Abstract
Background
Since the beginning of the pandemic, contact tracing has been one of the most relevant issues to understand SARS-CoV-2 transmission dynamics and, in this context, the analysis of quasispecies may turn out to be a useful tool for outbreak investigations. Analysis of the intra-host single nucleotide variants (iSNVs) found in the nsp2, ORF3, and ORF7 genes of SARS-CoV-2 was conducted in order to correctly identify virus transmission chain among patients hospitalized in Brescia Civic Hospital.
Methods
During the period between August and October 2023, 13 nasopharyngeal specimens, collected from patients admitted to Brescia Civic Hospital, were tested for SARS-CoV-2 positivity and molecularly characterized. Firstly, a phylogenetic analysis was performed to evaluate if they were epidemiologically linked and, then, the Beta-binomial method was used to estimate the transmission bottleneck size (Nb) and quantify the number of viral particles transmitted from one individual (donor) to another (recipient).
Results
According to the molecular characterization of specimens, we identified two transmission clusters in the cardiology unit: the first cluster concerned patients tested positive for the HV.1/EG.5.1.6 lineage, while the second cluster concerned patients tested positive for the FL.10.1 lineage. Moreover, evaluating the bottleneck size, we were able to solve SARS-CoV-2 transmission chain among infected patients.
Conclusion
Our method shows that it is possible to conduct a tracing study using a genomic approach based on iSNVs analysis.
Keywords: SARS-CoV-2, Hospital outbreak, Quasispecies, WGS, Traceability
Background
During COVID-19 pandemic, contact tracing in association with molecular testing turned out to be fundamental in order to contain virus spreading and interrupt transmission chains [1]. In this regard, Next Generation Sequencing (NGS) technology has been crucial since it allowed tracking of SARS-CoV-2 transmission between individuals and proved useful in transforming high-resolution genomic epidemiology into a tool for public health surveillance.
Although the latest SARS-CoV-2 variants may cause self-limited infection in the community, they can still induce serious, even life threatening, diseases in fragile patients and/or immunocompromised hosts [2]. Nosocomial SARS-CoV-2 outbreaks have been constantly reported up to date, and the several preventive measures adopted were unable to completely contrast virus spreading [3]. Indeed, SARS-CoV-2 outbreaks are difficult to recognize and control due to its high infectivity and the wide range of clinical manifestations of the infection [4]. Thus, understanding by genomic characterization the transmission chain in specific clinical units hosting patients at high risk for severe COVID-19 outcomes may help to develop ad hoc strategies to prevent and/or contain viral spreading.
Molecular tracing of SARS-CoV-2 is usually performed by analyzing consensus sequences that reveal the sequence of the dominant viral population without taking into account intra-host genetic variability [5]. Over the last year, the significant sequence similarity between SARS-CoV-2 variants has made it even more challenging to reconstruct transmission chains by consensus sequence analysis only.
When infecting a host, the RNA virus population is not represented by a single dominating sequence but rather consists of an ensemble of closely related sequences, referred to as viral quasispecies [6–8]. Quasispecies analysis has been described to be a useful instrument for accurately tracking person-to-person transmission for different RNA viruses as human immunodeficiency virus (HIV) and human hepatitis C virus (HCV). SARS-CoV-2 exhibits a remarkable propensity to mutate with an estimated mutation rate of roughly 8.0-9.8 × 10− 4 substitutions per site per year [9]. The capability of SARS-CoV-2 to give rise to quasispecies was assessed by a number of studies showing that SARS-CoV-2 infection exhibits intra-host genetic heterogeneity [10–13]. Therefore, molecular tracing and analysis of quasispecies, in particular, may turn out to be a more useful tool than consensus sequences for finely tuning SARS-CoV-2 spread during an outbreak [14].
In a previous work we showed that a more sophisticated genomic approach, which relies on SARS-CoV-2 quasispecies analysis, can be applied to accurately trace contacts [7]. Specifically, our method takes into consideration both the distribution of intra-host single nucleotide variants (iSNVs) along the SARS-CoV-2 genome and the bottleneck dimension, which defines how many viral particles can be transmitted from one individual to another. Here, we document a SARS-CoV-2 outbreak occurred in the cardiology unit of the Brescia Civic hospital from August to September 2023 and the application of our method of monitoring iSNVs to solve the virus transmission chains in a hospital setting.
