Abstract
The composition and function of animal gut microbiota are shaped by various factors, among which diet is one of the major factors. Diet is affected by seasonal shifts and geographical differences, which in turn impact the host’s nutritional levels. To adapt to these environmental changes, the gut microbiome often produces matching responses. Understanding the relationships among the environment, diet, host and the gut microbiome is helpful for exploring the environmental adaptation of wildlife. Here, we chose wild sika deer (Cervus nippon), which is composed natural allopatric populations, to explore how the environment shapes the gut microbiome and affects the relationship between microbiota composition and function and the mutual adaptation of the seasonal living environment to seasonal dietary changes. To this purpose we used DNA metabarcoding, 16S RNA gene amplification sequencing, metagenomic shotgun sequencing and nutritional analyses to comprehensively examine the relationships among the forage plant, nutrient status and host gut microbiome. Our analyses showed spatiotemporal differences in diet between the Tiebu and Hunchun regions, which ultimately led to varying intakes of protein, cellulose, and soluble sugar. The microbiome composition and function showed unique characteristics in each group, and significant differences were detected at the gene level for the protein absorption and metabolism pathway, the carbohydrate metabolic absorption pathway, and cellulase enzyme function, which are related to nutrition. We also found differences in the pathogenic bacteria and resistance mechanisms genes of the gut microbiota in different groups. Our results showed that the gut microbiome of allopatric populations adapts to changes in food composition and nutrition in different seasons and areas to help the host cope with spatiotemporal changes in the living environment. At the same time, varying levels of human activity can have potential health impacts on wild animals.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42523-024-00362-z.
Keywords: Gut microbiome, Mixed-fed herbivore, Sika deer (Cervus nippon), Coevolution, High-throughput sequencing, DNA metabarcoding, Metagenomic shotgun sequencing
Introduction
Animals often adopt flexible survival strategies to face a variety of survival challenges, such as diseases, energy and nutritional deficiencies and external interference [17, 55, 62, 64]. The gut microbiota, as a plastic entity, can respond to different environmental factors and it is critical in the adaptation process between the host and the environment [11]. As host symbionts, the gut microbiota members change more quickly and can be regulated in more ways than the host genome, thus resulting in novel evolution of the holobiont to adapt to specific environmental conditions [86]. Thus, studying the potential connections between host habitat and the gut microbiome can advance the understanding of host-gut microbe coevolution and might provide useful information for wildlife conservation [1, 51].
The composition and function of gut microbiota can be affected by many factors, such as the host’s phylogeny, physiological state, living environment, eating habits and social structure [5, 12, 38, 54, 65], and the gut microbiota responds to environmental changes to help the host adapt by influencing nutrient uptake, immune health, and physiological metabolism [30, 31, 35, 37, 81]. There is a great amount of evidence showing that environmental differences shape the gut microbiome in animals [13, 15, 51]. For example, declining sea ice levels divide polar bear population into “onshore bears” and “offshore bears”, and the different habitat preferences of polar bears affect the gut microbiota diversity, with “onshore bears” having greater diversity than “offshore bears” [72]. The gut microbiota of urban coyotes (Canis latrans) is rich in Streptococcus and Enterococcus, but that of rural coyotes is rich in Fusobacteria, Sutterella, and Anaerobiospirillum, which are associated with the different body conditions of the coyotes [63]. Studies have shown that large-scale geographical differences can also lead to changes in the gut microbiome [56, 71]. Among the six different Chinese rhesus macaque populations, the Tibetan population show a greater abundance of Firmicutes and a lower abundance of Bacteroidetes than the other geographical groups to help the host adapt to the high-altitude environment [84]. A study mapped beta diversity in the gut microbiota of 136 pairs of animals across the Americas by comparing the gut microbiota of sympatric and allopatric mammalian populations. The findings revealed that each group displayed a unique gut microbiota composition [52]. Even in the same area, seasonal shifts can lead to changes in habitat and diet, which can affect the community of the gut microbiome [69].
Diet is often a crucial determinant of the gut microbiome in different environments [16, 21]. Dietary changes affect nutrient access and lead to changes in the gut microbiome structure [22]. Animals adjust their eating habits according to changes in food resources, and they often consuming a wider variety of food and nutrients during food-rich seasons but relatively limited choices in the withering season, which further affects the microbial community in the body [78]. For example, the gut microbiome of wild black howler monkeys (Alouatta pigra), depending on its composition and activity, provides additional energy and nutrients to compensate for changes in diet caused by seasonal changes [2]. The functions of the gut microbiome, which has diverse metabolic pathways associated with cellulose degradation and short-chain fatty acid (SCFA) production, of the skywalker hoolock gibbon (Hoolock tianxing) were enriched in the high-leaf period [42]. Additionally, in herbivores, the gut microbiome can help the host digest indigestible nutrients and toxic substances [37, 57]. Human interference with habitats can alter the composition of gut microbiome in animals, potentially impacting their health. Habitats with varying levels of human interference can influence animals’ access to food, thereby affecting their gut microbial diversity [51]. Wild animal species that are more sensitive to human activities are also more susceptible to gut microbiota [3].
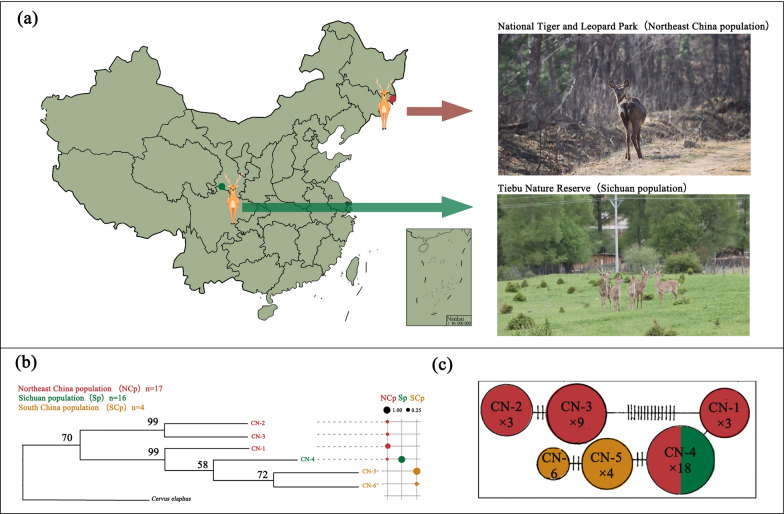
Researchers have generally focused on seasonal shifts or geographical scales to explore the response of the gut microbiome to the environment, while few studies have comprehensively considered spatiotemporal differences. Studies on large-scale geographical differences in ungulates have focused mainly on livestock [6, 25, 48]. In the case for endangered ungulates, researchers have generally focused on the differences between captive and wild populations [14, 20, 47, 70, 79]. Only a few studies have explored spatiotemporal changes in the gut microbiome of large herbivores within short geographical distances [16, 68]. Based on previous studies, we selected sika deer as a model species to explore the spatiotemporal response of the gut microbiome of endangered herbivores to the environment. Sika deer (Cervus nippon) is a mixed-diet herbivore that changes food choices according to changes in the environment [32]. In China, wild sika deer have undergone a long history of differentiation into several wild populations living in completely different habitats [27]. Liu et al. [45] sequenced 351 base pairs (bp) of the mitochondrial control region from 37 sika deer collected from Jilin (Northeast China population), Sichuan (Sichuan population), Anhui and Jiangxi (South China population), revealing significant population subdivision among sika deer in Mainland China. There has been considerable geographical isolation among these populations, limiting the potential for gene exchange. However, different populations of sika deer in Mainland China cannot be classified into evolutionarily significant units (ESUs). Sichuan population shared haplotype with Northeast China population (Fig. 1b, c).
Fig. 1.
