11(1):1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.P Songia E Branchetti A Parolari V Myasoedova G Ferrari F Alamanni 2016 Mitral valve endothelial cells secrete osteoprotegerin during endothelial mesenchymal transition J Mol Cell Cardiol 98 48 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gould RA, Butcher JT. Isolation of valvular endothelial cells. J Vis Exp. 2010;(46):2158. 10.3791/2158. [DOI] [PMC free article] [PubMed]
- 25.S Shu M Fu X Chen N Zhang R Zhao Y Chang 2022 Cellular Landscapes of Nondiseased Human Cardiac Valves From End-Stage Heart Failure-Explanted Heart Arterioscler Thromb Vasc Biol 42 12 1429 1446 [DOI] [PubMed] [Google Scholar]
- 26.K Xu S Xie Y Huang T Zhou M Liu P Zhu 2020 Cell-Type Transcriptome Atlas of Human Aortic Valves Reveal Cell Heterogeneity and Endothelial to Mesenchymal Transition Involved in Calcific Aortic Valve Disease Arterioscler Thromb Vasc Biol 40 12 2910 2921 [DOI] [PubMed] [Google Scholar]
- 27.T Lyu Y Liu B Li R Xu J Guo D Zhu 2023 Single-cell transcriptomics reveals cellular heterogeneity and macrophage-to-mesenchymal transition in bicuspid calcific aortic valve disease Biol Direct 18 1 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D Crean EP Murphy 2021 Targeting NR4A Nuclear Receptors to Control Stromal Cell Inflammation, Metabolism, Angiogenesis, and Tumorigenesis Front Cell Dev Biol 9 589770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.EE Puscheck AO Awonuga Y Yang Z Jiang DA Rappolee 2015 Molecular biology of the stress response in the early embryo and its stem cells Adv Exp Med Biol 843 77 128 [DOI] [PubMed] [Google Scholar]
- 30.EJ Nestler 2015 ∆FosB: a transcriptional regulator of stress and antidepressant responses Eur J Pharmacol 753 66 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C Rippe S Albinsson G Guron H Nilsson K Swärd 2019 Targeting transcriptional control of soluble guanylyl cyclase via NOTCH for prevention of cardiovascular disease Acta Physiol (Oxf) 225 1 e13094 [DOI] [PubMed] [Google Scholar]
- 32.JL Kielczewski YP Jarajapu EL McFarland J Cai A Afzal S Li Calzi 2009 Insulin-like growth factor binding protein-3 mediates vascular repair by enhancing nitric oxide generation Circ Res 105 9 897 905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K Bosse CP Hans N Zhao SN Koenig N Huang A Guggilam 2013 Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease J Mol Cell Cardiol 60 27 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Dong N, Hui H, Wang Y, Liu F, Xu L, et al. Endothelial cell-derived tetrahydrobiopterin prevents aortic valve calcification. Eur Heart J. 2022;43(17):1652–64. [DOI] [PubMed] [Google Scholar]
- 35.Chen YT, Wang J, Wee AS, Yong QW, Tay EL, Woo CC, et al. Differential MicroRNA Expression Profile in Myxomatous Mitral Valve Prolapse and Fibroelastic Deficiency Valves. Int J Mol Sci. 2016;17(5):753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LE Dupuis L Doucette AK Rice AE Lancaster MG Berger S Chakravarti 2016 Development of myotendinous-like junctions that anchor cardiac valves requires fibromodulin and lumican Developmental dynamics : an official publication of the American Association of Anatomists 245 10 1029 1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.W Witke AH Sharpe JH Hartwig T Azuma TP Stossel DJ Kwiatkowski 1995 Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin Cell 81 1 41 51 [DOI] [PubMed] [Google Scholar]
- 38.Goldsmith EC, Bradshaw AD, Spinale FG. Cellular mechanisms of tissue fibrosis. 2. Contributory pathways leading to myocardial fibrosis: moving beyond collagen expression. Am J Physiol Cell Physiol. 