Methods
Study participants
According to the guidelines of the Brescia Civic Hospital (Brescia, Lombardy, Italy), upon admission, patients are molecularly tested for SARS-CoV-2 using the STANDARD™ M10 SARS-CoV-2 test (SD Biosensor). During hospitalization, patients presenting with respiratory symptoms are monitored for SARS-CoV-2 positivity through antigen screening. During the period between August and October 2023, a total of 13 nasopharyngeal specimens were collected from hospitalized patients for the molecular confirmation of SARS-CoV-2 positivity using FLOQSwabs (Copan).
NGS and data analysis
Total RNA was extracted from 200 µL of nasopharyngeal swabs using QIAamp DSP Virus Kit® (Qiagen), according to the manufacturer’s instructions. Virus genomes were sequenced using the Paragon Genomics’ CleanPlex multiplex PCR Research and Surveillance Panel, as previously described [7]. Raw data were checked for quality using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then analyzed with the specifically designed software SOPHiA GENETICS’ SARS-CoV-2 Panel (SOPHiA GENETICS). A consensus sequence was reconstructed by mapping the reads to the SARS-CoV-2 reference sequence NC_045512 using Bowtie2 in high sensitivity-local mode. Lineage assessment was conducted using the Phylogenetic Assignment of Named Global Outbreak LINeages tool (Pangolin).
Phylogenetic analysis
A phylogenetic analysis was performed with the 13 sequences obtained in the study and 2331 sequences collected in Italy from different Omicron lineages (BA.1, BA.2, BA.5 and XBB) between November 2022 and October 2023. To ensure a globally representative subset of the Italian data, we used a previously described method called subsampler (https://github.com/andersonbrito/subsampler), which randomly selects sequences per country based on case counts, considering geographical, temporal, and epidemiological factors. We aligned all sequences using the ViralMSA tool and performed phylogenetic analysis using IQ-TREE2 [15] with the maximum likelihood approach. To obtain a dated tree topology, we used TreeTime [16] excluding outlier sequences and assuming a constant mean rate of 8.0 × 10− 4 nucleotide substitutions per site per year.
Quasi-species analysis
The variant calling was carried out by the Variant Finder Tool (Geneious ® software version 11.1.5; Biomat- ters Ltd) using a minimum variant frequency of 0 and default parameters for maximum variant p value (10− 6), as previously described [7]. To minimize false discoveries, a stringent approach to evaluate the presence of quasispecies spectrum in the patients’ samples was applied. iSNVs were identified as follows: (i) sequencing coverage of paired-end mapped reads > 100, (ii) at least four reads supporting the nucleotide substitution, (iii) minor allele frequency ≥ 1%. Thereafter, the Beta-binomial method was used to estimate the transmission bottleneck size (Nb) and quantify the number of viral particles transmitted from one individual (donor) to another (recipient). As previously described [7], the distribution of iSNVs found throughout the whole genome were visualized by a cluster heatmap obtained with the Python Matplotlib package (v.3.5.2). In detail, when evaluating the nsp2, ORF3 and ORF7 genes only, the Euclidean metric was applied for clustering using Pearson correlation coefficients.