Basic information about wild sika deer. a The study area of wild sika deer in China, namely, Tiebu Nature Reserve (Sichuan population) and Hunchun Nature Reserve (Northeast China population). The photo shows the differences in living environments between the two populations. b Phylogenetic relationship of sika deer mtDNA control region haplotypes reconstructed on the basis of the maximum likelihood algorithm with C. elaphus as the outgroup. The bubble plot represents the frequency with which haplotypes appear in the group, Northeast China population (red), Sichuan population (green), South China population (yellow), Figure is modified from Liu et al. [45]. c Minimum spanning network for haplotypes. The circle size is proportional to the number of individuals bearing that haplotype. Nucleotide transitions and transversions are indicated by dashes respectively (Only one transition between haplotypes is not marked), Figure is modified from Liu et al. [45]
We chose the Sichuan population and the Northeast China population as our research subjects. These two populations are located in distinct protected areas, each facing different habitat conditions and levels of human interference. This makes them ideal subjects for studying the adaptation of the gut microbiome of large wild animals to their living environments. We hypothesized that differences in habitat and phenology would influence diet composition, and that the gut microbiome function of wild sika deer would change across regions due to differences in food nutrition. Furthermore, we anticipated that the pathogens might differ based on the extent of human interference. To verify the above assumptions, we combined 16S RNA gene amplification sequencing, metabarcoding technology, metagenomic shotgun sequencing, and nutritional analyses to explore the response of the gut microbiome composition and function to the environment of sika deer.
Materials and methods
Study areas, animals and sampling
Our study area was in the Tiebu Nature Reserve (102°56′ ~ 103°10′E, 34°00′ ~ 34°11′N) in Ruoergai County, Sichuan Province, and National Tiger and Leopard Park (30°17′ ~ 131°14′E, 42°24′ ~ 43°28′N) in Hunchun city, Jilin Province (Fig. 1a). The population of wild sika deer in the Sichuan region is the largest population in China, inhabiting high-elevation environments that are complex, mostly mountain scrub meadows, forest scrub meadows, subalpine scrub meadows, valley scrub meadows and forest edge farmland habitats [26]. With respect to wild sika deer in Northeast China, the Hunchun region has a coastal mid-temperate maritime monsoon climate, and at lower altitudes, broad-leaved forest is the main vegetation type [77]. The Tiebu Nature Reserve is located in the pastoral area, providing greater opportunities for sika deer to come into contact with humans. In contrast, the Hunchun region of the National Tiger and Leopard Park, located along the border, is sparsely populated due to management policies (Fig. 1a).
We collected faecal samples during the green grass period (July to October) and the withered grass period (November to January) from 2020 to 2021. During the collection period, sika deer moved in groups. To avoid repetition during sample collection, we examined the activity areas of the different sika deer groups. The groups could be distinguished by the characteristics of the antlers and the white spots of the coats. After observing the defecation of the animals, we collected samples. Faecal samples with different morphological characteristics were collected from different groups, and when multiple faecal samples were present within a close range, only one faecal sample was collected. Sterile disposable PE gloves and sterile sampling bags were used to collect the samples, and only fresh samples were collected for identification based on their moist surfaces. All samples were transported and stored in dry ice and frozen at − 40 °C. PCR was used for sex detection, and different sex samples from one group were selected for this study to ensure that the collected samples did not come from one individual [76].
Diet analyses
DNA extraction and DNA amplification
35 faecal samples (green season group included female 12 and male 3; withered season group included female 12 and male 8) from the Tiebu region and 37 faecal samples (green season group included female 16 and male 4; withered season group included female 11 and male 6) from the Hunchun region were used to diet analyses. Total DNA was extracted from the faecal samples using a commercial reagent kit for the extraction of faecal DNA, namely, the MP Biomedicals SPIEasy DNA Kit for Faeces (MP Biomedicals). The DNA extracts were recovered in a total volume of 150 μL. We selected the psbCL primers to amplify chloroplast genomic fragments (Supporting Information Table 1). These primers have been shown to have high taxonomic coverage and discriminative capacity for vascular plants and have been applied to the study of animal eating habits [60, 74, 75].
The PCR parameters were as follows: predenaturation at 95 °C for 5 min; 45 cycles at 95 °C for 30 s, 45 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 10 min [75]. The follow-up experiments were carried out with only PCR products of the correct size and at a suitable concentration. The library was constructed and sequenced on the Illumina MiSeq platform (Shanghai Majorbio Biopharm Technology Co., Ltd.).
Processing of sequence data and sequence analysis
Sequence processing was completed on the Majorbio Cloud Platform (www.majorbio.com) [59]. We used fastp (version 0.19.6) to quality control, FLASH (version 1.2.7) to merge reads, and the DADA2 method to denoise the data and obtain the ASV sequence [10]. Considering the differences in plant species in different regions, we processed and analysed data samples from the Tiebu and Hunchun regions separately. The effective ASV sequences obtained were subjected to a BLASTN search (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi) to compare each ASV sequence with the available sequences in nt/nr database, and species identification was carried out according to sequence similarity. For plant sequence identification, we referred to the steps of Xiong et al. [75] and made several adjustments: (1) we considered only the alignment results with coverage and percentage of identity greater than 90%; (2) when the query sequence matched the single species sequence in the database and the consistency was ≥ 99% and the species distribution conformed to the result, the query was assigned to the species; (3) when a query matched more than one species sequence and the consistency was ≥ 90% and the distribution of multiple species was consistent with the results, the optimal results were retained according to the order of e values; (4) when a query matched more than one species sequence and the consistency was ≥ 90% and the distribution of multiple species was not consistent with the results, according to the optimal matching results, the classification unit of the upper level was recorded; and (5) considering the limited record of species distribution and information on bryophytes, identification of bryophytes was performed at the phylum level.
Nutritional analyses
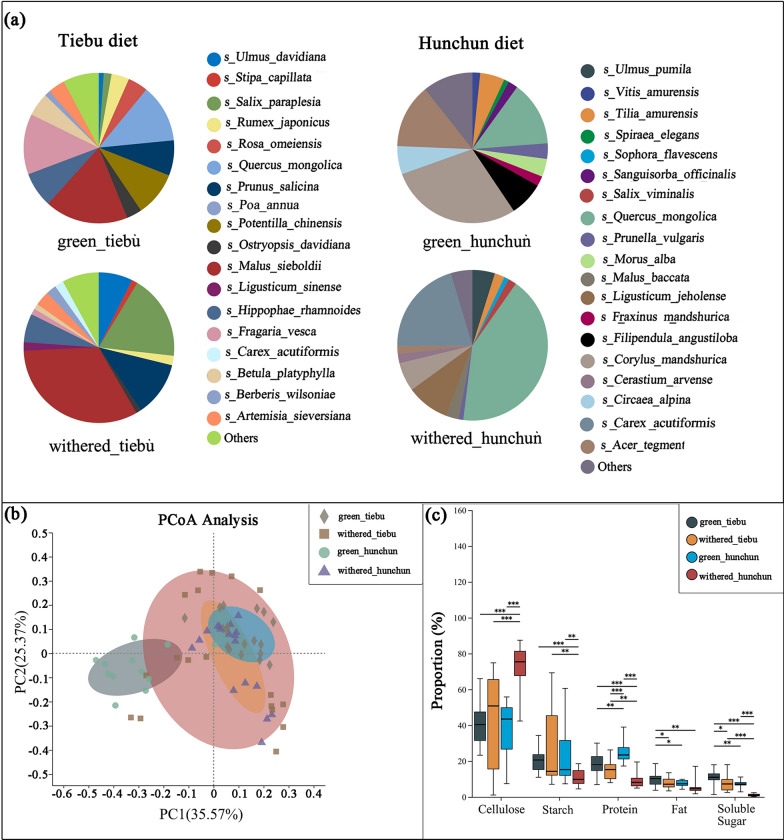
According to the morphophysiological feeding types of Cervidae, sika deer are considered to be an opportunistic adaptable selective [28]. Due to genetic and environmental influences, sika deer are often showed elastic in food habits. They showed browser type, intermediate type, and grazer type [50]. During the grass green period in the Tiebu and Hunchun areas, the sika deer showed a browser type. During the withered grass period in both areas, sika deer showed tend to intermediate type. Whether they showed browser type or intermediate type in food habits, the sika deer’s food choices need to meet its own energy needs. Main component of energy for ruminants is carbohydrate, protein, and lipid [67]. Therefore, we chose to measure protein, fat, starch, cellulose, soluble sugar in the main diet of sika deer as a nutritional evaluation. Based on the results of DNA metabarcoding, we selected plants that accounted for more than 10% of the food composition for nutritional composition determination (Fig. 2a, Supporting Information Table 2). Plant samples were collected from the Tiebu and Hunchun regions during the same phenological period from 2021 to 2022. Six samples of different plants of each type were collected. Herbs were collected from 6 different sites. Fresh plant leaves were stored and transported on dry ice and frozen at − 20 °C.