2013;304(5):C393-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.PE Bartko JP Dal-Bianco JL Guerrero J Beaudoin C Szymanski DH Kim 2017 Effect of Losartan on Mitral Valve Changes After Myocardial Infarction J Am Coll Cardiol 70 10 1232 1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.AC Liu VR Joag AI Gotlieb 2007 The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology Am J Pathol 171 5 1407 1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S Guo E Zhang B Zhang Q Liu Z Meng Z Li 2022 Identification of Key Non-coding RNAs and Transcription Factors in Calcific Aortic Valve Disease Frontiers in cardiovascular medicine 9 826744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S Aibar CB González-Blas T Moerman VA Huynh-Thu H Imrichova G Hulselmans 2017 SCENIC: single-cell regulatory network inference and clustering Nat Methods 14 11 1083 1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.F Fang AJ Shangguan K Kelly J Wei K Gruner B Ye 2013 Early growth response 3 (Egr-3) is induced by transforming growth factor-β and regulates fibrogenic responses Am J Pathol 183 4 1197 1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.J Cao Y Ni H Zhang X Ning X Qi 2022 Inhibition of Kruppel-like factor 7 attenuates cell proliferation and inflammation of fibroblast-like synoviocytes in rheumatoid arthritis through nuclear factor κB and mitogen-activated protein kinase signaling pathway Exp Anim 71 3 356 367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R Wan S Long S Ma P Yan Z Li K Xu 2024 NR2F2 alleviates pulmonary fibrosis by inhibition of epithelial cell senescence Respir Res 25 1 154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.K Gaspar G Kukova E Bunemann BA Buhren E Sonkoly AG Szollosi 2013 The chemokine receptor CCR3 participates in tissue remodeling during atopic skin inflammation J Dermatol Sci 71 1 12 21 [DOI] [PubMed] [Google Scholar]
- 47.X Yu Z Song L Rao Q Tu J Zhou Y Yin 2020 Synergistic induction of CCL5, CXCL9 and CXCL10 by IFN-γ and NLRs ligands on human fibroblast-like synoviocytes-A potential immunopathological mechanism for joint inflammation in rheumatoid arthritis Int Immunopharmacol 82 106356 [DOI] [PubMed] [Google Scholar]
- 48.F Rahimi K Hsu Y Endoh CL Geczy 2005 FGF-2, IL-1beta and TGF-beta regulate fibroblast expression of S100A8 Febs j 272 11 2811 2827 [DOI] [PubMed] [Google Scholar]
- 49.Jaiswal A, Rehman R, Dutta J, Singh S, Ray A, Shridhar M, et al. Cellular Distribution of Secreted Phospholipase A2 in Lungs of IPF Patients and Its Inhibition in Bleomycin-Induced Pulmonary Fibrosis in Mice. Cells. 2023;12(7):1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.I Ninou C Magkrioti V Aidinis 2018 Autotaxin in Pathophysiology and Pulmonary Fibrosis Front Med (Lausanne) 5 180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A Purohit A Sadanandam P Myneni RK Singh 2014 Semaphorin 5A mediated cellular navigation: connecting nervous system and cancer Biochim Biophys Acta 1846 2 485 493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.PS Connell AF Azimuddin SE Kim F Ramirez MS Jackson SH Little 2016 Regurgitation Hemodynamics Alone Cause Mitral Valve Remodeling Characteristic of Clinical Disease States In Vitro Ann Biomed Eng 44 4 954 967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.PS Connell DP Vekilov CM Diaz SE Kim KJ Grande-Allen 2018 Eliminating Regurgitation Reduces Fibrotic Remodeling of Functional Mitral Regurgitation Conditioned Valves Ann Biomed Eng 46 5 670 683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.S Ghatak S Misra RA Moreno-Rodrigue VC Hascall GW Leone RR Markwald 2019 Periostin/β1integrin interaction regulates p21-activated kinases in valvular interstitial cell survival and in actin cytoskeleton reorganization Biochim Biophys Acta Gen Subj 1863 5 813 829 [DOI] [PubMed] [Google Scholar]
- 55.Wang N, Wu W, Qiang C, Ma N, Wu K, Liu D, et al. Protective Role of Collectin 11 in a Mouse Model of Rheumatoid Arthritis. Arthritis Rheumatol. 2021;73(8):1430–40. 10.1002/art.41696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A Oikonomopoulos KI Sereti F Conyers M Bauer A Liao J Guan 2011 Wnt signaling exerts an antiproliferative effect on adult cardiac progenitor cells through IGFBP3 Circ Res 109 12 1363 1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.E Ghelfi CW Yu H Elmasri M Terwelp CG Lee V Bhandari 2013 Fatty acid binding protein 4 regulates VEGF-induced airway angiogenesis and inflammation in a transgenic mouse model: implications for asthma Am J Pathol 182 4 1425 1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S Pacini V Carnicelli L Trombi M Montali R Fazzi E Lazzarini 2010 Constitutive expression of pluripotency-associated genes in mesodermal progenitor cells (MPCs) PLoS ONE 5 3 