Results
Characteristics of the study participants
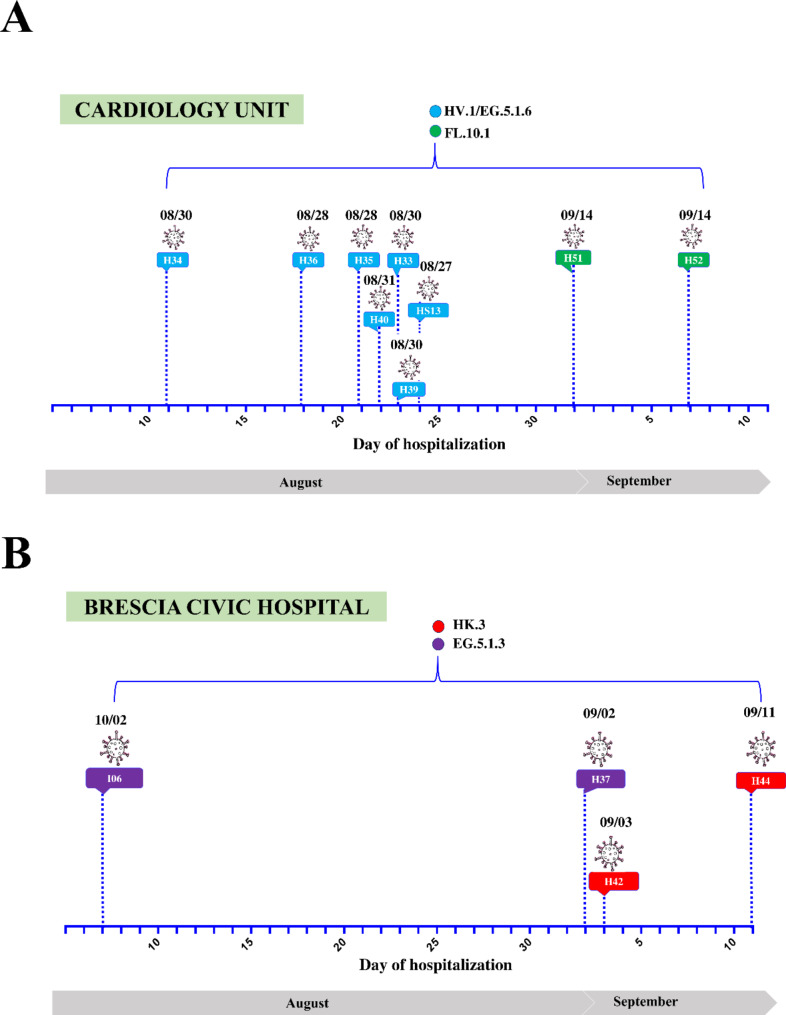
Following the onset of respiratory symptoms, nine patients (HS13, H33, H34, H35, H36, H39, H40, H51, and H52) who were hospitalized in the cardiology unit of the Brescia Civic Hospital between late August and September 2023, tested positive for SARS-CoV-2 (Fig. 1A). Since all patients were negative for SARS-CoV-2 infection based on molecular diagnostic testing at the time of admission, it is likely that they acquired the virus during hospitalization. In order to define the root source of the outbreak, whole genome sequencing (WGS) analysis was performed on nasopharyngeal specimens. In particular, WGS revealed that patients were infected with the following Omicron sub-lineages: HV.1 (H33, H34, H35, H36, H39, H40), EG.5.1.6 (HS13), and FL.10.1 (H51, H52) (Fig. 1A).
Fig. 1.
Timeline of hospitalization and SARS-CoV-2 positivity of patients’ hospitalized in the (A) cardiology unit or (B) other units of Brescia Civic Hospital. SARS-CoV-2 lineages are highlighted in different colors. indicates the day when each patient tested positive for SARS-CoV-2 infection
To establish whether the cases found in the cardiology ward represent a specific and confined nosocomial outbreak, patients (H37, H42, H44, I06) hospitalized in four different hospital’s units (emergency medicine unit, emergency room, general medicine unit and neuropsychiatry unit) were also included in the study as a control group. Differently from the patients belonging to the cardiology ward, they were infected with Omicron sub-lineages belonging to HK.3 (H42, H44), and EG.5.1.3 (H37, I06) lineages (Fig. 1B).
Epidemiological analysis
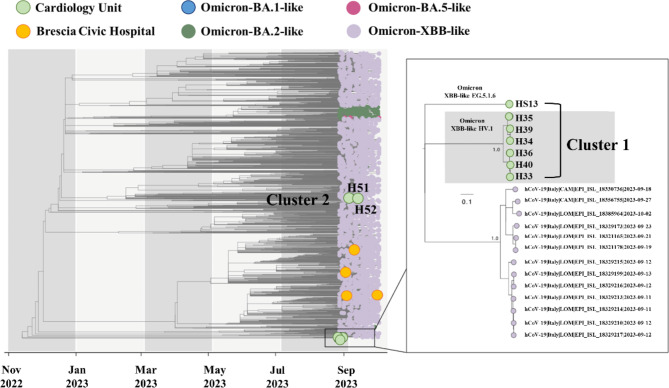
To clarify the transmission events leading to the outbreak and to understand if there was a common source of infection, we inferred a time-stamped phylogenetic tree placing samples collected from the cardiology unit within a larger context of SARS-CoV-2 genetic variations. In addition to our cases, representative sequences from Omicron variants circulating in Italy between November 2022 and October 2023 were evaluated. As shown in Fig. 2, the analysis revealed that consensus sequences collected from patients HS13, H33, H34, H35, H36, H39, and H40 sited close together on the phylogenetic tree (Cluster 1), while patients H51 and H52 formed a small distinct cluster (Cluster 2). Of note, the consensus sequence of patient HS13, in spite of belonging to the cardiology HV.1 group (Cluster 1), tested positive for a distinct SARS-CoV-2 Omicron variant (EG.5.1.6) and was located on a different branch of the tree. On the other hand, the control group patients H37, H42, H44, and I06 (yellow circles) were scattered across the phylogenetic tree (Fig. 2).
