Abstract
Purpose
Fetal nucleated red blood cells (fNRBCs) in the peripheral blood of pregnant women contain comprehensive fetal genetic information, making them an ideal target for non-invasive prenatal diagnosis (NIPD). However, challenges in identifying, enriching, and detecting fNRBCs limit their diagnostic potential.
Methods
To overcome these obstacles, we developed a novel biomimetic chip, replicating the micro-nano structure of red rose petals on polydimethylsiloxane (PDMS). The surface was modified with gelatin nanoparticles (GNPs) and affinity antibodies to enhance cell adhesion and facilitate specific cell identification. We subsequently investigated the chip’s characteristics, along with its in vitro capture and release system, and conducted further experiments using peripheral blood samples from pregnant women.
Results
In the cell line capture and release assay, the chip achieved a cell capture efficiency of 90.4%. Following metalloproteinase-9 (MMP-9) enzymatic degradation, the release efficiency was 84.08%, with cell viability at 85.97%. Notably, fNRBCs can be captured from the peripheral blood of pregnant women as early as 7 weeks of gestation. We used these fNRBCs to diagnose a case of single-gene disease and instances of chromosomal aneuploidies, yielding results consistent with those obtained from amniotic fluid punctures.
Conclusion
This novel chip not only enables efficient enrichment of fNRBCs for NIPD but also extends the diagnostic window for genetic and developmental disorders to as early as 7 weeks of gestation, potentially allowing for earlier interventions. By improving the accuracy and reliability of NIPD, this technology could reduce reliance on invasive diagnostic techniques, offering a new pathway for diagnosing fetal genetic conditions in clinical practice.
Keywords: non-invasive prenatal diagnosis, fetal nucleated red blood cells, nanostructure microchip, chromosomal aneuploidy, monogenic disease
Introduction
Birth defects are a significant cause of early newborn mortality and chronic illness1, primarily attributed to chromosomal or genetic abnormalities.2 These conditions place a heavy burden on affected families and society, highlighting the importance of prenatal testing in reducing the incidence of birth defects. Traditional prenatal diagnostic methods, such as chorionic villus sampling and amniocentesis,3 rely on the analysis of fetal materials. However, these invasive procedures carry an increased risk of miscarriage and impose psychological stress on pregnant women.4 Furthermore, these techniques are unsuitable for women with blood type incompatibilities, infections, or bleeding disorders.5–7 As a result, the development of non-invasive prenatal diagnostic (NIPD) methods has become a key area of research.
Currently, fetal chromosomal and genetic abnormalities are screened using cell-free fetal DNA (cffDNA).8 However, because cffDNA originates from the placenta, it can be affected by placental mosaicism, leading to discordant results.9,10 Additionally, due to the fragmented nature of cffDNA, complex mutations in homologous genomic regions may not be detectable.11 By contrast, circulating fetal cells in maternal blood and exfoliated fetal cells in the maternal vagina—primarily fetal nucleated red blood cells (fNRBCs) and trophoblast cells—offer greater potential for NIPD.12 Although trophoblast cells also originate from the placenta, they contain a complete fetal genome and have shown significant promise in clinical research.13,14 This study focuses on fNRBCs as the target cells, primarily because they are of fetal origin and thus avoid inconsistencies caused by placental mosaicism. fNRBCs offer several advantages for prenatal diagnosis: (1) They are continuously present in maternal blood throughout pregnancy;15 (2) After delivery, they are cleared from the maternal circulation, so cells from previous pregnancies do not interfere with results;16 (3) They carry the complete fetal genetic profile, making them ideal for diagnosing chromosomal and genetic disorders; (4) Their numbers are unaffected by maternal body mass index (BMI);17 and (5) They possess specific markers that facilitate cell sorting.18,19 Despite these advantages, fNRBCs are scarce in maternal circulation, driving research efforts to develop highly sensitive and specific enrichment methods for downstream analysis.
To date, various techniques for separating fNRBCs have been explored, including density gradient centrifugation20 (DGC), fluorescence-activated cell sorting21 (FACS), and magnetic activated cell sorting22 (MACS). DGC typically yields cells with low purity, while FACS requires advanced equipment and significant volumes of maternal peripheral blood. In MACS, magnetic particles can remain attached to the cells, potentially interfering with subsequent analyses. Consequently, there is ongoing research into chip technology for fNRBCs isolation. The microfluidic chip23 technique sorts cells by implementing multiple reaction protocols on the chip based on the cells’ characteristics. However, the complex material production processes and cumbersome experimental protocols hinder its widespread use for fNRBCs sorting. Recently, researchers have developed nanomaterials with multivalent activities and high affinities, capable of enriching fNRBCs by modifying the surfaces of nanoscale chips and microspheres.24–28 The construction and modification of these nanomaterials can significantly enhance capture efficiency. Nevertheless, non-specific adsorption of other cells poses a considerable challenge to the complete isolation and identification of fNRBCs.
Nanostructured biological surfaces facilitate numerous cellular functions, including recognition, adhesion,29 proliferation, and interaction.30 Previous studies have shown that micro-nanostructures can dramatically enhance cell capture ability compared to traditional plate substrates by strengthening local interactions between the substrate material and the filopodia and microvilli on the cell surface.31 Furthermore, micro-nanostructures increase the surface area available for antibody binding, thereby enhancing the rate of antibody attachment to membrane receptors and improving cell capture effectiveness. Additionally, these structures can create a more favorable environment for cell survival. This suggests that biological interfaces for target cells may be developed by integrating surface chemistry with nanostructures to promote efficient cell-material interactions. Although some studies have synthesized multi-layer composite materials using chemical techniques,32–34 these processes tend to be complex. Nature offers a powerful source of inspiration for materials design, having produced many sophisticated multi-layer composite structures with remarkable performance, enabling various complex functions35—such as cellulose, chitin,36 lotus leaves,37 and rose petals. Drawing from these biomimetic hierarchical designs, materials scientists have developed various sustainable, multi-level composite nanostructure materials using simplified preparation processes.
Prior studies have examined the microcapillaries and nanoscale epidermal folds on the surfaces of red rose petals. The integration of micro-nanostructures enhances cell adhesion to the surface, while the micropapillary system conforms to cell curvature, facilitating cell accommodation.38 With a micropapillary diameter of approximately 20–30 μm—comparable to that of fNRBCs—red rose petals promote cell attachment.39 Consequently, we selected red rose petals as the template for this investigation. Additionally, the nanoscale folds of the microstructure encourage the expansion of cell pseudopodia. Polydimethylsiloxane (PDMS) was chosen as the substrate due to its non-toxic nature, ease of shaping, and mechanical properties akin to the extracellular matrix. Gelatin nanoparticles (GNPs), a natural protein characterized by excellent biocompatibility, high cell affinity, and biodegradability, were utilized to create multifunctional nanofacials for fNRBCs isolation. The GNPs-based nanointerface rapidly responds to the matrix metalloproteinase-9 (MMP-9) enzyme, enabling partial enzymatic degradation of the GNPs layer on the chip, which facilitates the release of adhered cells.40
In this study, we present a novel non-invasive, cell-based prenatal diagnostic system designed to provide comprehensive fetal samples for the detection of fetal chromosomes and genetic diseases. We developed a bionic micro-nano hierarchical composite structure chip by modifying GNPs and biotinylated anti-CD147 antibodies on a concave PDMS scaffold, referred to as GNPs-rPDMS. Leveraging the complementary effects of physical terrain interactions and molecular recognition, this chip effectively captures fNRBCs. We conducted a series of studies to evaluate the cell capture and release capabilities of the chip, as well as the diagnostic potential of peripheral blood fNRBCs in pregnant women. To provide a visual summary of our experimental workflow, we present a flowchart in Figure 1. We believe this study will provide valuable guidance for the enrichment and analysis of fNRBCs, offering novel insights into non-invasive prenatal diagnosis.
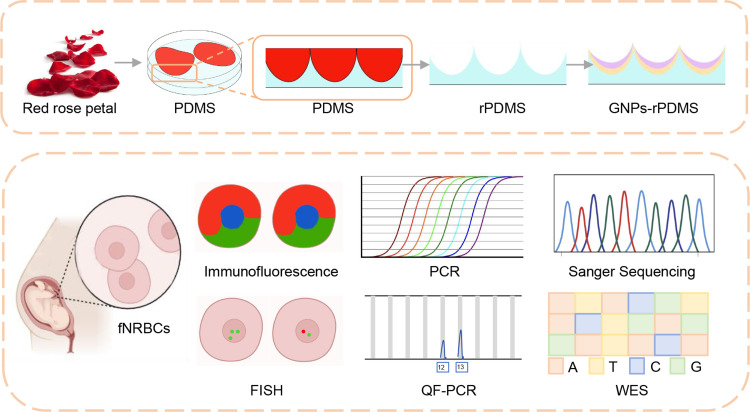
Figure 1.