Fig. 2.
Differences in the nutritional status of wild sika deer in different regions and during different periods. a The diet composition of wild sika deer in different regions and from different periods (those plants accounting for less than 1% of the deer diets were combined in the Others category). b The results of PCoA of plant nutrition. c The Mann–Whitney U test results for the different nutrient components. *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001
Fresh plants were dried at 65 °C, ground over a 50-mesh screen, and then placed into an envelope. We weighed approximately 1 g of plant sample for crude protein and crude fat analysis. A 0.2–0.5 g sample was used for soluble sugar, starch, and cellulose content analysis. Crude protein was measured with the Kjeldahl nitrogen method; crude fat was measured with the Soxhlet extraction method; and soluble sugar, starches and cellulose were measured with the anthrone colorimetric method; measurements were performed using commercial kits (Solarbio).
Considering the difference in the proportion of plants in the diet, the nutrient content of each sample was calculated using the following formula:
Pi: the percentage of plants in the diet, Di: original concentration of nutrients.
All nutrient contents were based on corrected data. Differences in nutritional content were identified using the Mann–Whitney U test, and post hoc tests were performed using the Welch-uncorrect method. We also performed a principal coordinate analysis (PCoA) of the five nutrient components of 11 plant parts from different spatiotemporal in main diet of groups using the Bray‒Curtis distance. PCoA is usually used to find the main impact indicators in evaluating plant nutrition [39]. Additionally, it shows the similarities and differences in nutrient composition among plants [41]. In order to better see the differences between groups, we conducted the analysis based on groups. The importance of each nutrient component in the grouping was determined by a random forest model, and the data were standardized using the relative abundance [41]. We considered P < 0.05 to indicate statistical significance. The contents of the original nutrients in the major foods are shown in Supporting Information Table 2.
Microbiome analyses
Gut microbiota community analyses
20 faecal samples (green season group included female 7 and male 3; withered season group included female 5 and male 5) from the Tiebu region and 16 faecal samples (green season group included female 4 and male 4; withered season group included female 4 and male 4) from the Hunchun region were used to gut microbiota community analyses. The primers 338F and 806R (Supporting Information Table 1) for the 16S rRNA gene were selected to amplify the variable region of bacterial V3-V4. The PCR parameters were as follows: predenaturation for 3 min at 95 °C; 27 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s; and a final extension for 10 min at 72 °C. The follow-up experiments were carried out only under the conditions of the correct size and suitable concentration of PCR products. The library was constructed and sequenced on the Illumina MiSeq platform (Shanghai Majorbio Biopharm Technology Co., Ltd.). We used fastp (v0.19.6) to quality control, FLASH (version 1.2.7) to merge reads, and the DADA2 method to denoise the data, obtain the ASV sequence, and flatten it according to the minimum number of sequences [10]. The effective ASV sequences were annotated by the Bayes method, and the annotation database used was the silva138/16s_bacteria database. The classification confidence of the species annotation method was 0.7. We focused on the differences in the microbial community composition between the green-grass period and the withered-grass period, calculated the α diversity and β diversity of the microbial community in the two periods, and analysis of similarities (ANOSIM) was used to judge whether the difference between groups was significant. The Mann‒Whitney U test was used to test the difference in genus-level species composition between the green-grass period and the withered period.
Metagenomic analyses
18 faecal samples (green season group included female 7 and male 3; withered season group included female 4 and male 4) from the Tiebu region and 13 faecal samples (green season group included female 3 and male 3; withered season group included female 4 and male 3) from the Hunchun region were used to Metagenomic analyses. Metagenomic sequencing was performed with fastp software for quality control. Reads with a length of less than 50 bp and an average weight less than 20 after mass shearing and reads containing N bases were removed, while high-quality paired-end reads and single-end reads were retained. A single-Megahit strategy was adopted for short-segment sequence assembly. Using the succinct de Bruijn graph method, the stitching parameters were iteratively stitched from small k-mers to large k-mers. MetaGene was used to predict ORFs in contigs in the splices, and genes with nucleic acid lengths greater than or equal to 100 bp were selected and translated into amino acid sequences to obtain a statistical table of gene prediction results for each sample.
CD-HIT software was used for clustering (default parameters: 90% identity and 90% coverage), and the longest gene of each class was taken as the representative sequence to construct the nonredundant gene set [19]. Using SOAPaligner software, the high-quality reads of each sample were compared with the nonredundant gene set (default parameter: 95% identity) to collect information on the abundance of genes in the corresponding sample [43]. Diamond software was used to compare the sequences of nonredundant gene sets with various databases to obtain species and functional annotations, in which the BLASTP parameter was set as an E-value ≤ 1e-5 [8, 9]. In this study, the gene sets were compared and annotated according to the NR database to obtain the species information. The species annotation method was the best hit method. The microbial functional annotation results were analysed using the KEGG database [36]. The pathogenic bacteria annotation results were analysed using the PHI database (Pathogen Host Interactions Database) [66]. The CARD database (Comprehensive Antibiotic Resistance Database) was used to annotation the resistance mechanism gene [34]. The Kruskal Wallis H test was used to test the difference groups, post hoc tests were used with the Welch-uncorrect method, and the confidence interval was 0.95. The Mann‒Whitney U test as used to test in two groups. We also used DESeq2 analysis for the gut microbiota function. The threshold for multiple differences in gene expression was established at a factor of 2.We considered P < 0.05 to indicate statistical significance. We used Benjamini–Hochberg multiple correction method. Differences in pathogenic bacteria among the groups were analyzed using LEfSe differential discriminant analysis, with a linear discriminant analysis (LDA) threshold set at greater than 3. Microbiome sequence processing and analyses were performed on the online Majorbio Cloud Platform (www.majorbio.com) [59].
Results
Diet composition
A total of 35 faecal samples from the Tiebu region were successfully typed at the psbCL locus. A total of 531,199 sequence reads were obtained, from which we identified 213 ASVs. Among them, 197 ASVs met the identification criteria, and a total of 107 plant taxa were identified, including 43 families, 93 genera, and 105 species. In the green-grass period, 81 plant taxa were identified, including 30 families, 76 genera, and 79 species. In the withered grass period, 78 plant taxa were identified, including 31 families, 73 genera, and 77 species. We chose to rarefy our sampling depth at 9449 sequences (per sample) to equalize the sampling depth across all samples in the Tiebu region. Based on the mean sequence calculation, we found that the plants that accounted for more than 10% of the diets were Malus sieboldii (17.83%), Fragaria vesca (13.01%), and Quercus mongolica (12.81%) during the green grass period and Malus sieboldii (32.64%), Salix paraplesia (18.19%), and Prunus salicina (11.76%) during the withered grass period (Fig. 2a).
A total of 37 faecal samples were successfully collected at the psbCL locus in the Hunchun region. A total of 577,710 sequence reads were obtained, from which we obtained 264 ASVs. Among them, 156 ASVs met the identification criteria, and a total of 92 plant taxa were identified, including 45 families, 86 genera, and 91 species. In the green-grass period, 80 plant taxa were identified, including 41 families, 75 genera, and 79 species. In the withered grass period, 45 plant taxa were identified, including 26 families, 43 genera, and 45 species. We chose to rarefy our sampling depth at 10,112 sequences (per sample) to equalize the sampling depth across all the samples in the Hunchun region. Based on the mean sequence calculation, we found that the plants that accounted for more than 10% of the diets were Corylus mandshurica (28.96%), Quercus mongolic (13.81%), and Acer tegmentosum (13.70%) in the green grass period and Quercus mongolic (42.20%) and Carex dispalata (20.64%) in the withered grass period (Fig. 2a).