Fig. 2.
Time-resolved maximum-likelihood tree of SARS-CoV-2 including 13 whole genomes obtained from the hospitalized patients considered for this study in addition with a set of representative Italian data (n = 2331) collected between November 2022 and October 2023. The genomes are colored according to the SARS-CoV-2 lineages. In addition, the Clusters 1 and 2, that include patients admitted to the cardiology unit identified in this study, are highlighted
Due to substantial sequence similarity among SARS-CoV-2 Omicron variants, the phylogenetic tree obtained with the consensus sequences, could not accurately determine the virus transmission chain among patients and, so, could not clearly solve the hospital outbreak.
Definition of virus transmission through bottleneck size evaluation
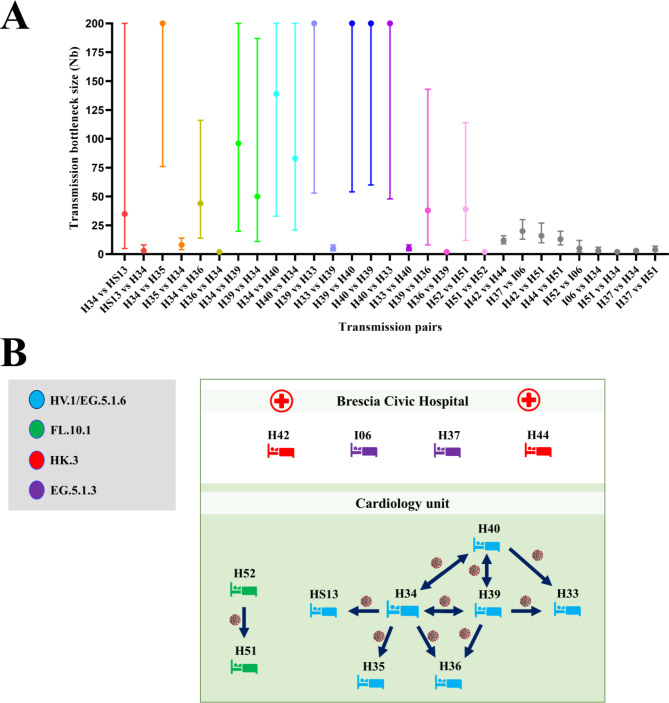
Estimating the number of transmitted viral particles is crucial to track epidemiologically linked SARS-CoV‐2 infections [7]. The Beta Binomial inference method was used to calculate the bottleneck sizes and, together with the dates of hospitalization and samples collection, was taken into account in order to determine the transmission directionality among the SARS-CoV-2 positive patients. We performed 106 bottleneck size estimations, pairing randomly the patients and testing each patient both as a donor and recipient. H34, as shown in Fig. 1A, was the first patient admitted to the cardiology unit, so we speculated that the outbreak may have started with this patient. Then, each hospitalized subject was evaluated as donor or recipient in a hypothetical transmission chain according to the bottleneck dimension. This analysis (Fig. 3A) clearly showed the directionality of the viral transmission for some pairs, for example for H34-HS13, H34-H35, H34-H36, H39-H33, H40-H33, H39-H36, and H52-H51, while other subjects (H34, H39, H40), if coupled, displayed similar results in both directions, making it difficult to establish who is the donor and who is the recipient. Taking into consideration both bottleneck sizes and date of hospital admission, we reconstructed a hypothetical chain of viral transmission among the cardiology unit patients (Fig. 3B). In particular, a wide bottleneck was found when H34 was coupled with all the other patients from Cluster 1, supporting our hypothesis that this patient was the cluster’s index case. However, considering the bottleneck dimension, we hypothesized that patient H33 most likely acquired the infection via H39 (H39-H33 range: 53–200 Nb). Determining the H34-H39-H40 transmission chain was a challenging issue, due to a wide bottleneck for all the possible donor-recipient pairs (H34-H39 range: 20–200 Nb vs. H39-H34 range: 11–187 Nb; H34-H40 range: 33–200 Nb vs. H40-H34 range: 21–200 Nb, H40-H39 range: 60–200 Nb vs. H39-H40 range: 54–200 Nb). Although we do not exclude a possible transmission between H39 and H40, considering both the bottleneck dimension and the date of hospitalization/positivity, it is more likely that they both contracted the virus from subject H34. Concerning Cluster 2, both H51 and H52 tested positive for SARS-CoV-2 on September 14th. However, the bottleneck analysis suggests that H52 was the donor and H51 the recipient (H52-H51 range: 12–114 Nb vs. H51-H52 range: 2–4 Nb). Furthermore, a narrow bottleneck dimension was found with control patients (H37, H42, H44, I06), confirming the hypothesis that the SARS-CoV-2 cases detected in the cardiology department represent a confined nosocomial outbreak.