Overview of the preparation, characterization, and application of GNPs-rPDMS for fetal cell isolation and genetic diagnosis. The red rose petal structure is replicated onto polydimethylsiloxane (PDMS) to create reverse PDMS (rPDMS). This surface is further modified with gelatin nanoparticles (GNPs) to enhance cell capture efficiency. Fetal nucleated red blood cells (fNRBCs) are identified through immunofluorescence and fluorescence in situ hybridization (FISH). The enriched cells are applied for FISH, quantitative fluorescence polymerase chain reaction (QF-PCR), Sanger sequencing, and whole-exome sequencing (WES) to enable genetic diagnosis.
Material and Methods
Materials
The polydimethylsiloxane (PDMS) and curing agent were obtained from Momentive (Waterford, NY, USA). Lymphocyte separation medium (LSM), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), (3-aminopropyl) triethoxysilane (APTES), paraformaldehyde (PFA), Triton X-100, bovine serum albumin (BSA), gelatin (Type B, 225 Bloom), glutaraldehyde solution, and matrix metalloproteinase-9 (MMP-9) were sourced from Sigma-Aldrich (St. Louis, MO, USA). Anti-fluorescence quenching edge sealing solution (containing 4.6-diamidino-2-phenylindole dihydrochloride) was purchased from Beyotime Biotechnology (Shanghai, China). Streptavidin was acquired from Invitrogen (Carlsbad, CA, USA), while biotinylated anti-CD147 was obtained from R&D Systems (Minneapolis, MN, USA). Phycoerythrin (PE)-mouse anti-human fetal hemoglobin antibody was procured from BD Biosciences (Franklin Lakes, NJ, USA), and fluorescein isothiocyanate (FITC)-anti-human GPA antibody and rhGM-CSF were obtained from Proteintech (Wuhan, China). PRMI-1640 and 100× penicillin/streptomycin were sourced from Procell (Wuhan, China). Fetal bovine serum was purchased from Biological Industries (Kibbutz Beit Haemek, Israel). Fluorescein diacetate (FDA) and propidium iodide (PI) were obtained from Solarbio (Beijing, China). The MALBAC single-cell whole genome amplification kit was obtained from Yikon Genomics (Suzhou, China), while the Short Tandem Repeat detection kit and the fluorescence in situ hybridization detection kit were both acquired from Microread (Beijing, China) and LBP (Guangzhou, China), respectively.
Synthesis and Activation of GNPs
To prepare the gelatin solution,41 0.625 g of Gelatin Type B was dissolved in 12.5 mL of deionized DI water at 50 °C. The solution was kept heated while 12.5 mL of acetone was added and stirred. After allowing the mixture to settle, the supernatant was removed, and an additional 12.5 mL of DI water was added to redissolve the gelatin at 50 °C. The pH was adjusted to 10, and acetone was added until the solution turned turbid. Subsequently, 1 mL of 2.5% glutaraldehyde was added slowly, and the mixture was stirred at room temperature for 16 hours. The gelatin nanoparticles (GNPs) were purified by centrifugation three times, redispersed in DI water, and stored at 4 °C.
Before use, the GNPs were thoroughly washed with DI water and 0.1 M MES solution, then activated in a 0.1 M MES solution containing EDC (4 mg/mL) and NHS (6 mg/mL) for 30 minutes.
Preparation of GNPs-rPDMS
The red roses (Rosa carolina) used in this study were procured from a local florist in Wuhan, China, as cut flowers, which were immediately transported to the laboratory for subsequent experiments. No specific permissions were required for the purchase of these commercially available plants, commonly cultivated for decorative purposes. In this study, the reverse pattern of red rose petals was replicated on PDMS. The PDMS elastomer and curing agent were mixed in a 10:1 ratio, degassed, and poured over fresh red rose petals. After curing at 70 °C for 2 hours, the red rose petals were completely removed.38 The resulting chip was washed and baked at 80 °C for 1 hour. Next, the chip was treated with O2 plasma for 3 minutes and immersed in a 5% APTES solution in ethanol at room temperature for 1 hour, followed by three washes with ethanol. Subsequently, the activated GNP solution (2 mg/mL) was incubated on the chip for 1.5 hours, allowing GNPs to modify the chip surface. Finally, the chip was treated with 50 μg/mL streptavidin in PBS solution at 4 °C.
Before use, the prepared chip was washed three times with PBS at room temperature, and surface moisture was slightly removed. Subsequently, the surface was modified with biotinylated anti-CD147 antibody for 2 hours. Following the cleaning of the uncombined antibody, the experiment commenced.
Characterization of GNPs-rPDMS
The size of the GNPs was measured using dynamic light scattering (DLS; Malvern Zetasizer Nano 266 ZS90) and scanning electron microscopy (SEM; Zeiss, 264 Sigma 300). The zeta potential of GNPs was also determined by DLS, while SEM provided insights into the morphology of GNPs-rPDMS.
TF-1 Cell Culture, Capture and Release Assays
The nucleated erythrocyte cell line (359057, TF-1) was obtained from BNCC. TF-1 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2.5 ng/mL rhGM-CSF, under an atmosphere of 5% CO2 at 37°C.
For the cell capture assay, 104 cells were stained with FDA and placed on the chip surface. After varying incubation times, non-specifically adsorbed cells were washed away with PBS. The cells were then photographed under a fluorescence microscope (IX-81, Olympus, Japan) and counted using Image-Pro Plus.
In the cell release assay, the chip was placed in a 12-well plate, and 0.1 mg/mL MMP-942 was incubated on the chip surface for different durations to degrade GNPs. The resulting suspension was gently shaken and removed. Cells were stained with FDA and PI for 5 minutes and observed under a fluorescence microscope to evaluate cell activity. The calculation formula applied is as follows:
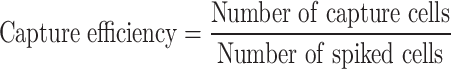 |
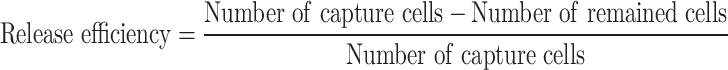 |
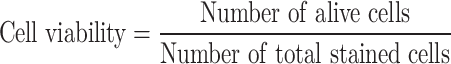 |
fNRBCs Capture, Immunofluorescence Staining and Release Assays
This study was conducted in accordance with the Declaration of Helsinki and approved by the Medical Ethics Committee of Zhongnan Hospital of Wuhan University (ethical number: 2021127). Informed consent was obtained from all participants, including pregnant women undergoing routine medical testing at the hospital. Each participant received a thorough explanation of the study, and written informed consent was obtained before commencement. The consent process was conducted by trained research staff in a private setting, allowing participants to ask questions and seek clarifications. Blood samples were collected as part of routine medical examinations, with no additional procedures or burdens imposed on the participants. To ensure confidentiality and privacy, all personal identifiers were removed from the dataset, and samples were assigned unique identifiers. Access to the data was restricted to authorized personnel only, and all data were securely stored in accordance with the policies of Zhongnan Hospital of Wuhan University. Blood samples were collected from 28 pregnant women at 5–24 weeks of gestation (n = 28), with each woman providing 2 mL of blood in a K2-EDTA tube. Samples were stored at 4°C, transported to the laboratory in frozen packaging, and processed within 6 hours. The pre-processing of blood samples adhered to the LSM protocol. An equal volume of PBS was mixed with the blood, resulting in the formation of a distinct red-white interface on the surface of the LSM. Following centrifugation at 1500 rpm for 30 minutes, four liquid layers were obtained, with the second layer comprising the mononuclear cells containing fNRBCs. The cell suspension was then applied to a chip modified with a biotinylated anti-CD147 antibody. Capture and release assays for fNRBCs were conducted under optimal conditions established through assays in cell lines. After 60 minutes of capture, the cells were fixed with 4% PFA, permeabilized with 0.5% Triton X-100, and blocked with 5% BSA. The chip was then incubated with a fluorescent antibody diluent containing PE-labelled anti-human fetal hemoglobin, FITC-labelled anti-GPA, and Cy5-labelled anti-CD45 for 2 hours. DAPI, combined with an anti-fluorescent quenching agent, was applied for 10 minutes. Before downstream analysis, samples were incubated with 0.1 mg/mL MMP-9 for 30 minutes and stored at −20 °C, to be processed within 1 month of collection.
