Abstract
Background
Circular RNAs (circRNAs) are a novel class of endogenous non-coding RNA molecules in eukaryotes, involved in many essential biological processes. However, their role in allergic rhinitis (AR) has not been extensively studied.
Methods
The expression levels of hsa_circRNA_100791 were measured using qRT-PCR in peripheral blood mononuclear cells (PBMCs) and nasal mucosa from AR patients. The biological function of hsa_circRNA_100791 in AR was investigated through RNA-seq and a series of in vitro experiments. Western blotting, luciferase reporter assays, and rescue experiments were conducted to elucidate the molecular mechanisms underlying hsa_circRNA_100791. Additionally, a mouse model was used to assess the functional role of hsa_circRNA_100791 in vivo.
Results
Upregulation of hsa_circRNA_100791 was observed in both PBMCs and nasal mucosa of AR patients. In vitro, increased expression of hsa_circRNA_100791 promoted the production of pro-inflammatory mediators (IL-1β, IL-4, IL-5, IL-6, IL-8, IL-13, IL-17, IL-18, IL-33, TNF-α, and NF-κB) and inhibited IL-2 and IFN-γ. Conversely, knockdown of hsa_circRNA_100791 both in vitro and in vivo alleviated AR symptoms, reduced pro-inflammatory mediators, and enhanced IL-2 and IFN-γ levels. Mechanistically, we found hsa_circRNA_100791 contributing to the pathological processes of AR, which upregulate TRIM13 via sponging miR-487b-5p.
Conclusion
Our study demonstrated that hsa_circRNA_100791 mitigates the inhibitory effect of miR-487b-5p on Trim13 by directly binding to miR-487b-5p. This interaction regulates the expression of inflammatory factors and facilitates AR. Thus, hsa_circRNA_100791 could be a promising new therapeutic target for AR.
Keywords: allergic rhinitis, hsa_circ_100791, miR-487b-5p, Trim13, diagnosis, inflammation, NF-κB
Introduction
AR is one of the most common chronic airway diseases, characterized by IgE-mediated type 2 inflammation in response to inhaled allergens. Clinically, AR presents with nasal congestion, itching, sneezing, and rhinorrhea.1,2 During acute allergic reactions, inflammatory mediators primarily released by mast cells (eg histamine and leukotrienes) are responsible for the development of AR symptoms. Additionally, activated allergen-specific TH2 cells secrete large amounts of IL-4, IL-5, and IL-13, which contribute to the enhancement and persistence of type 2 mucosal inflammation in AR.1–3 However, the specific molecular and cellular mechanisms underlying these processes are not yet fully understood.
MicroRNAs (miRNAs) are small non-coding RNA molecules that can decrease gene expression either by degrading mRNA or by binding to the 3ʹ-untranslated region of target mRNAs to inhibit translation.4 Several studies have suggested that miRNAs play a role in the pathological processes of AR.5–7 For instance, miR-133b has been reported to alleviate allergic symptoms in a mouse model of AR.7 Additionally, miR-487b has been shown to mitigate AR in an OVA-induced animal model by inhibiting the IL-33/ST2 signaling pathway, and reduced expression of miR-487b can lead to increased production of pro-inflammatory cytokines such as IL-6 and TNF-α.5 Furthermore, miR-487b is known to play a significant role in allergic airway inflammation.8 Despite these insights, the precise molecular mechanisms underlying these pathological events in AR still require further investigation.
Circular RNAs (circRNAs) are a novel class of endogenous non-coding RNA molecules found in eukaryotes. They are formed by the back-splicing of exons from precursor mRNA (pre-mRNA) transcripts, resulting in a covalently closed loop structure that lacks the 5ʹ–3ʹ polarity and 3ʹ polyA tail typical of linear RNAs.9,10 Unlike traditional linear RNAs, circRNAs are resistant to degradation by RNA exonucleases, which contributes to their stability.9–11 With advances in biotechnology, particularly RNA sequencing and bioinformatics, a significant number of circRNAs have been identified across various organisms. Increasing evidence suggests that abnormal circRNA expression plays a crucial role in allergic disorders such as asthma12–14 and AR.15,16 Emerging research also indicates that circRNAs can function as competitive endogenous RNAs (ceRNAs), regulating the expression of miRNAs and their target genes.9–11 Despite these findings, the role and mechanisms of circRNAs in AR remain largely unexplored.
Tripartite motif-containing protein 13 (TRIM13), an E3 ubiquitin ligase, plays a key role in the progression of various diseases. TRIM13 overexpression can inhibit HCC progression and attenuated alveolar epithelial cell injury in COPD.17 Inhibiting TRIM13 expression suppressed MMC’s inflammation and proliferation by inhibiting the activation of the NF-κB pathway.18 TRIM13 overexpression was proposed to promote activity of NF-κB via p65.18
To identify differentially expressed circRNAs in AR, we conducted circRNA sequencing on peripheral blood samples from AR patients and healthy controls through KangChen Bio-tech (Shanghai, China).19 Notably, we observed that hsa_circ_100791 (also known as hsa_circ_0004851 in circBase, http://circrna.org) was overexpressed in the peripheral blood samples from AR patients compared to healthy controls. Bioinformatics analysis suggested a potential interaction between hsa_circ_100791 and miR-487b. However, the precise function and underlying mechanism of hsa_circ_100791 in the pathological processes of AR remain unclear. This study aims to investigate the biomolecular role of hsa_circ_100791 in the progression and development of AR, with the goal of providing a scientific basis for its potential use in AR prevention and treatment.
Methods and Materials
Patients
Biopsies of nasal mucosal from the inferior nasal turbinate of 23 patients with AR and 21 healthy controls were collected from the Shandong Provincial ENT Hospital, Cheeloo College of Medicine, Shandong University. Patients were diagnosed as AR based on combined assessment of medical history, symptoms and positive skin prick test against specific allergen.1 We also collected the venous blood from 41 AR patients and 37 healthy controls to isolate the peripheral blood mononuclear cells (PBMCs). The measure of isolating the PBMC from venous blood is according to the manufacturer’s instructions (Solarbio, China). All participants were provided informed consent. All participants also signed the informed consent. The study protocol was approved by The Institutional Review Boards of Shandong Provincial ENT Hospital, Cheeloo College of Medicine, Shandong University. The detailed information of study participants is described in Tables 1 and 2.
Table 1.
Summary of the Clinical Features for Nasal Mucosal Tissues of Study Participants
| Parameters | Control Subjects | Allergic Rhinitis | |
|---|---|---|---|
| Sample sizes | 21 | 23 | |
| Age, years (mean ± SD) | 44.33 ± 3.847 | 40.09 ±2.635 | p=0.34 |
| Gender (M/F) | 13/8 | 17/6 |
Table 2.
Summary of the Clinical Features for PBMC of Study Participants
| Parameters | Control Subjects | Allergic Rhinitis | |
|---|---|---|---|
| Sample sizes | 37 | 41 | |
| Age, years (mean ± SD) | 29.00 ±1.314 | 26.66 ±1.539 | p=0.256 |
| Gender (M/F) | 20/17 | 17/24 |
Abbreviation: PBMC, Peripheral blood mononuclear cell.
Cell Culture and Cell Transfection
HNEpC, human nasal epithelial cell line, were obtained from Cellbio (Cat. No. CBR-130634), and maintained in RPIM-1640 (Gibco, USA) medium supplemented with 10% fetal bovine serum (FBS, Gibco, USA) in a humidified atmosphere with 5% CO2 at 37°C.
The recombinant vectors expressing hsa_circ_100791, small interfering RNAs of hsa_circ_100791 (si-circ100791), and their corresponding negative controls were purchased from GenePharm (Shanghai, China). We listed the siRNA sequences as following: 5ʹ-AACAGCUUCAAACAGGUUCTT-3ʹ. MiR-487b-5p inhibitors, miR-487b-5p mimics, and their corresponding negative controls were synthesized from RiboBio (Guangzhou, China). HNEpCs were planked in 6-well plates with 1×105 cell density per well and cultured for 24 hours. Then, transiently transfected with designated vector or sequences for 48h or 72h using Lipofectamine 3000 (Invitrogen, USA) according to manufacturer’s instructions. RNA expression levels after transfection verified by qRT-PCR.
RNA Sequencing (RNA-Seq) Analysis, GO and KEGG Pathways Enrichment Analysis
We extracted the total RNA from HNEpCs, which transfected with si- circ100791 and negative control (n=3). The NanoDrop ND-1000 (NanoDrop Thermo) were used to evaluate the concentration of RNA. To enrich and purificate the mRNA, we used oligo dT beads to treat the mRNA. The RNA sequencing, GO, KEGG pathways enrichment and other bioinformatics analysis were carried out by BGI Tech (Shenzheng, China).19 The GO and KEGG pathway enrichment analyses were conducted using the BGI online analysis platform (https://report.bgi.com/).