e9861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.LA Wylie KP Mouillesseaux DC Chong VL Bautch 2018 Developmental SMAD6 loss leads to blood vessel hemorrhage and disrupted endothelial cell junctions Dev Biol 442 2 199 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.N Froese B Kattih A Breitbart A Grund R Geffers JD Molkentin 2011 GATA6 promotes angiogenic function and survival in endothelial cells by suppression of autocrine transforming growth factor beta/activin receptor-like kinase 5 signaling J Biol Chem 286 7 5680 5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.C Sturtzel K Lipnik R Hofer-Warbinek J Testori B Ebner J Seigner 2018 FOXF1 Mediates Endothelial Progenitor Functions and Regulates Vascular Sprouting Front Bioeng Biotechnol 6 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.L Wu KK Wary S Revskoy X Gao K Tsang YA Komarova 2015 Histone Demethylases KDM4A and KDM4C Regulate Differentiation of Embryonic Stem Cells to Endothelial Cells Stem Cell Reports 5 1 10 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.AB Jaykumar S Plumber DM Barry D Binns C Wichaidit M Grzemska 2022 WNK1 collaborates with TGF-β in endothelial cell junction turnover and angiogenesis Proc Natl Acad Sci U S A 119 30 e2203743119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O Marsit MA Clavel A Paquin V Deschênes S Hadjadj I Sénéchal-Dumais 2022 Effects of Cyproheptadine on Mitral Valve Remodeling and Regurgitation After Myocardial Infarction J Am Coll Cardiol 80 5 500 510 [DOI] [PubMed] [Google Scholar]
- 65.H Ma L Xie L Zhang X Yin H Jiang X Xie 2018 Activated hepatic stellate cells promote epithelial-to-mesenchymal transition in hepatocellular carcinoma through transglutaminase 2-induced pseudohypoxia Commun Biol 1 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.WW Hwang-Verslues WH Kuo PH Chang CC Pan HH Wang ST Tsai 2009 Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers PLoS ONE 4 12 e8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.B Yang S Xia X Ye W Jing B Wu 2021 MiR-379-5p targets microsomal glutathione transferase 1 (MGST1) to regulate human glioma in cell proliferation, migration and invasion and epithelial-mesenchymal transition (EMT) Biochem Biophys Res Commun 568 8 14 [DOI] [PubMed] [Google Scholar]
- 68.Q Tang AJ McNair K Phadwal VE Macrae BM Corcoran 2022 The Role of Transforming Growth Factor-β Signaling in Myxomatous Mitral Valve Degeneration Frontiers in cardiovascular medicine 9 872288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.JJ Silbiger 2013 Novel pathogenetic mechanisms and structural adaptations in ischemic mitral regurgitation Journal of the American Society of Echocardiography : official publication of the American Society of Echocardiography 26 10 1107 1117 [DOI] [PubMed] [Google Scholar]
- 70.SJ Conway JD Molkentin 2008 Periostin as a heterofunctional regulator of cardiac development and disease Curr Genomics 9 8 548 555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.X Chen Q Zhang Q Zhang 2022 Predicting potential biomarkers and immune infiltration characteristics in heart failure Math Biosci Eng 19 9 8671 8688 [DOI] [PubMed] [Google Scholar]
- 72.MS Poetsch A Strano K Guan 2020 Role of Leptin in Cardiovascular Diseases Front Endocrinol (Lausanne) 11 354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.JM Pilewski L Liu AC Henry AV Knauer CA Feghali-Bostwick 2005 Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition Am J Pathol 166 2 399 407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.W Chen W Zhao A Yang A Xu H Wang M Cong 2017 Integrated analysis of microRNA and gene expression profiles reveals a functional regulatory module associated with liver fibrosis Gene 636 87 95 [DOI] [PubMed] [Google Scholar]