Fig. 3.
(A) Maximum likelihood estimation for the mean transmission bottleneck size among paired-patients. Values represented as colored dots correspond to the hypothesized linked pairs, while the gray dots correspond to pairs that include at least one control patient. (B) Schematic representation of patients’ interaction across the epidemiological cluster. Arrows indicate the hypothesis of transmission direction. The subjects in the study are colored according to the SARS-CoV-2 lineages
Intrahost single nucleotide variants characterization and cluster analysis for genomic tracing
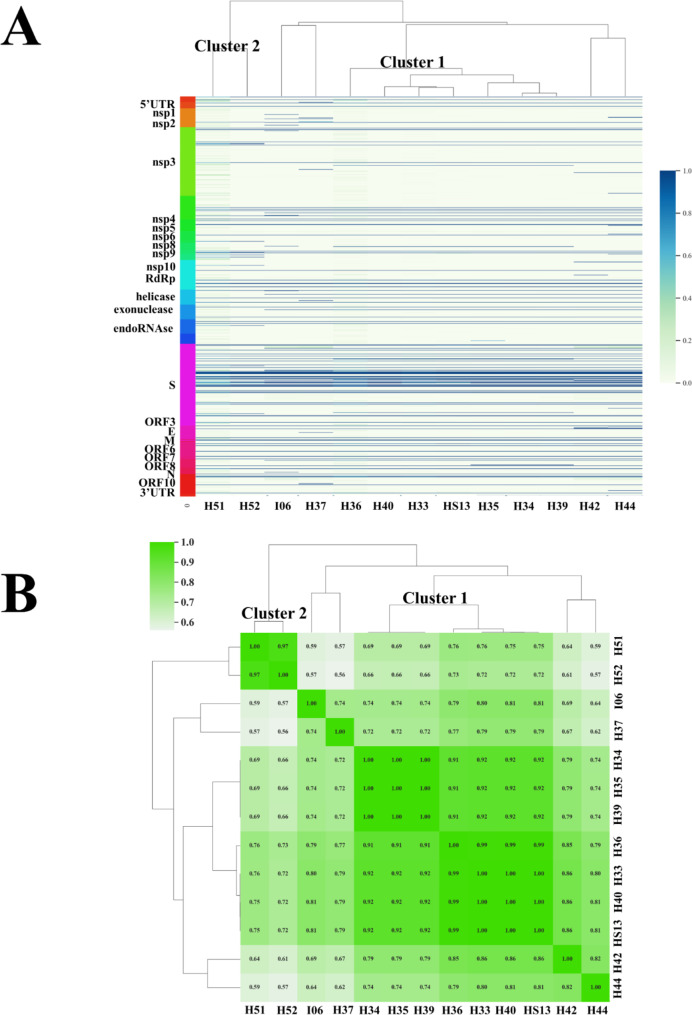
To further confirm the presence of two distinct clusters among the hospitalized patients in the cardiology unit, the Euclidean clustering method was performed to analyze the iSNVs found in the SARS-CoV-2 genome, as previously described [7]. As shown in Fig. 4A, the variations of iSNVs in terms of frequency and distribution along the whole genome displayed useful patterns to comprehend the viral transmission chains among patients. Data plotted in the heatmap (Fig. 4A) highlighted the close position of patients HS13, H33, H34, H35, H36, H39, H40 in the central branch and the localization of patients H51 and H52 in the same left branch, confirming the presence in the cardiology unit of the Cluster 1 and 2. Furthermore, Fig. 4A documents how patients I06, H37, H42, and H44, admitted to Brescia Civil Hospital in the same period but in different wards, were placed on distinct branches, separated from the clusters identified in the cardiology unit.