Fluorescence in situ Hybridization Analysis
FISH analysis was performed using a prenatal chromosome number detection kit. The fNRBCs were fixed onto slides with a mixture of methanol and acetic acid (3:1), digested with 0.01% pepsin for 10 minutes at 37°C, and dehydrated through a gradient of ethanol (70%, 85%, and 100%). Following dehydration, fluorescent probes specific for CSP 18/X/Y or CSP 13/21 were applied to the slides. Hybridization occurred at 85°C for 2 minutes, followed by a 2-hour incubation at 45°C in a humidified chamber. After hybridization, slides were washed with 2x SSC buffer at 45°C and dehydrated through the ethanol series. DAPI was used for nuclear counterstaining. The cells were observed and photographed under a fluorescence microscope, and hybridization signals were analyzed for chromosomal abnormalities.
Detection of Short Tandem Repeat Loci Using Quantitative Fluorescence Polymerase Chain Reaction
Whole genome amplification43 (WGA) of fNRBCs was conducted using the MALBAC WGA kit. Maternal genomic DNA (gDNA) was extracted from peripheral blood using a TIANGEN genomic DNA kit. The concentration and quality of the DNA were measured with a NanoDrop 2000 (Thermo Scientific, US). Twenty-one human STR loci from maternal DNA and fNRBCs were analyzed: D19S433, D5S818, D6S1043, AMEL, D3S1358, D7S820, D16S539, CSF1PO, Penta D, D2S441, D8S1179, vWA, TPOX, Penta E, TH01, D12S391, D2S1338, FGA, D21S11, D18S51, and D13S317. PCR products were analyzed using an ABI 3730xl Genetic Analyzer (Thermo Fisher Scientific, USA), with results identified using GeneMapper 4.2 (Applied Biosystems Inc., Thermo Fisher Scientific).
Sanger Sequencing
For Sanger sequencing, the genotypes of 10 random single nucleotide polymorphism (SNP) loci from 10 chromosomes and one pathogenic SNP locus from chromosome 16 were determined. Gene amplification was performed using an ABI PCR thermocycler with 50 ng of WGA product. The amplified products were purified through 1% agarose gel electrophoresis and analyzed using an ABI Genetic Analyzer 3730xl.
Whole-Exome Sequencing Analysis of Samples
Whole-exome sequences (WES) were efficiently enriched from genomic DNA using the Agilent liquid capture system, and the DNA libraries were sequenced on the Illumina platform. Valid sequencing data were then mapped to the reference genome (Hg19).
Statistical Analysis
All experimental data were statistically analyzed using GraphPad Prism 9.0 software. Data are presented as mean ± standard deviation (mean ± SD). Each experiment was repeated at least three times to ensure the reliability of the results. For the comparison of capture efficiency, release efficiency, and cell viability under different incubation times, a one-way analysis of variance (ANOVA) was conducted, followed by Tukey’s post hoc test for pairwise comparisons. This statistical approach was used to determine whether there were significant differences between experimental groups, with a significance level set at p < 0.05.
Results
Characterization and Optimization of GNPs-rPDMS
The mechanism of cell capture and release is illustrated in Figure 2. The chip’s biomimetic red rose petal surface is grooved to a size that maximizes contact with cells, enhancing capture efficiency. The incorporation of GNPs increases the substrate’s structural hierarchy and roughness. Their nanoscale dimensions complement the microvilli and filamentous pseudopods on the cell surface, promoting adhesion. The chips were modified with a biotinylated anti-CD147 antibody for the identification and capture of fNRBCs.24 Moreover, MMP-9 was employed to dissolve GNPs, facilitating the release of fNRBCs from the GNPs-rPDMS.28,44
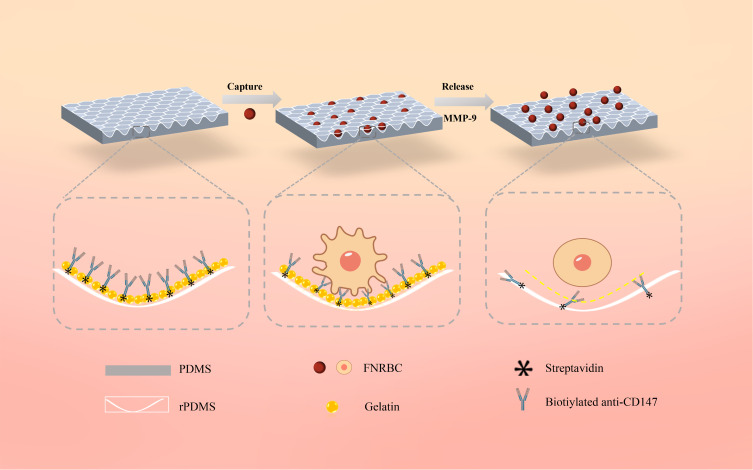
Figure 2.
Schematic illustration of the fetal cell separation and release mechanism utilizing GNPs-rPDMS technology. The red rose petal morphology is replicated onto PDMS, which, adorned with GNPs, forms a concave structure to furnish an expanded array of binding sites for the capture antibody (anti-CD147), thereby amplifying the affinity between fNRBCs and the substrate. Post cell capture, the GNPs are selectively degraded by an MMP-9 solution, facilitating the isolation and retrieval of intact cells.
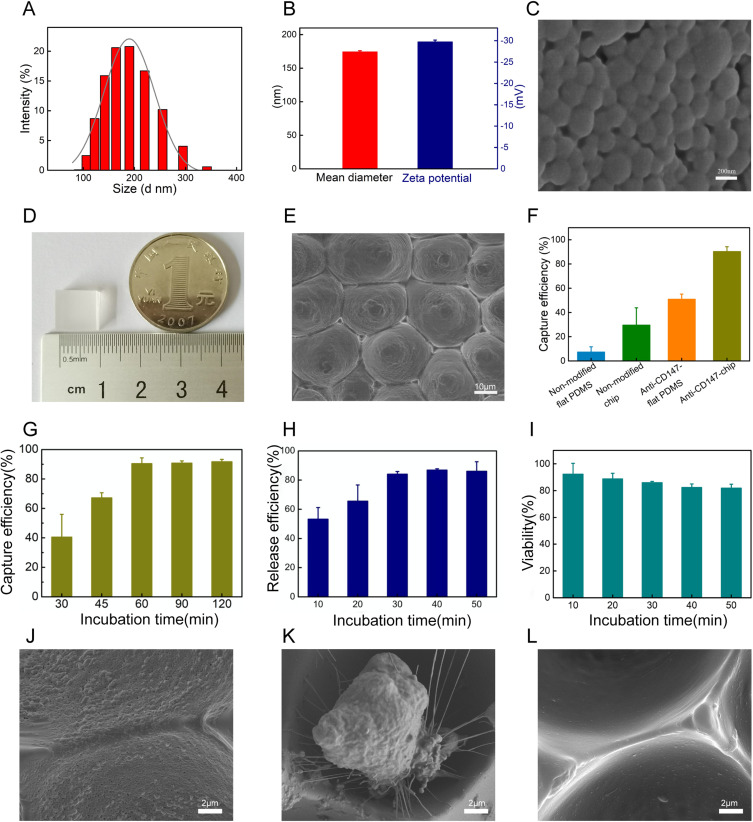
In this study, a cell enrichment system was developed using soft lithography technology. The assembly and performance of the chip are influenced by various parameters, including the size and stability of GNPs, as well as capture and release times. Dynamic light scattering (DLS) results provided detailed data on the diameter distribution of GNPs (Figure 3A), revealing an average size of approximately 174.3 nm and a zeta potential of –29.7 mV, confirming their stability (Figure 3B). The morphology of the GNPs was visualized through scanning electron microscopy (SEM) (Figure 3C), demonstrating that their structure met the experimental requirements.45,46 Additionally, photographs of the GNPs-rPDMS chip provided an overview of its physical dimensions (Figure 3D). The transparent characteristics of the captured cells enhance analysis. Due to the high biocompatibility and enzymatic breakdown capabilities of GNPs, they were modified on the chip surface to improve cell-substrate affinity and facilitate the release of target cells. As shown in Figure 3E, the SEM images, the GNPs-rPDMS exhibited densely arranged cavities on the chip surface with diameters of approximately 20–30 μm, indicating the successful acquisition of a biomimetic red rose petal structure.47 The rough and granular surface of each groove confirms the attachment of GNPs.