- 75.SE Adamson R Griffiths R Moravec S Senthivinayagam G Montgomery W Chen 2016 Disabled homolog 2 controls macrophage phenotypic polarization and adipose tissue inflammation J Clin Investig 126 4 1311 1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.M Liu Z Tong C Ding F Luo S Wu C Wu 2020 Transcription factor c-Maf is a checkpoint that programs macrophages in lung cancer J Clin Investig 130 4 2081 2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Y Bi J Chen F Hu J Liu M Li L Zhao 2019 M2 Macrophages as a Potential Target for Antiatherosclerosis Treatment Neural Plast 2019 6724903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Y Zhang X Shi J Han W Peng Z Fang Y Zhou 2021 Convallatoxin Promotes M2 Macrophage Polarization to Attenuate Atherosclerosis Through PPARγ-Integrin α(v)β(5) Signaling Pathway Drug Des Devel Ther 15 803 812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.K Yan Y Wang Y Lu Z Yan 2021 Coexpressed Genes That Promote the Infiltration of M2 Macrophages in Melanoma Can Evaluate the Prognosis and Immunotherapy Outcome J Immunol Res 2021 6664791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.D Jia S Chen P Bai C Luo J Liu A Sun 2022 Cardiac Resident Macrophage-Derived Legumain Improves Cardiac Repair by Promoting Clearance and Degradation of Apoptotic Cardiomyocytes After Myocardial Infarction Circulation 145 20 1542 1556 [DOI] [PubMed] [Google Scholar]
- 81.C Meng G Liu H Mu M Zhou S Zhang Y Xu 2015 Amphiregulin may be a new biomarker of classically activated macrophages Biochem Biophys Res Commun 466 3 393 399 [DOI] [PubMed] [Google Scholar]
- 82.Clayton SA, Daley KK, MacDonald L, Fernandez-Vizarra E, Bottegoni G, O’Neil JD, et al. Inflammation causes remodeling of mitochondrial cytochrome c oxidase mediated by the bifunctional gene C15orf48. Sci Adv. 2021;7(50):eabl5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.J Li M Zhou JQ Feng SM Hong SY Yang LX Zhi 2022 Bulk RNA Sequencing With Integrated Single-Cell RNA Sequencing Identifies BCL2A1 as a Potential Diagnostic and Prognostic Biomarker for Sepsis Front Public Health 10 937303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.K Budhachandra RK Brojen Singh GI Menon 2008 Microtubule dynamics regulated by stathmin Comput Biol Chem 32 2 141 144 [DOI] [PubMed] [Google Scholar]
- 85.T Tian M Bu X Chen L Ding Y Yang J Han 2021 The ZATT-TOP2A-PICH Axis Drives Extensive Replication Fork Reversal to Promote Genome Stability Mol Cell 81 1 198 211.e6 [DOI] [PubMed] [Google Scholar]
- 86.JA Han JY Kim JI Kim 2014 Analysis of gene expression in cyclooxygenase-2-overexpressed human osteosarcoma cell lines Genomics Inform 12 4 247 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.H Yanai T Ban Z Wang MK Choi T Kawamura H Negishi 2009 HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses Nature 462 7269 99 103 [DOI] [PubMed] [Google Scholar]
- 88.Li X, Kolling FW, Aridgides D, Mellinger D, Ashare A, Jakubzick CV. ScRNA-seq expression of IFI27 and APOC2 identifies four alveolar macrophage superclusters in healthy BALF. Life Sci Alliance. 2022;5(11):e202201458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.SS Bohlson SD O'Conner HJ Hulsebus MM Ho DA Fraser 2014 Complement, c1q, and c1q-related molecules regulate macrophage polarization Front Immunol 5 402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.K Wang Q Zheng X Liu B Geng N Dong J Shi 2022 Identifying hub genes of calcific aortic valve disease and revealing the immune infiltration landscape based on multiple WGCNA and single-cell sequence analysis Front Immunol 13 1035285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.S Yang H Zhao W Xiao L Shao C Zhao P Sun 2022 Extracellular vesicle-packaged miR-181c-5p from epithelial ovarian cancer cells promotes M2 polarization of tumor-associated macrophages via the KAT2B/HOXA10 axis J Gene Med 24 10 e3446 [DOI] [PubMed] [Google Scholar]
- 92.AR Pinto JW Godwin A Chandran L Hersey A Ilinykh R Debuque 2014 Age-related changes in tissue macrophages precede cardiac functional impairment Aging (Albany NY) 6 5 399 413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.J Wang X Zhang M Long M Yuan J Yin W Luo 2023 Macrophage-derived GPNMB trapped by fibrotic extracellular matrix promotes pulmonary fibrosis Commun Biol 6 1 136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leung GA, Cool T, Valencia CH, Worthington A, Beaudin AE, Forsberg EC. The lymphoid-associated interleukin 7 receptor (IL7R) regulates tissue-resident macrophage development. Development (Cambridge, England). 