Nutritional analyses of food components
PCoA showed that there was a greater difference in nutrient availability between the green-grass period and the withered-grass period in the Hunchun region than in the Tiebu region (Fig. 2b). The results of the random forest model showed that proteins, cellulose and soluble sugars were the nutritional components with major differences among the groups (Supporting Information Fig. 1). The Mann–Whitney U test showed that there was a significant difference between the groups (Fig. 2c). We focused on the differences in protein, cellulose and soluble sugar levels, which are the major factors influencing nutrition (Supporting Information Fig. 2). There was a significant difference in the protein content between the green-grass period and the withered-grass period in Hunchun (P < 0.001), and the protein content was greater during the green-grass period. However, there was no significant difference in the Tiebu region. There was significantly more protein in the green grass period in Hunchun than in Tiebu (P < 0.01). However, in the withered grass period, the protein content in Tiebu was significantly greater than that in Hunchun (P < 0.01). The cellulose content in the withered grass period in Hunchun was significantly greater than that in Tiebu (P < 0.001), and that in the Hunchun in the withered grass period was significantly greater than that in the Hunchun in the green grass period (P < 0.001). The soluble sugar content in Tiebu during both the withered grass period (P < 0.001) and the green grass period (P < 0.01) was greater than that in Hunchun. The soluble sugar content in Tiebu during the green-grass period was greater than that in Tiebu during the withered-grass period (P < 0.05), and similar results were obtained for Hunchun (P < 0.001). The differences in fat and starch between the groups were also analysed (Supporting Information Fig. 2).
Gut microbiota structure
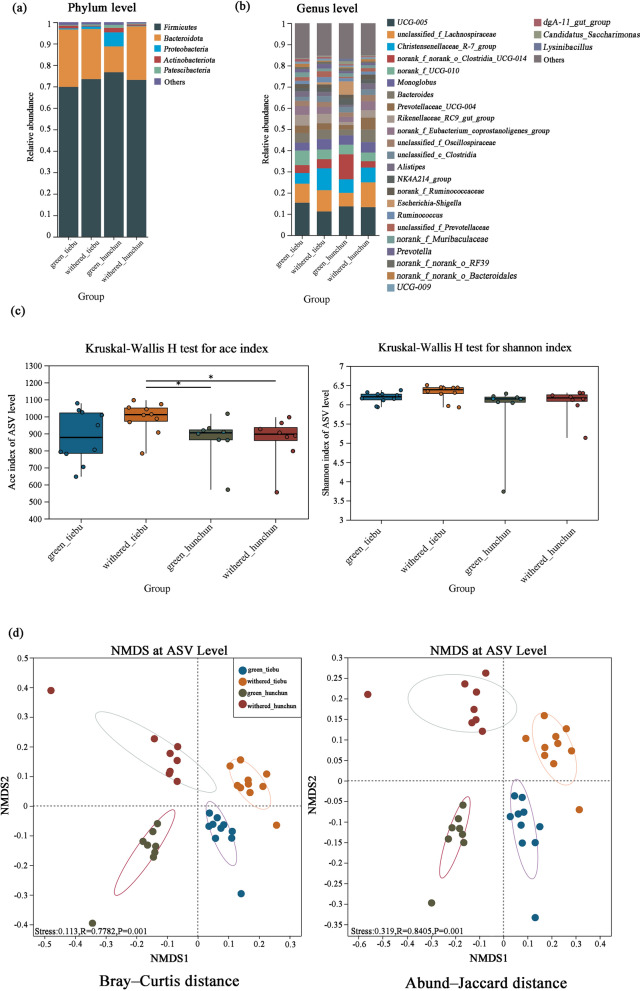
A total of 36 faecal samples were successfully amplified. A total of 1,902,222 sequence reads were obtained, from which we identified 8692 ASVs, 302 genera, 140 families, 31 classes, and 19 phyla. We chose to rarefy our sampling depth at 18,909 sequences (per sample) to equalize the sampling depth across all samples. The major phyla of all the groups were Firmicutes and Bacteroidetes. Firmicutes was most abundant in Hunchun in the green grass period (73.69%), moderately abundant in Hunchun in the withered grass period (73.11%) and Tiebu in the withered grass period (73.49%), and least abundant in Tiebu in the green grass period (69.84%). Bacteroidetes was most abundant in Tiebu in the green grass period (26.74%), moderately abundant in Tiebu in the withered grass period (23.30%) and Hunchun in the withered grass period (24.97%), and least abundant in Hunchun in the green grass period (12.02%) (Fig. 3a). Oscillospiraceae and Lachnospiraceae were the major families of the microbiome in every group (Supporting Information Fig. 3). At the genus level, UCG-005, unclassified_f_Lachnospiraceae, Christensenellaceae_R-7_group, norank_f_norank_o_Clostridia_UCG-014, norank_f_UCG-010, and Monoglobus were the major taxa (Fig. 3b).
Fig. 3.
Gut microbiota community and diversity dynamics. a The dominant phyla with less than 0.01% abundance were combined into the Others category. b The dominant genera with less than 0.01% abundance were merged into the Others category. c Alpha diversity. d NMDS analysis of beta diversity
The Tiebu withered grass group had the greatest alpha diversity, as revealed by the observed ASVs, the ACE index and the Shannon index (Fig. 2c). The ACE index was greater in the Tiebu withered grass group than in the Hunchun withered grass group (P ≤ 0.05). There was no significant difference in the Shannon index between the groups (Fig. 3c). NMDS analysis of beta diversity confirmed that the differences of microbiome in each group, Bray‒Curtis distance (ANOSIM: R = 0.7782, stress < 0.02, P < 0.005; Fig. 3d) and Abund‒Jaccard distance (ANOSIM: R = 0.8405, stress < 0.02, P < 0.005; Fig. 3d). We also performed ANOSIM during the withered grass and green grass periods in the two regions based on the Abund‒Jaccard index and found that the difference between the green grass period and the withered grass period in the Hunchun (ANOSIM: R = 0.77, P < 0.005) area was greater than that in Tiebu (ANOSIM: R = 0.71, P < 0.005). ANOSIM of the Abund‒Jaccard index results between different regions in the same season revealed that the difference between the two regions during the green grass period (ANOSIM: R = 0.92, P < 0.005) was greater than that during the withered grass period (ANOSIM: R = 0.81, P < 0.005) (Fig. 3e). The Mann–Whitney U test revealed that the abundances of 64 genera were significantly different between the Hunchun and Tiebu regions during the green grass period, while those of 45 genera were significantly different between the two regions during the withered grass period.
Gut microbiome function
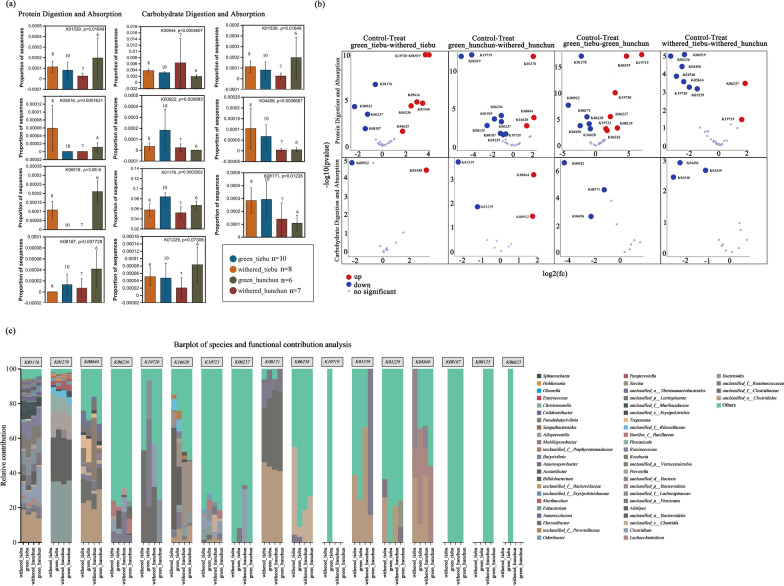
The nutritional differences between the spatiotemporal groups were considered. We focused on two metabolic pathways (protein digestion and absorption pathway: ko04974 and carbohydrate digestion and absorption pathway: ko04973) and the gut microbial genes encoding cellulose-related enzymes. In the protein digestion and absorption pathway, the genes K01539, K05616, K06519, and K08187 were significantly different according to both the Kruskal Wallis H test and DESeq2 analysis (Fig. 4a, b). In the carbohydrate digestion and absorption pathway, the genes K00844, K00922, K01539, K04456, and K08171 also exhibited significant differential expression in both the Kruskal Wallis H test and DESeq2 analysis (Fig. 4a, b). The composition of the bacterial community encoding these genes is shown in Fig. 4c.