Figure 3.
Characterization and optimization of the GNPs-rPDMS (A and B) DLS analysis of GNPs indicated an average size of 175 nm, with a zeta potential of –29.7 mV. (C) Representative SEM image of GNPs (scale bar = 200 nm). (D) Photograph of GNPs-rPDMS. (E) SEM image of GNPs-rPDMS showing the micro-nano structure resembling a red rose petal (scale bar = 10 μm). (F) Cell capture efficiency comparison between flat PDMS and GNPs-rPDMS with or without CD147. (G) Capture efficiency of GNPs-rPDMS at different incubation times. (H) Release efficiency of GNPs-rPDMS under varying MMP-9 incubation times. (I) Viability of released TF-1 cells after incubation with MMP-9 for different times. (J) SEM image of GNPs-rPDMS before cell incubation (scale bar = 2 μm). (K) SEM image of captured cells on GNPs-rPDMS (scale bar = 2 μm). (L) SEM image of GNPs-rPDMS after MMP-9 incubation (scale bar = 2 μm).
We investigated whether the concave, multi-layer structure of the chip could enhance cell capture performance. The effects of different surface modifications on the capture efficiency of TF-1 cells using the anti-CD147 modified micro-nano chip (GNPs-rPDMS), anti-CD147 modified flat PDMS, non-modified micro-nano chip, and non-modified flat PDMS are summarized in Figures 3F and Figure S1. The anti-CD147 antibody-modified GNPs-rPDMS exhibited significantly higher capture ability than the other configurations. These results underscore the critical roles of both the multi-layer structure and antibody modification in cell recognition and capture. The multi-layer architecture, inspired by red rose petals and integrated with GNPs, provided a rough surface and an optimal environment for cell interaction. These features enhance the interaction between cells and the GNPs-rPDMS surface.
High capture and release efficiency, along with the viability of released cells, are essential for downstream analyses. To determine the optimal cell capture time, TF-1 cells were incubated with the chip for 30, 45, 60, 90, and 120 minutes. Capture efficiency peaked at 90.4% after 60 minutes and did not significantly increase with prolonged incubation (one-way ANOVA, p > 0.05, Figure 3G). Consequently, we selected 60 minutes as the optimal capture time for subsequent experiments. We then evaluated the effects of incubation time on cell release under MMP-9 treatment, with captured TF-1 cells incubated for 10, 20, 30, 40, and 50 minutes. At 30 minutes, capture efficiency was 84.08%, reaching a plateau (Figure 3H). Statistical analysis (one-way ANOVA) showed no significant increase beyond 30 minutes (p > 0.05), indicating that 30 minutes is an effective incubation time for optimal cell release. To assess the impact of the capture and release processes on cell viability, TF-1 cells were pre-stained with FDA/PI prior to incubation. As shown in Figure 3I, cell viability was 85.97% after 30 minutes of incubation, with the error bars indicating standard deviation (SD) values across three replicates. The statistical comparison confirmed that cell viability remained above 80% across all tested conditions, suggesting satisfactory biocompatibility of the GNPs-rPDMS. Figure S2 displays live/dead fluorescence images of TF-1 cells on the chip surface, demonstrating that nearly all captured cells could be released and maintained high viability after MMP-9 treatment.
Further observations of the captured cells and chip surface morphology were conducted using SEM. The insets in Figure 3J reveal that the GNPs-rPDMS surface is uniformly coated with GNPs prior to cell release. The captured TF-1 cells exhibited a spread morphology with numerous pseudopodia (Figure 3K). Following MMP-9 enzymatic degradation, the cells were released, and the GNPs were digested (Figure 3L). Collectively, our findings demonstrate that the hierarchical micro-nano structure of GNPs-rPDMS enhances the binding affinity and adhesion of cells to the chip, enabling specific capture and release of CD147-expressing cells, which can be utilized for enriching fNRBCs in non-invasive prenatal diagnosis.
Isolation and Fetal Origin Identification of fNRBCs from Maternal Blood
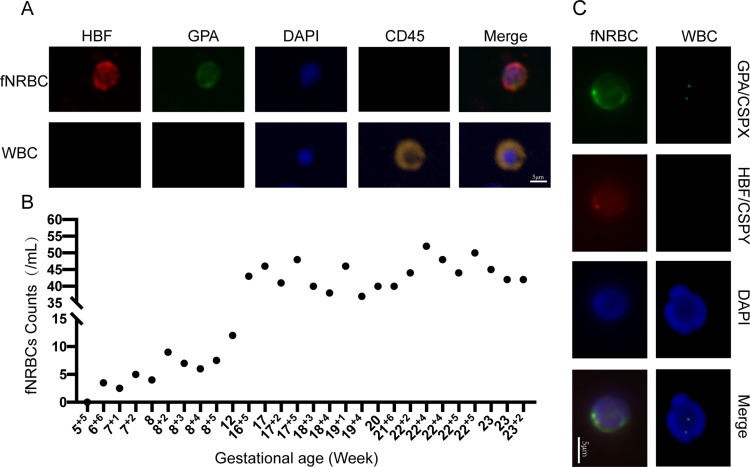
To assess the clinical feasibility of our novel chip, we enriched fNRBCs from the peripheral blood samples of pregnant women. fNRBCs were specifically stained with anti-GPA and anti-HBF markers,48 while maternal white blood cells were stained with anti-CD45. The presence of nuclei was confirmed using DAPI staining. Fluorescence imaging revealed that fNRBCs (HBF+/GPA+/CD45−) were successfully captured on the chip, whereas maternal leukocytes (GPA−/HBF−/CD45+) were nonspecifically adsorbed (Figure 4A). Statistical analysis of 28 blood samples, summarized in Figure 4B and Table S1, demonstrated successful collection of fNRBCs in most samples. The GNPs-rPDMS technology captured cells as early as 7 weeks of gestation, highlighting its excellent potential for early prenatal diagnosis. Additionally, fNRBCs were collected from 7 to 23 gestational weeks, prior to 16 weeks, the number of enriched cells per milliliter ranged from 2.5 to 12, with a median of 6. Between 16 and 24 weeks, the count increased to 37–52 cells per milliliter, with a median of 43.5, confirming the versatility of GNPs-rPDMS.
Figure 4.
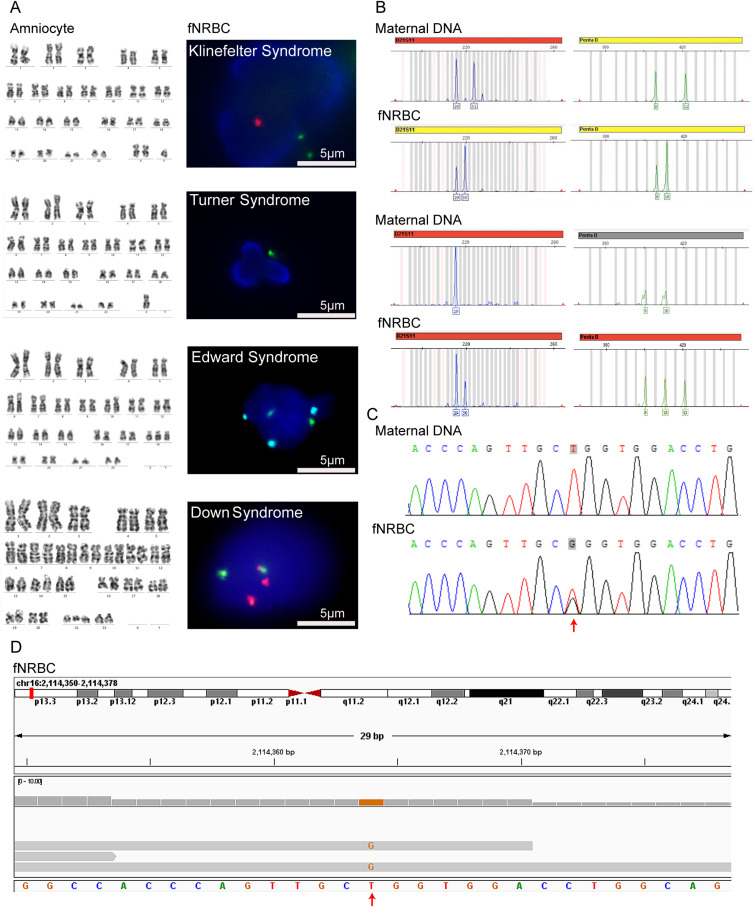
(A) Four-color immunofluorescence images of captured fNRBCs and maternal leukocytes (scale bar = 5 μm). (B) Counts of captured fNRBCs from 28 maternal blood samples. (C) Identification of fetal origin of captured fNRBCs using FISH combined with three-color immunofluorescence (scale bar = 5 μm).