2019;146(14):dev176180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.V Konduri D Oyewole-Said J Vazquez-Perez SA Weldon MM Halpert JM Levitt 2020 CD8(+)CD161(+) T-Cells: Cytotoxic Memory Cells With High Therapeutic Potential Front Immunol 11 613204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Y Zhou J Yuan Y Pan Y Fei X Qiu N Hu 2009 T cell CD40LG gene expression and the production of IgG by autologous B cells in systemic lupus erythematosus Clin Immunol 132 3 362 370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.K Kokubo K Hirahara M Kiuchi K Tsuji Y Shimada Y Sonobe 2023 Thioredoxin-interacting protein is essential for memory T cell formation via the regulation of the redox metabolism Proc Natl Acad Sci U S A 120 2 e2218345120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.SS Ng RF Labastida De J Yan D Corvino I Das P Zhang 2020 The NK cell granule protein NKG7 regulates cytotoxic granule exocytosis and inflammation Nat Immunol 21 10 1205 1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.B Bengsch T Ohtani RS Herati N Bovenschen KM Chang EJ Wherry 2018 Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns J Immunol Methods 453 3 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.JM Rice A Zweifach MA Lynes 2016 Metallothionein regulates intracellular zinc signaling during CD4(+) T cell activation BMC Immunol 17 1 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.S Choi X Liu Z Pan 2018 Zinc deficiency and cellular oxidative stress: prognostic implications in cardiovascular diseases Acta Pharmacol Sin 39 7 1120 1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.CY Yeh CT Shun YM Kuo CJ Jung SC Hsieh YL Chiu 2015 Activated human valvular interstitial cells sustain interleukin-17 production to recruit neutrophils in infective endocarditis Infect Immun 83 6 2202 2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xian S, Chen A, Wu X, Lu C, Wu Y, Huang F, et al. Activation of activin/Smad2 and 3 signaling pathway and the potential involvement of endothelial-mesenchymal transition in the valvular damage due to rheumatic heart disease. Mol Med Rep. 2021;23(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.L Fan J Wang P Deng Y Wang A Zhang M Yang 2022 Foxhead box D1 promotes the partial epithelial-to-mesenchymal transition of laryngeal squamous cell carcinoma cells via transcriptionally activating the expression of zinc finger protein 532 Bioengineered 13 2 3057 3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Z Wang M Fu Y Li 2020 miR-142-5p and miR-212-5p cooperatively inhibit the proliferation and collagen formation of cardiac fibroblasts by regulating c-Myc/TP53INP1 Can J Physiol Pharmacol 98 5 314 323 [DOI] [PubMed] [Google Scholar]
- 106.X Luo Y Xu Z Zhong P Xiang X Wu A Chong 2022 miR-8485 alleviates the injury of cardiomyocytes through TP53INP1 J Biochem Mol Toxicol 36 10 e23159 [DOI] [PubMed] [Google Scholar]
- 107.G Huang L An M Fan M Zhang B Chen M Zhu 2020 Potential role of full-length and nonfull-length progranulin in affecting aortic valve calcification J Mol Cell Cardiol 141 93 104 [DOI] [PubMed] [Google Scholar]
- 108.LA Meier JL Auger BJ Engelson HM Cowan ER Breed MI Gonzalez-Torres 2018 CD301b/MGL2(+) Mononuclear Phagocytes Orchestrate Autoimmune Cardiac Valve Inflammation and Fibrosis Circulation 137 23 2478 2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.W Zhao Z Wang Z Sun S Wang M Wu L Zheng 2017 Lentivirus-mediated overexpression of CD97/ADGRE5 reverses dysregulated high glucose-induced endothelial cell migration Mol Med Rep 15 5 3048 3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.IV Subramanian BC Fernandes T Robinson J Koening KS Lapara S Ramakrishnan 2009 AAV-2-mediated expression of IGF-1 in skeletal myoblasts stimulates angiogenesis and cell survival J Cardiovasc Transl Res 2 1 81 92 [DOI] [PubMed] [Google Scholar]
- 111.N Cheng DM Brantley H Liu Q Lin M Enriquez N Gale 2002 Blockade of EphA receptor tyrosine kinase activation inhibits vascular endothelial cell growth factor-induced angiogenesis Mol Cancer Res 1 1 2 11 [PubMed] [Google Scholar]
- 112.A Freywald N Sharfe CD Miller C Rashotte CM Roifman 2006 EphA receptors inhibit anti-CD3-induced apoptosis in thymocytes J Immunol 176 7 4066 4074 [DOI] [PubMed] [Google Scholar]