Animals Modeling
We purchased 8-week-old female BALB/c mice (n=24) from Jinan Pengyue Laboratory Animal Breeding Company. Mice were randomly divided into control (n=6), AR (n=6), AR+ circ-negative control (n=6), and AR+si-circ100791 (n=6) groups. The control group were treated with PBS, AR group were treated with ovalbumin (OVA) and aluminum hydroxide (Al (OH)3), AR+ circ- negative control group were treated with AR-induced and tail vein injection the circRNA- siRNA negative control, and another group were treated with AR-induced and tail vein injection the siRNA of hsa_circ_100791. The mice were housed at 20±1°C and were maintained at a 12-h: 12-h dark-light cycle and could free access to standard chow and water. The animal experiments of this study were conducted the basis of the Animal Ethics guidelines of Shandong Provincial ENT Hospital, Cheeloo College of Medicine, Shandong University and approved by the hospital.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was extracted from HNEpCs or tissues using Trizol reagent (Invitrogen, CA) following the manufacturer’s instructions and about 0.5µg RNA as a model for reverse transcription according to protocol of Thermo Fisher Scientific RT kit (USA). The mRNA expression of miR-487b-5p, hsa_circRNA_100791, CAPRIN1, Trim13, IL-1β, IL-4, IL-5, IL-6, IL-8, IL-13, IL-17, IL-18, IL-33, TNF-α, IL-2 and IFN-γ were determined by qRT-PCR analysis, which was performed using SYBR Green Premix Ex TaqTM (Takara, Japan) in the light of protocol. The relative expression level of miRNA and genes was normalized to that of internal control U6 and GAPDH by using 2−ΔΔCt cycle threshold method. Primers used were listed in Table 3.
Table 3.
The Sequences of the qRT-PCR Gene Primers Were as Follows
| Primer | Assay |
|---|---|
| GAPDH | F: 5ʹ-GCACCGTCAAGGCTGAGAAC |
| R: 5ʹ-TGGTGAAGACGCCAGTGGA | |
| circ-100791 | F: 5ʹ-ACCAGAGCTTTTCTAGTCAGCC |
| R: 5ʹ-TTCTGTGGTCGTTGCTCTGT | |
| IL-1β | F: 5ʹ-GGGCCTCAAGGAAAAGAATC |
| R: 5ʹ-TTCTGCTTGAGAGGTGCTGA | |
| IL-2 | F: 5ʹ-ATCCCAAACTCACCAGGATGC |
| R: 5ʹ-AGATGTTTCAGTTCTGTGGCCT | |
| IL-4 | F: 5ʹ-TTCCTGAAACGGCTCGACAG |
| R: 5ʹ-CGTACTCTGGTTGGCTTCCT | |
| IL-5 | F: 5ʹ-TCGAACTCTGCTGATAGCCA |
| R: 5ʹ-CTCCAGTGTGCCTATTCCCT | |
| IL-6 | F: 5ʹ-TGAGGAGACTTGCCTGGTGAA |
| R: 5ʹ-CAGCTCTGGCTTGTTCCTCAC | |
| IL-8 | F: 5ʹ-CTCTGTGTGAAGGTGCAGTTTT |
| R: 5ʹ-GTTTTCCTTGGGGTCCAGACA | |
| IL-13 | F: 5ʹ-AGGAGGGTTAGGGAGGGGTA |
| R: 5ʹ-GGGCACCCACTGTAATGCTA | |
| IL-17 | F: 5ʹ-TCCATCTCATAGCAGGCACA |
| R: 5ʹ-GTCTTCCCAGGAGTCATCGT | |
| IL-18 | F: 5ʹ-TGACCAAGGAAATCGGCCTC |
| R: 5ʹ-GGTCCGGGGTGCATTATCTCT | |
| TNF-α | F: 5ʹ-GGGCAGGTCTACTTTGGGAT |
| R: 5ʹ-AGGTTGAGGGTGTCTGAAGG | |
| IL-33 | F: 5ʹ-TCCACAGCAAAGTGGAAGAACA |
| R: 5ʹ-TGGCCTTCTGTTGGGATTTTCC | |
| IFN-γ | F: 5ʹ-AAGTGATGGCTGAACTGTCG |
| R: 5ʹ-TACTGGGATGCTCTTCGACC | |
| CAPRIN1 | F: 5ʹ-ACAAGTGAGGGGTACACAGC |
| R: 5ʹ-TTGCTTGTCCCAGCCTGAAA | |
| Trim13 | F: 5ʹ-ATCCTTCTGCTTGGCCTTGT |
| R: 5ʹ-GACAGCCTTTCCAAGTTGCC | |
| trim13(Mus) | F: 5ʹ- TTGATGACCCCCGAGTGTTG |
| R: 5ʹ- AGCTGAGGTTTCCTTACGGC | |
| GAPDH(Mus) | F: 5ʹ- TGTGTCCGTCGTGGATCTGA |
| R: 5ʹ- TTGCTGTTGAAGTCGCAGGAG |
RNase R Treatment
RNase R can digest almost all linear RNA molecules, but it is not easy to digest circular RNA, so can enrich circular RNAs (circRNAs). The 2.5µg total RNA was isolated from HNEpCs using Trizol reagent and was either treated or untreated with 10U RNase R in 2μL 10× Reaction Buffer, incubated at 37°C for 30 min, following at 70°C for 10 min to inactivate the enzyme.
Western Blotting
The total protein was extracted from cells using the Radio Immunoprecipitation Assay (RIPA) lysis buffer containing protease inhibitor and phenylmethanesulfonyl fluoride (Solarbio, China) followed to determine the protein concentration using bicinchoninic acid (BCA) protein analysis. 20μg of protein were separated by 8–16% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride membrane (PVDF, Millipore, USA) by electroblotting, and then blocked with 5% skim milk in Tris-Buffered Saline tween (TBST) buffer for 2h at room temperature. The membranes were incubated by primary antibodies overnight at 4°C: Trim13 (1: 1000; ab34847, Abcam, UK), IL-4(1: 1000; ab62351, Abcam, UK), IL-5(1: 1000; 310D1A14, Invitrogen, USA), IL-13(1: 1000; ab133353, Abcam, UK)p-p65(1: 1000; ab131109, Abcam, UK), p65(1:1000; ab16502, Abcam, UK) and β-actin (1:3000, Beyotime, China), then washed with TBST buffer and incubated with HRP-conjugated secondary antibodies (Proteintech, USA) for 1h at room temperature. The images were determined by chemiluminescence system (Bio-Rad, USA) and normalized to those of β-actin.
Hematoxylin-Eosin (H-E) Staining
Briefly, Fresh nasal mucosa were immediately fixed by 4% formaldehyde. Then, the samples were embedded in paraffin and were cut into 4-µm serial sections. To evaluated eosinophil infiltration, the sections were stained by the hematoxylin and eosin, and visualized by under high-power fields (400× magnification) microscope and photographed.
Immunohistochemistry (IHC)
The nasal mucosa tissues were fixed, paraffin-embedded and cut into 4-µm serial sections. IHC experiment were performed on serial sections. Primary antibodies against Trim13 (1:200; ab34847, Abcam, UK) was used. Then, all sections visualized by under high-power fields (400× magnification) microscope and validated by two experienced pathologists.
Immunofluorescence (IF) Staining
Paraffin tissue sections were incubated with primary antibodies against Trim13 (1:150; ab34847, Abcam, UK) at 4°C overnight, then incubation with Alexa Fluor 594 conjugated secondary antibodies (Life Technologies, Carlsbad, CA, USA) for 1h at room temperature. The SlowFade Gold anti-fade reagent with 4ʹ6-diamidino-2-phenylindole (DAPI; Life Technologies Inc., USA) was used to mounted the slides. All sections visualized by under high-power fields (400× magnification) microscope.
Fluorescence in situ Hybridization (FISH) Assay
The location of hsa_circ_100791 and miR-487b-5p were performed by the probes, which were respectively labelled by Cy3 and 488 (Genepharma, China). After hsa_circ_100791 and miR-487b-5p probes were incubated with probes at 37°C for 15h, and the nuclei were stained using DAPI, and signal were captured by confocal microscope.
Dual Luciferase Reporter Assay
The 3ʹ-untranslated region (3ʹ-UTR) of hsa_circ_100791 or Trim13 contain the targeted site of miR-487b-5p. We synthesized luciferase reporter plasmid of pMIRGLO- circRNA-100791 (circRNA-WT), which contain the targeted site of miR-487b-5p in 3ʹ-untranslated region (3ʹ-UTR) of hsa_circ_100791 “ATAACCA”. Simultaneously we constructed luciferase reporter plasmid of pMIRGLO -Trim13 (Trim13-WT), which contain the targeted site of miR-487b-5p in 3ʹ-untranslated region (3ʹ-UTR) of Trim13 “ATAACCA”. Then, we change the binding site of miR-487b-5p in hsa_circ_100791 or Trim 13 to construct the mutated vector (circRNA-MUT or Trim13-MUT). The HNEpCs were transfected with miR-487b-5p inhibitors or mimics, circRNA-WT or circRNA-MUT, Trim13-WT or Trim13-MUT, and the corresponding negative controls with lipofectamine 3000. After 48h transfection, Cell’s luciferase activities were measured by using dual luciferase reporter assay kit (Promega, Massachusetts, USA), and normalized to renilla luciferase activity.
Statistics
Data were expressed as mean ± standard error of the mean. Statistical analysis was performed with unpaired Student’s t-test or F-test in two groups and analyzed by GraphPad. Prism. v 8.0 software. The chi-square test was used statistics the age difference and sex ratio between two groups. p-value < 0.05 was taken to indicate statistical significance. (*p < 0.05, **p < 0.01, ***p < 0.001).