To further distinguish the released fNRBCs, we employed specific protein expression and genotyping analyses for fetal origin identification. Immunofluorescence staining with HBF and GPA antibodies, combined with X/Y chromosome FISH assays, validated the fetal origin of the cells49 (Figure 4C). Male fNRBCs were identified by two FISH signals (X/Y chromosomes) in the nucleus, along with positive expressions of HBF, GPA, and DAPI. By contrast, maternal white blood cells were stained with X/X chromosomes and DAPI. These results affirm the reliability of GNPs-rPDMS in identifying fNRBCs. While the Y chromosome staining method provides clear visualization of fNRBCs, it is limited by gender specificity. Consequently, we incorporated genotyping analyses, including STR50,51 profiling and SNP52,53 genotyping, to further ascertain the fetal origin of the captured cells.
In the STR analysis, we utilized maternal DNA and WGA products from released fNRBCs to target 20 autosomal loci and 1 sex chromosome locus. The STR results indicated that the released cells could be traced back to maternal DNA STR fingerprints, confirming fetal-maternal relationships (Figure 5A). Notably, no instances of three or more alleles were detected, indicating that the collected target cells were pure fetal cells. Furthermore, SNP profiles for these cells included 10 SNPs on 10 human autosomes, with a minor allele frequency (MAF) ranging from 0.27 to 0.4954 (Figure 5B). Distinct genotypes at several loci further demonstrated the different origins of the two cell types. Overall, the genotyping results reinforce that the cells enriched by GNPs-rPDMS were indeed fNRBCs.
Figure 5.
(A) STR analysis confirming the fetal origin of captured fNRBCs. (B) SNP analysis confirming the fetal origin of captured fNRBCs.
Prenatal Diagnosis of Chromosomal Aneuploidies and Single-Gene Disease
We confirmed that GNPs-rPDMS can effectively capture and release fNRBCs, verifying the fetal origin of the released cells. Subsequently, we validated the clinical application of GNPs-rPDMS for diagnosing prenatal chromosomal aneuploidies and single-gene diseases. Maternal peripheral blood samples were collected from pregnant women identified as high-risk for chromosomal aneuploidy through cffDNA-based non-invasive prenatal testing55 or fetal structural abnormalities identified through ultrasound. Following the enrichment of fNRBCs, FISH and QF-PCR assays were utilized to analyze fetal genetic information, facilitating the diagnosis of fetal chromosome aneuploidy.
In the FISH examination, fNRBCs were incubated with fluorescent probes targeting chromosomes 21, 13, 18, X, and Y to diagnose conditions such as Down syndrome, Edwards syndrome, Klinefelter syndrome, and Turner syndrome (Figure 6A). The diagnostic results were consistent with the karyotype obtained from amniocentesis. These findings indicate that the isolated fetal cells possess sufficient genetic information for diagnosing fetal chromosomal disorders using FISH techniques. In the QF-PCR examination, autosomal loci were amplified for fetal identification, while the No. 21 chromosome loci were amplified to assess chromosomal aneuploidy. The volume ratio of results for the target chromosomes was consistent with trisomy (Figure 6B). This method identified two cases of Down syndrome, and the number of replications in autosomes confirmed the fetal-maternal relationships (Figure S3). Additionally, the results aligned with the karyotype and copy number variation sequencing (CNV-seq) findings from amniocentesis. QF-PCR requires only 1 µg of DNA to perform a multi-locus test, making it particularly suitable for fNRBCs, which may be present in sparse numbers and exhibit low DNA concentration. Thus, our findings demonstrate that fNRBCs isolated using GNPs-rPDMS can be effectively employed for non-invasive prenatal diagnosis of chromosomal aneuploidy.
Figure 6.
(A) Representative images for diagnosing fetal chromosome aneuploidy using FISH analysis of isolated fNRBCs from maternal peripheral blood. Red fluorescence represents chromosomes 21 and Y, green represents chromosomes 13 and X, and cyan represents chromosome 18 (scale bar = 5 μm). All results were consistent with amniocentesis karyotype results. (B) Representative images for diagnosing fetal trisomy 21 using QF-PCR analysis of isolated fNRBCs from maternal peripheral blood. (C) Sequencing results of candidate fNRBCs from a pregnancy affected by Bourneville disease. The results suggested that the fetus harbors a heterozygous c.1535T > G mutation in the TSC2 gene, while the mother is unaffected. The arrow indicates the location of the variant. (D) WES of candidate fNRBCs in the above cases suggested the presence of the c.1535T > G mutation in the same gene.
In a typical case, a 20-week pregnant woman underwent fetal echocardiography, revealing multiple solid lesions in the left and right ventricles (possible rhabdomyomas) and tricuspid regurgitation. Following amniocentesis, CNV-seq and WES were performed according to ACOG guidelines. The sequencing results identified the NM_000548.3:c.1535T>G (p.Leu512Arg) mutation in the TSC2 gene,56 associated with nodular sclerosis type 2, with no family history of the condition. We collected maternal peripheral blood and enriched fNRBCs by incubating the samples with GNPs-rPDMS. STR analysis confirmed the fetal origin of the cells (Figure S4). Site-specific amplification products were subsequently analyzed using Sanger sequencing. As shown in Figure 6C, the mother exhibited a T/T genotype, while the fetus displayed a heterozygous T/G genotype. Further exploration using WES on the target fNRBCs yielded an average effective data volume of 88.04%, a Q20 score of 93.97%, and an average error rate of 0.04%. The average raw data generated was 9.02 Gb. These results demonstrate high-quality sequencing data that meet analytical requirements.57,58 The sequencing depth for chromosome 16 reached 58.66x, with an average coverage of 87.5%. Figure 6D illustrates the T>G mutation at the same locus of the TSC2 gene, corroborating the WES results from amniotic fluid samples. This confirms that fNRBCs captured by GNPs-rPDMS can be effectively utilized for diagnosing point mutations associated with monogenic diseases.
Discussion
The technological advancements of this study include the enrichment and identification of rare fNRBCs, yielding high-purity fetal DNA. This is achieved through genotyping and specific protein staining to confirm fetal origin. We also analyzed the feasibility of using cell-based tests for detecting chromosomal disorders and monogenic diseases. Compared with previous studies, this research employs a bioinspired structure modeled after red rose petals, resulting in lower costs and a simpler fabrication process. The ability to collect sufficient quantities of high-quality rare cells for genetic analysis is crucial, while achieving comparable capture efficiency. The GNPs-rPDMS chip boasts several superior characteristics: (1) a surface-specific micro-nano multi-layered structure that increases antibody binding sites and enhances cell adhesion; (2) excellent light transmission for effective cell detection; (3) the capability to release captured fNRBCs via MMP-9 digestion with minimal cellular impairment; and (4) the versatility of released fNRBCs for various subsequent analyses.
Utilizing chips with micro-nano structural differences on their surfaces, combined with specific antibodies, represents a strategic approach for the rapid screening of rare cells. Micrometer-sized chips that match the scale of cell surfaces, in conjunction with GNPs to enhance cell adhesion and facilitate release, render these chips both simple and effective in differentiating fNRBCs from maternal cells. Subsequent studies confirmed the accurate source identification of cells, achieving high precision in distinguishing fetal from maternal cells. With this advanced chip technology, we successfully identified between 2.5 and 52 target cells per milliliter of blood in 96% of the samples, as early as 7 weeks into gestation. This indicates that chromosomal or genetic testing of the fetus can be performed as early as 7 weeks of pregnancy, preceding the gestational age at which invasive prenatal diagnostic procedures or screening are currently employed in clinical practice. This advancement enables both pregnant women and clinicians to gain earlier insights into fetal health, facilitating improved pregnancy monitoring and management, and potentially allowing for early interventions or treatment measures in the first trimester. Notably, the only exception was a sample from a woman at 5 weeks of gestation. This discrepancy may result from the early gestational age, leading to an insufficient quantity of fNRBCs in the maternal peripheral blood for effective analysis. This non-invasive and highly accurate approach is particularly effective for enriching fNRBCs and yields high-quality molecular analysis outcomes. A limitation of this study is the sample size of 28 cases in the counting section, expanding this in future studies will facilitate a more comprehensive investigation of the clinical feasibility of this chip, which is a key direction for future research. Establishing the fetal origin of presumed cells is essential for their use in prenatal testing. We observed positive HBF and GPA expression in enriched cells. Additionally, during pregnancies with male fetuses, Y chromosome signals were positively identified in isolated cells through FISH analysis. Collectively, these findings indicate that the isolated target cells originate from the fetus. Beyond mere staining, employing STR and SNP genotyping of these cells represents the gold standard for confirming fetal origin.