Fig. 4.
Genes with significant differences in the protein absorption and metabolism pathways and the carbohydrate metabolic absorption pathway. a The Kruskal Wallis H test of difference groups. *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001. b DESeq2 analysis of the groups in pairs. c The abundances of the top 50 major contributing bacteria at the genus level (KO on ko04974 pathway and ko04973 pathway)
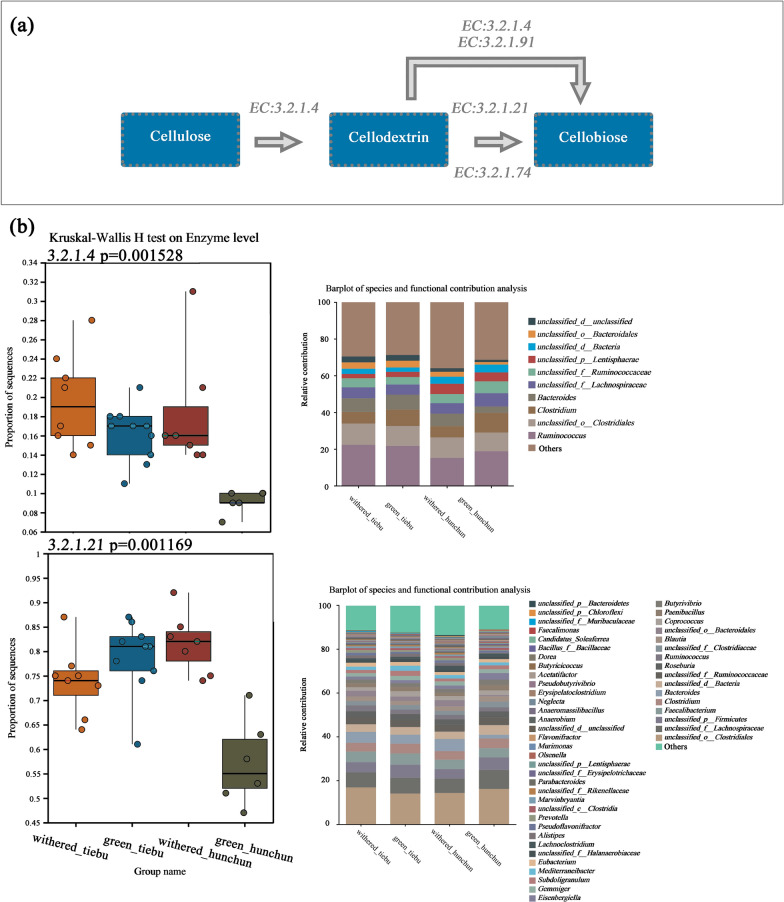
In terms of cellulase enzyme function, we observed significant differences in the levels of endoglucanase (EC:3.2.1.4) and beta-glucosidase (EC:3.2.1.21) between the groups (Fig. 5b). The differences for each pair of groups are illustrated in Supporting Information Fig. 4. There were more genes encoding endoglucanase in the Hunchun withered grass group than in the Hunchun green grass group (P < 0.01). The gene encoding endoglucanase was also more abundant in the Tiebu green grass group than in the Hunchun green grass group (P < 0.001). The beta-glucosidase activity in the Hunchun withered grass group was greater than that in the Hunchun green grass group (P < 0.001), that in the Hunchun withered grass group was greater than that in the Tiebu withered grass group (P < 0.05), and that in the Tiebu green grass group was greater than that in the Hunchun green grass group (P < 0.001). For cellulose 1,4-beta-cellobiosidase (EC:3.2.1.91), there were no significant differences between the groups (P > 0.05).
Fig. 5.
Spatiotemporal differences in the cellulase enzyme function of the gut microbiome. a Cellulose metabolic pathway. b Functional differences in enzymes related to cellulose metabolism and the abundances of the top 10 major contributing bacteria at the genus level
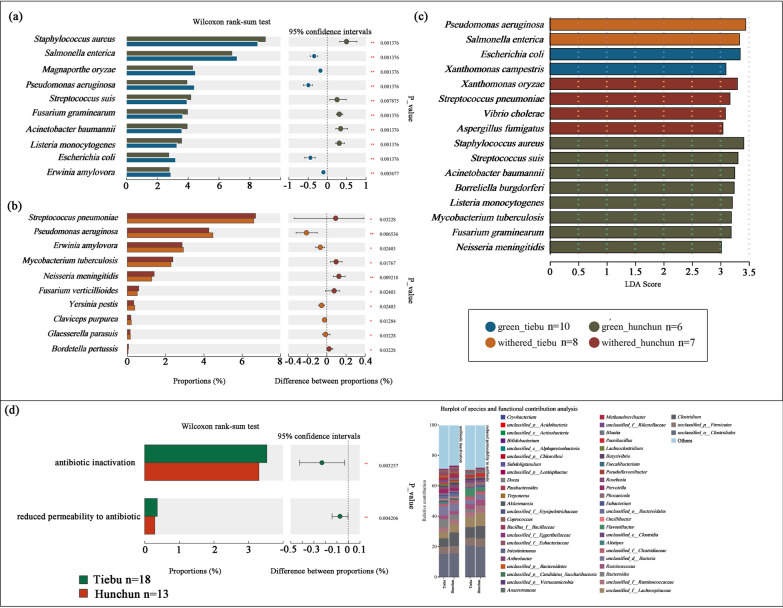
LEfSe differential discriminant analysis revealed that Pseudomonas aeruginosa and Salmonella enterica were enriched in the Tiebu withered grass group. Escherichia coli and Xanthomonas campestris were found to be enriched in the Tiebu green grass group. In the Hunchun withered grass group, Xanthomonas oryzae, Streptococcus pneumoniae, Vibrio cholerae, and Aspergillus fumigatus were identified as enriched species. The Hunchun green grass group exhibited enrichment of Staphylococcus aureus, Streptococcus suis, Acinetobacter baumannii, Borreliella burgdorferi, Listeria monocytogenes, Mycobacterium tuberculosis, Fusarium graminearum, and Neisseria meningitidis (Fig. 6c). The differences between the Tiebu and the Hunchun during the green grass and dry grass periods were shown in Fig. 6a and b. There were differences in drug resistance mechanisms between Tiebu group and Hunchun group. Tiebu group was enriched on the antibiotic inactivation and reduced permeability to antibiotic genes (P < 0.005) (Fig. 6d).
Fig. 6.
Spatiotemporal differences in pathogenic bacteria. a The Mann‒Whitney U test of grass period between Tiebu and Hunchun. *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001. b The Mann‒Whitney U test of withered period between Tiebu and Hunchun. *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001. c Differences in pathogenic bacteria among the groups were analyzed using LEfSe differential discriminant analysis. d Differences in resistance mechanism between Tiebu group and Hunchun group, and the abundances of the top major contributing bacteria at the genus level
Discussion
Firmicutes and Bacteroides were the major components of the gut microbiota at the phylum level in wild and captive sika deer in Northeast China. At the genus level, UCG-005, UCG-010, Christensenellaceae_R-7_group, Bacteroides, UCG-013 and other genera constitute the dominant bacteria in the gut microbiota of Sika deer [24]. Our results on the gut microbiota community composition in the two regions are similar to those of the above studies. The composition of the gut microbiome is affected by phylogeny, and allopatric populations of the same species often exhibit commonality in the gut microbiome, which is the result of the coevolution of the mammalian gut microbiome with its host [80]. However, different geographical locations can change the dynamic composition of the gut microbiota community, and biogeography can increase the diversity of the gut microbiota [4, 44]. The gut microbial communities of nearby hosts are more similar to those of nearby hosts than to those of distant hosts [23, 58]. Our PCoA showed that the compositions of gut microbiota communities in the same region were similar even in different phenological periods. Environments can lead to changes in the microbiome of animals. The habitats of sika deer in the Tiebu and Hunchun regions differ significantly. The plant compositions in these two areas are entirely distinct, resulting in varying food sources available for sika deer. Additionally, there are considerable altitude differences and varying levels of human interference between the two regions, which may influence the structure and function of their microbial communities.