Result
Increased Expression Levels of Hsa_circ_100791 in AR Patients and Generated from the CAPRIN1 Gene
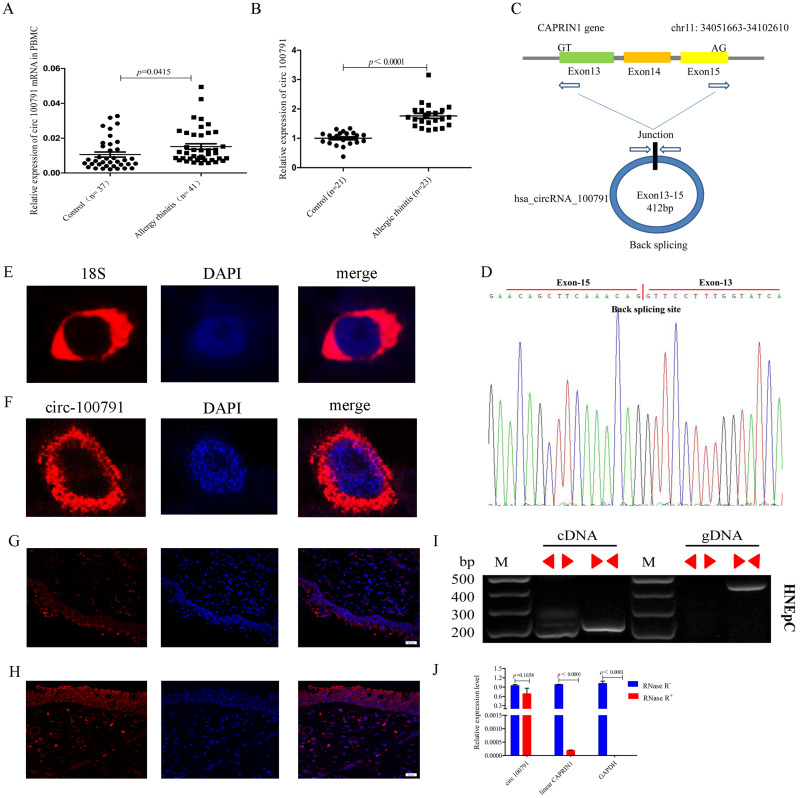
The expression of hsa_circRNA_100791 was significantly elevated (p=0.0415) in peripheral blood mononuclear cells (PBMCs) from 41 AR patients compared to 37 healthy controls (Figure 1A). A similar increase (p<0.0001) was observed in the nasal mucosa of AR patients (Figure 1B). This circRNA is derived from the CAPRIN1 gene, formed by the head-to-tail splicing of exons 13, 14, and 15, located at chr11:34051663–34102610, and it comprises 412 nucleotides (Figure 1C and D).
Figure 1.
Hsa_circ_100791 expression was increased in AR patients compared to healthy controls. (A and B) hsa_circ_100791 expression was elevated in both PBMCs and nasal mucosa from AR patients compared to healthy controls. (C) Schematic diagram illustrating that hsa_circ_100791 is generated from exons 13, 14, and 15 of the CAPRIN1 gene. (D) Sanger sequencing of hsa_circ_100791 PCR products, with the red arrow indicating the head-to-tail splicing site. (E and F) FISH analysis showing abundant expression of 18S and hsa_circ_100791 in the cytoplasm of human nasal epithelial cells (HNEpCs). Nuclei were stained with DAPI, and 18S and hsa_circ_100791 were labeled with Cy3. (G and H) FISH analysis demonstrating the abundant expression of hsa_circ_100791 in the cytoplasm of nasal mucosa from both healthy controls and AR patients. (I) PCR amplification of hsa_circ_100791 using convergent and divergent primers on cDNA and genomic DNA from HNEpCs. Results indicate successful amplification of hsa_circ_100791 with divergent primers using cDNA, but not with genomic DNA. (J) qRT-PCR analysis of RNA extracted from HNEpCs treated with RNase R, showing that hsa_circ_100791 is more stable than linear CAPRIN1 and GAPDH. Data are presented as mean ± SEM.
Abbreviations: AR, allergic rhinitis; PBMC, peripheral blood mononuclear cell; FISH, fluorescence in situ hybridization; HNEpCs, human nasal epithelial cell line; SEM, standard error of the mean.
We designed two sets of primers: convergent and divergent primers for hsa_circ_100791. PCR and agarose gel electrophoresis results showed that hsa_circ_100791 was successfully amplified by divergent primers using cDNA but not genomic DNA (Figure 1I). Fluorescence in situ hybridization (FISH) analysis revealed that hsa_circ_100791 was abundantly expressed in the cytoplasm of human nasal epithelial cells (HNEpCs) and in the nasal mucosa of both healthy controls and AR patients (Figure 1E–H). Additionally, qRT-PCR demonstrated that hsa_circ_100791 is more resistant to degradation by RNase R compared to the linear CAPRIN1 mRNA, indicating its greater stability (Figure 1J).
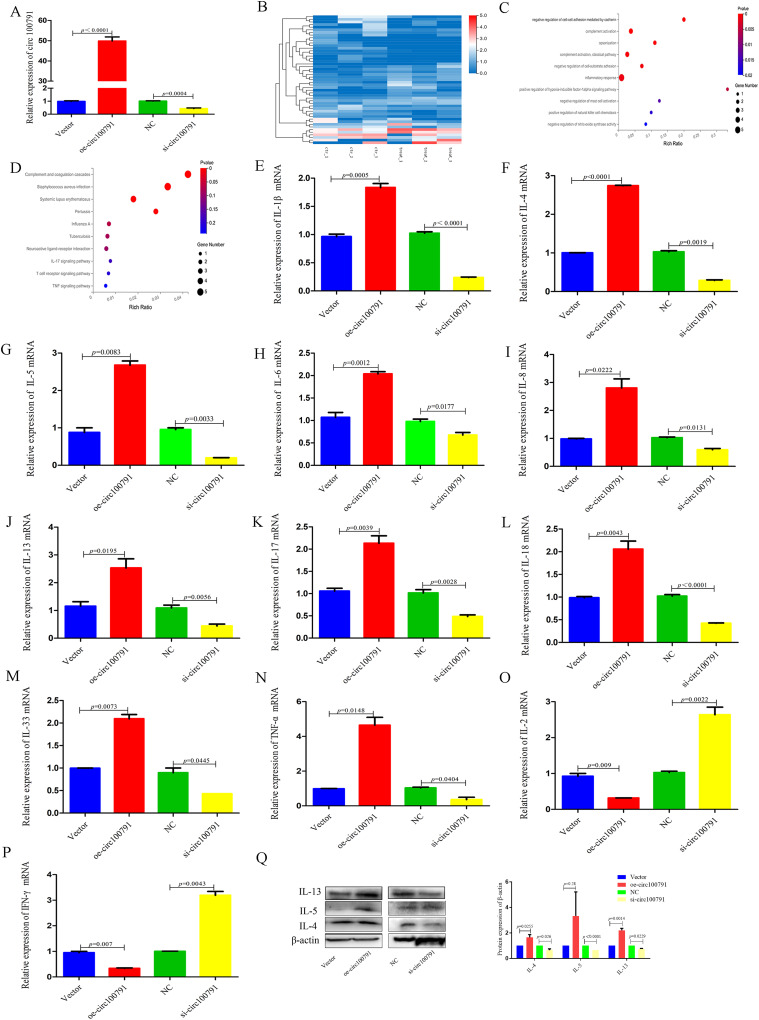
Hsa_circ_100791 Regulates the Expression of Inflammatory Factors in HNEpCs
The qRT-PCR confirmed the successful transfection of the hsa_circ_100791 overexpression vector and siRNA into HNEpCs (Figure 2A). In the si-circ-100791 group, 49 differentially expressed genes (DEGs) were identified compared to negative controls (Figure 2B). Gene Ontology (GO) analysis of these DEGs revealed their involvement in inflammatory responses, the negative regulation of mast cell activation and nitric oxide synthase activity, and the positive regulation of the hypoxia-inducible factor-1 alpha (HIF-1α) signaling pathway, among other biological processes (Figure 2C). KEGG pathway analysis of the DEGs highlighted their association with systemic lupus erythematosus, the IL-17 signaling pathway, the T cell receptor signaling pathway, and the TNF signaling pathway (Figure 2D).
Figure 2.
Hsa_circ_100791 regulates the expression of inflammatory factors in HNEpCs. (A) qRT-PCR confirmed the transfection efficiency of the hsa_circ_100791 overexpression vector or siRNA in HNEpCs. (B) The heatmap shows the differentially expressed genes between si-circ-100791 groups and NC groups, as analyzed by RNA-seq. (C) The biological process of gene ontology (GO) analysis of DEGs related to hsa_circ_100791 knockdown. (D) KEGG analysis of the DEGs related to hsa_circ_100791 knockdown. (E–N) qRT-PCR analysis shows the mRNA expression level of IL-1β, IL-4, IL-5, IL-6, IL-8, IL-13, IL-17, IL-18, IL-33 and TNF-α was significantly enhanced in hsa_circ_100791 overexpression group and obviously reduced in hsa_circ_100791 knockdown group. (O and P) qRT-PCR analysis shows the mRNA expression level of IL-2, IFN-γ was significantly reduced in hsa_circ_100791 overexpression group and obviously enhanced in hsa_circ_100791 knockdown group. (Q) Western blot analysis further revealed that the protein expression levels of IL-4, IL-5 and IL-13 was enhanced in hsa_circ_100791 overexpression group and reduced in hsa_circ_100791 knockdown group. Data were presented as mean ± SEM. N= 3 for each group.