Currently, only a few studies have investigated the fetal origin of fNRBCs through STR.26 Most research has focused on fetal trophoblast cells,14,59,60 likely due to the scarcity of fNRBCs in peripheral blood, complicating their separation. The successful detection of STR and SNP in rare fNRBCs in our study supports the efficacy of GNPs-rPDMS in enriching fetal cells, enabling the extraction of precise fetal genetic information. Given the high purity of fetal cells, the accuracy of fetal genotyping is comparable to invasive strategies and potentially superior to that of cffDNA.
Presently, prenatal diagnostic methods for fetal chromosomal or genetic diseases still predominantly rely on invasive procedures. Some studies have attempted to diagnose monogenic diseases using cffDNA,61 however, its fragmented nature poses limitations. By contrast, fetal cells provide advantages due to the high integrity of pure fetal DNA. Our approach utilizes a simple chip to isolate rare fNRBCs and employs established clinical genetic analysis techniques such as FISH,62 QF-PCR,63 Sanger sequencing,64 and WES, to explore the potential for diagnosing other genetic disorders based on these cells. The use of fNRBCs for NIPD of monogenic diseases is rare, and only preliminary attempts have been made in this study. Notably, this is the first study to utilize fNRBCs isolated from peripheral blood to diagnose fetal Bourneville disease. Further research is necessary to investigate the potential of this cell-based test for diagnosing other genetic disorders.
Currently, commercially available cell-based NIPD primarily employs ARCEDI’s method of sorting fetal trophoblast cells, which are then used for research on chromosomal aneuploidies, microdeletions, and monogenic diseases.65,66 However, this study did not extend the enriched fNRBCs to microdeletion or duplication testing, representing a limitation. While the turnaround time and gestational weeks for obtaining results are comparable between both methods, ARCEDI requires a larger blood volume. Despite its higher cost, ARCEDI’s two-step sorting reduces non-specific cell adsorption, highlighting an area where we can improve.
Several important limitations must be acknowledged in our study. For instance, we detected only two reads in the target region, likely due to sample deterioration, low concentration, or randomness in the WES.67–69 Furthermore, before our diagnostic platform can be clinically implemented, additional studies with larger sample sizes are necessary, including a more diverse cohort of patients with various genetic anomalies. Additionally, the current DNA amplification method is fragmentary, making comparative analyses of DNA quality unfeasible. Therefore, a more robust WGA technique may be required to obtain higher-quality fetal DNA, suitable for extensive molecular analyses. Current research indicates that fNRBCs are released in greater quantities in conditions such as fetal cardiac malformations70 or maternal complications such as preeclampsia.71 However, whether fNRBCs undergo specific expression changes under pathological conditions remains unclear and warrants further investigation. Future research should explore RNA sequencing or other molecular biology approaches to analyze enriched fNRBCs and assess their potential clinical applications.
In summary, the GNPs-rPDMS method presented here offers a non-invasive means of obtaining valuable molecular information from fetuses at risk for chromosomal or monogenic diseases. The chip exhibits high accuracy in enriching rare fNRBCs. Compared with invasive procedures, this cell-based enrichment and detection approach maintains high diagnostic accuracy without posing safety risks to the fetus, positioning it as a promising candidate for clinical use. Our findings suggest that fNRBC enrichment from maternal peripheral blood holds substantial promise for identifying fetuses with congenital defects.
Conclusion
The biomimetic micro-nano hierarchical composite structure chip, GNPs-rPDMS, effectively enriches fNRBCs from the peripheral blood of pregnant women, providing a novel non-invasive prenatal diagnostic method. This study aims to address the limitations of NIPD and offer new insights into the genetic and developmental disorders of fetuses.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 82001644) and Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University (Grant No. ZNJC202008).
Disclosure
The author(s) report no conflicts of interest in this work.
References
- 1.Yue W, Zhang E, Liu R, et al. The China Birth Cohort Study (CBCS). Eur J Epidemiol. 2022;37(3):295–304. doi: 10.1007/s10654-021-00831-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehtonen L, Gimeno A, Parra-Llorca A, et al. Early Neonatal Death: a Challenge Worldwide. Semin Fetal Neonatal Med. 2017;22(3):153–160. doi: 10.1016/j.siny.2017.02.006 [DOI] [PubMed] [Google Scholar]
- 3.Alfirevic Z, Navaratnam K, Mujezinovic F. Amniocentesis and Chorionic Villus Sampling for Prenatal Diagnosis. Cochrane Database Syst Rev. 2017;9(9):CD003252. doi: 10.1002/14651858.CD003252.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durand M-A, Stiel M, Boivin J, et al. Information and Decision Support Needs of Parents Considering Amniocentesis: interviews with Pregnant Women and Health Professionals. Health Expect Int J Public Particip Health Care Health Policy. 2010;13:125–138. doi: 10.1111/j.1369-7625.2009.00544.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du X, Zhang L, Liu Z, et al. Risk of Mother-to-Child Transmission after Amniocentesis in Pregnant Women with Hepatitis B Virus: a Retrospective Cohort Study. Am J Obstet Gynecol. 2024;230(2):249.e1–249.e8. doi: 10.1016/j.ajog.2023.07.032 [DOI] [PubMed] [Google Scholar]
- 6.Bakker M, Birnie E, Robles de Medina P, et al. Total Pregnancy Loss after Chorionic Villus Sampling and Amniocentesis: a Cohort Study. Ultrasound Obstet Gynecol. 2017;49(5):599–606. doi: 10.1002/uog.15986 [DOI] [PubMed] [Google Scholar]
- 7.Han Z, Zhang Y, Bai X, et al. Mother-to-Child Transmission of Hepatitis B Virus after Amniocentesis: a Retrospective Matched Cohort Study. Prenat Diagn. 2019;39(6):431–440. doi: 10.1002/pd.5452 [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Li J, Saucier JB, et al. Non-Invasive Prenatal Sequencing for Multiple Mendelian Monogenic Disorders Using Circulating Cell-Free Fetal DNA. Nat Med. 2019;25(3):439–447. doi: 10.1038/s41591-018-0334-x [DOI] [PubMed] [Google Scholar]
- 9.Bianchi DW, Chiu RWK. Sequencing of Circulating Cell-Free DNA during Pregnancy. N Engl J Med. 2018;379(5):464–473. doi: 10.1056/NEJMra1705345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosner M, Kolbe T, Hengstschläger M. Fetomaternal Microchimerism and Genetic Diagnosis: on the Origins of Fetal Cells and Cell-Free Fetal DNA in the Pregnant Woman. Mutat Res. 2021;788:108399. doi: 10.1016/j.mrrev.2021.108399 [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Li J, Chen S, et al. Genetic Deconvolution of Fetal and Maternal Cell-Free DNA in Maternal Plasma Enables next-Generation Non-Invasive Prenatal Screening. Cell Discov. 2022;8:109. doi: 10.1038/s41421-022-00457-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rezaei M, Winter M, Zander-Fox D, et al. A Reappraisal of Circulating Fetal Cell Noninvasive Prenatal Testing. Trends Biotechnol. 2019;37(6):632–644. doi: 10.1016/j.tibtech.2018.11.001 [DOI] [PubMed] [Google Scholar]