- 113.P Ranjan R Kumari SK Goswami J Li H Pal Z Suleiman 2021 Myofibroblast-Derived Exosome Induce Cardiac Endothelial Cell Dysfunction Frontiers in cardiovascular medicine 8 676267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.MR Chastney JRW Conway J Ivaska 2021 Integrin adhesion complexes Curr Biol 31 10 R536 R542 [DOI] [PubMed] [Google Scholar]
- 115.M Kashiwazaki T Tanaka H Kanda Y Ebisuno D Izawa N Fukuma 2003 A high endothelial venule-expressing promiscuous chemokine receptor DARC can bind inflammatory, but not lymphoid, chemokines and is dispensable for lymphocyte homing under physiological conditions Int Immunol 15 10 1219 1227 [DOI] [PubMed] [Google Scholar]
- 116.H Elmasri E Ghelfi CW Yu S Traphagen M Cernadas H Cao 2012 Endothelial cell-fatty acid binding protein 4 promotes angiogenesis: role of stem cell factor/c-kit pathway Angiogenesis 15 3 457 468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.J Jin Z Zhang S Zhang X Chen Z Chen P Hu 2018 Fatty acid binding protein 4 promotes epithelial-mesenchymal transition in cervical squamous cell carcinoma through AKT/GSK3β/Snail signaling pathway Mol Cell Endocrinol 461 155 164 [DOI] [PubMed] [Google Scholar]
- 118.MK Rausch FA Tibayan DC Miller E Kuhl 2012 Evidence of adaptive mitral leaflet growth J Mech Behav Biomed Mater 15 208 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.JP Dal-Bianco E Aikawa J Bischoff JL Guerrero MD Handschumacher S Sullivan 2009 Active adaptation of the tethered mitral valve: insights into a compensatory mechanism for functional mitral regurgitation Circulation 120 4 334 342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.H Elmasri C Karaaslan Y Teper E Ghelfi M Weng TA Ince 2009 Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells FASEB journal : official publication of the Federation of American Societies for Experimental Biology 23 11 3865 3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.H Yamauchi EN Feins NV Vasilyev S Shimada D Zurakowski PJ Nido Del 2013 Creation of nonischemic functional mitral regurgitation by annular dilatation and nonplanar modification in a chronic in vivo swine model Circulation 128 11 Suppl 1 S263 S270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Information of patients enrolled in the study. Table S2. Quality control results. Figure S1. Histological examination (HE staining) of mitral valve corresponding to each patient. Figure S2. Quality control for scRNA-seq datasets. Figure S3. Identification of valve cells. Figure S4. Potential effects of gender on the cellular landscape of mitral regurgitation valves. Figure S5. Potential effects of age on the cellular landscape of mitral regurgitation valves. Figure S6. VIC subclusters helps the valve change its structure to accommodate hemodynamic changes. Figure S7. Supplementary figures on VEC analysis. Figure S8. Supplementary figures on myeloid analysis. Figure S9. Supplementary figures on Lymphocyte analysis. Figure S10. Supplementary figures on function of FABP4 in VEC.
Additional file 5: VIC_TF and Target_network diagram.
Additional file 6: The highly expressed genes in each VIC subpopulation (VIC_top5DEGs_regulon).
Additional file 7: DEGs between the NC and MR groups in VIC (VIC DESeq2 DEG).
Additional file 10: VEC_TF and Target_network diagram.
Additional file 11: The highly expressed genes in each VEC subpopulation (VEC_top5DEGs_regulon).
Additional file 12: DEGs between the NC and MR groups in VEC (VEC DESeq2 DEG).
Additional file 13: Myeloid cluster DEGs.
Additional file 14: Myeloid_cluster_regulon.
Additional file 15: Myeloid_cluster_TF and Target_network diagram.
Additional file 16: DEGs between the NC and MR groups in Myeloid_cluster (Myeloid DESeq2 DEG).
Additional file 17: Lymphocyte cluster DEGs.
Additional file 18: Lymphocyte cluster_regulon.
Additional file 19: Lymphocyte_cluster_TF and Target_network diagram.
Additional file 20: DEGs between the NC and MR groups in Lymphocyte cluster (Lymphocyte DESeq2 DEG).
Additional file 21: The original, uncropped gels.
Data Availability Statement
Sequencing data are available at Genome Sequence Archive (accession no. HRA005416), and processed gene expression data are deposited in Gene Expression Omnibus (accession no. GSE241947).