Abbreviations: HNEpCs, human nasal epithelial cell line; AR, allergic rhinitis; NC, negative control; RNA-seq, RNA sequencing; GO, gene ontology; DEGs, Differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes; SEM, standard error of mean.
The mRNA expression levels of pro-inflammatory cytokines, including IL-1β, IL-4, IL-5, IL-6, IL-8, IL-13, IL-17, IL-18, IL-33, and TNF-α, were significantly higher in the hsa_circ_100791 overexpression group compared to the control group (p=0.0005, p<0.0001, p=0.0083, p=0.0012, p=0.0222, p=0.0195, p=0.0039, p=0.0043, p=0.0073, p=0.0148). In contrast, the mRNA expression of these pro-inflammatory cytokines was significantly reduced in the hsa_circ_100791 knockdown group compared to the negative control group (p<0.0001, p=0.0019, p=0.0033, p=0.0091, p=0.0131, p=0.0056, p=0.0028, p<0.0001, p=0.0445, p=0.0404) (Figure 2E–N).
Additionally, mRNA expression levels of IL-2 and IFN-γ were significantly reduced in the hsa_circ_100791 overexpression group (p=0.009, p=0.007) and markedly increased in the hsa_circ_100791 knockdown group (p=0.0022, p=0.0043) (Figure 2O and P). Western blot analysis further demonstrated that overexpression of hsa_circ_100791 increased the protein levels of IL-4, IL-5 and IL-13, whereas these levels were decreased in the hsa_circ_100791 knockdown group (Figure 2Q).
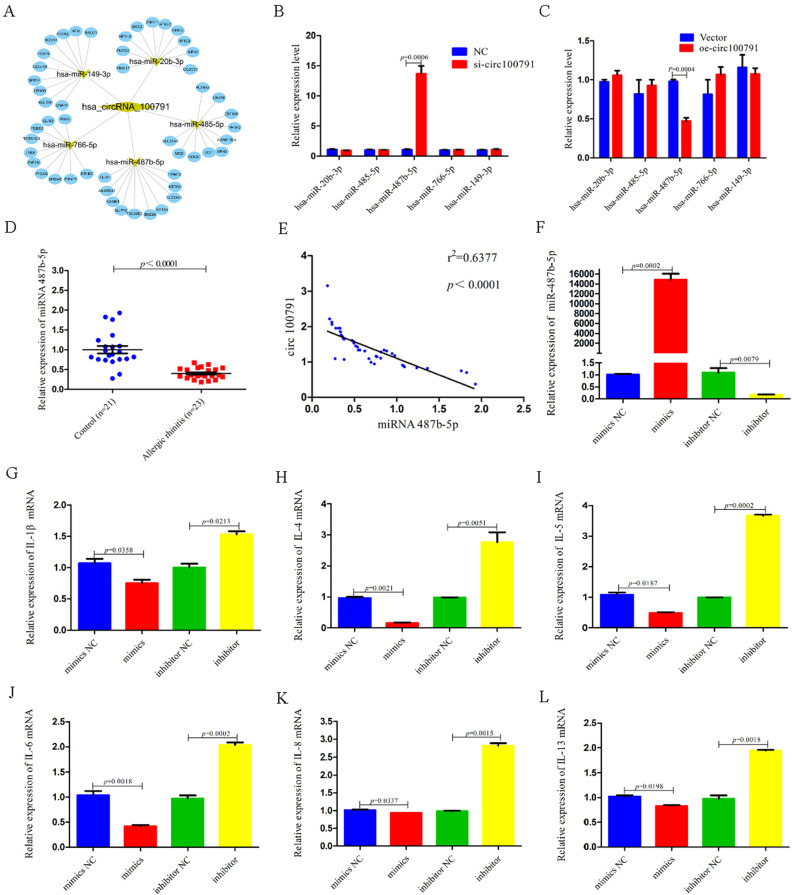
MiR-487b-5p is Lowly Expressed in AR Patients and Mediates Expression of Inflammatory Cytokines
We identified five candidate miRNAs with potential binding sites for hsa_circ_100791 using online bioinformatics tools TargetScan (http://www.targetscan.org/vert_72/) and miRanda (http://sanderlab.org/tools/micrornas.html) (Figure 3A). Among these candidates, only miR-487b-5p showed significant changes: its expression was markedly increased (p=0.0006) in hsa_circ_100791 knockdown cells and significantly reduced (p=0.0004) in hsa_circ_100791 overexpression cells (Figure 3B and C). Furthermore, miR-487b-5p levels were notably decreased (p<0.0001) in the nasal mucosa of AR patients (Figure 3D). A statistical analysis revealed a strong negative correlation (p<0.0001, r2=0.6377) between hsa_circ_100791 and miR-487b-5p in nasal mucosal tissues (Figure 3E).
Figure 3.
Continued.
Figure 3.
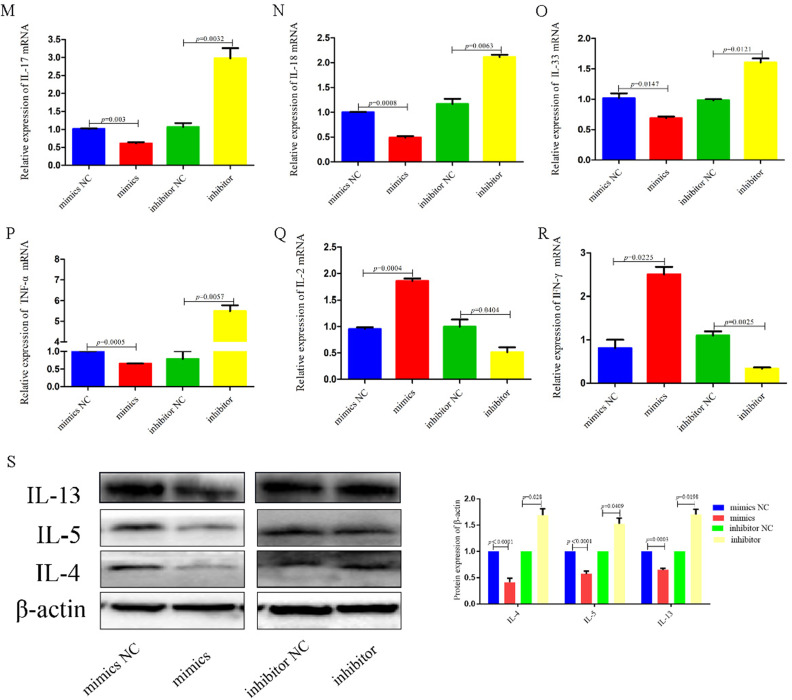
MiR-487b-5p is low expression in AR patients and mediates expression of inflammatory cytokines. (A) The 5 candidate miRNAs have binding sites for hsa_circ_100791. (B and C) qRT-PCR analysis shows the expression level of 5 candidate miRNAs after being transfected with si-hsa_circ_100791 and NC or hsa_circ_100791 overexpression and control vector into HNEpCs. (D and E) qRT-PCR analysis shows that the miR-487b-5p was decreased in nasal mucosa of the AR patients. And statistical analysis indicated that negative correlation between hsa_circ_100791 and miR-487b-5p. (F) The qRT-PCR verified transfection efficiency of transfected the miR-487b-5p mimics or inhibitor into HNEpCs. (G–P) qRT-PCR analysis shows the mRNA expression level of IL-1β, IL-4, IL-5, IL-6, IL-8, IL-13, IL-17, IL-18, IL-33 and TNF-α was significantly reduced in miR-487b-5p mimics group, but obviously enhanced in miR-487b-5p inhibitor group. (Q and R) qRT-PCR analysis shows the mRNA expression level of IL-2, IFN-γ significantly enhanced in miR-487b-5p mimics group, but obviously reduced in miR-487b-5p inhibitor group. (S) Western blot analysis further revealed that the protein expression levels of IL-4, IL-5 and IL-13 was significantly reduced in miR-487b-5p mimics group, but obviously enhanced in miR-487b-5p inhibitor group. Data were presented as mean ± SEM. N= 3 for each group.
Abbreviation: NC, negative control.
To confirm the functional role of miR-487b-5p, we assessed the transfection efficiency of miR-487b-5p mimics and inhibitors in HNEpCs using qRT-PCR (Figure 3F). The mRNA expression levels of proinflammatory cytokines IL-1β, IL-4, IL-5, IL-6, IL-8, IL-13, IL-17, IL-18, IL-33, and TNF-α were significantly reduced (p=0.0358, p=0.0021, p=0.0187, p=0.0018, p=0.0337, p=0.0198, p=0.003, p=0.0008, p=0.0147, p=0.0005) in cells transfected with miR-487b-5p mimics. Conversely, these cytokines showed significant increases (p=0.0213, p=0.0051, p=0.0002, p=0.0002, p=0.0015, p=0.0018, p=0.0032, p=0.0063, p=0.0121, p=0.0057) in cells treated with miR-487b-5p inhibitors (Figure 3G–P). Additionally, the mRNA levels of IL-2 and IFN-γ were significantly enhanced (p=0.0004 and p=0.0225) in the miR-487b-5p mimics group, while they were markedly reduced (p=0.0404 and p=0.0025) in the miR-487b-5p inhibitor group (Figure 3Q and R). Western blot analysis further confirmed that increased expression of miR-487b-5p led to a notable reduction (p<0.0001 for both IL-4 and IL-5) in the protein levels of IL-4, IL-5 and IL-13. Conversely, decreased miR-487b-5p expression resulted in a significant increase (p=0.028 and p=0.0409) in these protein levels (Figure 3S).