- 13.Hatt L, Ravn K, Dahl Jeppesen L, et al. How Does Cell-Based Non-Invasive Prenatal Test (NIPT) Perform against Chorionic Villus Sampling and Cell-Free NIPT in Detecting Trisomies and Copy Number Variations? A Clinical Study from Denmark. Prenat Diagn. 2023;43(7):854–864. doi: 10.1002/pd.6387 [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Situ B, Huang L, et al. Nondestructive Identification of Rare Trophoblastic Cells by Endoplasmic Reticulum Staining for Noninvasive Prenatal Testing of Monogenic Diseases. Adv Sci. 2020;7(7):1903354. doi: 10.1002/advs.201903354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Cheng L, Wei X, et al. High-Throughput Isolation of Fetal Nucleated Red Blood Cells by Multifunctional Microsphere-Assisted Inertial Microfluidics. Biomed Microdevices. 2020;22(4):75. doi: 10.1007/s10544-020-00531-2 [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Yang Y, Li X, et al. Frequency-Enhanced Transferrin Receptor Antibody-Labelled Microfluidic Chip (FETAL-Chip) Enables Efficient Enrichment of Circulating Nucleated Red Blood Cells for Non-Invasive Prenatal Diagnosis. Lab Chip. 2018;18(18):2749–2756. doi: 10.1039/c8lc00650d [DOI] [PubMed] [Google Scholar]
- 17.Kruckow S, Schelde P, Hatt L, et al. Does Maternal Body Mass Index Affect the Quantity of Circulating Fetal Cells Available to Use for Cell-Based Noninvasive Prenatal Test in High-Risk Pregnancies? Fetal Diagn Ther. 2019;45(5):353–356. doi: 10.1159/000492028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Wu Z, Sutlive J, et al. Noninvasive Prenatal Diagnosis Targeting Fetal Nucleated Red Blood Cells. J Nanobiotechnology. 2022;20(1):546. doi: 10.1186/s12951-022-01749-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karsten U, Butschak G, Stahn R, et al. A Novel Series of Anti-Human Glycophorin A (CD235a) Antibodies Defining Five Extra- and Intracellular Epitopes. Int Immunopharmacol. 2010;10(11):1354–1360. doi: 10.1016/j.intimp.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 20.Kwon KH, Jeon YJ, Hwang HS, et al. A High Yield of Fetal Nucleated Red Blood Cells Isolated Using Optimal Osmolality and a Double-Density Gradient System. Prenat Diagn. 2007;27(13):1245–1250. doi: 10.1002/pd.1888 [DOI] [PubMed] [Google Scholar]
- 21.de Goede OM, Lavoie PM, Robinson WP. Robinson WP Cord Blood Hematopoietic Cells from Preterm Infants Display Altered DNA Methylation Patterns. Clin Clin Epigenet. 2017;9(1):39. doi: 10.1186/s13148-017-0339-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemescu D, Constantinescu D, Gorduza V, et al. Comparison between Paramagnetic and CD71 Magnetic Activated Cell Sorting of Fetal Nucleated Red Blood Cells from the Maternal Blood. J Clin Lab Anal. 2020;34(9):e23420. doi: 10.1002/jcla.23420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumo T, Tomizawa Y, Kita M, et al. Concentration and Extraction Chip of Fetal Nucleated Red Blood Cells (NRBCs) by Micro Gap with Diaphragm for Fetal DNA Diagnosis from Maternal Blood. Microsyst Technol. 2017;23(12):5351–5355. doi: 10.1007/s00542-017-3309-9 [DOI] [Google Scholar]
- 24.Feng C, He Z, Cai B, et al. Non-Invasive Prenatal Diagnosis of Chromosomal Aneuploidies and Microdeletion Syndrome Using Fetal Nucleated Red Blood Cells Isolated by Nanostructure Microchips. Theranostics. 2018;8(5):1301–1311. doi: 10.7150/thno.21979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Li N, Cai B, et al. A Biocompatible Nanofibers-Based Microchip for Isolation and Nondestructive Release of Fetal Nucleated Red Blood Cells. Adv Mater Interfaces. 2020;7(23):2001028. doi: 10.1002/admi.202001028 [DOI] [Google Scholar]
- 26.Zhang Q, Zhang K, Guo Y, et al. The Isolation and Analysis of Fetal Nucleated Red Blood Cells Using Multifunctional Microbeads with a Nanostructured Coating toward Early Noninvasive Prenatal Diagnostics. J Mater Chem B. 2021;9(13):3047–3054. doi: 10.1039/D1TB00005E [DOI] [PubMed] [Google Scholar]
- 27.Xu S, Wu L, Qin Y, et al. Sequential Ensemble-Decision Aliquot Ranking Isolation and Fluorescence In Situ Hybridization Identification of Rare Cells from Blood by Using Concentrated Peripheral Blood Mononuclear Cells. Anal Chem. 2021;93(6):3196–3201. doi: 10.1021/acs.analchem.0c04629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei X, Cai B, Chen K, et al. Enhanced Isolation and Release of Fetal Nucleated Red Blood Cells Using Multifunctional Nanoparticle-Based Microfluidic Device for Non-Invasive Prenatal Diagnostics. Sens Actuators B Chem. 2019;281:131–138. doi: 10.1016/j.snb.2018.10.027 [DOI] [Google Scholar]
- 29.Liu Q, Zheng S, Ye K, et al. Cell Migration Regulated by RGD Nanospacing and Enhanced under Moderate Cell Adhesion on Biomaterials. Biomaterials. 2020;263:120327. doi: 10.1016/j.biomaterials.2020.120327 [DOI] [PubMed] [Google Scholar]
- 30.Yin H, Liu W, Huang Y, et al. Surface Epitaxial Crystallization-Directed Nanotopography for Accelerating Preosteoblast Proliferation and Osteogenic Differentiation. ACS Appl Mater Interfaces. 2019;11(46):42956–42963. doi: 10.1021/acsami.9b14800 [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Cui H, Zhang P, et al. Efficient Capture of Cancer Cells by Their Replicated Surfaces Reveals Multiscale Topographic Interactions Coupled with Molecular Recognition. ACS Appl Mater Interfaces. 2017;9(12):10537–10543. doi: 10.1021/acsami.7b01147 [DOI] [PubMed] [Google Scholar]
- 32.Alagoz AS, Cansizoglu H, Karabacak T. Karabacak T Hierarchical Multi-Diameter Single-Crystal Silicon Nanowires by Successive Wet Chemical Etching. J Nanosci Nanotechnol. 2017;17(4):2857–2860. doi: 10.1166/jnn.2017.13856 [DOI] [PubMed] [Google Scholar]
- 33.Kim K, Mun S, Jang M, et al. Thermoelectric Properties of Ni/Ge-Multilayer-Laminated Silicon. Appl Phys A. 2021;127(1):50. doi: 10.1007/s00339-020-04200-2 [DOI] [Google Scholar]
- 34.Li Q, Li F, Li Y, et al. Hydrogen Induced Etching Features of Wrinkled Graphene Domains. Nanomaterials. 2019;9(7):930. doi: 10.3390/nano9070930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Martinez FJ, Jin K, Barreiro DL, et al. The Rise of Hierarchical Nanostructured Materials from Renewable Sources: learning from Nature. ACS Nano. 2018;12(8):7425–7433. doi: 10.1021/acsnano.8b04379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabritius H-O, Ziegler A, Friák M, et al. Functional Adaptation of Crustacean Exoskeletal Elements through Structural and Compositional Diversity: a Combined Experimental and Theoretical Study. Bioinspiration & Biomimetics. 2016;11(5):055006. doi: 10.1088/1748-3190/11/5/055006 [DOI] [PubMed] [Google Scholar]
- 37.Ye W, Tang J, Wang Y, et al. Hierarchically Structured Carbon Materials Derived from Lotus Leaves as Efficient Electrocatalyst for Microbial Energy Harvesting. Sci Total Environ. 2019;666:865–874. doi: 10.1016/j.scitotenv.2019.02.300 [DOI] [PubMed] [Google Scholar]
- 38.Dou X, Li P, Jiang S, et al. Bioinspired Hierarchically Structured Surfaces for Efficient Capture and Release of Circulating Tumor Cells. ACS Appl Mater Interfaces. 2017;9(10):8508–8518. doi: 10.1021/acsami.6b16202 [DOI] [PubMed] [Google Scholar]
- 39.Li P, Dou X, Feng C, et al. Enhanced Cell Adhesion on a Bio-Inspired Hierarchically Structured Polyester Modified with Gelatin-Methacrylate. Biomater Sci. 2018;6(4):785–792. doi: 10.1039/c7bm00991g [DOI] [PubMed] [Google Scholar]
- 40.Wei X, Chen K, Cai B, et al. An Acoustic Droplet-Induced Enzyme Responsive Platform for the Capture and On-Demand Release of Single Circulating Tumor Cells. ACS Appl Mater Interfaces. 2019;11(44):41118–41126. doi: 10.1021/acsami.9b16566 [DOI] [PubMed] [Google Scholar]