Fig. 4.
(A) Cluster heatmap built up with the iSNVs found in the whole SARS-CoV-2 genome, based on the similarity between samples. (B) Cluster heatmap constructed with iSNVs identified in the nsp2, ORF3, and ORF7 genes. Colors are defined according to the scale
In order to validate our method, which considers the quasispecies found in a small portion of the SARS-CoV-2 genome, we conducted the same analysis taking into account the frequency and distribution of the iSNVs found in the nsp2, ORF3, and ORF7 genes only [7]. The Pearson correlation coefficients were used to evaluate the iSNVs variations and the results were plotted in a heatmap (Fig. 4B). As obtained with the whole genome, the iSNVs found in these 3 well conserved SARS-CoV-2 genes correctly identify transmission chains for both Cluster 1 and Cluster 2. In details, patients HS13, H33, H34, H35, H36, H39, H40 (Cluster 1) were plotted together in the central portion of the heatmap, while H51 and H52 formed a small distinct cluster (Cluster 2) in the left portion. Once again, our analysis confirmed that the iSNVs found in these genes can be utilized as a peculiar signature for contact tracing when the consensus sequences are identical.
Discussion
Here we documented that a contact tracing study can be conducted by applying a genomic approach based on iSNVs analysis. Our molecular method allowed us to precisely reconstruct the transmission chains of a SARS-CoV-2 outbreak in a hospital setting, and highlight the role of genomic sequencing in establishing a direct transmission. Of note, traceability of SARS-CoV-2 transmission was possible by limiting the analysis of iSNVs to only three well conserved genes, namely nsp2, ORF3, and ORF7. This has several advantages such as the speed of results and the easy implementation to different SARS-CoV-2 variants.
Since the beginning of the COVID-19 pandemic, contact tracing has been one of the most relevant issues to understand SARS-CoV-2 transmission dynamics and trace person-to-person contacts, which is critical to understand the epidemiology of community transmission. Up to date, sequencing by consensus analysis was found to be a useful tool to rule out direct transmission if significant differences are detected in the viral sequences. Indeed, if genomes are almost identical, as occurs in the more recent circulating SARS-CoV-2 variants, it is challenging to establish a possible route of spread.
The existence of quasispecies, which are thought to be a viral evolution strategy, attests the intra-host genetic variation of SARS-CoV-2 [17]. When the virus interacts with the host’s immune system, quasispecies evolve carrying iSNVs, as well documented for other RNA viruses such as HIV and HCV [18]. Interestingly, during the acute phase of infection in the human host, viral population can grow up to 1014 virions but, when the virus spreads, bottleneck events occur and viral particles that are actually transmitted are estimated to be from one to a few virions only [19]. Since iSNVs are generated overtime in SARS-CoV-2 infected patients, they can be transmitted among acutely-infected hosts [20]. Therefore, the presence of specific iSNVs in the transmission bottleneck determines how much of the viral diversity generated in one host passes to another, making it possible to prove transmission with a degree of certainty.
We have recently proposed a NGS-based method to trace the transmission chains among acutely infected individuals by analyzing iSNVs, and in particular those originated in three well-conserved viral genes (nsp2, ORF3 and ORF7) [7]. With our method we identified two transmission clusters in the cardiology unit of Brescia Civic hospital: the first one concerned patients tested positive for HV.1/EG.5.1.6 lineage, while the second one concerned patients tested positive for FL.10.1 lineage. To establish transmission events, we generated a time-stamped phylogenetic tree, together with representative Italian sequences from Omicron variants, confirming the presence of two small distinct clusters. However, we were unable to accurately determine the virus transmission chain by analyzing the consensus sequences only. Whereas, the evaluation of the bottleneck dimension for 106 random couples proved crucial to assess each hospitalized patient as either a donor or a recipient in the transmission chain. This analysis together with the evaluation of the frequency and distribution of iSNVs allowed us to clearly solve the hospital outbreak.