MiR-487b-5p Mediates Expression of Inflammatory Cytokines of HNEpCs in vitro by Targeting TRIM13
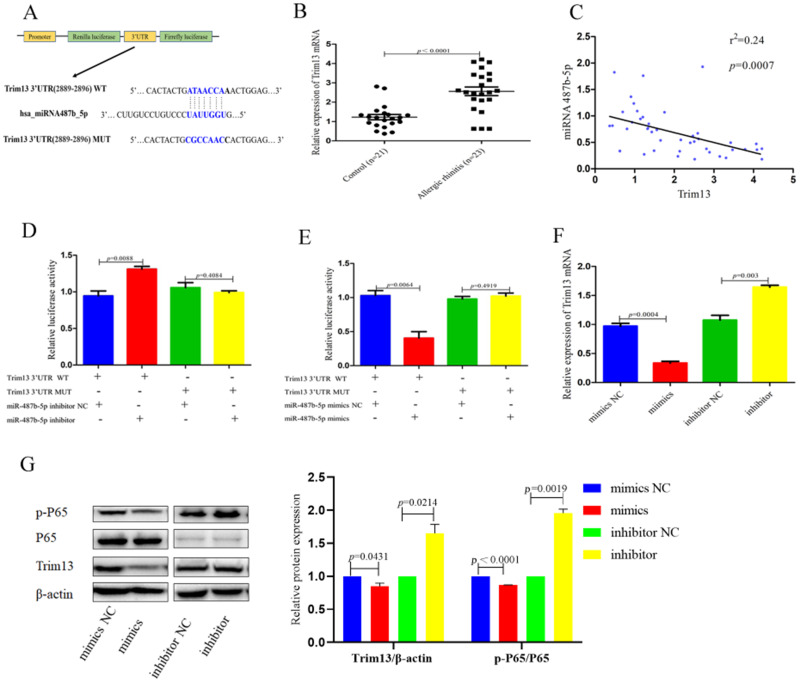
We measured the mRNA expression level of Trim13 in the nasal mucosa of AR patients and found a significant upregulation of Trim13 (p<0.0001) compared to controls (Figure 4B). Notably, there was a negative correlation between TRIM13 mRNA levels and miR-487b-5p expression (p=0.0007, r2=0.24) in these tissue samples (Figure 4C).
Figure 4.
MiR-487b-5p induces allergic responses of HNEpCs in vitro by targeting TRIM13. (A) The sequences of 3ʹUTR for TRIM13, which are complementary to miR-487b-5p seed sequence. WT represent wild-type and Mut represent mutant sequences. (B and C) qRT-PCR analysis shows that the expression of TRIM13 was increased in nasal mucosa of the AR patients (n=23) compared with nasal mucosal tissues of healthy controls (n=21). And statistical analysis indicated that negative correlation between TRIM13 and miR-487b-5p in nasal mucosal tissues. (D and E) The luciferase reporter assays results demonstrated that the luciferase activity of TRIM13-wild-type version significantly increased when miR-487b-5p knockdown in HNEpCs, while decreased when miR-487b-5p mimics in HNEpCs. However, the luciferase activity of Trim13- mutant version was unchanged. (F) qRT-PCR analysis result showed that the expression mRNA level of TRIM13 was significantly reduced in miR-487b-5p mimics group, but obviously enhanced in miR-487b-5p inhibitor group. (G) Western blot analysis result showed that the expression protein level of Trim13, p65 and p-p65 was significantly reduced in miR-487b-5p mimics group, but obviously enhanced in miR-487b-5p inhibitor group. Data were presented as mean ± SEM. Data are pooled from 3 independent experiments.
Abbreviations: HNEpCs, human nasal epithelial cell line; UTR, untranslated region; AR, allergic rhinitis; SEM, standard error of mean.
Further analysis showed that both mRNA and protein levels of TRIM13 were significantly reduced (p=0.0004, p=0.0431) in cells transfected with miR-487b-5p mimics, while they were markedly increased (p=0.003, p=0.0214) in cells treated with miR-487b-5p inhibitors (Figure 4F and G). The phosphorylation of p65 (p-p65) was significantly reduced (p<0.0001) in the miR-487b-5p mimics group and significantly increased (p=0.0019) in the miR-487b-5p inhibitor group (Figure 4G).
We cloned both the wild-type and mutant versions of the 3ʹ untranslated region (3ʹUTR) of TRIM13 into the pMIRGLO vector (Figure 4A). The luciferase assay results demonstrated that the luciferase activity of the TRIM13wild-type reporter significantly increased (p=0.0088) upon knockdown of miR-487b-5p in HNEpCs, while it significantly decreased (p=0.0064) upon overexpression of miR-487b-5p. In contrast, the luciferase activity of the TRIM13 mutant version remained unchanged regardless of whether miR-487b-5p was inhibited or mimicked (Figure 4D and E).
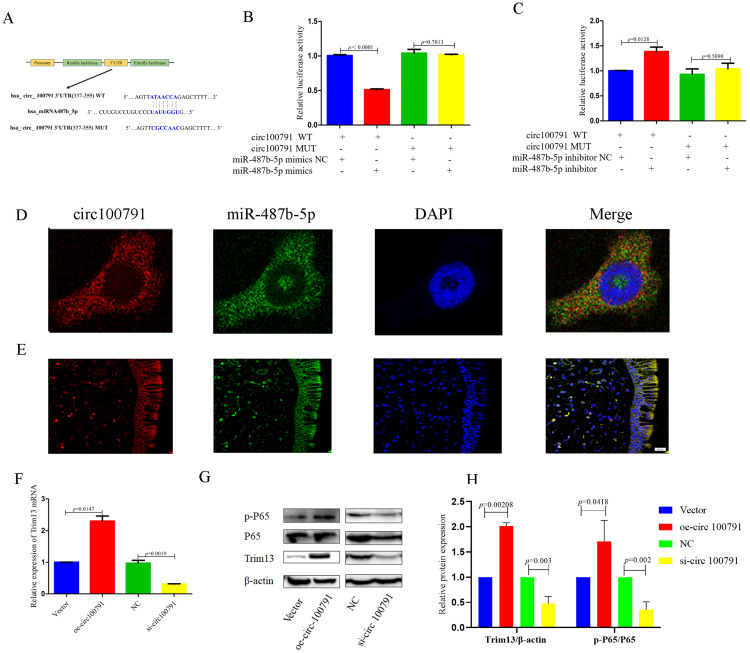
Hsa_circ_100791 Acts as a Sponge for miR-487b-5p to Regulate TRIM13 in HNEpCs
Bioinformatics analysis using TargetScan and miRanda predicted that miR-487b-5p could target hsa_circ_100791 (Figure 5A). The wild-type version of the reporter construct, which includes the complete binding sites for miR-487b-5p within the hsa_circ_100791 sequence, showed a significant reduction in luciferase activity (p<0.0001) upon transfection with miR-487b-5p mimics in HNEpCs. Conversely, luciferase activity was significantly increased (p=0.0128) when miR-487b-5p was knocked down. In contrast, the luciferase activity of the mutant version, which lacks these binding sites, did not show significant changes (Figure 5B and C).
Figure 5.
Hsa_circ_100791 acts as a sponge for miR-487b-5p to regulate TRIM13 in HNEpCs. (A) The databases predicted the sequences of hsa_circ_100791, which are complementary to miR-487b-5p seed sequence. (B and C) The luciferase reporter assays results showed that the luciferase activity of wild-type version significantly reduced when miR-487b-5p mimics in HNEpCs, while enhanced when miR-487b-5p knockdown in HNEpCs. But, the luciferase activity of mutant version was obviously different. (D and E) FISH assay revealed that hsa_circ_100791 (red) localized with miR-487b-5p (green) in the cytoplasm. Nuclei were stained with DAPI. Hsa_circ_100791 and miR-487b-5p were labeled with Cy3 and FAM, respectively. (F) qRT-PCR analysis result showed that the expression mRNA level of TRIM13 was significantly enhanced in hsa_circ_100791 overexpression group, but obviously reduced in hsa_circ_100791 knockdown group. (G and H) Western blot analysis result showed that the expression protein level of Trim13, p65 and p-p65 was significantly increased in hsa_circ_100791 overexpression group, whereas decreased in hsa_circ_100791 knockdown group. Data were presented as mean ± SEM. N=3 for each group.
Abbreviations: FISH, Fluorescence in situ hybridization; SEM, standard error of mean.