Among the influencing factors, forage habits greatly influence the gut microbiome [68]. Gut microbiota community of herbivores is highly species specific and is affected by phylogeny, and the dominant flora of the gut microbiome of the same species are usually similar, but diet type is a factor affecting the variation in the gut microbiome structure [61]. DNA barcoding revealed that there were more species in the diet of sika deer in Tiebu in the withered period than in Hunchun in the withered period, and the ACE index of the gut microbiota was greater in the Tiebu withered grass group than in the Hunchun withered grass group. The ACE index reflects the community richness of the gut microbiota community; the greater the ACE index is, the greater the number of species in the community. A study revealed that generalist herbivore gut microbiota diversity is affected by dietary species richness and that the alpha diversity of the gut microbiota increases linearly with dietary species richness [40]. According to the feeding characteristics and morphological and structural changes in the stomach, ruminants are divided into three nutrient adaptation types: grazer, browser and mixed-fed [29]. As mixed feeders, sika deer can change their diet according to phenological changes. During the green grass period, sika deer in the two regions could obtain rich food, so there was no significant difference in the alpha diversity of the gut microbiota; however, during the withering grass period, the Tiebu area had a more complex habitat than did the Hunchun area. For beta diversity, there is separation between different groups. However, the difference between the two periods in Hunchun groups is greater than that in the Tiebu groups. During the withering grass period, sika deer in the Tiebu area could obtain more kinds of food, resulting in more abundant microbial species compared with those in the Hunchun area. The difference in food availability between the two phenological periods in Hunchun may be the reason for the result. In addition, the altitude difference between the two regions may also affect the alpha diversity. The Tiebu region is located on the border of the Qinghai-Tibet Plateau, belongs to the plateau area, whereas the Hunchun region is located in a plain area. Studies have shown that the alpha diversity of animal gut microbiota is higher at elevated altitudes [83].
Because of the differences in the types of major foods consumed, there are also differences in the nutritional intake of wild sika deer between the two regions. Many studies have shown the effects of food nutrition on the microbial community and microbial function. For example, porcupines (Erethizon dorsatum), which eat lignified plant material, have relatively high proportions of genes encoding cellulose-degrading enzymes [18]. In this study, we focused on crude protein, soluble sugars and cellulose. There were significant differences in crude protein content between the different groups, and the crude protein content in the green grass period was generally greater than that in the withered grass period in the two regions. The crude protein content in Hunchun during the green-grass period was significantly greater than that in the other groups. In terms of gut microbiota function, we found that the K01539, K06519, and K08187 genes were significantly enriched in the Hunchun green grass group, while the K01539, K05616, and K06519 genes were enriched in the Tiebu withered grass group, which is similar to the overall trend of differences in nutrition between the groups. The K05616, K06519, and K08187 genes all participate in alanine, aspartate and glutamate metabolism. The K05616, K06519, and K08187 genes belong to the solute carrier family. K01539 encodes sodium/potassium-transporting ATPase subunit alpha [36]. The major amino acids absorbed in ruminant intestines are microbial proteins (MCPs), rumen undegradable proteins (RUPs) and amino acids derived from the digestion of endogenous proteins. The gut absorbs large amounts of free amino acids; this process depends mainly on transport against the concentration gradient and depends on the pump transport system for Na+ and other ions [46]. Atkinson [7] reported that with increasing RUP levels, the content of amino acids entering the small intestine increases; therefore, a greater RUP content in the diet increases the absorption of amino acids in the small intestine. There is no way to determine the specific content of RUPs in crude protein, but the genes related to protein digestion and absorption pathway digestion in the microbiome are consistent with the change trend of crude protein among different groups. The differential expression of genes encoding components involved in amino acid transport are likely adaptations of the microbiome to differences in protein content in food during different seasons.
The results of the carbohydrate digestion and absorption pathway showed that there were differences in the abundance of related genes, such as K00844, which encodes hexokinase, which significantly increased during the withering grass period in the Hunchun region. Hexokinase (HK) is one of the rate-limiting enzymes in the glycolytic pathway and catalyses the phosphorylation of hexose to glucose-6-phosphate. Glucose-6-phosphate is an important intermediate in the process of glucose metabolism; it generates pyruvate and enters the tricarboxylic acid cycle, which is important for regulating energy metabolism [73]. During the withering period, the proportion of cellulose, which accounted for the largest proportion of nutrient components, increased significantly. In the cellulose metabolism pathway, the end product of cellulose metabolism is D-glucose. The increase in the content of hexokinase genes may be attributed to the increase in reactive substrates caused by metabolites, and the body needs more energy to cope with the harsh winter environment during the withered grass season. The above results may indicate that in the face of different environmental differences, the gut microbiome of wild sika deer responds faster at the genetic level to help it adapt to different nutritional differences at different times and in different locations.
Endoglucanase (EC3.2.1.4), a cellulose-degrading enzyme, can act on the amorphous region in cellulose, and it can randomly hydrolyse β-1,4-glucoside bonds to decompose long-chain cellulose molecules into fibrodextrin, fibrodisose and glucose. An exonuclease (EC3.2.1.91) can act on the ends of cellulose molecules and can degrade crystalline cellulose and hydrolysate β-1,4-glucoside bonds. β-Glucosidase (EC3.2.1.2) can further decompose cellodisose, cellotriose and other low-molecular-weight dextrins to glucose. The cellulase abundance increased significantly during the withering grass period in both regions, which was consistent with the winter diet. The difference between the withered and green grass periods in the Hunchun region was greater than that in the Tiebu region and consistent with the differences in the local environment. Compared with that in the Tiebu area, the vegetation composition in the Hunchun area is simpler, consisting mainly of deciduous broad-leaved forest. Every winter, the variety of food decreased, the amount of available food for wild sika deer decreased, and only Cyperaceae was consumed as a staple food. Through these experiments, we found that Cyperaceae has a high cellulose content. Changes in enzyme function can well reflect the interactions between the gut microbiome and diet. For example, the level of amylase activity in the digestive system of giant pandas increases, and giant pandas have a good ability to digest this nutrient, which is also reflected in the function of the gut microbiome [81, 85].
We also looked at differences in pathogen bacteria and drug resistance mechanisms genes in groups. Human activities can promote the growth of Escherichia coli in the environment [49]. Escherichia coli was enriched in the Tiebu group, it may because Tiebu region is a pastoral area, the sika deer has more opportunity to contact with human. Salmonella enterica and Escherichia coli, both of which have Bos and humans as hosts, are enriched in the Tiebu region, suggesting a potential risk of disease transmission [66]. In fact, we have observed close contact between sika deer and livestock in Tiebu area several times. Hunchun area due to its status as a border control zone and few residents in the core area, we have not observed sika contact with human or livestocks (Supporting Information Fig. 5). LefSe analysis showed that more pathogenic bacteria were significantly enriched in the Hunchun area. It may be due to differences in vector organisms. Ticks, which serve as vectors for multiple pathogens and viruses, are more diverse in the Hunchun area compared to the Tiebu area [53, 82]. Additionally, it is important to note that the Tiebu region exhibits significantly more genes associated with antibiotic resistance mechanisms than the Hunchun region, a discrepancy likely attributable to human activities [33].
In conclusion, this is the first study to comprehensively consider the effects of both seasonal and geographical factors on the gut microbiome of wild sika deer in different regions. The seasonal changes in food availability affect the nutrient intake of sika deer, leading to differences in the gut microbiome composition and function. At the same time, human activities pose potential health risks to animals.
Supplementary Information
Acknowledgements
Thanks to Haitao Yang, Qiang Dai, Baowei Zhang, Chengcheng Zhang, Tiebu Nature Reserve and National Tiger and Leopard Park for their assistance in collecting samples.