Concerning the study’s limitations, firstly the analysis was carried out on 13 samples only. Therefore, in order to determine whether our method can reconstruct the transmission chain even in a more comprehensive clinical context, it would be necessary to apply it to a larger number of samples. Secondly, patients included in the study were hospitalized between August and October 2023, when the COVID-19 pandemic had already been declared over and, in Italy, SARS-CoV-2 circulated with a low incidence [21]. For this reason, it would be important to confirm whether our approach can be used to track infections even during the most critical phases of the pandemic. At the same time, with the goal of monitoring the spread of novel variants that harbor concerning new mutations, we cannot rule out that, in the next future, further analysis will highlight additional mutational patterns and genes which may define a more accurate strategy for contact tracing.
In consideration of the decreasing costs of genome sequencing, our method represents a promptly available, accurate, and standardized surveillance strategy to adequately support SARS-CoV-2 infection prevention programs in hospital settings and hospices, where population, who has increased risk of acquiring a serious, or even life threatening infection, is hosted.
Conclusions
To date, despite all the preventive measures that have been implemented in order to restrict the nosocomial virus’ circulation, patients can still contract COVID-19 in hospitals. In addition, global health security has also been threatened by the emergence of new human viral diseases affecting the respiratory tract, such as the highly pathogenic avian influenza viruses [22]. Thus, implementing new approaches to track viral spreading could help to improve surveillance programs and disease control efforts, and shape the response to future pandemic threats. In recent years, the NGS technology has turned out to be convenient for laboratories in terms of cost and turnaround time, making real-time surveillance a possible strategy to prevent the spread of pathogens and infectious diseases. At the same time, systematic molecular surveillance allows characterization of quasispecies, pathogenic variants, and possible recombinant strains. In particular, characterizing viral quasispecies might provide insights into the routes of transmission and selection of newly emerging viral strains and, as shown in this work, also offers genetic fingerprints that could be a valuable tool for a molecularly contact tracing approach.
Acknowledgements
The authors are grateful to all individuals who participated in this study. The authors also gratefully acknowledge all data contributors, that is, the authors and their originating laboratories responsible for obtaining the specimens, and their submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
Author contributions
SM: Study design, Data collection, Roles/Writing – original draft. MG: Data analysis. AR: Data analysis. MB: Data collection. MD: Data collection. FC: Conceptualization, Supervision. MC: Conceptualization, Supervision. AC: Study design, Conceptualization, Supervision Writing - review & editing.
Funding
Not applicable.
Data availability
Sequences obtained in this study have been deposited in Global Initiative on Sharing All Influenza Data (GISAID) database (accession numbers: EPI_ISL_19025631; EPI_ISL_19025632; EPI_ISL_19025633; EPI_ISL_19025634; EPI_ISL_19025635; EPI_ISL_19025636; EPI_ISL_19025637; EPI_ISL_19025638; EPI_ISL_19025639; EPI_ISL_19025640; EPI_ISL_19025641; EPI_ISL_19025642; EPI_ISL_19025643).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thomas Craig KJ, Rizvi R, Willis VC, Kassler WJ, Jackson GP. Effectiveness of Contact Tracing for Viral Disease Mitigation and Suppression: Evidence-Based Review. JMIR Public Health Surveill. 2021;7(10):e32468. 10.2196/32468. Published 2021 Oct 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caillard S, Benotmane I, Gautier Vargas G, Perrin P, Fafi-Kremer S. SARS-CoV-2 viral dynamics in immunocompromised patients. Am J Transpl. 2021;21(4):1667–9. 10.1111/ajt.16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darby J, Falco C. Infection Control and the Need for Family-/Child-Centered Care. Healthcare-Associated Infections Child. 2018. 10.1007/978-3-319-98122-2_4. 57–79. Published 2018 Jul 16. [Google Scholar]