- 41.Coester CJ, Langer K, van Briesen H, et al. Gelatin Nanoparticles by Two Step Desolvation--A New Preparation Method, Surface Modifications and Cell Uptake. J Microencapsul. 2000;17(2):187–193. doi: 10.1080/026520400288427 [DOI] [PubMed] [Google Scholar]
- 42.Wei X, Ao Z, Cheng L, et al. Highly Sensitive and Rapid Isolation of Fetal Nucleated Red Blood Cells with Microbead-Based Selective Sedimentation for Non-Invasive Prenatal Diagnostics. Nanotechnology. 2018;29(43):434001. doi: 10.1088/1361-6528/aad8c4 [DOI] [PubMed] [Google Scholar]
- 43.Huang L, Ma F, Chapman A, et al. Single-Cell Whole-Genome Amplification and Sequencing: methodology and Applications. Annu Rev Genomics Hum Genet. 2015;16(1):79–102. doi: 10.1146/annurev-genom-090413-025352 [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Gao F, Li L, et al. Gelatin–Mesoporous Silica Nanoparticles as Matrix Metalloproteinases-Degradable Drug Delivery Systems in Vivo. Microporous Mesoporous Mater. 2013;182:165–172. doi: 10.1016/j.micromeso.2013.08.050 [DOI] [Google Scholar]
- 45.Wang S, Wan Y, Liu Y. Liu Y Effects of Nanopillar Array Diameter and Spacing on Cancer Cell Capture and Cell Behaviors. Nanoscale. 2014;6(21):12482–12489. doi: 10.1039/c4nr02854f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W, Weng S, Zhang F, et al. Nanoroughened Surfaces for Efficient Capture of Circulating Tumor Cells without Using Capture Antibodies. ACS Nano. 2013;7(1):566–575. doi: 10.1021/nn304719q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo R, Yu Y, Zeng J, et al. Biomimicking Topographic Elastomeric Petals (E-Petals) for Omnidirectional Stretchable and Printable Electronics. Adv Sci. 2015;2(3):1400021. doi: 10.1002/advs.201400021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bianchi DW, Zickwolf GK, Yih MC, et al. Erythroid-Specific Antibodies Enhance Detection of Fetal Nucleated Erythrocytes in Maternal Blood. Prenat Diagn. 1993;13(4):293–300. doi: 10.1002/pd.1970130408 [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Cheng L, Sun Y, et al. Enhanced Isolation of Fetal Nucleated Red Blood Cells by Enythrocyte-Leukocyte Hybrid Membrane-Coated Magnetic Nanoparticles for Noninvasive Pregnant Diagnostics. Anal Chem. 2021;93(2):1033–1042. doi: 10.1021/acs.analchem.0c03933 [DOI] [PubMed] [Google Scholar]
- 50.Wright SE, Todd PK. Todd PK Native Functions of Short Tandem Repeats. eLife. 2023;12:e84043. doi: 10.7554/eLife.84043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mo S, Liu Y, Wang S, et al. Exploring the Efficacy of Paternity and Kinship Testing Based on Single Nucleotide Polymorphisms. Forensic Sci Int Genet. 2016;22:161–168. doi: 10.1016/j.fsigen.2016.02.012 [DOI] [PubMed] [Google Scholar]
- 52.Flanagan SP, Jones AG. The Future of Parentage Analysis: from Microsatellites to SNPs and Beyond. Mol Ecol. 2019;28(3):544–567. doi: 10.1111/mec.14988 [DOI] [PubMed] [Google Scholar]
- 53.Brumfield RT, Beerli P, Nickerson DA, et al. The Utility of Single Nucleotide Polymorphisms in Inferences of Population History. Trends Ecol Evol. 2003;18(5):249–256. doi: 10.1016/S0169-5347(03)00018-1 [DOI] [Google Scholar]
- 54.Tam JCW, Chan YM, Tsang SY, et al. Noninvasive Prenatal Paternity Testing by Means of SNP-Based Targeted Sequencing. Prenat Diagn. 2020;40(4):497–506. doi: 10.1002/pd.5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gadsbøll K, Petersen OB, Gatinois V, et al. Current Use of Noninvasive Prenatal Testing in Europe, Australia and the USA: a Graphical Presentation. Acta Obstet Gynecol Scand. 2020;99(6):722–730. doi: 10.1111/aogs.13841 [DOI] [PubMed] [Google Scholar]
- 56.Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG Clinical Laboratory Standards for Next-Generation Sequencing. Genet Med. 2013;15(9):733–747. doi: 10.1038/gim.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santani A, Simen BB, Briggs M, et al. Designing and Implementing NGS Tests for Inherited Disorders: a Practical Framework with Step-by-Step Guidance for Clinical Laboratories. J Mol Diagn JMD. 2019;21(3):369–374. doi: 10.1016/j.jmoldx.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 58.Sathirapongsasuti JF, Lee H, Horst BAJ, et al. Exome Sequencing-Based Copy-Number Variation and Loss of Heterozygosity Detection: exomeCNV. Bioinforma Oxf Engl. 2011;27(19):2648–2654. doi: 10.1093/bioinformatics/btr462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou S, Chen J-F, Song M, et al. Imprinted NanoVelcro Microchips for Isolation and Characterization of Circulating Fetal Trophoblasts: toward Noninvasive Prenatal Diagnostics. ACS Nano. 2017;11(8):8167–8177. doi: 10.1021/acsnano.7b03073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain CV, Kadam L, van Dijk M, et al. Fetal Genome Profiling at 5 weeks of Gestation after Noninvasive Isolation of Trophoblast Cells from the Endocervical Canal. Sci Transl Med. 2016;8(363):363re4. doi: 10.1126/scitranslmed.aah4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen M, Lu S, Lai ZF, et al. Targeted Sequencing of Maternal Plasma for Haplotype-Based Non-Invasive Prenatal Testing of Spinal Muscular Atrophy. Ultrasound Obstet Gynecol. 2017;49(6):799–802. doi: 10.1002/uog.15947 [DOI] [PubMed] [Google Scholar]
- 62.Liu S, Wang J, Zhang R, et al. Expert Consensus on Prenatal Fluorescence in Situ Hybridization. Chin J Med Genet. 2020;37:918–923. doi: 10.3760/cma.j.cn511374-20190430-00218 [DOI] [PubMed] [Google Scholar]
- 63.Scott F, Murphy K, Carey L, et al. Prenatal Diagnosis Using Combined Quantitative Fluorescent Polymerase Chain Reaction and Array Comparative Genomic Hybridization Analysis as a First-Line Test: results from over 1000 Consecutive Cases. Ultrasound Obstet Gynecol. 2013;41(5):500–507. doi: 10.1002/uog.12429 [DOI] [PubMed] [Google Scholar]
- 64.Seroussi E. Seroussi E Estimating Copy-Number Proportions: the Comeback of Sanger Sequencing. Genes. 2021;12(2):283. doi: 10.3390/genes12020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeppesen LD, Lildballe DL, Hatt L, et al. Noninvasive Prenatal Screening for Cystic Fibrosis Using Circulating Trophoblasts: detection of the 50 Most Common Disease-Causing Variants. Prenat Diagn. 2023;43(1):3–13. doi: 10.1002/pd.6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kølvraa S, Singh R, Normand EA, et al. Genome-Wide Copy Number Analysis on DNA from Fetal Cells Isolated from the Blood of Pregnant Women. Prenat Diagn. 2016;36(12):1127–1134. doi: 10.1002/pd.4948 [DOI] [PubMed] [Google Scholar]
- 67.Eldomery MK, Coban-Akdemir Z, Harel T, et al. Lessons Learned from Additional Research Analyses of Unsolved Clinical Exome Cases. Genome Med. 2017;9(1):26. doi: 10.1186/s13073-017-0412-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.LaDuca H, Farwell KD, Vuong H, et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PLoS One. 2017;12(2):e0170843. doi: 10.1371/journal.pone.0170843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Zhang D, Zhao X, et al. Exploration of a Novel Noninvasive Prenatal Testing Approach for Monogenic Disorders Based on Fetal Nucleated Red Blood Cells. Clin Chem. 2023;69(12):1396–1408. doi: 10.1093/clinchem/hvad165 [DOI] [PubMed] [Google Scholar]
- 70.Tseng SY, Gao Z, Kalfa TA, et al. Altered Erythropoiesis in Newborns with Congenital Heart Disease. Pediatr Res. 2022;91(3):606–611. doi: 10.1038/s41390-021-01370-4 [DOI] [PubMed] [Google Scholar]
- 71.Em H, Aj E, van der Z M, et al. The Impact of Early- and Late-Onset Preeclampsia on Umbilical Cord Blood Cell Populations. J Reprod Immunol. 2016;116:81–85. doi: 10.1016/j.jri.2016.05.002 [DOI] [PubMed] [Google Scholar]