Author contributions
Y.N. conceived and designed the study; Y.L. and Y.N. secured funding; X.M., K.L., W.W., W.J., J.L., Y.C. and Y.M. collected the samples; X.M., X.H. and Y.G. carried out the laboratory work; X.M., M.L. and Y.L. conducted the analysis; X.M. wrote the manuscript; all authors revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32225033, 32071496, 32100399) and the Ministry of Science and Technology of China (No. 2022YFF1301500).
Availability of data and materials
Data is provided within the manuscript or supplementary information files.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Amato KR. Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microb Sci Med. 2013;1:10–29. [Google Scholar]
- 2.Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, et al. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouattapigra). Microb Ecol. 2014;69:434–43. [DOI] [PubMed] [Google Scholar]
- 3.Amato KR, Kuthyar S, Ekanayake-Weber M, Salmi R, Snyder-Mackler N, Wijayathunga L, et al. Gut microbiome, diet, and conservation of endangered langurs in Sri Lanka. Biotropica. 2020;00:1–10. [Google Scholar]
- 4.Andersen-Ranberg E, Barnes C, Rasmussen L, Salgado-Flores A, Grøndahl C, Mosbacher J, et al. A comparative study on the faecal bacterial community and potential zoonotic bacteria of muskoxen (Ovibosmoschatus) in Northeast Greenland, Northwest Greenland and Norway. Microorganisms. 2018;6:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antwis RE, Edwards KL, Unwin B, Walker SL, Shultz S. Rare gut microbiota associated with breeding success, hormone metabolites and ovarian cycle phase in the critically endangered eastern black rhino. Microbiome. 2019;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aricha H, Simujide H, Wang C, Zhang J, Lv W, Jimisi X, et al. Comparative analysis of fecal microbiota of grazing Mongolian cattle from different regions in Inner Mongolia, China. Animals. 2021;11:1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkinson RL, Toone CD, Ludden PA. Effects of supplemental ruminally degradable protein versus increasing amounts of supplemental ruminally undegradable protein on site and extent of digestion and ruminal characteristics in lambs fed low-quality forage1. J Anim Sci. 2007;85:3322–30. [DOI] [PubMed] [Google Scholar]
- 8.Buchfink B, Reuter K, Drost H-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18:366–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12(59–60):9. [DOI] [PubMed] [Google Scholar]
- 10.Callahan B, McMurdie P, Rosen M, Han A, Johnson A, Holmes S. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candela M, Biagi E, Maccaferri S, Turroni S, Brigidi P. Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol. 2012;20:385–91. [DOI] [PubMed] [Google Scholar]
- 12.Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C-W, Huang B-H, Lin S-M, Huang C-L, Liao P-C. Changes of diet and dominant intestinal microbes in farmland frogs. BMC Microbiol. 2016;16:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi X, Gao H, Wu G, Qin W, Song P, Wang L, et al. Comparison of gut microbiota diversity between wild and captive bharals (Pseudoisnayaur). BMC Vet Re. 2019;15:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, et al. Captivity humanizes the primate microbiome. Proc Natl Acad Sci. 2016;113:10376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl S-A, Hudler M, Windisch W, Bolduan C, Brugger D, König A. High fibre selection by roe deer (Capreoluscapreolus): evidence of ruminal microbiome adaption to seasonal and geographical differences in nutrient composition. Anim Prod Sci. 2020;60:1303–14. [Google Scholar]
- 17.DeCandia AL, Brenner LJ, King JL, vonHoldt BM. Ear mite infection is associated with altered microbial communities in genetically depauperate Santa Catalina Island foxes (Urocyonlittoralis catalinae). Mol Ecol. 2020;29:1463–75. [DOI] [PubMed] [Google Scholar]
- 18.Finlayson-Trick ECL, Getz LJ, Slaine PD, Thornbury M, Lamoureux E, Cook J, et al. Taxonomic differences of gut microbiomes drive cellulolytic enzymatic potential within hind-gut fermenting mammals. PLoS ONE. 2017;12:e0189404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao H, Chi X, Qin W, Wang L, Song P, Cai Z, et al. Comparison of the gut microbiota composition between the wild and captive Tibetan wild ass (Equuskiang). J Appl Microbiol. 2019;126:1869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobet A, Mest L, Perennou M, Dittami SM, Caralp C, Coulombet C, et al. Seasonal and algal diet-driven patterns of the digestive microbiota of the European abalone Haliotistuberculata, a generalist marine herbivore. Microbiome. 2018;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene LK, Blanco MB, Rambeloson E, Graubics K, Fanelli B, Colwell RR, et al. Gut microbiota of frugo-folivorous sifakas across environments. Anim Microb. 2021;3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths SM, Antwis RE, Lenzi L, Lucaci A, Behringer DC, Butler MJ, et al. Host genetics and geography influence microbiome composition in the sponge Ircinia campana. J Anim Ecol. 2019;88:1684–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan Y, Yang H, Han S, Feng L, Wang T, Ge J. Comparison of the gut microbiota composition between wild and captive sika deer (Cervusnippon hortulorum) from feces by high-throughput sequencing. AMB Express. 2017;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo R, Zhang W, Shen W, Zhang G, Xie T, Li L, et al. Analysis of gut microbiota in chinese donkey in different regions using metagenomic sequencing. BMC Genom. 2023;24:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y. Study on the food habits of Sichuan sika deer (Cervusnippon Sichuanicus). Sichuan Teach Coll Nat. 2001;22:112–9. [Google Scholar]
- 27.Guo Y, Zheng H. On the geological distribution, taxonomic status of species and evolutionary history of sika deer in China. Acta Theriologica Sinica. 2000;3:168–79. [Google Scholar]
- 28.Hofmann RR. Digestive physiology of deer. Royal Soc N Z Bull. 1985;22:393–407. [Google Scholar]
- 29.Hofmann RR, Stewart DRM. Grazer of browser: a classification based on the stomach-structure and feeding habits of East African ruminants. Mammalia. 1972;36:226–40. [Google Scholar]
- 30.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. [DOI] [PubMed] [Google Scholar]
- 32.Hume ID. Digestive strategies of mammals. Acta Zool Sinica. 2002;48:1–19. [Google Scholar]
- 33.Ishibashi S, Sumiyama D, Kanazawa T, Murata K. Prevalence of antimicrobial-resistant Escherichiacoli in endangered Okinawarail (Gallirallusokinawae) inhabiting areas around a livestock farm. Vet Med Sci. 2019;00:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia B, Raphenya RA, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. (2016) [DOI] [PMC free article] [PubMed]
- 35.Just S, Mondot S, Ecker J, Wegner K, Rath E, Gau L, et al. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome. 2018;6:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohl KD, Weiss RB, Cox J, Dale C, Denise Dearing M, van Dam N. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol Lett. 2014;17:1238–46. [DOI] [PubMed] [Google Scholar]
- 38.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Li Z, Xing Y. Analysis on nutrients component of sixteen functional plants leaves in Daxing’an Mountains. For Eng. 2019;35(1):29–35. [Google Scholar]
- 40.Li G, Shi C, Song Y, Chu H, Zhang Z, Faust K. The role transition of dietary species richness in modulating the gut microbial assembly and postweaning performance of a generalist herbivore. mSystems. 2021;6:e00979-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G, Yin B, Li J, Wang J, Wei W, Bolnick DI, Wan X, et al. Host-microbiota interaction helps to explain the bottom-up effects of climate change on a small rodent species. ISME J. 2020;14:1795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Fei HL, Luo ZH, Gao SM, Wang PD, Lan LY, et al. Gut microbiome responds compositionally and functionally to the seasonal diet variations in wild gibbons. NPJ Biofilms Microb. 2023;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. [DOI] [PubMed] [Google Scholar]
- 44.Linnenbrink M, Wang J, Hardouin EA, Künzel S, Metzler D, Baines JF. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22:1904–16. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, Yang G, Wei F, Li M, Hu J. Sequence variability of the mitochondrial DNA control region and population genetic structure of sika deers (Cervusnippon) in China. Acta Zool Sinica. 2003;49(1):53–60. [Google Scholar]
- 46.Liu J. Nutritional physiology of ruminants. Beijing: China Agriculture Press; 2019. [Google Scholar]
- 47.Liu J, Liang X, Liu Y. Comparison of the gut microbiota composition between captive and wild roe deer. BioRxiv. 2019;11:831222. [Google Scholar]
- 48.Liu W, Wang Q, Song J, Xin J, Zhang S, Lei Y, et al. Comparison of gut microbiota of yaks from different geographical regions. Front Microbiol. 2021;12:666940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo Q, Tong F, Tao X, Cao W, Fu L, Zhu H. Effects of tourism disturbance on the habitat and water quality for Andriasdavidianus in Zhangjiajie, Hunan, China. Chin J Appl Ecol. 2019;30:2101–8. [DOI] [PubMed] [Google Scholar]
- 50.McCullough., D. R., Takatsuki, S. & Kaji., K. (2009). Geographical variations in food habits of sika deer: the northern grazer vs. the southern browser. In: Sika deer biology & management of native & introduced populations, pp. 231–2.