- 4.Kannangara CI, Seetulsingh P, Foley J, Bennett G, Carter T. Investigation and management of an outbreak of COVID-19 infection in an acute admission unit in a District General Hospital: lessons learnt. Infect Prev Pract. 2021;3(3):100156. 10.1016/j.infpip.2021.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader W, Delerce J, Aherfi S, La Scola B, Colson P. Quasispecies Analysis of SARS-CoV-2 of 15 Different Lineages during the First Year of the Pandemic Prompts Scratching under the Surface of Consensus Genome Sequences. Int J Mol Sci. 2022;23(24):15658. 10.3390/ijms232415658. Published 2022 Dec 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun F, Wang X, Tan S, et al. SARS-CoV-2 Quasispecies Provides an Advantage Mutation Pool for the Epidemic Variants. Microbiol Spectr. 2021;9(1):e0026121. 10.1128/Spectrum.00261-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messali S, Rondina A, Giovanetti M, et al. Traceability of SARS-CoV-2 transmission through quasispecies analysis. J Med Virol. 2023;95(6):e28848. 10.1002/jmv.28848. [DOI] [PubMed] [Google Scholar]
- 8.Jary A, Leducq V, Malet I, et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin Microbiol Infect. 2020;26(11):1560e. 1-1560.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasco N, Damaggio G, Esposito L, Colonna V, Vitagliano L. A comprehensive analysis of SARS-CoV-2 missense mutations indicates that all possible amino acid replacements in the viral proteins occurred within the first two-and-a-half years of the pandemic. Int J Biol Macromol. 2024;266(Pt 1):131054. 10.1016/j.ijbiomac.2024.131054. [DOI] [PubMed] [Google Scholar]
- 10.Lythgoe KA, Hall M, Ferretti L, et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372(6539):eabg0821. 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Wang D, Zhang L, et al. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021;13(1):30. 10.1186/s13073-021-00847-5. Published 2021 Feb 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karamitros T, Papadopoulou G, Bousali M, Mexias A, Tsiodras S, Mentis A. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies. J Clin Virol. 2020;131:104585. 10.1016/j.jcv.2020.104585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caccuri F, Messali S, Bortolotti D, et al. Competition for dominance within replicating quasispecies during prolonged SARS-CoV-2 infection in an immunocompromised host. Virus Evol. 2022;8(1):veac042. 10.1093/ve/veac042. Published 2022 May 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houwaart T, Belhaj S, Tawalbeh E, et al. Integrated genomic surveillance enables tracing of person-to-person SARS-CoV-2 transmission chains during community transmission and reveals extensive onward transmission of travel-imported infections, Germany, June to July 2021. Euro Surveill. 2022;27(43):2101089. 10.2807/1560-7917.ES.2022.27.43.2101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minh BQ, Schmidt HA, Chernomor O et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era [published correction appears in Mol Biol Evol. 2020;37(8):2461. doi: 10.1093/molbev/msaa131]. Mol Biol Evol. 2020;37(5):1530–1534. 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed]
- 16.Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4(1):vex042. 10.1093/ve/vex042. Published 2018 Jan 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao R, Zu W, Liu Y, et al. Quasispecies of SARS-CoV-2 revealed by single nucleotide polymorphisms (SNPs) analysis. Virulence. 2021;12(1):1209–26. 10.1080/21505594.2021.1911477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skums P, Zelikovsky A, Singh R, et al. QUENTIN: reconstruction of disease transmissions from viral quasispecies genomic data. Bioinformatics. 2018;34(1):163–70. 10.1093/bioinformatics/btx402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev. 2012;76(2):159–216. 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messali S, Bugatti A, Filippini F, Caruso A, Caccuri F. Emergence of S gene-based quasispecies explains an optimal adaptation of Omicron BA.5 subvariant in the immunocompetent vaccinated human host. J Med Virol. 2023;95(1):e28167. 10.1002/jmv.28167. [DOI] [PubMed] [Google Scholar]
- 21.Sclavi L, Bertelli M, Messali S, Caruso A, Caccuri F. Epidemiology and molecular characterization of respiratory viruses at the end of COVID-19 pandemic in Lombardy, Northern Italy. New Microbiol. 2024;47(1):80–7. [PubMed] [Google Scholar]
- 22.Zhang Z, Lei Z. The Alarming Situation of Highly Pathogenic Avian Influenza Viruses in 2019–2023. Glob Med Genet. 2024;11(3):200–13. 10.1055/s-0044-1788039. Published 2024 Jun 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences obtained in this study have been deposited in Global Initiative on Sharing All Influenza Data (GISAID) database (accession numbers: EPI_ISL_19025631; EPI_ISL_19025632; EPI_ISL_19025633; EPI_ISL_19025634; EPI_ISL_19025635; EPI_ISL_19025636; EPI_ISL_19025637; EPI_ISL_19025638; EPI_ISL_19025639; EPI_ISL_19025640; EPI_ISL_19025641; EPI_ISL_19025642; EPI_ISL_19025643).