Fluorescence in situ hybridization (FISH) assays demonstrated that hsa_circ_100791 (red) co-localized with miR-487b-5p (green) in the cytoplasm of both HNEpCs and nasal mucosal tissue (Figure 5D and E).
Additionally, the mRNA and protein levels of TRIM13, a direct target gene of miR-487b-5p, were significantly increased (p=0.0147, p=0.002) upon overexpression of hsa_circ_100791, while they were significantly decreased (p=0.0019, p=0.003) in the hsa_circ_100791 knockdown group. Furthermore, the expression of phosphorylated p65 (p-p65) was significantly increased (p=0.0418) in the hsa_circ_100791 overexpression group, but markedly decreased (p=0.002) in the hsa_circ_100791 knockdown group (Figure 5F–H).
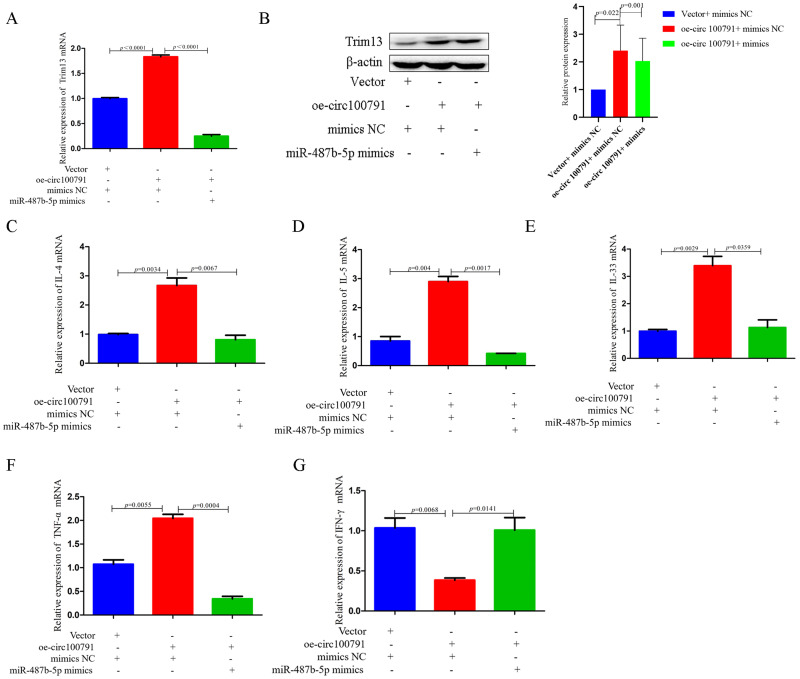
Hsa_circ_100791 Exerts Biological Functions in AR by Regulating the Expression of miR-487b-5p
QRT-PCR and Western blot analyses revealed that the expression of Trim13 was significantly decreased (p<0.0001, p=0.001) in HNEpCs co-transfected with both the hsa_circ_100791 overexpression vector and miR-487b-5p mimics, compared to cells transfected with the hsa_circ_100791 vector alone (Figure 6A and B).
Figure 6.
Hsa_circ_100791 exerts biological functions in AR by regulating the expression of miR-487b-5p. (A) qRT-PCR analysis of the expression protein level of TRIM13 in HNEpCs co-transfected with vector+ mimics NC or oe- circ-100791 + mimics NC or oe- circ-100791 + miR-487b-5p mimics. (B) Western blot analysis of the expression protein level of Trim13 in HNEpCs co-transfected with vector+ mimics NC or oe- circ-100791 + mimics NC or oe- circ-100791 + miR-487b-5p mimics. (C–G) qRT-PCR analysis of the expression protein level of IL-4, IL-5, IL-33, TNF-α and IFN-γ in HNEpCs co-transfected with vector+ mimics NC or oe- circ-100791 + mimics NC or oe- circ-100791 + miR-487b-5p mimics. Data were presented as mean ± SEM. N=3 for each group.
Abbreviations: AR, allergic rhinitis; NC, negative control; oe- circ-100791, overexpression circ-100791; SEM, standard error of mean.
Additionally, the levels of proinflammatory cytokines IL-4, IL-5, IL-33, and TNF-α were partially reduced in HNEpCs co-transfected with the hsa_circ_100791 overexpression vector and miR-487b-5p mimics, relative to those in cells transfected with the hsa_circ_100791 vector alone (Figure 6C–F). In contrast, IFN-γ levels were elevated in the co-transfected group compared to the group transfected with the hsa_circ_100791 vector alone (Figure 6G).
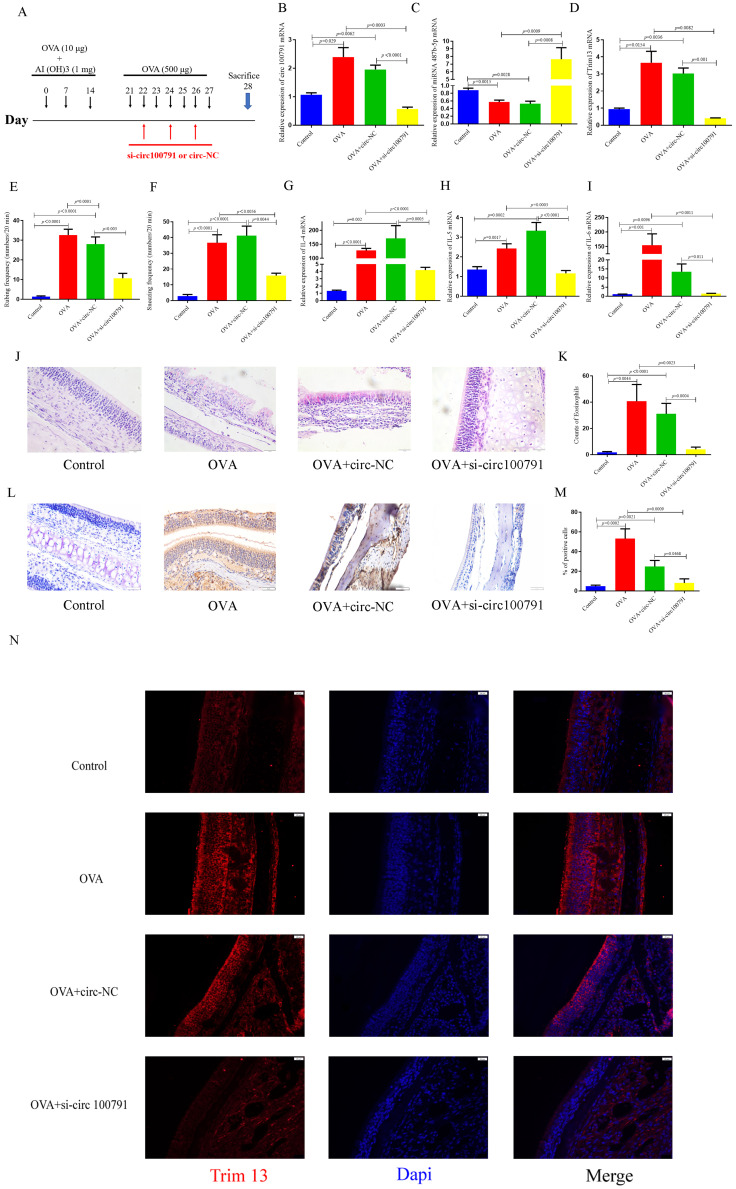
Knockdown Hsa_circ_100791 Increased the Expression of miR-487b-5p and Reduced the Nasal Symptoms of AR in OVA-Induced AR Mice
We conducted tail vein injections with circ-NC and si-circ100791 in AR mice (Figure 7A). The expression of hsa_circ_100791 was significantly increased (p=0.029) in the OVA group compared to the control group, and it was markedly decreased (p=0.0003) in the OVA+si-circ100791 group compared to the OVA group (Figure 7B). Conversely, miR-487b-5p expression was significantly reduced (p=0.0015) in the OVA group compared to the control group, and it was notably enhanced (p=0.0009) in the OVA+si-circ100791 group compared to the OVA group (Figure 7C).
Figure 7.
Knockdown hsa_circ_100791 increase the expression of miR-487b-5p and reduced the nasal symptoms of AR in OVA-induced AR mice. (A) Schematic diagram of mouse model. (B–D) qRT-PCR analysis of the expression of hsa_circ_100791, miR-487b-5p and TRIM13 in the OVA group or the OVA+ si-circ100791 or the OVA+ circ-NC group. (E and F) The frequencies of nose rubbing and sneezing were record in the OVA group or the OVA+ si-circ100791 or the OVA+ circ-NC group. (G–I) qRT-PCR analysis of the expression of IL-4, IL-5 and IL-6 in the OVA group or the OVA+ si-circ100791 or the OVA+ circ-NC group. (J and K) HE analysis of the number of eosinophils in the OVA group or the OVA+ si-circ100791 or the OVA+ circ-NC group. (L and M) IHC analysis of the expression protein level of Trim13 in the OVA group or the OVA+ si-circ100791 or the OVA+ circ-NC group. (N) IF analysis showed that the Trim13 colocalized in the cytoplasm and the expression protein level of Trim13 in the OVA group or the OVA+ si-circ100791 or the OVA+ circ-NC group. Data were presented as mean ± SEM. N=6 for each group.
Abbreviations: AR, allergic rhinitis; OVA, ovalbumin; NC, negative control; HE, hematoxylin-eosin; SEM, standard error of mean.