- 51.McManus N, Holmes SM, Louis EE, Johnson SE, Baden AL, Amato KR. The gut microbiome as an indicator of habitat disturbance in a Critically Endangered lemur. BMC Ecol Evol. 2021;21:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc Natl Acad Sci. 2017;114:13768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mu C, Zhao N, Luo G, Ma C, Tian Y, Guo J, et al. Report on main vectors surveillance at Hunchun port. Chin Front Health Quar. 2015;38:30–3. [Google Scholar]
- 54.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nie Y, Speakman JR, Wu Q, Zhang C, Hu Y, Xia M, et al. Exceptionally low daily energy expenditure in the bamboo-eating giant panda. Science. 2015;349:171–4. [DOI] [PubMed] [Google Scholar]
- 56.Pan B, Han X, Yu K, Sun H, Mu R, Lian CA. Geographical distance, host evolutionary history and diet drive gut microbiome diversity of fish across the Yellow River. Mol Ecol. 2022;32:1183–96. [DOI] [PubMed] [Google Scholar]
- 57.Pennisi E. How do gut microbes help herbivores? Counting the ways. Science. 2017;355:236–236. [DOI] [PubMed] [Google Scholar]
- 58.Ren T, Boutin S, Humphries MM, Dantzer B, Gorrell JC, Coltman DW, et al. Seasonal, spatial, and maternal effects on gut microbiome in wild red squirrels. Microbiome. 2017;5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren Y, Yu G, Shi C, Liu L, Guo Q, Han C, et al. Majorbio cloud: a one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta. 2022;1:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 2011;39:e145–e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rojas CA, Ramírez-Barahona S, Holekamp KE, Theis KR. Host phylogeny and host ecology structure the mammalian gut microbiota at different taxonomic scales. Anim Microb. 2021;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommer F, Ståhlman M, Ilkayeva O, Arnemo JM, Kindberg J, Josefsson J, et al. The gut microbiota modulates energy metabolism in the hibernating brown bear Ursusarctos. Cell Rep. 2016;14:1655–61. [DOI] [PubMed] [Google Scholar]
- 63.Sugden S, Sanderson D, Ford K, Stein LY, St Clair CC. An altered microbiome in urban coyotes mediates relationships between anthropogenic diet and poor health. Sci Rep. 2020;10:22207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tilman D, Clark M, Williams DR, Kimmel K, Polasky S, Packer C. Future threats to biodiversity and pathways to their prevention. Nature. 2017;546:73–81. [DOI] [PubMed] [Google Scholar]
- 65.Trosvik P, de Muinck EJ, Rueness EK, Fashing PJ, Beierschmitt EC, Callingham KR, et al. Multilevel social structure and diet shape the gut microbiota of the gelada monkey, the only grazing primate. Microbiome. 2018;6:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urban M, Cuzick A, Seager J, Wood V, Rutherford K, Venkatesh SY, et al. PHI-base in 2022: a multi-species phenotype database for pathogen-host interactions. Nucleic Acids Res. 2022;50:D837–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Soest PJ. Nutritional ecology of the ruminant. 2nd ed. Ithaca: Cornell University Press; 1994. [Google Scholar]
- 68.Wang L, Ding J, Yang Z, Chen H, Yao R, Dai Q, et al. Père David’s deer gut microbiome changes across captive and translocated populations: implications for conservation. Evol Appl. 2019;12:622–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Huang G, Hou R, Qi D, Wu Q, Nie Y, et al. Multi-omics reveals the positive leverage of plant secondary metabolites on the gut microbiota in a non-model mammal. Microbiome. 2021;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Wu X, Shang Y, Gao Y, Li Y, Wei Q, et al. High-altitude drives the convergent evolution of alpha diversity and indicator microbiota in the gut microbiomes of ungulates. Front Microbiol. 2022a;13:953234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Zhang C, Li G, Yi X. The influence of species identity and geographic locations on gut microbiota of small rodents. Front Microbiol. 2022b;13:983660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watson SE, Hauffe HC, Bull MJ, Atwood TC, McKinney MA, Pindo M, et al. Global change-driven use of onshore habitat impacts polar bear faecal microbiota. ISME J. 2019;13:2916–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson JE. An introduction to the isoenzymes of mammalian hexokinase types I-III. Biochem Soc Trans. 1997;25:103–7. [DOI] [PubMed] [Google Scholar]
- 74.Wu F, Zhu D, Wen P, Tang Z, Bao L, Guan Y, et al. Domestic cattle in a national park restricting the sika deer due to diet overlap. Animals. 2023;13:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiong M, Shao X, Long Y, Bu H, Zhang D, Wang D, et al. Molecular analysis of vertebrates and plants in scats of leopard cats (Prionailurusbengalensis) in southwest China. J Mammal. 2016;97:1054–64. [Google Scholar]
- 76.Yamazaki S, Motoi Y, Nagai K, Ishinazaka T, Asano M, Suzuki M. Sex determination of sika deer (Cervusnippon yesoensis) using nested PCR from feces collected in the field. J Vet Med Sci. 2011;73:1611–6. [DOI] [PubMed] [Google Scholar]
- 77.Yang H, Xie B, Han S, Wang T, Feng L. Seasonal spatial pattern of abundance in manchurian sika deer and influcing factors in Hunchun Nationsl nature reserve, Jilin province. J Beijing Normal Univ (Nat Sci). 2018;54:498–505. [Google Scholar]
- 78.You Z, Deng J, Liu J, Fu J, Xiong H, Luo W, et al. Seasonal variations in the composition and diversity of gut microbiota in white-lipped deer (Cervusalbirostris). PeerJ. 2022;10:e13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang B, Shi M, Xu S, Zhang H, Li Y, Hu D. Analysis on changes and influencing factors of the intestinal microbiota of alpine musk deer between the place of origin and migration. Animals. 2023;13:3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, Gao H, Jiang F, Liu D, Hou Y, Chi X, et al. Comparative analysis of gut microbial composition and functions in Przewalski’s gazelle (Procapraprzewalskii) from various habitats. Front Microbiol. 2022;13:913358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang W, Liu W, Hou R, Zhang L, Schmitz-Esser S, Sun H, et al. Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose. ISME J. 2018;12:1319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y. Molecular detection and phylogenetic analysis of some insect-borne pathogen in Ruoergai county of Sichuan province. Chengdu: Southwest Minzu University; 2022. [Google Scholar]
- 83.Zhao J, Yao Y, Dong M, Xiao H, Xiong Y, Yang S, et al. Diet and high altitude strongly drive convergent adaptation of gut microbiota in wild macaques, humans, and dogs to high altitude environments. Front Microbiol. 2023;14:1067240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao J, Yao Y, Li D, Xu H, Wu J, Wen A, et al. Characterization of the gut microbiota in six geographical populations of Chinese Rhesus Macaques (Macacamulatta), implying an adaptation to high-altitude environment. Microb Ecol. 2018;76:565–77. [DOI] [PubMed] [Google Scholar]
- 85.Zheng Y-C, Fei L-S, Li F, Niu L-L, Zhang Z-H. Analysis of digestive enzyme activities in the digestive tract of giant pandas. Sichuan J Zoolgy. 2009;28:397–400. [Google Scholar]
- 86.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.