TRIM13 mRNA and protein levels were markedly upregulated (p=0.0154, p=0.0002) in the OVA group compared to the control group. In contrast, these levels were significantly downregulated (p=0.0082, p=0.0009) in the OVA+si-circ100791 group compared to the OVA group (Figure 7D and L–N).
The mRNA expression of IL-4, IL-5, and IL-6 was significantly elevated (p<0.0001, p=0.0017, p=0.001) in the OVA group compared to the control group, and these levels were notably decreased (p<0.0001, p=0.0003, p=0.0011) in the OVA+si-circ100791 group compared to the OVA group (Figure 7G–I).
The frequencies of nose rubbing and sneezing were significantly increased (p<0.0001, p<0.0001) in the OVA group compared to the control group, and these frequencies were markedly decreased (p=0.0001, p=0.0056) in the OVA+si-circ100791 group compared to the OVA group (Figure 7E and F).
Additionally, HE staining revealed a significant increase (p=0.0044) in the number of eosinophils in the OVA group compared to the control group. The number of eosinophils was significantly decreased (p=0.0023) in the OVA+si-circ100791 group compared to the OVA+circ-NC group (Figure 7J and K).
Discussion
AR is a prevalent allergic upper respiratory condition characterized primarily by type 2 (Th2) inflammation, which involves the excessive release of chemokines and cytokines, such as IL-4, IL-5, and IL-13, in the nasal mucosa.1,2,20 Emerging evidence highlights the significant role of circular RNAs (circRNAs) in autoimmune diseases, both in diagnosis and therapy, by regulating gene expression.16,21–23 For example, circRNAs have been implicated in systemic lupus erythematosus22 and rheumatoid arthritis.23 Most research on circRNAs has concentrated on their functions in immune cells, particularly neutrophils and macrophages.24 Furthermore, circRNAs are closely linked to various biological pathways in peripheral blood mononuclear cells (PBMCs), including “Cytokine-cytokine receptor interaction”, “Chemokine signaling pathway”, and “Bacterial invasion of epithelial cells”.25,26 Additionally, circRNAs are associated with antibacterial immune responses; for instance, Huang et al25 identified hsa_circ_001937 as playing a role in tuberculosis by regulating the NF-κB signaling pathway, suggesting its potential as a diagnostic marker for the disease. CircHIPK3 promotes Th2 differentiation via the miR-495/GATA-3 pathway.16 circZNF652 enhances goblet cell metaplasia by targeting the miR-452-5p/JAK2 signaling pathway in allergic airway epithelia.27 circRAPGEF5 competitively binds to miR-27a-3p, regulating TXNIP levels and inhibiting the progression of renal cell carcinoma.28 circTP63 promotes lung squamous cell carcinoma progression through the miR-873-3p/FOXM1 pathway.29 These findings underscore the diverse roles of circRNAs in regulating various biological processes and highlight their potential as therapeutic targets.
In this study, we confirmed, in line with previous research, that hsa_circ_100791 is generated by the head-to-tail splicing of exons 13, 14, and 15 of the CAPRIN1 gene.9,30,31 This head-to-tail splicing is the result of reverse splicing of cDNA, not gDNA. Hsa_circ_100791 is more stable than linear RNA due to its resistance to degradation by RNase R. We found that hsa_circ_100791 was significantly upregulated in patients with AR compared to healthy controls. These results suggest a close relationship between the overexpression of hsa_circ_100791 and the development of AR. Our findings also indicate that increased levels of hsa_circ_100791 can enhance the expression of pro-inflammatory mediators and reduce the expression of Th1-related inflammatory cytokines in vitro. Conversely, silencing hsa_circ_100791 expression can partially alleviate AR symptoms and the type 2 inflammatory process both in vivo and in vitro. Thus, our study highlights the crucial role of hsa_circ_100791 in promoting type 2 inflammation associated with AR.
Research has indicated that exon-derived circRNAs are primarily located in the cytoplasm, where they function as miRNA sponges to regulate downstream target genes and exert various biological effects. For instance, CircPVT1 promotes the expression of IGF2BP1, KRAS, and HMGA2 by sponging the let-7 family, thus preventing cellular senescence.32 Similarly, circSLC8A1 regulates PTEN expression by sponging miR-130b/miR-494 in bladder cancer,33 and circ-MBOAT2 counteracts the inhibitory effects of miR-433-3p on its target gene GOT1 in pancreatic cancer.34
In our study, FISH assays demonstrated that hsa_circ_100791 predominantly and stably exists in the cytoplasm of HNEpCs. Using online bioinformatics tools such as TargetScan (http://www.targetscan.org/vert_72/) and miRanda (http://sanderlab.org/tools/micrornas.html), we identified five candidate miRNAs with potential binding sites for hsa_circ_100791. qRT-PCR results confirmed that the expression of miR-487b-5p was significantly altered following transfection with either a specific small interference RNA targeting hsa_circ_100791 or a hsa_circ_100791 overexpression vector in HNEpCs. Statistical analysis revealed a negative correlation between the expression levels of hsa_circ_100791 and miR-487b-5p in AR tissues. Further validation through dual luciferase reporter assays and FISH experiments confirmed the interaction and co-localization of hsa_circ_100791 and miR-487b-5p in HNEpCs. These findings suggest that hsa_circ_100791 regulates miR-487b-5p expression by acting as a sponge.
Studies have shown that miR-487b can suppress the expression of proinflammatory cytokines and alleviate the pathological changes associated with AR.5 To explore the biological functions and mechanisms of miR-487b-5p, we transfected HNEpCs with miR-487b-5p mimics or inhibitors. Our results revealed that miR-487b-5p is downregulated in AR patients. Additionally, miR-487b-5p significantly reduced the expression of proinflammatory cytokines (including IL-1β, IL-4, IL-5, IL-6, IL-8, IL-13, IL-17, IL-18, IL-33, and TNF-α) and enhanced the expression of IL-2 and IFN-γ, thereby mitigating inflammation in AR.
Since miRNAs regulate target gene expression by binding to the 3ʹ UTR of these genes, we used the online bioinformatics tool miRDB to identify Trim13 as a potential target of miR-487b-5p. Our findings indicate that Trim13 is upregulated in AR patients and negatively correlates with miR-487b-5p levels. Moreover, the expression of Trim13 was significantly altered following the introduction of miR-487b-5p mimics or inhibitors in HNEpCs. These results suggest that miR-487b-5p plays a critical role in inhibiting the inflammatory response in AR by targeting Trim13. This was further supported by our qRT-PCR, Western blotting, and dual-luciferase reporter assay experiments.
TRIM13, a RING-type E3 ubiquitin ligase, is known to enhance the activity of NF-κB, thereby amplifying immune responses.35 Previous studies have indicated that TRIM13 positively regulates NF-κB activation. For instance, NF-κB and its p65 subunit have been found to be upregulated in AR nasal mucosa tissues, where activated NF-κB contributes to the pathogenesis and progression of AR.36 In our study, we observed that the expression levels of both p65 and phosphorylated p65 (p-p65) were significantly reduced in the miR-487b-5p mimics group and the hsa_circ_100791 knockdown group. Conversely, these levels were notably increased in the miR-487b-5p inhibitor group and the hsa_circ_100791 overexpression group. The expression of p-p65 being unexpectedly higher than that of total p65 in Figure 4G. One possible explanation for this observation could be related to the dynamic regulation of p65 phosphorylation in response to the Lipo2000 transfection of miRNA NC and inhibitor sequences used in our experiments. Phosphorylation may occur rapidly following stimulation, leading to a transient increase in p-p65 levels compared to total p65. Additionally, variations in protein stability or degradation rates could also contribute to the observed discrepancy.
To assess the role of miR-487b-5p in the inflammatory response induced by hsa_circ_100791, we conducted a rescue experiment in vitro. Our findings demonstrated that upregulation of miR-487b-5p could partially mitigate the inflammatory effects caused by hsa_circ_100791 overexpression in AR. These results suggest that hsa_circ_100791 promotes AR inflammation by sponging miR-487b-5p, thereby modulating TRIM13 expression.
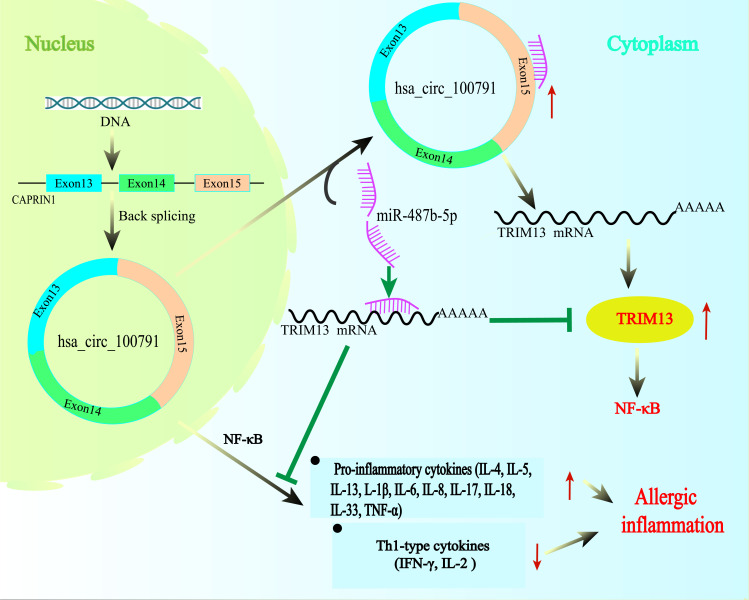
In summary, our study reveals that hsa_circ_100791 is significantly upregulated in AR patients. Elevated levels of hsa_circ_100791 enhance the expression of several inflammatory cytokines (including IL-1β, IL-4, IL-5, IL-6, IL-13, IL-17, IL-18, IL-33, TNF-α) and NF-κB, while suppressing Th1-related cytokines (IL-2 and IFN-γ) in HNEpCs. Furthermore, hsa_circ_100791 counteracts the inhibitory effects of miR-487b-5p on Trim13 by directly binding to miR-487b-5p, thereby significantly influencing the expression of inflammatory factors and contributing to the inflammatory response in AR (Figure 8). Our study not only elucidates the molecular mechanism by which hsa_circ_100791 regulates AR pathogenesis but also identifies potential new targets for future therapeutic interventions.
Figure 8.
Schematic show that the mechanism of hsa_circ_100791 promote the progression of allergic rhinitis. Hsa_circ_100791 could increase the expression of TRIM13 by sponging miR-487b-5p, and consequently activate the NF-κB signaling pathway, and upregulates the expression of pro-inflammatory factors (IL-4, IL-5 and IL-13, IL-1β, IL-6, IL-8, IL-17, IL-18, IL-33 and TNF-α), inhibits the expression of Th1-related inflammatory factors (IL-2 and IFN-γ), and ultimately promotes the the inflammatory response of AR.
Funding Statement
This work was supported by the National Nature Science Foundation of China (82371117; Li Shi) and Key R & D Program of Shandong Province (2018CXGC1214; Li Shi), the National Nature Science Foundation of China (82101195; Xiaoxue Zi).
Ethics Statement
We confirm that our study complies with the Declaration of Helsinki, and all procedures involving human participants were approved by the relevant ethical review board.
Disclosure
The authors declared that no compete of interest exists.
References
- 1.Bousquet J, Anto JM, Bachert C, et al. Allergic rhinitis. Nat Rev Dis Primers. 2020;6:95. doi: 10.1038/s41572-020-00227-0 [DOI] [PubMed] [Google Scholar]
- 2.Brożek JL, Bousquet J, Agache I, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-2016 revision. J Allergy Clin Immunol. 2017;140:950–958. doi: 10.1016/j.jaci.2017.03.050 [DOI] [PubMed] [Google Scholar]
- 3.Chang CJ, Yang Y-H, Liang Y-C, et al. A novel phycobiliprotein alleviates allergic airway inflammation by modulating immune responses. Am J Respir Crit Care Med. 2011;183:15–25. doi: 10.1164/rccm.201001-0009OC [DOI] [PubMed] [Google Scholar]
- 4.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 5.Liu HC, Liao Y, Liu CQ. miR-487b mitigates allergic rhinitis through inhibition of the IL-33/ST2 signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:8076–8083. doi: 10.26355/eurrev_201812_16497 [DOI] [PubMed] [Google Scholar]
- 6.Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016;137:1423–1432. doi: 10.1016/j.jaci.2016.01.029 [DOI] [PubMed] [Google Scholar]
- 7.Xiao L, Jiang L, Hu Q, Li Y. MicroRNA-133b ameliorates allergic inflammation and symptom in murine model of allergic rhinitis by targeting Nlrp3. Cell Physiol Biochem. 2017;42:901–912. doi: 10.1159/000478645 [DOI] [PubMed] [Google Scholar]
- 8.Yamazumi Y, Sasaki O, Imamura M, et al. The RNA binding protein Mex-3B is required for IL-33 induction in the development of allergic airway inflammation. Cell Rep. 2016;16:2456–2471. doi: 10.1016/j.celrep.2016.07.062 [DOI] [PubMed] [Google Scholar]
- 9.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 10.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 12.Huang Z, Fu B, Qi X, et al. Diagnostic and therapeutic value of Hsa_circ_0002594 for T helper 2-mediated allergic asthma. Int Arch Allergy Immunol. 2020;16:1–11. [DOI] [PubMed] [Google Scholar]
- 13.Huang Z, Cao Y, Zhou M, et al. Hsa_circ_0005519 increases IL-13/IL-6 by regulating hsa-let-7a-5p in CD4+ T cells to affect asthma. Clin Exp Allergy. 2019;49:1116–1127. doi: 10.1111/cea.13445 [DOI] [PubMed] [Google Scholar]
- 14.Zeng H, Gao H, Zhang M, et al. Atractylon treatment attenuates pulmonary fibrosis via regulation of the mmu_circ_0000981/miR-211-5p/TGFBR2 axis in an ovalbumin-induced asthma mouse model. Inflammation. 2021;44:1856–1864. doi: 10.1007/s10753-021-01463-6 [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Wang P, Chen D, Xu Z, Yang L. circARRDC3 contributes to interleukin‑13‑induced inflammatory cytokine and mucus production in nasal epithelial cells via the miR‑375/KLF4 axis. Mol Med Rep. 2021;23:141. doi: 10.3892/mmr.2020.11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Wang X, Wang Y, Zhao Y. The regulatory network among CircHIPK3, LncGAS5, and miR-495 promotes Th2 differentiation in allergic rhinitis. Cell Death Dis. 2020;11:216. doi: 10.1038/s41419-020-2394-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Y, Li C, Wang Z, et al. TRIM13 reduces damage to alveolar epithelial cells in COPD by inhibiting endoplasmic reticulum stress-induced ER-Phagy. Lung. 2024;202:821–830. doi: 10.1007/s00408-024-00753-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatchi EM, Poalas K, Cordeiro N, N’Debi M, Gavard J, Bidère N. Participation of the E3-ligase TRIM13 in NF-κB p65 activation and NFAT-dependent activation of c-Rel upon T-cell receptor engagement. Int J Biochem Cell Biol. 2014;54:217–222. doi: 10.1016/j.biocel.2014.07.012 [DOI] [PubMed] [Google Scholar]
- 19.Jin P, Sun K, Zhang Q, et al. Peripheral circular RNA profiling from patients with allergic rhinitis identified hsa_circRNA_404013 as a potential diagnostic biomarker. Int Arch Allergy Immunol. 2022;183(10):1078–1088. PMID: 35882184. doi: 10.1159/000525791 [DOI] [PubMed] [Google Scholar]
- 20.Breiteneder H, Peng Y-Q, Agache I, et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy. 2020;75:3039–3068. doi: 10.1111/all.14582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Xiao X, He S, Qiao Y, Ma S. Altered circular RNA expression profiles in an ovalbumin-induced murine model of allergic rhinitis. Allergol Immunopathol. 2021;49:94–103. doi: 10.15586/aei.v49i2.33 [DOI] [PubMed] [Google Scholar]
- 22.Li LJ, Zhu Z-W, Zhao W, et al. Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology. 2018;155:137–149. doi: 10.1111/imm.12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep. 2017;16:8029–8036. doi: 10.3892/mmr.2017.7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang Y, Li X, Zhang M, Lv K. Microarray analysis of circular RNA expression patterns in polarized macrophages. Int J Mol Med. 2017;39:373–379. doi: 10.3892/ijmm.2017.2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian Z, Liu H, Li M, et al. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine. 2018;27:18–26. doi: 10.1016/j.ebiom.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang ZK, Yao F-Y, Xu J-Q, et al. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell Physiol Biochem. 2018;45:1230–1240. doi: 10.1159/000487454 [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Xu C, Cai Y, et al. CircZNF652 promotes the goblet cell metaplasia by targeting the miR-452-5p/JAK2 signaling pathway in allergic airway epithelia. J Allergy Clin Immunol. 2022;150(1):192–203. PMID: 35120971. doi: 10.1016/j.jaci.2021.10.041 [DOI] [PubMed] [Google Scholar]
- 28.Chen Q, Liu T, Bao Y, et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77. doi: 10.1016/j.canlet.2019.10.017 [DOI] [PubMed] [Google Scholar]
- 29.Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. doi: 10.1038/s41467-019-11162-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maass PG, Glažar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med. 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 32.Panda AC, Grammatikakis I, Kim KM, et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017;45:4021–4035. doi: 10.1093/nar/gkw1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Liu K, Cui J, et al. Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. J Exp Clin Cancer Res. 2021;40:124. doi: 10.1186/s13046-021-01894-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang B, Baek SH. Trim13 potentiates toll-like receptor 2-mediated nuclear factor κB activation via K29-linked polyubiquitination of tumor necrosis factor receptor-associated factor 6. Mol Pharmacol. 2017;91:307–316. doi: 10.1124/mol.116.106716 [DOI] [PubMed] [Google Scholar]
- 36.Wang SZ, Ma FM, Zhao JD. Expressions of nuclear factor-kappa B p50 and p65 and their significance in the up-regulation of intercellular cell adhesion molecule-1 mRNA in the nasal mucosa of allergic rhinitis patients. Eur Arch Otorhinolaryngol. 2013;270:1329–1334. doi: 10.1007/s00405-012-2136-y [DOI] [PubMed] [Google Scholar]